Abstract
Membrane transport is a fundamental means to control basic cellular processes such as apoptosis, inflammation, and neurodegeneration and is mediated by a number of transporters, pumps, and channels. Accumulating evidence over the last half century has shown that a type of so-called “large-pore channel” exists in various tissues and organs in gap-junctional and non-gap-junctional forms in order to flow not only ions but also metabolites such as ATP. They are formed by a number of protein families with little or no evolutionary linkages including connexin, innexin, pannexin, leucine-rich repeat-containing 8 (LRRC8), and calcium homeostasis modulator (CALHM). This review summarizes the history and concept of large-pore channels starting from connexin gap junction channels to the more recent developments in innexin, pannexin, LRRC8, and CALHM. We describe structural and functional features of large-pore channels that are crucial for their diverse functions on the basis of available structures.
Keywords: Large-pore channel, gap junction, hemichannel, connexin, innexin, pannexin, CALHM, LRRC8, SWELL1, ATP, metabolites, osmolytes, neurotransmitters, voltage-gating, x-ray crystallography, cryo-EM, electrophysiology
Graphical Abstract

Large-pore channels in early days
Connexin gap junction channel and hemichannel
While the majority of channels on the cell surface are designed to selectively permeate ions such as potassium, sodium, and chloride to underpin cellular homeostasis, there are also channels that are permeable to larger molecules with greater pore diameters than the selective ion channels. The first example is the discovery that connexin gap junction channels are permeable to not only ions but also larger molecules such as ATP, demonstrating that they are effective molecular machines for exchanging metabolites between bridged cells [1–3]. Gap junction channels mediate intercellular communication between cells and thereby make tissues a functional syncytium. Interestingly, later studies suggested the presence of connexin channels which do not exist as gap junctions but as a hemichannel in Xenopus oocytes heterologously expressing Cx43 [4] or catfish retina [5]. Other studies showed that some connexin hemichannels can release ATP from cells and that the released ATP molecules bind purinergic receptors to induce calcium transients in adjacent cells [6, 7]. This cell-to-cell spread of a calcium transient, deemed a calcium wave, was originally discovered to involve a gap junction mechanism [8]. However, the above studies showed that it can also occur in non-junctional cells via ATP release by the connexin hemichannel and the subsequent purinergic signaling. While the existence of large-pore connexin channels as gap junctions has been well accepted for a long time, the concept of connexin hemichannels was substantially slower to develop until the field reached a consensus that opening of connexin hemi-channels occurs by lowering of extracellular Ca2+ and membrane depolarization [9–11]. That is, the extracellularly exposed large-pore connexin channel can control channel opening and closing within the physiological concentrations of extracellular Ca2+ to prevent uncontrolled leakage of ions and metabolites such as ATP from the cytoplasm.
There are more than twenty connexin isoforms identified to date [12]. These different connexin isoforms can associate in many different combinations to give rise to diverse properties such as gating and permeability [13]. They can form a homotypic gap junction (assembly of two identical hemichannels) or a heterotopic gap junction (assembly of different hemichannels). Furthermore, the hemichannels can form heteromeric assembly containing different connexin isoforms (Fig. 1a). Of the connexin isoforms identified so far, Cx26, Cx32, Cx43, Cx46, and Cx50 have been shown to exist as hemichannels that efflux ATP when they are heterologously expressed in cell cultures [6, 14, 15]. These studies further confirmed the formation of connexin-based ATP permeating large-pore channels on cell surfaces. Ion flux of some of these channels have been shown to be regulated by voltage, pH, and extracellular Ca2+ [9–11, 16]. Later studies showed that voltage-sensing is mainly mediated by the amino terminal (N-terminal) region which faces the channel pore [17–20]. The calcium sensing motif has been delineated to be in the extracellular domain [11, 21] as well as in the pore [22, 23]. In this review, we define large-pore channel as those shown to be permeable not only to small ions (e.g. Na+, Ca2+, K+, Cl−, etc.) but also larger molecules such as ATP with an estimated pore size larger than ~14 Å [2]. This is in stark contrast to sodium, calcium and potassium selective channels with the pore diameters of 3.5, 6, and 3 Å, respectively [24–26].
Fig. 1. Connexin gap junction channels.
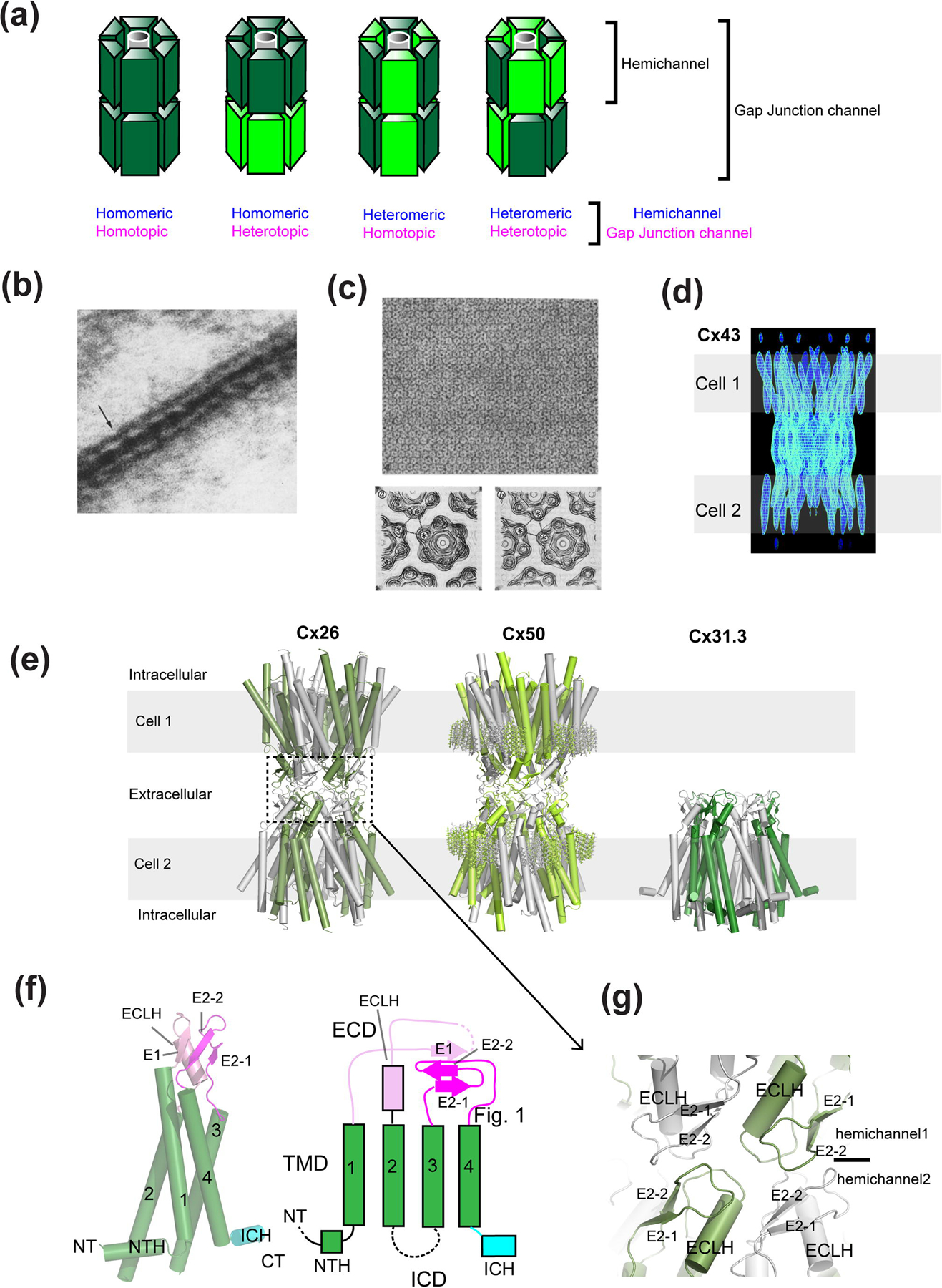
(a) Various types of connexin hemichannels can assemble to form distinct gap junction channels. Connexin can form homo- or hetero-hexameric hemichannels where they form homotypic or heterotypic gap junctions. Note that the pattern of heteromeric and heterotopic protomer arrangements are arbitrary since they have not been determined experimentally. (b) Electron microscopic observation of the gap junction in Mauthner cell synapses. (c) Isolated gap junction sheets in a 2D crystalline array viewed from the ‘top’ (top panel). Maps of the gap junction channels from image analysis in the presence (left) and absence (right) of Ca2+. (d) Maps calculated from cryo-electron crystallography showing rod-like structures representing transmembrane helices. (e) X-ray crystallographic structure of Cx26 (left, PDB code: 2ZW3) and cryo-EM structures of Cx50 (middle, PDB code: 7JJP) and Cx31.3 hemichannel (right, PDB code: 6L3T). Sticks in the Cx50 structure represent resolved lipid molecules at the extracellular leaflet of the plausible lipid bilayer. Gray belts represent the approximate location of the lipid bilayers. Note that the Cx50 model was built from the averaged cryo-EM map of native Cx46/50. (f) A monomer of Cx31.1 and the schematic presentation of the structural topology. Note that in Cx26 and Cx50, the beta strand in the TM1-2 loop and ICH are missing or not resolved. (g) The gap junction formation occurs through interactions of the extracellular domain (ECD). Shown here is the interface of the Cx26 gap junction channel. The images in panel b, c, and d have been adapted from the article by Robertson [30], Unwin and Zampighi [35], Unwin and Ennis [36], and Unger et al. [37] Copyright was obtained from the journals, J. Cell Biology, Nature, and Science.
Connexin large-pore channels viewed from cellular and structural biological perspectives
Gap junctions were discovered in myocardium and nerve by electrophysiology detecting the fast electrical transmission properties between adjacent cells [27–29]. From the cellular/structural biological perspective, electron microscopy revealed the hexagonally shaped ultrastructure at the club ending of Mauthner cell synapses from goldfish brains [30] (Fig. 1b). Since then, the prototypical large-pore channels formed by proteins, which we know as connexins, have been visualized and measured by a series of structural biological efforts initially by electron microscopic and low angle X-ray diffraction analyses of native tissues, which form natural crystal arrays [31–34]. Gap junctions have been effective specimens for structural biology because of their unique ultrastructure that tethers cells and their ability to form hexagonal paracrystalline arrays on the membranes or two-dimensional (2D) crystals. More extensive analysis of electron micrographs collected from negative-stained 2D crystals of gap junctions at multiple angles allowed observation of two hexagonal units forming gap junctions in the 1980s [35, 36] (Fig. 1c).
The major breakthrough in 1999 was the first clear observation of some secondary structural elements of the Cx43 gap junction channel using cryoelectron diffraction from vitrified 2D crystals [37]. The electron diffraction reached resolutions of 7.5 Å and 21 Å in the membrane plane and vertical directions, respectively. With this accomplishment, the field further accepted the view that the connexin gap junction channels are formed by tethering of two hexameric hemichannels at the extracellular region of the opposing channels. Importantly, this electron crystallographic structure observed 24 rods of density per hemichannel (48 per gap junction channel) representing transmembrane helices, thereby confirming that each connexin subunit contains four transmembrane domains (Fig. 1d). At this point, issues with sample heterogeneity were overcome by the establishment of the stably transfected BHK cell-line of CX43 [38]. Remarkably, the membrane fraction already contained small 2D crystals. The size and crystal packing of these crystals were further improved by extraction of an enriched membrane fraction by a mild treatment with Tween20. In a later study on the Cx26 mutant, Met34Ala, proteins were expressed by the baculovirus insect cell system followed by detergent extraction, affinity purification, and reconstitution into liposomes for 2D crystallization [39]. The subsequent cryoelectron crystallography observed some extra density in the middle of the hexameric pore, which appeared to plug the channel [39]. The amino terminal sequences that precede the first predicted transmembrane helix in the Cx32/Cx26 gap junction channel were previously shown to control voltage sensitivity [18]; thus, the authors predicted the amino-terminal motif to be a part of the ‘plug’-like density. Also importantly, the above studies showed that the secondary and tertiary structures were similar between Cx26 and Cx43 indicating the structural conservation within the connexin family.
The major advance in 2009 is represented by the 3.5 Å structure of Cx26 gap junction channel by X-ray crystallography where the quality of experimental density was sufficiently high to model most of the amino acids except for those in the cytoplasmic loop and the carboxyl terminal residues, allowing the field to assess the molecular basis of gap junction functions in substantially finer detail [40] (Fig. 1e). A number of high-resolution structures of connexins by X-ray crystallography and single-particle electron-cryomicroscopy (cryo-EM) are available today (Fig. 1e, Table 1). The crystal structure of Cx26 gap junction channel was assumed to be in the ‘open’ conformation based on the fact that the ‘plug’-like density observed in the previous cryoelectron crystallographic study [39] was not present. This structural study observed and confirmed the presence of four transmembrane helices per subunit and also an extra helix at the N-terminal region looping back toward the pore from the cytoplasmic side (Fig. 1f). In most of the structures available to date, the intracellular domain (ICD) is not well resolved. The crystal structure also revealed the specific modes of interactions of the extracellular loops between opposing Cx26 hemichannels; thereby, showing the mechanism of gap junction formation at the molecular level for the first time (Fig. 1g). Importantly, this crystal structure clearly showed that the size of the narrowest region of the pore is almost wide enough (~13.5 Å) to permit ATP permeation (Fig. 2a). In other words, it would take only a minor protein movement to open the channel pore sufficiently wide (~ 14 Å) for permeation of ATP and other molecules up to 1,000 Daltons [2]. Overall, the crystal structure solidified the view that connexin channels are permeable to large metabolites. A later X-ray crystallographic study on Cx26 in the presence of calcium showed that there is no robust structural change between the calcium-bound and calcium-free forms but rather the differences are in the side chain orientations of acidic residues in the extracellular regions (one binding site per subunit) (Fig. 2b) [41]. Molecular dynamics simulations demonstrated that calcium binding results in a positively charged electrostatic barrier that interferes with cations or disrupt electrostatic networks of charged residues at the extracellular loops and that the physical gate may located deeper in the pore [21, 41]. Therefore, the Ca2+ mediated gating is controlled by electrostatic but not steric factors. And more recently, by implementing single-particle cryo-EM, it was demonstrated that Cx26 gap junction channels at low pH (pH 6.4) can open and close physically by dynamic movement of the N-terminal region (Fig. 2c) [42]. Therefore, the series of structural studies showed that channel inhibition by Ca2+ and protons occurs through distinct mechanisms.
Table 1.
Available structures of Connexin in 2021
| Structure | Functional state* | PDB code | Species | Oligomeric state | Solubilization conditions | Freezing buffer conditions | Reference |
|---|---|---|---|---|---|---|---|
| Cx46 (Cx46/50) 3.4 Å | Open | 6MHQ | Ovis aries | 6; gap junction 12-mer | TBS + 2% DDM | Amphipol A8-35; 20 mM HEPES pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA | Myers et al., 2018 |
| Cx50 (Cx46/50) 3.4 Å | Open | 6MHY | Ovis aries | 6; gap junction 12-mer | TBS + 2% DDM | Amphipol A8-35; 20 mM HEPES pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA | Myers et al., 2018 |
| Cx31.3 2.63 Å | Open | 6L3V | Homo sapiens | 6 | 20 mM CAPS pH 10.5, 250 mM NaCl, 2 mM βME, 1 mM EDTA, 10% glycerol, 0.4% LMNG | 20 mM HEPES pH 7.5, 250 mM NaCl, 2 mM βME, 0.004% LMNG | Lee et al., 2020 |
| Cx31.3 + Ca2+ 2.53 Å | Open | 6L3U | Homo sapiens | 6 | 20 mM CAPS pH 10.5, 250 mM NaCl, 2 mM βME, 1 mM EDTA, 10% glycerol, 0.4% LMNG | 20 mM HEPES pH 7.5, 250 mM NaCl, 2 mM βME, 0.004% LMNG, 50 mM Ca2+ | Lee et al., 2020 |
| Cx31.3 R15G mutant 2.34 Å | Open | 6L3T | Homo sapiens | 6 | 20 mM CAPS pH 10.5, 250 mM NaCl, 2 mM βME, 1 mM EDTA, 10% glycerol, 0.4% LMNG | 20 mM HEPES pH 7.5, 250 mM NaCl, 2 mM βME, 0.004% LMNG | Lee et al., 2020 |
| Cx46 (lipid class 3) 2.5 Å | Open | 7JN1 | Ovis aries | 6; gap junction 12-mer | 10 mM Tris, 2 mM EDTA, 2 mM EGTA, 1% DM | MSP1E1 + DMPC lipid nanodiscs; 20 mM HEPES pH 7.4, 150 mM NaCl | Flores et al, 2020 |
| Cx46 (lipid class 2) 2.5 Å | Open | 7JN0 | Ovis aries | 6; gap junction 12-mer | 10 mM Tris, 2 mM EDTA, 2 mM EGTA, 1% DM | MSP1E1 + DMPC lipid nanodiscs; 20 mM HEPES pH 7.4, 150 mM NaCl | Flores et al, 2020 |
| Cx46 (lipid class 1) 2.5 Å | Open | 7JMD | Ovis aries | 6; gap junction 12-mer | 10 mM Tris, 2 mM EDTA, 2 mM EGTA, 1% DM | MSP1E1 + DMPC lipid nanodiscs; 20 mM HEPES pH 7.4, 150 mM NaCl | Flores et al, 2020 |
| Cx50 1.94 Å | Open | 7JJP | Ovis aries | 6; gap junction 12-mer | 10 mM Tris, 2 mM EDTA, 2 mM EGTA, 1% DM | MSP1E1 + DMPC lipid nanodiscs; 20 mM HEPES pH 7.4, 150 mM NaCl | Flores et al, 2020 |
| Cx46 1.9 Å | Open | 7JKC | Ovis aries | 6; gap junction 12-mer | 10 mM Tris, 2 mM EDTA, 2 mM EGTA, 1% DM | MSP1E1 + DMPC lipid nanodiscs; 20 mM HEPES pH 7.4, 150 mM NaCl | Flores et al, 2020 |
| Cx50 (lipid class 1) 2.5 Å | Open | 7JLW | Ovis aries | 6; gap junction 12-mer | 10 mM Tris, 2 mM EDTA, 2 mM EGTA, 1% DM | MSP1E1 + DMPC lipid nanodiscs; 20 mM HEPES pH 7.4, 150 mM NaCl | Flores et al, 2020 |
| Cx50 (lipid class 2) 2.5 Å | Open | 7JM9 | Ovis aries | 6; gap junction 12-mer | 10 mM Tris, 2 mM EDTA, 2 mM EGTA, 1% DM | MSP1E1 + DMPC lipid nanodiscs; 20 mM HEPES pH 7.4, 150 mM NaCl | Flores et al, 2020 |
| Cx50 (lipid class 3) 2.5 Å | Open | 7JMC | Ovis aries | 6; gap junction 12-mer | 10 mM Tris, 2 mM EDTA, 2 mM EGTA, 1% DM | MSP1E1 + DMPC lipid nanodiscs; 20 mM HEPES pH 7.4, 150 mM NaCl | Flores et al, 2020 |
| Cx26 3.5 Å | Open | 2ZW3 | Homo sapiens | 6; gap junction 12-mer | 10 mM CAPS pH 10.5, 1 M NaCl, 10 mM DTT, 1–1.5% DDM | Final SEC buffer: 10 mM HEPES pH 7.5, 200 mM NaCl, 2 mM DTT, 0.01% UDM | Maeda et al, 2009 |
| Cx26 3.8 Å | Open | 5ERA | Homo sapiens | 6; gap junction 12-mer | 50 mM HEPES pH 7.5, 300 mM NaCl, 10 mM imidazole, 2% glycerol, 1% DM | Final SEC buffer: 100 mM HEPES pH 7, 1 M NaCl, 2.5% glycerol, 0.02% FA-3 | Bennett et al, 2016 |
| Cx26 + Ca2+ 3.28 Å | Closed | 5ER7 | Homo sapiens | 6; gap junction 12-mer | 50 mM HEPES pH 7.5, 300 mM NaCl, 10 mM imidazole, 2% glycerol, 1% DM | Final SEC buffer: 100 mM HEPES pH 7, 1 M NaCl, 2.5% glycerol, 0.02% FA-3 | Bennett et al, 2016 |
| Cx26 (neutral pH) 4 Å | Open | 6UVR | Homo sapiens | 6; gap junction 12-mer | 50 mM HEPES pH 7.5, 300 mM NaCl, 10 mM imidazole, 2% glycerol, 1% DM | Amphipol A8-35; 50 mM HEPES pH 7.5, 300 mM NaCl | Khan et al, 2020 |
| Cx26 (low pH) 7.5 Å | Closed | 6UVT | Homo sapiens | 6; gap junction 12-mer | 50 mM HEPES pH 7.5, 300 mM NaCl, 10 mM imidazole, 2% glycerol, 1% DM | Amphipol A8-35; 12.5 mM HEPES, 37.5 mM MES pH 6.4, 125 mM NaCl | Khan et al, 2020 |
| Cx26 (low pH) 4.2 Å | Open | 6UVS | Homo sapiens | 6; gap junction 12-mer | 50 mM HEPES pH 7.5, 300 mM NaCl, 10 mM imidazole, 2% glycerol, 1% DM | Amphipol A8-35; 12.5 mM HEPES, 37.5 mM MES pH 6.4, 125 mM NaCl | Khan et al, 2020 |
| Cx26 M34A 6 Å | Closed | 3IZ1 | Homo sapiens | 6; gap junction 12-mer | 1 M NaCl, 10 mM HEPES pH 7.5, 2% DDM, 0.005% NaN3 | 2D crystallography in DM/POPC, 10 mM MES pH 5.8, 100 mM NaCl, 50 mM MgCl2, 5 mM CaCl2, 2 mM DTT, 0.1 mM CBX, 0.005% NaN3, 1% glycerol | Oshima et al, 2011 |
| Cx26 M34A Del2-7 10 Å | Closed | 3IZ2 | Homo sapiens | 6; gap junction 12-mer | 1 M NaCl, 10 mM HEPES pH 7.5, 2% DDM, 0.005% NaN3 | 2D crystallography in DM/POPC, 10 mM MES pH 5.8, 100 mM NaCl, 50 mM MgCl2, 5 mM CaCl2, 2 mM DTT, 0.1 mM CBX, 0.005% NaN3, 1% glycerol | Oshima et al, 2011 |
| Cx26 M34A 10 Å | n/a | n/a | Homo sapiens | 6; gap junction 12-mer | 1 M NaCl, 10 mM HEPES pH 7.5, 2% DDM, 0.005% NaN3 | 2D crystallography in DM/POPC, 10 mM MES pH 5.8, 100 mM NaCl, 50 mM MgCl2, 5 mM CaCl2, 2 mM DTT, 0.1 mM CBX, 0.005% NaN3, 1% glycerol | Oshima et al, 2007 |
Fig. 2. Pore sizes of connexin channels.
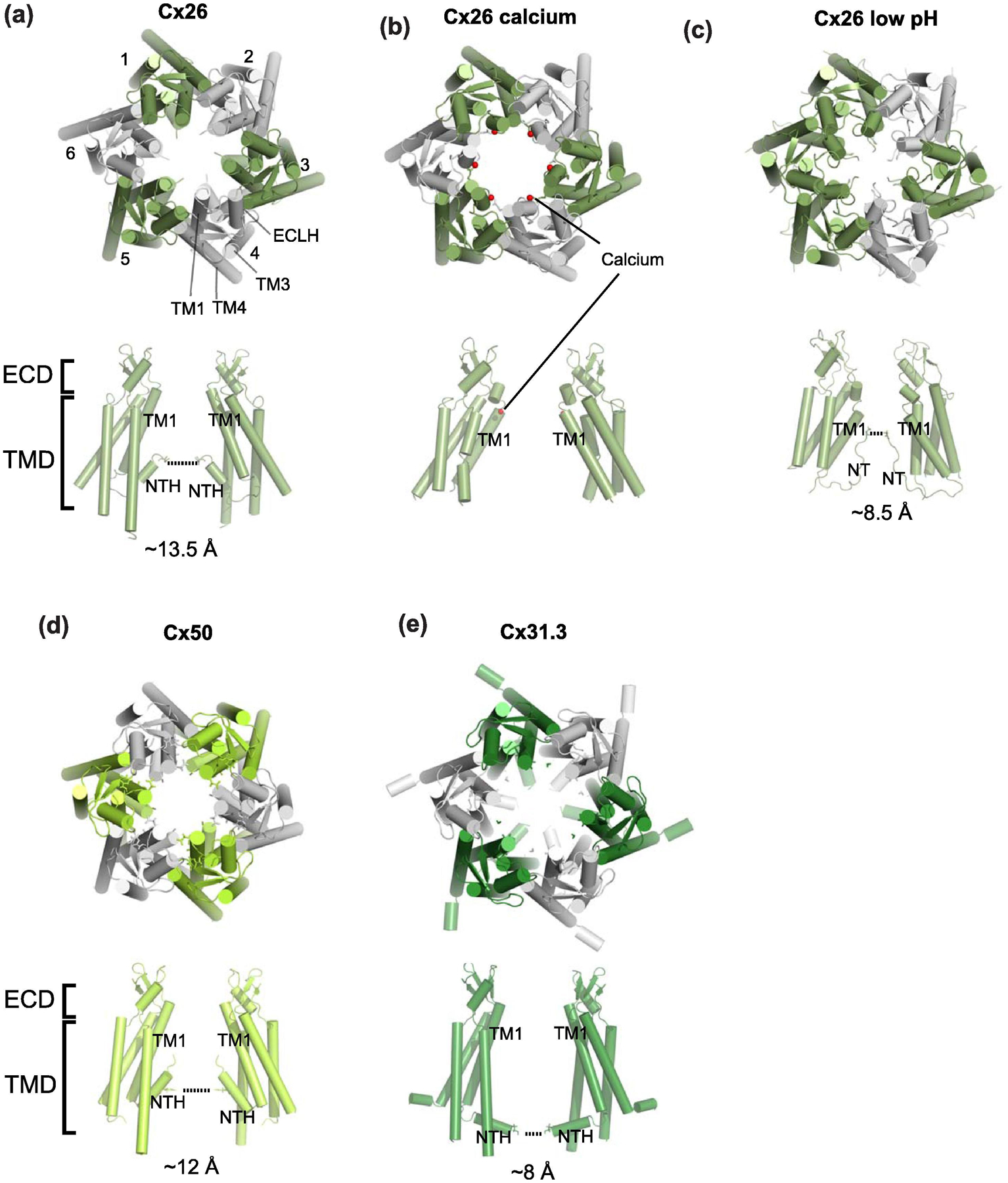
(a-e) The views of hemichannels from the extracellular side (upper panels) and from the side (lower panels). Only two opposing subunits are shown for clarity. The locations of the narrowest constrictions at the N-terminal regions are highlighted with dotted lines except for Cx26 calcium (panel b; calcium in red spheres) where the N-terminal ends are not well resolved. At low pH, the N-terminal region narrows down (panel c). Also note that the orientations of NTHs in Cx31.3 (panel e) are different from the rest. Cx26 low pH in the closed conformation (PDB code: 6UVT) is modelled based on a 7.5 Å cryo-EM map. Cx26 calcium (PDB code: 5ER7) is from the X-ray crystallographic data at 3.28 Å.
In addition to Cx26, high-resolution structures of other connexin channels, including Cx46/50 and Cx31.3/GJC3 became available, by single-particle cryo-EM [43–45] (Fig. 1e). The Cx46/50 heteromeric gap junction was isolated and purified from eye lens of lamb and sheep [43, 44]. The authors suggest that the hemichannels contain both Cx46 and Cx50 by showing that the N-terminal residues (Gly2) from both subunits, which are located within the pore, can be tethered by short cross-linkers [44]. During the single-particle analysis, D6 symmetry (two-fold symmetric dimer of the two hexameric connexin channels) was imposed; thus, the unique cryo-EM density which distinguishes Cx46 and Cx50 from each other were averaged out. Consequently, the pattern of heteromeric subunit arrangement in this gap junction channel was unresolved. Nevertheless, the resolution of the most recent 3D reconstruction reached as high as 1.9 Å, which marks the most highly resolved structure amongst all of the large-pore channels [43]. This stems from the fact that Cx46 and Cx50 are highly similar in sequence (80% identity and 88% similarity) and structure. This high-resolution structural analysis resolved ordered water molecules and lipid molecules of the extracellular leaflet of the bilayer, which stabilize the channel architecture (Fig. 1e). Furthermore, this structure is suggested to be in an open conformation and has a pore diameter size of ~12 Å or larger (Fig. 2d). It is interesting to note that a recent cryo-EM study on the connexin hemichannel, Cx31.3/GJC3 (Fig. 1e and 2e) [45] measured the pore size to be 8 Å, which is not sufficiently large for ATP to permeate implying a difference in pore size among different members of connexin channels. It is expected that as more structures of connexin channels become available, the common and different features between different connexin isoforms should become clearer. Important to reemphasize is the fact that in all of the studies so far, the connexin hemichannels and gap junctions have been shown as strictly hexameric and dodecameric (dimer of hexamers), respectively. Also, every connexin subunit has been shown to have the same membrane topology containing four transmembrane helices per protomer, thus far. Overall, the structural studies on the connexin channels over the last half century set a foundation to study newer members of large-pore channels discussed in the later sections.
Overview of non-connexin large-pore channel members
Since the discovery of connexin gap junction channels and hemichannels, a number of functionally related large-pore channels have been identified. The non-connexin members of the large-pore channel family today include innexin, pannexin, LRRC8, and CALHM, which play distinct biological roles. Here we provide an overview of these large-pore channel members (Fig. 3).
Fig. 3. Overview of large-pore channels.
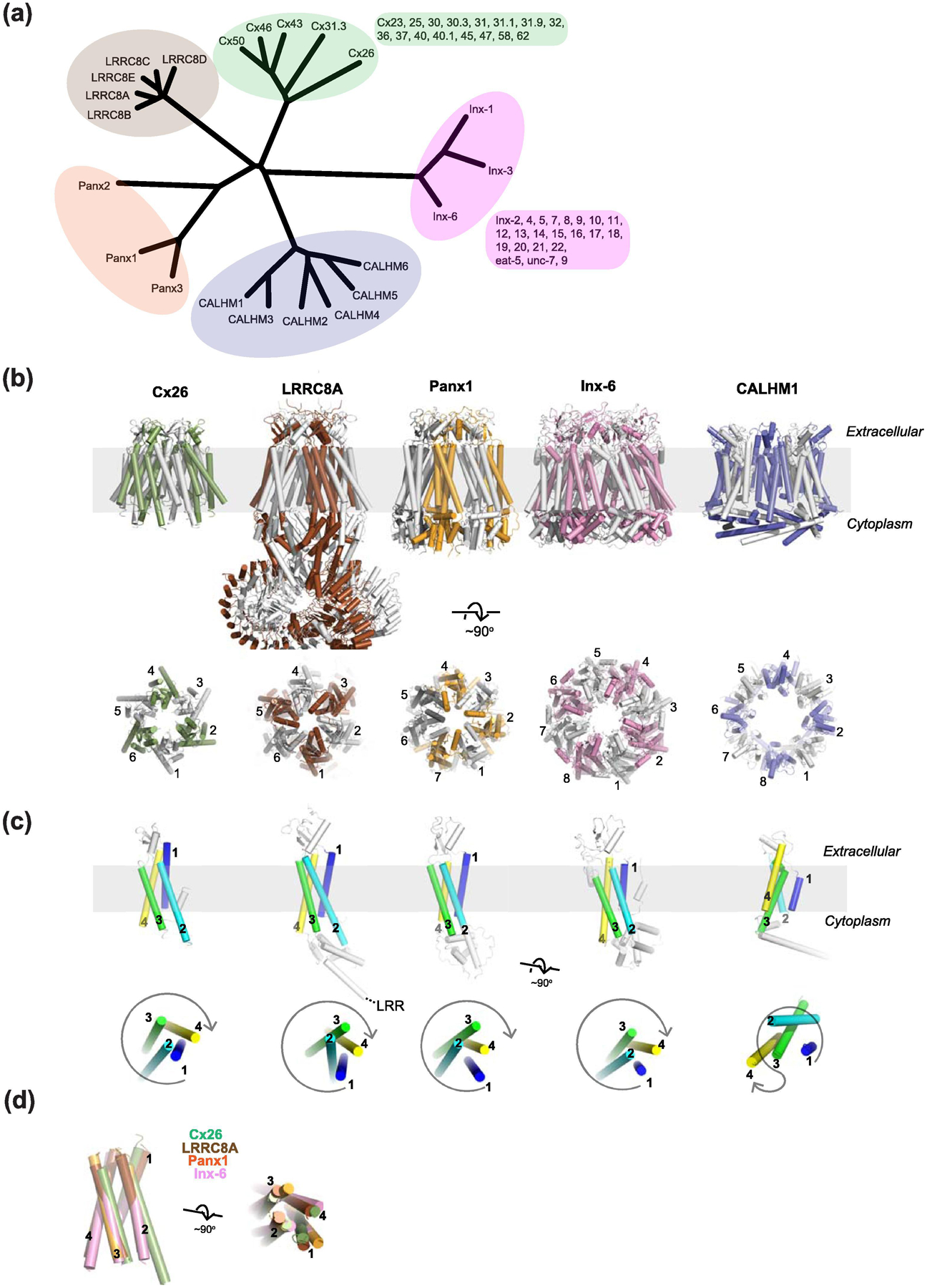
(a) Phylogenic tree all members of LRRC8, pannexin, and CALHM and some representative members of connexin and innexin families. Other family members of connexin and innexin are listed next to the tree. (b) Representative structures from different large-pore channels from the side (upper panels) and the top (lower panels) of the membrane plane. The PDB codes of Cx26, LRRC8A, Panx1, Inx6, and CALHM1 shown here are 2ZW3, 6G9O, 6VD7, 5H1Q, and 6VAM, respectively. Only one hemichannel of the Cx26 gap junction structure is shown here for clear comparison with others. The numbering in the lower panel is to show different oligomeric states. (c) The monomeric subunits of Cx26, LRRC8A, Panx1, Inx-6, and CALHM1 viewed from the side (upper panel) and from the top (lower panel) of the membrane plane where the first, second, third, and fourth transmembrane helices are colored blue, cyan, green, and yellow, respectively. (d) Superposition of the transmembrane region of Cx26, LRRC8A, Panx1, and Inx6 from the side (left) and the top (right) showing the similar helical orientation. The transmembrane helices of CALHM1 cannot be superimposed and therefore was excluded here.
First, the observation of gap junctions in invertebrate tissues by electron microscopy [46] led to a logical prediction that similar proteins to connexins likely exist also in invertebrates. Forward genetic screens for disrupting locomotion in Caenorhabditis elegans (C.elegans) led to the eventual identification of a gene encoding an invertebrate gap junction protein, innexin. Innexin, like connexin, forms hemichannels that release ATP from cells [47–49].
Pannexin was discovered by a sequence-based search for a second family of gap junction proteins conserved in vertebrates and invertebrates [50–52]. It was later shown that pannexin does not form a gap junction channel when properly glycosylated but exists as a cell surface channel with its extracellular domain exposed to the exterior environment [53–56]. Pannexin mediates release of ATP like connexin and innexin hemichannels [57]. Importantly, the pannexin-mediated ATP release acts as a ‘find-me’ signal of macrophage recruitment for inflammation and cell death, the biological role of which is distinct from that of connexin and innexin [58, 59].
LRRC8 was discovered to be responsible for Volume-Regulated Anion Channel (VRAC) activity in 2014 through whole genome small interfering RNA (siRNA) screens [60, 61]. The paradigm of VRAC current induced by hypotonic cell swelling was known since the 1980s but delineating its molecular entity was difficult since there are high endogenous levels of VRAC in almost all cells types. The obligatory subunit, LRRC8A (or SWELL1), was discovered through siRNA screening [60, 61]. The related subunits, LRRC8s (B-E), were shown to form heteromers with LRRC8A to generate the VRAC current. The LRRC8 channels allow the passage of not only halide ions (Cl− in the physiological condition) but also larger molecules such as taurine and even neurotransmitters such as glutamate and GABA [62].
CALHM was identified as a potential risk factor for late-onset Alzheimer’s disease by a genome analysis in 2008 [63]. CALHMs have no sequence homology to any of the known large-pore channels mentioned above. Indeed, CALHM1 was initially considered to have a similar motif to N-methyl-D-aspartate receptors (NMDARs) for mediating preferred permeability to Ca2+ [63]. However, later secondary structural and functional analyses showed that CALHM1 forms a hemichannel capable of conducting anions and cations as well as ATP in a voltage dependent manner [64–66]. ATP efflux through CALHM1 on type-II gustatory cells was shown to induce purinergic signaling for taste perception [65]. It eventually became clear with X-ray crystallographic structures and cryo-EM structures that CALHM1 and NMDARs do not have common structural motifs [67–69].
Despite little or no sequence homology, the large-pore channel members above were predicted to have similar transmembrane topologies with four transmembrane helices. The extensive structural biological efforts on all of the large-pore channel members in recent years revealed that connexin, innexin, pannexin, and LRRC8 channels have four transmembrane helices arranged in a highly similar manner. Furthermore, their extracellular and intracellular domains, excluding the LRR in the LRRC8 channels, have similar protein folds (Fig. 3b–d). The outliers are CALHMs, which have an unrelated pattern of transmembrane helical arrangement as well as unique protein folds in the extracellular and intracellular domains (Fig. 3b–d). The most striking structural observation of the large-pore channels is that the oligomeric states are variable. Connexin (Cx26, 43, 46/50, and 31.3), LRRC8 (A and D), innexin (Inx-6), pannexin (Panx1), and CALHM1 are hexameric, hexameric, octameric, heptameric, and octameric, respectively (Fig. 3b). As discussed later, the CALHM members can form oligomers ranging from 8-mers to 13-mers. Such variability in oligomeric states within the same channel family is unprecedented and is unique to CALHMs.
Mitochondrial channels such as the voltage-dependent anion channel (VDAC) form beta-barrel-based large pore channels that are permeable to ions and ATP, but their functions and cellular locations are not comparable to the large-pore channels found on the cell surface discussed here and thus will be outside the scope of this review. VDAC channels are discussed extensively in other reviews [70, 71].
In the following sections, we describe the most recent insights into the structural and functional understanding of large-pore channels including innexin, LRRC8, pannexin and CALHM, which function as voltage-gated channels for ions and metabolites. The progress of these studies has been highly facilitated by the recent development of cryo-EM methods. However, it is our opinion that numerical estimation of resolution does not necessarily correspond to the quality of maps at sites of interest. It is therefore a good practice to inspect the models and the quality of corresponding cryo-EM density (e.g., is cryo-EM density sufficiently resolved to confidently place molecular models?) using software such as UCSF Chimera and Coot [72, 73]. In all of the structures presented in this review, densities in the N-terminal domains including N-terminal helix (NTH) is generally poorly resolved although some studies decided to placed molecular models into them. Thus, the validity of molecular models around the region must be carefully assessed when designing structure-based experiments.
Innexin
Innexin channels form invertebrate gap junctions
Invertebrate gap junction formation involves a gene family called innexin (Inx) that has no sequence homology to the vertebrate connexin family. Genes with similar sequences to connexin are not found in any invertebrate species. However, later studies showed that distant homologues of innexin exist in vertebrates and are now known as pannexins [50, 51]. The innexin genes were first discovered in C. elegans as unc-7 [74] and in Drosophila melanogaster as Shaking-B [75, 76] through forward genetic screens. Neuronal coupling in the Giant Fiber System (GFS) in Drosophila was hampered in the Shaking-B mutant (originally called Passover mutant) [77] while the unc-7 mutant in C.elegans resulted in severe locomotion defects [74]. A similar gene, eat-5, was shown to disrupt synchronized pharyngeal muscle contractions, which resulted in altered eating behaviors [78]. Later studies isolated and determined the sequence of unc-7 and Shaking-B [79–81]. These studies allowed prediction of transmembrane topology, which suggested that unc-7 and Shaking-B encode similar proteins with similar transmembrane topology, and that these proteins have little or no sequence homology to connexin. The related genes in Drosophila and C.elegans were eventually named innexins [82]. Finally, expression of Shaking-B proteins in Xenopus oocytes showed pairing of oocytes and generation of transjunctional voltage as measured using a double voltage clamp procedure [83]. Thus, this study convincingly demonstrated that Shaking-B proteins (or innexin proteins) form gap junction channels. Inx-3 from C.elegans was later expressed in Xenopus oocytes and shown to form gap junction channels [84].
Innexins have been identified in all invertebrate phyla with the exception of sponges and echinoderms [85, 86]. For example, the C. elegans genome has 25 innexin coding genes together with multiple splice variants. As in the case for other large-pore channels, innexin channels can be activated by voltage and are functionally similar to connexin channels [47, 87]. They form not only gap junctions but also hemichannels. These currents are regulated by pH and intracellular calcium [47]. The innexin gap junctions, similar to connexin gap junctions, can form heterotypic channels that mediate rectification [88]. Finally, innexin channels have also been demonstrated to release ATP when overexpressed in Xenopus oocytes [47]. Both ion and ATP flux can be blocked by carbenoxolone (CBX) as in the case of other large-pore family members except CALHM. Overall, innexin channels possess many similar functional and pharmacological properties to other large-pore channels which indicates that invertebrates and vertebrates utilize similar molecular mechanisms to mediate intercellular metabolite exchange and electrical synaptic transmission.
Structural biology of Innexin
It took close to two decades for the field to determine the structure of an innexin channel. High resolution structures of C.elegans Inx-6 gap junction channels and hemichannels became available by single particle cryo-EM in 2016 (Fig. 4, Table 2). The initial structures were solved in detergent [89] and later in lipid nanodiscs [90]. Protein samples prepared for these studies was done using a method called GraDeR [91], where glycerol gradient centrifugation is used to remove free detergent from the protein-micelle complex to facilitate high quality cryo-EM imaging. This GraDeR preparation at 500 mM NaCl contained a mixture of gap junction channels and hemichannels, and structures of the innexin hemichannel and gap junction were independently obtained from the same dataset at 3.3 Å and 3.6 Å, respectively [89] (Fig. 4a). This structural study clearly showed that the Inx-6 hemichannels and gap junction channels are assembled as octamers (8-mers) and hexadecamers (16-mers), respectively. Gap junction channels are formed by interactions at the extracellular domains as in the case of connexins (Fig. 4a). Overall, the 8-meric assembly of the Inx-6 hemichannel was unlike the 6-meric assembly predicted based on the presumed structural similarity to the connexin hemichannel.
Fig. 4. Structures of Inx-6 gap junction channel and hemichannel.
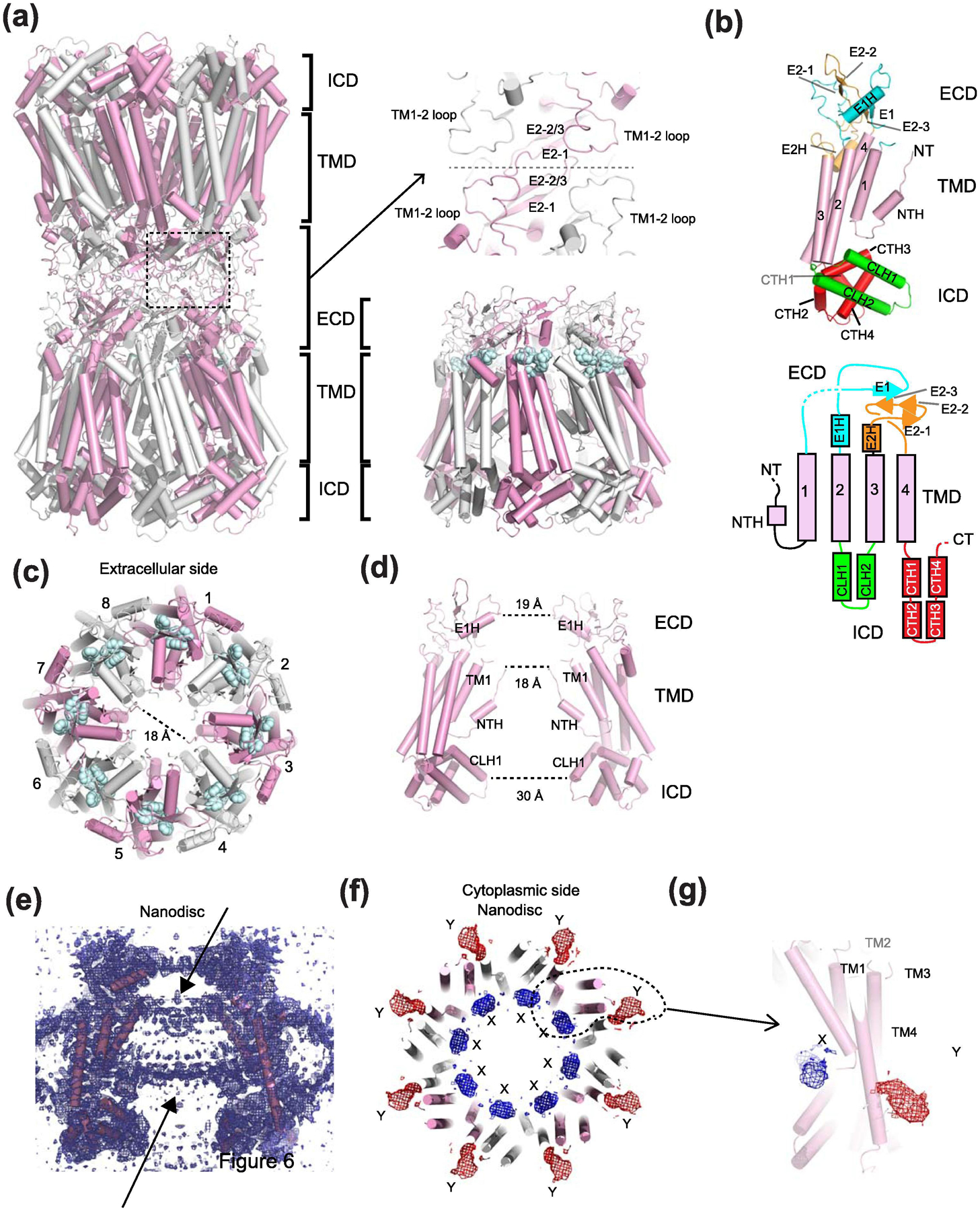
(a) The Inx-6 gap junction channel (left) and hemichannel (right) with subunits colored in alternating pink and white. The domain layers are partitioned into the transmembrane domain (TMD), the extracellular domain (ECD), and the intracellular domain (ICD). The two hemichannels interact at the ECD. Spheres in pale cyan are a conserved sequence (YYQWV) in the innexin family. (b) A monomer of Inx-6 and the schematic presentation of the structural topology. (c) Inx-6 hemichannel viewed from the extracellular side showing the octameric arrangement. The dotted line (18 Å) is drawn between Ala7 of the opposing subunits. (d) Side view showing two opposing subunits. Distances of narrow constrictions are measured between E1Hs, NTHs, and CLH1s. (e) The side view of cryo-EM density illustrating the presence of lipid-like density in Inx-6 in nanodiscs. (f) In addition to the lipid-like density, two amorphous densities, X (blue mesh) and Y (red mesh), are observed. (g) Close-up view of a single protomer and the locations of the X and Y densities. The X and Y densities are not present in the structure of the NTH deleted construct (residue 2–19 deletion).
Table 2.
Available structures of Innexin in 2021
| Structure | Functional state | PDB code | Species | Oligomeric state | Solubilization conditions | Freezing buffer conditions | Reference |
|---|---|---|---|---|---|---|---|
| Inx-6 3.3 Å | Open | 5H1Q | Caenorhab ditis elegans | 8 | TBS + 2% DDM | GraDeR; 10 mM Tris pH 7.5, 500 mM NaCl, trace LMNG | Oshima et al., 2016 |
| Inx-6 gap junction 3.6 Å | Open | 5H1R | Caenorhab ditis elegans | 8; head-to-head gap junction 16-mer | TBS + 2% DDM | GraDeR; 10 mM Tris pH 7.5, 500 mM NaCl, trace LMNG | Oshima et al., 2016 |
| Inx-6 3.8 Å | Open | 6KFF | Caenorhab ditis elegans | 8 | TBS + 2% DDM | MSP2N2 + POPC nanodiscs; 10 mM Tris pH 7.5, 150 mM NaCl | Burendei et al, 2020 |
| Inx-6 3.8 Å | Open | 6KFG | Caenorhab ditis elegans | 8 | TBS + 2% DDM | GraDeR; 10 mM Tris pH 7.5, 50 mM NaCl, trace LMNG | Burendei et al, 2020 |
| Inx-6 ΔN 3.6 Å | Open | 6KFH | Caenorhab ditis elegans | 8 | TBS + 2% DDM | MSP2N2 + POPC nanodiscs; 10 mM Tris pH 7.5, 150 mM NaCl | Burendei et al, 2020 |
| Inx-6 ΔN 10 Å | n/a | n/a | Caenorhab ditis elegans | 8; head-to-head gap junction 16-mer | TBS + 2% DDM | 2D electron crystallography | Oshima et al, 2016 |
The Inx-6 protomer has structural similarity to the connexin protomer despite no sequence homology. Like connexin, there are four transmembrane helices and the N-terminus has a short helix (NTH) facing the pore (Fig. 4b). The N-terminal residues have been known to play important roles in voltage sensitivity of connexin, pannexin, LRRC8, and CALHM channels. Given the structural similarity between innexin and other large-pore channel family members except CALHM, it is reasonable to predict that the N-terminal domain of innexin plays a role in voltage-sensitive activation. Indeed, truncation of the N-terminus has been shown to abolish voltage-dependent activity of Inx-6 channel [90].
The positions of the two disulfide bond pairs in the extracellular domain are similar to two of the three conserved disulfide bond pairs in connexin. However, the intracellular domain (ICD) of innexin is structurally well-defined and contains six helices which are distinct from connexin. There are two helices between TM2 and 3 (CLH1 and 2) and four short helices after TM4 (CTH1 to 4), which interact with each other to form the ICD that is similar to pannexin and LRRC8 channels as discussed later. No such features are observed in Cx26, Cx31.3, and Cx46/50 channels and CALHM channels. The ICD in Inx6 is structured like a dome (cytoplasmic dome) that interacts and controls the orientation of the NTH that is critically involved in voltage sensing.
In the context of the octameric hemichannel the narrowest constriction of the permeation pathway is in the NTH region where the diameter is measured to be 18 Å (Fig. 4c–d). Although the functional state of the cryo-EM structures is not clearly defined, this diameter is sufficiently wide to permeate ATP and perhaps other metabolites, which implies an open state. The ICD has a wide diameter, and thus, would not serve as a filter for substrates and ions. The constriction at the ECD around the helix in the first extracellular loop (E1H) (Fig. 4d) is nearly as narrow as the NTH region. Interestingly, the equivalent helix and the loop in the ECDs of LRRC8 and pannexin form narrow constrictions that serve as the selectivity filter for anions and possibly large metabolites as discussed later. The shorter diameters in the equivalent regions in LRRC8 and pannexin channels stem from their lower number of oligomeric states (6-mer for LRRC8 and 7-mer for pannexin).
An electron crystallography study at low resolution (10 Å in a horizontal plane) on lipid reconstituted 2D crystals of the Inx-6 gap junction clearly shows density in the middle of the cavity that appears to be plugging the pore [92]. This observation was followed up recently by the high resolution single particle cryo-EM analysis of Inx-6 in lipid nanodic in a buffer containing 150 mM NaCl [90]. The sample prepared in the above condition existed mostly as hemichannels and the central cavity contained plausible density for lipids. No such density was observed in the cryo-EM structure solved in minimal detergent [89]. The more pronounced lipid-like density was also observed in the structure of CALHM2 [67] obtained in a lipid nanodisc as well as CALHM4 in detergent [93] as discussed later. Although premature at this point, the above observation in Inx-6 may imply a potential role for lipids in gating that may be a common paradigm among the large-pore channels.
In the nanodisc reconstituted Inx-6 structure, no clear density for the NTH was observed [90] (Fig. 4e–g). This is in stark contrast to the structure solved in detergent where the NTH region was resolved and clearly present around the central pore region making contact with TM1 [89]. Instead, in the Inx-6 structure in nanodiscs, unresolved density was observed in two locations (termed X and Y densities), one around the lipid density close to the pore (density X) and the other toward the outside of the pore extending from TM1 and passing TM4 (density Y) (Fig. 4f and g). No such density was observed in the NTH deleted construct (Δ2–19) prepared in the same lipid nanodisc condition, supporting the rather bold hypothesis that these densities represent the NTHs [90]. Given that the N-terminal region undergoes such a robust conformational alteration it would be necessary to understand how it may regulate function of the Inx-6 channel.
Overall, the series of Inx-6 structures above provide the field with important blueprints to study and compare the functions of innexin with the other large-pore channels.
LRRC8 channels
LRRC8 as major component of Volume-Regulated Anion Channels
It is an absolute requirement that cells counteract osmotic swelling and shrinkage to maintain homeostasis. Mammalian cells regulate their volume by fluxing ions (Na+, K+, and Cl−) and osmolytes across the membrane to generate an osmotic gradient and facilitate influx and efflux of water. Cell swelling occurs during hypotonic stress, but a consequent restoration to normal volume occurs through a process called regulatory volume decrease (RVD). A part of this mechanism has been suggested to be mediated by swelling-activated K+ channels and by efflux of K+ and Cl− through K+/Cl− cotransporters [94, 95]. Swell-induced anion-selective currents were first observed in human lymphocytes as early as the 1980s [96]. This current is mediated mainly by Cl- (ICl, swell) in physiological conditions and also small organic osmolytes including taurine, glutamate, inositol and ATP [60, 61, 97–100] and most recently CGMP [101]. However, there have been reports indicating that the ATP release may be independent of the VRAC activity [102, 103] and that ATP rather blocks VRAC currents with minimal permeability [104]. The ICl, swell current has halide selectivity in the order of I− > Cl− > Br− > F−, has moderate outward rectification, requires intracellular ATP for activation, and inactivates at positive membrane potentials [105–108].
Despite decades of efforts, the molecular identity of the channels that mediate the ICl, swell currents, volume-regulated anion channels (VRAC), remained enigmatic. Bestrophin-1 in Drosophila cells was shown to mediate ICl, swell currents but its murine orthologues do not [109]. Remarkably in 2014, two groups independently identified leucine-rich repeat-containing protein 8A (LRRC8A or SWELL1) to be the major component for VRAC by a genome-wide small interfering RNA (siRNA) screen. In this assay, swelling induced I− flux was measured as fluorescence quenching of I−-sensitive yellow fluorescent proteins [60, 61]. Subsequent assays using electrophysiology on Hela cells and CD4+ T lymphocytes, where LRRC8A expression was suppressed by siRNA, verified LRRC8A as the major VRAC component. However, transfection of LRRC8A alone into HEK293 cells with the LRRC8A−/− background gives substantially smaller currents with stronger outward rectification than the normal VRAC current. The VRAC current can only be generated when LRRC8A forms a heteromeric assembly with LRRC8C, D or E [60]. Heterologous co-expression of LRRC8A and LRRC8B in Lrrc8−/− cells does not yield hypotonicity-induced VRAC activation [60, 110]. Instead, overexpression of LRRC8B was shown to mediate modest Ca2+ leak from the endoplasmic reticulum (ER) [110]. The heteromeric assembly between LRRC8A and LRRC8B-E has been verified by co-immunoprecipitations [60, 111]. Reconstitution of the purified heteromeric LRRC8 complexes into artificial droplets of lipid bilayers confirmed that they directly elicit anionic current in response to osmolality changes [111]. Interestingly ATP was not required for channel activation in this system [111] perhaps indicating that the previously measured effect in the cellular context involves other factors such as ATP-dependent channel regulation by interacting proteins. Also important is the observation that decreased ionic strength but not mechanical tension is the driving force for channel opening [111]. However, it is worth mentioning that the field has not reached a consensus on the activation mechanism. There remains a possibility that LRRC8 channels could be sensing mechanical forces by interactions between the LRR region and cellular elements such as the cytoskeleton. Furthermore, a recent study showed that pharmacological manipulation of diacylglycerol and protein kinase D, but not reduced intracellular ionic strength, activates VRAC [112].
Three or more subunits, with LRRC8A being an obligatory subunit, can be contained in the LRRC8 heteromeric channel [60, 111, 113]. The subunit composition controls substrate selectivity, inactivation kinetics, and single channel conductance. A wide variety of substrates have been shown to permeate through the LRRC8 channels including the osmolytes taurine and myo-inositol and also glutamate, aspartate, gamma-aminobutyric acid (GABA), and D-serine, which are effluxed from glial cells and can serve as potential neurotransmitters [114]. Release of these organic osmolytes and metabolites is mediated more effectively in the LRRC8A/D complex compared to others. More recently, the LRRC8A/C/E heteromeric channel was shown to flux 2’3’-cyclic-GMP-AMP (cGAMP), a paracrine innate immune messenger [101]. Besides volume-regulation, LRRC8 channels have been shown to play roles in apoptosis and uptake of cancer drug and antibiotics [60, 61, 97, 98, 111, 113, 115]. There have been some reports on ATP release from LRRC8 channels but it may occur only in an extreme hypotonic condition. Overall, the remarkable identification of the major VRAC component as LRRC8 was accomplished only recently. The LRRC8 family comprised of five subunits shows highly sophisticated differences in functions conferred by a number of different subunit combination. More substrates, subtype-specificity, and different modes of functional regulation may be discovered in the near future.
Structural biology of LRRC8 channels
A number of structures of the full length LRRC8A from mouse and human became available since 2018 by single particle cryo-EM (Fig. 5, Table 3) [116–119]. To facilitate complete modeling, X-ray crystallography was implemented on the isolated LRR domain to provide the 1.8 Å structure [116]. Furthermore, a 3D reconstruction map of LRRC8A/C heteromeric channel was obtained at 7.9 Å, the resolution of which was not sufficient to distinguish A and C subunits, therefore, stoichiometry and the pattern of subunit arrangement were not defined [116]. The structures were obtained from protein samples in detergent (digitonin) [116, 118, 119] and in lipid nanodiscs [117]. One of the structures of LRRC8A in nanodiscs was in complex with a channel blocker (4-[(2-Butyl-6,7-dichloro-2-cyclopentyl-2,3-dihydro-1-oxo-1H-inden-5-yl)oxy]butanoic acid) (DCPIB) [117]. Most recently, a cryo-EM structure of LRRC8D became available [120].
Fig. 5. Structure of homo-hexameric LRRC8A.
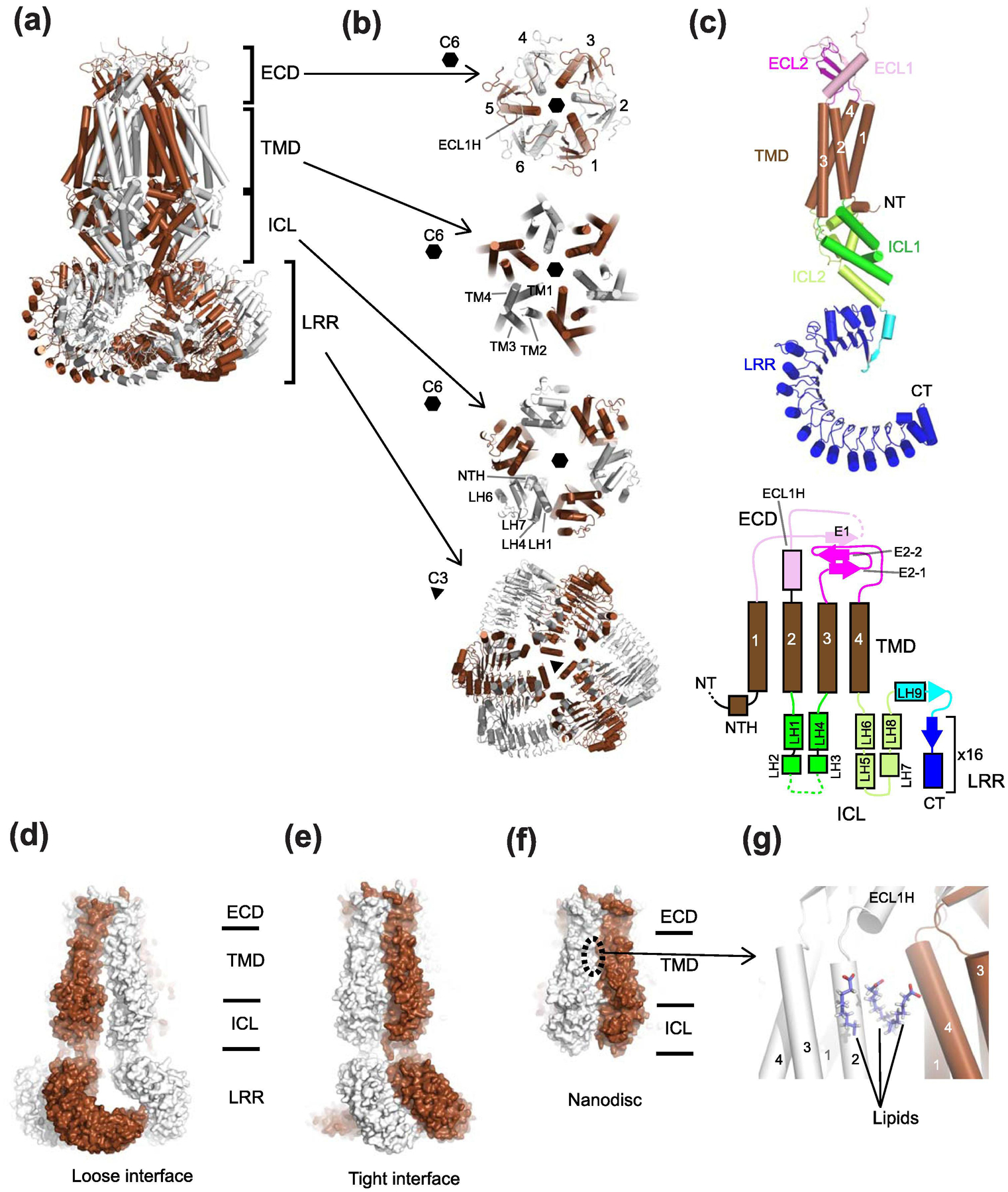
(a) Shown here is the structure of mouse LRRC8A (PDB code: 6G9O) where alternate subunits are colored brown and white. The side view shows four layers, ECD, TMD, ICL, and LRR. (b) Top views of LRRC8A in different layers. ECD, TMD, and ICL have C6 or pseudo-C6 symmetry whereas the LRR layer has C3 symmetry. (c) The structure of the LRRC8A protomer. The color code of the structure (left panel) is the same as the one for the schematic topology presentation (right panel). (d-e) The structure of human LRRC8A in detergent refined using C3 symmetry (PDB code: 5ZSU) where two distinct interfaces, ‘loose interface’ (panel d) and ‘tight interface’ (panel e) are observed. The gap in the ‘loose interface’ is more pronounced in the LRR layer than the ICL and TMD layers. (f) The structure of mouse LRRC8A in lipid nanodisc (PDB: 6NZZ) refined with C6 symmetry. In this study, the LRR layer was not modeled due to disordered density. The extent of inter-subunit packing is between those of ‘loose interface’ and ‘tight interface’ in panel d and e. (g) In this lipid nanodisc sample, the six inter-subunit interfaces in the TMD equally contain three molecules of phospholipids per interface, which strengthen the subunit-subunit interactions.
Table 3.
Available structures of LRRC8 in 2021
| Structure | Functional state | PDB code | Species | Oligomeric state | Solubilization conditions | Freezing buffer conditions | Reference |
|---|---|---|---|---|---|---|---|
| LRRC8A 4.4 Å | Not commented on | 6DJB | Homo sapiens | hexamer | 20 mM Tris pH 8, 150 mM NaCl, 1% DMNG, 2 mg/mL iodoacetamide, and EDTA-free protease inhibitor cocktail | 20 mM Tris pH 8, 150 mM NaCl, 0.05% digitonin, and EDTA-free protease inhibitor cocktail | Kefauver et al., 2018 |
| LRRC8A 4.25 Å | Suggestion that the channel may be open but authors state they can not precisely define open/cosed states on the basis of this structure | 5ZSU | Homo sapiens | hexamer | 50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM DTT, 1% digitonin | 50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM DTT, 0.1% digitonin | Kasuya et al., 2018 |
| LRRC8A 4.18 Å | Apo-LRRC8A in a constricted state | 6O00 | Mus musculus | hexamer | A 10%/2% solution of DDM/CHS was dissolved and clarified by bath sonication in 200 mM HEPES pH 8 prior to addition to the solubilisation buffer. 50 mM HEPES, 150 mM KCl, 1 mM EDTA, 1% DDM, 0.2% CHS, final pH 7.4. | 20 mM HEPES, 150 mM KCl, 1 mM EDTA pH 7.4, POPC, MSP2N2 | Kern et al., 2019 |
| LRRC8A-refined against 5.01 Å map | Authors suggest that the high salt conditions in the freezing buffer favor a closed conformation | 6G9L | Mus musculus | hexamer | 25 mM Tris-HCl, pH 8.5, 250 mM NaCl, 3% digitonin,50 μg ml−1 DNase, 50 μg ml−1 RNase A and protease inhibitors | 25 mM Tris-HCl pH 8.5, 250 mM NaCl, 0.12% digitonin | Deneka and Sawicka et al., 2018 |
| LRRC8A-refined against 4.25 Å map | Authors suggest that the high salt conditions in the freezing buffer favor a closed conformation | 6G9O | Mus musculus | hexamer | 25 mM Tris-HCl, pH 8.5, 250 mM NaCl, 3% digitonin,50 μg ml−1 DNase, 50 μg ml−1 RNase A and protease inhibitors | 25 mM Tris-HCl pH 8.5, 250 mM NaCl, 0.12% digitonin | Deneka and Sawicka et al., 2018 |
| LRRC8A-pore domain 3.66 Å | Authors suggest that the high salt conditions in the freezing buffer favor a closed conformation | 6G8Z | Mus musculus | hexamer | 25 mM Tris-HCl, pH 8.5, 250 mM NaCl, 3% digitonin,50 μg ml−1 DNase, 50 μg ml−1 RNase A and protease inhibitors | 25 mM Tris-HCl pH 8.5, 250 mM NaCl, 0.12% digitonin | Deneka and Sawicka et al., 2018 |
| LRRC8A-LRR domain 1.80 Å (X-ray structure) | n/a | 6FNW | Mus musculus | protomer | 10 mM Tris pH 9.4, 200 mM NaCl, 2% DDM, 50 μg ml−1 DNase, protease inhibitors | The protein was polished on SEC in 10 mM Tris pH 9.4, 200 mM NaCl, 0.1% CHAPS and then supplemented with 0.5% CHAPS and 1 mM tris(2-carboxyethyl)phosphine prior to crystallization in 0.1 M Bis-Tris propane pH 8.5, 0.2 M sodium malonate and 20% PEG3350 | Deneka and Sawicka et al., 2018 |
| LRRC8A-DCPIB 3.21 Å | Constricted state | 6NZW | Mus musculus | hexamer | A 10%/2% solution of DDM/CHS was dissolved and clarified by bath sonication in 200 mM HEPES pH 8 prior to addition to the solubilisation buffer. 50 mM HEPES, 150 mM KCl, 1 mM EDTA, 1% DDM, 0.2% CHS, final pH 7.4. | 20 mM HEPES, 150 mM KCl, 1 mM EDTA pH 7.4, POPC, MSP1E3D1. 100 μM of DCPIB was added prior to freezing. | Kern et al., 2019 |
| LRRC8A-DCPIB 3.32 Å | Expanded state | 6NZZ | Mus musculus | hexamer | A 10%/2% solution of DDM/CHS was dissolved and clarified by bath sonication in 200 mM HEPES pH 8 prior to addition to the solubilisation buffer. 50 mM HEPES, 150 mM KCl, 1 mM EDTA, 1% DDM, 0.2% CHS, final pH 7.4. | 20 mM HEPES, 150 mM KCl, 1 mM EDTA pH 7.4, POPC, MSP1E3D1. 100 μM of DCPIB was added prior to freezing. | Kern et al., 2019 |
| LRRC8D 4.36 Å | Undetermined. Some suggestion that the channel may be in an open conformation | 6M04 | Homo sapiens | hexamer | 50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM DTT, 1% digitonin | 50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM DTT, and 0.1% digitonin | Nakamura et al., 2020 |
| LRRC8A/C heteromer | n/a | n/a (7.94 Å map) | Mus musculus | hexamer | 25 mM Tris-HCl, pH 8.5, 250 mM NaCl, 3% digitonin,50 μg ml−1 DNase, 50 μg ml−1 RNase A and protease inhibitors | 25 mM Tris-HCl pH 8.5, 250 mM NaCl, 0.12% digitonin | Deneka and Sawicka et al., 2018 |
Structural analyses of LRRC8A, LRRC8A/C, and LRRC8D clearly showed that they all form hexamers. The structure of the heteromeric LRRC8A/C represents the most physiological channel assembly and, despite the limited 7.9 Å resolution, clearly showed similar hexameric subunit arrangement as in the homomeric LRRC8A and LRRC8D structures. Thus, a number of important insights into general functions of LRRC8 can be gained through the high resolution structures of LRRC8A and LRRC8D.
LRRC8 has four domain layers: the extracellular domain (ECD), transmembrane domain (TMD), intracellular linker (ICL), and LRR with leucine-rich repeats (16 for LRRC8A and 15 for LRRC8D) (Fig. 5). The LRRC8A and LRRC8D protomers share a similar transmembrane helical arrangement to connexin, innexin, and pannexin with the amino-terminal and the carboxy-terminal ends located in the cytoplasm (Fig. 5c). The carboxy-terminal region of LRRC8 contains the LRR domain. The amino-terminal end is disordered prior to Pro15 in the LRRC8A structures, but was visible in the LRRC8D structure up to Phe2, and contains a helix and a loop which extends toward the inner pore [120]. ECD and ICL of LRRC8 contains a similar set of secondary structures to pannexin and innexin but differs in their arrangements. The formation of a narrow constriction at ECD is a feature also observed in pannexin despite the two having different oligomeric states (six for LRRC8 and seven for pannexin, as discussed below). Such defined structure in ECD was not observed in other large-pore channel members. It is worth noting that the structural similarity was predicted by sequence analyses previously [121]. Remarkably, both pannexin and LRRC8 channels show anion selectivity, which is facilitated by their ECDs with narrow constrictions. However, as discussed later, the anion selectivity in pannexin and LRRC8 involves residues different in chemical nature.
Subunit arrangement and symmetry mismatch in LRRC8
In general, the LRRC8A hexamers in digitonin have six-fold or pseudo six-fold symmetry in the ECD, TMD, and ICL whereas the LRR layer has C3 symmetry formulated by a trimer-of-dimers assembly (Fig. 5b). The LRRC8A hexamer in a nanodisc has C6 symmetry in the ECD, TMD, and ICL layers and the highly heterogeneous LRR layer that was not well resolved by single particle cryo-EM. Deneka et al prepared mouse LRRC8A sample in detergent, applied overall C3 symmetry in the refinement, and further applied C6 symmetry locally to the pore region containing ECD, TMD, and ICL during refinement [116]. Kasuya et al and Kefauver et al prepared human LRRC8A and mouse LRRC8A in detergent, respectively, and applied C3 symmetry [118, 119]. In all cases, there are two modes of inter-subunit interactions in the LRR layer where the tight interaction is within the dimer unit and the loose interaction is at the trimer-of-dimers interface (Fig. 5d and e). This difference in the LRR layer is translated to the TMD region resulting in tight and loose packing although the difference is smaller [118, 119]. Thus, the ICL, TMD, and ECD layers can be regarded as pseudo-C6. Interestingly, the equivalent subunit interfaces are equally filled with clearly resolved lipid molecules when the LRRRC8A sample is prepared in lipid nanodiscs (Fig. 5f and g) [117]. The binding of lipids equally glues the six subunits together and promotes the subunit arrangement in the C6 symmetry but not pseudo-C6 symmetry. In the C3 symmetric structures, the loose TMD interface is suggested to be filled with digitonin-like molecules whereas the tight interface is too narrow to accommodate any lipid or detergent molecule. Kern et al saw weak density in the LRR layer but decided not to impose any symmetry, unlike other studies. Despite some differences in the patterns of assembly, it is fair to conclude from these high quality studies that the LRRC8A channel assembly involves a major symmetry mismatch between the LRR layer and the rest of the molecule. Symmetry mismatch is observed in other channels, including tetrameric ionotropic glutamate receptors. In these structures, the extracellular domains are arranged in C2 symmetry in a dimer-of-dimers arrangement whereas the transmembrane channel is arranged in C4 or pseudo-C4 symmetry especially around the pore-forming M3 helices [68, 69, 122, 123].
The cryo-EM study on LRRC8D in detergent showed overall C2 symmetry, unlike the C3 symmetry of LRRC8A. This difference is dictated by the arrangement of the LRR domains that are assembled as a dimer of trimers in LRRC8D, not a trimer of dimers as in LRRC8A. The ECD, TMD, and ICL layers appear to have pseudo-C6 symmetry, which implies that symmetry mismatch may be a common feature among the LRRC8 family members. The structure refined with C2 symmetry showed three different modes of inter-subunit interactions: tight, regular, and loose, most pronounced at the LRR layer and less so at the TMD layer. However, it should be noted that the ECD, TMD, and ICL layers of LRRC8D may have C6 symmetry and the LRR layer might not contain symmetry if the protein sample is prepared in nanodiscs as in the case of LRRC8A. At this point, it would be beneficial for the field to focus on defining the mode of inter-subunit interactions of the physiological heteromeric LRRC8 channels (e.g., LRC8A/C, LRRCA/D, etc.) at high resolution. Such an accomplishment would likely facilitate a more definitive understanding of functions such as channel gating from the perspective of subunit arrangement. Major alterations in patterns of inter-subunit interactions involving every domain are expected to occur in mediating channel activities as inferred by conformational heterogeneity of the LRR domains.
Critical structural features and functions in LRRC8A
The structures of homomeric LRRC8A at high resolution provided important insights into functions, which are applicable to understanding the more physiological heteromeric LRRC8 channels [116–119]. The LRRC8A homomeric channels have been demonstrated to have channel activity [116, 117, 119] at a substantially lower level compared to that of heteromeric LRRC8 channels (LRRC8A/B-E). At this point, perhaps wisely, none of the structural studies made any clear statement about the functional states of their respective structures. All of the cryo-EM structures are obtained in the presence of 150 mM NaCl [118–120], 150 mM KCl [117], or 250 mM NaCl [116]; thus, one could argue that the channels are in the closed state. Capturing the open channel state may require execution of structural biology on LRRC8-reconstituted liposomes under conditions that mimic cell swelling.
In all of the available structures of LRRC8 channels, the architecture of TMDs, ECD, and ICL are similar to each other. Apart from the LRR domains that are unique to the LRRC8 family, perhaps the most obvious feature that is different from connexin and innexin is the narrow constriction created by ECD, which appears to serve as a selectivity filter for ions and substrates. In all of the LRRC8A structures, the narrowest region of ECD measures approximately ~7–8 Å in diameter at Arg103 that is located on the E1H in the first extracellular loop (Fig. 6a–b). In this region, Arg103, His104, and Lys51 are organized to confer an overall positive net charge that likely contributes to the anion selectivity. Among the residues, Arg103 at the narrowest constriction plays the most critical role in anion selectivity. The equivalent residues of this arginine are Arg, Leu, Phe, and Phe for LRRC8B, C, D, and E, respectively. For example, LRRC8A(Arg103Ala)/LRRC8C (Ala and Leu residues in the constriction) lowers Cl− selectivity and confers measurable Na+ permeability [116]. LRRC8A Arg103Phe/LRRC8C (six Phe in the constriction) showed reduced reversal potential compared to wildtype LRRC8A/LRRC8C again indicating that Arg103 is critically involved in Cl− selectivity [118]. Interestingly, the mutant, LRRC8A/LRRC8C (Leu105Arg) containing six arginine residues in the narrow constriction show comparable channel activity to wildtype LRRC8A/LRRC8C indicating that the low activity for LRRC8A homomers does not attribute to the presence of the six arginine residues [116]. Considering that LRRC8A homomers are expressed on the cell surface at a level similar to LRRC8A/LRRC8C, there should be an unexplored mechanism for the requirement for heteromeric assembly to mediate native-like VRAC current [116]. A recent study showed that the intracellular loop between TM2 and TM3 of LRRC8A and the first extracellular loop between TM1 and TM2 of LRRC8C, D, and E are critical for VRAC activity [124]. In this work, the authors demonstrated that replacing the first extracellular loop of LRRC8A with that of LRRC8C-E or replacing the intracellular loop of LRRC8C-E with that of LRRC8A is sufficient for forming homomeric LRRC8 channels with the VRAC activity. The interpretation of the above result will likely be revealed as the field obtains heteromeric LRRC8 (A/C-E) channel structures in various functional states.
Fig. 6. Narrow constriction at ECD of LRRC8A.
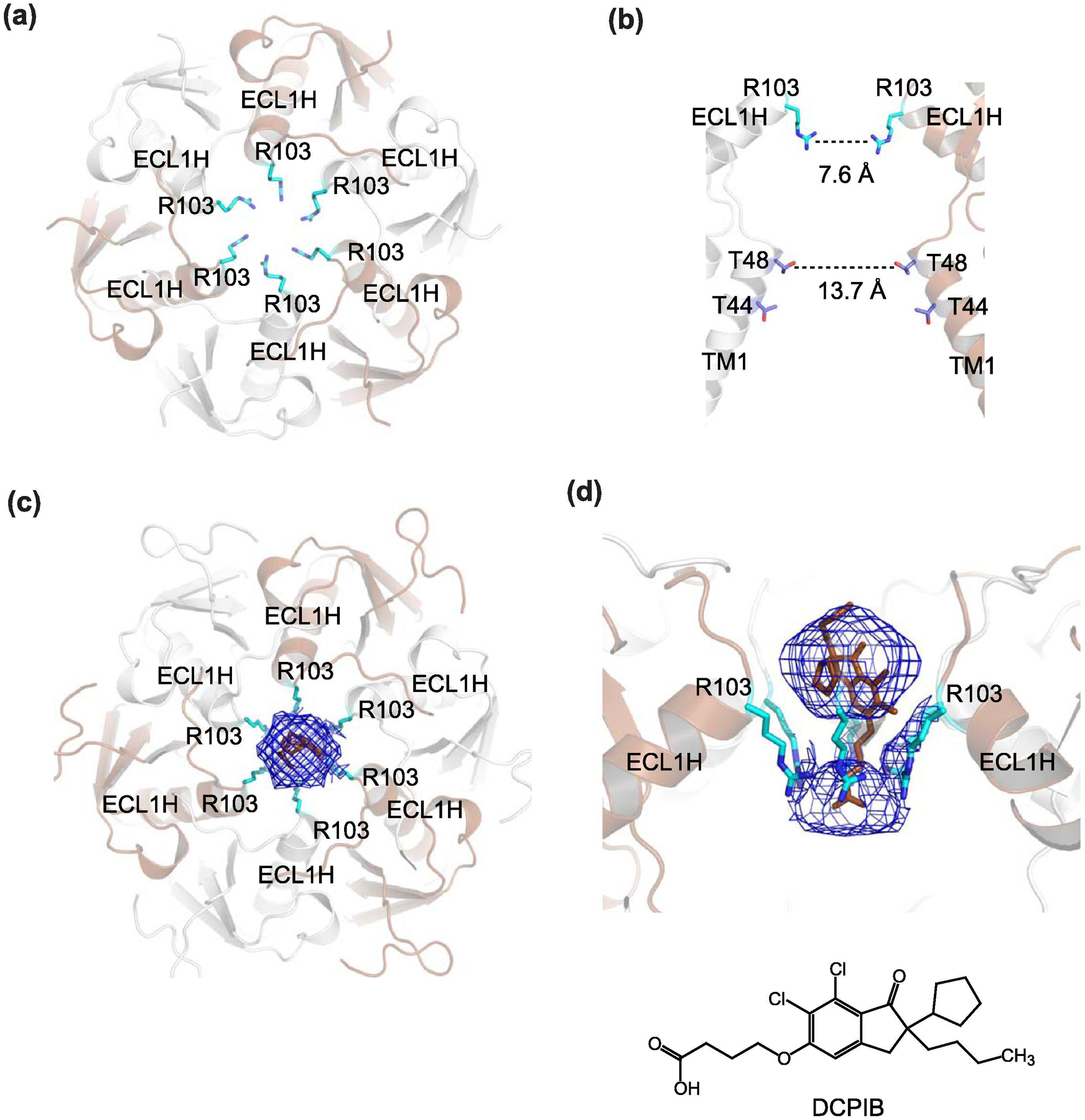
(a) The structure of human LRRC8A (PDB code: 5ZSU) looking ‘down’ the ECD. The narrow constriction is formed by six arginine residues (Arg103, cyan sticks), which are located on ECL1Hs. (b) The side view of the narrow constriction. Here only the two subunits opposite each other are shown for clarity. The Arg103 ring has a diameter of 7.6 Å whereas the second narrowest constriction with a diameter of 13.7 Å is created by Thr48 on TM1. (c and d) The narrow constriction in ECD is a binding site for a channel blocker, DCPIB (brown stick). Shown in mesh is the cryo-EM density likely representing DCPIB viewed from the top (c) and side (d). DCPIB density plugs ECD constriction around Arg103. The LRRC8A-DCPIB structure shown here is in the expanded conformation (PDB code: 6NZZ)
In addition to the anion selectivity, the ECD motif has been shown to be a binding site for a channel blocker, DCPIB (Fig. 6c–d). The LRRC8A sample containing DCPIB in cryo-EM showed a bulky density on the C6 symmetrical axis while the equivalent sample without DCPIB (apo-state) did not [117]. Intrinsic to a compound sitting on a symmetrical axis is a technical difficulty in resolving the density. DCPIB likely binds to the ECD motif in multiple orientations and possibly with various conformations resulting in undefined density even without imposed symmetry in refinement [117]. Nevertheless, this ECD constriction likely accommodates only one DCPIB molecule with the butanoic acid moiety possibly near Arg103. The binding of DCPIB physically plugs the ECD constriction like a cork in a bottle. LRRC8 channels are also known to be blocked by ATP (1 mM) and this ATP blockade is abolished in LRRC8A Arg103Phe/LRRC8C (six Phe in the constriction) indicating that ATP likely binds to this region and the positive charge is necessary for binding of a negatively charged ATP molecule [118]. In summary, the narrow ECD constriction is the filter for anion selectivity and the binding site for channel blocker compounds. Other methods such as molecular dynamics simulations may be useful in gaining deeper insights into the binding mode. As will be discussed in the later section, the narrow constriction at ECD is the hallmark structural feature of pannexin (Panx1) and serves as the anion selectivity filter and the binding site for channel blockers via different chemical mechanisms.
Beneath the ECD is the second narrowing (~10–15 Å diameter) created by Thr44 and Thr48 from the first TMD. Consistent with the structural notion that this narrowing is a portion of the permeation pathway, the substituted cysteine accessibility (SCAM) experiment on Thr44 downregulated current levels [61]. The TMD region is hydrophobic and becomes hydrophilic toward ICL. The N-terminal ends are disordered in all of the reported LRRC8A cryo-EM structures. These N-terminal residues are known to alter conductance, ion permeability, and inactivation gating [118, 125]. For example, modification of Thr5Cys by SCAM suppresses the channel activity indicating that this region is also a part of the channel pore as in the case of Cx26 connexin channel and possibly for other large-pore channel members [118]. Furthermore, the Thr5Arg and Glu6Cys mutation increased iodide selectivity over chloride, again indicating the critical functional role of the N-terminal ends [118, 125]. Finally, Kern et al observed two conformations, which they called ‘expanded’ and ‘constricted’, within the TMD and ICL layers in the context of the C6 symmetrical arrangement. Although premature at the moment, this minor change may be a clue in understanding how channel gating occurs.
Critical structural features for functional of LRRC8D
One of the hallmark functions of the LRRC8A/LRRC8D channel is that it is more permeable to osmolytes than other subunit combinations of LRRC8. The structure of LRRC8D homomers provided some speculation about this important functional feature (Fig. 7) [120]. Despite a dimer-of-trimer subunit arrangement with the overall C2 symmetry in LRRC8D, the regions containing ECD, TMD, and ICL have pseudo-C6 symmetry as in LRRC8A (Fig. 7a–c). The ECD is narrowly constricted in LRRC8D but with a slightly wider diameter (11.5 Å) than LRRC8A (7.6–9.6 Å) (Fig. 7d–e) [120]. The equivalent residue to Arg103 in LRRC8A is Phe143 in LRRC8D and together they may also form a constriction wider in diameter than LRRC8A/LRRC8B, C, or E). Indeed, the LRRC8D Phe143Arg mutant shows comparable anion channel activity to wildtype LRRC8A/D, but not glutamate permeation, indicating that placement of Phe143 is critical for permeation of osmolytes or at least glutamate [120]. It would be meaningful to test the effect of this mutant on other osmolytes (e.g. taurine) and on the anti-cancer drug cisplatin. While it is intuitive that LRRC8B and LRRC8C harboring arginine and leucine at the equivalent position to Phe143 have less permeability to osmolytes than LRRC8D, this is not the case for LRRC8E, which contains phenylalanine at the same position. While the narrow constriction at ECD may be one of the hotspots for determining osmolyte permeability, there may likely be other unknown factors for such function. Furthermore, while the diameter of the ECD narrow constriction may be wider in an LRRC8D-containing channel, the diameter shown in the structure is not sufficiently wide for permeation of molecules such as glutamate, GABA, and taurine. Nevertheless, the field should be reminded that LRRC8D homomers do not form functional channels and that clearer answers to the above question needs to be pursued via structural and functional studies on heteromeric LRRC8 channels. As discussed later, the structures of pannexin have a similar issue where the channels are wide enough for Cl− but not ATP to pass through the similar ECD narrow constriction.
Fig. 7. Cryo-EM structure of homo-hexameric LRRC8D.
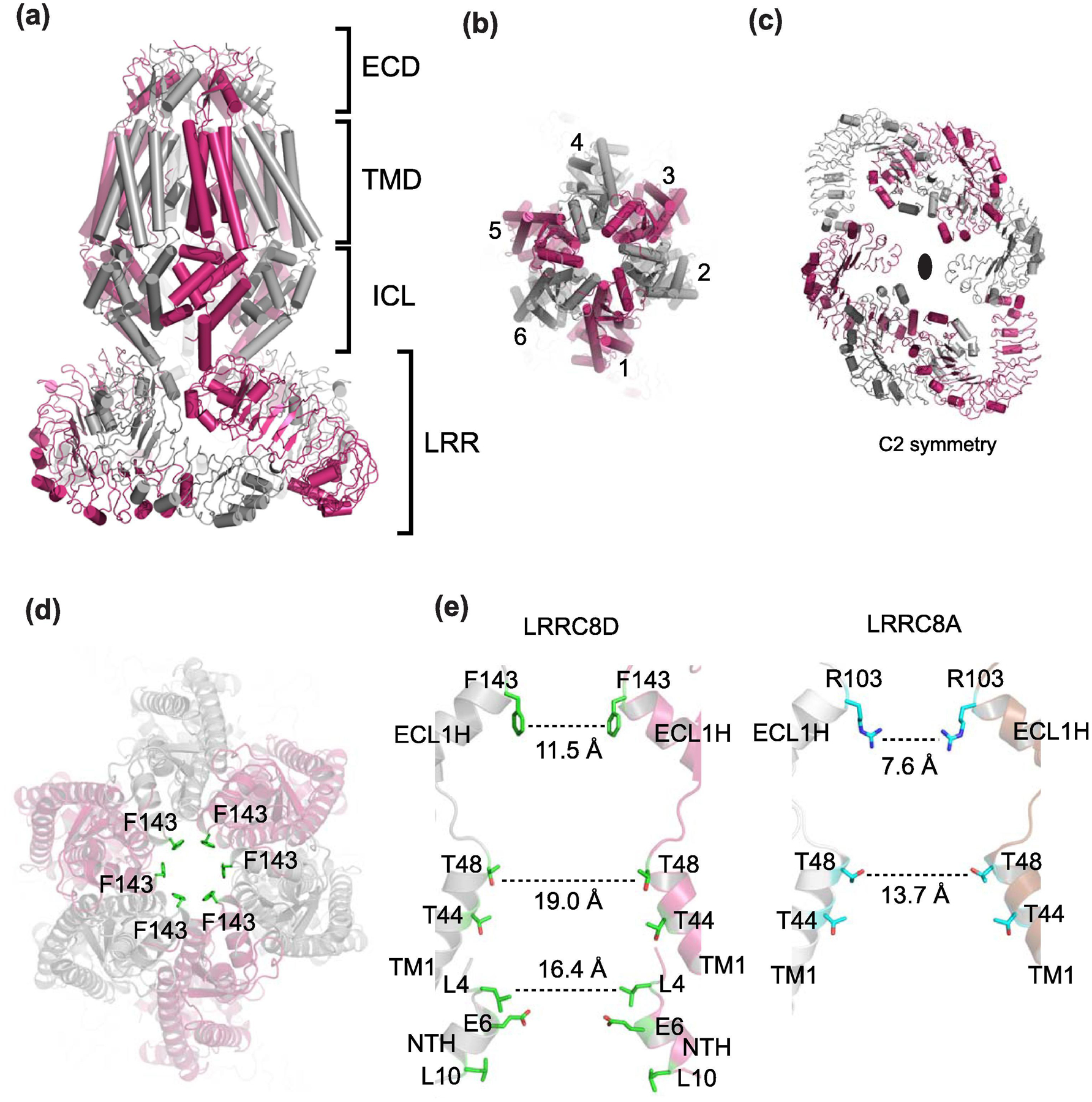
(a) Shown here is the structure of human LRRC8D (PDB code: 6M04) where alternate subunits are colored magenta and gray. The side view shows four layers, ECD, TMD, ICL, and LRR as in LRRC8A. (b) Top view without the LRR layer showing pseudo-C6 symmetry. (c) Top view of LRR showing C2 symmetry. (d) Top view showing the narrow constriction at ECD formed by Phe143 (green sticks). (e) Side view of human LRRC8D (left) and human LRRC8A (right). Here only the two subunits opposite to each other are shown for clarity. The diameters of the constriction sites at the ECD and TMD are wider in LRRC8D than LRRCA.
Pannexin channels
Discovery and general overview
The pannexin family was first reported in 2000 in a search for gap junction proteins conserved between vertebrates and invertebrates [50]. Using degenerate primers designed based on the innexin sequence, innexin-like DNAs were PCR amplified from mollusc and flatworm cDNA libraries. BLAST searches revealed that the isolated DNAs encoded proteins with weak homology to human proteins of unknown functions. Subsequent sequence analyses suggested that these human proteins contain four transmembrane helices and four conserved cysteines in the plausible extracellular region. These features are reminiscent of connexins, though no significant sequence homology exists between these two classes of membrane proteins [50, 126]. Under the assumption that these newly discovered proteins were vertebrate versions of innexins, they were dubbed “pannexins,” meaning “universal innexins.” The initial characterization using paired Xenopus oocytes supported the idea that pannexins can form gap-junctions [52]. However, later evidence suggests that glycosylation at the extracellular domain prevents this channel family from forming gap-junctions [53–56, 127].
The pannexin family comprises three different subtypes (Panx1-3). Panx1 and 2 are found in nearly all tissue types [128–132]. Expression of Panx3 is restricted to specialized areas such as skin, cartilage, heart, and osteoblast cells [133, 134]. Studies using Panx1 knockdown and knockout animals revealed that this subtype plays important roles in various pathophysiological events, such as pain sensation, blood pressure regulation, inflammasome activation, neurodegeneration, and epilepsy [135–143]. Much less is known about Panx2 and 3, though Panx2 has been linked with diabetes, focal cortical dysplasia-associated epilepsy, and neurodegeneration [137, 144, 145] and Panx3 is implicated in differentiation of keratinocytes and osteoblasts [134, 146]. Pannexins have also been shown to form heteromeric channels in vitro; however, the physiological relevance remains elusive [53, 147].
A number of important subjects are unresolved in the pannexin field. In particular, activation and inhibition mechanisms of pannexins remain controversial. This topic is elegantly covered in recent reviews [148, 149]. In the last two years, we and five other groups independently reported high-resolution cryo-EM structures of Panx1 channels (Fig. 8, Table 4) [127, 150–154]. The rapid succession of these publications reflects the growing interest in pannexin biology and the need for structural studies of pannexins to answer fundamental questions in the field. The ensemble of published structures illuminate previously unknown features of this channel, including a novel and unpredicted subunit stoichiometry, elements of the pore that comprise the permeation pathway, amino acids that control ion selectivity, and a potential mechanism for channel inhibition by carbenoxolone (CBX). Here we comprehensively summarize the findings presented in the Panx1 structures and highlight similarities and differences between the reported structures.
Fig. 8. Overall topology and extracellular gate of Panx1.

(a) Side view of the structure of human Panx1 (PDB: 6WBF). Panx1 harbors extracellular domain (ECD), transmembrane domain (TMD), and intracellular domain (ICD) layers. Sticks in the ECD and TMD represent N-linked glycosylation and bound lipids, respectively. (b) Topology of a Panx1 protomer viewed from within the plane of the membrane (left) and the schematic presentation of the structural topology (right). Disordered segments of the protein, including the intracellular loop and C-terminus, are represented by a dashed line. Caspase-7 cleaves off the last forty seven residues (scissors). (c) Close-up view of the extracellular constriction where Trp74 and Arg75 are shown as sticks (left panel). The ~9 Å diameter constriction is formed by Trp74 located on the N-terminal end of EH1. Cross-sectional view of the apo structure showing distance between Trp74 and Arg75 from neighboring protomers and potential cation-pi interactions (right panel).
Table 4.
Available structures of Pannexin in 2021
| Structure | Functional states | PDB code | Species | Oligomeric state | Solubilization conditions | Freezing buffer conditions | Reference |
|---|---|---|---|---|---|---|---|
| Panx1 3.38 Å | Open | 6UZY | Xenopus laevis | 7 | TBS + 1% LMNG | 20 mM Tris pH 8.0, 150 NaCl, 40 uM GDN | Deng et al., 2020 |
| Panx1 3.77 Å | Open | 6V6D | Human | 7 | TBS + 1% LMNG | 20 mM Tris pH 8.0, 150 NaCl, 40 uM GDN | Deng et al, 2020 |
| Panx1 4.10 Å | Open | 6M66 | Human | 7 | HBS + 2% DMNG | 50 mM HEPES pH 7.5, 150 mM NaCl, 0.05 mM LMNG | Jin et al., 2020 |
| Panx1 (EE) 3.60 Å | Open | 6M67 | Human | 7 | HBS + 2% DMNG | 50 mM HEPES pH 7.5, 150 mM NaCl, 0.05 mM LMNG | Jin et al, 2019 |
| Panx1 (EE + CBX) 4.60 Å | Inhibited | 6M68 | human | 7 | HBS + 2% DMNG | 50 mM HEPES pH 7.5, 150 mM NaCl, 0.05 mM LMNG | Jin et al., 2020 |
| Panx1 ΔLC 3.02 Å | Closed | 6VD7 | Xenopus laevis | 7 | PBS + 1% C12E8 | 20 mM Tris pH 8.0, 150 NaCl, 1 mM EDTA, MSP2N2, soybean polar lipids | Michalski et al., 2020 |
| Panx1 3.10 Å | Open | 6LTO | Human | 7 | TBS + 2% DDM | 25 mM Tris pH 8.0, 150 NaCl, 0.01% LMNG | Mou et al., 2020 |
| Panx1-cleaved 3.10 Å | Open | 6LTN | Human | 7 | TBS + 2% DDM | 25 mM Tris pH 8.0, 150 NaCl, 0.01% LMNG | Mou et al, 2019 |
| Panx1 3.20 Å | Closed | 6M02 | Human | 7 | TBS + 1% digitonin | 20 mM Tris pH 8.0, 150 NaCl, 0.1% digitonin | Qu et al, 2019 |
| Panx1 2.83 Å | Closed | 6WBF | Human | 7 | TBS + 1% GDN | TBS + 0.01% GDN | Ruan et al, 2020 |
| Panx1-cleaved 2.97 Å | Open | 6WBG | Human | 7 | TBS + 1% GDN | TBS + 0.01% GDN | Ruan et al, 2020 |
| Panx1-cleaved +CBX 4.39 Å | Inhibited | 6WBI | Human | 7 | TBS + 1% GDN | TBS + 0.01% GDN | Ruan et al, 2020 |
| Panx1 N255A hemichannel 2.86 Å | n/a | 6WBM | Human | 7 | TBS + 1% GDN | TBS + 0.01% GDN | Ruan et al, 2020 |
| Panx1 N255A Gap junction 2.83 Å | n/a | 6WBN | Human | 7; head-to-head gap junction 14-mer | TBS + 1% GDN | TBS + 0.01% GDN | Ruan et al, 2020 |
| Panx1 ΔN ΔC 6.01 Å | n/a | 6WBK | Human | 7 | TBS + 1% GDN | TBS + 0.01% GDN | Ruan et al, 2020 |
| Panx1 ΔN ΔC +CBX 5.13 Å | Inhibited | 6WBL | Human | 7 | TBS + 1% GDN | TBS + 0.01% GDN | Ruan et al, 2020 |
Panx1 proteins used for the cryo-EM studies
To date, there are 16 Panx1 structures deposited into the Protein Data Bank (PDB). Resolutions of these structures range from 2.8 to 6.0 Å. The Panx1 structures were obtained from proteins purified from three different expression hosts (insect, mammalian, or yeast cells) and of two different protein orthologues (human and Xenopus laevis (66% identical to human)). Despite such variations, all structures adopt a similar conformation; protomers of Panx1 from each group can be superimposed with small differences. This highlights that Panx1 structures are conserved among species and various recombinant systems. Biochemically, Panx1 proteins subjected to cryo-EM were obtained with similar strategies (Fig. 8, Table 4). All proteins were purified with C-terminal affinity tags with or without GFP/mRuby. This C-terminal tagging strategy is in line with a previous study demonstrating that modifications to the N-terminus could alter Panx1 channel activity [155]. Our group used MSP2N2 soybean polar lipid nanodiscs to image Panx1 samples, whereas others used detergents such as lauryl maltose neopentyl glycol (LMNG), digitonin, or glyco-diosgenin (GDN). Nevertheless, Panx1 appears to behave similarly in all the above listed conditions.
Similarities and Differences between the available Panx1 Structures
All Panx1 structures revealed that the channels assemble as heptamers with seven-fold symmetry down the axis of the pore (Fig. 8 and 9). This oligomeric state was consistent among protein samples in detergents and lipid nanodiscs, suggesting that the heptameric assembly is the dominant form of Panx1 channels. This is in contrast to the hexameric assembly suggested based off western blot, photo bleaching, concatemeric channels, and negative-stain EM [156–158]. However, distinguishing between hexamer and heptamer using such techniques is technically challenging because a small experimental error or variation could influence the interpretation of the data. The structure of each Panx1 protomer contains four transmembrane helices linked together by extracellular and intracellular domains (ECD and ICD) and cytoplasmic-oriented N- and C-termini (Fig. 8b). The arrangement of the four transmembrane helices are similar to connexin, innexin, and LRRC8 channels despite almost no sequence homology, and, these channels all share some structural similarity in their extracellular and intracellular domains. In particular, the Panx1 ECD and ICD are similar to that of innexin and LRRC8 channels (excluding the LRR domain of LRRC8). However, there is no structural similarity between Panx1 and CALHM in any domain layer, as described above (Fig. 3).
Fig. 9. Features of Panx1 structures.
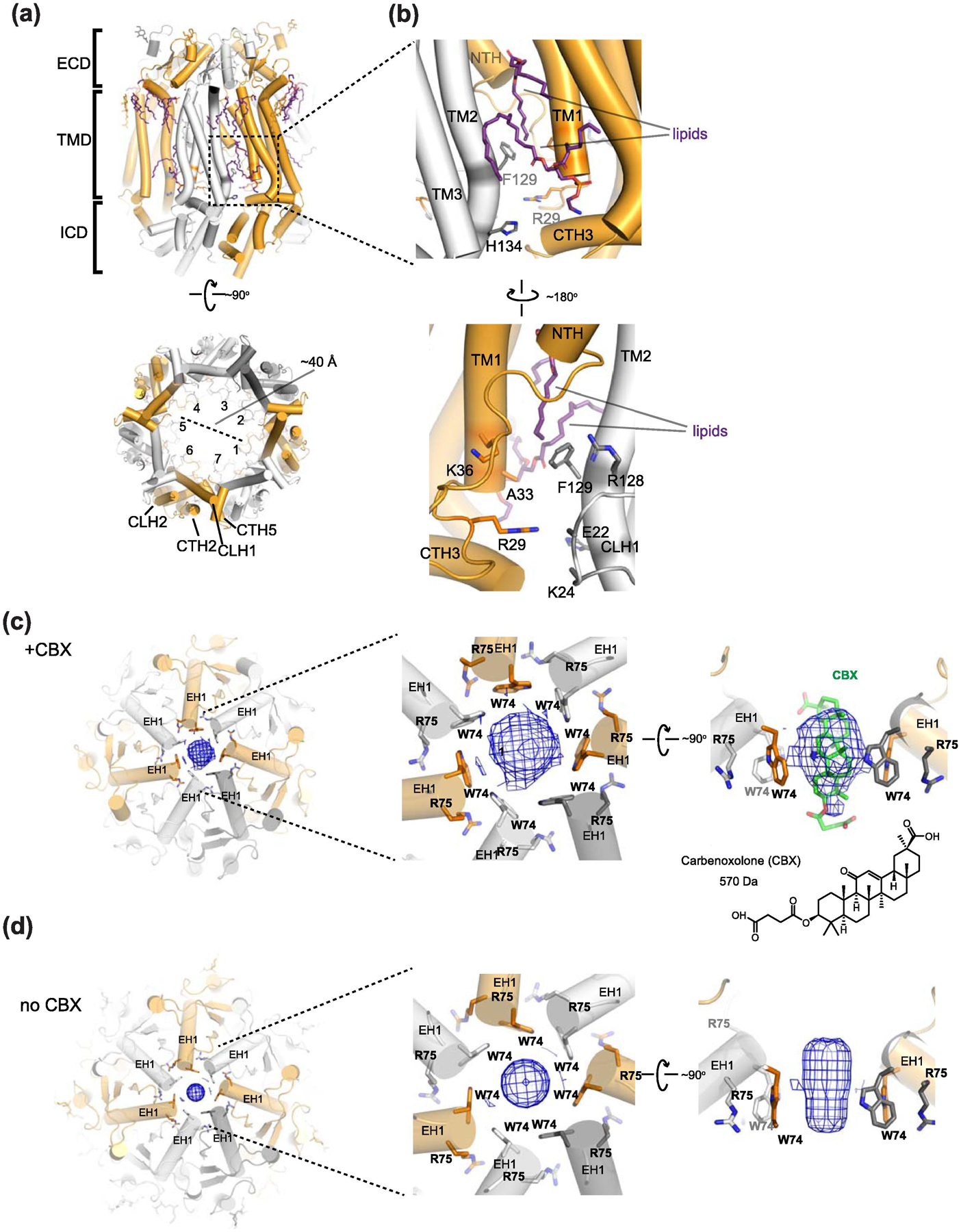
(a) The human Panx1 structure (PDB: 6WBG) viewed from the side of the membrane plane (top) and the cytoplasm (bottom). The cytoplasmic face of the channel pore is wide open with the measured diameter of ~40 Å. Lipids, N-linked glycosylation, and the side tunnel residues are shown in sticks (top). The dotted square is the location of the side tunnel. (b) Close-up views of the side tunnel at the inter-protomer interfaces viewed from the outside (top) and inside (bottom) of the central pore. The side tunnel residues are located on TM1, TM2, and the loop between TM1 and NTH. The ordered lipids (purple sticks) are located at the entrance of the side tunnel. (c-d) Top-down view of the extracellular face of caspase-7 cleaved human Panx1 with CBX (‘+CBX’; left and middle panel c) (PDB: 6WBG) and without CBX (no CBX, left and middle panel d) (PDB: 6WBI). Note that densities (blue mesh) in the ECD narrow constriction are observed in both no CBX and +CBX structures. Cross-sectional view of the ‘+CBX’ structure (right panel c). Side view of the density in no CBX showing clear density around Trp74 (right panel d). Note that the density is not sufficiently resolved to determine the presence of CBX although the molecular model (green sticks in panel c) was arbitrarily placed in the published PDB coordinates.
The permeation pathway of Panx1 has a wide cytoplasmic vestibule with a diameter of 30–40 Å, which gradually narrows to a ~9 Å constriction formed by Trp74 located in the first extracellular domain (Fig. 8c). Cation-pi interactions between Arg75 and Trp74 from neighboring subunits confer a net positive charge in the central pore, which presumably facilitates anion selectivity of Panx1 (Fig. 8c). Indeed, mutations in Trp74 and Arg75 alter the selectivity of Panx1. For example, the Arg75Ala mutation diminished chloride selectivity while a charge reversal mutant, Arg75Glu, facilitated sodium permeability [127, 152]. While these positions appear to define selectivity of the ion conducting form of Panx1, it would be interesting to examine whether residues within this constriction also affect selectivity for larger molecules such as ATP.
Globally, protomers of Panx1 reported from each publication can be aligned with only minor differences, especially within the transmembrane and extracellular domains along with the helical portion of the intracellular domain. However, there are differences in the conformations of the N-terminal domains. Situated within the channel’s wide cytoplasmic vestibule, four structural studies show a partially resolved N-terminus oriented towards and extending into the cytoplasm [150, 152–154]. Ruan et al modeled the N-terminus funneling into a constriction of the permeation pathway oriented towards the extracellular gate [127]. Notably, the other EM maps of the human Panx1 (e.g. EMD-21071) also showed similar, albeit weak, helix-like densities around the same positions. This pore-lining arrangement of the N-terminal helix is consistent with other large pore channels such as connexins and LRRC8. However, the EM densities of the N-terminal helix and the linker between this helix and the first transmembrane helix are weak and it is challenging to accurately build a model for this region. Interestingly, the EM maps of the frog Panx1 lack such helix-like density around the permeation pathway [150, 152]. Differences between these orientations might be attributed to a highly flexible and mobile N-terminus.
The C-terminus of Panx1
One unique and well-established gating mode of Panx1 involves the caspase-mediated cleavage of the C-terminal domain [59, 157, 159–161]. During apoptosis or pyroptosis, activated-caspases cleave a specific motif within the C-terminus, liberating the channel from its autoinhibitory C-terminal domain and allowing release of ATP [59, 157, 159] (Fig. 8b). This mode of Panx1 activation is “quantized,” meaning that the channel conductance and the open probability increase in a stepwise manner as each of the seven C-termini gets cleaved individually [157]. Recently, the Bayliss group unambiguously demonstrated the caspase-mediated release of the C-terminal autoinhibitory region and subsequent ATP release using an elegant in vitro reconstitution system [161]. However, the mechanism of C-terminal region dependent autoinhibition remains unclear. While it is alluring to consider that the C-terminus plugs the pore under basal conditions, such a mechanism is unlikely because a pannexin channel lacking the autoinhibitory region can be closed (e.g. Δ365–426) [162]. Perhaps it is possible that the C-terminal autoinhibitory region prevents the movement of another part of the channel (e.g. intracellular loop or N-terminus), which may directly control channel opening and closing.
Ideally, comparison of the full-length and the C-terminally truncated Panx1 would be instructive to understand the mechanism of autoinhibition. Unfortunately, however, none of the currently available full-length Panx1 maps shows interpretable density beyond the caspase cleavage site. Though Ruan et al claim that the full-length map obtained without imposing C7 averaging (“apoPANX(WT)”) showed a cloud of pore-covering density that disappeared in the map obtained from a caspase-treated Panx1 (“PANX1(ΔCTT)”), questions remain as to how such a loosely structured region can regulate channel gating in a quantized manner [127]. It is important for the field to carefully inspect the “C-terminal cloud” once this map becomes available at EMDB. In a different study, Mou et al modeled a C-terminal helix (residues 410–424) in a density within the pore [153]. The pore-lining C-terminus is supported by crosslinking and SCAM analyses, which suggest that the very C-terminus is sensitive to MTS reagents and may form part of the permeation pathway [160, 163]. However, this model needs further verification not only because the connectivity was invisible, but also because the equivalent density was interpreted as the N-terminal helix by Ruan et al [127].
Both Mou et al and Ruan et al reported nearly indistinguishable models of Panx1 before and after caspase cleavage (RMSD < 0.8 Å), suggesting that there was no global conformational change upon truncation of the C-terminal residues. At least the permeation pathway seemed unchanged after the removal of the C-terminus. Likewise, the map of the full-length frog Panx1 lacked density for the C-terminal region and was indistinguishable with the truncated version (RMSD = 0.49 Å) [150, 152]. Together, these structures suggest that liberating the C-terminus does not result in a detectable global conformational change, at least under the tested biochemical and cryo-EM conditions. As discussed below, this makes it challenging to assign this channel state as “open” or “closed.”
Are they open or closed?
All presented structures of Panx1 appear to adopt the same conformation, with the narrowest constriction of ~9 Å formed by Trp74 (Fig. 8). This permeation pathway is large enough for hydrated anions to go through, which indicates that all presented structures may represent an open conformation. However, there is general disagreement about whether this conformation represents an open or closed state of the channel, with three reports suggesting open [127, 150, 153], one suggesting closed [154], and two with no affirmative statement [151, 152]. The general confusion surrounding this is difficult to mediate as experimental data can support both hypotheses. Here, we broadly summarize the evidence and arguments that support whether the current Panx1 structures represent open or closed channel states.
On the basis of the published literature, the most straightforward experimental conditions that promote Panx1 opening is the inclusion of stimulating ions like Ca2+ and K+ [54, 57, 158]. As reported by Ruan et al, however, Ca2+ and K+ had no effect on Panx1 conformation [127]. This is corroborated by other reports in which K+ was incapable of activating Panx1 channels [148, 151]. These data indicate that Ca2+ and K+ may not directly act on Panx1 to promote channel opening.
Another straightforward means for trapping Panx1 in its active form is to remove the autoinhibitory C-terminus [161]. Single channel recordings of the C-terminally cleaved Panx1 dwell in both open and closed states, suggesting that the conformation of Panx1 is not necessarily rigid even when liberated from its C-terminus, and that the channel has the ability to gate when lacking this domain [158, 164–166]. This explains why removing the C-terminus by proteolysis or genetic truncations did not alter the overall conformation of Panx1 as observed under the cryo-EM conditions tested; the current structures of the full-length and truncated channel could both be representing either the closed or open conformations. It is possible that alternate conformations of Panx1 are transient and difficult to capture by 3D classification in single particle cryo-EM analyses or require additional factors (i.e. phosphorylation).
Our frog Panx1 structure was obtained using a construct that lacks the C-terminal region and a part of the intracellular loop [152]. This construct was non-conductive in HEK293 cells even under strong depolarization, suggesting it is incapable of conducting ions. Curiously, it adopted the same pore conformation as the full-length structures, arguing that all Panx1 models must represent a closed channel conformation. This may also indicate a role for the unstructured intracellular region in channel gating, which is consistent with reports of tyrosine phosphorylation in this domain promoting channel opening [167, 168].
As an ATP release channel, it is expected that an open-channel conformation would necessarily have a pore diameter equal-to or larger than the size of ATP. Given that the extended ATP is ~14 Å in length, one would predict this molecule is too large to pass through a 9 Å diameter pore. Additionally, recent metabolomics data indicate that Panx1 is also capable of permeating 20+ different metabolites [169]. These studies indicate that the current Panx1 structures do not represent a conformation in which ATP and other metabolites can permeate. As mentioned above, C-terminally cleaved Panx1 fluctuates between open and closed states, suggesting that the channel gate is not necessarily constricted to the pore size reported in the cryo-EM structures regardless of whether it is an open or closed conformation. It is possible that ATP and other metabolites permeate through an open state not yet captured by cryo-EM 3D reconstruction.
Ruan et al proposed a novel mechanism in which small ions such as chloride enter the pore through side “tunnels” (Fig. 9a) [127]. This mechanism suggests that while the C-terminus remains intact, chloride ions can partially dehydrate and transit through small openings at the subunit interface formed by TM1, CTH3, TM2, and CLH1 (Fig. 9b). While these tunnels are too small for larger molecules like ATP to permeate, once the C-terminus gets cleaved by caspases, the lower part of the main pore becomes accessible to the cytoplasm and available for larger molecules to access the pore. This mechanism was supported by their electrophysiology experiments where enlarging the most constricted region by mutating Arg29 to Ala increased the current density. The authors also claimed that the linker harboring Arg29 is flexible and acted as a “gate” based on molecular dynamics simulations. However, this mechanism needs further and rigorous verifications. First, it is unclear how the unstructured C-termini can completely plug the wide (40 Å in diameter) mouth of the cytoplasmic side of the pore (Fig. 9a) without having an aperture as small as the side tunnels. While a similar mechanism has been proposed for other channels such as P2X receptors, the cytoplasmic plug is well-ordered and readily visible in those structures [170, 171]. Second, right next to the proposed basic tunnel, there is a similarly-sized acidic opening near Glu22. If the tunnel constricted by Arg29 is large enough for small anions to permeate, would not small cations permeate through the opening next door as well? Third, the side tunnels are situated near the modeled lipids (Fig. 9b). It is possible that these openings may not exist in the native environment where the lipid bilayer may cover the side tunnels. Fourth, Panx1-mediated ATP release has been reported with channels activated via α1-adrenergic receptor stimulation [138]. Because the Panx1 C-terminus is intact under these conditions, the cytoplasmic entry to the permeation pathway should be completely blocked if the proposed mechanism is correct. Would ATP permeate through the side tunnels under these conditions? Besides ATP, molecules larger than chloride, such as methanesulfonate or thiocyanate have also been shown to permeate Panx1 with an intact C-terminus [166, 172]. And last, the mutagenesis and truncation studies by Ruan et al could be interpreted in a way that the N-terminal region plays important roles in Panx1 channel gating, as previously suggested [155]. Additional experiments are needed to fully flesh out mechanisms surrounding these side-tunnels.
In summary, whether the published structures of Panx1 represent a closed or open state remains unclear. There is currently no structure representing an undoubtedly closed or open state. Alternate conformations may be transient, may require activating stimuli that are difficult to manipulate biochemically, or may involve domains of the protein that are inherently disordered and flexible.
Potential mechanism of action of CBX-dependent Panx1 inhibition
CBX is a nonselective Panx1 inhibitor that blocks other large-pore channels including connexin, innexin, and LRRC8 [47, 53, 173]. Nevertheless, it has been the most popular compound for studying Panx1 function due to its relatively high potency (IC50 value of ~1 μM). An exhaustive mutational analysis previously identified Trp74 as a determinant which governs inhibition by CBX, suggesting this residue may form a crucial part of the CBX binding site, although other residues have also been postulated to be involved [174].
Two groups reported maps of Panx1 obtained in the presence of CBX [127, 151]. In both cases, a blobby cryo-EM density occupies the cross-sectional space encircled by Trp74, at the narrowest constriction of the permeation pathway located in the extracellular region (Fig. 9c). However, inspection of other deposited Panx1 cryo-EM maps under similar contour levels revealed that the majority of maps contain a pronounced central density encircled by Trp74 even when CBX was not included in the sample (Fig. 9d). One possible explanation is that some endogenous cellular metabolites during recombinant expression may have been carried over and that these ‘apo’ structures actually represent the ‘blocked’ state. Interestingly, the Panx1 sample prepared in nanodiscs (Table 4) has little or no density in the middle suggesting that detergents may be occupying this site. It is challenging to obtain high quality data to resolve the mechanism of CBX inhibition because CBX may have a number of distinct modes of interaction at the central pore. Furthermore, the density only covers ~50% of the arbitrarily placed CBX molecule in the PDB coordinate; thus, the actual occupancy of CBX may be low (Fig. 9d). Overall, while CBX may block the channel by plugging the narrow constriction around Trp74, the structural evidence for this mechanism is insufficient.
The potential channel ‘plugging’ mechanism of Panx1-CBX is similar to the inhibition of LRRC8A/VRAC by the compound DCPIB (Fig. 6c and d) [117]. Curiously, other Panx1 inhibitors, including probenecid, glybenclamide, and ATP, have also been shown to act through a mechanism involving Trp74 and nearby residues, [174, 175]. This might suggest a common mechanism of inhibition where blocker molecules have a tendency to plug the Panx1 pore and prevent the passage of ions. Alternatively, CBX and other inhibitors may bind somewhere else and prevent the movement important for channel gating. Obtaining a higher resolution structure of Panx1 complexed with an inhibitor without imposing C7-averaging may reveal finer details of chemistry that dictates binding.
CALHM channels
Discovery of CALHM and CALHM family
The human calhm1 gene was originally identified as a gene located on the Alzheimer’s disease (AD) locus on 10q24.33 and whose transcript is preferentially expressed in the hippocampus [63]. It was suggested that a single nucleotide polymorphism that results in the expression of a Pro86Leu mutant is linked to late onset AD and shown to promote accumulation of Aβ peptide when over-expressed in the SwAPP695-N2a cell line [63]. There are conflicting reports that show no association between the CALHM1 Pro86Leu substitution to AD [176–179]. In addition, there is a report indicating that the Pro86Leu mutation could result in an earlier age-of-onset for Alzheimer’s disease particularly in carriers of the APOE ε4 allele [180]. Other reported rare variants linked to AD include CALHM1 Arg154His [181, 182].
The CALHM protein family comprises six members in vertebrates (CALHM1-6) and one in C. elegans (CLHM-1). CALHM1 and CLHM-1 have been shown to form voltage-gated multimeric channels permeable to small ions with weak selectivity (Ca2+ ≫ Na+ ≈ K + > Cl−) and larger ions such as ATP [64, 65, 183]. The ion channel activity of CALHM1 is inhibited by ruthenium red (RuR), Gd3+, Zn2+, and 2-APB, and a physiological concentration of extracellular Ca2+ (~1.8 mM), and mildly potentiated by niflumic acid [64]. The narrowest point of the CALHM1 channel was measured to be 14.2 Å based on the permeability of differently-sized fluorescence dyes [184]. CALHM1, therefore, is predicted to form a large-pore channel like connexin, pannexin, innexin, and LRRC8. Thus far, CALHM1, CALHM1/3 heteromers, and CLHM-1 have been consistently demonstrated to form voltage-dependent ion channels that are regulated by extracellular calcium [63, 64, 183, 185, 186]. Similar calcium dependent ion channel activity has been reported for CALHM2 [187] although the majority of studies report no ion channel activity for CALHM2 [67, 93, 185]. Furthermore, CALHM4 and 6 have been shown not to have apparent voltage-gated or calcium gated ion channel activity [93]; however, there are implications that they are involved in biological processes. For example, CALHM6 is involved in interferon gamma production by NK cells [188] and CALHM4 has recently been shown to be highly expressed in placental tissue [93]. No functional insight or biological role of CALHM5 is known at this point. More studies need to be conducted to explore specific functions of CALHM2, 4, 5, and 6. For example, there is a possibility that activation of these proteins may require other unidentified factors such as ligands, lipids, post-translational modifications, and proteolytic cleavage for their channel activities.
CALHMs and purinergic signaling
Soon after its discovery, CALHM1 was reported to be expressed in primate taste buds [189]. This was followed by a breakthrough study that demonstrated the expression of CALHM1 in type II gustatory cells and the role of CALHM1 proteins in perception of sweet, bitter and umami tastes by effluxing ATP and mediating subsequent purinergic signaling [65]. Binding of a tastant to a G protein-coupled receptor (GPCR) in the type II cells triggers a signaling cascade resulting in the activation of phospholipase Cβ2 and the production of inositol 1,4,5-triphosphate (IP3) that binds to the type 3 IP3 receptors expressed in the endoplasmic reticulum (ER). The subsequent increase in intracellular Ca2+ activates TRPM5 channels in the plasma membrane leading to depolarization [190, 191], which in turn promotes CALHM1-mediated ATP release. The transmitted ATP activates the afferent gustatory neurons via binding of ATP-gated ion channels, P2X2/3 receptors [192]. Later studies showed that the CALHM1/3 heteromeric channel functions as an ATP release channel for GPCR-independent sensing of sodium taste [193–195] as well as potentially for sensing of fat taste mediated by CD36 [196]. Further studies suggested that the CALHM1/3 heteromer is the dominant form in taste buds, as co-expression of both CALHM1 and 3, but not expression of individual isoforms, mimicked the electrophysiological responses of these cells [185].
CALHM1 has also been reported as a component of ATP release machinery in the bladder [197] and airway epithelial cells in the lung [198]. Although the ion channel activities of CALHM1 have been well-characterized in the brain [63, 182], the physiological relevance of CALHM1-mediated ATP release in brain functions remains elusive. There is one study reporting that conditional knock-out of CALHM2 in astrocytes decreased spine density, long term potentiation, and depression-like behaviour, which were rescued by ATP replenishment [199]. CALHM2 has been shown to be permeable to ATP by a cell-based ATP detection assay, although not as robustly as CALHM1 [200]. It is also interesting to note that CALHM1 has been shown to be co-localized with mitochondria in one study, indicating the possibility that the major source of ATP efflux may come from mitochondria rather than the cytoplasm [201].
Architecture of CALHM proteins
Until recently, the structures of CALHM family proteins were unknown. Furthermore, the lack of sequence similarity to other proteins with known structures severely limited our mechanistic understanding of how this protein family functions. Within the past year, a veritable explosion of cryo-EM data resulted in twenty structures representing CALHM1, 2, 4, 6, several chimeras of CALHM1-2, and CLHM1, under various buffer conditions, being deposited into the PDB [67, 93, 187, 200, 202–204] (Fig. 10–11, Table 5). These studies provided the fundamental molecular blueprint to answer functional questions, yet also created a context and foundation for more questions.
Fig. 10. Basic architecture of CALHM.
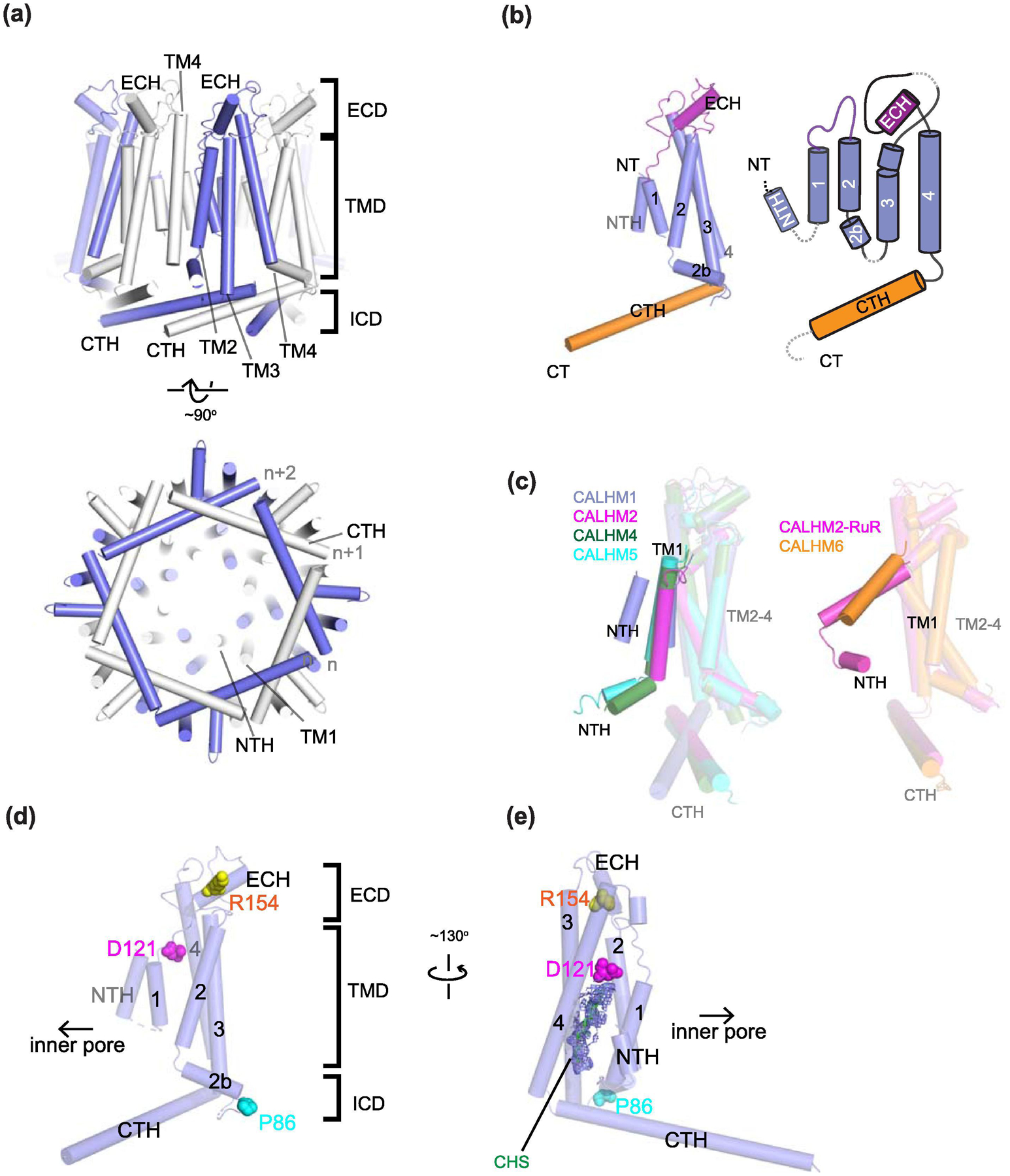
(a) Structure of CALHM1 from zebrafish (PDB ID: 6LYG) as an example. The structure is viewed from the plane of the membrane (upper panel) and the cytoplasm (lower panel). CALHM has an extracellular domain (ECD), a transmembrane domain (TMD) and an intracellular domain (ICD). Note that TMD2 and TMD4 in neighboring subunits form contacts. In the cytoplasm, the CTHs from a neighbor (n+1) and a neighbor of the neighbor (n+2) subunits interact with each other to stabilize the octameric arrangement. (b) Topology of a CALHM subunit. Each subunit contains four transmembrane helices (TM1-4), the N-terminal helix (NTH) in the middle of the central cavity, and ~40 residue long cytoplasmic helix (CTH). The remainder of the C-terminal structures are omitted for clarity. (c) Superposition of CALHM1 (slate), CALHM2 (magenta), CALHM4 (green), and CALHM5 (cyan) protomers (left panel) and of CALHM2 bound to RuR (magenta) and CALHM6 (orange) protomers (right panel) to illustrate that the arrangement of TM1 and NTH (highlighted) are different between the two groups. The NTH arrangement in CALHM1 differs from that in CALHM4 and 5 (left panel). The arrangement of TM2-4 is similar in all CALHMs. (d) CALHM1 viewed from the extracellular region to show locations of Asp121 (magenta sphere) on TM3, Arg154 (yellow sphere) in the extracellular domain, and Pro86 (cyan sphere) in the cytoplasmic loop between TM2 and 3. The residue numbers are based on human CALHM1. (e) Density likely representing cholesteryl hemisuccinate (CHS; green stick and density in slate mesh) is present around Asp121 in the structure of CALHM1 from killifish (PDB code: 6LMT).
Fig. 11. Oligomeric assembly of CALHM proteins.
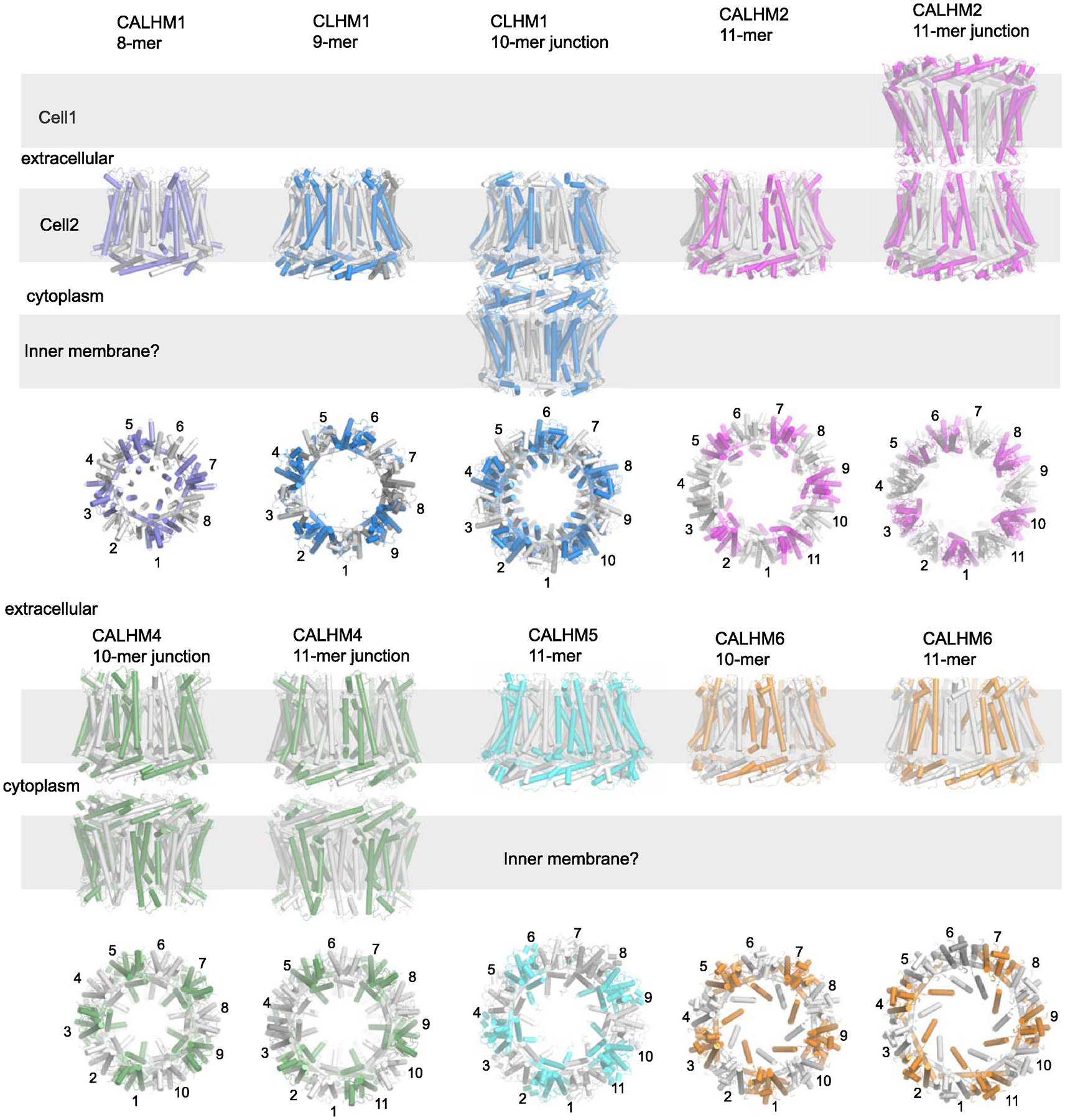
Structures of CALHM1, 2, 4, 5, and 6 and CLHM1 to illustrate variable numbers of oligomeric assembly. The PDB IDs used here are 6VAM, 6LMV, 6LOM, 6VAK, 6VAI, 6YTK, 6YTL, 6YTV, and 6YTX for CALHM1, CLHM1 (9-mer), CLHM1 (10-mer junction), CALHM2, CALHM2 (gap junction), CALHM4 (10-mer junction), CALHM4 (11-mer junction), CALHM6 (10-mer junction), and CALHM6 (11-mer junction), respectively.
Table 5.
Available structures of CALHM family proteins in 2021
| Structure | Functional state | PDB code | Species | Oligomeric state | Solubilization conditions | Freezing buffer conditions | Reference |
|---|---|---|---|---|---|---|---|
| CALHM1 2.66 Å map | open | 6LMT | (Oryzias latipes) Killifish | 8 | 1% DDM + 0.2% CHS | 50 mM Tris pH 8.0, 150 mM NaCl, 2 mM DTT, 0.05% GDN | Demura et al., 2020 |
| CALHM1 3.10 Å map | active | 6LYG | Danio rerio (Zebrafi sh) | 8 | 1% DDM + 0.2% CHS. Washed afterwards in 0.025% DDM and 0.005% CHS | 20 mM Tris-HCl pH 7.5, 200 mM NaCl, 0.0063% GDN | Ren et al, 2020 |
| CALHM1 3.63 Å map | Likely open | 6VAM | Chicken (Gallus gallus) | 8 | 1% C12E8, 1 mM EDTA, then washed in 0.01% C12E8, 1mM EDTA | 20 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 1 mM EDTA, MSP2N2, soybean polar lipid | Syrjanen et al., 2020 |
| CALHM2 3.30 Å map | Active or open | 6UIV | Human | 11 | 1% digitonin | 20 mM Tris-HCl pH 8.0,150 mM NaCl, 0.1% digitonin, EDTA | Choi et al, 2019 |
| CALHM2 3.40 Å map | open | 6LMU | Human | 11 | 1% DDM + 0.2% CHS, then washed in 0.06% digitonin | 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM DTT, PMAL-C8 | Demura et al., 2020 |
| CALHM2 3.48 Å map | n/a | 6VAK | Human | 11 | 1% C12E8, 1 mM EDTA, then washed in 0.01% C12E8, 1mM EDTA | 20 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 1 mM EDTA, MSP2N2, soybean polar lipid | Syrjanen et al., 2020 |
| CALHM2 3.68 Å map | n/a | 6VAI | Human | Dimer of undecamer (gap junctions form with extracellular domains in head-to-head contact.) | 1% C12E8, 1 mM CaCl2, then washed in 0.01% C12E8, 1 mM CaCl2 | 20 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 1 mM CaCl2, MSP2N2, soybean polar lipid | Syrjanen et al., 2020 |
| CALHM2-RuR 2.70 Å map | inhibited | 6UIW | Human | 11 | 1% digitonin | 20 mM Tris-HCl pH 8.0,150 mM NaCl, 0.1% digitonin, 1.5 mM RuR | Choi et al, 2019 |
| CALHM2-gap-choi 3.50 Å map | possibly inhibited | 6UIX | Human | Dimer of undecamer (gap junction with extracellular domains in head-to-head contact) | 1% digitonin | 20 mM Tris-HCl pH 8.0,150 mM NaCl, 0.1% digitonin, 1mM EDTA | Choi et al, 2019 |
| CALHM2 | n/a | n/a | Human | 11 and 12. Primarily 11. Also dimers of undecamers and dimers of dodecamers. From the 2D classes, it appears that the gap junctions form with extracellular domains in apposed contact. | 1% LMNG, 0.5mM CaCl2 | 10 mM HEPES pH 7.6, 150 mM NaCl, 2 mM CaCl2, 50 μM GDN | Drozdzyk, Sawicka et al, 2020 |
| CALHM4 4.24 Å map | n/a | 6YTO | Human | Dimer of decamer (gap junction with cytoplasmic domains in tail-to-tail contact) | 1% LMNG, 0.5mM CaCl2 | 10 mM HEPES pH 7.6, 150 mM NaCl, 2 mM CaCl2, 50 μM GDN | Drozdzyk, Sawicka et al, 2020 |
| CALHM4 4.07 Å map | n/a | 6YTK | Human | Dimer of decamer (gap junction with cytoplasmic domains in tail-to-tail contact) | 1% LMNG, 0.5mM CaCl2, replaced by 5 mM EGTA after protein extraction. | 10 mM HEPES pH 7.6, 150 mM NaCl, 2 EGTA, 50 μM GDN | Drozdzyk, Sawicka et al, 2020 |
| CALHM4 3.69 Å map | n/a | n/a but map deposited in the EMD (as an additional map under 10919) | Higher resolution map of an undecameric channel from a localized reconstruction | 1% LMNG, 0.5mM CaCl2, replaced by 5 mM EGTA after protein extraction. | 10 mM HEPES pH 7.6, 150 mM NaCl, 2 EGTA, 50 μM GDN | Drozdzyk, Sawicka et al, 2020 | |
| CALHM4 4.07 Å map | n/a | n/a but map deposited in the EMD (as an additional map under 10920) | Higher resolution map of an decameric channel from a localized reconstruction | 1% LMNG, 0.5mM CaCl2 | 10 mM HEPES pH 7.6, 150 mM NaCl, 2 mM CaCl2, 50 μM GDN | Drozdzyk, Sawicka et al, 2020 | |
| CALHM4 3.92 Å map | n/a | n/a but map deposited in the EMD (as an additional map under 10921) | Higher resolution map of an undecameric channel from a localized reconstruction | 1% LMNG, 0.5mM CaCl2 | 10 mM HEPES pH 7.6, 150 mM NaCl, 2 mM CaCl2, 50 μM GDN | Drozdzyk, Sawicka et al, 2020 | |
| CALHM4 4.02 Å map | n/a | 6YTQ | Human | Dimer of undecamer (gap junction with cytoplasmic domains in tail-to-tail contact) | 1% LMNG, 0.5mM CaCl2 | 10 mM HEPES pH 7.6, 150 mM NaCl, 2 mM CaCl2, 50 μM GDN | Drozdzyk, Sawicka et al, 2020 |
| CALHM4 3.82 Å map | n/a | 6YTL | Human | Dimer of decamer (gap junction with cytoplasmic domains in tail-to-tail contact) | 1% LMNG, 0.5mM CaCl2 replaced by 5 mM EGTA after protein extraction. | 10 mM HEPES pH 7.6, 150 mM NaCl, 2 mM EGTA, 50 μM GDN | Drozdzyk, Sawicka et al, 2020 |
| CALHM5 2.80 Å map | 7D61 | Human | 11 | 1% DDM supplemented with 0.1% CHS | 25 mM HEPES pH 7.5, 150 mM NaCl, EDTA MSP1E3D1 nanodisc protein and soybean polar lipid |
Liu et al, 2020 | |
| CALHM5 2.94 Å map | 7D65 | Human | 11 | 1% DDM supplemented with 0.1% CHS | 25 mM HEPES pH 7.5, 150 mM NaCl, Ca2+ MSP1E3D1 nanodisc protein and soybean polar lipid |
Liu et al, 2020 | |
| CALHM5 2.61 Å map | 7D60 | Human | 11 | 1% DDM supplemented with 0.1% CHS | 25 mM HEPES pH 7.5, 150 mM NaCl, ruthenium red MSP1E3D1 nanodisc protein and soybean polar lipid |
Liu et al, 2020 | |
| CALHM5 | n/a | n/a (2D classes) | Human | 10, 11, 12, 13 | 1% DDM supplemented with 0.1% CHS | In detergent | Liu et al, 2020 |
| CALHM6 4.39 Å map | n/a | 6YTV | Human | 10 | 1% LMNG, 0.5mM CaCl2 | 10 mM HEPES pH 7.6, 150 mM NaCl, 2 mM CaCl2, 50 μM GDN | Drozdzyk, Sawicka et al, 2020 |
| CALHM6 6.23 Å map | n/a | 6YTX | Human | 11 | 1% LMNG, 0.5mM CaCl2 | 10 mM HEPES pH 7.6, 150 mM NaCl, 2 mM CaCl2, 50 μM GDN | Drozdzyk, Sawicka et al, 2020 |
| CLHM1 3.60 Å map | n/a | 6LMV | C.elegans | 9 | 1.5% DDM + 0.15% CHS, then buffer exchanged into 0.06% GDN by SEC | 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM DTT, PMAL-C8 | Demura et al., 2020 |
| CLHM1 3.73 Å map | open | 6LOM | C.elegans | Dimer of decamer (gap junction with cytoplasmic domains in tail-to-tail contact) | The cell lysate was supplemented with 2 μg/ml iodoacetamide to block cysteine residues. 1% LMNG | 50 mM HEPES pH 7.5, 150 mM NaCl, 0.02% LMNG, 8 μM BPY | Yang et al, 2020 |
| CLHM1 | n/a | n/a | C.elegans | 9, 10, 11 hemichannels in the presence of the GFP tag. 9,10,11 dimers (“gap junctions”) in the absence of the GFP tag | The cell lysate was supplemented with 2 μg/ml iodoacetamide to block cysteine residues. 1% LMNG | 50 mM HEPES pH 7.5, 150 mM NaCl, 0.02% LMNG, (+/−8 μM BPY) | Yang et al, 2020 |
| Chimera of CALHM1-2 3.40 Å map | n/a | 6LMW | Killifish/human | 8 | 1% DDM + 0.2% CHS, then washed in 0.05% GDN | 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM DTT, PMAL-C8 | Demura et al., 2020 |
| Chimera of CALHM 1-2 3.40 Å map | n/a | 6LMX | Killifish/human | 9 | 1% DDM + 0.2% CHS, then buffer exchanged into 0.05% GDN by SEC | 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM DTT, PMAL-C8 | Demura et al., 2020 |
| Chimera CALHM1/2 3.87 Å map | n/a | 6VAL | Chicken/human | 11 | 1% C12E8, 1 mM EDTA, then washed in 0.01% C12E8, 1mM EDTA | 20 mM HEPES-NaOH pH 7.5, 200 mM NaCl, 1 mM EDTA, MSP2N2, soybean polar lipid | Syrjanen et al., 2020 |
All CALHM structures show common features. The protomer from all CALHMs has four transmembrane helices, with the first transmembrane helix (TM1) extending from the cytoplasm toward the extracellular side and facing the central pore, followed by three more transmembrane helices (TM2-4). Immediately after TM4 is a ~40 residue long cytoplasmic C-terminal helix (CTH), which makes contacts with the CTHs of an adjacent protomer (n+1) as well as a next to next (n+2) protomer (Fig. 10a). Viewed from the cytoplasm, the CTHs point counter clockwise and form an arrangement that resembles overlapping blades in an iris-like camera shutter. After this long CTH, there are an additional 70–100 residues. Our secondary structural analysis suggests that the C-terminal tail has short stretches of helices connected by loops. Indeed, in CALHM2 structures, it has been possible to model ~50 residues after the CTH. The TM1-TM2 loops are extracellularly exposed, and approximately 40 amino acids between TM3 and TM4 form an extracellular region containing small stretches of loops and a helix (ECH) (Fig. 10b). In addition, CALHM1 [184] and perhaps other CALHM members have been predicted to have a short N-terminal helix (NTH) prior to TMD1 as in other large-pore channels. As in connexin [20], pannexin [155], LRRC8 [120], and innexin [90] channels, the N-terminal region has been shown to play important roles in voltage dependent gating of CALHM1 [186]. The NTH has been partially modelled in the RuR-bound structure of CALHM2 (PDB ID: 6UIW), the killifish CALHM1 structure (PDB ID: 6LMT), the zebrafish CALHM1 structure (PDB ID: 6LYG), the CALHM4 structures in the presence and absence of calcium (PDB IDs: 6YTK and 6YTO), the CALHM5 structures in the presence of EDTA, Ca2+ and rubidium red (PDB IDs: 7D61, 7D65, 7D60), and one of the CLHM1 structures (PBD ID: 6LOM). However, the N-terminal ends of TM1 and the NTH region are not nearly as well resolved as TM2-4 in all of the reported cryo-EM structures, which leaves some ambiguity in the molecular models in this region. Perhaps the most well resolved NTHs are the ones from CALHM4 (EMD-10917/PDB-6YTK and EMD-10919/PDB-6YTL) and the zebrafish CALHM1 (EMD-30016/PDB-6LYG) even though the density in EMD-30016 is broken between TM1 and the plausible NTH, making the precise registering of residues difficult. Nevertheless, the structures show that the NTH in CALHM4 extends toward the pore center [93] (the green subunit in Fig. 10c) whereas the one in zebrafish CALHM1 loops back vertically to interact with TM1 [202] (the slate subunit in Fig. 10c).
Overall, the helical orientations of TM2-4 are similar between CALHM1, 2, 4, 5 and 6. TM1 can take distinct orientations depending on the isoform and buffer conditions (Fig. 10c). As described above, the TM helical orientation in CALHM proteins is distinct from the rest of the large-pore channels including connexin, pannexin, LRRC8, and innexin (Fig. 3c) implying that CALHM family may have taken a completely different route in molecular evolution.
Oligomeric assembly patterns differ within CALHM family
A surprising structural feature of the CALHM family members is that their oligomeric states are variable, ranging from 8-mer to 13-mer. In Figure 11, we display the most dominant oligomeric states of each CALHM, which ranges from 8-mer to 11-mer. CALHM1 from vertebrate species (chicken, killifish, and zebrafish) assemble as 8-mers [67, 200, 202] whereas the C.elegans CLHM1 has been reported to assemble as a 9-mer, 10-mer, and 11-mer [200, 203]. All of CALHM2 hemichannel structures 3D reconstructed to date have shown 11-meric assembly [67, 187, 200]. There is also evidence from 2D classification that a 12-meric assembly also exists for CALHM2 [93]. CALHM4 and CALHM6 hemichannels have been shown to exist as 10-mers and 11-mers [93]. Most recently, the 11-meric CALHM5 hemichannel structure has been reported [204]. CALHM5 proteins have also been shown to exist as 10-mers, 12-mers and 13-mers as assessed by 2D classification [204].
The variable oligomeric states alter the overall size of the CALHM proteins and importantly the pore size. The smallest CALHM, the 8-meric CALHM1, has a pore radius of around 20 Å, the width of which is sufficient for ATP permeation. The sizes of the plausible pores in the other CALHM proteins (CALHM2, 4, 5, and 6) are substantially larger than CALHM1, but whether the central pore-like structure is a permeation pathway or is blocked by factors such as lipids in the membrane or other proteins remain to be determined as discussed later.
A main locus of inter-protomer interaction for CALHM proteins is the ~40 residue long CTHs that are unique to CALHM among large-pore channel families. As mentioned above, a CTH from one subunit interacts with that from the next (n+1) and the next-to-next (n+2) subunits in the cytoplasmic region (Fig. 10a and 12). The only prominent interactions within the TMs occur between TM2 and 4 (Fig. 10a and 12b–g). Indeed, the orientations of the long cytoplasmic CTHs relative to TM4 appear to be the major determinant of oligomeric state. The CTH orientation is controlled by the nature of the inter CTH interactions and the positioning of a proline residue that introduces a kink in the TM4-CTH loop (Fig. 12b–g). The proline position in the TM4-CTH loop is conserved among all vertebrate CALHM1s, and the three structures show that the CTHs orient in a similar way to favor the 8-meric assembly [67, 200, 202] (Fig. 12b). The different proline positions in the TM4-CTH loops of the 8-meric CALHM1 and 11-meric CALHM2 introduces kinks at distinct positions (Fig. 12d). Syrjanen et al experimentally validated the above observation by swapping the C-terminal domain (CTD) including the TM4-CTH loop of CALHM1 with that of CALHM2 and showing that this CALHM1/2 chimera oligomerized as an 11-mer (Fig. 12a; CALHM1-2 chimera 11-mer) [67]. Consistent with this result, Demura et al swapped the CTD excluding the TM4-CTH loop and showed that their CALHM1-2 chimera oligomerized as an 8-mer and 9-mer but not as an 11-mer (Fig. 12a; CALHM1-2 chimera 9-mer) [200]. Most of the residues involved in inter-protomer interactions between TM2 and TM4 are conserved in CALHM1 and the CALHM1-2 chimeras (8–11-mers) although there are slight differences in knobs-into-holes packing of the two helices (Fig. 12b–g). This indicates that the TM interactions are permissive to changes in oligomeric assembly [67].
Fig. 12. Subunit interactions and CTH orientations in CALHM proteins.
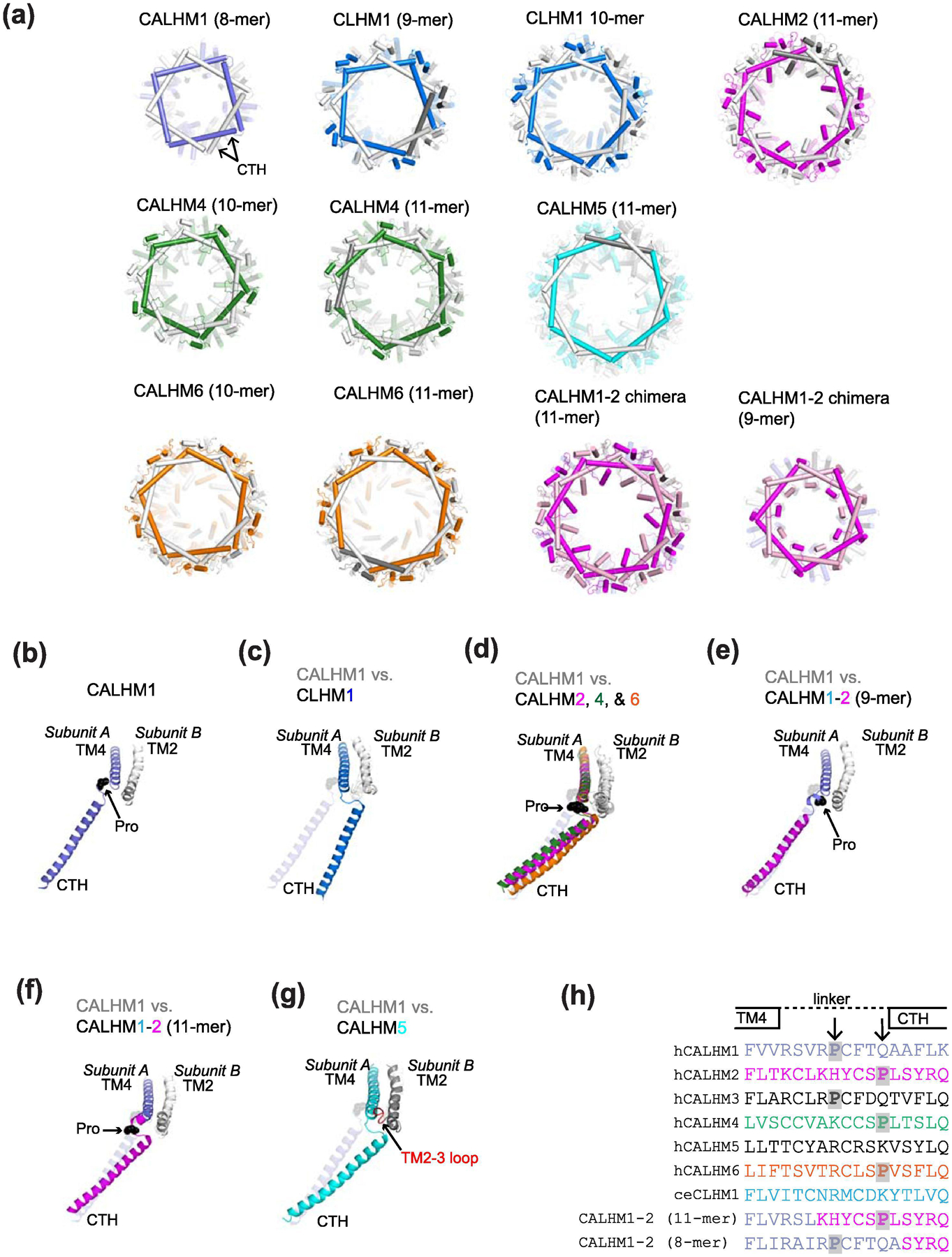
(a) Structures of CALHM proteins viewed from the cytoplasm where the C-terminal domains containing CTHs are highlighted. The PDB IDs used here are 6VAM, 6LMV, 6LOM, 6VAK, 6YTK, 6YTL, 7D61, 6YTV, 6YTX, 6VAL, and 6LMX for CALHM1, CLHM1 (9-mer), CLHM1 (10-mer junction), CALHM2, CALHM4 (10-mer junction), CALHM4 (11-mer junction), CALHM5 (11-mer), CALHM6 (10-mer junction), CALHM6 (11-mer junction), CALHM1-2 chimera (11-mer), and CALHM1-2 chimera (9-mer), respectively. (b) TM4-CTH orientation for CALHM1 viewed from the cytoplasm. The critical Pro residue in the TM4-CTH loop is shown as a black sphere. TM2 from the neighboring subunit (Subunit B) is shown. (c-g) The TM4-CTH orientations of CLHM1 (c), CALHM2, 4, and 6 (d), CALHM1-2 (9-mer) chimera (e), CALH1-2 (11-mer) chimera (f), and CALHM5 (g) in comparison with CALHM1. The TM2-3 loop from the neighboring subunit in CALHM5 is shown in red. (h) Sequence alignment of the TM4-CTH linker region. For clarity, sequences from the human orthologues are shown for CALHM1-6. The critical prolines are highlighted by gray shades. Note that the C.elegans (ce) CLHM1 and CALHM5 do not have proline residues in the TM4-CTH linker. CALHM1-2 (11-mer; chicken CALHM1 and human CALHM2 chimera) and CALHM1-2 (9-merl killifish CALHM1 and human CALHM2 chimera) have distinct construct designs which place the critical proline residues at different positions.
Together, the above work indicated that the TM4-CTH loop is indeed a locus for controlling the oligomeric state at least for CALHM1 and 2. It is important to note that the proline position in the TM4-CTH loop of CALHM4 and 6 is similar to CALHM2 but different from CALHM1 (Fig. 12d and h). Consistently, the oligomeric state of CALHM4 and 6 have been revealed as a mixture of 10-meric and 11-meric [93] where their TM4-CTH loops are in the orientation more similar to CALHM2 than CALHM1 (Fig. 12a and d). Furthermore, we note that there is no proline residue within the TM4-CTH loops of C. elegans CLHM1 and CALHM5 (Fig. 12c, g, and h). The CLHM1 structures illustrate that the oligomeric states could vary from 9- to 11-mer [200, 203] whereas those for CALHM5 could vary from 10-mer to 13-mer (Liu et al, in press 2020). In the case of CALHM5, the TM2-3 loop from the adjacent subunit uniquely interacts with the TM4-CTH loop so that the N-terminal end of the CTH is pulled toward TM2 (Fig. 12g). This results in the CALHM5 TM4-CTH orientation to be similar to CALHM2, 4, and 6. It is important to note that the TM2-3 loops of the CALHM isoforms have low sequence homology. Overall, CALHM family members form various oligomeric assemblies with different numbers of subunits, which are controlled by the nature of the CTHs, the TM4-CTH loops, and the TM2-3 loop.
In addition to the various oligomeric assemblies, some CALHM members have been shown to form gap junction channel-like assemblies. Specifically, CALHM2 forms a head-to-head gap junction channel-like assembly where the extracellular loops are forming contacts as observed under cryo-EM [67, 93, 187, 200]. Interestingly, the 10-meric CLHM1 from C.elegans and both the 10-meric and the 11-meric CALHM4 form tail-to-tail junctions where the cytoplasmic domains mediate interactions [93, 203] (Fig. 11). This tail-to-tail junction may potentially serve as a pathway to connect the ER and the extracellular environment if they express on the plasma and ER membrane. It would be intriguing to validate the physiological relevance of the tail-to-tail junction in the cellular context by a cutting-edge technology such as in situ electron cryotomography (cryo-ET). It is worth reminding that the head-to-head gap junction channel-like assembly and the tail-to-tail junction of the above CALHM proteins have never been observed in intact cells; thus, their physiological relevance is inconclusive at this point.
Overall, it can be concluded that CALHM family members contain a molecular blueprint to allow themselves to assemble with different subunit numbers. This variability in assembly patterns is in contrast to other connexin (Cx26, 31.3, 43, and 46/50) and LRRC8 (A and D) large-pore channel families where they strictly assemble as hexamers or a dimer of hexamers for hemichannels and gap junctions, respectively [35–37, 39–44, 116, 119, 120]. It would be interesting to see if a similar diversity in oligomeric states to CALHM exists in innexin and pannexin.
Also important to note is the fact that CALHM1 and CALHM3 proteins assemble to form channels with different functional properties from the homomeric CALHM1 [185]. On the basis of sequence alignment of the TM4-CTH loop (Fig. 12h), CALHM1 and CALHM3 are predicted to have the critical proline residue at the equivalent position within the TM4-CTH loop; thus, it is expected that the CALHM1/3 channel assembles in a similar manner to the homomeric CALHM1 channel. Much remains to be observed experimentally, including possibilities of other patterns of heteromeric assemblies involving other CALHM members. For example, could other subunits such as CALHM2, 4, 5, and 6 form hetero oligomers with each other or with CALHM1 and 3? It would also be important to correlate patterns of oligomeric states in CALHMs to functions.
Ion permeation, gating motif, and AD mutations in CALHM1
The basic functions of CALHM proteins are voltage-gating and calcium regulation of ion and ATP permeation. These functions are observed in CALHM1 and CALHM1/3 heteromers, possibly in CALHM2 and not in CALHM4, 5, and 6 as mentioned above. By far, the most well characterized CALHM member is CALHM1 and all of the CALHM1 structures to date have been obtained in the absence of calcium, thus, they likely represent the open channel. There are a few functional insights that can be gained from the structures. First is the location of the N-terminus including NTH that have been shown to be involved in voltage-sensitivity in CALHM1 and the C.elegans CLHM1 [183, 186]. Removal of the N-terminal residues between positions 2 and 19 alters voltage-sensitivity although the ion channel activity remains intact [186]. This is somewhat consistent with the structure showing that the NTH is interacting with the pore-lining TM1 and that the TM1 orientation may change in the absence of the NTH, thereby affecting but not disrupting channel function [202]. However, this speculation is based on the assumption that the permeation pathway lies in the middle of the center of the 8-fold assembly. Ma et al identified the conserved Asp121 in human CALHM1 as critical for calcium sensitivity and ion permeation by site-directed mutagenesis (Asp121Arg), which abrogated calcium sensitive ion conductance [64]. The structural studies of CALHM1 showed that this critical aspartate residue is located in TM3 (Fig. 10d); however, is not involved in forming the central pore [67, 200, 202]. This residue is also in close proximity to non-proteinaceous density modelled as cholesterol hemisuccinate (CHS) in the killifish CALHM1 structure (Fig. 10e) [200, 202]. Whether the ion conducting pathway is in the central cavity or in the region around Asp121 in each subunit remains an open question and requires more studies to be resolved. Overall, the structural loci for the permeation pathway and the calcium regulated voltage-gate are yet to be definitively determined. The field will benefit from a CALHM1 structure in a closed-channel conformation to unambiguously delineate the regulatory motifs mentioned above.
The CALHM1 structures located point mutations linked to AD and provided potential explanations to their functional roles. The two AD mutants, CALHM1 Pro86Leu [63] and CALHM1 Arg154His [205] were shown to increase the secretion level of amyloid beta (1–40 and 1–42) [63], however, no significant change in ion channel activity from the wild-type was reported in a later study [64]. The structural studies located the Pro86Leu in the cytoplasmic loop between TM2 and TM3 and the Arg154His in the extracellular loop between TM3 and 4 (Fig. 10d). These locations are far away from the central pore and are consistent with no change in the ion channel activity. Much needs to be explored to understand how these mutations result in the increased level of amyloid beta. For example, could these mutants affect plausible cellular signaling cascades mediated by CALHM1? The available CALHM1 structures should be fully utilized to address basic functional questions including permeation pathways for ions and ATP, and the voltage-sensing mechanism.
Conformations and functional states in CALHM2, 4, 5, and 6
The series of CALHM structures captured several protein conformations even though they are not clearly linked to functions at this point. All of the structures of CALHM1, the isoform that definitively shows voltage-gated currents and ATP flux activities, are in the similar open conformation. Thus, the structure-based gating mechanism of CALHM1 channels is not understood. Interestingly, distinct conformations in the NTH and TM1 regions were observed between CALHM4, CALHM5 and CALHM6 and between CALHM2 and CALHM2 containing RuR, indicating they are generally mobile in CALHMs.
The structures of CALHM4 and CALHM6 were determined in the presence and absence of calcium even though channel activity or calcium regulation have not been shown in these CALHM members [93]. Consistently, the authors did not observe any noticeable differences between CALHM4 structures in the presence and absence of calcium [93]. Indeed, the modelled TM1 region and NTH (residues 4–37) of decameric CALHM4 in the presence and absence of calcium overlay almost perfectly with an RMSD of 0.209Å. The limitation of resolution for the CALHM6 structures disallowed detailed comparisons between the calcium and no calcium conditions. However, a comparison of CALHM4 and 6 structures clearly showed the major difference in the orientation of TM1. In CALHM4, TM1 lines up and interacts with TM3 whereas in CALHM6, TM1 bends toward the central pore without interacting with TM3. The conformations of CALHM4 and CALHM6 were called cylindrical pore and conical pore conformations, respectively [93] (Fig. 13a–g). In the CALHM4 cylindrical conformation, ambiguous density likely representing lipids was observed in the central cavity whereas in the CALHM6 conical conformation, no such density was observed, possibly hinting a potential role of lipids accompanied by the conformation of TM1 [93].
Fig. 13. Configurations of TMD1 and NTH and assessment of cryo-EM density.
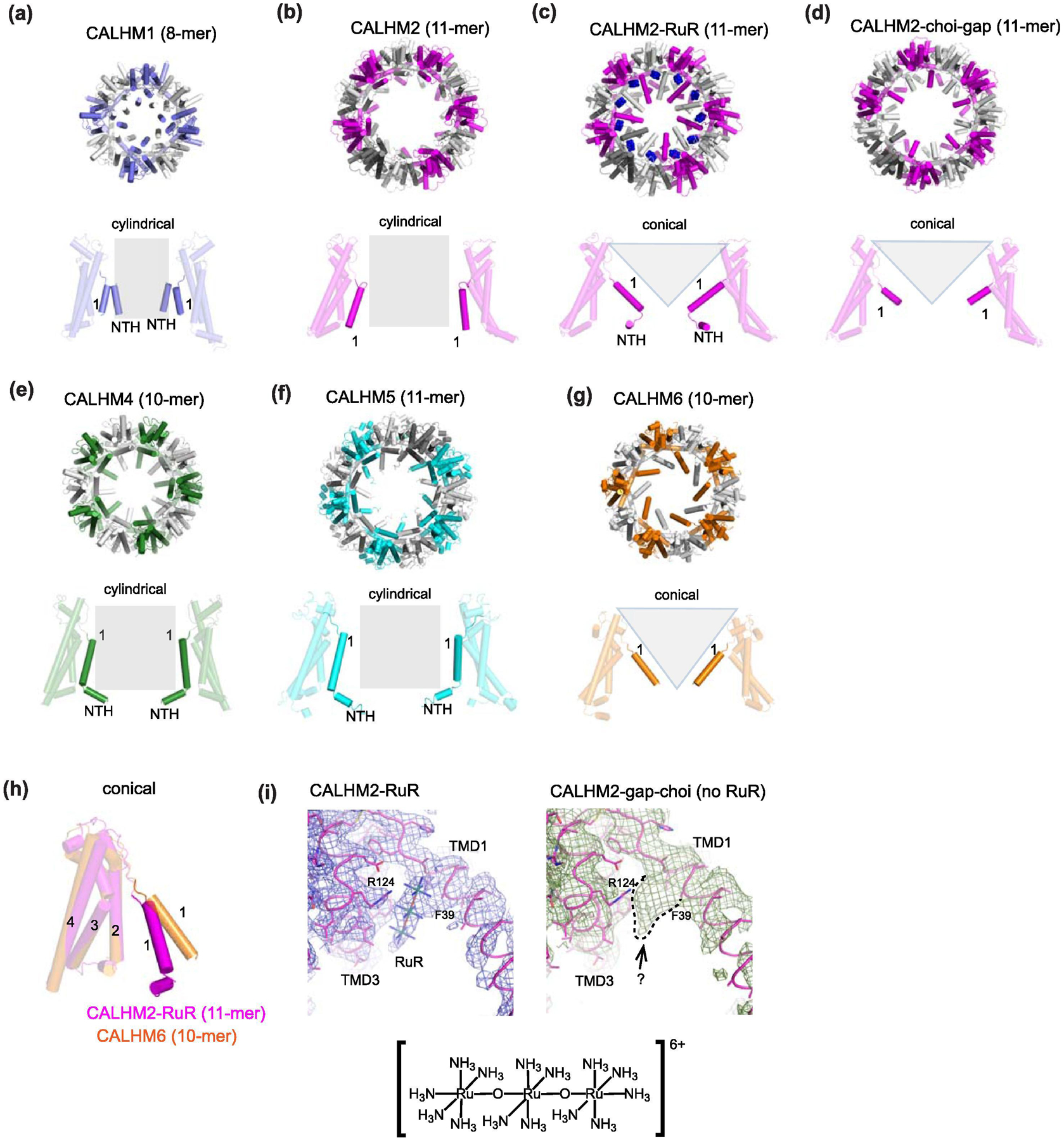
(a-g) Structures of CALHM1 (a), CALHM2 (in EDTA) (b), CALHM2-RuR (c), CALHM2-gap (CALHM2-gap-choi) (d), CALHM4 (10-mer) (e), CALHM5 (f), and CALHM6 (10-mer) (g) viewed from the extracellular side (upper panels) and the plane of the membrane (lower panels). The orientations of TMD1 (1) and NTH can be classified as cylindrical (CALHM1, 2, 4, and 5) and conical (CALHM2-RuR, CALHM2-gap, and CALHM6). Note that CALHM2-gap from Syrjanen et al has the cylindrical conformation. In panel c, putative binding of RuR molecules are shown in blue spheres). (h) Comparison of conical conformations between CALHM2-RuR and CALHM6 (10-mer). Here, TM2-4 are superposed to highlight different orientations of TM1s. (i) Cryo-EM density (blue mesh) around the extracellular side of TM1 and 3 of CALHM2-RuR (left) and CALHM2-gap-choi (right). RuR is in stick (PDB code: 6UIW). A similar density (green mesh) in the equivalent region in CALHM2-gap-choi is surrounded by dotted lines. Note that CALHM2-gap-choi does not contain RuR.
Whether CALHM2 forms functional channels or not has been controversial. While Choi et al reports activity as assessed by patch-clamp electrophysiology on tsA201 cells in the absence of calcium (in the presence of EGTA) [187], other groups have not been able to observe the equivalent CALHM2 mediated currents in Xenopus oocytes and HEK293 cells [67, 93, 185]. Thus, interpretation of structures with respect to functions requires caution until a consensus on functional properties of CALHM2 is reached in the field.
As described above, CALHM2 can exist as an undecamer or a dimer of undecamers under cryo-EM conditions. The overall architectures of the undecameric channels in the presence of EDTA [67, 187] (CALHM2) or DTT [200] from three groups are similar to each other although the samples were prepared in detergent [187], amphipol [200], and lipid nanodisc [67]. In CALHM2, TM1 lines up with TM3 in the similar manner to the cylindrical conformation in CALHM4 [93] (Fig. 13b). The largest structural change was reported in the structures of CALHM2 in the presence of RuR and EDTA (CALHM2-RuR) and in a dimer of undecamers in the absence of RuR and in the presence of EDTA by Choi et al (CALHM2-choi-gap) [187] where the TM1 swings toward the center of the central pore in a manner similar to the conical conformation in CALHM6 [93] but with some differences in directionality of the TM1s (Fig. 13c and d). In contrast the CALHM2 gap junction reported by Syrjanen et al (CALHM2-syr-gap) was in a cylindrical conformation similar to CALHM1, CALHM2, CALHM4, and CALHM5 (Fig. 13a, b, e, and f). The large structural change in CALHM2-RuR and CALHM2-choi-gap was observed mostly in the area of TM1 and NTH while little or no change was seen in the TM2-4 and the CTD regions (Fig. 13c). The CALHM2-RuR and CALHM2-EDTA structures excluding the TM1 and NTH have an RMSD of 1.122 Å. Choi et al observed a density at a region at the ‘top’ of the TM1 and TM3, which they assigned as the RuR molecule (Fig. 13i). The RuR model was placed to nestle into the extracellular side or the ‘top’ of TM1, surrounded by Glu37, Phe39, Ala43, Trp117, Arg124, and Glu126. The authors performed whole cell patch-clamp electrophysiology in which CALHM2 constructs were heterologously expressed in tsA201 cells, voltage-clamped at −60 mV, and detected current after complete removal of calcium by EGTA [187]. The wild-type CALHM2 channel is inhibited by calcium and mostly inhibited by RuR whereas the Glu37Arg mutant is still inhibited by calcium, but the extent of RuR inhibition was lessened. In addition, the current amplitude is decreased compared to the wild-type. They also showed that the Arg10Ala mutant in the NTH or removal of the entire NTH (2–20) distant from the RuR binding site can alter RuR inhibition and suggested that NTHs may be the channel gate.
Another observation by Choi et al is that their CALHM2 gap junction structure, CALHM2-choi-gap, obtained in the presence of EDTA and absence of RuR showed the similar orientation of TM1 to the one in CALHM2-RuR. Curiously, CALHM2-choi-gap has cryo-EM density at a similar spot where RuR was modelled in CALHM2-RuR (Fig. 13i). This observation suggests that the density in CALHM2-RuR may represent something other than RuR. Additionally, it may have resulted from alternative protein conformations around the region. The density in this area does not have sufficient features to unambiguously model RuR (Fig. 13i). Indeed, the molecular model of RuR placed in this coordinate has a fully extended (180°) Ru-O-Ru bond angle, the conformation of which is not favorable. The crystal structure of RuR showed that one of the two Ru-O-Ru bond angles is at 171.5° [206]. RuR is a channel blocker targeting several ion channel families including TRP channels [207], ryodinine channels [208], Piezo channels [209], calcium mitochondrial uniporter [210] and two-pore-domain potassium channels [211]. RuR can inhibit these channels through different mechanisms. For example, recent X-ray crystallographic work on K2P two-pore-domain potassium channels showed that RuR binds to a negatively charged pocket located between the extracellular CAP domain and the top of the pore [212]. In TrpV2, four RuR molecules were shown to bind to its selectivity filter by high resolution cryo-EM [213]. In both cases, densities were sufficiently resolved to model RuR molecules with the Ru-O-Ru bond angles of ~170°. Despite some wariness regarding the validity of RuR binding to CALHM2, the series of structural studies indicated that TM1 in CALHM is mobile. As mentioned above, much remains to be studied on the voltage-gated ion channel activity of CALHM2 channels, not least because of conflicting data on its activity or lack thereof [67, 93, 185].
Permeation of ATP through CALHM
All of the estimates of the diameter of the central pore in the published CALHM1 structure in a presumed open state fall between a range of ~16–20 Å, which is sufficiently large to conduct ATP. Indeed, if an extreme assumption is made that the substantially larger central pores of CALHM2, 4, 5, and 6 are hollow with no obstruction by possible presence of lipids, they should also be permeable to ATP and perhaps even larger metabolites.
The ATP release by CALHM1 was first reported by Taruno et al. as a source of the long sought purinergic signalling mechanism in type-II gustatory cells [65]. Taruno et al also established a luciferin-luciferase-based ATP detection assay on CALHM1 expressing HeLa cells. This assay confirmed that CALHM1 indeed releases ATP and that the release can be inhibited by extracellular calcium and RuR [65]. The similar ATP release was observed on HeLa cells co-expressing CALHM1 and CALHM3 [185]. By implementing the same assay, Demura et al. observed ATP permeation by killifish CALHM1, a CALHM1/2 chimera, C.elegans CLHM1 and human CALHM2 [200]. Perhaps noteworthy is that the kinetics of ATP release for all but human CALHM1 are similar and that they are substantially slower compared to human CALHM1. That is, the reported ATP flux by CALHM1 or CALHM1/3 appear to plateau 30–40 min after removal of calcium whereas that by killifish CALHM1, a CALHM1-2 chimera, C.elegans CLHM1 and human CALHM2 almost linearly increases even 60 min after calcium removal. The authors also performed the ATP assays with N-terminally truncated killifish CALHM1 (ΔN11 and ΔN19) with an intent to widen the pore size. These truncated constructs show higher levels of ATP release compared to the wildtype construct with an assumption that the N-terminal truncation did not upregulate expression level [200]. An equivalent truncation experiment has not been conducted on human CALHM1 or CALHM2 at this point. Furthermore, the ATP assay has not been reported on CALHM4, 5, and 6, thus far. The potential ATP permeation of CALHM2 is paradoxical to the studies that report no conductivity of ions, which are substantially smaller in size than ATP. Functions of CALHM2 may be more sophisticated than voltage- and/or calcium-dependent permeation and may require other factors such as ligands, unique membrane curvature, and interacting proteins. It would be important to definitively determine if ATP can be conducted through all of the CALHM family members. Although laborious, a more direct assay such as ATP depletion from liposomes packaged with ATP and reconstituted with CALHM proteins as demonstrated by Demura et al on killifish CALHM1 protein may be essential to answer outstanding questions on this issue.
CALHM and lipids in central pore?
One major question related to large-pore CALHM channels is how they might regulate their channel activity. While the pore diameter of the most well characterized CALHM1 is comparable to connexin and innexin larger-pore channels, the pores of CALHM2, 4, 5, and 6 are substantially larger if the central pore-like structures are assumed to be hollow. This poses major challenges in regulating their functions unless there are means to close the channels at resting states. Indeed, none of the pore diameters are narrow enough to have hydrophobic gates whose maximum allowable diameter is calculated to be ~12 Å in molecular simulations [214]. Some CALHM studies address possibilities that lipids may be involved in regulation.
There are broadly two manners of lipid-CALHM association. Firstly, some of the CALHM structures exhibit ordered lipid-like density in between the transmembrane helices. For example, the killifish CALHM1 structure has lipid-like density in the cleft between TM3, TM4, and TM2’ (where TM2’ belongs to an adjacent subunit) as well as in the interface between TM3 and TM4. Demura et al., modeled cholesteryl hemisuccinate, which was added during detergent solubilization, into these densities. Although not modeled, discrete molecules of lipid-like density are present in the zebrafish CALHM1 structure, as well as some of the CALHM2 structures [67, 187, 202]. Secondly, in addition to forming sequence specific interactions with the CALHM proteins, lipid-like density also appears to be present in the central pore in some instances.
For instance, the cryo-EM map of CALHM2, 4 and 5 in the cylindrical conformation [67, 93, 204] have densities in the central pore that likely is attributable to lipids whereas CALHM6 in conical conformation does not [93]. It is possible that the conformation of TM1 plays a role in lipid distribution in the central pore and hence regulates the plausible channel functions. It should be noted that the lipid-like densities in the central pores are amorphous unlike the specific lipid binding sites between the TM helices; thus, there remains a possibility that they may represent molecules other than lipids. Nevertheless, one study conducted molecular dynamics simulation to suggest that the central pore of CALHM2 could stably accommodate lipids in the bilayer configuration during the time course of the simulation, whereas the smaller octameric CALHM1 pore could not [67]. This implies that the size of the central pore which is controlled by TM1 conformations and oligomeric states may be the key determinant for the presence of lipids. Further experimental demonstration may be required for establishing this proposed lipid-gating mechanism. If CALHM2, 4, and 5 conduct ions and metabolites, questions remain regarding mechanisms by which these lipids are removed or rearranged to allow permeation.
Conclusion and outlooks
Unveiling the structures of connexin, innexin, LRRC8, pannexin, and CALHM channels has been the long-awaited cornerstone for understanding their functions. The recent structures provide unprecedented opportunities to assess functional questions within each family and in comparison with each other.
Though we introduced connexin as the most classic large-pore channel member, it is only a little over a decade since the first atomic resolution structure became available. Structural analysis of connexin gap junction channels at different conditions (e.g. low pH calcium, calcium free, etc.) in recent years facilitated our mechanistic understanding of functional regulations. Moving forward, there are several fundamental questions to be resolved. The field has not captured connexin channel structures in the presence of channel blockers such as carbenoxolone (CBX); thus, the mechanism and structural motifs responsible for channel blockade is not precisely understood. Perhaps the least understood is the pattern of protomer arrangement and stoichiometry of heteromeric connexin channels. The studies on the Cx46/50 gap junction channel [43, 44] could not distinguish Cx46 from Cx50 due to high similarity between these two isoforms. Furthermore, assessment of heterotopic gap junction assembly and function is an important remaining question. With the on-going development of biochemical and structural biological tools especially in cryo-EM, the above questions may be resolved in the near future. Effective heteromeric assembly may be mediated by a newly developed expression method such as EarlyBac [215, 216] and distinction of different Cx subunits may be achieved by specifically labeling them with antibodies (Fab fragments) as done in the study of hetero-petameric nicotinic acetylcholine receptors [217].
Innexin gap junction channels were perhaps the least studied large-pore channel member. Nevertheless, the recent breakthrough studies unveiled that innexin gap junction channels are composed of two 8-mer hemichannels. Innexin channels are indeed more similar in structure to LRRC8 and pannexin channels than connexin. Some intriguing questions remain to be answered in the field. For example, the mechanism of heterotopic innexin gap junction formation and rectification remain largely unknown. Functional states of the Inx-6 hemichannel and gap junction channel will need to be clearly defined. Capturing open and closed structures will likely facilitate a clearer mechanistic understanding of innexin channel gating. And finally, it would be beneficial for the field to delineate the binding site of channel blockers such as CBX.
Discovery of LRRC8 channels as the major component of VRAC in 2014 was a major milestone. Indeed, this opened up the new era to study molecular aspects of VRACs. The recent cryo-EM studies by several groups revealed a hexameric assembly but with symmetry mismatch between the pore components (ECD, TMD, and ICL) and the LRR domain. While the pore components have C6 symmetry or pseudo-C6 symmetry, the LRR domain has C3 and C2 symmetry in LRRC8A and LRRC8D, respectively. As the field is relatively new at this stage, much remains to be understood of the structure-function relationship of LRRC8 channels. Perhaps the two major missions are: 1) to obtain high resolution structures of heteromeric LRRC8 channels; and 2) to obtain the heteromeric LRRC8 channel structures that capture different functional states including a fully open channel for osmolyte permeation. Additionally, structural analysis of even more sophisticated tri-heteromeric channels such as LRRC8A/C/E may explain specific pharmacology including permeation of cGAMP. The fundamental question related to the activation mechanism still remains. Does the activation merely require lowering of ionic strength as previously shown in droplet bilayer experiments? [111]. Or does it involve membrane tension in cells? What is the role of the LRR domains, which have been shown to play a role in mechanosensing [218]? A mouse with the truncated LRR domain in LRRC8A resulting from a frameshift mutation (ébouriffé mouse) has a similar phenotype to the LRRC8A-null mouse [219]. Achieving the above goals in addition to the current insights will provide mechanistic schemes for this intriguing large-pore channel family.
Structural studies of Panx1 revealed unpredicted insights into the heptameric arrangement of the channel, novel structural similarities and differences to other large-pore channels, pore-lining and ion selectivity residues, and a potential mechanism for CBX inhibition. These fundamental discoveries represent only the beginning of pannexins viewed from a structural biology perspective, and future studies will likely focus on many questions yet to be resolved regarding the pannexin family. Are there additional conformations of Panx1, and can they be captured by cryo-EM? Would the narrow constriction (~9 Å diameter) of the central pore robustly widen to permeate ATP? How are signaling events (i.e. phosphorylation) transmitted into opening of the channel? Do Panx2 and Panx3 have similar structures and subunit stoichiometries to Panx1 and, if so, might structures of these putative channels help infer their physiological function? Answers to these questions will help guide the immediate future of the pannexin field.
Structural studies of CALHM revealed almost no similarity to other large-pore channel members, surprisingly variable patterns of oligomeric assembly ranging from 8- to 12-mers, potential presence of lipids in the central pore-like architecture in some CALHMs, and possible mechanisms of channel gating and inhibition. Perhaps even more so than pannexin, the structural studies raised more functional questions than answers. Do the two major orientations of TM1, conical and cylindrical, represent closed and open channels, respectively? Are the wide central-pores of CALHM2, 4, and 6 formed by 10- to 12-meric assemblies filled with lipids or do they work as extreme large-pore channels? How does CALHM1 close its gate? Can CALHM1 and 2 definitively permeate ATP? Is ATP the major metabolite that is effluxed, or are there others? What permeates through CALHM2, 4, 5, and 6 given that they are not permeable to ions? The CALHM1/3 heteromer appears to represent the ATP release channel in type II taste buds; can other CALHM proteins also form heteromeric complexes with each other? If so, do they form functional ion channels? Are there other proteins that interact with the cytoplasmic domains of CALHM family members? Do CALHM gap junction channels or tail-tail gap junction channels exist in vivo? The exciting new technology such as in situ cryo-ET using focused ion beam (FIB) milled cellular samples may be applied to answer this question. Nevertheless, the available CALHM structures set an excellent starting point to address the above questions.
Overall, the recent structural and functional studies represent major advances in the broad field of large-pore channels. In general, more structures with different functional states and subunit combinations, functional studies using conventional electrophysiology and cutting-edge biochemistry to identify novel activities and mechanisms, and perhaps application of techniques such as high-speed atomic force microscopy may contribute to further progress this exciting field.
Highlights.
Large-pore channels can permeate ions and metabolites such as ATP
Structures are available for connexin, innexin, pannexin, LRRC8, and CALHM
Connexin, innexin, pannexin, and LRRC8 are structurally related but not CALHM
CALHMs form oligomers with various protomer numbers
Acknowledgement
We thanks Drs. J. Contreras and R. Dutzler for valuable comments on this manuscript. This work was supported by NIH (NS111745, NS113632 and MH085926 to H.F., F32MH121061 to K.M., and GM114379 to T.K.), Charles Revson Senior Fellowship in Biomedical Science (to J.L.S.), Robertson funds at Cold Spring Harbor Laboratory, Doug Fox Alzheimer’s fund, Austin’s purpose, Heartfelt Wing Alzheimer’s fund, and the Gertrude and Louis Feil Family Trust (to H.F.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- [1].Bevans CG, Kordel M, Rhee SK, Harris AL. Isoform composition of connexin channels determines selectivity among second messengers and uncharged molecules. J Biol Chem. 1998;273:2808–16. [DOI] [PubMed] [Google Scholar]
- [2].Goldberg GS, Lampe PD, Nicholson BJ. Selective transfer of endogenous metabolites through gap junctions composed of different connexins. Nat Cell Biol. 1999;1:457–9. [DOI] [PubMed] [Google Scholar]
- [3].Goodenough DA, Paul DL. Gap Junctions. 2019.
- [4].Paul DL, Ebihara L, Takemoto LJ, Swenson KI, Goodenough DA. Connexin46, a novel lens gap junction protein, induces voltage-gated currents in nonjunctional plasma membrane of Xenopus oocytes. J Cell Biol. 1991;115:1077–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].DeVries SH, Schwartz EA. Hemi-gap-junction channels in solitary horizontal cells of the catfish retina. J Physiol. 1992;445:201–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cotrina ML, Lin JH, Alves-Rodrigues A, Liu S, Li J, Azmi-Ghadimi H, et al. Connexins regulate calcium signaling by controlling ATP release. Proc Natl Acad Sci U S A. 1998;95:15735–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Osipchuk Y, Cahalan M. Cell-to-cell spread of calcium signals mediated by ATP receptors in mast cells. Nature. 1992;359:241–4. [DOI] [PubMed] [Google Scholar]
- [8].Sanderson MJ, Charles AC, Dirksen ER. Mechanical stimulation and intercellular communication increases intracellular Ca2+ in epithelial cells. Cell Regul. 1990;1:585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Contreras JE, Saez JC, Bukauskas FF, Bennett MV. Gating and regulation of connexin 43 (Cx43) hemichannels. Proc Natl Acad Sci U S A. 2003;100:11388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li H, Liu TF, Lazrak A, Peracchia C, Goldberg GS, Lampe PD, et al. Properties and regulation of gap junctional hemichannels in the plasma membranes of cultured cells. J Cell Biol. 1996;134:1019–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gomez-Hernandez JM, de Miguel M, Larrosa B, Gonzalez D, Barrio LC. Molecular basis of calcium regulation in connexin-32 hemichannels. Proc Natl Acad Sci U S A. 2003;100:16030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Goodenough DA, Paul DL. Gap junctions. Cold Spring Harb Perspect Biol. 2009;1:a002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kumar NM, Gilula NB. The Gap Junction Communication Channel. Cell. 1996;84:381–8. [DOI] [PubMed] [Google Scholar]
- [14].Arcuino G, Lin JH, Takano T, Liu C, Jiang L, Gao Q, et al. Intercellular calcium signaling mediated by point-source burst release of ATP. Proc Natl Acad Sci U S A. 2002;99:9840–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kang J, Kang N, Lovatt D, Torres A, Zhao Z, Lin J, et al. Connexin 43 hemichannels are permeable to ATP. J Neurosci. 2008;28:4702–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ebihara L, Steiner E. Properties of a nonjunctional current expressed from a rat connexin46 cDNA in Xenopus oocytes. J Gen Physiol. 1993;102:59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rubin JB, Verselis VK, Bennett MV, Bargiello TA. A domain substitution procedure and its use to analyze voltage dependence of homotypic gap junctions formed by connexins 26 and 32. Proc Natl Acad Sci U S A. 1992;89:3820–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Verselis VK, Ginter CS, Bargiello TA. Opposite voltage gating polarities of two closely related connexins. Nature. 1994;368:348–51. [DOI] [PubMed] [Google Scholar]
- [19].Oh S, Verselis VK, Bargiello TA. Charges dispersed over the permeation pathway determine the charge selectivity and conductance of a Cx32 chimeric hemichannel. J Physiol. 2008;586:2445–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Purnick PE, Oh S, Abrams CK, Verselis VK, Bargiello TA. Reversal of the gating polarity of gap junctions by negative charge substitutions in the N-terminus of connexin 32. Biophys J. 2000;79:2403–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lopez W, Ramachandran J, Alsamarah A, Luo Y, Harris AL, Contreras JE. Mechanism of gating by calcium in connexin hemichannels. Proc Natl Acad Sci U S A. 2016;113:E7986–E95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pfahnl A, Dahl G. Localization of a voltage gate in connexin46 gap junction hemichannels. Biophys J. 1998;75:2323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Puljung MC, Berthoud VM, Beyer EC, Hanck DA. Polyvalent cations constitute the voltage gating particle in human connexin37 hemichannels. J Gen Physiol. 2004;124:587–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hille B The Permeability of the Sodium Channel to Organic Cations in Myelinated Nerve. Journal of General Physiology. 1971;58:599–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McCleskey EW, Almers W. The Ca channel in skeletal muscle is a large pore. Proceedings of the National Academy of Sciences. 1985;82:7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim DM, Nimigean CM. Voltage-Gated Potassium Channels: A Structural Examination of Selectivity and Gating. Cold Spring Harb Perspect Biol. 2016;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Weidmann S The electrical constants of Purkinje fibres. J Physiol. 1952;118:348–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Furshpan EJ, Potter DD. Mechanism of nerve-impulse transmission at a crayfish synapse. Nature. 1957;180:342–3. [DOI] [PubMed] [Google Scholar]
- [29].Furshpan EJ, Potter DD. Transmission at the giant motor synapses of the crayfish. J Physiol. 1959;145:289–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Robertson JD. The Occurrence of a Subunit Pattern in the Unit Membranes of Club Endings in Mauthner Cell Synapses in Goldfish Brains. J Cell Biol. 1963;19:201–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Benedetti EL, Emmelot P. Hexagonal array of subunits in tight junctions separated from isolated rat liver plasma membranes. J Cell Biol. 1968;38:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].McNutt NS, Weinstein RS. The ultrastructure of the nexus. A correlated thin-section and freeze-cleave study. J Cell Biol. 1970;47:666–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Caspar DL, Goodenough DA, Makowski L, Phillips WC. Gap junction structures. I. Correlated electron microscopy and x-ray diffraction. J Cell Biol. 1977;74:605–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Makowski L, Caspar DL, Phillips WC, Goodenough DA. Gap junction structures. II. Analysis of the x-ray diffraction data. J Cell Biol. 1977;74:629–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Unwin PN, Zampighi G. Structure of the junction between communicating cells. Nature. 1980;283:545–9. [DOI] [PubMed] [Google Scholar]
- [36].Unwin PN, Ennis PD. Two configurations of a channel-forming membrane protein. Nature. 1984;307:609–13. [DOI] [PubMed] [Google Scholar]
- [37].Unger VM, Kumar NM, Gilula NB, Yeager M. Three-dimensional structure of a recombinant gap junction membrane channel. Science. 1999;283:1176–80. [DOI] [PubMed] [Google Scholar]
- [38].Unger VM, Kumar NM, Gilula NB, Yeager M. Projection structure of a gap junction membrane channel at 7 A resolution. Nat Struct Biol. 1997;4:39–43. [DOI] [PubMed] [Google Scholar]
- [39].Oshima A, Tani K, Hiroaki Y, Fujiyoshi Y, Sosinsky GE. Three-dimensional structure of a human connexin26 gap junction channel reveals a plug in the vestibule. Proc Natl Acad Sci U S A. 2007;104:10034–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, et al. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 2009;458:597–602. [DOI] [PubMed] [Google Scholar]
- [41].Bennett BC, Purdy MD, Baker KA, Acharya C, McIntire WE, Stevens RC, et al. An electrostatic mechanism for Ca2+-mediated regulation of gap junction channels. Nature Communications. 2016;7:8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Khan AK, Jagielnicki M, McIntire WE, Purdy MD, Dharmarajan V, Griffin PR, et al. A Steric “Ball-and-Chain” Mechanism for pH-Mediated Regulation of Gap Junction Channels. Cell Reports. 2020;31:107482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Flores JA, Haddad BG, Dolan KA, Myers JB, Yoshioka CC, Copperman J, et al. Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 A. Nat Commun. 2020;11:4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Myers JB, Haddad BG, O’Neill SE, Chorev DS, Yoshioka CC, Robinson CV, et al. Structure of native lens connexin 46/50 intercellular channels by cryo-EM. Nature. 2018;564:372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lee HJ, Jeong H, Hyun J, Ryu B, Park K, Lim HH, et al. Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel. Sci Adv. 2020;6:eaba4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Flower NE. Invertebrate gap junctions. Journal of Cell Science. 1977;25:163. [DOI] [PubMed] [Google Scholar]
- [47].Bao L, Samuels S, Locovei S, Macagno ER, Muller KJ, Dahl G. Innexins form two types of channels. FEBS Lett. 2007;581:5703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Samuels SE, Lipitz JB, Dahl G, Muller KJ. Neuroglial ATP release through innexin channels controls microglial cell movement to a nerve injury. J Gen Physiol. 2010;136:425–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dahl G ATP release through pannexon channels. Philos Trans R Soc Lond B Biol Sci. 2015;370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Panchin Y, Kelmanson I, Matz M, Lukyanov K, Usman N, Lukyanov S. A ubiquitous family of putative gap junction molecules. Curr Biol. 2000;10:R473–4. [DOI] [PubMed] [Google Scholar]
- [51].Baranova A, Ivanov D, Petrash N, Pestova A, Skoblov M, Kelmanson I, et al. The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics. 2004;83:706–16. [DOI] [PubMed] [Google Scholar]
- [52].Bruzzone R, Hormuzdi SG, Barbe MT, Herb A, Monyer H. Pannexins, a family of gap junction proteins expressed in brain. Proc Natl Acad Sci U S A. 2003;100:13644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bruzzone R, Barbe MT, Jakob NJ, Monyer H. Pharmacological properties of homomeric and heteromeric pannexin hemichannels expressed in Xenopus oocytes. Journal of Neurochemistry. 2005;92:1033–43. [DOI] [PubMed] [Google Scholar]
- [54].Locovei S, Bao L, Dahl G. Pannexin 1 in erythrocytes: function without a gap. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7655–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].MacVicar BA, Thompson RJ. Non-junction functions of pannexin-1 channels. Trends in Neurosciences. 2010;33:93–102. [DOI] [PubMed] [Google Scholar]
- [56].Sosinsky GE, Boassa D, Dermietzel R, Duffy HS, Laird DW, MacVicar BA, et al. Pannexin channels are not gap junction hemichannels. Channels. 2011;5:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bao L, Locovei S, Dahl G. Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Letters. 2004;572:65–8. [DOI] [PubMed] [Google Scholar]
- [58].Qu Y, Misaghi S, Newton K, Gilmour LL, Louie S, Cupp JE, et al. Pannexin-1 is required for ATP release during apoptosis but not for inflammasome activation. J Immunol. 2011;186:6553–61. [DOI] [PubMed] [Google Scholar]
- [59].Chekeni FB, Elliott MR, Sandilos JK, Walk SF, Kinchen JM, Lazarowski ER, et al. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature. 2010;467:863–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Voss FK, Ullrich F, Munch J, Lazarow K, Lutter D, Mah N, et al. Identification of LRRC8 heteromers as an essential component of the volume-regulated anion channel VRAC. Science. 2014;344:634–8. [DOI] [PubMed] [Google Scholar]
- [61].Qiu Z, Dubin AE, Mathur J, Tu B, Reddy K, Miraglia LJ, et al. SWELL1, a plasma membrane protein, is an essential component of volume-regulated anion channel. Cell. 2014;157:447–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jentsch TJ. VRACs and other ion channels and transporters in the regulation of cell volume and beyond. Nat Rev Mol Cell Biol. 2016;17:293–307. [DOI] [PubMed] [Google Scholar]
- [63].Dreses-Werringloer U, Lambert JC, Vingtdeux V, Zhao H, Vais H, Siebert A, et al. A polymorphism in CALHM1 influences Ca2+ homeostasis, Abeta levels, and Alzheimer’s disease risk. Cell. 2008;133:1149–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ma Z, Siebert AP, Cheung KH, Lee RJ, Johnson B, Cohen AS, et al. Calcium homeostasis modulator 1 (CALHM1) is the pore-forming subunit of an ion channel that mediates extracellular Ca2+ regulation of neuronal excitability. Proc Natl Acad Sci U S A. 2012;109:E1963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Taruno A, Vingtdeux V, Ohmoto M, Ma Z, Dvoryanchikov G, Li A, et al. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature. 2013;495:223–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Siebert AP, Ma Z, Grevet JD, Demuro A, Parker I, Foskett JK. Structural and functional similarities of calcium homeostasis modulator 1 (CALHM1) ion channel with connexins, pannexins, and innexins. J Biol Chem. 2013;288:6140–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Syrjanen JL, Michalski K, Chou TH, Grant T, Rao S, Simorowski N, et al. Structure and assembly of calcium homeostasis modulator proteins. Nat Struct Mol Biol. 2020;27:150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344:992–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lee CH, Lu W, Michel JC, Goehring A, Du J, Song X, et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Rostovtseva T, Colombini M. VDAC channels mediate and gate the flow of ATP: implications for the regulation of mitochondrial function. Biophys J. 1997;72:1954–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zeth K, Zachariae U. Ten Years of High Resolution Structural Research on the Voltage Dependent Anion Channel (VDAC)-Recent Developments and Future Directions. Front Physiol. 2018;9:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. [DOI] [PubMed] [Google Scholar]
- [73].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–12. [DOI] [PubMed] [Google Scholar]
- [74].Brenner S THE GENETICS OF CAENORHABDITIS ELEGANS. Genetics. 1974;77:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Homyk T, Szidonya J, Suzuki DT. Behavioral mutants of Drosophila melanogaster. Molecular and General Genetics MGG. 1980;177:553–65. [DOI] [PubMed] [Google Scholar]
- [76].Thomas JB, Wyman RJ. Mutations altering synaptic connectivity between identified neurons in Drosophila. The Journal of Neuroscience. 1984;4:530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sun Y-A, Wyman RJ. Passover eliminates gap junctional communication between neurons of the giant fiber system in Drosophila. Journal of Neurobiology. 1996;30:340–8. [DOI] [PubMed] [Google Scholar]
- [78].Avery L The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133:897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Starich TA, Herman RK, Shaw JE. Molecular and genetic analysis of unc-7, a Caenorhabditis elegans gene required for coordinated locomotion. Genetics. 1993;133:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Watanabe T, Kankel DR. Molecular cloning and analysis of l(1)ogre, a locus of Drosophila melanogaster with prominent effects on the postembryonic development of the central nervous system. Genetics. 1990;126:1033–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Krishnan SN, Frei E, Swain GP, Wyman RJ. Passover: A gene required for synaptic connectivity in the giant fiber system of Drosophila. Cell. 1993;73:967–77. [DOI] [PubMed] [Google Scholar]
- [82].Phelan P, Bacon JP, Davies JA, Stebbings LA, Todman MG, Avery L, et al. Innexins: a family of invertebrate gap-junction proteins. Trends Genet. 1998;14:348–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Phelan P, Stebbings LA, Baines RA, Bacon JP, Davies JA, Ford C. Drosophila Shaking-B protein forms gap junctions in paired Xenopus oocytes. Nature. 1998;391:181–4. [DOI] [PubMed] [Google Scholar]
- [84].Landesman Y, White TW, Starich TA, Shaw JE, Goodenough DA, Paul DL. Innexin-3 forms connexin-like intercellular channels. Journal of Cell Science. 1999;112:2391. [DOI] [PubMed] [Google Scholar]
- [85].Phelan P Innexins: members of an evolutionarily conserved family of gap-junction proteins. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2005;1711:225–45. [DOI] [PubMed] [Google Scholar]
- [86].Hasegawa DK, Turnbull MW. Recent findings in evolution and function of insect innexins. FEBS Letters. 2014;588:1403–10. [DOI] [PubMed] [Google Scholar]
- [87].Dahl G, Muller KJ. Innexin and pannexin channels and their signaling. FEBS Lett. 2014;588:1396–402. [DOI] [PubMed] [Google Scholar]
- [88].Phelan P, Goulding LA, Tam JLY, Allen MJ, Dawber RJ, Davies JA, et al. Molecular Mechanism of Rectification at Identified Electrical Synapses in the Drosophila Giant Fiber System. Current Biology. 2008;18:1955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Oshima A, Tani K, Fujiyoshi Y, Martínez AD, Acuña R, Figueroa V, et al. Atomic structure of the innexin-6 gap junction channel determined by cryo-EM. Nature Communications. 2016;7:13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Burendei B, Shinozaki R, Watanabe M, Terada T, Tani K, Fujiyoshi Y, et al. Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids. Sci Adv. 2020;6:eaax3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Hauer F, Gerle C, Fischer N, Oshima A, Shinzawa-Itoh K, Shimada S, et al. GraDeR: Membrane Protein Complex Preparation for Single-Particle Cryo-EM. Structure. 2015;23:1769–75. [DOI] [PubMed] [Google Scholar]
- [92].Oshima A, Matsuzawa T, Murata K, Tani K, Fujiyoshi Y. Hexadecameric structure of an invertebrate gap junction channel. J Mol Biol. 2016;428:1227–36. [DOI] [PubMed] [Google Scholar]
- [93].Drozdzyk K, Sawicka M, Bahamonde-Santos MI, Jonas Z, Deneka D, Albrecht C, et al. Cryo-EM structures and functional properties of CALHM channels of the human placenta. Elife. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hoffmann EK, Lambert IH, Pedersen SF. Physiology of Cell Volume Regulation in Vertebrates. Physiological Reviews. 2009;89:193–277. [DOI] [PubMed] [Google Scholar]
- [95].Lang F, Busch GL, Ritter M, VÖLkl H, Waldegger S, Gulbins E, et al. Functional Significance of Cell Volume Regulatory Mechanisms. Physiological Reviews. 1998;78:247–306. [DOI] [PubMed] [Google Scholar]
- [96].Grinstein S, Clarke CA, Dupre A, Rothstein A. Volume-induced increase of anion permeability in human lymphocytes. Journal of General Physiology. 1982;80:801–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Planells-Cases R, Lutter D, Guyader C, Gerhards NM, Ullrich F, Elger DA, et al. Subunit composition of VRAC channels determines substrate specificity and cellular resistance to Pt-based anti-cancer drugs. EMBO J. 2015;34:2993–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Hyzinski-García MC, Rudkouskaya A, Mongin AA. LRRC8A protein is indispensable for swelling-activated and ATP-induced release of excitatory amino acids in rat astrocytes. The Journal of Physiology. 2014;592:4855–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hisadome K, Koyama T, Kimura C, Droogmans G, Ito Y, Oike M. Volume-regulated anion channels serve as an auto/paracrine nucleotide release pathway in aortic endothelial cells. J Gen Physiol. 2002;119:511–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Jackson PS, Strange K. Volume-sensitive anion channels mediate swelling-activated inositol and taurine efflux. Am J Physiol. 1993;265:C1489–500. [DOI] [PubMed] [Google Scholar]
- [101].Lahey LJ, Mardjuki RE, Wen X, Hess GT, Ritchie C, Carozza JA, et al. LRRC8A:C/E Heteromeric Channels Are Ubiquitous Transporters of cGAMP. Mol Cell. 2020;80:578–91 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hazama A, Shimizu T, Ando-Akatsuka Y, Hayashi S, Tanaka S, Maeno E, et al. Swelling-induced, CFTR-independent ATP release from a human epithelial cell line: lack of correlation with volume-sensitive cl(−) channels. J Gen Physiol. 1999;114:525–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Hazama A, Fan HT, Abdullaev I, Maeno E, Tanaka S, Ando-Akatsuka Y, et al. Swelling-activated, cystic fibrosis transmembrane conductance regulator-augmented ATP release and Cl- conductances in murine C127 cells. J Physiol. 2000;523 Pt 1:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Sabirov RZ, Dutta AK, Okada Y. Volume-dependent ATP-conductive large-conductance anion channel as a pathway for swelling-induced ATP release. J Gen Physiol. 2001;118:251–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Tsumura T, Oiki S, Ueda S, Okuma M, Okada Y. Sensitivity of volume-sensitive Cl-conductance in human epithelial cells to extracellular nucleotides. Am J Physiol-Cell Ph. 1996;271:C1872–C8. [DOI] [PubMed] [Google Scholar]
- [106].Worrell RT, Butt AG, Cliff WH, Frizzell RA. A volume-sensitive chloride conductance in human colonic cell line T84. Am J Physiol-Cell Ph. 1989;256:C1111–C9. [DOI] [PubMed] [Google Scholar]
- [107].Jackson PS, Morrison R, Strange K. The volume-sensitive organic osmolyte-anion channel VSOAC is regulated by nonhydrolytic ATP binding. Am J Physiol-Cell Ph. 1994;267:C1203–C9. [DOI] [PubMed] [Google Scholar]
- [108].Oike M, Droogmans G, Nilius B. The volume-activated chloride current in human endothelial cells depends on intracellular ATP. Pflügers Archiv. 1994;427:184–6. [DOI] [PubMed] [Google Scholar]
- [109].Stotz SC, Clapham DE. Anion-sensitive fluorophore identifies the Drosophila swell-activated chloride channel in a genome-wide RNA interference screen. PLoS One. 2012;7:e46865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Ghosh A, Khandelwal N, Kumar A, Bera AK. Leucine-rich repeat-containing 8B protein is associated with the endoplasmic reticulum Ca2+ leak in HEK293 cells. Journal of Cell Science. 2017;130:3818. [DOI] [PubMed] [Google Scholar]
- [111].Syeda R, Qiu Z, Dubin AE, Murthy SE, Florendo MN, Mason DE, et al. LRRC8 Proteins Form Volume-Regulated Anion Channels that Sense Ionic Strength. Cell. 2016;164:499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].König B, Hao Y, Schwartz S, Plested AJR, Stauber T. A FRET sensor of C-terminal movement reveals VRAC activation by plasma membrane DAG signaling rather than ionic strength. eLife. 2019;8:e45421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Lutter D, Ullrich F, Lueck JC, Kempa S, Jentsch TJ. Selective transport of neurotransmitters and modulators by distinct volume-regulated LRRC8 anion channels. J Cell Sci. 2017;130:1122–33. [DOI] [PubMed] [Google Scholar]
- [114].Gradogna A, Gavazzo P, Boccaccio A, Pusch M. Subunit-dependent oxidative stress sensitivity of LRRC8 volume-regulated anion channels. The Journal of Physiology. 2017;595:6719–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Lee CC, Freinkman E, Sabatini DM, Ploegh HL. The Protein Synthesis Inhibitor Blasticidin S Enters Mammalian Cells via Leucine-rich Repeat-containing Protein 8D. Journal of Biological Chemistry. 2014;289:17124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Deneka D, Sawicka M, Lam AKM, Paulino C, Dutzler R. Structure of a volume-regulated anion channel of the LRRC8 family. Nature. 2018;558:254–9. [DOI] [PubMed] [Google Scholar]
- [117].Kern DM, Oh S, Hite RK, Brohawn SG. Cryo-EM structures of the DCPIB- inhibited volume-regulated anion channel LRRC8A in lipid nanodiscs. eLife. 2019;8:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Kefauver JM, Saotome K, Dubin AE, Pallesen J, Cottrell CA, Cahalan SM, et al. Structure of the human volume regulated anion channel. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Kasuya G, Nakane T, Yokoyama T, Jia Y, Inoue M, Watanabe K, et al. Cryo-EM structures of the human volume-regulated anion channel LRRC8. Nat Struct Mol Biol. 2018;25:797–804. [DOI] [PubMed] [Google Scholar]
- [120].Nakamura R, Numata T, Kasuya G, Yokoyama T, Nishizawa T, Kusakizako T, et al. Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation. Commun Biol. 2020;3:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Abascal F, Zardoya R. LRRC8 proteins share a common ancestor with pannexins, and may form hexameric channels involved in cell-cell communication. Bioessays. 2012;34:551–60. [DOI] [PubMed] [Google Scholar]
- [122].Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Karakas E, Regan MC, Furukawa H. Emerging structural insights into the function of ionotropic glutamate receptors. Trends in biochemical sciences. 2015;40:328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Yamada T, Strange K. Intracellular and extracellular loops of LRRC8 are essential for volume-regulated anion channel function. Journal of General Physiology. 2018;150:1003–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zhou P, Polovitskaya MM, Jentsch TJ. LRRC8 N termini influence pore properties and gating of volume-regulated anion channels (VRACs). J Biol Chem. 2018;293:13440–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Panchin YV. Evolution of gap junction proteins--the pannexin alternative. J Exp Biol. 2005;208:1415–9. [DOI] [PubMed] [Google Scholar]
- [127].Ruan Z, Orozco IJ, Du J, Lü W. Structures of human pannexin 1 reveal ion pathways and mechanism of gating. Nature. 2020;584:646–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Ray A, Zoidl G, Weickert S, Wahle P, Dermietzel R. Site-specific and developmental expression of pannexin1 in the mouse nervous system. European Journal of Neuroscience. 2005;21:3277–90. [DOI] [PubMed] [Google Scholar]
- [129].Tachikawa M, Kaneko Y, Ohtsuki S, Uchida Y, Watanabe M, Ohtsuka H, et al. Targeted Proteomics-Based Quantitative Protein Atlas of Pannexin and Connexin Subtypes in Mouse and Human Tissues and Cancer Cell Lines. Journal of Pharmaceutical Sciences. 2020;109:1161–8. [DOI] [PubMed] [Google Scholar]
- [130].Vasseur ML, Lelowski J, Bechberger JF, Sin WC, Naus CC. Pannexin 2 protein expression is not restricted to the CNS. Frontiers in Cellular Neuroscience. 2014;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Vogt A, Hormuzdi SG, Monyer H. Pannexin1 and Pannexin2 expression in the developing and mature rat brain. Molecular Brain Research. 2005;141:113–20. [DOI] [PubMed] [Google Scholar]
- [132].Wang XH, Streeter M, Liu YP, Zhao HB. Identification and characterization of pannexin expression in the mammalian cochlea. Journal of Comparative Neurology. 2009;512:336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Bond SR, Lau A, Penuela S, Sampaio AV, Underhill TM, Laird DW, et al. Pannexin 3 is a novel target for Runx2, expressed by osteoblasts and mature growth plate chondrocytes. Journal of Bone and Mineral Research. 2011;26:2911–22. [DOI] [PubMed] [Google Scholar]
- [134].Ishikawa M, Iwamoto T, Nakamura T, Doyle A, Fukumoto S, Yamada Y. Pannexin 3 functions as an ER Ca 2+ channel, hemichannel, and gap junction to promote osteoblast differentiation. Journal of Cell Biology. 2011;193:1257–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Aquilino MS, Whyte-Fagundes P, Zoidl G, Carlen PL. Pannexin-1 channels in epilepsy. Neuroscience Letters. 2019;695:71–5. [DOI] [PubMed] [Google Scholar]
- [136].Bargiotas P, Krenz A, Monyer H, Schwaninger M. Functional outcome of pannexin-deficient mice after cerebral ischemia. Channels. 2012;6:453–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Bargiotas P, Krenz A, Hormuzdi SG, Ridder DA, Herb A, Barakat W, et al. Pannexins in ischemia-induced neurodegeneration. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20772–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Billaud M, Chiu YH, Lohman AW, Parpaite T, Butcher JT, Mutchler SM, et al. A molecular signature in the pannexin1 intracellular loop confers channel activation by the α1 adrenoreceptor in smooth muscle cells. Science Signaling. 2015;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].DeLalio LJ, Keller AS, Chen J, Boyce AKJ, Artamonov MV, Askew-Page HR, et al. Interaction between pannexin 1 and caveolin-1 in smooth muscle can regulate blood pressure. Arteriosclerosis, Thrombosis, and Vascular Biology. 2018;38:2065–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Good ME, Chiu YH, Poon IKH, Medina CB, Butcher JT, Mendu SK, et al. Pannexin 1 channels as an unexpected new target of the anti-hypertensive drug spironolactone. Circulation Research. 2018;122:606–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Silverman WR, de Rivero Vaccari JP, Locovei S, Qiu F, Carlsson SK, Scemes E, et al. The pannexin 1 channel activates the inflammasome in neurons and astrocytes. Journal of Biological Chemistry. 2009;284:18143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Spray DC, Hanani M. Gap junctions, pannexins and pain. Neuroscience Letters. 2019;695:46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Weaver JL, Arandjelovic S, Brown G, Mendu SK, Schappe MS, Buckley MW, et al. Hematopoietic pannexin 1 function is critical for neuropathic pain. Scientific Reports. 2017;7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Li S, Zang Z, He J, Chen X, Yu S, Pei Y, et al. Expression of pannexin 1 and 2 in cortical lesions from intractable epilepsy patients with focal cortical dysplasia. Oncotarget. 2017;8:6883–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Berchtold LA, Miani M, Diep TA, Madsen AN, Cigliola V, Colli M, et al. Pannexin-2-deficiency sensitizes pancreatic β-cells to cytokine-induced apoptosis in vitro and impairs glucose tolerance in vivo. Molecular and Cellular Endocrinology. 2017;448:108–21. [DOI] [PubMed] [Google Scholar]
- [146].Celetti SJ, Cowan KN, Penuela S, Shao Q, Churko J, Laird DW. Implications of pannexin 1 and pannexin 3 for keratinocyte differentiation. Journal of Cell Science. 2010;123:1363. [DOI] [PubMed] [Google Scholar]
- [147].Penuela S, Bhalla R, Nag K, Laird DW. Glycosylation Regulates Pannexin Intermixing and Cellular Localization. Molecular Biology of the Cell. 2009;20:4313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Chiu YH, Schappe MS, Desai BN, Bayliss DA. Revisiting multimodal activation and channel properties of Pannexin 1. Journal of General Physiology. 2018;150:19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Dahl G The Pannexin1 membrane channel: distinct conformations and functions. FEBS Letters. 2018;592:3201–9. [DOI] [PubMed] [Google Scholar]
- [150].Deng Z, He Z, Maksaev G, Bitter RM, Rau M, Fitzpatrick JAJ, et al. Cryo-EM structures of the ATP release channel pannexin 1. Nature Structural and Molecular Biology. 2020;27:373–81. [DOI] [PubMed] [Google Scholar]
- [151].Jin Q, Zhang B, Zheng X, Li N, Xu L, Xie Y, et al. Cryo-EM structures of human pannexin 1 channel. Cell Research. 2020;30:449–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Michalski K, Syrjanen JL, Henze E, Kumpf J, Furukawa H, Kawate T. The Cryo-EM structure of pannexin 1 reveals unique motifs for ion selection and inhibition. Elife. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Mou L, Ke M, Song M, Shan Y, Xiao Q, Liu Q, et al. Structural basis for gating mechanism of Pannexin 1 channel. Cell Research. 2020;30:452–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Qu R, Dong L, Zhang J, Yu X, Wang L, Zhu S. Cryo-EM structure of human heptameric Pannexin 1 channel. Cell Research. 2020;30:446–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Michalski K, Henze E, Nguyen P, Lynch P, Kawate T. The weak voltage dependence of pannexin 1 channels can be tuned by N-terminal modifications. J Gen Physiol. 2018;150:1758–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Boassa D, Ambrosi C, Qiu F, Dahl G, Gaietta G, Sosinsky G. Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. Journal of Biological Chemistry. 2007;282:31733–43. [DOI] [PubMed] [Google Scholar]
- [157].Chiu YH, Jin X, Medina CB, Leonhardt SA, Kiessling V, Bennett BC, et al. A quantized mechanism for activation of pannexin channels. Nature Communications. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Wang J, Ambrosi C, Qiu F, Jackson DG, Sosinsky G, Dahl G. The membrane protein Pannexin1 forms two open-channel conformations depending on the mode of activation. Science Signaling. 2014;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Yang D, He Y, Muñoz-Planillo R, Liu Q, Núñez G. Caspase-11 Requires the Pannexin-1 Channel and the Purinergic P2X7 Pore to Mediate Pyroptosis and Endotoxic Shock. Immunity. 2015;43:923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Sandilos JK, Chiu YH, Chekeni FB, Armstrong AJ, Walk SF, Ravichandran KS, et al. Pannexin 1, an ATP release channel, is activated by caspase cleavage of its pore-associated C-terminal autoinhibitory region. Journal of Biological Chemistry. 2012;287:11303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Narahari AK, Kreutzberger AJB, Gaete PS, Chiu Y-H, Leonhardt SA, Medina CB, et al. ATP and large signaling metabolites flux through caspase-activated Pannexin 1 channels. eLife. 2021;10:e64787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Dourado M, Wong E, Hackos DH. Pannexin-1 Is Blocked by Its C-Terminus through a Delocalized Non-Specific Interaction Surface. PLOS ONE. 2014;9:e99596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Wang J, Dahl G. SCAM analysis of Panx1 suggests a peculiar pore structure. Journal of General Physiology. 2010;136:515–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Chiu YH, Ravichandran KS, Bayliss DA. Intrinsic properties and regulation of Pannexin 1 channel. Channels. 2014;8:103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Ma W, Compan V, Zheng W, Martin E, North RA, Verkhratsky A, et al. Pannexin 1 forms an anion-selective channel. Pflugers Archiv European Journal of Physiology. 2012;463:585–92. [DOI] [PubMed] [Google Scholar]
- [166].Nomura T, Taruno A, Shiraishi M, Nakahari T, Inui T, Sokabe M, et al. Current-direction/amplitude-dependent single channel gating kinetics of mouse pannexin 1 channel: A new concept for gating kinetics. Scientific Reports. 2017;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].DeLalio LJ, Billaud M, Ruddiman CA, Johnstone SR, Butcher JT, Wolpe AG, et al. Constitutive SRC-mediated phosphorylation of pannexin 1 at tyrosine 198 occurs at the plasma membrane. Journal of Biological Chemistry. 2019;294:6940–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Weilinger NL, Tang PL, Thompson RJ. Anoxia-Induced NMDA Receptor Activation Opens Pannexin Channels via Src Family Kinases. J Neurosci. 2012;32:12579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Medina CB, Mehrotra P, Arandjelovic S, Perry JSA, Guo Y, Morioka S, et al. Metabolites released from apoptotic cells act as tissue messengers. Nature. 2020;580:130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].McCarthy AE, Yoshioka C, Mansoor SE. Full-Length P2X7 Structures Reveal How Palmitoylation Prevents Channel Desensitization. Cell. 2019;179:659–70.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Mansoor SE, Lü W, Oosterheert W, Shekhar M, Tajkhorshid E, Gouaux E. X-ray structures define human P2X3 receptor gating cycle and antagonist action. Nature. 2016;538:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Romanov RA, Bystrova MF, Rogachevskaya OA, Sadovnikov VB, Shestopalov VI, Kolesnikov SS. The ATP permeability of pannexin 1 channels in a heterologous system and in mammalian taste cells is dispensable. Journal of Cell Science. 2012;125:5514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Benfenati V, Caprini M, Nicchia GP, Rossi A, Dovizio M, Cervetto C, et al. Carbenoxolone inhibits volume-regulated anion conductance in cultured rat cortical astroglia. Channels. 2009;3. [DOI] [PubMed] [Google Scholar]
- [174].Michalski K, Kawate T. Carbenoxolone inhibits Pannexin1 channels through interactions in the first extracellular loop. J Gen Physiol. 2016;147:165–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Qiu F, Dahl G. A permeant regulating its permeation pore: inhibition of pannexin 1 channels by ATP. Am J Physiol-Cell Ph. 2009;296:C250–C5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Beecham GW, Schnetz-Boutaud N, Haines JL, Pericak-Vance MA. CALHM1 polymorphism is not associated with late-onset Alzheimer disease. Ann Hum Genet. 2009;73:379–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Minster RL, Demirci FY, DeKosky ST, Kamboh MI. No association between CALHM1 variation and risk of Alzheimer disease. Hum Mutat. 2009;30:E566–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Sleegers K, Brouwers N, Bettens K, Engelborghs S, van Miegroet H, De Deyn PP, et al. No association between CALHM1 and risk for Alzheimer dementia in a Belgian population. Hum Mutat. 2009;30:E570–4. [DOI] [PubMed] [Google Scholar]
- [179].Nacmias B, Tedde A, Bagnoli S, Lucenteforte E, Cellini E, Piaceri I, et al. Lack of implication for CALHM1 P86L common variation in Italian patients with early and late onset Alzheimer’s disease. J Alzheimers Dis. 2010;20:37–41. [DOI] [PubMed] [Google Scholar]
- [180].Lambert JC, Sleegers K, Gonzalez-Perez A, Ingelsson M, Beecham GW, Hiltunen M, et al. The CALHM1 P86L polymorphism is a genetic modifier of age at onset in Alzheimer’s disease: a meta-analysis study. J Alzheimers Dis. 2010;22:247–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Rubio-Moscardo F, Seto-Salvia N, Pera M, Bosch-Morato M, Plata C, Belbin O, et al. Rare variants in calcium homeostasis modulator 1 (CALHM1) found in early onset Alzheimer’s disease patients alter calcium homeostasis. PLoS One. 2013;8:e74203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Vingtdeux V, Chang EH, Frattini SA, Zhao H, Chandakkar P, Adrien L, et al. CALHM1 deficiency impairs cerebral neuron activity and memory flexibility in mice. Sci Rep. 2016;6:24250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Tanis JE, Ma Z, Krajacic P, He L, Foskett JK, Lamitina T. CLHM-1 is a functionally conserved and conditionally toxic Ca2+-permeable ion channel in Caenorhabditis elegans. J Neurosci. 2013;33:12275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Siebert AP, Ma Z, Grevet JD, Demuro A, Parker I, Foskett JK. Structural and functional similarities of calcium homeostasis modulator 1 (CALHM1) ion channel with connexins, pannexins, and innexins. Journal of Biological Chemistry. 2013;288:6140–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Ma Z, Taruno A, Ohmoto M, Jyotaki M, Lim JC, Miyazaki H, et al. CALHM3 Is Essential for Rapid Ion Channel-Mediated Purinergic Neurotransmission of GPCR-Mediated Tastes. Neuron. 2018;98:547–61 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Tanis JE, Ma ZM, Foskett JK. The NH2 terminus regulates voltage-dependent gating of CALHM ion channels. Am J Physiol-Cell Ph. 2017;313:C173–C86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Choi W, Clemente N, Sun W, Du J, Lu W. The structures and gating mechanism of human calcium homeostasis modulator 2. Nature. 2019;576:163–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Kasamatsu J, Azuma M, Oshiumi H, Morioka Y, Okabe M, Ebihara T, et al. INAM plays a critical role in IFN-gamma production by NK cells interacting with polyinosinic-polycytidylic acid-stimulated accessory cells. J Immunol. 2014;193:5199–207. [DOI] [PubMed] [Google Scholar]
- [189].Moyer BD, Hevezi P, Gao N, Lu M, Kalabat D, Soto H, et al. Expression of genes encoding multi-transmembrane proteins in specific primate taste cell populations. PLoS One. 2009;4:e7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Huang YA, Roper SD. Intracellular Ca(2+) and TRPM5-mediated membrane depolarization produce ATP secretion from taste receptor cells. J Physiol. 2010;588:2343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Zhang Z, Zhao Z, Margolskee R, Liman E. The transduction channel TRPM5 is gated by intracellular calcium in taste cells. J Neurosci. 2007;27:5777–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Finger TE, Danilova V, Barrows J, Bartel DL, Vigers AJ, Stone L, et al. ATP signaling is crucial for communication from taste buds to gustatory nerves. Science. 2005;310:1495–9. [DOI] [PubMed] [Google Scholar]
- [193].Tordoff MG, Ellis HT, Aleman TR, Downing A, Marambaud P, Foskett JK, et al. Salty taste deficits in CALHM1 knockout mice. Chem Senses. 2014;39:515–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Bigiani A Calcium Homeostasis Modulator 1-Like Currents in Rat Fungiform Taste Cells Expressing Amiloride-Sensitive Sodium Currents. Chem Senses. 2017;42:343–59. [DOI] [PubMed] [Google Scholar]
- [195].Nomura K, Nakanishi M, Ishidate F, Iwata K, Taruno A. All-Electrical Ca(2+)-Independent Signal Transduction Mediates Attractive Sodium Taste in Taste Buds. Neuron. 2020;106:816–29 e6. [DOI] [PubMed] [Google Scholar]
- [196].Sclafani A, Ackroff K. Greater reductions in fat preferences in CALHM1 than CD36 knockout mice. Am J Physiol Regul Integr Comp Physiol. 2018;315:R576–R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Sana-Ur-Rehman H, Markus I, Moore KH, Mansfield KJ, Liu L. Expression and localization of pannexin-1 and CALHM1 in porcine bladder and their involvement in modulating ATP release. Am J Physiol Regul Integr Comp Physiol. 2017;312:R763–R72. [DOI] [PubMed] [Google Scholar]
- [198].Workman AD, Carey RM, Chen B, Saunders CJ, Marambaud P, Mitchell CH, et al. CALHM1-Mediated ATP Release and Ciliary Beat Frequency Modulation in Nasal Epithelial Cells. Sci Rep. 2017;7:6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Jun M, Xiaolong Q, Chaojuan Y, Ruiyuan P, Shukun W, Junbing W, et al. Calhm2 governs astrocytic ATP releasing in the development of depression-like behaviors. Mol Psychiatry. 2018;23:1091. [DOI] [PubMed] [Google Scholar]
- [200].Demura K, Kusakizako T, Shihoya W, Hiraizumi M, Nomura K, Shimada H, et al. Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies. Sci Adv. 2020;6:eaba8105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Romanov RA, Lasher RS, High B, Savidge LE, Lawson A, Rogachevskaja OA, et al. Chemical synapses without synaptic vesicles: Purinergic neurotransmission through a CALHM1 channel-mitochondrial signaling complex. Sci Signal. 2018;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Ren Y, Wen T, Xi Z, Li S, Lu J, Zhang X, et al. Cryo-EM structure of the calcium homeostasis modulator 1 channel. Sci Adv. 2020;6:eaba8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Yang W, Wang Y, Guo J, He L, Zhou Y, Zheng H, et al. Cryo-electron microscopy structure of CLHM1 ion channel from Caenorhabditis elegans. Protein Sci. 2020;29:1803–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Liu J, Wan F, Jin Q, Li X, Bhat EA, Guo J, et al. Cryo-EM structures of human calcium homeostasis modulator 5. Cell Discovery. 2020;6:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Vingtdeux V, Tanis JE, Chandakkar P, Zhao H, Dreses-Werringloer U, Campagne F, et al. Effect of the CALHM1 G330D and R154H human variants on the control of cytosolic Ca2+ and Abeta levels. PLoS One. 2014;9:e112484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].de C.T. Carrondo MAAF, Griffith WP, Hall JP, Skapski AC. X-ray structure of [Ru3 O2 (NH3)14]6+, cation of the cytological reagent ruthenium red. Biochimica et Biophysica Acta (BBA) - General Subjects. 1980;627:332–4. [DOI] [PubMed] [Google Scholar]
- [207].Voets T, Prenen J, Vriens J, Watanabe H, Janssens A, Wissenbach U, et al. Molecular Determinants of Permeation through the Cation Channel TRPV4*. Journal of Biological Chemistry. 2002;277:33704–10. [DOI] [PubMed] [Google Scholar]
- [208].Smith JS, Imagawa T, Ma J, Fill M, Campbell KP, Coronado R. Purified ryanodine receptor from rabbit skeletal muscle is the calcium-release channel of sarcoplasmic reticulum. Journal of General Physiology. 1988;92:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Coste B, Xiao B, Santos JS, Syeda R, Grandl J, Spencer KS, et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature. 2012;483:176–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Moore CL. Specific inhibition of mitochondrial Ca++ transport by ruthenium red. Biochemical and Biophysical Research Communications. 1971;42:298–305. [DOI] [PubMed] [Google Scholar]
- [211].Czirják G, Enyedi P. Ruthenium red inhibits TASK-3 potassium channel by interconnecting glutamate 70 of the two subunits. Molecular pharmacology. 2003;63:646–52. [DOI] [PubMed] [Google Scholar]
- [212].Pope L, Lolicato M, Minor DL. Polynuclear Ruthenium Amines Inhibit K2P Channels via a “Finger in the Dam” Mechanism. Cell Chemical Biology. 2020;27:511–24.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Protopopova AD, Pumroy RA, de la Roche J, Haug FM, Sousa BB, Gallo PN, et al. TRPV2 interaction with small molecules and lipids revealed by cryo-EM. bioRxiv. 2020:2020.08.10.242008. [Google Scholar]
- [214].Beckstein O, Sansom MS. Liquid-vapor oscillations of water in hydrophobic nanopores. Proc Natl Acad Sci U S A. 2003;100:7063–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Furukawa H, Simorowski N, Michalski K. Effective production of oligomeric membrane proteins by EarlyBac-insect cell system. Methods Enzymol. 2021;in press:in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Chou TH, Tajima N, Romero-Hernandez A, Furukawa H. Structural Basis of Functional Transitions in Mammalian NMDA Receptors. Cell. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Walsh RM, Jr., Roh SH, Gharpure A, Morales-Perez CL, Teng J, Hibbs RE. Structural principles of distinct assemblies of the human alpha4beta2 nicotinic receptor. Nature. 2018;557:261–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Ju L, Chen Y, Xue L, Du X, Zhu C. Cooperative unfolding of distinctive mechanoreceptor domains transduces force into signals. eLife. 2016;5:e15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Platt CD, Chou J, Houlihan P, Badran YR, Kumar L, Bainter W, et al. Leucine-rich repeat containing 8A (LRRC8A)–dependent volume-regulated anion channel activity is dispensable for T-cell development and function. Journal of Allergy and Clinical Immunology. 2017;140:1651–9.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]


