Abstract
The COVID-19 pandemic has paralyzed the global economy and resulted in millions of deaths globally. People with co-morbidities like obesity, diabetes and hypertension are at an increased risk for severe COVID-19 illness. This is of overwhelming concern because 42% of Americans are obese, 30% are pre-diabetic and 9.4% have clinical diabetes. Here, we investigated the effect of obesity on disease severity following SARS-CoV-2 infection using a well-established mouse model of diet-induced obesity. Diet-induced obese and lean control C57BL/6 N mice, transduced for ACE2 expression using replication-defective adenovirus, were infected with SARS-CoV-2, and monitored for lung pathology, viral titers, and cytokine expression. No significant differences in tissue pathology or viral replication was observed between AdV transduced lean and obese groups, infected with SARS-CoV-2, but certain cytokines were expressed more significantly in infected obese mice compared to the lean ones. Notably, significant weight loss was observed in obese mice treated with the adenovirus vector, independent of SARS-CoV-2 infection, suggesting an obesity-dependent morbidity induced by the vector. These data indicate that the adenovirus-transduced mouse model of SARS-CoV-2 infection, as described here and elsewhere, may be inappropriate for nutrition studies.
Keywords: COVID-19, Diet-induced obesity, hACE2, Replication-defective adenovirus, Tissue, Pathology
1. Introduction
Coronavirus disease-2019 (COVID-19) is the third pandemic in the 21st century caused by a novel coronavirus, after severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 (Lee et al., 2003; Peiris et al., 2003; Jia et al., 2005; de Wit et al., 2016) and the Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 (Alagaili et al., 2014; Memish et al., 2013; Zaki et al., 2012). COVID-19 is responsible for ∼191 million confirmed cases, with over 4 million deaths globally, and ∼33 million confirmed cases and ∼600,000 deaths in the U.S. alone, as of July 21st, 2021 (Available from: https://covid19.who.int/region/amro/country/us). COVID-19 is characterized by fever and respiratory symptoms, which can progress to more severe and fatal disease (Guan et al., 2020). Infection with SARS-CoV-2, the causative agent of COVID-19, is more likely to cause critical illness or deaths in the elderly, immunocompromised and individuals with co-morbidities such as obesity, diabetes, and hypertension (Zhou et al., 2020; Haimovich et al., 2020; de Lusignan et al., 2020; Petrilli et al., 2020).
Obesity and diabetes have been identified as risk factors for poor outcomes in other major pandemics involving respiratory tract infections like MERS (Hui et al., 2018) and influenza (Van Kerkhove et al., 2011; O'Brien et al., 2012; Cocoros et al., 2014). Like COVID-19, obesity too has become a pandemic, affecting 12.5% of people globally and approximately 42% of Americans (National diabetes statist, 2020). Obesity is the most frequently observed component of metabolic syndrome, a condition that clusters abdominal obesity, dyslipidemia (abnormally high blood lipid levels), hyperglycemia (increased blood sugar levels) and hypertension (increased blood pressure) (Engin, 2017). Experiments using diet-induced obese (DIO) mice to study the pathogenesis of influenza A virus have shown not only are obese mice more susceptible to infection (Smith et al., 2007) but that they also promote the emergence of more virulent strains of the virus (Honce et al., 2020). To study the impact of obesity on SARS-CoV-2 pathogenesis and evolution, a similar mouse model is required; however, mice are not susceptible to infection with SARS-CoV-2 due to the lack of human angiotensin converting enzyme 2 (hACE2) receptor, required for entry of the virus (Letko et al., 2020; Wan et al., 2020). Several strategies have been developed to circumvent this issue: transgenic mice expressing hACE2 (hACE2-Tg) (McCray et al., 2007; Yang et al., 2007; Yoshikawa et al., 2009; Bao et al., 2020), mouse-adapted SARS-CoV-2 strains (Dinnon et al., 2020; Leist et al., 2020), and transduction with adenovirus vectors expressing hACE2 (AdV-hACE2) (Hassan et al., 2020; Sun et al., 2020). Recently it was shown that the SARS-CoV-2 variants containing the N501Y spike mutation could naturally use mouse ACE2 receptors for entry (Rathnasinghe et al., 2021), making this another suitable model for SARS-CoV-2 studies. The AdV-hACE2 approach has several advantages: it can be used with different strains of mice, is easy to manipulate, causes little pathology to wild-type (WT) mice, and allows for the use of WT SARS-CoV-2 (Hassan et al., 2020; Sun et al., 2020).
In this study, we sought to investigate the effect of obesity on disease outcome following SARS-CoV-2 infection using an adenovirus-transduced mouse model. Therefore, for our studies we used DIO and lean mice that were transduced with AdV-hACE2 followed by SARS-CoV-2 infection and monitored for morbidity, mortality, virus replication and cytokine expression. Surprisingly, obese, but not lean, mice inoculated with the replication-defective AdV-hACE2 lost significant weight independent of SARS-CoV-2 infection, suggesting an obesity-dependent morbidity induced by the AdV-hACE2 vector. SARS-CoV-2 replication occurred in the lungs of both lean and obese mice, though, we did not observe differences in terms of disease outcomes, tissue pathology, or viral replication. Accordingly, due to the confounding influence of AdV-hACE2 on SARS-CoV-2 infection, we could not determine obesity's impact on SARS-CoV-2 infection. These data underscore the importance of selecting an appropriate model for nutrition studies and suggest that researchers should exercise caution when using the AdV-hACE2 model described here and elsewhere to study SARS-CoV-2. Notably, it is possible that with optimization, for example reducing the titer of the vector or using other strains of mice, the AdV-hACE2 model may be appropriate. Further studies using transgenic mice, mouse-adapted strains (Dinnon et al., 2020; Leist et al., 2020), or other animals such as hamsters (Sia et al., 2020; Lee et al., 2020) or ferrets (Kim et al., 2020; Kutter et al., 2021) should also be conducted to explore the relationship between COVID-19 and metabolic co-morbidities.
2. Materials and methods
2.1. Cells and viruses
Vero E6 cells were obtained from American Type Culture Collection (ATCC; CRL-1586) and were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 5% fetal bovine serum (FBS), 1 mg/mL gentamicin, 1% non-essential amino acids (NEAA) and 25 mM HEPES buffer in an incubator at 37 °C with 5% CO2.
The replication-defective adenovirus encoding human ACE2 (AdV-hACE2) was originally prepared by the University of Iowa and Washington University in St. Louis, as reported previously (Jia et al., 2005). The viral stocks were propagated using 293A cells (ThermoFisher Catalog #R70507) and purified by cesium chloride gradient centrifugation prior to titering, also in 293A cells, as previously described (Calhoun et al., 2020). SARS-CoV-2 strain 2019 n-CoV/USA_WA1/2020 was obtained from BEI Resources (Catalog # NR-52281) (Rogers et al., 2020) and was propagated in Vero E6 cells to prepare stocks; infectious titers were determined by plaque assays on the same cell line.
2.2. Mice and diets
C57BL/6 N male and female mice were obtained from Charles River Laboratories at three to four weeks of age and allowed to acclimatize for a week before initiating diets. Mice were housed in groups of five per cage and maintained at ambient temperature with ad libitum supply of food and water, except for overnight food deprivation (12–14 h) prior to blood glucose measurements. All animal handling protocols were approved by the Institutional Animal Care and Use Committee (Protocol #20–060) at Virginia Tech.
All diets used for the study were obtained from Research Diets (New Brunswick, NJ, USA). Twenty mice (10 males and 10 females) were placed on a low-fat diet with 10% kcal fat (LFD; D12450K) and 20 (10 males and 10 females) on a high-fat diet with 60% kcal fat (HFD; D12492). Throughout the manuscript, we will refer to the groups as follows: lean (low fat diet) or obese (high fat diet). The mice were kept on these diets for 18–20 weeks before infections, and the same diets were continued until the end of the experiment. Table S1 displays the caloric information of these diets.
2.3. Fasting glucose levels
Two weeks prior to transduction by the adenovirus (16 weeks after diet initiation), mice were deprived of food overnight (12–14 h), blood was collected by submandibular bleed and glucose levels were measured by Abox glucose monitoring kit.
2.4. Mouse infections
Mice were moved to a BSL-3 facility 24 h prior to SARS-CoV-2 infections. For primary infections with the AdV-hACE2, mice were anesthetized with ketamine and xylazine (90 mg/kg and 5 mg/kg respectively) intraperitoneally and then inoculated intranasally with 108 plaque-forming units (PFUs) of the virus in 50 μL of Roswell Park Memorial Institute (RPMI)-1640 media with no additives. Five days post-adenovirus inoculation, the mice received 105 PFUs of SARS-CoV-2 intranasally under ketamine and xylazine anesthesia, or RPMI-1640 media alone for mock-infected groups. Six infected and four control mice from each group were sacrificed at days four and ten after SARS-CoV-2 infection. Mice were weighed daily post-SARS-CoV-2 infection and monitored for visual symptoms of the disease. Blood was collected via submandibular bleeds (∼200 μL per mouse), serum was collected via centrifugation at 5000×g for 10 min, transferred to fresh tubes and stored at −80 °C.
2.5. Histopathology and organ titration
Tissues for viral titration were harvested aseptically in 2 mL tubes containing a 5 mm stainless steel bead, RPMI-1640, 10 mM HEPES and 1% FBS (herein referred to as the “tissue diluent”), to a final concentration of 10% weight by volume. Tissues were homogenized in a Tissuelyser II (Qiagen) at 30 cycles per second for 2 min and then centrifuged at 5000×g for 10 min. Plaque assays were performed on the clarified supernatant. Briefly, Vero E6 cells at 100% confluency were inoculated with 50 μL of the serially diluted samples and incubated at 37 °C in 5% CO2 for 1 h. Overlay media containing 0.6% tragacanth gum, 1x MEM (minimum essential media), 20 mM HEPES and 4% FBS were then added, and the plates were allowed to incubate for two days for plaque formation.
For histopathology, the tissues harvested from mice were fixed in 4% paraformaldehyde for at least one week. The Virginia Tech Animal Laboratory Services (ViTALS) performed paraffin embedding, sectioning and hematoxylin-eosin staining, and a board-certified pathologist scored the slides in a blinded manner.
2.6. RNA extraction
A section of the infected lung tissues (4 dpi) was collected in 0.5 mL of TRIzol LS reagent (ThermoFisher) in 2 mL tubes with a 5 mm bead. Lung tissues were homogenized using a TissueLyser II (Qiagen) at 30 cycles per second for 2 min and stored at −80 °C. Samples were thawed and total RNA was extracted using the manufacturer's protocol for TRIzol extraction.
2.7. Reverse transcription quantitative PCR (RT-qPCR)
To quantify SARS-CoV-2 genomes, the 2019-nCoV RUO kit from Integrated DNA Technologies (IDT, Leuven, Belgium) was utilized. N2 combined primer-probe mix from the kit was used (Table S2) with Quantabio qScript XLT-One-Step RT-qPCR ToughMix (2X). The Bio-Rad CFX-96 (Hercules, CA, USA) was used for RT-qPCRs with the following conditions: 50 °C for 10 min for reverse transcription, 95 °C for 3 min for initial denaturation and polymerase activation, followed by 45 cycles of 95 °C for 10 s for denaturation, and 60 °C for 30 s for annealing/extension. For generating the standard curve, we used N gene RNA generated by in vitro transcription and used ten-fold serial dilutions to the point where no genome was detectable by qPCR. The virus concentration was calculated by fitting the Cq values of the samples to the standard curve and expressed in terms of N-gene copies/mL.
RT-qPCR was also performed on RNA extracted from the 4 dpi lung samples with NEB Luna Universal One-Step RT-qPCR Kit with SYBR-Green (NEB, Ipswich, MA, USA) to quantify cytokines. Primers were obtained from IDT and are listed in Table S2. The conditions for the reactions in a Bio-Rad CFX-96 were: 50 °C for 10 min for reverse transcription, 95 °C for 1 min for initial denaturation and polymerase activation, followed by 45 cycles of 95 °C for 10 s for denaturation, and 60 °C for 30 s for annealing/extension, followed by a melt curve. The samples were normalized with mouse GAPDH as the reference gene with respect to mock-infected groups (from the corresponding diet) as a control. The fold change between the SARS-CoV-2 infected lean and obese mice was calculated using the Delta Delta Ct (ΔΔCt) method of relative quantification (Livak and Schmittgen, 2001).
2.8. Statistical analysis
Statistical analysis was done using GraphPad Prism 9 (GraphPad Software, San Diego, CA, USA). Details of the statistical technique used for analyses are provided in the figure legends. The level of significance has been determined by the following p-values: p = 0.1234 (ns); p = 0.0332 (*), p = 0.0021(**), p = 0.0002 (***), p < 0.0001 (****). The error bars represent standard deviation (SD) from the mean, in all the figures and the dotted lines denote the limit of detection (L.O.D.). All statistical analyses were performed on data after testing them for normality using the Shapiro-Wilk test.
3. Results and discussion
3.1. Wild type C57BL/6 N mice fed a high fat diet had increased weight gain and blood glucose levels
The proportion of COVID-19 patients requiring intensive care was previously reported to be directly proportional to the body-mass index (BMI), being highest in patients with BMI ≥35 (morbidly obese) (Simonnet et al., 2020). Obesity has also been correlated with severe outcomes during the H1N1 influenza A epidemic (Cocoros et al., 2014; Bassetti et al., 2011; Balanzat et al., 2012; Huttunen and Syrjänen, 2013). Furthermore, DIO mice had significantly higher mortality rates when infected with influenza A virus (Smith et al., 2007) and more severe disease following chikungunya virus (CHIKV) (Weger-Lucarelli et al., 2019) and dengue virus (DENV) infection (Chuong et al., 2020). We sought to use a similar DIO mouse model to investigate whether obese mice infected with SARS-CoV-2 exhibit worse disease outcomes as compared to their lean counterparts.
We observed a significant weight gain in high-fat diet (HFD)-fed mice compared to the low-fat diet (LFD)-fed groups starting 7 weeks after the initiation of diets (p-value <0.0001) (Fig. 1 A). Weights were analyzed on the combined data for males and females on LFD and HFD. Separate analysis for males and females is presented in Supplementary Fig. S1. To assess other metabolic parameters, we measured overnight fasting blood glucose levels after 9 and 16 weeks of the initiation of diets. We tested only 10 mice per group at 9 weeks to reduce stress on the mice. Hyperglycemia, defined as overnight fasting glucose levels of ≥150 mg/dL, was not significantly different between the lean and obese groups at 9 weeks (p-value = 0.2344) (Fig. 1B). By 16 weeks, however, there was a significant difference between HFD and LFD groups (p-value = 0.0001) (Fig. 1C). This 15 to 20-week diet regimen produced a mouse model that mirrored obesity and hyperglycemia in humans and, therefore, was appropriate for infection studies.
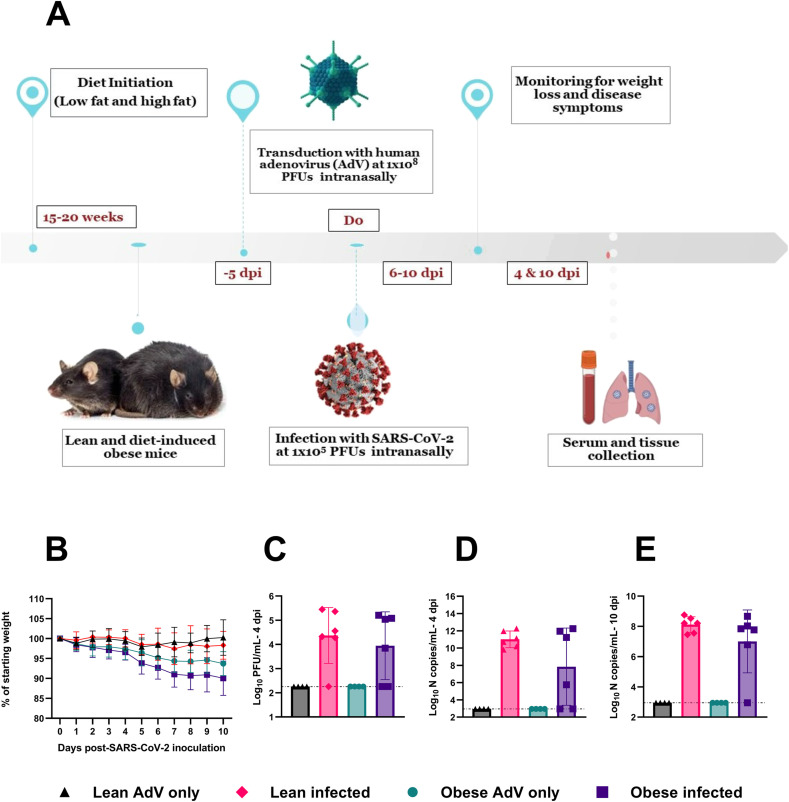
Fig. 1.
Metabolic characterization of mice fed low fat (lean) and high fat (obese) diets.
A) Four-week-old C57BL/6 N mice (20/group) were fed a high-fat diet (60% fat) or a low-fat diet (10% fat) for 15–20 weeks. Weights were measured weekly after initiating the diets (****p < 0.0001, 2way-ANOVA with Sidak's multiple comparisons test). B) Blood glucose levels were measured following 9 weeks of feeding (n = 10) (p = 0.2344, Mann-Whitney test). C) Glucose levels were measured after 16 weeks of feeding (n = 20) (***p = 0.0001, Mann-Whitney test). The error bars indicate standard deviation (SD) from mean.
3.2. Adenovirus-transduction caused morbidity in obese mice, independent of SARS-CoV-2 infection
Since mouse ACE2 receptors do not allow for WT SARS-CoV-2 infection, we first transduced mice with a replication-defective adenovirus encoding for human ACE2 (AdV-hACE2) intranasally to achieve hACE2 expression in the lungs and render mice susceptible to infection. We then inoculated the mice with SARS-CoV-2 or diluent five days post-transduction, as previously described (Hassan et al., 2020; Sun et al., 2020) (Fig. 2 A). Surprisingly, the obese mice inoculated only with AdV-hACE2 showed significant weight loss, similar to those infected with both AdV-hACE2 and SARS-CoV-2 (Fig. 2B). No weight loss was observed in lean mice treated with AdV-hACE2 or AdV-hACE2 and SARS-CoV-2. We found a significant weight loss in obese-AdV only mice compared to the lean-AdV only group starting from days 5 (p-value = 0.0010) through day 9 post-AdV transduction (p-value = 0.0242). The weights of the mice starting from the day of AdV inoculation in both percent weight loss and grams are presented in Supplementary Fig. S2. Collectively, these data suggested that the AdV-hACE2 vector at the dose used here, produced morbidity in obese mice. However, we proceeded to test other parameters to determine if differences could be found between lean and obese mice in terms of viral replication, immune response, or lung pathology characteristic of SARS-CoV-2 infection.
Fig. 2.
Timeline of the study and disease progression in lean and obese mice post-SARS-CoV-2 infection.
To measure viral replication, a subset of the mice was euthanized on days 4 and 10 post-SARS-CoV-2 inoculation and infectious virus was quantified in the lung homogenates. As expected, mice receiving only AdV-hACE2 tested negative for SARS-CoV-2 (Fig. 2C). Infectious lung viral titers between lean and obese infected groups were similar (p-value = 0.7377) at 4 days post-SARS-CoV-2 infection, and 1 lean and 2 obese mice were negative for infectious virus. We next used RT-qPCR against the N-gene as a more sensitive means to detect viral RNA. At 4 days post-SARS-CoV-2 inoculation, all lean mice were positive for the N-gene, while both infectious-virus-negative obese mice were negative, likely indicating a lack of productive infection. In Fig. 2E, we present RT-qPCR data for the N-gene for lungs collected at 10 days post-SARS-CoV-2 infection; all 6 lean mice and 5 of 6 obese mice tested positive for SARS-CoV-2 N-gene, and all the mice were negative for infectious virus tested by plaque assays (data not shown). No differences were observed between lean and obese groups in N-gene copies/mL at either timepoint. This finding is consistent with previous studies that found no significant difference in viral titers at peak viral replication between lean and obese mice infected with influenza virus (Smith et al., 2007), several alphaviruses (Weger-Lucarelli et al., 2019) or dengue virus (Chuong et al., 2020).
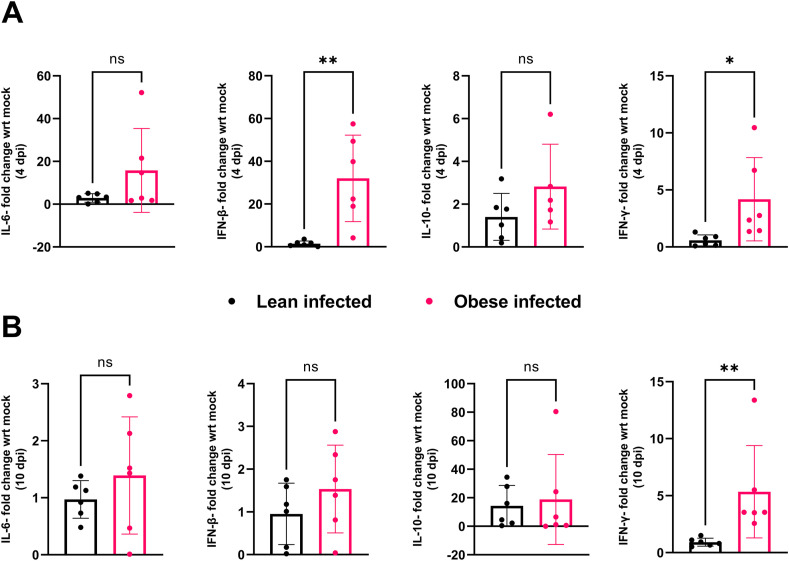
Severe COVID-19 disease is characterized by low levels of type I interferons (IFN-α and β) and over-production of inflammatory cytokines like interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α) (Hadjadj et al., 2020; Huang et al., 2020; Mehta et al., 2020; Wen et al., 2020; Chua et al., 2020). Moreover, invasive mechanical ventilation (IMV) was associated with severe obesity in patients infected with SARS-CoV-2, irrespective of age, sex, diabetes, and hypertension (Simonnet et al., 2020). Experiments using obese mice to study the pathogenesis of influenza A virus, another important respiratory virus, showed reduced expression of type I interferons (IFNs), delayed expression of pro-inflammatory cytokines and chemokines (Smith et al., 2007), severe lung pathology, and increased viral spread, leading to increased mortality (O'Brien et al., 2012). We thus sought to determine the contribution of obesity to the expression of several important cytokines and lung pathology following SARS-CoV-2 infection. For cytokine testing, RNA was extracted from lungs collected 4 days post-SARS-CoV-2 infection, and the expression of IL-6, IL-10, IFN-β and IFNγ was measured by RT-qPCR. The relative expression of these cytokines was calculated by the ΔΔCt method, with hACE2-AdV infected groups as control and mouse GAPDH as the reference gene. We selected these cytokines since they have previously been identified as predictors of severe COVID-19 disease outcome (Han et al., 2020; Bastard et al., 2020; Gadotti et al., 2020). We observed significantly higher expression of IFN-β (**p-value = 0.0042) and IFNγ (*p-value = 0.0376) in SARS-CoV-2 infected obese mice compared to their lean infected counterparts at 4 dpi (Fig. 3 A). At 10 dpi, there was no significant difference in the expression of IL-6, IL-10 and IFN-β, but IFNγ was significantly upregulated in obese mice infected with SARS-CoV-2 compared to lean infected mice (**p-value = 0.0022) (Fig. 3B). Previous studies have shown IFN-β and IFNγ to be potent inhibitors of SARS-CoV-2 (Busnadiego et al., 2020), which might explain why some obese mice did not become infected following SARS-CoV-2 infection.
Fig. 3.
Cytokine mRNA expression.
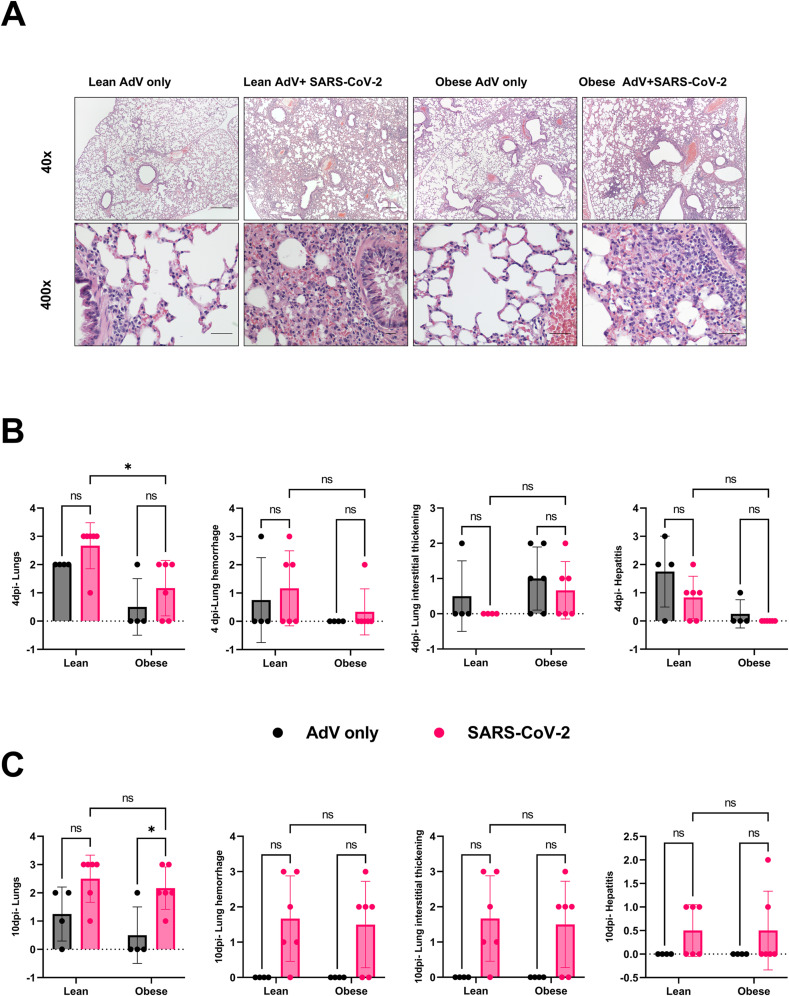
We also examined lungs, liver, and heart for histopathological lesions. Lungs were evaluated and scored based on evidence of interstitial inflammation, intra-alveolar hemorrhage, and peribronchiolar and perivascular lymphoid hyperplasia. There was no evidence of interstitial or alveolar septal necrosis or hyaline membrane formation. Livers were evaluated for evidence of lipidosis and single cell necrosis. The hearts were evaluated for evidence of myocardial necrosis and inflammation. Representative lung images collected at 4 dpi are presented in Fig. 4 A. The lung histopathology scores were similar for obese mice inoculated with both AdV-hACE2 and SARS-CoV-2 compared to their AdV-hACE2 only transduced counterparts at 4 dpi (Fig. 4B) but trended towards being more severe for the SARS-CoV-2 infected mice at 10 dpi (Fig. 4C). Lung pathology scores were lower for SARS-CoV-2 infected lean mice compared to their obese counterparts at 4 dpi (p-value = 0.03) but were similar at 10 dpi (p-value>0.9999). We hypothesize that the early difference was due to IFN-β production in infected obese mice controlling replication, and thus disease, but that lower IFN-β later in infection enabled SARS-CoV-2 replication and produced the resulting lung pathology. No differences in the histopathology scores for heart and liver were observed between the groups. Taken together, no major differences were observed between the two groups in tissue pathology; however, these data are likely confounded by the morbidity observed in AdV-hACE2 alone treated obese mice.
Fig. 4.
Tissue histopathology post-SARS-CoV-2 infection.
The mechanisms underlying the morbidity observed in AdV-hACE2 transduced obese mice are currently unclear. Future work using a control or empty AdV will enable parsing out of vector-related mechanisms vs. ectopic expression of hACE2 will be informative. Replication-defective adenoviruses have been used in many gene therapy applications (Crystal, 1995; Thacker et al., 2009) and are known to activate both innate and adaptive immune responses in humans (Juillard et al., 1995; Yang et al., 1995; Adesanya et al., 1996; Worgall et al., 1997). Furthermore, they have been shown to cause hepatotoxicity in mice due to induction of TNF-α in the liver (Engler et al., 2004; Varnavski et al., 2005) and dose-dependent morbidity in non-human primates (Schnell et al., 2001). Accordingly, we posit that obese hosts may be more permissive to disease following transduction with this dose of AdV-hACE2, possibly due to an overzealous immune response to the vector and SARS-CoV-2, as shown by increased interferon gene expression. The data presented here suggest some caution should be taken when using adenovirus vectors in broader populations. Notably, the race for developing a vaccine against SARS-CoV-2 at pandemic speed (Lurie et al., 2020) has renewed the interest of researchers in using adenovirus vectors, which makes our findings worth considering.
3.3. Limitations of the study
The major limitation of our study was the use of the adenovirus vector for transduction of mice with the hACE2 receptor. Inoculation of obese mice with adenovirus vector at this dose caused weight loss, confounding our ability to assess obesity's impact on SARS-CoV-2 infection in mice, the main objective of our study. We did not test a lower dose of the AdV-hACE2 vector, however, which may have provided enough ACE2 expression without any off-target effects. Another limitation of this study was the lack of a true mock-treated group, with no AdV-hACE2 or SARS-CoV-2 infection or an AdV with a control transgene, in the absence of which we cannot quantify the impact of the adenovirus vector versus hACE2 transgene on the obese host. Moreover, these differences could be specific to C57BL/6 N mice and we might observe different results with other mouse strains; though, we were constrained to use C57BL/6 N mice as they are sensitive to DIO and are an ideal model for human obesity (West et al., 1992). Another major limitation was not testing a wide range of cytokines; thus, we cannot make generalized claims about inflammation. Despite these constraints, this study provides useful information to the field relating to SARS-CoV-2 mouse models and advises caution when considering the AdV-hACE2 model as laid out here and elsewhere for nutrition studies.
4. Conclusion
The increased risk of grave illness and fatalities in obese people post-SARS-CoV-2 infection underscores the importance of having an effective animal model to study the mechanisms underlying worsened disease outcomes. Here, we generated DIO mice and used a previously established adenoviral-transduction model to render them susceptible to SARS-CoV-2 infection. Our results demonstrate that although adenovirus-transduction sensitized obese and lean mice to SARS-CoV-2 infection in their lungs, it also induced morbidity in obese mice. Therefore, this model was not appropriate for studying the relationship between underlying co-morbidities like obesity or diabetes and severe COVID-19. However, optimization of the viral dose, the strain of mice used, and other parameters may reveal the utility of the AdV-hACE2 system for nutrition studies with SARS-CoV-2. Future studies could also benefit from using another mouse model, for example, hACE2 transgenic mice or a mouse-adapted strain of SARS-CoV-2.
A) 18-20-week-old male and female C57BL/6 N mice received 108 PFUs of hACE2-AdV intranasally (day −5) and then 105 PFUs SARS-CoV-2 or viral diluent intranasally (day 0). B) Weights were measured daily after SARS-CoV-2 inoculation. Percent weight change was compared to the baseline weight on the day of SARS-CoV-2 infection (2way-ANOVA). C) Infectious viral titer in the lungs of SARS-CoV-2 infected lean and obese mice was determined at 4-dpi (p-value = 0.7377, Ordinary One-way ANOVA). The dotted horizontal line denotes the assay Limit of Detection (L.O.D) set at 2.255 Log10 PFUs/mL. D-E) Viral N-copies/mL from RNA extracted from lungs of mice at 4-dpi (D) and 10-dpi (E) measured by RT-qPCR (p-value at 4-dpi = 0.8241; p-value at 10-dpi = 0.6052, Ordinary One-way ANOVA). N copies and L.O.D. were calculated using the standard curve generated from N-gene RNA standard. The dotted horizontal line denotes the L.O.D. set at 2.947 Log10 N copies/mL. The study was conducted as two independent experiments with n = 6 for SARS-CoV-2 infected groups and n = 4 for only AdV-transduced controls. From study 1, two mice each from infected and uninfected groups were euthanized at 4-dpi and remaining at 10-dpi. In study 2, four mice from infected and two from uninfected groups were euthanized at 4-dpi and remaining at 10-dpi. Results are indicative of data combined from these duplicate independent studies.
Fold change in gene expression of cytokines, IL-6, IFN-β, IL-10 and IFN-γ at 4-days (A) and 10-days (B) post-infection, between SARS-CoV-2 infected lean and obese mice normalized to the respective AdV-only groups and GAPDH, as determined by RT-qPCR. No significant difference in the expression of IL-6 or IL-10 was observed at either 4 or 10-dpi. Fold change in between infected lean and obese mice was compared at 4-dpi (**p-value for IFN-β = 0.0042; *p-value for IFN-γ = 0.0376, unpaired t-test) and 10-dpi (**p-value for IFN-β = 0.2824, unpaired t-test; **p-value for IFN-γ = 0.0022, Mann-Whitney test). The study was conducted as two independent experiments with n = 6 for SARS-CoV-2 infected groups and n = 4 for only AdV-transduced controls. From study 1, two mice each from infected and uninfected groups were euthanized at 4-dpi and remaining at 10-dpi. In study 2, four mice from infected and two from uninfected groups were euthanized at 4-dpi and remaining at 10-dpi. Results are indicative of data combined from these duplicate independent studies.
A) Histopathology slides of the lungs of mice transduced only with AdV-hACE2 and those infected with AdV+SARS-CoV-2 at 10 days-post-SARS-CoV-2 infection. Images at the top row show a lower magnification (40×, scale bars = 500 μm) and the bottom row shows high magnification (400×), scale bars = 50 μm). Each image is representative of a group of 6 mice for SARS-CoV-2 infected groups and 4 for AdV-only groups. B–C) Histopathology scores of lungs and liver 4 days (B) and 10 days (C) post-SARS-CoV-2 infection. The tissue pathology was compared between the AdV-only groups and the respective SARS-CoV-2 infected groups (lean-AdV vs lean SARS-CoV-2; obese-AdV vs obese SARS-CoV-2; and between lean and obese SARS-CoV-2) at 4-dpi between SARS-CoV-2 infected lean and obese mice (*p-value = 0.0308, 2way ANOVA) and 10-dpi between obese-AdV only and obese-SARS-CoV-2 groups (*p-value = 0.0404, 2W-ANOVA). The study was conducted as two independent experiments with n = 6 for SARS-CoV-2 infected groups and n = 4 for only AdV-transduced controls. From study 1, two mice each from infected and uninfected groups were euthanized at 4-dpi and remaining at 10-dpi. In study 2, four mice from infected and two from uninfected groups were euthanized at 4-dpi and remaining at 10-dpi. Results are indicative of data combined from these duplicate independent studies.
Authors’ contributions
Study concept and design: PR, JWL. Data curation: PR, CC. Formal analysis: PR, JWL. Funding acquisition: JWL. Investigation: PR, CC. Methodology: PR, CC, JWL. Drafting of the manuscript: PR, JWL. Critical revision of the manuscript: all the authors. Statistical analysis: PR, JWL. Histopathology analysis: TLR. All authors have read and approved the final manuscript.
Funding
This research was funded my an Internal Research Competition (IRC) seed grant awarded to JWL by the VA-MD College of Veterinary Medicine.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Ethics approval and consent to participate
All animal handling protocols were approved by the Institutional Animal Care and Use Committee (Protocol #20–060) at Virginia Tech. Consent to participate “not applicable” for this study.
Consent for publication
Not applicable.
CRediT authorship contribution statement
Pallavi Rai: Conceptualization, and design, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Statistical analysis, Writing – review & editing. Christina Chuong: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. Tanya LeRoith: Histopathology analysis, Critical revision of the manuscript. James W. Smyth: Critical revision of the manuscript. Julia Panov: Critical revision of the manuscript. Moshe Levi: Critical revision of the manuscript. Kylene Kehn-Hall: Critical revision of the manuscript. Nisha K. Duggal: Critical revision of the manuscript. James-Weger Lucarelli: Conceptualization, Methodology, Validation, Statistical analysis, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2021.08.014.
Abbreviations
- COVID-19
Coronavirus disease 2019
- DIO
diet-induced obesity
- ACE2
Angiotensin-converting enzyme 2
- SARS-CoV
severe acute respiratory syndrome coronavirus
- SARS-CoV-2
severe acute respiratory syndrome coronavirus-2
- MERS
Middle East respiratory syndrome coronavirus
- hACE2
human angiotensin-converting enzyme
- hACE2-Tg
transgenic mice expressing human ACE2
- AdV-hACE2
adenovirus vectors expressing hACE2
- DMEM
Dulbecco's Modified Eagle's Medium
- FBS
fetal bovine serum
- NEAA
non-essential amino acids
- LFD
low-fat diet;
- HFD
high-fat diet;
- BSL-3
biosafety level-3
- PFUs
plaque forming units
- RPMI
Roswell Park Memorial Institute
- ViTALS
Virginia Tech Animal Laboratory Services
- RT-qPCR
reverse transcription quantitative polymerase chain reaction
- IL-6
interleukin-6
- IFN-β
interferon- β;
- L.O.D.
limit of detection
- BMI
body-mass index
- CHIKV
chikungunya virus
- DENV
dengue virus
- IFNs
interferons
- TNF-α
tumor necrosis factor-α.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adesanya M.R., et al. Immediate inflammatory responses to adenovirus-mediated gene transfer in rat salivary glands. Hum. Gene Ther. 1996;7(9):1085–1093. doi: 10.1089/hum.1996.7.9-1085. [DOI] [PubMed] [Google Scholar]
- Alagaili A.N., et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio. 2014;5(2):e00884–14. doi: 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanzat A.M., et al. An analysis of 332 fatalities infected with pandemic 2009 influenza A (H1N1) in Argentina. PloS One. 2012;7(4) doi: 10.1371/journal.pone.0033670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Bassetti M., et al. Risk factors for severe complications of the novel influenza A (H1N1): analysis of patients hospitalized in Italy. Clin. Microbiol. Infect. 2011;17(2):247–250. doi: 10.1111/j.1469-0691.2010.03275.x. [DOI] [PubMed] [Google Scholar]
- Bastard P., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370(6515) doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busnadiego I., et al. Antiviral activity of type I, II, and III interferons counterbalances ACE2 inducibility and restricts SARS-CoV-2. mBio. 2020;11(5) doi: 10.1128/mBio.01928-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun P.J., et al. Adenovirus targets transcriptional and posttranslational mechanisms to limit gap junction function. Faseb. J. 2020;34(7):9694–9712. doi: 10.1096/fj.202000667R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua R.L., et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- Chuong C., et al. Nutritional status impacts dengue virus infection in mice. BMC Biol. 2020;18(1):106. doi: 10.1186/s12915-020-00828-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocoros N.M., et al. Obesity as a risk factor for severe influenza-like illness. Influenza and other respiratory viruses. 2014;8(1):25–32. doi: 10.1111/irv.12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crystal R.G. Transfer of genes to humans: early lessons and obstacles to success. Science. 1995;270(5235):404–410. doi: 10.1126/science.270.5235.404. [DOI] [PubMed] [Google Scholar]
- Dinnon K.H., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586(7830):560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin A. The definition and prevalence of obesity and metabolic syndrome. Adv. Exp. Med. Biol. 2017;960:1–17. doi: 10.1007/978-3-319-48382-5_1. [DOI] [PubMed] [Google Scholar]
- Engler H., et al. Acute hepatotoxicity of oncolytic adenoviruses in mouse models is associated with expression of wild-type E1a and induction of TNF-α. Virology. 2004;328(1):52–61. doi: 10.1016/j.virol.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Gadotti A.C., et al. IFN-γ is an independent risk factor associated with mortality in patients with moderate and severe COVID-19 infection. Virus Res. 2020;289:198171. doi: 10.1016/j.virusres.2020.198171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich A.D., et al. Development and validation of the quick COVID-19 severity index: a prognostic tool for early clinical decompensation. Ann. Emerg. Med. 2020;76(4):442–453. doi: 10.1016/j.annemergmed.2020.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microb. Infect. 2020;9(1):1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182(3):744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honce R., et al. Obesity-related microenvironment promotes emergence of virulent influenza virus strains. mBio. 2020;11(2) doi: 10.1128/mBio.03341-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S., et al. Middle East respiratory syndrome coronavirus: risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect. Dis. 2018;18(8):e217–e227. doi: 10.1016/S1473-3099(18)30127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen R., Syrjänen J. Obesity and the risk and outcome of infection. Int. J. Obes. 2013;37(3):333–340. doi: 10.1038/ijo.2012.62. [DOI] [PubMed] [Google Scholar]
- Jia H.P., et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79(23):14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juillard V., et al. Long-term humoral and cellular immunity induced by a single immunization with replication-defective adenovirus recombinant vector. Eur. J. Immunol. 1995;25(12):3467–3473. doi: 10.1002/eji.1830251239. [DOI] [PubMed] [Google Scholar]
- Van Kerkhove M.D., et al. Risk factors for severe outcomes following 2009 influenza A (H1N1) infection: a global pooled analysis. PLoS Med. 2011;8(7) doi: 10.1371/journal.pmed.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.I., et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27(5):704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter J.S., et al. SARS-CoV and SARS-CoV-2 are transmitted through the air between ferrets over more than one meter distance. Nat. Commun. 2021;12:1653. doi: 10.1038/s41467-021-21918-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- Lee A.C.-Y., et al. Oral SARS-CoV-2 inoculation establishes subclinical respiratory Infection with virus Shedding in golden Syrian hamsters. Cell reports. Medicine. 2020;1(7):100121. doi: 10.1016/j.xcrm.2020.100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist S.R., et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183(4):1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lurie N., et al. Developing covid-19 vaccines at pandemic speed. N. Engl. J. Med. 2020;382(21):1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- de Lusignan S., et al. Risk factors for SARS-CoV-2 among patients in the oxford royal college of general practitioners research and surveillance centre primary care network: a cross-sectional study. Lancet Infect. Dis. 2020;20(9):1034–1042. doi: 10.1016/S1473-3099(20)30371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81(2):813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;19(11):1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National diabetes statistics report . 2020. Estimates of Diabetes and its Burden in the United States. [Google Scholar]
- O'Brien K.B., et al. Impaired wound healing predisposes obese mice to severe influenza virus infection. J. Infect. Dis. 2012;205(2):252–261. doi: 10.1093/infdis/jir729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrilli C.M., et al. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. Br. Med. J. 2020;369:m1966. doi: 10.1136/bmj.m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnasinghe R., et al. medRxiv; 2021. The N501Y Mutation in SARS-CoV-2 Spike Leads to Morbidity in Obese and Aged Mice and Is Neutralized by Convalescent and Post-vaccination Human Sera. [Google Scholar]
- Rogers T.F., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369(6506):956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell M.A., et al. Activation of innate immunity in nonhuman primates following intraportal administration of adenoviral vectors. Mol. Ther. 2001;3(5 Pt 1):708–722. doi: 10.1006/mthe.2001.0330. [DOI] [PubMed] [Google Scholar]
- Sia S.F., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583(7818):834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonnet A., et al. High prevalence of obesity in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity. 2020;28(7):1195–1199. doi: 10.1002/oby.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.G., et al. Diet-induced obese mice have increased mortality and altered immune responses when infected with influenza virus. J. Nutr. 2007;137(5):1236–1243. doi: 10.1093/jn/137.5.1236. [DOI] [PubMed] [Google Scholar]
- Sun J., et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182(3):734–743. doi: 10.1016/j.cell.2020.06.010. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker E.E., et al. A genetically engineered adenovirus vector targeted to CD40 mediates transduction of canine dendritic cells and promotes antigen-specific immune responses in vivo. Vaccine. 2009;27(50):7116–7124. doi: 10.1016/j.vaccine.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnavski A.N., et al. Evaluation of toxicity from high-dose systemic administration of recombinant adenovirus vector in vector-naive and pre-immunized mice. Gene Ther. 2005;12(5):427–436. doi: 10.1038/sj.gt.3302347. [DOI] [PubMed] [Google Scholar]
- Wan Y., et al. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger-Lucarelli J., et al. Host nutritional status affects alphavirus virulence, transmission, and evolution. PLoS Pathog. 2019;15(11) doi: 10.1371/journal.ppat.1008089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West D.B., et al. Dietary obesity in nine inbred mouse strains. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1992;262(6):R1025–R1032. doi: 10.1152/ajpregu.1992.262.6.R1025. [DOI] [PubMed] [Google Scholar]
- de Wit E., et al. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worgall S., et al. Innate immune mechanisms dominate elimination of adenoviral vectors following in vivo administration. Hum. Gene Ther. 1997;8(1):37–44. doi: 10.1089/hum.1997.8.1-37. [DOI] [PubMed] [Google Scholar]
- Yang Y., et al. Cellular and humoral immune responses to viral antigens create barriers to lung-directed gene therapy with recombinant adenoviruses. J. Virol. 1995;69(4):2004–2015. doi: 10.1128/jvi.69.4.2004-2015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.H., et al. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 2007;57(5):450–459. [PubMed] [Google Scholar]
- Yoshikawa N., et al. Differential virological and immunological outcome of severe acute respiratory syndrome coronavirus infection in susceptible and resistant transgenic mice expressing human angiotensin-converting enzyme 2. J. Virol. 2009;83(11):5451–5465. doi: 10.1128/JVI.02272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files].