Abstract
The STAT (signal transducer and activator of transcription) signaling pathway is activated by a large number of cytokines and growth factors. We sought to design a conditionally active STAT that could not only provide insight into basic questions about STAT function but also serve as a powerful tool to determine the precise biological role of STATs. To this end, we have developed a conditionally active STAT by fusing STATs with the ligand-binding domain of the estrogen receptor (ER). We have demonstrated that the resulting STAT-ER chimeras are estrogen-inducible transcription factors that retain the functional and biochemical characteristics of the cognate wild-type STATs. In addition, these tools have allowed us to evaluate separately the contribution of tyrosine phosphorylation and dimerization to STAT function. We have for the first time provided experimental data supporting the model that the only apparent role of STAT tyrosine phosphorylation is to drive dimerization, as dimerization alone is sufficient to unmask a latent STAT nuclear localization sequence and induce nuclear translocation, sequence-specific DNA binding, and transcriptional activity.
The JAK (Janus kinase)/STAT (signal transducer and activator of transcription) pathway, a recently discovered signaling pathway utilized by many cytokines and growth factors, was first elucidated in the context of interferon (IFN) signaling (11). It was later discovered that a large number of cytokines and growth factors, including most if not all of those that act through the cytokine receptor superfamily, activate overlapping sets of STAT family members, often in addition to activating other signaling pathways (11). IFN-γ signaling remains, however, a canonical example (2, 56). IFN-γ binding mediates IFN-γ receptor chain aggregation, which activates two cytoplasmic tyrosine kinases belonging to the JAK family, Jak1 and Jak2, that associate with the cytoplasmic face of the IFN-γ receptor chains. Upon receptor oligomerization, the JAKs phosphorylate each other and Tyr440 of the IFN-γ receptor α chain. Then Stat1, a latent cytoplasmic transcription factor that is a member of the STAT gene family, is recruited via its Src homology 2 domain (SH2 domain) to the phosphorylated Tyr440 of the receptor, whereupon Stat1 is itself phosphorylated by the JAKs on a specific tyrosyl residue, Tyr701. Phosphorylation triggers Stat1 homodimerization via the reciprocal binding of the SH2 domain of one Stat1 monomer with the phosphotyrosyl tail of the other Stat1 monomer in a head-to-tail interaction. It is thought that phosphorylation is the sole trigger for dimerization. Although it has been hypothesized that dimerization (and not tyrosine phosphorylation per se) in turn triggers nuclear translocation, there are no data that clearly demonstrate this. Indeed, this hypothesis has been challenged by recent studies on Stat5 activation by prolactin, as it has been reported that Stat5 tyrosine phosphorylation and Stat5 nuclear localization are controlled by different pathways that can be separated by prolactin receptor truncation (1). In any event, once in the nucleus, Stat1 homodimers bind to a distinct DNA element, the IFN-γ activation site found in the promoters of IFN-γ-regulated genes, thereby activating their transcription. Although sequence-specific DNA binding by STATs is thought to result from dimerization and not to be intrinsic to the tyrosine phosphorylation itself, the recent crystal structure of truncated, homodimeric Stat1 bound to DNA shows that the phosphate-binding loop of the Stat1 SH2 domain seems to communicate directly with a critical portion of the STAT DNA-binding domain (7). This has led to speculation that the phospho group on Tyr701 may play a more direct role in sequence-specific DNA binding than previously thought (7). Accordingly, a reagent that could separate STAT tyrosine phosphorylation from STAT dimerization would help shed light on the precise role of tyrosine phosphorylation in several aspects of STAT function.
Cytokine-activated receptors usually mediate the simultaneous activation of multiple signaling pathways (21). Determining the contribution of each of these signaling pathways to the eventual phenotypic outcome is a challenging problem and might help illuminate how cytokines direct different genetic or phenotypic programs in different cell types. Many approaches have been used to determine the specific contribution of STAT activation to overall cytokine action; these include receptor mutations (8, 9, 13, 14, 16, 33, 36, 42, 47, 49, 52, 57, 61, 67), dominant-negative STATs (20, 32, 35, 38, 40, 65), and the generation of STAT-deficient mice (11, 59). These approaches have all yielded important but limited information by providing data that address only what happens when a certain pathway or activity is lacking. Indeed, receptor truncations and mutations are rather blunt instruments, and the precise elimination of pathways emanating from a given receptor is more the exception than the rule (reference 59 and references cited therein). Similarly, the use of dominant-negative STAT constructs in several cases has yielded confusing or contradictory data (38, 59, 65). Results with STAT knockout mice have tended to be less ambiguous, though such experiments also have caveats. For example, in addition to their role in cytokine signal transduction, STATs may have unanticipated roles in the regulation of genes not directly associated with their role in cytokine signal transduction, as has been shown in Stat1-deficient fibroblasts, which lack the constitutive expression of certain caspases (22). Such phenomena can thus complicate the interpretation of the phenotype of STAT-deficient mice. For these reasons, we sought a system that would allow us to activate a specific STAT in the absence of interference from other pathways.
To design a conditionally active STAT, we chose the estrogen receptor (ER) ligand-binding domain (LBD), a heterologous, ligand-inducible dimerization domain, to serve as an inducible driver of STAT dimerization. The ER LBD, as defined by amino acids 282 to 595, is a domain responsible for binding ER ligands (23). In addition, the LBD also includes a dimerization domain, a transcriptional activation function called AF-2 (15), and an estrogen-regulated inactivation function (43). This inactivation function is portable (44), as it has been possible to render constitutively active enzymes and transcription factors estrogen dependent by fusion with the ER LBD (26). We have applied this system to the STAT transcription factors, which lack constitutive transcriptional activity, and have constructed chimeric STAT proteins whose activity is regulated by estrogen even in the absence of tyrosine phosphorylation. These tools have allowed us to evaluate separately the contribution of tyrosine phosphorylation and dimerization to STAT function. We have for the first time provided experimental data supporting the model that the only apparent role of STAT tyrosine phosphorylation is to drive dimerization, as dimerization alone is sufficient to unmask a latent STAT nuclear localization sequence and induce nuclear translocation, sequence-specific DNA binding, and transcriptional activity.
MATERIALS AND METHODS
Reagents.
Human IFN-γ was obtained from Genentech (South San Francisco, Calif.), and human interleukin-4 (IL-4) was obtained from R&D Systems (Minneapolis, Minn.). β-Estradiol (estrogen; E2) and 4-hydroxytamoxifen (hereafter referred to as tamoxifen) were obtained from Sigma (St. Louis, Mo.). ICI-182,870 and the ERE-tk-luc reporter were gifts from K. Marschke (Ligand Pharmaceuticals). The IRF-1x4-tk-luc reporter has been described previously (51), and the mGɛx4-pGL2 reporter, which contains four copies of the overlapping C/EBP and STAT-binding element from the murine germ line ɛ promoter in the context of pGL-2 (Promega, Madison, Wis.), was a gift from C. Lowe (Ligand). The natural ICAM-luc reporter construct was a gift from E. Delorme (Ligand).
Cells and cell culture.
HepG2 (human hepatoma) cells were obtained from the American Type Culture Collection and grown in Eagle minimal essential medium supplemented with fetal bovine serum (FBS; 10%, vol/vol). Parental 2fTGH (human fibrosarcoma) and derivative U3A (G. Stark, Cleveland Clinic) (39) cells were grown in Dulbecco modified Eagle medium supplemented with FBS (10%, vol/vol). Cos7 (simian kidney) cells were obtained from the American Type Culture Collection and grown in Dulbecco modified Eagle medium supplemented with FBS (10%, vol/vol). Cytokines were used at concentrations of 5 (IFN-γ) and 10 (IL-4) ng/ml. β-Estradiol, tamoxifen, and ICI-182,870 were used at 1 μM unless otherwise noted. In all cases, phenol red-free culture medium and charcoal-adsorbed FBS were used to avoid constitutive activation of the ER LBD by the weak ER agonist phenol red or estrogens typically present in untreated FBS.
Plasmid construction. (i) C-terminal fusion of Stat1 with the ER LBD (Stat1-ER).
The Stat1 cDNA was amplified by PCR from pMNC91 (39) (gift from J. E. Darnell, Jr., Rockefeller University), using 5′-KpnI (5′-CGCGCGGTACCATGTCTCAGTGGTACGAACTTCAGCAGCTT [sense])- and 3′-NotI (5′-CCGTCGTTCACGCGGCCGCTACTGTGTTCATCATACTGTCGAA [antisense])-containing primers (underlining indicates restriction enzyme sites). The ER LBD sequence encoding amino acids 282 to 595 was amplified by PCR from pER2 (gift from E. Allegretto, Ligand), using 5′-NotI ( 5′ - GCCCATCACACAC TGGCGGCCGCGTC TGC TGGAGACATGAGAGCT [sense])- and 3′-ApaI (5′-CCGTCGTTGGGCCCTCAGGATCCGACTGTGGCAGGGAAACCCTCT [antisense])-containing primers. The PCR products were digested as indicated and cloned into the KpnI/ApaI sites of pcDNA3.1(+) (Invitrogen, Carlsbad, Calif.), generating pStat1ER-3.1.
(ii) N-terminal fusion of the ER LBD with Stat1 (ER-Stat1).
The ER LBD was amplified by PCR from pER2 by using 5′-KpnI (5′-GCCCATGGTACCATGTCTGCTGGAGACATGAGAGCT [sense])- and 3′-BamHI (5′-CCGTCGTTGGATCCGACTGTGGCAGGGAAACCCTCT [antisense])-containing primers. The Stat1 cDNA was amplified by PCR from pMNC91 by using 5′-BamHI (5′-CGCGCGGATCCATGTCTCAGTGGTACGAACTTCAGCAGCTT [sense])- and 3′-ApaI (5′-CCGTCGTTGGGCCCTCATACTGTGTTCATCATACTGTCGAA [antisense])-containing primers. The PCR products were digested as indicated and cloned into the KpnI/ApaI sites of pcDNA3(+), generating pERStat1-3.0.
(iii) C-terminal fusion of Stat6 with the ER LBD (Stat6-ER).
The murine Stat6 cDNA was amplified by PCR from pRK5-Stat6 (48) (gift from J. N. Ihle, St. Jude Children’s Research Hospital), using 5′-NheI (5′-GCCCATCACGCTAGCGCCCATATGTCTCTGTGGGGCCTAATTTCCAAG [sense])- and 3′-NotI (5′-GCCCATCTCGAGGCGGCCGCCCAGCTGGGGTTGGTCCTTAGGTC [antisense])-containing primers. The NotI-ApaI ER LBD fragment from pStat1ER-3.1 was cloned into the NotI/ApaI sites of pcDNA3.1(+), resulting in pERLBD-3.1. The Stat6 PCR fragment was ligated with pERLBD-3.1 after both had been digested with NheI and NotI, generating pStat6ER-3.1.
(iv) Stat1 Ser727-Ala mutant in the Stat1-ER chimera.
Oligonucleotides comprising the unique Stat1 XbaI-EcoRI fragment were synthesized by substituting GCC (Ala) for the codon corresponding to Ser727. The oligonucleotides were then cloned into the XbaI/EcoRI sites of pStat1ER-3.1. All PCR-generated fragments were fully sequenced to verify that the sequences were correct. Plasmid pMNCStat1Ser727 Ala (69) was a gift from J. E. Darnell, Jr.
Cos7 cell transfections, preparation of nuclear extracts, EMSAs, and immunoprecipitations.
Cos7 cells were transfected in 10-cm-diameter plates by using LipofectAMINE (Life Technologies, Gaithersburg, Md.), using the manufacturer’s protocol. Nuclear extracts were prepared and electrophoretic mobility shift assays (EMSAs) were performed as described elsewhere (51). The EMSA probes were formed by annealing oligonucleotides with the sequence (50) 5′-GATCGATTTCCCGGAAATC-3′ (leaving 5′-GATC overhangs). Immunoprecipitations and immunoblotting were performed as described elsewhere (60), using an antibody directed against the N terminus of Stat1 (Transduction Laboratories, Lexington, Ky.), antiphosphotyrosine antibody 4G10 (Upstate Biotechnology, Lake Placid, N.Y.) or an anti-ER LBD antibody (Chemicon, Temecula, Calif.).
Analysis of gene induction.
Total RNA from transfected Cos7 cells was obtained by using an RNeasy kit (Qiagen, Valencia, Calif.), and cDNA was synthesized by using a Superscript cDNA kit (Life Technologies), using the manufacturers’ protocols. Total RNA (2 μg) and oligo(dT) primers (0.5 μg) were used in the reverse transcription (RT) reaction.
To analyze IRF-1 (IFN regulatory factor 1) gene induction, a fragment was amplified by PCR with 5′-TGGGCCCCTCTTATTCCTCTA-3′ (sense) and 5′-TCTGGGGTCACTGGTCTGTTC-3′ (antisense) primers, using standard conditions. To analyze cIITA gene induction, a fragment was amplified by PCR with 5′-ACGCCCACCATCCCATTCAGT-3′ (sense) and 5′-CCCTCTCACCGCCCCATTAGT-3′ (antisense) primers. For the PCRs, 1/25 of the RT reaction, 5 ng each of sense and antisense gene-specific primers, and 2.5 μCi of [α-33P]dATP were used. The PCR products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 4 to 20% acrylamide–Tris-borate-EDTA gels (Novex, San Diego, Calif.). The gels were exposed to X-ray film (Kodak X-Omat). Radioactive bands were quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Transient luciferase reporter assays.
HepG2 and U3A cells were transfected by calcium phosphate coprecipitation with the reporter, expression construct, and a control plasmid expressing β-galactosidase (β-Gal) as described previously (51). ER ligands or cytokines were then added, and the cells harvested after the indicated times. Cells were lysed, and luciferase and β-Gal activities determined by using standard techniques. For each sample, the normalized response was determined by dividing relative light units measured in a luciferase assay with the β-Gal activity in the same lysate. The fold induction (induced/untreated) was calculated by using the averaged normalized responses from three independent experiments.
RESULTS
Construction and activity of Stat1-ER chimeras.
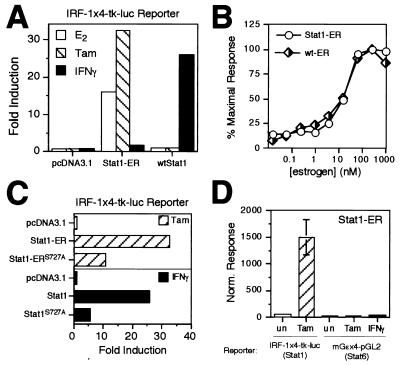
To construct a conditionally active Stat1, fusion proteins were generated by joining the ER LBD to either the amino or the carboxy terminus of Stat1 (Fig. 1). To test whether dimerization is sufficient to activate all STAT functions, we tested whether the chimeric Stat1-ER proteins could be activated by ER ligands such as β-estradiol (estrogen) or tamoxifen, which should cause ER LBD dimerization and, potentially, activation of the Stat1 chimeras. Constructs driving expression of wild-type Stat1 (wtStat1), ER-Stat1 (ER LBD fused to the N terminus of Stat1), or Stat1-ER (ER LBD fused to the C terminus of Stat1) were cotransfected with a Stat1-responsive reporter (IRF-1x4-tk-luc) into the U3A derivative of the 2fTGH cell line. The U3A cells lack endogenous Stat1 and are completely unresponsive to IFN-γ (39). Cells were then either left untreated or treated with estrogen, tamoxifen, or IFN-γ (optimum treatment times were 20 h for estrogen and tamoxifen and 5 h for IFN-γ). As shown in Fig. 2A, when wtStat1 was cotransfected with reporter, IFN-γ activated the reporter 20-fold over background whereas estrogen and tamoxifen had no effect. When the ER-Stat1 construct was cotransfected with reporter, none of the treatments resulted in reporter activation (data not shown). In contrast, when the Stat1-ER construct was cotransfected with the reporter, estrogen or tamoxifen treatment resulted in strong reporter activation, comparable to that induced by IFN-γ when wtStat1 is cotransfected. In U3A cells, IFN-γ activated the reporter marginally (less than 2.5-fold) when Stat1-ER was present, possibly due to a low-level ability of Stat1-ER to couple to the IFN-γ receptor. As shown in Fig. 2B, estrogen activated the Stat1-ER and wild-type ER with equivalent potency, as measured by luciferase reporter assays using Stat1-response- and estrogen-response-element-driven promoters, respectively. This supports the notion that Stat1-ER chimera activity is mediated by the ER LBD. Indeed, the pure ER antagonist ICI-182,870 (27) weakly (two- to threefold) induced chimera activity on the IRF-1x4-tk-luc reporter and antagonized estrogen action on the chimera down to the low level of activity exhibited by ICI-182,870 itself (data not shown), further demonstrating that estrogen-induced transcriptional activity of the chimera is ER LBD dependent. The observed low level of apparent dimerization activity of ICI-182,870 is consistent with studies of the ER LBD in yeast (66).
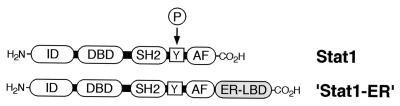
FIG. 1.
Schematic of Stat1 domain structure and Stat1-ER chimera. Domains shown include the interaction domain (ID), which is responsible for interaction with other transcription factors including other STATs, the DNA-binding domain (DBD), the SH2 domain (SH2), which mediates STAT dimerization, the tyrosine that is phosphorylated upon cytokine stimulation (Y), the transcriptional activation function (AF) and, for the chimeric construct, the ER LBD (ER-LBD).
FIG. 2.
(A) Activation of a Stat1-responsive reporter by the Stat1-ER chimera in Stat1-deficient U3A cells. U3A cells were transfected with an empty expression vector (pcDNA3.1) or an expression vector for either Stat1-ER or wtStat1 plus the Stat1-responsive reporter IRF-1x4-tk-luc and a β-Gal control plasmid. After transfection, cells were treated with estrogen (E2; 20 h), tamoxifen (Tam; 20 h) or IFN-γ (5 h), lysed, and assayed for luciferase and β-Gal activities. The normalized responses were determined by dividing relative light units measured in a luciferase assay with the β-Gal activity in the same lysate, and the fold induction (induced/untreated) was calculated by using the averaged normalized responses from three independent experiments. (B) HepG2 cells were transfected with the IRF-1x4-tk-luc reporter and an expression vector for Stat1-ER (circles) or the estrogen-responsive ERE-tk-luc reporter and an expression vector for wild-type ER (diamonds). Both sets included a β-Gal control plasmid. Cells were treated with the indicated concentrations of estrogen (20 h), lysed, and assayed for luciferase and β-Gal activities. The normalized responses are expressed as a percentage of the maximal normalized response for each treatment and are presented as the mean of three independent experiments. (C) Effect of Stat1 Ser727 Ala mutation on transcriptional activity of Stat1-ER and wtStat1 in Stat1-deficient U3A cells. (D) DNA sequence specificity of the Stat1-ER chimera. Stat1-deficient U3A cells were transfected with an expression vector for Stat1-ER and either a Stat1-responsive reporter (IRF-1x4-tk-luc) or a Stat6-selective reporter (mGɛx4-pGL2) together with a β-Gal control plasmid. Treatments and calculations were performed as described for panel A. un, untreated; Tam, tamoxifen treated.
Characterization of the transcriptional activation function of the Stat1-ER chimera.
Having established that the Stat1-ER chimera could activate a Stat1-inducible reporter in an estrogen-dependent fashion, we determined whether the transcriptional activity of the Stat1-ER chimera retained a Stat1-like character or whether it instead was dominated by the heterologous transcriptional activation function (AF-2) contributed by the ER LBD domain (15). This was an important issue to resolve if we expected to use the chimera to drive relevant, STAT-specific biological responses. Estrogen is a full ER agonist and can activate AF-2 in appropriate cellular contexts; however, tamoxifen, though able to induce ER LBD dimerization, has been previously shown to be unable to activate the ER’s AF-2 regardless of cellular context and in contexts where the ER LBD has been attached to heterologous DNA-binding domains (4, 27–31, 34, 63, 68). Therefore, this question was already answered by the experiment shown in Fig. 2A, which showed that tamoxifen is equal or better in efficacy than the full agonist estradiol itself with the Stat1-ER chimera. If the AF-2 were dominant in the context of the chimera, tamoxifen would have been less efficacious than estrogen in the reporter assay. Thus, estrogen (in this cellular context) and tamoxifen serve only to drive dimerization of the Stat1-ER chimera. Because of the importance of this issue, we further analyzed the chimera by making a point mutation of Stat1-Ser727 to Ala, a mutation that has been previously shown to reduce the transcriptional activity of Stat1 by approximately 80% in U3A cells (69). If the ER LBD AF-2 dominated the transcriptional activity of the chimera, we would expect to see little functional effect of this mutation. Instead, as shown in Fig. 2C, the Stat1-Ser727 Ala mutation, in the context of either the Stat1-ER chimera or Stat1 itself, reduces transcriptional activity similarly (70 or 80%, respectively). Thus, as assessed by two different measures, the ER LBD AF-2 does not contribute significantly to the activity of the Stat1-ER chimera, and the chimera, and the chimera retains the transactivation properties of Stat1 itself.
DNA sequence specificity of the Stat1-ER chimera.
For the STAT-ER chimera concept to be useful in examining STAT-driven biological responses, the DNA binding specificity of the cognate STAT protein must be retained. We therefore tested whether the Stat1-ER chimera retained its DNA sequence binding specificity by assessing the ability of the chimera to activate a Stat1-responsive reporter versus its ability to activate the mGɛx4-pGL2 reporter, which is selective for Stat6 (12, 51). In U3A cells, IL-4 strongly activates Stat6 (data not shown). As shown in Fig. 2D, the Stat1-ER chimera is able to activate the IRF-1x4-tk-luc reporter in U3A cells but is unable to activate the mGɛx4-pGL2 reporter. It should be noted that IL-4 can activate the mGɛx4-pGL2 reporter very strongly in these cells, as shown below (see Fig. 5). We further showed that a reporter driven by a 0.7-kb fragment of the natural promoter from the ICAM-1 gene, whose regulation by IFN-γ/Stat1 has been shown to be mediated by a Stat1-binding site (25), is also activated by the Stat1-ER chimera (data not shown). Thus, in two different contexts, the Stat1-ER chimera retains a DNA sequence and promoter specificity characteristic of Stat1.
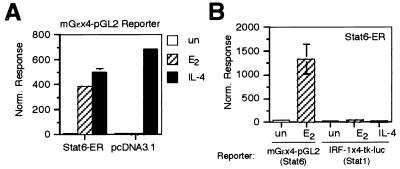
FIG. 5.
(A) Activation of a Stat6-responsive reporter by the Stat6-ER chimera. U3A cells were transfected with the Stat6-selective mGɛx4-tk-luc reporter and either an empty expression vector (pcDNA3.1) or an expression vector for Stat6-ER plus a β-Gal control plasmid. Cells were left untreated or treated with estrogen (E2; 20 h) or IL-4 (5 h), lysed, and assayed for luciferase and β-Gal activities. (B) DNA sequence selectivity of the Stat6-ER chimera. U3A cells were transfected with an expression vector for Stat6-ER and either a Stat1-responsive reporter (IRF-1x4-tk-luc) or a Stat6-selective reporter (mGɛx4-pGL2) together with a β-Gal-expressing control plasmid. After transfection, cells were left untreated or treated with estrogen (E2; 20 h) or IL-4 (5 h), lysed, and assayed for luciferase and β-Gal activities. The normalized responses were calculated by dividing the luciferase value by the β-Gal value for each transfection. The data are presented as the mean of at least three independent experiments.
Biochemical characterization of the Stat1-ER chimera.
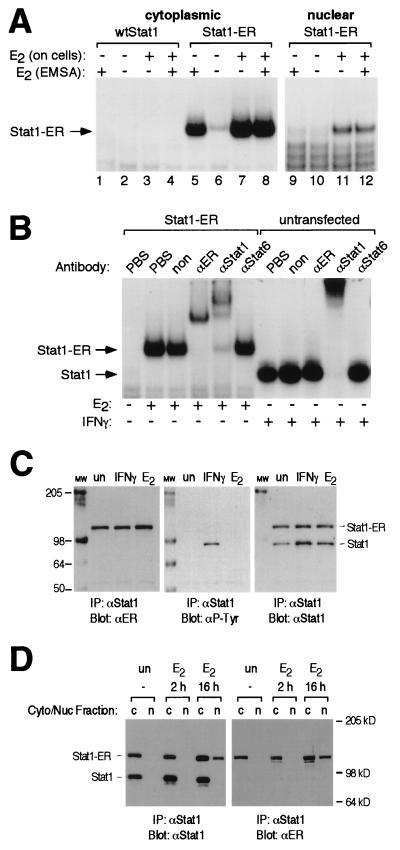
We next sought to determine whether estrogen or tamoxifen could induce nuclear translocation and in vitro DNA binding as a corollary to the reporter assays. We expressed Stat1-ER in Cos7 cells, made cell extracts, and, to assess DNA binding, subjected the extracts to EMSAs using a synthetic STAT-binding DNA element as probe (50). As shown in Fig. 3A (lanes 1 to 4), cytoplasmic extract from cells that were mock transfected produced no shifted complex with or without estrogen, while cytoplasmic extracts from Stat1-ER-transfected cells that were treated with estrogen prior to cell lysis generated a specific complex (lane 7) that was not further enhanced by addition of estrogen to the EMSA reaction (lane 8). Using antibody supershift experiments (Fig. 3B), we determined that the induced complex was made up of the Stat1-ER. Extracts of Stat1-ER-transfected cells that were not pretreated with estrogen (lane 6) produced a faint band that comigrated with the complex in lanes 7 and 8. Interestingly, addition of estrogen directly to the EMSA reaction containing untreated, Stat1-ER-transfected cell extract (lane 5) induced a strong complex that comigrated with the complexes in lanes 7 and 8. Thus, the unliganded Stat1-ER chimera could be induced by estrogen to dimerize and bind DNA in a cell-free, in vitro system.
FIG. 3.
(A) In vitro DNA binding of the Stat1-ER chimera. Cytoplasmic and nuclear extracts were prepared from Cos7 cells that had been either transfected with an expression vector driving the expression of Stat1-ER or left untransfected. Prior to the preparation of extracts, cells had either been left untreated or treated with estrogen (E2; 4 h). Extracts were analyzed by EMSA. In the indicated lanes, 1 μM estrogen (E2) was added directly to the EMSA reaction for 30 min. (B) Identification of shift complexes. Cytoplasmic extracts were prepared from Stat1-ER-transfected or untransfected Cos7 cells that had been treated as indicated. Extracts were incubated with phosphate-buffered saline (PBS), an irrelevant antibody (non), or antibodies specific for ER, Stat1, or Stat6 (αER, αStat1, or αStat6) and subjected to EMSA. The bands corresponding to the Stat1-ER chimera and wtStat1 are indicated. (C) Estrogen does not induce tyrosine phosphorylation of Stat1-ER. Whole-cell extracts from Stat1-ER-transfected Cos7 cells left untreated (un), treated with estrogen (E2; 4 h), or treated with IFN-γ (15 min) were immunoprecipitated (IP) with a Stat1 antibody. After resolution by SDS-PAGE and transfer to nitrocellulose, proteins were detected with the indicated antibodies (αP-Tyr, antiphosphotyrosine). The mobilities of molecular weight markers (lane MW; positions indicated in kilodaltons) and the bands corresponding to the Stat1-ER chimera and wtStat1 are indicated. (D) Estrogen induces nuclear translocation of the Stat1-ER chimera. Cytoplasmic (Cyto; lanes c) and nuclear (Nuc; lanes n) extracts prepared from Stat1-ER-transfected Cos7 cells left untreated (un) or treated with estrogen (E2; 2 h and 16 h) were immunoprecipitated with a Stat1 antibody. After resolution by SDS-PAGE and transfer to nitrocellulose, proteins were detected with the indicated antibodies. The mobilities of molecular weight markers and the bands corresponding to the Stat1-ER chimera and wtStat1 are indicated.
Using the Cos7 cell expression system, we next examined the ability of estrogen to induce nuclear translocation of the chimera. Cos7 nuclear extracts were prepared and subjected to EMSA analysis (Fig. 3A, lanes 9 to 12). Since estrogen treatment of nuclear extracts from untreated cells did not induce Stat1-ER DNA binding in the EMSA (compare lanes 9 and 10 with lanes 5 and 6 in Fig. 3A), we conclude that Stat1-ER did not preexist in the nuclei of untreated cells. Accordingly, Stat1-ER DNA binding activity appeared in the nucleus only after estrogen treatment of cells (lanes 11 and 12). To examine the localization of the Stat1-ER chimera more directly, immunoprecipitation and Western blotting experiments were performed on cytoplasmic and nuclear cell extracts prepared from Cos7 cell transfectants following treatment with estrogen (Fig. 3D). As shown in Fig. 3D, immunoprecipitation with a Stat1 antibody followed by Western blotting with either a Stat1 or an ER LBD antibody demonstrated that the Stat1-ER chimera appears in the nucleus only after estrogen treatment for 16 h. In this experiment, wtStat1 serves as an internal control for the cleanliness of the nuclear fractionation and also as an internal control protein whose localization is not affected by estrogen. Taken together, these data demonstrate that estrogen treatment induces translocation of latent Stat1-ER to the nucleus in much the same manner as cytokine treatment does for wtStat1.
To rule out the possibility that the mechanism by which estrogen induces specific DNA binding by the chimera is through an unanticipated, estrogen-induced tyrosine phosphorylation of Stat1-ER, extracts from Cos7 cells that had been transfected with Stat1-ER expression constructs were examined for tyrosyl-phosphorylated Stat1-ER. Examination of proteins immunoprecipitated with a Stat1-specific antibody and blotted with an antiphosphotyrosine antibody showed that estrogen treatment did not result in the tyrosyl phosphorylation of endogenous Stat1 or the Stat1-ER chimera (Fig. 3C).
Activation of endogenous Stat1-responsive genes by the Stat1-ER chimera.
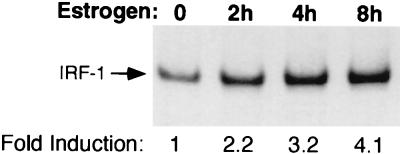
IRF-1 is a prototypic IFN-γ/Stat1-responsive gene that is thought to be involved in the antiproliferative and antiviral effects of IFNs (45, 55, 58). We thus determined whether the estrogen-activated Stat1-ER chimera could transcriptionally induce the endogenous IRF-1 gene. Cos7 cells were transfected with the Stat1-ER construct and were treated with estrogen for 0 to 8 h. As shown in Fig. 4, IRF-1 message levels, as assessed by RT-PCR, were increased roughly fourfold after 8 h. In the absence of transfected Stat1-ER, estrogen had no effect on IRF-1 levels, while IFN-γ induced IRF-1 levels roughly 24-fold (data not shown). The modest fourfold induction of the IRF-1 gene underestimates the true induction on a transfected-cell basis since the transfection efficiency in the experiment was approximately 15%, as judged by β-Gal staining of a parallel transfection. If one corrects for transfection efficiency, the true induction by estrogen/Stat1-ER is roughly 27-fold, which compares favorably with that by IFN-γ. The cIITA gene, a gene responsible for IFN-γ regulation of class II major histocompatibility complex genes that is also known to be IFN-γ/Stat1-regulated (37, 46), was activated in a similar manner by the Stat1-ER chimera after estrogen treatment (data not shown).
FIG. 4.
Induction of the endogenous IRF-1 gene by Stat1-ER in response to estrogen. Cos7 cells were transfected with an expression vector for Stat1-ER and treated for the indicated time with estrogen. Total RNA was prepared and subjected to RT-PCR analysis using specific primers for the IRF-1 gene. The PCR products were analyzed by PAGE, exposed to film (shown), and quantified by PhosphorImager analysis to give the fold inductions indicated (after normalization from parallel analyses of the housekeeping gene GAPDH).
Applicability to other STATs.
The general applicability of using ER LBD fusions to generate conditionally active STATs was tested by using Stat6, which, of the seven STAT proteins, is arguably the least related (only 22% identical) to Stat1 (19). Also, since Stat1 has only 49 amino acids following the tyrosyl residue that becomes phosphorylated, while Stat6 has 149 amino acids subsequent to the analogous residue, a C-terminally fused ER LBD would likely be placed in a quite different three-dimensional location relative to the rest of the protein in Stat6 versus Stat1. For these reasons, we felt that applying the ER LBD fusion method to Stat6 would provide the most stringent test as to whether the method could be applied to other STATs. As shown in Fig. 5A, a Stat6-ER chimera constructed analogously to the Stat1-ER chimera activates the IL-4/Stat6-responsive mGɛx4-pGL2 reporter 44-fold in response to estrogen, almost as strongly as endogenous Stat6 in response to IL-4 itself (58-fold). Furthermore, the Stat6-ER chimera retained the promoter specificity characteristic of wtStat6 and was unable to activate a Stat1-responsive reporter (Fig. 5B). To generalize the concept further, we have recently shown that similarly constructed Stat5A-ER and Stat5B-ER chimeras are also activated by estrogen in an analogous fashion (17).
DISCUSSION
We sought to design a conditionally active STAT that could serve as a powerful tool not only to provide insight into basic questions about STAT function but also to determine the precise biological role of STATs. To this end, we constructed a chimeric STAT protein with the ER LBD, a heterologous, inducible dimerization domain.
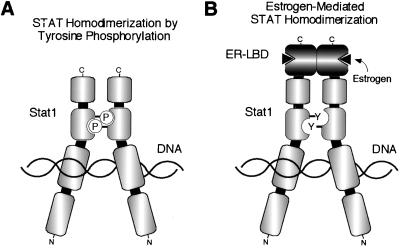
We have demonstrated that fusing the ER LBD with the C terminus of Stat1 results in a chimeric protein that is a novel cytokine-independent, estrogen-regulated transcriptional switch. Conversely, when the ER LBD is localized to the N terminus of Stat1, the chimera has no detectable transcriptional activity, in agreement with the recently published crystal structures of truncated Stat1 and Stat3 homodimers bound to DNA (3, 7), which show that the N termini of the STATs, modeled into the context of the dimer, are located quite far apart from each other, while the C termini are very close to each other (Fig. 6). For wtStat1, DNA binding has previously been shown to require dimerization (54). Accordingly, our data demonstrate that the ER LBD mediates the dimerization of the chimera in response to ER ligands like estrogen and tamoxifen, perhaps best demonstrated by the fact that the unliganded Stat1-ER chimera can be induced to bind a Stat1 DNA binding site in vitro by direct addition of estrogen. The estrogen-activated Stat1-ER chimera retains the DNA binding specificity and transcriptional activation properties of wtStat1 without any significant interference from the ER LBD. Furthermore, we have shown that the Stat1-ER chimera is capable of activating a natural IFN-γ/Stat1-responsive promoter from the ICAM-1 gene as well as endogenous, IFN-γ/Stat1-responsive genes such as IRF-1 and cIITA. These genes are each driven by promoters that have different constellations of required transcription factor binding sites surrounding the STAT-binding sites, and the ability of the Stat1-ER chimera to activate them faithfully is likely due to the fact that the major transcription factor interaction domain of Stat1 at the N terminus (18, 64, 70, 72) is unhindered in the chimera (Fig. 1 and 6). Based on all of these data, one might therefore extrapolate that this reagent will faithfully reproduce the phenotypic consequences of cytokine-activated Stat1. In addition, we have demonstrated the generality of the system by successfully applying it to four of the seven known mammalian STATs (Stat1, Stat5A, Stat5B, and Stat6), suggesting that STAT-ER chimeras will be broadly applicable to the study of STAT biology.
FIG. 6.
(A) Model of the tyrosine-phosphorylated Stat1 homodimer bound to DNA (7). (B) Model depicting the possible dimerization and DNA binding mode of the estrogen-activated Stat1-ER LBD chimera. Separate domains are indicated by the individual segments (Fig. 1). P, phosphorylated tyrosyl residue; Y, free tyrosyl residue.
Importantly, use of STAT-ER chimeras has allowed us to cleanly separate dimerization from tyrosine phosphorylation, and our data provide the first experimental support for the hypothesis that tyrosine phosphorylation serves simply as a dimerization trigger and is not intrinsically responsible for the other aspects of STAT activation and function. Because the ER LBD that we used provides an estrogen-regulated dimerization domain that lacks the key nuclear localization signals that are present in wild-type ER (43, 71), our results indicate that dimerization alone is sufficient to unmask the latent nuclear localization sequence of Stat1 and activate its (i) nuclear translocation, (ii) sequence-specific DNA binding, and (iii) transcriptional activation function. In contrast, it has recently been reported that STAT nuclear translocation is governed by a receptor-triggered pathway that is separate from STAT activation (1); however, our data clearly demonstrate that a second cytokine receptor-mediated pathway is not required for nuclear translocation. Finally, our data argue against an obligate role for the interaction between the phospho group on Tyr701 and the phosphate-binding loop of the STAT SH2 domain in orienting a key STAT DNA-binding helix, which leaves open an interesting structural question (7).
To extend the utility of this system to possible in vivo applications, a way of eliminating the uncontrolled activation of the ER LBD by endogenous estrogen must be found. Capitalizing on the finding that the G521R point mutant of the ER LBD renders the ER LBD insensitive to estrogen but preserves responsiveness to the synthetic ligand tamoxifen (10, 24), we have introduced this mutation into the Stat1-ER and Stat6-ER chimeras and have found these constructs to be insensitive to estrogen while retaining full inducibility by tamoxifen (17). We thus have tools that can be used both in vitro and in vivo to study STAT signaling.
Recently, there was a report of a coumermycin-inducible Stat3 that was generated by constructing a Stat3-gyrase B fusion (41). The Stat3-gyrase B construct was able to activate a Stat3-responsive reporter 2.5-fold in response to coumermycin, and the authors were able to demonstrate partial efficacy in inhibiting IL-10-induced proliferation. Effects of coumermycin on nuclear translocation of the Stat3-gyrase B fusion protein or on endogenous, Stat3-responsive genes were not reported, nor were functional data on gyrase B fusions with other STATs presented. We therefore feel that the STAT-ER design concept has clear utility and promises to provide a more general approach to the study of all STAT proteins. Of particular interest is the suspected role of activated Stat3 and Stat5 in cellular transformation (5, 6, 53, 62, 73), which may be conveniently studied using these methods.
In summary, the STAT-ER chimeras not only represent a novel, inducible method to specifically control gene expression but should prove useful in teasing apart the contribution of STATs to cytokine-induced phenotypes alone or in conjunction with other signaling pathways.
ACKNOWLEDGMENTS
We thank J. Miner and P. Lamb for helpful discussions. We also gratefully acknowledge C. Lowe, E. Delorme, G. Stark, I. Kerr, J. Darnell, J. Ihle, K. Marschke, and E. Allegretto for useful reagents.
ADDENDUM IN PROOF
A recent paper by Kamogawa et al. (J. Immunol. 161:1074–1077, 1998) reported the construction of a Stat6-ER chimera similar to one of the constructs described here. Kamogawa et al. obtained results consistent with ours in that they demonstrated a three- to fourfold induction of a Stat6-responsive luciferase reporter and up-regulation of CD23 surface expression by Stat6-ER in their system in response to tamoxifen.
REFERENCES
- 1.Ali S, Ali S. Prolactin receptor regulates Stat5 tyrosine phosphorylation and nuclear translocation by two separate pathways. J Biol Chem. 1998;273:7709–7716. doi: 10.1074/jbc.273.13.7709. [DOI] [PubMed] [Google Scholar]
- 2.Bach E A, Aguet M, Schreiber R D. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 3.Becker S, Groner B, Muller C W. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 4.Berry M, Metzger D, Chambon P. Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J. 1990;9:2811–2818. doi: 10.1002/j.1460-2075.1990.tb07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bromberg J F, Horvath C M, Besser D, Lathem W W, Darnell J E., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlesso N, Frank D A, Griffin J D. Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl. J Exp Med. 1996;183:811–820. doi: 10.1084/jem.183.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell J E, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 8.Chin H, Nakamura N, Kamiyama R, Miyasaka N, Ihle J N, Miura O. Physical and functional interactions between Stat5 and the tyrosine-phosphorylated receptors for erythropoietin and interleukin-3. Blood. 1996;88:4415–4425. [PubMed] [Google Scholar]
- 9.Chretien S, Varlet P, Verdier F, Gobert S, Cartron J P, Gisselbrecht S, Mayeux P, Lacombe C. Erythropoietin-induced erythroid differentiation of the human erythroleukemia cell line TF-1 correlates with impaired STAT5 activation. EMBO J. 1996;15:4174–4181. [PMC free article] [PubMed] [Google Scholar]
- 10.Danielian P S, White R, Hoare S A, Fawell S E, Parker M G. Identification of residues in the estrogen receptor that confer differential sensitivity to estrogen and hydroxytamoxifen. Mol Endocrinol. 1993;7:232–240. doi: 10.1210/mend.7.2.8469236. [DOI] [PubMed] [Google Scholar]
- 11.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 12.Delphin S, Stavnezer J. Characterization of an interleukin 4 (IL-4) responsive region in the immunoglobulin heavy chain germline epsilon promoter: regulation by NF- IL-4, a C/EBP family member and NF-kappa B/p50. J Exp Med. 1995;181:181–192. doi: 10.1084/jem.181.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drachman J G, Kaushansky K. Dissecting the thrombopoietin receptor: functional elements of the Mpl cytoplasmic domain. Proc Natl Acad Sci USA. 1997;94:2350–2355. doi: 10.1073/pnas.94.6.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gobert S, Porteu F, Pallu S, Muller O, Sabbah M, Dusanter-Fourt I, Courtois G, Lacombe C, Gisselbrecht S, Mayeux P. Tyrosine phosphorylation of the erythropoietin receptor: role for differentiation and mitogenic signal transduction. Blood. 1995;86:598–606. [PubMed] [Google Scholar]
- 15.Gronemeyer H. Transcription activation by estrogen and progesterone receptors. Annu Rev Genet. 1991;25:89–123. doi: 10.1146/annurev.ge.25.120191.000513. [DOI] [PubMed] [Google Scholar]
- 16.Gurney A L, Wong S C, Henzel W J, de Sauvage F J. Distinct regions of c-Mpl cytoplasmic domain are coupled to the JAK- STAT signal transduction pathway and Shc phosphorylation. Proc Natl Acad Sci USA. 1995;92:5292–5296. doi: 10.1073/pnas.92.12.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haslam, J. A., L. H. Milocco, and H. M. Seidel. Unpublished data.
- 18.Horvath C M, Stark G R, Kerr I M, Darnell J E., Jr Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol Cell Biol. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou J, Schindler U, Henzel W J, Ho T C, Brasseur M, McKnight S L. An interleukin-4-induced transcription factor: IL-4 Stat. Science. 1994;265:1701–1706. doi: 10.1126/science.8085155. [DOI] [PubMed] [Google Scholar]
- 20.Kaptein A, Paillard V, Saunders M. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. J Biol Chem. 1996;271:5961–5964. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- 21.Kishimoto T, Taga T, Akira S. Cytokine signal transduction. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 22.Kumar A, Commane M, Flickinger T W, Horvath C M, Stark G R. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 23.Kumar V, Green S, Stack G, Berry M, Jin J R, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- 24.Littlewood T D, Hancock D C, Danielian P S, Parker M G, Evan G I. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Look D C, Pelletier M R, Holtzman M J. Selective interaction of a subset of interferon-gamma response element- binding proteins with the intercellular adhesion molecule-1 (ICAM-1) gene promoter controls the pattern of expression on epithelial cells. J Biol Chem. 1994;269:8952–8958. [PubMed] [Google Scholar]
- 26.Mattioni T, Louvion J F, Picard D. Regulation of protein activities by fusion to steroid binding domains. Methods Cell Biol. 1994;43:335–352. doi: 10.1016/s0091-679x(08)60611-1. [DOI] [PubMed] [Google Scholar]
- 27.McDonnell D P, Clemm D L, Hermann T, Goldman M E, Pike J W. Analysis of estrogen receptor function in vitro reveals three distinct classes of antiestrogens. Mol Endocrinol. 1995;9:659–669. doi: 10.1210/mend.9.6.8592512. [DOI] [PubMed] [Google Scholar]
- 28.McInerney E M, Katzenellenbogen B S. Different regions in activation function-1 of the human estrogen receptor required for antiestrogen- and estradiol-dependent transcription activation. J Biol Chem. 1996;271:24172–24178. doi: 10.1074/jbc.271.39.24172. [DOI] [PubMed] [Google Scholar]
- 29.McInerney E M, Weis K E, Sun J, Mosselman S, Katzenellenbogen B S. Transcription activation by the human estrogen receptor subtype beta (ER beta) studied with ER beta and ER alpha receptor chimeras. Endocrinology. 1998;139:4513–4522. doi: 10.1210/endo.139.11.6298. [DOI] [PubMed] [Google Scholar]
- 30.Metzger D, Ali S, Bornert J M, Chambon P. Characterization of the amino-terminal transcriptional activation function of the human estrogen receptor in animal and yeast cells. J Biol Chem. 1995;270:9535–9542. doi: 10.1074/jbc.270.16.9535. [DOI] [PubMed] [Google Scholar]
- 31.Metzger D, Losson R, Bornert J M, Lemoine Y, Chambon P. Promoter specificity of the two transcriptional activation functions of the human oestrogen receptor in yeast. Nucleic Acids Res. 1992;20:2813–2817. doi: 10.1093/nar/20.11.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, Akira S. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci USA. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miura Y, Miura O, Ihle J N, Aoki N. Activation of the mitogen-activated protein kinase pathway by the erythropoietin receptor. J Biol Chem. 1994;269:29962–29969. [PubMed] [Google Scholar]
- 34.Montano M M, Muller V, Trobaugh A, Katzenellenbogen B S. The carboxy-terminal F domain of the human estrogen receptor: role in the transcriptional activity of the receptor and the effectiveness of antiestrogens as estrogen antagonists. Mol Endocrinol. 1995;9:814–825. doi: 10.1210/mend.9.7.7476965. [DOI] [PubMed] [Google Scholar]
- 35.Moriggl R, Gouilleux-Gruart V, Jahne R, Berchtold S, Gartmann C, Liu X, Hennighausen L, Sotiropoulos A, Groner B, Gouilleux F. Deletion of the carboxyl-terminal transactivation domain of MGF-Stat5 results in sustained DNA binding and a dominant negative phenotype. Mol Cell Biol. 1996;16:5691–5700. doi: 10.1128/mcb.16.10.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morita H, Tahara T, Matsumoto A, Kato T, Miyazaki H, Ohashi H. Functional analysis of the cytoplasmic domain of the human Mpl receptor for tyrosine-phosphorylation of the signaling molecules, proliferation and differentiation. FEBS Lett. 1996;395:228–234. doi: 10.1016/0014-5793(96)01047-2. [DOI] [PubMed] [Google Scholar]
- 37.Muhlethaler-Mottet A, Di Berardino W, Otten L A, Mach B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- 38.Muli A L, Wakao H, Kinoshita T, Kitamura T, Miyajima A. Suppression of interleukin-3-induced gene expression by a C-terminal truncated Stat5: role of Stat5 in proliferation. EMBO J. 1996;15:2425–2433. [PMC free article] [PubMed] [Google Scholar]
- 39.Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell J E, Jr, Stark G R, Kerr I M. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 41.O’Farrell A M, Liu Y, Moore K W, Mui A L. IL-10 inhibits macrophage activation and proliferation by distinct signaling mechanisms: evidence for Stat3-dependent and -independent pathways. EMBO J. 1998;17:1006–1018. doi: 10.1093/emboj/17.4.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pernis A, Witthuhn B, Keegan A D, Nelms K, Garfein E, Ihle J N, Paul W E, Pierce J H, Rothman P. Interleukin 4 signals through two related pathways. Proc Natl Acad Sci USA. 1995;92:7971–7975. doi: 10.1073/pnas.92.17.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Picard D, Kumar V, Chambon P, Yamamoto K R. Signal transduction by steroid hormones: nuclear localization is differentially regulated in estrogen and glucocorticoid receptors. Cell Regul. 1990;1:291–299. doi: 10.1091/mbc.1.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Picard D, Salser S J, Yamamoto K R. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell. 1988;54:1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- 45.Pine R, Canova A, Schindler C. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF- 1 promoter to mediate induction by IFN alpha and IFN gamma, and is likely to autoregulate the p91 gene. EMBO J. 1994;13:158–167. doi: 10.1002/j.1460-2075.1994.tb06245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piskurich J F, Wang Y, Linhoff M W, White L C, Ting J P. Identification of distinct regions of 5′ flanking DNA that mediate constitutive, IFN-gamma, STAT1, and TGF-beta-regulated expression of the class II transactivator gene. J Immunol. 1998;160:233–240. [PubMed] [Google Scholar]
- 47.Porteu F, Rouyez M C, Cocault L, Benit L, Charon M, Picard F, Gisselbrecht S, Souyri M, Dusanter-Fourt I. Functional regions of the mouse thrombopoietin receptor cytoplasmic domain: evidence for a critical region which is involved in differentiation and can be complemented by erythropoietin. Mol Cell Biol. 1996;16:2473–2482. doi: 10.1128/mcb.16.5.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quelle F W, Shimoda K, Thierfelder W, Fischer C, Kim A, Ruben S M, Cleveland J L, Pierce J H, Keegan A D, Nelms K, Paul W E, Ihle J N. Cloning of murine Stat6 and human Stat6, Stat proteins that are tyrosine phosphorylated in responses to IL-4 and IL-3 but are not required for mitogenesis. Mol Cell Biol. 1995;15:3336–3343. doi: 10.1128/mcb.15.6.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quelle F W, Wang D, Nosaka T, Thierfelder W E, Stravopodis D, Weinstein Y, Ihle J N. Erythropoietin induces activation of Stat5 through association with specific tyrosines on the receptor that are not required for a mitogenic response. Mol Cell Biol. 1996;16:1622–1631. doi: 10.1128/mcb.16.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raz R, Durbin J E, Levy D E. Acute phase response factor and additional members of the interferon-stimulated gene factor 3 family integrate diverse signals from cytokines, interferons, and growth factors. J Biol Chem. 1994;269:24391–24395. [PubMed] [Google Scholar]
- 51.Seidel H M, Milocco L H, Lamb P, Darnell J E, Jr, Stein R B, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seldin D C, Leder P. Mutational analysis of a critical signaling domain of the human interleukin 4 receptor. Proc Natl Acad Sci USA. 1994;91:2140–2144. doi: 10.1073/pnas.91.6.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shuai K, Halpern J, ten Hoeve J, Rao X, Sawyers C L. Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia. Oncogene. 1996;13:247–254. [PubMed] [Google Scholar]
- 54.Shuai K, Horvath C M, Huang L H, Qureshi S A, Cowburn D, Darnell J E., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 55.Sims S H, Cha Y, Romine M F, Gao P Q, Gottlieb K, Deisseroth A B. A novel interferon-inducible domain: structural and functional analysis of the human interferon regulatory factor 1 gene promoter. Mol Cell Biol. 1993;13:690–702. doi: 10.1128/mcb.13.1.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 57.Takatoku M, Kametaka M, Shimizu R, Miura Y, Komatsu N. Identification of functional domains of the human thrombopoietin receptor required for growth and differentiation of megakaryocytic cells. J Biol Chem. 1997;272:7259–7263. doi: 10.1074/jbc.272.11.7259. [DOI] [PubMed] [Google Scholar]
- 58.Taniguchi T, Lamphier M S, Tanaka N. IRF-1: the transcription factor linking the interferon response and oncogenesis. Biochim Biophys Acta. 1997;1333:M9–M17. doi: 10.1016/s0304-419x(97)00014-0. [DOI] [PubMed] [Google Scholar]
- 59.Teglund S, McKay C, Schuetz E, van Deursen J M, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle J N. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 60.Tian S S, Lamb P, Seidel H M, Stein R B, Rosen J. Rapid activation of the STAT3 transcription factor by granulocyte colony-stimulating factor. Blood. 1994;84:1760–1764. [PubMed] [Google Scholar]
- 61.Tian S S, Tapley P, Sincich C, Stein R B, Rosen J, Lamb P. Multiple signaling pathways induced by granulocyte colony-stimulating factor involving activation of JAKs, STAT5, and/or STAT3 are required for regulation of three distinct classes of immediate early genes. Blood. 1996;88:4435–4444. [PubMed] [Google Scholar]
- 62.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot R P, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tzukerman M T, Esty A, Santiso-Mere D, Danielian P, Parker M G, Stein R B, Pike J W, McDonnell D P. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- 64.Vinkemeier U, Cohen S L, Moarefi I, Chait B T, Kuriyan J, Darnell J E., Jr DNA binding in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 65.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle J N. Naturally occurring dominant negative variants of Stat5. Mol Cell Biol. 1996;16:6141–6148. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H, Peters G A, Zeng X, Tang M, Ip W, Khan S A. Yeast two-hybrid system demonstrates that estrogen receptor dimerization is ligand-dependent in vivo. J Biol Chem. 1995;270:23322–23329. doi: 10.1074/jbc.270.40.23322. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y D, Wong K, Wood W I. Intracellular tyrosine residues of the human growth hormone receptor are not required for the signaling of proliferation or Jak-STAT activation. J Biol Chem. 1995;270:7021–7024. doi: 10.1074/jbc.270.13.7021. [DOI] [PubMed] [Google Scholar]
- 68.Webster N J, Green S, Jin J R, Chambon P. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell. 1988;54:199–207. doi: 10.1016/0092-8674(88)90552-1. [DOI] [PubMed] [Google Scholar]
- 69.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 70.Xu X, Sun Y L, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 71.Ylikomi T, Bocquel M T, Berry M, Gronemeyer H, Chambon P. Cooperation of proto-signals for nuclear accumulation of estrogen and progesterone receptors. EMBO J. 1992;11:3681–3694. doi: 10.1002/j.1460-2075.1992.tb05453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Q, Nowak I, Vonderheid E C, Rook A H, Kadin M E, Nowell P C, Shaw L M, Wasik M A. Activation of Jak/STAT proteins involved in signal transduction pathway mediated by receptor for interleukin 2 in malignant T lymphocytes derived from cutaneous anaplastic large T-cell lymphoma and Sezary syndrome. Proc Natl Acad Sci USA. 1996;93:9148–9153. doi: 10.1073/pnas.93.17.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]