Abstract
Spermine oxidase (SMOX) catalyzes the oxidation of spermine to spermidine. Observational studies have reported SMOX as a source of reactive oxygen species associated with cancer, implying that inhibition of SMOX could be a target for chemoprevention. Here we test causality of SMOX levels with cancer risk using a Mendelian randomization analysis. We performed a GWAS of spermidine/spermine ratio to identify genetic variants associated with regulation of SMOX activity. Replication analysis was performed in two datasets of SMOX gene expression. We then did a Mendelian randomization analysis by testing the association between the SMOX genetic instrument and neuroblastoma, gastric, lung, breast, prostate, and colorectal cancers using GWAS summary statistics. GWAS of spermidine/spermine ratio identified SMOX locus (P = 1.34 × 10–49) explaining 32% of the variance. The lead SNP rs1741315 was also associated with SMOX gene expression in newborns (P = 8.48 × 10–28) and adults (P = 2.748 × 10–8) explaining 37% and 6% of the variance, respectively. Genetically determined SMOX activity was not associated with neuroblastoma, gastric, lung, breast, prostate nor colorectal cancer (P > 0.05). A PheWAS of rs1741315 did not reveal any relevant associations. Common genetic variation in the SMOX gene was strongly associated with SMOX activity in newborns, and less strongly in adults. Genetic down-regulation of SMOX was not significantly associated with lower odds of neuroblastoma, gastric, lung, breast, prostate and colorectal cancer. These results may inform studies of SMOX inhibition as a target for chemoprevention.
Subject terms: Cancer, Computational biology and bioinformatics, Genetics, Genetic association study, Medical genetics
Introduction
Although polyamines are essential for normal cell growth and development1,2, dysregulation of polyamine metabolism is involved in tumorigenesis3, and hence recognized as a potential target for chemotherapy and chemoprevention4. Already in the 1960s, ornithine decarboxylase (ODC), the first rate-limiting enzyme in polyamine biosynthesis, was demonstrated to be at high levels in human cancer specimens5. Moreover, a genetic variant in ODC, regulating its enzymatic activity, confirmed the role of this enzyme in human colon cancer risk6,7, and ODC levels have been shown to be elevated in human skin, breast and prostate cancer specimens8–10. Other polyamine metabolic enzymes have not yet been reported to be associated with tumorigenesis in humans.
Spermine oxidase (SMOX), a member of the mammalian polyamine catabolic pathway, encodes an enzyme with cytoplasmic and nuclear expression in most tissues11, that catalyzes the oxidation of spermine to spermidine with the production of hydrogen peroxide (H2O2) and 3-aminopropanal (3-AP)12,13. SMOX has been reported as a source of induced reactive oxygen species (ROS) associated with neuroblastoma, gastric, lung, breast, prostate and colon cancers14–20, implying that inhibition of SMOX could be a target for chemoprevention3. However, so far, studies of SMOX inhibition have been observational in their nature with no direct inference on causation14–20. Genome-wide association studies (GWAS) of SMOX activity are also lacking. A GWAS could potentially detect genetic determinants of SMOX activity that might serve as instruments to assess the causality of SMOX on cancer using a Mendelian randomization framework21.
To our knowledge, there has not been any Mendelian randomization study assessing the potential causal relationship between SMOX activity and risk of any type of cancer. In this study, we present the first GWAS of spermidine/spermine ratio as a proxy for SMOX activity. We then use the genetic determinants of SMOX activity to perform Mendelian randomization analysis to assess causal relationships between SMOX activity and risk of neuroblastoma, gastric, lung, breast, prostate and colon cancer. In addition, we perform a phenome-wide association study (PheWAS) to query possible deleterious effect of altered SMOX activity through all disease categories and to test possible pleiotropic effects.
Methods
Study population
The GWAS of spermidine/spermine ratio reflecting SMOX activity, was based on dried blood spot samples taken during routine newborn screening from 534 individuals of Danish ancestry, recruited in a matched case–control study of infantile hypertrophic pyloric stenosis (IHPS), as previously described22,23 (SSI-IHPS cohort). In addition, dried blood spot samples of 262 newborns of Danish ancestry were included for expression quantitative trait locus (eQTL) analysis (PSYCH-twin cohort). These 262 newborns were taken at random from each of 262 monozygotic twin pairs discordant for later diagnosis of psychiatric disorders as previously described24. Of these 262 newborns, 235 were included for methylation quantitative trait loci (mQTL) analysis. Furthermore, 671 whole blood samples taken from adults of the Estonian Genome Center University of Tartu (EGCUT) cohort25 were also used for eQTL (N = 508)26,27 and mQTL (N = 305; overlap with gene expression dataset 141 samples) lookups. The Danish Scientific Ethics Committee, the Danish Data Protection Agency and the Danish Neonatal Screening Biobank Steering Committee approved the PSYCH-twins study as well as the GWAS of metabolites in newborns. Usage of EGCUT RNA-seq dataset was approved by Estonian Committee on Bioethics and Human Research, protocol nr. 1.1–12/624, data extraction nr. N26.
The study also included GWAS summary statistics from four cancer-specific consortia cohorts: The ELLIPSE consortium (prostate cancer)28 with 79,194 cases and 61,112 controls; the BCAC consortium (breast cancer)29 with 62,533 cases and 60,976 controls; the TRICL consortium (lung cancer)30 with 29,266 cases and 56,450 controls, and the North American-based Children’s Oncology Group (neuroblastoma)31 with 2,101 cases and 4,202 controls. GWAS summary statistics for gastric and colorectal cancers relied on population-based cohorts: UK Biobank with 5,693 colorectal cancer cases and 386,740 controls32, and the BioBank Japan Project with 6,563 gastric cancer cases and 195,745 controls33. Analyses were restricted to individuals of European ancestry, except for gastric cancer due to the lack of publicly available European gastric cancer GWAS data.
Patients or the public were not involved in the design, or conduct, or reporting, or dissemination plans of our research.
Metabolite measurements and GWAS
The metabolites spermidine and spermine were quantified in whole blood of dried blood spots using the AbsoluteIDQ® p400 Kit (Biocrates Life Sciences AG, Innsbruck, Austria) (Supplementary Table S3, Figures S10-S12). The AbsoluteIDQ® p400 Kit allows quantification of 408 metabolites which were described in our previous study23. In our previous study23, we quantified 148 of the 408 metabolites, from which spermidine and spermine measurements were also used for this study. Briefly, since all samples were run sequentially, the only batch effect we noticed was plate effect, for which we corrected this by dividing the raw concentration values of all samples by a plate-specific correction factor23. We also did metabolite filtering by selecting the 148 metabolites that were above the limit of detection (LOD) in at least 80% of the samples and had a coefficient of variation of the replicates below 25%23. In this study we performed genome-wide association scans of all these 148 metabolites and their biologically relevant ratios (e.g. direct substrate and product of an enzyme), including spermidine to spermine ratio, which had the most significant genome-wide hit of all the metabolites and ratios tested. Our GWAS p-value threshold, correcting for the 148 metabolites tested, is P < 5 × 10–8/148 = 3.4 × 10–10. For computing time issues, the GWAS analysis was first implemented in PLINK34 and then repeated in R35 only for the genome-wide significant variants. The PLINK34 and R35 analyses were done using the following linear model: spermidine/spermine inverse normally transformed concentration ratios ~ SNP + sex + YOB + GA + Parity + IHPS. YOB pertains to year of birth, GA as gestational age in weeks, Parity as number of completed pregnancies, and IHPS as case/control status. Of note, the GWAS analysis was also repeated in R35 in order to obtain the variance explained (R2) of the genome-wide significant variants and to allow YOB to be coded as a factor. As described before22, the SSI-IHPS cohort was comprised of Danish ancestry and unrelated individuals of which DNA samples were array-genotyped with the Illumina Multi-Ethnic Global_v2_A2 array, and after genotyping QC, unobserved genotypes were imputed from the Haplotype Reference Consortium panel36. Altogether, 6,846,507 SNPs with minor allele frequency > = 1% and imputation info score > = 0.8 were used in the analysis. Being unrelated and of Danish ancestry was determined from the genome-wide SNP array data as previously described22, using the kinship and principal component analysis, respectively. The GWAS summary statistics of spermidine/spermine ratio of the SSI-IHPS newborn cohort is available here: danishnationalbiobank.com/gwas.
SMOX eQTL and mQTL identification
To replicate the genetic association with SMOX activity at the gene expression level, eQTL analysis was performed in whole blood from two additional data sets of 262 newborns (a subset of the PSYCH-twin cohort) and 508 adults (a subset of the EGCUT cohort) that had previously been RNA-sequenced and GWAS-genotyped24,26,27. Preprocessing of the EGCUT expression data is described elsewhere26. Briefly, in EGCUT, gene expression matrix was normalized by trimmed mean of M values (TMM)37, log2 transformed, gene expression values were centered and scaled, and genetic PCs were regressed out26. In addition, in order to remove non-genetic variance from the data, up to 20 gene expression-based PCs which were not associated with genetics were regressed out from the residuals of the expression data. Finally, inverse normal transformation was applied on the residuals from the previous step. In order to detect a possible age-dependent eQTL effect, the adult cohort (EGCUT) was stratified into different age groups (10-years intervals). Formal interaction analysis was also performed, in R35, between donor age at blood draw and rs1741315 genotype on SMOX expression.
Moreover, 235 out of 262 individuals from the newborn PSYCH-twin cohort24 also had genome-wide methylation array data from the Infinium HumanMethylation450 BeadChip (450 k) array, as previously described24. For this study, all samples were filtered by Call Rate > 0.99 (for detection P < 0.01 and bead count > 2) and median methylation and unmethylation intensity signal > 2000 to ensure proper sample measurement quality. All probes (CpGs) were filtered by Call Frequency > 0.99 (for detection P < 0.01 and bead count > 2) to ensure sufficient probe representation across most samples. The data was normalized with the noob background dye correction method after sample filtering38, implemented in the R35 package minfi. For all the 480 CpG sites within 1 Mb of the SMOX gene, we tested their methylation fraction against the lead (most significant) SMOX SNP. The analysis was implemented in R35 as the following linear model: CpG methylation fraction ~ lead SMOX SNP + sex + YOB + GA + BW (birth weight). An association was deemed significant if P-value < 0.05/480 = 1.04 × 10–4 to correct for the number of CpGs tested. Furthermore, 305 individuals from the adult EGCUT cohort also had genome-wide methylation array data from the Infinium HumanMethylation450 BeadChip (450 k) array. Briefly, methylation data was normalized according to the GoDMC (http://www.godmc.org.uk/) pipeline39 using functional normalization method as implemented in the R package meffil40. In step 1, we adjusted normalized methylation proportion for age, sex, predicted cell counts, predicted smoking and genetic PCs. Predicted smoking was performed as the methylation fraction of known CpG sites that are well correlated with smoking status41. In step 2, PCA was applied on the residuals from the previous step, and top non-genetic PCs were regressed out. To be able to estimate the SNPxAge effect on methylation, inverse normally transformed methylation proportions were regressed against covariates from step 1 and methylation PCs from step 2.
Variance explained, Mendelian randomization, and PheWAS analyses
The lead SNP located on chromosome 20q12 (in cis with SMOX) being genome-wide significantly (P < 5 × 10–8) associated with spermidine/spermine ratio was used as the genetic instrument in the Mendelian randomization analyses. Variance explained by the genetic determinant of SMOX activity was calculated as the adjusted R2 of lm function in R35 implemented as spermidine/spermine inverse normally transformed concentrations ratio ~ SNP, or normalized and inverse normally transformed gene expression value of SMOX ~ SNP. The Wald ratio method42, implemented in the R35 package MendelianRandomization (v0.3.0) was used to test whether the association between SMOX genotype and incident cancers could be caused by differences in SMOX activity (in the SSI-IHPS newborn cohort) or SMOX gene expression (in the EGCUT adult cohort). To confirm the results of the MR Wald ratio test, we also used another implementation of this method from the TwoSampleMR (v0.5.6) R35 package. The primary analysis measured the association between genetically determined SMOX activity and the risk of neuroblastoma, gastric, lung, breast, prostate and colorectal cancers. The PhenoScanner bioinformatic tool43, a curated database of publicly available results from large-scale genetic association studies, was used for the PheWAS analysis and detect possible concurrent risk factors of the investigated cancer types (pleiotropy). The PhenoScanner database was queried from the MendelianRandomization R23 package. In our main analyses we tested six key outcomes, namely neuroblastoma, gastric, lung, breast, prostate and colorectal cancers. A P-value of less than 0.05/6 (number of cancers tested) = 0.008 was considered statistically significant. Of note, Table S1 contains rs1741315 allele frequencies for reference populations relevant to our study.
To study the possible causal effects of reticulocyte count, lymphocyte count, and hemoglobin concentration on the cancers tested, MR-Base44 platform was used with default settings (accessed 29 April 2021). Briefly, genome-wide significant hits of GWAS of reticulocyte count45, lymphocyte count45 and hemoglobin concentration45 were used as genetic instruments (clumping was used to prune SNPS for LD) of those exposures to test their causal role on the six cancers tested.
All methods were carried out in accordance with relevant guidelines and regulations.
Ethical approval and consent to participate
The Danish Scientific Ethics Committee, the Danish Data Protection Agency and the Danish Neonatal Screening Biobank Steering Committee approved the PSYCH-twins study as well as the GWAS of metabolites in newborns. The Danish Scientific Ethics Committee granted exemption from obtaining informed consent from participants as this research project was based on genotyping samples from biobank material (H-4-2013-055). Usage of EGCUT RNA-seq dataset was approved by Estonian Committee on Bioethics and Human Research, protocol nr. 1.1-12/624, data extraction nr. N26. Patients or the public were not involved in the design, or conduct, or reporting, or dissemination plans of our research.
Consent for publication
All authors consent this study for publication.
Results
Participant characteristics
A total of 534 newborn participants (SSI-IHPS cohort) were included in the GWAS of spermidine/spermine ratio to find the genetic instrument of SMOX activity (Methods). An additional 262 newborns (PSYCH-twin cohort) and 508 adults (EGCUT cohort) were included to confirm the genetic instrument of SMOX activity at the gene expression level (Table 1 and Methods). A total of 950,575 participants, including 79,194 participants who had prostate cancer, 62,533 with breast cancer, 29,266 with lung cancer, 5,693 with colorectal cancer, 6,563 with gastric cancer and 2,101 with neuroblastoma were included as the cancer outcome cohorts (Table 2 and Methods). Demographic characteristics of each cohort are described in the original studies22–24,26,28–30,32,33.
Table 1.
SNP rs1741315 associations with SMOX activity, SMOX gene expression and cg07472708 methylation fraction.
| Pos | rsID | Cohort | Analysis | N | EAF | Beta | SE | P Value | R2 |
|---|---|---|---|---|---|---|---|---|---|
| 20:4,155,948 | rs1741315 | Newborns (SSI-IHPS) | SMOX activity | 534 | 0.458 | − 0.757 | 0.046 | 1.34 × 10–49 | 0.318 |
| Newborns (PSYCH-twins) | SMOX expr | 262 | 0.471 | − 0.830 | 0.062 | 2.47 × 10–30 | 0.366 | ||
| cg07472708 | 235 | − 0.012 | 0.003 | 4.63 × 10–05 | 0.005 | ||||
| Adults (EGCUT) | SMOX expr | 508 | 0.452 | − 0.366 | 0.065 | 2.75 × 10–08 | 0.057 | ||
| cg07472708 | 305 | − 0.062 | 0.069 | 0.371 | 0.002 |
SMOX activity and SMOX gene expression estimates were based on inverse normally transformed values (Methods). SNP rs1741315 ‘A’ allele is the effect allele. Pos, genomic position of rs1741315 (hg19); rsID, variant identifier; EAF, effect allele frequency; N, sample size; Beta, effect estimate; SE, standard error; R2, variance explained of rs1741315 on SMOX activity, SMOX gene expression and cg07472708 methylation fraction. An increased spermidine/spermine ratio represents increased SMOX activity.
Table 2.
Associations between genetically predicted SMOX activity from SSI-IHPS newborn cohort (SSI-IHPS instrument) and SMOX expression from the EGCUT adult cohort (EGCUT instrument), based on SNP rs1741315, and six site-specific cancers (prostate, breast, lung, colorectal and neuroblastoma).
| Outcome | Cases, No | Controls, No | EAF | SSI-IHPS instrument | EGCUT instrument | ||
|---|---|---|---|---|---|---|---|
| Odds ratio (95% CI) | P | Odds ratio (95% CI) | P | ||||
| Prostate cancer | 79,194 | 61,112 | 0.430 | 0.98 (0.96–1.00) | 0.046 | 0.96 (0.92–1.00) | 0.046 |
| Breast cancer | 62,533 | 60,976 | 0.434 | 0.99 (0.96–1.02) | 0.465 | 0.98 (0.93–1.03) | 0.465 |
| Lung cancer | 29,266 | 56,450 | 0.442 | 0.97 (0.94–1.00) | 0.084 | 0.95 (0.89–1.01) | 0.084 |
| Colorectal cancer | 5,693 | 386,740 | 0.439 | 0.96 (0.91–1.01) | 0.097 | 0.92 (0.83–1.02) | 0.097 |
| Gastric cancer | 6,563 | 195,745 | 0.361 | 1.02 (0.97–1.07) | 0.543 | 1.03 (0.93–1.14) | 0.543 |
| Neuroblastoma | 2,101 | 4,202 | 0.412 | 1.07 (0.97–1.19) | 0.182 | 1.16 (0.93–1.44) | 0.182 |
All estimations were based on the Wald ratio method, with A being the effect allele. EAF, effect allele frequency in each cohort (cases and controls combined); P, p-value. An increased spermidine/spermine ratio represents increased SMOX activity.
SMOX activity GWAS
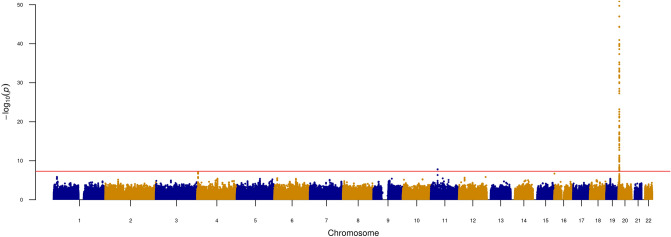
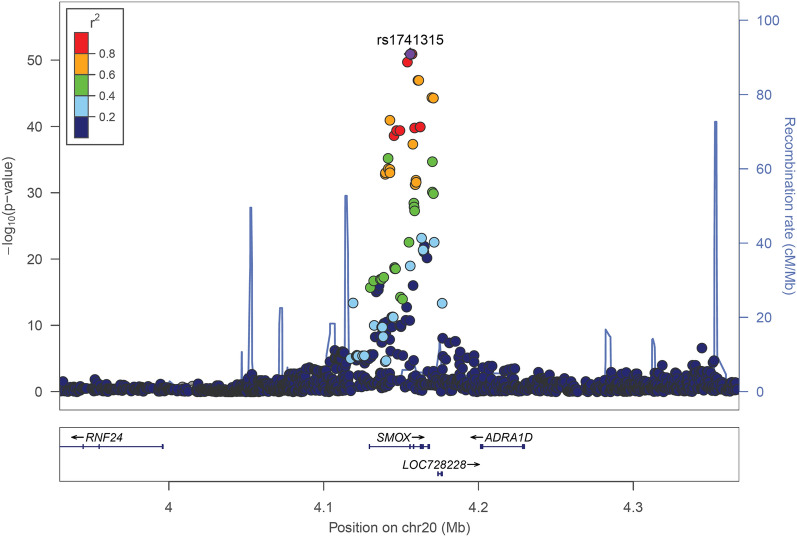
We performed a GWAS of spermidine/spermine ratio, with one locus reaching genome-wide significance, after correction for the 148 metabolites tested (P < 5 × 10–8/148 = 3.4 × 10–10) (Fig. 1, Methods). This locus was on chromosome 20q12, with the most significant SNP rs1741315 (P = 1.34 × 10–49) being an intronic variant in the SMOX gene (Fig. 2). This lead cis-acting SNP explained 32% of the variance in the spermidine/spermine ratio (Figure S9, Table 1), which is regarded as a powerful genetic instrument of SMOX activity for the Mendelian randomization analysis. Of note, there was no sex-difference in SMOX activity (Wilcoxon rank sum test P = 0.10, Figure S8).
Figure 1.
Genetic determinants of SMOX activity. Manhattan plot of the genome-wide association study (GWAS) of spermidine/spermine ratio (534 samples; SSI-IHPS cohort).
Figure 2.
Regional association plot of the new genome-wide significant loci for spermidine/spermine ratio at the SMOX locus. Color-coded linkage disequilibrium (LD) is shown for the top SNP rs1741315 (LD determined with 1000Genomes EUR population). The x-axis represents the genomic region (hg19 assembly) surrounding 200 kb of SMOX gene, while the y-axis represents the strength of the association in − log10(P-value).
SMOX eQTL and mQTL analyses
To assess effects of rs1741315 on SMOX activity at the gene expression level, we performed eQTL analyses in two additional cohorts. In a newborn cohort of 262 individuals (PSYCH-twin)24 rs1741315 was associated with SMOX gene expression (P = 2.47 × 10–30) explaining 37% of the variance (Table 1). In a cohort of 508 adults (EGCUT)26, rs1741315 was also associated with SMOX gene expression (eQTL P = 2.75 × 10–8) explaining 6% of the variance (Table 1, Figure S1). In the eQTLGen Consortium (https://www.eqtlgen.org/), which the EGCUT cohort is a part of, rs1741315 is also associated with SMOX gene expression (eQTL P = 3.27 × 10–310). Of note, rs1741315 was array-genotyped in all cohorts, in contrast to being just imputed. We stratified the adult cohort (EGCUT) into different age groups (10-years intervals) in order to detect a possible age-dependent eQTL effect in early vs. late adulthood (Figure S2), but found no significant interaction between donor age at blood draw and rs1741315 genotype (P = 0.50) on SMOX expression (Figure S3). Due to some missing values in the data, this age-stratified analysis was performed on 494 out of the total 508 adults. Moreover, 235 out of 262 individuals from the newborn PSYCH-twin cohort24 had genome-wide methylation array data (Methods). Zooming in on CpGs at the SMOX locus, we found rs1741315 to be associated with the methylation proportion of the CpG cg07472708 (mQTL P = 4.63 × 10–5) explaining 0.5% of the variance in methylation proportion for this CpG (Table 1). Contrary to the newborn data, we did not find rs1741315 to be associated with the methylation proportion of the CpG cg07472708 (mQTL P = 0.371) in 305 individuals from the adult EGCUT cohort who also had genome-wide methylation array data (Figure S4, Table 1 and Methods). Similar to the eQTL data, we also stratified the adult cohort (EGCUT) into different age groups (10-years intervals) in order to detect a possible age-dependent mQTL effect (Figure S5), but found no significant interaction between donor age at blood draw and rs1741315 genotype (P = 0.48) on cg07472708 methylation levels (Figure S6). The CpG cg07472708 is located in the same SMOX intron as rs1741315 and is under a region of transcription factor binding site for the transcription factor GATA-1 (Methods). It should be noted that the 50-mer probe containing cg07472708 overlaps with rs1741317 (37 bp apart from the C nucleotide of cg07472708), a high LD (r2 = 0.996) SNP with our lead rs1741315. This could lead to a technical artifact from poorer measurement conditions in those individuals carrying the minor allele, so the results from mQTL analysis should be interpreted with caution.
PheWAS association of SMOX genetic instrument
One of the assumptions of Mendelian randomization is that genetic variants should only affect the outcome through their effect on the risk factors, i.e. no pleiotropic effects46,47. To test if this assumption holds, we checked the association of the SMOX genetic instrument rs1741315 against the PhenoScanner database of genotype–phenotype associations43 at P < 1 × 10–5 (to correct for the 2998 traits/diseases tested in the database). With rs1741315 ‘A’ allele being the effect allele, we detected reticulocyte count (β = -0.150; 95% CI: -0.157, -0.143; P = 4.12 × 10–285), lymphocyte count (β = 0.016; 95% CI: 0.009, 0.023; P = 6.44 × 10–6) and hemoglobin concentration (β = 0.016; 95% CI: 0.009, 0.023; P = 8.05 × 10–6) as traits associated with this variant45. One potential way to test and adjust for pleiotropic effects through associations with these blood traits would be by multivariable Mendelian randomization analysis48. However, such analysis is only possible for multi-SNP genetic instruments, which we did not have. Moreover, running MR using genetics instruments for reticulocyte count45, lymphocyte count45, and hemoglobin concentration45 as exposures for cancer, provided no evidence of these potential confounders being causal for the cancers tested (Table S4, Methods).
Association of SMOX genetic instrument with cancer risk
By using the effect estimate form the SSI-IHPS newborn cohort, we found little evidence that the genetic instrument of SMOX activity, rs1741315, was associated with any of the five non-pediatric cancers evaluated (Table 2). Although genetically lower levels of SMOX were associated with slightly lower risk of prostate cancer at P = 0.047, this finding should be interpreted with caution as it becomes non-significant when correcting for the six different cancers tested.
Since the SMOX genetic instrument, rs1741315, explained 5 to 6 times more variance of SMOX expression in newborns than in adults, we used MR to test the causal association between SMOX expression and neuroblastoma, a pediatric cancer in which SMOX has been reported to play a role19,20. Genetically lower levels of SMOX did not associate with lower risk of developing neuroblastoma31 (OR = 0.95; 95% CI:0.88, 1.03; P = 0.182) (Table 2). We repeated the MR analysis for all 6 cancers tested with the effect estimates from the EGCUT adult cohort, and similar non-significant associations were obtained as for the SSI-IHPS newborn cohort (Table 2).
Discussion
We conducted the first GWAS of spermidine/spermine ratio to identify genetic variants regulating spermine oxidase (SMOX) activity. Variants in the SMOX gene explained a large proportion of the variance in this ratio in newborns and were eQTLs for SMOX expression in newborns as well as adults. We did not find genetically determined SMOX activity to be associated with risk of either pediatric (neuroblastoma) or adult cancers (gastric, lung, breast, prostate, and colorectal cancer).
Observational studies have reported elevated SMOX levels in gastric, lung, breast, prostate, and colorectal cancers14–18, and it has been hypothesized that SMOX inhibition could be an effective target for chemoprevention3. Our results do not indicate that genetically driven variation in SMOX activity plays a major role in cancer risk. Several factors and limitations of our study might explain this discrepancy between the observational epidemiological evidence14–18 and our Mendelian randomization results.
First, our results indicate that the genetic instrumental variable more accurately reflects SMOX activity in infancy, compared with later life, where most cancers occur. This variability in the degree to which SMOX expression is genetically regulated over the life course might violate an important MR assumption of monotonicity or homogeneity49. However, we also used the estimate of SMOX activity from the EGCUT adult cohort for the MR analysis and found similar results as when using the estimate of SMOX activity from the newborn cohort (Table 2).
Second, the positive association between observed SMOX levels and cancer in observational studies might be driven by reverse causation, in which cancer in an individual could cause SMOX to be elevated due to inflammation.
Third, elevated SMOX levels in cancer could also be due to environmental factors not captured by genetics, e.g. short-term pharmacological changes, induced by SMOX inhibitors. Our genetic instrument was developed based on normal range SMOX activity data, thus additional genetic variants might play a role in aberrant expression of this enzyme.
Fourth, despite we have corrected for disease status, half the samples from the discovery SSI-IHPS cohort were selected to be IHPS cases and are therefore not representative of the general population. However, the association of rs1741315 with spermidine/spermine ratio did not differ between IHPS cases and controls (Figure S7).
Fifth, the instrumental variable SNP is also associated with hematological traits45 that could potentially affect cancer risk. To assess whether pleiotropic effects drove our results, we performed a PheWAS analysis, with no diseases or non-hematological traits detected. We detected reticulocyte count, lymphocyte count and hemoglobin concentration associated with rs1741315. However, it is unclear if and how the variability in those blood cell phenotypes could affect the cancer predisposition. Moreover, MR analysis of those hematological traits as exposure, revealed no evidence that these potential blood trait confounders are causal for the cancers tested. Therefore, there was no reason to conclude that pleiotropy would affect our Mendelian Randomization analyses.
In addition, the GWAS of gastric cancer was performed on a different ancestry (Japanese, East Asian) than the SMOX GWAS (Danish, Western Northern European), which could have biased our MR estimates. However, the top correlated variants of rs1741315 in Europeans are also the same as in the Japanese population (Table S2), which means the MR estimate bias should be negligible.
Finally, SMOX activity and expression levels were measured in blood in this study, compared to cancer tissue cells in the observational studies.
Our study had important strengths. Several previous studies which examined SMOX activity and cancer risk were susceptible to recall bias, confounding and reverse causation13–18,21 none of which are concerns of Mendelian randomization studies21. Furthermore, using spermidine to spermine ratio to measure SMOX activity was here validated by the orthogonal measurement of SMOX gene expression and its genetic regulation. In addition, the fact that our genetic instrument explained a sizeable proportion of the variance of SMOX activity together with the fact that we used summary statistics from the largest meta-analyses of primary GWAS of these cancer types to date28–33 are factors enhancing the statistical power to detect causal effects50.
Conclusions
In conclusion, lifelong genetic exposure to low levels of SMOX was not associated with lower cancer risk. However, this finding cannot be assumed to represent a short-term strong drug inhibition of SMOX, thus establishing causality would require a randomized clinical trial. To better interpret the complexity of the relationship between SMOX activity and cancer, future studies should effectively distinguish newborn versus adulthood SMOX activity and short versus long exposures. This study is the first to use Mendelian randomization to assess the possible benefits of SMOX inhibitors on cancer risk, paving the way for other polyamine catabolic pathway enzymes to be tested with the same methodology.
Supplementary Information
Acknowledgements
This research has also been conducted using the UK Biobank Resource (project ID 31063) available from the Pan-UKB team. Furthermore, we acknowledge the Genetic Associations and Mechanisms in Oncology (GAME-ON)/Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE) Consortium, the Transdisciplinary Research Into Cancer of the Lung (TRICL) consortium, the Breast Cancer Association Consortium (BCAC), and the North American-based Children’s Oncology Group. We further thank all the participants, staff and researchers of the Danish National Biobank, Estonian, Japan and UK biobanks, along with the cancer case-control cohorts above described for their contribution to this research.
Abbreviations
- SMOX
Spermine oxidase
- GWAS
Genome-wide association study
- eQTL
Expression quantitative trait locus
- mQTL
Methylation quantitative trait locus
- PheWAS
Phenome-wide association study
- ODC
Ornithine decarboxylase
- H2O2
Hydrogen peroxide
- 3-AP
3-Aminopropanal
- ROS
Reactive oxygen species
- IHPS
Infantile hypertrophic pyloric stenosis
- EGCUT
Estonian Genome Center University of Tartu
- YOB
Year of birth
- GA
Gestational age
- RNA-seq
RNA sequencing
- TMM
Trimmed mean of M
- MDS
Multidimensional scaling
- PC
Principal component
- BW
Birth weight
- bp
Base pairs
- LD
Linkage disequilibrium
Author contributions
J.F. conceptualized, designed the study, coordinated data collection and carried out the initial analyses and drafted the initial manuscript. V.Y., U.V., C.S.H., S.K., J.C. and V.K. collected data and also carried initial analyses. L.S., F.G., T.E., A.B., M.M., T.M.W., D.M.H., L.M., J.B.G., A.S.C. conceptualized and designed the study, and critically reviewed the manuscript for important intellectual content. B.F. conceptualized, designed the study, coordinated and supervised data collection and analysis, and critically reviewed the manuscript for important intellectual content. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Funding
This study received partial support from the Danish Medical Reseach council (DFF 4004-00512), the Novo Nordisk Foundation (NNF18OC0053228) and the Oak Foundation (OCAY-18-598). It was conducted using the Danish National Biobank resource, supported by the Novo Nordisk Foundation, grant number 2010-11-12 and 2009-07-28. We would also like to acknowledge iPSYCH and The Lundbeck Foundation for their QTL validation datasets. The iPSYCH (The Lundbeck Foundation Initiative for Integrative Psychiatric Research) team acknowledges funding from The Lundbeck Foundation (grant numbers R102-A9118 and R155-2014-1724). This research has also been conducted using the UK Biobank Resource (project ID 31063) available from the Pan-UKB team. This work has also been supported by the European Regional Development Fund and the programme Mobilitas Pluss (MOBTP108), SP1GI18045T, No. 2014-2020.4.01.15-0012 GENTRANSMED and 2014-2020.4.01.16-0125. This study was also funded by EU H2020 grant 692145, Estonian Research Council Grant PUT1660 and PRG687. Data analyzes with Estonian datasets were carried out in part in the High-Performance Computing Center of University of Tartu.
Competing interests
JF works for Novo Nordisk A/S since January 2021. No other author has a competing interest regarding this study.
Data availability
Supporting data is available in Supplementary Material and on request.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
João Fadista, Email: jpsf@ssi.dk.
Bjarke Feenstra, Email: fee@ssi.dk.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-97069-x.
References
- 1.Pegg AE. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Can. Res. 1988;48(4):759–774. [PubMed] [Google Scholar]
- 2.Gerner EW, Meyskens FL., Jr Polyamines and cancer: Old molecules, new understanding. Nat. Rev. Cancer. 2004;4(10):781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 3.Casero RA, Jr, Murray Stewart T, Pegg AE. Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat. Rev. Cancer. 2018;18(11):681–695. doi: 10.1038/s41568-018-0050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 2007;6(5):373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 5.Russell D, Snyder SH. Amine synthesis in rapidly growing tissues: ornithine decarboxylase activity in regenerating rat liver, chick embryo, and various tumors. Proc. Natl. Acad. Sci. U.S.A. 1968;60(4):1420–1427. doi: 10.1073/pnas.60.4.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez ME, et al. Pronounced reduction in adenoma recurrence associated with aspirin use and a polymorphism in the ornithine decarboxylase gene. Proc. Natl. Acad. Sci. U.S.A. 2003;100(13):7859–7864. doi: 10.1073/pnas.1332465100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubner RA, et al. Ornithine decarboxylase G316A genotype is prognostic for colorectal adenoma recurrence and predicts efficacy of aspirin chemoprevention. Clin. Cancer Res. 2008;14(8):2303–2309. doi: 10.1158/1078-0432.CCR-07-4599. [DOI] [PubMed] [Google Scholar]
- 8.Gupta S, et al. Chemoprevention of prostate carcinogenesis by alpha- difluoromethylornithine in TRAMP mice. Can. Res. 2000;60(18):5125–5133. [PubMed] [Google Scholar]
- 9.Elmets CA, Athar M. Targeting ornithine decarboxylase for the prevention of nonmelanoma skin cancer in humans. Cancer Prev. Res. (Phila Pa). 2010;3(1):8–11. doi: 10.1158/1940-6207.CAPR-09-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manni A, et al. Prognostic influence on survival of increased ornithine decarboxylase activity in human breast cancer. Clin. Cancer Res. 1996;2(11):1901–1906. [PubMed] [Google Scholar]
- 11.Uhlén M, et al. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Devereux W, Woster PM, Stewart TM, Hacker A, Casero RA., Jr Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res. 2001;61(14):5370–5373. [PubMed] [Google Scholar]
- 13.Vujcic S, Diegelman P, Bacchi CJ, Kramer DL, Porter CW. Identification and characterization of a novel flavin-containing spermine oxidase of mammalian cell origin. Biochem. J. 2002;367(Pt 3):665–675. doi: 10.1042/bj20020720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babbar N, Casero RA., Jr Tumor necrosis factor-alpha increases reactive oxygen species by inducing spermine oxidase in human lung epithelial cells: A potential mechanism for inflammation-induced carcinogenesis. Cancer Res. 2006;66(23):11125–11130. doi: 10.1158/0008-5472.CAN-06-3174. [DOI] [PubMed] [Google Scholar]
- 15.Goodwin AC, et al. Increased spermine oxidase expression in human prostate cancer and prostatic intraepithelial neoplasia tissues. Prostate. 2008;68:766–772. doi: 10.1002/pros.20735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaturvedi R, et al. Spermine oxidase mediates the gastric cancer risk associated with Helicobacter pylori CagA. Gastroenterology. 2011;141:1696–1708. doi: 10.1053/j.gastro.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin AC, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl Acad. Sci. USA. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pledgie A, et al. Spermine oxidase SMO(PAOh1), not N1-acetylpolyamine oxidase PAO, is the primary source of cytotoxic H2O2 in polyamine analogue-treated human breast cancer cell lines. J. Biol. Chem. 2005;280:39843–39851. doi: 10.1074/jbc.M508177200. [DOI] [PubMed] [Google Scholar]
- 19.Amendola R, et al. Direct oxidative DNA damage, apoptosis and radio sensitivity by spermine oxidase activities in mouse neuroblastoma cells. Biochem. Biophys. Acta. 2005;1755(1):15–24. doi: 10.1016/j.bbcan.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Fratini E, et al. Link between spermine oxidase and apoptosis antagonizing transcription factor: A new pathway in neuroblastoma. Int. J. Oncol. 2019;55(5):1149–1156. doi: 10.3892/ijo.2019.4878. [DOI] [PubMed] [Google Scholar]
- 21.Ebrahim S, Davey Smith G. Mendelian randomization: can genetic epidemiology help redress the failures of observational epidemiology? Hum. Genet. 2008;123:15–33. doi: 10.1007/s00439-007-0448-6. [DOI] [PubMed] [Google Scholar]
- 22.Fadista J, et al. Genome-wide meta-analysis identifies BARX1 and EML4-MTA3 as new loci associated with infantile hypertrophic pyloric stenosis. Hum. Mol. Genet. 2019;28(2):332–340. doi: 10.1093/hmg/ddy347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fadista J, Skotte L, Courraud J, Geller F, Gørtz S, Wohlfahrt J, Melbye M, Cohen AS, Feenstra B. Integrating genetics with newborn metabolomics in infantile hypertrophic pyloric stenosis. Metabolomics. 2021;17(1):7. doi: 10.1007/s11306-020-01763-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen, C.S., et al. Elevated DNA methylation gestational age is associated with the risk of later bipolar disorder and anorexia nervosa in twins. medRxiv 2020.07.16.20155010
- 25.Leitsalu L, et al. Cohort profile: Estonian biobank of the estonian genome center University of Tartu. Int. J. Epidemiol. 2015;44(4):1137–1147. doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- 26.Võsa U, et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. 2018. bioRxiv 447367.
- 27.Lepik K, et al. C-reactive protein upregulates the whole blood expression of CD59 - an integrative analysis. PLoS Comput. Biol. 2017;13(9):e1005766. doi: 10.1371/journal.pcbi.1005766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schumacher FR, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018;50(7):928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michailidou K, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 2015;47(4):373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKay JD, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat. Genet. 2017;49(7):1126–1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDaniel LD, et al. Common variants upstream of MLF1 at 3q25 and within CPZ at 4p16 associated with neuroblastoma. PLoS Genet. 2017;13(5):e1006787. doi: 10.1371/journal.pgen.1006787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan-UKB team. https://pan.ukbb.broadinstitute.org. 2020.
- 33.Ishigaki K, et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat. Genet. 2020;52(7):669–679. doi: 10.1038/s41588-020-0640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;25(4):7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/ (2020).
- 36.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Triche TJ, Jr, et al. Low-level processing of illumina infinium DNA methylation BeadArrays. Nucleic Acids Res. 2013;41(7):e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min JL, et al. Genomic and phenomic insights from an atlas of genetic effects on DNA methylation. medRxiv 2020.09.01.20180406. [DOI] [PMC free article] [PubMed]
- 40.Min JL, et al. Meffil: Efficient normalization and analysis of very large DNA methylation datasets. Bioinformatics. 2018;34(23):3983–3989. doi: 10.1093/bioinformatics/bty476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wahl A, et al. IgG glycosylation and DNA methylation are interconnected with smoking. Biochim. Biophys. Acta Gen. Subj. 2018;1862(3):637–648. doi: 10.1016/j.bbagen.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Stephen B, Butterworth AS, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamat MA, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35(22):4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;30(7):e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Astle WJ, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167(5):1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.VanderWeele TJ, Tchetgen Tchetgen EJ, Cornelis M, Kraft P. Methodological challenges in mendelian randomization. Epidemiology. 2014;25:427–435. doi: 10.1097/EDE.0000000000000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas DC, Conti DV. Commentary: The concept of ‘Mendelian randomization’. Int. J. Epidemiol. 2004;33:21–25. doi: 10.1093/ije/dyh048. [DOI] [PubMed] [Google Scholar]
- 48.Stephen B, Thompson SG. Multivariable mendelian randomization: The use of pleiotropic genetic variants to estimate causal effects. Am. J. Epidemiol. 2015;181(4):251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017;32(5):377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int. J. Epidemiol. 2014;43:922–929. doi: 10.1093/ije/dyu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supporting data is available in Supplementary Material and on request.