Abstract
We report here that the Rad51 recombinase is cleaved in mammalian cells during the induction of apoptosis by ionizing radiation (IR) exposure. The results demonstrate that IR induces Rad51 cleavage by a caspase-dependent mechanism. Further support for involvement of caspases is provided by the finding that IR-induced proteolysis of Rad51 is inhibited by Ac-DEVD-CHO. In vitro studies show that Rad51 is cleaved by caspase 3 at a DVLD/N site. Stable expression of a Rad51 mutant in which the aspartic acid residues were mutated to alanines (AVLA/N) confirmed that the DVLD/N site is responsible for the cleavage of Rad51 in IR-induced apoptosis. The functional significance of Rad51 proteolysis is supported by the finding that, unlike intact Rad51, the N- and C-terminal cleavage products fail to exhibit recombinase activity. In cells, overexpression of the Rad51(D-A) mutant had no effect on activation of caspase 3 but did abrogate in part the apoptotic response to IR exposure. We conclude that proteolytic inactivation of Rad51 by a caspase-mediated mechanism contributes to the cell death response induced by DNA damage.
The response of eukaryotic cells to ionizing radiation (IR) and other DNA-damaging agents includes the induction of apoptosis. The available evidence indicates that IR induces DNA lesions by direct interaction with DNA or through the formation of reactive oxygen intermediates (27). However, the basis by which cells recognize DNA damage and transduce this information into signals that regulate events such as induction of apoptosis remains unclear.
Direct evidence for the activation of caspases in the induction of apoptosis comes from studies with peptide inhibitors (2, 52, 54), the cowpox virus protein CrmA (57), and the baculovirus protein p35 (9). Overexpression of CrmA inhibits the induction of apoptosis in diverse settings, including activation of the Fas receptor and treatment with tumor necrosis factor alpha (TNF) (21, 41, 71). Similarly, the p35 protein functions as an inhibitor of caspases and blocks apoptosis in insect and mammalian cells (10, 14, 15, 55). The recent finding that IR-induced apoptosis involves activation of a CrmA-insensitive pathway has supported the existence of apoptotic signals that are distinct from those activated by Fas and TNF (17). In this context, caspase 3 is inhibited by p35 but not CrmA in vitro, and IR-induced activation of caspase 3, like the induction of apoptosis, involves a p35-sensitive, CrmA-insensitive pathway (17). Whereas caspase 3 is activated by IR, as well as Fas ligand and TNF, these findings are explained by involvement of a CrmA-sensitive caspase in the Fas- and TNF-induced, but not the IR-induced, cascade. IR-induced activation of caspase 3 is associated with the proteolytic cleavage of poly(ADP-ribose) polymerase (PARP) (29, 36, 47), DNA-PK (11), protein kinase Cδ (20, 25), and protein kinase Cθ (18). The activation of caspase 3 and the subsequent substrate cleavage in irradiated cells are regulated by members of the Bcl-2/Bcl-xl family (17, 20). Bcl-2 and Bcl-xL block the release of cytochrome c from the mitochondria of cells exposed to IR and other agents (30, 32, 33, 78). Whereas cytochrome c is not released from the mitochondria of cells induced to undergo apoptosis with Fas ligand (13), this event upstream to activation of caspase 3 (37) and the insensitivity of IR-induced caspase-3 activity to CrmA (17) support distinct apoptotic signals in Fas- and IR-treated cells.
The present work provides support for the involvement of the Rad51 protein in IR-induced apoptosis. The RecA protein in Escherichia coli mediates recombinational repair of DNA double-strand breaks by initiating pairing and strand exchange between homologous DNAs (62). The identification of structural homologs of RecA in yeast, Xenopus laevis, mouse, and human cells has supported conservation of similar repair functions (8, 43, 49, 60, 61). ScRad51, the RecA homolog in Saccharomyces cerevisiae, is a member of the Rad52 epistasis group required for genetic recombination and the repair of IR-induced DNA double-strand breaks (61). ScRad51 converts DNA double-strand breaks to recombinatorial intermediates, and these breaks accumulate in rad51 mutants (61). In vitro, ScRad51 catalyzes DNA strand exchange in a reaction that is dependent on ATP and the single-strand binding factor replication protein A (67). Other studies indicate that ScRad52 functions as a cofactor for the Rad51 recombinase (46, 63, 66). Human Rad51 (HsRad51) also binds DNA, promotes ATP-dependent homologous pairing and strand transfer reactions in vitro, and interacts with Rad52 (4, 5, 26). These findings have suggested that Rad51 also plays a role in recombinatorial repair in mammalian cells. In this context, decreased expression of Rad51 in mouse cells confers sensitivity to IR-induced DNA lesions (69). Other studies in mice have demonstrated that targeted disruption of the rad51 gene results in an embryonic lethal phenotype (38, 73). Mutant embryos arrest early in development and exhibit increased apoptosis (38). Recent studies in chicken cells have further demonstrated that Rad51 functions in the repair of spontaneously occurring chromosome breaks in proliferating cells (64). These findings and the failure to generate viable rad51−/− embryonic stem cells (73) have indicated that Rad51 function is essential for cell viability.
In the present studies, we show that Rad51 is cleaved at a DVLD/N site by a caspase-mediated mechanism in IR-induced, but not in TNF-induced, apoptosis. Cleavage of Rad51 to N- and C-terminal fragments abrogates Rad51 recombinase activity. Importantly, expression of a caspase-resistant Rad51 protects in part against IR-induced apoptosis. These findings support a role for the Rad51 protein in cell death mechanisms induced by DNA damage.
MATERIALS AND METHODS
Cell culture.
U-937 cells, Rat1/myc cells, HeLa cells, MCF-7 cells, and 293 embryonal kidney cells were grown as described earlier (31, 79). U2-OS cells (American Type Culture Collection) were grown in McCoy’s 5A medium supplemented with 10% fetal bovine serum. Irradiation was performed by using a Gammacell 1000 (Atomic Energy of Canada) with a 137Cs source emitting at a fixed dose of 0.21 Gy min−1 as determined by dosimetry. The DNA content was assessed by staining ethanol-fixed cells with propidium iodide and monitoring with a FACScan (Becton Dickinson). Numbers of cells with sub-G1 DNA content were determined with a MODFIT LT program (Verity Software House, Topsham, Maine). Statistical analysis of the data was performed with Statview (BrainPower, Inc., Calabasas, Calif.). Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were performed with ApopTag fluorescein (Oncor).
Immunoblot analyses.
Cell lysates were prepared as described earlier (81) in lysis buffer containing 1% Nonidet P-40. Proteins were separated in 8 or 15% polyacrylamide gels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transblotted to nitrocellulose. Immunoblot analysis was performed with the following antibodies: anti-caspase 3 (anti-CPP32; Transduction Laboratories), anti-caspase 7 (Transduction Laboratories), anti-PARP (UBI), anti-Rad51, anti-PCNA (Santa Cruz), anti-green fluorescent protein (anti-GFP; Clontech) or anti-Flag (M2; Eastman Kodak).
Generation of recombinant Rad51 proteins and protease cleavage assays.
A Rad51 (D184A and D187A) mutant was generated in two steps by overlapping primer extension and verified by DNA sequencing. Vectors expressing Flag-Rad51, Flag-Rad51(D-A), Flag-Rad51(1–187), or Flag-Rad51(188–339) were prepared by subcloning PCR-generated HsRad51 into a Flag-tagged pcDNA3 vector. The identity of the subcloned HsRad51 was confirmed by restriction enzyme digestion and DNA sequencing. The wild-type or mutant Rad51 proteins were expressed in 293 cells and purified by using anti-Flag immunoaffinity columns. The purified proteins were incubated with 2.5 μg of purified recombinant caspase 3 or caspase 7 (70) per ml in 50 mM HEPES (pH 7.5), 10% glycerol, 2.5 mM dithiothreitol, and 0.24 mM EDTA at room temperature for 30 min. Cleavage reactions were also performed at 37°C for 90 min in the presence of 5 μg of lysate from cells prepared 3 h after IR exposure. The IR-treated cell lysate was depleted of caspase 3 by two rounds of immunoprecipitation with anti-caspase 3 antibody and was reconstituted by the addition of recombinant caspase 3. The reaction products were analyzed by electrophoresis (SDS–15% PAGE) transferred to nitrocellulose membranes, and immunoblotted with anti-Rad51 antibody.
DNA binding assays.
Full-length, N-terminal (amino acid 1 to 187), and C-terminal (amino acid 188 to 339) Rad51 proteins were labeled with [35S]methionine during synthesis in coupled transcription and translation reactions (Promega, Madison, Wis.). The labeled proteins were incubated with single-stranded DNA (ssDNA) cellulose beads for 30 min at 4°C. The beads were extensively washed, boiled in loading dye, resolved by SDS-PAGE, and analyzed by autoradiography.
Cell transfections.
The wild-type HsRad51 and the (D-A) mutant of HsRad51 [Rad51(D-A)] were subcloned into the pEGFP-C1 vector (Clontech). The vectors were transiently transfected into Rat1/myc or U2-OS cells by the calcium phosphate method. HeLa cells stably expressing Flag-tagged wild-type Rad51 or the Rad51(D-A) mutant were prepared by selection in the presence of neomycin.
Homologous recombination assays.
Wild-type, Rad51(D-A) mutant, N-terminal (positions 1 to 187), or C-terminal (positions 188 to 339) Rad51 proteins were transiently (GFP-tagged) or stably (Flag-tagged) expressed in FSH cells, and homologous recombination rates were assayed as described previously (42).
RESULTS
Rad51 is cleaved during IR-induced apoptosis.
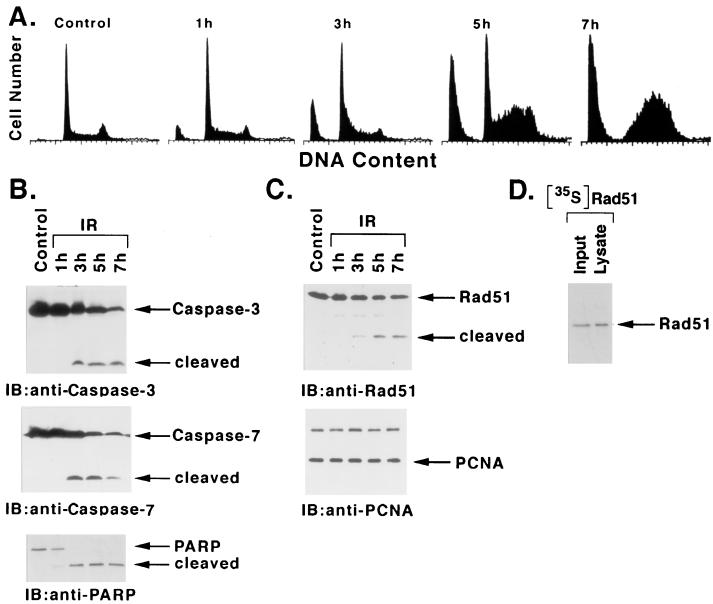
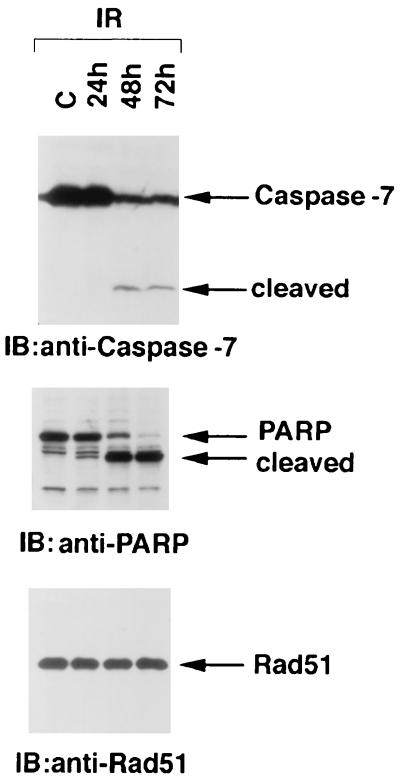
To assess the temporal effects of IR on the induction of apoptosis, irradiated U-937 cells were monitored for the accumulation of sub-G1 DNA content (45). There was little effect at 1 h after IR exposure; however, sub-G1 DNA was apparent by 3 h and exhibited further increases at 5 to 7 h (Fig. 1A). This induction of sub-G1 DNA was temporally associated with the activation of caspase 3 and caspase 7. Lysates from irradiated cells exhibited cleavage of the caspase 3 proenzyme to its active subunits (23, 47) at 3 h, and further activation was detectable at 5 to 7 h (Fig. 1B). Similar findings were obtained for the activation of caspase 7 (Fig. 1B). In concert with these findings, caspase-dependent cleavage of PARP (47) exhibited a similar pattern (Fig. 1B). Significantly, immunoblot analysis of the lysates with anti-Rad51 demonstrated cleavage of the intact 36-kDa Rad51 to a 21-kDa fragment (Fig. 1C). Similar patterns of Rad51 cleavage were observed in U-937 cells induced to undergo apoptosis by exposure to 5 or 10 Gy of IR (data not shown). The IR-induced cleavage of Rad51 exhibited kinetics similar to those for caspase 3 and caspase 7 activation and appearance of apoptotic cells with the sub-G1 DNA content. In contrast, there was no detectable cleavage of proliferating cell nuclear antigen (PCNA) in IR-induced apoptosis (Fig. 1C). As a control, 35S-labeled Rad51 prepared by in vitro transcription or translation was added to cells prior to the lysis procedure. The finding that exogenous [35S]Rad51 is not cleaved during preparation of the lysate supports specific cleavage of endogenous Rad51 as part of the apoptotic response (Fig. 1D).
FIG. 1.
Rad51 is cleaved during IR-induced apoptosis. U-937 cells were treated with 20 Gy IR and harvested at the indicated times. (A) DNA content was assessed by flow cytometry after ethanol-fixed cells were stained with propidium iodide. (B and C) Cell lysates were subjected to immunoblot analysis with anti-caspase 3, anti-caspase 7, anti-PARP, anti-Rad51, or anti-PCNA. (D) U-937 cells treated with 20 Gy of IR were harvested at 5 h. [35S]Rad51 was added to the cells just prior to lysis at 4°C. Input [35S]Rad51 and lysate were analyzed by SDS-PAGE and autoradiography.
IR induces Rad51 cleavage by a CrmA-insensitive, p35-sensitive pathway.
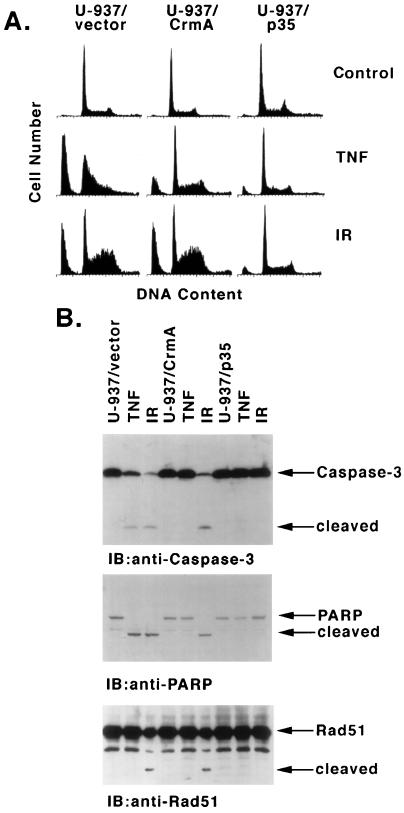
The temporal association between caspase 3/caspase 7 activation and Rad51 proteolysis in irradiated cells suggested the involvement of a caspase-mediated mechanism in Rad51 cleavage. U-937 cell clones that overexpress CrmA or p35 (17) were assayed to determine whether Rad51 cleavage is sensitive to inhibitors of caspases. For purposes of comparison, the cells were treated with TNF or IR. Whereas TNF treatment of U-937 cells stably transfected with the empty vector produced accumulation of sub-G1 DNA, TNF-induced apoptosis was inhibited in U-937/CrmA and U-937/p35 cells (Fig. 2A). These findings confirm that CrmA is functional in the U-937/CrmA cells. In contrast, while apoptosis induced by IR was insensitive to the expression of CrmA, the finding that p35 blocks the appearance of sub-G1 DNA in irradiated cells supports the involvement of caspases. In concert with these results, CrmA blocked TNF-induced, but not IR-induced, activation of caspase 3 and proteolytic cleavage of PARP, while p35 inhibited these events in both TNF- and IR-treated cells (Fig. 2B). Although there was no detectable cleavage of Rad51 in the TNF-treated cells, IR-induced cleavage of Rad51, like that for PARP, was insensitive to CrmA and inhibited by p35 expression (Fig. 2B). Of note, in certain preparations the anti-Rad51 antibody reacts with a protein that migrates faster than intact Rad51 and is not dependent on caspase activation. Nonetheless, the results support cleavage of Rad51 by a caspase-mediated mechanism which is activated during IR- and not TNF-induced apoptosis.
FIG. 2.
IR induces Rad51 cleavage by a CrmA-insensitive, p35-sensitive mechanism. U-937 cells stably expressing an empty vector, CrmA, or p35 were treated with 30 ng of TNF per ml for 6 h or with 20 Gy of IR and then harvested at 7 h. (A) DNA content was assessed by flow cytometry. (B) Cell lysates were subjected to immunoblotting analysis with anti-caspase 3, anti-PARP, or anti-Rad51.
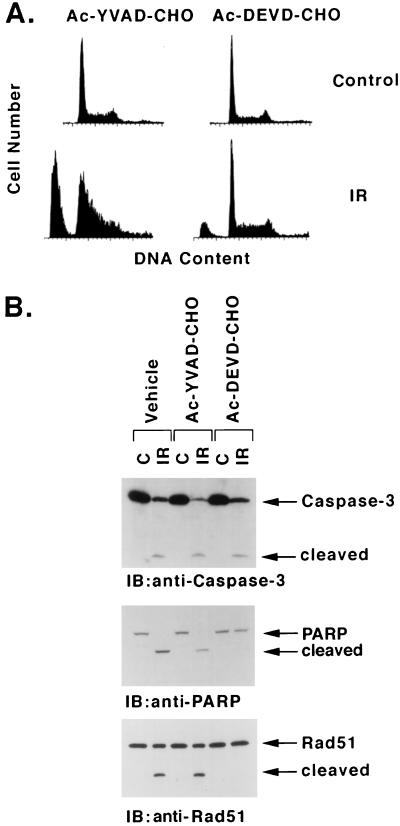
To provide further support for the involvement of a caspase, cells were treated with the tetrapeptide aldehydes acetyl-DEVD-CHO (Ac-DEVD-CHO) and Ac-YVAD-CHO. The peptide component of Ac-DEVD-CHO is that found in the S4-S1 sites of caspase 3 substrates, and the aldehyde is a potent inhibitor of this protease. By contrast, Ac-YVAD-CHO preferentially inhibits caspase 1 and related subfamily members (48). Preincubation of U-937 cells with Ac-DEVD-CHO, and not Ac-YVAD-CHO, inhibited IR-induced accumulation of sub-G1 DNA content (Fig. 3A). Ac-DEVD-CHO had little effect on IR-induced activation of caspase 3 but blocked cleavage of PARP (Fig. 3B). Importantly, Ac-DEVD-CHO, but not Ac-YVAD-CHO, functioned as an inhibitor of Rad51 cleavage (Fig. 3B). These findings and those obtained with the p35 inhibitor suggest that Rad51 is a substrate for a caspase sensitive to Ac-DEVD-CHO in IR-induced apoptosis.
FIG. 3.
Inhibition of IR-induced apoptosis and Rad51 cleavage by Ac-DEVD-CHO. U-937 cells were preincubated with 100 μM Ac-YVAD-CHO or Ac-DEVD-CHO for 30 min, treated with 20 Gy of IR, and then harvested at 7 h. (A) DNA content was assessed by flow cytometry. (B) Cell lysates were subjected to immunoblot analysis with anti-caspase 3, anti-PARP, or anti-Rad51.
Rad51 is cleaved by caspase 3 in vitro.
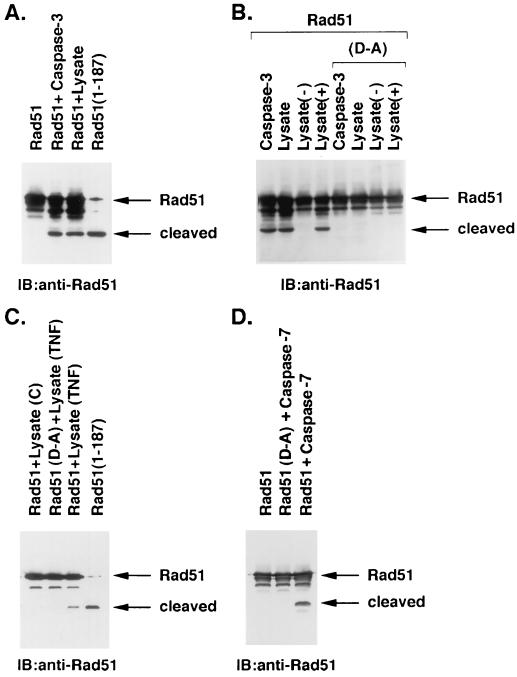
As deduced from synthetic peptide substrates, caspase 3 and caspase 7 exhibit an S4–S1 subsite preference with a DXXD consensus (70, 72). A DVLD/N site is present in mouse and human Rad51 at amino acids 184 to 188 (60). To determine whether Rad51 is cleaved by caspase 3, Flag-tagged Rad51 was expressed in 293 cells and purified by using immunoaffinity columns. The purified Flag-tagged Rad51 was incubated with recombinant caspase 3, and the products were subjected to immunoblotting with anti-Rad51 antibody. Caspase 3 cleaved Rad51 to a 21-kDa fragment (Fig. 4A). Similar results were obtained when the purified Flag-Rad51 was incubated with lysates from IR-treated U-937 cells that exhibit activation of caspase 3 (Fig. 4A). The 21-kDa cleavage fragment of Rad51 exhibited an electrophoretic mobility similar to that of a recombinant N-terminal fragment of Rad51 that extends to the DVLD187 site (Fig. 4A). Whereas these findings provided support for caspase-3-mediated cleavage of Rad51 at the DVLD/N site, we generated a Rad51 protein with the S4 to S1 aspartic acids mutated to alanines (AVLA/N). Compared to wild-type Rad51, the Rad51(D-A) mutant was more resistant to cleavage by recombinant caspase 3 and lysates from IR-induced apoptotic U-937 cells (Fig. 4B). To confirm the involvement of caspase 3, we depleted caspase 3 from the U-937 cell lysate with an anti-caspase 3 antibody. The findings that the immunodepleted lysate fails to cleave wild-type Rad51 and that reconstitution with recombinant caspase 3 restores the activity provided additional support for a caspase 3-mediated mechanism of Rad51 cleavage (Fig. 4B). Moreover, resistance of the Rad51(D-A) mutant to cleavage by caspase 3 and by lysates from TNF-treated cells (Fig. 4C) confirms the involvement of the DVLD/N site. To determine whether Rad51 is cleaved by other executioner caspases, we incubated purified Rad51 with recombinant caspase 7. The results demonstrate that caspase 7 cleaves Rad51 and not Rad51(D-A) (Fig. 4D). These findings demonstrate that caspase 7, like caspase 3, cleaves Rad51 at the DVLD/N site in vitro. Thus, to assess the role of caspase 3 and/or caspase 7 in the cleavage of Rad51 in vivo, studies were performed on MCF-7 cells that are caspase 3 deficient (28). IR treatment of MCF-7 cells was associated with activation of caspase 7 and the cleavage of PARP (Fig. 5). In contrast, there was no detectable cleavage of Rad51 (Fig. 5). These results provide support for the cleavage of Rad51 by caspase 3, and not caspase 7, in irradiated cells. The discrepancy between the cleavage of Rad51 by caspase 7 in vitro, but not in irradiated cells, may be related to subcellular localization of caspase 7 to the mitochondria and endoplasmic reticulum (12).
FIG. 4.
Recombinant Rad51 is cleaved at a DVLD/N site by caspase 3 in vitro. (A) Flag-tagged Rad51 was overexpressed in 293 cells and purified by immunoaffinity column. The purified Rad51 was incubated with recombinant caspase 3 (2.5 μg/ml) for 30 min at room temperature or lysate from U-937 cells treated with 20 Gy of IR and harvested at 3 h. The reaction products were subjected to immunoblotting with anti-Rad51. The recombinant N-terminal Rad51(1–187) fragment was purified from 293 cells and included as a control. The small amount of full-length Rad51 in this lane is a contaminant present after partial purification of the recombinant Rad51(1–187) from 293 cell lysates. (B) Flag-tagged wild-type Rad51 and the DVLD/N-to-AVLA/N mutant, designated (D-A), were overexpressed in 293 cells and purified by using an immunoaffinity column. Purified Rad51 and the Rad51(D-A) mutant were incubated with recombinant caspase 3 and lysates from IR-treated U-937 cells. The cell lysate was depleted of caspase 3 by two immunoprecipitations with anti-caspase 3 [lysate (−)]. Reconstitution of this lysate was achieved by the addition of 2.5 μg of recombinant caspase 3 per ml [lysate (+)]. The reaction products were analyzed by immunoblotting with anti-Rad51. (C) Purified Rad51 was incubated with lysates from control (lane 1) and TNF-treated (lane 3; 30 ng/ml for 6 h) U-937 cells. Purified Rad51(D-A) was incubated with lysate from TNF-treated cells (lane 2). The reaction products were analyzed by immunoblotting with anti-Rad51. The recombinant Rad51(1–187) fragment was included as a control (lane 4). (D) Purified Rad51 and the Rad51(D-A) mutant were incubated with recombinant caspase 7. The reaction products were analyzed by immunoblotting with anti-Rad51.
FIG. 5.
IR-induced cleavage of Rad51 is abrogated in caspase 3-deficient MCF-7 cells. Lysates were prepared from control (C) MCF-7 cells and, at the indicated times after exposure to 20 Gy of IR, were subjected to immunoblot analysis with anti-caspase 7, anti-PARP, or anti-Rad51.
Rad51 is cleaved at the DVLD/N site in vivo.
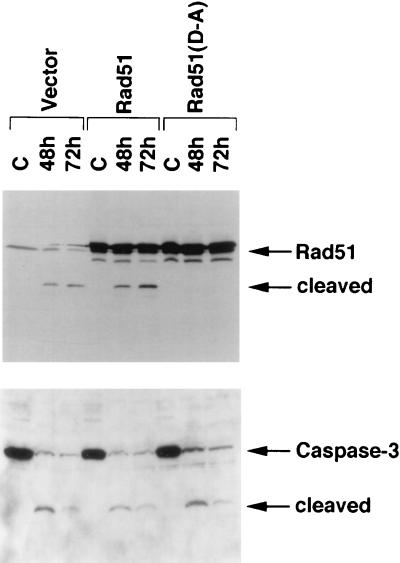
To determine whether Rad51 is cleaved at the DVLD/N site in cells induced to undergo apoptosis, we stably expressed wild-type Rad51 and the Rad51(D-A) mutant in HeLa cells. Immunoblot analysis of the transfectants demonstrated increased reactivity with anti-Rad51 antibody compared to cells transfected with the empty vector (Fig. 6, upper panel). Irradiation of cells expressing the empty vector resulted in the cleavage of Rad51 and the activation of caspase 3 (Fig. 6, upper panel). Similar findings were obtained in cells overexpressing the wild-type Rad51 protein (Fig. 6, upper panel). In contrast and in concert with the in vitro findings, there was no detectable cleavage of the Rad51(D-A) mutant (Fig. 6, upper panel). The activation of caspase 3, however, was comparable in HeLa cells expressing the empty vector, wild-type Rad51, or Rad51(D-A) (Fig. 6, lower panel). These results demonstrate that Rad51 is cleaved at the DVLD/N site in irradiated cells.
FIG. 6.
Rad51 is cleaved at the DVLD/N site in vivo. Wild-type Rad51 and the Rad51(D-A) mutant were stably expressed in HeLa cells. Control HeLa cells expressed the empty vector. The HeLa cell transfectants were treated with 20 Gy of IR and harvested at the indicated times. Cell lysates were subjected to immunoblotting with anti-Rad51 or anti-caspase 3.
Functional role of Rad51 cleavage.
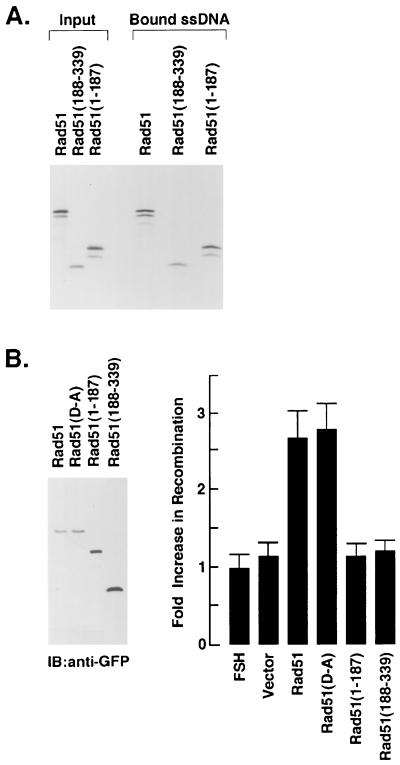
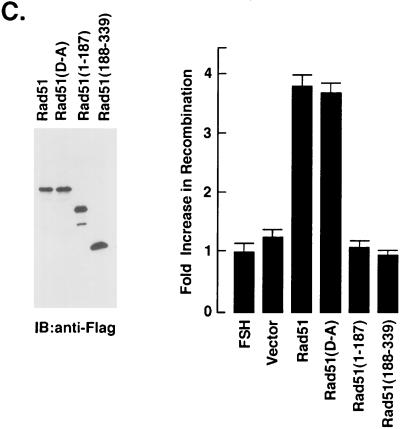
Rad51 binds ssDNA to form nucleoprotein filaments that undergo pairing with duplex DNA (6, 68). To assess the effects of cleavage on the binding of Rad51 to ssDNA, we prepared N- and C-terminal fragments that correspond to those generated by cleavage at the DVLD/N site (Fig. 7A). As shown previously (6, 49), full-length Rad51 binds to ssDNA (Fig. 7A). Similar results were obtained with the N-terminal Rad51(1–187) and C-terminal Rad51(188–339) fragments (Fig. 7A). In contrast, there was no detectable binding of these proteins to cellulose beads devoid of ssDNA (data not shown). Whereas it is unlikely that proteins from the reticulocyte lysate contribute to the binding of the Rad51 fragments, these findings suggest that the cleavage of Rad51 at the DVLD/N site has little if any effect on the direct binding of Rad51 to ssDNA. To assess the effects of cleavage of Rad51 recombinase activity, we assayed the homologous recombination between lacZ chromosomal direct repeats in FSH cells (42). Transient expression of both wild-type Rad51 and the Rad51(D-A) mutant tagged with GFP (Fig. 7B, left panel) stimulated greater recombination compared to that obtained with transfection of the empty vector (Fig. 7B, right panel). In contrast, there was little if any stimulation of homologous recombination as a result of transiently expressing the GFP-tagged Rad51(1–187) or Rad51(188–339) fragments (Fig. 7B). To confirm the effects of cleavage on Rad51 recombinase activity, FSH cells were stably transfected with Flag-tagged Rad51, the Rad51(D-A) mutant, or the Rad51 fragments. Immunoblot analysis of the transfectants with anti-Flag antibody confirmed the overexpression of the Rad51 proteins (Fig. 7C, left panel). As shown in the transient-transfection studies, recombination was stimulated by wild-type Rad51 and the Rad51(D-A) mutant but not by the N- or C-terminal fragments (Fig. 7C, right panel). These findings indicate that cleavage of Rad51 by caspase 3 results in the abrogation of Rad51 recombinase activity.
FIG. 7.
Cleavage of Rad51 at the DVLD/N site abrogates Rad51 function in homologous recombination. (A) Full-length Rad51, the N-terminal Rad51(1–187) fragment and the C-terminal Rad51(188–339) fragment were synthesized by in vitro transcription and translation in the presence of [35S]methionine. The products were incubated with ssDNA-conjugated cellulose beads for 30 min. The beads were extensively washed and boiled in loading dye. Input proteins (not bound to ssDNA) and proteins eluted from the ssDNA-conjugated cellulose beads were subjected to SDS-PAGE and autoradiography. (B) GFP-tagged Rad51 (full length), Rad51(D-A), Rad51(1–187) and Rad51(188–339) were transiently overexpressed in FSH cells for 72 h. The transfectants were subjected to immunoblot analysis with anti-GFP (left panel). Homologous recombination was assessed by determining the β-galactosidase activity of GFP-positive cells. Results are expressed as the fold increase (mean ± the standard deviation [SD] of three experiments, each performed in duplicate) in recombination compared to that obtained in the FSH cells expressing the GFP-empty vectors (right panel). (C) FSH cells were stably transfected to express Flag (vector), Flag-tagged Rad51, Flag-tagged Rad51(D-A), Flag-tagged Rad51(1–187), or Flag-tagged Rad51(188–339). Transfectants were subjected to immunoblot analysis with anti-Flag (left panel). The fold increase in recombination was assessed by comparing the β-galactosidase activity to that obtained in the FSH cells expressing the Flag-empty vector. The results are expressed as the mean ± the SD of three determinations, each performed in duplicate (right panel).
Involvement of Rad51 cleavage in apoptosis.
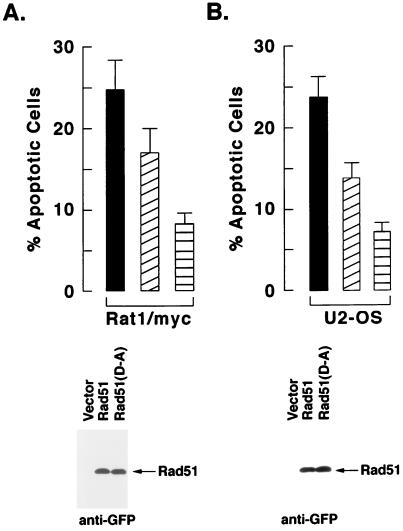
Whereas cells deficient in Rad51 exhibit an increased sensitivity to IR (69), we asked whether the cleavage of Rad51 has a functional role in the induction of apoptosis. Rat1/myc cells were transiently transfected with GFP vectors expressing wild-type or mutant Rad51. After 24 h, the transfected cells were exposed to IR and incubated for an additional 72 h, and the GFP-positive cells were analyzed for sub-G1 DNA content. Compared to cells transfected with the empty vector, the overexpression of wild-type Rad51 partially inhibited the induction of apoptosis (Fig. 8A). Importantly, overexpression of the Rad51(D-A) mutant was more effective than wild-type Rad51 in protecting Rat1/myc cells against IR-induced apoptosis (Fig. 8A). Similar results were obtained with U2-OS cells transiently transfected with Rad51 or Rad51(D-A) and treated with IR (Fig. 8B). These findings indicate that the protective effects of Rad51(D-A) against IR-induced apoptosis are not cell type specific.
FIG. 8.
Transient overexpression of the Rad51(D-A) mutant confers resistance to IR-induced apoptosis of Rat1/myc and U2-OS cells. GFP-tagged wild-type Rad51 or the Rad51(D-A) mutant were transiently transfected into Rat1/myc (A) and U2-OS (B) cells. As controls, cells were transfected with the GFP-expressing empty vector. The cells were treated with 20 Gy of IR at 24 h posttransfection and then incubated for an additional 72 h. The GFP-positive cells were sorted and analyzed for DNA content by flow cytometry (upper panels) or for GFP-tagged Rad51 by immunoblotting with anti-GFP (lower panel). The flow cytometery results are expressed as the percentage (mean ± the standard deviation from two independent experiments, each performed in triplicate) of cells with sub-G1 DNA content. Cells were transfected with the empty vector (solid bars), wild-type Rad51 (diagonal lined bar), or the Rad51(D-A) mutant (horizontal lined bar).
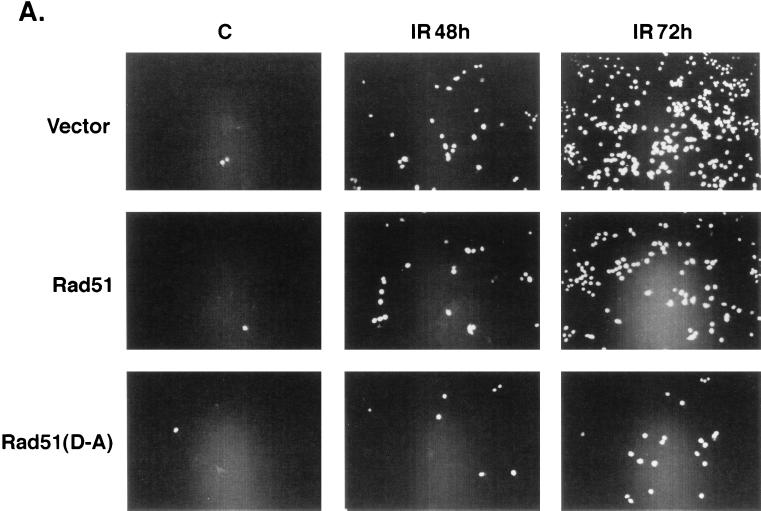
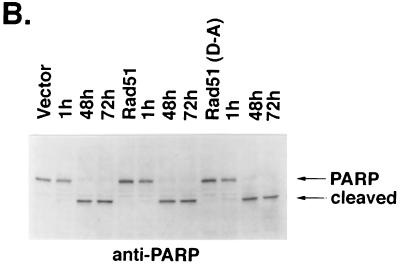
Other studies were performed with HeLa cell clones that stably express the Flag-tagged wild-type or mutant Rad51 (Fig. 9A). Assessment of IR-induced apoptosis demonstrated that, whereas wild-type Rad51 inhibited the response, mutant Rad51(D-A) was more effective in blocking the appearance of cells positive for sub-G1 DNA content (Fig. 9B). In contrast, overexpression of Rad51 or Rad51(D-A) had no apparent effect on the extent of apoptosis induced by exposure to TNF (Fig. 9C). To confirm these findings, the HeLa cell transfectants were exposed to IR and then assayed for TUNEL staining. The results demonstrate that, as shown for the induction of sub-G1 DNA, overexpression of Rad51 and, in particular, the Rad51(D-A) mutant decreased the percentage of cells that exhibit TUNEL positivity (Fig. 10A). Whereas HeLa cells overexpressing Rad51 failed to exhibit defects in the cleavage of procaspase 3 by IR exposure (Fig. 6), caspase-3-mediated cleavage of PARP was assessed to confirm the activation of caspase 3. The results demonstrate that overexpression of wild-type Rad51 or the Rad51(D-A) mutant has little if any effect on IR-induced PARP cleavage (Fig. 10B). These findings demonstrate that overexpression of wild-type Rad51 and particularly the Rad51(D-A) mutant confers resistance to DNA fragmentation, and not caspase 3 activation, associated with the induction of apoptosis by IR exposure.
FIG. 9.
Stable expression of the Rad51(D-A) mutant confers resistance to IR-induced apoptosis of HeLa cells. HeLa cells were stably transfected to express the Flag-empty vector (solid bars), Flag-tagged Rad51 (diagonal lined bars), or Flag-tagged Rad51(D-A) (horizontal lined bars). (A) Clones (a and b) selected from each of two independent transfections were subjected to immunoblot analysis with anti-Flag. The transfectants were treated with 20 Gy of IR and harvested at the indicated times (B) or were treated with TNF for 12 h (C). The cells were analyzed for DNA content by flow cytometry. The results are expressed as the percentage (mean ± standard deviation from three separate experiments) of cells with sub-G1 DNA content.
FIG. 10.
Expression of Rad51 confers resistance to IR-induced DNA degradation and not caspase 3 activation. (A) HeLa cells stably expressing Flag-empty vector, Flag-tagged Rad51, or Flag-tagged Rad51(D-A) were exposed to 20 Gy of IR, fixed at 48 or 72 h, and subjected to TUNEL staining. (B) The HeLa cell transfectants were exposed to 20 Gy of IR and harvested at the indicated times. Cell lysates were subjected to immunoblotting with anti-PARP.
DISCUSSION
Functional significance of caspase-mediated proteolysis to apoptosis.
Members of the caspase family of cysteine proteases have been identified as key effectors responsible for the cleavage of proteins during the induction of apoptosis. Certain proteins inactivated as a result of caspase cleavage, such as DNA-PK and the retinoblastoma tumor suppressor, are not essential for cell viability. PARP, a protein involved in DNA repair and the suppression of an apoptotic endonuclease (39), is also cleaved during apoptosis (29, 36); however, the significance of this event is uncertain, since PARP-deficient cells exhibit an intact apoptotic response to DNA-damaging agents (75). Substrates that are activated as a consequence of caspase-induced cleavage include protein kinase Cδ (PKCδ) (19, 20), PKCθ (18), p21-activated kinase 2 (PAK2) (58), cytosolic phospholipase A2 (76), sterol regulatory binding proteins (51), the 45-kDa subunit of DNA fragmentation factor (40), and PITSLRE kinase α2-1 (7). Although expression of the cleaved fragments of PKCδ, PKCθ, or PAK2 induces certain characteristics of apoptosis (18, 25, 58), the available evidence is insufficient to assess the role of these activated products in cell death. Conversely, certain substrates that exhibit functional roles for caspase-mediated cleavage in the apoptotic response have been identified. A caspase-activated DNase that degrades DNA during apoptosis has recently been identified (22). Other work has shown that caspase-mediated cleavage of nuclear lamin and the actin regulatory protein gelsolin contributes to changes characteristic of apoptosis (34, 50, 56). Collectively, these findings have thus resulted in the identification of diverse proteins that are activated or inactivated by caspases and thereby contribute to the apoptotic program.
Rad51 is cleaved by a member of the caspase family.
The present results support cleavage of Rad51 by a caspase-mediated mechanism in IR-induced apoptosis. In this regard, Rad51 cleavage in irradiated cells is blocked by expression of the baculovirus p35 protein. p35 inhibits caspases 1, 2, 3, and 4 in vitro (9, 77) and blocks apoptosis induced by DNA-damaging agents (16) and diverse other stimuli (74). In contrast to p35, CrmA inhibits caspases 1 and 8 at nM concentrations in vitro (82) and blocks TNF-induced, but not DNA damage-induced, apoptosis (16, 17). In concert with these findings, CrmA had no effect on cleavage of Rad51 in irradiated cells.
The present findings also demonstrate that Rad51 is cleaved at a DVLD/N site by caspase 3 in vitro. Mutation of the aspartic acid residues to alanines (AVLA/N) blocked caspase-3-mediated cleavage of Rad51 in vitro. The finding that expression of the Rad51(D-A) mutant in cells also blocks IR-induced cleavage provided support for the physiologic importance of the DVLD/N site. After completion of these studies, other work which demonstrates that Rad51 is cleaved during the apoptosis of T lymphocytes was published (24). In contrast to our results, the Rad51 cleavage site was not mapped and Rad51 was not cleaved by caspase 3 (24). Thus, Rad51 may be subject to cleavage by different mechanisms. Whereas caspases 2, 3, and 7 cleave at DXXD sites (70, 72), any of these proteases may be responsible for cleavage of Rad51 in cells. For example, PARP cleavage has been attributed to caspase 3 in in vitro studies (29, 36), yet cells deficient in caspase 3 effectively cleave PARP during the induction of apoptosis (35). In contrast, the finding that Rad51 is not cleaved in caspase 3-deficient MCF-7 cells after IR treatment provides support for a caspase 3-dependent mechanism. Of interest is the present finding that Rad51 is not cleaved during TNF-induced apoptosis, yet lysates from TNF-treated cells mediate Rad51 cleavage. Whereas caspase 3 is activated by both TNF and IR treatment, cleavage of Rad51 may be dependent on a posttranslational modification that occurs during DNA damage-induced signaling and results in the subcellular accessibility of Rad51 to caspase 3-mediated proteolysis. In this context, recent studies have demonstrated that Rad51 is phosphorylated in IR-treated cells by the proapoptotic c-Abl tyrosine kinase and that c-Abl-mediated phosphorylation of Rad51 abrogates the binding of Rad51 to DNA (80).
Role for Rad51 in cell survival.
RecA in E. coli and ScRad51 in S. cerevisiae are essential for recombinational DNA repair. In yeast cells, rad51 mutants are sensitive to IR and defective in DNA damage-induced mitotic recombination (61). Also, ScRad51 expression is induced in response to IR and other DNA-damaging agents (1, 3, 61). Whereas yeast cells deficient in Rad51 are viable, disruption of the rad51 gene in mice results in an early arrest of embryonic development (38, 73). These findings indicate that Rad51 performs an essential function in mammalian and not in yeast cells, yet both rad51−/− cell types are defective in cell proliferation, exhibit increased radiosensitivity, and display chromosome loss (38, 44, 73). Antisense inhibition of Rad51 expression in mouse cells has confirmed an essential role for this protein in cell proliferation and in the repair of IR-induced DNA lesions (69). Other studies in Rad51−/− chicken cells expressing a human Rad51 transgene confirm a role for Rad51 in the repair of chromosome breaks that accumulate during proliferation (64). Mammalian Rad51 binds to ssDNA and thereby promotes homologous pairing and strand exchange in vitro (4). The present studies demonstrate that cleavage of Rad51 at the DVLD/N site generates N- and C-terminal fragments that retain binding to ssDNA. However, in a cell-based model of homologous recombination, the results demonstrate that cleavage at the DVLD/N site abrogates the Rad51 recombinase activity. Taken together, these findings support a model in which caspase-mediated cleavage of Rad51 in irradiated cells inhibits Rad51 activity and thereby recombinational repair. Proteolytic cleavage of Rad51 in the induction of apoptosis could also affect an essential function of Rad51 that involves interactions with other proteins such as p53 (65) or BRCA1 (59).
Caspase-mediated cleavage of Rad51 is a function of the apoptotic response.
The present results further demonstrate that transient overexpression of wild-type Rad51 and particularly the Rad51(D-A) mutant inhibits IR-induced apoptosis. These findings could be explained by the requirement of a critical level of Rad51 that is essential for recombinational repair of IR-induced DNA double-strand breaks and/or cell survival. In this context, our findings demonstrate that cells overexpressing Rad51 exhibit increased levels of recombinational activity. While it is perhaps unexpected that the level of homologous recombination is stimulated by overexpression of only Rad51, other studies have shown that mammalian cells overexpressing Rad52 exhibit increased homologous recombination and double-stranded DNA break repair (53). Taken together with our results and more recent studies demonstrating that Rad52 stimulates the function of Rad51 (5, 46, 63, 66), these findings suggest that overexpression of either component, i.e., Rad51 or Rad52, stimulates the level of homologous recombination and thereby the repair of IR-induced lesions. Alternatively, the present results do not exclude the possibility that overexpression of Rad51 interferes with DNA metabolism and results in DNA lesions that are repaired by recombination. In addition, given findings that Rad51 functions as a polymer in recombinational repair, our results do not exclude the potential for Rad51 cleavage products to be functionally active in the context of a mixed polymer with intact Rad51 that persists after IR treatment.
Activation of caspase 3 was similar in cells expressing wild-type or mutant Rad51 compared to those expressing the empty vector. These findings indicate that the caspase pathway is activated in response to IR-induced DNA damage in cells transfected to express wild-type or mutant Rad51. The demonstration that these cells also respond appropriately to IR with activation of c-Abl and the SAPK pathway (data not shown) supports an intact response to the DNA lesions induced by IR exposure. Moreover, the results suggest that cleavage of Rad51 and thereby inactivation of this protein may be involved in the cell death response. Compared to control cells, the transfectants expressing the wild-type protein exhibit increased levels of Rad51. If inactivation of Rad51 by proteolysis contributes to apoptotic cell death, then increased levels should be protective. Studies with the caspase-resistant Rad51 mutant support this hypothesis. Cells that express Rad51(D-A) were significantly more resistant to IR-induced apoptosis than cells that overexpress wild-type Rad51. Notably, overexpression of Rad51 or Rad51(D-A) could protect cells against apoptosis by the titration of caspases and/or other effectors of the cell death response. Thus, one could argue that Rad51 has little if any effect on apoptosis under physiological conditions. Nonetheless, the essential role for Rad51 in cell survival suggests that proteolytic inactivation of Rad51 could contribute in part to the induction of apoptosis.
ACKNOWLEDGMENTS
This investigation was supported by Public Health Service grant CA55241 awarded by the National Cancer Institute.
We thank Akira Shinohara for the anti-human Rad51 antibody and the human Rad51 cDNA and James Stringer for the FSH cells.
REFERENCES
- 1.Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semi-dominant suppressors of srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to prokaryotic RecA proteins. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong R C, Aja T, Xiang J, Gaur S, Krebs J F, Hoang K, Bai X, Korsmeyer S J, Karanewsky D S, Fritz L C, Tomaselli K J. Fas-induced activation of the cell death-related protease CPP32 is inhibited by Bcl-2 and by ICE family protease inhibitors. J Biol Chem. 1996;271:16850–16855. doi: 10.1074/jbc.271.28.16850. [DOI] [PubMed] [Google Scholar]
- 3.Basile G, Aker M, Mortimer R K. Nucleotide-sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol Cell Biol. 1992;12:3235–3246. doi: 10.1128/mcb.12.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann P, Benson F E, West S C. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 5.Benson F E, Baumann P, West S C. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 6.Benson F E, Stasiak A, West S C. Purification and characterisation of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyaert R, Kidd V J, Cornelis S, Van de Craen M, Denecker G, Lahti J M, Gururajan R, Vandenabeele P, Fiers W. Cleavage of PITSLRE kinases by ICE/CASP-1 and CPP32/CASP-3 during apoptosis induced by tumor necrosis factor. J Biol Chem. 1997;272:11694–11697. doi: 10.1074/jbc.272.18.11694. [DOI] [PubMed] [Google Scholar]
- 8.Bishop D K, Park D, Xu L Z, Keckner N. DMC1: a meiosis-specific yeast homolog of E. coli RecA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 9.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, Licari P, Mankovich J, Shi L, Greenberg A H, Miller L K, Wong W W. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 10.Cartier J L, Hershberger P A, Friesen P D. Suppression of apoptosis in insect cells stably transfected with baculovirus p35: dominant interference by N-terminal sequences p35(1–76) J Virol. 1994;68:7728–7737. doi: 10.1128/jvi.68.12.7728-7737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casciola-Rosen L, Nicholson D W, Chong T, Rowan K R, Thornberry N A, Miller D K, Rosen A. Apopain/CPP32 cleaves proteins that are essential for cellular repair: a fundamental principle of apoptotic death. J Exp Med. 1996;183:1957–1964. doi: 10.1084/jem.183.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandler J M, Cohen G M, MacFarlane M. Different subcellular distribution of caspase-3 and caspase-7 following Fas-induced apoptosis in mouse liver. J Biol Chem. 1998;273:10815–10818. doi: 10.1074/jbc.273.18.10815. [DOI] [PubMed] [Google Scholar]
- 13.Chauhan D, Pandey P, Ogata A, Teoh G, Krett N, Halgren R, Rosen S, Kufe D, Kharbanda S, Anderson K. Cytochrome-C dependent and independent induction of apoptosis in multiple myeloma cells. J Biol Chem. 1997;272:29995–29997. doi: 10.1074/jbc.272.48.29995. [DOI] [PubMed] [Google Scholar]
- 14.Clem R J, Fechheimer M, Miller L K. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 15.Clem R J, Miller L K. Control of programmed cell death by the baculovirus genes p35 and iap. Mol Cell Biol. 1994;14:5212–5222. doi: 10.1128/mcb.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta R, Banach D, Kojima H, Talanian R V, Alnemri E S, Wong W W, Kufe D. Activation of the CPP32 protease in apoptosis induced by 1-β-d-arabinofuranosylcytosine and other DNA damaging agents. Blood. 1996;88:1936–1943. [PubMed] [Google Scholar]
- 17.Datta R, Kojima H, Banach D, Bump N J, Talanian R V, Alnemri E S, Weichselbaum R R, Wong W W, Kufe D W. Activation of a CrmA-insensitive, p35-sensitive, pathway in ionizing radiation-induced apoptosis. J Biol Chem. 1997;272:1965–1969. doi: 10.1074/jbc.272.3.1965. [DOI] [PubMed] [Google Scholar]
- 18.Datta R, Kojima H, Yoshida K, Kufe D. Caspase-3 mediated cleavage of protein kinase Cθ in induction of apoptosis. J Biol Chem. 1997;272:20317–20320. doi: 10.1074/jbc.272.33.20317. [DOI] [PubMed] [Google Scholar]
- 19.Emoto Y, Kisaki H, Manome Y, Kharbanda S, Kufe D. Activation of protein kinase Cδ in human myeloid leukemia cells treated with 1-β-d-arabinofuranosylcytosine. Blood. 1996;87:1990–1996. [PubMed] [Google Scholar]
- 20.Emoto Y, Manome G, Meinhardt G, Kisaki H, Kharbanda S, Robertson M, Ghayur T, Wong W W, Kamen R, Weichselbaum R, Kufe D. Proteolytic activation of protein kinase Cδ by an ICE-like protease in apoptotic cells. EMBO J. 1995;14:6148–6156. doi: 10.1002/j.1460-2075.1995.tb00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enari M, Hug H, Nagata S. Involvement of an ICE-like protease in Fas-mediated apoptosis. Nature. 1995;375:78–81. doi: 10.1038/375078a0. [DOI] [PubMed] [Google Scholar]
- 22.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwanatsu A, Natata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 23.Fernandes-Alnemri T, Litwack G, Alnemri E S. CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1 beta-converting enzyme. J Biol Chem. 1994;269:30761–30764. [PubMed] [Google Scholar]
- 24.Flygare J, Armstrong R C, Wennborg A, Orsan S, Hellgren D. Proteolytic cleavage of HsRad51 during apoptosis. FEBS Lett. 1998;427:247–251. doi: 10.1016/s0014-5793(98)00433-5. [DOI] [PubMed] [Google Scholar]
- 25.Ghayur T, Hugunin M, Talanian R V, Ratnofsky S, Quinlan C, Emoto Y, Pandey P, Datta R, Kharbanda S, Allen H, Kamen R, Wong W, Kufe D. Proteolytic activation of protein kinase Cδ by an ICE/CED 3-like protease induces characteristics of apoptosis. J Exp Med. 1996;184:2399–2404. doi: 10.1084/jem.184.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta R C, Bazemore L R, Golub E I, Radding C M. Activities of human recombination protein Rad51. Proc Natl Acad Sci USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall, E. J. (ed.). Radiobiology for the radiologist, p. 17–38. Lippincott, Philadelphia, Pa.
- 28.Janicke R U, Sprengart M L, Wati M R, Porter A G. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J Biol Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 29.Kaufmann S H, Desnoyers S, Ottaviano Y, Davidson N E, Poirier G G. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 30.Kharbanda S, Pandey P, Schofield L, Roncinske R, Bharti A, Yuan Z-M, Weichselbaum R, Nalin C, Kufe D. Role for Bcl-xL as an inhibitor of cytosolic cytochrome C accumulation in apoptosis. Proc Natl Acad Sci USA. 1997;94:6939–6942. doi: 10.1073/pnas.94.13.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kharbanda S, Ren R, Pandey P, Shafman T D, Feller S M, Weichselbaum R R, Kufe D W. Activation of the c-Abl tyrosine kinase in the stress response to DNA-damaging agents. Nature. 1995;376:785–788. doi: 10.1038/376785a0. [DOI] [PubMed] [Google Scholar]
- 32.Kim C N, Wang X, Huang Y, Ibrado A M, Liu L, Fang G, Bhalla K. Overexpression of Bcl-xL inhibits araC-induced mitochondrial loss of cytochrome c and other perturbations that activate the molecular cascade of apoptosis. Cancer Res. 1997;57:3115–3120. [PubMed] [Google Scholar]
- 33.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 34.Kothakota S, Azuma T, Reinhard C, Klippel A, Tang J, Chu K, McGarry T J, Kirschner M W, Koths K, Kwiatkowski D J, Williams L T. Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science. 1997;278:294–298. doi: 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- 35.Kuida K, Zheng T S, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell R A. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 36.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 37.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 38.Lim D S, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindahl T, Satoh M S, Poirier G G, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 40.Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 41.Los M, van de Craen M, Penning L C, Schenk H, Westendorp M, Baeuerle P A, Droge W, Krammer P H, Fiers W, Schulze-Osthoff K. Requirement of an ICE-CED-3 protease for Fas/APO-1-mediated apoptosis. Nature. 1995;375:81–83. doi: 10.1038/375081a0. [DOI] [PubMed] [Google Scholar]
- 42.Ludwig D L, Stringer J R. Spontaneous and induced homologous recombination between lacZ chromosomal direct repeats in CV-1 cells. Somat Cell Mol Genet. 1994;20:11–25. doi: 10.1007/BF02257482. [DOI] [PubMed] [Google Scholar]
- 43.Maeshima K, Morimatsu K, Shinohara A, Horii T. Rad51 homologs in Xenopus laevis: two distinct genes are highly expressed in ovary and testis. Gene. 1995;160:195–200. doi: 10.1016/0378-1119(95)00148-y. [DOI] [PubMed] [Google Scholar]
- 44.Malkova A, Ivanov E L, Haber J E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCloskey T W, Oyaizu N, Coronesi M, Pahwa S. Use of a flow cytometric assay to quantitate apoptosis in human lymphocytes. Clin Immunol Immunopathol. 1994;71:14–18. doi: 10.1006/clin.1994.1045. [DOI] [PubMed] [Google Scholar]
- 46.New J H, Sugiyama T, Zaitseva E, Kowalczykowski S C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 47.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, Munday N A, Raju S M, Smulson M E, Yamin T-T, Yu V L, Miller D K. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 48.Nicholson D W, Thornberry N A. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 49.Ogawa T, Yu X, Shinohara A, Egelman E H. Similarity of the yeast Rad51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 50.Ohtsu M, Sakai N, Fujita H, Kashiwagi M, Gasa S, Shimuzu S, Eguchi Y, Tsujimoto Y, Sakiyama Y, Kobayashi K, Kuzumaki N. Inhibition of apoptosis by the actin-regulatory protein gelsolin. EMBO J. 1997;16:4650–4656. doi: 10.1093/emboj/16.15.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pai J T, Brown M S, Goldstein J L. Purification and cDNA cloning of a second apoptosis-related cysteine protease that cleaves and activates sterol regulatory element binding proteins. Proc Natl Acad Sci USA. 1996;93:5437–5442. doi: 10.1073/pnas.93.11.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park D S, Stefanis L, Yan C Y I, Farinelli S E, Greene L A. Ordering the cell death pathway. J Biol Chem. 1996;271:21898–21905. doi: 10.1074/jbc.271.36.21898. [DOI] [PubMed] [Google Scholar]
- 53.Park M S. Expression of human RAD52 confers resistance to ionizing radiation in mammalian cells. J Biol Chem. 1995;270:15467–15470. doi: 10.1074/jbc.270.26.15467. [DOI] [PubMed] [Google Scholar]
- 54.Pronk G J, Ramer K, Amiri P, Williams L T. Requirement of an ICE-like protease for induction of apoptosis and ceramide generation by REAPER. Science. 1996;271:808–810. doi: 10.1126/science.271.5250.808. [DOI] [PubMed] [Google Scholar]
- 55.Rabizadeh S D, LaCount D J, Friensen P D, Bredesen D E. Expression of the baculovirus p35 gene inhibits mammalian neural cell death. J Neurochem. 1993;61:2318–2321. doi: 10.1111/j.1471-4159.1993.tb07477.x. [DOI] [PubMed] [Google Scholar]
- 56.Rao L, Perez D, White E. Lamin proteolysis facilitates nuclear events during apoptosis. J Cell Biol. 1996;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ray C A, Black R A, Kronheim S R, Greenstreet T A, Sleath P R, Salvesen G S, Pickup D J. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1β-converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 58.Rudel T, Bokoch G M. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 59.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 60.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 61.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in Saccharomyces cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 62.Shinohara A, Ogawa T. Homologous recombination and the roles of double-strand breaks. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 63.Shinohara A, Ogawa T. Stimulation by Rad51 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 64.Sonoda E, Sasaki M S, Buerstedde J-M, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stürzbecher H-W, Donzelmann B, Henning W, Knippschild U, Buchhop S. p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 66.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 67.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 68.Sung P, Robberson D L. DNA strand exchange mediated by a Rad51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 69.Taki T, Ohnishi T, Yamamoto A, Hiraga S, Arita N, Izumoto S, Hayakawa T, Morita T. Antisense inhibition of the RAD51 enhances radiosensitivity. Biochem Biophys Res Commun. 1996;223:434–438. doi: 10.1006/bbrc.1996.0911. [DOI] [PubMed] [Google Scholar]
- 70.Talanian R V, Quinlan C, Trautz S, Hackett M C, Mankovich J A, Banach D, Ghayur T, Brady K D, Wong W W. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 71.Tewari M, Dixit V M. Fas- and tumor necrosis factor-induced apoptosis is inhibited by the poxvirus crmA gene product. J Biol Chem. 1995;270:3255–3260. doi: 10.1074/jbc.270.7.3255. [DOI] [PubMed] [Google Scholar]
- 72.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, Chapman K T, Nicholson D W. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 73.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Villa P, Kaufmann S H, Earnshaw W C. Caspases and caspase inhibitors. Trends Biochem Sci. 1997;22:388–393. doi: 10.1016/s0968-0004(97)01107-9. [DOI] [PubMed] [Google Scholar]
- 75.Wang Z-Q, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner E F. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 76.Wissing D, Mouritzen H, Egeblad M, Poirier G G, Jaattela M. Involvement of caspase-dependent activation of cystolic phospholipase A2 in tumor necrosis factor-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:5073–5077. doi: 10.1073/pnas.94.10.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xue D, Horwitz H R. Inhibition of the Caenorhabditis elegans cell-death protease CED-3 by a CED-3 cleavage site in baculovirus p35 protein. Nature. 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 78.Yang J, Liu X, Bhalla K, Kaekyung Kim C, Ibrado A M, Cai J, Peng T-I, Jones D P, Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 79.Yuan Z-M, Huang Y, Ishiko T, Kharbanda S, Weichselbaum R, Kufe D. Regulation of DNA damage-induced apoptosis by the c-Abl tyrosine kinase. Proc Natl Acad Sci USA. 1997;94:1437–1440. doi: 10.1073/pnas.94.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan Z M, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kharbanda S, Sung P, Shinohara A, Weichselbaum R, Kufe D. Regulation of Rad51 function by c-Abl in response to DNA damage. J Biol Chem. 1998;273:3799–3802. doi: 10.1074/jbc.273.7.3799. [DOI] [PubMed] [Google Scholar]
- 81.Yuan Z M, Huang Y, Whang Y, Sawyers C, Weichselbaum R, Kharbanda S, Kufe D. Role for the c-Abl tyrosine kinase in the growth arrest response to DNA damage. Nature. 1996;382:272–274. doi: 10.1038/382272a0. [DOI] [PubMed] [Google Scholar]
- 82.Zhou Q, Snipar S, Orth K, Muzio M, Dixit V M, Salvesen G S. Target protease specificity of the viral Serpin CrmA. J Biol Chem. 1997;272:7797–7800. doi: 10.1074/jbc.272.12.7797. [DOI] [PubMed] [Google Scholar]