Abstract
The irreversible oxidation of cysteine residues can be prevented by protein S-thiolation, in which protein -SH groups form mixed disulfides with low-molecular-weight thiols such as glutathione. We report here the identification of glyceraldehyde-3-phosphate dehydrogenase as the major target of protein S-thiolation following treatment with hydrogen peroxide in the yeast Saccharomyces cerevisiae. Our studies reveal that this process is tightly regulated, since, surprisingly, despite a high degree of sequence homology (98% similarity and 96% identity), the Tdh3 but not the Tdh2 isoenzyme was S-thiolated. The glyceraldehyde-3-phosphate dehydrogenase enzyme activity of both the Tdh2 and Tdh3 isoenzymes was decreased following exposure to H2O2, but only Tdh3 activity was restored within a 2-h recovery period. This indicates that the inhibition of the S-thiolated Tdh3 polypeptide was readily reversible. Moreover, mutants lacking TDH3 were sensitive to a challenge with a lethal dose of H2O2, indicating that the S-thiolated Tdh3 polypeptide is required for survival during conditions of oxidative stress. In contrast, a requirement for the nonthiolated Tdh2 polypeptide was found during exposure to continuous low levels of oxidants, conditions where the Tdh3 polypeptide would be S-thiolated and hence inactivated. We propose a model in which both enzymes are required during conditions of oxidative stress but play complementary roles depending on their ability to undergo S-thiolation.
Sulfhydryl groups (-SH) play a remarkably broad range of roles in the cell, since the redox status of cysteine residues is involved in both the structure and function of numerous enzymes, receptors, and transcription factors. However, cysteine residues are among the most easily oxidized residues in proteins, and the oxidation of -SH groups is one of the earliest observable events during reactive oxygen species (ROS)-mediated damage (6, 12). The oxidation of -SH groups can result in the formation of protein disulfides (S-S) through two protein cysteines or mixed disulfides between protein-SH groups and a number of low-molecular-weight thiols (protein S-thiolation). While the role of disulfide bond formation in protein folding has been well characterized (see, for example, reference 23), its role during conditions of oxidative stress is unclear. In contrast, protein S-thiolation has been proposed to serve an antioxidant function by preventing the irreversible oxidation of cysteine residues to higher oxidation states (e.g., sulfenic acid) following exposure to ROS (10, 47).
Modification of proteins by S-thiolation does not require an enzymatic activity and has been proposed to occur either by the reaction of partially oxidized protein sulfhydryls (thiyl radical or sulfenic acid intermediates) with thiols such as cysteine or glutathione (GSH) or by thiol-disulfide exchange reactions with the oxidized disulfide form of GSH (GSSG) (47). To provide a defense against conditions of oxidative stress, this modification must be reversible. Dethiolation can occur via direct reduction by GSH. In addition, dethiolation can be catalyzed by glutaredoxin, thioredoxin, and protein disulfide isomerase, but glutaredoxin appears to be the most efficient dethiolase based on in vitro experiments (29). The range of proteins that can be modified by S-thiolation has been determined in studies with human endothelial cells and murine macrophages (40, 41). A few proteins that are S-thiolated in response to oxidants, including carbonic anhydrase III (4), actin (3), creatine kinase (8), glycogen phosphorylase b (37) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (39, 42), have been positively identified in mammalian cells. The physiological relevance of the modification of these proteins is unclear. There do not appear to be any unifying structural or functional features apart from the relative abundance of these proteins in mammalian cells.
The intracellular milieu is usually a reducing environment due to a high concentrations (millimolar) of the low-molecular-weight thiol GSH, which is maintained in the cytosol predominantly in a reduced form (24, 35). GSH is a ubiquitous tripeptide (γ-glutamylcysteinylglycine) that has proposed functions in many cellular processes, including the detoxification of various carcinogens, mutagens, and ROS (34, 44). Studies with the model eukaryote Saccharomyces cerevisiae have led to a better understanding of the role of this peptide in cellular metabolism and have shown that GSH is an essential metabolite that is required as a reductant during normal growth conditions (reviewed in reference 16). This essential function may be the removal of toxic intermediates which accumulate during the course of normal cell metabolism (5, 18). In addition, yeast strains lacking GSH, or altered in their GSH redox state, are sensitive to conditions of oxidative stress induced by hydroperoxides and the superoxide anion, as well as to the toxic products of lipid peroxidation (14, 15, 17, 19, 27, 48). This sensitivity to ROS presumably reflects the antioxidant capacity of GSH both as a free-radical scavenger and as an electron donor for glutathione peroxidases and transferases (16, 34).
In this present study, we demonstrate that protein S-thiolation occurs in yeast and examine the role of GSH in the process. GAPDH is identified as the most abundant S-thiolated protein during conditions of oxidative stress, but, surprisingly, only the Tdh3 and not the Tdh2 GAPDH isoenzyme was modified. We present evidence that both enzymes are required for survival during conditions of oxidative stress but play complementary roles depending on their ability to undergo S-thiolation.
MATERIALS AND METHODS
Yeast strains and media.
The S. cerevisiae strains used in this study are described in Table 1. The strains were grown in rich YEPD medium (2% [wt/vol] glucose, 2% [wt/vol] Bacto Peptone, 1% [wt/vol] yeast extract) or minimal SD medium (0.17% [wt/vol] yeast nitrogen base without amino acids, 5% [wt/vol] ammonium sulfate, 2% [wt/vol] glucose) supplemented with appropriate amino acids and bases: 2 mM leucine, 4 mM isoleucine, 1 mM valine, 0.3 mM histidine, 0.4 mM tryptophan, 0.15 mM adenine, and 0.2 mM uracil. The media were solidified by the addition of 2% (wt/vol) agar.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| CY4 | MATa ura3-52 leu2-3 leu2-112 trp1-1 ade2-1 his3-11 can1-100 | 17 |
| CY7 | As in strain CY4 but glr::TRP1 | 15 |
| CY97 | As in strain CY4 but gsh2::HIS3 | 18 |
| JL3 | As in strain CY4 but gsh1::LEU2 | J.-C. Lee |
| LM1 | MATa leu2-3 leu2-112 his3-Δ1 ura3-52 trp1-289 tdh1::URA3 | 33 |
| LM5 | MATa leu2-3 leu2-112 his3-Δ1 ura3-52 trp1-289 tdh2::URA3 | 33 |
| LM17 | MATα leu2-3 leu2-112 his3-Δ1 ura3-52 ade1-101 trp1-289 tdh3::URA3 | 33 |
| LM7 | MATα leu2-3 leu2-112 ura3-52 trp1-289 tdh1::URA3 tdh2::URA3 | 33 |
| LM28 | MATa leu2-3 leu2-112 his3-Δ1 ura3-52 trp1-289 tdh1::URA3 tdh3::URA3 | 33 |
| S173-6B | MATa leu2-3 leu2-112 his3-Δ1 ura3-52 trp1-289 | 33 |
| S173-29A | MATα leu2-3 leu2-112 his3-Δ1 ade1-101 trp1-289 | 33 |
Sensitivity to oxidants.
Dose-response curves were generated by growing cells to the exponential phase (1 × 107 to 2 × 107 cells/ml) in SD medium at 30°C and treating them with 4 mM H2O2 for 1 h. Aliquots of cells were diluted into fresh YEPD medium at 20-min intervals and plated in triplicate on YEPD plates to obtain viable counts after 3 days of growth. Sensitivity to H2O2, tert-butyl hydroperoxide, and diamide was determined by spotting strains onto YEPD plates containing various concentrations of oxidants (18). Cells were grown to stationary phase in YEPD, and 10-μl aliquots of each culture diluted to an absorbance at 600 nm of 2.0 were spotted onto appropriate plates. Sensitivity was determined by comparison of growth to that of the wild-type strain after 3 days.
Analysis of protein S-thiolation.
The analysis of S-thiolation in yeast was adapted from methods described for mammalian cells (43, 46). Yeast cells were grown to exponential phase (1 × 107 to 2 × 107 cells/ml) in minimal SD medium and treated with 50 μg of cycloheximide per ml for 15 min to completely inhibit cytoplasmic protein synthesis (9). The cells were incubated with a final concentration of 0.51 nM l-[35S]cysteine for 1 h to facilitate labelling of the intracellular pool of low-molecular-weight sulfhydryls. They were then washed with SD medium and resuspended in SD medium supplemented with the necessary auxotrophic requirements, as well as H2O2 where desired. Cell extracts were prepared in 20 mM sodium phosphate buffer (pH 7.4) containing 100 mM phenylmethylsulfnonyl fluoride by breaking cells with glass beads, using a Minibead Beater (Biospec Scientific), for 30 s at 4°C. Parallel extracts were prepared in buffer containing either 50 mM N-ethylmaleimide (NEM) to prevent thiolation during the sample preparation or 25 mM dithiothreitol (DTT) to reduce any S-thiolated proteins. To quantify protein S-thiolation, aliquots of cell extracts were precipitated on Whatman GF/C glass microfiber filters with 10% trichloroacetic acid. Radioactive incorporation was measured by scintillation counting, and S-thiolation is expressed as the difference between the NEM- and DTT-treated extracts (cpm per microgram of protein).
Autoradiography, following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), was used to investigate the range of proteins that were S-thiolated. Cell extracts (10 to 50 μg), diluted in Laemmli buffer, were boiled (3 min), adjusted to 40 mM iodoacetamide, and electrophoresed on 4 to 15% polyacrylamide SDS-PAGE minigels (Bio-Rad). For autoradiography, the gels were stained in 0.125% (wt/vol) Coomassie blue–30% (vol/vol) methanol–10% (vol/vol) acetic acid, pretreated with En3Hance (NEN-Life Sciences) as specified by the following manufacturer, and dried before being exposed to film (Hyperfilm N; Amersham-Pharmacia).
HPLC and electrochemical detection of low-molecular-weight sulfhydryls.
The preparation of yeast cell extracts for analysis of low-molecular-weight sulfhydryls was based on that described for tissue extracts (20). Cell pellets were broken in ice-cold 0.6 N perchloric acid containing 2 mM EDTA by vortexing with glass beads. Following cell breakage, proteins were precipitated by incubating on ice for 15 min and collected by centrifugation in a microcentrifuge for 15 min at 4°C. The resulting supernatants were diluted in a mobile phase before being separated by reverse-phase chromatography on a C18 column (5 μm; 4.5 by 250 mm; Shperisorb octadecyl silane 2) under isocratic conditions with 0.1 M KH2PO4–0.35% (vol/vol) acetonitrile (13) at a flow rate of 0.9 ml/min. To identify the range of low-molecular-weight thiols involved in protein S-thiolation, the protein precipitates were treated with 25 mM DTT at 37°C for 20 min and the resulting supernatants were analyzed by high-performance liquid chromatography (HPLC). Compounds were identified based on known sulfhydryl standards including GSH, γ-Glu-Cys, and cysteine, which were measured by electrochemical detection with a Coulochem II (ESA) detector (E = + 550 mV).
GAPDH enzyme assays.
GAPDH activity was measured by the method of McAlister and Holland (32) and is expressed as micromoles of NADH formed per minute per microgram of protein.
Western blot analysis.
Protein extracts (20 μg/lane) were electrophoresed under reducing conditions on 4 to 15% polyacrylamide gradient SDS-PAGE minigels and electroblotted onto a polyvinylidene difluoride membrane (NEN Research Products, Boston, Mass.). The blot was incubated in 2 μg of anti-GAPDH monoclonal antibody (MAB374; Chemicon International Inc., Temecula, Calif.) per ml. Bound antibody was visualized by chemiluminescence (Renaissance; NEN Research Products) following incubation of the blot in rabbit anti-mouse immunoglobulin-horseradish peroxidase conjugate (1/2,000 dilution; DakoPatts, Carpinteria, Calif.).
RESULTS
Characterization of protein S-thiolation in yeast.
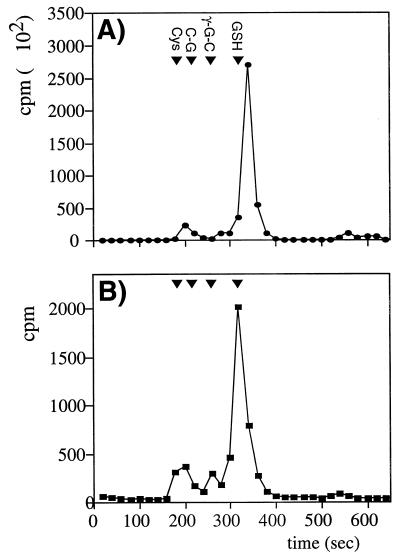
We first established that the intracellular pool of sulfhydryls in yeast could be radiolabelled by incubation with l-[35S]cysteine as described in Materials and Methods. Low-molecular-weight sulfhydryls were separated by HPLC, and radioactivity was analyzed to determine the fate of the radiolabel (Fig. 1A). The major peak comigrated with GSH, accounting for greater than 75% of the radioactivity detected. In addition, two smaller peaks that comigrated with cysteine and γ-glutamylcysteine were detected.
FIG. 1.
HPLC separation of low-molecular-weight sulfhydryls. (A) Sulfhydryls were separated by HPLC (see Materials and Methods) following radiolabelling of the intracellular pool with [35S]cysteine. The retention times of the known standards cysteine (Cys), cysteinylglycine (C-G), γ-glutamylcysteine (γ-G-C), and glutathione (GSH) are shown. (B) Sulfhydryls bound to proteins by S-thiolation were released by treatment with DTT and separated by HPLC.
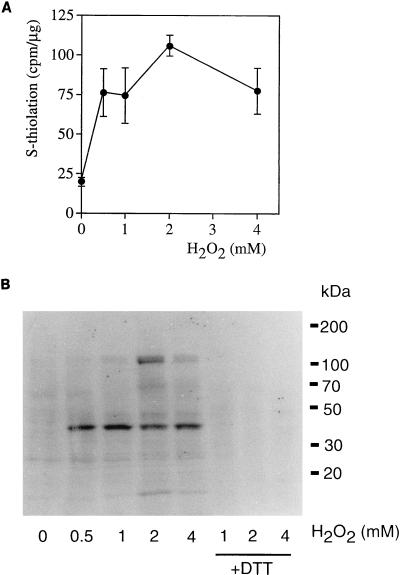
To characterize the effect of oxidative stress on protein S-thiolation, a dose-response curve to H2O2 was generated by challenging labeled cells with H2O2 at concentrations between 0.5 and 4 mM for 30 min (Fig. 2A). Cell extracts were prepared in the presence of either NEM to prevent S-thiolation during the extraction procedure or DTT to reduce any S-thiolated proteins, and the level of S-thiolation was quantified as the difference between the NEM- and DTT-prepared extracts. The basal level of S-thiolation was very low in unstressed cells but increased at all concentrations of H2O2 tested (Fig. 2A). The maximum level of S-thiolation was observed at 2 mM H2O2, which was chosen for all subsequent thiolation experiments. To determine the number and range of proteins that are S-thiolated, total-cell extracts were examined by SDS-PAGE and autoradiography. As expected from the radioactive counts, few S-thiolated proteins were detected in unstressed cells (Fig. 2B) whereas several proteins (e.g., of 45, 70, and 120 kDa) were detected following treatments with H2O2, including a prominent protein at approximately 38 kDa. Radioactivity incorporation was confirmed to occur as a result of S-thiolation since it was reversed by treatment with the reducing agent DTT (Fig. 2B, lanes +DTT).
FIG. 2.
Protein S-thiolation following treatment with hydrogen peroxide. Following radiolabelling of the intracellular pool of low-molecular-weight sulfhydryls, cells were treated with various concentrations of H2O2 for 30 min. (A) Protein S-thiolation increased at all concentrations of H2O2 tested. (B) Proteins were separated by SDS-PAGE and analyzed by autoradiography. The cell extracts challenged with 1, 2, and 4 mM H2O2 were treated with 25 mM DTT to reduce S-thiolated proteins (lanes +DTT). Molecular mass markers are indicated in kilodaltons.
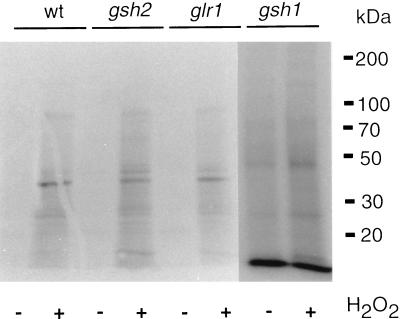
To determine the role of GSH in protein S-thiolation, we examined yeast strains in which GSH1, encoding γ-glutamylcysteine synthetase, GSH2, encoding glutathione synthetase, or GLR1, encoding glutathione reductase, was deleted. The gsh1 mutant is completely devoid of GSH, whereas, the gsh2 mutant lacks GSH but can still make the dipeptide γ-glutamylcysteine (γ-Glu-Cys) intermediate in GSH synthesis (18). The glr1 mutant contains a lowered GSH redox ratio (GSH-to-GSSG ratio) due to the inability to recycle oxidized GSSG to the reduced GSH form (15). A different pattern of protein S-thiolation was observed in the gsh1 mutant compared to the wild-type strain, with several novel proteins labelled during both stressed and nonstressed conditions (Fig. 3). In contrast, the pattern of protein S-thiolation in the glr1 and gsh2 mutants was similar to that in the wild-type strain. The role of GSH was further analyzed by determining the range of low-molecular-weight sulfhydryls that could participate in protein S-thiolation. The proteins were treated with the reducing agent DTT following the S-thiolation reaction, and released thiols were examined by HPLC analysis (Fig. 1B). A major peak corresponding to GSH was detected, indicating that GSH is the main thiol compound that undergoes S-thiolation. In addition, smaller peaks corresponding to cysteine and γ-Glu-Cys were detected. Having established the range and extent of protein S-thiolation, we turned our attention to the identification of the most prominent, 38-kDa, protein detected.
FIG. 3.
Protein S-thiolation in mutants affected in GSH metabolism. Protein S-thiolation was analyzed following treatment with 2 mM H2O2 for 30 min in the wild-type (wt) strain (CY4) and strains with GSH1 (γ-glutamylcysteine synthetase), GSH2 (glutathione synthetase), and GLR1 (glutathione reductase) deleted.
Identification of GAPDH as the major S-thiolated protein in yeast.
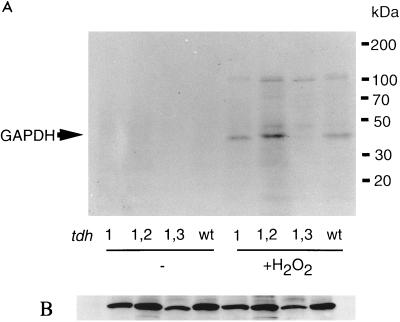
Given the size of the 38-kDa protein and the fact that GAPDH has been identified as a target of S-thiolation in human endothelial cells and blood monocytes (39, 42), we investigated whether the 38-kDa yeast protein is GAPDH. Three unlinked genes (TDH1, TDH2, and TDH3) encode isoenzymes of GAPDH in yeast (33). Tdh2 and Tdh3 are highly homologous (98% similarity, 96% identity), whereas Tdh1 is somewhat less conserved (94% similarity and 88% identity to Tdh3). Strains lacking both TDH2 and TDH3 are inviable, indicating that at least one of these genes must be present for normal cell growth (33). Therefore, we examined protein S-thiolation in mutants with TDH1, TDH1 and TDH2, or TDH1 and TDH3 deleted. The 38-kDa S-thiolated protein was detected in strains that contain Tdh3 (tdh1 tdh2) but not in strains that contain Tdh2 (tdh1 tdh3), indicating that Tdh3 is the 38-kDa S-thiolated protein (Fig. 4A). We confirmed the identity of the 38-kDa protein as GAPDH by immunoprecipitation with an anti-GAPDH antibody (data not shown). This result is surprising given the high degree of sequence homology between Tdh2 and Tdh3. In addition to the overall sequence conservation, the yeast GAPDH proteins contain two active-site cysteine residues in a conserved pentapeptide (C-T-T-N-C), which could serve as possible sites for S-thiolation following an oxidative stress. This difference in protein S-thiolation could not be accounted for by differences in the relative levels of the Tdh2 and Tdh3 polypeptides (Fig. 4B) (32). In addition, the Western blot analysis in Fig. 4B demonstrates that treatment with cycloheximide or H2O2 did not affect the levels of the various Tdh polypeptides. To determine whether S-thiolation of Tdh3 but not Tdh2 is physiologically relevant, we next examined the effects of oxidative stress on mutants lacking GAPDH isoenzymes.
FIG. 4.
Protein S-thiolation in tdh mutants. (A) Protein S-thiolation was analyzed following treatment with 2 mM H2O2 for 30 min in the wild-type (wt) strain (6B) and strains with TDH1, TDH1 and TDH2, or TDH1 and TDH3, all encoding GAPDH, deleted. (B) Western blot analysis probed with anti-GAPDH antibody.
Sensitivity of tdh mutants to oxidants.
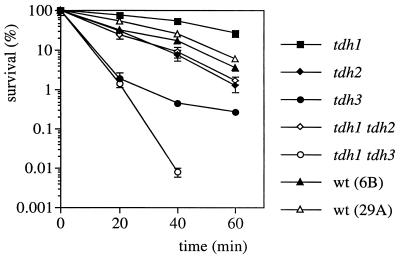
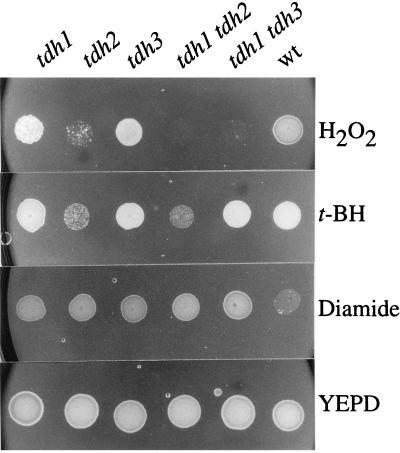
If S-thiolation of Tdh3 serves to protect the GAPDH enzyme from irreversible oxidation, we reasoned that strains lacking TDH3 should be sensitive to oxidants. This was tested by treating the various mutant strains with 4 mM H2O2 for 1 h while monitoring cell viability (Fig. 5). As predicted, strains lacking TDH3 (tdh3 and tdh1 tdh3 strains) were extremely sensitive to H2O2 compared to the wild-type controls. The strain lacking both TDH1 and TDH3 was the most sensitive, with no survivors detected after 40 min of treatment. In this experiment, the H2O2 stress was relieved by diluting the cells into YEPD medium before plating to determine viable counts. We next examined the effect of a constant exposure to oxidants by growing the strains to stationary phase and spotting them onto plates containing concentrations of H2O2 (4.5 mM) or tert-butyl hydroperoxide (2.5 mM) that allowed the wild-type control to grow (Fig. 6). We reasoned that strains containing Tdh3 as the major GAPDH enzyme (tdh2 and tdh1 tdh2 strains) would be unable to grow if S-thiolation inhibits the activity of Tdh3. In agreement with this prediction, the growth of the tdh1 and tdh3 mutants (contain Tdh2) was unaffected in the presence of hydroperoxides whereas the tdh2 and tdh1 tdh2 mutants did not grow. Similarly, the tdh1 tdh3 double mutant was able to grow in the presence of tert-butyl hydroperoxide but, in contrast, was unable to grow in the presence of H2O2, indicating that Tdh2 may also play some role during continued exposure to H2O2 stress. In addition, none of the tdh mutants were sensitive to the thiol oxidant diamide (Fig. 6), in agreement with the finding that it does not promote protein S-thiolation (data not shown). However, all of the tdh mutants were more resistant to diamide than were the wild-type control, similar to the findings that mutants altered in sulfhydryl regulation, including the GSH, glutaredoxin, and thioredoxin systems, are also resistant to this thiol oxidant (18, 31, 36). We next examined the effect of H2O2 stress on GAPDH enzyme activity.
FIG. 5.
Dose-response curves of tdh mutants to hydrogen peroxide. The wild-type (wt) strains 6B and 29A and strains with TDH1, TDH2, TDH3, TDH1 and TDH2, or TDH1 and TDH3 deleted were grown to exponential phase in SD medium and treated with 4 mM H2O2 for 1 h. The cells were diluted and plated in triplicate onto YEPD medium to monitor cell viability at 20-min intervals. Percent survival is expressed relative to the untreated control cultures (100%).
FIG. 6.
Sensitivity of tdh mutants to a continued exposure to oxidants. Sensitivity to H2O2 (4.5 mM), tert-butyl hydroperoxide (t-BH) (2.5 mM), and diamide (3 mM) was determined by spotting strains onto YEPD plates containing the appropriate oxidants. Cultures of the wild type (wt) (6B) and mutants with TDH1, TDH2, TDH3, TDH1 and TDH2, or TDH1 and TDH3 deleted were grown to stationary phase in YEPD and diluted to an absorbance at 600 nm of 2.0, and 10-μl aliquots were spotted onto appropriate plates. The plates were incubated at 30°C for 2 days before growth was scored.
Inhibition and recovery of GAPDH enzyme activity following exposure to H2O2 stress.
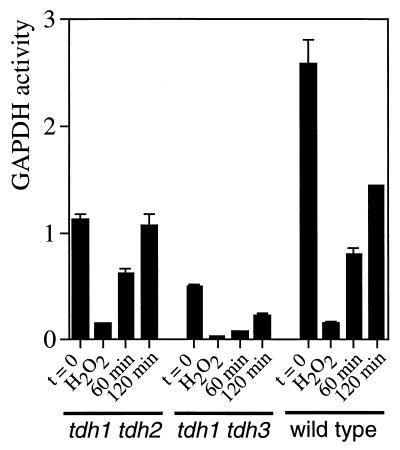
The tdh1 tdh2, tdh1 tdh3, and wild-type strains were grown to exponential phase and treated with 2 mM H2O2 for 30 min before GAPDH activity was determined (Fig. 7). Prior to the peroxide treatment, the tdh1 tdh2 and tdh1 tdh3 mutants were found to contain approximately 44 and 19% of the GAPDH activity found in the wild-type strain. Following treatment with H2O2, GAPDH activity was inhibited to approximately 6 to 13% of its initial activity in all three strains tested. This indicates that both the Tdh2 and Tdh3 GAPDH isoenzymes are extremely sensitive to inactivation by oxidation. However, this may either arise due to irreversible oxidation or due to S-thiolation of the enzyme active site, both of which would inhibit GAPDH activity. To distinguish between these possibilities, cells were transferred into fresh SD medium to monitor the recovery of GAPDH activity once the H2O2 stress was relieved. The tdh1 tdh2 mutant, which contains the S-thiolatible Tdh3 isoenzyme, recovered GAPDH activity to 55 and 95% of the starting activity following a 1- and 2-h growth period, respectively. In contrast, the tdh1 tdh3 mutant recovered only 45% activity after 2 h. The wild-type strain contained 56% activity following the 2-h recovery period, presumably reflecting the presence of both the Tdh2 and Tdh3 polypeptides.
FIG. 7.
Inhibition and recovery of GAPDH enzyme activity following exposure to H2O2 stress. GAPDH enzyme activity (μmoles per minute per microgram) was determined in the wild-type strain (6B) and in tdh1 tdh2 and tdh1 tdh3 mutants grown to exponential phase in SD medium (t = 0). Cells were treated with 2 mM H2O2 for 30 min, and aliquots were taken to determine GAPDH activity (H2O2). The cells were washed and resuspended in fresh SD medium to monitor the recovery of GAPDH activity for 60 or 120 min.
DISCUSSION
Hydrogen peroxide is a ubiquitous molecule that is formed as a product of many oxidase-mediated reactions and of auto-oxidation of hemoproteins and flavoproteins (38). As well as being both freely diffusible and reactive itself, H2O2 must be removed from cells to avoid formation of the highly reactive hydroxyl radical. Detoxification can be mediated by catalases, which catalyze its breakdown into H2O and O2, and by glutathione peroxidases, which utilize GSH as a reductant. In yeast, GSH-mediated reactions appear to be the primary means of detoxifying this ROS, although catalases appear to provide an overlapping defense system during particular growth conditions (16, 26). In addition to the oxidant sensitivity of gsh1 and glr1 mutants (15, 17), H2O2 was found to increase the levels of oxidized (GSSG), protein-bound (GSSP), and extracellular GSH at the expense of intracellular GSH (16a). In the present study, we have shown that the levels of GSSP are increased due to modification of specific target proteins and that Tdh3 is the major S-thiolated protein.
Most of the S-thiolated proteins detected were formed due to reaction with GSH, since this was the most abundant sulfhydryl released by reaction with DTT, and the gsh1 mutant, which is totally devoid of GSH (17), contained an altered pattern of S-thiolation. Interestingly, the dipeptide γ-Glu-Cys may be able to replace GSH in the S-thiolation reaction, since the pattern observed in the gsh2 mutant was similar to that seen in the wild type. We have previously shown that the gsh2 mutant is devoid of GSH but contains elevated levels of the dipeptide γ-Glu-Gys, which can substitute for GSH as an antioxidant in yeast (18). The fact that the wild-type pattern of S-thiolation was found in the glr1 mutant, which contains elevated levels of GSSG (15, 19), argues against a mechanism that proceeds via a thiol-disulfide exchange reaction with the oxidized disulfide form of GSH (GSSG). Similarly, the 2 mM H2O2 treatment used for these experiments did not cause any increase in the level of GSSG (data not shown).
GAPDH was identified as the most abundant S-thiolated protein in yeast following a challenge with H2O2. Previous studies with mammalian cells have also identified GAPDH as a target of S-thiolation, but the studies presented here show for the first time that this is a regulated process and that it affects survival during conditions of oxidative stress. GAPDH is a glycolytic enzyme that is active as a tetramer of identical 37-kDa subunits catalyzing the conversion of glyceraldehyde-3-phosphate to 1,3-diphosphoglycerate. It has been extensively characterized in S. cerevisiae, in which it is one of the most abundant soluble proteins and is encoded by three unlinked genes designated TDH1, TDH2, and TDH3 (1, 22). McAlister and Holland reported the isolation of yeast mutants lacking combinations of the GAPDH isoenzymes (33). None of the TDH genes were individually essential for cell viability, but a functional copy of either TDH2 or TDH3 was required since tdh2 tdh3 double mutants were inviable. These results implied that Tdh1 may perform a function distinct from that of Tdh2 or Tdh3, since Tdh1 alone was unable to support growth. Both the Tdh2 and Tdh3 polypeptides are able to form catalytically active homotetramers, but the apparent Vmax of the Tdh3 homotetramer is approximately two- to threefold lower than that of the Tdh2 homotetramer (32). Interestingly, two forms of the Tdh3 polypeptide that differ in their isoelectric charge were detected by two-dimensional PAGE and may represent catalytically different forms of the GAPDH tetramer (32). The two forms of Tdh3 polypeptide did not arise as a result of differential phosphorylation, and it is tempting to speculate that they arose due to differences in protein S-thiolation. Therefore, the relatively low Vmax of Tdh3 may be due to the S-thiolated and hence inactivated form of Tdh3.
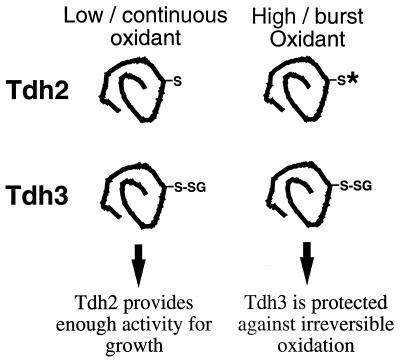
Given that the Tdh2 and Tdh3 polypeptides have 98% homology and are functionally redundant for glycolysis, it is surprising that they differ in protein S-thiolation. Both Tdh2 and Tdh3 GAPDH enzyme activities were inactivated by exposure to H2O2. However, Tdh3 (thiolated) activity was recovered within 2 h after the removal of the H2O2 stress, suggesting that the oxidative inactivation was reversible. In contrast, Tdh2 (nonthiolated) activity was restored to only 45% of the initial activity during the 2-h recovery period. The S-thiolation of Tdh3 appears to be physiologically relevant since strains lacking TDH3 were hypersensitive to a lethal dose of H2O2 compared to the wild type and tdh2 strains. Thus, S-thiolation, and hence protection of Tdh3 against irreversible oxidation, was required for survival following a challenge with H2O2. However, the nonthiolated Tdh2 polypeptide is also necessary during exposure to oxidative-stress conditions, since it was required for growth during the continued presence of oxidants. Our model to explain these results postulates that during a prolonged exposure to low levels of oxidants, the continued S-thiolation of Tdh3 would negatively affect cell growth, but during such conditions, enough of the Tdh2 enzyme may avoid oxidation to provide the necessary GAPDH activity for growth. At higher concentrations of oxidants, which would irreversibly oxidize the Tdh2 polypeptide, the Tdh3 polypeptide could be protected by S-thiolation until such time as the oxidative pressure is removed (Fig. 8).
FIG. 8.
Model for the oxidant sensitivity of tdh mutants. During continuous or low exposure to oxidants, Tdh3 is S-thiolated (-SG), inhibiting GAPDH activity. Under these conditions, Tdh2 must provide enough GAPDH activity for growth. Following an exposure to high concentrations of oxidant, Tdh2 is irreversibly oxidized (*). Tdh3 is protected by protein S-thiolation until such time as the oxidant pressure is removed and the cells can resume growth.
In mammalian cells, the glycolytic synthesis of ATP is inactivated by conditions of oxidative stress, mainly at the level of GAPDH, due to three independent effects (7, 25): (i) direct inactivation of the enzyme active site, (ii) a decrease in the cofactor NAD, and (iii) a shift in the cytosolic pH from the enzyme optima (7, 25). Thus, protection of the GAPDH active site by S-thiolation would protect against oxidative inactivation of this crucial glycolytic enzyme. In addition, it has been proposed that blocking glycolysis at the GAPDH step would be beneficial during conditions of oxidative stress since it would result in an increased flux of glucose equivalents through the pentose phosphate pathway, leading to the generation of NADPH (39, 42). Such NADPH could provide reducing power for antioxidant enzymes including catalases and the GSH-GSH peroxidase system (21, 30). Evidence is also accumulating that GAPDH functions in processes unrelated to glycolysis, including, DNA repair and replication, translational control of gene expression, and endocytosis and in a plasma membrane oxidoreductase complex (reviewed in references 2 and 45). It is unknown whether S-thiolation of GAPDH affects its activity in these various processes.
In addition to its role in protecting individual protein -SH groups from oxidation, and given the high concentrations (millimolar) of sulfhydryl groups in the cell, thiolation has been suggested to serve as a general antioxidant defense system analogous to free-radical scavengers (47). However, this seems unlikely from the present data, since the tdh3 mutant, lacking the most abundant S-thiolated protein, did not lead to increases in the level of thiolation of existing proteins or the appearance of new S-thiolated proteins. Rather, the consistent pattern of labelling indicates that S-thiolation is a highly controlled form of posttranslational modification resulting from an oxidant challenge. This modification may serve to protect the cysteine residues of particular proteins from oxidation or, alternatively, may provide a novel form of protein regulation. Two recent reports have indicated a role for reversible thiolation in the regulation of enzyme activity. First, S-thiolation was implicated in the regulation of the HIV-1 protease under the control of glutaredoxin (11); second, S-thiolation was shown to regulate the E1 and E2 ubiquitin-conjugating enzymes in bovine retina cells (28). Here, we have demonstrated that the activity of GAPDH in protection against oxidative stress is regulated by protein S-thiolation and that this process is physiologically important for survival during conditions of oxidative stress.
ACKNOWLEDGMENTS
We thank Michael Holland for the GAPDH mutants and Daniel Gonzalbo for the anti-GAPDH antibody.
REFERENCES
- 1.Boucherie H, Bataille N, Fitch I T, Perrot M, Tuite M F. Differential synthesis of glyceraldehyde-3-phosphate dehydrogenase polypeptides in stressed yeast cells. FEMS Microbiol Lett. 1995;125:127–134. doi: 10.1111/j.1574-6968.1995.tb07348.x. [DOI] [PubMed] [Google Scholar]
- 2.Bulliard C, Zurbiggen R, Tornare J, Faty M, Dastoor Z, Dreyer J-L. Purification of a dichlorophenol-indophenol oxidoreductase from rat and bovine synaptic membranes: tight complex association of a glyceraldehyde-3-phosphate dehydrogenase isoform, TOAD64, enolase-γ and aldolase C. Biochem J. 1997;324:555–563. doi: 10.1042/bj3240555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chai Y-C, Hendrich S, Thomas A J. S-thiolation of individual neutrophil proteins including actin by stimulation of the respiratory burst: evidence against a role for glutathione disulphide. Arch Biochem Biophys. 1994;310:273–281. doi: 10.1006/abbi.1994.1167. [DOI] [PubMed] [Google Scholar]
- 4.Chai Y-C, Jung C-H, Lii C K, Ashraf S S, Hendrich S, Wolf B, Sies H, Thomas J A. Identification of an S-thiolated rat liver protein as carbonic anhydrase III: characterization of S-thiolation and dethiolation reactions. Arch Biochem Biophys. 1991;284:270–278. doi: 10.1016/0003-9861(91)90295-t. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri B, Ingavale S, Bachhawat A K. apd1+, a gene required for red pigment formation in ade6 mutants of Schizosaccharomyces pombe, encodes an enzyme required for glutathione biosynthesis: a role for glutathione and a glutathione-conjugate pump. Genetics. 1997;145:75–83. doi: 10.1093/genetics/145.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coan C, Ji J-Y, Hideg K, Mehlhorn R J. Protein sulfhydryls are protected from irreversible oxidation by conversion to mixed disulfides. Arch Biochem Biophys. 1992;295:369–378. doi: 10.1016/0003-9861(92)90530-a. [DOI] [PubMed] [Google Scholar]
- 7.Cochrane C G. Cellular injury by oxidants. Am J Med. 1991;91(Suppl. 3C):23S–30S. doi: 10.1016/0002-9343(91)90280-b. [DOI] [PubMed] [Google Scholar]
- 8.Collison M, Thomas J A. S-thiolation of cytoplasmic cardiac creatine kinase in heart cells treated with diamide. Biochim Biophys Acta. 1987;928:121–129. doi: 10.1016/0167-4889(87)90112-1. [DOI] [PubMed] [Google Scholar]
- 9.Cooper T G, Bossinger J. Selective inhibition of protein synthesis initiation in Saccharomyces cerevisiae by low concentrations of cycloheximide. J Biol Chem. 1976;251:7278–7286. [PubMed] [Google Scholar]
- 10.Cotgreave I A, Gerdes R G. Recent trends in glutathione biochemistry-glutathione-protein interactions: a molecular link between oxidative stress and cell proliferation? Biochem Biophys Res Commun. 1998;242:1–9. doi: 10.1006/bbrc.1997.7812. [DOI] [PubMed] [Google Scholar]
- 11.Davis D A, Newcomb F M, Starke D W, Ott D E, Mieyal J J, Yarchoan R. Thioltransferase (glutaredoxin) is detected within HIV-1 and can regulate the activity of glutathionylated HIV-1 protease in vitro. J Biol Chem. 1997;272:25935–25940. doi: 10.1074/jbc.272.41.25935. [DOI] [PubMed] [Google Scholar]
- 12.Dean R T, Fu S, Stocker R, Davies M J. Biochemistry and pathology of radical-mediated protein oxidation. Biochem J. 1997;324:1–18. doi: 10.1042/bj3240001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudman N P B, Guo X W, Crooks R, Xie L J, Silverberg J S. Assay of plasma homocysteine—light sensitivity of the fluorescent 7-benzo-2-oxa-1,3,diazole-4-sulfonic acid derivative, and use of appropriate calibrators. Clin Chem. 1996;42:2028–2032. [PubMed] [Google Scholar]
- 14.Evans M V, Turton H E, Grant C M, Dawes I W. Toxicity of linoleic acid hydroperoxide to Saccharomyces cerevisiae: Involvement of a respiration-related process for maximal sensitivity and adaptive response. J Bacteriol. 1997;180:483–490. doi: 10.1128/jb.180.3.483-490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant C M, Collinson L P, Roe J-H, Dawes I W. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol Microbiol. 1996;21:171–179. doi: 10.1046/j.1365-2958.1996.6351340.x. [DOI] [PubMed] [Google Scholar]
- 16.Grant C M, Dawes I W. Synthesis and role of glutathione in protection against oxidative stress in yeast. Redox Rep. 1996;2:223–229. doi: 10.1080/13510002.1996.11747054. [DOI] [PubMed] [Google Scholar]
- 16a.Grant C M, Perrone G, Dawes I W. Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;253:893–898. doi: 10.1006/bbrc.1998.9864. [DOI] [PubMed] [Google Scholar]
- 17.Grant C M, MacIver F H, Dawes I W. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr Genet. 1996;29:511–515. doi: 10.1007/BF02426954. [DOI] [PubMed] [Google Scholar]
- 18.Grant C M, MacIver F H, Dawes I W. Glutathione synthetase is dispensable for growth under both normal and oxidative stress conditions in the yeast Saccharomyces cerevisiae due to an accumulation of the dipeptide gamma-glutamylcysteine. Mol Biol Cell. 1997;8:1699–1707. doi: 10.1091/mbc.8.9.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant C M, MacIver F M, Dawes I W. Stationary phase induction of GLR1 expression is mediated by the yAP-1 transcriptional protein in Saccharomyces cerevisiae. Mol Microbiol. 1996;22:739–746. doi: 10.1046/j.1365-2958.1996.d01-1727.x. [DOI] [PubMed] [Google Scholar]
- 20.Harvey P R C, Ilson R G, Strasberg S M. The simultaneous determination of oxidized and reduced glutathione in liver tissues by ion pairing reverse phase high performance liquid chromatography with a coulometric electrochemical detector. Clin Chim Acta. 1989;180:203–212. doi: 10.1016/0009-8981(89)90001-6. [DOI] [PubMed] [Google Scholar]
- 21.Hillar A, Nicholls P, Switala J, Loewen P C. NADPH binding and control of catalase compound II formation: comparison of bovine, yeast, and Escherichia coli enzymes. Biochem J. 1994;300:531–539. doi: 10.1042/bj3000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holland J P, Holland M J. Structural comparison of two non-tandemly repeated glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1980;255:2596–2605. [PubMed] [Google Scholar]
- 23.Huppa J B, Ploegh H L. The eS-Sence of -SH in the ER. Cell. 1998;92:145–148. doi: 10.1016/s0092-8674(00)80907-1. [DOI] [PubMed] [Google Scholar]
- 24.Hwang C, Sinskey A J, Lodish H F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 25.Hyslop P A, Hinshaw D B, Halsey W A, Schraufstatter I U, Sauerheber R D, Spragg R G, Jackson J H, Cochrane C G. Mechanisms of oxidant-mediated injury. J Biol Chem. 1988;263:1665–1675. [PubMed] [Google Scholar]
- 26.Izawa S, Inoue Y, Kimura A. Importance of catalase in the adaptive response to hydrogen peroxide: analysis of acatalasaemic Saccharomyces cerevisae. Biochem J. 1996;320:61–67. doi: 10.1042/bj3200061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izawa S, Inoue Y, Kimura A. Oxidative stress in yeast: effect of glutathione on adaption to hydrogen peroxide stress in Saccharomyces cerevisiae. FEBS Lett. 1995;368:73–76. doi: 10.1016/0014-5793(95)00603-7. [DOI] [PubMed] [Google Scholar]
- 28.Jahngen-Hodge J, Obin M S, Gong X, Shang F, Nowell T R, Gong J, Abasi H, Blumberg J, Taylor A. Regulation of ubiquitin-conjugating enzymes by glutathione following oxidative stress. J Biol Chem. 1997;272:28218–28226. doi: 10.1074/jbc.272.45.28218. [DOI] [PubMed] [Google Scholar]
- 29.Jung C-H, Thomas J A. S-Glutathiolated hepatocyte proteins and insulin disulfides as substrates for reduction by glutaredoxin, thioredoxin, protein disulfide isomerase, and glutathione. Arch Biochem Biophys. 1996;335:61–72. doi: 10.1006/abbi.1996.0482. [DOI] [PubMed] [Google Scholar]
- 30.Kirkman H N, Galiano S, Gaetani G F. The function of catalase-bound NADPH. J Biol Chem. 1987;262:660–666. [PubMed] [Google Scholar]
- 31.Luikenhuis S, Dawes I W, Grant C M. The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol Biol Cell. 1997;9:1081–1091. doi: 10.1091/mbc.9.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McAlister L, Holland M J. Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1985;260:15019–15027. [PubMed] [Google Scholar]
- 33.McAlister L, Holland M J. Isolation and characterization of yeast strains carrying mutations in the glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1985;260:15013–15018. [PubMed] [Google Scholar]
- 34.Meister A. Glutathione metabolism and its selective modification. J Biol Chem. 1988;263:17205–17208. [PubMed] [Google Scholar]
- 35.Meister A, Anderson M E. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 36.Muller E G D. A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol Biol Cell. 1996;7:1805–1813. doi: 10.1091/mbc.7.11.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park E M, Thomas J A. S-thiolation of creatine kinase and glycogen phosphorylase b initiated by partially reduced oxygen species. Biochem Biophys Acta. 1988;964:151–160. doi: 10.1016/0304-4165(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 38.Penninckx M J, Elskens M T. Metabolism and functions of glutathione in micro-organisms. Adv Microb Physiol. 1993;34:239–301. doi: 10.1016/s0065-2911(08)60031-4. [DOI] [PubMed] [Google Scholar]
- 39.Ravichandran V, Seres T, Moriguchi T, Thomas J A, Johnston R B. S-thiolation of glyceraldehyde-3-phosphate dehydrogenase induced by the phagocytosis-associated respiratory burst in blood monocytes. J Biol Chem. 1994;269:25010–25015. [PubMed] [Google Scholar]
- 40.Rokutan K, Thomas J A, Johnson R B. Phagocytosis and stimulation of the respiratory burst by phorbol diester initiate S-thiolation of specific proteins in macrophages. J Immunol. 1991;147:260–264. [PubMed] [Google Scholar]
- 41.Schuppe I, Moldeus P, Cotgreave I A. Protein-specific S-thiolation in human endothelial cells during oxidative stress. Biochem Pharmacol. 1992;44:1757–1764. doi: 10.1016/0006-2952(92)90069-u. [DOI] [PubMed] [Google Scholar]
- 42.Schuppe-Koistinen I, Moldeus P, Bergman T, Cotgreave I A. S-thiolation of human endothelial cell glyceraldehyde-3-phosphate dehydrogenase after hydrogen peroxide treatment. Eur J Biochem. 1994;221:1033–1037. doi: 10.1111/j.1432-1033.1994.tb18821.x. [DOI] [PubMed] [Google Scholar]
- 43.Seres T, Ravichandran V, Moriguchi T, Rokutan K, Thomas J A, Johnston R B. Protein S-thiolation and dethiolation during the respiratory burst in human monocytes. J Immunol. 1996;156:1973–1980. [PubMed] [Google Scholar]
- 44.Sies H. Strategies of antioxidant defense. Eur J Biochem. 1993;215:213–219. doi: 10.1111/j.1432-1033.1993.tb18025.x. [DOI] [PubMed] [Google Scholar]
- 45.Sirover M A. Emerging new functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. Life Sci. 1996;58:2271–2277. doi: 10.1016/0024-3205(96)00123-3. [DOI] [PubMed] [Google Scholar]
- 46.Thomas J A, Chai Y-C, Jung C-H. Protein S-thiolation and dethiolation. Methods Enzymol. 1994;233:385–395. doi: 10.1016/s0076-6879(94)33045-x. [DOI] [PubMed] [Google Scholar]
- 47.Thomas J A, Poland B, Honzatko R. Protein sulphydryls and their role in the antioxidant function of protein S-thiolation. Arch Biochem Biophys. 1995;319:1–9. doi: 10.1006/abbi.1995.1261. [DOI] [PubMed] [Google Scholar]
- 48.Turton H E, Dawes I W, Grant C M. Saccharomyces cerevisiae exhibits a yAP-1-mediated adaptive response to malondialdehyde. J Bacteriol. 1997;179:1096–1101. doi: 10.1128/jb.179.4.1096-1101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]