Abstract
Chemoresistance is a major cause of treatment failure in pancreatic cancer (PC). It has been demonstrated that epithelial-to-mesenchymal transition (EMT) is closely related to drug resistance in PC; however, the underlying mechanisms are not yet fully understood. Recently found evidence has suggested that nuclear-enriched abundant transcript 1 (NEAT1) is involved in the development of chemoresistance. However, the role and mechanism of NEAT1 in PC gemcitabine resistance remain unknown. In the present study, we first established two independent gemcitabine-resistant (GR) PC cell lines, PANC-1/GR and SW1990/GR. We found that GR cells displayed markedly enhanced migration and invasion abilities, decreased expression of E-cadherin, and upregulation of N-cadherin, Vimentin, Snail, ZEB1, and ZEB2. Our findings suggested that downregulation of NEAT1 enhanced the sensitivity of GR cells to gemcitabine by reversing the EMT process. Mechanistically, NEAT1 mediates ZEB2 mRNA expression through sponging miR-506-3p. Downregulation of NEAT1 can reverse the EMT process of GR PC cells by reducing the expression of ZEB2, thus enhancing the sensitivity of GR PC cells to gemcitabine. These findings were further confirmed in a nude mouse xenograft model. Taken together, downregulation of NEAT1 sensitized the GR PC cells to gemcitabine through modulation of the miR-506-3p/ZEB2/EMT axis. These results provide the novel evidence for understanding the function and molecular mechanism of NEAT1, and a new direction for improving the chemotherapeutic effects in PC.
Keywords: NEAT1, pancreatic cancer, gemcitabine, drug resistance, epithelial-to-mesenchymal transition
Introduction
Pancreatic cancer (PC) is one of the most aggressive types of cancer with an approximate 5-year survival rate of 9%; it represents the fourth leading cause of cancer death [1,2]. Since majority of the patients are diagnosed at a later or metastatic stage, only less than 20% of PCs are resectable [3]. Currently, chemotherapy is still one of the indispensable adjuvant methods for the treatment of PC. Since 1997, gemcitabine has been the first-line treatment choice for patients with locally advanced and metastatic PC [4]. However, with intrinsic or acquired drug resistance, PC has proven to be highly resistant to chemotherapy in clinical practice, which poses a significant challenge for the treatment of PC [5]. Hence, there is an urgent need to explore the mechanism of gemcitabine resistance, which could help us find a promising strategy for treatment of PC.
Epithelial-to-mesenchymal transition (EMT), a common feature of various types of tumors, allows the epithelial cells to acquire the mesenchymal phenotype, resulting in enhanced migration and invasion capacities [6]. Emerging evidence has demonstrated that this process may also be involved in the acquisition of chemoresistance in cancer cells [7,8]. For example, the transcription factor Twist1, one of the key EMT regulatory factors, has been found to be resistant to 5-fluorouracil in breast cancer cells [9]. Furthermore, Yang et al. [10] confirmed that EMT could induce gemcitabine resistance in PC, and that reversing EMT in PC cells enhances the sensitivity of the cells to gemcitabine. These studies provide a new direction for improving the effect of cancer chemotherapy. To investigate the development mechanism of gemcitabine resistance in PC, the present study established two independent gemcitabine-resistant (GR) PC cell lines, PANC-1/GR and SW1990/GR. We found that GR cells displayed markedly enhanced migration and invasion abilities, decreased expression of E-cadherin, and upregulation of N-cadherin, Vimentin, Snail, ZEB1, and ZEB2.
Long non-coding RNAs (lncRNAs) are generally defined as RNA transcripts longer than 200 nucleotides without evident protein coding functions [11]. It is well known that lncRNAs function as competing endogenous RNAs (ceRNAs) to protect mRNAs by competing for targeting the microRNAs (miRNAs), and play significant roles in a wide range of biological processes [12]. Accumulating evidence has revealed that lncRNAs could be crucial players in the progression and chemoresistance of various cancers [13-15]. Nuclear-enriched abundant transcript 1 (NEAT1), a newly identified nuclear-restricted lncRNA, is associated with malignant biological behavior in a variety of human cancers [16,17]. Recently, Feng et al. has demonstrated that NEAT1 facilitates PC growth and metastasis through stabilizing ELF3 mRNA [18]. Our previous study showed that NEAT1 is upregulated in PC tissues and cell lines, and is closely correlated with the progression of PC and a poor overall survival [19]. However, the role of NEAT1 in PC gemcitabine resistance still has not been investigated. We, for the first time, found that NEAT1 was upregulated in GR cells and associated with drug resistance of the cells. Furthermore, both in vivo and in vitro, we validated that downregulation of NEAT1 enhanced the sensitivity of PANC-1/GR and SW1990/GR cells to gemcitabine through modulation of the miR-506-3p/ZEB2/EMT axis. Our data provide the novel evidence for understanding the function and molecular mechanism of NEAT1, and for reversing chemotherapy resistance in patients with PC.
Materials and methods
Cell lines and culture conditions
Human PC cell lines (SW1990 and PANC-1), obtained from the American Type Culture Collection (ATCC; Rockville, USA), were cultured as described previously [19]. GR PC cells (PANC-1/GR and SW1990/GR) were selected from their respective parental cells that were exposed to gradually increasing concentrations of gemcitabine (Sigma-Aldrich, St. Louis, USA) for more than 6 months. These cells were then maintained in high-glucose Dulbecco’s modified Eagle medium (DMEM; Invitrogen, Waltham, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, USA) in a humidified atmosphere containing 5% carbon dioxide at 37°C.
MTT assay
A total of 5,000 cells were plated in 96-well plates for overnight incubation and then treated with different concentrations of gemcitabine for 48 h. According to the manufacturer’s instructions, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT; Sigma-Aldrich) assay was performed to determine the cell viability. SPSS statistical software (version 24.0; Chicago, USA) was used to calculate the concentration of gemcitabine required for 50% growth inhibition (IC50). All assays were performed thrice independently.
Transwell assay (cell migration and invasion)
The migration and invasion capacities of the cells were detected using the Transwell assay, performed according to the manufacturer’s instructions, as described previously [20]. Briefly, in the invasion assay, 5 × 104 cells were added to the transwell chambers precoated with MatrigelTM (BD Biosciences, San Jose, CA, USA). Then, the chambers were placed in a 24-well plate (Corning, New York, USA) and incubated for 24 h. After fixing with 4% paraformaldehyde and staining with 0.1% crystal violet, the number of invading cells was counted using a microscope (Olympus, Tokyo, Japan) in five randomly chosen fields. The same steps were performed in the cell migration assay, except that MatrigelTM was not used.
RNA extraction and quantitative RT-PCR assay
Total RNA was isolated from the cell lines using Trizol reagent (Invitrogen, Thermo Fisher Scientific, Inc.), then reverse transcribed using the PrimeScript RT reagent kit or the PrimeScript miRNA cDNA Synthesis Kit (TaKaRa, Dalian, China), according to the manufacturer’s instructions. Then, qPCR was performed using SYBR Premix Ex Taq II kit (Takara) or Taqman miRNA kit (Applied Biosystems, Foster City, CA), following the manufacturer’s instructions. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 were regarded as the internal controls, and the relative gene expression was determined using the 2-ΔΔCt method. The primer sequences used are listed in Table 1.
Table 1.
qPCR primers used in this study
| Genes | Primers | Sequences (5’-3’) |
|---|---|---|
| NEAT1 | Forward | TGGCTAGCTCAGGGCTTCAG |
| Reverse | TCTCCTTGCCAAGCTTCCTT | |
| miR-506-3p | Forward | GCCACCACCATCAGCCATAC |
| Reverse | GCACATTACTCTACTCAGAAGGG | |
| E-cadherin | Forward | GACAACAAGCCCGAATT |
| Reverse | GGAAACTCTCTCGGTCCA | |
| N-cadherin | Forward | GTATCCGGTCCGATCTGCA |
| Reverse | ATAGTCCTGCTCACCACCAC | |
| Vimentin | Forward | ATTCCACTTTGCGTTCAAGG |
| Reverse | CTTCAGAGAGAGGAAGCCGA | |
| Snail | Forward | CTTCCAGCAGCCCTACGACCA |
| Reverse | GCCCAGGCTGAGGTACTCC | |
| ZEB1 | Forward | AAGTGGCGGTAGATGGTAATGT |
| Reverse | AAGGAAGACTGATGGCTGAAAT | |
| ZEB2 | Forward | ACCAGCGGAAACAAGGAT |
| Reverse | TTTATGTCGCAGAAGGGAAC | |
| GAPDH | Forward | CATCACCATCTTCCAGGAGCG |
| Reverse | TGACCTTGCCCACAGCCTTG | |
| U6 | Forward | GCAGGAGGTCTTCACAGAGT |
| Reverse | TCTAGAGGAGAAGCTGGGGT |
Western blotting
The cells were lysed using radioimmunoprecipitation assay buffer (RIPA buffer) (Beyotime, Guangzhou, China) containing protease inhibitors. Protein concentrations were detected using a bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL, USA). Protein samples were separated using 10% SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and then incubated with primary antibodies. The antibodies against E-cadherin (#14472, 1:1000), N-cadherin (#13116, 1:1000), vimentin (#5741, 1:1000), Snail (#3879, 1:1000), ZEB1 (#3396, 1:1000), and ZEB2 (#97885, 1:1000) were obtained from Cell Signaling Technology (Danvers, MA, USA) and the anti-GAPDH antibody (ab8245, 1:2000) was obtained from Abcam (Cambridge, MA, USA). All the antibodies were used according to the manufacturers’ instructions.
Transfection of NEAT1 short hairpin RNA
For knockdown of lncRNA NEAT1, three different specific shRNAs (sh-NEAT1-1, sh-NEAT1-2, sh-NEAT1-3) were synthesized in a plasmid vector (GenePharma, Shanghai, China), and then incorporated into the lentivirus (GV118) vector to get Lv-sh-NEAT1. Hsa-miR-506-3p mimics and inhibitors along with their corresponding negative controls were prepared by Research-Bio Co., Ltd (Shanghai, China). Special siRNA against ZEB2 (si-ZEB2), ZEB2 overexpression plasmid (ZEB2-pcDNA3.1-EGFP) and their respective negative control vector were purchased from GenePharma (Shanghai, China). Cells were transfected using Lipofectamine 2000 (Invitrogen) when PANC-1/GR and SW1990/GR cells at the logarithmic growth phase. The transfection efficiency was confirmed using qRT-PCR. In the specified group, cells were exposed to 10 ng/mL TGF-β (R&D Systems, Minneapolis, USA) 24 h after transfection and then allowed to incubate at 37°C for 48 h. For generation of stably transfected cells, the cells were treated with 2 μg/mL puromycin (Sigma-Aldrich) for two weeks, following which the GFP-positive cells were selected for subsequent assays.
Cell apoptosis assay
Cell apoptosis assay was performed using the Annexin V/PI staining kit (Sungene Biotech, Tianjing, China), as described previously [20]. In brief, cells (1 × 105) were harvested for each assay, washed with cold PBS, and resuspended in 200 mL binding buffer. Then incubated with Annexin V-fluorescein isothiocyanate (5 µl) for 10 min and propidium iodide (PI) for 5 min at room temperature in the dark. Finally, the samples were analyzed by flow cytometry (BD Biosciences, San Jose, CA, USA) within 1 h.
Dual-luciferase reporter assay
The online software StarBase3.0 (http://starbase.sysu.edu.cn/) was used to predict the binding sites of NEAT1 to mi-506-3p. ZEB2 was a potential target of miR-506-3p by bioinformatical prediction tools, TargetScan 7.2 (http://www.targetscan.org/vert_72/). The fragment from NEAT1 or 3’UTR of ZEB2 containing the predicted binding sites of miR-506-3p were amplified using PCR, and cloned into the XhoI and XbaI sites of the downstream of Firefly luciferase gene in the pmirGLO vector (Promega, Madison, USA) to generate luciferase reporter vectors NEAT1-wt and ZEB2-3’UTR-wt, respectively. To test the binding specificity, the corresponding mutant type NEAT1 or 3’UTR of ZEB2 lacking the binding sites of miR-506-3p was also cloned into the pmirGLO vector to form the reporter vectors, NEAT1-mut and ZEB2-3’UTR-mut, respectively. The PANC-1/GR or SW1990/GR cells were seeded into 24-well plates and co-transfected with either 50 nM miR-506-3p mimics or NC using Lipofectamine 2000, together with 100 ng of the indicated luciferase reporter vector. After 48 h, luciferase activities in the transfected cells were detected using the Dual Luciferase® Reporter Assay kit (Promega, Madison, USA), according to the manufacturer’s protocol. The firefly luciferase activity was measured and normalized based on Renilla luciferase activity.
In vivo experiments
Female BALB/c nude mice (five-week-old) were purchased from Hunan SJA Laboratory Animal Co. Ltd., China. PANC-1/GR cells (1 × 107 cells per mice) stably expressed sh-NEAT1-1 or corresponding controls were subcutaneously injected into the mice. We allowed one week for the formation of palpable subcutaneous tumors and then intraperitoneally injected the mice with gemcitabine (50 mg/kg) or vehicle weekly. All experimental nude mice were randomly separated into different groups as follows: (1) sh-NC + vehicle; (2) sh-NEAT1-1 + vehicle; (3) sh-NC + Gemcitabine; (4) sh-NEAT1-1 + Gemcitabine. Tumor sizes were recorded weekly after gemcitabine treatment using the formula: width2 × length/2. Mice were sacrificed at the last measurement, the tumors were weighed, and subsequent assays were performed. All the animal experiments were performed in accordance with the experimental animal use guidelines of the National Institutes of Health and approved by the Ethics Committee for Animal Experiments of the Second Affiliated Hospital of Nanchang University.
Transferase dUTP nick end-labeling assay
In situ, cell apoptosis was detected using a One Step Transferase dUTP nick end-labeling (TUNEL) apoptosis assay kit (Beyotime, Shanghai, China) on formalin-fixed, paraffin-embedded tissues of nude mouse xenografts, according to the manufacturer’s instructions. The tissues were then photographed using an inverted fluorescence microscope (Nikon, Tokyo, Japan).
Statistical analysis
All results have been expressed as mean ± SD and analyzed using SPSS statistical software (version 24.0; Chicago, USA). Statistical analysis was performed using two-tailed Student’s t-test or one-way ANOVA to identify significant differences between two or multiple groups. All assays were performed independently three times. P≤0.05 was considered statistically significant.
Results
Establishment of PANC-1/GR and SW1990/GR cell lines
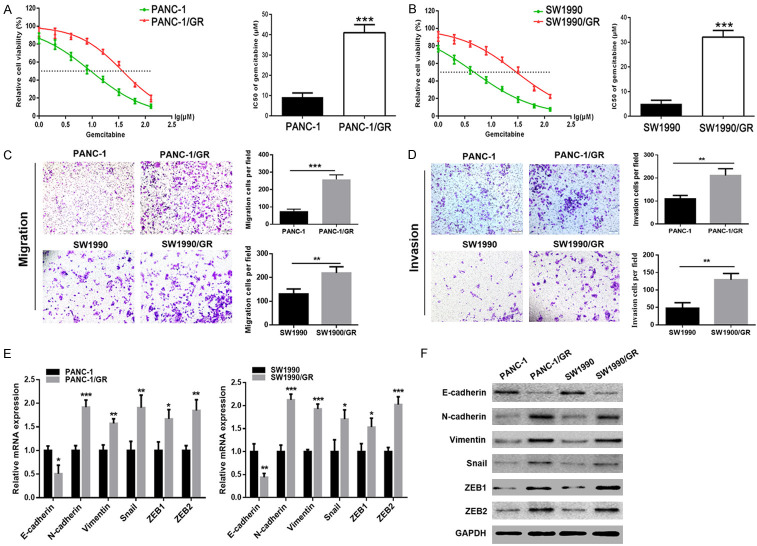
To determine the underlying mechanism for chemoresistance in PC, we first established two independent GR PC cell lines, PANC-1/GR and SW1990/GR. MTT assay was performed to verify whether PANC-1/GR and SW1990/GR cells were gemcitabine-resistant, followed by the calculation of IC50 values of gemcitabine in each cell line. As shown in Figure 1A and 1B, there was a significant increase in the IC50 values of gemcitabine in PANC-1/GR and SW1990/GR cells, compared to their respective parental cells. These findings demonstrated that PANC-1/GR and SW1990/GR cells exhibited chemoresistance to gemcitabine, and thus, these two GR PC cell lines were continuously cultured for the following study.
Figure 1.
PANC-1/GR and SW1990/GR cells displayed an EMT phenotype. A, B. MTT assay was used to determine the cytotoxicity of gemcitabine to PANC-1, PANC-1/GR, SW1990, and SW1990/GR cells. IC50 values of gemcitabine in each cell line were calculated. C, D. Transwell assay was performed to determine the cell migration and invasion capacities of GR cells (PANC-1/GR and SW1990/GR) and their respective parental cells. E, F. The mRNA and protein levels of EMT molecular markers including E-cadherin, N-cadherin, Vimentin, Snail, ZEB1, and ZEB2 were detected using qRT-PCR and western blot analysis, respectively. At least three independent experiments were performed in each group and mean ± SD was used to represent the final result. *P<0.05, **P<0.01, ***P<0.001.
GR cells acquired EMT capability and showed EMT characteristics
In addition to the role of EMT in promoting tumor invasion, emerging evidence demonstrates that this process may also be involved in the acquisition of the chemosensitivity in cancer cells. In the present study, we performed Transwell assay to investigate the cell migration and invasion abilities of PANC-1/GR and SW1990/GR cells. GR cells had markedly enhanced migration ability, compared to their respective parental cells (Figure 1C), and there was a parallel upward trend in their invasive ability (Figure 1D). Then, the expression of EMT molecular markers in GR cells, including E-cadherin, N-cadherin, Vimentin, Snail, ZEB1, and ZEB2, were examined using qRT-PCR and western blotting. Compared to their respective parental cells, both the mRNA and protein levels of N-cadherin, Vimentin, Snail, ZEB1, and ZEB2 were found to be upregulated, whereas E-cadherin was found to be downregulated in GR cells (Figure 1E, 1F). These results further suggested that PANC-1/GR and SW1990/GR cells acquired EMT capability.
Expression of NEAT1 was upregulated in GR cell lines
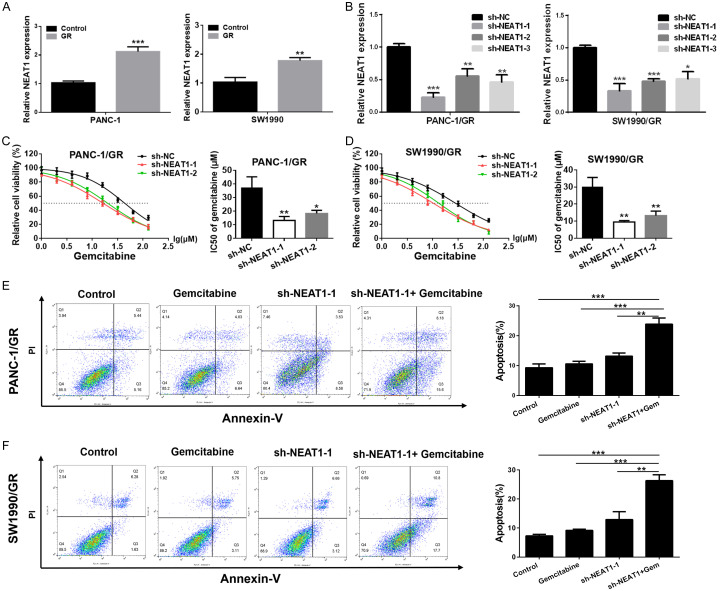
Our previous study has shown that NEAT1 is upregulated in PC tissues and cell lines, and is closely correlated with the progression of PC and poor overall survival. To explore whether NEAT1 is involved in chemoresistance of PC cells, we first detected the expression level of NEAT1 in PANC-1/GR, SW1990/GR cells and their respective parental cells using qRT-PCR. As shown in Figure 2A, elevated expression of NEAT1 was observed in GR cells compared to their respective parental cells, suggesting that the chemoresistance of GR cells could partly be due to the upregulation of NEAT1.
Figure 2.
Downregulation of NEAT1 enhanced the cytotoxicity of gemcitabine to SW1990/GR and PANC-1/GR cells. A. Relative expression of NEAT1 in GR cells (PANC-1/GR and SW1990/GR) and their respective parental cells. B. Relative expression of NEAT1 in GR cells after transfection with NEAT1 shRNA or negative control shRNA. C, D. MTT assay was used to determine the cytotoxicity of gemcitabine to GR cells after transfection with NEAT1 shRNA or negative control shRNA. IC50 values of gemcitabine in each group were calculated. E, F. The cell apoptosis in GR cells (PANC-1/GR and SW1990/GR) after NEAT1 knockdown or/and gemcitabine treatment was detected using flow cytometry analysis (gemcitabine: 5 μM, 48 h). At least three independent experiments were performed in each group and mean ± SD was used to represent the final result. *P<0.05, **P<0.01, ***P<0.001.
Downregulation of NEAT1 sensitized GR cells to gemcitabine
To further understand the relationship between NEAT1 and PC gemcitabine resistance, we silenced NEAT1 expression in PANC-1/GR and SW1990/GR cells using NEAT1 shRNA (sh-NEAT1-1, sh-NEAT1-2 and sh-NEAT1-3). As shown in Figure 2B, PANC-1/GR and SW1990/GR cells transfected with NEAT1 shRNA displayed a significantly decreased expression level of NEAT1 as compared to the control group. When NEAT1 was knocked down, PANC-1/GR and SW1990/GR cells showed more susceptibility to gemcitabine, with a significant decline in the IC50 value for gemcitabine (Figure 2C, 2D). Then, we investigated the effects of NEAT1 knockdown on gemcitabine induced cell apoptosis. Our results showed that the gemcitabine induced apoptosis was dramatically reinforced in GR cells treated with a combination of NEAT1 shRNA and gemcitabine compared to cells treated with gemcitabine or NEAT1 shRNA alone (Figure 2E, 2F). These data collectively indicated that knockdown of NEAT1 could significantly enhance the sensitivity of PANC-1/GR and SW1990/GR cells to gemcitabine.
Downregulation of NEAT1 enhanced the sensitivity of GR cells to gemcitabine by reversing the EMT process
To explore the relationship between NEAT1 and EMT in PC, we performed a Transwell assay to investigate the cell migration and invasion abilities of PANC-1/GR and SW1990/GR cells upon knockdown of NEAT1 expression. As shown in Figure 3A, cells transfected with NEAT1 shRNA presented a significantly impaired migration ability than those in the control group, and there was a parallel downward trend in their invasive ability (Figure 3B). Correspondingly, both mRNA and protein levels of N-cadherin, Vimentin, Snail, ZEB1, and ZEB2 were downregulated in the NEAT1 shRNA group, while E-cadherin was upregulated (Figure 3C, 3D). Further, to determine whether NEAT1 regulates GR in PANC-1/GR and SW1990/GR cells through EMT, we downregulated the expression of NEAT1 in PANC-1/GR and SW1990/GR cells, and then stimulated the cells using an inducer of EMT (TGF-β) to observe the effect on gemcitabine sensitivity. First, we observed that the NEAT1 shRNA-induced impairment in the migration and invasion abilities of PANC-1/GR and SW1990/GR cells was partly reversed upon TGF-β treatment (Figure 4A, 4B). Our data also showed that the NEAT1 shRNA-mediated enhancement in the cytotoxicity of gemcitabine to GR cells was partially offset by TGF-β (Figure 4C). Meanwhile, the increase in gemcitabine-induced apoptosis mediated by NEAT1 shRNA was partly offset by TGF-β (Figure 4D). These findings suggested that downregulation of NEAT1 enhanced the sensitivity of PANC-1/GR and SW1990/GR cells to gemcitabine by reversing the EMT process.
Figure 3.
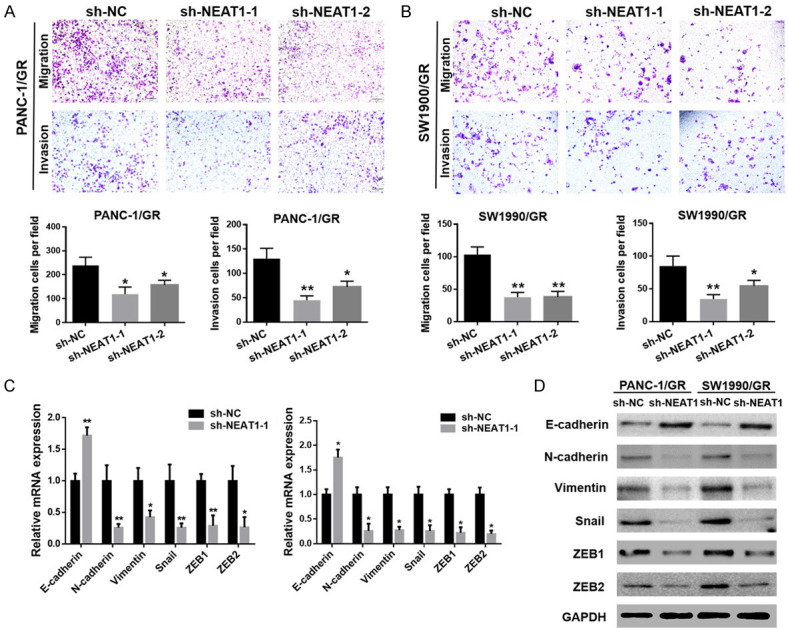
Downregulation of NEAT1 reversed the EMT process in SW1990/GR and PANC-1/GR cells. A, B. Transwell assay was performed to determine the cell migration and invasion capacities of the GR cells after transfection with NEAT1 shRNA or negative control shRNA. C, D. The mRNA and protein levels of EMT molecular markers including E-cadherin, N-cadherin, Vimentin, Snail, ZEB1, and ZEB2 in GR cells after transfection with NEAT1 shRNA or negative control shRNA were detected using qRT-PCR and western blot analysis, respectively. At least three independent experiments were performed in each group and mean ± SD was used to represent the final result. *P<0.05, **P<0.01.
Figure 4.
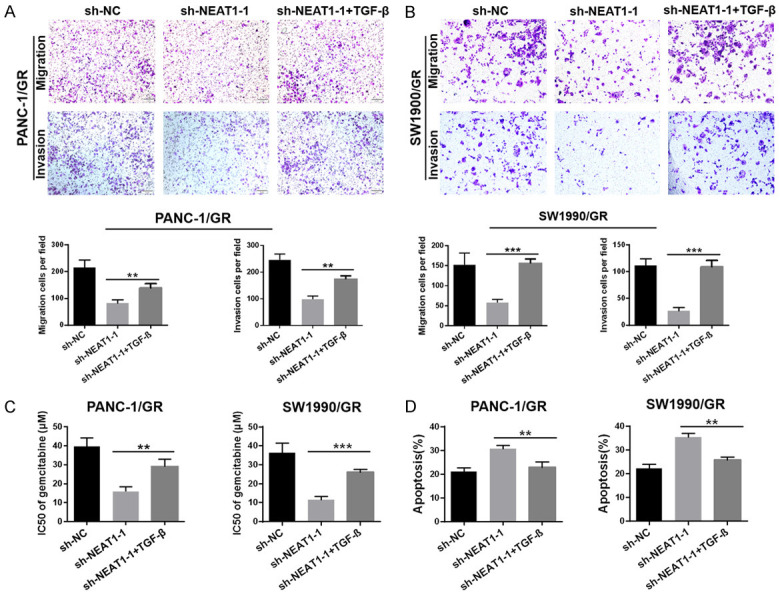
Downregulation of NEAT1 enhanced the sensitivity of SW1990/GR and PANC-1/GR cells to gemcitabine by reversing the EMT process. A, B. Transwell assay was performed to determine the cell migration and invasion capacities of GR cells after NEAT1 knockdown combined with TGF-β treatment or not. C. MTT assay was used to determine the cytotoxicity of gemcitabine to GR cells after NEAT1 knockdown combined with TGF-β treatment or not. IC50 values of gemcitabine in each group were calculated. D. Flow cytometry analysis was used to determine the gemcitabine-induced cell apoptosis in GR cells after NEAT1 knockdown combined with TGF-β treatment or not. Cells in each group were exposed to 5 μM gemcitabine for 48 h. At least three independent experiments were performed in each group and mean ± SD was used to represent the final result. *P<0.05, **P<0.01, ***P<0.001.
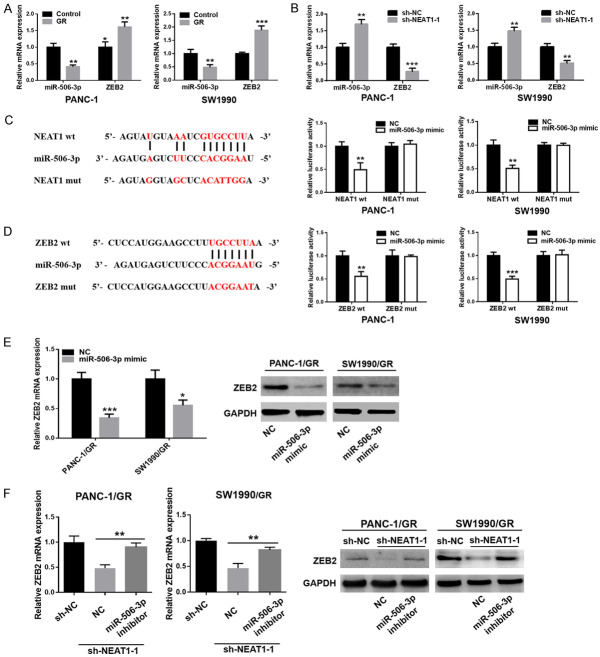
NEAT1 regulated ZEB2 expression by sponging miR-506-3p
LncRNAs are known to function as ceRNAs to protect mRNAs by competing for their targeting miRNAs. In the present study, we observed that the expression of miR-506-3p was downregulated, while the expression of ZEB2 was upregulated in PANC-1/GR and SW1990/GR cells (Figure 5A). Furthermore, downregulation of NEAT1 in GR cells could upregulate the expression of miR-506-3p, accompanied by a decrease of ZEB2 expression (Figure 5B). Bioinformatics prediction tools showed that miR-506-3p could directly bind to NEAT1 and ZEB2 (Figure 5C, 5D). Dual-luciferase assays confirmed that the miR-506-3p mimic significantly decreased the luciferase activity of the reporter with wild-type NEAT1-3’UTR but did not affect the activity of the mutant vector in GR cells (Figure 5C). Similarly, the results indicated a significant reduction in luciferase activities after co-transfection of miR-506-3p mimic and wild-type ZEB2 reporter vector, but not mutant ZEB2 (Figure 5D). Ectopic expression of miR-506-3p also remarkably decreased ZEB2 mRNA and protein expression in GR cells (Figure 5E). More importantly, the reduced mRNA and protein level of ZEB2, induced by NEAT1 knockdown in GR cells, was partly restored upon introduction of the miR-506-3p inhibitor (Figure 5F). These data strongly indicated that NEAT1 regulated ZEB2 expression by sponging miR-506-3p in GR cells.
Figure 5.
NEAT1 regulated ZEB2 expression by sponging miR-506-3p in SW1990/GR and PANC-1/GR cells. A. Relative expression of miR-506-3p and ZEB2 in GR cells (PANC-1/GR and SW1990/GR) and their respective parental cells. B. Relative expression of miR-506-3p and ZEB2 in GR cells after transfection with NEAT1 shRNA or negative control shRNA. C, D. The predicted binding sites of miR-506-3p in the 3’-UTR of NEAT1 or ZEB2 were determined using Starbase v3.0 or TargetScan. Luciferase reporter assays showed that miR-506-3p overexpression significantly suppressed the activity of the reporter containing wild-type NEAT1 or wild-type ZEB2 in GR cells (PANC-1/GR and SW1990/GR). E. The mRNA and protein levels of ZEB2 in GR cells after miR-506-3p overexpression were detected using qRT-PCR and western blot analysis, respectively. F. qRT-PCR and western blot analysis of the expression of ZEB2 in GR cells transfected with NEAT1 shRNA in the presence of miR-506-3p inhibitor or NC. At least three independent experiments were performed in each group and mean ± SD was used to represent the final result. *P<0.05, **P<0.01, ***P<0.001.
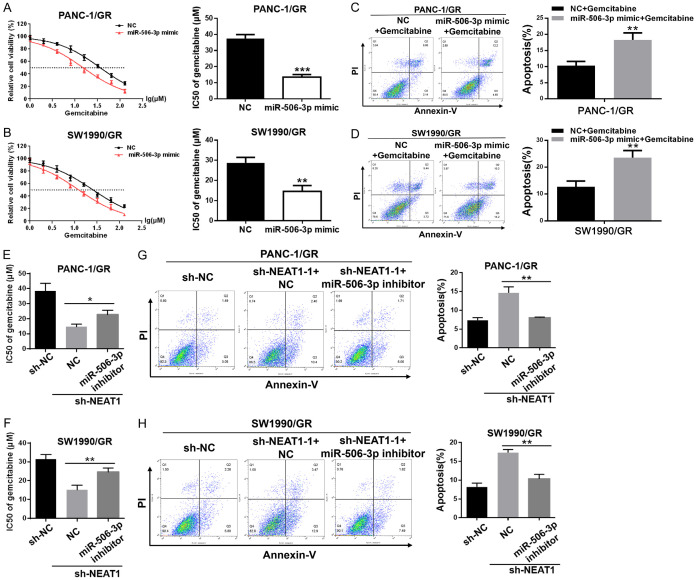
The function of NEAT1 in GR cells was dependent on miR-506-3p and ZEB2
The MTT assay data showed that miR-506-3p mimic sensitized PANC-1/GR and SW1990/GR cells to gemcitabine, with a significant decline in the IC50 value for gemcitabine (Figure 6A, 6B). Furthermore, miR-506-3p mimic significantly increased the number of cells undergoing gemcitabine-induced apoptosis in PANC-1/GR and SW1990/GR cells (Figure 6C, 6D). The rescue experiments showed that miR-506-3p inhibitor restored the resistance of GR cells transfected with NEAT1 shRNA to gemcitabine, with a significant increase in the IC50 value for gemcitabine (Figure 6E, 6F) and a decease on gemcitabine induced cell apoptosis (Figure 6G, 6H). Similarly, MTT assay data showed that ZEB2 knockdown sensitized PC GR cells to gemcitabine, with a significant decline in the IC50 value for gemcitabine (Figure S1A, S1B). Furthermore, ZEB2 knockdown significantly increased the number of cells undergoing gemcitabine-induced apoptosis in PC GR cells (Figure S1C, S1D). The rescue experiments showed that ZEB2 overexpression restored the resistance of GR cells transfected with NEAT1 shRNA to gemcitabine (Figure S1E-H). Taken together, these data suggested that the function of NEAT1 in GR cells was at least partly dependent on miR-506-3p and ZEB2.
Figure 6.
The function of NEAT1 in SW1990/GR and PANC-1/GR cells was dependent on miR-506-3p. A, B. The MTT assay was used to determine the cytotoxicity of gemcitabine to GR cells after transfection with miR-506-3p mimic or NC. IC50 values of gemcitabine in each group were calculated. C, D. Flow cytometry analysis of gemcitabine induced cell apoptosis in GR cells after transfection with miR-506-3p mimic or NC. Cells in each group were exposed to 5 μM gemcitabine for 48 h. E, F. MTT assay was used to determine the cytotoxicity of gemcitabine to GR cells transfected with NEAT1 shRNA in the presence of miR-506-3p inhibitor or NC. IC50 values of gemcitabine in each group were calculated. G, H. Flow cytometry analysis of the gemcitabine-induced cell apoptosis in GR cells transfected with NEAT1 shRNA in the presence of miR-506-3p inhibitor or NC. Cells in each group were exposed to 5 μM gemcitabine for 48 h. At least three independent experiments were performed in each group and mean ± SD was used to represent the final result. *P<0.05, **P<0.01, ***P<0.001.
Downregulation of NEAT1 improved the chemotherapeutic effect of gemcitabine in nude mouse xenograft models
To evaluate the influence of NEAT1 inhibition on the efficacy of chemotherapy, in vivo, we performed studies using mouse xenograft models. We observed that xenografts treated with a combination of NEAT1 shRNA and gemcitabine grew at the slowest rate, with the slowest tumor volumes and weights, as compared to the other three groups (Figure 7A-C). As shown in Figure 7D, there was a significant decrease in the mRNA expression level of NEAT1 in NEAT1 shRNA cells-derived xenografts, while the expression of miR-506-3p was upregulated. Correspondingly, compared to those in the control group, both mRNA and protein levels of N-cadherin, Vimentin, Snail, ZEB1, and ZEB2 were downregulated in NEAT1 shRNA cells-derived xenografts, while E-cadherin was upregulated (Figure 7E, 7F). Moreover, TUNEL analysis of apoptotic cell numbers further confirmed the improved chemotherapeutic effect of gemcitabine in tumors with NEAT1 inhibition (Figure 7G). Collectively, our xenograft studies provide further evidence that downregulation of NEAT1 enhances the chemotherapeutic effect of gemcitabine, likely by inhibiting the EMT process.
Figure 7.
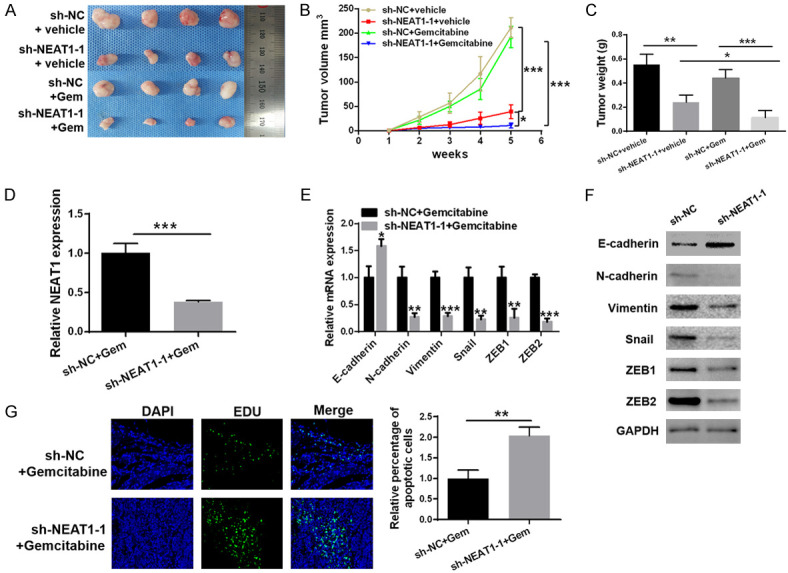
Downregulation of NEAT1 improved the chemotherapeutic effect of gemcitabine in nude mouse xenograft models. A. Representative images of xenograft tumors in each group, including sh-NC + vehicle, sh-NEAT1-1 + vehicle, sh-NC + Gemcitabine and sh-NEAT1-1 + Gemcitabine. B, C. Tumor growth curves and tumor weights of each group. D. qRT-PCR analysis of the expression of NEAT1 in the two tumor xenografts groups. E, F. qRT-PCR and western blot analysis of the expression of E-cadherin, N-cadherin, Vimentin, Snail, ZEB1, and ZEB2 in the two tumor xenografts groups. G. TUNEL staining images for apoptosis in the two tumor xenografts groups. At least three independent experiments were performed in each group and mean ± SD was used to represent the final result. *P<0.05, **P<0.01, ***P<0.001.
Discussion
Since 1997, the FDA has used gemcitabine as a first-line drug for the treatment of PC [4]. Although chemotherapy has improved the prognosis of PC patients to some extent, the death rate is still on the rise [5]. The main reason for chemotherapy failure in case of PC is chemotherapy resistance, either intrinsic or acquired drug resistance [21]. Resistance to gemcitabine represents a clinical and scientific challenge for PC patients. EMT is a process where the epithelial cell phenotype transitions to a mesenchymal cell phenotype, resulting in enhanced migration and invasion capacities, which play an important role in tumorigenesis and evolution [6]. Interestingly, recent research studies have suggested that EMT is one of the key factors for acquisition of chemoresistance in cancer cells [7,8]. A wealth of evidence has confirmed that EMT could induce gemcitabine resistance in PC [22,23]. In the present study, we established two independent GR PC cell lines, PANC-1/GR and SW1990/GR. Our study consistently demonstrated that GR cells exhibited stronger cell migration and invasion abilities than their respective parental cells. Moreover, GR cells gradually lost expression of the epithelial marker E-cadherin, while the expression levels of mesenchymal markers N-cadherin, Vimentin, Slug, ZEB1, and ZEB2 increased. These findings confirmed that chemoresistance is closely associated with the EMT process.
A growing body of studies has indicated that the dysregulation of lncRNAs contributes to chemoresistance [24,25]. NEAT1, a newly identified nuclear-restricted lncRNA, has been reported to be upregulated, and thus identified as a potential therapeutic target in many cancers, including PC [19], gastric cancer [26], liver cancer [27], colon cancer [28], esophageal cancer [29], etc. Recently, new evidence has suggested that NEAT1 is involved in the development of drug resistance. NEAT1 has been found to be upregulated in a paclitaxel-resistant non-small cell lung carcinoma (NSCLC) cell line, where it contributes to paclitaxel-resistance [30]. Parasramka et al. [31] indicated that exogenous regulation of NEAT1 expression alters the tumor cell phenotype and modulates the sensitivity to gemcitabine in cholangiocarcinoma. In the present study, elevated expression of NEAT1 was observed in GR cells. Knockdown of NEAT1 significantly enhanced the sensitivity of GR cells to gemcitabine. Furthermore, downregulation of NEAT1 improved the chemotherapeutic effect of gemcitabine in nude mouse xenograft models. These findings indicated that the chemoresistance of GR cells could partly be due to the upregulation of NEAT1.
Multiple studies have revealed that lncRNAs play a critical role in the regulation of drug resistance-mediated EMT [32]. Yao et al. [14] reported that the lncRNA NONHSAT101069 is upregulated in breast cancer tissues and promotes epirubicin resistance via regulation of Twist1. Gao et al. [33] indicated that the downregulation of lncRNA H19 elevates tamoxifen sensitivity by inhibiting the Wnt pathway and EMT process in tamoxifen-resistant breast cancer cells. The expression of lncRNA LEIGC has been found to be significantly lower in human gastric cancers; the upregulation of LEIGC enhances the sensitivity of gastric cancer cells to 5-fluorouracil, whereas downregulation of LEIGC has the opposite effect. Moreover, LEIGC functions by inhibiting the EMT process in gastric cancer [34]. Consistently, we found that downregulation of NEAT1 reversed the EMT process in GR cells. Furthermore, the NEAT1 shRNA-induced enhanced cytotoxicity of gemcitabine to GR cells was partially offset by an inducer of EMT (TGF-β). These findings suggested that downregulation of NEAT1 enhances the chemotherapeutic effect of gemcitabine, most likely by inhibiting the EMT process.
It is well known lncRNAs function as ceRNAs to protect mRNAs by competing for their targeting miRNAs. For example, Liu et al. [35] demonstrated that the lncRNA growth arrest-specific 5 (GAS5) suppresses gemcitabine resistance in PC by regulating the miR-221/SOCS3 pathway, which mediates the EMT process. lncRNA LINC00346 promotes PC growth and gemcitabine resistance by sponging miR-188-3p to derepress BRD4 expression [36]. Likewise, NEAT1 has a critical role in cancers by acting as a sponge of miRNAs. Zhou et al. [37] reported that NEAT1 promotes cell proliferation and invasion in liver cancer by regulating miRNA-22-3p/akt2. ZEB2, a key EMT regulatory factor, plays its role in the development of a variety of tumors by inhibiting the expression of E-cadherin and promoting EMT [38]. Duan et al. [39] found that miR-203 inhibits EMT and enhances the chemosensitivity of lung cancer by targeting ZEB2. LncRNA ATB induces EMT and invasion of HCC cells in vitro and in vivo by upregulating its target gene ZEB2 through competitive binding with miR-200 [40]. In the present study, the luciferase reporter assay confirmed that ZEB2 was a direct target of miR-506-3p and that NEAT1 regulated ZEB2 expression by sponging miR-506-3p in GR cells. Furthermore, the rescue experiments demonstrated that the function of NEAT1 in GR cells was dependent on miR-506-3p and ZEB2. Collectively, these findings indicated that downregulation of lncRNA NEAT1 sensitized GR cells to gemcitabine through modulation of the miR-506-3p/ZEB2/EMT axis.
In summary, the present study provides convincing evidence that GR PC cells displayed an EMT phenotype. Furthermore, we identified the lncRNA, NEAT1, as a key modulator of PC chemoresistance for the first time. Downregulation of NEAT1 enhanced the sensitivity of GR cells to gemcitabine by reversing the EMT process. Mechanistically, NEAT1 mediates ZEB2 mRNA expression through sponging miR-506-3p. Our findings suggested that downregulation of NEAT1 sensitized the GR PC cells to gemcitabine through modulation of the miR-506-3p/ZEB2/EMT axis (Figure 8). These results provide the novel evidence for understanding the function and molecular mechanism of NEAT1, and a new direction for improving the chemotherapeutic effects in PC.
Figure 8.
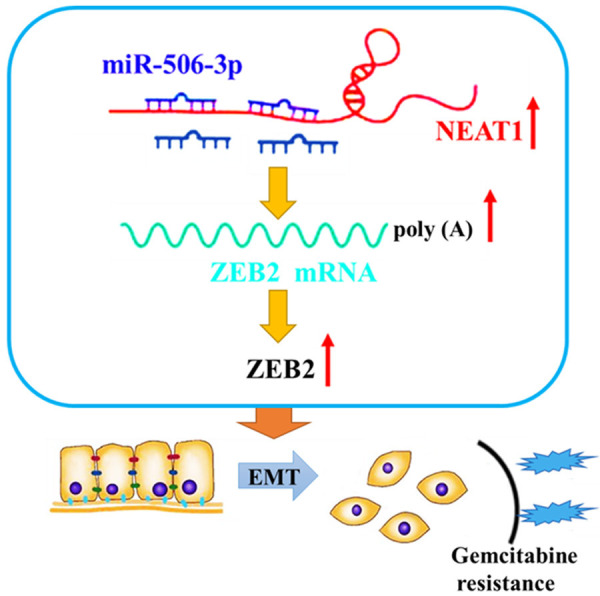
The schematic illustration of the potential molecular mechanism of NEAT1 as a key regulator in PC chemoresistance. NEAT1 was remarkably upregulated in gemcitabine-resistant pancreatic cancer cells and regulated ZEB2 expression by sponging miR-506-3p. Downregulation of NEAT1 sensitizes gemcitabine-resistant pancreatic cancer cells to gemcitabine through modulation of the miR-506-3p/ZEB2/EMT axis.
Acknowledgements
The current study was supported by the National Natural Science Foundation of China (81860530), the Key Research and Development Program of Jiangxi Province (20171BBG70123), the Natural Science Foundation of Jiangxi Province (20202BABL206017) and the Jiangxi Province Graduate Innovation Special Fund Project (YC2019-B007).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol. 2010;7:163–172. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Oettle H. Progress in the knowledge and treatment of advanced pancreatic cancer: from benchside to bedside. Cancer Treat Rev. 2014;40:1039–1047. doi: 10.1016/j.ctrv.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Lee KH, Kim MK, Kim YH, Ryoo BY, Lim HY, Song HS, Kim HK, Lee MA, Im SA, Chang HM. Gemcitabine and oxaliplatin combination as first-line treatment for advanced pancreatic cancer: a multicenter phase II study. Cancer Chemother Pharmacol. 2009;64:317–325. doi: 10.1007/s00280-008-0873-9. [DOI] [PubMed] [Google Scholar]
- 5.Custodio A, Puente J, Sastre J, Dã Az-Rubio E. Second-line therapy for advanced pancreatic cancer: a review of the literature and future directions. Cancer Treat Rev. 2009;35:676–684. doi: 10.1016/j.ctrv.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11:28–39. doi: 10.1002/1878-0261.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, Lebleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El RT, Ryu S, Troeger J. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nuti SV, Mor G, Li P, Yin G. TWIST and ovarian cancer stem cells: implications for chemoresistance and metastasis. Oncotarget. 2014;5:7260–7271. doi: 10.18632/oncotarget.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang RM, Zhan M, Xu SW, Long MM, Yang LH, Chen W, Huang S, Liu Q, Zhou J, Zhu J. miR-3656 expression enhances the chemosensitivity of pancreatic cancer to gemcitabine through modulation of the RHOF/EMT axis. Cell Death Dis. 2017;8:e3129. doi: 10.1038/cddis.2017.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 12.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 13.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao N, Fu Y, Chen L, Liu Z, He J, Zhu Y, Xia T, Wang S. Long non-coding RNA NONHSAT101069 promotes epirubicin resistance, migration, and invasion of breast cancer cells through NONHSAT101069/miR-129-5p/Twist1 axis. Oncogene. 2019;38:7216–7233. doi: 10.1038/s41388-019-0904-5. [DOI] [PubMed] [Google Scholar]
- 15.Shu D, Xu Y, Chen W. Knockdown of lncRNA BLACAT1 reverses the resistance of afatinib to non-small cell lung cancer via modulating STAT3 signalling. J Drug Target. 2020;28:300–306. doi: 10.1080/1061186X.2019.1650368. [DOI] [PubMed] [Google Scholar]
- 16.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Y, Gao L, Cui G, Cao Y. LncRNA NEAT1 facilitates pancreatic cancer growth and metastasis through stabilizing ELF3 mRNA. Am J Cancer Res. 2020;10:237–248. [PMC free article] [PubMed] [Google Scholar]
- 19.Huang B, Liu C, Wu Q, Zhang J, Min Q, Sheng T, Wang X, Zou Y. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem Biophys Res Commun. 2017;482:828. doi: 10.1016/j.bbrc.2016.11.120. [DOI] [PubMed] [Google Scholar]
- 20.Fu X, Zhu Y, Zheng B, Zou Y, Wang C, Wu P, Wang J, Chen H, Du P, Liang B. KIFC1, a novel potential prognostic factor and therapeutic target in hepatocellular carcinoma. Int J Oncol. 2018;52:1912–1922. doi: 10.3892/ijo.2018.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Sousa Cavalcante L, Monteiro G. Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur J Pharmacol. 2014;741:8–16. doi: 10.1016/j.ejphar.2014.07.041. [DOI] [PubMed] [Google Scholar]
- 22.Bera A, Freeman JW, Venkatasubbarao K, Zhao S. Abstract LB-22: gemcitabine resistance is associated with EMT phenotype along with expression of cancer stem cell markers and a specific miRNA profile in the human pancreatic cancer cell line BxPC3. Cancer Res. 2012;72:22. [Google Scholar]
- 23.Wang R. Gemcitabine resistance is associated with epithelial-mesenchymal transition and induction of HIF-1α in pancreatic cancer cells. Curr Cancer Drug Tar. 2014;14:407–17. doi: 10.2174/1568009614666140226114015. [DOI] [PubMed] [Google Scholar]
- 24.Ayers D, Vandesompele J. Influence of microRNAs and long non-coding RNAs in cancer chemoresistance. Genes (Basel) 2017;8:95. doi: 10.3390/genes8030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang L, Zeng L, Chu J, Xu P, Lv M, Xu J, Wen J, Li W, Wang L, Wu X. Chemoresistance-related long non-coding RNA expression profiles in human breast cancer cells. Mol Med Rep. 2018;18:243–253. doi: 10.3892/mmr.2018.8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571–1579. doi: 10.1007/s00432-016-2152-1. [DOI] [PubMed] [Google Scholar]
- 27.Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong M, Dang Y, Feng Z, Chen G. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol. 2015;8:5395–402. [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F, Zhuo C, Zheng Y, Li B, Wang Z. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer. 2015;14:191. doi: 10.1186/s12943-015-0455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Kong J, Ma Z, Gao S, Feng X. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am J Cancer Res. 2015;5:2808–2815. [PMC free article] [PubMed] [Google Scholar]
- 30.Li B, Gu W, Zhu X. NEAT1 mediates paclitaxel-resistance of non-small cell of lung cancer through activation of Akt/mTOR signalling pathway. J Drug Target. 2019;27:1061–1067. doi: 10.1080/1061186X.2019.1585437. [DOI] [PubMed] [Google Scholar]
- 31.Parasramka M, Yan IK, Wang X, Nguyen P, Matsuda A, Maji S, Foye C, Asmann Y, Patel T. BAP1 dependent expression of long non-coding RNA NEAT-1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol Cancer. 2017;16:22. doi: 10.1186/s12943-017-0587-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia H, Hui KM. Mechanism of cancer drug resistance and the involvement of noncoding RNAs. Curr Med Chem. 2014;21:3029–41. doi: 10.2174/0929867321666140414101939. [DOI] [PubMed] [Google Scholar]
- 33.Gao H, Hao G, Sun Y, Li L, Wang Y. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. Onco Targets Ther. 2018;11:8001–8012. doi: 10.2147/OTT.S172379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Y, Ye J, Wu D, Wu P, Chen Z, Chen J, Gao S, Huang J. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. BMC Cancer. 2014;14:932. doi: 10.1186/1471-2407-14-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu B, Wu S, Ma J, Yan S, Xiao Z, Wan L, Zhang F, Shang M, Mao A. lncRNA GAS5 Reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol Ther Nucleic Acids. 2018;13:472–482. doi: 10.1016/j.omtn.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi W, Zhang C, Ning Z, Hua Y, Li Y, Chen L, Liu L, Chen Z, Meng Z. Long non-coding RNA LINC00346 promotes pancreatic cancer growth and gemcitabine resistance by sponging miR-188-3p to derepress BRD4 expression. J Exp Clin Cancer Res. 2019;38:60. doi: 10.1186/s13046-019-1055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou X, Wang X, Zhou Y, Cheng L, Zhang Y, Zhang Y. Long noncoding RNA NEAT1 promotes cell proliferation and invasion and suppresses apoptosis in hepatocellular carcinoma by regulating miRNA-22-3p/akt2 in vitro and in vivo. Onco Targets Ther. 2019;12:8991–9004. doi: 10.2147/OTT.S224521. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Ahn SM, Cha JY, Kim J, Kim D, Trang HT, Kim YM, Cho YH, Park D, Hong S. Smad3 regulates E-cadherin via miRNA-200 pathway. Oncogene. 2012;31:3051–3059. doi: 10.1038/onc.2011.484. [DOI] [PubMed] [Google Scholar]
- 39.Duan X, Fu Z, Gao L, Zhou J, Deng X, Luo X, Fang W, Luo R. Direct interaction between miR-203 and ZEB2 suppresses epithelial-mesenchymal transition signaling and reduces lung adenocarcinoma chemoresistance. Acta Biochim Biophys Sin (Shanghai) 2016;48:1042–1049. doi: 10.1093/abbs/gmw099. [DOI] [PubMed] [Google Scholar]
- 40.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.