ABSTRACT
T cells must recognize pathogen-derived peptides bound to major histocompatibility complexes (MHCs) in order to initiate a cell-mediated immune response against an infection, or to support the development of high-affinity antibody responses. Identifying antigens presented on MHCs by infected cells and professional antigen-presenting cells (APCs) during infection may therefore provide a route toward developing new vaccines. Peptides bound to MHCs can be identified at whole-proteome scale using mass spectrometry—a technique referred to as “immunopeptidomics.” This technique has emerged as a powerful tool for identifying potential vaccine targets in the context of many infectious diseases. In this review, we discuss the contributions immunopeptidomic studies have made to understanding antigen presentation and T cell priming in the context of infection and the potential for immunopeptidomics to inform the development of vaccines to address pressing global health problems in infectious disease.
KEYWORDS: host-pathogen interactions, immunology, immunopeptidomics, infectious disease, mass spectrometry, proteomics, vaccines
INTRODUCTION
Recognition of pathogen-specific peptide-major histocompatibility complexes (pMHCs) is required for naive T cells to become activated and proliferate, and for effector T cells to recognize infected cells and carry out effector functions such as cytokine secretion and lysis of infected cells that ultimately lead to control of an infection. Antigen-specific T cell responses are also required for B cells to undergo affinity maturation and produce high-affinity antibodies (1). Identifying antigens that can be presented on MHCs and recognized by T cells is therefore essential for understanding natural immunity to infection and developing effective vaccines.
The pMHC repertoire is highly complex within any given individual, and highly variable among individuals. MHC class I and class II present peptides for recognition by CD8+ and CD4+ T cells, respectively, and are loaded with peptides via distinct pathways of antigen processing that have been reviewed in detail elsewhere (2, 3). MHC molecules are highly genetically polymorphic, and different alleles preferentially bind different sets of peptide sequences. Three distinct human leukocyte antigen (HLA) loci encode the alpha chain of MHC class I (HLA-A, HLA-B, and HLA-C), for up to six alleles in a given individual. Similarly, three loci each encode an alpha chain and a beta chain of MHC class II (HLA-DR, HLA-DQ, and HLA-DP) (4). Given the diversity and variability of the immunopeptidome, whole-proteome-scale approaches are needed to comprehensively identify pathogen-derived antigens presented on MHCs.
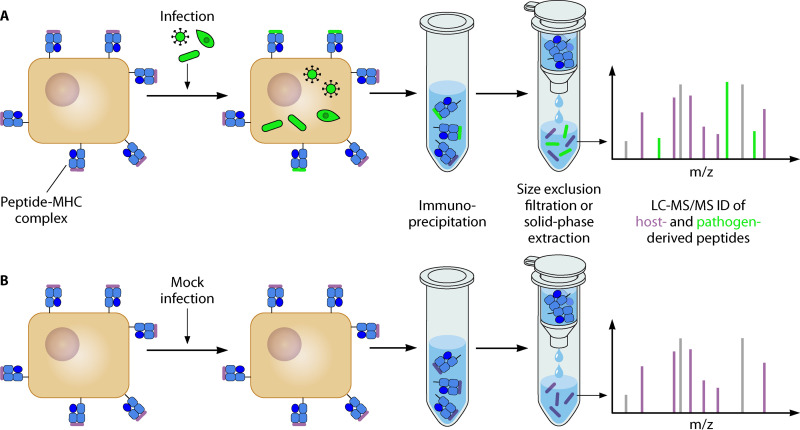
Mass spectrometry (MS) provides a means of experimentally identifying the repertoire of peptide antigens presented on MHCs by cells infected with a pathogen of interest. In a typical immunopeptidomic workflow, MHC-peptide complexes are immunoprecipitated (IP) from a population of cells of interest using an HLA-allele-specific antibody or pan-HLA antibody. Peptides are released by acid elution, MHCs are separated from peptides by size exclusion or solid-phase extraction, and eluted peptides are analyzed by liquid chromatography coupled to tandem MS (LC-MS/MS) (Fig. 1) (5, 6). Mass spectra are then searched against a database of possible peptides that could be derived from the host and/or pathogen proteomes to identify pathogen-derived peptides presented on MHCs.
FIG 1.
Schematic representation of an immunopeptidomic workflow to identify pathogen-derived peptides in infected cells. Infected cells and mock-infected control cells are separately lysed, lysate is subjected to immunoprecipitation with an MHC-specific antibody, and peptides are eluted in acid and separated from MHC proteins using size filtration or solid-phase extraction. Finally, the eluted peptides are analyzed by LC-MS/MS, and mass spectra are searched against a database that includes the proteomes of both the pathogen and the host to identify MHC-associated peptides. Putative pathogen-derived peptides identified in the samples from infected cells can be looked for in the data from the mock-infected control to identify and eliminate some false positives.
MS analyses of MHC peptides can be carried out in two distinct modes. In data-dependent mode, peptides are detected and prioritized for tandem mass spectrum acquisition based on their abundance, without prior knowledge of the composition of the sample, enabling discovery of novel vaccine targets. In targeted mode, a predetermined list of targets is selected for acquisition, enabling studies of the mechanism and kinetics of presentation of known antigens by providing reliable detection and quantification across multiple samples.
Here, we review the use of MS-based immunopeptidomics to study antigen presentation in infectious disease. These studies reveal basic insights into the biology of antigen presentation, provide data sets for training predictive models of antigen presentation, and guide the rational selection of vaccine targets. We go on to discuss opportunities for further study in this area, technical pitfalls that remain to be overcome, and the potential for immunopeptidomics to inform vaccine development.
ANTIGEN PRESENTATION IN MODEL SYSTEMS
Vaccinia virus infection has served as a model system for research on the basic biology of antigen presentation in viral infection. Vaccinia virus is a large, enveloped poxvirus and a relative of variola virus (the etiologic agent of smallpox). It was widely used as a vaccine against smallpox before the eradication of the disease, and some variants such as Modified Vaccinia Ankara (MVA) are being used experimentally as viral vectors for delivery of vaccine antigens (7–9). Although the immunopeptidome of vaccinia virus could provide insight into the mechanism by which it provides protection against smallpox (10), its use in basic studies of antigen presentation is due primarily to its attractiveness as a well-characterized, tractable model of viral infection.
An immunopeptidomic study of vaccinia virus-infected cells assessed whether most viral peptides presented by MHCs are immunogenic or if a large proportion are immunologically silent (11). The authors identified 170 viral peptides bound to MHC class I, over 80% of which could be recognized by T cells isolated from vaccinia virus-infected mice (11). The authors argue that their data represent evidence that most viral peptides presented on MHCs are immunogenic, but it is unclear to what extent this conclusion can be generalized beyond vaccinia virus.
Vaccinia virus infection has also been used as a model to study the kinetics of antigen presentation during viral infection and the relationship between pMHC abundance and immunogenicity. Croft et al. (12) used a targeted MS approach to monitor presentation of eight known vaccinia virus peptides in infected mouse B cells over time. They found that peptide presentation occurred almost immediately after initiation of viral gene expression, consistent with cotranslational antigen processing (13, 14). They also found that comparably immunogenic peptides can vary widely in their abundance in the MHC repertoire, suggesting that pMHC abundance may not strongly correlate with immunogenicity. Using targeted MS to monitor antigen presentation over time could be useful in selecting vaccine antigens presented during different stages of infection (15).
COMPUTATIONAL MODEL BUILDING
Computational methods for predicting peptides likely to be presented on MHCs were first trained using data from in vitro binding assays (16–19), but recently developed models incorporate large MS-based immunopeptidomic data sets as training data, resulting in improvements in performance (20–24). The high-throughput nature of in silico predictions enables such analyses to cover a much wider range of HLA alleles. Computational methods can also select sets of epitopes that maximize coverage of a population with a given set of HLA allele frequencies (25), which will aid in the design of broadly protective vaccines.
Immunopeptidomic data sets have facilitated the development of models that explicitly take into account factors beyond MHC binding affinity alone, such as gene expression level, protein subcellular localization, and predicted protease cleavage sites (22–24, 26). These modeling approaches could be applied to learn pathogen-specific rules for antigen presentation, such as whether secreted bacterial proteins are preferentially presented on MHCs, relative to proteins in the pathogen’s cytosol (27). Existing data sets of pathogen-derived MHC peptides are small, but highly data-efficient machine learning methods (28) may help build pathogen-specific models from limited numbers of training examples.
Rapid computational predictions can inform efforts to develop vaccines for emerging infectious diseases sooner than experimental data. Multiple computational analyses of predicted T cell epitopes in the SARS-CoV-2 proteome were published early in the pandemic (29, 30), whereas experimental data identifying MHC peptides derived from SARS-CoV-2 were not published until months later (31, 32). Obtaining accurate predictions of T cell epitopes could therefore be critical to rapidly developing vaccines against future emerging pathogens.
VACCINE TARGET DISCOVERY
The direct discovery of vaccine targets is one of the most exciting applications of immunopeptidomics in infectious disease. Whereas T cell responses contribute to protective immunity against many globally important infectious diseases, most rationally designed vaccines licensed to date have been designed to elicit protective immunity primarily through humoral immune responses (33). Immunopeptidomics can provide a systematic, rational basis on which to select targets for vaccines that elicit protective cell-mediated immunity, which may be essential in order to develop highly effective vaccines against pathogens for which none yet exist.
Developing effective vaccines against intracellular bacteria has historically proven challenging, in part due to a lack of knowledge of protective T cell antigens (34). Immunopeptidomic analyses have helped to address this problem by identifying protective T cell epitopes of Mycobacterium tuberculosis—the causative agent of tuberculosis (35)—and Chlamydia trachomatis—the causative agent of chlamydia (36) and trachoma (37). M. tuberculosis-derived peptides have been identified that are presented on MHC class I (38) and on the noncanonical class Ib MHC molecule HLA-E (39) by M. tuberculosis-infected human cells, as well as MHC class I and II peptides presented by human macrophages infected with the bacillus Calmette-Guérin (BCG) vaccine strain (40). Administering these antigens to mice using the ChAdOx1 viral vector in a prime-boost regimen with BCG reduced M. tuberculosis bacterial burden in the lung significantly more than did BCG alone (40). Similarly, nine antigens identified in an immunopetidomic analysis of C. trachomatis-infected cells (41) were recognized by T cells of infected mice and reduced vaginal shedding of C. trachomatis when delivered as a recombinant protein vaccine (42). These findings suggest that immunopeptidomics can help advance the development of effective vaccines against intracellular bacterial pathogens, though the clinical utility of these vaccine targets has yet to be tested in humans.
Like intracellular bacteria, eukaryotic parasites have proven difficult to vaccinate against, with only one vaccine currently licensed (43). Mou et al. (44) used MS to identify peptides presented on MHC class II by murine macrophages infected with Leishmania major, a parasite that causes leishmaniasis (45). They identified an immunodominant epitope of phosphoenolpyruvate carboxykinase (PEPCK) that reduced the parasite burden in a mouse model of leishmaniasis when administered as a peptide vaccine or DNA vaccine. These results demonstrate the potential for immunopeptidomics to aid in the development of vaccines against intracellular parasites.
Immunopeptidomics may help identify viral T cell epitopes that vaccines can target to confer lasting protection in the face of viral evasion of antibody-mediated immunity. A high frequency of escape mutations (46) has made it difficult to develop vaccines that elicit antibody-mediated protection against human immunodeficiency virus (HIV)—the causative agent of AIDS (47). However, some individuals naturally control HIV infection in a manner associated with CD8+ T cell responses, suggesting T cells can contribute to durable immunity against HIV (48–51). Several research groups have used immunopeptidomic methods to identify HIV-derived epitopes presented on MHC class I by infected CD4+ T cells (52–56). Some of these studies specifically isolated peptides bound to HLA alleles associated with improved control of HIV infection (53, 55), thereby identifying antigenic targets that may be associated with durable protection. Many of these epitopes elicited gamma interferon (IFN-γ) production in T cells of HIV-positive patients, validating their relevance to T cell responses against HIV. Including additional viral T cell epitopes in future HIV vaccine candidates could help mitigate the effects of viral escape mutations, but this concept has not been tested in animal models or humans.
INFECTION AND AUTOIMMUNITY
Some infections are known to be associated with the onset of autoimmune disease, but the mechanisms underlying this relationship are not fully understood (57). Immunopeptidomics offers a way to systematically identify pathogen-derived MHC peptides that can trigger self-reactive T cell responses, leading to autoimmunity. For example, Alvarez-Navarro et al. (58) used immunopeptidomics to investigate why Chlamydia trachomatis infection is associated with reactive arthritis in individuals carrying HLA-B alleles of the HLA-B*27 group (59). They identified three C. trachomatis peptides presented by HLA-B*27 that had high sequence similarity to endogenous human peptides and were predicted to adopt similar conformations when bound to HLA-B*27. T cells that respond to these C. trachomatis peptides might cross-react with endogenous peptides, leading to autoimmunity. Similarly, Wang et al. (60) used immunopeptidomics to reveal a mechanism that may explain why the HLA-DR*15 haplotype and Epstein-Barr virus (EBV) infection are both associated with increased risk of multiple sclerosis. They identified peptides presented by HLA-DR*15 derived from the neuronal marker RASGRP2 and found that T cells specific for these peptides cross-reacted with peptides derived from EBV. These results suggest that EBV infection may lead to T cell responses that cross-react with neuronal markers in individuals expressing HLA-DR*15, leading to neurodegeneration. Cross-reactive epitopes identified in immunopeptidomic studies could in principle be targeted by therapies designed to induce epitope-specific immune tolerance (61, 62) and mitigate autoimmunity associated with infection.
FUTURE DIRECTIONS AND OUTSTANDING CHALLENGES
Immunopeptidomics can elucidate the basic biology and mechanisms of antigen presentation in infectious disease, improve predictive modeling of pathogen-derived T cell epitopes, draw connections between infection and autoimmunity, and identify promising vaccine targets. The discovery of new T cell antigens would aid in the development of vaccines against several globally important pathogens. For example, liver-resident CD8+ T cells are known to be a strong correlate of immunity to malaria (63, 64), but only a few T cell antigens presented in liver-stage malaria have been identified (65, 66). T cell responses are also thought to be important for immunity to bacterial pathogens with a rapidly growing incidence of antibiotic resistance, such as Salmonella spp. (67) and Staphylococcus aureus (68), as well as parasites such as Trypanosoma cruzi, which causes Chagas disease (69). Some of these protective T cell responses could target ligands that standard immunopeptidomic workflows do not detect, such as noncanonical translation products (70) or small molecules presented by the HLA-like molecules CD1 (71) and MR1 (72), but methods are being developed to analyze these unconventional epitopes by MS as well (70, 73, 74). The use of immunopeptidomics to comprehensively identify T cell antigens could therefore help combat both long-standing global health problems and emerging crises.
Although immunopeptidomics can provide valuable information about the repertoire of pathogen-derived MHC ligands at a whole-proteome scale, it currently has at least three important limitations, enumerated in Table 1. Some of these limitations can be overcome by combining MS-based immunopeptidomics with other complementary techniques, and some may be overcome through ongoing method development.
TABLE 1.
Three significant limitations of current immunopeptidomic methods
| Limitation of immunopeptidomics | Implications | Relevant references | |
|---|---|---|---|
| 1 | Not all peptides presented on MHCs are immunogenic | Targets identified by immunopeptidomics should ideally be further screened for T cell recognition and immunogenicity using other assays. | 75 – 79 |
| 2 | Cell-to-cell heterogeneity in antigen presentation cannot be resolved | Some contributions to the MS signal in an immunopeptidomic study may come from uninfected bystander cells, and a mixture of different cell types cannot be deconvoluted to learn which types are important for antigen presentation. Presentation of specific antigens can be probed at a single-cell level using pMHC-specific antibodies or biotin labels transferred between cells. | 80 – 85 |
| 3 | Large amounts of sample input are required | Studies in primary cells or with pathogens that cannot be cultured in large quantities are currently difficult to carry out. Microfluidic methods and other low-volume sample-handling techniques may help overcome these limitations. Targeted MS analyses may have higher sensitivity and require lower input. | 6, 86 |
Determining whether a peptide discovered through immunopeptidomics is immunogenic (problem 1) generally requires combining immunopeptidomics with techniques that can directly measure a T cell response against a given peptide of interest. Traditionally, this has been done by measuring production of IFN-γ by T cells specific for each epitope of interest (40). T cells specific for a pMHC complex of interest can also be stained using pMHC tetramers (75). More recently, a suite of methods for high-throughput measurement of T cell receptor (TCR)-pMHC interactions has been developed (76). Computational models have also been developed to predict the immunogenicity of putative T cell epitopes (77–79), which may help prioritize hits identified in an immunopeptidomic experiment for development as potential vaccine targets.
Cell-to-cell heterogeneity in antigen presentation (problem 2) may be consequential for immunity to some infections. For example, certain antigens of Mycobacterium tuberculosis can be presented by both infected cells and uninfected bystander cells (80, 81), while other antigens may be specific to infected cells. An antigen presented by uninfected bystander cells could potentially cause off-target lysis of bystander cells by cytotoxic T cells if targeted by a vaccine and/or might be presented at a lower level on infected cells than immunopeptidomic data would suggest, leading to less effective targeting of infected cells. Antibodies that recognize specific peptide-MHC complexes (82, 83) can enable measurements of antigen presentation at the single-cell level using flow cytometry or microscopy. Interactions between T cells and antigen-presenting cells (APCs) can also be tracked using a transferable biotin label (84, 85), providing a single-cell readout of pMHC-TCR interactions. These methods can complement immunopeptidomics by probing antigen presentation in a heterogeneous population of cells.
Whereas recent immunopeptidomic studies in cancer immunology have used sample input on the order of 106 to 107 cells (6), sample input on the order of 108 to 109 cells (problem 3) is often required to detect pathogen-derived peptides. The use of microfluidic devices or other low-volume sample handling techniques (86) could help reduce input requirements dramatically while retaining high sensitivity. Serial immunoprecipitation has already been successfully used to isolate both MHC class I and class II peptides from the same sample, further conserving input material (40). Lower sample input requirements could enable immunopeptidomic studies on infected primary cells or ex vivo samples from animals infected with a pathogen of interest or studies of pathogens that are impossible to culture in large quantities, such as the liver stage of malaria parasites (Plasmodium spp.).
Combining computational modeling with targeted mass spectrometry approaches could provide another avenue toward reducing sample input requirements (problem 3). Targeted MS analyses can detect MHC-associated peptides with greater sensitivity than data-dependent analyses, potentially reducing sample input requirements and increasing the likelihood of detecting peptides that would not be detectable in a data-dependent analysis. Accurate predictive models of peptide presentation on MHCs could identify promising candidates to experimentally validate using highly sensitive targeted MS workflows, and these experimental results could in turn be used for further model refinement.
Future improvements in immunopeptidomic methods may also focus on overcoming technical pitfalls that can result in inaccurate identification of MHC peptides by MS. In one instance, for example, HIV-derived peptides previously identified as MHC class I ligands (52) were shown to nonspecifically copurify with host membrane proteins (87). This result highlights the importance of proper controls in immunopeptidomic studies. Ambiguities in peptide identification may also be a source of uncertainty. It has recently been proposed that posttranslationally spliced peptides (88–91) and peptides derived from noncanonical translation products (92, 93) contribute substantially to the immunopeptidome. However, matching mass spectra against an expanded search space that includes spliced peptides and noncanonical translation products may increase the risk of false positives (70, 94). The false-discovery rate associated with these identifications must be carefully estimated to obtain accurate results (70).
Immunopeptidomics has already been used to design vaccines that improve control of infection in mouse models of chlamydia (42), leishmaniasis (44), and tuberculosis (40). As the field continues to advance, it is likely that immunopeptidomics and/or computational models trained on immunopeptidomic data will provide a rapid, sensitive, and systematic means of identifying vaccine targets in many human pathogens. Combining immunopeptidomics with other next-generation immunoassays and existing preclinical and clinical vaccine development platforms has the potential to have a transformative impact on global health.
ACKNOWLEDGMENTS
We acknowledge helpful, informative conversations with Lauren Stopfer and Cameron Flower.
O.K.L. and B.D.B. are supported by funding from the Ragon Institute of MGH and Harvard and the MIT Department of Biological Engineering, and by a grant from the National Institutes of Health (R21DE026582, R01A1022553, R01AR073252). O.K.L. and F.M.W. are supported by a grant from the National Cancer Institute (U54 CA210180).
Contributor Information
Forest M. White, Email: fwhite@mit.edu.
Bryan D. Bryson, Email: bryand@mit.edu.
Aleksandra Nita-Lazar, NIAID, NIH.
REFERENCES
- 1.Mitchison NA. 2004. T-cell–B-cell cooperation. Nat Rev Immunol 4:308–312. doi: 10.1038/nri1334. [DOI] [PubMed] [Google Scholar]
- 2.Blum JS, Wearsch PA, Cresswell P. 2013. Pathways of antigen processing. Annu Rev Immunol 31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruz FM, Colbert JD, Merino E, Kriegsman BA, Rock KL. 2017. The biology and underlying mechanisms of cross-presentation of exogenous antigens on MHC-I molecules. Annu Rev Immunol 35:149–176. doi: 10.1146/annurev-immunol-041015-055254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dendrou CA, Petersen J, Rossjohn J, Fugger L. 2018. HLA variation and disease. Nat Rev Immunol 18:325–339. doi: 10.1038/nri.2017.143. [DOI] [PubMed] [Google Scholar]
- 5.Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, Cox AL, Appella E, Engelhard VH. 1992. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255:1261–1264. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 6.Stopfer LE, Mesfin JM, Joughin BA, Lauffenburger DA, White FM. 2020. Multiplexed relative and absolute quantitative immunopeptidomics reveals MHC I repertoire alterations induced by CDK4/6 inhibition. Nat Commun 11:2760. doi: 10.1038/s41467-020-16588-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panicali D, Davis SW, Weinberg RL, Paoletti E. 1983. Construction of live vaccines by using genetically engineered poxviruses: biological activity of recombinant vaccinia virus expressing influenza virus hemagglutinin. Proc Natl Acad Sci U S A 80:5364–5368. doi: 10.1073/pnas.80.17.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tameris MD, Hatherill M, Landry BS, Scriba TJ, Snowden MA, Lockhart S, Shea JE, McClain JB, Hussey GD, Hanekom WA, Mahomed H, McShane H. 2013. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet 381:1021–1028. doi: 10.1016/S0140-6736(13)60177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ternette N, Block PD, Sánchez-Bernabéu Á, Borthwick N, Pappalardo E, Abdul-Jawad S, Ondondo B, Charles PD, Dorrell L, Kessler BM, Hanke T. 2015. Early kinetics of the HLA class I-associated peptidome of MVA.HIVconsv-infected cells. J Virol 89:5760–5771. doi: 10.1128/JVI.03627-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson KL, Ovsyannikova IG, Mason CJ, Bergen HR, III, Poland GA. 2009. Discovery of naturally processed and HLA-presented class I peptides from vaccinia virus infection using mass spectrometry for vaccine development. Vaccine 28:38–47. doi: 10.1016/j.vaccine.2009.09.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croft NP, Smith SA, Pickering J, Sidney J, Peters B, Faridi P, Witney MJ, Sebastian P, Flesch IEA, Heading SL, Sette A, La Gruta NL, Purcell AW, Tscharke DC. 2019. Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc Natl Acad Sci U S A 116:3112–3117. doi: 10.1073/pnas.1815239116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croft NP, Smith SA, Wong YC, Tan CT, Dudek NL, Flesch IEA, Lin LCW, Tscharke DC, Purcell AW. 2013. Kinetics of antigen expression and epitope presentation during virus infection. PLoS Pathog 9:e1003129. doi: 10.1371/journal.ppat.1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yewdell JW, Antón LC, Bennink JR. 1996. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J Immunol 157:1823–1826. [PubMed] [Google Scholar]
- 14.Trentini DB, Pecoraro M, Tiwary S, Cox J, Mann M, Hipp MS, Hartl FU. 2020. Role for ribosome-associated quality control in sampling proteins for MHC class I-mediated antigen presentation. Proc Natl Acad Sci U S A 117:4099–4108. doi: 10.1073/pnas.1914401117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen SG, Zak DE, Xu G, Ford JC, Marshall EE, Malouli D, Gilbride RM, Hughes CM, Ventura AB, Ainslie E, Randall KT, Selseth AN, Rundstrom P, Herlache L, Lewis MS, Park H, Planer SL, Turner JM, Fischer M, Armstrong C, Zweig RC, Valvo J, Braun JM, Shankar S, Lu L, Sylwester AW, Legasse AW, Messerle M, Jarvis MA, Amon LM, Aderem A, Alter G, Laddy DJ, Stone M, Bonavia A, Evans TG, Axthelm MK, Früh K, Edlefsen PT, Picker LJ. 2018. Prevention of tuberculosis in rhesus macaques by a cytomegalovirus-based vaccine. Nat Med 24:130–143. doi: 10.1038/nm.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sylvester-Hvid C, Kristensen N, Blicher T, Ferré H, Lauemøller SL, Wolf XA, Lamberth K, Nissen MH, Pedersen LØ, Buus S. 2002. Establishment of a quantitative ELISA capable of determining peptide-MHC class I interaction. Tissue Antigens 59:251–258. doi: 10.1034/j.1399-0039.2002.590402.x. [DOI] [PubMed] [Google Scholar]
- 17.Peters B, Bui H-H, Frankild S, Nielson M, Lundegaard C, Kostem E, Basch D, Lamberth K, Harndahl M, Fleri W, Wilson SS, Sidney J, Lund O, Buus S, Sette A. 2006. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Comput Biol 2:e65. doi: 10.1371/journal.pcbi.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidney J, Southwood S, Oseroff C, Guercio M, Sette A, Grey HM. 2001. Measurement of MHC/peptide interactions by gel filtration. Curr Protoc Immunol Chapter 18:Unit 18.3. doi: 10.1002/0471142735.im1803s31. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, Røder G, Peters B, Sette A, Lund O, Buus S. 2007. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. 2017. NetMHCpan-4.0: improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol 199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. 2020. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abelin JG, Keskin DB, Sarkizova S, Hartigan CR, Zhang W, Sidney J, Stevens J, Lane W, Zhang GL, Eisenhaure TM, Clauser KR, Hacohen N, Rooney MS, Carr SA, Wu CJ. 2017. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46:315–326. doi: 10.1016/j.immuni.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abelin JG, Harjanto D, Malloy M, Suri P, Colson T, Goulding SP, Creech AL, Serrano LR, Nasir G, Nasrullah Y, McGann CD, Velez D, Ting YS, Poran A, Rothenberg DA, Chhangawala S, Rubinsteyn A, Hammerbacher J, Gaynor RB, Fritsch EF, Greshock J, Oslund RC, Barthelme D, Addona TA, Arieta CM, Rooney MS. 2019. Defining HLA-II ligand processing and binding rules with mass spectrometry enhances cancer epitope prediction. Immunity 51:766–779.e17. doi: 10.1016/j.immuni.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Sarkizova S, Klaeger S, Le PM, Li LW, Oliveira G, Keshishian H, Hartigan CR, Zhang W, Braun DA, Ligon KL, Bachireddy P, Zervantonakis IK, Rosenbluth JM, Ouspenskaia T, Law T, Justesen S, Stevens J, Lane WJ, Eisenhaure T, Lan Zhang G, Clauser KR, Hacohen N, Carr SA, Wu CJ, Keskin DB. 2020. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat Biotechnol 38:199–209. doi: 10.1038/s41587-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu G, Carter B, Bricken T, Jain S, Viard M, Carrington M, Gifford DK. 2020. Computationally optimized SARS-CoV-2 MHC class I and II vaccine formulations predicted to target human haplotype distributions. Cell Syst 11:131–144.e6. doi: 10.1016/j.cels.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Donnell TJ, Rubinsteyn A, Laserson U. 2020. MHCflurry 2.0: improved pan-allele prediction of MHC class I-presented peptides by incorporating antigen processing. Cell Syst 11:42–48.e7. doi: 10.1016/j.cels.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Woodworth JS, Fortune SM, Behar SM. 2008. Bacterial protein secretion is required for priming of CD8+ T cells specific for the Mycobacterium tuberculosis antigen CFP10. Infect Immun 76:4199–4205. doi: 10.1128/IAI.00307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hie B, Bryson BD, Berger B. 2020. Leveraging uncertainty in machine learning accelerates biological discovery and design. Cell Syst 11:461–477.e9. doi: 10.1016/j.cels.2020.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. 2020. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiyotani K, Toyoshima Y, Nemoto K, Nakamura Y. 2020. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J Hum Genet 65:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knierman MD, Lannan MB, Spindler LJ, McMillian CL, Konrad RJ, Siegel RW. 2020. The human leukocyte antigen class II immunopeptidome of the SARS-CoV-2 spike glycoprotein. Cell Rep 33:108454. doi: 10.1016/j.celrep.2020.108454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weingarten-Gabbay S, Klaeger S, Sarkizova S, Pearlman LR, Chen D-Y, Gallagher KME, Bauer MR, Taylor HB, Dunn WA, Tarr C, Sidney J, Rachimi S, Conway HL, Katsis K, Wang Y, Leistritz-Edwards D, Durkin MR, Tomkins-Tinch CH, Finkel Y, Nachshon A, Gentili M, Rivera KD, Carulli IP, Chea VA, Chandrashekar A, Bozkus CC, Carrington M, MGH COVID-19 Collection & Processing Team, Bhardwaj N, Barouch DH, Sette A, Maus MV, Rice CM, Clauser KR, Keskin DB, Pregibon DC, Hacohen N, Carr SA, Abelin JG, Saeed M, Sabeti PC. 2021. Profiling SARS-CoV-2 HLA-I peptidome reveals T cell epitopes from out-of-frame ORFs. Cell doi: 10.1016/j.cell.2021.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson HL, Amara RR. 2005. T cell vaccines for microbial infections. Nat Med 11:S25–S32. doi: 10.1038/nm1212. [DOI] [PubMed] [Google Scholar]
- 34.Titball RW. 2008. Vaccines against intracellular bacterial pathogens. Drug Discov Today 13:596–600. doi: 10.1016/j.drudis.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. 2020. Global tuberculosis report 2020. World Health Organization, Geneva, Switzerland. https://www.who.int/publications/i/item/9789240013131.
- 36.World Health Organization. 2018. Report on global sexually transmitted infection surveillance, 2018. World Health Organization, Geneva, Switzerland. https://apps.who.int/iris/bitstream/handle/10665/277258/9789241565691-eng.pdf?ua=1.
- 37.Emerson PM, Hooper PJ, Sarah V. 2017. Progress and projections in the program to eliminate trachoma. PLoS Negl Trop Dis 11:e0005402. doi: 10.1371/journal.pntd.0005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flyer DC, Ramakrishna V, Miller C, Myers H, McDaniel M, Root K, Flournoy C, Engelhard VH, Canaday DH, Marto JA, Ross MM, Hunt DF, Shabanowitz J, White FM. 2002. Identification by mass spectrometry of CD8+-T-cell Mycobacterium tuberculosis epitopes within the Rv0341 gene product. Infect Immun 70:2926–2932. doi: 10.1128/IAI.70.6.2926-2932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMurtrey C, Harriff MJ, Swarbrick GM, Duncan A, Cansler M, Null M, Bardet W, Jackson KW, Lewinsohn DA, Hildebrand W, Lewinsohn DM. 2017. T cell recognition of Mycobacterium tuberculosis peptides presented by HLA-E derived from infected human cells. PLoS One 12:e0188288. doi: 10.1371/journal.pone.0188288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bettencourt P, Müller J, Nicastri A, Cantillon D, Madhavan M, Charles PD, Fotso CB, Wittenberg R, Bull N, Pinpathomrat N, Waddell SJ, Stylianou E, Hill AVS, Ternette N, McShane H. 2020. Identification of antigens presented by MHC for vaccines against tuberculosis. NPJ Vaccines 5:2. doi: 10.1038/s41541-019-0148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karunakaran KP, Rey-Ladino J, Stoynov N, Berg K, Shen C, Jiang X, Gabel BR, Yu H, Foster LJ, Brunham RC. 2008. Immunoproteomic discovery of novel T cell antigens from the obligate intracellular pathogen Chlamydia. J Immunol 180:2459–2465. doi: 10.4049/jimmunol.180.4.2459. [DOI] [PubMed] [Google Scholar]
- 42.Karunakaran KP, Yu H, Foster LJ, Brunham RC. 2016. Using MHC molecules to define a Chlamydia T cell vaccine. Methods Mol Biol 1403:419–432. doi: 10.1007/978-1-4939-3387-7_23. [DOI] [PubMed] [Google Scholar]
- 43.The RTS,S Clinical Trials Partnership. 2012. A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. N Engl J Med 367:2284–2295. doi: 10.1056/nejmoa1208394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mou Z, Li J, Boussoffara T, Kishi H, Hamana H, Ezzati P, Hu C, Yi W, Liu D, Khadem F, Okwor I, Jia P, Shitaoka K, Wang S, Ndao M, Petersen C, Chen J, Rafati S, Louzir H, Muraguchi A, Wilkins JA, Uzonna JE. 2015. Identification of broadly conserved cross-species protective Leishmania antigen and its responding CD4+ T cells. Sci Transl Med 7:310ra167. doi: 10.1126/scitranslmed.aac5477. [DOI] [PubMed] [Google Scholar]
- 45.Kaye P, Scott P. 2011. Leishmaniasis: complexity at the host-pathogen interface. Nat Rev Microbiol 9:604–615. doi: 10.1038/nrmicro2608. [DOI] [PubMed] [Google Scholar]
- 46.Parren PI, Moore JP, Burton DR, Sattentau QJ. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13(Suppl A):S137–S162. [PubMed] [Google Scholar]
- 47.World Health Organization. 2019. Progress report on HIV, viral hepatitis and sexually transmitted infections, 2019. World Health Organization, Geneva, Switzerland. https://apps.who.int/iris/bitstream/handle/10665/324797/WHO-CDS-HIV-19.7-eng.pdf?ua=1.
- 48.Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M, Ehler L, Metcalf J, Liu S, Connors M. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol 3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 49.Vaccari M, Trindade CJ, Venzon D, Zanetti M, Franchini G. 2005. Vaccine-induced CD8 + central memory T cells in protection from simian AIDS. J Immunol 175:3502–3507. doi: 10.4049/jimmunol.175.6.3502. [DOI] [PubMed] [Google Scholar]
- 50.Gillespie GMA, Kaul R, Dong T, Yang H-B, Rostron T, Bwayo JJ, Kiama P, Peto T, Plummer FA, McMichael AJ, Rowland-Jones SL. 2002. Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. AIDS 16:961–972. doi: 10.1097/00002030-200205030-00002. [DOI] [PubMed] [Google Scholar]
- 51.Collins DR, Gaiha GD, Walker BD. 2020. CD8+ T cells in HIV control, cure and prevention. Nat Rev Immunol 20:471–482. doi: 10.1038/s41577-020-0274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yaciuk JC, Skaley M, Bardet W, Schafer F, Mojsilovic D, Cate S, Stewart CJ, McMurtrey C, Jackson KW, Buchli R, Olvera A, Cedeño S, Plana M, Mothe B, Brander C, West JT, Hildebrand WH. 2014. Direct interrogation of viral peptides presented by the class I HLA of HIV-infected T cells. J Virol 88:12992–13004. doi: 10.1128/JVI.01914-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chikata T, Paes W, Akahoshi T, Partridge T, Murakoshi H, Gatanaga H, Ternette N, Oka S, Borrow P, Takiguchi M. 2019. Identification of immunodominant HIV-1 epitopes presented by HLA-C*12:02, a protective allele, using an immunopeptidomics approach. J Virol 93:e00634-19. doi: 10.1128/JVI.00634-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ternette N, Yang H, Partridge T, Llano A, Cedeño S, Fischer R, Charles PD, Dudek NL, Mothe B, Crespo M, Fischer WM, Korber BTM, Nielsen M, Borrow P, Purcell AW, Brander C, Dorrell L, Kessler BM, Hanke T. 2016. Defining the HLA class I-associated viral antigen repertoire from HIV-1-infected human cells. Eur J Immunol 46:60–69. doi: 10.1002/eji.201545890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramarathinam SH, Gras S, Alcantara S, Yeung AWS, Mifsud NA, Sonza S, Illing PT, Glaros EN, Center RJ, Thomas SR, Kent SJ, Ternette N, Purcell DFJ, Rossjohn J, Purcell AW. 2018. Identification of native and posttranslationally modified HLA-B*57:01-restricted HIV envelope derived epitopes using immunoproteomics. Proteomics 18:1700253. doi: 10.1002/pmic.201700253. [DOI] [PubMed] [Google Scholar]
- 56.Rucevic M, Kourjian G, Boucau J, Blatnik R, Garcia Bertran W, Berberich MJ, Walker BD, Riemer AB, Le Gall S. 2016. Analysis of major histocompatibility complex-bound HIV peptides identified from various cell types reveals common nested peptides and novel T cell responses. J Virol 90:8605–8620. doi: 10.1128/JVI.00599-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Münz C, Lünemann JD, Getts MT, Miller SD. 2009. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat Rev Immunol 9:246–258. doi: 10.1038/nri2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alvarez-Navarro C, Cragnolini JJ, Santos HG, Dos Barnea E, Admon A, Morreale A, De Castro JAL. 2013. Novel HLA-B27-restricted epitopes from Chlamydia trachomatis generated upon endogenous processing of bacterial proteins suggest a role of molecular mimicry in reactive arthritis. J Biol Chem 288:25810–25825. doi: 10.1074/jbc.M113.493247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colmegna I, Cuchacovich R, Espinoza L. 2004. HLA-B27-associated reactive arthritis: pathogenetic and clinical considerations. Clin Microbiol Rev 17:348–369. doi: 10.1128/CMR.17.2.348-369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Jelcic I, Mühlenbruch L, Haunerdinger V, Toussaint NC, Zhao Y, Cruciani C, Faigle W, Naghavian R, Foege M, Binder TMC, Eiermann T, Opitz L, Fuentes-Font L, Reynolds R, Kwok WW, Nguyen JT, Lee J-H, Lutterotti A, Münz C, Rammensee H-G, Hauri-Hohl M, Sospedra M, Stevanovic S, Martin R. 2020. HLA-DR15 molecules jointly shape an autoreactive T cell repertoire in multiple sclerosis. Cell 183:1264–1281.e20. doi: 10.1016/j.cell.2020.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maldonado RA, LaMothe RA, Ferrari JD, Zhang A-H, Rossi RJ, Kolte PN, Griset AP, O’Neil C, Altreuter DH, Browning E, Johnston L, Farokhzad OC, Langer R, Scott DW, von Andrian UH, Kishimoto TK. 2015. Polymeric synthetic nanoparticles for the induction of antigen-specific immunological tolerance. Proc Natl Acad Sci U S A 112:E156–E165. doi: 10.1073/pnas.1408686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hunter Z, McCarthy DP, Yap WT, Harp CT, Getts DR, Shea LD, Miller SD. 2014. A biodegradable nanoparticle platform for the induction of antigen-specific immune tolerance for treatment of autoimmune disease. ACS Nano 8:2148–2160. doi: 10.1021/nn405033r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Villarino N, Schmidt N. 2013. CD8+ T cell responses to plasmodium and intracellular parasites. Curr Immunol Rev 9:169–178. doi: 10.2174/1573395509666131126232327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holz LE, Fernandez-Ruiz D, Heath WR. 2016. Protective immunity to liver-stage malaria. Clin Transl Immunol 5:e105. doi: 10.1038/cti.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Good MF, Pombo D, Quakyi IA, Riley EM, Houghten RA, Menon A, Alling DW, Berzofsky JA, Miller LH. 1988. Human T-cell recognition of the circumsporozoite protein of Plasmodium falciparum: immunodominant T-cell domains map to the polymorphic regions of the molecule. Proc Natl Acad Sci U S A 85:1199–1203. doi: 10.1073/pnas.85.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valencia-Hernandez AM, Ng WY, Ghazanfari N, Ghilas S, de Menezes MN, Holz LE, Huang C, English K, Naung M, Tan PS, Tullett KM, Steiner TM, Enders MH, Beattie L, Chua YC, Jones CM, Cozijnsen A, Mollard V, Cai Y, Bowen DG, Purcell AW, La Gruta NL, Villadangos JA, de Koning-Ward T, Barry AE, Barchet W, Cockburn IA, McFadden GI, Gras S, Lahoud MH, Bertolino P, Schittenhelm RB, Caminschi I, Heath WR, Fernandez-Ruiz D. 2020. A natural peptide antigen within the plasmodium ribosomal protein RPL6 confers liver TRM cell-mediated immunity against malaria in mice. Cell Host Microbe 27:950–962.e7. doi: 10.1016/j.chom.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 67.de Wit J, Souwer Y, Jorritsma T, Bos HK, ten Brinke A, Neefjes J, Marieke van Ham S. 2010. Antigen-specific B cells reactivate an effective cytotoxic T cell response against phagocytosed Salmonella through cross-presentation. PLoS One 5:e13016. doi: 10.1371/journal.pone.0013016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clegg J, Soldaini E, Bagnoli F, McLoughlin RM. 2021. Targeting skin-resident memory T cells via vaccination to combat Staphylococcus aureus infections. Trends Immunol 42:6–17. doi: 10.1016/j.it.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 69.Ward AI, Lewis MD, Taylor MC, Kelly JM. 2021. Incomplete recruitment of protective T cells facilitates Trypanosoma cruzi persistence in the mouse colon. bioRxiv doi: 10.1101/2021.02.26.433032. [DOI] [PMC free article] [PubMed]
- 70.Chong C, Müller M, Pak H, Harnett D, Huber F, Grun D, Leleu M, Auger A, Arnaud M, Stevenson BJ, Michaux J, Bilic I, Hirsekorn A, Calviello L, Simó-Riudalbas L, Planet E, Lubiński J, Bryśkiewicz M, Wiznerowicz M, Xenarios I, Zhang L, Trono D, Harari A, Ohler U, Coukos G, Bassani-Sternberg M. 2020. Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat Commun 11:1293. doi: 10.1038/s41467-020-14968-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barral DC, Brenner MB. 2007. CD1 antigen presentation: how it works. Nat Rev Immunol 7:929–941. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 72.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O’Hair RAJ, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 73.Yuan W, Kang S, Evans JE, Cresswell P. 2009. Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J Immunol 182:4784–4791. doi: 10.4049/jimmunol.0803981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harriff MJ, McMurtrey C, Froyd CA, Jin H, Cansler M, Null M, Worley A, Meermeier EW, Swarbrick G, Nilsen A, Lewinsohn DA, Hildebrand W, Adams EJ, Lewinsohn DM. 2018. MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci Immunol 3:eaao2556. doi: 10.1126/sciimmunol.aao2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bentzen AK, Marquard AM, Lyngaa R, Saini SK, Ramskov S, Donia M, Such L, Furness AJS, McGranahan N, Rosenthal R, Straten PT, Szallasi Z, Svane IM, Swanton C, Quezada SA, Jakobsen SN, Eklund AC, Hadrup SR. 2016. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat Biotechnol 34:1037–1045. doi: 10.1038/nbt.3662. [DOI] [PubMed] [Google Scholar]
- 76.Joglekar AV, Li G. 2020. T cell antigen discovery. Nat Methods doi: 10.1038/s41592-020-0867-z. [DOI] [PubMed] [Google Scholar]
- 77.Calis JJA, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, Sette A, Keşmir C, Peters B. 2013. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol 9:e1003266. doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Riley TP, Keller GLJ, Smith AR, Davancaze LM, Arbuiso AG, Devlin JR, Baker BM. 2019. Structure based prediction of neoantigen immunogenicity. Front Immunol 10:2047. doi: 10.3389/fimmu.2019.02047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu J, Wang W, Zhang J, Zhou B, Zhao W, Su Z, Gu X, Wu J, Zhou Z, Chen S. 2019. DeepHLApan: a deep learning approach for neoantigen prediction considering both HLA-peptide binding and immunogenicity. Front Immunol 10:2559. doi: 10.3389/fimmu.2019.02559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Srivastava S, Grace PS, Ernst JD. 2016. Antigen export reduces antigen presentation and limits T cell control of M. tuberculosis. Cell Host Microbe 19:44–54. doi: 10.1016/j.chom.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Srivastava S, Ernst JD. 2014. Cell-to-cell transfer of M. tuberculosis antigens optimizes CD4 T cell priming. Cell Host Microbe 15:741–752. doi: 10.1016/j.chom.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saini NK, Baena A, Ng TW, Venkataswamy MM, Kennedy SC, Kunnath-Velayudhan S, Carreño LJ, Xu J, Chan J, Larsen MH, Jacobs WR, Porcelli SA. 2016. Suppression of autophagy and antigen presentation by Mycobacterium tuberculosis PE-PGRS47. Nat Microbiol 1:16133. doi: 10.1038/nmicrobiol.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ortega PA, Silva-Miranda M, Torres-Larios A, Campos-Chávez E, Franken KCLCM, Ottenhoff THM, Ivanyi J, Espitia C. 2020. Selection of a single domain antibody, specific for an HLA-bound epitope of the mycobacterial Ag85B antigen. Front Immunol 11:577815. doi: 10.3389/fimmu.2020.577815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li G, Bethune MT, Wong S, Joglekar AV, Leonard MT, Wang JK, Kim JT, Cheng D, Peng S, Zaretsky JM, Su Y, Luo Y, Heath JR, Ribas A, Witte ON, Baltimore D. 2019. T cell antigen discovery via trogocytosis. Nat Methods 16:183–190. doi: 10.1038/s41592-018-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu Z, Li JP, Chen M, Wu M, Shi Y, Li W, Teijaro JR, Wu P. 2020. Detecting tumor antigen-specific T cells via interaction-dependent fucosyl-biotinylation. Cell 183:1117–1133.e19. doi: 10.1016/j.cell.2020.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Furlan C, Dirks RAM, Thomas PC, Jones RC, Wang J, Lynch M, Marks H, Vermeulen M. 2019. Miniaturised interaction proteomics on a microfluidic platform with ultra-low input requirements. Nat Commun 10:1525–1528. doi: 10.1038/s41467-019-09533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Partridge T, Nicastri A, Kliszczak AE, Yindom LM, Kessler BM, Ternette N, Borrow P. 2018. Discrimination between human leukocyte antigen class I-bound and co-purified HIV-derived peptides in immunopeptidomics workflows. Front Immunol 9:912–919. doi: 10.3389/fimmu.2018.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mishto M, Liepe J. 2017. Post-translational peptide splicing and T cell responses. Trends Immunol 38:904–915. doi: 10.1016/j.it.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 89.Liepe J, Marino F, Sidney J, Jeko A, Bunting DE, Sette A, Kloetzel PM, Stumpf MPH, Heck AJR, Mishto M. 2016. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354:354–358. doi: 10.1126/science.aaf4384. [DOI] [PubMed] [Google Scholar]
- 90.Faridi P, Li C, Ramarathinam SH, Illing PT, Mifsud NA, Ayala R, Song J, Gearing LJ, Hertzog PJ, Ternette N, Rossjohn J, Croft NP, Purcell AW. 2018. A subset of HLA-I peptides are not genomically templated: evidence for cis- and trans-spliced peptide ligands. Sci Immunol 3:eaar3947. doi: 10.1126/sciimmunol.aar3947. [DOI] [PubMed] [Google Scholar]
- 91.Platteel ACM, Mishto M, Textoris-Taube K, Keller C, Liepe J, Busch DH, Kloetzel PM, Sijts AJAM. 2016. CD8+ T cells of Listeria monocytogenes-infected mice recognize both linear and spliced proteasome products. Eur J Immunol 46:1109–1118. doi: 10.1002/eji.201545989. [DOI] [PubMed] [Google Scholar]
- 92.Laumont CM, Vincent K, Hesnard L, Audemard É, Bonneil É, Laverdure J-P, Gendron P, Courcelles M, Hardy M-P, Côté C, Durette C, St-Pierre C, Benhammadi M, Lanoix J, Vobecky S, Haddad E, Lemieux S, Thibault P, Perreault C. 2018. Noncoding regions are the main source of targetable tumor-specific antigens. Sci Transl Med 10:eaau5516. doi: 10.1126/scitranslmed.aau5516. [DOI] [PubMed] [Google Scholar]
- 93.Ruiz Cuevas MV, Hardy M-P, Hollý J, Bonneil É, Durette C, Courcelles M, Lanoix J, Côté C, Staudt LM, Lemieux S, Thibault P, Perreault C, Yewdell JW. 2021. Most non-canonical proteins uniquely populate the proteome or immunopeptidome. Cell Rep 34:108815. doi: 10.1016/j.celrep.2021.108815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rolfs Z, Müller M, Shortreed MR, Smith LM, Bassani-Sternberg M. 2019. Comment on “A subset of HLA-I peptides are not genomically templated: evidence for cis- and trans-spliced peptide ligands.” Sci Immunol 4:eaaw1622. doi: 10.1126/sciimmunol.aaw1622. [DOI] [PMC free article] [PubMed] [Google Scholar]