Abstract
The ubiquitously expressed bromodomain-containing protein 4 (BRD4) is an epigenetic reader, which recruits transcriptional regulatory complexes to acetylated chromatin. Because of its role in enhancing proliferation, BRD4 has become a therapeutic target in oncology, as the inhibition of this protein leads to the reduction of the growth of many tumours. Even though BRD4 is more and more studied, its mechanism of action has not been fully described yet. Therefore, we aimed at generating a cellular reporter system to monitor BRD4 inhibition. Such reporter can be potentially used in high throughput chemical and genetic screenings, in order to uncover new possible BRD4 functional pathways. The deeper understanding of the mechanism of action of BRD4 activity will certainly help in developing new therapy strategies for those cancers so called BRD4-dependent.
Keywords: Cellular chromatin reporters, Epigenetic, Chromatin reorganization, Heterochromatinization, BRD4
Background
Research in the epigenetic field has recently highlighted the central role of BRD4 in cancer progression. BRD4 is an acetyl-lysine reader of the BET (bromodomain and extraterminal domain) family ( Dey et al., 2003 ; Filippakopoulos et al., 2012 ; Wang et al., 2012 ) able to bind to acetylated histones at promoter and enhancer regions ( Dey et al., 2003 ; Filippakopoulos et al., 2012 ; Nagarajan et al., 2014 ). The mechanism of action of this epigenetic reader consists in the activation of gene promoters and enhancers by recruiting several transcription factors, cofactors and RNA polymerase II (RNApol II), which results in modulating, mostly enhancing, the transcription of certain target genes. The BRD4-histone module has been described to play a key role regulating cell cycle progression ( Dey et al., 2003 ; Wu and Chiang, 2007; Yang et al., 2008 ; Devaiah and Singer, 2013) and genomic structure and stability (Wu and Chiang, 2007; Floyd et al., 2013 ); for those reasons, BRD4 has frequently been associated with cancer development and progression ( Yang et al., 2008 ; Zuber et al., 2011 ; Nagarajan et al., 2014 ; Wu et al., 2015 ).
Chromatin reporter cell lines have been already developed in order to identify modulators of position effect variegation ( Tchasovnikarova et al., 2015 ) or to discover new chromatin-targeting compound ( Johnson et al., 2008 ; Best et al., 2011 ; Wang et al., 2013 ). In contrast to previous approaches, we wanted to develop a protocol for the generation of a reporter cell line able to monitor the BRD4-dependent heterochromatization of a generic reporter. To achieve that, we used a common retroviral vector carrying an RFP (Red Fluorescent Protein) gene, and selected clones that integrated it in fully repressed genomic regions specifically reactivated by BRD4 inhibition. The haploid nature of the cell line used (KBM7 [ Andersson et al., 1995 ]), makes the reporter easily amenable not only to chemical screens, but also to genetic screens. Both methods can be used for the identification of new BRD4 direct and functional partners, and results from these approaches will provide further insights into BRD4 biology.
Materials and Reagents
Pipette tips
6-well plates, tissue culture treated (Corning, Costar®, catalog number: 3506)
15 ml Falcon® conical centrifuge tube (Corning, Falcon®, catalog number: 352196)
0.45 μm syringe filters (VWR, catalog number: 514-8021)
24-well plates, tissue culture treated (STARLAB INTERNATIONAL, catalog number: CC7682-7524)
96-well plates, tissue culture treated (Corning, catalog number: 3598)
Viewplate-96 black, optically clear bottom, tissue culture treated, sterile, 96-Well with lid (PerkinElmer, catalog number: 6005182)
10 cm plates, tissue culture treated (Corning, catalog number: 430167)
293T cell line (ATCC, catalog number: CRL-3216)
KBM7 cell line (Chronic Myeloid Leukaemia) (Horizon Discovery, catalog number: C628)
Fluorescent reporter vector (LZRS-RFP-ires-ZEO retroviral vector, a gift from S. Nijman Lab, Ludwig Cancer Research, Oxford)
Packaging vector (e.g., pCMV-Gag-Pol retroviral vector, Addgene, catalog number: 14887; pCMV-VSV-G envelope vector, Addgene, catalog number: 8454)
DMEM media (Thermo Fisher Scientific, GibcoTM, catalog number: 41965039)
Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10500064)
Lipofectamine 2000 (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11668019)
Opti-MEM
IMDM media (Thermo Fisher Scientific, GibcoTM, catalog number: 21980032)
Phosphate-buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14190094)
(S)-JQ1 (MedChemExpress, catalog number: HY-13030)
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418)
Equipment
Pipettes (Gilson)
Cell culture centrifuge (Sigma Laborzentrifugen, model: 3-18K, catalog number: 10290)
FACS (BD, BD Bioscience, model: BD FACSCALIBUR)
FACS sorter (BD, BD Biosciences, model: BD FACSAria)
Fluorescence microscope (PerkinElmer, model: HH12000000)
Cell culture hood (Thermo Fisher Scientific, model: HerasafeTM KS, catalog number: 51022515)
Cell culture incubator (Eppendorf, model: Galaxy® 170 R, catalog number: CO170R-230-1000)
Procedure
-
Virus preparation
293T cells are seeded at 200,000 cells/ml and grown until 60% confluence in DMEM plus 10% FBS in 6-well plates. When they reach such confluence rate (usually the day after), they are transfected using Lipofectamine 2000, according to manufacturer’s instructions, in order to produce the virus needed for the reporter generation (see details below). In order to get optimal virus production, it is important to use 293T cells at low passage number, and which have never been overconfluent.
For the LZRS-RFP-ires-ZEO retrovirus production, 1.5 μg of LZRS-RFP-ires-ZEO retroviral vector were used in combination with 1.5 μg of packaging vectors (pCMV-Gag-Pol retroviral vector 8:1 pCMV-VSV-G envelope vector). Briefly (for 1 well), vectors are diluted and gently mixed in 250 μl of Opti-MEM. 5 μl of Lipofectamine 2000 are diluted in 250 μl of Opti-MEM and incubate for 5 min at RT (room temperature). After the 5-min incubation, the vector dilution and the Lipofectamine 2000 dilution are combined and gently mixed. This suspension is incubated for 20 min at RT and then added to the cell media drop by drop. Finally, the plate is mixed gently by rocking back and forth in order to distribute homogenously the transfection solution.
-
Virus harvest is done at 30 and 48 h post transfection by collecting the media, centrifuging it in 15 ml Falcon conical centrifuge tubes at 1,200 × g for 5 min at RT and then filtering it through 0.45 μm syringe filters into new 15 ml Falcon conical centrifuge tubes (RT).
Note: At this point the virus can be used for titration and cell infection (steps 2 and 3) or stored at -80 °C.
-
Virus titration
KBM7 cells are seeded in a 6-well plate in IMDM plus 10% FBS at 2 x 106 cells/ml (1 ml/well), and then infected using different ratios of media/virus. The virus is resuspended in 1 ml of IMDM plus 10% FBS. In this way, the final cell concentration is of 106 cells/ml.
The ratio typically tested were: 1/100, 1/40, 1/20, 1/10, 1/4 and 1/2 (virus volume vs. cell volume).
5 μg/ml (final concentration) of Polybrene is added to each well by pipetting it in the cell media, once the virus has been added (Polybrene stock solution is 5 mg/ml).
24 and 48 h post infection cells are washed in PBS and the media is replaced with fresh aliquots (2 ml/well). Cells of each well are resuspended once in 5 ml of PBS (RT) and then centrifuged at 1,200 × g for 5 min at RT.
-
48 h post infection, FACS analysis is performed in order to check the percentage of infection of each well/ratio. A concentration of virus giving between 25% and 40% of cell infection (25-40% of RFP positive cells) is chosen to be used for the reporter generation (therefore ensuring that likely no more than one infection event could happen per cell). LZRS-RFP-ires-ZEO infection.
KBM7 cells are seeded in 6-well plates in IMDM plus 10% FBS at 106 cells/ml (30 x 106 cells are used in total).
KBM7 cells are treated with 0.5 μM (S)-JQ1 for 18 h and then infected with the LZRS-RFP-ires-ZEO retrovirus (using the virus concentration determined in step 2).
Media is changed after 24 and 48 h post infection; (S)-JQ1 is kept in the culture media at 0.5 μM.
-
First sorting (pool)
RFP-positive cells are sorted in presence of 0.5 μM (S)-JQ1 (the whole population is sorted: cells number is at this stage approximately 120 x 106).
-
Second sorting (single cells)
(S)-JQ1 is removed from the media and after 30 h the RFP negative population is sorted into single cell clones. Approximately 10 96-well plates are filled; the rest of cell is frozen in IMDM media plus 10% FBS and 10% DMSO.
Note: Usually for the sorting procedure cells must be washed at least once in 5 ml of PBS (RT), centrifuged at 1,200 x g for 5 min at RT and resuspended in 0.5 ml of PBS (maximum cell concentration allowed is 30 x 106 cell/ml); we recommend to add 10% of cell media to the resuspension in order to avoid cell-cluster formation, especially during the first sorting.
-
Single clone amplification and reporter selection
Outgrowing clones (about 10 days after step 5) are amplified and frozen in aliquots of 10 x 106 cells in IMDM media plus 10% FBS and 10% DMSO. 20,000 cells of each clone are seeded in 96-well plates (Viewplate-96 black, optically clear bottom) and treated with (S)-JQ1 for 24 h.
Cells are imaged using the Operetta high-content imaging system (20x objective and non-confocal mode) (Figure 1). A clone is selected as reporter if after (S)-JQ1 treatment is able to express RFP.
Figure 1. (S)-JQ1 treatment activates RFP expression in KBM7 reporter cells.
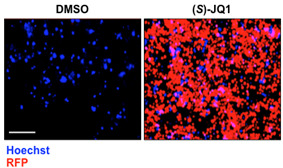
Examples of live cell imaging pictures of KBM7 reporter cells treated with 0.5 μM of (S)-JQ1 for 24 h; equal amount of DMSO was added as control (scale bar = 100 μm).
Notes
All the cells used in this protocol were grown in a cell culture incubator at 37 °C and 5% CO2.
KBM7 freezing media: IMDM, 20% FCS, 10% DMSO. Cells are gradually frozen at -80 °C and then passed to liquid nitrogen.
LZRS-RFP-ires-ZEO vector can be replaced with any other fluorescent reporter vector, either retro or lentiviral.
(S)-JQ1 treatment can be replaced with any other epigenetic or not epigenetic drug treatment in order to generate reporters for the specific enzyme of interest.
KBM7 cells can be replaced with any other cell line of interest. We have chosen this cell line because they are haploid and therefore allows unambiguous monoallelic genetic configurations.
Acknowledgments
Research in the Kubicek laboratory is supported by the Austrian Federal Ministry of Science, Research and Economy; the National Foundation for Research, Technology, and Development; the Marie Curie Career Integration Grant EPICAL; and the JDRF. Sara Sdelci acknowledges support by JDRF postdoctoral fellowship 3-PDF-2014-206-A-N ‘Reprogramming by Loss of Function’.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Andersson B. S., Collins V. P., Kurzrock R., Larkin D. W., Childs C., Ost A., Cork A., Trujillo J. M., Freireich E. J., Siciliano M. J. and et al.(1995). KBM-7, a human myeloid leukemia cell line with double Philadelphia chromosomes lacking normal c-ABL and BCR transcripts. Leukemia 9(12): 2100-2108. [PubMed] [Google Scholar]
- 2.Best A. M., Chang J., Dull A. B., Beutler J. A. and Martinez E. D.(2011). Identification of four potential epigenetic modulators from the NCI structural diversity library using a cell-based assay. J Biomed Biotechnol 2011: 868095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devaiah B. N. and Singer D. S.(2013). Two faces of BRD4: mitotic bookmark and transcriptional lynchpin. Transcription 4(1): 13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dey A., Chitsaz F., Abbasi A., Misteli T. and Ozato K.(2003). The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A 100(15): 8758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J. P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Muller S., Pawson T., Gingras A. C., Arrowsmith C. H. and Knapp S.(2012). Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149(1): 214-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Floyd S. R., Pacold M. E., Huang Q., Clarke S. M., Lam F. C., Cannell I. G., Bryson B. D., Rameseder J., Lee M. J., Blake E. J., Fydrych A., Ho R., Greenberger B. A., Chen G. C., Maffa A., Del Rosario A. M., Root D. E., Carpenter A. E., Hahn W. C., Sabatini D. M., Chen C. C., White F. M., Bradner J. E. and Yaffe M. B.(2013). The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 498(7453): 246-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson R. L., Huang W., Jadhav A., Austin C. P., Inglese J. and Martinez E. D.(2008). A quantitative high-throughput screen identifies potential epigenetic modulators of gene expression. Anal Biochem 375(2): 237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagarajan S., Hossan T., Alawi M., Najafova Z., Indenbirken D., Bedi U., Taipaleenmaki H., Ben-Batalla I., Scheller M., Loges S., Knapp S., Hesse E., Chiang C. M., Grundhoff A. and Johnsen S. A.(2014). Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep 8(2): 460-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tchasovnikarova I. A., Timms R. T., Matheson N. J., Wals K., Antrobus R., Gottgens B., Dougan G., Dawson M. A. and Lehner P. J.(2015). GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 348(6242): 1481-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L., Chang J., Varghese D., Dellinger M., Kumar S., Best A. M., Ruiz J., Bruick R., Pena-Llopis S., Xu J., Babinski D. J., Frantz D. E., Brekken R. A., Quinn A. M., Simeonov A., Easmon J. and Martinez E. D.(2013). A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat Commun 4: 2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R., Li Q., Helfer C. M., Jiao J. and You J.(2012). Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J Biol Chem 287(14): 10738-10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu S. Y. and Chiang C. M.(2007). The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem 282(18): 13141-13145. [DOI] [PubMed] [Google Scholar]
- 13.Wu T., Pinto H. B., Kamikawa Y. F. and Donohoe M. E.(2015). The BET family member BRD4 interacts with OCT4 and regulates pluripotency gene expression. Stem Cell Reports 4(3): 390-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z., He N. and Zhou Q.(2008). Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol 28(3): 967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuber J., Shi J., Wang E., Rappaport A. R., Herrmann H., Sison E. A., Magoon D., Qi J., Blatt K., Wunderlich M., Taylor M. J., Johns C., Chicas A., Mulloy J. C., Kogan S. C., Brown P., Valent P., Bradner J. E., Lowe S. W. and Vakoc C. R.(2011). RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478(7370): 524-528. [DOI] [PMC free article] [PubMed] [Google Scholar]


