Abstract
The cytosine analog 5-azacytidine (5-AzaC) is a demethylating agent that is also known to induce mutagenesis in mammalian cells. In this study, the mutagenic potential of this drug was tested in the G10 and G12 transgenic Chinese hamster cell lines, which have a single bacterial gpt gene integrated into the genome at different sites, with its expression driven by a simian virus 40 (SV40) promoter. We show that the mutation frequencies following a 48-h exposure to different concentrations of 5-AzaC were 10 to 20 times higher than those of any of the other numerous mutagens that have been tested in the G10-G12 system. Moreover, the mutation frequencies were much higher in the G10 cell line than in the G12 cells. Detailed molecular analysis of the 6-thioguanine (6-TG)-resistant variants demonstrated that transgene silencing by de novo DNA methylation and increased chromatin condensation in the SV40 promoter was the major factor responsible for this high level of 6-TG resistance. As would be expected, exposure to 5-AzaC lowered the overall genomic DNA methylation levels, but it unexpectedly caused hypermethylation and increased chromatin condensation of the transgene in both the G10 and G12 cell lines. These results provide the first evidence that 5-AzaC may also induce transgene-specific DNA methylation, a phenomenon that can further be used for the elucidation of the mechanism that controls silencing of foreign DNA.
Transfection of exogenous marker genes into mammalian cells and integration of those genes into the mammalian genome facilitate the study of factors affecting mutagenesis and gene expression (25). G10 and G12 are two transgenic gpt+ hprt− V79 cell lines that have been studied extensively and represent an important set of mammalian cell lines for mutagenesis studies (19, 20). The bacterial xanthine guanine phosphoribosyltransferase enzyme is functionally analogous to the endogenous mammalian hypoxanthine phosphoribosyltransferase. The G10 cell line is more susceptible to spontaneous and induced mutagenicity than is the G12 cell line. Most of the spontaneous mutations and mutations induced by X ray or bleomycin were characterized as deletions in the G10 cell line (21). This was explained by the genomic mapping data that showed partial duplication of gpt-flanking plasmid sequences in G10 but not in G12 cells (21). These sequences could facilitate homologous recombination, resulting in the observed high deletion frequencies (21). Additionally, it is known that a single copy of the gpt gene was integrated into each of these cell lines on different chromosomes (22). In the G12 cell line, the gpt gene was integrated on chromosome 1, near the telomere and adjacent to a dense region of heterochromatin, while in the G10 cell line, the gpt gene was integrated on chromosome 6, distant from any heterochromatin.
Unusual results were obtained for the highly carcinogenic nickel compounds (22). Nickel subsulfide and crystalline nickel sulfide were only weakly mutagenic in G10 cells (15), but they induced extremely high (10−3) frequencies of 6-thioguanine (6-TG) resistance in the G12 cells (22). It was shown that the mechanism of the 6-TG resistance involved chromatin condensation and silencing of the gpt gene by de novo DNA methylation. The localization of the gpt sequence in the vicinity of a dense heterochromatin area in G12 cells has been proposed to explain the selective sensitivity of this sequence to nickel (22). Similar DNA methylation silencing of transgenes have been reported for gpt sequences in transfected human cell lines as a result of UV or ethyl methanesulfonate treatment (7, 23).
For evaluation of the mutagenic and epigenetic potential of other carcinogens, a combined G10-G12 screening may be informative. In this study, we used the G10 and G12 cell lines to analyze the mutagenic effects of 5-azacytidine (5-AzaC). 5-AzaC is a cytosine analog that when incorporated into DNA caused extensive demethylation of 5-methylcytosine. This was due to covalent binding of DNA methyltransferase to 5-AzaC in DNA (14, 31) and a subsequent reduction of the enzyme activity in the cell, as well as the incorporation of a nonmethylating site, such as 5-AzaC, in place of the normal base cytosine. The demethylation activity of this drug induces muscle cell differentiation in CH3 10T1/2 or 3T3 cells (34). Treatment of mammalian cell lines with 5-AzaC or its deoxyribose congener 5-Aza-2′-deoxycytidine has resulted in a variety of altered phenotypes, including changes in chromosome structure, gene expression, and cellular morphology (29, 32), and in the induction of apoptosis (18). Numerous investigations have described the reactivation of de novo-methylated silenced genes by 5-Aza-CR or 5-Aza-2′-deoxycytidine. These genes include loci on the inactive X chromosome (13), the VHL gene (9), the E-cadherin gene (35), the estrogen receptor gene (27), and the p16 gene (2). Silenced transgenes are also reactivated after treatment with this drug (22, 23).
The adducts formed between DNA methyltransferase and genomic DNA with a 5-Aza substitution (14) can sterically inhibit DNA replication, transcription, and DNA repair and may play a role in 5-Aza-induced mutagenesis in mammalian cells (1, 3, 24). 5-AzaC is also known to be a mutagen in Escherichia coli (6), Salmonella typhimurium (3, 30), and Saccharomyces cerevisiae (36).
Here we describe the molecular analysis of 6-TGr G10 and G12 variants induced by treatment with 5-AzaC and surprisingly show that in most cases the inactivation of the gpt gene was correlated with higher DNA methylation levels of CpG islands and increased chromatin condensation in the gpt regions in both cell lines. Meanwhile, as expected, the overall genomic DNA methylation levels in the variants were lower than those in the original untreated cells. This finding may allow new approaches for the isolation and characterization of specific regulators that inhibit transgene expression and point to the importance of DNA hypermethylation as a mechanism that silences foreign DNA.
MATERIALS AND METHODS
Cell culture conditions.
The gpt+ (xanthine guanine phosphoribosyltransferase gene) transgenic G10 and G12 cells were cultured in F-12 medium (Life Technologies, Inc., GIBCO BRL, Grand Island, N.Y.) supplemented with 5% fetal bovine serum (GIBCO) and 1% penicillin-streptomycin (GIBCO) at 37°C in a humid 5% CO2 atmosphere. In order to maintain a low spontaneous-mutation frequency, G10 and G12 cultures were supplemented with hypoxanthine-aminopterin-thymidine, and fresh cultures were defrosted every 6 weeks. One day prior to the beginning of each mutagenesis experiment, the cells were removed from hypoxanthine-aminopterin-thymidine selection.
5-AzaC mutagenesis assay.
A total of 5 × 105 G10 or G12 cells were seeded into 80-cm2 tissue culture flasks and after 4 to 6 h were exposed to various concentrations of 5-AzaC for 2 days. After the treatment was removed, the cells were rinsed with saline A twice and incubated in F-12 medium for a 7-day expression period. Cytotoxicity was determined for each treatment by plating 400 cells in each of three 6-cm dishes and determining the clonal survival relative to untreated controls. Following the expression period, 2 × 106 mutagenized cells per treatment were reseeded at a maximum cell density of 2 × 105 cells/100-mm dish into F-12 containing 10 μg of freshly prepared 6-TG per ml for 10 days. The reseeding plating efficiency in nonselective medium (F-12) was determined for mutation frequency calculations after 7 days of growth without selection. The number of mutant colonies growing in the selection medium following a correction for the number of clonable cells was used to calculate the mutation frequency. Thus, the mutation frequency refers to the number of cells that survived the 5-AzaC treatment. Unstained mutant colonies were individually isolated and characterized.
Deletion screen by PCR amplification of coding sequences.
The gpt gene was amplified by the PCR method as previously described (22). The amplification reaction mixtures (100 μl) contained 1 μg of genomic DNA (G10, G12, and 6-TGr clones), 100 pmol of amplification primers 5′-AACACTTTTTAAGCCGTAGATAAA and 5′-TATTGTAACCCGCCTGAAGTTAAA (these primers hybridize 18 bases before and 39 bases after the gpt coding region, respectively), 200 μM deoxynucleoside triphosphate, and 2.5 U of Taq polymerase in a 50 mM KCl–10 mM Tris (pH 8.0; 1.5 mM MgCl2–0.01% [wt/vol] gelatin) buffer. Amplification (typically 30 cycles) was performed in Perkin-Elmer thermal cycler. The resultant PCR products were then separated on 1.2% agarose gels to screen for any tentative deletions.
Genomic methylation level.
A modification of the methyl-accepting assay (17) was used to determine the methylation level of DNA isolated from G10, G12, and 6-TGr cells. DNA (200 ng) was incubated with 4 U of SssI methylases (New England Biolabs) in the presence of 1.5 μM S-adenosyl-l-[methyl-3H]methionine and 1.5 μM nonradioactive S-adenosylmethionine. The reaction mixtures (20 μl), in the manufacturer’s buffer containing 0.1 μg of RNase A, were incubated at 37°C for 4 h. The reactions were terminated by adding 300 μl of stop solution (1% sodium dodecyl sulfate, 2 mM EDTA, 5% 2-propanol, 125 mM NaCl, 1 mg of proteinase K per ml, 0.25 mg of carrier DNA per ml) for 1 h at 37°C. The DNA was extracted with phenol-chloroform and ethanol precipitated. The recovered DNA was resuspended in 30 μl of 0.3 M NaOH and incubated for 30 min at 37°C. DNA was spotted on Whatman GF/C filter discs, dried, and then washed five times with 5% (wt/vol) trichloroacetic acid followed by 70% (vol/vol) ethanol. Filters were placed in scintillation vials and incubated for 1 h at 60°C with 500 μl of 0.5 M perchloric acid. Then 5 ml of scintillation cocktail was added and the 3H incorporation was determined by a Beckman liquid scintillation counter. Higher levels of [3H]methyl group incorporated into DNA indicated lower levels of genomic DNA methylation, but when less [3H]methyl group was incorporated, a higher level of genomic DNA methylation was indicated (17).
DNA methylation studies with methylation-sensitive restriction enzymes.
Ten micrograms of DNA from G10 and G12 cells and 6-TGr 5-AzaC G10 and G12 variants were first digested with the restriction endonuclease EcoRV (5 U/μg of genomic DNA) and then extracted with phenol-chloroform and ethanol precipitated. This digestion released a 1.7-kb genomic fragment in G12 DNA and a 5.3-kb genomic fragment in G10 DNA. This DNA fragment was digested again with 5 U of the methylation-sensitive restriction endonuclease HpaII (5′-CCGG-3′; U.S. Biochemicals) or HaeII (5′-PuGCGCPy-3′; New England Biolabs) per μg or with the insensitive isoschizomer MspI. The digested DNA was fractionated on agarose gels, blotted onto nylon membranes (Nytran; Schleicher & Schuell), and hybridized with an appropriate radiolabeled probe.
Bisulfite genomic sequencing for the detection of 5-methylcytosine.
Bisulfite genomic sequencing was performed as described by Clark et al. (5) with the modifications described by Singal et al. (32). The primers for the PCR amplification (5′-ACATAAATCTACAACATATCCCAAATAACA/GATA and 5′-ATGTAAAGTATGTATTTTAATTAGTTAGTAATTA) were constructed after the bisulfite conversion reaction had been taken into account. (The sequence of the unmodified sense strand for which these primers were constructed is depicted in Fig. 5.) Direct sequencing of PCR-amplified product (408 bp) was performed with an automated DNA sequencer (Genomis Inc.).
FIG. 5.
Map of 408 bp from the SV40 early promoter region flanking the gpt coding region in G10 and G12 transgenic cell lines. CpG dinucleotides are underlined. The numbers above the CpG sites correspond to the methylation map data in Table 3. Bold numbers represent the sites that were methylated or partially methylated in the genomes of five out of the eight variants that were examined. The arrows indicate the sequences used for the primer design. (The primers were constructed after the bisulfite conversion reaction had been taken into account.)
DNase I sensitivity assay.
The procedure for the isolation of nuclei was reported previously (22). A total of 5 × 105 nuclei in DNase I buffer (10 mM Tris-Cl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 100 μM CaCl2) were treated with increasing amounts (0, 0.5, 1, 2, and 10 U) of DNase I (Boehringer) in a reaction volume of 200 μl for 30 min at 25°C. The reactions were terminated by adding equal volume of stop solution (1% sodium dodecyl sulfate, 0.1 M NaCl, 50 mM Tris-Cl [pH 8.0], and 10 mM EDTA) containing 1 mg of proteinase K per ml and incubated at 55°C for 2 h. The DNA was extracted with phenol-chloroform and ethanol precipitated. The gpt gene was amplified by PCR (50 ng/reaction; 30 cycles) with the same primers that were described above for the deletion screen. The PCR products were separated on 1.2% agarose gel and stained with ethidium bromide (EtBr).
RESULTS
High mutation frequencies induced by 5-AzaC in G10 and G12 cells.
Dose-response studies for 5-AzaC were performed on the G10 and G12 cell lines (Fig. 1). The cells were grown for 48 h in the presence of the drug, and then the survival rates and mutation frequencies of the gpt gene (6-TGr) were determined as described in Materials and Methods. The concentration of 5-AzaC necessary to inhibit growth by 50% was 2 μM for both cell lines. The mutation frequencies were very high (approximately 10 to 20 times higher) compared to those of other mutagens tested in the G10 and G12 cell lines (22); however, there were dissimilarities between the two cell lines. The mutation frequency in the G10 cell line was much higher than in the G12 cell line. This level of mutagenesis (4 × 10−2) is the highest level ever reported for this cell line. The differences between the cell lines may be explained by the different localizations of the gpt gene.
FIG. 1.
Mutation frequency and cytotoxicity of 5-AzaC in G10 and G12 cells. The cells were exposed to various concentrations of 5-AzaC for 48 h and then incubated for a 7-day expression period. The selection for gpt− cells was then done in a medium containing 6-TG (10 μg/ml) for 10 days. The data represent the median values (for mutation frequencies) and the means ± standard deviations (for survivals) of three to eight determinations. Filled symbols and open symbols represent the mutation frequency and percent survival, respectively. The spontaneous mutation frequencies of the G10 (▴) and G12 (●) cells were 100 and 30 per 106 surviving cells, respectively.
Northern and deletion analysis revealed no transcript but intact transgene existence in most of the 6-TGr cell lines.
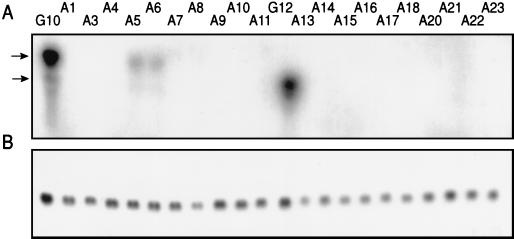
We examined gpt-specific mRNA levels and also gpt genomic sequences in several G10- and G12-derived cell lines (summarized in Table 1). These and all mutant clones described here were independently isolated from individually treated populations of cells. As clearly seen in the Northern blot (Fig. 2A), only the wild-type G10 and G12 cells accumulate gpt transcript. Note, however, that in two of the G10 variants very low levels of expression of gpt transcripts were observed (variants A5 and A6 [Fig. 2A]).
TABLE 1.
Summary of molecular characterization of 5-AzaC-induced 6-TGr variants of G10 and G12 cell lines
| Cell line | No. (%) of variants
|
|||
|---|---|---|---|---|
| Tested | Without transcripts | With transcripts | With deletions of gpt | |
| G10 | 23 | 20 (87) | 1 (4.3) | 2 (8.7) |
| G12 | 32 | 28 (87.6) | 3 (9.3) | 1 (3.1) |
| Total | 55 | 48 (87.2) | 4 (7.3) | 3 (5.5) |
FIG. 2.
Analysis of gpt transcription in several 6-TG-resistant G10 and G12 variants. (A) Northern blot analysis of gpt transcription. Fourteen micrograms of total RNA per cell line was fractionated on 1% agarose gels and transferred to a nylon membrane. The membrane was hybridized with a 32P-labeled gpt probe that was generated by random primer labeling of a 561-bp PCR product of the gpt coding region. The arrows indicate the expected size of the gpt transcript in the G10 (top arrow) and G12 (bottom arrow) parental lines. (B) To control for gel-loading differences, the membranes were stripped and rehybridized with a GAPDH probe.
The results of the PCR analysis that was done in order to determine the presence or absence of the gpt sequence are shown in Fig. 3. The frequency of deletion mutants is very low in both cell lines (Table 1). This resembled the low level of deletion that was found for the 6-TGr G12-derived cell line induced by nickel (22). However, the mutagenic spectrum found in other studies showed that transgene deletions occurred in 20% of the spontaneous G12 mutants and in about 50% of the X-ray- and bleomycin-induced G12 mutants (21). The levels were even higher (up to 95%) in G10 mutant cells (21), which were much more highly prone to gpt deletion than G12 cells. The lower deletion level (less than the spontaneous frequency) obtained with 5-AzaC demonstrated a unique mechanism activated by this drug, which silences these transgenes.
FIG. 3.
Deletion screen by PCR amplification of the gpt coding sequence in G12-derived (A) and G10-derived (B) 6-TG-resistant cell lines. PCR products were separated on 1.2% agarose gels and stained with EtBr. The expected 561-bp PCR product is clearly visible in the control G10 and G12 cell lines and most of the examined variants. No products are shown in the negative control reaction (no template DNA) (lane N). M, molecular weight markers.
Lower genomic methylation levels in the 5-AzaC-induced variants.
The overall methylation levels were checked for a number of the 5-AzaC G12-derived variants and compared to those of the original G12 cell line. The genomic methylation levels, as expected, decreased as a result of the 5-AzaC treatment (Table 2). These low genomic methylation levels were maintained for many cell cycles because the cells were selected for 6-TGr before isolation and genomic DNA methylation patterns were known to be inherited.
TABLE 2.
Genomic DNA methylation levels in 5-AzaC-induced 6-TGr variants of the G12 cell line
| Cell line | Genomic DNA methyl-ation level (% of control)a |
|---|---|
| G12 (control) | 100 |
| G12 6-TGr | |
| A13 | 59.4 ± 4.7 |
| A14 | 68.9 ± 1.9 |
| A15 | 80.10 ± 2.5 |
| A16 | 47.11 ± 4 |
| A18 | 50.4 ± 2.6 |
Methylation levels of genomic DNA were measured from duplicate samples by a methyl-accepting assay with CpG methylase (SssI) and calculated as follows: {1 + [(control − treated)/control]} × 100.
Evidence for DNA methylation in 5-AzaC-induced variants.
As shown in Table 1, 87% of all the 5-AzaC variants did not accumulate gpt transcript despite having an intact transgene. In order to check if the transgene expression had been silenced by mechanisms involving DNA methylation, we performed two different assays. Digestion of genomic DNA with a methylation-sensitive restriction enzyme followed by Southern analysis of the gpt region is shown in Fig. 4. Figure 4A shows digestion of genomic DNA from 6-TGr G12-derived cell lines with the methylation-sensitive restriction enzyme HaeII. There is one cutting site in the 5′ flanking region of the integrated gpt target sequence in the G12 cell line. When G12 genomic DNA was digested with EcoRV and HaeII, a 1.2-kb fragment would have been obtained if both enzymes cut, or a 1.7-kb fragment would have appeared if the HaeII site was methylated, but only EcoRV cut the DNA (Fig. 4D). The appearance of both bands in almost all the cell lines examined (except A18 and A19) demonstrated partial methylation of this site. The digestion of control G12 DNA yielded the lower band only. The results for the G10-derived cell lines, digested with the methylation-sensitive enzyme HpaII and the enzyme EcoRV, are shown in Fig. 4B. There are three restriction sites for methylation-sensitive HpaII restriction enzyme in this transgene sequence (Fig. 4D). When the DNA was completely methylated, a 5.3-kb fragment would have been observed. If the DNA was not methylated, 5.04- and 0.15-kb fragments (and also 0.07- and 0.04-kb fragments that are not detected with the blotting conditions used) would result from the double digest. Also in this case, the appearance of intermediate bands (0.22 and 0.26 kb) together with the lower band (for variants A2, A3, A4, A7, A8, A9, A10, A11, and A12) indicated increased methylation. These intermediate bands are not seen in the G10 control. In the A5 and A6 variants, we could not detect intermediate bands. In these same cell lines, low transcription levels were detected by the Northern analysis (Fig. 2).
FIG. 4.
Methylation of the gpt gene and its flanking sequence in G12 (A) and G10 (B and C) 5-AzaC-induced 6-TG-resistant cell lines. Ten micrograms of EcoRV-digested DNA was further digested with the methylation-sensitive restriction enzyme HaeII (A) or HpaII (B). Lane C contains EcoRV-digested DNA not subjected to digestion with the other enzymes. (C) Digestion of G10 and the A2, A3, and A7 5-AzaC variants with the methylation-sensitive enzyme HpaII and the methylation-insensitive isoschizomer MspI. To better resolve the bands, the DNA was cut with both EcoRV and HindIII, and fragments were separated in a 1.7% agarose gel (6 h at 55 V). The DNA was transferred to a membrane in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). (D) Restriction of the gpt gene on the genomic map of G12 and G10 cell lines. The fragments were separated on agarose gels (1% [A], 1.7% [B], and 1.7% [C]), transferred to nylon membranes, and then hybridized with a 32P-labeled gpt probe. The variants are identified as in Fig. 2. H1, H2, H3, and H4 are the first, second, third, and fourth HpaII sites, respectively.
Figure 4C compares the digestion of the G10 and three G10 5-AzaC variants (A2, A3, and A7) with the methylation-sensitive HpaII and the methylation-insensitive isoschizomer MspI. (The DNA was first digested with EcoRV and HindIII, yielding lower molecular weights than in Fig. 4B, in which the DNA was cut with only EcoRV.) A darker 470-bp band in the HpaII cut than that in the MspI cut indicated some partial methylation at the first HpaII site in wild-type G10 (Fig. 4D). However, there were additional differences in the 5-AzaC-induced variants of G10 between MspI and HpaII cutting with respect to the presence of the 150-bp fragment in the MspI lane and not in the HpaII lane in each variant, indicating more methylation in the variants at the first and second HpaII sites (Fig. 4D). Additionally, there was less intensity of the 320-bp band in the variants cut with the HpaII than in the same DNA cut with MspI, indicating more methylation at the first HpaII site (Fig. 4D). There was also greater intensity of the 470- and 540-bp fragments with HpaII cutting than with MspI cutting in the variants (i.e., a 470-bp fragment would be present if the first HpaII site was methylated, a 540-bp fragment would be present if the second HpaII site was methylated, and a 580-bp fragment would be present if the third site was methylated [Fig. 4D]).
The second assay performed was methylation analysis of the promoter region in different 6-TGr cells by the sodium bisulfite sequencing method (5, 32). The region examined included 408 bp of the simian virus 40 (SV40) early promoter region, which was located 5′ to the E. coli gpt gene in these transgenic cell lines. This region contained 18 CpG sites (Fig. 5). The results for eight independent 6-TGr cell lines and two control cell lines are summarized in Table 3. Since direct sequence of the PCR product was performed, the data represent a population average (5). Table 3 shows that the amount of sites methylated in the different cell lines varies. Note that many sites are only partially methylated. However, almost all the cell lines had some level of methylation in the specific region tested, excluding those with some gpt expression, A5 and A6 variants, and the wild-type G12. The A5 and A6 cell lines showed either no methylation or partially methylated sites, respectively. This correlated with the low levels of transcription and undetectable methylation after digestion with methylation-sensitive restriction enzymes (Fig. 4).
TABLE 3.
CpG-methylated, nonmethylated, and partially methylated sites in the SV40 early promoter region of 5-AzaC-induced 6-TGr variants derived from the G10 and G12 cell linesa
| Site no.b | Methylationc in:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| G10 variants
|
G12 variants
|
G12 | N37d | |||||||
| A2 | A3 | A5 | A6 | A7 | A14 | A15 | A23 | |||
| 1 | − | + | − | ND | − | ND | − | − | − | − |
| 2 | − | ± | − | ND | − | ND | − | − | − | − |
| 3 | ± | + | − | − | ± | − | − | ± | ± | ± |
| 4 | + | + | − | − | ± | − | − | ± | − | ± |
| 5 | + | + | − | − | + | − | ± | + | − | + |
| 6 | ± | + | − | − | ± | − | + | ± | − | ± |
| 7 | + | + | − | − | ± | ± | ± | ± | − | + |
| 8 | ± | ± | − | − | − | − | − | ± | − | ± |
| 9 | ± | ± | − | − | − | − | − | ± | − | ± |
| 10 | + | + | − | − | ± | − | ± | + | − | + |
| 11 | ± | ± | − | − | − | − | − | − | − | + |
| 12 | ± | ± | − | − | − | − | − | ± | − | + |
| 13 | ± | + | − | − | ± | − | ± | + | − | + |
| 14 | + | + | − | − | ± | − | + | + | − | + |
| 15 | + | + | − | − | + | ± | + | + | − | + |
| 16 | + | ± | − | − | ± | − | ± | + | ND | + |
| 17 | + | + | − | − | ± | − | ± | + | − | + |
| 18 | + | + | − | ± | ± | − | ± | ± | − | + |
Data were obtained by the sodium bisulfite genomic sequencing technique (5, 32) of independent cell lines. Direct PCR sequencing was done with an automated DNA sequencer to obtain a methylation map from the population average. Partial methylation at any site was observed by the presence of both a cytosine and a thymidine residue at the same sequencing position. Cytosines that are not associated with CpG dinucleotides have been converted to thymidines in all the samples.
The sites in this table correspond to the CpG sites marked in Fig. 5.
+, methylation; −, no methylation; ±, partial methylation.
N37 is a nickel-induced 6-TGr G12 variant, which was shown to be highly methylated in the gpt locus (22).
The 10 common sites that were methylated or partially methylated in five out of eight variants are marked on Fig. 5. Each of these genomic methylation patterns resulted in transgene silencing, and all were induced by the demethylation drug, 5-AzaC.
Reversion assays with 3 μM 5-AzaC were performed in the A3, A11, A13, and A21 cell lines, as described in detail previously (22), in order to check if the transgene was still potentially functional and inactivated only by its epigenetic status. The high reversion frequencies (6 × 10−4, 10 × 10−4, 60 × 10−4, and 500 × 10−4, respectively) further emphasized the correlation between the methylation state and the silencing of the transgene. The control cell lines that were used for these assays were the N37 nickel-induced variant, which was shown to be highly methylated in the gpt locus and gave high reversion frequencies (6 × 10−4), and the N126 cell line, in which the gpt gene is mutated. In this cell line no reversion was observed with 5-AzaC treatment (22). The spontaneous reversion rates for methylated variants in these experiments were on the order of 3 × 10−6.
Changes in chromatin structure in the gpt locus of the 5-AzaC-induced variants.
To study whether the observed changes in DNA methylation were correlated with changes in the chromatin structure in the gpt locus, DNase I sensitivity was examined in isolated nuclei from different variant cell lines. As shown in Fig. 6, the variant clones of both G12 and G10 exhibited marked resistance to increasing concentrations of DNase I compared with that in the parental G10 and G12 cell lines. The N37 cell line was used as a positive control (Table 3). These data indicated a more condensed chromatin structure of the transgene, induced as a consequence of 5-AzaC treatment.
FIG. 6.
Resistance of 5-AzaC-induced 6TG-resistant clones to DNase I. Nuclei isolated from G12, N37, A13, A14, and A15 cells (A) and G10, A2, A6, and A7 cells (B) were treated with 0, 0.5, 1, 2, and 10 U of DNase I. PCRs were performed on 50 ng of DNase I-digested DNA. PCR products were separated on 1.2% agarose gels and stained with EtBr. The expected gpt product is 561 bp. M, molecular weight markers.
DISCUSSION
Our results showed that treatment of G10 and G12 transgenic cell lines with 5-AzaC resulted in an unusually high frequency of 6-TG resistant variants (1.5 × 10−2 to 4.5 × 10−2). Other studies (for examples, see references 24 and 33) showed 5-AzaC-induced 6-TG and TFT resistance in AS52 (33) and L5178Y cell lines (24). In those studies the mutation frequency levels were lower than the results presented here. This was probably due to the shorter 5-AzaC treatment periods that were used in the two mentioned studies (4 to 5 h compared to 48 h here). However, 5-AzaC is more effective at lowering DNA methylation with longer treatment periods (i.e., at least one cell cycle), and this is usually the way cells are treated with this drug.
Detailed molecular analysis of the variants demonstrated that transgene silencing by de novo DNA methylation was the major factor responsible for this high level of 6-TG resistance. Transgene silencing is a well-known phenomenon (7, 28), but it was unexpectedly observed after treatment with the classical demethylation agent, 5-AzaC. Transgenes are probably recognized in cells by the specific bacterial or viral sequences that they contain and their different structures in the chromosomal environment (11, 28), and by some unknown mechanism, 5-AzaC caused these transgenes to become silenced by inducing de novo DNA methylation and increased chromatin condensation. It is important to emphasize that the variant cells were isolated after drug selection. As a result of 5-AzaC treatment for 2 days, the transgene was probably demethylated. Then the cells were grown in regular medium for a 7-day expression period (10 to 14 cell divisions). After this period, the majority of the nuclei did not contain 5-AzaC (12). When the cells were transferred to the selection media (6-TG), the selection was toward cells that could inactivate the transgene. It is possible that part of the 6-TGr cell lines was a result of mutation (like the A5 and A6 cell lines and also the A18 cell line), but the high frequency of variants obtained supports a more general process of gene inactivation by methylation of cytosines in DNA. An equivalent model for silencing and reactivation of recombinant viral genes has recently been suggested (4). According to this model, a host protein or protein complex binds to viral sequences and recruits a histone deacetylase to the site. The enzyme deacetylates histones H3 and H4, resulting in a more condensed chromatin structure and inhibition of transgene transcription. A direct relationship between DNA methylation and histone deacetylation was shown by Nan et al. (26). It is possible that the same events happened in our system, due to the selection conditions, leading to DNA methylation and chromatin condensation of the gpt locus (16).
The reason for the higher frequency of G10 variants, compared to G12 variants, may be the different chromosomal localizations of the gpt gene. In the G10 cells it was localized in a euchromatic region on chromosome 6, which might be more susceptible to protein binding and modifying enzymes, such as DNA methyltransferase, than the gpt locus in G12 cells, which was located in close proximity to a condensed heterochromatic region near the telomere on chromosome 1.
The methylation maps of the different variants are not equal, but among the 18 tested sites, there were 10 sites (Table 3) completely or partially methylated in five out of the eight cell lines examined. It was already demonstrated that the discrimination between methylated and unmethylated alleles may be attributed to differences in methylation of a very short and specific region (8, 10); thus, it is possible that methylation of only a few critical sites was sufficient to silence the gpt gene. To emphasize the relation between the transgene silencing and methylation, reactivation of the transgene silenced by 5-AzaC was performed with 5-AzaC again. The high reversion frequencies suggested additional evidence for the epigenetic effect induced by 5-AzaC.
The results we report here demonstrated that under specific conditions 5-AzaC may induce transgene silencing by DNA methylation and chromatin condensation. Isolation of the specific regulatory protein(s) that enhanced transgene silencing and the elucidation of its expression pattern may be a useful tool for a controlled transgene inactivation process. Further study of this phenomenon may help treat disease involving viral integration and assist us in designing better strategies for gene therapy.
ACKNOWLEDGMENTS
We thank Tomasz Kluz for performing the experiments shown in Fig. 4C and for excellent technical assistance. We also thank Konstantin Salnikow for his consultation during the course of this project.
This work was supported by grants ES05512 and ES00260 from the National Institute of Environmental Health Sciences and grant CA16037 from the National Cancer Institute.
REFERENCES
- 1.Amacher D, Turner G. The mutagenicity of 5-azacytidine and other inhibitors of replicative DNA synthesis in the L5178Y mouse lymphoma cell. Mutat Res. 1987;176:123–131. doi: 10.1016/0027-5107(87)90259-4. [DOI] [PubMed] [Google Scholar]
- 2.Bender C M, Pao M M, Jones P A. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 3.Call K, Jensen J, Liber H, Thilly W. Studies of mutagenicity and clastogenicity of 5-azacytidine in human lymphoblasts and Salmonella typhimurium. Mutat Res. 1986;160:249–257. doi: 10.1016/0027-5107(86)90135-1. [DOI] [PubMed] [Google Scholar]
- 4.Chen W Y, Bailey E C, McCune S L, Dong J-Y, Townes T M. Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc Natl Acad Sci USA. 1997;94:5798–5803. doi: 10.1073/pnas.94.11.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark S J, Harrison J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman S. The effect of 5-azacytidine on E. coli DNA methylase. Biochem Biophys Res Commun. 1979;89:1328–1333. doi: 10.1016/0006-291x(79)92154-5. [DOI] [PubMed] [Google Scholar]
- 7.Gebara M M, Drevon C, Harcourt S A, Steingrimsdottir H, James M R, Burke J F, Arlett C F, Lehmann A R. Inactivation of a transfected gene in human fibroblasts can occur by deletion, amplification, phenotypic switching, or methylation. Mol Cell Biol. 1987;7:1459–1464. doi: 10.1128/mcb.7.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D-S, Gnarra J R, Linehan W M, Baylin S B. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J-P J, Davidson N E, Sidransky D, Baylin S B. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 11.Jahner D, Stuhlmann H, Stewart C L, Harbers K, Lohler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 12.Jones P A, Taylor S M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 13.Jones P A, Taylor S M, Mohandas T, Shapiro L J. Cell cycle-specific reactivation of an inactive X-chromosome locus by 5-azadeoxycytidine. Proc Natl Acad Sci USA. 1982;79:1215–1219. doi: 10.1073/pnas.79.4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jutterman R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kargacin B, Klein C B, Costa M. Mutagenic responses of nickel oxides and nickel sulfides in Chinese hamster V79 cell lines at the xanthine guanine phosphoribosyl transferase locus. Mutat Res. 1993;300:63–72. doi: 10.1016/0165-1218(93)90141-y. [DOI] [PubMed] [Google Scholar]
- 16.Kass S U, Pruss D, Wolfe A P. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 17.Kautiainen T L, Jones P A. DNA methyl-transferase levels in tumorigenic and nontumorigenic cells in culture. J Biol Chem. 1986;261:1594–1598. [PubMed] [Google Scholar]
- 18.Kizaki H, Ohnishi Y, Azuma Y, Mizuno Y, Ohsaka F. 1-β-d-Arabinosylcytosine and 5-azacytidine induce internucleosomal DNA fragmentation and cell death in thymocytes. Immunopharmacology. 1992;24:219–227. doi: 10.1016/0162-3109(92)90077-p. [DOI] [PubMed] [Google Scholar]
- 19.Klein C B, Rossman T G. Transgenic Chinese hamster V79 cell lines which exhibit variable levels of gpt mutagenesis. Environ Mol Mutagen. 1990;16:1–12. doi: 10.1002/em.2850160102. [DOI] [PubMed] [Google Scholar]
- 20.Klein C B, Su L, Rossman T G, Snow E T. Transgenic gpt+ V79 cell lines differ in their mutagenic response to clastogens. Mutat Res. 1994;304:217–228. doi: 10.1016/0027-5107(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 21.Klein C B, Su L, Singh J, Snow E T. Characterization of gpt deletion mutations in transgenic Chinese hamster cell lines. Environ Mol Mutagen. 1997;30:418–428. [PubMed] [Google Scholar]
- 22.Lee Y-W, Klein C B, Kargacin B, Salnikow K, Kitahara J, Dowjat K, Zhitkovich A, Christie N T, Costa M. Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: a new model for epigenetic carcinogens. Mol Cell Biol. 1995;15:2547–2557. doi: 10.1128/mcb.15.5.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehmann A R, Arlett C F, Harcourt S A, Steingrimsdottir H, Gebara M M. Mutagenic treatments result in inactivation of expression of a transfected bacterial gene integrated into a human cell line. Mutat Res. 1989;220:255–262. doi: 10.1016/0165-1110(89)90029-8. [DOI] [PubMed] [Google Scholar]
- 24.McGregor D B, Brown A G, Cattanach P, Shepherd W, Riach C, Daston D S, Caspary W J. TFT and 6-TG resistance of mouse lymphoma cells to analogs of azacytidine. Carcinogenesis. 1989;10:2003–2008. doi: 10.1093/carcin/10.11.2003. [DOI] [PubMed] [Google Scholar]
- 25.Mulligan R C, Berg P. Expression of a bacterial gene in mammalian cells. Science. 1980;209:1422–1427. doi: 10.1126/science.6251549. [DOI] [PubMed] [Google Scholar]
- 26.Nan X, Ng H-H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 27.Ottaviano Y L, Issa J-P, Parl F F, Smith H S, Baylin S B, Davidson N E. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994;54:2552–2555. [PubMed] [Google Scholar]
- 28.Palmer T D, Rosman G J, Osborne W R A, Miller D A. Genetically modified skin fibroblasts persist long after transplantation but gradually inactivate introduced genes. Proc Natl Acad Sci USA. 1991;88:1330–1334. doi: 10.1073/pnas.88.4.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parrow V C, Alestrom P, Gautvik K M. 5-Azacytidine induced alterations in the GH12C1 cells: effects on cellular morphology, chromosome structure, DNA and protein synthesis. J Cell Sci. 1989;93:533–543. doi: 10.1242/jcs.93.3.533. [DOI] [PubMed] [Google Scholar]
- 30.Podger D M. Mutagenicity of 5-azacytidine in Salmonella typhimurium. Mutat Res. 1983;121:1–6. doi: 10.1016/0165-7992(83)90079-9. [DOI] [PubMed] [Google Scholar]
- 31.Santi D V, Norment A, Garrett C E. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singal R, Ferris R, Little J A, Wang S Z, Ginder G D. Methylation of the minimal promoter of an embryonic globin gene silences transcription in primary erythroid cells. Proc Natl Acad Sci USA. 1997;94:13724–13729. doi: 10.1073/pnas.94.25.13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spencer D L, Caspary W J, Hines K C, Tindall K R. 5-Azacytidine-induced 6-thioguanine resistance at the gpt locus in AS52 cells: cellular response. Environ Mol Mutagen. 1996;28:100–106. doi: 10.1002/(SICI)1098-2280(1996)28:2<100::AID-EM5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 34.Taylor S M, Jones P A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 35.Yoshiura K, Kanai Y, Ochiari A, Shimoyama Y, Sugimura T, Hirohashi S. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci USA. 1995;92:7416–7419. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmermann F K, Scheel I. Genetic effects of 5-azacytidine in Saccharomyces cerevisiae. Mutat Res. 1984;139:21–24. doi: 10.1016/0165-7992(84)90116-7. [DOI] [PubMed] [Google Scholar]