ABSTRACT
The overuse and misuse of antibiotics in clinical settings and in food production have been linked to the increased prevalence and spread of antimicrobial resistance (AR). Consequently, public health and consumer concerns have resulted in a remarkable reduction in antibiotics used for food animal production. However, there are no data on the effectiveness of antibiotic removal in reducing AR shared through horizontal gene transfer (HGT). In this study, we used neonatal broiler chicks and Salmonella enterica serovar Heidelberg, a model food pathogen, to test if chicks raised antibiotic free harbor transferable AR. We challenged chicks with an antibiotic-susceptible S. Heidelberg strain using various routes of inoculation and determined if S. Heidelberg isolates recovered carried plasmids conferring AR. We used antimicrobial susceptibility testing and whole-genome sequencing (WGS) to show that chicks grown without antibiotics harbored an antimicrobial resistant S. Heidelberg population at 14 days after challenge and chicks challenged orally acquired AR at a higher rate than chicks inoculated via the cloaca. Using 16S rRNA gene sequencing, we found that S. Heidelberg infection perturbed the microbiota of broiler chicks, and we used metagenomics and WGS to confirm that a commensal Escherichia coli population was the main reservoir of an IncI1 plasmid acquired by S. Heidelberg. The carriage of this IncI1 plasmid posed no fitness cost to S. Heidelberg but increased its fitness when exposed to acidic pH in vitro. These results suggest that HGT of plasmids carrying AR shaped the evolution of S. Heidelberg and that antibiotic use reduction alone is insufficient to limit antibiotic resistance transfer from commensal bacteria to Salmonella enterica.
IMPORTANCE The reported increase in antibiotic-resistant bacteria in humans has resulted in a major shift away from antibiotic use in food animal production. This shift has been driven by the assumption that removing antibiotics will select for antibiotic susceptible bacterial taxa, which in turn will allow the currently available antibiotic arsenal to be more effective. This change in practice has highlighted new questions that need to be answered to assess the effectiveness of antibiotic removal in reducing the spread of antibiotic resistance bacteria. This research demonstrates that antibiotic-susceptible Salmonella enterica serovar Heidelberg strains can acquire multidrug resistance from commensal bacteria present in the gut of neonatal broiler chicks, even in the absence of antibiotic selection. We demonstrate that exposure to acidic pH drove the horizontal transfer of antimicrobial resistance plasmids and suggest that simply removing antibiotics from food animal production might not be sufficient to limit the spread of antimicrobial resistance.
KEYWORDS: antimicrobial resistance, horizontal gene transfer, Salmonella enterica
INTRODUCTION
Antimicrobial resistance (AR) spread is a worldwide health challenge (1, 2). A major aspect of this challenge stems from the ability of microbes to share their genetic material through horizontal gene transfer (HGT) (3). Exposure to antibiotics can select for a microbiota of commensals and pathogens that can withstand antibiotic selective pressure, create a reservoir of resistance genes that are acquired by susceptible taxa, and allow for their survival and proliferation (3). The overuse and misuse of antibiotics in a clinical setting (4) and in food production (5) have been shown to result in such selective environments contributing to the increased prevalence and spread of AR. These public health concerns have resulted in a major shift away from antibiotic use for food production in developed North American and European countries. This change has been driven by the assumption that removing this selective pressure will stop HGT of AR and allow the currently available antibiotic arsenal to be effective for a longer period (6–9). This shift in food production practices has highlighted new questions that need to be answered to understand and assess the effectiveness of antibiotic removal in reducing the spread and prevalence of AR in foodborne pathogens.
To shed some light on this problem, we focused on broiler chickens, the most consumed meat in the United States. Broiler chicken production accounted for the largest reduction in medically and nonmedically important antibiotics sold (∼47% from 2016 to 2017) in the United States (10). We chose Salmonella enterica serovar Heidelberg as the model pathogen because of its promiscuity to plasmids carrying AR and because of its virulence properties (11–14). Infections associated with S. Heidelberg have been reported to be more invasive than other serovars, including Salmonella enterica serovar Enteritidis or Salmonella enterica serovar Typhimurium (15, 16). Therefore, our goal was to evaluate if an antibiotic-susceptible S. Heidelberg strain will acquire plasmid-borne AR from the microbiome of neonatal chicks raised antibiotic free and to identify the nonantibiotic selective pressures and mechanism that might drive such HGT events.
RESULTS
The route of challenge affected S. Heidelberg abundance in ceca and litter.
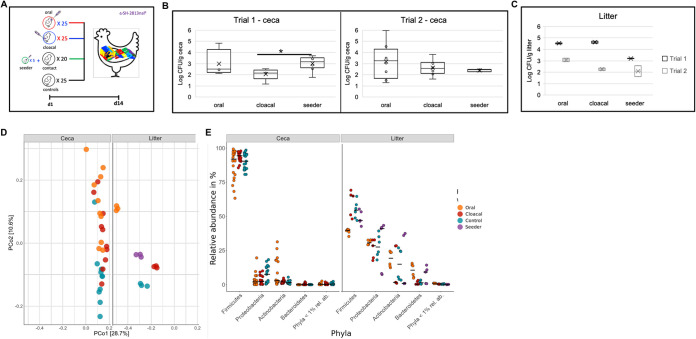
To determine if the route of exposure to S. Heidelberg affected intestinal and litter colonization, we challenged 150 1-day-old broiler chicks with a nalidixic (nal)-resistant strain of S. Heidelberg (SH-2813nalR) either through oral gavage, through cloacal inoculation, or by the seeder method (i.e., a few chicks [n = 5] were challenged orally and comingled with unchallenged chicks [n = 20]) (Fig. 1a). Chicks were raised for 14 days on fresh pine shavings in 4 separate floor pens (1.8-m length by 1.16-m width or 5.9-ft length by 3.8-ft width), including an unchallenged control group (n = 25). Chicks were not administered any medication or antibiotics for the duration of the study. The challenge experiments, namely, trial 1 and trial 2, were performed in September 2017 and April 2018, respectively, and we have published the rate that S. Heidelberg colonized the ceca (17).
FIG 1.
S. Heidelberg colonization changed the microbiome of broiler chicks (a) Experimental design for trial 1 and trial 2 conducted in September 2017 and April 2018, respectively. (b, c) Box plot of S. Heidelberg concentration in the ceca (n = 10 per trial) and litter (n = 2 per trial) 14 days after challenging chicks with S. Heidelberg through oral gavage, cloacal inoculation, or the seeder method (*, P < 0.05; Wilcoxon signed-rank test). (d) Principal-coordinate analysis of Bray-Curtis distances based on 16S rRNA gene libraries obtained from ceca and litter samples. Each point is a value from individual libraries with colors expressing the route used for S. Heidelberg challenge for respective samples. (e) Phylum‐level classification of 16S rRNA gene sequence reads in each cecum and litter sample grouped by route of S. Heidelberg challenge. Black bar represents the median.
S. Heidelberg colonization rates in the ceca were similar for trial 1 and 2 (test statistic for Wilcoxon signed-rank test [W] = 157, P = 0.90) (Fig. 1b), but chicks inoculated cloacally carried lower levels of S. Heidelberg than seeder chicks in trial 1 (W = 5, P = 0.03) (Fig. 1b). In the litter, S. Heidelberg levels were higher in trial 1 than those in trial 2 (W = 34, P = 0.009) for all treatments (Fig. 1c). The percentage of chicks that had S. Heidelberg detected in their ceca was 80% for the oral and seeder methods and 100% for the cloacal method during trial 1. For trial 2, 100% of the chicks in the oral and cloacal treatment were positive compared with 40% for the seeder group (n = 10, 10, and 15 chicks for oral, cloacal, and seeder treatments, respectively, in both trials). The ceca (n = 10 chicks) and litter of control chicks were negative for S. Heidelberg by culture; however, 1 litter sample was positive after a 24-h enrichment in buffered peptone water. Litter pH and moisture were higher for trial 1 (pH, 6.86 ± 0.27; moisture, 25% ± 0.03%) than those of trial 2 (pH, 6.54 ± 0.17; moisture, 21% ± 0.03%) (pH, W = 14, P = 0.11; moisture, W = 13, P = 0.18) (see Fig. S1 in the supplemental material).
Litter pH and moisture differed between trial 1 and 2. Litter was collected as grab samples (n = 7) and pooled into one whirl pak bag (n = 1 per pen). The pH of the litter was determined from the eluate made from a 1:5 dilution of litter in 1× PBS. Download FIG S1, TIF file, 0.1 MB (81.7KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
S. Heidelberg perturbed the ceca and litter microbiome in a challenge route-specific manner.
Next, we determined if S. Heidelberg challenge perturbed the microbiome of broiler chicks. We profiled the microbial community present in the ceca and litter using 16S rRNA gene amplicon sequencing. The bacterial alpha diversity was higher in the litter than in the ceca for the oral treatment for all alpha diversity measures (observed, Chao1, Shannon, and InvSimpson) examined (P < 0.01) (see Fig. S2a in the supplemental material). In addition, observed and Chao1 alpha diversity indices were higher for litter than ceca for cloacal and control treatments (P = 0.007). Contrastingly, the ceca from orally treated chicks had the lowest alpha diversity for all measures used compared with the control (P < 0.05).
S. Heidelberg challenge affected the diversity of bacterial species in ceca and litter. (a) Average alpha diversity indices of ceca and litter samples grouped by route of S. Heidelberg challenge. Box plots show the interquartile range (75th to 25th) of the data. Whiskers extend to the most extreme value within 1.5 × interquartile range, and dots are outliers beyond that range. (b) Heatmap of the top 50 most abundant amplicon sequence variants (ASVs) grouped by route of S. Heidelberg challenge and split by sample type (ceca and litter). Data are the average of ASV counts from replicate 16S rRNA gene libraries for each category. Download FIG S2, TIF file, 0.5 MB (510.1KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Beta diversity differed between the litter and ceca, and the route of S. Heidelberg challenge affected the beta diversity of the litter (F = 43.7, df = 11, P = 0.001) more than the ceca (F = 2.21, df = 29, P = 0.007) (Fig. 1d). Furthermore, cecal samples were more dispersed than litter samples. Firmicutes sp. dominated the cecal microbiome of broiler chicks (Fig. 1e) and Ruminococcaceae and Lachnospiraceae (class Clostridia) were the major families. Amplicon sequence variants (ASVs) matching Subdolinagrum, Lactobacillus, and Family_Lachnospiraceae were the most abundant ASVs in oral and cloacal chicks compared with controls in the ceca (Fig. S2b).
Firmicutes, Proteobacteria, Actinobacteria, and Bacteriodetes were the dominant phyla present in litter (Fig. 1e), and Escherichia/Shigella and Klebsiella were the most abundant ASVs in litter (Fig. S2b). Escherichia/Shigella was higher in cloacal and control chicks than in oral and seeder chicks, and Klebsiella was higher in control than in oral, cloacal, and seeder chicks (Escherichia/Shigella, χ2 = 9.9744, P = 0.018; Klebsiella, χ2 = 9.6667, P = 0.021). This result suggested that S. Heidelberg perturbed the bacterial community present in the litter and ceca of broiler chicks, especially members of the family Enterobacteriaceae.
Determining if S. Heidelberg acquired AR plasmid after broiler chicks were challenged.
To determine if S. Heidelberg acquired AR after 14 days of challenge, we used the National Antimicrobial Resistance Monitoring System Sensititre panel for Gram-negative bacteria to conduct antimicrobial susceptibility testing (AST). We performed AST on 250 S. Heidelberg isolates recovered from the ceca and litter of broiler chicks. SH-2813nalR carries a gyrA mutation for nal resistance; therefore, all isolates recovered were resistant to nal. S. Heidelberg also carries a chromosome-carried fosA7 gene that confers resistance to fosfomycin (18).
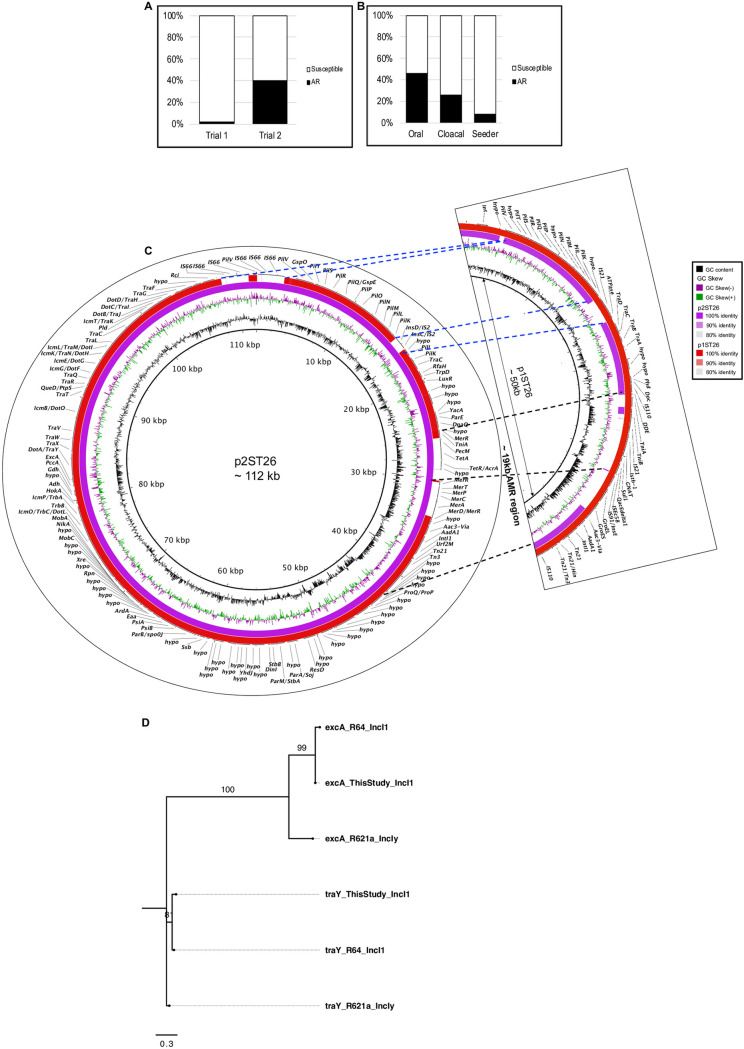
In trial 1, only 2% of S. Heidelberg isolates (n = 92) developed resistance to an antibiotic. These two isolates were resistant to gentamicin, streptomycin, and sulfisoxazole. For trial 2, 40% of the isolates (n = 158) developed resistance to at least 1 antibiotic (Fig. 2a) and >86% displayed resistance to gentamicin, streptomycin, and tetracycline (see Fig. S3a in the supplemental material). Chicks challenged orally carried a higher percentage (46%) of AR S. Heidelberg isolates than cloacally inoculated (24%) or seeder (8%) chicks (χ2 = 29.2, df = 2, P < 0.001) (Fig. 2b). A disk diffusion assay on a selected number of AR S. Heidelberg isolates from trial 2 (n = 4) revealed that these isolates were also resistance to tobramycin, netilmicin, and kanamycin (data not shown).
FIG 2.
S. Heidelberg acquired antibiotic-resistant plasmids from the microbiome of broiler chicks. (a) Percentage of S. Heidelberg isolates that acquired antibiotic resistance in trial 1 (n = 92) and trial 2 (n = 158) (b) Percentage of S. Heidelberg isolates in trial 2 that acquired antibiotic resistance grouped by the route the broiler chicks and were challenged (n = 54, 62, and 42 for oral, cloaca, and seeder treatment, respectively). (c) BLASTn alignment of the IncI1 plasmid acquired during trial 1 (p1ST26) and trial 2 (p2ST26); dashed black lines highlight mobile region carrying antimicrobial resistance genes, while blue dashed lines show other regions that are different between p1ST26 and p2ST26. (d) Maximum likelihood tree constructed using TraY and ExcA protein sequences from a representative IncI1 plasmid from this study, R64 (IncI1), and R621a (IncI1-gamma). The GTR model of nucleotide substitution and the GAMMA model of rate heterogeneity were used for sequence evolution prediction. Tree was rooted with the traY sequence of R621a.
Acquisition of IncI1 plasmid by S. Heidelberg conferred resistance to multiple antibiotics. Antimicrobial susceptibility was performed on S. Heidelberg isolates recovered from the ceca and litter of broiler chicks 14 days after challenge. (a) Percentage of S. Heidelberg isolates (n = 63) from trial 2 that acquired resistance to two or more antibiotics compared with isolate resistance to only 1 antibiotic. (b) Linear map of the ∼19-kb variable region carrying antimicrobial resistance genes, transposons, and integrase in IncI1-p1ST26 (∼102 kb). Download FIG S3, TIF file, 0.2 MB (202.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Next, we found the genetic determinants responsible for the acquired AR phenotype using whole-genome sequencing (WGS). We performed WGS on 69 S. Heidelberg isolates from trial 2 and 2 isolates from trial 1. We focused our resources on trial 2, for which AR acquisition was higher than trial 1. The isolates sequenced were either susceptible (n = 33) or carried resistance to at least one antibiotic in two antimicrobial drug classes (n = 38). The ancestral SH-2813nalR (isolate used for challenge) harbors an IncX1 plasmid, and carriage of this plasmid does not confer AR in S. Heidelberg. The IncX1 incompatibility (inc) region was not detected by PlasmidFinder (19) in 42 of the 71 isolates sequenced.
Thirty-nine percent of S. Heidelberg isolates acquired Col plasmids, but none carried a known antibiotic resistance gene (ARG). ARGs known to confer resistance to aminoglycosides [aadA1 and aac(3)-Via] and tetracyclines (tetA) were detected in all AR isolates from trial 2. The AR isolates from trial 1 harbored a sulfonamide resistance gene (sul1) in addition to aminoglycoside genes but no tet gene. The ARGs were found on a plasmid belonging to inc group IncI1 (Fig. 2c) and plasmid multilocus sequence type 26 (pST26). A change in AR profile was seen in four isolates from the seeder group; for these isolates, AST confirmed that they acquired AR but after WGS the AR determinants were not detected. Upon AST retest, these isolates were found to be susceptible. Acquired ARG was absent in four isolates with resistance to either tetracycline or streptomycin.
pST26 differed in its AR genetic context and genome architecture.
We questioned if the pST26s present in the sequenced S. Heidelberg isolates were similar or different in gene content or structure. To answer this question, we used hybrid assemblies (Illumina short reads and MinION or PacBio long reads) to achieve complete circular pST26 plasmids for two AR S. Heidelberg isolates recovered from trial 1 and trial 2. The pST26 plasmids from trial 1 (p1ST26) and trial 2 (p2ST26) were ∼112 kbp long and ∼87% identical (Fig. 2c). Both carried the atypical IncI1 backbone, including regions encoding replication, stability, leading, and conjugative transfer (20). To determine the IncI1-complex group, we used the classical traY and excA protein sequences of plasmids R64 (IncI1) and R621a (IncI1-gamma). A reconstructed tree confirmed that both plasmids belong to the IncI1 group (Fig. 2d). A 19.8-kb variable region encoding transposons and AR was found in both plasmids. For p1ST26, this region encoded aadA1, aac(3)-Via, sul1 and quaternary (quats) ammonium compound resistance (qacEΔ1) (Fig. 2c, Fig. S3b). In contrast, this region carried tetA and mer operons in addition to aadA1 and aac(3)-Via in p2ST26 (Fig. 2c).
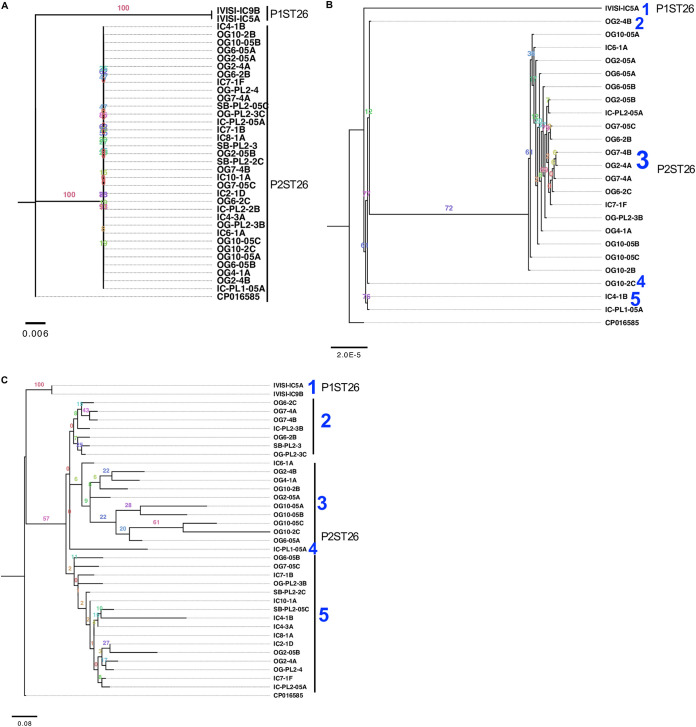
To investigate whether gene flux influenced IncI1 genome architecture, we used p1ST26 and p2ST26 as reference genomes for the rest of the AR isolates carrying IncI1. By aligning raw reads to complete pST26 genomes, we were able generate a consensus IncI1 plasmid contig for each isolate. A tree built with these plasmids resulted in two clades represented by p1ST26 and p2ST26 plasmids, respectively (Fig. 3a). Multiple alignment of protein sequences revealed that pST26 plasmids differed in the number of genes carried and gene alleles. This finding was pronounced for regions encoding AR, pilus/shufflon assembly proteins (pil), and IS66 family of transposases (see Data Set S1 in the supplemental material; Data Set S1 to S5are available in the Zenodo repository online at https://doi.org/10.5281/zenodo.4976002 and in the supplemental material). A pangenome analysis revealed that the pST26 from this study carried 70 core genes (genes present in ≥95% of the plasmids) and 90 accessory genes (genes present in <95% of the plasmids).
FIG 3.
Gene flux contributed to the diversity of IncI1 plasmids. (a to c) Maximum likelihood tree of pST26 IncI1 plasmids from this study (n = 36) constructed using complete plasmid DNA sequences (a), core genes (b), and accessory genes (c). The GTR+I, GTR, and JC+I models of nucleotide substitution and the GAMMA model of rate heterogeneity were used for sequence evolution prediction for a, b, and c, respectively (tree was rooted with the closest relative; GenBank CP016585), found through NCBI BLASTn search. Clade numbering was assigned arbitrarily to show the number of clades found. Numbers shown next to the branches represent the percentage of replicate trees where associated isolates cluster together based on ∼100 bootstrap replicates.
A tree reconstructed with the core genes and accessory genes divided the pST26 into 5 major groups (Fig. 3b and c). For both trees, the p1ST26 formed one clade and the p2ST26 made up the other clades. No single nucleotide polymorphisms (SNPs) were found between p2ST26s, but p1ST26 differed from p2ST26 with 21 SNPs and 2 insertions (Data Set S1, see Table S1 in the supplemental material). The insertions were present in only one p1ST26 plasmid. One insertion was found on tniA (a DDE-type integrase/transposase/recombinase protein) and the other was between tn3-like transposon and yadA. Inserted DNA showed significant homology to tniA present in E. coli plasmid EcPF5 (GenBank accession no. CP054237) and IS5057 present in E. coli plasmid pIOMTU792 (GenBank accession no. LC542972.1), respectively.
Mutations found in p1ST26 but absent in p2ST26. Download Table S1, DOCX file, 0.01 MB (14.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Determining if the acquisition of new genes affected the genome of S. Heidelberg.
We hypothesized that successful S. Heidelberg colonization will be dependent on gene flux (gain and loss of genes) and mutations. To test this hypothesis, we reconstructed a maximum likelihood (ML) tree based on the pangenome and mutations of S. Heidelberg strains recovered. The core genome (genes present in ≥95% of the strains) and accessory genome (genes present in <95% of the strains) was composed of 3,729 and 1,531 genes, respectively. A total of 91 new mutations (SNPs and indels) were on the chromosome of S. Heidelberg strains, and no single mutation was present in every strain. Rather, mutations were unique to individual strains or shared between 2 and 26 isolates (Data Set S2). Consequently, the core genome and SNP-based trees did not provide a clear division of strains based on plasmid acquired, and clades had low bootstrap support values (see Fig. S4a and b in the supplemental material).
Maximum likelihood tree reconstruction of S. Heidelberg isolates. Core genome (a) and SNP-based (b) maximum likelihood (ML) tree of S. Heidelberg isolates sequenced from trial 1 (n = 2) and trial 2 (n = 69). Isolates with duplicated SNPs/indels were removed before the SNP-based ML tree was reconstructed. All S. Heidelberg strains used for the core genome tree were assembled using Illumina short reads except OG8-05A, IC9B, and IC4-3A and SH2813-ancestral that were assembled using both Illumina short reads and PacBio or MinION long reads. The GTR and JC models of nucleotide substitution was used for core and SNP-based trees, respectively, and the GAMMA model of rate heterogeneity were used for sequence evolution prediction. Numbers shown next to the branches are the percentage of replicate trees where associated isolates cluster together based on ∼100 bootstrap replicates. The susceptible SH-2813 ancestor was used to root the for the core genome, while the SNP tree was rooted with the nalidixic-resistant S. Heidelberg ancestor (SH-2813nalR). (Nal, nalidixic acid; Gen, gentamicin; Str, streptomycin; Tet, tetracycline; Fis, sulfisoxazole; NA, not applicable)FIG S4, TIF file, 0.8 MB (824.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
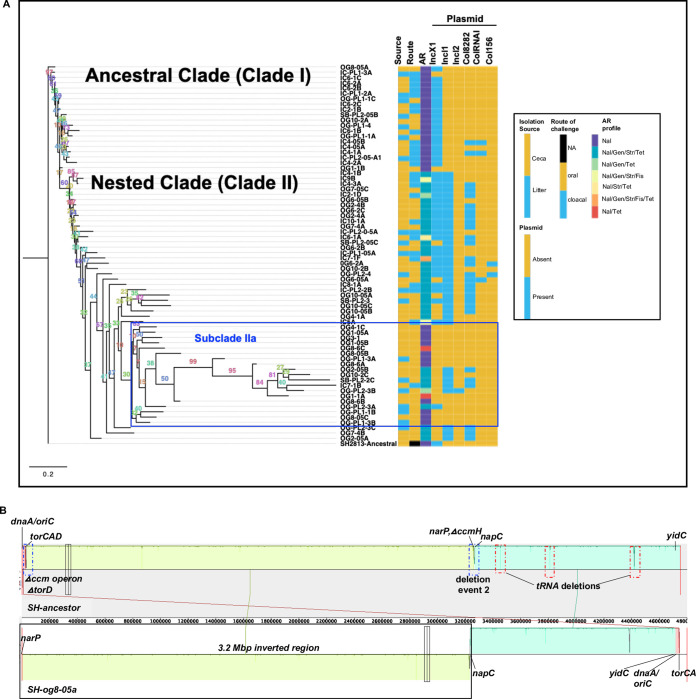
Contrastingly, the accessory genome tree grouped the strains based on AR phenotype, plasmid, and ARG carried (Fig. 4a). Acquisition of IncI (IncI1 and IncI2) and Col plasmids and the presence/absence of IncX1 plasmids defined the clades seen on the accessory genome tree. Susceptible strains carrying only IncX1 plasmids represented the ancestral clade and AR strains dominated nested clade II. Clade II strains carried only either IncI1 or IncI1 plus a Col plasmid or both were present with an IncX1 plasmid (Fig. 4a). A few susceptible and strains with no plasmid (n = 12) were nested within subclade IIa, and the genome of these strains showed signs of substantial gene loss (Fig. 4a, see Fig. S5a in the supplemental material). One S. Heidelberg strain (og8-05a) harbored an untypeable circular episome encoding uncharacterized proteins and two genes encoding NADH-quinone oxidoreductase (nuo) (∼22 kbp) (Fig. S5b) and an IncX1 plasmid with divergent protein sequences from the IncX1 present in the ancestor (Fig. S5c). Subclade IIa strains also had a higher number of contigs, higher misassemblies, and lower genome size than the rest of the strains sequenced for this study (test statistic for Kruskal-Wallis test [H] = 18.82, df =1, P < 0.001) (Fig. S5d to f).
FIG 4.
HGT and chromosome inversion changed the genome of S. Heidelberg. (a) Maximum likelihood tree constructed using accessory genes present in S. Heidelberg isolates (n = 72) recovered from the ceca and litter of chicks colonized with S. Heidelberg. The GTR model of nucleotide substitution and the GAMMA model of rate heterogeneity were used for sequence evolution prediction. Numbers shown next to the branches represent the percentage of replicate trees where associated isolates cluster together based on ∼100 bootstrap replicates. All S. Heidelberg strains were assembled using Illumina short reads except OG8-05A, IC9B, and IC4-3A and SH2813-ancestral that were assembled using both Illumina short reads and PacBio or MinION long reads. The tree was rooted with the ancestral susceptible S. Heidelberg strain. Clade numbers were assigned arbitrarily to ease discussion on isolates that acquired antibiotic resistance. A blue rectangular box highlights the subclade with susceptible strains nested within antibiotic resistance strains due to misassembly bias. (Nal, nalidixic acid; Gen, gentamicin; Str, streptomycin; Tet, tetracycline; Fis, sulfisoxazole; NA, not applicable). (b) Mauve visualization of the inverted genome of S. Heidelberg strain og8-05a. The chromosomal contigs of og8-05a were aligned and ordered to the complete chromosome of the S. Heidelberg ancestor. A colored similarity plot is shown for each genome, of which the height is proportional to the level of sequence identity in that region. When the similarity plot points downward, it shows an alignment to the reverse strand of the S. Heidelberg ancestor genome, i.e., inversion. A segment highlighted with solid black rectangular box denotes the inverted region in og8-05a, while dashed blue and red rectangular boxes denote segments with mutations.
Gene flux contributed to the diversity of S. Heidelberg. (a) Heatmap of genes present and absent in S. Heidelberg genomes determined by pangenome analysis. All S. Heidelberg strains were assembled using Illumina short reads except OG8-05A, IC9B, and IC4-3A and SH2813-ancestral that were assembled using both Illumina short reads and PacBio or MinION long reads. The top black rectangular box highlights S. Heidelberg strains that carried no plasmid, while the bottom rectangular box denotes strains with no plasmids but clustered with S. Heidelberg strains carrying IncI1. (b) Circular map of novel episome present in S. Heidelberg strain og8-05a. (c) BLASTn alignment of IncX1 present in S. Heidelberg ancestor and IncX1 present in S. Heidelberg strain og8-05a. (d to f) S. Heidelberg genome assembly statistics for S. Heidelberg strains with assembly error (n = 12; subclade IIa of ML tree reconstructed using accessory genes) (see Fig. 2a) compared with other S. Heidelberg strains (n = 59) (***P < 0.001; binomial logistic regression; error bar, standard deviation). Download FIG S5, TIF file, 1.3 MB (1.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To determine if erroneous assembly affected the clustering of this clade, we performed long read sequencing (PacBio) on one strain from this clade and used it in a hybrid approach with Illumina short reads. This procedure resulted in a partially complete chromosome (longest contig, 4.7 Mbp), but 30 noncircular contigs were not scaffolded. An alignment of the ordered contigs with the ancestor showed that the strain did not suffer significant gene loss as suggested through short or long read only assembly (see Table S2 in the supplemental material); however, a 3.2-Mbp chromosomal region was inverted (Fig. 4b). To confirm if chromosomal inversion influenced the assembly of these genomes, we aligned protein sequences of two complete circular S. Heidelberg chromosomes from trial 1 and 2 with the ancestor. This analysis showed that the isolate from trial 1 harbored an ∼2-Mbp chromosomal inversion (see Fig. S6a in the supplemental material).
Assembly report for a S. Heidelberg strain with sequencing bias. Download Table S2, DOCX file, 0.01 MB (13.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Chromosomal inversion reoriented the origin of replication of S. Heidelberg strains and positively skewed their GC. Complete chromosome contigs from three S. Heidelberg strains recovered from ceca during trial 1 (ic9b) and 2 (ic4-3a and og8-05a) were aligned and compared with the ancestor. (a) progressiveMauve alignment of S. Heidelberg strains ic4-3a and ic9b. A colored similarity plot is shown for each genome, of which the height is proportional to the level of sequence identity in that region. When the similarity plot points downward, it shows an alignment to the reverse strand of the genome of S. Heidelberg-ancestor, i.e., inversion. A segment highlighted with solid black rectangular box denotes the inverted 2-Mbp region in ic9b. A segment highlighted with a horizontal blue box denotes regions with relevant mutations. (b and c) GC skew in og8-05a and ic9b compared with S. Heidelberg-ancestor. The red arrow points toward reoriented oriC or ter, whereas red and blue arcs show the inverted region. The number of guanine and cytosine in inverted area was used for GC skew calculation. Dashed blue and red rectangular boxes denote segments with mutations. Download FIG S6, TIF file, 1.5 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Gene inversion changes the leading or lagging strand sequence to its reverse complement, thus altering the GC skew of the affected gene or genome from a positive value to a negative value or negative to a positive (21). Accordingly, we confirmed that there was a GC skew inversion in these regions (Fig. S6b and c). Gene inversions occur after rejoining DNA breaks and inverted genes can introduce sequencing bias and misassembly (22). Here, deletions affecting the cytochrome c maturation gene cluster (ccm) contributed to the inversion of genes in S. Heidelberg strain og8-05a (Fig. 4b, see Fig. S6b in the supplemental material). Furthermore, the oriC of og8-05a was reoriented compared with the ancestor. On the other hand, deletion of a putative YebC/PmpR transcriptional regulator and a deletion between cysK and cysZ led to the reorientation of oriC in strain ic9b (Fig. S6a and c).
The inverted region in these genomes encoded 34% to 60% of the 168 virulence genes present in the ancestor, including Salmonella pathogenicity islands 1 and 2, fimbrial and adherence genes, and type 1 and 2 secretion system proteins and prophages (Data Set S3; Fig. S6b and c). Gene inversions can increase the diversity and virulence of pathogens (21), and our bioinformatic analysis confirmed that these S. Heidelberg strains inverted their genomes. Nevertheless, in vivo testing will be required to confirm their virulence potential. Our results suggested that gene flux and homologous recombination shaped the genome of S. Heidelberg strains after they gained entry into the gut of broiler chicks.
Determining the commensal reservoir of IncI1 plasmids in broiler chicks.
The application of the proximity-ligation method (Hi-C) has improved the assembly of metagenomes and made it possible to detect plasmid-host associations (23). Therefore, we used Hi-C metagenomics to find the bacterial species carrying IncI1 plasmids in the microbiome of broiler chicks. First, we checked if Hi-C correctly assembled S. Heidelberg genome present in the ceca of challenged chicks. We selected two cecal samples from cloacal chicks from trial 2 (here referred to as Hi-IC-FL1 and Hi-IC-FL2). We chose cloacal chicks because all chicks in this group were colonized with S. Heidelberg compared to oral or seeder. The two chicks carried S. Heidelberg at ∼100 CFU/g of ceca.
A total of ∼125 million shotgun reads and ∼206 million Hi-C reads were obtained per sample. Taxonomic assignment of Hi-C metagenome-assembled genomes (MAGs) using Mash (24, 25) assigned 226 MAGs to a taxon for Hi-IC-FL1, while 87 MAGs were assigned a taxon for Hi-IC-FL2. One MAG from Hi-IC-FL1 and one from Hi-IC-FL2 were classified as Salmonella enterica serovar Cubana and S. Heidelberg, respectively. However, when we used a Salmonella serovar prediction tool (26) to validate the result from Mash, the MAGs (∼1.4- and 1.2-Mbp total contig size for Hi-IC-FL1 and Hi-IC-FL2, respectively) were not Salmonella sp. Furthermore, aligning these MAGs to a complete S. Heidelberg genome yielded poor alignment results (data not shown). CheckM also revealed that the Salmonella MAGs were incomplete for both Hi-IC-FL1 (42% completeness) and Hi-IC-FL2 (12.4% completeness). Similarly, taxonomic assignments performed with the Genome Taxonomy Database Toolkit (GTDB-Tk) (27) did not assign any MAG as Salmonella sp. Contrastingly, 23,610 ± 7,156 raw shotgun reads from each sample were classified as Salmonella enterica using the Kraken 2 database (28) (Fig. 5a). This result suggested that our metagenomic approach did not have the discriminatory power to identify the S. Heidelberg strain in the cecal samples.
FIG 5.
Proteobacteria was the main reservoir of IncI1 plasmids carried by broiler chickens. (a) Percentage of metagenomic reads assigned to the top 20 bacterial genera of the phylum Proteobacteria present in samples Hi-IC4 and Hi-IC6. Only reads assigned to phylum Proteobacteria (70.9% of the metagenomic reads) were used and zoomed in to show 0% to 1% of total reads within phylum Proteobacteria (96.7% of reads within phylum Proteobacteria were assigned to Komagataeibacter). (b and c) IncI1 contig hosts found by Hi-C contacts. IncI1-derived contigs (horizontal axis of heatmap) show specific Hi-C associations with metagenome-assembled genomes (MAGs) present in samples Hi-IC4 (a) and Hi-IC6 (b) (vertical axis of heatmap). MAGs are derived from Hi-C deconvolution of the metagenome assembly and placed into a bacterial phylogeny using Mash and CheckM. Cluster.10 in each sample is an Escherichia coli genome. In Hi-IC4, one MAG (cluster.82) representing an extremely fragmented and partially contaminated Escherichia coli genome is omitted for clarity (due to its contamination, this cluster was placed in Clostridia by CheckM). Heatmap values indicate transformed counts of Hi-C read contacts (indicating intracellular physical proximity of IncI1 contigs to those genomes). Heatmap values were pseudocounted to facilitate plotting of log-transformed data, including zeroes.
Next, we investigated if the MAGs harbored IncI1 plasmids. The IncI1 plasmid contigs detected in both samples were linked to E. coli MAGs (Fig. 5b and c; see Table S3 in the supplemental material) and multiple MAGs were classified as E. coli by Mash for both Hi-IC-FL1 (n = 3) and Hi-IC-FL2 (n = 12), and one MAG was ∼95% complete. Some bacterial MAGs assigned to the genus Firmicutes showed a low level of linkage to IncI1 (∼100× less compared than E. coli in Hi-IC-FL1). In addition, we found contigs (n = 151; mean contig size, 17,607 bp; smallest contig, 1,027 bp; largest contig, 282,389 bp) with protein sequences identical to pST26 in both Hi-IC-FL1 and Hi-IC-FL2 (Data Set S4). This result suggested that E. coli was the main reservoir of IncI1 plasmids.
Plasmids found in the cecal metagenome. Download Table S3, DOCX file, 0.01 MB (13KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Broiler chicks carried E. coli strains harboring pST26 IncI1 plasmids.
To confirm if broiler chicks from this study harbored E. coli populations that could serve as a reservoir of pST26 plasmids, we retrospectively screened the cecal contents of the broiler chicks used for Hi-C on CHROMagar supplemented with gentamicin and tetracycline and selected five colonies for whole-genome sequencing. As expected, we found pST26 in two strains (phylogroup F, multilocus sequence type [MLST] 6858), while the other three strains (phylogroup D, MLST 69) carried an IncI1 that harbored no ARG and an unknown MLST (Table 1). Next, we compared the pST26 in S. Heidelberg to that found in E. coli using complete plasmids. The p1ST26 and p2ST6 IncI1 plasmids of S. Heidelberg were 86.8% and 99.4% identical to pST26 present in E. coli. The main difference was in the region encoding AR and transposases in p1ST26 (see Fig. S7 in the supplemental material; Fig. S7 to S10 are available in Zenodo repository online at https://doi.org/10.5281/zenodo.4976002 and in the supplemental material).
TABLE 1.
Antibiotic-resistant E. coli genomes found in the ceca of broiler chicks
| Isolate identifier | Resistance phenotypea | Antibiotic resistance gene(s)b | Plasmidsc | Phylogroup | MLST |
|---|---|---|---|---|---|
| Ec-FL1-1X | Amp Gen Str Fis Tet | aac(3)-Iid aph(3′)-Ia aph(6)-Id aph(3″)-Ib blaTEM-1B tet (B) sul2 | IncF:B:A-, IncI1 | D | 69 |
| Ec-FL2-2X | Amp Gen Str Fis Tet | aac(3)-Iid aph(3′)-Ia aph(6)-Id aph(3″)-Ib blaTEM-1B tet (B) sul2 | IncF:B:A-, IncI1 | D | 69 |
| Ec-FL2-5X | Amp Gen Str Fis Tet | aac(3)-Iid aph(3′)-Ia aph(6)-Id aph(3″)-Ib blaTEM-1B tet (B) sul2 | IncF:B:A-, IncI1 | D | 69 |
| Ec-FL1-2Xd | Gen Str Tet | aadA1 aac(3)-Via tet(A) | IncF:B:A- IncI1 IncI2 Col8282 ColRNAI | F | 6858 |
| Ec-FL1-5Xd | Gen Str Tet | aadA1 aac(3)-Via tet(A) | IncF:B:A- IncI1 IncI2 Col8282 ColRNAI | F | 6858 |
Amp, ampicillin; Gen, gentamicin; Str, streptomycin; Tet, tetracycline; Fis, sulfisoxazole.
Boldface denotes antibiotic resistance genes and phenotype carried on the chromosome.
Illumina short reads and PacBio long reads were combined to assemble the plasmids of E. coli strains Ec-FL1-1X and Ec-FL1-2X.
E. coli isolates carrying IncI1-pST26 with >99% identical DNA sequence with p2ST26 acquired by S. Heidelberg.
pST26 carriage poses a variable fitness cost and benefit under selection pressure.
pST26 carried accessory genes for AR, metal resistance, or disinfectants (i.e., quaternary ammonium compounds). Furthermore, S. Heidelberg strains carrying pST26 differed by route of exposure, e.g., higher proportion in oral versus cloacal exposure. To this end, we questioned if any of these factors could exert a selective pressure for pST26. We did not administer antibiotics to broiler chicks throughout study; therefore, we did not consider antibiotic selective pressure. We detected metals, including nickel, chromium, copper, and zinc, in the starter feed in parts per million; and we found contaminants, such as arsenic, lead, and cadmium, in trace amounts (see Table S4 in the supplemental material). Disinfectants, including quaternary ammonium compounds, were used for cleaning broiler houses and equipment; thus, we considered disinfectant as another source of selective pressure in this study.
Concentration of metals and pollutants in starter feed. Download Table S4, DOCX file, 0.01 MB (13.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To determine their effect on S. Heidelberg fitness, we used phenotype MicroArray (PM) 96-well plates supplemented with selected metal chlorides and disinfectants at various concentrations to compare a p2ST26-carrying strain (here referred to as genR) to a susceptible evolved strain. There was no significant difference in metabolic activity for S. Heidelberg strain carrying genR compared with its susceptible counterpart for metals and disinfectant, but genR exhibited a higher metabolism for benzethonium chloride, chromate, tellurite, and zinc, whereas the susceptible strain showed higher metabolism for dequalinum chloride, chromium, and cesium (χ2 = 1.83, df = 1, P = 0.1762) (Fig. S8a and b).
Cox and colleagues have suggested that the acidic pH of the upper gastrointestinal (GI) tract was associated with the lower number of chicks positive for Salmonella and Campylobacter sp. after oral gavage than that after cloacal inoculation (29–31). Based on this premise, we hypothesized that S. Heidelberg strains carrying genR will survive better under exposure to acidic pH. To address this hypothesis, we first compared the survival of the strains under different pH levels (3.5 to 10) with or without nitrogen sources and amino acids using PM plates. An S. Heidelberg strain carrying genR showed lower metabolic activity than the susceptible strain at pH 3.5 to 4 (V-statistic for Wilcoxon signed-rank test [V] = 3; P = 0.5), at pH 4.5 with and without nitrogen sources (V = 599; P = 3.426e-06), and at pH 9.5 to 10 with and without sources of nitrogen (V = 714; P = 6.547e-07) (Fig. 6a).
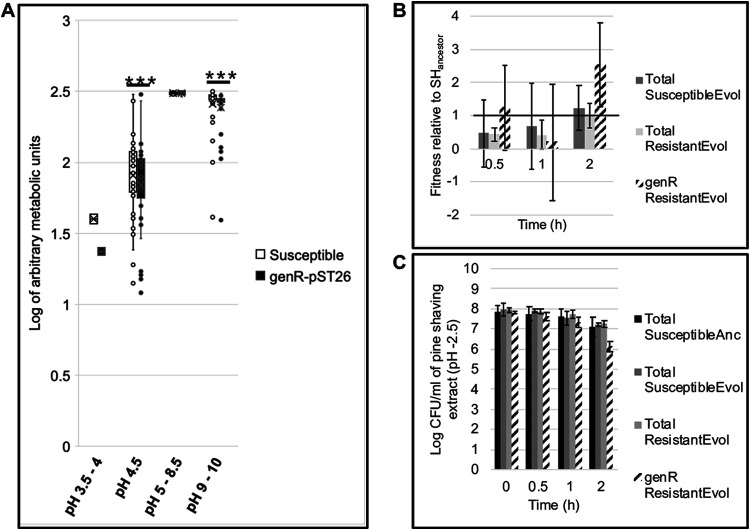
FIG 6.
S. Heidelberg exposure to acidic pH imposes negative frequency-dependent selection for IncI1. (a) Box plot comparing the metabolic activity of one evolved susceptible (no IncI1) and one genR (carries p2ST26) S. Heidelberg strain using phenotype microarray (PM) plates (Biolog Inc.). Preconfigured PM plates are composed of microtiter plates with one negative-control well and 95 wells prefilled with or without nitrogen and at pH of 3.5 to 10 (n = 1 plate per strain; the number of wells per plate for pH 3.5 to 4, 4.5, 5 to 8.5, and 9 to 10 was 2, 6, 37, and 38, respectively; error bars, standard deviation; ***, P < 0.001 (Wilcoxon signed-rank test). (b and c) Fitness and abundance of evolved susceptible and genR S. Heidelberg strains when exposed to pine shaving extract of pH 2.5. Each bar represents the average fitness or abundance (CFU per ml of pine shaving extract) of three individual populations that were established from three single bacterial colonies from three different strains (error bars, standard deviation; P > 0.05; Kruskal-Wallis rank-sum test). The horizontal line in b represents the fitness of three S. Heidelberg ancestor populations established from three single bacterial colonies. (Anc, ancestor; Evol, evolved, i.e., S. Heidelberg isolate recovered during in vivo experiment; genR, population resistant to gentamicin). Values of <1 show a reduction in fitness, while >1 indicates an increase in fitness.
Next, we compared the survival of the susceptible ancestor, susceptible evolved strains, and genR evolved strains (n = 3 strains for each population) in pine shaving extract (PSE) adjusted to a pH of 2.5. We acclimatized the bacterial population to PSE (pH 6.5) for 2 h before exposing it to acidified PSE (Fig. S8c). We screened for genR-carrying populations using the gentamicin resistance marker on p2ST26. We found that p2ST26 carriage produced higher variable fitness effects at all time points than the susceptible strains (Fig. 6b). For instance, two genR strains showed a decrease in fitness (values of <1 show a reduction in fitness [w], while a value of >1 indicates an increase in fitness) after 30 minutes in PSE, while one strain showed an increase (w = 2.7). After 2 h of exposure, there was no significant difference in fitness between susceptible and genR populations, but cells carrying genR exhibited higher fitness than their susceptible counterparts (χ2 = 4.35, df = 2, P = 0.113) (Fig. 6b). Similarly, there was no significant difference in the final population of susceptible and evolved strains; however, the genR strain had a lower population size (χ2 = 6.37, df = 3, P = 0.095) (Fig. 6c). This result suggested that p2ST26 carriage poses a variable fitness cost and benefit on the S. Heidelberg host when exposed to acidic pH.
DISCUSSION
In this study, neonatal Cobb 500 broiler chicks challenged with a susceptible S. Heidelberg strain carried susceptible and antimicrobial resistant S. Heidelberg populations 2 weeks after inoculation in their ceca and litter. This resistance phenotype was conferred by the acquisition of IncI1-pST26 plasmids. These plasmids are present in S. Heidelberg, S. Typhimurium, Salmonella enterica serovar Anatum, Salmonella enterica serovar Derby, Salmonella enterica serovar Schwarzengrund, Salmonella enterica serovar Saintpaul, and E. coli strains isolated from animal and human sources (Fig. S9). The closest IncI1 plasmids to pST26 from this study are pST26 present in E. coli (CP018625) and S. Typhimurium (CP027409). The plasmids carry aminoglycoside resistance genes with or without tetracycline, sulfonamide, and qac and mer resistance genes, making it an antibiotic/disinfectant/metal resistance plasmid.
The acquisition of pST26 was higher in chicks challenged orally than in chicks inoculated via the cloaca in trial 2. This route-specific rate of conjugation suggested that the selection for pST26 in our study was associated with the differential pH in the upper GI versus the lower GI tract. This finding was corroborated by in vitro experiments, where acidic pH reduced the metabolism of S. Heidelberg cells carrying p2ST26 compared with S. Heidelberg cells with no p2ST26. Furthermore, carriage of p2ST26 imposed a variable fitness effect on S. Heidelberg exposed to acidic pH. This type of acid-imposed selection for p2ST26 alludes to the “negative frequency-dependent selection hypothesis”—where the fitness of the population is higher when a plasmid is present at a low frequency in the population (32) and also supports the tenet that variability of fitness effects contributes to plasmid maintenance and persistence in bacterial communities (33).
The length of the GI tract and the route traveled could make it challenging for S. Heidelberg to colonize the ceca of chicks or acquire AR via HGT. When we measured the distance of the cloaca and esophagus to the ceca of a 49-day-old Cobb 500 broiler chicken (photo not shown), we estimated that S. Heidelberg will have to travel ∼20× farther to get to the ceca of chicks gavaged orally than chicks inoculated cloacally. It is plausible that this longer “residence” time in the upper GI tract allows more opportunities for S. Heidelberg to make contact and compete with other members of the chicken microbiome. In this study, such an event could also explain the higher transfer rate of p2ST26 to S. Heidelberg in chicks that were gavaged.
It is noteworthy that AR acquisition occurred at a lower rate during trial 1 than during trial 2. On one hand, the lower pH of the litter in trial 2 than that in trial 1 supports our acid-imposed selection hypothesis; alternatively, several other factors can be responsible for the observed difference, including the season we conducted the experiments. Trial 1 was done in the fall, and trial 2 was performed in the spring. The temperature outside for trial 1 was higher (average, 70°F; high, 90°F; low, 53°F) than trial 2 (average, 57°F; high, 81°F; low, 34°F) (www.timeanddate.com/weather/usa/athens/). The temperature and relative humidity inside a broiler house are correlated with the weather outside, and these environmental parameters are controlled by manipulating the ventilation system installed in the house. Broiler house ventilation rates have a direct effect on the moisture, pH, ammonia levels, and microbiome of the litter (34). The higher levels of S. Heidelberg in the litter from trial 1 than that from trial 2 suggest that high house temperature, litter moisture, and litter pH selected for an S. Heidelberg population that had no plasmid-borne AR. Lastly, we did not administer antibiotics to the 1-day-old chicks used for both trials and do not have the information to speculate on the antimicrobial practices of the hatchery or parent/breeder farms of the chicks. One or all of these factors could explain the differences between trials and indicate that further studies are needed before the results can be generalized.
The IncI1 complex is widespread in Enterobacteriaceae and has been reported to be the major driver of AR in Salmonella sp. and E. coli. Yet, our understanding of its mode of transfer and AR evolution has been limited to studies on its prevalence, mating experiments, and in vivo challenge with mice (20, 35, 36). In this study, IncI1 plasmid contigs found using metagenomics were linked to E. coli MAGs, and this result was corroborated by WGS of E. coli isolates. The E. coli isolates sequenced carried IncI1 plasmids that were similar to the pST26 acquired by S. Heidelberg. Fischer et al. (37) showed that E. coli populations in broiler chickens acquired β-lactam resistance after 4-days-old broiler chicks were gavaged with an E. coli strain carrying blaCTX-M-1 on an IncI1 plasmid. In a twin pair experiment, Hagbo et al. (38) revealed that IncI1 carrying blaCMY-2, ColE1, and P1 bacteriophage are often transferred between microbiota present in the preterm infant gut. Salmonella resistance to β-lactams in retail meat was shown to correlate with AR found in E. coli, and horizontal transfer was the hypothesis put forward (39). This study and others have demonstrated that IncI1 is the most prevalent plasmid carrying β-lactam in Enterobacteriaceae and that they are transferable from Salmonella to E. coli and vice versa.
The nutrient-niche hypothesis suggests that S. Heidelberg will be able to invade a new niche only if it can metabolize a growth-limiting resource or if it can outcompete a resident species and a vacant niche arises due to a new component in diet or by eliminating the competitor (40). Metagenomics revealed that the two most abundant bacterial phyla in the ceca and litter from this study were Firmicutes and Proteobacteria. Therefore, bacterial species belonging to these phyla are the resident bacteria with which S. Heidelberg will have to compete. The most abundant genera of Firmicutes found by aligning shotgun reads to the Kraken database were Staphylococcus, Lachnoclostridium, Streptococcus, Enterococcus, Faecalibacterium, and Lactobacillus. Firmicutes have been shown to be the dominant phylum in chicken ceca in earlier studies, with relative abundances usually well above >50% as found by 16S rRNA gene sequencing (41, 42). Comparable results have also been demonstrated with shotgun metagenomics (43–45). Interestingly, S. Heidelberg inoculation did not have an impact on the abundance of Firmicutes in this study. Contrastingly, we saw an effect of S. Heidelberg inoculation on Proteobacteria and was significantly associated with the genera Escherichia/Shigella and Klebsiella. The most abundant species of these genera found in ceca using shotgun metagenomics were E. coli, Shigella dysenteriae and Klebsiella pneumoniae.
Competition for oxygen was shown to be the limiting factor that contributed to the virulence of S. Enteritidis toward E. coli in neonatal chickens (46). Similarly, competition between S. Heidelberg strains and members of the family Enterobacteriaceae played a key role in selecting for antibiotic-resistant S. Heidelberg in our study. This conclusion was drawn from results showing that S. Heidelberg challenge perturbed the bacterial community of broiler chicks and S. Heidelberg acquired plasmids found only in E. coli populations. For S. Heidelberg and E. coli to coevolve and persist in the same new niche, a recombination of genes occurred and led to the emergence of new S. Heidelberg strains. In our study, S. Heidelberg strains carrying pST26 appeared after conjugating with E. coli populations. As expected, S. Heidelberg genomes were grouped into nine core genome MLST clusters, with eight clusters representing new strains (Fig. S10). The S. Heidelberg strains that acquired pST26 exhibited variable fitness under acid exposure experiments and are expected to be persist longer in the new niche. Therefore, we conclude that simply removing antibiotics from food animal production might not be sufficient to reverse the spread of antimicrobial resistance.
MATERIALS AND METHODS
S. Heidelberg inoculum preparation.
We have previously described the characteristics of the S. Heidelberg strain (SH-2813nalR) used for this study and how the inoculum was prepared (17). Briefly, the strain belongs to the multilocus sequence type 15; carries a 37-kb conjugative plasmid; and is resistant to erythromycin, tylosin, and fosfomycin. The strain was recovered from a broiler chicken carcass in 2013 and made resistant to 200 ppm of nalidixic acid (nalR) for selective enumeration. The nalR phenotype is conferred by a serine-to-tyrosine substitution at position 83 of DNA gyrase subunit A protein (GyrA). The S. Heidelberg inoculum was grown overnight in poultry litter extract, centrifuged, and resuspended in 1× phosphate-buffered saline (PBS). The resuspended cells were used as inocula. The genome of the ancestor to SH-2813nalR was sequenced using Illumina and PacBio sequencing to achieve a complete circular chromosome and plasmid (GenBank accession number CP066851). SH-2813nalR carries six mutations (Data Set S2), including the mutation affecting gyrA, that are not present in the ancestor.
Challenging broiler chicks with S. Heidelberg.
One-day-old Cobb 500 broiler chicks were purchased from a commercial hatchery in Cleveland, Georgia. Upon purchase, chicks were placed in plastic crates lined with brown paper and transported to the University of Georgia, Poultry Research Center (33.90693362620281, −83.37871698746522). One hundred chicks were either uninoculated (n = 25), gavaged orally (n = 25), or inoculated cloacally (n = 25) with a 100 μl-volume of S. Heidelberg inoculum (Fig. 1a). We included a seeder colonization method, whereby five gavaged chicks were mingled with 20 uninoculated chicks. Each inoculated chick received ∼106 colony forming units (CFU) of SH-2813nalR. Afterward, chicks were placed in floor pens at a stocking density of 0.65 m2/chick on fresh pine shaving litter. Broiler chicks were given water and feed ad libitum and raised antibiotic-free on starter diet for 2 weeks (starter feed was synthesized by the University of Georgia poultry research center feed mill). The concentration of metals and priority pollutants in feed was determined by the University of Georgia’s feed and environmental water laboratory (Athens, GA, USA) (see Table S4 in the supplemental material). Husbandry and management followed commercial broiler chicken industry guidelines. After 2 weeks, 10 chicks from oral, cloacal, and control groups and 15 chickens from seeder (5 seeder chicks and 10 uninoculated pen mate) were sacrificed to determine the extent of S. Heidelberg colonization in the ceca. The experiments were done in two trials conducted in September 2017 (trial 1) and April 2018 (trial 2). The study was approved by the University of Georgia Office of Animal Care and Use under Animal Use Protocol A2017 04-028-A2.
Cecal and litter bacteriological analyses.
Ceca were aseptically removed from the eviscera of 2-week-old broiler chicks, placed in a stomacher bag, and transported on ice to the U.S. National Poultry Research Center for analysis. Ceca were weighed, and buffered peptone water (BPW) (BD Difco, MD, USA) was added 3× volume to the weight (v/w) and stomached for 60 s. Serial dilutions were made and plated onto Brilliant green sulfa agar (BGS) (BD Difco, MD, USA) containing 200 ppm nal. In addition, a 10-μl inoculating loop was used to streak cecal slurry (cecal contents in BPW) onto xylose lysine tergitol-4 agar (BD Difco, Sparks, MD) supplemented with 4 ppm tetracycline for trial 1 and BGS supplemented with 32 ppm ampicillin (amp) or 32 ppm streptomycin for trial 2. All antibiotics were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise noted. Plates were incubated along with the cecal slurry for 24 h. All bacterial incubations were carried out at 37°C, unless otherwise noted. After incubation, colonies were manually counted, and serial dilution plates with 2 to 100 colonies were used for CFU per gram calculation. If no colonies appeared on serial dilution plates, a cecal slurry was streaked onto a new BGS plus nal plate and incubated overnight. After incubation, plates were examined for the presence/absence of Salmonella colonies.
Broiler chicken litter was collected as grab samples from 7 locations (4 corners of the pen and under the waterer) in each pen after chicks were removed. The litter samples were pooled, and 30 g was processed in duplicates from each pen as previously described (47). Serial dilutions of the litter slurry were made and plated onto BGS agar containing 200 ppm nal. Plates were incubated overnight, and colonies were manually counted and reported per gram litter dry weight. Litter pH and moisture were determined as described previously (47). We selected randomly 2 to 6 single colonies from BGS plates supplemented with nal from each ceca and litter sample and archived them in 30% Luria Bertani broth (LB) glycerol at −80°C. In addition, cecal slurry was saved at a 4:1 ratio in Luria Bertani broth (BD Difco, MD, USA) containing 30% glycerol at −80°C, whereas litter samples were stored in vacuum-sealed whirl pak bags at −20°C.
Antimicrobial susceptibility testing.
We performed antimicrobial susceptibility testing (AST) on 2 50 S. Heidelberg isolates recovered from the ceca and litter of broiler chicks following the National Antimicrobial Resistance Monitoring System (NARMS) protocol for Gram-negative bacteria. MICs for isolates were determined by broth microdilution using the Sensititre semiautomated antimicrobial susceptibility system (Thermo Fisher Scientific, Inc., MA, USA) and interpreted according to clinical and laboratory standards institute guidelines when available; otherwise, breakpoints established by NARMS were used. AST was also done on E. coli isolates (n = 5) recovered from the ceca of two broiler chicks challenged intracloacally with S. Heidelberg. The Kirby-Bauer disk diffusion assay for tobramycin, kanamycin, neomycin, and netilmicin was done on four gentamicin-resistant S. Heidelberg isolates as previously described (47).
DNA extraction.
DNA was extracted and purified from bacterial colonies using a FastDNA spin kit (MP Biomedicals, LLC, CA, USA), whereas 250 mg of cecal and litter was extracted with the Qiagen DNeasy power soil DNA kit (Hilden, Germany). The modifications we made to the manufacturer’s protocol for DNA extraction have been reported (47, 48).
16S rRNA gene sequence processing and analysis.
Cecal and litter DNA were used for bacterial community analysis through the sequencing of the V4 hypervariable region of the 16S rRNA gene of bacterial genomes. Sequencing was done using the paired‐end (2 × 250 bp) method on the Illumina MiSeq platform. Statistical analysis of microbial communities was performed in the R environment using the packages “phyloseq,” “Ampvis2,” and “vegan.” Alpha diversity indices were calculated using the function “estimate_richness” from phyloseq with a data set rarefied to the minimum sample size (8,740 sequences) (49). After assessing normal distribution by qqplots, histograms, and the Shapiro-Wilk normality test, the not normally distributed groups were compared using the Wilcoxon signed-rank test. Beta diversity was calculated with “amp_ordinate” in the ampvis2 package (50). Data have been Hellinger transformed, and ASVs that were not present in more than 0.1% relative abundance in any sample have been removed. The principal-coordinate analysis was based on Bray-Curtis distances. The permutational multivariate analysis of variance (PERMANOVA) was calculated with the “adonis” function and 5,000 permutations in vegan (51). All raw FASTQ reads for 16S rRNA gene sequences have been deposited under NCBI accession number PRJNA669215.
Whole-genome sequencing and processing.
Illumina short read sequencing was performed on DNA extracted from S. Heidelberg isolates recovered from ceca and litter. In addition, five E. coli isolates recovered from two cecal samples were sequenced. Libraries were prepared using either Nextera XT or Nextera DNA flex library preparation kits (Illumina, Inc., San Diego, CA) following the manufacturer’s protocol. Libraries were sequenced on the Illumina MiSeq platform with 150- or 250-bp paired-end reads. Additionally, five S. Heidelberg and two E. coli isolates were selected for long-read sequencing using the Sequel II system (PacBio Biosciences Inc.) or MinION device (Oxford Nanopore Technology).
Preparation and sequencing of long read libraries were done by next-generation sequencing core centers of University of Georgia and Colorado State University. MinION sequencing of samples was done using R9.5 chemistry on a 1D flowcell. Lima (https://github.com/PacificBiosciences/pbbioconda) was used to demultiplex barcoded PacBio samples and to convert the split BAM files into FASTQ format. BBMap reformat.sh (https://github.com/BioInfoTools/BBMap) was used to randomly subsample generated PacBio reads down to 200× coverage. Porechop v0.2.4 (https://github.com/rrwick/Porechop) was used to demultiplex barcoded MinION samples. Raw Illumina reads were trimmed using Trimmomatic v0.39 (52) (command line parameters, PE ILLUMINACLIP:NexteraPE-PE.fa:2:30:10:3 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:36).
Genome assembly, resistome characterization, and quality assessment of long reads were done using Reads2Resistome pipeline v1.1.1 (53). Reads2Resistome performed hybrid assemblies using either Illumina reads and PacBio or Illumina and MinION reads, using both Unicycler (54) and SPAdes (55) (see Data Set S5 for assembly statistics). In addition, hierarchical genome assembly process (HGAP) assembly was performed using the PacBio single-molecule real-time (SMRT) Link v9.0 analysis software suite using default settings. Assembly quality was assessed by QUAST v5.0.2 (56), using default settings, and genome annotation was done using Prokka (57), Rapid Annotation using Subsystem Technology (RAST) (58), and BlastKOALA (59). We confirmed that all S. Heidelberg isolates were Salmonella enterica serovar Heidelberg using Salmonella In Silico Typing Resource (SISTR) (26) and used ClermonTyping (60) and multilocus sequence typing (MLST) (61) for E. coli strains.
For resistome characterization, Reads2Resistome used ARG-ANNOT (62), the Comprehensive Antibiotic Resistance Database (63), MEGARes (64), AMRFinderPlus (65), PlasmidFinder (19), ResFinder (66), and VirulenceFinder database (67). For plasmid typing and IncI1 clonal complex determination, we used plasmid MLST (19). PHAST (68) was used to identify prophages present in chromosomal contigs, and the predicted prophage DNA sequence was annotated with RAST and BlastKOALA. progressiveMAUVE v1.1.1 (69) and MAFFT v1.4.0 (70) implemented in Geneious Prime v2020.0.1 were used for aligning and comparing sequences. In addition, a pangenome analysis of annotated assemblies was conducted with Roary (71). Phylogenetic trees based on the core genome and accessory genome were reconstructed using the maximum likelihood (ML) method implemented in RAxML-NG 2.0.0 (72). When computationally possible, the best model of sequence evolution predicted by jModelTest (73) was used for tree reconstruction; otherwise, the GTR+GAMMA model was implemented. Lastly, we used the bacterial genomes sequenced in the study to create a BLAST database that can be searched for sequences of interest in Geneious Prime.
Illumina short reads were assembled de novo into contigs using Unicycler v0.4.7 and characterized with bioinformatic tools described for long reads. Illumina reads were used to determine single nucleotide polymorphisms (SNPs) and indels present in S. Heidelberg isolates. Alignment of raw FASTQ reads to the genome of the S. Heidelberg ancestor was done using Burrows-Wheeler aligner (BWA) (74), and SNPs/indels were called using the Genome Analysis Toolkit (75) as described previously (48). Variant call format (VCF) files of identified SNPs/indels and the Linux/Unix shell script used have been deposited in Dryad Digital Repository online at https://doi.org/10.5061/dryad.4tmpg4f8d. The SNP-based ML tree was reconstructed by converting merged VCF files to the PHYLogeny Inference Package format using PGDSpider (76). Afterward, isolates with duplicated SNPs/indels were removed, and an ML tree was drawn using the Jukes-Cantor model of nucleotide substitution and GAMMA model of rate heterogeneity.
To determine the IncI1-pST26 consensus, first we used BWA (74) or Bowtie v 7.2.1 (77) (implemented in Geneious Prime) to align raw FASTQ files against a complete circular p1ST26 or p2ST26 and used the binary alignment map file generated for consensus determination in Geneious Prime. To identify identical IncI1-pST26 plasmids, we performed a BLAST search against the NCBI nonredundant database and selected the top 50 plasmids matching p2ST26 from this study. To construct a whole plasmid-based maximum likelihood tree, we downloaded the FASTA files for the top 50 plasmids from NCBI and used them for whole-genome alignment. All raw FASTQ reads for sequenced bacterial genomes are publicly available under NCBI accession numbers PRJNA683658, PRJNA684578, and PRJNA684580.
Hi-C and cecal metagenome library preparation and analysis.
A shotgun and Hi-C DNA library of two cecal samples was created using the Nextera XT library preparation kit and Phase Genomics (Seattle, WA) ProxiMeta Hi-C Microbiome kit following the manufacturer’s instructions. Library sequencing was performed by Novogene corporation (Sacramento, CA, USA) on the Illumina HiSeq platform using 150-bp paired-end reads. Two libraries were sequenced per HiSeq flow cell lane. Metagenomic FASTQ files were uploaded to the Phase Genomics cloud-based bioinformatics portal for subsequent analysis.
Shotgun reads were filtered and trimmed for quality using bbduk (78), normalized using bbtools (78), and then assembled with metaSPAdes (79) using default options. Hi-C reads were then aligned to the assembly following the Hi-C kit manufacturer’s recommendations (https://phasegenomics.github.io/2019/09/19/hic-alignment-and-qc.html). Briefly, reads were aligned using BWA-MEM with the -5SP and -t 8 options specified and all other options default. SAMBLASTER (80) was used to flag PCR duplicates. Alignments were then filtered with SAMtools (81) using the -F 2304 filtering flag to remove nonprimary and secondary alignments. Lastly, Hi-C read alignments to the assembly were filtered using Matlock (https://github.com/phasegenomics/matlock) with default options (removing all alignments with MAPQ of <20, edit distance greater than 5, and read duplicates). Metagenome deconvolution was performed with ProxiMeta (82, 83), resulting in the creation of putative genome and genome fragment clusters.
Metagenome-assembled genomes/clusters were assessed for quality using CheckM (84) and assigned preliminary taxonomic classifications with Mash (24) and GTDB-Tk (27) (database release 202). Quality-controlled shotgun reads were additionally classified using Kraken2 (v2.0.8 beta) (28). To identify the plasmids and ARGs present in metagenome-assembled genomes, we used Minimap2 (85) to align assembled metagenome contigs to the PlasmidFinder and ResFinder database. We used a 75% identity threshold for the alignment match. To search the metagenome for ARG and plasmids of interest, we used Geneious Prime to create a BLAST metagenome database. Metagenome contigs with significant homology (E value of <0.05 and at least 1,000 bp of aligned sequence) were considered contigs of the respective plasmid or ARG. Hi-C data were then used to link identified sequences to host genomes and genome fragments within ProxiMeta (23). Phylogenetic visualizations of Hi-C linkages used the placement of clusters in a large prokaryotic phylogeny as estimated by CheckM (84) and Mash (24). Shotgun and Hi-C reads are publicly available under NCBI accession number PRJNA688069.
Isolation of E. coli from ceca of broiler chicks.
To isolate E. coli, we retrospectively screened two cecal slurry samples from chicks challenged intracloacally on CHROMagar plates supplemented with gentamicin (8 ppm) and tetracycline (8 ppm). Frozen vials of cecal contents were thawed on ice and vortexed. For each sample, a 10-μl aliquot was struck for isolation, and a 100-μl aliquot was spread plated onto CHROMagar with relevant antibiotics. Plates were incubated overnight at 37°C. Blue-green and blue-cream colonies were counted as presumptive E. coli, and colonies were streaked again for isolation on CHROMagar supplemented with gentamicin and tetracycline. After incubation, 2 to 3 colonies from each sample were selected to represent different forms of blue-green/blue-cream colonies, streaked in parallel onto sheep blood agar (SBA) and eosin methylene Bbue (EMB) (Remel Inc., KS, USA), and incubated overnight. Growth on EMB was the characteristic green metallic sheen after incubation. Colonies from SBA were stored in 30% LB glycerol at −80°C and were used for AST and WGS.
Bioinformatic tools used for visualization of high-throughput sequence data.
The gene presence/absence heatmap, with gene presence/absence data obtained from Roary, was generated using the pheatmap v1.0.12, tidyverse v1.3.0, and viridis v0.5.1 packages in R v4.0.2. BLAST Ring Image Generator (86) was used for genome comparison visualization, including GC skew change, and Phandango (87) was used for visualizing phylogenetic trees with their associated metadata. PHYLOViZ 2.0 (88) was used to generate a minimum spanning tree of S. Heidelberg isolates from core genome information obtained through SISTR (i.e., wgMLST_330:complete-alleles, wgMLST_330:missing-alleles, wgMLST_330:partial-alleles, closest-public-genome-alleles-matching, and cgMLST_cluster_level). SnapGene was used for drawing linear maps of the plasmid, prophages, and other regions of interest in bacterial genomes. Geneious Prime and SnapGene were used to view MAFFT alignments of DNA and amino acid sequences.
Determining S. Heidelberg metabolism under selective pressure.
To determine if exposure to metals, disinfectants, or acidic pH poses a selection on S. Heidelberg strains carrying IncI1 plasmids, we used 96-well phenotype microarray (PM) MicroPlates (PM10, PM12B, PM13B, PM15B, and PM16A) (Biolog, Inc., Hayward CA) to compare the metabolic profile of one gentamicin-resistant strain harboring IncI1 and one susceptible strain. PM plates use cell respiration via NADH production to determine cell metabolic activity. If the phenotype is positive in a well, the cells respire actively, reducing a tetrazolium dye and forming a strong color (Biolog, Inc.). Briefly, S. Heidelberg strains were subcultured twice in universal growth agar (Biolog Universal Growth Medium + 5% sheep blood) for 24 h at 37°C. Cells were removed with a sterile swab and transferred to 16 ml inoculating fluid 0 (IF-0) to achieve a final turbidimetric transmission of 42% transmittance (42%T). Afterward, 15 ml of the 42%T cell suspension was transferred to 75 ml of IF-0+ dye to achieve a final transmission of 85%T before 600 μl of the cell suspension was transferred to 120 ml IF-10+ dye. The suspension was mixed, and each well in a microplate was inoculated with 100 μl of the suspension. After inoculation, microplates were covered with a sterile plastic film and monitored automatically for color development every 15 min for 48 h at 37°C using and an OmniLog reader for 48 h. To identify phenotypes, the kinetic curves of the gentamicin-resistant and -susceptible strains were compared using OmniLog PM software. For each PM plate, the respiratory unit for each well at 22 h was extracted and log10 transformed. All supplies used were purchased from Biolog, Inc.
Exposing S. Heidelberg to acidic pH.
To determine the fitness of evolved S. Heidelberg strains that acquired antibiotic resistance compared with that of evolved susceptible strains after exposure to acidic pH, we exposed gentamicin-resistant (n = 3) and -susceptible (n = 3) strains recovered from the litter of chicks gavaged with S. Heidelberg to acidified filter-sterilized pine shaving extract (PSE). PSE was prepared as described for poultry litter extract (47), using fresh pine shavings collected from the University of Georgia, Poultry Research Center, Athens, GA, USA. Three single colonies of each strain, including the ancestral SH-2813nalR, were selected from overnight cultures grown on sheep blood agar and transferred to a microcentrifuge tube containing 900 μl of PSE (pH 6.52), i.e., one tube per strain.
After transfer, tubes were vortexed, covered with a gas-permeable paper strip, and incubated at 41°C under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) for 2 h. After incubation, tubes were vortexed, and 100 μl of the suspension was transferred to a microcentrifuge tube containing 900 μl of PSE (pH of PSE was adjusted to 2.54 using 1 M HCl [Spectrum Chemical Mfg. Corp., CA, USA] and 1 M NaOH [Fisher Chemical, NJ, USA]) Afterward, tubes were vortexed and incubation was continued for another 2 h as was done for PSE at pH 6.5. To determine the S. Heidelberg population in PSE, one replicate tube per strain was removed and serially diluted in 1× PBS at time points 0 and 2 h for pH 6.5 and 0.5 and 1 and 2 h for pH 2.5. Serial dilutions were plated onto BGS agar plates supplemented with or without 16 ppm gentamicin. S. Heidelberg colonies were counted 18 to 24 h after incubation using a calibrated automated colony counter.
The fitness of each evolved S. Heidelberg population relative to the SH-2813nalR population was determined after 2 h at pH 2.54 as described by San Millan et al. (89):
where Wstrain is the fitness of the evolved susceptible (total susceptible) or gentamicin-resistant populations (total resistant and genR); Ninitial,evol and Nfinal,evol are the numbers of cells (in CFU) of the evolved susceptible or gentamicin-resistant population at time point 0 and 2 h after exposure to PSE; and Ninitial,anc and Nfinal,anc are the numbers of cells of SH-2813nalR population at time point 0 and 2 h.
Statistical analyses.
Continuous variables did not meet the assumption of a normal distribution; therefore, nonparametric testing for direct comparisons was performed using Wilcoxon rank-sum and signed-rank tests, and the Kruskal-Wallis rank-sum test was used for one-way analysis of variance tests. Furthermore, continuous variables were log-transformed before any statistical tests were performed. Statistical analyses were performed using R (v3.4.1).
Ethics statement.
All animal experiments were approved by the University of Georgia Office of Animal Care and Use under Animal Use Protocol A2017 04-028-A2.
Data availability.
All raw FASTQ reads, including short and long reads for sequenced bacterial genomes, are publicly available under NCBI accession numbers PRJNA683658, PRJNA684578, and PRJNA684580. Shotgun and Hi-C reads are publicly available under NCBI accession number PRJNA688069 and 16S rRNA gene sequences under NCBI accession number PRJNA669215. The whole-genome assemblies for S. Heidelberg strains SH-2813-ancestor and ic9b harboring p1ST26 have been made available under NCBI GenBank accession number CP066851 and DDBJ/ENA/GenBank JAEMHU000000000, respectively. The whole-genome assemblies for E. coli strain Ec-FL1-1X and Ec-FL1-2X carrying IncI1 are available under GenBank accession numbers CP066836 and JAFCXR000000000, respectively. Variant call format (VCF) files of identified SNPs/indels and the Linux/Unix shell script used have been deposited in Dryad Digital Repository online at https://doi.org/10.5061/dryad.4tmpg4f8d.
ACKNOWLEDGMENTS
We are grateful to Marlo Sommers, Jasmine Johnson, Carolina Hall, Jeromey Jackson, and Latoya Wiggins for their logistical and technical assistance. This work was supported by USDA Agricultural Research Service (project number 6040‐32000‐010‐00‐D), nonassistance cooperative agreement (58-6040-6-030) between USDA Agricultural Research Service and University of Georgia, Research Foundation, and research service agreement (58-6040-8-035) between USDA Agricultural Research Service and Colorado State University. M.O.P. was supported in part by grant R44AI150008 from NIAID to Phase Genomics. B.Z. was supported by the competence center. The competence center (FFoQSI) is funded by the Austrian ministries BMVIT and BMDW and the Austrian provinces Niederoesterreich, Upper Austria, and Vienna within the scope of COMET‐Competence Centers for Excellent Technologies. The Austrian Research Promotion Agency FFG handles the program COMET. This study was supported in part by resources and technical expertise from the Georgia Advanced Computing Resource Center, a partnership between the University of Georgia’s Office of the Vice President for Research and Office of the Vice President for Information Technology. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the USDA or the National Science Foundation, and any mention of products or trade names does not constitute recommendation for use.
M.O.P. was an employee of Phase Genomics during the performance of this work.
A.O., Z.A., K.C., and N.A.C. designed the study. A.O., G.Z., D.E.C., and C.R. performed live-broiler chicken studies. A.O., J.P.L., G.Z., S.H., and D.C. performed bacteriological analyses, antibiotic susceptibility testing, and DNA extraction. J.P.L., G.Z., and A.O. performed Illumina whole-genome sequencing. A.O. performed cecal shotgun and Hi-C library preparation. T.L. made 16S rRNA gene libraries and sequencing. B.Z. performed 16S rRNA bacterial community analysis and interpretation. K.I., A.H., and A.O. performed phenotype microarray analyses with the help of J.G. and E.L. A.O., Z.A., M.O.P., J.C.T., S.M.L., R.W., J.P.L., D.C., and L.L. performed bioinformatic analyses and data curation. K.H. and A.O. performed Salmonella Heidelberg fitness testing and analysis. A.O., J.P.L., and D.C. isolated multidrug-resistant E. coli donors from cecal contents with help from M.J.R. A.O., B.Z., and M.E.B. performed statistical analyses. A.O., Z.A., N.A.C., B.Z., R.W., M.O.P., S.M.L., K.I., G.Z., J.P.L., K.H., and L.L. drafted the manuscript, which was reviewed and edited by all authors. A.O., Z.A., S.E.A., L.C., and M.W. supervised the study.
Contributor Information
Adelumola Oladeinde, Email: ade.oladeinde@usda.gov.
Zaid Abdo, Email: zaid.abdo@colostate.edu.
Daniel Garrido, Pontificia Universidad Catolica de Chile.
REFERENCES
- 1.Frieri M, Kumar K, Boutin A. 2017. Antibiotic resistance. J Infect Public Health 10:369–378. doi: 10.1016/j.jiph.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Akova M. 2016. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence 7:252–266. doi: 10.1080/21505594.2016.1159366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Von Wintersdorff CJ, Penders J, Van Niekerk JM, Mills ND, Majumder S, Van Alphen LB, Savelkoul PH, Wolffs PF. 2016. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol 7:173. doi: 10.3389/fmicb.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ventola C. 2015. The antibiotic resistance crisis: part 1: causes and threats. PT 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 5.Angulo FJ, Baker NL, Olsen SJ, Anderson A, Barrett TJ. 2004. Antimicrobial use in agriculture: controlling the transfer of antimicrobial resistance to humans. Semin Pediatr Infect Dis 15:78–85. doi: 10.1053/j.spid.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Ventola CL. 2015. The antibiotic resistance crisis: part 2: management strategies and new agents. P T 40:344–352. [PMC free article] [PubMed] [Google Scholar]
- 7.Scott AM, Beller E, Glasziou P, Clark J, Ranakusuma RW, Byambasuren O, Bakhit M, Page SW, Trott D, Del Mar C. 2018. Is antimicrobial administration to food animals a direct threat to human health? A rapid systematic review. Int J Antimicrobial Agents 52:316–323. doi: 10.1016/j.ijantimicag.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Aidara-Kane A, Angulo FJ, Conly JM, Minato Y, Silbergeld EK, McEwen SA, Collignon PJ, WHO Guideline Development Group. 2018. World Health Organization (WHO) guidelines on use of medically important antimicrobials in food-producing animals. Antimicrob Resist Infect Control 7:7. doi: 10.1186/s13756-017-0294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang KL, Caffrey NP, Nóbrega DB, Cork SC, Ronksley PE, Barkema HW, Polachek AJ, Ganshorn H, Sharma N, Kellner JD, Ghali WA. 2017. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet Health 1:e316–e327. doi: 10.1016/S2542-5196(17)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karavolias J, Salois MJ, Baker KT, Watkins K. 2018. Raised without antibiotics: impact on animal welfare and implications for food policy. Transl Anim Sci 2:337–348. doi: 10.1093/tas/txy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bekal S, Berry C, Reimer AR, Van Domselaar G, Beaudry G, Fournier E, Doualla-Bell F, Levac E, Gaulin C, Ramsay D, Huot C, Walker M, Sieffert C, Tremblay C. 2016. Usefulness of high-quality core genome single-nucleotide variant analysis for subtyping the highly clonal and the most prevalent Salmonella enterica serovar Heidelberg clone in the context of outbreak investigations. J Clin Microbiol 54:289–295. doi: 10.1128/JCM.02200-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liakopoulos A, Geurts Y, Dierikx CM, Brouwer MS, Kant A, Wit B, Heymans R, van Pelt W, Mevius DJ. 2016. Extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg strains, the Netherlands. Emerg Infect Dis 22:1257–1261. doi: 10.3201/eid2207.151377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edirmanasinghe R, Finley R, Parmley EJ, Avery BP, Carson C, Bekal S, Golding G, Mulvey MR. 2017. A whole-genome sequencing approach to study cefoxitin-resistant Salmonella enterica serovar Heidelberg isolates from various sources. Antimicrob Agents Chemother 61:e01919-16. doi: 10.1128/AAC.01919-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gieraltowski L, Higa J, Peralta V, Green A, Schwensohn C, Rosen H, Libby T, Kissler B, Marsden-Haug N, Booth H, Kimura A, Grass J, Bicknese A, Tolar B, Defibaugh-Chávez S, Williams I, Wise M, Salmonella Heidelberg Investigation Team. 2016. National outbreak of multidrug resistant Salmonella Heidelberg infections linked to a single poultry company. PLoS One 11:e0162369. doi: 10.1371/journal.pone.0162369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakao JH, Talkington D, Bopp CA, Besser J, Sanchez ML, Guarisco J, Davidson SL, Warner C, McIntyre MG, Group JP, Comstock N, Xavier K, Pinsent TS, Brown J, Douglas JM, Gomez GA, Garrett NM, Carleton HA, Tolar B, Wise ME. 2018. Unusually high illness severity and short incubation periods in two foodborne outbreaks of Salmonella Heidelberg infections with potential coincident Staphylococcus aureus intoxication. Epidemiol Infect 146:19–27. doi: 10.1017/S0950268817002655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent C, Usongo V, Berry C, Tremblay DM, Moineau S, Yousfi K, Doualla-Bell F, Fournier E, Nadon C, Goodridge L, Bekal S. 2018. Comparison of advanced whole genome sequence-based methods to distinguish strains of Salmonella enterica serovar Heidelberg involved in foodborne outbreaks in Quebec. Food Microbiol 73:99–110. doi: 10.1016/j.fm.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Cox NA, Oladeinde AA, Cook KL, Zock GS, Berrang ME, Ritz CW, Hinton A. 2020. Research Note: evaluation of several inoculation procedures for colonization of day-old broiler chicks with Salmonella Heidelberg. Poult Sci 99:1615–1617. doi: 10.1016/j.psj.2019.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehman MA, Yin X, Persaud-Lachhman MG, Diarra MS. 2017. First detection of a fosfomycin resistance gene, fosA7, in Salmonella enterica serovar Heidelberg isolated from broiler chickens. Antimicrob Agents Chemother 61:e00410-17. doi: 10.1128/AAC.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carattoli A, Zankari E, Garcia-Fernandez A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carattoli A, Villa L, Fortini D, Garcia-Fernandez A. 2018. Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid S0147-619X(18)30046-5. doi: 10.1016/j.plasmid.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Merrikh CN, Merrikh H. 2018. Gene inversion potentiates bacterial evolvability and virulence. Nat Commun 9:4662. doi: 10.1038/s41467-018-07110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucas Lledo JI, Caceres M. 2013. On the power and the systematic biases of the detection of chromosomal inversions by paired-end genome sequencing. PLoS One 8:e61292. doi: 10.1371/journal.pone.0061292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stalder T, Press MO, Sullivan S, Liachko I, Top EM. 2019. Linking the resistome and plasmidome to the microbiome. ISME J 13:2437–2446. doi: 10.1038/s41396-019-0446-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ondov BD, Starrett GJ, Sappington A, Kostic A, Koren S, Buck CB, Phillippy AM. 2019. Mash Screen: high-throughput sequence containment estimation for genome discovery. Genome Biol 20:232. doi: 10.1186/s13059-019-1841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VP, Nash JH, Taboada EN. 2016. The Salmonella In Silico Typing Resource (SISTR): an open Web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox NA, Bailey JS, Blankenship LC, Meinersmann RJ, Stern NJ, McHan F. 1990. Fifty percent colonization dose for Salmonella Typhimurium administered orally and intracloacally to young broiler chicks. Poult Sci 69:1809–1812. doi: 10.3382/ps.0691809. [DOI] [PubMed] [Google Scholar]
- 30.Bailey JS, Cox NA, Cosby DE, Richardson LJ. 2005. Movement and persistence of Salmonella in broiler chickens following oral or intracloacal inoculation. J Food Prot 68:2698–2701. doi: 10.4315/0362-028x-68.12.2698. [DOI] [PubMed] [Google Scholar]
- 31.Cox N, Richardson L, Cosby D, Berrang M, Harrison M. 2016. Recovery of Campylobacter from external and internal spleen samples from baby broiler chicks following various routes of inoculation. J Food Saf 36:132–135. doi: 10.1111/jfs.12220. [DOI] [Google Scholar]
- 32.Dimitriu T, Medaney F, Amanatidou E, Forsyth J, Ellis RJ, Raymond B. 2019. Negative frequency dependent selection on plasmid carriage and low fitness costs maintain extended spectrum beta-lactamases in Escherichia coli. Sci Rep 9:17211. doi: 10.1038/s41598-019-53575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alonso-Del Valle A, Leon-Sampedro R, Rodriguez-Beltran J, DelaFuente J, Hernandez-Garcia M, Ruiz-Garbajosa P, Canton R, Pena-Miller R, San Millan A. 2021. Variability of plasmid fitness effects contributes to plasmid persistence in bacterial communities. Nat Commun 12:2653. doi: 10.1038/s41467-021-22849-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fairchild BD. 2009. Environmental factors to control when brooding chicks. The University of Georgia, Athens, Georgia. [Google Scholar]
- 35.Benz F, Huisman JS, Bakkeren E, Herter JA, Stadler T, Ackermann M, Diard M, Egli A, Hall AR, Hardt W-D, Bonhoeffer S. 2021. Plasmid- and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo. ISME J 15:862–878. doi: 10.1038/s41396-020-00819-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wotzka SY, Kreuzer M, Maier L, Arnoldini M, Nguyen BD, Brachmann AO, Berthold DL, Zünd M, Hausmann A, Bakkeren E, Hoces D, Gül E, Beutler M, Dolowschiak T, Zimmermann M, Fuhrer T, Moor K, Sauer U, Typas A, Piel J, Diard M, Macpherson AJ, Stecher B, Sunagawa S, Slack E, Hardt W-D. 2019. Escherichia coli limits Salmonella Typhimurium infections after diet shifts and fat-mediated microbiota perturbation in mice. Nat Microbiol 4:2164–2174. doi: 10.1038/s41564-019-0568-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer EAJ, Dierikx CM, van Essen-Zandbergen A, Mevius D, Stegeman A, Velkers FC, Klinkenberg D. 2019. Competition between Escherichia coli populations with and without plasmids carrying a gene encoding extended-spectrum beta-lactamase in the broiler chicken gut. Appl Environ Microbiol 85:e00892-19. doi: 10.1128/AEM.00892-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagbø M, Ravi A, Angell IL, Sunde M, Ludvigsen J, Diep DB, Foley SL, Vento M, Collado MC, Perez-Martinez G, Rudi K. 2020. Experimental support for multidrug resistance transfer potential in the preterm infant gut microbiota. Pediatr Res 88:57–65. doi: 10.1038/s41390-019-0491-8. [DOI] [PubMed] [Google Scholar]
- 39.Nyirabahizi E, Tyson GH, Dessai U, Zhao S, Kabera C, Crarey E, Womack N, Crews MK, Strain E, Tate H. 2020. Evaluation of Escherichia coli as an indicator for antimicrobial resistance in Salmonella recovered from the same food or animal ceca samples. Food Control 115:107280. doi: 10.1016/j.foodcont.2020.107280. [DOI] [Google Scholar]
- 40.Pereira FC, Berry D. 2017. Microbial nutrient niches in the gut. Environ Microbiol 19:1366–1378. doi: 10.1111/1462-2920.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohd Shaufi MA, Sieo CC, Chong CW, Gan HM, Ho YW. 2015. Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog 7:4. doi: 10.1186/s13099-015-0051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding J, Zhao L, Wang L, Zhao W, Zhai Z, Leng L, Wang Y, He C, Zhang Y, Zhang H, Li H, Meng H. 2016. Divergent selection-induced obesity alters the composition and functional pathways of chicken gut microbiota. Genet Sel Evol 48:93. doi: 10.1186/s12711-016-0270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qu A, Brulc JM, Wilson MK, Law BF, Theoret JR, Joens LA, Konkel ME, Angly F, Dinsdale EA, Edwards RA, Nelson KE, White BA. 2008. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS One 3:e2945. doi: 10.1371/journal.pone.0002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Danzeisen JL, Kim HB, Isaacson RE, Tu ZJ, Johnson TJ. 2011. Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS One 6:e27949. doi: 10.1371/journal.pone.0027949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang K, Hu Y, Wu S, Shi S. 2021. Comparative metagenomic analysis of chicken gut microbial community, function, and resistome to evaluate noninvasive and cecal sampling resources. Animals (Basel) 11:1718. doi: 10.3390/ani11061718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Litvak Y, Mon KKZ, Nguyen H, Chanthavixay G, Liou M, Velazquez EM, Kutter L, Alcantara MA, Byndloss MX, Tiffany CR, Walker GT, Faber F, Zhu Y, Bronner DN, Byndloss AJ, Tsolis RM, Zhou H, Bäumler AJ. 2019. Commensal Enterobacteriaceae protect against Salmonella colonization through oxygen competition. Cell Host Microbe 25:128–139.e125. doi: 10.1016/j.chom.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Oladeinde A, Cook K, Orlek A, Zock G, Herrington K, Cox N, Plumblee Lawrence J, Hall C. 2018. Hotspot mutations and ColE1 plasmids contribute to the fitness of Salmonella Heidelberg in poultry litter. PLoS One 13:e0202286. doi: 10.1371/journal.pone.0202286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oladeinde A, Cook K, Lakin SM, Woyda R, Abdo Z, Looft T, Herrington K, Zock G, Lawrence JP, Thomas JC, Beaudry MS, Glenn T. 2019. Horizontal gene transfer and acquired antibiotic resistance in Salmonella enterica serovar Heidelberg following in vitro incubation in broiler ceca. Appl Environ Microbiol 85:e01903-19. doi: 10.1128/AEM.01903-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersen KS, Kirkegaard RH, Karst SM, Albertsen M. 2018. ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. bioRxiv 10.1101/299537. [DOI]
- 51.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Peter S. 2019. vegan: community ecology package. R package version 2.5–6. https://CRAN.R-project.org/package=vegan.
- 52.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woyda RR, Oladeinde A, Abdo Z. 2020. Reads2Resistome: an adaptable and high-throughput whole-genome sequencing pipeline for bacterial resistome characterization. bioRxiv 10.1101/2020.05.18.102715. [DOI]
- 54.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antipov D, Korobeynikov A, McLean JS, Pevzner PA. 2016. hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics 32:1009–1015. doi: 10.1093/bioinformatics/btv688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 58.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 60.Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. 2018. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom 4:e000192. doi: 10.1099/mgen.0.000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lakin SM, Dean C, Noyes NR, Dettenwanger A, Ross AS, Doster E, Rovira P, Abdo Z, Jones KL, Ruiz J, Belk KE, Morley PS, Boucher C. 2017. MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res 45:D574–D580. doi: 10.1093/nar/gkw1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Tyson GH, Zhao S, Hsu CH, McDermott PF, et al. 2019. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu B, Zheng D, Jin Q, Chen L, Yang J. 2019. VFDB 2019: a comparative pathogenomic platform with an interactive Web interface. Nucleic Acids Res 47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kozlov A, Darriba D, Flouri T, Morel B, Stamatakis A. 2018. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. bioRxiv 10.1101/447110. [DOI] [PMC free article] [PubMed]
- 73.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lischer HE, Excoffier L. 2012. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28:298–299. doi: 10.1093/bioinformatics/btr642. [DOI] [PubMed] [Google Scholar]
- 77.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bushnell B, Rood J, Singer E. 2017. BBMerge—accurate paired shotgun read merging via overlap. PLoS One 12:e0185056. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Faust GG, Hall IM. 2014. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30:2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Press MO, Wiser AH, Kronenberg ZN, Langford KW, Shakya M, Lo C-C, Mueller KA, Sullivan ST, Chain PS, Liachko I. 2017. Hi-C deconvolution of a human gut microbiome yields high-quality draft genomes and reveals plasmid-genome interactions. bioRxiv 10.1101/198713. [DOI]
- 83.Stewart RD, Auffret MD, Warr A, Wiser AH, Press MO, Langford KW, Liachko I, Snelling TJ, Dewhurst RJ, Walker AW, Roehe R, Watson M. 2018. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat Commun 9:870. doi: 10.1038/s41467-018-03317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR. 2018. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34:292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nascimento M, Sousa A, Ramirez M, Francisco AP, Carrico JA, Vaz C. 2017. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 33:128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- 89.San Millan A, Heilbron K, MacLean RC. 2014. Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J 8:601–612. doi: 10.1038/ismej.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Litter pH and moisture differed between trial 1 and 2. Litter was collected as grab samples (n = 7) and pooled into one whirl pak bag (n = 1 per pen). The pH of the litter was determined from the eluate made from a 1:5 dilution of litter in 1× PBS. Download FIG S1, TIF file, 0.1 MB (81.7KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
S. Heidelberg challenge affected the diversity of bacterial species in ceca and litter. (a) Average alpha diversity indices of ceca and litter samples grouped by route of S. Heidelberg challenge. Box plots show the interquartile range (75th to 25th) of the data. Whiskers extend to the most extreme value within 1.5 × interquartile range, and dots are outliers beyond that range. (b) Heatmap of the top 50 most abundant amplicon sequence variants (ASVs) grouped by route of S. Heidelberg challenge and split by sample type (ceca and litter). Data are the average of ASV counts from replicate 16S rRNA gene libraries for each category. Download FIG S2, TIF file, 0.5 MB (510.1KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Acquisition of IncI1 plasmid by S. Heidelberg conferred resistance to multiple antibiotics. Antimicrobial susceptibility was performed on S. Heidelberg isolates recovered from the ceca and litter of broiler chicks 14 days after challenge. (a) Percentage of S. Heidelberg isolates (n = 63) from trial 2 that acquired resistance to two or more antibiotics compared with isolate resistance to only 1 antibiotic. (b) Linear map of the ∼19-kb variable region carrying antimicrobial resistance genes, transposons, and integrase in IncI1-p1ST26 (∼102 kb). Download FIG S3, TIF file, 0.2 MB (202.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Mutations found in p1ST26 but absent in p2ST26. Download Table S1, DOCX file, 0.01 MB (14.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Maximum likelihood tree reconstruction of S. Heidelberg isolates. Core genome (a) and SNP-based (b) maximum likelihood (ML) tree of S. Heidelberg isolates sequenced from trial 1 (n = 2) and trial 2 (n = 69). Isolates with duplicated SNPs/indels were removed before the SNP-based ML tree was reconstructed. All S. Heidelberg strains used for the core genome tree were assembled using Illumina short reads except OG8-05A, IC9B, and IC4-3A and SH2813-ancestral that were assembled using both Illumina short reads and PacBio or MinION long reads. The GTR and JC models of nucleotide substitution was used for core and SNP-based trees, respectively, and the GAMMA model of rate heterogeneity were used for sequence evolution prediction. Numbers shown next to the branches are the percentage of replicate trees where associated isolates cluster together based on ∼100 bootstrap replicates. The susceptible SH-2813 ancestor was used to root the for the core genome, while the SNP tree was rooted with the nalidixic-resistant S. Heidelberg ancestor (SH-2813nalR). (Nal, nalidixic acid; Gen, gentamicin; Str, streptomycin; Tet, tetracycline; Fis, sulfisoxazole; NA, not applicable)FIG S4, TIF file, 0.8 MB (824.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Gene flux contributed to the diversity of S. Heidelberg. (a) Heatmap of genes present and absent in S. Heidelberg genomes determined by pangenome analysis. All S. Heidelberg strains were assembled using Illumina short reads except OG8-05A, IC9B, and IC4-3A and SH2813-ancestral that were assembled using both Illumina short reads and PacBio or MinION long reads. The top black rectangular box highlights S. Heidelberg strains that carried no plasmid, while the bottom rectangular box denotes strains with no plasmids but clustered with S. Heidelberg strains carrying IncI1. (b) Circular map of novel episome present in S. Heidelberg strain og8-05a. (c) BLASTn alignment of IncX1 present in S. Heidelberg ancestor and IncX1 present in S. Heidelberg strain og8-05a. (d to f) S. Heidelberg genome assembly statistics for S. Heidelberg strains with assembly error (n = 12; subclade IIa of ML tree reconstructed using accessory genes) (see Fig. 2a) compared with other S. Heidelberg strains (n = 59) (***P < 0.001; binomial logistic regression; error bar, standard deviation). Download FIG S5, TIF file, 1.3 MB (1.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Assembly report for a S. Heidelberg strain with sequencing bias. Download Table S2, DOCX file, 0.01 MB (13.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Chromosomal inversion reoriented the origin of replication of S. Heidelberg strains and positively skewed their GC. Complete chromosome contigs from three S. Heidelberg strains recovered from ceca during trial 1 (ic9b) and 2 (ic4-3a and og8-05a) were aligned and compared with the ancestor. (a) progressiveMauve alignment of S. Heidelberg strains ic4-3a and ic9b. A colored similarity plot is shown for each genome, of which the height is proportional to the level of sequence identity in that region. When the similarity plot points downward, it shows an alignment to the reverse strand of the genome of S. Heidelberg-ancestor, i.e., inversion. A segment highlighted with solid black rectangular box denotes the inverted 2-Mbp region in ic9b. A segment highlighted with a horizontal blue box denotes regions with relevant mutations. (b and c) GC skew in og8-05a and ic9b compared with S. Heidelberg-ancestor. The red arrow points toward reoriented oriC or ter, whereas red and blue arcs show the inverted region. The number of guanine and cytosine in inverted area was used for GC skew calculation. Dashed blue and red rectangular boxes denote segments with mutations. Download FIG S6, TIF file, 1.5 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Plasmids found in the cecal metagenome. Download Table S3, DOCX file, 0.01 MB (13KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Concentration of metals and pollutants in starter feed. Download Table S4, DOCX file, 0.01 MB (13.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
All raw FASTQ reads, including short and long reads for sequenced bacterial genomes, are publicly available under NCBI accession numbers PRJNA683658, PRJNA684578, and PRJNA684580. Shotgun and Hi-C reads are publicly available under NCBI accession number PRJNA688069 and 16S rRNA gene sequences under NCBI accession number PRJNA669215. The whole-genome assemblies for S. Heidelberg strains SH-2813-ancestor and ic9b harboring p1ST26 have been made available under NCBI GenBank accession number CP066851 and DDBJ/ENA/GenBank JAEMHU000000000, respectively. The whole-genome assemblies for E. coli strain Ec-FL1-1X and Ec-FL1-2X carrying IncI1 are available under GenBank accession numbers CP066836 and JAFCXR000000000, respectively. Variant call format (VCF) files of identified SNPs/indels and the Linux/Unix shell script used have been deposited in Dryad Digital Repository online at https://doi.org/10.5061/dryad.4tmpg4f8d.