Abstract
The high-mobility-group I (HMGI) protein is a nonhistone component of active chromatin. In this work, we demonstrate that HMGI protein specifically binds to the AT-rich region of the murine beta interferon (IFN-β) promoter localized upstream of the murine virus-responsive element (VRE). Contrary to what has been described for the human promoter, HMGI protein did not specifically bind to the VRE of the murine IFN-β promoter. Stably transfected promoters carrying mutations on this HMGI binding site displayed delayed virus-induced kinetics of transcription. When integrated into chromatin, the mutated promoter remained repressed and never reached normal transcriptional activity. Such a phenomenon was not observed with transiently transfected promoters upon which chromatin was only partially reconstituted. Using UV footprinting, we show that the upstream AT-rich sequences of the murine IFN-β promoter constitute a preferential binding region for histone H1. Transfection with a plasmid carrying scaffold attachment regions as well as incubation with distamycin led to the derepression of the IFN-β promoter stably integrated into chromatin. In vitro, HMGI protein was able to displace histone H1 from the upstream AT-rich region of the wild-type promoter but not from the promoter carrying mutations on the upstream high-affinity HMGI binding site. Our results suggest that the binding of histone H1 to the upstream AT-rich region of the promoter might be partly responsible for the constitutive repression of the promoter. The displacement by HMGI protein of histone H1 could help to convert the IFN-β promoter from a repressed to an active state.
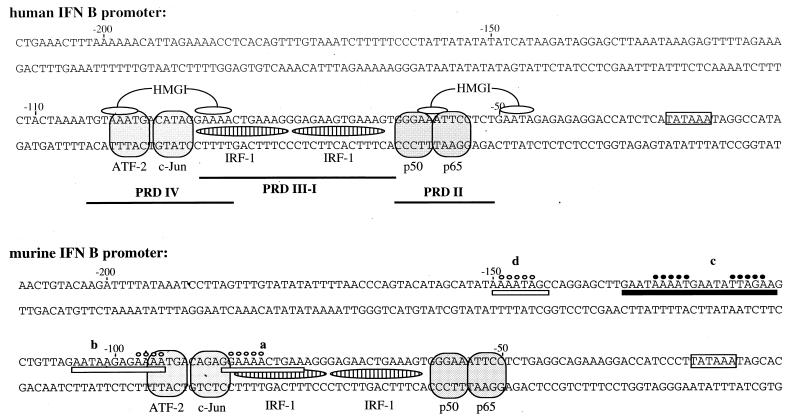
The activation of genes encoding type I interferons (alpha interferon [IFN-α] and IFN-β) constitutes the primary cellular defense against virus infections. The IFN-β gene is constitutively repressed in uninduced cells, and its transient activation can be achieved after virus infection. The virus-responsive element (VRE) corresponding to the minimal DNA sequence necessary for virus induction of the human IFN-β (huIFN-β) gene has been extensively studied over the last 10 years (24, 35, 37). It includes the sequence from −110 to −55, containing DNA binding sites for several transcription factors: NF-κB, which binds to positive regulatory domain II (PRDII, from −64 to −55); proteins belonging to the family of interferon regulatory factors (IRFs), which bind to positive regulatory domains I and III (PRDI and PRDIII, from −89 to −64); and activating transcription factor 2 (ATF-2/c-Jun), which binds to positive regulatory domain IV (PRDIV, from −104 to −86). More recently, CREB binding protein (CBP) and p300 have been described as coactivators participating in the synergistic transcriptional activation of the huIFN-β gene (29, 48). The sequences corresponding to the four positive regulatory domains, constituting the human VRE (huVRE), are extremely well conserved in the murine promoter (see Fig. 1). A murine VRE (muVRE), analogous to the huVRE, can thereby be considered present in the murine IFN-β (muIFN-β) promoter from positions −92 to −50.
FIG. 1.
DNA sequences of the huIFN-β and muIFN-β promoters spanning from the TATA box to position −210, as described by Vodjdani et al. (41) and Thanos and Maniatis (36). PRDI, PRDII, PRDIII, and PRDIV, corresponding to the huVRE and containing the DNA binding sites of transcription factors NF-κB (p50 and p65 subunits), IRF-1, and ATF-2/c-Jun and the binding sites of HMGI protein, are indicated as they have been determined on the human promoter. The regions (a, b, c, and d) of the murine promoter protected by HMGI protein during DNase I and hydroxyl radical footprinting reactions are indicated. Open bars indicate regions protected by HMGI protein during DNase I footprinting only in the absence of competitor DNA. Closed bars indicate regions that remained protected by HMGI protein during DNase I footprinting in the presence of competitor DNA. Open circles indicate bases protected by HMGI protein during hydroxyl radical footprinting only in the absence of competitor DNA. Closed circles indicate bases that remained protected by HMGI protein during hydroxyl radical footprinting in the presence of competitor DNA.
Binding sites for high-mobility-group I (HMGI) protein have been observed in the huIFN-β promoter region, one near the NF-κB binding site and the other near the ATF-2/c-Jun site. Mutations that affect these HMGI binding sites for PRDII or PRDIV have been shown to decrease virus induction of huIFN-β transcription (36, 47).
HMGI protein is a nonhistone component of active chromatin which has been demonstrated to contribute to the transcriptional regulation of several promoters, modulating DNA and chromatin structure (7) as well as DNA topology (2, 13). In most differentiated adult tissues, HMGI protein is not significantly expressed; in contrast, its expression is highly increased in embryonic undifferentiated cells as well as in proliferating and transformed cells (14, 46). Three “AT-hook” peptides in HMGI protein bind to short DNA AT-tract sequences (7, 47). A strong HMGI binding site contains at least two correctly spaced AT tracts (27). More recently, HMGI protein has been described as also being able to specifically bind to non-B DNA structures, such as cruciform DNA (13). With regard to the role of protein HMGI during transcriptional regulation of the huIFN-β gene, it was suggested that HMGI protein may act upon the huIFN-β promoter as an architectural protein necessary for the formation of a transcriptionally active multiprotein-DNA complex (38). In this work, we show that this is not the case for the binding of HMGI protein to the muIFN-β promoter.
Our footprinting data clearly indicate that there is only one high-affinity HMGI binding site in the murine promoter. It is located 5′ to the muVRE, between positions −133 and −114. When wild-type promoters and promoters with mutations in this high-affinity HMGI binding site were transiently transfected, the virus-induced kinetics of expression of the mutated promoters was delayed compared to that of the analogous wild-type promoters. Nevertheless, no significant difference was observed between the corresponding maximal virus-induced transcriptional activities of the wild-type and mutated promoter.
However, when these promoters were stably transfected, mutation of the high-affinity HMGI binding site in the region from positions −133 to −114 produced a much more dramatic effect. When integrated into chromatin, the mutated promoter not only displayed strongly delayed kinetics but also remained partly repressed and never reached virus-induced transcriptional activity equivalent to that of the wild-type promoter under the same conditions. The differences observed between the transiently and stably transfected promoters strongly suggest that the effect of mutations introduced into the high-affinity HMGI binding site could be linked to chromatin.
The mutated promoters lacking the upstream sequences from positions −330 to −150 did not display delayed virus-induced kinetics of transcriptional activation. UV footprints of histone H1 on supercoiled plasmids containing the muIFN-β promoter indicated preferential binding of histone H1 to the upstream region of the promoter from positions −220 to −110. This region corresponds to an AT-rich (76% AT content) sequence in the murine promoter as well as in the human promoter and has been described as a negative regulatory element responsible for the constitutive repression of the huIFN-β promoter (50). Distamycin, a drug which contacts the minor groove of AT-rich DNA sequences and which has been described as capable of specifically displacing histone H1 (17), activated the IFN-β promoter containing the upstream AT-rich sequence. Such a phenomenon was not observed with promoters which contained only the VRE sequence and which lacked the upstream AT-rich sequence. Histone H1 has been described as preferentially binding to AT-rich scaffold attachment regions (SAR) (17). Transfection of stably transfected cells with a plasmid carrying SAR sequences also derepressed the IFN-β promoter.
We discuss here a new role for HMGI protein, acting as a potential antirepressor of the muIFN-β promoter. Our results suggest that histone H1, bound to the AT-rich region of the IFN-β promoter, could participate in the repression of the promoter. In vitro, HMGI protein bound to its site from positions −133 to −114 was able to displace histone H1 from the upstream region of the promoter. The displacement of histone H1 by HMGI protein could lead to the conversion of the promoter from a repressed state to an active state.
MATERIALS AND METHODS
HMGI and H1 proteins.
Purification of HMGI protein was carried out as described by Reeves and Nissen (30). Full-length recombinant human HMGI protein was produced with plasmid pET3b-HMGI (a generous gift from E. Käs, LBME, Toulouse, France) carrying the complete human HMGI cDNA cloned between the NdeI and BamHI sites of expression vector pET3b. Plasmid pET3b-HMGI was transformed in Escherichia coli BL21pLysS, which was grown in Luria-Bertani medium supplemented with 100 μg of ampicillin and 40 μg of chloramphenicol per ml to an optical density at 600 nm of 0.5. At that stage, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM; 3 h later, the cells were collected by centrifugation. The pellet from 1 liter of culture was resuspended in 10 ml of phosphate-buffered saline–0.1% Triton–1 mM phenylmethylsulfonyl fluoride (PMSF). The bacteria were disrupted by two cycles in a French press at 20,000 lb/in2 and extracted with 5% perchloric acid. Acid-soluble proteins were precipitated from the extract with 25% trichloroacetic acid. For further purification, the proteins were applied to a CM52 cellulose column previously equilibrated with 10 bed volumes of buffer A (20 mM potassium phosphate [pH 7.4], 20 mM NaCl, 0.2 mM PMSF, 0.5 mM EDTA). Pure full-length HMGI protein was eluted in buffer A–150 to 200 mM NaCl. The purity of the protein was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Purified histone H1 was a generous gift from A. Prunell (IJM, Paris, France).
Plasmid constructions.
Plasmid pGCAT-C3 (see Fig. 2) was constructed by B. Soury in our laboratory by insertion of the 420-bp SacI-PstI muIFN-β promoter (positions −405 to +20) fragment (41) into the corresponding sites of plasmid vector pGCAT-C (11).
FIG. 2.
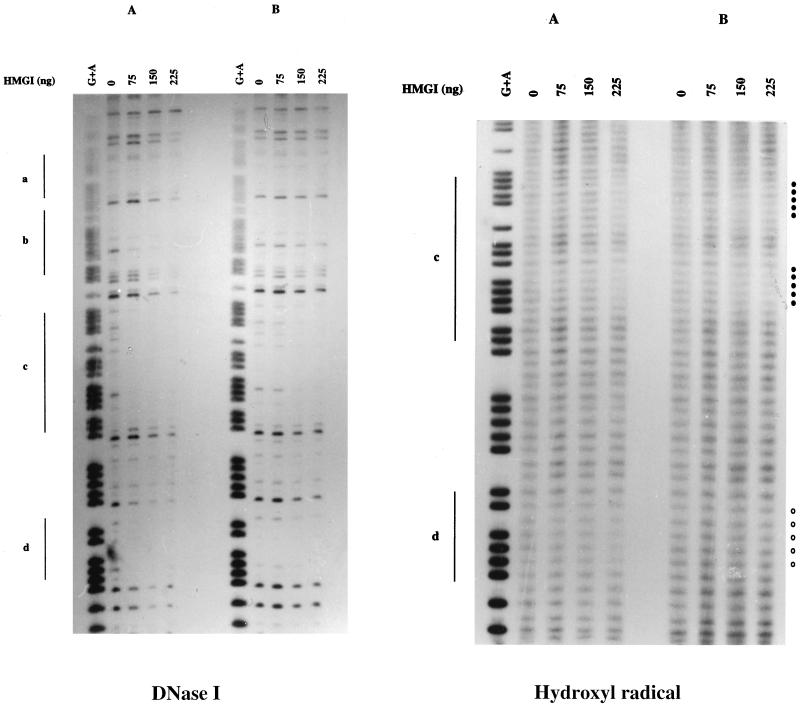
DNase I and hydroxyl radical footprinting reactions of the muIFN-β promoter carried out with HMGI protein in the presence (A) or absence (B) of 10 μg of nonradioactive sonicated salmon sperm DNA per ml as nonhomologous competitor DNA. The EcoRI-PvuII fragment of plasmid pGCAT-C3 containing the muIFN-β promoter DNA labelled at the 5′ end of the EcoRI site (position −162) was digested as described in Materials and Methods and analyzed on a 7 M urea–8% polyacrylamide gel. Lane G+A, specific G+A DNA sequencing reaction of the corresponding labelled DNA fragment. The regions protected by HMGI protein were as follows: a, −76 to −86; b, −94 to −101; c, −114 to −143; and d, −144 to −150. Closed circles indicate bases that remained protected by HMGI protein in the presence of competitor DNA. Open circles indicate bases that remained protected by HMGI protein only in the absence of competitor DNA.
The wild-type and mutated (mutI.g and mutI.cg) muIFN-β promoters (positions −330 to +20) were constructed by single or double PCR with plasmid pGCAT-C3 as a template. For all constructions, the PstI oligonucleotide (5′-AGCTACTCTGCAGGGCTTTTCAGTG-3′) was used as a 5′ primer, and the BamHI oligonucleotide (5′-CCCGGATCCTGGCAGTGAGAATGAT-3′) was used as a 3′ primer. The final PCR products were cleaved with PstI and BamHI and cloned in pBLCAT3 (26). All constructions were checked by nucleotide sequencing of the double-stranded DNA templates.
Plasmid pBLCAT3-muIFN-β wt, carrying the wild-type muIFN-β promoter fragment from −330 to +20, was constructed by single PCR with PstI and BamHI oligonucleotides as primers.
Plasmid pBLCAT3-muIFN-β mutI.g and pBLCAT3-muIFN-β mutI.cg, carrying the muIFN-β promoter fragment from positions −330 to +20 and with mutations in the AT tracts of the upstream HMGI binding site, were constructed by double PCR. For mutI.g, the primers were the PstI oligonucleotide and 5′-CTTCTAATATTCCTCTCATTCAAGC-3′ and the BamHI oligonucleotide and 5′-GCTTGAATGAGAGGAATATTAGAAG-3′. For mutI.cg, the primers were the PstI oligonucleotide and 5′-CTTCGCATCGTCCGTTTATTCAAGC-3′ and the BamHI oligonucleotide and 5′-GCTTGAATAAACGGACGATGCGAAG-3′. The corresponding PCR products were purified, annealed, and used in a new round of PCR.
Plasmid pBLCAT3-muIFN-β wt150 and pBLCAT3-muIFN-β mutI.cg150, carrying a short wild-type promoter fragment and the mutI.cg muIFN-β promoter fragment from positions −150 to +20, respectively, were constructed by single PCR with plasmids pBLCAT3-muIFN-β wt and pBLCAT3-muIFN-β mutI.cg, respectively, as templates and with the BamHI oligonucleotide and 5′-ATAGCCTGCAGCTTGAAT-3′.
Plasmid pBLCAT3-muIFN-β wt110, carrying the short wild-type muIFN-β promoter fragment from positions −110 to +20, was constructed by single PCR with plasmid pBLCAT3-muIFN-β wt as a template and with the BamHI oligonucleotide and 5′-AAAACTGCAGTGTTAGAATAAGAGAAAATG-3′.
Plasmid pSP64-34 (17) contains the 657-bp histone SAR fragment cloned in vector pSP64.
DNase I footprinting.
For the footprinting experiments with the coding strand, the muIFN-β promoter fragment was isolated from 20 μg of plasmid pGCAT-C3 after digestion with EcoRI. After dephosphorylation with alkaline phosphatase, the fragment was 5′ end labelled with T4 polynucleotide kinase and further digested with PvuII. The 379-bp EcoRI-PvuII fragment (5 to 10 ng), 5′ end labelled at the EcoRI site, was incubated with various amounts of recombinant HMGI protein in a 100-μl volume of HMGI binding buffer containing 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 50 mM NaCl in the presence or absence of 10 μg of nonradioactive, sonicated salmon sperm DNA per ml for 10 min at room temperature. The digestion was carried out as previously described (4).
Hydroxyl radical footprinting.
The EcoRI-PvuII fragment isolated from plasmid pGCAT-C3 and 5′ end labelled at the EcoRI site was incubated with various amounts of recombinant HMGI protein in 100 μl of HMGI binding buffer in the presence or absence of 10 μg of nonradioactive, sonicated salmon sperm DNA per ml for 10 min at room temperature. The reaction was carried out in the presence of 2 μl of a freshly prepared solution of 20 mM Fe(II)–25 mM EDTA, 2 μl of a freshly prepared solution of 200 mM sodium ascorbate, and 2 μl of 6% hydrogen peroxide as previously described (5).
UV footprinting.
Various amounts of histone H1 were incubated with 750 ng of sonicated salmon sperm DNA in 50 μl of H1 binding buffer containing 10 mM Tris-HCl (pH 8.0), 12.5 mM NaCl, and 1 mM EDTA for 10 min at room temperature. Then, 500 ng of supercoiled plasmid pBLCAT3-muIFN-β wt or pBLCAT3-muIFN-β wt150 was added to the reaction mixture, and incubation was continued for 30 min at room temperature. Unless indicated otherwise, the samples were irradiated at 254 nm at room temperature with a manual Mineralight multiband UV 254- to 366-nm lamp (model UVGL-58). The lamp was preheated for 15 min before irradiation. The irradiated or nonirradiated DNA was denatured by incubation for 5 min with 5 μl of 2 M NaOH–2 mM EDTA. After precipitation, the DNA was annealed with the corresponding 5′-end-labelled primer and submitted to an elongation reaction with 1 U of T7 DNA polymerase and 125 μM cold deoxynucleoside triphosphate mixture in a final volume of 60 μl of reaction buffer containing 40 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 50 mM NaCl, and 1 mM dithiothreitol. After 30 min at 37°C, the DNA products were ethanol precipitated and loaded onto a denaturing 7 M urea–8% polyacrylamide sequencing gel buffered with Tris-borate-EDTA.
Gel binding assays.
Gel retardation experiments were done with 8% polyacrylamide gels (acrylamide-bisacrylamide, 38:2) by incubation of various amounts of HMGI protein with 5′-end-labelled wild-type, mutI.g, or mutI.cg double-stranded DNA probes for 10 min at room temperature in 20 μl of HMGI binding buffer in the presence of 250 ng of nonradioactive, sonicated salmon sperm DNA. The gels were dried and autoradiographed. The 5′-to-3′ sequences of the oligonucleotides used to prepare the corresponding DNA probes were as follows: (i) for the wild-type probe, GCTTGAATAAAATGAATATTAGAAG (coding strand) and CTTCTAATATTCATTTTATTCAAGC (noncoding strand); (ii) for the mutI.g probe, GCTTGAATGAGAGGAATATTAGAAG (coding strand) and CTTCTAATATTCCTCTCATTCAAGC (noncoding strand) and (iii) for the mutI.cg probe, GCTTGAATAAACGGACGATGCGAAG (coding strand) and CTTCTGCATCGTCCGTTTATTCAAGC (noncoding strand).
Transient DNA transfection, virus induction, and CAT assays.
L929 and HeLa S3 cells (six-well tissue culture plates; 200,000 cells/well) seeded in Dulbecco’s modified Eagle’s medium supplemented with antibiotics, l-glutamine, nonessential amino acids, and 10% fetal calf serum were transfected at 50% confluence by the calcium phosphate precipitation method with 1 μg of reporter plasmid. L929 cells were glycerol shocked with 10% glycerol for 1 min. Virus induction was carried out with Newcastle disease virus (NDV) as previously described (9). Mock-induced cells were treated as described above except that no NDV was added. The cells were harvested at different times postinduction, and chloramphenicol acetyltransferase (CAT) was assayed as previously described (25). Transfection efficiency was normalized with β-galactosidase as previously described when necessary (25). In each experiment, a given reporter plasmid was transfected, NDV induced, or mock induced in duplicate. Two different clones of each plasmid were tested in transient transfection assays. The results presented in Fig. 4A and B correspond to the averages of three independent experiments.
FIG. 4.
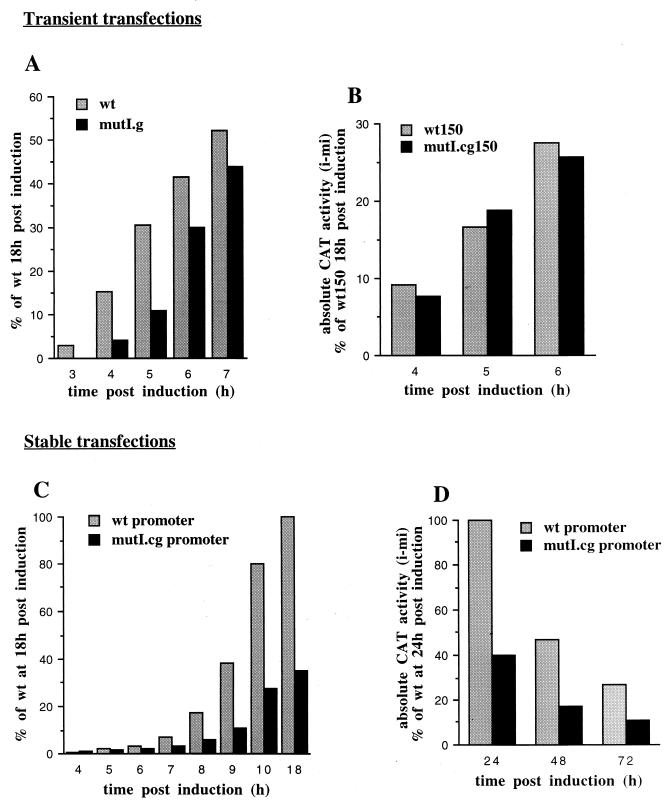
Kinetics of virus-induced transcriptional activation of the wild-type (wt) and mutI.cg muIFN-β promoters transiently or stably transfected. (A) HeLa cells were transiently transfected with the muIFN-β promoter (positions −330 to +20), either wild type (pBLCAT3-muIFN-β wt) or mutated on the upstream HMGI binding site (pBLCAT3-muIFN-β mutI.g) and fused to the CAT gene. (B) Kinetics of virus-induced transcriptional activation of short wild-type (pBLCAT3-muIFN-β wt150) and mutI.cg150 (pBLCAT3-muIFN-β mutI.cg150) muIFN-β promoters (positions −150 to +20) transiently transfected into HeLa cells. (C and D) Kinetics of virus-induced transcriptional activation of wild-type and mutI.cg muIFN-β promoters stably transfected into L929 cells. L929 cells were stably transfected with the muIFN-β promoter (positions −330 to +20), either wild type or mutated on the upstream HMGI binding site (mutI.cg) and fused to the CAT gene. For all experiments, the cells were induced (i) or mock induced (mi) with NDV. The absolute CAT activities (induced minus mock induced) of the cells collected at different times postinduction were measured as indicated in Materials and Methods. The corresponding values are expressed as a percentage of the CAT activity reached by the wild-type promoter at 18 h postinduction (24 h for panel D), which we have considered to correspond to 100%. Except for panel D, the values shown correspond to the average values obtained from three independent experiments, with duplicate determinations for each induced and mock-induced point.
Cell lines.
To construct the corresponding cell lines, wild-type or mutI.cg reporter plasmids were cotransfected with plasmid pCB6 carrying resistance to Geneticin (1). L929 cells (5 × 105 cells/100-mm dish) seeded in Dulbecco’s modified Eagle’s medium supplemented with antibiotics, l-glutamine, nonessential amino acids, and 10% fetal calf serum were transfected at 50% confluence by the calcium phosphate precipitation method with 10 μg of plasmid pBLCAT3-muIFN-β wt or pBLCAT3-muIFN-β mutI.cg and 10 μg of plasmid pCB6. Four hours after transfection, the cells were glycerol shocked with 10% glycerol for 1 min and washed three times with phosphate-buffered saline. The transfected cells were then selected in medium containing G418 (600 μg/ml; GIBCO) for 3 weeks. Clones were isolated, propagated, and tested for NDV-induced CAT activity during several passages of the cells. Except for the wt110 clone (see Fig. 7A), an average of 10 positive clones were pooled and frozen. The data for some experiments (see Fig. 4C and D) were obtained with freshly thawed cells that were passaged once. One day prior to virus induction, the cells were split among six-well plates (200,000 cells/well) containing medium without G418. The cells were virus induced, harvested, and assayed for CAT activity as previously described. Mock-induced cells were treated as described above except that no NDV was added. In each experiment, NDV inductions or mock inductions were carried out in duplicate. The results for these experiments (see Fig. 4C and D) correspond to the averages of three independent experiments.
FIG. 7.
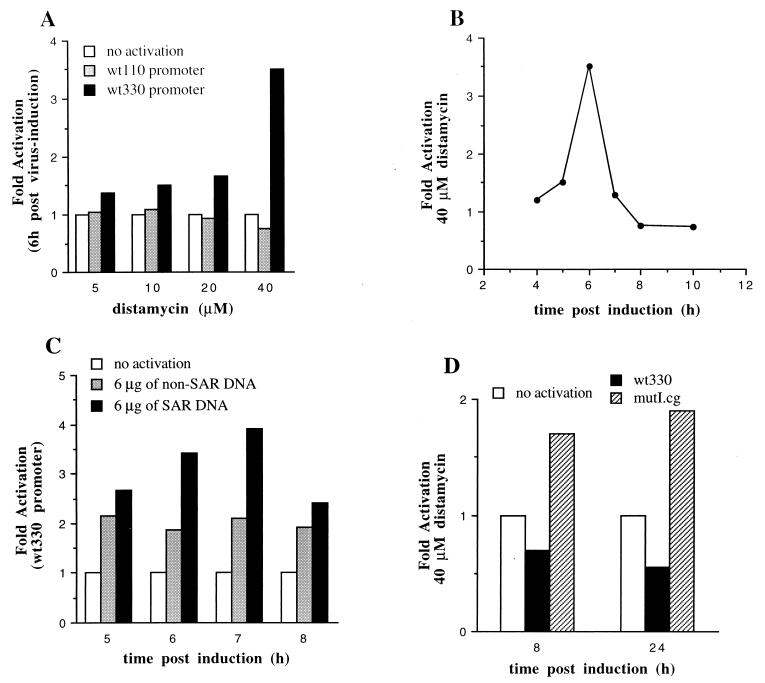
Effect of distamycin or SAR DNA upon the virus-induced transcriptional activation of the muIFN-β promoter. (A) Stably transfected cells containing the wt330 or wt110 IFN-β promoter were incubated in the absence or presence of increasing amounts of distamycin A. The cells were collected 6 h after NDV infection and tested for their corresponding CAT activities. (B) Stably transfected cells containing the wt330 IFN-β promoter were incubated in the presence of 40 μM distamycin. (C) Stably transfected cells containing the wt330 IFN-β promoter were transfected with 6 μg of plasmid pSP64-34 containing SAR DNA or 6 μg of plasmid pGEM4 containing vector non-SAR DNA. (D) Stably transfected wt330 or mutI.cg cell lines were incubated in the presence of 40 μM distamycin. In panels A, B, and D, the fold activation corresponds to the absolute CAT activity measured in the presence of distamycin divided by the absolute CAT activity measured in the absence of distamycin. In panel C, the fold activation corresponds to the absolute CAT activity measured after transfection divided by the absolute CAT activity measured in the absence of transfection. The values correspond to the average values obtained from two independent experiments.
Distamycin A (Sigma) at 5 mg/ml in 96% ethanol was freshly diluted in Dulbecco’s modified Eagle’s medium to final concentrations ranging from 5 to 40 μM. Cells were incubated in the presence of distamycin 20 h before virus infection and for various times after virus infection.
The wild-type cell line was transfected as previously described for L929 cells with 6 μg of plasmid pSP64-34 or pGEM4. The results of these experiments (see Fig. 7C) are the averages of two independent transfections with each point determined in duplicate.
RESULTS
High-affinity binding of the HMGI protein to the upstream AT-rich region of the IFN-β promoter.
Comparative analyses of the human and murine promoter sequences within the minimal DNA region necessary for the virus-induced expression of the IFN-β gene indicate that the sequences of NF-κB, IRF, and ATF-2/c-Jun binding sites present on the human promoter are conserved on the murine promoter (Fig. 1). In contrast, the HMGI binding site positioned near PRDII on the human promoter is disrupted in the murine promoter, since one of the two AT tracts interacting with HMGI protein is missing from the sequence of the murine promoter. Binding of HMGI protein to the human PRDII region has been described as essential for the synergistic transcriptional activation of the huIFN-β promoter (20).
A series of DNase I footprinting experiments was carried out in order to investigate the binding of HMGI protein to the muIFN-β promoter. The results obtained with a DNA fragment labelled at position −162 on the 5′ end of the coding strand are shown in Fig. 2. In the absence of competitor DNA (Fig. 2A), four protected DNA elements were localized on the murine promoter, as follows (Fig. 1): a, from −86 to −76; b, from −105 to −94; c, from −133 to −114; and d, from −150 to −143. Protection of a and b corresponds to the binding of HMGI protein to PRDIV. This binding is identical to that described for HMGI protein on human PRDIV. Protection of c and d is located on the region 5′ to murine PRDIV and has not been described yet for any IFN-β promoter. Sonicated salmon sperm DNA was added to the reaction mixture as competitor DNA in order to detect, from among the different HMGI binding sites, the one(s) for which the protein displayed a higher affinity and therefore the one(s) more likely to have functional significance. In the presence of competitor DNA (Fig. 2B), the only DNA region that remained protected by HMGI protein was the region corresponding to the c sequence. The d sequence remained weakly protected in the presence of competitor DNA, and the protection previously observed for PRDIV (a and b sequences) completely disappeared under these conditions.
Even in the absence of competitor DNA, we never observed on the coding strand protection by HMGI protein of the murine PRDII region.
We also carried out HMGI protein footprinting analysis of the noncoding strand (data not shown). This analysis confirmed the results previously obtained for the coding strand. However, only partial HMGI protein protection of the murine PRDII sequence corresponding to the region spanning from positions −60 to −54 could be observed on the noncoding strand. This protection immediately disappeared when competitor DNA was added to the reaction mixture.
Hydroxyl radical footprinting analysis was carried out in order to determine more precisely the DNA bases contacted by HMGI protein in region c. The results shown in Fig. 2 indicate that the bases directly protected by HMGI protein in this region constitute two AT tracts separated by 5 bp and corresponding to the sequence GAATAAAATGAATATTAGAAG, with the underlined bases being those directly protected by HMGI protein during hydroxyl radical footprinting.
These results obtained with DNase I and hydroxyl radical footprinting indicated (i) the presence of a high-affinity HMGI binding site on the upstream AT-rich region of the muIFN-β promoter and (ii) that the two HMGI binding sites previously described on the huIFN-β promoter as being necessary for the correct transcriptional activation of this gene were nonspecific HMGI binding sites on the murine promoter. It was therefore, interesting to investigate if the high-affinity HMGI binding site present in the regions from positions −133 to −114 (c) played a role during virus-induced transcriptional activation of the murine gene. We therefore disrupted this HMGI binding site by introducing mutations on the AT tracts directly protected by HMGI protein. The corresponding mutated sequences were as follows: mutI.g, 5′-GCTTGAATgAgAgGAATATTAGAAG-3′; and mutI.cg, 5′-GCTTGAATAAAcgGAcgATgcGAAG (with the mutated bases being shown in lowercase letters).
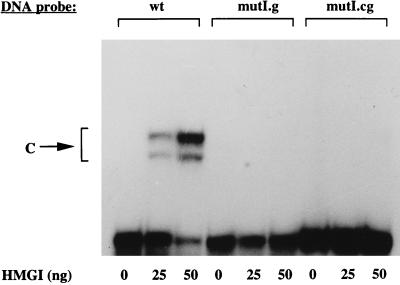
Gel retardation experiments with HMGI protein and either the wild-type site or the mutated sites (mutI.g and mutI.cg) were performed in order to confirm that HMGI protein had no affinity for any of the mutated sequences. As shown in Fig. 3, protein-DNA complexes were obtained only when HMGI protein was incubated with the wild-type sequence. It is also interesting to note that the complexes formed between HMGI protein and the wild-type probe systematically migrated as two distinct bands, a phenomenon that we have recurrently observed with different protein preparations as well as with different short DNA probes. This result may indicate the formation of two different protein-DNA complexes: a first, less retarded complex with one HMGI protein molecule per DNA probe bound to the two AT tracts present on the DNA probe, and a second, more retarded complex with two HMGI protein molecules per DNA probe, each HMGI protein molecule bound to only one of the two AT tracts. Nevertheless, no complex was observed under these conditions with the mutI.g or mutI.cg sequences, confirming, therefore, that HMGI protein had no affinity for these mutated sequences.
FIG. 3.
Autoradiograph of an 8% polyacrylamide gel showing protein-DNA complexes (C) between the indicated amounts of HMGI protein and the wild-type (wt) or mutated (mutI.g or mutI.cg) 5′-end-labelled double-stranded DNA sequences of the upstream HMGI binding site. The incubation reaction was carried out as described in Materials and Methods in the presence of 12.5 μg of nonradioactive sonicated salmon sperm DNA per ml as nonhomologous competitor DNA.
Transient transfection of muIFN-β promoters with mutations in the upstream high-affinity HMGI binding site shows delayed kinetics of virus-induced transcriptional activation.
The wild-type or mutated murine promoters were cloned in plasmid pBLCAT3 upstream of the CAT gene (see Materials and Methods). The resulting plasmids were transiently transfected into human HeLa cells or into murine L929 cells. The transfected cells were induced with NDV and collected 18 h after NDV induction. As shown in Table 1, 18 h postinduction no significant difference was detected between the wild-type and mutated promoters.
TABLE 1.
Virus-induced CAT activities of transiently transfected wild-type and mutI.g promoters (18 h postinduction)a
| Cell type | Promoter | CAT activity (cpm/h/mg) normalized with β-galactosidase | Inducibility |
|---|---|---|---|
| HeLa | Wild type | 171,211 ± 9,333 | 61-fold |
| mutI.g | 132,804 ± 34,565 | 57-fold | |
| L929 | Wild type | 45,910 ± 14,267 | ND |
| mutI.g | 52,590 ± 7,881 | ND |
ND, not determined. Overall low levels of CAT activities in the absence of NDV made the values for mock-induced cells undetectable in L929 cells.
Cells transiently transfected with wild-type or mutI.g promoters were also collected at short times after virus induction, and the corresponding CAT activities were measured. From the final CAT activities obtained after virus induction, we subtracted the mock-induced CAT activities present in cells not induced by the virus. We called the resulting activities the absolute CAT activities. Figure 4A shows the CAT activities obtained at different times after NDV induction of HeLa cells. The values are expressed as percentages of the absolute CAT activity reached by the wild-type promoter 18 h postinduction.
The results of the transient transfection experiments shown in Fig. 4A indicate that the kinetics of the virus-induced transcriptional activation of the mutI.g promoter are delayed compared to those of the wild-type promoter. Transcription from the wild-type promoter was detectable starting at 3 h postinduction, whereas at this time transcription from the mutI.g promoter was undetectable. Four hours postinduction, the activity of the mutI.g promoter was four times weaker than the activity of the wild-type promoter. This difference between the transcriptional activities of the mutated and wild-type promoters slowly diminished with time. At 5 h postinduction, the mutI.g promoter was three times weaker, and at 6 h, the difference was only 1.4 times.
The general pattern of the kinetics of induction obtained when the mock-induced values were not subtracted from the final CAT activities is very similar to that obtained when we considered the absolute CAT activities. This result indicated that the alterations correlated with the mutations introduced in the upstream HMGI binding site are clearly not related to the mock-induced values. The same experiments were carried out with murine L929 cells. The results obtained were analogous to those obtained with HeLa cells, except that lower expression was observed. The same phenomenon was also observed when equivalent transfections were performed with the murine promoter carrying mutI.cg mutations (data not shown).
During transient transfections, the mutI.g and mutI.cg mutations delayed the kinetics of virus-induced transcription but did not prevent the promoters from reaching transcriptional activities equivalent to that of the wild-type promoter.
We constructed short IFN-β promoters spanning from positions −150 to +20 (as opposed to promoters spanning from positions −330 to +20 [described before]), either wild type (wt150) or mutated on the upstream high-affinity HMGI binding site (mutI.cg150). Region c, corresponding to the high-affinity HMGI binding site, and the muVRE region are entirely contained on the sequences of these short promoters. However, only a small part of the upstream AT-rich region is contained on these sequences.
The wt150 and mutI.cg150 promoters were transiently transfected, cells were virus induced as previously described, and the corresponding absolute CAT activities were measured at different times postinduction. No significant difference in the kinetics of virus-induced transcriptional activation was observed between the wt150 and mutI.cg150 promoters. The wt150 and mutI.cg150 promoters were transcriptionally active independent of the mutation introduced in the HMGI binding site, even at short times after virus induction (Fig. 4B).
No delayed kinetics were observed for the mutI.cg150 promoter, suggesting that the role of this high-affinity HMGI binding site is accomplished via the sequences positioned 5′ of the muVRE. These sequences have been described as being responsible for the constitutive repression of the promoter (50). The binding of HMGI protein to position −130 therefore could influence the derepression of the promoter.
Stably transfected mutI.cg muIFN-β promoters are unable to reach normal virus-induced transcriptional activation.
Promoter derepression often requires chromatin remodelling (23). Chromatin is correctly reconstituted on stably integrated DNA templates, while it is incompletely organized on transiently transfected DNAs (21, 34). We therefore analyzed the kinetics of virus-induced transcriptional activation of the wild-type and mutated muIFN-β promoters after stable integration into chromatin and compared them with the kinetics obtained with transiently transfected promoters.
Stably transfected wild-type or mutI.cg murine L929 cell lines were obtained and pooled as described in Materials and Methods. The virus induction kinetics of the wild-type or mutI.cg pooled cell lines were tested with freshly thawed cells that were passaged once. Figure 4C shows the average of three independent analyses of the kinetics of induction. The effect of the mutI.cg mutation on the kinetics of virus-induced transcriptional activation is much more dramatic for the stably transfected promoters. Even 9 and 10 h postinduction, the mutI.cg pooled cell lines remained partially repressed, displaying reduced transcriptional activity. Contrary to the results obtained during transient transfections, the stably transfected mutated promoter never reached virus-induced transcriptional activation equivalent to that obtained with the wild-type promoter.
The induction of the IFN-β promoter is only transient, since a feedback inhibitory mechanism blocks the virus-induced transcription of the gene (42). Ten hours postinduction, the promoter has almost reached its maximum activity. The difference observed between the wild-type and mutI.cg cell lines at 10 h persisted at 18 h postinduction and remained long after 18 h (Fig. 4D), with the CAT activities of the wild-type and mutI.cg cell lines gradually diminishing due to the gradual degradation of the corresponding CAT mRNAs.
No significant difference was observed for the corresponding mock-induced CAT activities of the wild-type and mutI.cg promoters, at early as well as later times after induction. The values obtained in the absence of NDV induction consistently remained very low, equivalent to the values obtained under the same conditions with nontransfected L929 cells. We were therefore unable to really examine the constitutive expression of either the wild-type or the mutI.cg pooled cell lines.
The results shown in Fig. 4C confirmed the delayed virus-induced kinetics of transcriptional activation previously observed for the mutI.cg promoter. Up to 6 h postinduction, both promoters displayed very low transcriptional capacities, with no strong difference between the wild-type and mutI.cg promoters. Starting at 7 h after virus infection, the CAT activity under the control of the wild-type promoter started to be frankly induced, doubling every hour until it approached its maximum 10 h postinduction. On the contrary, the CAT activity under the control of the mutI.cg promoter started to be frankly induced only 9 h postinduction. At 10 h postinduction, when the mutI.cg promoter approached its maximum activity, it still remained partly repressed and was therefore unable to reach transcriptional activation equivalent to that of the wild-type promoter.
It is also interesting to compare in Fig. 4 the kinetics for the virus-induced transcriptional activation of the transiently transfected wild-type promoter with the kinetics for the same promoter stably integrated into chromatin. The transiently transfected wild-type promoter reached, 7 h after virus infection, at least 50% of its maximal transcriptional activity (Fig. 4A). However, when stably transfected, the wild-type promoter reached, at the same time, no more than 7% of its maximal transcriptional activity (Fig. 4C). The repression of the promoter when integrated into chromatin is clearly more strongly established.
In vitro UV footprints of supercoiled muIFN-β DNA show preferential binding of histone H1 to the upstream AT-rich region of the promoter.
The data obtained with wt150 and mutI.cg150 strongly suggested that HMGI protein bound to position −130 accomplished its role via the sequences located 5′ to position −150. The sequences of muIFN-β as well as huIFN-β promoters positioned 5′ to the corresponding VRE and spanning from positions −220 to −110 are highly AT rich (Fig. 1). For the huIFN-β promoter, this region has been described as a negative regulatory domain (50).
Preferential binding of histone H1 has been described for the AT-rich DNA sequences of the SAR (17) as well as the AT-rich flanking sequences of the oocyte-type 5S RNA gene of Xenopus laevis (39), leading to the specific repression of the Xenopus oocyte 5S rRNA gene (6, 16). In order to determine if histone H1 could also preferentially bind to the AT-rich DNA sequence present on the upstream region of the IFN-β promoter, we carried out a series of photofootprinting analyses of the muIFN-β promoter in the presence and absence of histone H1.
Becker and Wang (3) were the first to describe UV radiation as a way to detect protein-DNA contacts. UV irradiation of DNA with a 254-nm light induces a series of photoproducts. The spectrum of these photoproducts is affected when protein-DNA complexes are formed. The differences in UV reactivity between irradiated naked DNA and irradiated protein-bound DNA can be detected during primer extension reactions, since the UV-induced photoproducts are DNA polymerase arrest sites (32). Subsequent analysis of the primer extension reaction products in a denaturing gel gives rise to a footprint.
During UV irradiation, the degree of supercoiling of the irradiated DNA is preserved. This is an advantageous characteristic when one is searching for histone H1-DNA interactions, since histone H1 has been described as binding with a higher affinity to supercoiled DNA than to relaxed DNA (15).
Before UV irradiation, increasing amounts of histone H1 were incubated with different supercoiled plasmids containing the muIFN-β promoter sequence spanning from positions −330 to +20 (wt330), positions −150 to +20 (wt150), or positions −110 to +20 (wt110) (Fig. 5). UV irradiation of naked DNA as well as protein-bound DNA was carried out as described in Materials and Methods. After irradiation, the DNA was denatured and annealed with the corresponding 5′-end-labelled primers indicated in Fig. 6. After annealing, an elongation reaction was carried out with T7 DNA polymerase in order to detect the UV photoproducts formed after UV irradiation of the naked plasmid as well as the plasmid previously incubated with histone H1.
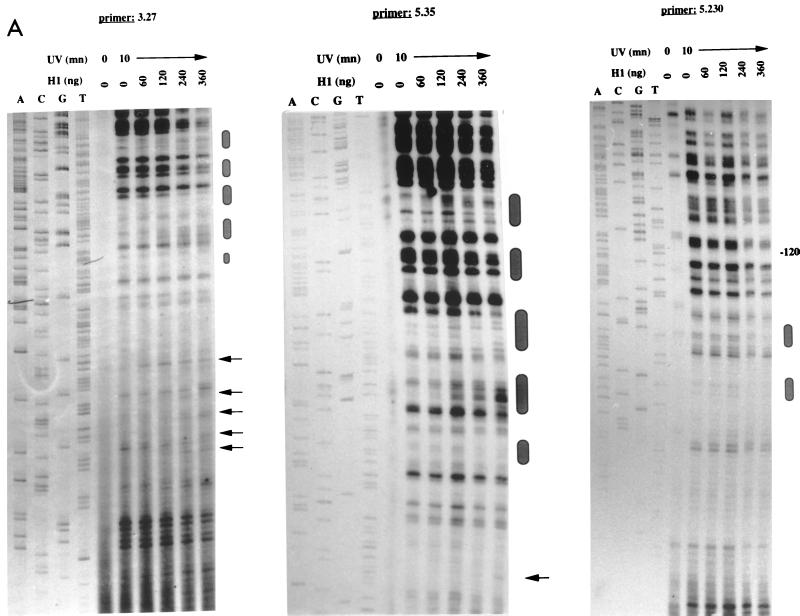
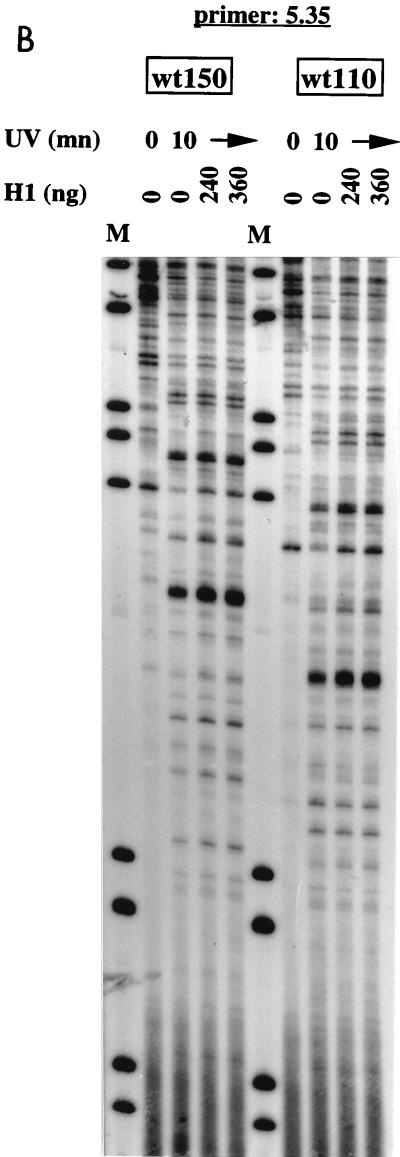
FIG. 5.
UV footprinting reactions of histone H1 with supercoiled plasmids containing the long IFN-β promoter wt330 (positions −330 to +20) (A) or the short IFN-β promoters wt150 (positions −150 to +20) and wt110 (positions −110 to +20). The footprinting reactions were carried out as described in Materials and Methods. In panel A, A, C, G, and T correspond to a standard sequencing reaction carried out with the indicated primers. In panel B, M corresponds to molecular size standards (pBR322 digested with AluI) of 57, 63, 100, 136, 257, 403, and 521 bp. From the 5′ end of the indicated primers to the top of the gels, approximately 300 bp is shown in each panel. In panel A, arrows and ovals indicate weak and strong isolated histone H1-induced UV photoproduct modifications, respectively, and −120 indicates a muIFN-β promoter position. mn, minute.
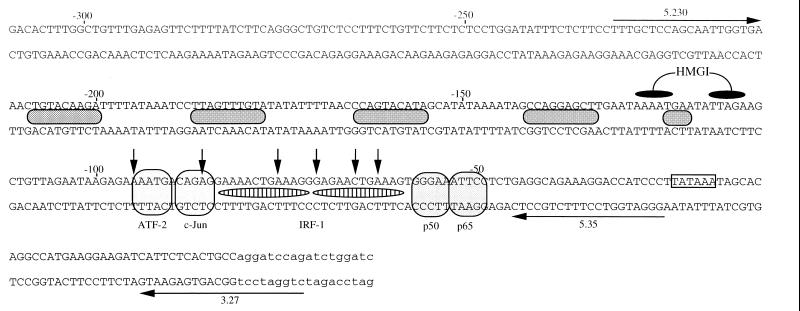
FIG. 6.
DNA sequence of muIFN-β promoter. The positions of the different primers used during T7 elongation reactions and the regions of histone H1-induced modifications during UV footprinting are indicated. Black ovals indicate specific HMGI binding sites. Horizontal arrows indicate 5′-to-3′-oriented primers. Shaded ovals between the sequences indicate regions of strong histone H1-induced UV photoproduct modifications. Vertical arrows indicate weak isolated histone H1-induced UV photoproduct modifications.
As shown in Fig. 5A, the spectrum of photoproducts obtained with the wt330 plasmid was modified in the presence of increasing amounts of histone H1. The modifications detected on the coding strand with primers 3.27 and 5.35 can be grouped in a series of specific regions which are all located in the upstream AT-rich sequence of the promoter (Fig. 6). No specific modifications were observed with primer 5.230 on the region from position −120 to the TATA box, which corresponds to the muVRE region (Fig. 5A). With this primer, we just observed on the VRE region a general inhibition of the T7 extension reaction, which was also observed with the other primers and which corresponds to T7 elongation being slowed down by histone H1 binding to the promoter.
Using primer 5.35, we carried out UV footprint analyses of the wt150 and wt110 plasmids in the presence of histone H1. These plasmids are identical to the wt330 plasmid, except that they lack the upstream AT-rich region. The extension reactions shown in Fig. 5B were carried out over 300 bp like those shown in Fig. 5A. No modifications of the spectrum of photoproducts were induced by histone H1 in either the promoter DNA sequence or the vector sequence DNA on wt150 or wt110 plasmids (Fig. 5B). This result confirmed that the UV footprints shown in Fig. 5A represent the specific in vitro binding of histone H1 to the upstream AT-rich region of the IFN-β promoter rather than that they are a consequence of random, nonspecific binding of histone H1.
Distamycin A increases the virus-induced transcriptional activation of the wt330 IFN-β promoter.
In order to further examine the eventual association of histone H1 with the upstream AT-rich region of the promoter, we used the drug distamycin A, which interacts with the minor groove of AT-rich DNA sequences and which has been described as selectively displacing histone H1 from oligo(dA) · oligo(dT) runs (17, 23). The stably transfected wt330 cell line was incubated with different concentrations of distamycin before and after virus induction as described in Materials and Methods. As shown in Fig. 7A, distamycin activated in a dose-dependent manner IFN-β promoter activity after virus infection. At 6 h postinduction, the strongest activation was obtained with 40 μM distamycin. The same experiment was carried out with a stably transfected short wt110 IFN-β promoter. Distamycin had no activating effect on this promoter, which lacked the upstream AT-rich region (Fig. 7A). On the contrary, distamycin had a slight inhibitory effect on this short promoter, since the fold activation dropped to 0.76 at 40 μM distamycin. The inhibition of transcription by distamycin is a general phenomenon usually observed with this antiproliferation drug.
The fold activation induced by distamycin varied at different times after virus infection (Fig. 7B). The strongest activation was observed in the presence of 40 μM distamycin at 6 h postinduction. At this time, the stably transfected wt330 promoter is, in the absence of distamycin, still highly repressed (Fig. 4C). At 10 h after virus infection, when the promoter is fully active, distamycin had no activating effect. On the contrary, a slight inhibitory effect was observed.
Overall, these results are consistent with our in vitro data indicating a selective interaction of histone H1 with the upstream AT-rich region of the IFN-β promoter.
Overproduction of AT-rich SAR DNA increases the virus-induced transcriptional activation of the stably transfected wt330 IFN-β promoter.
Histone H1 has been described to preferentially bind to SAR (17). We expected, then, that overproduction of SAR sequences should compete for histone H1 binding to the upstream AT-rich region of the IFN-β promoter. We therefore transfected the wt330 cell line with plasmids containing either SAR DNA (pSP64-34) or just vector DNA (pGEM4).
Transfection with plasmid pSP64-34, leading to an overproduction of SAR sequences inside the cell, resulted in an increase in virus-induced transcriptional activation (Fig. 7C). As with distamycin, the effect of the overproduction of SAR sequences was not constant at different times after virus infection. The highest fold activation was observed at 6 and 7 h postinduction, that is, before the promoter is fully active.
The plasmid containing just vector DNA was also able to activate the promoter, but to a much lesser extent and with no significant variation in the kinetics of induction.
In vitro HMGI protein is capable of displacing histone H1 from the upstream AT-rich region of the muIFN-β promoter.
With regard to the binding of histone H1 to SAR, it has been shown that HMGI protein can compete with histone H1 for binding to AT-rich DNA sequences and displace histone H1 (49). Our next question, then, was whether HMGI protein can displace histone H1 from the AT-rich region of the muIFN-β promoter.
In order to study this issue, we took advantage of the fact that when bound to DNA, histone H1 causes T7 DNA polymerase arrest even in the absence of UV irradiation.
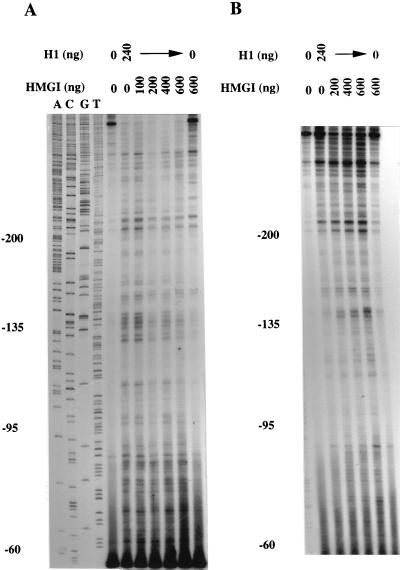
The plasmid containing the wild-type promoter region spanning from positions −330 to +20 was incubated with a fixed amount of histone H1 in the absence or presence of increasing amounts of HMGI protein. The naked or protein-bound plasmids were annealed with primer 5.35 and submitted to an elongation reaction with T7 DNA polymerase. Figure 8A shows that the intensity of the sites of arrest caused by the presence of histone H1 diminished as increasing amounts of HMGI protein were added to the reaction mixture. This phenomenon was not homogeneous over the entire promoter, since no effect linked to the presence of HMGI protein could be observed on the promoter region extending from the 5′ end of the primer to position −95. On the contrary, HMGI protein displaced histone H1 from the promoter region spanning from positions −95 to −200, this result being most visible for the region from positions −135 to −200 of the muIFN-β promoter.
FIG. 8.
Competition experiments. Supercoiled plasmids containing the wild-type (A) or mutI.cg IFN-β promoter (B) (positions −330 to +20) were incubated with 0 or 240 ng of histone H1 in the absence or presence of the indicated amounts of HMGI protein. Incubation was carried out as for the UV footprinting reactions, except that the plasmids were not UV irradiated. HMGI protein was added at the same time as the plasmids. The plasmids were denatured, annealed with primer 5.35, and elongated with T7 DNA polymerase. In panel A, A, C, G, and T indicate a standard sequencing reaction. Numbers at the left of panels indicate positions on the muIFN-β promoter.
We then wanted to investigate if the binding of HMGI protein to its −130 binding site was necessary for this protein to displace histone H1. In order to do this, the same experiment as that shown in Fig. 8A was carried out with the plasmid containing the mutI.cg promoter sequence, that is, the sequence spanning from positions −330 to +20 but carrying a mutation on the upstream high-affinity HMGI binding site. As shown in Fig. 8B, HMGI protein was unable to displace histone H1 from the upstream AT-rich region of the mutI.cg promoter. This result indicated that correct binding of HMGI protein to its upstream high-affinity binding site was necessary for this protein to be able to displace histone H1 in vitro.
The fact that in vitro HMGI protein is able to displace histone H1 is, of course, not proof that in vivo it will. Nevertheless, if this were the case, we would expect distamycin to compensate for the lack of binding of HMGI protein. In order to test this idea, we measured the effect of distamycin upon the mutI.cg cell line. As was observed with the wt330 cell line, distamycin was also able to increase the virus-induced transcriptional activation of the mutI.cg promoter (data not shown). However, contrary to the results obtained with the wild-type cell line, the effect of distamycin upon the mutI.cg promoter persisted for up to 24 h after virus infection (Fig. 7D), so that distamycin was able to partly compensate for the lack of binding of HMGI protein to position −130 of the IFN-β promoter.
DISCUSSION
In this work, we have examined the binding of HMGI protein and histone H1 to the promoter region of the muIFN-β gene. Concerning HMGI protein, we describe a new, specific HMGI binding site on the upstream AT-rich region of the muIFN-β promoter. This site is located outside the VRE in the region from positions −133 to −114 and has not been described for any other IFN-β promoter. The bases that appeared directly protected by HMGI protein in this site constituted two AT tracts (of 5 bases each) separated by five nonprotected bases and therefore facing the same side of the DNA helix. From what is known concerning the mode of binding of HMGI protein to DNA (7, 27), the presence of two AT tracts facing the same side of the helix, which could be contacted by two HMGI protein AT-hook peptides, would account for the high-affinity binding of HMGI protein to this particular site. Besides, weaker HMGI binding sites were observed upstream and downstream of the region from positions −133 to −114. These weaker sites may contribute to the stabilization of the binding of HMGI protein to the upstream region of the promoter, since intermolecular cooperativity has been previously described for the binding of two molecules of HMGI protein (47).
This site present on the region from positions −133 to −114 of the promoter was the only one that remained protected by HMGI protein in the presence of sonicated salmon sperm DNA used as random, nonspecific competitor DNA. As a matter of fact, we show in this work that the HMGI binding site next to the NF-κB binding site on the PRDII region of the human promoter is nonspecific on the muIFN-β promoter. These results have also been obtained with gel retardation assays (31). In the murine PRDII promoter region, only one AT tract is present; there are two AT tracts in the same region of the human promoter (Fig. 1). Yie et al. (47) have reported that mutations introduced in one of the two AT tracts present in the human PRDII promoter region strongly reduced the affinity of HMGI protein for this site. Indeed, when we introduced into the murine PRDII promoter region the “missing” second AT tract, we observed that the affinity of HMGI protein for this reconstituted site became equivalent to that for the corresponding site in the human promoter (30a). Overall, these results indicate that it is the lack of one of the two AT tracts that renders this HMGI binding site very weak and almost nonexistent in the murine promoter.
The HMGI binding site present in the PRDIV region of the huIFN-β promoter is conserved in the murine promoter (Fig. 1). Nevertheless, during our footprint experiments, this site appeared weakly specific, since it immediately disappeared when sonicated salmon sperm DNA was used as random, nonspecific competitor DNA. The two AT tracts present in the PRDIV HMGI binding site are separated by 8 bp instead of 5 bp and therefore are not positioned on the same side of the DNA helix. This fact can probably account, at least in part, for the weak binding of HMGI protein to the PRDIV region. Besides, in the human promoter, the binding of HMGI protein to the PRDIV is stabilized by the HMGI protein molecule bound to the PRDII region (47). It is possible, therefore, that the absence of correct binding of HMGI protein to the murine PRDII region also reduces the affinity of HMGI protein for the murine PRDIV region.
In order to assess the role of the binding of HMGI protein to position −130 during virus induction of the muIFN-β promoter, we have compared the virus-induced transcriptional capacity of a wild-type promoter and promoters with mutations in the upstream HMGI binding site (mutI.g and mutI.cg) during transient as well as stable transfections.
During transient transfection experiments, the mutated promoters displayed delayed virus-induced kinetics of transcription, but 7 h after virus infection, the wild-type and mutated promoters reached similar transcriptional capacities. This result suggested that HMGI protein bound to upstream position −130 could play a role during promoter derepression. This observation was confirmed by the lack of difference observed between the wild-type promoter and the short mutated promoter (mutI.cg150), which contained the entire muVRE region plus the wild-type (wt150) or mutated (mutI.cg150) HMGI binding site but which lacked upstream positions −330 to −150. Both promoters displayed the same virus-induced kinetics of transcriptional activation, suggesting that the effect that was correlated with the mutation introduced in the HMGI binding site was accomplished via the sequences which have been previously described for the huIFN-β promoter as being responsible for the constitutive repression of the promoter. These results corroborate our hypothesis of the potential role of HMGI protein during promoter derepression.
The integration of the mutated promoter spanning from positions −330 to +20 into a chromatin context enhanced the in vivo effect correlated with the loss of HMGI protein binding to its upstream high-affinity binding site that we observed in vitro. Unlike the results obtained with the transiently transfected promoter, the mutI.cg cell line never reached virus-induced transcriptional activity equivalent to that displayed by the wild-type cell line (Fig. 4C and D). Chromatin is only partially reconstituted on transiently transfected plasmids, whereas it is fully reconstituted on DNA templates when they are integrated into the genome (34). The difference that we observed between transiently and stably transfected IFN-β promoters is therefore probably a consequence of chromatin which has been fully reconstituted only on stably transfected muIFN-β promoters. The role of chromatin structures during transcriptional control is an extensively documented and fully established phenomenon (recently reviewed in references 43 and 45). The results shown in Fig. 4 suggest that chromatin might play a role during the establishment of the repressed state of the promoter. As a matter of fact, it is interesting to note that the stably transfected wild-type promoter reached, 7 h after virus infection, no more than 7% of its maximal transcriptional activity (Fig. 4C), whereas when transiently transfected, it reached, at the same time, about 50% of its maximal transcriptional activity (Fig. 4A). The repression of the promoter clearly appears more strongly established after the promoter is integrated into chromatin. Recently published data demonstrate that a transcription complex containing IRF-3/CBP/p300 positively regulates the virus-induced transcriptional activation of the huIFN-β gene (48). Histone acetylase activity is associated with CBP/p300. The participation of histone acetylase during the virus-inducible activation of IFN-β is in agreement with our observation that chromatin could participate in the regulation of virus-induced IFN-β gene expression.
The transiently transfected mutI.cg150 promoter, which lacked the upstream AT-rich region, did not display retarded kinetics (Fig. 4B), indicating that the phenotype associated with the mutations introduced in the HMGI binding site was mediated through this region of the promoter. Using UV footprints, we show in this work that in vitro histone H1 can preferentially bind to the AT-rich region of a promoter localized immediately upstream of the muVRE. This interaction appeared specific, since in the absence of the AT-rich region, no histone H1 protection could be observed either in promoter DNA or in vector DNA (Fig. 5B).
Distamycin has been described as a drug that can specifically displace histone H1 from oligo(dA) · oligo(dT) runs (17, 33). In the case of the wild-type muIFN-β promoter integrated into chromatin, distamycin was able to activate the promoter after virus infection but only if the promoter carried the upstream AT-rich region (Fig. 7A). This result suggested that in vivo histone H1 could also bind to this particular sequence and, by doing so, could be at least partly responsible for promoter repression. It was interesting to observe that distamycin had no affect 10 h after virus infection, when the promoter was fully active (Fig. 7B). This result could be due to the fact that 10 h after induction, when the promoter is fully active, histone H1 might no longer interact with the promoter AT-rich DNA sequence.
A specific interaction of histone H1 with SAR DNA sequences has been described (17). The overproduction of SAR sequences activated the virus-induced transcriptional activation of the muIFN-β promoter to a higher extent than the overproduction of non-SAR vector DNA (Fig. 7C). This finding corroborated the results obtained with distamycin, since they also suggested that histone H1 might interact with the IFN-β promoter and, by doing so, might contribute at least partly to its repression.
Histone H1 is an interesting protein which has been considered, until recently, to generally repress transcription by preventing nucleosome mobility and contributing to higher-order chromatin structures. Nevertheless, the results recently obtained with histone H1-knockout strains revealed a specific role for histone H1 in transcription, histone H1 being able to activate or repress a subset of genes (recently reviewed in references 12 and 44). Some of the genes directly affected by histone H1 have characteristics similar to those that we have described in this work. A recent work shows that the effect of dephosphorylated histone H1 upon the inactivation of the mouse mammary tumor virus promoter is observed with the stably transfected promoter assembled as chromatin but not with the transiently transfected promoter (22). Also, it has been shown that the histone H1-mediated repression of oocyte-type 5S rRNA genes is dependent upon the binding of histone H1 to the AT-rich flanking sequences of these genes (19, 39). More recent work suggests that the specific transcriptional repressor effect of histone H1 on the Xenopus oocyte 5S rRNA gene is linked to the presence of an AT-rich sequence 3′ to the gene as well as to the binding of histone H1 to the 3′ end of the nucleosome core, blocking the access to a key promoter element (33). For Xenopus embryos, Vermaak et al. have described the globular domain of histone H1 as being sufficient to direct this specific gene repression (40).
In this work, we describe data suggesting that the binding of histone H1 to the upstream AT-rich region of the muIFN-β promoter is at least partly responsible for the repression of the IFN-β gene. Our hypothesis, then, is that HMGI protein bound to the region from positions −133 to −114 would be able to regulate promoter derepression by displacing histone H1 from the upstream AT-rich region of the promoter. At the moment, two observations support this hypothesis. The first one is that in vitro HMGI protein is indeed capable of displacing histone H1 from the AT-rich region of the promoter but only in the presence of a wild-type HMGI binding site. The second one is that distamycin, which specifically displaces histone H1, is able to partly compensate for the lack of binding of HMGI protein to its −130 site. Nevertheless, until further confirmation, this remains a work hypothesis.
The linkage of the displacement of histone H1 to gene derepression has been previously described with several systems. For the binding of histone H1 to SAR, it was shown that HMGI protein can compete with histone H1 for binding to these AT-rich DNA sequences, displace histone H1, and derepress a T7 promoter (49). Upstream binding factor 3 has also been described as capable of displacing histone H1. As a matter of fact, upstream binding factor 3, a high-mobility-group-like protein, displaces and causes complete dissociation of histone H1 from rRNA gene enhancers associated with nucleosomes (18). Ding et al. have demonstrated that histone H1 can be a functional target of high-mobility-group protein 14 in both transcriptional enhancement and chromatin decompaction (10). Hepatocyte nuclear factor 3 has a configuration very similar to that of histone H1 and, like histone H1, has nucleosome positioning properties (28). This factor has also been described as capable of replacing histone H1 in the chromatin of the mouse serum albumin enhancer (8).
Genomic footprinting done before and after virus infection with the wild-type and mutI.cg cell lines should help us to understand the mechanisms of histone H1, HMGI protein, and IFN-β promoter derepression. Meanwhile, it is interesting to recall here a set of experiments previously described by Zinn and Maniatis (51). They carried out a comparative study of the DNase I-hypersensitive sites present on the huIFN-β promoter before and after poly(I)-poly(C) induction. They observed that promoter induction leads to a remarkable change in the pattern of the hypersensitive sites present on the upstream promoter region previously described as a negative regulatory element (50). They suggested that a factor bound to the region from positions −167 to −94 could be responsible for the constitutive repression of the huIFN-β promoter, its dissociation after induction being necessary for the activation of the promoter to take place. This factor could be histone H1 either alone or associated with core histones. Immunoprecipitation experiments with anti-histone H1 antibodies as well as nucleosome reconstitution experiments in the presence and absence of histone H1 should help clarify this issue.
Khadake and Rao (19), using circular dichroism spectroscopy, have shown that the binding of histone H1 to the AT-rich sequences of the SAR results in chromatin condensation. This condensation is three- to fourfold higher than that brought about by histone H1 on a random DNA fragment. Such an effect of histone H1 on the muIFN-β promoter could prevent the binding of transactivation factors to the muVRE, resulting in a repressed state of the promoter. Displacement of histone H1 by HMGI protein would help convert the promoter from a repressed state to an active state, allowing recruitment of the different transcription factors to the VRE region.
We have described a strong HMGI binding site at the 3′ boundary of the AT-rich region on the murine promoter. It is interesting to note that the murine DNA sequence corresponding to this site (5′-AATAAAATGAATATTAGAAG-3′) is strongly conserved in the human promoter (5′-TAAATAAAGAGTTTTAGAAA-3′) being located at positions −134 to −115, at the 3′ boundary of the AT-rich region of the human promoter. Considering not only the role of the 110-bp promoter sequence immediately flanking the TATA box but also the role of the upstream regions of the promoter has allowed us to enlarge our view of the transcriptional regulation of the IFN-β gene, unmasking the role of HMGI protein during promoter derepression and implicating histone H1 as a potential repressor of this gene.
ACKNOWLEDGMENTS
We are grateful to Emmanuel Käs for the gift of plasmid pET3b-HMGI and fruitful discussions as well as to Ariel Prunel for the gift of purified histone H1. We thank Pascale Debey for critical reading of the manuscript, S. Chusterman and S. Navarro for discussions and encouragement, and Eugenio Prieto for photographic work.
This work was supported by the Centre de la Recherche Scientifique and by grants from the Association pour la Recherche sur le Cancer (contract 1042) and the Federation Nationale des Groupements des Entreprises Francçaises et Monégasques dans la Lutte contre le Cancer.
REFERENCES
- 1.Andersson S, Davis D L, Dahlbäck H, Jörnvall H, Russell D W. Cloning, structure and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 2.Bagga R, Emerson B M. An HMGI/Y-containing repressor complex and supercoiled DNA topology are critical for long-range enhancer-dependent transcription in vitro. Genes Dev. 1997;11:629–639. doi: 10.1101/gad.11.5.629. [DOI] [PubMed] [Google Scholar]
- 3.Becker M M, Wang J C. Use of light for footprinting DNA in vivo. Nature. 1984;309:682–687. doi: 10.1038/309682a0. [DOI] [PubMed] [Google Scholar]
- 4.Bonnefoy E, Rouvière-Yaniv J. HU, the major histone-like protein of E. coli, modulates the binding of IHF to oriC. EMBO J. 1992;11:4489–4496. doi: 10.1002/j.1460-2075.1992.tb05550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnefoy E, Takahashi M, Rouvière-Yaniv J. DNA-binding parameters of the HU protein of Escherichia coli to cruciform DNA. J Mol Biol. 1994;242:116–129. doi: 10.1006/jmbi.1994.1563. [DOI] [PubMed] [Google Scholar]
- 6.Bouvet P, Dimitrov S, Wolffe A P. Specific regulation of Xenopus chromosomal 5S RNA gene transcription in vivo by histone H1. Genes Dev. 1994;8:1147–1159. doi: 10.1101/gad.8.10.1147. [DOI] [PubMed] [Google Scholar]
- 7.Bustin M, Reeves R. High-mobility-group proteins: architectural components that facilitate chromatin function. Prog Nucleic Acids Res. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 8.Cirillo L A, McPherson C E, Bossard P, Stevens K, Cherian S, Shim E Y, Clark K L, Burley S K, Zaret K S. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Civas A, Dion M, Vodjdani G, Doly J. Repression of the murine interferon alpha 11 gene: identification of negatively acting sequences. Nucleic Acids Res. 1991;19:4497–4502. doi: 10.1093/nar/19.16.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding H-F, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol Cell Biol. 1997;17:5843–5853. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frebourg T, Brison O. Plasmid vectors with multiple cloning sites and cat-reporter gene for promoter cloning and analysis in animal cells. Gene. 1988;65:315–318. doi: 10.1016/0378-1119(88)90468-4. [DOI] [PubMed] [Google Scholar]
- 12.Hartzog G A, Winston F. Nucleosomes and transcription: recent lessons from genetics. Curr Opin Genet Dev. 1997;7:192–198. doi: 10.1016/s0959-437x(97)80128-1. [DOI] [PubMed] [Google Scholar]
- 13.Hill D A, Reeves R. Competition between HMG-I(Y), HMG-1 and histone H1 on four-way junction DNA. Nucleic Acids Res. 1997;25:3523–3531. doi: 10.1093/nar/25.17.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holth L T, Thorlacius A E, Reeves R. Effects of epidermial growth factor and estrogen on the regulation of the HMG-I/Y gene in human mammary epithelial cell lines. DNA Cell Biol. 1997;16:1299–1309. doi: 10.1089/dna.1997.16.1299. [DOI] [PubMed] [Google Scholar]
- 15.Ivanchenko M, Zlanatova J, van Holde K. Histone H1 preferentially binds to superhelical DNA molecules of higher compaction. Biophys J. 1997;72:1388–1395. doi: 10.1016/S0006-3495(97)78785-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kandolf H. The H1A histone variant is an in vivo repressor of oocyte-type 5S gene transcription in Xenopus laevis embryos. Proc Natl Acad Sci USA. 1994;91:7257–7261. doi: 10.1073/pnas.91.15.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Käs E, Izaurralde E, Laemmli U K. Specific inhibition of DNA binding to nuclear scaffolds and H1 by distamycin. The role of oligo (dA) · (dT) tracts. J Mol Biol. 1989;210:587–599. doi: 10.1016/0022-2836(89)90134-4. [DOI] [PubMed] [Google Scholar]
- 18.Kermechiev M, Workman J L, Pikaard C S. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol Cell Biol. 1997;17:5833–5842. doi: 10.1128/mcb.17.10.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khadake J R, Rao M R S. Preferential condensation of SAR-DNA by histone H1 and its SPKK containing octapeptide repeat motif. FEBS Lett. 1997;400:193–196. doi: 10.1016/s0014-5793(96)01393-2. [DOI] [PubMed] [Google Scholar]
- 20.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 21.Lee H-Y, Archer T K. Nucleosome-mediated disruption of transcription factor-chromatin initiation complex at the mouse mammary tumor virus long terminal repeat in vivo. Mol Cell Biol. 1994;14:32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H-Y, Archer T K. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 1998;17:1454–1466. doi: 10.1093/emboj/17.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, Wrange O, Eriksson P. The role of chromatin in transcriptional regulation. Int J Biochem Cell Biol. 1997;29:731–742. doi: 10.1016/s1357-2725(97)00016-2. [DOI] [PubMed] [Google Scholar]
- 24.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez S, Reeves R, Island M-L, Bandu M-T, Christeff N, Doly J, Navarro S. Silencer activity in the interferon-A gene promoters. J Biol Chem. 1997;272:22788–22799. doi: 10.1074/jbc.272.36.22788. [DOI] [PubMed] [Google Scholar]
- 26.Luckow B, Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory element. Nucleic Acids Res. 1987;15:5490–5494. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maher J F, Nathans D. Multivalent DNA-binding properties of the HMGI proteins. Proc Natl Acad Sci USA. 1996;93:6716–6720. doi: 10.1073/pnas.93.13.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McPherson C E, Horowitz R, Woodcock C L, Jiang C, Zaret K. Nucleosome positioning properties of the albumin transcriptional enhancer. Nucleic Acids Res. 1996;24:397–404. doi: 10.1093/nar/24.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 30.Reeves R, Nissen M S. Interaction of high mobility group-I(Y) nonhistone proteins with nucleosome core particles. J Biol Chem. 1993;268:21137–21146. [PubMed] [Google Scholar]
- 30a.Robbe, K. Unpublished results.
- 31.Robbe K, Bonnefoy E. Non-B-DNA structures on the interferon-β promoter? Biochimie. 1998;80:665–671. doi: 10.1016/s0300-9084(99)80020-0. [DOI] [PubMed] [Google Scholar]
- 32.Selleck S B, Majors J. Photofootprinting in vivo detects transcription-dependent changes in yeast TATA boxes. Nature. 1987;325:173–177. doi: 10.1038/325173a0. [DOI] [PubMed] [Google Scholar]
- 33.Sera T, Wolffe A P. Role of histone H1 as an architectural determinant of chromatin structure and as a specific repressor of transcription on Xenopus oocyte 5S rRNA genes. Mol Cell Biol. 1998;18:3668–3680. doi: 10.1128/mcb.18.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith C L, Hager G L. Transcriptional regulation of mammalian genes in vivo. A tale of two templates. J Biol Chem. 1997;272:27493–27496. doi: 10.1074/jbc.272.44.27493. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka N, Taniguchi T. Cytokine gene regulation: regulatory cis-elements and DNA binding factors involved in the interferon system. Adv Immunol. 1992;52:263–281. doi: 10.1016/s0065-2776(08)60877-9. [DOI] [PubMed] [Google Scholar]
- 36.Thanos D, Maniatis T. The high mobility group protein HMGI(Y) is required for NF-κB-dependent virus induction of the human IFNβ gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 37.Thanos D, Du W, Maniatis T. The high-mobility group protein HMGI(Y) is an essential structural component of the virus-inducible enhancer complex. Cold Spring Harbor Symp Quant Biol. 1993;58:73–81. doi: 10.1101/sqb.1993.058.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 39.Tomaszewski R, Jerzmanowski A. The AT-rich flanks of the oocyte-type 5S RNA gene of Xenopus laevis act as a strong local signal for histone H1-mediated chromatin reorganization in vitro. Nucleic Acids Res. 1997;25:458–465. doi: 10.1093/nar/25.3.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vermaak D, Steinbach O C, Dimitrov S, Rupp R A W, Wolffe A P. The globular domain of histone H1 is sufficient to direct specific gene repression in early Xenopus embryos. Curr Biol. 1998;8:533–536. doi: 10.1016/s0960-9822(98)70206-4. [DOI] [PubMed] [Google Scholar]
- 41.Vodjdani G, Coulombel C, Doly J. Structure and characterization of a murine chromosomal fragment containing the interferon β gene. J Mol Biol. 1988;204:221–231. doi: 10.1016/0022-2836(88)90571-2. [DOI] [PubMed] [Google Scholar]
- 42.Whittemore L A, Maniatis T. Postinduction repression of the β-interferon gene is mediated through two positive regulatory domains. Proc Natl Acad Sci USA. 1990;87:7799–7803. doi: 10.1073/pnas.87.20.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolffe A P. Histones, nucleosomes and the roles of chromatin structure in transcriptional control. Biochem Soc Trans. 1997;25:354–358. doi: 10.1042/bst0250354. [DOI] [PubMed] [Google Scholar]
- 44.Wolffe A P. Histone H1. Int J Biochem Cell Biol. 1997;29:1463–1466. doi: 10.1016/s1357-2725(97)00026-5. [DOI] [PubMed] [Google Scholar]
- 45.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 46.Wunderlich, V., and M. Böttger. High-mobility group proteins and cancer—an emerging link. J. Cancer Res. Clin. Oncol. 123:133–140. [DOI] [PMC free article] [PubMed]
- 47.Yie J, Liang S, Merika M, Thanos D. Intra- and intermolecular cooperative binding of high-mobility-group I(Y) to the beta interferon promoter. Mol Cell Biol. 1997;17:3649–3662. doi: 10.1128/mcb.17.7.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao K, Käs E, Gonzalez E, Laemmli U K. SAR-dependent mobilization of histone H1 by HMG-I(Y) in vitro: HMG-I(Y) is enriched in H1-depleted chromatin. EMBO J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zinn K, DiMaio D, Maniatis T. Identification of two distinct regulatory regions adjacent to the human β-interferon gene. Cell. 1983;34:865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]
- 51.Zinn K, Maniatis T. Detection of factors that interact with the human β-interferon regulatory region in vivo by DNAase I footprinting. Cell. 1986;45:611–618. doi: 10.1016/0092-8674(86)90293-x. [DOI] [PubMed] [Google Scholar]