Abstract
Background and Objectives:
Escherichia coli is a Gram-negative organism causing mild to severe infections, with a wide spectrum range of organs involved. The study aimed to describe antibiotics susceptibility of E. coli from clinical specimens from October 11, 2019 to September 11, 2020.
Materials and Methods:
Study was conducted retrospectively in a private microbiology laboratory in Mataram Indonesia. Period of study divided as two groups after WHO declared COVID-19 as pandemic by March 11, 2020; group A including the specimen related to September 2019 to March 11th 2020 and group B including the specimens related to March 11th 2020 to September 2020. All clinical specimens were subjected to identify E. coli isolates and their antibiotics susceptibility using WHO-NET 5.6 version.
Results:
Totally, 148 E. coli isolates were found in group A and 62 isolates in group B. Prevalence of extended-spectrum beta lactamase (ESBL)- producing E. coli in group A was 50% and in group B was 20.9% with significantly difference (p<0.05). There was an increase in susceptibility to 10/16 antibiotics; where 3 antibiotics ofloxacin, aztreonam, and fosfomycin were significant (p<0.05). There was a significant decrease in susceptibility to the antibiotics piperacillin (p=0.012), amoxicillin (p=0.002), cefadroxil (p=0.036) and ampicillin (p=0.036). Type of infections between two groups: musculoskeletal infections, pneumonia, urinary tract infections and sepsis were not significant.
Conclusion:
Reduced number of E. coli isolates between two groups with decrease of ESBL-producing E. coli contribute in dynamics of antibiotics susceptibility. The longer period of analysis is needed to be done, due to the ongoing COVID-19 pandemic.
Keywords: Antimicrobial resistance, COVID-19 pandemic, Escherichia coli
INTRODUCTION
Antimicrobial resistance (AMR) is a complex health problem globally. The complexity is described as the cross boundary of AMR between hosts and interspecies within microorganisms. Resistance itself means a change in the response of pathogens to antibiotics which results in ineffective antibiotics, which begins early when antibiotic is used (1, 2).
Escherichia coli is Gram negative bacterium causing urinary tract infection, enteritis, soft tissue infection, septicaemia and other types of clinical infections. They can spread rapidly in community and hospital settings (3–5) and may be multi-drug resistant and capable of breaking down commonly used antibiotics. The extended-spectrum beta lactamase (ESBL) producing strains of E. coli is one of the important forms of multi-drug resistant phenotypes and categorized as a serious threat by CDC (3). ESBL-producing E. coli has experienced increased resistance to a wide variety of penicillins and cephalosporins (6).
There has been an increasing trend in antimicrobial resistance and mortality caused by E. coli infections. Between 1997–2011, in the US recorded ESBL- E. coli incidence from 0 per 10.000 patient-days to 16.4 per 10.000 patient-days discharged from hospitals (7). The incidence of bacteremia with E. coli in the UK has increased from 67.0 cases per 100,000 population in 2015 to 78.0 cases per 100,000 population in 2019 (8). In Indonesia, the prevalence of ESBL-producing E. coli in 4 major hospitals has increased from 29% in 2005 to 52% in 2012 (9, 10).
COVID-19 pandemic that has hit the world has affected various aspects of life, socio-economic, health policy, healthcare system, including the impact on bacterial patterns and antibiotic sensitivity (11, 12). The way of COVID-19 affected antimicrobial resistance comes from inappropriate antibiotics prescribing, shifting in antimicrobial stewardship programs, disruption in health services, and the wide use of environmental and personal disinfection (11). The resistance pattern of E. coli is interesting to study because of the following reasons: i) It is common Gram-negative microorganisms both as intestinal flora and hospital pathogen, ii) it causes a broad spectrum of infections. This study aimed to describe antibiotic susceptibility of E. coli isolates associated with two occasions, before and during the COVID-19 pandemic.
MATERIALS AND METHODS
Specimens.
Specimens consisting of blood, sputum, pus and urine were received in the microbiology laboratory of a private hospital in Mataram, Indonesia. This is a referral laboratory and gives services to surrounding private and public hospitals and private practices.
Cultures and antibiotic susceptibility tests.
Cultures and antibiotics susceptibility tests were performed according to Clinical and Laboratory Standard Institute (CLSI, 2018) (13). Microorganism identification was determined using the API20E biochemical identification system (BioMerieux France). The susceptibility of microorganisms to antibiotics was assessed using a disc diffusion method. The antibiotics susceptibility data was analyzed using WHO-NET Version 5.6 program.
Antibiotics.
Our laboratory uses 21 antibiotics for Gram negative antibiotics panels, but only 16 were consistently available (stock shortage). The following antibiotic discs were used: penicillin (P) 10 U, amoxicillin (AMX) 20 μg, cefixime (CFM) 5 μg, ciprofloxacin (CIP) 5 μg, cefadroxil (CFR) 30 μg, amikacin (AMK) 30 μg, ceftriaxone (CRO) 30 μg, ampicillin (AMP) 10 μg, amoxicillin/clavulanic acid (AMC) 20/10 μg, cefuroxime (CXM) 30 μg, cefoperazone/sulbactam (CFP) 75/30 μg, levofloxacin (LVX) 5 μg, fosfomycin (FOS) 50 μg, ofloxacin (OFX) 5 μg, piperacillin (PRL) 100 μg and aztreonam (ATM) 30 μg.
RESULTS
There were 308 Gram negative isolates in group A and 243 Gram negative isolates in group B. E. coli decreases proportionally in both groups from 48% in group A to 25.5% in group B (Table 1).
Table 1.
Gram negatives microorganisms isolated from group A and B
| Group A N (%) | Group B N (%) | |
|---|---|---|
| Enterobacter aerogenes | 0 (0) | 10 (4.11) |
| Enterobacter cloacae | 74 (24) | 51 (20.9) |
| Escherichia coli | 148 (48) | 62 (25.5) |
| Klebsiella pneumoniae | 42 (13.6) | 21 (8.6) |
| Pseudomonas aeruginosa | 21 (6.8) | 80 (32.9) |
| Serratia marcescens | 10 (3.2) | 11 (4.5) |
| Stenotrophomonas maltophilia | 0 (0) | 1 (0.4) |
| Other | 13 (4.2) | 7 (2.8) |
| Total | 308 | 243 |
Antibiotic susceptibility.
Susceptibility to most of antibiotics (10/16) in group B was higher than group A; these antibiotics were cefoperazone/sulbactam, cefuroxime, ceftriaxone, cefixime, aztreonam, amikacin, ciprofloxacin, ofloxacin, levofloxacin and fosfomycin. Five antibiotics were decreased in susceptibility: ampicillin, amoxicillin, piperacillin, amoxicillin/clavulanic acid and cefadroxil. In both groups, all isolates were resistant to penicillin (Table 2).
Table 2.
Prevalence of susceptible E. coli isolates in each group. NA=not applicable
| No. of susceptible E. coli isolates in group A (%) n = 148 | No. of susceptible E. coli isolates in group B (%) n = 62 | P value | |
|---|---|---|---|
| Penicillin | 0 | 0 | NA |
| Ampicillin | 11 (7.4) | 0 | 0.036 |
| Amoxicillin | 18 (12.2) | 0 | 0.002 |
| Piperacillin | 13 (8.7) | 0 | 0.012 |
| Amoxicillin/clavulanic acid | 28 (18.6) | 9 (14.5) | 0.553 |
| Cefadroxil | 11 (7.4) | 0 | 0.036 |
| Cefoperazone/sulbactam | 101 (68.2) | 44 (70.9) | 0.746 |
| Cefuroxime | 20 (13.5) | 9 (14.5) | 0.829 |
| Ceftriaxone | 32 (21.6) | 18 (29) | 0.287 |
| Cefixime | 46 (31.1) | 21 (33.9) | 0.746 |
| Aztreonam | 56 (37.8) | 41 (66.1) | 0.000 |
| Amikacin | 86 (58.1) | 44 (70.9) | 0.088 |
| Ciprofloxacin | 63 (42.6) | 35 (56.4) | 0.071 |
| Levofloxacin | 83 (56.1) | 38 (61.3) | 0.542 |
| Ofloxacin | 81 (54.7) | 50 (80.6) | 0.000 |
| Fosfomycin | 104 (70.3) | 53 (85.5) | 0.023 |
Among increased susceptibility antibiotics, three antibiotics fosfomycin, ofloxacin and aztreonam were significant (p<0.05). While in decreased susceptibility antibiotics, four antibiotics ampicillin, amoxicillin, piperacillin and cefadroxil were significant, while amoxicillin/clavulanic acid was not significant.
Phenotypic ESBL producers.
Extended spectrum beta lactamase (ESBL)-producing E. coli was phenotypically identified. There were 74/148 (50%) ES-BL-producing E. coli in group A and 13/62 (20.9%) in group B, which showed there was a significant decrease (p=0.000).
Type of infections.
Type of infections in group A, musculoskeletal 68/148 (45.9%), pneumonia 32/148 (21.6%), urinary tract infection 44/148 (29.7%) and sepsis 4/148 (2.7%). In group B soft tissue infection 27/62 (43.5%), pneumonia 20/62 (32.3%), urinary tract infection 15/62 (24.1%), sepsis 0. Chi-square test of both groups: musculoskeletal infection (p=0.764), pneumonia (p=0.116), urinary tract infection (p=0.502) and sepsis (p=0.322). All kinds of infection were not significant.
DISCUSSION
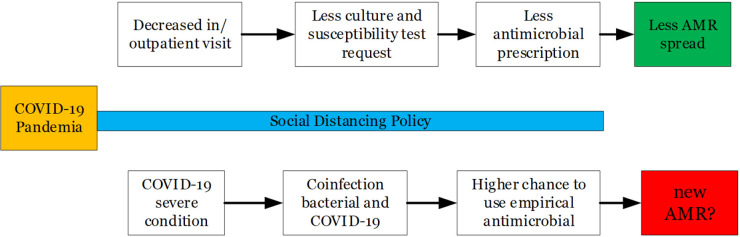
The prevalence of extended spectrum beta-lactamase-producing E. coli has increased worldwide, with the highest rate in South Asia (11,5%) and Turkey (10.9%) before and after 2010 (14). The time separation before and during COVID-19 pandemic is important because the introduction of the social distancing policy simultaneously worldwide will automatically limit people’s visits to the hospital, leading to a decrease in the number of requests for culture and susceptibility tests to the microbiology laboratory.
The actual impact of COVID-19 pandemic is an ongoing issue. Van Duin et al. (2020) stated there are two sides to debate regarding how COVID-19 affected antimicrobial resistance profile: increase or decrease antimicrobial resistance event. The decreased incidence of AMR is due to a direct impact of reduced outpatient / inpatient visits due to the implementation of social distancing policies carried out by the government. Increasing infection and prevention control practice in community and hospital will reduce the spread of resistance (15).
Broad spectrum antibiotics are often used in a case of suspected secondary bacterial infection in COVID-19 patients, thus affecting unnecessary prescribing. The most common type of infection is pneumonia, followed by blood-stream and urinary tract infection (16).
This study describes the incidence of decreased antimicrobial resistance as a direct result of the implementation of the social distancing policy implemented by the government in the first 6 months of the COVID-19 pandemic (Fig. 1).
Fig. 1.
COVID-19 and antimicrobial resistance. AMR=antimicrobial resistance. Developed according to reference 11.
Overall number of Gram-negative isolates were reduced between two groups, corresponding to reduced number outpatients/inpatients admission to the hospital (Table 1). Antibiotics susceptibility is dynamically changed between two groups, with the majority of antibiotics increase in susceptibility. This phenomenon is likely correlated with reduced consumption of antibiotics, similarly to restrict use of antibiotics is applied, then susceptibility tends to increase (17, 18). Increased number of aztreonam susceptible isolates seems to correspond to decreased number of ESBL-E. coli between two groups. Fosfomycin as one of drugs of choice for urinary tract infection (UTI) usage tends to decrease as lower number of UTI cases in group B, that has outcome in increasing its susceptibility.
Type of infections dominance between two groups showed difference in sequence, in group A, soft tissue infection, UTI, pneumonia and sepsis while in group B, pneumonia replace UTI position as runner up and no sepsis cases. This type is differed from elsewhere that urinary tract infections dominance the clinical samples (14, 19, 20). The number of pneumonia cases shifting from third in group A to second place in group B is not confirmed by the number of pulmonary manifestations of COVID-19, that is still needed for further investigation.
Collignon and Beggs (15) stated there were difference between developed and developing countries in resistance spread during COVID-19 pandemic, developed countries tends to decrease spread of resistance, while in developing countries with poor control on the spread of bacteria will increase, but in our study shows tendency resistance spread to decrease within given period.
The limitation of our study is the short period of time and limited area is not sufficient to draw a big picture impact of COVID-19 pandemic to E. coli susceptibility pattern. The subject in group B had no further investigation of SARS-CoV-2 infection status. The azithromycin antibiotics were not evaluated in this study.
CONCLUSION
Total E. coli isolates and ESBL-producing E. coli were in decline during COVID-19 pandemic. This phenomenon contributes to the dynamic of antibiotic susceptibility between two groups. The longer period of analysis is needed to be done, due to the ongoing COVID-19 pandemic.
REFERENCES
- 1.WHO . Fact sheet Antibiotic Resistance. 2020. Available at: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance
- 2.Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist 2018;11:1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC . Antibiotic Resistance Threats in the United States 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf
- 4.Breijyeh Z, Jubeh B, Karaman R.Resistance of Gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 2020; 25:1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allocati N, Masulli M, Alexeyev MF, Di Ilio C.Escherichia coli in Europe: an overview. Int J Environ Res Public Health 2013;10:6235–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ventola CL.The antibiotic resistance crisis: part 1: causes and threats. P T 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 7.McDanel J, Schweizer M, Crabb V, Nelson R, Samore M, Khader K, et al. Incidence of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella infections in the United States: A systematic literature review. Infect Control Hosp Epidemiol 2017; 38:1209–1215. [DOI] [PubMed] [Google Scholar]
- 8.Public Health England (2020). English surveillance programme for antimicrobial utilisation and resistance (ESPAUR) 2015 to 2019.
- 9.Hadi U, Kuntaman, Qiptiyah M, Paraton H.Problem ofantibiotic use and antimicrobial resistance in Indonesia: are we really making progress? IJTID 2013; 4: 5–8. [Google Scholar]
- 10.Hayati Z, Rizal S, Putri R.Isolation of extended-spectrum β-lactamase (ESBL) producing Escherichia coli and Klebsiella pneumiae from Dr. Zainoel Abidin General Hospital, Aceh. Int J Trop Vet Biomed Res 2019;4:16–22. [Google Scholar]
- 11.van Duin D, Barlow G, Nathwani D.The impact of the COVID-19 pandemic on antimicrobial resistance: a debate. JAC Antimicrob Resist 2020; 2: dlaa053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Getahun H, Smith I, Trivedi K, Paulin S, Balkhy HH.Tackling antimicrobial resistance in the COVID-19 pandemic. Bull World Health Organ 2020;98:442–442A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI Supplement M100. Wayne PA: Clinical and Laboratory Institute. [Google Scholar]
- 14.Lee DS, Lee SJ, Choe HS.Community-acquired urinary tract infection by Escherichia coli in the era of antibiotic resistance. Biomed Res Int 2018;2018: 7656752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collignon P, Beggs JJ.CON: COVID-19 will not result in increased antimicrobial resistance prevalence. JAC Antimicrob Resist 2020; 2: dlaa051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clancy CJ, Buehrle DJ, Nguyen MH.PRO: The COVID-19 pandemic will result in increased antimicrobial resistance rates. JAC Antimicrob Resist 2020; 2: dlaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fair RJ, Tor Y.Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem 2014; 6: 25–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manyi-Loh C, Mamphweli S, Meyer E, Okoh A.Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 2018; 23:795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kibret M, Abera B.Antimicrobial susceptibility patterns of E. coli from clinical sources in northeast Ethiopia. Afr Health Sci 2011; 11(Suppl 1): S40–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulkarni SR, Peerapur BV, Sailesh KS.Isolation and antibiotic susceptibility pattern of Escherichia coli from urinary tract infections in a tertiary care hospital of North Eastern Karnataka. J Nat Sci Biol Med 2017; 8:176–180. [DOI] [PMC free article] [PubMed] [Google Scholar]