Abstract
Bacteria of the genus Streptomyces have a linear chromosome, with a core region and two ‘arms’. During their complex life cycle, these bacteria develop multi-genomic hyphae that differentiate into chains of exospores that carry a single copy of the genome. Sporulation-associated cell division requires chromosome segregation and compaction. Here, we show that the arms of Streptomyces venezuelae chromosomes are spatially separated at entry to sporulation, but during sporogenic cell division they are closely aligned with the core region. Arm proximity is imposed by segregation protein ParB and condensin SMC. Moreover, the chromosomal terminal regions are organized into distinct domains by the Streptomyces-specific HU-family protein HupS. Thus, as seen in eukaryotes, there is substantial chromosomal remodelling during the Streptomyces life cycle, with the chromosome undergoing rearrangements from an ‘open’ to a ‘closed’ conformation.
Subject terms: Chromosomes, Bacterial development, Bacterial genetics, Cellular microbiology
Streptomyces bacteria have a linear chromosome and a complex life cycle, including development of multi-genomic hyphae that differentiate into mono-genomic exospores. Here, Szafran et al. show that the chromosome of Streptomyces venezuelae undergoes substantial remodelling during sporulation, from an ‘open’ to a ‘closed’ conformation.
Introduction
Chromosomal organisation differs notably among the domains of life, and even among bacteria, the chromosomal arrangement is not uniform. While linear eukaryotic chromosomes undergo dramatic condensation before their segregation, the compaction of usually circular bacterial chromosomes remains relatively constant throughout the cell cycle1–3. To date, chromosome conformation capture methods have been applied in only a few model bacterial species, revealing that bacterial chromosomes adopt one of two distinct conformation patterns. Either both halves of the circular chromosome (replichores) align closely (as in Bacillus subtilis4,5, Caulobacter crescentus6, Corynebacterium glutamicum7, Mycoplasma pneumoniae8, and chromosome II of Vibrio cholerae9) or they tend to adopt an ‘open’ conformation, which lacks close contacts between both chromosomal arms (as in Escherichia coli10 and V. cholerae chromosome I9). Cytological studies have shown that chromosome organisation may alter depending on the bacterial growth phase or developmental stage1,11–13. While the most prominent changes in bacterial chromosome organisation can be observed in the extensive DNA compaction that occurs during sporulation of Bacillus spp.14 and Streptomyces spp.13, the chromosome conformation during this process has not been elucidated.
The folding of bacterial genomes requires a strictly controlled interplay between several factors, including macromolecular crowding, transcription and the activity of various dedicated DNA-organising proteins (topoisomerases, condensins and nucleoid-associated proteins (NAPs))10,15,16. By non-specific DNA binding, NAPs induce its bending, bridging or wrapping to govern the local organisation and global compaction of bacterial chromosomes1,10,16. Since the subcellular repertoire of NAPs is dependent on growth conditions, changes in their supply can account for the fluctuation of chromosome compaction in response to environmental clues. Furthermore, the repertoire of NAPs also varies between bacterial species, with some (such as HU homologues) being widespread while others are less conserved13,17,18. By contrast, condensins (SMC-ScpAB complex found in most phyla, or the MukBEF complex found in the gammaproteobacteria including E. coli) are among the most conserved DNA-organising proteins identified in all domains of life19–21. According to a recently proposed model, the primary activity of SMC is the extrusion of large DNA loops22,23. SMC complexes were shown to move along bacterial chromosomes from the centrally positioned origin of replication (oriC), leading to gradual chromosome compaction and parallel alignment of both replichores5,6,24,25. Importantly, although the alignment of two chromosomal arms was not seen in E. coli, the MukBEF complex is still responsible for long-range DNA interactions10,26.
In B. subtilis, C. glutamicum and C. crescentus, the loading of SMC in the vicinity of the oriC were shown to be dependent on the chromosome segregation protein ParB4,6,7,27,28. Binding to a number of parS sequences, ParB forms a large nucleoprotein complex (segrosome), that interacts with ParA, an ATPase bound non-specifically to nucleoid29,30. Interaction with ParB triggers ATP hydrolysis and nucleoid release by ParA, generating a concentration gradient that drives segrosome separation31–33. Segrosome formation was shown to be a prerequisite for chromosomal arm alignment in Gram-positive bacteria with circular chromosomes5,7,28. There have been no reports on the 3D organisation of linear bacterial chromosomes to date.
Streptomyces spp. are among a few bacterial genera that possess linear chromosome. They are Gram-positive mycelial soil actinobacteria, which are highly valued producers of antibiotics and other clinically important metabolites. Notably, the production of these secondary metabolites is closely correlated with Streptomyces unique and complex life cycle, which encompasses vegetative growth and sporulation, as well as exploratory growth in some species34,35. During the vegetative stage, Streptomyces grow as branched hyphae composed of elongated cell compartments, each containing multiple copies of nonseparated and largely uncondensed chromosomes1,3,36. Under stress conditions, sporogenic hyphae develop, and their rapid elongation is accompanied by extensive replication of chromosomes37. When hyphae stop extending, the multiple chromosomes undergo massive compaction and segregation to generate unigenomic spores13. These processes are accompanied by multiple synchronised cell divisions initiated by FtsZ, which assembles into a ladder of regularly spaced Z-rings38,39. Chromosome segregation is driven by ParA and ParB proteins, whereas nucleoid compaction is induced by the concerted action of SMC and sporulation-specific NAPs that include HupS - one of the two Streptomyces HU homologues, structurally unique to actinobacteria owing to the presence of a histone-like domain40–44. Thus, while in unicellular bacteria chromosome segregation occurs during ongoing replication, during Streptomyces sporulation, intensive chromosome replication is temporally separated from their compaction and segregation, with the latter two processes occurring only during sporogenic cell division.
While Streptomyces chromosomes are among the largest bacterial chromosomes, our knowledge concerning their organisation is scarce. These linear chromosomes are flanked with terminal inverted repeats (TIRs) which encompass palindromes that form telomeric secondary structures. Replication of telomeres is mediated by a covalently bound telomere-associated complex of terminal proteins (TPs)45,46. The elimination of TPs results in circularisation of the chromosome47,48. Interestingly, the length of TIRs varies for different Streptomyces species, ranging from less than a few hundred nucleotides (in S. avermitilis49), to 200 kbp (in S. ambofaciens50) or even over 600 kbp (in S. collinus51), while in S. venezuelae it was determined to be about 1.5 kbp52. Limited cytological evidence suggests that during vegetative growth, both ends of the linear chromosome colocalize, and the oriC region of the apical chromosome is positioned at the tip-proximal edge of the nucleoid53,54. The visualisation of the nucleoid and ParB-marked oriC regions in sporogenic hyphae showed that oriCs are positioned centrally within pre-spore compartments, but did not elucidate chromosome architecture during sporulation39,54.
In this study, we investigated if S. venezuelae chromosome organisation changes during sporulation. Using the chromosome conformation capture method (Hi-C), we observed a dramatic rearrangement of chromosome structure associated with sporogenic development. Moreover, by combining the Hi-C with chromatin immunoprecipitation (ChIP) and fluorescence microscopy techniques, we investigated the contribution of ParB, SMC and HupS (the sporulation-dedicated HU homologue) to global nucleoid compaction. We demonstrated the substantial rearrangement of the S. venezuelae chromosomes which undertake compact folded conformation during sporogenic cell division.
Results
Hi-C reveals alignment of chromosomal arms extending to terminal domains during S. venezuelae sporogenic cell division
During Streptomyces sporulation, multi-genomic hyphae undergo considerable morphological changes, turning into a chains of spores through multiple cell divisions. This event is linked to the inhibition of DNA replication followed by synchronised compaction and segregation of tens of chromosomes37,39,55–57. To explore whether the increasing chromosome compaction during sporulation is reflected in overall changes in chromosome organisation, we optimised Hi-C6,58,59 to map DNA contact frequency across the S. venezuelae chromosome (Fig. 1 and Supplementary Fig. 1).
Fig. 1. Spatial organisation of the S. venezuelae linear chromosome during sporulation.
A The S. venezuelae (wild-type ftsZ-ypet derivative, MD100) growth curve (in 5 ml culture), performed in n = 3 independent experiments. The mean optical density values and standard deviation (whiskers) are shown on the scheme. The critical time points of sporogenic development are marked with arrows: growth arrest and cessation of DNA replication (red), the appearance of FtsZ ladders (green) and the spore formation (blue). The normalised FtsZ-YPet fluorescence (a marker of a synchronous cell division) [U/µg] and the relative oriC/arm ratio (a marker of DNA replication; n = 3 independent experiments: mean oriC/arm values as well as calculated standard deviations (whiskers) are shown on the diagram) are shown as insets in the plot, with X axes corresponding to the main plot X axis. The oriC/arm ratio at the 26 h of growth was set as 1.0. The right panel shows the representative visualisation of condensing nucleoids (n = 250 analysed nucleoids per a single time point; DNA stained with 7-AAD) and FtsZ-YPet at 22 h of growth; scale bar 2 µm. B The normalised Hi-C contact map obtained for the wild-type (ftsZ-ypet derivative, MD100) after 22 h of growth (in 5 ml culture). The dotted pink lines mark the position of the oriC region. The dotted black lines mark the positions of the left (LTD) and right (RTD) terminal domains. The boundaries of the LTD and RTD are marked directly on the Hi-C contact map with the orange dotted lines. The contact range within the secondary diagonal axis is marked with black dots. C Top panel: the normalised Hi-C contact maps obtained for the wild-type (ftsZ-ypet derivative, MD100) strain growing for 13, 15, 17 and 25 h (in 5 ml culture). Bottom panel: the differential Hi-C maps in the logarithmic scale (log2) comparing the contact enrichment at 22 h of growth (red) versus 13, 15, 17 and 25 h of growth (blue). The X and Y axes indicate chromosomal coordinates binned in 30 kbp. The right panel shows the model of chromosome rearrangement in the course of sporulation. D Top panel: the normalised Hi-C contact maps generated for parA and parB mutants (ftsZ-ypet derivatives MD011 and MD021, respectively) growing for 22 h (in 5 ml culture). Bottom panel: the differential Hi-C maps in the logarithmic scale (log2) comparing the contact enrichment in the wild-type strain (red) versus the mutant strain (blue). The right panel shows the model of chromosome organisation in the parA and parB mutants.
First, using ftsZ-ypet strain, and having confirmed that production of FtsZ-Ypet fusion protein does not affect S. venezuelae growth (Supplementary Fig. 2A), we determined the time points at which critical sporulation events took place, namely growth arrest and cessation of DNA replication, formation of the FtsZ ladder, and appearance of spores (Fig. 1A). After 13–14 h of growth (under 5 ml culture conditions), we observed vegetative growth inhibition. Slowed hyphal growth correlated with a reduction in the oriC/arm ratio corresponding to decreased DNA replication (Fig. 1A and Supplementary Fig. 3A), indicating entry into a developmental stage that we named the pre-sporulation phase, and which lasted until 22 h of growth (~8 h). During this phase, the DNA replication rate transiently increased (in 19–21 h), and this increased replication rate coincided with a slight increase in the optical density of the culture, which could be explained by the rapid growth of sporogenic hyphae (Fig. 1A). A subsequent drop in the replication rate was accompanied by an increase in the FtsZ-YPet level (21–23 h) and assembly of FtsZ-YPet into ladders of Z-rings (22 h) (Supplementary Fig. 4A). At this time point, the spore chains entered the maturation phase; during this phase, chromosome compaction was shown to gradually increase, achieving significant condensation at 25 h (Supplementary Fig. 4B), while the FtsZ-YPet level rapidly decreased and the hyphae fragmented. This stage ended when immature spores could be detected (25–26 h) (Fig. 1A and Supplementary Fig. 4A). Thus, we established that 22 h of S. venezuelae culture is the point at which multi-genomic hyphae undergo synchronised cell division to form spores, replication of chromosomes ceases, while their condensation increases, which would be expected to minimise interchromosomal contacts. Based on these findings, we set out to establish the chromosome conformation at this time point.
Hi-C mapping of the chromosomal contacts in S. venezuelae sporogenic hyphae (22 h) undergoing cell division indicated the presence of two diagonal axes crossing perpendicularly in the proximity of the centrally located oriC region (Fig. 1B). The primary diagonal corresponds to the contact frequency between neighbouring chromosomal loci. Based on the contact range (extended in the proximity of chromosome termini) and principal component analysis (PCA1), three distinct chromosome regions were distinguished: the core region that encompasses 4.4 Mbp (position: 1.9–6.3 Mbp) around the centrally positioned oriC and two well-organised domains flanking the chromosome core, which we named the left and right terminal domains (LTD and RTD, respectively), with each measuring ~1.5–2 Mbp in size (Fig. 1B and Supplementary Fig. 5). The secondary diagonal, which results from the interarm interactions, was prominent in the region of ~2.0–2.2 Mbp in each direction from the oriC region and severely diminished within terminal domains (Fig. 1B). Additionally, the Hi-C contact map showed some diffuse signals at the termini proximal fragments of the secondary diagonal, suggesting the existence of spatial proximity but also somewhat longer range contacts at the termini of the linear S. venezuelae chromosome (Fig. 1B and Supplementary Fig. 3B).
In summary, these results indicate that at the time of S. venezuelae synchronised cell division, the linear chromosomes are folded with both arms closely aligned within the core region, while the terminal regions form distinct domains.
Arms proximity increases during sporulation and requires segregation protein activity
Having established chromosomal contact maps during S. venezuelae sporogenic cell division, we set out to characterise the changes in chromosomal arrangement during sporogenic development. To this end, we obtained normalised Hi-C contact maps for the wild-type strain during the vegetative stage at the entry into the pre-sporulation phase when growth slows and DNA replication decreases (13 h) and during the pre-sporulation phase when the chromosomes are still not compacted (15 and 17 h), as well as during spore maturation (25 h), which is when chromosome compaction reaches a maximum (Fig. 1A and Supplementary Fig. 4B).
At the entry into the pre-sporulation phase (13 h), only the primary diagonal was clearly visible, whereas the secondary diagonal was barely detectable across the core and terminal domains, indicating limited contact between the chromosomal arms during vegetative growth (Fig. 1C). During the pre-sporulation phase, the signals of the secondary diagonal were enhanced and extended at distances of up to 1 Mbp from the oriC region at 15 and 17 h of growth (Fig. 1C). Finally, in the maturing spores (25 h), we observed almost complete arm alignment, similar to that detected at 22 h (Fig. 1C). This result suggests that although nucleoid compaction was still underway during the spores maturation phase (Supplementary Fig. 4B), the global chromosome organisation did not change critically after that established during cell division (Fig. 1C). Since the contact maps show various chromosomal arrangements within the hyphae of S. venezuelae, we cannot estimate the timing of the process for a single chromosome. However, taking into account synchronous S. venezuelae development, we can conclude that the average arm cohesion gradually increases during the pre-sporulation phase.
In model bacteria whose chromosomal arms are in close proximity, their positioning is directed by ParB-dependent SMC loading within the oriC region4,7,28. During Streptomyces sporogenic cell division, coincident with the time of observed folding of chromosomes with proximal arms, ParB has been shown to bind numerous parS scattered in the proximity of oriC (Supplementary Fig. 3B), assembling into regularly spaced complexes that position oriC regions along the hyphae57. At the same time, ParA accumulates in sporogenic hyphae and facilitates the efficient formation and segregation of ParB complexes39,60. Elimination of S. venezuelae segregation proteins disturbs aerial hyphal development and chromosome segregation39 but does not affect the culture growth rate (Supplementary Fig. 2B). To verify whether segregation proteins have a role in the organisation of S. venezuelae chromosomes, we obtained Hi-C maps of chromosomal contacts in sporogenic hyphae of parA or parB mutants.
The Hi-C map generated for the parB mutant showed the complete disappearance of the secondary diagonal axis, confirming the expected role of the ParB complex in facilitating interarm contacts (Fig. 1D). The same lack of a secondary diagonal was observed for the parB mutant at each time point in the S. venezuelae life cycle (Supplementary Fig. 6A). Deletion of parA also had a detectable influence on arm alignment, although it varied among the samples analysed (Fig. 1D and Supplementary Fig. 6B). The strength of the signals at the secondary diagonal was either significantly or only slightly lowered in the parA mutant, as indicated by the differential Hi-C map in comparison to the wild-type strain (Fig. 1D and Supplementary Fig. 6B). While the effect of parA or parB deletions was predominantly manifested by the partial or complete disappearance of the interarm contacts, respectively, their influence on the local chromosome structure, including LTD and RTD folding, was marginal (Fig. 1D and Supplementary Fig. 5).
Taken together, these results indicate that chromosomal arm alignment progresses during pre-sporogenic and sporogenic hyphal development (Fig. 1C, scheme) and that this chromosomal rearrangement is dependent on ParB (Fig. 1D, scheme).
SMC- and HupS-induced chromosome compaction permits regularly spaced cell division
In many bacteria, ParB impacts chromosome organisation by mediating the loading of SMC, which promotes global chromosome compaction. Prior studies using S. coelicolor showed that the elimination of either SMC or HupS, a Streptomyces-specific sporulation-associated NAP, affected nucleoid compaction in spores40,42,61. We hypothesised that SMC would be responsible for ParB-induced chromosomal arm alignment but that since ParB elimination did not influence short-range chromosome interactions, other DNA-organising factors, such as HupS, may also contribute to chromosome compaction in sporogenic hyphae.
The disruption of either the hupS or smc gene in S. venezuelae did not significantly affect the growth rate or differentiation of these bacteria; however, a double hupS smc mutant grew slower than wild-type or either of the single mutants (Supplementary Fig. 7A, inset and Supplementary Fig. 7B). Interestingly, the elimination of hupS or, to a lesser extent, smc increased spore size, while the double hupS smc deletion led to more pronounced disturbances in spore size (Fig. 2A, B). Normal spore length was restored in the single mutants by complementation with in trans-delivered hupS-FLAG or smc-FLAG genes (Fig. 2B).
Fig. 2. Phenotypic effects of smc, hupS, and double smc/hupS deletions.
A Scanning electron micrographs showing sporulating hyphae and spores of the wild-type and hupS, smc and hupS smc double mutant (ftsZ-ypet derivatives, MD100, TM005, TM004 and TM006, respectively) (representative of 10 images). Elongated spores are marked with a yellow arrowhead. Scale bar: 10 µm. B Box plot analysis of the spore length distribution in the S. venezuelae wild-type (light green), hupS (yellow), smc (blue) and hupS smc (pink) (AK200, TM010, TM003, respectively), as well as in hupS and smc mutants complemented with in trans-delivered hupS-flag (orange) and smc-flag genes (dark blue) (TM015 and TM019, respectively) (300 spores were analysed for each strain). Boxplots show median with first and third quartile while the lower and upper ‘whiskers’ extend to the value no further than 1.5 * IQR (interquartile range) from the ‘hinge’. The statistical significance between strains determined by a one-way anova with a Games–Howell post-hoc test (two-sided) is marked with asterisks: p-value ≤0.05 (*), ≤0.01 (**) and ≤0.001 (***). p-values: wild-type-hupS: <0.0001, wild-type-smc: 0.0242, wild-type-hupS smc: <0.0001, wild-type-hupS; hupS-FLAG: 0.0625, wild-type-smc; smc-FLAG: 0.3871, hupS-smc: 0.0089, hupS-hupS smc: 0.1241, smc-hupS smc: <0.0001. C Examples of time-lapse images (representative of 10 repetitions) showing visualisation of nucleoids (marked with HupA-mCherry fusion) and Z-rings (visualised by FtsZ-YPet fusion) in the wild-type background (ftsZ-ypet, hupA-mCherry derivative, TM011) hupS, smc and hupS smc double mutant background (ftsZ-ypet, hupA-mCherry derivative, TM013, TM012 and TM014, respectively). An abnormal hyphal fragment is marked with a yellow arrowhead. Scale bar: 5 µm. D Box plot analysis of the nucleoid area (visualised by HupA-mCherry fusion) in the wild-type (light green, 262 spores), hupS (yellow, 232 spores), smc (blue, 220 spores) and hupS smc (pink, 218 spores) double mutant (ftsZ-ypet, hupA-mcherry derivative, MD100, TM011, TM013, TM012, TM014, respectively). Boxplots show median with first and third quartile while the lower and upper ‘whiskers’ extend to the value no further than 1.5 * IQR (interquartile range) from the ‘hinge’. The statistical significance between strains determined by Wilcoxon rank-sum test (two-sided) with Holm method used for multiple comparisons is marked with asterisks: p-value ≤0.05 (*), ≤0.01 (**) and ≤0.001 (***). p-values: wild-type-hupS: <2e−16, wild-type-smc: <2e−16, wild-type-hupS smc: <2e−16, hupS-smc: 0.9082, hupS-hupS smc: 0.0028, smc-hupS smc: 0.0039. E Kymographs showing the intensity of Z-ring fluorescence along the representative hyphae in the wild-type and hupS, smc and hupS smc double mutant (ftsZ-ypet derivatives, MD100, TM005, TM004, TM006, respectively) during maturation. F The variation (standard deviation) of FtsZ-Ypet (Z-rings) fluorescence during spore maturation of strains calculated for 25–30 hyphae of the wild-type (light green), hupS (yellow), smc (blue) and hupS smc (pink) double mutant (ftsZ-ypet derivatives, TM005, TM004, TM006, respectively). Fluorescence was measured starting from 30 min before growth arrest until spore formation. Points represent values for each hyphae, while lines show the result of Local Polynomial Regression Fitting (loess) with 95% confidence intervals (shaded area).
Next, to visualise nucleoid compaction and segregation during cell division, we used fluorescence microscopy to observe sporogenic hyphal development of smc and/or hupS mutant strains labelled with HupA-mCherry and FtsZ-YPet (Fig. 2C and Supplementary Fig. 7C, Supplementary Movies 1–4). None of the mutant strains formed anucleate spores, but all of them showed significantly increased nucleoid areas, in particular the double hupS smc mutant, where notably enlarged spores could be observed (Fig. 2A–D). The increased nucleoid area was accompanied by increased distances between Z-rings (Supplementary Fig. 7D). The largest fraction was found for Z-rings spaced at increased distances (longer than 1.3 μm) in the double hupS smc mutant and in the smc mutant (Supplementary Fig. 7D). Additionally, the intensity of Z-rings in single smc or the double mutant was more varied along the hyphae than in the wild-type strain, indicating their lowered stability (Fig. 2E, F). The disturbed positioning and stability of Z-rings explain the presence of elongated spores in mutant strains.
In summary, both SMC and HupS contribute to chromosome compaction in spores. The decreased chromosome compaction in the mutant strains correlated with decreased Z-ring stability and their aberrant positioning which, in all studied mutants, resulted in increased distance between septa and elongated spores.
SMC and HupS collaborate during sporogenic chromosome rearrangement
Having confirmed that both SMC and HupS mediate global chromosome compaction during S. venezuelae sporulation, we investigated how both proteins contribute to chromosomal contacts during sporulation-associated chromosome rearrangement.
The Hi-C contact map obtained for the smc mutant at sporogenic cell division (22 h) showed the complete disappearance of the secondary diagonal (similar to the map obtained for the parB mutant), indicating abolished chromosomal arm interactions (Figs. 1D and 3A). In contrast, the elimination of smc resulted in an increase in short-range DNA contact frequency (<200 kbp) along the primary diagonal, whereas in the parB mutant, the short-range DNA contacts were only slightly affected, suggesting an additional role of SMCs in chromosome organisation (Figs. 1D, 3A and Supplementary Fig. 8A). Indeed, while nucleoid compaction was previously noted to be affected by parB deletion39, the measurements of the nucleoid area confirmed higher chromosome compaction in parB than in smc mutant strains (Supplementary Fig. 8B).
Fig. 3. Influence of SMC and HupS on chromosome organisation.
A Top panels: the normalised Hi-C contact maps obtained for the wild-type and smc, hupS and smc hupS double mutants (ftsZ-ypet derivatives, MD100, TM005, TM004, TM006, respectively) growing for 22 h or for 25 h (only in the case of the double hupS smc mutant) (in 5 ml cultures). Bottom panels: the corresponding differential contact maps in the logarithmic scale (log2) comparing the contact enrichment in the wild-type strain (red) versus the mutant strains (blue) are shown below each Hi-C contact map. B The number of FLAG-SMC binding sites along the chromosome of the wild-type (blue) (TM017) or parB mutant background (red) (KP4F4) determined by ChIP-Seq analysis at the time of sporogenic cell division (14 h of 50 ml culture growth). The number of FLAG-SMC binding sites (250 bp) was determined by normr analysis averaged over 0.5 Mbp sliding window every 1000 bp with a loess model fit. Points below the plot show positions of individual sites coloured according to their FDR (false discovery rate) values (yellow less significant, dark purple more significant). C The number of HupS-FLAG binding sites along the chromosome in the wild-type background (hupS deletion complemented with hupS-FLAG delivered in trans, TM015) determined by ChIP-Seq analysis at the time of sporogenic cell division (14 h of 50 ml culture growth). The number of HupS-FLAG binding sites was determined by differential analysis of the HupS-FLAG and wild-type strains using edgeR and averaged over 0.5 Mbp sliding window every 1000 bp. Smooth line shows loess model fit. The points below the plot show positions of individual sites coloured according to their FDR values (yellow less significant, dark purple more significant). D Heatmaps showing the number of reads for all 307 regions bound by HupS-FLAG in the wild-type background (TM015), smc mutant background (TM016) and negative control (the wild-type strain). The reads were normalised by the glmQL model from the edgeR package. For each region, position 0 is the position with the maximum number of reads. Regions are sorted according to their logFC values. E Comparison of HupS-FLAG binding in the wild-type (blue) and smc deletion background (red). The lines show mean values of reads (for 69 bp long regions) with 95% confidence intervals for all 307 sites differentially bound by HupS-FLAG protein. For all sites, 1000 bp long fragments were extracted from the chromosome centred around the best position as determined by edgeR analysis.
To confirm that ParB promotes the interaction of SMC with DNA, we used ChIP-seq to analyse FLAG-SMC binding in the wild-type background and in the parB mutant. First we assessed how the developmental time points for 50-ml culture conditions used in our ChIP-seq experiments corresponded to those in the 5 ml culture previously described for Hi-C experiments (Fig. 1A and Supplementary Fig. 9A). At the 12 h of ‘ChIP-seq culture’, DNA replication decreased (corresponding to 15–17 h of ‘Hi-C culture’), while at 14 h of ‘ChIP-seq culture’, DNA compaction and the formation of regularly spaced Z-rings were clearly detected (Supplementary Fig. 9B) (corresponding to 22 h in ‘Hi-C culture’ conditions), indicating sporogenic cell division and chromosome segregation. Notably, the level of FLAG-SMC remained constant during sporogenic development (Supplementary Fig. 10A) and the N-terminal FLAG-SMC fusion did not affect the function of the protein (Supplementary Fig. 10B).
Analysis of DNA fragments bound by FLAG-SMC at the time of sporogenic cell division did not identify any specific binding sites for FLAG-SMC, suggesting a lack of sequence preference. However, quantification of the detected SMC binding sites along the chromosome (using an algorithm dedicated to non-specific DNA binders) showed that their frequency increased up to twofold in the chromosomal core region (Fig. 3B and Supplementary Fig. 11A). The increased frequency of SMC binding overlapped with the secondary diagonal identified in Hi-C experiments (Fig. 3A, B and Supplementary Fig. 11B). SMC binding was severely diminished in the parB mutant (Fig. 3B), demonstrating that the ParB complex is a prerequisite for SMC loading.
The Hi-C contact map generated for the hupS mutant at the time sporogenic cell division (at the 22 h) revealed the presence of both diagonals. However, the identified local signals within both diagonals were more dispersed than in the wild-type, and diagonals were visibly broadened (Fig. 3A and Supplementary Fig. 6C). Moreover, along the primary diagonal, a less clear distinction between the core region and terminal domains could be observed. The effect of hupS deletion, while spreading over the whole chromosome compaction, was most prominent within terminal domains, especially within RTD (Fig. 3A and Supplementary Fig. 5).
To further investigate the involvement of HupS in the organisation of terminal domains and to test the cooperation between HupS and SMC, we analysed HupS-FLAG binding in the complemented hupS and double hupS smc deletion strains at the time of cell division. In contrast to SMC, HupS-FLAG showed low sequence binding preference, and we identified 307 regions differentially bound by HupS-FLAG with a mean width of 221 bp (Fig. 3C, D and Supplementary Fig. 12A). The analysis of the number of HupS-FLAG binding sites exhibited a significant increase in the terminal domains, especially the right terminal domain, but HupS binding was also detected in the core region of the chromosome (Fig. 3C). The enhanced binding of HupS observed at chromosomal termini is consistent with the lowered number of chromosomal contacts in RTD observed in the Hi-C maps obtained for the hupS mutant.
Further analysis of ChIP-seq data performed using two independent methods (rGADEM and MEME) identified the motif recognised by HupS-FLAG (Supplementary Fig. 12B). Although the motif was observed to be uniformly widespread along the chromosome, and was not required for HupS binding, it was enriched in HupS binding sites versus random regions, with an increased probability for occurrence in the middle of the HupS-bound regions (confirmed with CentriMo) (Supplementary Figs. 12B–E). Furthermore, in vitro studies showed that immobilised GST-HupS-bound digested cosmid DNA with no sequence specificity (Supplementary Fig. 12F). Interestingly, ChIP-seq analysis of HupS-FLAG binding in the absence of SMC (hupS smc deletion complemented with hupS-FLAG) showed a lower number of reads for all HupS-FLAG binding sites, indicating that SMC enhanced the HupS-DNA interaction (Fig. 3D, E). This observation could not be explained by the lowered level of HupS-FLAG in the smc deletion background (Supplementary Fig. 10C). We also noted marginal effect of hupS deletion on chromosome organisation at the early pre-sporulation phase, as indicated by the Hi-C contact map for the hupS mutant at this time point (Supplementary Fig. 10D), which corresponded with the lower HupS-FLAG level during the vegetative growth than during sporulation42.
Since our ChIP-seq analysis indicated cooperation between HupS and SMC binding and that the double hupS smc deletion most severely affected chromosome compaction, we investigated how the double deletion affects chromosomal arrangement. The Hi-C contact map generated for the double hupS smc mutant at the time of sporogenic cell division (22 h) showed the combination of effects observed independently for hupS and smc single mutations, namely, the disruption of arm alignment and the destabilization of local DNA contacts (Fig. 3A). Thus, while long-range interarm interactions are facilitated by SMC, short-range compaction is governed by HupS, and the effect of the elimination of both proteins is additive.
In summary, our Hi-C and ChIP-seq analyses of smc and hupS mutants showed their contribution to global chromosome compaction. While SMC binds in the chromosomal core and is responsible for the close contact of chromosome arms, HupS binds preferentially in terminal domains and organises them. Cooperation between both proteins is manifested by SMC enhancing HupS binding to DNA. Elimination of both proteins disturbs global chromosome organisation.
Discussion
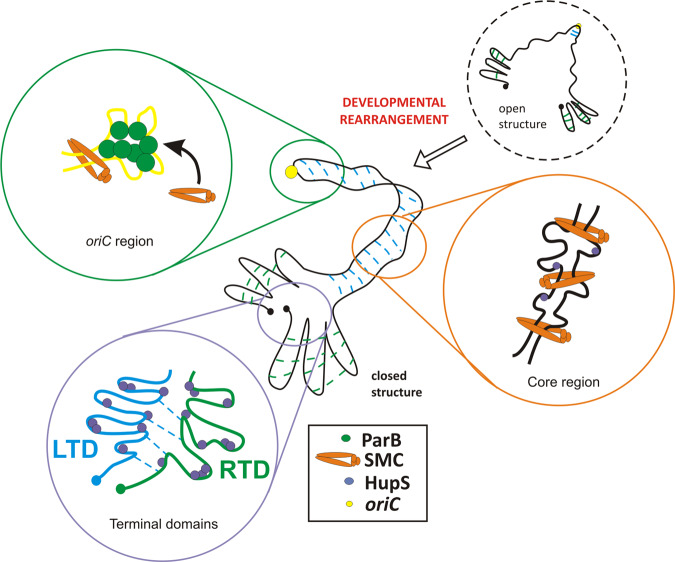
In this study, we provide a complex picture of the dynamic rearrangement of a linear bacterial chromosome. Hi-C mapping of the ~8.2 Mbp S. venezuelae chromosome during sporogenic cell division revealed chromosomal folding with interarm contacts, which extend ~4 Mbp around the oriC region. Arms proximity is diminished at the independently folded 1.5 and 2 Mbp terminal domains LTD and RTD, respectively (Fig. 4). The independent folding of core and terminal domain corroborates with distinguished by genomic analyses regions of Streptomyces chromosome: core that contains housekeeping genes and exhibits high synteny and 1.5–2 Mbp long arms with low evolutionary conservation62. Further analysis of chromosomal contacts shows spatial proximity of both chromosomal termini, which is consistent with the findings of previous cytological studies53. Folding of the Streptomyces linear chromosome into core and terminal domains was also independently demonstrated for the S. ambofaciens63.
Fig. 4. Model of spatial rearrangement of the S. venezuelae chromosome during sporogenic development showing the contribution of ParB (green), SMC (orange) and HupS (blue) to chromosomal arm alignment and organisation of terminal domains (TLD and RTD).
Blue lines indicated intrachromosomal contacts, oriC region is marked with a yellow circle.
Our Hi-C data disclosed substantial rearrangement of chromosomal conformation occurring during S. venezuelae sporulation. At the end of vegetative growth, at the entrance to the sporulation phase contacts between the chromosomal arms are scarce. After this time point, chromosomal arms become increasingly aligned, reaching a maximum at the time of initiation of sporogenic cell division (Z-ring formation) which is also the time point when chromosome segregation is initiated39. Thus, during Streptomyces differentiation, changes in hyphal morphology, gene expression, chromosome replication and mode of cell division13,34 are accompanied by rearrangement of chromosomes from an extended conformation during vegetative growth to almost fully folded and compacted structures.
Our data demonstrate that in S. venezuelae, both ParB and SMC are essential for imposing chromosomal arms proximity, reinforcing previous work describing ParB-dependent SMC loading onto the chromosome. Importantly, in S. venezuelae, SMC binding is enriched within the core region of the chromosome, matching the higher frequency of interarm contacts in this region. SMC was previously shown to be involved in chromosome compaction during S. coelicolor sporulation40,61. Interestingly, parB deletion was also observed to decrease chromosome condensation at the time of cell division39. This observation may be explained by the lack of SMC loading in absence of ParB, which was also reported for the other model bacteria4,6,7,9. The increase in long-range chromosomal contacts induced by SMC in the chromosomes of S. venezuelae and other bacteria corroborates the proposed model of action in which SMC zips DNA regions when translocating from the loading site4.
While the increase in arm alignment during sporulation63 cannot be explained by increased SMC level during sporulation, the parAB genes were both earlier shown to be transcriptionally induced in Streptomyces sporogenic hyphae56,64. ParB binds in the proximity of the oriC regions of all chromosomes throughout the whole life cycle but complexes formed during vegetative growth and at the early stages of sporogenic development are not as massive as the regularly spaced segrosomes that accompany sporogenic-associated cell division60. Notably, the Streptomyces chromosome contains an unusually high number of ParB binding sites (16–20), indicating a complex architecture of the segrosome. Since interarm contacts occur during cell division, a characteristic spatial architecture of sporogenic segrosomes is implied to be a prerequisite for SMC loading. The significance of the ParB complex architecture for SMC loading has been established by studies of ParB mutants with impaired spreading or bridging in B. subtilis and in C. glutamicum7,27. Accordingly, our observations suggest that during Streptomyces differentiation ParB complexes spatially rearrange to increase their capability of SMC loading and initiating chromosomal folding with proximal arms.
What factors could induce the reorganisation of ParB complexes during Streptomyces sporulation? ParB has recently been shown capable of binding to and hydrolysing CTP, which affects the formation of segrosomes and appears to be conserved among ParB homologues65–67. While not yet confirmed in Streptomyces, CTP binding and hydrolysis may be involved in the rearrangement of ParB complexes on chromosomes in sporogenic hyphae. The formation of ParB complexes has also been shown to be associated with sporulation-dependent deacetylation of ParB in S. coelicolor68. Furthermore, segrosome rearrangement may also result from the increased transcriptional activity of the parAB operon. ParA accumulates in sporogenic cells, promoting the formation and regular distribution of the segrosomes39,60. In other bacteria (B. subtilis, V. cholerae, and M. smegmatis) the elimination of ParA also decreased ParB binding69–71. Indeed, our Hi-C mapping showed that in the absence of ParA, interarm interactions are somewhat diminished, reinforcing the role played by ParA in executing ParB architecture. Finally, since the transcriptional activity was proven to have a considerable impact on chromosomal arm alignment5,63,72, sporulation-associated changes in transcription within the chromosomal core may account for the development of interarm contacts.
Our study has established that although HupS promotes contacts all over the chromosome, it is particularly involved in the organisation of the terminal domains. HupS exhibited binding sequence preference in vivo, but no sequence specificity when binding linear DNA fragments. Thus, we infer that specific DNA structural features may account for enhanced HupS binding in particular regions. Reduced binding of HupS in the absence of SMC could be explained by preferential binding by HupS to structures such as loops, generated by SMC. This notion corroborates earlier suggestions that HU homologues could stabilise DNA plectonemic loops, increasing short-range contacts6. E. coli studies also suggested cooperation between condensin MukB and HU homologues in maintaining long-range chromosomal contacts10. In fact, in E. coli, condensin activity was suggested to be increased by DNA structures induced by HU protein10. Previous analysis of S. coelicolor showed that HupS deletion resulted in chromosome decompaction in sporogenic hyphae42. Our Hi-C analyses have established that both SMC and HupS contribute to chromosome compaction during sporulation, and the double hupS smc deletion has the strongest effect on chromosomal conformation, affecting both interarm and short-range chromosomal contacts (Fig. 4).
Although elimination of SMC and HupS disturbed the S. venezuelae chromosome structure, the phenotype of the double mutant was still rather mild and manifested predominantly in increased nucleoid volume and formation of elongated spores. While a mild phenotype for an smc mutant was also described in C. glutamicum, in most bacteria (e.g., B. subtilis. C. crescentus and S. aureus), smc deletion results in either chromosome decompaction and/or segregation defects and often led to the formation of elongated cells7,21,73–75. Interestingly, condensins have been suggested to be particularly important for chromosome segregation in bacteria lacking the complete parABS system (such as E. coli)76–78. The variation of spore size observed in S. venezuelae smc mutants correlates with irregular placement and lowered stability of Z-rings, suggesting that decreased chromosome compaction affects Z-ring formation and/or stability.
In conclusion, we have established the first maps of chromosomal contacts for a linear bacterial chromosome. The S. venezuelae chromosome conformation undergoes extensive rearrangement during sporulation. Similar chromosomal rearrangements have been observed during the transition from the vegetative to stationary phase of growth in S. ambofaciens63. DNA-organising proteins HupS and SMC contribute to chromosome compaction, and they also affect cell division. Although roles for HU and SMC have been established in numerous bacteria genera, in Streptomyces, their function is seemingly adjusted to meet the requirements of compacting the large linear chromosomes. Owing to the unique developmental life cycle of Streptomyces spp., in which chromosome replication and segregation are spatiotemporally separated, the transition between the particular stages of the life cycle requires chromosomal rearrangement (Fig. 4) on a similar scale and complexity to those seen for chromosomes during the eukaryotic cell cycle.
Methods
Bacterial strains and plasmids
The S. venezuelae and E. coli strains utilised in the study are listed in Supplementary Tables 1 and 2 respectively, and the plasmids and oligonucleotides are listed in Supplementary Tables 3 and 4, respectively. DNA manipulations were performed by standard protocols79. Culture conditions, antibiotic concentrations, transformation and conjugation methods followed the commonly employed procedures for E. coli79 and Streptomyces80.
To construct unmarked smc deletion S. venezueale strain (TM010), first apramycin resistance cassette was inserted into the chromosome to replace smc gene using λ Red recombination based PCR-targeting protocol81. First the cassette encompassing apra gene and oriT was amplified on the template of pIJ773 using primers psmc_Fw and psmc_Rv and used to transform BW25113 carrying cosmid Sv-3-B07 and λ RED plasmid, pIJ790. The resulting cosmid Sv-3-B07Δsmc::apra was used for conjugation into wild-type S. venezuelae. The AprR exconjugants were screened for the loss of KanR, indicating a double-crossover allelic exchange of the smc locus. The obtained colonies were verified by sequencing of the PCR products obtained using the chromosomal DNA as the template and the primers psv_smc_spr_Fw and psv_smc_spr_Rv. The obtained strain TM001 was next used for further modifications.
To obtain unmarked smc deletion strain, FLP recombinase, that recognises FRT sites flanking apra cassette, was used. To this end, DH5α containing pCP20 plasmid, that encodes yeast FLP recombinase, were transformed with the cosmid Sv-3-B07 Δsmc::apra. KanR and AprR resistant transformants were incubated at 42 °C and next clones sensitive to apramycin were selected. After verification of the scar sequence generated in smc locus, the obtained Sv-3-B07 Δsmc::scar cosmid was modified, using PCR-targeting protocol, by insertion of apra-oriT cassette in the bla locus in the SuperCos. The cassette was amplified at the template of pIJ773 plasmid using primers pblaP1 and pblaP2. The resulting cosmid Sv-3-B07 Δsmc::scar was used for conjugation into S. venezuelae smc::apra (TM001) strain. The AprR, KanR exconjugates were next screened for the loss of KanR AprR, indicating a double-crossover allelic exchange of the smc locus. The obtained strain TM010 was verified by sequencing of the PCR products obtained using the chromosomal DNA as the template and the primers psv_smc_spr_Fw and psv_smc_spr_Rv.
Strains producing FLAG-SMC: FLAG-smc (TM017) and parB FLAG-smc (KP4F4) were constructed by homologous recombination and modification of smc gene in its native chromosomal locus. First the apramycin resistance cassette flanked with the oligonucleotide encoding FLAG was amplified on the template of pIJ773, using papra_FLAG_Fw and papra_FLAG_Rv primers that contained NdeI restriction sites and used to transform BW25113 carrying cosmid Sv-3-B07 and λ RED plasmid, pIJ790. Next, the obtained cosmid Sv-3-B07 apra-oriT- FLAG-smc was digested with NdeI and religated to remove apra-oriT from the upstream region of smc gene. The obtained cosmid was verified using PCR with primers psmc_promoter_Fw and psmc_NFLAG_Rv and then modified, using PCR-targeting protocol, by insertion of apra-oriT cassette in the bla locus in the SuperCos. The cassette was amplified at the template of pIJ773 plasmid using primers pblaP1 and pblaP2. The resulting disrupted cosmid Sv-3-B07 FLAG-smc was used for conjugation into S. venezuelae smc::apra (TM001) strain. The AprR, KanR exconjugates were next screened for the loss of KanR AprR, indicating a double-crossover allelic exchange of the smc locus. The obtained strain TM017 was verified by sequencing of the PCR products obtained with the primers psmc_promoter_Fw and psmc_NFLAG_Rv on the template of the chromosomal DNA as well as Western Blotting with the anti-FLAG M2 antibody (Merck).
To obtain parB FLAG-smc strain (KP4F4) cosmid Sv-4-A98ΔparB::apra was used for conjugation into S. venezuelae FLAG-smc (TM017) strain. The AprR exconjugants were screened for the loss of KanR, indicating a double-crossover allelic exchange of the parAB locus.
S. venezuelae hupS deletion strains (hupS (AKO200), hupS smc (TM003) and hupS FLAG-smc (TM018)) were constructed using λ Red recombination PCR-targeting method81 by the introduction of apra resistance cassette in the hupS locus. First, the cassette encompassing apra gene and oriT was amplified on the template of pIJ773 using primers phupS_Fw and phupS_Fw and used to transform BW25113 carrying cosmid Sv-5-D08 and λ RED plasmid, pIJ790. The resulting cosmid Sv-5-D08ΔhupS::apra was used for conjugation into the wild-type, smc::scar (TM010) and to FLAG-smc (TM017) strains. The AprR exconjugants were screened for the loss of KanR, indicating a double-crossover allelic exchange in the hupS locus. The obtained hupS (AKO200), hupS smc (TM003) and hupS FLAG-smc (TM018) strains were verified by sequencing of the PCR products obtained using the chromosomal DNA as the template and the primers phupSspr_Fw and phupSspr_Rv.
To construct hupS-FLAG strains (hupS-FLAG (TM015) and hupS-FLAG smc (TM016)) plasmid pIJ10770-hupS-FLAG, that contained hupS gene under the control the native hupS promoter, was used. The oligonucleotide encoding FLAG was amplified using primers PpSS170_xhoI_FLAG_Fw and PpSS170-eco32I_FLAG_Rv that included restriction sites XhoI and Eco32I and cloned into pIJ10770 plasmid (HygR, kind gift from dr. Susan Schlimpert, John Innes Centre, Norwich, UK) delivering pIJ10770-FLAG. hupS gene with its native promoter was amplified on the template of cosmid Sv-5-D08 using primers PpSS170_hupS_FLAG_Fw and PhupS-tocherry-Rv. The fragment containing hupS gene was cloned to pIJ10770-FLAG using restriction sites Acc65I and XhoI. The obtained plasmid pIJ10770-hupS-FLAG was used for conjugation to hupS (AKO200) and hupS smc (TM003) strains delivering hupS-FLAG (TM015) and hupS-FLAG smc (TM016). Strains were verified by sequencing of the PCR products obtained using primers ppSS_spr_Fw and ppSS_spr_Rv on the template of chromosomal DNA, and western blotting with anti-FLAG M2 antibody (Merck).
To construct the S. venezuelae strains expressing ftsZ-ypet the obtained strains TM010, AKO200 and TM003 were used for conjugation with the E. coli carrying pKF351 ftsZ-ypet apra (to TM010) or pKF351 ftsZ-ypet hyg (to AKO200 and TM003) plasmid. smc ftsZ-ypet (TM004 AprR), hupS ftsZ-ypet (TM005, HygR) and hupS smc ftsZ-ypet (TM006, HygR) were verified by PCR with pftszypet_fw and pftszypet_Rv and fluorescence microscopy.
To construct the S. venezuelae strains expressing ftsZ-ypet and hupA-mcherry, we used plasmid pSS172hupA-mcherry (hygR, kind gift from dr. Susan Schlimpert, John Innes Centre, Norwich, UK) that contained hupA-mCherry under the control of the native hupA promoter. Plasmid pSS172hupA-mCherry was introduced to ftsZ-ypet (in the wild-type background, MD100, AprR) and smc ftsZ-ypet (TM004, AprR) strain generating ftsZ-ypet hupA-mcherry (TM011, AprR, HygR) and smc ftsZ-ypet hupA-mCherry (TM012, AprR, HygR), respectively. Strains hupS ftsZ-ypet, hupA-mCherry (TM013, AprR, HygR SpecR) and hupS smc ftsZ-ypet hupA-mCherry (TM014, AprR, HygR SpecR) were constructed by subsequent delivery of pKF351 ftsZ-ypet spec and pSS172hupA-mCherry to hupS (AKO200) and hupS smc (TM003). The obtained strains were verified by PCR with PpSS_spr_Fw and PpSS_spr_Rv primers using chromosomal DNA as template and by using fluorescence microscopy.
Genetic complementation of Δsmc::scar (TM010) was performed by delivering smc-FLAG in trans. To this end the smc gene was amplified using cosmid DNA Sv-3B07 as template, using primers psmc_poczatek_Fw and psmc_exp_Rv, next PCR product was cloned into pGEM-T-Easy vector, to which also double-stranded oligonucleotide encoding FLAG was inserted using BamHI and Mph1103I restriction enzymes. The promoter region of smc was amplified using Sv-3-B07 cosmid DNA as template and primers psmc_promoter_Fw and psmc_promoter_Rv and cloned directly into pMS83 using KpnI and NdeI. Next, fragment encoding SMC-FLAG was cloned from pGEM-TEasy-smc-FLAG to pMS83-psmc yielding pMS83-smc-FLAG, which was used for conjugation to Δsmc::scar (TM010) yielding TM019 (Δsmc::scar; pMS83-smc-FLAG). The construct was verified by sequencing of PCR products obtained using the chromosomal TM019 as template and primers psmc_spr_Fw and psmc_spr_Rv; as well as psmc_inter_Fw and psmc_NFLAG_Rv,. and then by western blotting.
S. venezuelae cultures and growth rate analysis
To obtain the standard growth curves comparing the growth rate of S. venezuelae strains, a Bioscreen C instrument (Growth Curves US) was used. Cultures (at least three independent cultures for the same strain) in liquid MYM medium (300 µl) were set up by inoculation with 6 × 103 colony-forming units (CFUs) of S. venezuelae spores. The cultures grew for 48 h at 30 °C under the ‘medium’ speed and shaking amplitude settings, and their growth was monitored by optical density measurement (OD600) every 20 min. The data were collected using BioScreener 3.0.0 software.
The Hi-C cultures were set up as follows: 5 ml of MYM medium was inoculated with 108 CFUs, and the cultures were incubated for 6–26 h at 30 °C with shaking (180 rpm). For the ‘5 ml culture’ growth curve depiction, the optical density (OD600) of the culture diluted (1:10) with MYM medium was measured in 1-h intervals.
Preparation of Hi-C libraries and data analysis
For the preparation of S. venezuelae Hi-C contact maps, 5 ml cultures were established as described above. Each S. venezuelae strain and/or developmental stage was analysed in at least 2 experimental replicates. The cultures were incubated for 13 to 25 h at 30 °C with shaking (180 rpm). After this step, the cultures were cross-linked with 1% formaldehyde (Sigma Aldrich) for 7 min and 30 s and blocked with 0.2 M glycine for 15 min, both steps performed at 30 °C with shaking (180 rpm). One millilitre of the cross-linked culture was centrifuged (1 min, 2400 g at room temperature). The mycelium pellet was washed twice with 1.2 ml of TE buffer (10 mM Tris-HCl pH = 8.0, 1 mM EDTA) and resuspended in 0.6 ml of TE buffer. Two vials, each containing 25 µl of the resuspended mycelium, were independently processed according to the protocol below (a modification of the protocol described by Le et al.82). The cells were lysed using 0.5 µl of ReadyLyse solution (Invitrogen) at 37 °C for 30 min followed by the addition of 1.25 µl of 5% SDS solution (Sigma Aldrich) and incubated at room temperature for 15 min. After cell lysis, the reaction was supplemented with 5 µl of 10% Triton X-100 (Sigma Aldrich), 5 µl of 10× reaction buffer 3 (New England Biolabs) and 10.75 µl of DNase-free water (Invitrogen) and subsequently incubated for 15 min at room temperature. The chromosomal DNA was digested with 2.25 µl of BglII restriction enzyme (New England Biolabs; 50,000 U/ml) for 2 h and 30 min at 37 °C followed by the addition of 0.25 µl of BglII (New England Biolabs; 50,000 U/ml) and incubation for another 30 min at 37 °C. 50 µl of the BglII-digested reaction was cooled down on ice and next, the generated sticky ends were labelled with biotin-14-dATP (Invitrogen) by addition of 0.9 µl of each: 2 mM dGTP, dTTP and dCTP (New England Biolabs) as well as 4.5 µl of 0.4 mM biotin-14-dATP (Invitrogen), 1.6 µl of ultrapure water and 1.2 µl of Klenow large fragment (New England Biolabs; 5000 U/ml). The reaction mixtures were incubated for 45 min at room temperature and subsequently stopped with 3 µl of 5% SDS (Sigma Aldrich). In the next step, the filled-in DNA ends were ligated by addition to the reaction mixture: 75 µl of 10% Triton X-100, 100 µl of 10× T4 ligation buffer (New England Biolabs), 2.5 µl of bovine serum albumin (New England Biolabs; 10 mg/ml) and 800 µl ultrapure water. The reaction mixtures were incubated for 15 min on ice, subsequently supplemented with 3 µl of T4 DNA ligase (New England Biolabs, 2,000,000 U/ml), and incubated overnight in ice-bath. Next day, the ligation reaction was stopped by the addition of 20 µl of 0.5 M EDTA pH = 8.0 (Invitrogen) and 2.5 µl of proteinase K (New England Biolabs, 20 mg/ml) followed by incubation at 65 °C for 6–8 h. Next, the samples were extracted twice with one volume of phenol/chloroform/isoamyl alcohol (25:24:1) pH = 8.0 (Sigma Aldrich). The water phase containing DNA was next mixed with one volume of isopropanol (Sigma Aldrich) supplemented with 3 µl of GlycoBlue co-precipitant (ThermoFisher Scientific) and incubated overnight at −80 °C. Next, two parallelly processed samples containing precipitated DNA were combined and resuspended in ultrapure water in final volume 60 µl. Next, the DNA was sonicated to reach DNA in average size 200–500 bp, resolved in 1% agarose gel and purified using Qiaquick Gel Extraction Kit (Qiagen). The shared DNA was eluted with 50 µl of ultrapure water. To repair DNA ends after fragmentation, 50 µl of the shared DNA from the previous step was supplemented with 10 µl of 10× T4 DNA ligase buffer (New England Biolabs), 2.5 µl of 10 mM dNTPs (New England Biolabs), 28.75 µl of ultrapure water, 4 µl of T4 DNA polymerase (New England Biolabs, 3000 U/ml), 4 µl of T4 polynucleotide kinase (New England Biolabs, 1000 U/ml) and 0.75 µl of Klenow large fragment (New England Biolabs, 5000 U/ml), and followed by incubation at room temperature for 30 min. After the reaction, DNA was purified using MinElute Reaction CleanUp Kit (Qiagen). From the column, DNA was eluted with 30 µl of ultrapure water. In the next step, 3′-adenine-overhangs were added to the repaired DNA by incubation of 30 µl of the reaction from the step above for 45 min at room temperature with 4 µl of 10× reaction buffer 2 (New England Biolabs), 4 µl of 2 mM dATP (New England Biolabs) and 3 µl of Klenow 3′–5′ exo- (New England Biolabs, 5000 U/ml). Next, DNA was purified using MinElute Reaction CleanUp Kit (Qiagen) and eluted with 15 µl of ultrapure water. The NEBNext adaptors (5 µl) (New England Biolabs) were subsequently ligated according to the manufacture’s protocol. In the next step, the biotin-labelled DNA was purified from non-labelled DNA using DynaBead MyOne Streptavidin C1 magnetic beads (Invitrogen). First, 25 µl of magnetic beads were washed twice with 200 µl of NTB buffer (5 mM Tris-HCl pH 8.0, 0.5 mM EDTA, 1 M NaCl) and twice with 200 µl of ultrapure water. Finally, the magnetic beads were resuspended in 10 µl of ultrapure water and transferred to the NEBNext adaptors ligation mixture (from the earlier step). The pulled-down beads containing biotinylated DNA were washed twice with 200 µl of NBT buffer, twice with 200 µl of ultrapure water and subsequently resuspended with 12 µl of ultrapure water 1.2 µl of beads resuspension was used immediately as a template for Hi-C library preparation as well as NEBNext Index and NEBNext Universal PCR primers (New England Biolabs). The DNA was amplified using Phusion DNA polymerase enzyme (ThermoFisher Scientific) according to the manufacturer’s protocol. The number of PCR amplification cycles was limited to 18. The amplified Hi-C libraries were purified from a 1% agarose gel, and their concentrations were measured using a Qubit dsDNA HS Assay Kit (ThermoFisher Scientific) in OptiPlate-96 White (Perkin Elmer) with 490-nm and 520-nm excitation and emission wavelengths, respectively. The agarose-purified Hi-C libraries were diluted to a 10 nM concentration, pooled together, and sequenced using Illumina NextSeq500 (2 × 75 bp; paired reads), as offered by the Fasteris SA (Plan-les-Ouates, Switzerland) sequencing facility. Sequencing data were collected and pre-processed by Fasteris SA using the software as below: NextSeq Control Software 4.0.1.41, RTA 2.11.3 and bcl2fastq2.17 v2.17.1.14.
Complete data processing was performed using the Galaxy HiCExplorer web server83. The paired reads were first trimmed using Trimmomatic (version 0.36.5) and subsequently filtered using PRINSEQ (version 0.20.4) to remove reads with more than 90% GC content. The pre-processed reads were mapped independently to the S. venezuelae chromosome using Bowtie2 (version 2.3.4.3; with the -sensitive -local presets) (Supplementary Fig. 1A). The Hi-C matrix was generated using hicBuildMatrix (version 2.1.4.0) with a bin size of 10,000 bp. In the next step, three neighbouring bins were merged using hicMergeMatrixBins (version 3.3.1.0) and corrected using the hicCorrectMatrix tool (version 3.3.1.0) based on Imakaev’s iterative correction algorithm84 (with the presets as follows: number of iterations: 500; skip diagonal counts: true (a standard pipeline) to highlight secondary diagonal contacts, or false to highlight primary diagonal contacts (used only for wild-type 22 h; presented in Fig. 1B); remove bins of low coverage: −1.5; remove bins of high coverage: 5.0). The threshold values for the low and high data coverage were adjusted based on the diagnostic plot generated by the hicCorrectMatrix tool (version 3.3.1.0). The corrected Hi-C matrixes were normalised and compared using hicCompareMatrixes (version 3.4.3.0, presets as below: log2ratio), yielding differential Hi-C contact maps. The differential maps were plotted using hicPlotMatrix (version 3.4.3.0; with masked bin removal and seismic pallets of colours). To prepare the Hi-C contact map, the corrected matrixes were normalised in the 0 to 1 range using hicNormalize (version 3.4.3.0; settings: set values below the threshold to zero: value 0) and subsequently plotted using hicPlotMatrix (version 3.4.3.0; with the removal of the masked bins), yielding 30-kbp-resolution Hi-C heatmaps. The threshold values (min/max) were manually adjusted for each Hi-C contact map independently, and the min and max values oscillated in the ranges of 0.05–0.1 and 0.3–0.4, respectively. The corrected matrixes served as the input files for the multiple pairwise matrix comparison using hicCorrelate (version 3.6 + galaxy0). The Hi-C contact matrixes were compared and grouped based on the calculated Pearson correlation coefficients (Supplementary Fig. 1B). To identify the LTD and RTD domain organisation, principal component analysis (PCA1) was performed using hicPCA (version 3.6 + galaxy0). The generated bedgraph files were extracted and smoothed using the moving average, and the PCA1 scores for each 30 kbp bin were plotted against the S. venezuelae chromosome position. The domain boundaries were identified based on the position of the X axis intersection.
Chromatin immunoprecipitation combined with next-generation sequencing (ChIP-seq)
For chromatin immunoprecipitation, S. venezuelae cultures (three independent cultures for each analysed strain) were grown in 50 ml liquid MYM medium supplemented with Trace Element Solution (TES, 0.2× culture volume)80 at 30 °C with shaking (ChIP-seq cultures). Cultures were inoculated with 7 × 108 CFUs. After 14 h of growth, the cultures were cross-linked with 1% formaldehyde for 30 min and blocked with 125 mM glycine. Next, the cultures were washed twice with PBS buffer, and the pellet obtained from half of the culture volume was resuspended in 750 μl of lysis buffer (10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 14 mg/ml lysozyme, protease inhibitor (Pierce)) and incubated at 37 °C for 1 h. Next, 200 μl of zircon beads (0.1 mm, BioSpec products) were added to the samples, which were further disrupted using a FastPrep-24 Classic Instrument (MP Biomedicals; 2 × 45 s cycles at 6 m/s speed with 5-min breaks, during which the lysates were incubated on ice). Next, 750 μl IP buffer (50 mM Tris-HCl pH 8.0, 250 mM NaCl, 0.8% Triton X-100, protease inhibitor (Pierce)) was added, and the samples were sonicated to shear DNA into fragments ranging from 300 to 500 bp (verified by agarose gel electrophoresis). The samples were centrifuged, 25 μl of the supernatant was stored to be used as control samples (input), and the remaining supernatant was used for chromatin immunoprecipitation and mixed with magnetic beads (60 μl) coated with anti-FLAG M2 antibody (Merck). Immunoprecipitation was performed overnight at 4 °C. Next, the magnetic beads were washed twice with IP buffer, once with IP2 buffer (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 0.8% Triton X-100, protease inhibitors (Pierce)), and once with TE buffer (Tris-HCl, pH 7.6, 10 mM EDTA 10 mM). DNA was released by overnight incubation of beads resuspended in IP elution buffer (50 mM Tris-HCl pH 7.6, 10 mM EDTA, 1% SDS) at 65 °C. The IP elution buffer was also added to the ‘input’ samples, which were treated further as immunoprecipitated samples. Next, the samples were centrifuged, and proteinase K (Roche) was added to the supernatants to a final concentration of 100 µg/ml followed by incubation for 90 min at 55 °C. DNA was extracted with phenol and chloroform and subsequently precipitated overnight with ethanol. The precipitated DNA was dissolved in nuclease-free water (10 µl). The concentration of the DNA was quantified using a Qubit dsDNA HS Assay Kit (ThermoFisher Scientific). DNA sequencing was performed by Fasteris SA (Switzerland) using the Illumina ChIP-Seq TruSeq protocol, which included quality control, library preparation and sequencing from both ends (2 × 75 bp) of amplified fragments. Sequencing data for HupS-FLAG ChIP-Seq experiment were collected and pre-processed by Fasteris SA using software: HiSeq Control Software 4.0.1.41, RTA 2.7.7, bcl2fastq2.17v2.17.1.14. Sequencing data for FLAG-SMC ChIP-Seq experiment were collected and pre-processed by Fasteris SA using software: NovaSeq Control Software 1.6.0, RTA 3.4.4, bcl2fastq2.20 v2.20.0.422.
The complete bioinformatic analysis was performed using the R packages edgeR (3.30.3), normr (1.14.0), rGADEM (2.36) and csaw (1.22.1), as well as the MACS2 (2.2.7.1) and MEME (5.1.0) programmes. HupS-FLAG binding was analysed using an earlier published protocol85. The mapping of ChIP-seq data was performed using the Bowtie2 tool (version 2.3.5.186,87). The successfully mapped reads were subsequently sorted using samtools (version 1.10)88. The total number of mapped reads was above 106 on average. Regions differentially bound by HupS-FLAG were identified using the R packages csaw and edgeR89–92, which also normalised the data, as described earlier, as follows85. The mapped reads were counted within a 69-bp-long sliding window with a 23-bp slide. Peaks were filtered using the local method from the edgeR package, which compares the number of reads in the region to the background computed in the surrounding window of 2000 bp. Only regions with log2-fold change above 2.0 were utilised for further analysis. Each window was tested using the QL F-test. Regions that were less than 100 bp apart were merged, and the combined p-value for each was calculated. Only regions with false discovery rate (FDR) values below the 0.05 threshold were considered to be differentially bound. The identified regions were further confirmed by analysis with the MASC2 programme using the broad option93. The R packages rGADEM and MEME Suite were used to search for probable HupS binding sites94. FLAG-SMC bound regions were analysed using the R package normr using Input ChIP-seq data as a control. Reads were counted in 250-bp-long regions, and a region was considered to be bound by FLAG-SMCs if the FDR value compared to the Input value was below the 0.05 threshold and if it was not found to be significant in a control strain not producing FLAG-SMC protein95.
Quantification of the oriC/arm ratio
To estimate the oriC/arm ratio, chromosomal DNA was extracted from S. venezuelae 5 ml cultures (as for Hi-C) growing for 13–26 h or from 50 ml culture (as for ChIP-seq) growing for 8 to 16 h. One millilitre of each culture was centrifuged (1 min at 2400 g and at room temperature). The supernatant was discarded, and the mycelium was used for the subsequent isolation of chromosomal DNA with application a Genomic Mini AX Streptomyces kit (A&A Biotechnology) according to the manufacturer’s protocol. The purified DNA was dissolved in 50 µl of DNase-free water (Invitrogen). The DNA concentration was measured at 260 nm and subsequently diluted to a final concentration of 1 ng/µl.
qPCR was performed using Power Up SYBR Green Master Mix (Applied Biosystems) with 2 ng of chromosomal DNA serving as a template for the reaction and oligonucleotides complemented to the oriC region (oriC) (gyr2_Fd, gyr2_Rv) or to the right arm termini (arm) (arg3_Fd, arg3_Rv) (Supplementary Table 4). The changes in the oriC/arm ratio were calculated using the comparative ΔΔCt method, with the arm region being set as the endogenous control. The oriC/arm ratio was estimated as 1 in the culture (5 ml) growing for 26 h, corresponding to the appearance of spore chains.
Light microscopy
The analyses of spore sizes were performed using brightfield microscopy. Spores from 72-h plate cultures (MYM medium without antibiotics) were transferred to microscopy slides. The samples were observed using a Zeiss Microscope (Axio Imager M1), and images were analysed using Fiji software (ImageJ 2.0.0 or ImageJ 1.53c).
For the visualisation of Z-rings, strains expressing ftsZ-ypet as the second copy of the ftsZ gene were used (the modification did not affect the growth rate, Supplementary Fig. 2A). For the snapshot analysis of S. venezuelae development in liquid, 5 µl from the 5 ml culture (as for Hi-C) growing for 18–26 h or from the 50-ml culture (as for ChIP-Seq) growing for 8–16 h was spread on a coverslip and fixed by washing twice with absolute methanol. For the snapshot analysis of nucleoid areas, strains were cultured on coverslips inserted in MM agar plates supplemented with 1% mannitol80, and after 22–24 h of growth, they were fixed with 2.8% formaldehyde in PBS. The nucleoids were subsequently stained for 5 min at room temperature with 7-amino-actinomycin D (1 mg/ml 7-AAD in DMSO, ThermoFisher Scientific) diluted 1:400 in PBS buffer. In the next step, the mycelium was washed twice with PBS buffer, and the coverslip was mounted using 50% glycerol in PBS buffer. Microscopic observation of FtsZ-YPET and DNA stained with 7-AAD was performed using a Zeiss Microscope (Axio Imager M1). The images were analysed with dedicated software (Axio Vision Rel software). To calculate the nucleoid separation, the distance between the two neighbouring DNA-free areas was measured for 250 nucleoids. The boxplots showing the length of nucleoid-covered areas were constructed using the Plotly tool (plotly.com).
Time-lapse fluorescence microscopy was performed using CellAsic Onix system according to a previously described protocol55 as follows: B04A microfluidic plates (ONIX; CellASIC) were washed with MYM medium, next 100 μl of spores were loaded into microfluidic plate at pressure 4 psi repeatedly for 2–10 s, dependently on spore suspension density. Cultures were started by perfusing the spores with MYM medium at constant flow rate (at pressure 3 psi and temperature 30 °C). For growth rate analysis, the extension of the hyphae emerging from spores was measured within 1 h. For sporulation analyses, after 3 h of preincubation, spent MYM, derived from 40 h flask culture and filter-sterilised, was applied to flow channel. The images were taken every 10 min for ~20 h. The DeltaVision Elite Ultra High-Resolution microscope equipped with a DV Elite CoolSnap camera. Resolve3D softWoRX- Acquire Version:6.1.1 software, and ×100, 1.46 NA, DIC lens was used. Samples were exposed for 50 ms at 5% transmission in the DIC channel and for 80 ms at 50% transmission in the YFP and mCherry channels. The time-lapse movies were analysed using ImageJ Fiji software. For nucleoid visualisation in time-lapse microscopy, strains expressing hupA-mCherry as an additional copy of the hupA gene were used. Nucleoid area was measured using HupA-mCherry as the marker at the time of disassembly of Z-rings (visualised using FtsZ-Ypet fusion), which was ~120 min after their appearance and hyphal growth cessation. Analysis of the HupA-mCherry-labelled chromosome images was based on determining the area and Feret diameter (the maximal distance between two points) of nucleoid fluorescence using the Versatile Wand Tool with 8-connected mode and manually adjusted value tolerance. The distances between Z-rings were measured 60 min after hyphal growth cessation for ~300 Z-rings. The fluorescence intensity of the Z-rings was collected along and averaged across the hyphae with a line selection tool whose width was adjusted to the width of the measured hyphae. The fluorescence intensity of each Z-ring in 25–30 hyphae of each strain was subsequently determined by analysing the data with RStudio using a custom shiny application findpeaks based on the Peaks R package. Tracking of Z-rings in the time-lapse images was performed using the nearest neighbour algorithm. Only Z-rings observed for more than 90 min were used in the analysis. The data from each time point were combined to show how the standard deviation of fluorescence intensity changed during hyphal development. Representative hyphae of each strain were selected, the FtsZ-YPet fluorescence intensity and Z-ring position were determined using the script described above, and its variability was visualised in the form of a kymograph with a custom script based on the ggplot2 package. Statistical analysis was performed using the R programme, for data preparation dplyr 1.0.2 was used. Since the variances in the analysed samples were not homogenous, one-way ANOVA with a two-sided Games–Howell post-hoc test was used to assess the differences between sample means. This test compares the difference between each pair of means with an appropriate adjustment for multiple testing.
Scanning electron microscopy
Streptomyces samples were mounted on an aluminium stub using Tissue TekR OCT (optimal cutting temperature) compound (BDH Laboratory Supplies, Poole, England). The stub was then immediately plunged into liquid nitrogen slush at approximately −210 °C to cryo-preserve the material. The sample was transferred onto the cryo-stage of an ALTO 2500 cryo-transfer system (Gatan, Oxford, UK) attached to an FEI Nova NanoSEM 450 (FEI, Eindhoven, The Netherlands). Sublimation of surface frost was performed at −95 °C for ~3 min before sputter coating the sample with platinum for 150 sat 10 mA, at colder than −110 °C. After sputter-coating, the sample was moved onto the cryo-stage in the main chamber of the microscope, held at −125 °C. The sample was imaged at 3 kV and digital TIFF files were stored.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was funded by the Polish National Science Centre: HARMONIA grant 2016/22/NZ1/00122 (to M.J.S.) and OPUS grant 2018/31/B/NZ1/00614 (to D.J.). We are grateful to Susan Schlimpert for sharing pIJ10770 and pSS172. We thank the Bioimaging facility of the John Innes Centre, supported by a core capability grant from BBSRC, for use of their resources. We thank Jolanta Zakrzewska-Czerwińska for discussions and comments on the manuscript. T.B.K.L. acknowledges the Royal Society University Research Fellowship (UF140053) and BBSRC (BBS/E/J/000PR9791) for funding.
Source data
Author contributions
Conceptualisation and experiment design (M.J.S., D.J.). Methodology including strains construction (T.M., A.Z., A.K.-O., K.P.), epifluorescence microscopy observation and data analysis (T.M., M.J.S., K.P., A.S.); electron microscopy observations (K.F.); introduction to the Hi-C technique (T.B.K.L.); Hi-C libraries preparation and data analysis (M.J.S.); ChIP-Seq libraries preparation and data analysis (T.M., K.P., A.S.); western blots and in vitro DNA binding assays (T.M., K.P., J.D.); growth curves (M.J.S.); ori/arm ratio analyses (M.J.S.). Figures preparation and writing the manuscript (M.J.S., D.J., A.S.); manuscript revision and discussion (T.B.K.L., A.S.).
Data availability
The Hi-C data generated in this study (shown in Figs. 1B–D, 3A and Supplementary Figs. 6A–C, 10C) have been deposited in the ArrayExpress database (EMBL-EBI) under accession code E-MTAB-9810. The raw ChIP-Seq data, as well as the processed data generated in this study (shown in Fig. 3C), have been deposited in the ArrayExpress database (EMBL-EBI) under accession code E-MTAB-9821. The raw ChIP-Seq data, as well as the processed data generated in this study (shown in Fig. 3B), have been deposited in the ArrayExpress database (EMBL-EBI) under accession code E-MTAB-9822. The microscopy data are available in Figshare as follows: data shown in Fig. 2A: [10.6084/m9.figshare.15042900], the microscopy images used for Fig. 2B: [10.6084/m9.figshare.15043329], and for Supplementary Fig. 8B. [10.6084/m9.figshare.15043359], the time-lapse movies used for Fig. 2E, F and Supplementary Fig. 7D: [10.6084/m9.figshare.15042801] and for Supplementary Fig. 7A, inset: [10.6084/m9.figshare.15043338]. The images used for Supplementary Fig. 4B: [10.6084/m9.figshare.15042921]. Source data are provided with this paper.
Code availability
The code used to determine FtsZ foci positions in time-lapse fluorescence profiles: https://github.com/astrzalka/findpeaks.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review informationNature Communications thanks Martial Marbouty and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/16/2021
A Correction to this paper has been published: 10.1038/s41467-021-25897-6
Contributor Information
Marcin J. Szafran, Email: marcin.szafran@uwr.edu.pl
Dagmara Jakimowicz, Email: dagmara.jakimowicz@uwr.edu.pl.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-25461-2.
References
- 1.Badrinarayanan A, Le TBK, Laub MT. Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Biol. 2015;31:171–199. doi: 10.1146/annurev-cellbio-100814-125211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Montero Llopis P, Rudner DZ. Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 2013;14:191–203. doi: 10.1038/nrg3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Rudner DZ. Spatial organization of bacterial chromosomes. Curr. Opin. Microbiol. 2014;22:66–72. doi: 10.1016/j.mib.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, et al. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 2015;29:1661–1675. doi: 10.1101/gad.265876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Brandão HB, Le TBK, Laub MT, Rudner DZ. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science. 2017;355:524–527. doi: 10.1126/science.aai8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le TBK, Imakaev MV, Mirny LA, Laub MT. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böhm K, et al. Chromosome organization by a conserved condensin-ParB system in the actinobacterium Corynebacterium glutamicum. Nat. Commun. 2020;11:1485. doi: 10.1038/s41467-020-15238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trussart M, et al. Defined chromosome structure in the genome-reduced bacterium Mycoplasma pneumoniae. Nat. Commun. 2017;8:14665. doi: 10.1038/ncomms14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Val M-E, et al. A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae. Sci. Adv. 2016;2:e1501914. doi: 10.1126/sciadv.1501914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lioy VS, et al. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell. 2018;172:771–783.e18. doi: 10.1016/j.cell.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 11.Remesh SG, et al. Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling. Nat. Commun. 2020;11:2905. doi: 10.1038/s41467-020-16724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Montero Llopis P, Rudner DZ. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc. Natl Acad. Sci. USA. 2014;111:12877. doi: 10.1073/pnas.1407461111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szafran, M. J., Jakimowicz, D. & Elliot, M. A. Compaction and control - the role of chromosome organizing proteins in Streptomyces. FEMS Microbiol. Rev.10.1093/femsre/fuaa028 (2020). [DOI] [PMC free article] [PubMed]
- 14.Errington J. Septation and chromosome segregation during sporulation in Bacillus subtilis. Curr. Opin. Microbiol. 2001;4:660–666. doi: 10.1016/s1369-5274(01)00266-1. [DOI] [PubMed] [Google Scholar]
- 15.Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dame, R. T., Rashid, F.-Z. M. & Grainger, D. C. Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat. Rev. Genet.10.1038/s41576-019-0185-4 (2019). [DOI] [PubMed]
- 17.Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Li G-W, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobbe N, Heck MMS. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol. Biol. Evol. 2004;21:332–347. doi: 10.1093/molbev/msh023. [DOI] [PubMed] [Google Scholar]
- 20.Gligoris T, Löwe J. Structural insights into ring formation of cohesin and related Smc complexes. Trends Cell Biol. 2016;26:680–693. doi: 10.1016/j.tcb.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nolivos S, Sherratt D. The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol. Rev. 2014;38:380–392. doi: 10.1111/1574-6976.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganji M, et al. Real-time imaging of DNA loop extrusion by condensin. Science. 2018;360:102–105. doi: 10.1126/science.aar7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banigan EJ, Mirny LA. Loop extrusion: theory meets single-molecule experiments. Curr. Opin. Cell Biol. 2020;64:124–138. doi: 10.1016/j.ceb.2020.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Marko JF, De Los Rios P, Barducci A, Gruber S. DNA-segment-capture model for loop extrusion by structural maintenance of chromosome (SMC) protein complexes. Nucleic Acids Res. 2019;47:6956–6972. doi: 10.1093/nar/gkz497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terakawa T, et al. The condensin complex is a mechanochemical motor that translocates along DNA. Science. 2017;358:672–676. doi: 10.1126/science.aan6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mäkelä J, Sherratt D. SMC complexes organize the bacterial chromosome by lengthwise compaction. Curr. Genet. 2020 doi: 10.1007/s00294-020-01076-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawalek A, Wawrzyniak P, Bartosik AA, Jagura-Burdzy G. Rules and exceptions: the role of chromosomal ParB in DNA segregation and other cellular processes. Microorganisms. 2020;8:105. doi: 10.3390/microorganisms8010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonard TA, Butler PJ, Löwe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer-a conserved biological switch. EMBO J. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surovtsev IV, Jacobs-Wagner C. Subcellular organization: a critical feature of bacterial cell replication. Cell. 2018;172:1271–1293. doi: 10.1016/j.cell.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim HC, et al. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. eLife. 2014;3:e02758. doi: 10.7554/eLife.02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surovtsev IV, Campos M, Jacobs-Wagner C. DNA-relay mechanism is sufficient to explain ParA-dependent intracellular transport and patterning of single and multiple cargos. Proc. Natl Acad. Sci. USA. 2016;113:E7268–E7276. doi: 10.1073/pnas.1616118113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flardh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 35.Jones SE, Elliot MA. ‘Exploring’ the regulation of Streptomyces growth and development. Curr. Opin. Microbiol. 2018;42:25–30. doi: 10.1016/j.mib.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Hopwood DA, Glauert AM. Observations on the chromatinic bodies of Streptomycescoelicolor. J. Biophys. Biochem. Cytol. 1960;8:257–265. doi: 10.1083/jcb.8.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruban-Ośmiałowska B, Jakimowicz D, Smulczyk-Krawczyszyn A, Chater KF, Zakrzewska-Czerwińska J. Replisome localization in vegetative and aerial hyphae of Streptomycescoelicolor. J. Bacteriol. 2006;188:7311–7316. doi: 10.1128/JB.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwedock J, McCormick JR, Angert ER, Nodwell JR, Losick R. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomycescoelicolor. Mol. Microbiol. 1997;25:847–858. doi: 10.1111/j.1365-2958.1997.mmi507.x. [DOI] [PubMed] [Google Scholar]
- 39.Donczew M, et al. ParA and ParB coordinate chromosome segregation with cell elongation and division during Streptomyces sporulation. Open Biol. 2016;6:150263. doi: 10.1098/rsob.150263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kois A, Swiatek M, Jakimowicz D, Zakrzewska-Czerwińska J. SMC protein-dependent chromosome condensation during aerial hyphal development in Streptomyces. J. Bacteriol. 2009;191:310–319. doi: 10.1128/JB.00513-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swiercz JP, Nanji T, Gloyd M, Guarné A, Elliot MA. A novel nucleoid-associated protein specific to the actinobacteria. Nucleic Acids Res. 2013;41:4171–4184. doi: 10.1093/nar/gkt095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salerno P, et al. One of the two genes encoding nucleoid-associated HU proteins in Streptomycescoelicolor is developmentally regulated and specifically involved in spore maturation. J. Bacteriol. 2009;191:6489–6500. doi: 10.1128/JB.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Facey PD, et al. Streptomycescoelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol. Microbiol. 2009;73:1186–1202. doi: 10.1111/j.1365-2958.2009.06848.x. [DOI] [PubMed] [Google Scholar]
- 44.Kim HJ, Calcutt MJ, Schmidt FJ, Chater KF. Partitioning of the linear chromosome during sporulation of Streptomycescoelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 2000;182:1313–1320. doi: 10.1128/jb.182.5.1313-1320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang C-C, Tseng S-M, Chen CW. Telomere-associated proteins add deoxynucleotides to terminal proteins during replication of the telomeres of linear chromosomes and plasmids in Streptomyces. Nucleic Acids Res. 2015;43:6373–6383. doi: 10.1093/nar/gkv302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang C-C, et al. The terminal proteins of linear Streptomyces chromosomes and plasmids: a novel class of replication priming proteins. Mol. Microbiol. 2002;43:297–305. [PubMed] [Google Scholar]
- 47.Bao K, Cohen SN. Recruitment of terminal protein to the ends of Streptomyces linear plasmids and chromosomes by a novel telomere-binding protein essential for linear DNA replication. Genes Dev. 2003;17:774–785. doi: 10.1101/gad.1060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang C-C, Tseng S-M, Pan H-Y, Huang C-H, Chen CW. Telomere associated primase Tap repairs truncated telomeres of Streptomyces. Nucleic Acids Res. 2017;45:5838–5849. doi: 10.1093/nar/gkx189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Omura S, et al. Genome sequence of an industrial microorganism Streptomycesavermitilis: deducing the ability of producing secondary metabolites. Proc. Natl Acad. Sci. USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leblond P, et al. The unstable region of Streptomyces ambofaciens includes 210 kb terminal inverted repeats flanking the extremities of the linear chromosomal DNA. Mol. Microbiol. 1996;19:261–271. doi: 10.1046/j.1365-2958.1996.366894.x. [DOI] [PubMed] [Google Scholar]
- 51.Rückert C, et al. Complete genome sequence of the kirromycin producer Streptomyces collinus Tü 365 consisting of a linear chromosome and two linear plasmids. J. Biotechnol. 2013;168:739–740. doi: 10.1016/j.jbiotec.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 52.Gomez-Escribano JP, et al. Streptomyces venezuelae NRRL B-65442: genome sequence of a model strain used to study morphological differentiation in filamentous actinobacteria. J. Ind. Microbiol. Biotechnol. 2021 doi: 10.1093/jimb/kuab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang MC, Losick R. Cytological evidence for association of the ends of the linear chromosome in Streptomycescoelicolor. J. Bacteriol. 2001;183:5180–5186. doi: 10.1128/JB.183.17.5180-5186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kois-Ostrowska A, et al. Unique function of the bacterial chromosome segregation machinery in apically growing Streptomyces - targeting the chromosome to new hyphal tubes and its anchorage at the tips. PLoS Genet. 2016;12:e1006488. doi: 10.1371/journal.pgen.1006488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlimpert, S., Flärdh, K. & Buttner, M. Fluorescence time-lapse imaging of the complete S. venezuelae life cycle using a microfluidic device. J. Vis. Exp.10.3791/53863 (2016). [DOI] [PMC free article] [PubMed]
- 56.Bush MJ, Bibb MJ, Chandra G, Findlay KC, Buttner MJ. Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. mBio. 2013;4:e00684–00613. doi: 10.1128/mBio.00684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jakimowicz D, Gust B, Zakrzewska-Czerwinska J, Chater KF. Developmental-stage-specific assembly of ParB complexes in Streptomycescoelicolor hyphae. J. Bacteriol. 2005;187:3572–3580. doi: 10.1128/JB.187.10.3572-3580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Berkum, N. L. et al. Hi-C: a method to study the three-dimensional architecture of genomes. J. Vis. Exp.10.3791/1869 (2010). [DOI] [PMC free article] [PubMed]
- 59.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jakimowicz D, Zydek P, Kois A, Zakrzewska-Czerwińska J, Chater KF. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol. Microbiol. 2007;65:625–641. doi: 10.1111/j.1365-2958.2007.05815.x. [DOI] [PubMed] [Google Scholar]
- 61.Dedrick RM, Wildschutte H, McCormick JR. Genetic interactions of smc, ftsK, and parB genes in Streptomycescoelicolor and their developmental genome segregation phenotypes. J. Bacteriol. 2009;191:320–332. doi: 10.1128/JB.00858-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bentley SD, et al. Complete genome sequence of the model actinomycete Streptomycescoelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 63.Lioy, V. S. et al. Dynamics of the compartmentalized Streptomyces chromosome during metabolic differentiation. Nat. Commun.10.1038/s41467-021-25462-1 (2021). [DOI] [PMC free article] [PubMed]
- 64.Jakimowicz D, Mouz S, Zakrzewska-Czerwinska J, Chater KF. Developmental control of a parAB promoter leads to formation of sporulation-associated ParB complexes in Streptomycescoelicolor. J. Bacteriol. 2006;188:1710–1720. doi: 10.1128/JB.188.5.1710-1720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soh Y-M, et al. Self-organization of parS centromeres by the ParB CTP hydrolase. Science. 2019;366:1129–1133. doi: 10.1126/science.aay3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jalal AS, Tran NT, Le TB. ParB spreading on DNA requires cytidine triphosphate in vitro. eLife. 2020;9:e53515. doi: 10.7554/eLife.53515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Osorio-Valeriano M, et al. ParB-type DNA segregation proteins are CTP-dependent molecular switches. Cell. 2019;179:1512–1524.e15. doi: 10.1016/j.cell.2019.11.015. [DOI] [PubMed] [Google Scholar]
- 68.Li P, Zhang H, Zhao G-P, Zhao W. Deacetylation enhances ParB-DNA interactions affecting chromosome segregation in Streptomycescoelicolor. Nucleic Acids Res. 2020;48:4902–4914. doi: 10.1093/nar/gkaa245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ginda K, et al. ParA of Mycobacterium smegmatis co-ordinates chromosome segregation with the cell cycle and interacts with the polar growth determinant DivIVA. Mol. Microbiol. 2013;87:998–1012. doi: 10.1111/mmi.12146. [DOI] [PubMed] [Google Scholar]
- 70.Baek JH, Rajagopala SV, Chattoraj DK. Chromosome segregation proteins of Vibrio cholerae as transcription regulators. mBio. 2014;5:e01061–01014. doi: 10.1128/mBio.01061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Breier AM, Grossman AD. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 2007;64:703–718. doi: 10.1111/j.1365-2958.2007.05690.x. [DOI] [PubMed] [Google Scholar]
- 72.Tran NT, Laub MT, Le TBK. SMC progressively aligns chromosomal arms in caulobacter crescentus but is antagonized by convergent transcription. Cell Rep. 2017;20:2057–2071. doi: 10.1016/j.celrep.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu W, Herbert S, Graumann PL, Götz F. Contribution of SMC (structural maintenance of chromosomes) and SpoIIIE to chromosome segregation in Staphylococci. J. Bacteriol. 2010;192:4067. doi: 10.1128/JB.00010-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl Acad. Sci. USA. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rybenkov VV, Herrera V, Petrushenko ZM, Zhao H. MukBEF, a chromosomal organizer. J. Mol. Microbiol. Biotechnol. 2014;24:371–383. doi: 10.1159/000369099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol. Microbiol. 2007;65:1485–1492. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Niki H, Jaffé A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sambrook, J. & Russell, D. Molecular Cloning: A Laboratory Manual 3rd edn (Cold Spring Harbor Laboratory Press, 2001).
- 80.Kieser, T., Bibb, M., Buttner, M., Chater, K. & Hopwood, D. Practica Streptomyces Genetics (John Innes Foundation).
- 81.Gust B, et al. Red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 2004;54:107–128. doi: 10.1016/S0065-2164(04)54004-2. [DOI] [PubMed] [Google Scholar]
- 82.Le TBK. Chromosome conformation capture with deep sequencing to study the roles of the structural maintenance of chromosomes complex in vivo. Methods Mol. Biol. 2019;2004:105–118. doi: 10.1007/978-1-4939-9520-2_9. [DOI] [PubMed] [Google Scholar]
- 83.Wolff J, et al. Galaxy HiCExplorer: a web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 2018;46:W11–W16. doi: 10.1093/nar/gky504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lun ATL, Smyth GK. From reads to regions: a Bioconductor workflow to detect differential binding in ChIP-seq data. F1000Research. 2015;4:1080. doi: 10.12688/f1000research.7016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Langmead B, Wilks C, Antonescu V, Charles R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics. 2019;35:421–432. doi: 10.1093/bioinformatics/bty648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lun ATL, Smyth GK. csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 2016;44:e45. doi: 10.1093/nar/gkv1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lun ATL, Smyth GK. De novo detection of differentially bound regions for ChIP-seq data using peaks and windows: controlling error rates correctly. Nucleic Acids Res. 2014;42:e95. doi: 10.1093/nar/gku351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Helmuth, J. et al. normR: Regime enrichment calling for ChIP-seq data. Preprint at bioRxiv10.1101/082263 (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The Hi-C data generated in this study (shown in Figs. 1B–D, 3A and Supplementary Figs. 6A–C, 10C) have been deposited in the ArrayExpress database (EMBL-EBI) under accession code E-MTAB-9810. The raw ChIP-Seq data, as well as the processed data generated in this study (shown in Fig. 3C), have been deposited in the ArrayExpress database (EMBL-EBI) under accession code E-MTAB-9821. The raw ChIP-Seq data, as well as the processed data generated in this study (shown in Fig. 3B), have been deposited in the ArrayExpress database (EMBL-EBI) under accession code E-MTAB-9822. The microscopy data are available in Figshare as follows: data shown in Fig. 2A: [10.6084/m9.figshare.15042900], the microscopy images used for Fig. 2B: [10.6084/m9.figshare.15043329], and for Supplementary Fig. 8B. [10.6084/m9.figshare.15043359], the time-lapse movies used for Fig. 2E, F and Supplementary Fig. 7D: [10.6084/m9.figshare.15042801] and for Supplementary Fig. 7A, inset: [10.6084/m9.figshare.15043338]. The images used for Supplementary Fig. 4B: [10.6084/m9.figshare.15042921]. Source data are provided with this paper.
The code used to determine FtsZ foci positions in time-lapse fluorescence profiles: https://github.com/astrzalka/findpeaks.