Abstract
Interleukin-2 (IL-2) responsiveness of T lymphocytes is controlled through transcription of the IL-2 receptor (IL-2R) α subunit by antigen and by IL-2 itself. IL-2 induces IL-2Rα transcription via an IL-2-responsive enhancer (IL-2rE), whose activity depends on the cooperative binding of IL-2-induced STAT5 to two sites and of constitutively active Elf-1 to a third one. Here we describe the changes in IL-2rE chromatin that occur in normal T lymphocytes upon activation of IL-2Rα expression. In cells induced to transiently express IL-2Rα with concanavalin A (which mimics antigen), none of the IL-2rE sites is occupied despite the presence of Elf-1 and STAT1, which bind to the IL-2rE in vitro. The two STAT binding sites are occupied rapidly upon IL-2 stimulation, concomitantly with STAT5 activation. Occupation of the Elf-1 binding site is delayed, although Elf-1 concentration and binding activity are not modified by IL-2. Digestion of T-cell chromatin with DNase I and micrococcal nuclease shows that IL-2 induces the appearance of nuclease-hypersensitive sites flanking the IL-2rE. Thus IL-2, in addition to activating STAT5, appears to regulate IL-2Rα transcription by making IL-2Rα chromatin accessible to transcription factors.
Interleukin-2 (IL-2) is the principal growth factor for antigen-activated T lymphocytes. It promotes T-cell proliferation by binding to a high-affinity receptor composed of three transmembrane proteins, the α, β, and γc chains (43). The γc chain is shared with the receptors for IL-2, -4, -7, -9, and -15 (22, 33, 50, 54, 55) and is constitutively expressed in mature T cells and their thymic precursors (8, 32, 48). IL-2 receptor β (IL-2Rβ) is present on a subpopulation of resting T cells (51, 62). β and γc chains combine to form an intermediate-affinity IL-2R that can transmit signals (47, 49), but cannot stimulate the proliferation of normal T lymphocytes (7, 38, 59). The α chain is undetectable on resting T cells. Its expression is triggered by antigen (53), a stimulus that can be mimicked by lectins such as concanavalin A (ConA) (31) or by antibodies against the T-cell receptor (TCR) (20). These signals also result in secretion of IL-2, which increases and prolongs IL-2Rα expression (4, 15, 39), thus acting as a positive feedback regulator of its own high-affinity receptor.
IL-2Rα gene expression is regulated mostly through changes in its rate of transcription (13, 34, 52). In transgenic mice bearing a reporter gene under the control of 2.6 kb of 5′ flanking region of the murine IL-2Rα gene, transgene expression is restricted to lymphoid organs (60). In T cells, the transgene can be induced by ConA and IL-2 with kinetics very similar to those of the endogenous gene. The responses of both the human and mouse genes to signals from the TCR depend on cis-acting elements in the promoter-proximal region (1, 5, 10, 28, 61). By transient transfection of IL-2Rα mutant promoter constructs into a rodent T-cell line, PC60, we identified a 51-nucleotide (nt) enhancer necessary and sufficient for the response of the IL-2Rα gene to IL-2 1.3 kb upstream of the transcription start site (61). Subsequently the homologue of this IL-2-responsive enhancer (IL-2rE) was found at around −4 kb in the human IL-2Rα gene (6, 29, 36). The mouse enhancer consists of three separate elements: two binding sites for signal transducers and activators of transcription (STAT) and a consensus motif for Ets family members. In PC60 cells, the STAT motifs (sites I and II) and the Ets binding site (site III) are all required for the response of the enhancer to IL-2. Recently, we have shown that IL-2 stimulation of IL-2rE activity depends on the cooperative binding of two IL-2-induced STAT5 dimers to sites I and II (42). This is concordant with the finding that, in mice lacking STAT5A, IL-2-induced IL-2Rα expression is severely reduced (46). Site II overlaps with a recognition sequence for GATA proteins, but this motif does not appear to be important for IL-2rE function (42). Elf-1, a member of the Ets family of proteins, contributes to IL-2rE activity by binding to site III (58). Elf-1 binding activity to site III is constitutive in PC60 cells.
There is growing evidence that chromatin structure participates in gene regulation, partly by modulating access of transcription factors to DNA (3, 17, 65). Gene activation is often accompanied by perturbation or disruption of the nucleosomes that occupy cis-acting elements. Such an opening of chromatin is generally reflected in a change in its sensitivity to nucleases. Indeed, in mouse T lymphocytes, activation of IL-2Rα expression results in the appearance of a DNase I-hypersensitive site in, or close to, the IL-2rE (60). Induction of DNase I-hypersensitive sites by IL-2 has also been described in the B-cell-specific enhancer of the immunoglobulin J chain gene (30).
Here we present the results of an analysis, by in vivo footprinting and nuclease digestion, of the changes in IL-2rE chromatin during activation of IL-2Rα expression in normal mouse T lymphocytes. We find that protein-DNA interactions on all three elements of the IL-2rE depend on stimulation by IL-2, although Elf-1 binding activity is present in resting T lymphocytes and is not affected by IL-2. Stimulation with ConA, which induces activation of STAT1 but not STAT5, does not result in the occupation of the STAT binding sites in the IL-2rE, even though both site I and site II can bind STAT1 in vitro. Stimulation of ConA-activated cells with IL-2 leads to the appearance of micrococcal nuclease hypersensitive sites flanking the IL-2rE, indicating that IL-2 induces translational positioning of nucleosomes at the borders of the IL-2rE. Our data show that IL-2 regulates the accessibility of the IL-2rE to the transcription factors which control its enhancer activity and that IL-2rE chromatin configuration participates in the regulation of IL-2Rα transcription by IL-2.
MATERIALS AND METHODS
Cell preparation and culture.
Cells from spleens of 3- to 6-month-old DBA/2 mice (Harlan) were prepared by gentle homogenization. To enrich for T cells, B cells and accessory cells were removed by adherence to nylon wool (9). When high purity was required, T cells were purified by incubating cell preparations with anti-Thy1.2 monoclonal antibody (MAb [mouse CD90]) coupled to magnetic microbeads (MACS-r; Miltenyi Biotec GmbH, Bergisch-Gladbach, Germany), followed by two to three rounds of sorting with a magnet cell separation system (MACS-r; Miltenyi Biotec GmbH). Cells were used either directly (resting T lymphocytes) or cultured at 2 × 106 cells/ml for the indicated time with ConA (2.5 μg/ml; Sigma) and human recombinant IL-2 (100 U/ml; kindly provided by Glaxo, Geneva, Switzerland) or ConA with antimouse IL-2 MAb S4B6.1 (66) and antimouse IL-2Rα MAb 5A2 (44). Culture medium was Dulbecco’s modified Eagle’s medium containing 10 mM HEPES, 50 μM 2-mercaptoethanol, and 5% fetal calf serum. For some experiments, IL-2-responsive cells were obtained by stimulating lymphocytes for 24 h with ConA in the presence of the IL-2 blocking antibodies. Cells were then washed in medium containing an excess of IL-2 and subsequently cultured for the indicated time with 1,000 U of IL-2 per ml. Splenic T lymphocytes from 3-month-old gamma interferon receptor (IFN-γR)-deficient mice (eS/eV/129 background) and wild-type eS/eV/129 mice (generously provided by Jacques Louis, Lausanne, Switzerland) were enriched by passage through nylon wool and cultured with ConA and IL-2 as mentioned above. Bone marrow cells were obtained by flushing femur and tibia bones with ice-cold phosphate-buffered saline (PBS) through a 26-gauge needle.
Flow cytometry.
For immunofluorescence analysis of IL-2Rα surface expression, a biotinylated rat anti-mouse IL-2Rα MAb (PC61) (37) was used together with either fluorescein isothiocyanate (FITC)-conjugated anti-CD4 (GK1.5) or anti-CD8 (53.6.7) antibodies alone or with a mixture of both antibodies. PE-conjugated streptavidin (Caltag Laboratories, San Francisco, Calif.) was used as the second-step reagent. The purity of T cells after nylon wool or MACS purification was assessed by staining with FITC-conjugated anti-CD4 plus anti-CD8 antibodies. Samples were analyzed with a FACScan flow cytometer (Becton Dickinson, California) by using the LYSIS II program.
In vivo footprinting analysis.
For dimethyl sulfate (DMS) treatment, 1.5 × 107 cells were resuspended in 1 ml of culture medium, and 40 μl of 0.5 M HEPES (pH 7.5) containing 2.5% DMS (Fluka) was added. The reaction was stopped after 1 min by washing twice with ice-cold PBS containing 2% 2-mercaptoethanol. Cells were lysed in presence of 0.5 mg of proteinase K per ml, and the genomic DNA was isolated. As a naked DNA control, 50 μg of genomic DNA isolated from mouse liver were incubated with 0.5% DMS for 30 s in 10 mM Tris-HCl (pH 7.5). Piperidine (Fluka) cleavage was performed as described in reference 40.
Ligation-mediated PCR (LMPCR) was used to visualize DMS-dependent cleavage sites. The procedure described in reference 19 was used, with the following changes. (i) Annealing of the first primer to genomic DNA was performed at 61°C. (ii) PCR amplification was done through 22 cycles of 1 min at 95°C, 2 min at 64°C, and 3 min at 76°C. Five seconds was added to the 76°C step at each cycle, with the exception of the last cycle, where extension proceeded for 10 min. (iii) End labeling was carried out in a thermal cycler (two cycles of 1 min at 95°C, 2 min at 68°C, and 10 min at 76°C). Reaction mixtures were separated on a 6% polyacrylamide sequencing gel. The primers used for LMPCR of the lower strand were 5′-ACCACCTTCTACTGTTAGAAAGAGC-3′ (primer 1), 5′-CAAAGGCTCACCTCTACCCTAAGAGG-3′ (primer 2), 5′-AAGGCTCACCTCTAACCTAAGAGGAGGC-3′ (primer 3), or 5′-CATAACGTTCTCTTTGCTAAGCGTC-3′ (primer 1′), 5′-TCCCGCTCTTCTTCATCATGATCACTG-3′ (primer 2′), and 5′-CCGCTCTTCTTCATCATGATCACTGGGC-3′ (primer 3′). The unidirectional linker was formed by hybridization of 5′-GCAGTGACTCGAGAGTACTGAGCTC-3′ (linker 1) with 5′-GAGCTCAGTAC-3′ (linker 2).
Protein extracts, electrophoretic mobility shift assays, and Western blots.
Nuclear extracts from fresh and cultured cells were prepared essentially as described previously (56), in the presence of protease inhibitors (1 μg of leupeptin per ml, 1 μg of aprotinin per ml, 1 mM phenylmethylsulfonyl fluoride [PMSF]) and phosphatase inhibitors (25 mM β-glycerol phosphate, 0.1 mM Na3VO3). For STAT proteins, binding reactions were performed in a final volume of 20 μl in binding buffer [10 mM Tris-HCl (pH 7.5), 20 mM KCl, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol (DTT), 1 mg of bovine serum albumin per ml, 0.1 μg of poly(dI-dC) and 1 μg of sonicated salmon sperm DNA] containing 0.5 to 5 μg of nuclear extract and 2 × 104 to 3 × 104 cpm of end-labeled probe. For Elf-1, binding reactions were performed as described previously (58). Reaction mixtures were incubated for 15 min on ice and separated on a 5% nondenaturing polyacrylamide gel in 0.3× Tris-borate buffer. Oligonucleotide probes have been described previously (42, 58).
For Western blots, 5 μg of denatured nuclear extract was separated on a sodium dodecyl sulfate-polyacrylamide (7.5%) gel electrophoresis (SDS-PAGE) gel. Proteins were transferred to nitrocellulose membrane (Amersham Life Science), and Elf-1 was detected with a 1/500 dilution of anti-Elf-1 antibodies (C-20; Santa Cruz Biotechnology, Santa Cruz, Calif.) by the ECL (enhanced chemiluminescence) system (Amersham Life Science) with horseradish peroxidase-coupled goat anti-rabbit antibodies (Jackson ImmunoResearch Laboratories).
DNase I and MNase digestion.
DNase I treatment of cells was performed as described previously (60). For micrococcal nuclease (MNase) digestion, 5 × 107 to 6 × 107 cells were washed with PBS and resuspended in 15 ml of ice-cold solution 1 (15 mM Tris-HCl [pH 7.4], 0.2 mM spermine, 0.5 mM spermidine, 2 mM K-EDTA, 80 mM KCl, 0.5 mM EDTA, 1% thiodiethylene glycol, 1 mM DTT, 0.05% Nonidet P-40) containing freshly added protease inhibitors (0.4 mM PMSF, 3 μg of aprotinin per ml, 0.5 μg of leupeptin per ml, 1 μg of pepstatin per ml). Cells were submitted to 10 strokes of a tight-fitting pestle in a Dounce homogenizer. Nuclei were pelleted by centrifugation for 5 min at 1,000 × g with the brake off, resuspended in 7.5 ml of solution 1 containing the protease inhibitors and 20% glycerol, and Dounce homogenized again (four or five strokes). After centrifugation, the pellet was resuspended in 2 ml of ice-cold solution 2 (7.5 mM Tris-HCl [pH 7.4], 0.1 mM spermine, 0.25 mM spermidine, 40 mM KCl, 5% glycerol, 1% thiodiethylene glycol, 1 mM DTT, 5 mM MgCl2, 1 mM CaCl2) containing the protease inhibitors. Nuclei were aliquoted to 1 × 107 to 2 × 107/tube, and samples were incubated with MNase for 5 min at 25°C. Digestion was stopped by the addition of 3 volumes of SDS buffer (25 mM Tris-HCl, [pH 8.0], 10 mM EDTA, 200 mM NaCl, 0.4% SDS) and 0.5 mg of proteinase K per ml.
Southern blotting.
Forty to fifty micrograms of DNA was digested to completion with the indicated restriction enzymes and electrophoresed in 1 to 1.5% agarose gels in Tris-borate buffer at 40 V. DNA was transferred by capillarity to nylon membranes (Appligene Oncor, Illkirch, France) with 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Probes were prepared with the random priming kit supplied by Boehringer Mannheim, by using IL-2Rα PCR fragments (probe 8 runs from nt −539 to +58 and probe 3 runs from nt −586 to −286 from the transcription start site) as templates. Membranes were hybridized in Church’s buffer at 68°C.
RESULTS
ConA-induced, IL-2-independent IL-2Rα expression does not require activation of STAT proteins.
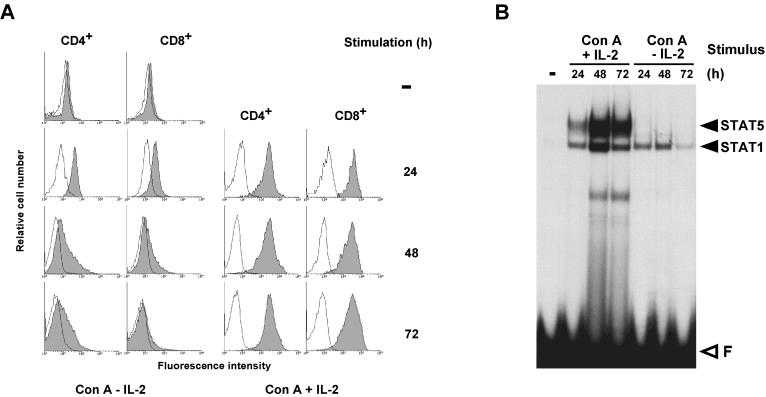
Previously, we showed that ConA or anti-TCR antibodies induce transient IL-2Rα expression on mouse spleen T lymphocytes in the absence of IL-2 stimulation (reference 60 and our unpublished observations). Figure 1A confirms this result and shows, in addition, that both CD4+ and CD8+ T cells are homogeneously IL-2Rα+ after 24 h of culture in ConA only (i.e., in the presence of a mixture of antimouse IL-2 and antimouse IL-2Rα antibodies that prevent auto- or paracrine stimulation by IL-2). In both populations, IL-2Rα expression drops to very low levels during the next 48 h unless the cells are stimulated with IL-2.
FIG. 1.
IL-2 stimulates IL-2Rα expression on CD4+ and CD8+ T lymphocytes. Nylon wool-purified T lymphocytes (>70% CD4+ or CD8+) from mouse spleens were used fresh or after culture for the indicated times either with ConA in the presence of MAbs against mouse IL-2 (S4B6.1) and the IL-2R (5A2) that block auto- or paracrine IL-2 stimulation or with ConA and human IL-2. (A) IL-2Rα expression on CD4+ or CD8+ cells was monitored by FACS analysis. (B) Nuclear extracts from 2 × 105 cells were incubated for 15 min with a probe containing the STAT motif of the FcγRI gene and separated on a nondenaturing polyacrylamide gel. The identity of the proteins forming specific retarded complexes was determined by supershift experiments with the appropriate antibodies (not shown).
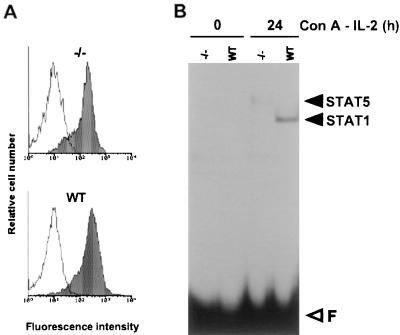
ConA triggers strong activation of STAT1, whereas STAT5 DNA binding is induced by IL-2 (Fig. 1B). Since autocrine activation of STAT1 by IFN-γ following T-cell activation has been reported (21), it seemed likely that ConA stimulated STAT1 via IFN-γ. In order to confirm the origin of STAT1 induction and to assess its role in IL-2Rα expression, we monitored ConA-induced IL-2Rα expression and STAT1 activation in T lymphocytes from IFN-γR-deficient mice (26). Figure 2A shows that, in these cells, IL-2Rα expression after 1 day of stimulation with ConA is completely normal despite the absence of STAT1 and STAT5 (Fig. 2B). These experiments demonstrate that the ConA-induced wave of IL-2Rα expression is independent of STAT1 or STAT5 activation. These observations confirm the biphasic model of the regulation of IL-2Rα transcription based on transient transfection experiments, according to which antigen (or ConA) triggers IL-2Rα expression through cis-acting elements in the promoter-proximal region while IL-2 enhances transcription through the distal IL-2rE (45, 61).
FIG. 2.
ConA induces IL-2Rα expression in absence of active STAT factors. T lymphocytes from IFN-γR-deficient (−/−) or wild-type (WT) mice were stimulated for 24 h with ConA in the presence of the IL-2-blocking MAbs. (A) Flow cytometric analysis of IL-2Rα expression on T cells. (B) Bandshift assay with 3 μg of nuclear extracts incubated with the FcγRI probe for 15 min.
STAT1 does not play a role in the IL-2-dependent phase of IL-2Rα expression.
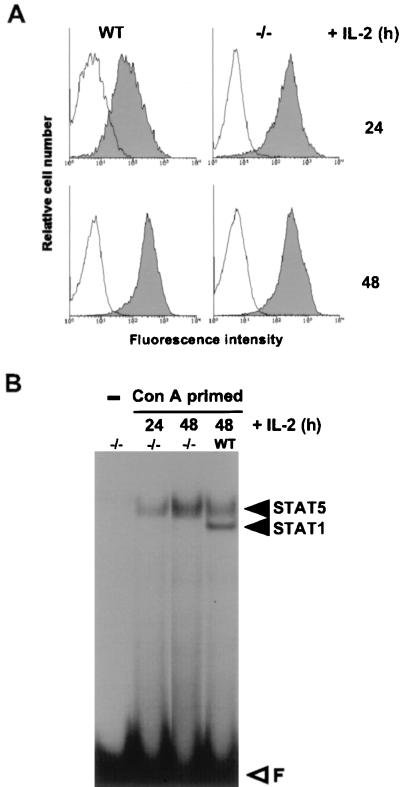
Figure 3A shows that IL-2Rα expression during 72 h of stimulation with ConA and IL-2 in cells from IFN-γR-deficient mice is indistinguishable from that in wild-type cells of the same genetic background. At no time during this period did IFN-γR-deficient cells contain detectable STAT1 activity (Fig. 3B). This indicates that STAT1 is not necessary for IL-2-induced IL-2Rα expression, nor does it act as a negative regulator. This is consistent with the finding that IFN-γ stimulation of the rodent T-cell line which we used to map the IL-2rE does not induce IL-2Rα expression (results not shown). Thus the antiproliferative effect of IFN-γ on T cells (11) is not a consequence of reduced IL-2Rα expression.
FIG. 3.
IL-2-induced expression of IL-2Rα is normal in IFN-γR-deficient mice. T lymphocytes from IFN-γR-deficient (−/−) or wild-type (WT) mice were stimulated for 24 h with ConA in the presence of the IL-2-blocking MAbs (ConA primed), and IL-2 was then added for the indicated times. (A) FACS analysis of IL-2Rα expression on T cells. (B) Bandshift assay with 3 μg of nuclear extracts incubated with the FcγRI probe for 15 min.
In vivo footprinting experiments confirm the role of the IL-2rE in IL-2-induced IL-2Rα expression.
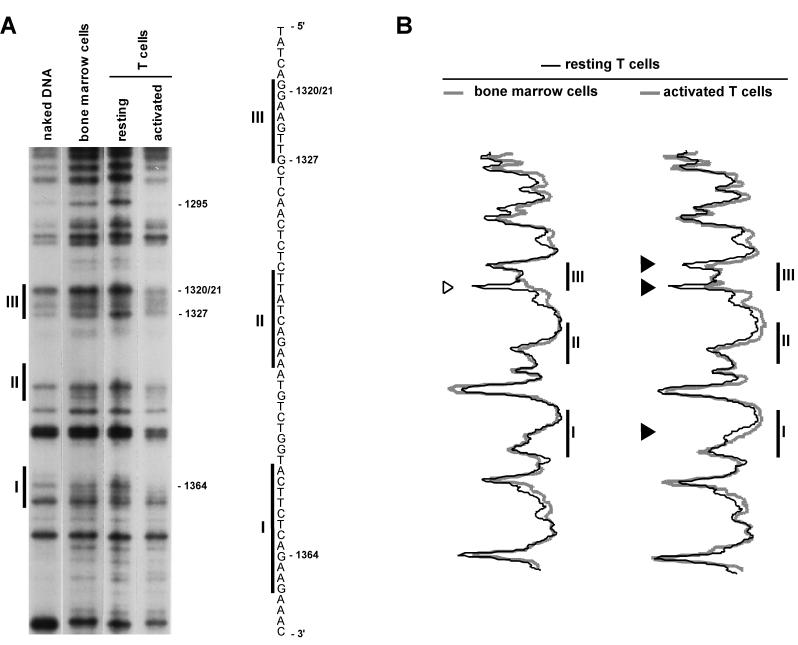
Changes in protein-chromatin interactions taking place in the IL-2rE were analyzed by DMS footprinting. DMS specifically methylates guanine and, to a lesser extent, adenine residues of DNA (40). The binding of a protein to its target site modulates the accessibility of DMS to DNA by either protecting nucleotides in the binding site from or by rendering neighboring residues more sensitive to methylation. We compared the methylation pattern on IL-2rE chromatin from resting T cells and cells activated with ConA and IL-2 with that of the IL-2Rα-negative bone marrow cells (Fig. 4). Bone marrow chromatin does not show any significant protection of nucleotides in the IL-2rE compared to naked DNA. The enhanced methylation of adenine bases in living cells compared to naked DNA has also been observed by others (23) and might be due to the differences existing between the composition of cell nuclei and the buffer in which naked DNA is resuspended for the DMS reaction. In the chromatin of resting T cells, we reproducibly observed increased methylation of guanine residue −1327 located at the 5′ boundary of site III. This is unlikely to be due to Elf-1, the factor that mediates the contribution of site III to IL-2rE activity, since in vitro binding analysis indicates that Elf-1 protects the G doublet in the core of the Ets motif (27). Upon activation with ConA and IL-2, G −1364, which is part of the STAT5 consensus motif (3′-AAGN3CTT-5′) (25, 57) in site I, and G −1327 and the doublet −1320/−1321 in site III are partially protected from methylation, corroborating the previous mapping of the IL-2-responsive elements in the IL-2rE. In vivo footprinting of the human IL-2rE has also shown IL-2-dependent protection of site I (36). In some experiments, IL-2 also induces a weak reduction in methylation of G −1344 in site II (see Fig. 6). That this protection is not always seen may reflect the low affinity of STAT5 for site II (42). We also detected partial protection of G −1293 in activated T cells. This suggests that an additional protein may bind to this site. However, since this residue lies outside the IL-2rE, in a region which is not required for IL-2rE activity (61), the protein is unlikely to play an important role in IL-2Rα regulation.
FIG. 4.
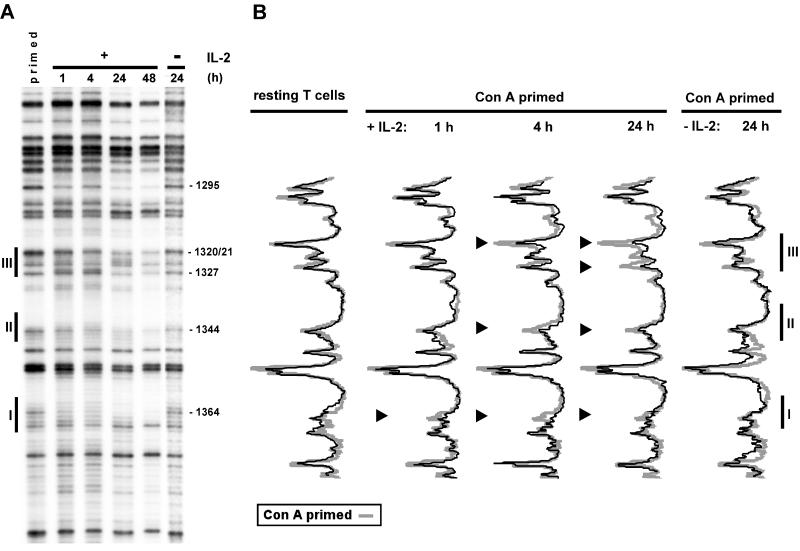
T-lymphocyte activation results in genomic footprints in the IL-2rE. LMPCR was performed with in vitro-methylated liver DNA or in vivo-methylated DNA from bone marrow cells, resting T lymphocytes, and T cells stimulated with ConA and IL-2 for 72 h. (A) Guanine sequence of the lower strand. The positions of the three sites that constitute the IL-2rE are shown to the left. The positions of the guanine residues, the methylation of which varies, are indicated on the right. The autoradiogram is representative of four independent experiments. (B) Histogram representation of the sequences shown in the gel presented in panel A. The gel was scanned with a phosphorimager, and histograms of the different lanes were overlaid. Each peak in the histogram corresponds to a band in the gel. Solid arrowheads indicate guanine bases which are partially protected in activated T cells. The open arrowhead points to a guanine residue at the border of site III with increased sensitivity to DMS in resting T cells.
FIG. 6.
IL-2-induced in vivo protection of IL-2rE sites follows a temporal pattern. (A) In vivo footprinting analysis was performed with ConA-primed cells or primed cells cultured further with or without IL-2 for the indicated times. (B) The phosphorimager histograms of the footprints obtained with the different populations are compared to the one from ConA-primed cells. The results shown are representative of three experiments. Arrowheads indicate partial protection of guanine bases. Nucleotide positions are marked to the right of panel A.
No major changes in the sensitivity to DMS were detected in the upper strand of the IL-2rE (data not shown). The G −1360 residue that is part of the palindromic STAT motif in site I is very poorly methylated on naked DNA and is insensitive to methylation in vivo. Intriguingly, a similar insensitivity has been observed for the homologous G residue (G −4175) in the human gene (36). Site II differs from the STAT consensus motif in that it contains a T instead of a G in position −1340.
We did not observe any protection of the G −1342 residue in the GATA consensus motif that overlaps with site II (data not shown).
Although more than 95% of the activated cells were expressing IL-2Rα at their surface, protections were only partial. The most plausible explanation for this is that binding of STAT5 and Elf-1 to the IL-2rE is not strong enough to prevent access of DMS completely or that the IL-2rE sites are not occupied all the time.
Occupation of site III depends on stimulation with IL-2.
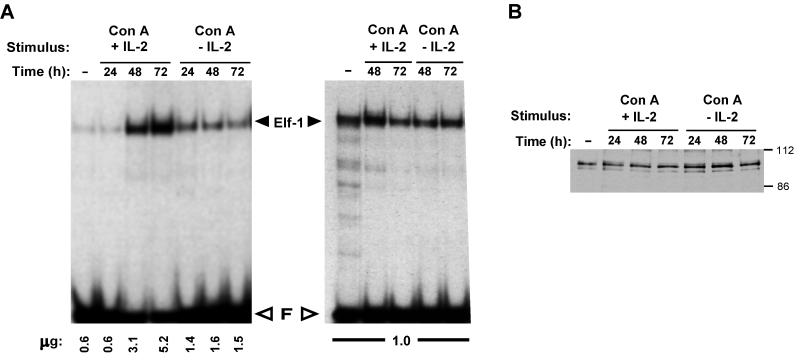
From the studies with PC60 cells, we had proposed that Elf-1 was constitutively bound to the IL-2rE. The observation that occupation of all three sites of the IL-2rE depends on stimulation of T cells indicates that this model is too simple. One explanation for this apparent inconsistency could be that activation of normal T cells, in contrast to stimulation of PC60, results in an increase in nuclear Elf-1 concentration. Figure 5 shows that this is not the case. Although the absolute amount of Elf-1 binding activity per cell is augmented with activation (Fig. 5A, left panel), the relative concentration of this activity compared to total nuclear protein (Fig. 5A, right panel) remains constant, as does the concentration of Elf-1 protein (Fig. 5B). Activation of T cells affects neither the electrophoretic mobility of Elf-1–site III complexes or denatured Elf-1 protein, nor does it result in an alteration of the affinity of Elf-1 for site III as measured by in vitro assays (data not shown).
FIG. 5.
Elf-1 DNA binding activity is not modified upon T-cell activation. (A) Nuclear extracts from 2 × 105 cells (left panel) or a constant amount of protein of the same extracts (right panel) was incubated with a labeled oligonucleotide containing site III, and the complexes formed were fractionated on a nondenaturing gel. The amount of protein per sample is indicated on the bottom of each lane. The identity of the Elf-1 complex was confirmed by supershift analysis with an anti-Elf-1 antibody (not shown) and has been fully documented (58). (B) Five micrograms of nuclear extract was fractionated on a denaturing gel, and the Western blot was probed with anti-Elf-1 antibodies. Molecular mass markers (in kilodaltons) are indicated on the right.
In order to distinguish whether the appearance of the footprint on site III was the result of ConA activation or depended on stimulation by IL-2, we compared the chromatin of T lymphocytes activated with ConA alone with that of cells stimulated with ConA for 24 h (ConA primed), and then subsequently cultured with IL-2 for different times (Fig. 6). Stimulation with ConA only for 24 or 48 h does not result in any detectable changes in the IL-2rE methylation pattern. IL-2 stimulation of ConA-primed cells leads, as expected, to a rapid reduction of the sensitivity of sites I and II to DMS, correlating with IL-2-induced apparition of active STAT5 (data not shown). Protection in site I is observed as soon as 1 h after IL-2 addition and is maintained throughout stimulation, as is nuclear STAT5 activity (Fig. 1B). The persistence of active STAT5 in response to IL-2 was also observed in PC60 cells and paralleled IL-2rα expression and IL-2rE-driven reporter gene activity (42, 61). Continuous STAT5 activation in T cells can be explained by the fact that IL-2Rα expression and STAT5 activation are regulated by a positive feedback loop. Protection of the G doublet in the core of site III also depends on IL-2 stimulation, but this response is slower than the changes in the STAT motifs. In vivo changes in site III are first detected 4 h after addition of IL-2, but maximal protection is observed only after 24 h of culture in the presence of IL-2.
Taken together, these results suggest that the accessibility of site III to Elf-1 in vivo depends on changes in the IL-2rE chromatin that are induced by IL-2.
STAT1 does not occupy sites I and II in ConA-primed cells.
Nuclei of T cells activated with ConA for 24 h contain substantial amounts of active STAT1 (Fig. 1B). Although STAT1 can specifically bind to sites I and II, as shown by the ability of wild-type but not mutated IL-2rE to compete for STAT1-FcγRI complex formation (data not shown), these sites are not protected against DMS methylation in cells stimulated with ConA only (Fig. 6). Comparison of the affinities of STAT1 and STAT5 for the IL-2rE shows that both proteins bind with similar efficiency to the latter (data not shown). Thus, the lack of occupation of the STAT motifs in the IL-2rE in chromatin of ConA-activated cells is not due to a lower avidity of STAT1 for the IL-2rE. Since we observe occupation of the STAT sites very early after addition of IL-2 to primed cells, at a time when the amounts of STAT1 and STAT5 DNA binding activity are similar, it is unlikely that, in ConA-primed cells, the IL-2rE is not occupied because the STAT1 concentration is too low. These data rather suggest that the IL-2rE in ConA-activated cells is not accessible to STAT1.
IL-2 induces positioning of nucleosomes around the IL-2rE.
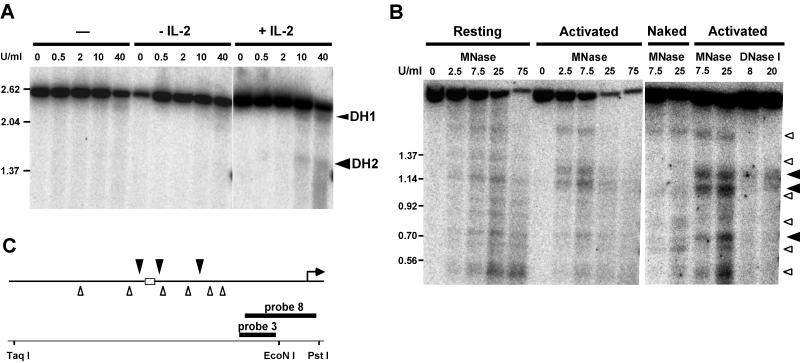
Previously we described the appearance of a DNase I-hypersensitive site (DH2) at the IL-2rE in T cells activated with ConA and IL-2 (60). Figure 7A shows that this site is not detectable in cells stimulated with ConA alone but appears after stimulation with IL-2. This confirms our hypothesis that IL-2 induces a modification of IL-2rE chromatin, making it accessible for transcription factors.
FIG. 7.
IL-2 induces the appearance of DNase I and MNase sites flanking the IL-2rE. (A) DNase I-treated chromatin from ConA-primed cells or primed cells induced further with or without IL-2 for 24 h was digested with TaqI and PstI and electrophoresed in 1% agarose gels. DNase I-hypersensitive (DH) sites were visualized by hybridization with probe 8. DH1 is a lymphocyte-specific constitutive site that maps close to the transcription start site (60). DH2 is located at the IL-2rE. Size markers are indicated on the left (in kilobases). (B) MNase cleavage of naked DNA and chromatin from resting T cells or cells stimulated for 72 h with ConA and IL-2. DNA was digested with TaqI and EcoNI and electrophoresed in a 1.5% agarose gel. The DNase I-treated samples from activated cells in the right panel were digested with the same restriction enzymes. Blots were hybridized with probe 3. Solid arrowheads indicate chromatin-specific cleavage sites in activated cells. Open arrowheads indicate the cutting sites in naked DNA. (C) Summary of MNase analysis. The IL-2rE is represented as a hatched box. Arrowheads (as in panel B) show mean cleavage positions of the MNase cuts deduced from two experiments.
By probing IL-2rE chromatin with MNase, we observed a number of cuts in resting T cells (Fig. 7B). Most of these were also observed when naked DNA was digested with MNase and can be attributed to the sequence preferences of this nuclease. Thus, in resting T cells, there appears to be little or no translational positioning of nucleosomes over the IL-2rE. In fully activated cells, however, MNase cuts IL-2rE chromatin preferentially at positions flanking the IL-2rE core (which contains sites I, II, and III). An additional site appears downstream of the IL-2rE. Most of the cuts observed in resting cells and naked DNA are no longer detectable. Digestion of the same population of activated T cells with DNase I shows that the region between the two MNase-hypersensitive sites, i.e., the IL-2rE itself, is hypersensitive to DNase I.
Thus, stimulation of IL-2Rα gene transcription results in the translational positioning of nucleosomes at the borders of the IL-2rE sequence and in the opening of the IL-2rE chromatin.
DISCUSSION
Our results reveal that the function of the IL-2-responsive enhancer in the IL-2Rα gene depends not only on the presence of previously identified transcription factors, but also on IL-2-induced changes in IL-2rE chromatin. We show that IL-2 controls the accessibility of the binding site for Elf-1 that is required for enhancer activity.
We confirm the two-stage model of the regulation of IL-2Rα gene transcription during T-cell activation (45). According to this model, antigen or ConA stimulates a transient peak of IL-2Rα transcription through promoter-proximal elements. In a second phase, IL-2 increases the level of IL-2Rα transcription by activating STAT5, which binds to an IL-2-responsive enhancer conserved in mice and humans. Here we show that ConA stimulation of IL-2Rα transcription in normal T lymphocytes is independent of STAT activation and does not involve the IL-2rE. IL-2 induces genomic footprints in the IL-2rE, confirming the function of the three distinct cis-acting elements in the IL-2rE mapped by transient transfection in lymphoid cell lines (29, 36, 61). In the chromatin of mouse T lymphocytes stimulated to express the high-affinity receptor chain by IL-2, all three sites within the 51-nt IL-2rE are occupied. From transient transfection experiments, we concluded that the two more distal sites control IL-2 responsiveness by binding to STAT5 (42), whereas Elf-1 contributes to enhancer activity by binding to the third site (58). The in vivo footprints shown here are completely consistent with this interpretation. In both site I and site II, the consensus STAT binding motif is protected. The absence of protection of the GATA consensus motif that overlaps with site II supports our previous results that argued against a role of GATA proteins in IL-2rE function (42). Protection of the G doublet and the 5′ flanking G residue in site III by Elf-1 is predicted by methylation interference analysis of the in vitro interaction between Elf-1 and its binding site (27, 28). The enhanced sensitivity to methylation 5′ of site III specifically observed in the chromatin of nonactivated T lymphocytes raises the possibility that a protein may bind at or near site III in resting T cells, although such a factor has not yet been detected by in vitro binding experiments.
Occupation of the STAT binding sites in the IL-2rE requires stimulation with IL-2 and is concomitant with STAT5 activation. That IL-2 stimulation is essential for prolonged IL-2Rα expression is reflected in the persistence of activated STAT5 and in vivo footprints in the IL-2rE. Occupation of the Elf-1 binding site also depends on IL-2 stimulation. This is surprising, because Elf-1 DNA binding activity is present in the nuclei of unstimulated T cells, and IL-2 does not stimulate an increase in the nuclear concentration or in the in vitro binding affinity of the protein for the IL-2rE, nor does it induce any detectable changes in Elf-1 electrophoretic mobility in native or denaturing gels. Binding of Elf-1 to certain sites has been shown to depend on cooperation with other transcription factors that are activated by stimuli which mimic antigen triggering (63). This is clearly not the case for site III, which matches very closely the highest-affinity sites for mouse and human Elf-1 obtained in site-selection experiments (12, 27) and is identical in 7 of 8 nt to the Ets binding site, which regulates transcription of the terminal deoxytransferase gene (16). Elf-1 binding to this sequence is also independent of activation signals. We have failed to find any evidence for interactions between Elf-1 and STAT5 in in vitro binding assays or in transient transfection experiments (references 42 and 58 and our unpublished results).
The simplest hypothesis for the delayed occupation of site III by Elf-1 is suggested by the comparison of the accessibility of the IL-2rE to nucleases in resting, ConA-primed and fully activated cells. IL-2, but not ConA, induces substantial changes in IL-2rE chromatin. Our data indicate that IL-2 triggers both an opening of IL-2rE chromatin (that accounts for the sensitivity of the IL-2rE to DNase I) and the translational positioning of nucleosomes at the borders of the IL-2rE (resulting in MNase-hypersensitive sites). Thus, it appears likely that accessibility of site III to Elf-1 depends on the IL-2-induced opening of the IL-2rE chromatin. Elucidation of the molecular events and the role of STAT5 in the IL-2-induced chromatin changes will require different experimental approaches.
Finally, our data also suggest that sites I and II in IL-2rE chromatin are accessible to STAT5 but not to STAT1. If confirmed, this finding would add a new level at which the specificity of cytokine-induced gene expression may be controlled and provide a possible explanation for the conclusion from the analysis of STAT1-deficient mice that the physiological role of STAT1 is restricted to transduction of IFN responses (14, 41), although STAT1 is activated by many other cytokines (18, 24, 35, 64).
In summary, while the experiments described here provide a clear-cut confirmation of the previously formulated model of the regulation of IL-2Rα gene expression during T-cell activation, they indicate that changes in chromatin also play an important role in IL-2-stimulated gene transcription.
ACKNOWLEDGMENTS
We are grateful to Glaxo, Geneva, Switzerland, for the generous gift of IL-2. Jacques Louis, WHO, IRTC, Biochemistry Institute, Lausanne, Switzerland, kindly provided IFN-γR-deficient mice. Anne-Lise Peitrequin, Ludwig Institute, Lausanne, supplied us with the antibodies for the FACS analyses. We thank Jovan Mirkovitch, Jean Imbert, and Michèle Algarté for helpful technical advice and Véronique Imbert, Friedrich Beermann, and Moira Cockell for valuable discussions and advice during the preparation of the manuscript.
This work was supported, in part, by grants from the Swiss National Science Foundation, the Swiss Cancer League, and the Swiss Federal Office of Science and Technology.
REFERENCES
- 1.Algarté M, Lécine P, Costello R, Plet A, Olive D, Imbert J. In vivo regulation of interleukin-2 receptor α gene transcription by the coordinated binding of constitutive and inducible factors in human primary T cells. EMBO J. 1995;14:5060–5072. doi: 10.1002/j.1460-2075.1995.tb00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartl S, Taplick J, Lagger G, Khier H, Kuchler K, Seiser C. Identification of mouse histone deacetylase 1, as a growth factor-inducible gene. Mol Cell Biol. 1997;17:5033–5043. doi: 10.1128/mcb.17.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beato M, Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25:3559–3563. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bismuth G, Moreau J L, Sommé G, Duphot M, Dautry Varsat A, Robb R J, Thèze J. Regulation of interleukin-2 (IL-2) receptor expression: IL-2 as an inducing signal for the expression of its own receptor on a murine T helper cell line. Eur J Immunol. 1985;15:723–727. doi: 10.1002/eji.1830150716. [DOI] [PubMed] [Google Scholar]
- 5.Böhnlein E, Ballard D W, Bogerd H, Peffer N J, Lowenthal J W, Greene W C. Induction of interleukin-2 receptor α gene expression is regulated by post-translational activation of kappaB specific DNA binding proteins. J Biol Chem. 1989;264:8475–8478. [PubMed] [Google Scholar]
- 6.Bucher P, Corthésy P, Imbert J, Nabholz M. A conserved IL-2 responsive enhancer in the IL-2Rα gene. Immunobiology. 1997;197:133–140. doi: 10.1016/s0171-2985(97)80034-4. [DOI] [PubMed] [Google Scholar]
- 7.Cantrell D A, Smith K A. The interleukin-2 T cell system: a new cell growth model. Science. 1984;224:1312–1316. doi: 10.1126/science.6427923. [DOI] [PubMed] [Google Scholar]
- 8.Cao X, Kozak C A, Liu Y-J, Noguchi M, O’Connell E, Leonard W J. Characterization of cDNAs encoding the murine interleukin-2 receptor (IL-2R) γ chain: chromosomal mapping and tissue specificity of IL-2R gamma chain expression. Proc Natl Acad Sci USA. 1993;90:8464–8468. doi: 10.1073/pnas.90.18.8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W. Current protocols in Immunology. Boston, Mass: Greene Publishing Associates; 1996. pp. 3.2.1–3.2.4. [Google Scholar]
- 10.Cross S L, Halden N F, Lenardo M J, Leonard W J. Functionally distinct NF-kappaB binding sites in the immunoglobulin kappa and IL-2 receptor α chain genes. Science. 1989;244:466–469. doi: 10.1126/science.2497520. [DOI] [PubMed] [Google Scholar]
- 11.Dalton D K, Pitts-Meek S, Keshav S, Figari I S, Bradley A, Stewart T A. Multiple defects of immune cell function in mice with disrupted interferon-γ genes. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 12.Davis J N, Roussel M F. Cloning and expression of the murine Elf-1 cDNA. Gene. 1996;171:265–269. doi: 10.1016/0378-1119(96)00013-3. [DOI] [PubMed] [Google Scholar]
- 13.Depper J M, Leonard W J, Drogula C, Krönke M, Waldmann T A, Greene W C. Interleukin-2 (IL-2) augments transcription of the IL-2 receptor gene. Proc Natl Acad Sci USA. 1985;82:4230–4234. doi: 10.1073/pnas.82.12.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Targeted disruption of the mouse STAT1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 15.Erard F, Corthésy P, Smith K A, Fiers W, Conzelmann A, Nabholz M. Characterization of soluble factors that induce the cytolytic activity and the expression of T cell growth factor receptors of a T cell hybrid. J Exp Med. 1984;160:584–599. doi: 10.1084/jem.160.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst P, Hahm K, Trinh L, Davis J N, Roussel M F, Turck C W, Smale S T. A potential role for Elf-1 in terminal transferase gene regulation. Mol Cell Biol. 1996;16:6121–6131. doi: 10.1128/mcb.16.11.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V. Chromatin structure and gene expression. Proc Natl Acad Sci USA. 1996;93:9384–9388. doi: 10.1073/pnas.93.18.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu X-Y, Zhang J-J. Transcription factor p91 interacts with the epidermal growth factor receptor and mediates activation of the c-fos gene promoter. Cell. 1993;74:1135–1145. doi: 10.1016/0092-8674(93)90734-8. [DOI] [PubMed] [Google Scholar]
- 19.Garrity P, Wold B. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc Natl Acad Sci USA. 1992;89:1021–1025. doi: 10.1073/pnas.89.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geppert T D, Lipsky P E. Accessory cell independent proliferation of human T4 cells stimulated by immobilized monoclonal antibodies to CD3. J Immunol. 1987;138:1660–1666. [PubMed] [Google Scholar]
- 21.Girdlestone J, Wing M. Autocrine activation by interferon-γ of STAT factor following T cell activation. Eur J Immunol. 1996;26:704–709. doi: 10.1002/eji.1830260329. [DOI] [PubMed] [Google Scholar]
- 22.Giri J G, Ahdieh M, Eisenman J, Shanebeck K, Grabstein K, Kumaki S, Namen A, Park L S, Cosman D, Anderson D. Utilization of the β and γ chains of the IL-2 receptor by the novel cytokine IL-15. EMBO J. 1994;13:2822–2830. doi: 10.1002/j.1460-2075.1994.tb06576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grange T, Rigaud G, Bertrand E, Fromont-Racine M, Espinas M L, Roux J, Pictet R. In vivo footprinting of the interaction of proteins with DNA and RNA. Adv Mol Cell Biol. 1997;21:73–109. doi: 10.1006/meth.1996.0401. [DOI] [PubMed] [Google Scholar]
- 24.Gronowski A M, Zhong Z, Wen Z, Thomas M J, Darnell J E, Jr, Rotwein P. In vivo growth hormone treatment rapidly stimulates the tyrosine phosphorylation and activation of STAT3. Mol Endocrinol. 1995;9:171–177. doi: 10.1210/mend.9.2.7776967. [DOI] [PubMed] [Google Scholar]
- 25.Horvath C M, Wen Z, Darnell J E., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 26.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel R M, Aguet M. Immune response in mice that lack the interferon-γ receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 27.John S, Marais R, Child R, Light Y, Leonard W J. Importance of low affinity Elf-1 sites in the regulation of lymphoid-specific inducible gene expression. J Exp Med. 1996;183:743–750. doi: 10.1084/jem.183.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.John S, Reeves R B, Lin J-X, Child R, Leiden J M, Thompson C B, Leonard W J. Regulation of cell-type-specific interleukin-2 receptor α-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF-κB family proteins. Mol Cell Biol. 1995;15:1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.John S, Robbins C M, Leonard W J. An IL-2 response element in the human IL-2 receptor α chain promoter is a composite element that binds STAT5, Elf-1, HMG-I (Y) and a GATA family protein. EMBO J. 1996;15:5627–5635. [PMC free article] [PubMed] [Google Scholar]
- 30.Kang C-J, Sheridan C, Koshland M E. A stage-specific enhancer of immunoglobulin J chain gene is induced by IL-2 in a presecretor B cell stage. Immunity. 1998;8:285–295. doi: 10.1016/s1074-7613(00)80534-8. [DOI] [PubMed] [Google Scholar]
- 31.Kimura A, Ersson B. Activation of T lymphocytes by lectins and carbohydrate-oxidizing reagents viewed as an immunological recognition of cell surface modifications seen in the context of “self” major histocompatibility complex antigens. Eur J Immunol. 1981;11:475–483. doi: 10.1002/eji.1830110607. [DOI] [PubMed] [Google Scholar]
- 32.Kondo M, Ohashi Y, Tada K, Nakamura M, Sugamura K. Expression of the mouse interleukin-2 receptor γ chain in various cell populations of the thymus and spleen. Eur J Immunol. 1994;24:2026–2030. doi: 10.1002/eji.1830240914. [DOI] [PubMed] [Google Scholar]
- 33.Kondo M, Takeshita T, Ishii N, Nakamura M, Watanabe S, Arai K-I, Sugamura K. Sharing of the interleukin-2 (IL-2) receptor γ chain between receptors for IL-2 and IL-4. Science. 1993;262:1874–1877. doi: 10.1126/science.8266076. [DOI] [PubMed] [Google Scholar]
- 34.Krönke M, Leonard W J, Depper J M, Greene W C. Sequential expression of genes involved in human T lymphocyte growth and differentiation. J Exp Med. 1985;161:1593–1598. doi: 10.1084/jem.161.6.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larner A C, David M, Feldman G M, Igarashi K, Hackett R H, Webb D S A, Sweitzer S M, Petricoin III E F, Finbloom D S. Tyrosine phosphorylation of DNA binding proteins by multiple cytokines. Science. 1993;261:1730–1733. doi: 10.1126/science.8378773. [DOI] [PubMed] [Google Scholar]
- 36.Lécine P, Algarté M, Rameil P, Beadling C, Bucher P, Nabholz M, Imbert J. Elf-1 and Stat5 bind to a critical element in a new enhancer of the human interleukin-2 receptor α gene. Mol Cell Biol. 1996;16:6829–6840. doi: 10.1128/mcb.16.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowenthal J W, Corthésy P, Tougne C, Lees R, MacDonald H R, Nabholz M. High and low affinity IL-2 receptors: analysis by IL-2 dissociation rate and reactivity with monoclonal anti-receptor antibody PC61. J Immunol. 1985;135:3988–3994. [PubMed] [Google Scholar]
- 38.Lowenthal J W, Tougne C, MacDonald H R, Smith K A, Nabholz M. Antigenic stimulation regulates the expression of IL-2 receptors in a cytolytic T lymphocyte clone. J Immunol. 1985;134:931–939. [PubMed] [Google Scholar]
- 39.Malek T R, Ashwell J D. Interleukin-2 upregulates expression of its receptor on a T cell clone. J Exp Med. 1985;161:1575–1580. doi: 10.1084/jem.161.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 41.Meraz M A, White J M, Sheehan K C F, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, Carver-Moore K, DuBois R N, Clark R, Aguet M, Schreiber R D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–450. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 42.Meyer W K H, Reichenbach P, Schindler U, Soldaini E, Nabholz M. IL-2 regulates IL-2Rα transcription through interaction of STAT5 dimers on two low affinity binding sites. J Biol Chem. 1997;272:31821–31828. doi: 10.1074/jbc.272.50.31821. [DOI] [PubMed] [Google Scholar]
- 43.Minami Y, Kono T, Miyazaki T, Taniguchi T. The IL-2 receptor complex: its structure, function, and target genes. Annu Rev Immunol. 1993;11:245–268. doi: 10.1146/annurev.iy.11.040193.001333. [DOI] [PubMed] [Google Scholar]
- 44.Moreau J-L, Nabholz M, Diamantstein T, Malek T, Shevach E, Thèze J. Monoclonal antibodies identify three epitope clusters on the mouse p55 subunit of the interleukin-2 receptor: relationship to the interleukin-2 binding site. Eur J Immunol. 1987;17:929–935. doi: 10.1002/eji.1830170706. [DOI] [PubMed] [Google Scholar]
- 45.Nabholz M, Soldaini E, Sperisen P, Pla M, Wang S-M, MacDonald H R, Reichenbach P, Beermann F, Bucher P. The cis-acting elements controlling mouse IL-2Rα transcription. Immunobiology. 1995;193:259–262. doi: 10.1016/s0171-2985(11)80552-8. [DOI] [PubMed] [Google Scholar]
- 46.Nakajima H, Liu X-W, Wynshaw-Boris A, Rosenthal L A, Imada K, Finbloom D S, Hennighausen L, Leonard W J. An indirect effect of STAT5A in IL-2 induced proliferation: a critical role for STAT5A in IL-2 mediated IL-2 receptor α chain induction. Immunity. 1997;7:1–20. doi: 10.1016/s1074-7613(00)80389-1. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura Y, Russell S M, Mess S A, Friedmann M, Erdos M, Francois C, Jacques Y, Adelstein S, Leonard W J. Heterodimerization of the IL-2 receptor β and γ chain cytoplasmic domains is required for signaling. Nature (London) 1994;369:330–333. doi: 10.1038/369330a0. [DOI] [PubMed] [Google Scholar]
- 48.Nakarai T, Robertson M J, Streuli M, Wu Z, Ciardelli T L, Smith K A, Ritz J. Interleukin-2 receptor γ chain expression on resting and activated lymphoid cells. J Exp Med. 1994;180:241–251. doi: 10.1084/jem.180.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson B H, Lord J D, Greenberg P D. Cytoplasmic domains of the interleukin-2 receptor β and γ chains mediate the signal for T cell proliferation. Nature (London) 1994;369:333–336. doi: 10.1038/369333a0. [DOI] [PubMed] [Google Scholar]
- 50.Noguchi M, Nakamura Y, Russell S M, Ziegler S F, Tsang M, Cao X, Leonard W J. Interleukin-2 receptor γ chain: a functional component of the interleukin-7 receptor. Science. 1993;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- 51.Ohashi Y, Takeshita T, Nagata K, Mori S, Sugamura K. Differential expression of the IL-2 receptor subunits, p55 and p75 on various populations of primary peripheral blood mononuclear cells. J Immunol. 1989;143:3548–3555. [PubMed] [Google Scholar]
- 52.Plaetinck G, Combe M-C, Corthésy P, Sperisen P, Kanamori H, Honjo T, Nabholz M. Control of IL-2 receptor α expression by IL-1, tumor necrosis factor, and IL-2. Complex regulation via elements in the 5′ flanking region. J Immunol. 1990;145:3340–3347. [PubMed] [Google Scholar]
- 53.Robb R J, Munck A, Smith K A. T cell growth factor receptors. Quantitation, specificity, and biological relevance. J Exp Med. 1981;154:1455–1474. doi: 10.1084/jem.154.5.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russell S M, Johnston J A, Noguchi M, Kawamura M, Bacon C M, Friedmann M, Berg M, McVicar D W, Witthuhn B A, Silvennoinen O, Goldman A S, Schmalstieg F C, Ihle J N, O’Shea J, Leonard W J. Interaction of IL-2Rβ and γc chains with Jak1 and Jak3: implications for XSCID and XCID. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 55.Russell S M, Keegan A D, Harada N, Nakamura Y, Noguchi M, Leland P, Friedmann M C, Miyajima A, Puri R K, Paul W E, Leonard W J. Interleukin-2 receptor γ chain: a functional component of the interleukin-4 receptor. Science. 1993;262:1880–1883. doi: 10.1126/science.8266078. [DOI] [PubMed] [Google Scholar]
- 56.Schreiber E, Matthias P, Müller M M, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seidel H M, Milocco L H, Lamb P, Darnell J E, Stein R B, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serdobova I, Pla M, Reichenbach P, Sperisen P, Ghysdael J, Wilson A, Freeman J, Nabholz M. Elf-1 contributes to the function of the complex IL-2 responsive enhancer in the mouse IL-2 receptor α gene. J Exp Med. 1997;185:1211–1221. doi: 10.1084/jem.185.7.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soldaini E, MacDonald H R, Nabholz M. Minimal growth requirements of mature T lymphocytes: IL 1 and IL 6 increase growth rate but not plating efficiency of CD4 cells stimulated with anti-CD3 and IL-2. Eur J Immunol. 1992;22:1707–1711. doi: 10.1002/eji.1830220707. [DOI] [PubMed] [Google Scholar]
- 60.Soldaini E, Pla M, Beermann F, Espel E, Corthésy P, Barangé S, Waanders G A, MacDonald H R, Nabholz M. Mouse interleukin-2 receptor α gene expression: delimitation of cis-acting regulatory elements in transgenic mice and by mapping of DNase I hypersensitive sites. J Biol Chem. 1995;270:10733–10742. doi: 10.1074/jbc.270.18.10733. [DOI] [PubMed] [Google Scholar]
- 61.Sperisen P, Wang S M, Soldaini E, Pla M, Bucher P, Rusterholz C, Corthésy P, Reichenbach P, Nabholz M. Mouse interleukin-2 receptor α gene expression: interleukin-1 and interleukin-2 control transcription via distinct cis-acting elements. J Biol Chem. 1995;270:10743–10753. doi: 10.1074/jbc.270.18.10743. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka T, Tsudo M, Karasuyama H, Kitamura F, Kono T, Hatakeyama M, Taniguchi T, Miyasaka M. A novel monoclonal antibody against murine IL-2 receptor β chain. Characterization of receptor expression in normal lymphoid cells and EL-4 cells. J Immunol. 1991;147:2222–2228. [PubMed] [Google Scholar]
- 63.Wang C-Y, Bassuk A G, Boise L H, Thompson C B, Bravo R, Leiden J M. Activation of the granulocyte-macrophage colony-stimulating factor promoter in T cells requires cooperative binding of Elf-1 and AP-1 transcription factors. Mol Cell Biol. 1994;14:1153–1159. doi: 10.1128/mcb.14.2.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wen Z, Zhong Z, Darnell J J E. Maximal activation of transcription by STAT1 and STAT3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 65.Wolffe A P. Nucleosome positioning and modification: chromatin structures that potentiate transcription. Trends Biochem Sci. 1994;19:240–244. doi: 10.1016/0968-0004(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 66.Zurawski S M, Mosmann T R, Benedik M, Zurawski G. Alterations in the amino-terminal third of mouse interleukin-2. Effects on biological activity and immunoreactivity. J Immunol. 1986;137:3354–3360. [PubMed] [Google Scholar]