Abstract
Topoisomerase IIα has been a representative anti-cancer target for decades thanks to its functional necessity in highly proliferative cancer cells. As type of topoisomerase IIα targeting drugs, topoisomerase II poisons are frequently in clinical usage. However, topoisomerase II poisons result in crucial consequences resulted from mechanistically induced DNA toxicity. For this reason, it is needed to develop catalytic inhibitors of topoisomerase IIα through the alternative mechanism of enzymatic regulation. As a catalytic inhibitor of topoisomerase IIα, AK-I-191 was previously reported for its enzyme inhibitory activity. In this study, we clarified the mechanism of AK-I-191 and conducted various types of spectroscopic and biological evaluations for deeper understanding of its mechanism of action. Conclusively, AK-I-191 represented potent topoisomerase IIα inhibitory activity through binding to minor groove of DNA double helix and showed synergistic effects with tamoxifen in antiproliferative activity.
Keywords: Topoisomerase IIα, Topoisomerase catalytic inhibitor, DNA minor groove binder, Trifluoromethyl 2-(3-trifluoromethylphenyl)-4-(3-h, Halogenated 2, 4-diphenyl Indeno[1, 2-b]-pyridinol derivative
INTRODUCTION
DNA topoisomerases are essential enzymes that solve topological stress by cleavage and relegation of DNA strands during DNA replication, transcription, chromosomal segregation and condensation processes (Champoux, 2001). In human, topoisomerases I, IIα and IIβ are identified (Nitiss, 1998; Cortés et al., 2003; Capranico et al., 2017). Among them, only topoisomerase IIα expression changes depending on the proliferative phase of cells (Woessner et al., 1991; Isaacs et al., 1998; Delgado et al., 2018). Considering the highly proliferative nature of cancer cell and the essential role of topoisomerase IIα in cell proliferation, topoisomerase IIα has been regarded as one of the most important anticancer therapeutic targets. Topoisomerase IIα inhibitors are generally categorized into two groups, topoisomerase II poisons and catalytic inhibitors (Sehested and Jensen, 1996; Larsen et al., 2003; McClendon and Osheroff, 2007). The topoisomerase II poisons stabilize DNA-topoisomerase II cleavage complexes and inhibit the enzymatic activity (Baviskar et al., 2011). The topoisomerase II catalytic inhibitors attenuate enzymatic activity of topoisomerase without stabilization of DNA-enzyme cleavage complex. This difference in mechanism of action is quite important as the stabilized DNA-enzyme complex induced truncated DNAs which can result in undesired side effects. Most of clinically using medications are belong to the topoisomerase II poisons because of their potent activities. Despite of their potent enzyme inhibitory activities, topoisomerase IIα poisons have negative consequences in use such as secondary malignancy during medication (Winick et al., 1993). Topoisomerase II poisons have a high potential to form undesired double-stranded truncated DNAs, which are attributed to DNA toxicity and secondary malignancies. For this reason, topoisomerase IIα catalytic inhibitors has been considered as an alternative intervention to regulate the activity of topoisomerase IIα in development of anticancer agents (Wilstermann and Osheroff, 2003).
In recent years, there are several reports of catalytic inhibitors (Bau et al., 2014; Jeon et al., 2017; Park et al., 2019; Bergant Loboda et al., 2020). However, the catalytic inhibitors currently on the market are not as effective as the poison inhibitors (Larsen et al., 2003). We therefore aimed to develop catalytic inhibitors that were more effective and less damaging to DNA than poison inhibitors. Based on the previous study with trifluoromethylphenyl derivatives, we, in this study, biologically evaluated AK-I-191, one of the most potent compounds (Kadayat et al., 2019). As a result, AK-I-191 was proved to be a DNA minor groove binder with potent topoisomerase IIα inhibitory activity. AK-I-191 successfully induced apoptosis and showed synergistic effect in combination with tamoxifen in proliferation assay with T47D, luminal A subtype breast cancer cell line.
MATERIALS AND METHODS
Chemicals and reagents
A trifluoromethylphenyl derivative, AK-I-191, was synthesized as previously reported (Kadayat et al., 2019). All compounds were dissolved in dimethyl sulfoxide (DMSO) (Sigma Aldrich, St. Louis, MO, USA) to produce 100 mM stock solutions and stored at –20°C. GangNam-STAIN™ Protein Marker was ordered from INTRON Biotechnology (Seongnam, Korea). BSA™ protein assay kit was obtained from Thermo Fisher Scientific Inc. (Waltham, MA, USA). Recombinant human topoisomerase I and kinetoplast DNA were purchased from TopoGEN (Buena Vista, CO, USA), and human recombinant topoisomerase IIα was from USB Corp. (Cleveland, OH, USA) and Inspiralis (Norwich, UK). The pBR322 plasmid DNA was obtained from Thermo Fisher Scientific Inc. Camptothecin, etoposide, adriamycin, and ellipticin were purchased from Sigma Aldrich.
Cell culture
T47D (human breast cancer cells) was obtained from the Korea Cell Line Bank (Seoul, Korea). Cells were cultured in RPMI 1640 medium (HyClone Lab Inc., Logan, UT, USA) supplemented with 10% fetal bovine serum (Hyclone Laboratories Inc.) and 1% penicillin-streptomycin (Hyclone Laboratories Inc.) in an atmosphere of 5% CO2 and 37°C.
kDNA decatenation assay
The detailed assay protocol was followed the previous report (Park et al., 2019). Briefly, a mixture of 50 ng kinetoplast DNA (kDNA) (TopoGEN) and reaction buffer (10 mM Tris-HCl, pH 7.9, 50 mM KCl, 50 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 15 μg/mL bovine serum albumin, and 1 mM ATP) was prepared. The total concentration of the compound added to the mixture was 20 or 100 μM. Subsequently, 4 units of topoisomerase IIα was added and incubated at 37°C for 30 min. Stop buffer was added to stop topoisomerase IIα enzymatic activity and 2 μL proteinase K was added to the mixture to remove protein. After additional incubation at 55°C for 30 min, the mixture was loaded onto a 1.2% agarose gel and electrophoresed for 7 h at 30 V. The intensity of the DNA bands was measured by UV light using an AlphaImager™ (Alpha Innotech, San Leandro, CA, USA).
Cleavage complex assay
A mixture of pBR322 DNA (125 ng) and the reaction buffer (10 mM Tris-HCl, pH 7.9, 50 mM KCl, 50 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 15 μg/mL bovine serum albumin, and 1 mM ATP) were prepared. The total concentration of the test compound added to the mixture was 250 μM or 500 μM. Then, 3 units of topoisomerase IIα were added and incubated at 37°C for 30 min. After incubation, 2.5 μL stop buffer was added to stop topoisomerase IIα activity. To capture the DNA enzyme complex, 4 μL of proteinase K was added to the mixture, which was then centrifuged and incubated at 45°C for 30 min. The mixture was electrophoresed on a 1% agarose gel containing 0.5 μg/mL ethidium bromide (EtBr) for 6 h at 35 V. The intensity of the DNA bands was measured by UV light using an AlphaImager™ (Alpha Innotech).
Band depletion assay
T47D cells were seeded in 6 well cell culture plate (SPL, Pocheon, Korea) with 70% confluence. Cells were treated with etoposide or AK-I-191 in serum-free medium. Next, the cells were incubated for 2 h and harvested. The harvested cells were washed with PBS, etoposide or AK-I-191 was added again at the same concentration as above, and then incubated on ice for 15 min. The cells were centrifuged at 3,200 rpm for 3 min to separate the cells, and the separated cells were lysed with denaturing buffer (2% SDS, 62.5 mM Tris-HCl (pH 6.8), and 1 mM EDTA). After sonication at 20% power for 3 s, followed by centrifugation at 12,000 rpm for 20 min, the supernatant was subjected to western blotting on an 8% SDS-PAGE gel.
Western blot
Cells were lysed using RIPA buffer (Cell Signaling Technology, Danvers, MA, USA) containing 1 mM PMSF and 1% protease inhibitor cocktail. Lysate from each sample was loaded with a volume containing 35 μg of protein per well. Samples were run on 10-15% polyacrylamide gels and transferred to polyvinylidene fluoride membranes (Pall Life Science, Port Washington, NY, USA). Proteins were detected using primary antibodies against topoisomerase IIα (Cat# ab74715, Abcam, Cambridge, UK), α-tubulin (Cat# 2144, Cell Signaling Technology), cleaved PARP (Cat#9541, Cell Signaling Technology), Bcl-2 (Cat# 2876, Cell Signaling Technology), Bax (Cat# 2772, Cell Signaling Technology), and β-Actin (Cat# 4967, Cell Signaling Technology), followed by incubation with HRP–conjugated anti-mouse (Cat# GTX213111-01, GeneTex, Irvine, CA, USA; Cat# 7076, Cell Signaling Technology) or anti-rabbit antibodies (Cat# GTX213110-01, GeneTex; Cat# 7074, Cell Signaling Technology) and enhanced chemiluminescence (GE Healthcare Life Sciences, Little Chalfont, UK). The chemiluminescence was detected using an LAS-3000 (Fuji Photo Film Co., Tokyo, Japan), and blot images were analyzed using Multi-Gauge (Fuji Photo Film Co.).
Alkaline comet assay
The details of experiment have been previously reported (Shrestha et al., 2018). To briefly describe the current evaluation, T47D cells were seeded in 6 well cell culture plate (SPL) with 70% confluence. Cells were treated with 5 or 10 μM of AK-I-191 in serum free medium and incubated for 20 h at 37°C and humified condition. After harvested, cells were mixed with LM agarose and transferred to the CometSlide™ slides (Trevigen, Gaithersburg, MD, USA). The slides were incubated in the dark and placed in lysis solution. After incubation in the unwinding solution, the cells were electrophoresed. The slides were washed with distilled water and 70% alcohol. The slides were incubated at 42°C until dry, stained with SYBR Gold staining solution. Then, it was mounted using the Dako fluorescent mounting medium (Agilent, Santa Clara, CA, USA). The images were captured by using an Axio Observer Inverted Microscope (Carl Zeiss Co. Ltd., Jena, Germany). A minimum of 30 spots of T47D cells were randomly selected for imaging. The tail length of the cells represented the DNA damage and was calculated by using Komet 5.5 software (Kinetic Imaging Ltd, Liverpool, UK).
Titration of DNA with AK-I-191 measured by UV-visible spectroscopy
UV-visible spectra of calf-thymus DNA (ctDNA) (Sigma Aldrich) were recorded using Nano Drop ND-2000 (Thermo Fisher Scientific Inc.) in 10 mM Tris-HCl (pH 7.2) with or without adding AK-I-191 at various molar ratios of ctDNA to AK-I-191 as designated in figure legend. Spectra of ctDNA and ctDNA-AK-I-191 complex were measured in the wavelength range of 220-300 nm.
Hoechst displacement assay
Hoechst 33258 (Hoechst), well-known as a DNA minor groove binder, was used to decipher the binding mode of compound on interaction with DNA. ctDNA (30 μM) and Hoechst (30 μM) were dissolved in 10 mM Tris-HCl (pH 7.2). Later, increasing concentrations of AK-I-191 were added to Hoechst-DNA solution. Solution was excited at 360 nm and emission spectra were recorded in the range 400-600 nm. Quenching constant (Ksv) values of AK-I-191 in reduction of Hoechst fluorescence was analyzed through the Stern-Volmer equation. The fluorescence was measured using Microplate Multi-Reader Infinite M200 PRO (Tecan, Männnedorf, Switzerland).
KI quenching assay
KI quenching assay was conducted in the presence and absence of ctDNA (60 μM). Briefly, AK-I-191 (20 μM) was dissolved in 10 mM Tris-HCl (pH 7.2) and titrated with increasing concentrations of KI. AK-I-191 was excited at 360 nm and fluorescence emitted at 420 nm was recorded. Fluorescence of AK-I-191 quenched by increasing concentrations of KI was also assessed without ctDNA. Quenching constant (Ksv) values in the presence and absence of ctDNA was analyzed via the Stern-Volmer equation. The fluorescence was measured using Microplate Multi-Reader Infinite M200 PRO (Tecan).
Field analysis & molecular docking study
The three-dimensional docking study of DNA with AK-I-191 was performed with structures retrieved from PDB code, 2B0K. The Flare tool provided by Cresset Software (Cresset, Cambridgeshire, UK) was used for protein preparation, small molecule preparation, and the docking analysis. The docking experiment between AK-I-191 and DNA structure was conducted using active sites defined by the ligand in the PDB structure. Molecular docking simulations were performed using an algorithm provided by the Flare software package from Cresset.
Cell cycle analysis
T47D cells were seeded in 60 mm cell culture dish. After 70% confluence, the cells were treated with AK-I-191 either in a time-dependent or in a concentration-dependent manner in serum-free medium (RPMI 1640). The harvested cells were washed twice with PBS, and the cells were centrifuged at 3,200 rpm for 3 min. For fixation, 70% ethanol was added to the cell pellet and incubated overnight at –20°C. The fixed cells were centrifuged at 3,200 rpm for 3 min, resuspended with 500 μL of staining solution containing propidium iodide 50 mg/mL, RNase 0.5 mg/mL and Triton X-100 0.25 μL in PBS and incubated at 37°C for 25 min. The stained cells were transferred to a 5 mL test tube (SPL). The fluorescence was measured by using a Fluorescence Activated Cell Sorting (FACS)-Caliber flow cytometer (BD Biosciences, San Jose, CA, USA). A minimum of 10,000 cells were measured for each sample.
Annexin V-PI double staining
The experiments were conducted according to the manufacturer’s protocol. Briefly, T47D cells were seeded in 60 mm cell culture dish. After the cells reached 70% confluence, AK-I-191 was added at the final concentration of 5 or 10 μM and the cells were incubated for 36 h. Then, cells were washed twice with PBS, harvested by using trypsin, and subjected to centrifugation. The cell pellet was resuspended in 100 μL binding buffer. Then, annexin V-FITC and PI were added to the cell suspension and incubated for 20 min at room temperature. The stained sample was examined by using a FACS-Calibur flow cytometer (BD Biosciences). The analysis was performed by using the BD Cell Quest Pro software (BD Biosciences).
Human data analysis from public data sets
Kaplan–Meier survival graphs were generated from data available from KM Plotter (http://www.kmplotter.com) (Györffy et al., 2010). To assess the gene expression of TOP2A, data was analyzed using two probes of microarray, 201291_s_at and 201292_at.
Combination analysis
Prior to combined effects on anti-proliferative activity, cellular growth was assessed using WST reagent (EZ-Cytox, DoGenBio, Seoul, Korea). After 4 h starvation, compounds at the indicated concentrations were treated and then the cellular viability was measured using WST reagent and Microplate Multi-Reader Infinite M200 PRO (Tecan).
Combination index (CI) values were determined for drug-drug concentrations and effect levels (Fa, fraction affected; inhibition of cellular growth). CI values were calculated according to method of isobologram-combination index (Chou et al.) with CompuSyn software (ComboSyn Inc., Paramus, NJ, USA).
Statistical analysis
Data are presented as the mean ± SEM. Multiple comparisons were analyzed by one-way ANOVA followed by Tukey’s post hoc test. Differences were regarded significant when *p<0.05, **p<0.01, and ***p<0.001. Statistical analysis was carried out with GraphPad Prism version 6 (GraphPad Software Inc., La Jolla, CA, USA).
RESULTS
AK-I-191 is a topoisomerase IIα catalytic inhibitor
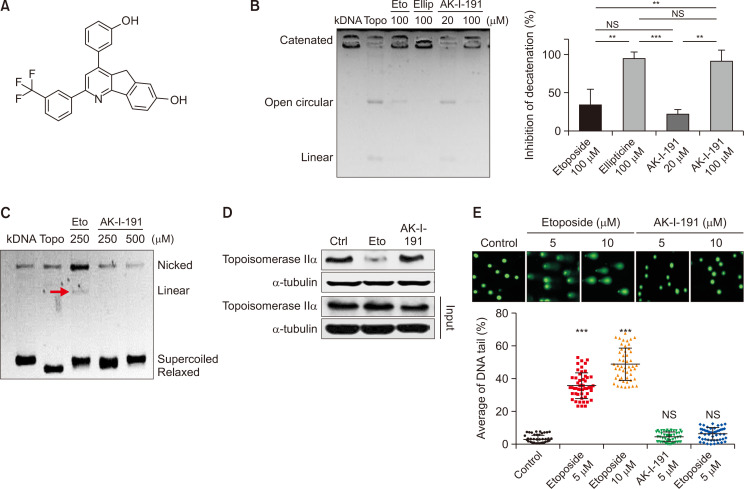
AK-I-191 was reported for its topoisomerase inhibition (Fig. 1A) (Kadayat et al., 2019). The enzyme inhibition of AK-I-191 was highly selective on topoisomerase IIα against topoisomerase I. Prior to the further investigation for clarification of mechanism of action, we confirmed its topoisomerase IIα inhibitory activity using kDNA decatenation assay (Fig. 1B). AK-I-191 successfully inhibited decatenating activity of topoisomerase IIα. To uncover the mechanism of the potent and selective topoisomerase IIα inhibition, cleavage complex assay was conducted (Fig. 1C). After enzymatic reaction with high units of topoisomerase IIα, DNA-enzyme complex was captured, enzyme digested, and loaded on to agarose gel. As a result, not only bands of supercoiled and relaxed DNA but also that of linear DNA were shown in topoisomerase II poison inhibitor-treated sample. Through the experiment, AK-I-191 was found to be a catalytic inhibitor of topoisomerase IIα rather than topoisomerase poison. The function as a topoisomerase IIα catalytic inhibitor was confirmed using the band depletion assay (Fig. 1D). Topoisomerase IIα enzyme covalently bound to DNA is precipitated during western blot sample preparation process in etoposide-treated cells. Unlike etoposide, AK-I-191 did not created topoisomerase IIα-DNA cleavage complex and represented band of the similar intensity to that of control cells. The secondary malignancy, a major side effect of topoisomerase poisons, is possibly due to the truncated DNA which is generated by formation of stable enzyme-DNA cleavage complex, leading to blocking re-ligation (Baviskar et al., 2011). For this reason, the formation of truncated DNA is a critical factor to evaluate the topoisomerase catalytic inhibitor and AK-I-191 was subjected to the alkaline comet assay (Fig. 1E). Alkaline comet assay shows truncation of DNA as DNA tails looks like comet. AK-I-191 did not show comet tail products, whereas etoposide produced high proportion of comet tails, which suggests that AK-I-191 inhibited topoisomerase IIα activity without generation of truncated DNA.
Fig. 1.
Mechanistic evaluation of AK-I-191 as a catalytic inhibitor of topoisomerase IIα. (A) Structure of AK-I-191. (B) kDNA decatenation assay of AK-I-191. Lane kDNA: kDNA only, Lane Topo: kDNA+topoisomerase IIα, Lane Eto: kDNA+topoisomerase IIα+etoposide, Lane Ellip: kDNA+topoisomerase IIα+ellipticine, Lane AK-I-191: kDNA+topoisomerase IIα+AK-I-191 (20 or 100 μM). The values measured in the gel image are depicted as a graph (right). (C) Cleavage complex assay. Supercoiled DNA (pBR322) was pre-incubated with DNA topoisomerase and each compound was added and additionally incubated. After the agarose gel electrophoresis, band of linear form of DNA was analyzed (red arrow). (D) Band depletion assay. Formation of topoisomerase IIα-DNA cleavage complex was assessed using band depletion assay. Cells treated with vehicle, Etoposide, or AK-I-191 were lysed with denaturing buffer and centrifugated. The supernatant was subjected to assessment of DNA-unbound topoisomerase IIα (upper panel). The whole cell lysate in the same experimental condition was utilized as input. (E) Alkaline comet assay. Comet slides were electrophoresed at high pH and stained with a SYBR gold solution. The comet tails were observed by fluorescence microscopy. Quantification of DNA tails was conducted using Komet 5.5 software. Intensities of comet tails versus comet heads reflected the extent of DNA breaks (n=50). ***p<0.001 and **p<0.01 are significantly different from control.
AK-I-191 is a minor groove binding catalytic inhibitor
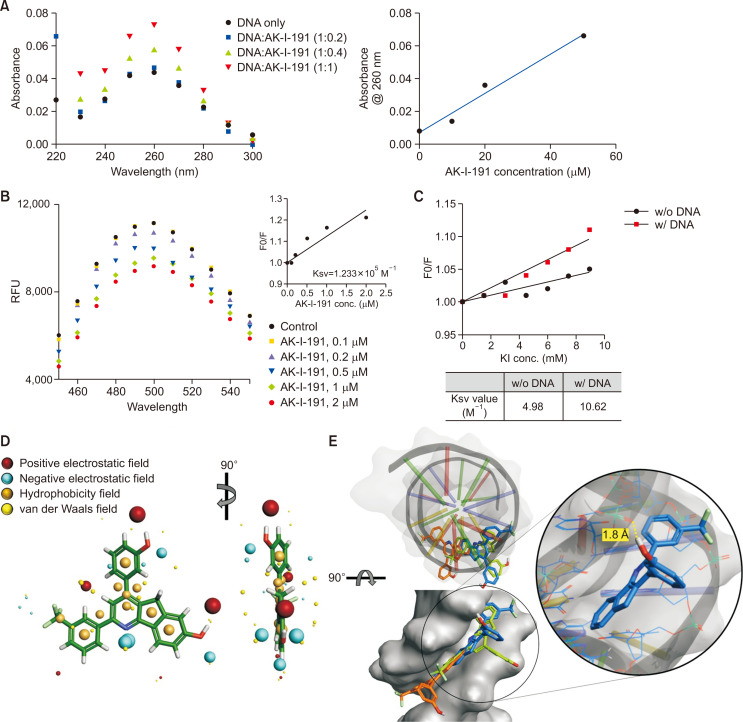
As a topoisomerase IIα catalytic inhibitor, mechanistic evaluation of AK-I-191 was conducted. First, we assessed its ability to interact with DNA (Fig. 2A). Absorbance of DNA with or without AK-I-191 was evaluated. As the concentration of the compound increased, the absorbance of DNA increased. The hyperchromicity in DNA absorbance induced by compound has been reported previously in the studies of DNA groove binding compounds (Rehman et al., 2014). To confirm the exact DNA binding mode, AK-I-191 was subjected to the Hoechst and EtBr displacement assays (Fig. 2B, Supplementary Fig. 1). As a result, AK-I-191 was found to be a groove binder by replacing the Hoechst from minor groove of DNA. In addition, KI quenching assay was conducted for further evaluation of DNA interacting mode of AK-I-191. By the iodide ion in KI solution, fluorescence signal of chemicals can be quenched. If the compound is DNA intercalator, the DNA molecule with negatively charged surface because of its phosphate groups can protect the compound from the access of iodide ion. For this reason, in KI quenching assay, DNA addition reduced the activity of quencher, iodide ion, on the intercalative compound. In contrast, fluorescence signal of AK-I-191 was quenched by iodide ion in both DNA present and absent conditions, which proves that AK-I-191 is a groove binder rather than an intercalator (Fig. 2C). The binding mode of AK-I-191 to DNA was further assessed by field analysis and molecular docking study. As shown in Fig. 2D, AK-I-191 possesses hydrophobic field-rich flat structure of phenyl indeno[1,2-b]pyridine ring. The docking system with the PDB structure (PDB code: 2B0K) was evaluated prior to docking analysis through calculating the RMSD of redocked original ligand (1.272 Å). Based on the docking results that all three poses of AK-I-191 possessing the lowest energy are located in the minor groove of DNA and the phenyl indeno[1,2-b]pyridine structure fits well into the minor groove of DNA with hydrophobic interaction (Fig. 2E), AK-I-191 was further validated as a minor groove binding catalytic inhibitor. The hydroxyl group on the phenyl ring of AK-I-191 was involved in hydrogen bonding with DNA (Fig. 2E), which could enhance the probability of AK-I-191 to function as a minor groove binder.
Fig. 2.
DNA minor groove binding of AK-I-191. (A) UV-visible absorption spectra of ctDNA (50 μM) in the presence of increasing concentrations of AK-I-191. Hyperchromism in UV/Vis absorbance of DNA was observed with increasing concentrations of AK-I-191, which confirmed the interaction between AK-I-191 and DNA. (B) Titration of AK-I-191 to the complex of ctDNA with Hoechst (minor groove binder) with monitoring fluorescence. The Hoechst-DNA complex was excited at 340 nm and emission spectra were recorded from 450 nm to 550 nm. Fluorescence intensity decreased with subsequent addition of AK-I-191. (C) KI quenching experiment. Stern-Volmer plot for fluorescence quenching of AK-I-191 (20 μM) by KI in the absence and presence of ctDNA (60 μM). The degree of quenching of AK-I-191 fluorescence intensity was measured by increasing concentrations of KI in the absence and presence of ctDNA and quenching constant was calculated in both the case. (D) Field analysis of AK-I-191. Positive electrostatic (red), negative electrostatic (cyan), hydrophobicity (orange), and van der Waals (yellow) fields were presented with the structure of AK-I-191. (E) Docking analysis were carried out with DNA (PDB ID: 2B0K) duplex of sequence d(CGCGAATTCGCG)2 using Flare program. The structure of DNA is shown as a ribbon diagram and grey surface. Three docking poses AK-I-191 with the lowest energy were shown as sticks and colored by blue, green, and orange. The lowest energy docking pose painted with blue is shown as a magnified view. Possible hydrogen bonding was highlighted with a yellow dashed line.
AK-I-191 induced apoptosis
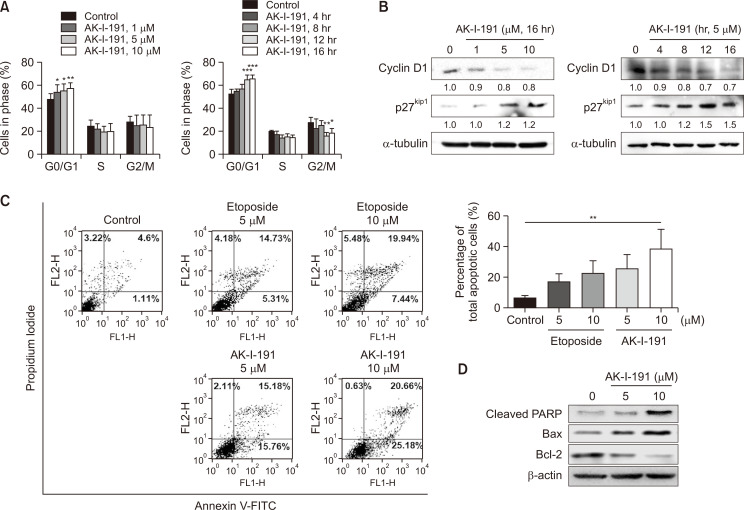
The anticancer effect of AK-I-191 was assessed previously by measuring anti-proliferative activity using dehydrogenase enzyme (Kadayat et al., 2019). To confirm the previous study and to evaluate the activity in depth, alteration of cellular population in each cell cycle was analyzed (Fig. 3A). As a result, cells treated with AK-I-191 were arrested in G1 phase in time- and concentration-dependent manner. The cell cycle arrest was also confirmed with the level of G1 checkpoint markers, cyclin D1 and p27kip1 (Fig. 3B). The decrease in cyclin D1 and the increase in p27kip1 confirmed that AK-I-191 induced G1 arrest (Baldin et al., 1993; Ray et al., 2009). The induction of apoptotic process was also assessed through annexin V and PI double staining (Fig. 3C). Flow cytometric analysis of double stained cells showed that AK-I-191 induced apoptosis to a greater extent than etoposide. The population of early apoptotic cells (annexin V-positive and PI-negative cells as indicated in the lower right box in Fig. 3C) after treatment of AK-I-191 increased in dose-dependent manner, and the extent of increase was higher that of etoposide. The population of late apoptotic cells or post-apoptotic necrosis-induced cells (annexin V-positive and PI-positive cells as shown in upper right box in Fig. 3C) increased dose-dependently by treatment of both etoposide and AK-I-191. The degree of increases by etoposide and AK-I-191 was similar to each other. In addition, apoptotic induction was confirmed by western blot analysis (Fig. 3D). The results showed that bcl-2, an anti-apoptotic protein, was decreased by treatment with AK-I-191 in a concentration-dependent manner, whereas the pro-apoptotic proteins, cleaved PARP (c-PAPP) and bax, were increased. Overall, AK-I-191 induced apoptosis in a concentration-dependent manner.
Fig. 3.
Apoptotic induction of AK-I-191. (A) Cell cycle analysis of AK-I-191-treated cells. G1 arrest was observed in AK-I-191 concentration- and time-dependent manner. ***p<0.001, **p<0.01 and *p<0.05 are significantly different from control without compound. (B) Western blot analysis of G1 checkpoint markers, cyclin D1 and p27kip1 in T47D cells after treatment of AK-I-191. (C) Apoptosis assessment using Annexin V-PI double staining. T47D cells were treated with varying concentrations of AK-I-191 for 36 h. Apoptotic cells were detected by flow cytometry and plotted as Annexin V-positive cells on the right side. (D) Assessment of Apoptotic induction with changes in protein levels. Protein levels of pro- and anti-apoptotic proteins, bcl-2, bax, and cleaved PARP, were analyzed using western blotting.
Topoisomerase IIα catalytic inhibition with AK-I-191 is a promising therapeutic strategy in luminal A subtype of breast cancer
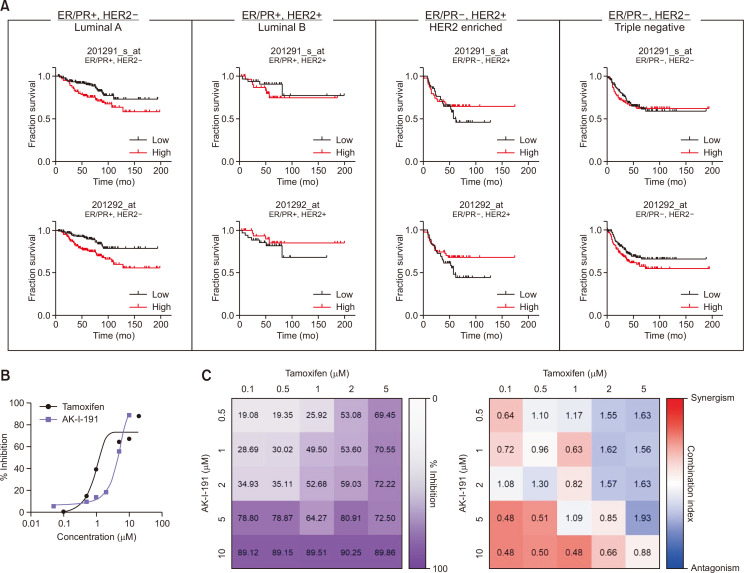
AK-I-191 had been reported to show the most potent anti-proliferative activity in T47D cells compared to other types of cancer cell lines. T47D cell line belongs to the luminal A subtype with ER/PR positive and HER2 negative phenotype. Based on the previous study, we assessed the correlation between topoisomerase IIα expression and prognosis in each subtypes of breast cancer from dataset of the Cancer Genome Atlas (TCGA). As a result, the expression level of TOP2A gene encoding topoisomerase IIα is closely related with clinical outcome of luminal A breast cancer patients (Fig. 4A, Supplementary Table 1). Unlike luminal A subtype, there was no significant correlations in luminal B, HER2 enriched, or triple negative breast cancer. Next, to determine whether AK-I-191 had a synergistic effect when combined with tamoxifen, a clinically used drug for luminal A breast cancer (Yu et al., 2019), cell viability was measured (Fig. 4B, 4C). The combination of tamoxifen and AK-I-191 resulted in additive and synergistic effects at lower concentrations of tamoxifen in T47D cells.
Fig. 4.
Antiproliferative activity of AK-I-191. (A) Correlation between TOP2A expression and survival of subtypes of breast cancer. Red and black lines indicate high and low expression of TOP2A gene, respectively. (B) Growth inhibitory activities of tamoxifen and AK-I-191 in T47D cell line. (C) Viability assays of T47D cells treated with either tamoxifen, AK-I-191, or combinations of both (n=3 experiments performed in triplicate, values indicate mean). Right panels: heatmap of combination indexes generated using CompuSyn Software. Heatmap was colored according to log(combination index), red is synergism (–1) and blue is antagonism (+1).
DISCUSSION
In this study, we confirmed the topoisomerase IIα inhibitory activity of AK-I-191 and uncovered its mechanism of action using systematic evaluation. Through DNA minor groove binding, AK-I-191 exerted anticancer effects in T47D luminal A subtype breast cancer cell line. AK-I-191 induced apoptosis and G0/G1 cell cycle arrest. In addition, AK-I-191 showed the synergistic effect in combination with tamoxifen, a drug clinically used to treat breast cancer patients with luminal A subtype.
Topoisomerase IIα has been regarded as an important target in cancer treatment due to its essential characteristics during cell division (Nitiss, 2009). As cancer cells are known to be highly proliferative, targeting topoisomerase IIα is a very effective way to attenuate growth of cancer. Topoisomerase II inhibitors in currently clinical use effectively inhibit the enzymatic activity and attenuate tumor progression, but have limitations. Secondary malignancy is one of the dire side effects of topoisomerase II poisons (Ezoe, 2012). The topoisomerase II inhibitors under clinical use are mostly topoisomerase II poisons which stabilize transiently formed topoisomerase II-DNA cleavage complex, block DNA re-ligation, and release DNA with double strand breaks (Pommier et al., 2010; Vollmer et al., 2010). It is certainly necessary to inhibit topoisomerase IIα without producing undesired double-strand DNA breaks to avoid significant side effects arising from the mechanism of drugs (Catanzaro et al., 2020). Catalytic inhibitor of topoisomerase IIα prevents the enzyme before DNA cleavage or in the last step of the catalytic cycle after re-ligation. (Baviskar et al., 2011) Most of catalytic inhibitors developed have tendency to show low potency. This is one of the reasons that the catalytic inhibitors of topoisomerase IIα are yet to be the first line therapeutics in cancer therapy. For this reason, we focused on discovery of topoisomerase IIα catalytic inhibitor with high potency. AK-I-191 exerted strong activity in topoisomerase IIα inhibition, anti-proliferation, and apoptotic induction. Its mode of action was systematically characterized. AK-I-191 inhibited topoisomerase IIα enzymatic activity through DNA minor groove binding with much stronger activity than etoposide, a topoisomerase IIα poison in clinical use.
AK-I-191 was identified as a DNA minor groove binder. Along with the experimental analysis of Hoechst assay, the mechanism of action was simulated in silico docking study. In molecular docking study, AK-I-191 fit in the minor groove of DNA. All the three poses with the lowest energy show that 2-(3-trifluoromethylphenyl)-5H-indeno[1,2-b]pyridin-7-ol structure was placed in the minor groove through hydrophobic interaction with the aromatic rings in purine or pyrimidine structure of DNA. Hydroxyl group of phenyl ring which positioned in the outside of DNA exhibits the possibility of hydrogen bonding with phosphate group of DNA.
Among several mechanisms of topoisomerase catalytic inhibitors, DNA interaction is one of the frequently considered mechanisms. As topoisomerases must interact with DNA double helix to perform their proper activity, it is reasonable to target DNA for inhibition of the enzymatic activity. There are two general modes of DNA binding that characterize the interaction between small molecular compounds and DNA for topoisomerase inhibition, DNA intercalation and major groove binding (Pindur et al., 2005; Gurova, 2009). Most of small molecular groove binders are usually crescent-shaped molecules with large molecular mass and therefore have low druglikeness (Palchaudhuri and Hergenrother, 2007). AK-I-191 with a molecular weight of 405.41 Da was estimated to have one candidate hydrogen donor, five hydrogen bond acceptors, and an log P (octanol-water partition coefficient) of 5.00. All the calculated molecular properties of AK-I-191 indicate good druglikeness based on the Lipinski’s rule of five (Lipinski et al., 2001).
Breast cancer is a type of cancer that is highly heterogeneous. There are four subtypes in clinically using classification; luminal A, luminal B, HER2-enriched, and triple negative breast cancers. One of the subtypes, luminal A is the most abundant subtype of breast cancer (Pandit et al., 2020). Among the four subtypes of breast cancer, only patients of luminal A subtype with high expression of TOP2A showed clear correlation with clinical outcome (Fig. 4A). The correlation between TOP2A expression and prognosis in luminal or ER-positive breast cancer had been reported (An et al., 2018). However, luminal B, ER-positive and HER2-positive, breast cancer did not show significant correlation in results of database analysis. Based on the previous reports and results of database analysis, targeting topoisomerase IIα as cancer treatment is possibly more effective in luminal A subtype of breast cancer than any other subtypes. Based on strong antiproliferative activity of AK-I-191 against T47D cells and the correlation of higher TOP2A expression with worse prognosis analyzed using breast cancer patient dataset, we confirmed the anticancer activity of AK-I-191 using the cell line of luminal A subtype. The anticancer activity of AK-I-191 was confirmed not only in the cellular topoisomerase activity assessment, but also in the apoptotic induction and cell cycle arrest. Moreover, combination effect of AK-I-191 with tamoxifen, a selective estrogen receptor modulator in clinical use, was assessed (Yu et al., 2019). With low concentration of tamoxifen, AK-I-191 exhibited additive and synergistic effects in cellular growth of T47D cells.
In conclusion, topoisomerase IIα is a highly beneficial therapeutic target in luminal A subtype of breast cancer and AK-I-191, a minor groove binding catalytic inhibitor of topoisomerase IIα, can be a new candidate of anticancer agent in combination with clinically used drugs, such as tamoxifen.
SUPPLEMENTARY MATERIALS
ACKNOWLEDGMENTS
This work was supported by grants from National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIT) (2018R1A5A2025286, 2017R1A2B2003944, and 2019R1I1A1A01050921).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interests.
REFERENCES
- An X., Xu F., Luo R., Zheng Q., Lu J., Yang Y., Qin T., Yuan Z., Shi Y., Jiang W., Wang S. The prognostic significance of topoisomerase II alpha protein in early stage luminal breast cancer. BMC Cancer. 2018;18:331. doi: 10.1186/s12885-018-4170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldin V., Lukas J., Marcote M. J., Pagano M., Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- Bau J. T., Kang Z., Austin C. A., Kurz E. U. Salicylate, a catalytic inhibitor of topoisomerase II, inhibits DNA cleavage and is selective for the α isoform. Mol. Pharmacol. 2014;85:198–207. doi: 10.1124/mol.113.088963. [DOI] [PubMed] [Google Scholar]
- Baviskar A. T., Madaan C., Preet R., Mohapatra P., Jain V., Agarwal A., Guchhait S. K., Kundu C. N., Banerjee U. C., Bharatam P. V. N-fused imidazoles as novel anticancer agents that inhibit catalytic activity of topoisomerase IIα and induce apoptosis in G1/S phase. J. Med. Chem. 2011;54:5013–5030. doi: 10.1021/jm200235u. [DOI] [PubMed] [Google Scholar]
- Bergant Loboda K., Janežič M., Štampar M., Žegura B., Filipič M., Perdih A. Substituted 4,5'-bithiazoles as catalytic inhibitors of human DNA topoisomerase IIα. J. Chem. Inf. Model. 2020;60:3662–3678. doi: 10.1021/acs.jcim.0c00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capranico G., Marinello J., Chillemi G. Type I DNA topoisomerases. J. Med. Chem. 2017;60:2169–2192. doi: 10.1021/acs.jmedchem.6b00966. [DOI] [PubMed] [Google Scholar]
- Catanzaro E., Betari N., Arencibia J. M., Montanari S., Sissi C., De Simone A., Vassura I., Santini A., Andrisano V., Tumiatti V., De Vivo M., Krysko D. V., Rocchi M. B. L., Fimognari C., Milelli A. Targeting topoisomerase II with trypthantrin derivatives: discovery of 7-((2-(dimethylamino)ethyl)amino)indolo[2,1-b]quinazoline-6,12-dione as an antiproliferative agent and to treat cancer. Eur. J. Med. Chem. 2020;202:112504. doi: 10.1016/j.ejmech.2020.112504. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Cortés F., Pastor N., Mateos S., Domínguez I. Roles of DNA topoisomerases in chromosome segregation and mitosis. Mutat. Res. 2003;543:59–66. doi: 10.1016/S1383-5742(02)00070-4. [DOI] [PubMed] [Google Scholar]
- Delgado J. L., Hsieh C. M., Chan N. L., Hiasa H. Topoisomerases as anticancer targets. Biochem. J. 2018;475:373–398. doi: 10.1042/BCJ20160583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezoe S. Secondary leukemia associated with the anti-cancer agent, etoposide, a topoisomerase II inhibitor. Int. J. Environ. Res. Public Health. 2012;9:2444–2453. doi: 10.3390/ijerph9072444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurova K. New hopes from old drugs: revisiting DNA-binding small molecules as anticancer agents. Future Oncol. 2009;5:1685–1704. doi: 10.2217/fon.09.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy B., Lanczky A., Eklund A. C., Denkert C., Budczies J., Li Q., Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- Isaacs R. J., Davies S. L., Sandri M. I., Redwood C., Wells N. J., Hickson I. D. Physiological regulation of eukaryotic topoisomerase II. Biochim. Biophys. Acta. 1998;1400:121–137. doi: 10.1016/S0167-4781(98)00131-6. [DOI] [PubMed] [Google Scholar]
- Jeon K. H., Park C., Kadayat T. M., Shrestha A., Lee E. S., Kwon Y. A novel indeno[1,2-b]pyridinone derivative, a DNA intercalative human topoisomerase IIα catalytic inhibitor, for caspase 3-independent anticancer activity. Chem. Commun. 2017;53:6864–6867. doi: 10.1039/C7CC02372C. [DOI] [PubMed] [Google Scholar]
- Kadayat T. M., Park S., Shrestha A., Jo H., Hwang S. Y., Katila P., Shrestha R., Nepal M. R., Noh K., Kim S. K., Koh W. S., Kim K. S., Jeon Y. H., Jeong T. C., Kwon Y., Lee E. S. Discovery and biological evaluations of halogenated 2,4-diphenyl Indeno[1,2-b]pyridinol derivatives as potent topoisomerase IIα-targeted chemotherapeutic agents for breast cancer. J. Med. Chem. 2019;62:8194–8234. doi: 10.1021/acs.jmedchem.9b00970. [DOI] [PubMed] [Google Scholar]
- Larsen A. K., Escargueil A. E., Skladanowski A. Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol. Ther. 2003;99:167–181. doi: 10.1016/S0163-7258(03)00058-5. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- McClendon A. K., Osheroff N. DNA topoisomerase II, genotoxicity, and cancer. Mutat. Res. 2007;623:83–97. doi: 10.1016/j.mrfmmm.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss J. L. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta. 1998;1400:63–81. doi: 10.1016/S0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- Nitiss J. L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchaudhuri R., Hergenrother P. J. DNA as a target for anticancer compounds: methods to determine the mode of binding and the mechanism of action. Curr. Opin. Biotechnol. 2007;18:497–503. doi: 10.1016/j.copbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Pandit P., Patil R., Palwe V., Gandhe S., Patil R., Nagarkar R. Prevalence of molecular subtypes of breast cancer: a single institutional experience of 2062 patients. Eur. J. Breast Health. 2020;16:39–43. doi: 10.5152/ejbh.2019.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Hwang S. Y., Shin J., Jo H., Na Y., Kwon Y. A chromenone analog as an ATP-competitive, DNA non-intercalative topoisomerase II catalytic inhibitor with preferences toward the alpha isoform. Chem. Commun. 2019;55:12857–12860. doi: 10.1039/C9CC05524J. [DOI] [PubMed] [Google Scholar]
- Pindur U., Jansen M., Lemster T. Advances in DNA-ligands with groove binding, intercalating and/or alkylating activity: chemistry, DNA-binding and biology. Curr. Med. Chem. 2005;12:2805–2847. doi: 10.2174/092986705774454698. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Leo E., Zhang H., Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., James M. K., Larochelle S., Fisher R. P., Blain S. W. p27Kip1 inhibits cyclin D-cyclin-dependent kinase 4 by two independent modes. Mol. Cell. Biol. 2009;29:986–999. doi: 10.1128/MCB.00898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman S. U., Yaseen Z., Husain M. A., Sarwar T., Ishqi H. M., Tabish M. Interaction of 6 mercaptopurine with calf thymus DNA--deciphering the binding mode and photoinduced DNA damage. PLoS ONE. 2014;9:e93913. doi: 10.1371/journal.pone.0093913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehested M., Jensen P. B. Mapping of DNA topoisomerase II poisons (etoposide, clerocidin) and catalytic inhibitors (aclarubicin, ICRF-187) to four distinct steps in the topoisomerase II catalytic cycle. Biochem. Pharmacol. 1996;51:879–886. doi: 10.1016/0006-2952(95)02241-4. [DOI] [PubMed] [Google Scholar]
- Shrestha A., Park S., Shin S., Man Kadayat T., Bist G., Katila P., Kwon Y., Lee E. S. Design, synthesis, biological evaluation, structure-activity relationship study, and mode of action of 2-phenol-4,6-dichlorophenyl-pyridines. Bioorg. Chem. 2018;79:1–18. doi: 10.1016/j.bioorg.2018.03.033. [DOI] [PubMed] [Google Scholar]
- Vollmer T., Stewart T., Baxter N. Mitoxantrone and cytotoxic drugs' mechanisms of action. Neurology. 2010;74 Suppl 1:S41–S46. doi: 10.1212/WNL.0b013e3181c97f5a. [DOI] [PubMed] [Google Scholar]
- Wilstermann A. M., Osheroff N. Stabilization of eukaryotic topoisomerase II-DNA cleavage complexes. Curr. Top. Med. Chem. 2003;3:321–338. doi: 10.2174/1568026033452519. [DOI] [PubMed] [Google Scholar]
- Winick N. J., McKenna R. W., Shuster J. J., Schneider N. R., Borowitz M. J., Bowman W. P., Jacaruso D., Kamen B. A., Buchanan G. R. Secondary acute myeloid leukemia in children with acute lymphoblastic leukemia treated with etoposide. J. Clin. Oncol. 1993;11:209–217. doi: 10.1200/JCO.1993.11.2.209. [DOI] [PubMed] [Google Scholar]
- Woessner R. D., Mattern M. R., Mirabelli C. K., Johnson R. K., Drake F. H. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- Yu N. Y., Iftimi A., Yau C., Tobin N. P., van 't Veer L., Hoadley K. A., Benz C. C., Nordenskjöld B., Fornander T., Stål O., Czene K., Esserman L. J., Lindström L. S. Assessment of long-term distant recurrence-free survival associated with tamoxifen therapy in postmenopausal patients with luminal a or luminal B breast cancer. JAMA Oncol. 2019;5:1304–1309. doi: 10.1001/jamaoncol.2019.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.