Abstract
We sought novel approaches to improve transfection efficiencies of microRNAs (miRNAs) in platelets, and to apply these approaches to investigate roles of miRNAs in regulating signal-activated protein translation and functional effects. We found that ex vivo human platelets support gymnosis - internalization of ectopic miRNAs following co-incubation in the absence of conventional transfection reagents or schemes - and subsequently incorporate transfected miRNA into ARGONAUTE2 (AGO2)-based RNA-induced silencing complexes (RISC). Thrombin/fibrinogen stimulation activated translation of miR-223–3p target SEPTIN2, which was suppressed by miR-223–3p transfection in an AGO2/RISC-dependent manner. Thrombin/fibrinogen-induced exosome and microvesicle generation was inhibited by miR-223–3p transfection, and this effect was reversed with a RISC inhibitor. Platelet gymnosis of naked miRNAs appeared to be mediated in part by endocytic pathways including clathrin-dependent and fluid-phase endocytosis and caveolae. These results demonstrate the ability of ex vivo platelets to internalize ectopic miRNAs by unassisted transfection, and utilize them to modulate signal-activated translation and platelet function. Our results identify new roles for miR-223–3p in extracellular vesicle generation in stimulated platelets. High efficiency gymnotic transfection of miRNAs in ex vivo platelets may be a broadly useful tool for exploring molecular genetic regulation of platelet function.
Keywords: miRNA, translation, mRNA, microvesicles, exosomes, transfection
Introduction
Anucleate platelets are highly enriched in microRNAs (miRNAs), 19–22 nucleotide non-coding RNAs which function as translational suppressors by multiple mechanisms collectively referred to as RNA inhibition (RNAi). Platelet miRNAs originate in the megakaryocyte nucleus as unprocessed pri-miRNA transcripts, which undergo initial processing and are exported to the megakaryocyte cytosol as short hairpin pre-miRNAs.1 Platelets inherit pre-miRNAs and mature miRNAs from megakaryocytes, and harbor all the protein machinery necessary to support RNAi.1 The endoribonuclease DICER1, expressed in megakaryocytes and platelets, removes the hairpin to generate mature double-stranded (ds) miRNA duplexes comprised of two putative functional miRNAs, the so-called −5p and −3p miRNA arms of the cognate pre-miRNA. The RNA-induced silencing complex (RISC) recruits functional miRNAs by ds miRNA docking into a binding pocket in the central RISC protein ARGONAUTE2 (AGO2), and selection of one of the miRNA strands to retain as the active strand. The active miRNA strand can then suppress translation by recruiting a cognate target mRNA to the binding pocket in AGO2 via seed site complementarity2. Platelets harbor AGO2-based RNAi machinery, but functional roles for miRNAs and RNAi in platelets are not well understood.1
Platelet miRNAs may play important roles in platelet function, but this has been difficult to test experimentally. Platelet activation following agonist exposure stimulates splicing of pre-mRNAs and translation of new protein from newly spliced as well as extant mature mRNAs, suggesting the possibility that miRNAs may modulate signal-activated translation.14,15 Individual variation in platelet miRNA expression levels in humans is associated with platelet reactivity,3, 4 and depletion of most miRNAs by platelet-specific deletion of Dicer1 in mice yields increased mRNA expression and platelet hyper-reactivity.5 However, these effects may reflect alterations in expression of proteins inherited from megakaryocytes, potentially obscuring functional roles of miRNAs in platelets. Hence, the ability to over-express or antagonize miRNAs ectopically in platelets could be valuable for both research and clinical applications. Transfection of miRNAs in ex vivo platelets has been achieved using coupling reagents such as liposome carriers which are standard reagents for transfection of nucleated cells; however, transfection efficiencies with these approaches have generally been low as reported by us and others, limiting their utility.6–8
Circulating platelets internalize plasma-borne RNAs including miRNAs; indeed, platelets are currently being investigated as repositories of RNA-based biomarker signatures for diseases including cancer.9 Recent studies identified microvesicles as a major delivery vehicle for platelet uptake of extracellular mRNAs derived from vascular cells including endothelium, contributing to platelet heterogeneity, and modulating platelet lifespan.10 However, in addition to vesicle-encapsulated miRNAs, many plasma miRNAs appear to be non-vesicular, protected from RNase degradation by encapsulation in AGO2-based RISC or association with other protein macromolecular complexes.11–13 Whether platelets can internalize non-vesicular extracellular miRNAs is not known. Moreover, various cell types have recently been shown to harbor the ability to internalize “naked” miRNAs not associated with protein complexes and without addition of transfection reagents in vitro, through a poorly understood process referred to as “gymnosis”, but this yet to be investigated in the context of platelets.14–16 Platelets can utilize small locked nucleic acid antagomiRs introduced by intravenous transfusion in mice, suggesting the possibility that platelets may employ gymnosis as a mechanism for uptake of short naked RNAs, although potential uptake in megakaryocytes and transfer to nascent platelets was not investigated in those studies.17,18, 19 The functional consequences of uptake of naked miRNAs in platelets remains to be characterized. In this study, we explored gymnosis as a putative mechanism and experimental approach of high efficiency miRNA transfection in platelets, and investigated effects on signal-activated protein translation and functional outcomes.
Methods
Materials
Mouse monoclonal anti-GP1bα (CD42b), goat anti-SEPTIN2, and rabbit anti-CD63 antibodies were from Santa Cruz Biotechnologies (Santa Cruz, CA, USA). Rabbit anti-AGO2 and anti-β-ACTIN were from Cell Signaling (Danvers, MA, USA). Rabbit anti-TC21 antibodies were from Abnova (Taipei City, Taiwan). Fluorophore-conjugated secondary antibodies and pre-immune IgG were from Jackson (West Grove, PA, USA). Streptavidin-conjugated AlexaFluor488 was from Jackson Immunoresearch (Pittsburgh, PA, USA). α-CD45- and α-CD235a-conjugated magnetic beads and MACS separation LD columns were from Miltenyi Biotec (Auburn, CA, USA). Oligonucleotides were from IDT (Coralville, IA, USA). hsa-miR-223–3p miRNA mimics harboring either a 3’ biotin tag on the active strand and a 3’ DY547 fluorophore on the passenger strand (rendered ineffective by 3’ diuridylation), or a 3’ DY547 fluorophore on the active strand only, or with no tags, and miRIDIAN negative control miRNA (C. elegans Cel-miR-67) were from Dharmacon (Lafayette, CO, USA). The miR-223–3p active strand sequence was 5’-ugucaguuugucaaauacccca; the inactive passenger strand sequence was 5’-ugggguauuugacaaacugaca. The Cel-miR-67 active strand sequence was 5’- ucacaaccuccuagaaagaguaga. DNA sequencing was provided by Genewiz (South Plainfield, NJ, USA). Methyl β-cyclodextrin and dynasore hydrate were from Sigma (St. Louis, MO, USA), and LY294002 was from LC Laboratories (Woburn, MA, USA). Thrombin and BCI-137 were from Millipore Sigma (Burlington, MA, USA) and plasminogen-depleted fibrinogen was from EMD Millipore (Danvers, MA, USA). Mitotracker Deep Red FM was from Invitrogen (Carlsbad, CA, USA).
Platelet preparation
Human blood was obtained by venipuncture from a pool of healthy volunteers in a one-sixth volume of acid-citrate-dextrose. Red blood cells were removed by centrifugation at 230×g for 20 min at room temperature. Platelet-rich plasma was recovered, and platelets were pelleted at 800×g for 10 min at room temperature. The platelet pellet was suspended in Tyrode’s buffer (138 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 3 mM NaH2PO4, 5 mM glucose, 10 mM Hepes, pH 7.4 in distilled, DNase/RNase-free water with 0.01 units/ml of apyrase added fresh) containing 1 mg/ml bovine serum albumin (BSA). Leukocyte depletion was achieved by incubation of platelet suspensions with α-CD45- and α-CD235a-conjugated magnetic microbeads at 10 μl each per 10 ml blood, for 15 min with mixing at 4°C, followed by bead extraction with MACS separation LD columns according to the manufacturer’s instructions. Leukocyte depletion was confirmed by RNA extraction from a sample of the pre- and post-depleted platelet supernatants, followed by cDNA synthesis and PCR for CD45 mRNA as described below; data were obtained only from platelet samples with no detectable CD45 expression. Approval for this study was obtained from the Institutional Review Board of Thomas Jefferson University. Written informed consent was obtained after the nature and possible consequences of the study were explained.
Platelet transfection and fluorometry
Platelets were kept in suspension in Tyrode’s buffer with or in some cases without BSA at 2 × 108/ml at room temperature. A stock solution of double-stranded miRNA mimic in distilled, DNase/RNase-free water was added directly to the platelet suspension at a final concentration of 200 nM and the mixture was kept at 37°C with gentle agitation for different times. For fluorometry studies, total mass of the miRNA mimic was calculated as the sum of the oligonucleotide mass (14.664 kDa). RNA concentrations after incubation with platelets were measured by optical density based on RNA fluorescence in a Qubit fluorometer (ThermoFisher Scientific, Waltham, MA, USA), and calculated using the oligonucleotide mass only. In some cases, transfected platelets were treated with 100 ng/ml of RNase III for 30 min followed by inhibition with Superase·In at 0.1 U/ml for 15 min, or miRNA mimic solutions were pre-treated with RNase III under the same conditions, all at room temperature. In some cases, platelets were first captured on coated surfaces prior to miRNA transfection. Platelets were washed in buffer before proceeding with further procedures.
Immunocytochemistry, fluorescence microscopy, and FACS
Platelet suspensions or platelet-depleted releasates were fixed in 0.5% formaldehyde and seeded for 16 hours at 4°C on coverslips coated with 0.01% v/v poly-L-lysine, or unfixed platelets were seeded for 1 h at 37°C on coverslips coated with 5 μg/ml fibronectin before being fixed and processed. Non-adherent material was washed away with PBS and coverslips were treated with 0.1% Triton X-100/PBS for 5 min, equilibrated in PBS, and processed for immunocytochemical staining by incubation with primary antibodies at a dilution of 1:200 plus 1:100 dilution normal donkey serum, or streptavidin-AlexaFluor488 at 1:100 concentration, overnight at 4°C. The next day, coverslips were washed 4x, then 1 mg/ml solutions of fluorescein isothiocyanate (FITC)-, Cy3- or Cy5-conjugated antibodies were applied at a dilution of 1:100 (FITC) or 1:2000 (Cy3/5), for 1 h at room temperature, washed, and mounted with Prolong™ Gold antifade reagent (Life Technologies Corporation, OR, USA). Images were acquired on an EVOS FL Auto microscope with a 60x/1.42 NA oil-immersion objective at room temperature with Pearl EVOS software, or a Zeiss LSM 780 microscope with a 63x/1.40 NA oil-immersion objective at room temperature utilizing a TCS SL confocal system running Zeiss ZEN 2011 SP7 FP3 (64bit) software. Post-acquisition image processing was performed using ImageJ and Adobe Photoshop. Operations included brightness/contrast adjustment to all pixels in the images, grouping of images, and automated particle counting. Background particle counts with secondary antibodies only were subtracted from total particle counts. For FACS, suspensions of fixed platelets were run on a BD LSRII flow cytometer recording 100,000 events. Data was analyzed with FlowJo version 7.6.5.
Neutravidin pulldown, immunoprecipitation and immunoblotting
Platelets were subject to crosslinking by exposure to 312 nm light on a Foto/PrepII UV transilluminator (Fotodyne, Hartland, WI, USA) for 2 min, rotated 90°, and then exposed again for 2 min. Platelets were collected, re-suspended in lysis buffer (10 mM Tris-Cl, pH 7.5, 100 mM NaCl, 2 mM MgOAc, 0.5% Nonidet P-40, and a cocktail of protease inhibitors [Roche, Indianapolis, IN, USA]) for 30 min on ice. Insoluble material and ribosomes were removed by ultracentrifugation at 30,000×g for 30 min at 4°C. For neutravidin pulldowns, supernatants were pre-cleared with agarose beads (Sigma) for 30 min at 4°C. Cleared lysates were incubated with neutravidin-coupled agarose beads (ThermoFisher Scientific) for 16 h at 4°C. For immunoprecipitations, supernatants were pre-cleared by incubation with Protein G-coupled agarose beads (Roche) for 30 minutes at 4°C. Cleared lysates were incubated with 5 μg of AGO2 antibodies for 16 h at 4°C, followed by antibody capture on Protein G-agarose beads for 1 h. Bead-bound complexes were pelleted by centrifugation, washed at least five times in lysis buffer, followed by RNA extraction and processing as described below. For all immunoblotting studies, platelet lysates were generated and normalized to total protein levels, and immunoblotting were performed as described.20
Nanoparticle tracking analysis
NT analysis was performed with a NanoSight NS300 fitted with a 488 nm laser (Malvern, Westborough, MA, USA). Filtered media or buffer (0.22 mm filter) was analyzed each time for background subtraction; typical counts were ~107 (i.e., ~1/100 of the particle counts in the samples). Final particle counts were derived from averaging the final concentrations with background subtracted, for six scans per sample, 30 sec per scan. Addition of 0.1% Triton X-100 caused immediate disappearance of event counts, indicating that the particle counts represented membranous structures (results not shown).
RNA extraction, cDNA synthesis, and PCR
RNA was extracted using TRIzol reagent according to the manufacturer’s instructions, except that 95% EtOH was used for RNA washes to preserve small RNAs. cDNA libraries were constructed using the NCode miRNA First-Strand cDNA Synthesis Kit (Life Technologies, Grand Island, NY, USA) according to the manufacturer’s instructions. Conventional PCR was carried out on an Eppendorf thermal cycler. Quantitative real-time PCR (qRT-PCR) analysis was performed using the LightCycler (Roche, Indianapolis, IN, USA) and the FastStart DNA Master SYBR Green I Kit, according to the manufacturer’s instructions. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a housekeeping control for qRT-PCR experiments for mRNAs, and hsa-miR-24–5p-1 was used as a housekeeping control for miRNAs. Primers used for conventional PCR and qRT-PCR amplification included the following: CD45-F: 5’-ACAGCCAGCACCTTTCCTAC; CD45-R: 5’-GTGCAGGTAAGGCAGCAGA; miR-223–3p-F: 5’-TGTCAGTTTGTCAAATACCCA; Cel-miR-67-F: 5’-TCACAACCTCCTAGAAAGAG; SEPT2-F: 5’-TAAACAGCCTATTCCTAACT; SEPT2-R: 5’- CCCCTCGCTCTTCAATTTC; P2RY12-F: 5’-GAAGACCACCAGGCCATTTAAAAC; P2RY12-R: 5’-GCCTGTTGGTCAGAATCATGTTAG; universal 3’ reverse primer from the NCode cDNA kit; GAPDH-F: 5’-CATGGCCTTCCGTGTTCCTA; GAPDH-R: 5′-CCTGCTTCACCACCTTCTTGAT; hsa-miR-24–5p-1-F: 5’-TGCCTACTGAGCTGATATCAGT. Single bands of predicted product size for each PCR reaction were confirmed by gel electrophoresis from all qRT-PCR samples. Gene expression levels relative to controls were determined using the 2-ΔΔCt method with housekeeping genes.
Statistical methods
Statistical testing described throughout was done using GraphPad Prism software (Graphpad, La Jolla, CA). In some cases, data were normalized to starting measurements to control for baseline variation and statistical analysis was performed prior to normalization. P < 0.05 was considered statistically significant. Values are expressed as mean ± s.e.m. unless otherwise stated. Experiments were repeated at least 3 times unless otherwise stated.
Results
Rapid gymnotic uptake of naked double-stranded miRNA mimic in ex vivo platelets unassisted by transfection reagents
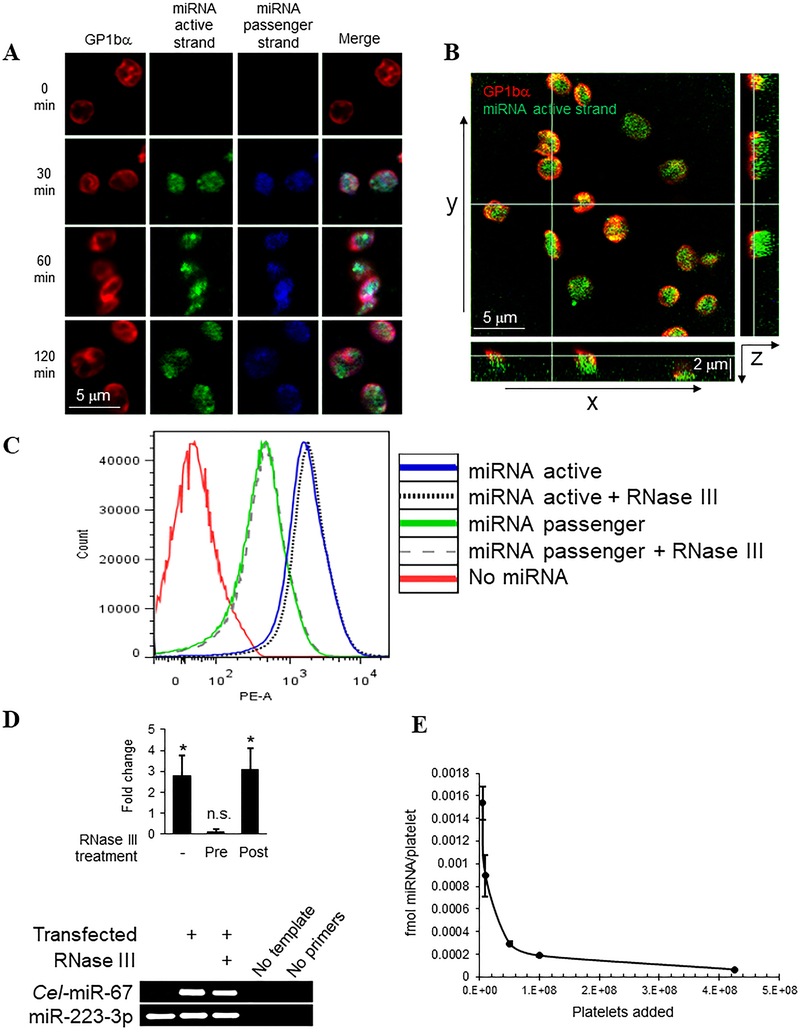
We considered whether platelets can support gymnosis of ds miRNAs ex vivo. miR-223 is enriched in platelets and apparently a major regulator of platelet function.4, 21, 22 Therefore we selected miR-223–3p as a model system to test platelet uptake and downstream effects, reasoning that stoichiometric increase in miR-223–3p would offset the homeostatic balance of target mRNA suppression in platelets and yield functional effects. We incubated human platelets at room temperature with ds miR-223–3p mimic harbouring a biotin tag at the 3’ end of the active strand and a DY547 fluorophore at the 3’ end of the passenger strand – rendered inactive by diuridylation at the 3’ end of this strand - directly in buffer containing 0.1% bovine serum albumin but without supplemental transfection reagents, and then assessed association and internalization of the ectopic ds miRNA. Both active and passenger miRNA strand labels were detected in the platelet cytosol as early as 30 minutes after direct co-incubation. Moreover, nearly all the captured platelets displayed internalized miRNA, indicating rapid, high efficiency transfection per platelet by direct exposure, unassisted by transfection reagents (Figure 1A, B).
Figure 1.
Rapid uptake of naked double-stranded miRNA mimic in ex vivo platelets unassisted by transfection reagents. Freshly isolated leukocyte-depleted platelets (2 × 108/ml, A-C) were suspended in Tyrode’s/BSA buffer and end-end-labelled DY547-miR-223–3p-biotin ds miRNA mimic was added to the platelet suspensions at a final concentration of 200 nM for the indicated times. (A) Platelets were fixed and captured on poly-L-lysine-coated coverslips, permeablized with Triton X-100 and stained with antibodies to GP1bα followed by Cy5-conjugated secondary antibodies to label platelet surfaces (red pseudocolor), and streptavidin-AlexaFluor488 to label the miR-223–3p active (top) strand (green). Confocal microscopy images were obtained including DY547 fluorescence indicating the passenger (bottom) miRNA strand (blue), and merged images are shown to the right for each panel. (B) Confocal z-stack image slice (0.2 μm thick) shown as a central selected xy plane of platelets treated for 60 min as in (A), with xz orthogonal section shown below and yz orthogonal section to the right. (C) Platelets were incubated with miRNA mimics, harboring a DY547 conjugated to the 3’ end of either the active or passenger strand, for 60 min followed in some cases by treatment with RNase III as indicated. Single platelets were analyzed for DY547 fluorescence by FACS using phycoerythrin (PE-A) detection. (D) Platelets were transfected with untagged miR-223–3p mimic or C. elegans Cel-miR-67 miRNA mimic as above, with either pre-treatment of miRNA mimic or post-transfection treatment of the platelets with RNase III, and total extracted RNA was poly(dA) tailed, converted to cDNA and processed for qRT-PCR (upper panel, shown as fold change miR-223–3p over untransfected) or conventional PCR (lower panel, “RNase III” indicates post-treatment) as indicated. *, p < 0.05; n.s., not significant. n = 5. (E) Serial dilutions of platelets (lowest platelet counts shown, 5×106, 1×107) were incubated for 60 min with 1 μM ds miRNA mimic in 100 μl volume of buffer, platelets were depleted by centrifugation, and non-sequestered total miRNA was measured by fluorometry in the supernatants. miRNA consumed per platelet was calculated from the differences from starting material to remaining fraction in each sample, shown ± s.e.m.
As a secondary approach to assess miRNA internalization and to quantify transfection efficiencies, we analysed transfected platelet populations by FACS, taking advantage of the DY547 fluorophore label on the passenger strand, coupled with addition of RNase III after miRNA co-incubation to degrade non-internalized ds RNA.23 We observed a marked rightward shift in mean fluorescence intensities of the entire population of single platelets in samples incubated with miRNA relative to un-transfected platelets. We observed a similar shift using the same miRNA mimic, but with a DY547 fluorophore at the 3’ end of the active strand only. The observed peak was even farther shifted to the right with this miRNA mimic than in platelets incubated with miRNA harboring DY547 anchored to the 3’ end of the passenger strand. These differences in mean fluorescence may reflect possible degradation of the passenger strand upon transfection; however, we cannot rule out the possibility of different transfection efficiencies of the ds miRNA mimics as a function of the strand placement of the fluorophore. Together these data indicate that ds miRNAs are internalized efficiently by the platelets on a per-platelet basis under these conditions (Figure 1C). RNase III treatment applied 60 min after miRNA transfection did not reduce the platelet fluorescence significantly for either the active or passenger strand, confirming that the increased fluorescence was derived from miRNA mimic internalized in the platelets (Figure 1C). Thus, ds miRNA was internalized in platelets under these conditions at close to 100% efficiency on a per-platelet basis.
We considered whether platelet miRNA uptake is a general phenomenon or specific to the miR-223–3p mimic. To investigate this question, and also to quantify the relative increase in miR-223–3p over endogenous levels after transfection, we incubated platelets with either miR-223–3p mimic or Cel-miR-67 mimic (based on C. elegans Cel-miR-67 which has minimal sequence identity with any known human miRNAs), both untagged, and assessed miRNA content in the treated platelets by qRT-PCR and conventional PCR. Direct incubation of platelets with miR-223–3p mimic increased the platelet load of this miRNA by 2.81-fold ± 0.98 over untreated platelets as determined by qRT-PCR, indicating significant stoichiometric increase in miR-223–3p over endogenous levels by this transfection method. In contrast, pre-treatment of the ds miRNA mimic with RNase III prior to transfection resulted in 0.14 ± 0.11-fold change in miR-223–3p in platelets, whereas we observed 3.08 ± 1.02-fold increase in miR-223–3p when platelets were treated with RNase III beginning 1 hr after transfection (Fig. 1D). Cel-miR-67 was undetectable in untreated platelets by qRT-PCR (not shown) or by conventional PCR, but increased dramatically in platelets upon co-incubation, demonstrating that miRNA uptake in platelets is not sequence-dependent. Both miRNA mimics were detected by conventional PCR in transfected platelets treated post-transfection with RNase III (Fig. 1D). To quantify the amount of ectopic miRNA that can be sequestered per platelet by direct co-incubation, we measured the concentration of miRNA remaining in buffer after incubation of 1 μM ds miRNA with serial dilutions of leukocyte-depleted platelets for 60 min, and used these observed values to calculate miRNA sequestration on a per platelet basis. As shown in Figure 1E, platelets sequestered ectopic ds miRNA with an observed maximal ds miRNA consumption of 0.00154 fmol/platelet under these conditions, at the lower concentration of 5×106 platelets in a total volume of 100 μl. Interestingly, the ability of platelets to sequester ectopic miRNA dropped exponentially as platelet concentration increased in the same volume of buffer. This result could indicate that platelet uptake of miRNA may be very rapid and saturating under these conditions, such that increased platelet number dilutes the availability of miRNA molecules per platelet. Alternatively, the increased platelet concentration may alter or interfere with the ability of the platelets to access the ectopic miRNA. Together, these results demonstrate unassisted transfection of ex vivo platelets by direct co-incubation with ectopic ds miRNA, i.e., gymnosis of ectopic miRNA by isolated platelets, at nearly 100% per-platelet efficiency and high stoichiometry.
Transfected naked miRNA is rapidly incorporated into RNA-induced silencing complexes in resting platelets
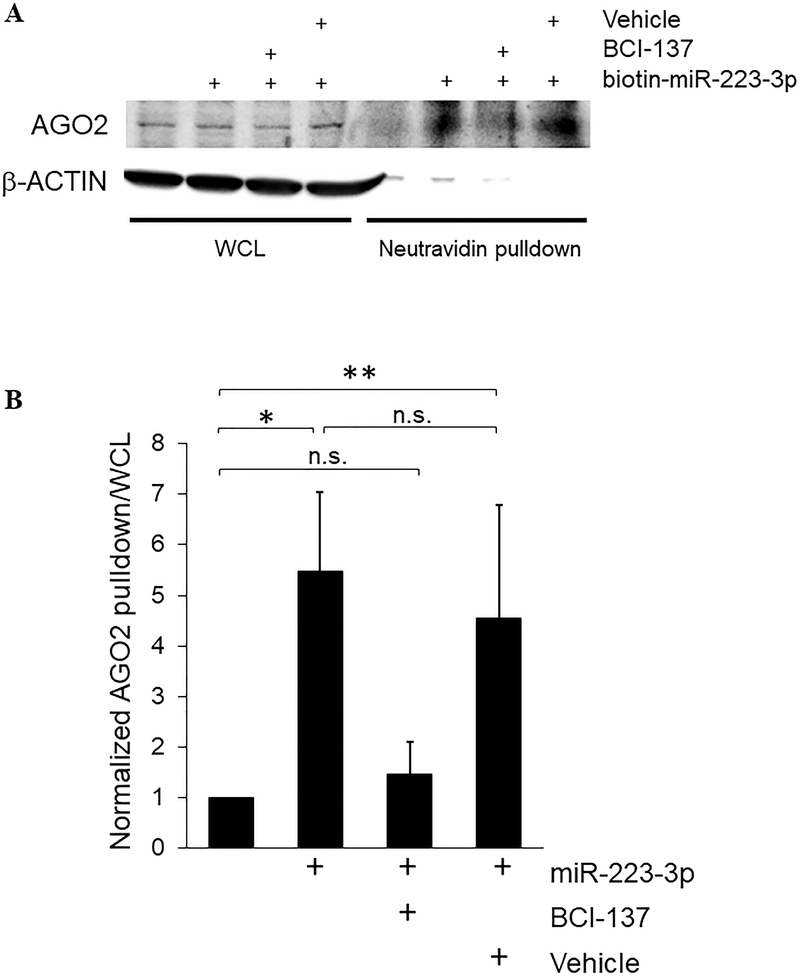
miRNA-mediated translational suppression requires docking of a miRNA in RISC, separation of the ds duplex and ejection of the passenger strand, creating a template for sequence-driven recruitment of mRNA targets of the cognate RISC-bound miRNA.24 We assessed incorporation of gymnotically transfected miRNA into platelet RISC by detection of AGO2 in biotin-miRNA pulldown fractions with specific antibodies.8, 25 AGO2 was detected in the biotin-miR-223–3p pulldown fraction after platelet transfection for 60 minutes, demonstrating rapid association of transfected miRNA with platelet RISC, as well as providing further confirmation of overall rapid uptake into the platelet cytosol. Pre-treatment of platelets with BCI-137, a small molecule inhibitor of AGO2/miRNA binding,26 reduced AGO2/miR-223–3p association, whereas the association was maintained in the presence of DMSO vehicle, further supporting direct association of the transfected miRNA with AGO2 complexes (Figure 2A,B). We evaluated co-precipitation in reverse using immunoprecipitation with AGO2 antibodies from platelets transfected with untagged miR-223–3p mimic, followed by RNA extraction and qRT-PCR.8 We observed a 3.3-fold ± 1.9 increase in miR-223–3p in AGO2 immunoprecipitate fractions in the transfected platelets compared to un-transfected platelets (n = 3, results not shown). Thus, unassisted ds miRNA transfection in platelets is associated with enrichment of the ectopic miRNA active strand in RISC.
Figure 2.
Transfected naked miRNA is rapidly incorporated into RNA-induced silencing complexes in resting platelets. Platelets at 2 × 108/ml were subject to UV crosslinking at the indicated times after co-incubation with ds miR-223–3p mimic, with 10 μM BCI-137 or equivalent volume of DMSO vehicle as shown, and lysed under non-denaturing conditions. RNA-protein complexes containing the biotin-miRNA mimic were enriched by pulldown with neutravidin beads, and proteins in the whole cell lysate (WCL) and bead pulldown fractions were subject to Western blotting with antibodies to AGO2, and β-actin as loading control. (A) Representative blot. (B) AGO2 pulldown/WCL ratios normalized to un-transfected samples, + s.e.m. *, p < 0.02; **, p < 0.05. n.s., not significant. n = 4.
Transfected naked miRNAs suppress signal-activated translation of target mRNAs in platelets
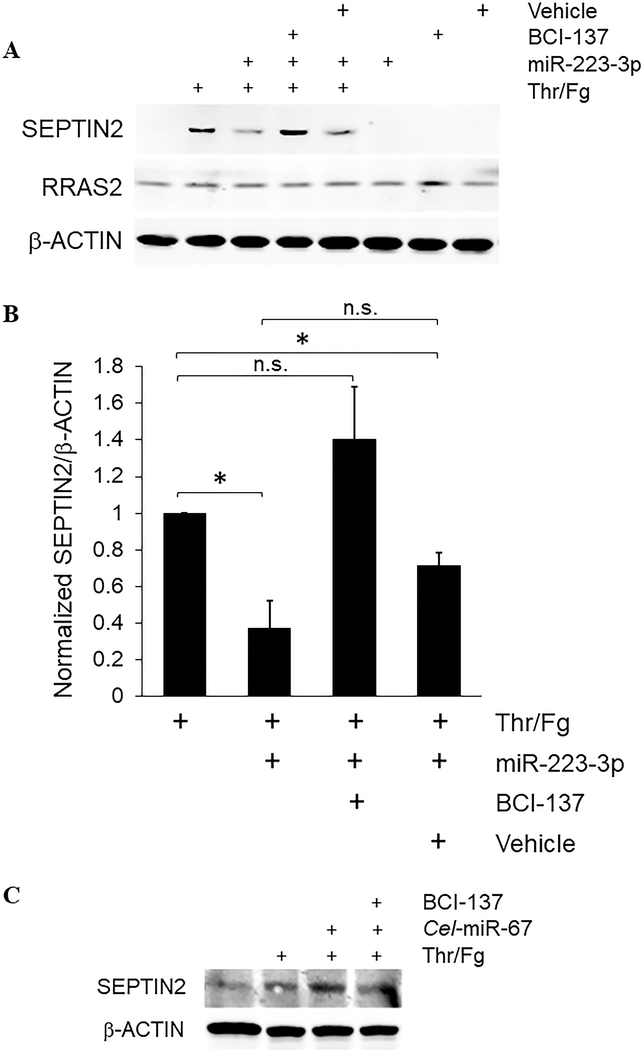
We considered whether ectopic over-expression of miR-223–3p by gymnotic transfection modulates signal-activated translation of target mRNAs in platelets. Established and predicted miR-223–3p target mRNAs coding for platelet-expressed proteins include SEPTIN2, SEPTIN6, P2RY12 encoding the P2Y12 ADP receptor, and RRAS2 (TC21) which we previously showed regulates glycoprotein (GP)VI pathway signalling and functional responses in platelets.1, 27–29 To investigate effects of miR-223–3p transfection on mRNA target translation in stimulated platelets, we used 0.5 U/ml thrombin combined with 100 μg/ml plasminogen-depleted fibrinogen as stimulus, since this treatment has been shown to induce signal-activated translation in platelets.18, 19 We incubated leukocyte-depleted platelets with miR-223–3p mimic for 2 hours, then stimulated platelets with thrombin and fibrinogen for an additional 18 hours in the continuous presence of the miRNA mimic. We tested several commercial P2Y12 antibodies but we were unable to identify reliable P2Y12 antibodies for western blotting (results not shown). RRAS2/TC21 expression was unaltered by platelet stimulation, and miR-223–3p transfection did not have any apparent effect on RRAS2 expression in stimulated or resting platelets. These data suggest that further study is needed to establish RRAS2 mRNA as a functional target of miR-223–3p, as predicted by sequence analysis. We observed low levels of SEPTIN2 protein in freshly isolated resting platelets, confirming previous proteomics and western blot identification of SEPTIN2 in human platelets, although the low levels we and others observed differ slightly from apparently higher levels of SEPTIN2 observed recently in unstimulated platelets after 2 days in storage.28, 30, 31 However, in freshly isolated platelets SEPTIN2 protein expression increased dramatically after thrombin/fibrinogen stimulation, indicating signal-activated translation of SEPTIN2 in platelets under these conditions. Interestingly, gymnotic transfection with miR-223–3p mimic strongly inhibited signal-activated SEPTIN2 protein increase induced by thrombin/fibrinogen stimulation (Figure 3A,B). To assess whether the inhibitory effect of transfected miR-223–3p on induced SEPTIN2 was due to RISC-mediated translational suppression, we pre-treated platelets with BCI-137 or with vehicle. RISC blockade with BCI-137 but not vehicle treatment prevented the ability of miR-223–3p to suppress signal-activated translation of SEPTIN2, indicating that the transfected miRNA functionally inhibited translation of the SEPTIN2 target mRNA via a RISC-mediated process. miR-223–3p mimic, BCI-137 or vehicle alone had no effect on expression of any of the proteins analysed in platelets not stimulated with thrombin and fibrinogen (Figure 3A,B). In contrast to miR-223–3p mimic, induced SEPTIN2 translation was sustained in platelets transfected with negative control miRNA Cel-miR-67 mimic, confirming specific suppression of induced SEPTIN2 translation by miR-223–3p over-expression (Figure 3C). Suppression of induced increase in SEPTIN2 protein levels was not due to direct RNAi via the miRNA mimic passenger strand which resembles (although is not identical to) miR-223–5p, due to diuridylation of this passenger strand which renders it inactive for RNAi; moreover, this miRNA strand does not have predicted seed recognition sites in the human SEPTIN2 mRNA, whereas there are two tandem predicted miR-223–3p seed recognition sites in the SEPTIN2 mRNA 3’-UTR.28 We did not observe significant changes to SEPTIN2 or P2RY12 mRNAs in miR-223–3p-transfected platelets by qRT-PCR (fold change = 1.11 ± 0.4 and 0.83 ± 0.3, respectively, n = 3, results not shown), suggesting blockade of induced increase in SEPTIN2 protein levels by over-expressed miR-223–3p primarily via mechanisms other than target mRNA degradation. Thus, miRNA transfected by gymnosis is functionally active and suppresses signal-induced translation of specific mRNA targets via RISC-mediated suppression of translation in ex vivo platelets.
Figure 3.
Transfected naked miRNAs suppress signal-activated translation of target mRNAs in platelets. Leukocyte-depleted platelets were suspended at 2 × 108/ml in buffer with or without 200 nM ds miR-223–3p, 10 μM BCI-137 or equivalent volume of DMSO (Vehicle) for 2 hours, then stimulated with 0.5 U/ml thrombin combined with 100 μg/ml plasminogen-depleted fibrinogen as indicated, for 18 hours at RT with gentle agitation. Platelet protein extracts were collected by precipitation with perchloric acid, separated by SDS-PAGE, and probed in immunoblots with antibodies to RRAS2, SEPTIN2, and β-actin as loading control. (A) Representative blots. (B) SEPTIN2/β-ACTIN ratios normalized to thrombin/fibrinogen-only samples, + s.e.m. *, p < 0.02. n.s., not significant. n = 4. (C) Representative blots of platelets transfected with Cel-miR-67 and treated as in (A). n = 3.
Platelet transfection with miR-223–3p suppresses extracellular vesicle release induced by thrombin/fibrinogen
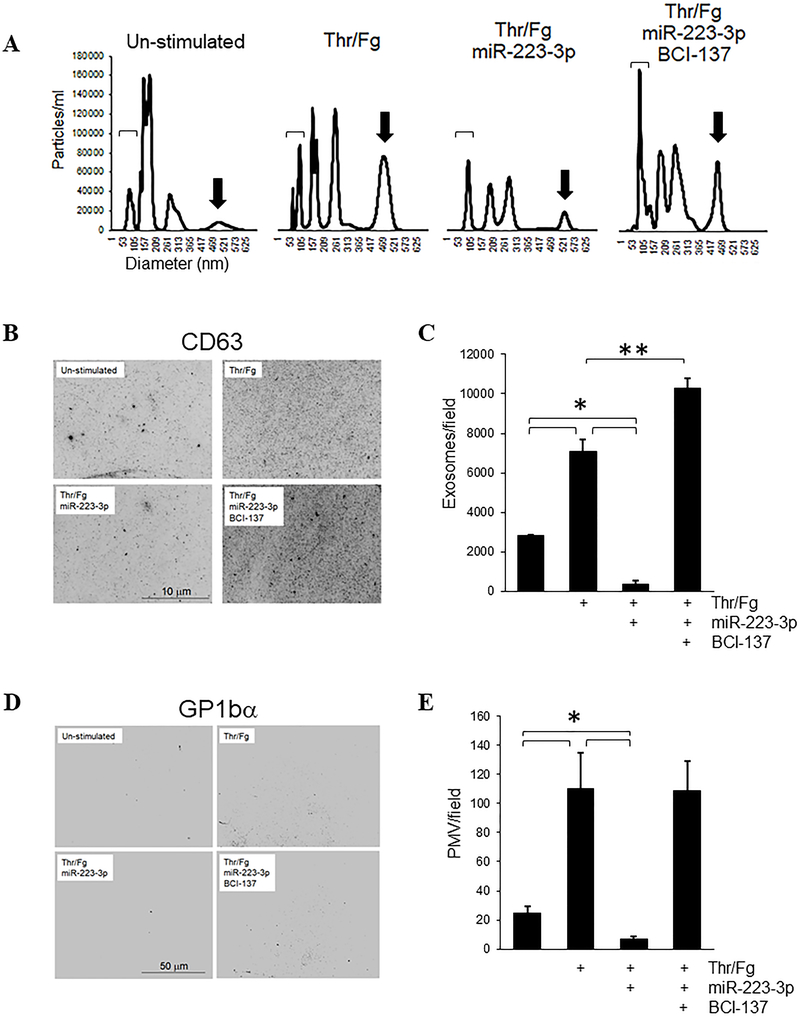
We investigated functional outcomes of miR-223–3p over-expression by gymnotic transfection in platelets stimulated with thrombin and fibrinogen, focusing on long-term outcomes reflecting modulation of signal-activated protein translation of SEPTIN2, and putatively other proteins, by ectopic miR-223–3p. Specific functions of SEPTIN2 in platelets are not known. SEPTIN2 has been associated with SNARE-dependent exocytosis which mediates exosome secretion and microvesicle release,32, 33 and with mitochondrial fission.34–36 Whereas α- and δ-granule secretion from platelets occurs rapidly after stimulation, platelet exosome secretion and microvesicle generation and release occur over much longer time frames that may permit regulation by signal-activated protein translation.37–39 We considered whether platelet exosome and/or microvesicle generation may be modulated by miR-223–3p. We assessed the exosome and microvesicle content in platelet releasates collected 18 hours after stimulation by nanoparticle tracking analysis (NTA). Platelets maintained with gentle agitation but without thrombin/fibrinogen released exosomes, identified as vesicles with diameter ranging from 10–120 nm (hereafter abbreviated PEX for platelet exosomes), and a heterogeneous cohort of platelet microvesicles (PMV). Thrombin/fibrinogen stimulation enhanced PEX release, as well as an increase in generation of PMV as predicted, as evidenced by a decrease in release of small diameter vesicles coupled with concomitant increased concentration of vesicles of larger diameter after stimulation.37 In contrast, gymnotic transfection with miR-223–3p mimic inhibited thrombin/fibrinogen-induced release of both PEX and PMV. This inhibition of PEX and PMV generation by ectopic miR-223–3p was reversed by pre-treatment with BCI-137. Pre-treatment with BCI-137 also substantially enhanced induced PEX generation relative to resting platelets and to stimulated platelets without BCI-137 (Figure 4A).
Figure 4.
Platelet transfection with miR-223–3p suppresses exosome and microvesicle release induced by thrombin/fibrinogen. Leukocyte-depleted platelets at 2 × 108/ml were stimulated and transfected with 200 nM miR-223–3p mimic +/− 10 μM BCI-137 as shown for 18 hours, then platelets were collected by centrifugation and discarded. (A) Platelet releasates in the resulting supernatants were analyzed by NTA. Representative NTA tracings are shown. Brackets indicate 50–100 nm exosomes; arrows indicate ~400–550 nm diameter PMV. (B) Particles in releasates were fixed and captured on poly-L-lysine-coated coverslips, and stained with α-CD63 and α-GP1bα antibodies. Shown are the central segments of representative micrographs of exosomes imaged by epifluorescence microscopy for CD63 staining. (C) Exosomes/field were counted from full-frame micrograph images as in (B) for at least 5 fields per sample, shown as mean values after background subtraction + s.e.m. (D) Representative images of the same captured material as in (C), stained with GP1bα antibodies to label PMV. (E) PMV/field were counted from images as in (D) for at least 5 fields per sample, shown + s.e.m. *, p < 0.01; **, p < 0.02.
To distinguish PEX and PMV on a basis other than diameter and to confirm that the observed differences in NTA events represented differences in specific cohorts of exosomes and PMV, we captured platelet-derived extracellular vesicles from the releasates on poly-L-lysine-coated coverslips, and immunostained with antibodies to markers for exosomes (CD63) and PMV (GP1bα). Reflecting the NTA results, thrombin/fibrinogen stimulation yielded a significant increase in CD63+ PEX and GP1bα+ PMV, whereas we observed a substantial reduction in both PEX and PMV in stimulated platelets that had been transfected with miR-223–3p relative to un-transfected platelets, and this suppressive effect of miR-223–3p was reversed by BCI-137 (Figure 4B–E). The increased exosome release with BCI-137 in stimulated platelets against a background of miR-223–3p over-expression, compared with stimulated platelets alone, suggests that other miRNAs in addition to miR-223–3p contribute to thrombin/fibrinogen-induced platelet exosome release. We note that due to the 200-nm limit of resolution of classical optical microscopy, the observed CD63+ puncta likely represent expanded fluorescence from point sources of individual exosomes but may also represent small aggregates of exosomes; however, we detected only background fluorescence in coverslips stained directly without exposure to platelet releasates or releasate-exposed coverslips with secondary antibody conjugates alone (results not shown; background subtracted in Figure 4C), confirming that the observed staining represents CD63+ platelet-derived material above background in the releasate samples (Figure 4B,C). Thus, ectopic over-expression of miRNAs by gymnotic transfection can modulate platelet function via RISC-mediated translational suppression. These data further demonstrate new functions for miR-223–3p in platelets: regulation of exosome and microvesicle release from thrombin/fibrinogen-stimulated platelets.
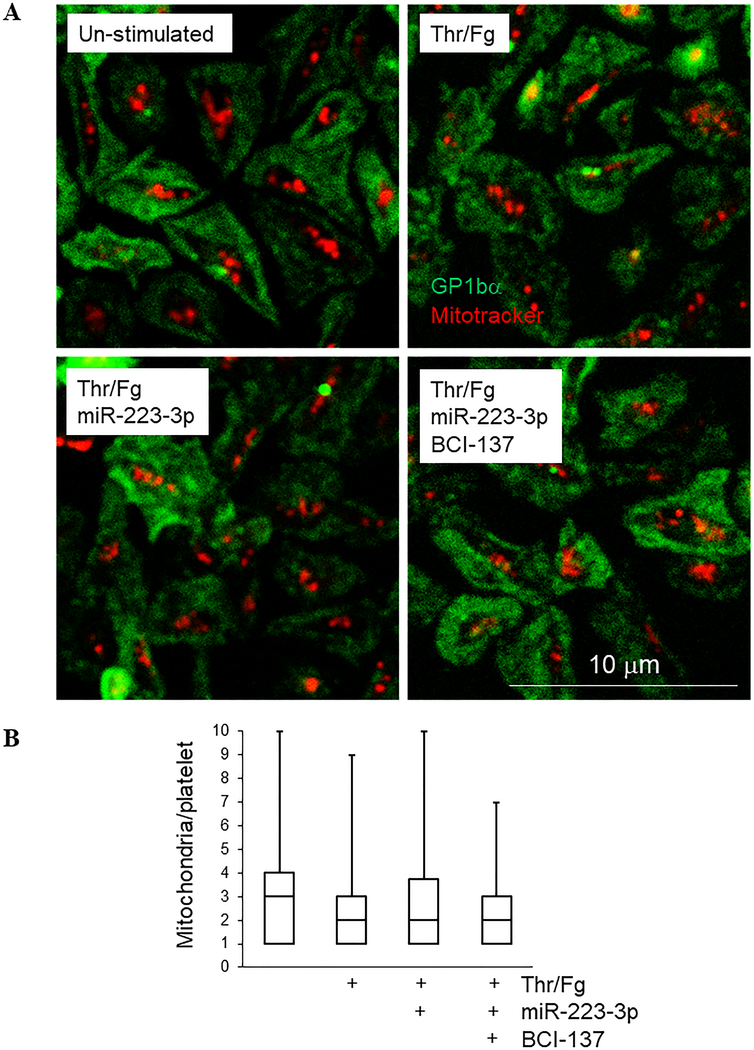
Mitochondrial count is not affected by miR-223–3p over-expression or RNAi inhibition in resting or stimulated platelets
Based on roles for SEPTIN2 in mitochondrial fission in nucleated cells, we considered whether miR-223–3p over-expression or RISC inhibition affects the number of mitochondria in resting and stimulated platelets. Imaging of platelet mitochondria with Mitotracker revealed the presence of ~2–8 mitochondria per platelet (median, 3–4) under resting conditions as anticipated, but we did not observe significant changes in platelet mitochondrial count under any of the conditions tested (Figure 5), consistent with recent indications that mitochondrial fission occurs during late megakaryopoiesis or early thrombopoiesis rather than as an ongoing or stimulated process in platelets.40 From these results we conclude that mitochondrial count is stable and not altered by thrombin/fibrinogen stimulation in platelets, and miRNA-mediated translational suppression does not modulate mitochondrial count in resting or stimulated platelets.
Figure 5.
Mitochondrial count is not affected by miR-223–3p over-expression or RNAi inhibition in resting or thrombin/fibrinogen-stimulated platelets. (A) Mitotracker-labelled platelets were captured on fibronectin-coated coverslips and allowed to spread for 60 minutes before being treated as shown for 18 hours, stained with antibodies to GP1bα to indicate platelet borders (green), then imaged by confocal fluorescence microscopy. Mitochondria are shown in red. (B) Mitochondria per platelet are shown + max values for at least 50 platelets per sample from (A).
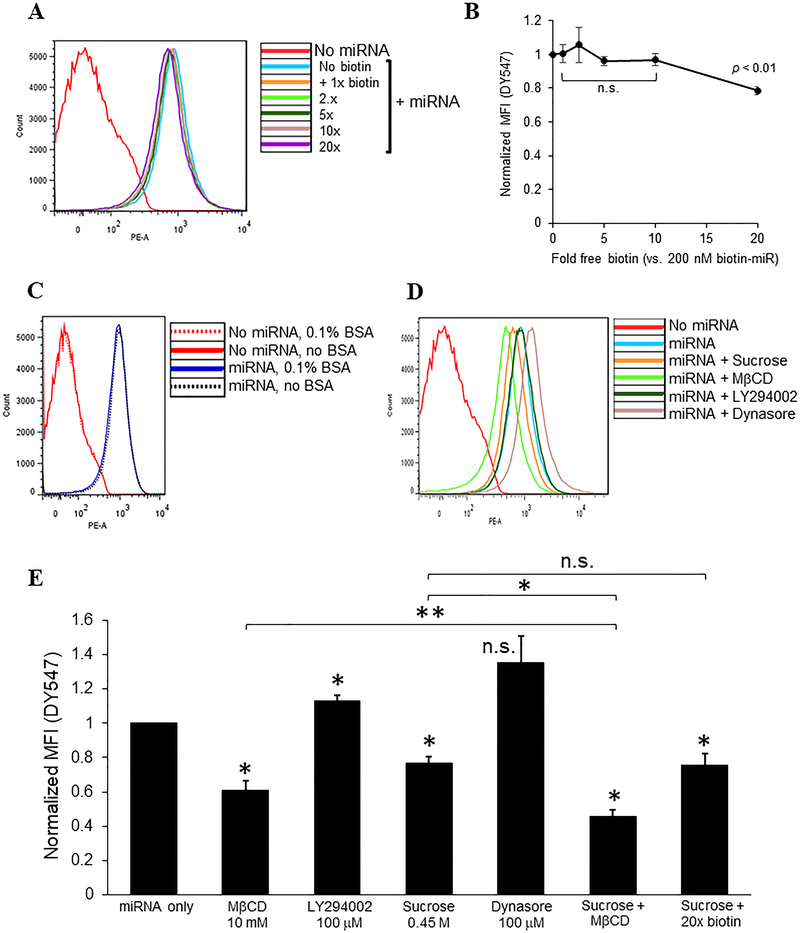
Gymnosis of naked miRNAs in platelets is partially mediated by caveolae and clathrin-dependent or fluid-phase endocytosis
We next examined cellular mechanisms for gymnosis of naked ds miRNAs in platelets. We first considered whether the linked biotin on the miRNA mimic mediates platelet association and subsequent uptake. We incubated platelets with excess free biotin prior to transfection and RNase A post-transfection treatment, and assessed miRNA uptake by flow cytometry. We observed minimal blockade of uptake of miRNA with excess biotin, by which blockade appeared to be dose-dependent but only reached significance at 20-fold molar excess competitive free biotin, with only 20% inhibition at the highest dose of competitor (Figure 6A, B). We conclude from these results that the biotin linker has a very minor role in uptake of free miRNA in ex vivo platelets. We next considered whether the albumin (0.1% bovine serum albumin) added to the Tyrode’s platelet buffer was involved in miRNA uptake. Platelet albumin is well-known to be internalized by circulating platelets through multiple mechanisms, and as a carrier protein, albumin could facilitate miRNA internalization.41 Surprisingly, platelets kept in Tyrode’s buffer with or without added albumin internalized ectopic naked miRNA to similar levels, indicating that platelet gymnosis of extracellular miRNA does not require exogenous albumin (Fig. 6C).
Figure 6.
Gymnosis of naked miRNAs in platelets is partially mediated by lipid rafts/caveolae and clathrin-dependent and fluid-phase endocytosis. Platelets at 2 × 108/ml were pre-treated with biotin or inhibitors for 30 min prior to addition of miR-223–3p mimic at 200 nM, incubated for 60 minutes in the continued presence of biotin or inhibitors, or in Tyrode’s buffer lacking 0.1% bovine serum album (BSA), washed and fixed, treated with RNase A followed by RNase inhibition, then analyzed by FACS for DY547 fluorescence. (A, C, D) Representative FACS histograms of DY547 fluorescence (PE-A) for platelets treated with (A) fold molar excess free biotin or (D) endocytic pathway inhibitors, or (C) in the presence or absence of BSA as indicated. (B, E) Normalized mean fluorescence intensities (MFI) from (A, D, respectively) after background subtraction of transfected platelets treated as indicated, ± s.e.m. *, p < 0.01 compared to miRNA only, or as indicated; **, p < 0.05. n.s., not significant. n = 4.
Gymnosis of extracellular RNA in nucleated cells has been proposed to involve endocytic engulfment, requiring subsequent translocation of engulfed RNA from the endo-lysosomal system into the cytosol.42 To identify putative endocytic mechanisms of miRNA gymnosis in platelets, we incubated platelets with specific inhibitors targeting lipid rafts/caveolae (methyl-β-cyclodextrin, MβCD), macropinocytosis (LY294002), clathrin-dependent endocytosis (hyperosmotic sucrose, which also inhibits fluid-phase endocytosis), or dynamin (dynasore) prior to and concurrent with miRNA transfection, and monitoring of internalized miRNA by flow cytometry.43–47 Treatment with sucrose or MβCD each caused partial inhibition of miRNA uptake in platelets, whereas LY294002 and dynasore had little effect. Combined treatment with sucrose and MβCD had an additive inhibitory effect over each inhibitor alone, whereas addition of 20-fold molar excess biotin in combination with sucrose did not yield further inhibition of uptake beyond sucrose treatment alone (Figure 6D,E). These results indicate that clathrin-dependent endocytosis and potentially fluid-phase endocytosis, and caveolae independently contribute to gymnotic ds miRNA uptake in ex vivo platelets, with apparently minimal roles for macropinocytosis.
Discussion
The major findings of this study are that human platelets are capable of internalizing naked double-stranded miRNAs ex vivo in the absence of supplemental transfection reagents, and platelets incorporate internalized naked miRNAs into RNA-induced silencing complexes and subsequently utilize the transfected miRNA to mediate RNA inhibition. We confirmed that internalized miRNAs can function to suppress signal-dependent translation in platelets, as evidenced by suppression of signal-activated SEPTIN2 expression using unassisted transfection of a miR-223–3p mimic, which in turn inhibited signal-activated release of extracellular vesicles in a RISC-dependent manner. MiRNA uptake was mediated by multiple independent mechanisms including clathrin-dependent and/or fluid-phase endocytosis and caveolae. Together, these results demonstrate the utility of gymnotic transfection to investigate functional roles of ectopic miRNAs in platelet protein expression and platelet cellular function.
Whereas miRNA transfection in platelets has been achieved in recent years by us and others using variations of traditional liposome-based transfection approaches, efficiencies were generally low, although functional effects could be observed in some cases even with these low efficiencies.6–8, 48 High efficiency gymnosis of miRNAs by platelets ex vivo offers improved utility as a simple and effective technique for molecular genetics studies of platelet function. Whether circulating platelets can support gymnosis of plasma miRNAs in vivo has not been investigated. However, naked miRNAs are unlikely to be prevalent in plasma due to RNase activity; plasma miRNAs are highly stable due primarily to protection from degradation by association with various plasma proteins such as HDL or extra-vesicular AGO2.12, 13, 49 Several groups have also transfused locked nucleic acid (LNA) antagomiRs, which are refractory to cleavage by plasma RNases due to the LNA linkages.15, 17 Therefore, it is possible that these oligonucleotides could be internalized directly as “free” oligonucleotides by circulating platelets, although whether the transfused antagomiRs remain unbound from other molecules or vesicles in plasma, or potential uptake in megakaryocytes and transfer to new platelets, was not investigated in those studies.
Gymnotic over-expression of miR-223–3p suppressed translation of SEPTIN2 in stimulated human platelets, identifying SEPTIN2 as a new target of miR-223–3p in platelets. Alterations in miRNA:mRNA target stoichiometry are well known to be sufficient to affect target suppression.50 Human platelets harbor an abundance of miR-223 (−5p and −3p),4, 51 and the −3p arm has recently been found to target SEPTIN2 in other cell types28, but a connection in platelets was not previously established. A recent quantitative proteomic screen of resting versus agonist-stimulated human platelets indicated moderate increase in total SEPTIN2 protein levels after 2 hr stimulation with thrombin receptor activating peptide (TRAP), which activates human platelets selectively via the PAR1 but not the PAR4 thrombin receptor.52 Our results showed substantial increase in SEPTIN2 protein levels after 18 hr stimulation with thrombin and fibrinogen. These differences in SEPTIN2 expression under each condition may indicate continuing translation of SEPTIN2 over time upon platelet stimulation, or it may be the case that PAR4 and/or αIIbβ3 integrin fibrinogen receptor ligation stimulates robust SEPTIN2 translation through receptor-specific signaling pathways, although this question requires further investigation. Precise roles for SEPTIN2 in platelets remain unknown, but may include regulation of extracellular vesicles via interactions with SNARE proteins.35
We found that miR-223–3p inhibited agonist-induced generation of platelet exosomes and microvesicles, in particular PMV ranging in diameter up to 550 nm (compared to the reported upper limit of 2000 nm).37 These findings are consistent with long-term effects and physiological functions of prolonged platelet stimulation and the extended time required to translate new protein in stimulated platelets, beyond their ability to respond rapidly to stimulation in a hemostatic context (e.g., to secrete granule contents, activate integrins, and crosslink fibrinogen), which cannot rely on the much slower process of translation. Regulation of stimulated exosome and microvesicle release in platelets are new functions for miR-223–3p; however, we cannot rule out indirect effects of long-term incubation of platelets with miRNA mimics on this or other long-term platelet responses. The importance of blood cell-derived miR-223 in various processes and pathologies has been established, such as in cancer, angiogenesis, liver disease, vascular injury, and recently platelet-derived miR-223 was associated with vascular injury repair, but specific contributions of miR-223–3p to generation of platelet extracellular vesicles in these contexts remains to be investigated.22, 53–58 Genomic deletion of miR-223 in mice had surprisingly little effect on platelet production or reactivity, although roles in platelet extracellular vesicle generation were not tested under those conditions.59 Further studies are needed to elucidate the functional roles of modulation of platelet extracellular vesicle release by loss or over-expression of miR-223–3p. As miR-223 contained in extracellular vesicles has been shown to regulate function in distal cells and this transfer is associated with various pathologies, it will also be important to consider packaging of miR-223–3p itself as well as other miRNAs in platelet exosomes and microvesicles in further studies.8, 60–63
Our results indicate that platelets can utilize multiple mechanisms for internalizing ectopic naked ds miRNA, with apparent contributions from various endocytic pathways spanning caveolae, clathrin-dependent and fluid-phase endocytosis. Platelets are well-known to support each of these endocytic pathways, which together provide platelets with both degrading and non-degrading mechanisms for internalizing plasma components.41, 64, 65 Although albumin can act as a carrier protein in many contexts and is internalized by circulating platelets via a combination of the above mechanisms, ex vivo platelets were able to take up ectopic miRNA even in the absence of bovine serum albumin in the buffer, suggesting that albumin is not required for this process.66 However, a role for platelet-associated albumin carried over from the donor plasma could not be ruled out.
While caveolae and clathrin-based endocytosis typically mediate internalization of receptor-bound molecules, the specific surface receptors that may mediate double-stranded miRNA uptake via platelet gymnosis remain to be determined. Platelets express various surface proteins involved in RNA binding and uptake such as toll-like receptors. Our results also leave open the possibility of a receptor-independent mechanism of ectopic miRNA via fluid-phase endocytosis (pinocytosis). It will be interesting in future studies to elucidate the precise molecular mechanisms of miRNA uptake by ex vivo platelets. Ectopic miRNAs internalized via endocytic pathways must subsequently leave the endosomal system to access RISC in the cytosol, a process referred to as endosomal escape.67, 68 Mechanisms underlying endosomal escape of ectopic naked miRNAs that may be endocytosed by platelets also remain to be determined.42, 69 A portion of the internalized miRNA appeared non-vesicular and diffuse in the platelet cytosol at the earliest time points after miRNA exposure (Fig. 1), suggesting the presence of internalized miRNA that is available for RISC loading. Whether this pool of cytosolic internalized miRNA originated in endocytic vesicles or was transferred directly to the cytosol from the extracellular space could not be determined. Ultimately, gymnotically internalized miRNA reached sufficient levels in the cytosol to be detected in RISC and to mediate RNAi, indicating that endosomal entrapment is not a technical barrier to efficient platelet transfection of miRNAs by this method.
The overall mechanisms and functions of RNAi in platelet biology remain to be elucidated. Gymnosis of naked miRNAs is a novel technique for transfecting extracellular miRNAs into platelets ex vivo, which can be used to test molecular and functional effects of platelet miRNAs. This method provides a simple, direct, high efficiency approach to suppress expression of target genes in platelets without the need for pharmacological or genomic manipulations. However, as each miRNA has many mRNA targets, careful interpretation of functional data following miRNA over-expression is always warranted. We propose that, as with all miRNA expression studies, ex vivo miRNA transfection should mainly be considered for testing specific roles of miRNAs in platelet gene expression and function. Although the studies here focused on human platelets, platelets from other species may be amenable to gymnotic transfection with miRNAs, which may support adoptive transfer-based functional assays to investigate roles of miRNAs in platelet function in vivo.
Acknowledgements
This work was supported by the National Institutes of Health under Grants R01HL137207 (L.E.G.) and R01HL144574 (P.M.). S.L., J.G.T.W. and X.C. performed experiments. S.L. and L.E.G. conceived the study and wrote the manuscript. P.M. provided critical reagents and scientific consultation. All authors read the manuscript and approved its contents.
Footnotes
Declaration of Interest Statement
The authors report no conflicts of interest.
References
- 1.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol 2009:16(9): 961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004:305(5689): 1437–1441. [DOI] [PubMed] [Google Scholar]
- 3.Nagalla S, Shaw C, Kong X, Kondkar AA, Edelstein LC, Ma L, Chen J, McKnight GS, Lopez JA, Yang L et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood 2011:117(19): 5189–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon LM, Edelstein LC, Nagalla S, Woodley AB, Chen ES, Kong X, Ma L, Fortina P, Kunapuli S, Holinstat M et al. Human platelet microRNA-mRNA networks associated with age and gender revealed by integrated plateletomics. Blood 2014:123(16): e37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rowley JW, Chappaz S, Corduan A, Chong MM, Campbell R, Khoury A, Manne BK, Wurtzel JG, Michael JV, Goldfinger LE et al. Dicer1 mediated miRNA processing shapes the mRNA profile and function of murine platelets. Blood 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pendaries C, Watson SP, Spalton JC. Methods for genetic modification of megakaryocytes and platelets. Platelets 2007:18(6): 393–408. [DOI] [PubMed] [Google Scholar]
- 7.Hong W, Kondkar AA, Nagalla S, Bergmeier W, Jin Y, Herman JH, Bray PF. Transfection of human platelets with short interfering RNA. Clin Transl Sci 2011:4(3): 180–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michael JV, Wurtzel JGT, Mao GF, Rao AK, Kolpakov MA, Sabri A, Hoffman NE, Rajan S, Tomar D, Madesh M et al. Platelet microparticles infiltrating solid tumors transfer miRNAs that suppress tumor growth. Blood 2017:130(5): 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Best MG, Vancura A, Wurdinger T. Platelet RNA as a circulating biomarker trove for cancer diagnostics. J Thromb Haemost 2017:15(7): 1295–1306. [DOI] [PubMed] [Google Scholar]
- 10.Clancy L, Beaulieu LM, Tanriverdi K, Freedman JE. The role of RNA uptake in platelet heterogeneity. Thromb Haemost 2017:117(5): 948–961. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008:18(10): 997–1006. [DOI] [PubMed] [Google Scholar]
- 12.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011:13(4): 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011:108(12): 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soifer HS, Koch T, Lai J, Hansen B, Hoeg A, Oerum H, Stein CA. Silencing of gene expression by gymnotic delivery of antisense oligonucleotides. Methods Mol Biol 2012:815(333–346. [DOI] [PubMed] [Google Scholar]
- 15.Stein CA, Hansen JB, Lai J, Wu S, Voskresenskiy A, Hog A, Worm J, Hedtjarn M, Souleimanian N, Miller P et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res 2010:38(1): e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi M, Contu VR, Kabuta C, Hase K, Fujiwara Y, Wada K, Kabuta T. SIDT2 mediates gymnosis, the uptake of naked single-stranded oligonucleotides into living cells. RNA Biol 2017:14(11): 1534–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Abraham S, Andre P, Edelstein LC, Shaw CA, Dangelmaier CA, Tsygankov AY, Kunapuli SP, Bray PF, McKenzie SE. Anti-miR-148a regulates platelet FcgammaRIIA signaling and decreases thrombosis in vivo in mice. Blood 2015:126(26): 2871–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weyrich AS, Dixon DA, Pabla R, Elstad MR, McIntyre TM, Prescott SM, Zimmerman GA. Signal-dependent translation of a regulatory protein, Bcl-3, in activated human platelets. Proc Natl Acad Sci U S A 1998:95(10): 5556–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmerman GA, Weyrich AS. Signal-dependent protein synthesis by activated platelets: new pathways to altered phenotype and function. Arterioscler Thromb Vasc Biol 2008:28(3): s17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wurtzel JG, Kumar P, Goldfinger LE. Palmitoylation regulates vesicular trafficking of R-Ras to membrane ruffles and effects on ruffling and cell spreading. Small GTPases 2012:3(3): 139–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elgheznawy A, Shi L, Hu J, Wittig I, Laban H, Pircher J, Mann A, Provost P, Randriamboavonjy V, Fleming I. Dicer cleavage by calpain determines platelet microRNA levels and function in diabetes. Circ Res 2015:117(2): 157–165. [DOI] [PubMed] [Google Scholar]
- 22.Shi R, Zhou X, Ji WJ, Zhang YY, Ma YQ, Zhang JQ, Li YM. The Emerging Role of miR-223 in Platelet Reactivity: Implications in Antiplatelet Therapy. Biomed Res Int 2015:2015(981841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev 2010:34(5): 883–923. [DOI] [PubMed] [Google Scholar]
- 24.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 2007:130(1): 101–112. [DOI] [PubMed] [Google Scholar]
- 25.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010:141(1): 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masciarelli S, Quaranta R, Iosue I, Colotti G, Padula F, Varchi G, Fazi F, Del Rio A. A small-molecule targeting the microRNA binding domain of argonaute 2 improves the retinoic acid differentiation response of the acute promyelocytic leukemia cell line NB4. ACS Chem Biol 2014:9(8): 1674–1679. [DOI] [PubMed] [Google Scholar]
- 27.Janapati S, Wurtzel JG, Dangelmaier C, Manne BK, Bhavanasi D, Kostyak JC, Kim S, Holinstat M, Kunapuli S, Goldfinger LE. TC21/RRas2 regulates GPVI-FcRg-mediated platelet activation and thrombus stability. Journal of Thrombosis and Haemostasis 2018:Accepted manuscript in press.( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chattopadhyay M, Dahiya N, Atreya C. MicroRNA-223 Regulates Septin-2 and Septin-6 in Stored Platelets. Microrna 2018:7(3): 223–228. [DOI] [PubMed] [Google Scholar]
- 29.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007:129(7): 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartsch I, Blaser S, Roseler S, Sandrock K, Busse A, Huber M, Rempp H, Lieber M, Horn J, Brendle C et al. Human endothelial and platelet septin SEPT11: cloning of novel variants and characterisation of interaction partners. Thromb Haemost 2010:104(6): 1201–1210. [DOI] [PubMed] [Google Scholar]
- 31.Trugilho MRO, Hottz ED, Brunoro GVF, Teixeira-Ferreira A, Carvalho PC, Salazar GA, Zimmerman GA, Bozza FA, Bozza PT, Perales J. Platelet proteome reveals novel pathways of platelet activation and platelet-mediated immunoregulation in dengue. PLoS Pathog 2017:13(5): e1006385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014:30(255–289. [DOI] [PubMed] [Google Scholar]
- 33.Somasundaram A, Taraska JW. Local protein dynamics during microvesicle exocytosis in neuroendocrine cells. Mol Biol Cell 2018:29(15): 1891–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silio V, Marques M, Cortes I, Zuluaga S, Carrera AC. A cascade involving p85, Cdc42 and septin 2 regulates cytokinesis. Biochem Soc Trans 2007:35(Pt 2): 222–224. [DOI] [PubMed] [Google Scholar]
- 35.Tokhtaeva E, Capri J, Marcus EA, Whitelegge JP, Khuzakhmetova V, Bukharaeva E, Deiss-Yehiely N, Dada LA, Sachs G, Fernandez-Salas E et al. Septin dynamics are essential for exocytosis. J Biol Chem 2015:290(9): 5280–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagliuso A, Tham TN, Stevens JK, Lagache T, Persson R, Salles A, Olivo-Marin JC, Oddos S, Spang A, Cossart P et al. A role for septin 2 in Drp1-mediated mitochondrial fission. EMBO Rep 2016:17(6): 858–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponomareva AA, Nevzorova TA, Mordakhanova ER, Andrianova IA, Rauova L, Litvinov RI, Weisel JW. Intracellular origin and ultrastructure of platelet-derived microparticles. J Thromb Haemost 2017:15(8): 1655–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duvernay M, Young S, Gailani D, Schoenecker J, Hamm HE. Protease-activated receptor (PAR) 1 and PAR4 differentially regulate factor V expression from human platelets. Mol Pharmacol 2013:83(4): 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ambrose AR, Alsahli MA, Kurmani SA, Goodall AH. Comparison of the release of microRNAs and extracellular vesicles from platelets in response to different agonists. Platelets 2018:29(5): 446–454. [DOI] [PubMed] [Google Scholar]
- 40.Tugolukova EA, Campbell RA, Hoerger KB, Schwertz H, Weyrich AS, Rowley JW. Mitochondrial Fission Protein Drp1 Regulates Megakaryocyte and Platelet Mitochondrial Morphology, Platelet Numbers, and Platelet Function. Blood 2017:130(455. [Google Scholar]
- 41.Banerjee M, Whiteheart SW. The ins and outs of endocytic trafficking in platelet functions. Curr Opin Hematol 2017:24(5): 467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen TA, Smith BRC, Elgass KD, Creed SJ, Cheung S, Tate MD, Belz GT, Wicks IP, Masters SL, Pang KC. SIDT1 Localizes to Endolysosomes and Mediates Double-Stranded RNA Transport into the Cytoplasm. J Immunol 2019:202(12): 3483–3492. [DOI] [PubMed] [Google Scholar]
- 43.Araki N, Johnson MT, Swanson JA. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J Cell Biol 1996:135(5): 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gousset K, Wolkers WF, Tsvetkova NM, Oliver AE, Field CL, Walker NJ, Crowe JH, Tablin F. Evidence for a physiological role for membrane rafts in human platelets. J Cell Physiol 2002:190(1): 117–128. [DOI] [PubMed] [Google Scholar]
- 45.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell 2006:10(6): 839–850. [DOI] [PubMed] [Google Scholar]
- 46.Guo S, Zhang X, Zheng M, Zhang X, Min C, Wang Z, Cheon SH, Oak MH, Nah SY, Kim KM. Selectivity of commonly used inhibitors of clathrin-mediated and caveolae-dependent endocytosis of G protein-coupled receptors. Biochim Biophys Acta 2015:1848(10 Pt A): 2101–2110. [DOI] [PubMed] [Google Scholar]
- 47.Wang S, Singh RD, Godin L, Pagano RE, Hubmayr RD. Endocytic response of type I alveolar epithelial cells to hypertonic stress. Am J Physiol Lung Cell Mol Physiol 2011:300(4): L560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu S, Huang H, Deng G, Xie Z, Ye Y, Guo R, Cai X, Hong J, Qian D, Zhou X et al. miR-326 targets antiapoptotic Bcl-xL and mediates apoptosis in human platelets. PLoS One 2015:10(4): e0122784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muth DC, Powell BH, Zhao Z, Witwer KW. miRNAs in platelet-poor blood plasma and purified RNA are highly stable: a confirmatory study. BMC Res Notes 2018:11(1): 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009:136(2): 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bray PF, McKenzie SE, Edelstein LC, Nagalla S, Delgrosso K, Ertel A, Kupper J, Jing Y, Londin E, Loher P et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genomics 2013:14(1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cimmino G, Tarallo R, Nassa G, De Filippo MR, Giurato G, Ravo M, Rizzo F, Conte S, Pellegrino G, Cirillo P et al. Activating stimuli induce platelet microRNA modulation and proteome reorganisation. Thromb Haemost 2015:114(1): 96–108. [DOI] [PubMed] [Google Scholar]
- 53.Bozec A, Zangari J, Butori-Pepino M, Ilie M, Lalvee S, Juhel T, Butori C, Brest P, Hofman P, Vouret-Craviari V. MiR-223–3p inhibits angiogenesis and promotes resistance to cetuximab in head and neck squamous cell carcinoma. Oncotarget 2017:8(34): 57174–57186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shan Z, Qin S, Li W, Wu W, Yang J, Chu M, Li X, Huo Y, Schaer GL, Wang S et al. An Endocrine Genetic Signal Between Blood Cells and Vascular Smooth Muscle Cells: Role of MicroRNA-223 in Smooth Muscle Function and Atherogenesis. J Am Coll Cardiol 2015:65(23): 2526–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao Y, Lin L, Li T, Yang J, Wei Y. The role of miRNA-223 in cancer: Function, diagnosis and therapy. Gene 2017:616(1–7. [DOI] [PubMed] [Google Scholar]
- 56.Ye D, Zhang T, Lou G, Liu Y. Role of miR-223 in the pathophysiology of liver diseases. Exp Mol Med 2018:50(9): 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng Z, Xia L, Fan X, Ostriker AC, Yarovinsky T, Su M, Zhang Y, Peng X, Xie Y, Pi L et al. Platelet-derived miR-223 promotes a phenotypic switch in arterial injury repair. J Clin Invest 2019:129(3): 1372–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldfinger LE, Edelstein LC. Horizontal RNA transfer goes deep: platelet consumption and microRNA utilization by vascular smooth muscle cells. J Thromb Haemost 2019:17(7): 1014–1017. [DOI] [PubMed] [Google Scholar]
- 59.Leierseder S, Petzold T, Zhang L, Loyer X, Massberg S, Engelhardt S. MiR-223 is dispensable for platelet production and function in mice. Thromb Haemost 2013:110(6): 1207–1214. [DOI] [PubMed] [Google Scholar]
- 60.Pan Y, Liang H, Liu H, Li D, Chen X, Li L, Zhang CY, Zen K. Platelet-secreted microRNA-223 promotes endothelial cell apoptosis induced by advanced glycation end products via targeting the insulin-like growth factor 1 receptor. J Immunol 2014:192(1): 437–446. [DOI] [PubMed] [Google Scholar]
- 61.Nian W, Ao X, Wu Y, Huang Y, Shao J, Wang Y, Chen Z, Chen F, Wang D. miR-223 functions as a potent tumor suppressor of the Lewis lung carcinoma cell line by targeting insulin-like growth factor-1 receptor and cyclin-dependent kinase 2. Oncol Lett 2013:6(2): 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y, Song Y, Huang J, Qu M, Zhang Y, Geng J, Zhang Z, Liu J, Yang GY. Increased Circulating Exosomal miRNA-223 Is Associated with Acute Ischemic Stroke. Front Neurol 2017:8(57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang H, Yan X, Pan Y, Wang Y, Wang N, Li L, Liu Y, Chen X, Zhang CY, Gu H et al. MicroRNA-223 delivered by platelet-derived microvesicles promotes lung cancer cell invasion via targeting tumor suppressor EPB41L3. Mol Cancer 2015:14(1): 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Behnke O Coated pits and vesicles transfer plasma components to platelet granules. Thromb Haemost 1989:62(2): 718–722. [PubMed] [Google Scholar]
- 65.Behnke O Degrading and non-degrading pathways in fluid-phase (non-adsorptive) endocytosis in human blood platelets. J Submicrosc Cytol Pathol 1992:24(2): 169–178. [PubMed] [Google Scholar]
- 66.Handagama PJ, Shuman MA, Bainton DF. Incorporation of intravenously injected albumin, immunoglobulin G, and fibrinogen in guinea pig megakaryocyte granules. Journal of Clinical Investigation 1989:84(73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ben-Shushan D, Markovsky E, Gibori H, Tiram G, Scomparin A, Satchi-Fainaro R. Overcoming obstacles in microRNA delivery towards improved cancer therapy. Drug Deliv Transl Res 2014:4(1): 38–49. [DOI] [PubMed] [Google Scholar]
- 68.Johannes L, Lucchino M. Current Challenges in Delivery and Cytosolic Translocation of Therapeutic RNAs. Nucleic Acid Ther 2018:28(3): 178–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen TA, Smith BRC, Tate MD, Belz GT, Barrios MH, Elgass KD, Weisman AS, Baker PJ, Preston SP, Whitehead L et al. SIDT2 Transports Extracellular dsRNA into the Cytoplasm for Innate Immune Recognition. Immunity 2017:47(3): 498–509 e496. [DOI] [PMC free article] [PubMed] [Google Scholar]