Abstract
Background:
The oral microbiome is a complex assembly of microbial species, whose constituents can tilt the balance towards progression of oral disease or sustained health. Recently we identified sex-specific differences in the salivary microbiome contained within caries-active and caries-free children. In this study, we sought to ascertain if adjunctive dental therapies, including povidone iodine and chlorhexidine, were effective in shifting the cariogenic microbiome from dysbiosis to non-cariogenic health.
Design:
We recruited young children (ages 2–12 years) to enter five enrollment groups, with each group (N = 9–30 participants/group) receiving caries restorative and/or adjunctive therapies, either singularly or in combination (OHSU IRB #6535). Saliva specimens were collected pre- and post-treatment (4–8 weeks) of caries preventive measures, and oral microbiota were identified using next generation sequencing (HOMINGS, Forsyth Institute, Cambridge, MA).
Results:
With the use of multi-dimensional scaling plots, support vector machine learning, odds ratio analysis, and other statistical methods, we have determined that treatment with povidone iodine can shift the composition of the salivary cariogenic microbiome to include higher proportions of aerobic microorganisms, such as Stentrophomonas maltophila, as well as non-cariogenic, anaerobic microorganisms including Poryphyromonas and Fusobacterium species.
Conclusion:
We have identified microorganisms that are associated with caries-active children and have determined that povidone iodine is an effective adjunctive therapy that has the potential to shift the composition of the cariogenic microbiome to one more closely aligned with non-cariogenic health.
Keywords: Salivary microbiome, Dental caries, Caries-active children, Microbiome shift, Stentrophomonas maltophila, Povidone iodine, Dysbiosis, Oral health
1. Introduction
The oral microbiome is a complex collection of microbial species, the balance of which contributes to the development of oral disease processes and health. Of the various microbial communities in humans, the oral microbiome is considered as one of the most constant over time [1–3]. Following administration of antibiotics, the oral microbiome exhibits greater resilience and eventual full recovery, whereas the gut microbiota tends to sustain long-term modifications [4]. Though the oral microbiome exhibits natural defenses to drastic shifts in microbial composition via the effects of saliva and other factors, it exhibits limited capability in recovering from disease states that result from the over-abundance of specific oral pathogens.
Dental caries is a common oral disease affecting children and adults, and is influenced by socio-economic, genetic, and microbiological risk factors, including disruption of the composition of oral microorganisms within the oral cavity [4–6]. Several adjunctive therapies have been applied to reduce the cariogenic microbial burden, including the use of povidone iodine and chlorhexidine treatment. Povidone iodine is a popular antimicrobial administered via mouth rinse, with a history of effective treatment for antisepsis and healing [7,8]. Povidone iodine promotes the release of free iodine from carrier complexes in aqueous medium [7,9,10], and has the ability to restore healthy microbial flora following bacterial infection, including bacterial vaginosis [11].
Chlorhexidine is also widely used as an adjunctive therapy for dental caries and exhibits both bacteriostatic and bactericidal effects. This antimicrobial mechanism of action targets the cell membrane and inhibits the attachment of biofilms to tooth surfaces [8,12]. Limitations of chlorhexidine include its cationic nature, making it incompatible with certain dentifrice constituents, preference for an alkaline pH, reduced activity in the presence of organic matter, and reversible tooth discoloration [8,13,14]. Acquired bacterial resistance has been observed in chlorhexidine treatment, likely due to alterations in bacterial membranes and prevention of chlorhexidine adsorption [10,15].
In our prior publication [16], we identified sex-specific differences in the salivary microbiome in children with dental caries. In this current work, using multidimensional scaling plots, machine learning and odds ratio analyses, we now assess the ability of adjunctive dental therapies to shift the salivary microbiota from dysbiosis to non-cariogenic health.
2. Materials and methods
2.1. Participant selection and enrollment groups
We recruited 85 children ages 2–12 years, either caries-active (N = 61) or caries-free (N = 24) from patients visiting OHSU pediatric dentistry clinics, or from an external practitioner site in Albany, Oregon. The inclusion and exclusion parameters for study participation were described previously [16]. Participants entered one of five enrollment groups as defined in Table 1.
Table 1.
Enrollment groups describing participant number, caries status and treatment.
| Group number | N value | Caries status | Treatment at Visit 1 | Treatment at Visit 2 |
|---|---|---|---|---|
| 1 | 18 | Caries-Active | Restorative | Fluoride |
| 2 | 18 | Caries-Active | Restorative + CHX | Fluoride |
| 3 | 25 | Caries-Active | Restorative + PI | Fluoride |
| 4 | 9 | Caries-Free | Fluoride | None |
| 5 | 15 | Caries-Free | CHX | Fluoride |
Caries status was assessed by clinical examination, DMFT indices and radio-graphs.
Caries-active participants had DMFT scores of 1–18 at enrollment (DMFT average = 8.6).
N value = number of participants.
CHX = 0.12 chlorhexidine.
PI = 10% povidone iodine (equivalent to Betadine).
Single application of PI was applied to the dentition, then immediately wiped away with gauze.
Restorative therapy is considered as standard-of-care for all caries-active participants.
Fluoride application was applied to all caries-active or caries-free participants. Visit 2 = 4–8 weeks following Visit 1.
Saliva specimens were collected at the beginning of each visit.
2.2. Specimen collection and identification of salivary microorganisms
Non-stimulated saliva specimens were collected from all enrolled participants at the beginning of Visit 1 and Visit 2. Because young children under the age of 5 years have general difficulty expelling saliva, we used non-stimulated saliva in all collections, either using a swab for children under the age of 5 years, or the expulsion of saliva (1 ml) from all other children. There were no stipulated conditions of food or drink intake, or teeth brushing, prior to the collection of specimens. Microbial genomic DNA from saliva [16,17] was applied in PCR using V3-V4 16S rDNA-specific primers and in next generation sequencing (NGS) (Forsyth Institute, Cambridge, MA).
2.3. Statistical analysis
The relative abundance of salivary microbial species was determined by enrollment group and visit number. As described previously [16], we used a zero-inflated negative binomial regression model by the zinb-wave and edgeR packages in R statistical language [18,19]. Two-sided p-values for testing relative abundance were adjusted by the false discovery rate (FDR) for multiple test correction [20]. In addition, multi-dimensional scaling plots were used to visualize the level of similarity between caries-active and caries-free data sets, based on the fold-changes of the log (cpm) values [21,22]. To predict caries status for 65 cases at Visit 2, we used the support vector machine (SVM) [23] to build a prediction rule for caries-active versus caries-free, using data from Visit 1 from all 85 enrolled participants as training data. Further analyses were conducted using the ratio of odds of caries-active over caries-free between Visit 1 and Visit 2; in this case, the odds that an adjunctive therapy within a specific individual restored the cariogenic salivary microbiome to one more representative of caries-free health.
3. Results
3.1. Changes in salivary microorganisms based on enrollment group, visit number, and age categories
Fig. 1 ranks the abundance of the top twenty salivary microorganisms found in caries-active children and caries-free children based on enrollment group and visit, either pre- or post-adjunctive treatment (also Table 2). Streptococci is the most abundant microorganism found in all enrollment groups, and with the exception of Group 2, appears to be diminished in abundance following treatment. Haemophilus parainfluenza is ranked as the second most abundant microorganism overall, and in Group 4, this microbial species appears to rise as the most abundant microorganism post-treatment. As described previously [16], Prevotella melaninogenica is the second most abundant microorganism in caries-active children and is the fourth most abundant in caries-free children, and appears to be significantly affected in abundance percentage, especially in Groups 2, 4 and 5, when comparing pre- and post-treatment numbers. In our prior publication (16) with the same participant cohort, we illustrated several significant microbial changes based on the sex of the participants. When examining two age categories – 2–5 years and 6–12 years – reflective of children with primary dentition versus mixed dentition respectively, we find that caries-active boys, ages 2–5, have a much higher prevalence of known cariogenic microorganisms, Streptococcus sobrinus and Streptococcus mutans, when compared to older caries-active boys, ages 6–12 (Table 3A). No statistically-significant fold changes in microbial species were identified when examining age category comparisons for caries-active girls, caries-free boys, or caries-free girls.
Fig. 1. Abundance of Top 20 Salivary Microorganisms Found in Caries-Active and Caries-Free Children.

Abundance of top 20 salivary microorganisms found in caries-active (Groups 1–3) and caries-free (Groups 4 and 5) children based on enrollment group (G1–G5) and visit number (V1-pre-treatment and V2-post-treatment).
Table 2.
Abundance of salivary microorganisms found in caries-active and caries-free children based on enrollment group and visit number.
| Probe ID | Microorganism | G1V1 | G1V2 | G2V1 | G2V2 | G3V1 | G3V2 | G4V1 | G4V2 | G5V1 | G5V2 | Average |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GP-081 | Streptococcus_Genus_probe_4 | 27.29% | 23.77% | 24.19% | 29.30% | 20.96% | 18.69% | 19.97% | 18.17% | 29.15% | 27.60% | 23.91% |
| HA-05 | Haemophilus_parainfluenzae | 6.69% | 7.04% | 6.49% | 4.60% | 7.81% | 9.43% | 16.64% | 23.45% | 6.68% | 5.63% | 9.45% |
| RO-03 | Rothia_mucilaginosa | 3.75% | 4.31% | 6.42% | 3.66% | 9.50% | 6.83% | 8.99% | 9.47% | 8.54% | 5.64% | 6.71% |
| PR-14 | Prevotella_melaninogenica | 10.27% | 8.92% | 6.73% | 2.21% | 9.02% | 7.10% | 2.74% | 7.26% | 7.31% | 3.53% | 6.51% |
| GP-060 | Neisseria_Genus_probe_2 | 4.07% | 6.71% | 5.84% | 9.04% | 4.39% | 3.07% | 9.30% | 8.88% | 2.72% | 5.17% | 5.92% |
| GE-02 | Gemella_haemolysans | 4.64% | 4.10% | 3.75% | 5.80% | 5.65% | 5.55% | 3.13% | 1.34% | 5.41% | 5.73% | 4.51% |
| GR-02 | Granulicatella_elegans | 5.60% | 2.40% | 3.56% | 4.58% | 3.91% | 4.51% | 1.68% | 0.54% | 4.16% | 6.22% | 3.72% |
| FU-10 | Fusobacterium_periodonticum | 2.07% | 3.57% | 1.24% | 2.44% | 3.64% | 3.51% | 4.06% | 3.17% | 2.66% | 1.96% | 2.83% |
| PO-09 | Porphyromonas_pasteri | 2.17% | 1.41% | 2.20% | 2.79% | 2.19% | 1.76% | 1.52% | 0.64% | 1.81% | 3.59% | 2.01% |
| NE-03 | Neisseria_flavescens | 1.61% | 2.15% | 3.46% | 0.68% | 2.19% | 1.84% | 2.96% | 0.04% | 1.76% | 2.03% | 1.87% |
| BE-02 | Bergeyella_sp_HOT_322 | 0.93% | 0.78% | 2.84% | 3.00% | 1.52% | 0.74% | 1.89% | 0.29% | 2.56% | 2.70% | 1.73% |
| GP-040 | Haemophilus_Genus_probe_3 | 2.72% | 0.97% | 2.35% | 2.00% | 1.29% | 1.68% | 0.83% | 0.95% | 1.24% | 1.87% | 1.59% |
| AL-04 | Alloprevotella_sp_HOT_473 | 0.84% | 0.61% | 2.17% | 3.35% | 0.76% | 1.17% | 0.26% | 0.77% | 1.05% | 3.43% | 1.44% |
| HA-07 | Haemophilus_pittmaniae | 0.43% | 0.63% | 1.11% | 1.05% | 1.35% | 0.70% | 1.84% | 2.34% | 1.32% | 1.24% | 1.20% |
| GR-01 | Granulicatella_adiacens | 0.83% | 1.37% | 1.26% | 1.29% | 1.16% | 0.99% | 1.37% | 1.20% | 1.55% | 0.94% | 1.20% |
| PR-79 | Prevotella_nanceiensis | 1.00% | 1.78% | 1.03% | 0.95% | 1.34% | 1.23% | 0.85% | 0.77% | 1.49% | 0.71% | 1.11% |
| ST-20 | Streptococcus_sanguinis | 1.07% | 1.04% | 1.52% | 1.74% | 0.57% | 0.60% | 0.90% | 1.41% | 0.96% | 0.70% | 1.05% |
| GP-038 | Fusobacterium_Genus_probe_4 | 0.78% | 0.92% | 0.88% | 0.73% | 1.01% | 0.93% | 1.49% | 1.51% | 0.78% | 1.00% | 1.00% |
| GP-089 | Veillonella_Genus_probe_2 | 0.86% | 1.70% | 1.27% | 1.58% | 0.66% | 0.77% | 1.01% | 1.09% | 0.60% | 0.51% | 1.00% |
| PO-24 | Porphyromonas_pasteri | 0.72% | 0.72% | 0.67% | 0.54% | 0.73% | 0.38% | 0.92% | 0.32% | 0.78% | 1.17% | 0.70% |
| GP-039 | Gemella_Genus_probe | 0.75% | 1.30% | 0.62% | 0.87% | 0.63% | 0.53% | 0.23% | 0.15% | 0.93% | 0.58% | 0.66% |
| GP-126 | Streptococcus_Genus_probe_1 | 0.57% | 0.54% | 0.67% | 0.85% | 0.49% | 0.38% | 0.36% | 0.43% | 0.35% | 1.72% | 0.64% |
| LA-29 | Lautropia_mirabilis | 0.30% | 0.90% | 0.27% | 0.88% | 0.32% | 0.33% | 0.69% | 1.18% | 0.87% | 0.43% | 0.62% |
| LE-06 | Leptotrichia_hongkongensis | 0.71% | 0.46% | 0.59% | 1.04% | 0.21% | 0.85% | 0.21% | 0.86% | 0.28% | 0.89% | 0.61% |
| PR-25 | Prevotella_salivae | 0.88% | 0.97% | 0.26% | 0.05% | 0.88% | 0.95% | 0.05% | 0.14% | 0.58% | 0.11% | 0.49% |
| GE-04 | Gemella_sanguinis | 0.21% | 0.20% | 0.30% | 0.25% | 0.56% | 0.36% | 0.85% | 0.97% | 0.56% | 0.33% | 0.46% |
| PR-09 | Prevotella_histicola | 0.70% | 0.70% | 0.45% | 0.14% | 1.13% | 0.51% | 0.07% | 0.16% | 0.30% | 0.10% | 0.43% |
| LE-16 | Leptotrichia_sp_HOT_417 | 0.44% | 0.47% | 0.42% | 0.38% | 0.74% | 1.08% | 0.06% | 0.22% | 0.23% | 0.16% | 0.42% |
| OR-01 | Oribacterium_sinus | 0.34% | 0.83% | 0.19% | 0.18% | 0.21% | 0.31% | 0.53% | 1.02% | 0.35% | 0.21% | 0.42% |
| VE-08 | Veillonella_sp_HOT_780 | 0.86% | 0.17% | 0.65% | 0.39% | 0.15% | 0.33% | 0.19% | 0.16% | 0.48% | 0.73% | 0.41% |
| VE-03 | Veillonella_dispar | 0.43% | 1.09% | 0.47% | 0.38% | 0.22% | 0.57% | 0.12% | 0.33% | 0.19% | 0.21% | 0.40% |
| NE-19 | Neisseria_subflava | 0.44% | 0.60% | 0.51% | 0.39% | 0.27% | 0.39% | 0.39% | 0.04% | 0.43% | 0.52% | 0.40% |
| LE-07 | Leptotrichia_shahii | 0.11% | 0.61% | 1.22% | 0.34% | 0.13% | 0.24% | 0.46% | 0.05% | 0.16% | 0.38% | 0.37% |
| GP-112 | Haemophilus_Genus_probe_2 | 0.24% | 0.10% | 0.50% | 0.51% | 0.34% | 0.27% | 0.27% | 0.34% | 0.24% | 0.47% | 0.33% |
| GP-099 | Aggregatibacter_Genus_probe_1 | 0.25% | 0.26% | 0.49% | 0.11% | 0.31% | 0.48% | 0.31% | 0.59% | 0.17% | 0.29% | 0.33% |
| VE-07 | Veillonella_rogosae | 0.15% | 0.55% | 0.27% | 0.17% | 0.21% | 0.19% | 0.53% | 0.45% | 0.55% | 0.17% | 0.32% |
| GP-019 | Campylobacter_Genus_probe_2 | 0.56% | 0.59% | 0.21% | 0.16% | 0.52% | 0.42% | 0.14% | 0.25% | 0.22% | 0.14% | 0.32% |
| GP-050 | Leptotrichia_Genus_probe_4 | 0.26% | 0.41% | 0.36% | 0.35% | 0.08% | 0.20% | 0.26% | 0.57% | 0.19% | 0.49% | 0.32% |
| GP-073 | Rothia_Genus_probe | 0.20% | 0.21% | 0.36% | 0.28% | 0.49% | 0.35% | 0.33% | 0.26% | 0.33% | 0.29% | 0.31% |
| RO-01 | Rothia_aeria | 0.32% | 0.32% | 0.34% | 0.68% | 0.40% | 0.31% | 0.25% | 0.08% | 0.18% | 0.20% | 0.31% |
| GP-080 | Staphylococcus_Genus_probe_3 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 3.04% | 0.00% | 0.01% | 0.00% | 0.00% | 0.31% |
| PR-22 | Prevotella_pallens | 0.65% | 0.66% | 0.22% | 0.07% | 0.49% | 0.27% | 0.05% | 0.18% | 0.20% | 0.09% | 0.29% |
| AB-01 | Abiotrophia_defectiva | 0.69% | 0.23% | 0.25% | 0.30% | 0.16% | 0.13% | 0.52% | 0.23% | 0.23% | 0.09% | 0.28% |
| VE-21 | Veillonella_atypica | 0.43% | 0.38% | 0.31% | 0.09% | 0.38% | 0.31% | 0.04% | 0.18% | 0.48% | 0.12% | 0.27% |
| CO-03 | Corynebacterium_matruchotii | 0.10% | 0.18% | 0.15% | 0.08% | 0.10% | 0.30% | 0.42% | 0.16% | 0.32% | 0.73% | 0.25% |
| RO-02 | Rothia_dentocariosa | 0.20% | 0.11% | 0.23% | 0.18% | 0.38% | 0.28% | 0.15% | 0.28% | 0.42% | 0.31% | 0.25% |
| AG-06 | Aggregatibacter_sp_HOT_513 | 0.29% | 0.39% | 0.14% | 0.25% | 0.54% | 0.44% | 0.11% | 0.00% | 0.03% | 0.34% | 0.25% |
| TM-13 | TM7[G-1]_sp_HOT_348 | 0.01% | 0.01% | 0.07% | 0.01% | 0.03% | 0.04% | 1.01% | 0.00% | 0.13% | 1.11% | 0.24% |
| NE-18 | Neisseria_oralis | 0.11% | 0.22% | 0.26% | 1.16% | 0.07% | 0.16% | 0.04% | 0.00% | 0.16% | 0.15% | 0.23% |
| VE-20 | Veillonella_atypica | 0.43% | 0.44% | 0.27% | 0.07% | 0.25% | 0.17% | 0.04% | 0.09% | 0.20% | 0.14% | 0.21% |
| CL-03 | Ruminococcaceae[G-1]_sp_HOT_075 | 0.62% | 0.34% | 0.15% | 0.09% | 0.15% | 0.12% | 0.09% | 0.22% | 0.06% | 0.03% | 0.19% |
| SE-05 | Selenomonas_noxia | 0.02% | 0.04% | 0.07% | 0.00% | 0.02% | 0.01% | 1.61% | 0.01% | 0.01% | 0.01% | 0.18% |
| AL-11 | Alloprevotella_sp_HOT_914 | 0.15% | 0.34% | 0.22% | 0.21% | 0.25% | 0.14% | 0.21% | 0.05% | 0.10% | 0.10% | 0.18% |
| GP-049 | Leptotrichia_Genus_probe_3 | 0.12% | 0.12% | 0.36% | 0.24% | 0.19% | 0.49% | 0.02% | 0.05% | 0.05% | 0.04% | 0.17% |
| GP-076 | Selenomonas_&_Centipeda_Genus_probe | 0.03% | 0.04% | 0.17% | 0.03% | 0.03% | 0.07% | 0.25% | 0.09% | 0.30% | 0.63% | 0.16% |
| LE-26 | Leptotrichia_sp_HOT_215 | 0.21% | 0.24% | 0.12% | 0.15% | 0.13% | 0.13% | 0.13% | 0.27% | 0.09% | 0.13% | 0.16% |
| GE-07 | Gemella_morbillorum | 0.10% | 0.12% | 0.09% | 0.09% | 0.39% | 0.32% | 0.08% | 0.08% | 0.16% | 0.15% | 0.16% |
| NE-08 | Neisseria_pharyngis | 0.55% | 0.07% | 0.17% | 0.23% | 0.12% | 0.24% | 0.00% | 0.04% | 0.09% | 0.04% | 0.15% |
| LE-13 | Leptotrichia_sp_HOT_221 | 0.47% | 0.18% | 0.12% | 0.24% | 0.14% | 0.20% | 0.06% | 0.02% | 0.04% | 0.08% | 0.15% |
| PR-18 | Prevotella_nigrescens | 0.09% | 0.12% | 0.06% | 0.19% | 0.34% | 0.17% | 0.23% | 0.29% | 0.03% | 0.02% | 0.15% |
| ST-29 | Stenotrophomonas_maltophilia | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 1.53% | 0.00% | 0.00% | 0.00% | 0.00% | 0.15% |
| GP-100 | Aggregatibacter_Genus_probe_2 | 0.16% | 0.05% | 0.09% | 0.27% | 0.18% | 0.17% | 0.01% | 0.29% | 0.09% | 0.14% | 0.15% |
| PR-51 | Prevotella_veroralis | 0.14% | 0.62% | 0.39% | 0.02% | 0.06% | 0.12% | 0.00% | 0.00% | 0.06% | 0.03% | 0.14% |
| GP-005 | Alloprevotella_Genus_probe | 0.14% | 0.18% | 0.17% | 0.17% | 0.15% | 0.09% | 0.08% | 0.13% | 0.14% | 0.14% | 0.14% |
| NE-16 | Neisseria_flavescens | 0.29% | 0.03% | 0.27% | 0.01% | 0.35% | 0.17% | 0.00% | 0.00% | 0.01% | 0.22% | 0.14% |
| LA-06 | Lachnoanaerobaculum_umeaense | 0.09% | 0.30% | 0.13% | 0.12% | 0.08% | 0.06% | 0.16% | 0.18% | 0.12% | 0.09% | 0.13% |
| GP-003 | Actinomyces_Genus_probe_3 | 0.05% | 0.04% | 0.08% | 0.18% | 0.06% | 0.19% | 0.03% | 0.37% | 0.10% | 0.22% | 0.13% |
| GP-004 | Actinomyces_Genus_probe_4 | 0.11% | 0.11% | 0.10% | 0.10% | 0.18% | 0.15% | 0.12% | 0.13% | 0.17% | 0.05% | 0.12% |
| LE-08 | Leptotrichia_sp_HOT_212 | 0.03% | 0.29% | 0.05% | 0.29% | 0.05% | 0.14% | 0.06% | 0.21% | 0.02% | 0.07% | 0.12% |
| TM-05 | TM7[G-1]_sp_HOT_352 | 0.11% | 0.22% | 0.06% | 0.04% | 0.26% | 0.18% | 0.06% | 0.03% | 0.20% | 0.04% | 0.12% |
| NE-02 | Neisseria_elongata | 0.09% | 0.15% | 0.08% | 0.08% | 0.05% | 0.36% | 0.16% | 0.14% | 0.05% | 0.04% | 0.12% |
| CA-37 | Catonella_morbi | 0.11% | 0.35% | 0.04% | 0.09% | 0.08% | 0.06% | 0.12% | 0.18% | 0.08% | 0.05% | 0.12% |
| GE-05 | Gemella_morbillorum | 0.07% | 0.07% | 0.13% | 0.10% | 0.15% | 0.22% | 0.13% | 0.06% | 0.04% | 0.16% | 0.11% |
| GP-067 | Porphyromonas_Genus_probe_2 | 0.13% | 0.19% | 0.11% | 0.13% | 0.08% | 0.15% | 0.03% | 0.03% | 0.10% | 0.19% | 0.11% |
| GP-096 | Fusobacterium_Genus_probe_3 | 0.07% | 0.12% | 0.16% | 0.08% | 0.05% | 0.15% | 0.10% | 0.16% | 0.08% | 0.14% | 0.11% |
| LE-22 | Leptotrichia_wadei | 0.12% | 0.03% | 0.12% | 0.04% | 0.09% | 0.52% | 0.14% | 0.02% | 0.01% | 0.02% | 0.11% |
| LE-11 | Leptotrichia_sp_HOT_218 | 0.18% | 0.03% | 0.38% | 0.09% | 0.11% | 0.23% | 0.00% | 0.00% | 0.07% | 0.02% | 0.11% |
| CA-12 | Capnocytophaga_leadbetteri | 0.10% | 0.15% | 0.06% | 0.04% | 0.05% | 0.15% | 0.18% | 0.20% | 0.04% | 0.09% | 0.11% |
| FU-12 | Fusobacterium_nucleatum_subsp_animalis | 0.08% | 0.21% | 0.09% | 0.06% | 0.33% | 0.21% | 0.02% | 0.04% | 0.01% | 0.01% | 0.11% |
| KI-03 | Kingella_oralis | 0.12% | 0.14% | 0.14% | 0.13% | 0.06% | 0.08% | 0.02% | 0.14% | 0.04% | 0.18% | 0.10% |
| PR-20 | Prevotella_oris | 0.05% | 0.10% | 0.06% | 0.11% | 0.17% | 0.20% | 0.15% | 0.03% | 0.05% | 0.04% | 0.10% |
| SO-Ol | Solobacterium_moorei | 0.13% | 0.17% | 0.07% | 0.07% | 0.10% | 0.06% | 0.03% | 0.15% | 0.12% | 0.05% | 0.09% |
| TA-02 | Tannerella_sp_HOT_286 | 0.03% | 0.09% | 0.08% | 0.03% | 0.02% | 0.05% | 0.12% | 0.15% | 0.21% | 0.16% | 0.09% |
| BA-03 | Bacteroidales[G-2]_sp_HOT_274 | 0.03% | 0.04% | 0.04% | 0.01% | 0.11% | 0.02% | 0.33% | 0.02% | 0.29% | 0.01% | 0.09% |
| CA-31 | Cardiobacterium_hominis | 0.04% | 0.09% | 0.08% | 0.06% | 0.02% | 0.20% | 0.04% | 0.21% | 0.06% | 0.10% | 0.09% |
| CA-30 | Capnocytophaga_sputigena | 0.05% | 0.09% | 0.08% | 0.13% | 0.04% | 0.13% | 0.14% | 0.12% | 0.05% | 0.05% | 0.09% |
| VE-06 | Veillonella_parvula | 0.06% | 0.08% | 0.26% | 0.32% | 0.03% | 0.05% | 0.02% | 0.03% | 0.02% | 0.02% | 0.09% |
| GE-03 | Gemella_morbillorum | 0.05% | 0.12% | 0.08% | 0.09% | 0.12% | 0.16% | 0.07% | 0.06% | 0.05% | 0.08% | 0.09% |
| PR-21 | Prevotella_oulorum | 0.04% | 0.04% | 0.16% | 0.02% | 0.10% | 0.29% | 0.05% | 0.04% | 0.02% | 0.04% | 0.08% |
| C002 | Corynebacterium_durum | 0.04% | 0.09% | 0.05% | 0.08% | 0.07% | 0.07% | 0.07% | 0.04% | 0.19% | 0.10% | 0.08% |
| PR-26 | Prevotella_scopos | 0.05% | 0.02% | 0.06% | 0.03% | 0.24% | 0.08% | 0.02% | 0.04% | 0.09% | 0.13% | 0.08% |
| SR-03 | SR1[G-1]_sp_HOT_875 | 0.03% | 0.13% | 0.02% | 0.08% | 0.11% | 0.01% | 0.11% | 0.01% | 0.20% | 0.06% | 0.08% |
| GP-113 | Kingella_Genus_probe_1 | 0.08% | 0.16% | 0.10% | 0.11% | 0.05% | 0.06% | 0.10% | 0.01% | 0.04% | 0.04% | 0.07% |
| GP-130 | Veillonella_Genus_probe_1 | 0.08% | 0.10% | 0.08% | 0.08% | 0.08% | 0.08% | 0.04% | 0.05% | 0.10% | 0.03% | 0.07% |
| PE-21 | Peptostreptococcus_stomatis | 0.07% | 0.13% | 0.06% | 0.05% | 0.12% | 0.06% | 0.05% | 0.07% | 0.08% | 0.04% | 0.07% |
| OR-06 | Oribacterium_asaccharolyticum | 0.13% | 0.13% | 0.03% | 0.03% | 0.09% | 0.11% | 0.02% | 0.03% | 0.09% | 0.04% | 0.07% |
| CA-04 | Campylobacter_gracilis | 0.03% | 0.06% | 0.10% | 0.03% | 0.05% | 0.14% | 0.04% | 0.02% | 0.10% | 0.14% | 0.07% |
| ME-01 | Megasphaera_micronuciformis | 0.20% | 0.16% | 0.07% | 0.01% | 0.07% | 0.06% | 0.01% | 0.01% | 0.10% | 0.01% | 0.07% |
| PR-70 | Prevotella_pallens | 0.24% | 0.23% | 0.04% | 0.00% | 0.07% | 0.03% | 0.01% | 0.01% | 0.05% | 0.02% | 0.07% |
| GP-083 | TM7_Genus_probe | 0.03% | 0.05% | 0.09% | 0.03% | 0.05% | 0.10% | 0.22% | 0.01% | 0.05% | 0.07% | 0.07% |
| PO-27 | Porphyromonas_sp_HOT_284 | 0.02% | 0.04% | 0.04% | 0.05% | 0.03% | 0.23% | 0.11% | 0.08% | 0.04% | 0.05% | 0.07% |
| CA-10 | Capnocytophaga_granulosa | 0.02% | 0.05% | 0.04% | 0.04% | 0.02% | 0.07% | 0.05% | 0.26% | 0.02% | 0.10% | 0.07% |
| AC-20 | Actinomyces_sp_HOT_172 | 0.08% | 0.06% | 0.02% | 0.02% | 0.14% | 0.11% | 0.05% | 0.02% | 0.15% | 0.01% | 0.07% |
| EU-03 | Peptostreptococcaceae[XI][G-1][Eubacterium]_sulci | 0.11% | 0.12% | 0.04% | 0.01% | 0.16% | 0.07% | 0.03% | 0.02% | 0.06% | 0.02% | 0.06% |
| GP-069 | Prevotella_Genus_probe_2 | 0.05% | 0.09% | 0.07% | 0.04% | 0.15% | 0.09% | 0.04% | 0.02% | 0.08% | 0.02% | 0.06% |
| AC-24 | Actinomyces_lingnae | 0.04% | 0.05% | 0.05% | 0.03% | 0.07% | 0.09% | 0.09% | 0.05% | 0.13% | 0.02% | 0.06% |
| LE-15 | Leptotrichia_sp_HOT_392 | 0.09% | 0.11% | 0.06% | 0.05% | 0.02% | 0.08% | 0.06% | 0.02% | 0.08% | 0.05% | 0.06% |
| AG-01 | Aggregatibacter_actinomycetemcomitans | 0.00% | 0.00% | 0.02% | 0.57% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.06% |
| ST-03 | Stenotrophomonas_maltophilia | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.59% | 0.00% | 0.00% | 0.00% | 0.00% | 0.06% |
| ST-15 | Streptococcus_mutans | 0.04% | 0.00% | 0.06% | 0.35% | 0.02% | 0.06% | 0.00% | 0.00% | 0.04% | 0.01% | 0.06% |
| FU-07 | Fusobacterium_nucleatum_subsp_nucleatum | 0.03% | 0.04% | 0.06% | 0.06% | 0.07% | 0.15% | 0.06% | 0.02% | 0.04% | 0.06% | 0.06% |
| EI-01 | Eikenella_corrodens | 0.06% | 0.05% | 0.05% | 0.04% | 0.02% | 0.22% | 0.03% | 0.02% | 0.02% | 0.04% | 0.06% |
| SE-04 | Selenomonas_noxia | 0.02% | 0.01% | 0.11% | 0.01% | 0.02% | 0.09% | 0.00% | 0.00% | 0.27% | 0.00% | 0.05% |
| CL-04 | Ruminococcaceae[G-2]_sp_HOT_085 | 0.11% | 0.06% | 0.04% | 0.02% | 0.12% | 0.06% | 0.02% | 0.06% | 0.03% | 0.01% | 0.05% |
| TM-01 | TM7[G-1]_sp_HOT_346 | 0.01% | 0.01% | 0.31% | 0.02% | 0.05% | 0.07% | 0.00% | 0.00% | 0.06% | 0.00% | 0.05% |
| CA-09 | Capnocytophaga_gingivalis | 0.03% | 0.06% | 0.01% | 0.03% | 0.03% | 0.02% | 0.07% | 0.16% | 0.02% | 0.08% | 0.05% |
| PR-39 | Prevotella_sp_HOT_309 | 0.09% | 0.09% | 0.04% | 0.03% | 0.09% | 0.05% | 0.01% | 0.03% | 0.04% | 0.04% | 0.05% |
| AG-03 | Aggregatibacter_paraphrophilus | 0.01% | 0.01% | 0.00% | 0.01% | 0.02% | 0.34% | 0.07% | 0.02% | 0.01% | 0.01% | 0.05% |
| ST-14 | Streptococcus_intermedius | 0.04% | 0.07% | 0.06% | 0.09% | 0.03% | 0.06% | 0.03% | 0.03% | 0.02% | 0.06% | 0.05% |
| GP-095 | Fusobacterium_Genus_probe_2 | 0.03% | 0.07% | 0.05% | 0.04% | 0.06% | 0.12% | 0.04% | 0.01% | 0.01% | 0.02% | 0.05% |
| OR-07 | Oribacterium_parvum | 0.08% | 0.10% | 0.03% | 0.01% | 0.03% | 0.03% | 0.04% | 0.05% | 0.06% | 0.02% | 0.04% |
| AG-09 | Aggregatibacter_paraphrophilus | 0.01% | 0.21% | 0.01% | 0.04% | 0.03% | 0.04% | 0.09% | 0.00% | 0.01% | 0.01% | 0.04% |
| PO-17 | Porphyromonas_catoniae | 0.02% | 0.09% | 0.02% | 0.03% | 0.01% | 0.02% | 0.01% | 0.01% | 0.01% | 0.22% | 0.04% |
| LE-19 | Leptotrichia_sp_HOT_498 | 0.03% | 0.02% | 0.16% | 0.02% | 0.03% | 0.13% | 0.00% | 0.00% | 0.01% | 0.00% | 0.04% |
| PR-80 | Prevotella_sp_HOT_942 | 0.13% | 0.00% | 0.05% | 0.04% | 0.03% | 0.04% | 0.00% | 0.00% | 0.06% | 0.04% | 0.04% |
| LE-04 | Leptotrichia_goodfellowii | 0.02% | 0.14% | 0.03% | 0.01% | 0.02% | 0.11% | 0.01% | 0.02% | 0.02% | 0.01% | 0.04% |
| PR-15 | Prevotella_micans | 0.00% | 0.01% | 0.03% | 0.01% | 0.01% | 0.00% | 0.33% | 0.00% | 0.00% | 0.00% | 0.04% |
| LA-01 | Lachnoanaerobaculum_orale | 0.10% | 0.02% | 0.04% | 0.04% | 0.06% | 0.03% | 0.01% | 0.04% | 0.04% | 0.01% | 0.04% |
| AC-36 | Actinomyces_naeslundii | 0.01% | 0.03% | 0.02% | 0.02% | 0.02% | 0.16% | 0.02% | 0.04% | 0.02% | 0.03% | 0.04% |
| GP-020 | Capnocytophaga_Genus_probe_2 | 0.02% | 0.03% | 0.04% | 0.03% | 0.02% | 0.05% | 0.07% | 0.02% | 0.03% | 0.07% | 0.04% |
| PO-26 | Porphyromonas_sp_HOT_930 | 0.17% | 0.02% | 0.10% | 0.04% | 0.01% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% | 0.04% |
| PR-46 | Prevotella_sp_HOT_472 | 0.02% | 0.06% | 0.02% | 0.01% | 0.04% | 0.14% | 0.01% | 0.02% | 0.02% | 0.03% | 0.04% |
| LA-28 | Lactococcus_lactis | 0.35% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.04% |
| GP-063 | Parvimonas_Genus_probe | 0.01% | 0.14% | 0.01% | 0.02% | 0.03% | 0.09% | 0.04% | 0.00% | 0.01% | 0.00% | 0.04% |
| CA-32 | Cardiobacterium_valvarum | 0.01% | 0.04% | 0.01% | 0.01% | 0.00% | 0.05% | 0.01% | 0.10% | 0.01% | 0.10% | 0.03% |
| AL-09 | Alloprevotella_tannerae | 0.02% | 0.04% | 0.03% | 0.06% | 0.15% | 0.03% | 0.00% | 0.00% | 0.01% | 0.00% | 0.03% |
| GP-118 | Moraxella_Genus_probe_1 | 0.00% | 0.00% | 0.33% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.03% |
| SR-02 | SR1[G-1]_sp_HOT_874 | 0.02% | 0.03% | 0.03% | 0.03% | 0.02% | 0.01% | 0.02% | 0.09% | 0.01% | 0.08% | 0.03% |
| GP-021 | Capnocytophaga_Genus_probe_3 | 0.03% | 0.04% | 0.03% | 0.03% | 0.02% | 0.04% | 0.04% | 0.06% | 0.01% | 0.03% | 0.03% |
| CA-17 | Capnocytophaga_sp_HOT_332 | 0.01% | 0.02% | 0.02% | 0.01% | 0.01% | 0.01% | 0.03% | 0.02% | 0.03% | 0.16% | 0.03% |
| LA-02 | Lachnoanaerobaculum_saburreum | 0.01% | 0.03% | 0.04% | 0.01% | 0.01% | 0.08% | 0.02% | 0.04% | 0.00% | 0.07% | 0.03% |
| GP-022 | Cardiobacterium_Genus_probe | 0.01% | 0.04% | 0.03% | 0.02% | 0.01% | 0.03% | 0.01% | 0.02% | 0.11% | 0.02% | 0.03% |
| GP-023 | Catonella_Genus_probe | 0.02% | 0.02% | 0.03% | 0.04% | 0.03% | 0.02% | 0.04% | 0.03% | 0.03% | 0.03% | 0.03% |
| ST-05 | Stomatobaculum_sp_HOT_097 | 0.02% | 0.05% | 0.01% | 0.02% | 0.03% | 0.02% | 0.04% | 0.07% | 0.03% | 0.02% | 0.03% |
| DI-01 | Dialister_invisus | 0.02% | 0.04% | 0.02% | 0.01% | 0.10% | 0.02% | 0.01% | 0.04% | 0.02% | 0.00% | 0.03% |
| AT-08 | Atopobium_parvulum | 0.00% | 0.02% | 0.02% | 0.00% | 0.04% | 0.08% | 0.00% | 0.03% | 0.08% | 0.00% | 0.03% |
| AC-35 | Actinomyces_graevenitzii | 0.05% | 0.01% | 0.04% | 0.01% | 0.06% | 0.04% | 0.02% | 0.02% | 0.03% | 0.01% | 0.03% |
| GP-127 | Streptococcus_Genus_probe_2 | 0.22% | 0.03% | 0.01% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.03% |
| SE-08 | Selenomonas_sp_HOT_136 | 0.15% | 0.04% | 0.01% | 0.00% | 0.02% | 0.01% | 0.00% | 0.00% | 0.02% | 0.01% | 0.03% |
| PR-27 | Prevotella_shahii | 0.02% | 0.09% | 0.01% | 0.01% | 0.03% | 0.02% | 0.05% | 0.01% | 0.03% | 0.01% | 0.03% |
| PE-28 | Peptostreptococcaceae[XI][G-7]_sp_HOT_922 | 0.00% | 0.12% | 0.01% | 0.00% | 0.01% | 0.00% | 0.04% | 0.06% | 0.01% | 0.01% | 0.03% |
| SI-01 | Simonsiella_muelleri | 0.07% | 0.02% | 0.00% | 0.06% | 0.02% | 0.02% | 0.02% | 0.00% | 0.01% | 0.04% | 0.03% |
| GP-025 | Corynebacterium_Genus_probe | 0.01% | 0.02% | 0.02% | 0.02% | 0.03% | 0.04% | 0.02% | 0.01% | 0.03% | 0.05% | 0.03% |
| LA-08 | Lachnospiraceae[G-2]_sp_HOT_096 | 0.09% | 0.03% | 0.01% | 0.00% | 0.03% | 0.07% | 0.00% | 0.00% | 0.00% | 0.01% | 0.03% |
| PO-19 | Porphyromonas_sp_HOT_275 | 0.02% | 0.04% | 0.03% | 0.02% | 0.02% | 0.02% | 0.04% | 0.00% | 0.04% | 0.02% | 0.02% |
| SR-01 | SR1[G-1]_sp_HOT_345 | 0.02% | 0.10% | 0.02% | 0.01% | 0.01% | 0.01% | 0.02% | 0.00% | 0.04% | 0.02% | 0.02% |
| LA-11 | Butyrivibrio_sp_HOT_455 | 0.10% | 0.02% | 0.01% | 0.01% | 0.06% | 0.02% | 0.00% | 0.01% | 0.01% | 0.01% | 0.02% |
| PR-06 | Prevotella_denticola | 0.02% | 0.01% | 0.01% | 0.00% | 0.17% | 0.01% | 0.00% | 0.01% | 0.01% | 0.00% | 0.02% |
| PO-03 | Porphyromonas_endodontalis | 0.08% | 0.00% | 0.00% | 0.03% | 0.11% | 0.01% | 0.00% | 0.00% | 0.01% | 0.00% | 0.02% |
| BE-04 | Bergeyella_sp_HOT_907 | 0.02% | 0.02% | 0.04% | 0.02% | 0.02% | 0.02% | 0.06% | 0.00% | 0.01% | 0.03% | 0.02% |
| BE-05 | Bergeyella_sp_HOT_931 | 0.01% | 0.02% | 0.02% | 0.03% | 0.02% | 0.01% | 0.01% | 0.00% | 0.03% | 0.09% | 0.02% |
| LA-31 | Lachnospiraceae[G-3]_sp_HOT_100 | 0.00% | 0.01% | 0.02% | 0.01% | 0.00% | 0.00% | 0.04% | 0.00% | 0.14% | 0.01% | 0.02% |
| TM-14 | TM7[G-1]_sp_HOT_952 | 0.01% | 0.01% | 0.02% | 0.01% | 0.01% | 0.02% | 0.04% | 0.00% | 0.02% | 0.09% | 0.02% |
| ST-30 | Stomatobaculum_longum | 0.06% | 0.04% | 0.02% | 0.01% | 0.02% | 0.04% | 0.00% | 0.00% | 0.03% | 0.01% | 0.02% |
| AC-03 | Actinobaculum_sp_HOT_183 | 0.00% | 0.01% | 0.10% | 0.04% | 0.01% | 0.06% | 0.01% | 0.00% | 0.00% | 0.00% | 0.02% |
| PR-42 | Prevotella_sp_HOT_317 | 0.01% | 0.03% | 0.03% | 0.01% | 0.01% | 0.02% | 0.08% | 0.02% | 0.01% | 0.02% | 0.02% |
| OR-08 | Oribacterium_sp_HOT_078 | 0.00% | 0.01% | 0.01% | 0.00% | 0.01% | 0.01% | 0.16% | 0.01% | 0.00% | 0.00% | 0.02% |
| TM-03 | TM7[G-1]_sp_HOT_348 | 0.00% | 0.01% | 0.05% | 0.01% | 0.01% | 0.12% | 0.02% | 0.00% | 0.01% | 0.00% | 0.02% |
| LE-12 | Leptotrichia_sp_HOT_219 | 0.01% | 0.01% | 0.01% | 0.01% | 0.01% | 0.06% | 0.03% | 0.01% | 0.04% | 0.02% | 0.02% |
| PO-16 | Porphyromonas_catoniae | 0.01% | 0.01% | 0.02% | 0.00% | 0.00% | 0.03% | 0.00% | 0.04% | 0.06% | 0.04% | 0.02% |
| PR-65 | Prevotella_intermedia | 0.03% | 0.06% | 0.05% | 0.02% | 0.02% | 0.00% | 0.00% | 0.00% | 0.01% | 0.01% | 0.02% |
| AL-03 | Alloprevotella_sp_HOT_308 | 0.03% | 0.04% | 0.01% | 0.01% | 0.03% | 0.03% | 0.00% | 0.01% | 0.03% | 0.02% | 0.02% |
| CA-01 | Campylobacter_concisus | 0.01% | 0.02% | 0.02% | 0.02% | 0.01% | 0.04% | 0.02% | 0.02% | 0.03% | 0.02% | 0.02% |
| KI-05 | Kingella_sp_HOT_012 | 0.02% | 0.04% | 0.01% | 0.00% | 0.00% | 0.01% | 0.01% | 0.07% | 0.00% | 0.03% | 0.02% |
| BE-03 | Bergeyella_sp_HOT_900 | 0.02% | 0.02% | 0.02% | 0.01% | 0.01% | 0.01% | 0.01% | 0.02% | 0.07% | 0.02% | 0.02% |
| AC-45 | Actinomyces_oris | 0.00% | 0.00% | 0.01% | 0.01% | 0.01% | 0.04% | 0.00% | 0.05% | 0.02% | 0.04% | 0.02% |
| TM-02 | TM7[G-1]_sp_HOT_347 | 0.01% | 0.02% | 0.01% | 0.01% | 0.00% | 0.02% | 0.02% | 0.06% | 0.01% | 0.02% | 0.02% |
| PR-23 | Prevotella_pleuritidis | 0.06% | 0.01% | 0.01% | 0.01% | 0.07% | 0.01% | 0.00% | 0.01% | 0.00% | 0.01% | 0.02% |
| GN-02 | GN02[G-1]_sp_HOT_872 | 0.01% | 0.01% | 0.01% | 0.02% | 0.01% | 0.02% | 0.01% | 0.05% | 0.00% | 0.05% | 0.02% |
| PR-37 | Prevotella_sp_HOT_305 | 0.04% | 0.02% | 0.01% | 0.00% | 0.05% | 0.02% | 0.00% | 0.00% | 0.02% | 0.01% | 0.02% |
| PA-03 | Parvimonas_micra | 0.01% | 0.03% | 0.02% | 0.01% | 0.04% | 0.02% | 0.01% | 0.01% | 0.01% | 0.00% | 0.02% |
| GP-057 | Moraxella_Genus_probe_2 | 0.01% | 0.00% | 0.14% | 0.00% | 0.01% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.02% |
| PR-78 | Prevotella_maculosa | 0.01% | 0.01% | 0.03% | 0.00% | 0.02% | 0.02% | 0.04% | 0.01% | 0.01% | 0.01% | 0.02% |
| PR-76 | Prevotella_aurantiaca | 0.00% | 0.00% | 0.00% | 0.01% | 0.02% | 0.00% | 0.11% | 0.00% | 0.00% | 0.00% | 0.01% |
| CA-36 | Capnocytophaga_sp_HOT_326 | 0.02% | 0.03% | 0.02% | 0.00% | 0.01% | 0.01% | 0.01% | 0.01% | 0.02% | 0.02% | 0.01% |
| TM-16 | TM7[G-6]_sp_HOT_870 | 0.03% | 0.01% | 0.02% | 0.01% | 0.02% | 0.01% | 0.02% | 0.00% | 0.01% | 0.01% | 0.01% |
| PE-17 | Peptostreptococcaceae[XI][G-7][Eubacterium]_yurii_subsp_schtitka | 0.00% | 0.10% | 0.00% | 0.00% | 0.01% | 0.00% | 0.01% | 0.00% | 0.00% | 0.01% | 0.01% |
| AL-02 | Alloprevotella_rava | 0.02% | 0.00% | 0.00% | 0.02% | 0.04% | 0.02% | 0.00% | 0.00% | 0.03% | 0.00% | 0.01% |
| LE-25 | Leptotrichia_sp_HOT_223 | 0.01% | 0.01% | 0.01% | 0.00% | 0.01% | 0.01% | 0.05% | 0.00% | 0.01% | 0.03% | 0.01% |
| SE-37 | Selenomonas_sp_HOT_126 | 0.00% | 0.00% | 0.09% | 0.00% | 0.00% | 0.01% | 0.01% | 0.00% | 0.01% | 0.00% | 0.01% |
| LA-09 | Lachnospiraceae[G-3]_sp_HOT_100 | 0.00% | 0.01% | 0.00% | 0.01% | 0.00% | 0.01% | 0.01% | 0.01% | 0.06% | 0.02% | 0.01% |
| PR-73 | Prevotella_sp_HOT_317 | 0.01% | 0.02% | 0.02% | 0.01% | 0.00% | 0.02% | 0.00% | 0.01% | 0.04% | 0.00% | 0.01% |
| PR-74 | Prevotella_sp_HOT_317 | 0.00% | 0.01% | 0.01% | 0.00% | 0.00% | 0.00% | 0.08% | 0.01% | 0.00% | 0.00% | 0.01% |
| VE-18 | Veillonella_denticariosi | 0.01% | 0.03% | 0.00% | 0.00% | 0.02% | 0.01% | 0.00% | 0.00% | 0.03% | 0.00% | 0.01% |
| AC-27 | Actinomyces_sp_HOT_448 | 0.00% | 0.00% | 0.01% | 0.00% | 0.00% | 0.10% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% |
| PR-71 | Prevotella_saccharolytica | 0.00% | 0.00% | 0.01% | 0.00% | 0.00% | 0.01% | 0.06% | 0.01% | 0.01% | 0.00% | 0.01% |
| AT-03 | Atopobium_rimae | 0.02% | 0.01% | 0.01% | 0.00% | 0.03% | 0.01% | 0.01% | 0.02% | 0.00% | 0.00% | 0.01% |
| TR-57 | Treponema_sp_HOT_231 | 0.00% | 0.01% | 0.01% | 0.01% | 0.02% | 0.00% | 0.01% | 0.00% | 0.05% | 0.00% | 0.01% |
| AC-11 | Actinomyces_massiliensis | 0.00% | 0.01% | 0.00% | 0.00% | 0.00% | 0.01% | 0.03% | 0.01% | 0.02% | 0.01% | 0.01% |
| GP-082 | Tannerella_Genus_probe | 0.01% | 0.01% | 0.01% | 0.00% | 0.01% | 0.00% | 0.01% | 0.01% | 0.00% | 0.05% | 0.01% |
| VE-05 | Veillonella_parvula | 0.01% | 0.01% | 0.01% | 0.01% | 0.01% | 0.02% | 0.00% | 0.01% | 0.01% | 0.01% | 0.01% |
| PO-22 | Porphyromonas_sp_HOT_277 | 0.04% | 0.01% | 0.01% | 0.00% | 0.02% | 0.02% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% |
| SE-22 | Selenomonas_dianae | 0.00% | 0.00% | 0.05% | 0.00% | 0.00% | 0.00% | 0.02% | 0.01% | 0.00% | 0.01% | 0.01% |
| LA-07 | Lachnospiraceae[G-2]_sp_HOT_088 | 0.00% | 0.00% | 0.01% | 0.02% | 0.00% | 0.03% | 0.02% | 0.01% | 0.00% | 0.00% | 0.01% |
| CA-21 | Capnocytophaga_sp_HOT_338 | 0.00% | 0.01% | 0.00% | 0.02% | 0.00% | 0.03% | 0.01% | 0.00% | 0.01% | 0.00% | 0.01% |
| CE-02 | Centipeda_periodontii | 0.00% | 0.00% | 0.03% | 0.00% | 0.00% | 0.00% | 0.06% | 0.00% | 0.00% | 0.00% | 0.01% |
| SE-18 | Selenomonas_sputigena | 0.01% | 0.00% | 0.03% | 0.00% | 0.00% | 0.01% | 0.01% | 0.02% | 0.00% | 0.00% | 0.01% |
| AC-18 | Actinomyces_sp_HOT_170 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% | 0.01% | 0.05% | 0.02% | 0.01% |
| GP-066 | Porphyromonas_Genus_probe_1 | 0.01% | 0.02% | 0.01% | 0.04% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% |
| GP-056 | Mogibacterium_Genus_probe | 0.01% | 0.01% | 0.03% | 0.00% | 0.01% | 0.01% | 0.00% | 0.01% | 0.01% | 0.00% | 0.01% |
| TM-09 | TM7[G-3]_sp_HOT_351 | 0.01% | 0.01% | 0.00% | 0.01% | 0.02% | 0.01% | 0.00% | 0.01% | 0.01% | 0.00% | 0.01% |
| GP-079 | SR1_Genus_probe | 0.01% | 0.02% | 0.01% | 0.01% | 0.01% | 0.00% | 0.01% | 0.01% | 0.01% | 0.02% | 0.01% |
| GP-009 | Atopobium_Genus_probe | 0.01% | 0.01% | 0.00% | 0.00% | 0.02% | 0.02% | 0.00% | 0.01% | 0.02% | 0.00% | 0.01% |
| ST-09 | Streptococcus_anginosus | 0.00% | 0.00% | 0.01% | 0.00% | 0.01% | 0.01% | 0.00% | 0.00% | 0.02% | 0.03% | 0.01% |
| AL-12 | Alloprevotella_sp_HOT_912 | 0.00% | 0.00% | 0.05% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% | 0.00% | 0.01% |
| PR-55 | Propionibacterium_propionicum | 0.01% | 0.01% | 0.01% | 0.01% | 0.01% | 0.02% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% |
| GP-072 | Pseudomonas_Genus_probe | 0.01% | 0.00% | 0.00% | 0.01% | 0.00% | 0.05% | 0.00% | 0.00% | 0.01% | 0.00% | 0.01% |
| AC-19 | Actinomyces_sp_HOT_171 | 0.00% | 0.03% | 0.00% | 0.00% | 0.00% | 0.01% | 0.00% | 0.00% | 0.01% | 0.01% | 0.01% |
| GP-111 | Haemophilus_Genus_probe_1 | 0.00% | 0.06% | 0.01% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% |
| PR-72 | Prevotella_saccharolytica | 0.00% | 0.01% | 0.01% | 0.00% | 0.00% | 0.01% | 0.01% | 0.00% | 0.03% | 0.00% | 0.01% |
| PR-45 | Prevotella_sp_HOT_443 | 0.00% | 0.03% | 0.00% | 0.00% | 0.02% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% |
| GP-128 | Streptococcus_Genus_probe_3 | 0.01% | 0.00% | 0.01% | 0.01% | 0.01% | 0.01% | 0.00% | 0.01% | 0.00% | 0.00% | 0.01% |
| PE-22 | Peptococcus_sp_HOT_167 | 0.00% | 0.02% | 0.01% | 0.01% | 0.01% | 0.00% | 0.00% | 0.01% | 0.00% | 0.00% | 0.01% |
| ST-10 | Streptococcus_constellatus | 0.01% | 0.01% | 0.01% | 0.01% | 0.01% | 0.01% | 0.00% | 0.00% | 0.01% | 0.00% | 0.01% |
| AC-21 | Actinomyces_sp_HOT_175 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% | 0.00% | 0.03% | 0.00% | 0.01% | 0.01% |
| LE-18 | Leptotrichia_sp_HOT_463 | 0.01% | 0.02% | 0.03% | 0.01% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% |
| AL-08 | Alloprevotella_sp_HOT_914 | 0.00% | 0.01% | 0.01% | 0.01% | 0.01% | 0.00% | 0.00% | 0.01% | 0.00% | 0.00% | 0.01% |
| JO-01 | Johnsonella_ignava | 0.00% | 0.01% | 0.00% | 0.00% | 0.01% | 0.02% | 0.02% | 0.00% | 0.00% | 0.00% | 0.01% |
| VE-02 | Veillonella_denticariosi | 0.02% | 0.01% | 0.01% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% | 0.01% | 0.01% |
| LE-21 | Leptotrichia_sp_HOT_879 | 0.01% | 0.01% | 0.00% | 0.00% | 0.01% | 0.01% | 0.01% | 0.01% | 0.00% | 0.01% | 0.01% |
| GP-047 | Lactobacillus_Genus_probe_4 | 0.06% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% |
| EU-04 | Peptostreptococcaceae[XI][G-9][Eubacterium]_brachy | 0.01% | 0.01% | 0.01% | 0.00% | 0.01% | 0.00% | 0.01% | 0.00% | 0.00% | 0.01% | 0.01% |
| LA-03 | Lachnoanaerobaculum_sp_HOT_083 | 0.00% | 0.04% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% | 0.00% | 0.00% | 0.00% | 0.01% |
| AC-22 | Actinomyces_sp_HOT_178 | 0.01% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% | 0.00% | 0.01% | 0.02% | 0.01% |
| NE-12 | Neisseria_sp_HOT_499 | 0.03% | 0.00% | 0.00% | 0.00% | 0.01% | 0.00% | 0.01% | 0.00% | 0.00% | 0.00% | 0.01% |
| TR-56 | Treponema_socranskii | 0.01% | 0.00% | 0.00% | 0.01% | 0.02% | 0.01% | 0.00% | 0.00% | 0.01% | 0.00% | 0.01% |
| SE-28 | Selenomonas_sp_HOT_137 | 0.00% | 0.00% | 0.02% | 0.00% | 0.00% | 0.01% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% |
| CA-06 | Campylobacter_sp_HOT_044 | 0.02% | 0.02% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.01% |
This table includes microorganisms with average measurable abundance levels >0.01%. Microorganism species (N =284) with average measurable abundance <0.01% were excluded from this table for space considerations.
Table 3.
Significant Differences in Salivary Microorganisms Found Between Specific Cohorts or Between Visits 1 and 2.
| A. Differences Found Between Caries-Active Boys, Ages 2–5 Years Versus Caries-Active Boys, Ages 6–12 Years | ||||
|---|---|---|---|---|
| Probe ID | Microorganism | Fold changes | p-value | FDR p-value |
| GP-057 | Moraxella_Genus_probe_2 | 301.4 | 0.0002 | 0.0231 |
| LE-11 | Leptotrichia_sp_HOT_218 | 83.8 | 0.0006 | 0.0358 |
| ST-15 | Streptococcus_mutans | −24.6 | 0.0000 | 0.0051 |
| PO-26 | Porphyromonas_sp_HOT_930 | −86.9 | 0.0000 | 0.0051 |
| ST-21 | Streptococcus_sobrinus | −129.0 | 0.0005 | 0.0339 |
| B. Differences Found Between Visits 1 and 2 for Group 3 | ||||
|---|---|---|---|---|
| Probe ID | Microorganism | Fold changes | p-value | FDR p-value |
| ST-29 | Stenotrophomonas_maltophilia | 3473.9 | 4.88E–08 | 4.85E–06 |
| GP-080 | Staphylococcus_Genus_probe_3 | 1001.8 | 1.45E–11 | 4.33E–09 |
| GP-072 | Pseudomonas_Genus_probe | 29.3 | 0.00207681 | 0.03998775 |
| EI-01 | Eikenella_corrodens | 18.6 | 9.66E–10 | 1.44E–07 |
| CA-32 | Cardiobacterium_valvarum | 11.4 | 0.00029569 | 0.01101428 |
| NE-02 | Neisseria_elongata | 10.2 | 1.00E–06 | 7.48E–05 |
| LE-04 | Leptotrichia_goodfellowii | 9.4 | 0.0001934 | 0.00960559 |
| AC-36 | Actinomyces_naeslundii | 6.4 | 0.00023007 | 0.00979436 |
| GP-063 | Parvimonas_Genus_probe | 6.2 | 0.00210945 | 0.03998775 |
| CA-31 | Cardiobacterium_hominis | 5.2 | 5.87E–05 | 0.0034991 |
| PO-27 | Porphyromonas_sp_HOT_284 | 5.1 | 0.00185532 | 0.03998775 |
| LE-06 | Leptotrichia_hongkongensis | 5.1 | 0.00061526 | 0.02037181 |
| GP-095 | Fusobacterium_Genus_probe_2 | 3.3 | 0.00171068 | 0.03998775 |
| VE-03 | Veillonella_dispar | 3.3 | 0.00123336 | 0.03341292 |
| GP-096 | Fusobacterium_Genus_probe_3 | 3.2 | 0.00102153 | 0.03044152 |
| GN-03 | GN02[G-2]_sp_HOT_873 | −17.4 | 0.00214699 | 0.03998775 |
| C. Differences Found Between Visits 1 and 2 for Group 5 | ||||
| Probe ID | Microorganism | Fold changes | p-value | FDR p-value |
| GP-022 | Cardiobacterium_Genus_probe | −10.6 | 0.00022085 | 0.02360636 |
| TM-09 | TM7[G-3]_sp_HOT_351 | −16.4 | 0.00023765 | 0.02360636 |
| SE-04 | Selenomonas_noxia | −135.7 | 4.52E–05 | 0.01348336 |
For Table 3A, microorganisms exhibiting positive value-fold changes indicate higher prevalence in caries-active boys, ages 6–12 years.
For Table 3A, microorganisms exhibiting negative value-fold changes indicate higher prevalence in caries-active boys, ages 2–5 years.
FDR = false discovery rate; FDR p-value is the more stringent statistical indicator and the one used to determine statistical-significance.
Group 3 is defined as the caries-active cohort treated with restorative therapy and povidone iodine.
For Table 3B, microorganisms exhibiting positive value-fold changes indicate higher prevalence following treatment.
For Table 3B, microorganisms exhibiting negative value-fold change indicates higher prevalence before treatment.
Group 5 is defined as the caries-free cohort treated with chlorhexidine.
For Table 3C, microorganisms exhibiting negative value-fold changes indicate higher prevalence before treatment compared to after treatment. No microorganisms exhibiting positive value-fold changes (indicating higher prevalence after treatment) were identified.
3.2. Evidence supporting efficacy of povidone iodine in shifting the microbiome profile from caries-active to caries-free
Multidimensional Scaling Plots.
Fig. 2A displays the relative position of each microbiome in a multi-dimensional scaling plot illustrating the general and independent clustering of caries-active (red) and caries-free (black) microbiomes at pre-treatment (Visit 1). Following adjunctive therapy (Fig. 2B) including povidone iodine (Group 3), we observe a shift of several microbiomes into the lower right quadrant of the multi-dimensional graph, or more closely aligned in the placement of microbiomes defined as caries-free in Visit 1. This is especially evident when microbiomes affected by povidone iodine in Visit 2 (as seen in Fig. 2B; Group 3) are overlaid directly upon microbiomes identified as caries-free (Groups 4 and 5) in Fig. 2A. Interestingly, many microbiomes from caries-free children treated with fluoride (Group 4) or chlorhexidine (Group 5) become more dispersed in the multi-dimensional scaling plot at Visit 2 (Fig. 2B). Fold changes were also assessed for each microbial species at pre-treatment versus post-treatment and then ranked from high positive values (indicating high prevalence at Visit 2; post-treatment) to low negative values (indicating high prevalence at Visit 1; pre-treatment). Microorganisms found in Groups 3 (caries-active; povidone iodine) and 5 (caries-free; chlorhexidine) were identified to undergo statistically-significant changes following adjunctive treatment (Table 3B and C). Microorganisms found to statistically increase in Group 3 following povidone iodine treatment include Stentrophomonas melophilia and Staphylococcus genus probe 3, as well as several other microorganisms found in periodontal disease, including Porphyromonas species HOT 284 and Fusobacterium genus probe 2 and probe 3 (Table 3B). In Group 5 (caries-free group), treatment with chlorhexidine results in statistical reduction of three microorganisms, including Cardiobacterium genus probe, TM7 (G-3) species HOT 351 and Selenomonas noxia (Table 3C).
Fig. 2. Multidimensional Scaling Plots of Microbiome Profiles of Participants at Visit 1 and Visit 2.
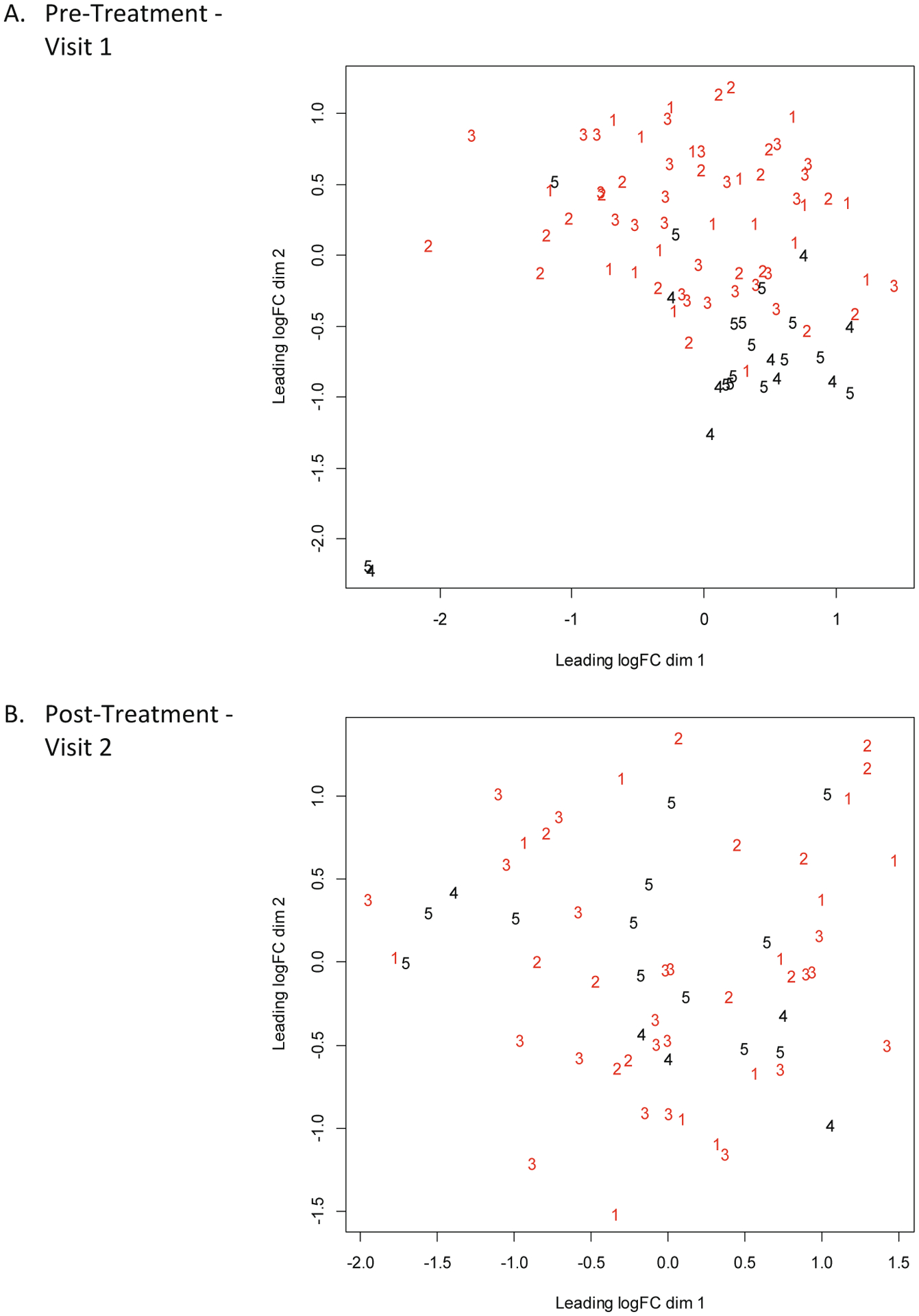
Multidimensional scaling plots of microbiome profiles of participants at Visit 1 (A) and Visit 2 (B). Numbers represent the treatment groups. Groups 1–3 are caries-active and Groups 4–5 are caries-free.
Support Vector Machine and Building Prediction Rule to Define Caries-Active and Caries-Free Individuals Based on Microbiome Profiles.
Data from untreated caries-active and caries-free individuals at Visit 1 (61 from Groups 1–3 and 24 from Groups 4 and 5) were used as training data with support vector machine (SVM) to define the microbiomes of caries-active and caries-free individuals. SVM with the default parameters and cost-tuning parameter of 0.89 generated a caries-active versus caries-free prediction rule with 100% accuracy. When applying this highly-conservative prediction rule to identify the caries-status of individuals following treatment, we identified zero out of 11 individuals in Group 1, one out of 12 individuals in Group 2, and four out of 21 individuals in Group 3 as caries-free based on microbiome profiles. Alternatively, for caries-free individuals in Groups 4 and 5, zero out of nine individuals in Group 4 and only two out of 12 individuals in Group 5 were reclassified as caries-active following adjunctive dental therapy.
Odds Ratio Analyses.
We also computed odd1 = probability of being caries-active at Visit 1 (pre-treatment) / probability of being caries-free at Visit 1 (pre-treatment), and similarly odd2 at Visit 2 (post-treatment) using the outputs from the SVM analysis. An odds ratio was computed as OR = odd1/odd2. If the log of the odds ratio is greater than zero, then odd2 > odd1, which implies that the odds of being caries-active is relatively larger at Visit 2. If the odd2 < odd1, then the odds of being caries-free becomes larger at Visit 2. Using this approach, we computed the ratios of odds for prediction of caries-active versus caries-free between Visits 1 and 2 for each of the five enrollment groups. Odds ratio analyses indicate that the microbiome profiles of seven out of nine individuals in Group 1, seven out of ten individuals in Group 2 and seventeen out of eighteen individuals in Group 3 had shifted microbiome profiles from caries-active to those more-typically identified as caries-free. Using the Group 5 caries-free control group, only one out of eleven individuals showed a reversal of microbiome profiles defined originally as caries-free to one observed as caries-active.
4. Discussion
4.1. Adjunctive dental therapies and anti-microbial activity
Adjunctive therapies, including povidone iodine or chlorhexidine, have been used to promote caries prevention and oral health. Regular applications of povidone iodine are especially useful in treating childhood caries and suppressing levels of S. mutans [10,24]. In addition to caries control, povidone iodine is known to be effective in managing periodontal disease and gingivitis, and results in the reduction of periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum [9,10]. In totality, the clinical evidence demonstrates povidone iodine as an excellent adjunctive antimicrobial agent due to its broad spectrum of microbicidal effects [25].
Chlorhexidine is an effective bacteriostatic agent with the ability to alter microbiota composition and metabolic activity, and can promote higher abundance of pathobionts associated with chronic inflammatory conditions and high lactate production [26]. Although chlorhexidine can decrease the pathogenic microbial burden, it is less efficacious than povidone iodine in altering subgingival microbial composition [8]. Additionally, chlorhexidine produces indiscriminate antimicrobial effects, producing widespread decreases in biomass and variety throughout the oral microbiome [27]. Chlorhexidine therapy also results in the development of microbial tolerance, allowing resistant species to survive and reduce therapeutic efficacy [10,28]. For our specific study, relative changes in the percent composition of specific microorganisms could be due to either decreases in absolute numbers of microorganisms directly affected by the adjunctive therapy, or indirectly due to concomitant percent increases of other microbial species that are more resistant to treatment. It is important to recognize that the starting microbial populations found in children within Groups 2 and 5, reflect conditions of disease versus health, respectively. These starting microbial populations are not identical and have several species differences which could account for the variability in fold changes observed between Groups 2 and 5. Streptococci is broad and composed of a multitude of individual species, which are different within caries-active versus caries-free children, and thus may be differentially affected by chlorhexidine treatment. For example, S. gordonii, typically associated with caries-free conditions, is more tolerant to chlorhexidine action [26] because it is a primary colonizer and inhabitant of the lower layers of the oral biofilm and thus more protected from chlorhexidine action.
4.2. Microbial dysbiosis and reversion with povidone iodine
The application of povidone iodine in the oral cavity is shown to have a variety of effects, from preventing dental caries and periodontal disease, to reducing instances of respiratory infections and preventing pneumonia [9,10,24,29,30–32]. Mouth rinses with povidone iodine results in reduced oropharyngeal levels of P. aeruginosa, S. aureus, and H. influenza, bacteria linked to chronic respiratory infections and pneumonia [10,31,32]. Povidone iodine therapy controls oxidative stress and promotes wound healing via anti-inflammatory effects, free-radical scavenging, and promotion of re-epithelialization [33,34].
4.3. Limitations
The primary limitation of our study was the restrictive number of caries-free children available in our University dental clinics, which are more dedicated towards treatment of advanced dental cases, including rampant childhood caries. This was compounded further by participant no-shows for Visit 2 in all enrollment groups, especially affecting the caries-free Groups 4 and 5. We also understand that restorative materials may have an anti-microbial effect, and that there may be a combinatorial effect on bacterial composition when using both restorative and adjunctive therapies. Due to the necessity of providing standard-of-care practices in treatment of caries-active participants, it was not possible to deploy an adjunctive therapy without restorative therapy, and acknowledge that the type of restoration (fissure sealant versus restoration of simple or complex cavity) may influence important qualities of the microbial population, including bacterial load and virulence potential. We also recognize that there are many additional variables that could influence dental caries that were not controlled in this study, including genetic, socioeconomic, dietary, ethnicity and cultural determinants. We understand that the statistical power of our study could have been increased with larger numbers of participants.
4.4. Conclusions and future applications
With the use of multiple lines of analyses, including the use of multidimensional scaling plots, support vector machine, and odds ratio determinations, we found that povidone iodine has the potential to shift the salivary cariogenic microbiome to one more typically observed in non-cariogenic health. We also compared individual microbial species, prior to and following povidone iodine therapy (Group 3), and identified fifteen microbial species that underwent statistically-significant fold changes in prevalence following therapy (Table 3B). None of the major cariogenic microbiological agents – S. mutans, S. sobrinus or S. wiggsiae – were prevalent following povidone iodine therapy, in contrast to higher prevalence of several periodontal microorganisms, including Porphyromonas and Fusobacterium (Table 3B). Interestingly, the use of povidone iodine dramatically increased the prevalence of Stentrophomonas maltophila, an uncommon aerobic microorganism typically related to health, but more recently identified as a multi-drug resistant opportunistic pathogen that are particularly high-risk among immunocompromised hospital patients [35]. Repeated exposure of povidone iodine over an extended period of time appears to be necessary to restore complete conditions of health [36] and we assume this also applies to the long-term restoration of the non-cariogenic microbiome. The percent increase of Stentophormonas maltophilia in Group 3 is presumably due to its resistance to povidone iodine and the subsequent reduction of other microorganisms within the overall microbial population. Due to the unexpected prevalence of S. maltophilia found to affect cystic fibrosis and other respiratory patients, we recommend limiting the use of povidone iodine in immunocompromised dental patients as well as other individuals exhibiting endocrine disorders such as hyperparathyroidism who may be adversely affected by additional iodine intake.
5. Declarations
5.1. Human participant approval and informed consent
The OHSU Institutional Review Board approved the use of human participants for this study (IRB Study Number 0006535). Parents or guardians of the participants in this study have provided written informed consent prior to specimen collection. For children older than seven years of age, written child assent was also obtained. This study was purely an observational, non-interventional study, where the application of fluoride was required for all dental patients in the provision of standard-of-care therapy. The range of 4–8 weeks between Visits 1 and 2 was recommended to provide flexibility in scheduling patient returns for Visit 2.
Acknowledgments
EH is a recipient of the Dean’s Student Research Fellowship Award from the OHSU School of Dentistry. EH, SO, BK, and MS are current students in the academic DMD degree program at the OHSU School of Dentistry. CL is the current Clinical Study Coordinator for our project. EW, JW, EP, TM, and CM receive salary support from the OHSU School of Dentistry and AF has an adjunct appointment at the School of Dentistry. This project and CM were also supported in part by NIH DE024317. KK is a collaborating independent investigator covered by OHSU FWA #161, who provided several caries-free study participants from his private dental practice in Albany, Oregon. JI, AJ, SS, JW, EW, BN, JC, and CC were former residents, staff members, or students who contributed in earlier phases of the project, and have started professional dental practice, obtained faculty appointments, or moved to other academic institutions. We also thank KK for establishing and supporting with Carifree (Albany, Oregon), the Bob Bowers Memorial Award for Student Research in Dental Caries. The Bob Bowers Memorial Award helps support dental students to conduct and present caries-related research at professional dental meetings and conferences. We thank the many other students, residents and staff who provided technical support in the earlier years of the project – including Paige Schmidt, Aisha Saradi, Manizha Rezayee, Tyler Horton, Courtney Levasa, Alyssa Avila, and Allen Yoshinaga. Special thanks are also extended to DMD students, Kareem Raslan and Ghazaleh Vargha, for their encouragement and support of their dentistry colleagues. We also thank Dr. Bruce Paster and the HOMINGS Laboratory for providing commentary on next generation sequencing procedures, and Drs. Tom Shearer, Bill Knight, Bob Steelman, John Engle, John Peterson, and Dena Fischer (NIDCR) for many discussions and support.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol 2014;15(7):R89. 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rasiah IA, Wong L, Anderson SA, Sissons CH. Variation in bacterial DGGE patterns from human saliva: over time, between individuals and in corresponding dental plaque microcosms. Arch Oral Biol 2005;50(9):779–87. 10.1016/j.archoralbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- [3].Zhou Y, Gao H, Mihindukulasuriya KA, La Rosa PS, Wylie KM, Vishnivetskaya T, et al. Biogeography of the ecosystems of the healthy human body. Genome Biol 2013;14(1):R1. 10.1186/gb-2013-14-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zaura E, Brandt BW, Teixeira DE, Mattos MJ, Buijs MJ, Caspers MP, et al. Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. MBio 2015;6(6):e01693–15. 10.1128/mBio.01693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shaffer JR, Wang X, McNeil DW, Weyant RJ, Crout R, Marazita ML. Genetic susceptibility to dental caries differs between the sexes: a family-based study. Caries Res 2015;49(2):133–40. 10.1159/000369103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hakan Ç, Dülgergil Ç, Dalli M, Hamidi MM. Early childhood caries update: a review of causes, diagnoses, and treatments. J Nat Sci Bio Med 2013;4(1):29–38. 10.4103/0976-9668.107257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kanagalingam J, Chopra A, Hong MH, Ibrahim W, Villaon A, Lin JC. Povidone-iodine for the management of oral mucositis during cancer therapy. Oncol Rev 2017;11(2):341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Slots J. Selection of antimicrobial agents in periodontal therapy. J Periodont Res 2002;37(5):389–98. 10.4081/oncol.2017.341. [DOI] [PubMed] [Google Scholar]
- [9].Hosaka Y, Saito A, Maeda R, Fukaya C, Morikawa S, Makino A, et al. Antibacterial activity of povidone-iodine against an artificial biofilm of Porphyromonas gingivalis and Fusobacterium nucleatum. Arch Oral Biol 2012;57(4):364–8. 10.1016/j.archoralbio.2011.09.005. [DOI] [PubMed] [Google Scholar]
- [10].Kanagalingam J, Feliciano R, Hah JH, Labib H, Le TA, Lin JC. Practical use of povidone-iodine antiseptic in the maintenance of oral health and in the prevention and treatment of common oropharyngeal infections. Int J Clin Pract 2015;69(11):1247–56. 10.1111/ijcp.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wewalka G, Stary A, Bosse B, Duerr HE, Reimer K. Efficacy of povidone-iodine vaginal suppositories in the treatment of bacterial vaginosis. Dermatology 2002; 204(Suppl. 1):79–85. 10.1159/000057731. [DOI] [PubMed] [Google Scholar]
- [12].Mohammadi Z, Abbott PV. The properties and applications of chlorhexidine in Endodontics. Int Endod J 2009;42(4):288–302. 10.1111/j.1365-2591.2008.01540.x. [DOI] [PubMed] [Google Scholar]
- [13].Jorgensen MG, Aalam A, Slots J. Periodontal antimicrobials – finding the right solutions. Int Dent J 2005;55(1):3–12. 10.1111/j.1875-595x.2005.tb00025.x. [DOI] [PubMed] [Google Scholar]
- [14].Yildirim A, Metzler P, Lübbers HT, Yildirim V. Chlorhexidine – history, mechanism and risks. Swiss Dent J 2015;125(7–8):830–1. [DOI] [PubMed] [Google Scholar]
- [15].Lachapelle J, Castel O, Casado AF, Leroy B, Micali G, Tennstedt D, et al. Antiseptics in the era of bacterial resistance: a focus on povidone iodine. Future Med 2013;10(5):579–92. 10.2217/CPR.13.50. [DOI] [Google Scholar]
- [16].Ortiz S, Herrman E, Lyashenko C, Purcell A, Raslan K, Khor B, et al. Sex-specific differences in the salivary microbiome of caries-active children. J Oral Microbiol 2019;11(1):1653124. 10.1080/20002297.2019.1653124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Forsyth A, Raslan K, Lyashenko C, Bona S, Snow M, Khor B, et al. Children with autism spectrum disorder: Pilot studies examining the salivary microbiome and implications for gut metabolism and social behavior. Hum Microbiome J 2019;15(2020). 10.1016/j.humic.2019.100066. [DOI] [Google Scholar]
- [18].Risso D, Perraudeau F, Gribkova S, Dudoit S, Vert JP. A general and flexible method for signal extraction from single-cell RNA-seq data. Nat Commun 2018;9 (1):284. 10.1038/s41467-017-02554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucl Acids Res 2012;40(10):4288–97. 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 1995;57(1):289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- [21].Mead A. Review of the development of multidimensional scaling methods. J Roy Stat Soc D-Stat 1992;41(1):27–39. 10.2307/2348634. [DOI] [Google Scholar]
- [22].Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl Acids Res 2015;43(7):e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;20(3):273–97. 10.1007/BF00994018. [DOI] [Google Scholar]
- [24].Simratvir M, Singh N, Chopra S, Thomas AM. Efficacy of 10% povidone iodine in children affected with early childhood caries: an in vivo study. J Clin Pediatr Dent 2010;34(3):233–8. 10.17796/jcpd.34.3.l552816527xtv122. [DOI] [PubMed] [Google Scholar]
- [25].Reilly C, Goettl M, Steinmetz M, Nikrad J, Jones RS. Short-term effects of povidone iodine and sodium fluoride therapy on plaque levels and microbiome diversity. Oral Dis 2016;22(2):155–61. 10.1111/odi.12407. [DOI] [PubMed] [Google Scholar]
- [26].Chatzigiannidou I, Teughels W, Van de Wiele T, Boon N. Oral biofilms exposure to chlorhexidine results in altered microbial composition and metabolic profile. Biofilms Microbiom 2020;6:13. 10.1038/s41522-020-0124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Al-Kamel A, Baraniya D, Al-Hajj WA, Halboub E, Abdulrab S, Chen T, et al. Subgingival microbiome of experimental gingivitis: shifts associated with the use of chlorhexidine and N-acetyl cysteine mouthwashes. J Oral Microbiol 2019;11(1):1608141. 10.1080/20002297.2019.1608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kaspar JR, Godwin MJ, Velsko IM, Richards VP, Burne RA. Spontaneously arising Streptococcus mutans variants with reduced susceptibility to chlorhexidine display genetic defects and diminished fitness. Antimicrob Agents Chemother 2019;63(7): e00161–219. 10.1128/AAC.00161-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Amin MS, Harrison RL, Benton TS, Roberts M, Weinstein P. Effect of povidone-iodine on Streptococcus mutans in children with extensive dental caries. Pediatr Dent 2004;26(1):5–10. [PubMed] [Google Scholar]
- [30].Evans A, Leishman SJ, Walsh LI, Seow WK. Inhibitory effects of antiseptic mouthrinses on Streptococcus mutans, Streptococcus sanguinis and Lactobacillus acidophilus. Aust Dent J 2015;60(2):247–54. 10.1111/adj.12312. [DOI] [PubMed] [Google Scholar]
- [31].Nagatake T, Ahmed K, Oishi K. Prevention of respiratory infections by povidone-iodine gargle. Dermatology 2002;204(Suppl. 1):32–6. 10.1159/000057722. [DOI] [PubMed] [Google Scholar]
- [32].Sakai M, Shimbo T, Omata K, Takahashi Y, Satomura K, Kitamura T, et al. Cost effectiveness of gargling for the prevention of upper respiratory tract infections. BMC Health Serv Res 2008;8:258. 10.1186/1472-6963-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Beukelman CJ, van den Berg AJ, Hoekstra MJ, Uhl R, Reimer K, Mueller S. Anti-inflammatory properties of a liposomal hydrogel with povidone-iodine (Repithel) for wound healing in vitro. Burns 2008;34(6):845–55. 10.1016/j.burns.2007.11.014. [DOI] [PubMed] [Google Scholar]
- [34].Vogt PM, Hauser J, Mueller S, Bosse B, Hopp M. Efficacy of conventional and liposomal povidone-iodine in infected mesh skin grafts: an exploratory study. Infect Dis Ther 2017;6(4):545–55. 10.1007/s40121-017-0172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Brooke J. Stentrophormonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 2012;25(1):1–41. 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jayabel JB, Mahesh R. Current state of topical antimicrobial therapy in management of early childhood caries. ISRN Dentistry 2014, Article ID762458, 10.1155/2014/762458. [DOI] [PMC free article] [PubMed] [Google Scholar]


