Abstract
Current estimates of COVID-19 prevalence are largely based on symptomatic, clinically diagnosed cases. The existence of a large number of undiagnosed infections hampers population-wide investigation of viral circulation. Here, we quantify the SARS-CoV-2 concentration and track its dynamics in wastewater at a major urban wastewater treatment facility in Massachusetts, between early January and May 2020. SARS-CoV-2 was first detected in wastewater on March 3. SARS-CoV-2 RNA concentrations in wastewater correlated with clinically diagnosed new COVID-19 cases, with the trends appearing 4–10 days earlier in wastewater than in clinical data. We inferred viral shedding dynamics by modeling wastewater viral load as a convolution of back-dated new clinical cases with the average population-level viral shedding function. The inferred viral shedding function showed an early peak, likely before symptom onset and clinical diagnosis, consistent with emerging clinical and experimental evidence. This finding suggests that SARS-CoV-2 concentrations in wastewater may be primarily driven by viral shedding early in infection. This work shows that longitudinal wastewater analysis can be used to identify trends in disease transmission in advance of clinical case reporting, and infer early viral shedding dynamics for newly infected individuals, which are difficult to capture in clinical investigations.
Keywords: SARS-CoV-2, Wastewater surveillance, Longitudinal, Viral shedding, Foreshadow, Convolution model
Graphical abstract
1. Introduction
The ongoing coronavirus disease (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has quickly become a global health crisis, with over 2 million confirmed cases in the US and 7.4 million worldwide as of June 11, 2020 (Johns Hopkins University Center for Systems Science and Engineering, 2020). Due to limited diagnostic capacity in many countries and high rates of asymptomatic individuals (Gandhi et al., 2020; Mizumoto et al., 2020), these numbers are considered to be underestimates of the true prevalence of infection (Kaashoek and Santillana, 2020; Lu et al., 2021).
Wastewater surveillance has been used to detect illicit drug consumption, human health-related biomarkers, and infectious diseases including SARS-CoV-2. It offers a complementary approach to clinical disease surveillance to track viral outbreak and circulation in the population (Daughton, 2018; Fuschi et al., 2021; Lodder and de R. Husman, 2020; Mao et al., 2020; Sims and Kasprzyk-Hordern, 2020). In addition, wastewater surveillance provides an unbiased sample of the infected population, including asymptomatic and pre-symptomatic individuals, those who are symptomatic but have not yet been clinically confirmed, and individuals who may have the disease but do not seek healthcare. Early work has shown that wastewater surveillance could detect SARS-CoV-2 before it became widespread in a population, and that viral levels in wastewater largely paralleled local increases in clinical cases of COVID-19 in Australia (Ahmed et al., 2020a), France (Wurtzer et al., 2020), the Netherlands (Medema et al., 2020), Spain (Orive et al., 2020; Randazzo et al., 2020), Turkey (Kocamemi et al., 2020), and Israel (Bar-Or et al., 2021). These studies highlight the potential of wastewater surveillance to provide early warning of emerging outbreaks. However, it remains largely unexplored how viral concentrations in wastewater can be used for quantitative epidemiological modeling, such as informing viral shedding dynamics for infected individuals that is challenging to capture clinically.
The Massachusetts wastewater treatment facility where samples were collected has two major influent streams, which are referred to as the “northern” and “southern” influents. The daily flow rates during the sampling period for the northern and southern influents ranged from 7.53 × 105 to 1.84 × 106 m3/d, and 4.26 × 105 to 1.18 × 106 m3/d, respectively. Together the two sewersheds represent approximately 2.25 million sewered individuals in Middlesex, Norfolk, and Suffolk counties. We previously reported the detection of SARS-CoV-2 in wastewater from this facility, and showed that viral concentrations were significantly higher than would be expected based on clinical cases alone (Wu et al., 2020a, Wu et al., 2021). Here, we use longitudinal sampling from January to mid-May 2020 (116 samples in total) to identify the virus's first appearance and spread, to infer viral shedding dynamics, and to investigate the relationship between wastewater SARS-CoV-2 concentrations, clinically reported COVID-19 cases, and statewide public health interventions.
2. Materials and methods
2.1. Sample collection and viral inactivation
The 24-h composite samples of raw sewage were collected from the Deer Island Wastewater Treatment Plant in Massachusetts. The January, February and early March samples were stored at 4 °C in the wastewater treatment facility before being transferred to the laboratory on ice in a second container. Those samples were received in two groups and processed on April 17 and 21, respectively, and the longest storage time at 4 °C was 97 d (January 14 to April 21) before analysis. Samples from mid-March to May were received in six groups in the order of time and processed as previously described (Wu et al., 2020a, Wu et al., 2020b). Their storage time at 4 °C varied from 1 to 34 d. Briefly, samples were pasteurized at 60 °C for 90 min to inactivate the virus. Previous studies showed that pasteurization has little influence on the detection of SARS-CoV-2 RNA copies, and PMMoV is highly resistance to heat inactivation (Auerswald et al., 2021; Pastorino et al., 2020; Shirasaki et al., 2020; Wang et al., 2020b). Raw sewage was then vacuum filtered through a 0.2 μm membrane (Millipore Sigma) to remove bacterial cells and debris. Filtrate was used for the viral enrichment with the methods described below.
2.2. Viral precipitation, RNA extraction, reverse transcription and quantitative PCR (Method I)
Two methods were used for RNA extraction, reverse transcription and quantitative PCR (Methods I and II). For Method I, the filtrate (40 ml) was mixed with 4 g of Polyethylene glycol 8000 (10% w/v, Millipore Sigma) and 0.9 g NaCl (0.3 M, Millipore Sigma) and centrifuged at 12,000 ×g at 4 °C for 2 h (or overnight centrifugation at 3200 ×g) to precipitate the viral particles. The viral pellet was then resuspended in 1.5 ml Trizol reagent (Cat# 15596026, Thermo Fisher Scientific) for RNA extraction. cDNA was synthesized by reverse transcription (RT) based on the manufacturer's protocol (M0368, New England Biosciences). Briefly, 10 μl of RNA was mixed with random hexamers and then incubated at 70 °C for 5 min and 4 °C for 3 min. After that, the RNA-hexamer mixture was mixed with 5× ProtoScript II buffer (5 μl), 0.1 M DTT (2.5 μl), ProtoScript II Reverse Transcriptase (200 U/μl. 1.25 μl), 10 mM dNTP (1.25 μl), RNase Inhibitor (40 U/μl, 0.5 μl), and Nuclease-free water (2.5 μl) to a total volume of 25 μl. The mixture was then incubated at 42 °C for 1 h and inactivated at 65 °C for 20 min.
The quantitative PCR (qPCR) was performed with TaqMan assay techniques. Briefly, the TaqMan® Fast Advanced Master Mix (4444557, ThermoFisher Scientific) was mixed with the primers and water, and then distributed into the 96-well PCR plate. 2 μl of cDNA template from RT was added into the plate, which was sealed with adhesive PCR plate seals (AB0558, ThermoFisher Scientific). The qPCR reaction was carried out for 48 cycles using Bio-Rad CFX96 Real-Time PCR Detection System based on the following program: polymerase activation (95 °C for 2 min), PCR (48 cycles, denature at 95 °C for 1 s, and anneal/extend at 55 °C for 30 s).
The primers (N1 and N2) and DNA standards of SARS-CoV-2 nucleocapsid (N) gene were used to quantify the genome copies of SARS-CoV-2 (Table S1, Ref. (CDC, 2020)). Briefly, Cq values for N1 or N2 primer sets were first converted to viral gene copies in the cDNA sample (copies per μl of cDNA) based on the calibration curves established with the positive control plasmid (Table S1). This concentration was further converted to viral gene copies per microliter of the wastewater sample by multiplying the dilution factor (volume of total cDNA ∗ total volume of RNA) / (volume of RNA used for reverse transcription ∗ starting volume of filtered wastewater sample). Two or three replicates were performed for each primer set, and mean values were reported. The MIQE (Minimum Information for Publication of Quantitative Real-Time PCR Experiments) checklist was also provided as supplementary information (Table S1).
Concentrations of fecal materials in wastewater are subject to wide fluctuations in daily sewage flow rates at the wastewater treatment plant. To correct for these fluctuations, we used pepper mild mottle virus (PMMoV), a positive-strand RNA virus prevalent in human feces (Hamza et al., 2019; Rosario et al., 2009; Zhang et al., 2005), as an internal reference for quantification. PMMoV is globally distributed and highly stable in the wastewater (Hamza et al., 2019; Kitajima et al., 2018; Rosario et al., 2009; Zhang et al., 2005). Our previous results showed that PMMoV is relatively stable across samples, and the data after PMMoV adjustment are much less noisy and match the upward trend of clinical COVID-19 cases (Wu et al., 2020a, Wu et al., 2020b). Briefly, we used qPCR to evaluate the PMMoV concentrations across the sewage samples in our time-series with the PMMoV primers and probe (Table S1, Ref. Zhang et al., 2005). Results showed that PMMoV is detectable and relatively stable between daily samples, with a standard deviation of 1.789 Ct in the 108 tested samples (Fig. S8).
To adjust the SARS-CoV-2 viral concentrations for each sample, we first calculated the deviation of its PMMoV copies from the median of PMMoV copies in all 108 samples: Deviation factor = 10^(k ∗ (sample Ct − median Ct)), where k = −0.2991 is the slope of the standard curve (amplification efficiency is 99.11% for this primer set as previously measured (Wu et al., 2020a, Wu et al., 2020b)). The SARS-CoV-2 viral concentration was then divided by this deviation.
In Method I, we processed 108 wastewater samples, in batches as they were received in the lab, from January 8 to May 5, 2020 (Fig. S1).
2.3. Viral precipitation, RNA extraction and RT-qPCR (Method II)
For Method I, raw wastewater samples were processed as they were received, in chronological groups – providing near-real time information on viral concentrations in sewage but raising the possibility of batch effects because of different reagent lots and personnel changes. Therefore, we processed 60 samples from March 3 to May 20 (including 8 new samples from May) with a second method (Method II) to allow all samples from a single influent to be processed together as a single batch. Before the reprocessing, these samples were stored at 4 °C and the longest storage time was about two and half months (March 4 to May 22). Compared to Method I, this method started with less volume of sewage samples and greatly shortened the experimental time. Fifteen milliliters of filtrate were first concentrated with 10 kDa Amicon Ultra Centrifugal Filter (Sigma, Cat# UFC9010) to 150–200 μl, which is further lysed with 600 μl AVL buffer (Qiagen, Cat# 19073) for RNA extraction (Qiagen RNeasy kit, Cat# 74182). The eluted RNA (3 μl) was immediately used for one-step RT-PCR with TaqMan™ Fast Virus 1-Step Master Mix (Thermofisher, Cat# 4444436) based on the following protocol: 50 °C 10 min for reverse transcription, 95 °C 20 s for RT inactivation and initial denaturation, and 48 cycles of denature (95 °C, 1 s) and anneal/extend (55 °C, 30 s).
Viral gene copies per ml of sewage were converted from the Ct values for N1 or N2 primer sets using the calibration curves (Table S1), and multiplied by the dilution factor (Total volume of RNA sample/Starting volume of filtered wastewater sample). Two technical replicates were performed for each primer set, and mean values were reported. No significant difference was found between the viral gene concentrations in the wastewater estimated from N1 or N2 primer sets (Student's t-test with a p-value 0.407). PMMoV was also quantified in the newly extracted samples, and SARS-CoV-2 RNA concentrations were adjusted by the corresponding PMMoV concentrations in the sample using the method described above. Data analysis was performed with both raw and PMMoV normalized viral concentrations to confirm agreement of observed trends. Viral genome copies per ml of wastewater sample were averaged from the N1 and N2 primer sets.
To assess the robustness of Method I and Method II, we evaluated the recovery rate using murine hepatitis virus (MHV, ATCC® VR-764), which is also an enveloped coronavirus like SARS-CoV-2. SARS-CoV-2 virus was not used due to safety reasons. Briefly, MHV was spiked into the wastewater and then concentrated by PEG8000 and Amicon filter in Method I and Method II, respectively. In parallel, the same amount of virus was directly lysed using Trizol reagent or AVL buffer in Method I and Method II, respectively. Viral RNA extraction, RT, and qPCR were performed by following the steps as described above using MHV-specific primers and probes (Table S1). Results indicate that the recovery rates of viral concentration in Method I and Method II were 58.09 ± 20.21% and 31.42 ± 2.59%, respectively. Using the same protocol, Ahmed et al. obtained recovery rates ranging from 26.7 to 65.7% (Ahmed et al., 2020b).
All data presented was obtained with Method II, unless specified otherwise. The longitudinal data generated with Method I are only presented in Figs. 1B, S1 and S9, showing high correspondence of viral concentrations obtained from the two methods. In Method II, we reprocessed 52 samples from March 3 to May 5, and 8 new samples from May 2 to May 20. In total, 116 samples were processed in this study from January 8 to May 20.
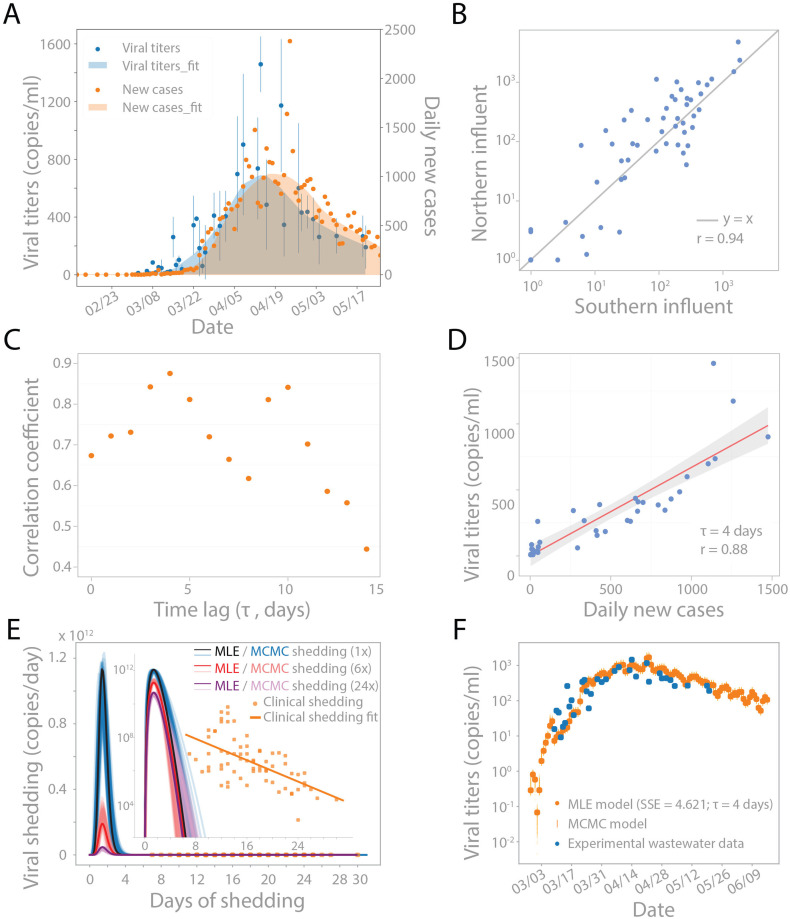
Fig. 1.
SARS-CoV-2 concentrations in wastewater correlate with new clinical cases, with a temporal offset. (A) Viral RNA concentrations in wastewater samples from March 3 to May 20 (blue dots) and new clinical cases from February 12 to May 20 in Norfolk, Suffolk, and Middlesex counties served by the wastewater treatment plant (orange dots). LOWESS smoothing was applied to show the trends for viral concentrations (shaded blue) and daily new cases (shaded orange). Viral concentrations were normalized by PMMoV concentrations in each sample, and the blue dot represents the mean of northern and southern viral concentrations. (B) Unsmoothed viral concentrations in the northern and southern influents are highly correlated and have similar magnitudes for all the data generated by Method I and II combined (see Materials and methods). Blue line represents y = x. Pearson's r = 0.94. (C) Linear correlation between unsmoothed viral concentrations in wastewater and unsmoothed daily new cases with different time lags from 0 to 14 days. Pearson's correlation coefficient is highest with a 4 day time lag. Grey bar highlighted the points with the 1st and 2nd highest correlation coefficients. (D) Viral concentrations correlate with daily new cases with a 4-d time lag. Red solid line is the linear regression fitting. Grey area: 95% confidence interval from standard error of the fitting. Pearson's r = 0.88. (E–F) Modeling wastewater concentrations as a convolution of new cases per day and virus shedding per day. (E) Beta function with optimal shape and scaling parameters (α, β, c) representing the average viral shedding function S(t) on a linear scale and log scale (inset). The shedding function was inferred using 1× reported cases (black), 6× reported cases (red), and 24× reported cases (purple) based on reports that true case numbers could be 6–24× higher than reported cases. Markov Chain Monte Carlo (MCMC) simulation was used to investigate the uncertainty landscape around the MLE shedding function, and 100 random MCMC results are shown in blue, pink, and lavender for 1×, 6×, and 24× reported cases, respectively. Clinically reported values provided by Wolfel et al. are added in orange for reference, with linear regression fit (Wölfel et al., 2020). (F) Viral concentrations based on the convolution model compared to viral concentrations observed in wastewater. Model output is the convolution of new cases per day I(t) and MLE shedding function S(t) from (E). 100 random MCMC simulation results of the shedding function were convolved with I(t) to illustrate the uncertainty around the MLE model results (orange vertical lines). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.4. Contemporaneous societal data collection and correlation analysis
Clinical case data from March 1, 2020 to May 20, 2020 from Norfolk, Suffolk, and Middlesex Counties was downloaded from Mass.gov (“Department of Public Health | Mass.gov,” 2020). We summed the clinical cases from each county to represent the cases in the sewershed of the wastewater treatment plant and calculated the new cases per day. We conducted locally weighted scatterplot smoothing (LOWESS) of wastewater concentrations (smoothing parameter = 0.4) and new clinical cases (smoothing parameter = 0.2) purely to show the qualitative trends in Fig. 1A. No further analysis was done with the smoothed data. LOWESS smoothing was done with statsmodels.nonparametric.smoothers_lowess.lowess in python 3.6.5 and statsmodels 0.9.0.
Pearson's correlation was calculated between unsmoothed wastewater data and unsmoothed clinical data. Higher correlations were seen when comparing wastewater data to daily new clinical cases than to cumulative cases (Figs. 1C and S2). Moreover, cumulative cases are monotonically increasing, and do not reflect the trend of wastewater viral concentrations over the time, and thus we decided to do further analysis with daily new clinical cases. Correlation analysis was done in R (3.5.0). p-Values for the correlations between viral concentrations and clinical cases were not provided because of the potential autocorrelations within the two datasets.
Influenza-like illness data reported to ILINet was downloaded from CDC FluView Portal (“U.S. Outpatient Influenza-like Illness Surveillance Network (ILINet): Overall Percentage of Visits for ILI | CDC,” n.d.). Data on hospitalizations, reported deaths, number of tests administered, and daily positive test rates in Massachusetts were downloaded from Mass.gov (“Department of Public Health | Mass.gov,” 2020). Public transit and cellular mobility data were downloaded from Massachusetts Bay Transportation Authority (Massachusetts Department of Transportation, 2020) and Citymapper (“Citymapper Mobility Index,” 2020), respectively. Supermarket visits data for Massachusetts was downloaded from SafeGraph (SafeGraph, 2020).
2.5. Social media data analysis
For analysis of emotional content, we used the dictionaries from LIWC2015 (Tausczik and Pennebaker, 2010), which are optimized to capture social and psychological states within written text. Our analysis focused specifically on health, anxiety, and death-related texts.
Facebook posts were collected by querying for posts containing the keyword “massachusetts” between January 1, 2020 and May 20, 2020 from the CrowdTangle API (CrowdTangle Team, 2020), resulting in 475,938 posts. 105,127 (22.1%) of the posts contained words within the LIWC categories of interest.
For analysis, the available text fields from the Facebook posts were pre-processed, and psychometrics were calculated using the prevalence of words in the LIWC2015 dictionaries within the corpus of posts for a given day. Counts of words related to anxiety, death, and health were retained for analysis. The prevalence of posts about anxiety, death, and health by day was calculated by summing the counts of word matches and dividing by the number of posts in that day.
2.6. Estimation of viral shedding function
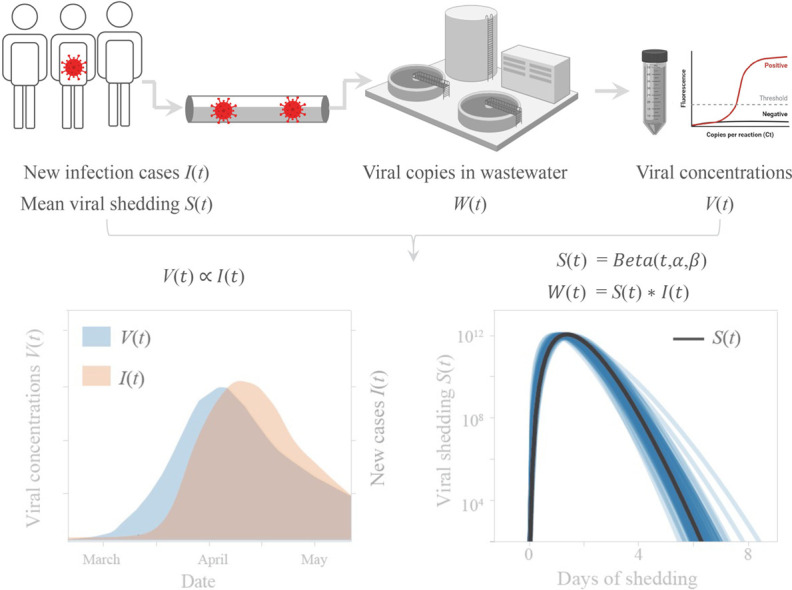
Wastewater data W(t) was modeled as a convolution of new clinical cases I(t) and S(t) describing the average viral shedding detectable in wastewater from infected individuals in the sewershed. We assume I(t) = z ∗ G(t), where G(t) represents government-reported new cases, while z represents the ratio between non-reported and reported cases. We chose z = 1, 6, 24 based on a CDC report that the estimated number of total infections based on serological testing could be 6–24× higher than reported cases (Havers et al., 2020).
We hypothesized that the viral shedding function S(t) could be fit by a beta distribution with parameters α, β, and scaling factor c. Beta and gamma distributions harbor a rich variety of shapes with only two parameters, and are widely employed to model viral shedding or viral load dynamics from clinical data (Ferretti et al., 2020; He et al., 2020; Huynh and Rong, 2012; Milbrath et al., 2013). Since the beta distribution is defined on the range [0, 1], we sampled the probability density function for points 1/30 apart as a proxy for viral shedding over 30 days. Time lag between wastewater data and clinical counts was denoted as parameter τ. Based on the results of the correlation analysis, we back-dated the clinical cases by τ = 4 days to get the new cases per day function I(t). The full model is:
| (1) |
| (2) |
| (3) |
The discrete convolution formula is defined as:
and beta probably density function is defined as: ,
where Γ is the gamma function .
We defined the score function as the sum of squared errors (SSE) between log10(observed copies/ml in wastewater) and log10((S ∗ I)[t]). We used the L-BFGS method in the scipy.optimize.minimize function to find parameters α, β, c of the beta distribution that minimized the SSE, which is equivalent to conducting maximum likelihood estimation under the assumption of normally distributed homoscedastic noise. For initial parameter guesses, we used a combination of α = [2, 20, 50, 100, 200], β = [2, 20, 50, 100, 200], and c = [0.01, 0.1], which gives a wide variety of starting shapes for the shedding function. Optimal S(t) was multiplied by an estimated wastewater volume of 1.36e6 m3 to obtain the total copies shed per day. This total viral shedding per day per individual does not consider person-to-person variation and can be regarded as the average shedding in the sewershed.
To investigate the relationship between the shape of the shedding function and the clinical time lag τ, we conducted the optimization for τ ranging from 0 to 10 days and reported the optimal shedding function for each τ. This analysis was performed for northern and southern influent data separately, as well as the average wastewater data. We also modeled the shedding function as a gamma distribution over [0, 30] and had similar results as with the beta distribution. All shedding estimation work was done with python 3.6.5, numpy 1.14.3, pandas 0.23.0, and scipy 1.1.0.
2.7. Markov Chain Monte Carlo (MCMC) simulation to quantify uncertainty in shedding model
MCMC simulation was performed to investigate the uncertainty landscape around the maximum likelihood estimation of the parameters for the viral shedding function. Briefly, we started at the maximum likelihood estimate for each parameter α, β, and c. We defined the transition function as a normal distribution centered around the previous parameters, with standard deviation (1, 1, 0.001) for α, β, and c, respectively. At each iteration, we selected a new set of parameters using the transition function and computed the log likelihood. New parameters were accepted if the log likelihood was higher. If the log likelihood was lower, we accepted the parameters with probability exp(−delta(SSE)/T), where we used T = 5 to tune the acceptance ratio. In 10,000 iterations, 1318 samples were accepted. We selected 100 random selected parameter sets and plot them in Fig. 1E and F to illustrate the uncertainty around the maximum likelihood estimate of the shedding function. MCMC simulation was done with python 3.6.5, numpy 1.14.3, pandas 0.23.0, and scipy 1.1.0.
3. Results and discussion
We first quantified SARS-CoV-2 RNA concentrations in 108 longitudinal wastewater samples from the wastewater treatment facility's northern and southern influents, from January 8 to May 5, 2020. Samples from March 3 to May 20 including eight new samples in May were processed a second time in late May, in a single batch for each sewershed, to complement initial quantification, which had been performed in chronological groups as samples were received (see Materials and methods).
3.1. Longitudinal wastewater sampling captures the emergence and spread of SARS-CoV-2 in the population
Viral concentrations in wastewater followed a trend similar to new clinical cases over the sampled time period (Fig. 1A). SARS-CoV-2 was not detected in either influent stream in January or February samples, and was first detected on March 3 (northern influent) and March 10 (southern influent), at a concentration of ≤15 copies per ml of wastewater (Figs. 1A and S1). Only two clinically confirmed COVID-19 cases had been reported in Massachusetts as of March 3, indicating viral detection in wastewater in the early stage of the local outbreak. SARS-CoV-2 concentrations remained relatively low (<90 copies per ml) in both influents until March 15, after which they increased exponentially. Viral concentrations appeared to rise in wastewater in advance of clinical cases. SARS-CoV-2 levels in wastewater began trending downward about a month later (April 13), while the peak and decline in new clinical cases occurred later, on April 24. Together, these qualitative trends suggested that wastewater viral concentrations during this time period might reflect disease incidence in advance of clinical reporting.
SARS-CoV-2 was consistently detected at comparable levels, and demonstrated similar dynamics, in the northern and southern influents (Fig. 1B).
3.2. Wastewater data correlates best with clinically confirmed new cases, with a temporal offset
Since SARS-CoV-2 was first detected in wastewater when only two cases were clinically confirmed, we hypothesized that the wastewater data included a significant undiagnosed, COVID-19 infected population. This difference between wastewater and clinical data could be due to underdiagnosis of asymptomatic or mildly symptomatic cases, limitations in clinical testing capacity, or a time delay between viral shedding and the onset of respiratory and other symptoms.
We tested the correlation of viral concentrations with new clinical cases and cumulative cases, allowing for a variable time lag (Figs. 1C/D and S2). The number of cumulative cases was considered because of reports that SARS-CoV-2 can be shed in feces for more than 20 days, in which case the long tail of shedding may contribute significant signal to wastewater (Wölfel et al., 2020; Wu et al., 2020b; Zheng et al., 2020). Higher correlations were seen when comparing wastewater data to daily new clinical cases than to cumulative cases (Figs. 1C and S2). Strong correlations (Pearson's r > 0.8) were observed when comparing new clinical cases back-dated by 3–5 and 9–10 days, and the maximum agreement between the two time series was observed for a time offset of 4 days (Pearson's r = 0.88, Fig. 1C/D). Similar results were found when each sewershed was considered individually (Fig. S3). This time lag between the wastewater signal and clinically reported cases is consistent with the typical 4–5 day incubation period from SARS-CoV-2 infection to symptom onset (Guan et al., 2020; Lauer et al., 2020; Li et al., 2020). Thus, wastewater surveillance could potentially be used to predict trends in new COVID-19 cases.
3.3. An inferred viral shedding function suggests an early burst of high viral shedding
The high correlation between wastewater viral concentrations and daily reports of new clinically confirmed COVID-19 cases (Fig. 1D) – combined with the time lag between the wastewater signal and clinical data – suggests that newly infected individuals contribute significant viral loads to the wastewater and that most of this shedding may occur early in infection, prior to the individual seeking healthcare and being tested. As a result, viral dynamics in wastewater could provide insight into early shedding, which would be challenging to capture clinically if it precedes patient presentation to the clinic.
We attempted to infer average population-level viral shedding dynamics observed in wastewater by comparing our longitudinal wastewater data with estimates of daily new cases. Wastewater viral RNA concentrations W(t) was modeled as a convolution of new clinical cases I(t) and a function S(t) describing the average viral shedding (whether from fecal, urine, or other unknown sources) detectable in wastewater from infected individuals in the sewershed (see Materials and methods).
Modeling revealed a short burst of viral shedding that peaked around day 2 of shedding and lasted for 3–4 days (Fig. 1E and F). This feature of the modeling was robust to different time lags (0–10 days) applied to data of daily new clinical cases, as well as the distribution used to fit the shedding function (both beta and gamma distributions were investigated, Fig. S4). This short burst of shedding was also seen after modeling each sewershed individually (Figs. S5–S6). Inferring the viral shedding function using values for z (estimated true number of infected individuals) reflecting 6× or 24× the reported number of new cases (Havers et al., 2020) resulted in a similar narrow-peaked viral shedding function peaking around day 2, but with a lower average magnitude per individual (Fig. 1E). The lower magnitude of viral shedding is expected because W(t) is defined as a convolution of I(t) and S(t); when I(t) increases, S(t) will decrease for a fixed W(t).
A limitation to this modeling method is that the trends inferred from Boston data early in the pandemic may be confounded by limited testing capacity. To address this limitation and validate our findings, we have repeated our analysis with wastewater and clinical new case data, collected over an additional 9 month period, from August 15, 2020 to May 13, 2021 (Massachusetts Water Resources Authority), after on-demand testing became readily available in Massachusetts (“Baker-Polito Administration Announces New Initiatives to Stop Spread of COVID-19 | Mass.gov,” n.d.). Using this second data set, we found a similar narrow-peaked viral shedding function during the early phase of infection (Fig. S7), replicating our initial findings. These independent models of the two waves of the COVID-19 pandemic showed similar results, improving our confidence that Boston municipal wastewater data reflects an early period of viral shedding, in alignment with clinical reports. As additional groups test this methodology it will be useful to compare results to understand the factors underpinning trends in different types of wastewater.
The early timing of the inferred shedding is consistent with reports that viral load and infectiousness may peak before symptom onset (Benefield et al., 2020; He et al., 2020; Wei, 2020), and that fecal and respiratory shedding may occur 3–5 days before symptom onset (Jones et al., 2020; Wang et al., 2020a, p. 138). Although the early peak of viral shedding in our model does not directly reflect patients' reported clinical courses, it is consistent with reports of abdominal pain, nausea, and diarrhea preceding onset of respiratory symptoms in COVID-19 patients (Cholankeril et al., 2020; Dane et al., 2020; Siegel et al., 2020), suggesting that individuals may shed SARS-CoV-2 virus early in this process. In several clinical studies, the earliest stool samples collected have been positive for viral RNA (day 3 post symptom onset (Wölfel et al., 2020); and day 0 of hospital admission (Chen et al., 2020, p. 2)), suggesting fecal shedding may start before individuals seek medical care. Our finding about the early peak shedding is also supported by observations in a transgenic mouse model with humanized ACE2 receptors, where viral concentrations in the intestines peaked at 1 day post infection (Bao et al., 2020). Moreover, Benefield et al. performed a pooled analysis of 66 clinical studies (1198 patients across 14 counties) reporting temporal viral load and shedding data, and found that SARS-CoV-2 viral load peaks prior to – not at – symptom onset (Benefield et al., 2020). Finally, a recent study modeled SARS-CoV-2 fecal shedding and found that hospitalized patients represent the tail end of shedding and their shedding levels cannot explain the high RNA concentrations observed in wastewater (Hoffmann and Alsing, 2021). This modeling result is consistent with ours and suggests the presence of more abundant shedding early in infection.
The inferred peak shedding was generally several orders of magnitude greater than typical values (104–109 copies per 200 g stool), which are measured after symptom onset and clinical testing (Zheng et al., 2020; Wölfel et al., 2020; Jones et al., 2020) (Fig. 1E). In a more recent study, Han et al. measured the viral RNA load in infected individuals in South Korea and found the median fecal RNA load was 107.68 (ranging from 104.1–1010.27) copies/ml (Han et al., 2020). Assuming one 200 g stool per day per individual, our peak shedding value is very similar to this report, illustrating the variability in the clinical literature.
We note that inadequate testing capacity, changing criteria for testing, and populations who do not seek medical care introduce uncertainty to the reported new cases. It should be noted, however, that a good model fit could be obtained even if a large fraction of cases is underreported. For example, if only half of cases are reported, then the total amount of shedding per individual would be inferred to be twice the actual value, even though the fit to the model would be equally good. Attempting to correct for these factors would decrease the magnitude of average viral shedding per person. On the other hand, factors such as viral RNA degradation in sewage lines and storage and recovery efficiency introduce uncertainty in the observed wastewater concentrations and attempting to correct for these factors would increase the magnitude of average viral shedding per person. Because these factors introduce sizable uncertainty to the true magnitude of shedding, we do not seek to challenge the absolute shedding number reported in the literature, but rather emphasize the timing- that wastewater signals may reflect shedding dynamics early in infection.
Together, our data suggest that SARS-CoV-2 levels in wastewater may be largely driven by an early burst of shedding, followed by a period of prolonged low-level shedding as reported in the literature (Chen et al., 2020; Hoffmann and Alsing, 2021; Wölfel et al., 2020; Zheng et al., 2020).
3.4. Wastewater SARS-CoV-2 RNA concentrations in the context of behavior and interventions
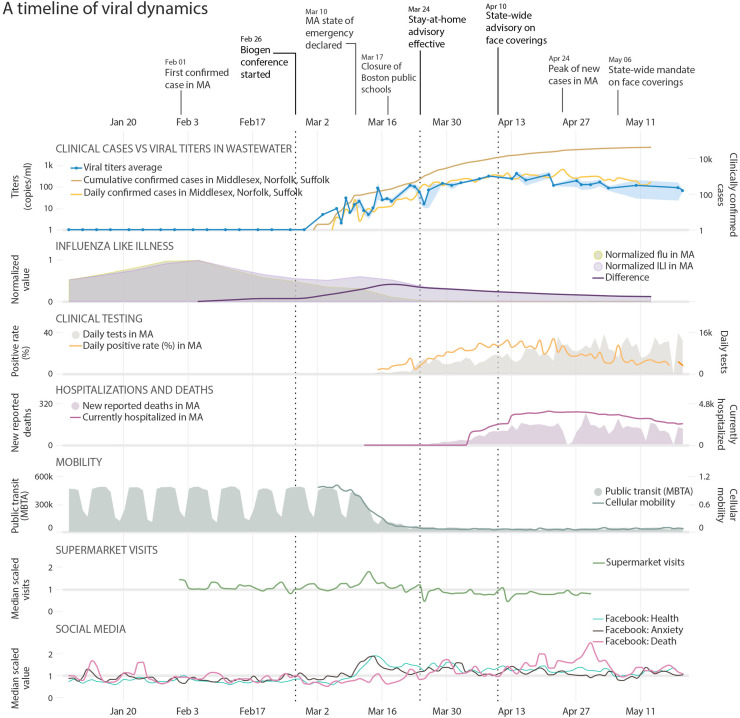
SARS-CoV-2 levels in wastewater began to increase exponentially after March 15, coinciding with a peak in reports to the US Centers for Disease Control and Prevention of “influenza-like illnesses” (ILI) in Massachusetts that were not caused by influenza (Fig. 2 , panel 3) – and which showed early dynamics similar to SARS-CoV-2 levels in wastewater, suggesting these ILIs may have been undiagnosed COVID-19 (Centers for Disease Control and Prevention, 2020; Lu et al., 2021; “U.S. Outpatient Influenza-like Illness Surveillance Network (ILINet): Overall Percentage of Visits for ILI | CDC,” n.d.). Wastewater concentrations of SARS-CoV-2 began to drop in mid-April (Fig. 1A), roughly one month after the state of emergency was declared (March 10, Fig. 2) and the statewide school closure (March 17; Fig. 2), and approximately three weeks after the Massachusetts stay-at-home advisory went into effect (March 24, Fig. 2) (“Information on the Outbreak of Coronavirus Disease 2019 (COVID-19) | Mass.gov,” n.d.). Public transport and cellular mobility data indicate that public movement began to decrease significantly ahead of the stay-at-home advisory, starting soon after the state of emergency was declared and the first school closures (“Citymapper Mobility Index,” 2020; Massachusetts Department of Transportation, 2020). The time between infection and forward transmission is approximately 4–6 days (Ferretti et al., 2020; He et al., 2020), suggesting that there were likely 4–5 additional cycles of infection after the implementation of the stay-at-home advisory before viral concentrations began to decline in wastewater.
Fig. 2.
A timeline of viral dynamics in the context of key events and clinical/behavioral data. Trends are plotted in the same time frame, from January 8 to May 20. Row 1: Timeline of COVID-19 pandemic and important events in MA. Row 2: Clinical Cases vs Viral concentrations in Wastewater: Viral concentrations in wastewater (blue line along the primary y-axis, shaded area represents the minimum/maximum of PMMoV-adjusted viral concentrations), daily (orange line) and cumulative (brown line) confirmed cases along the secondary y-axis. Influenza Like Illness: Visits for influenza-like illness (ILI, purple shading) and confirmed flu cases (light green shading), and the difference between the two after normalization (purple line), which shows a peak of non-flu ILI at March 18. Clinical Testing: Daily SARS-CoV-2 tests and positive rates in MA. Hospitalizations and Deaths: New reported COVID-19 related deaths and hospitalizations in MA. Mobility: Public transit and cellular mobility data. Supermarket Visits: Supermarket visits in MA (normalized by the median value). Social Media: Facebook posts with terms expressing “Health”, “Anxiety”, and “Death”. Dashed lines in all the panels represent the date of the Biogen conference, the stay-at-home advisory in MA, and the state-wide face covering advisory. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
What could explain the approximately month-long lag between decreased public mobility and the drop in population-wide viral levels as measured in wastewater? Several factors likely contributed. First, despite efforts to limit human-to-human contact, disease transmission could have remained high among especially at-risk populations such as essential workers and their families. Second, individuals infected before the stay-at-home order could have infected multiple family members or roommates in subsequent days, especially in areas with high numbers of inhabitants per household. Finally, despite the stay-at-home order, human-to-human interactions may still have occurred at a significant rate outside the household, in essential venues such as supermarkets and pharmacies. These early interactions may have been more likely to result in disease transmission because the Massachusetts advisory recommending face coverings in public was not announced until April 10, and the state order requiring face coverings did not go into effect until May 6 (“Department of Public Health | Mass.gov,” 2020). Social media indices of public anxiety showed a pronounced rise in anxiety around the time of the declaration of the state of emergency (CrowdTangle Team, 2020), followed shortly thereafter by a peak in Massachusetts supermarket traffic on March 13 (Fig. 2, panel 7–8; Ref. SafeGraph, 2020). This concentration of people into supermarkets – without masks, and before the institution of supermarket policies to limit the number of shoppers – might have contributed to the sharp rise in viral concentrations in wastewater (Fig. 1A) that began in the following days.
4. Uncertainties and future directions of wastewater surveillance
Although wastewater data should be relatively unbiased, relatively high day-to-day variation in our results suggests that they are noisy. On the other hand, clinical data may be less noisy but more prone to systematic bias due to constraints such as limited clinical testing capacity. Together, these independent data streams from clinical- and wastewater-based surveillance can provide a more complete picture of viral dynamics, and a new opportunity to explore the relationship between COVID-19 dynamics and concomitant public health interventions and behavioral changes. In this study, we infer a viral shedding function that can help to connect these two data streams in a quantitative fashion to allow for more accurate epidemiological modeling – and ultimately, more informed decision-making about how and when to intervene.
Several knowledge gaps introduce uncertainty to our modeling approaches. First, viral concentrations in wastewater are highly dependent on individual shedding rates, viral stability in wastewater, and flow rates of the influent. While we have noted that normalization with the human fecal indicator PMMoV helps reduce noise due to sampling time and flow, significant uncertainties remain regarding the consistency of viral shedding among COVID-19 patients (Chen et al., 2020; Zheng et al., 2020). Second, the viral shedding profile inferred by our modeling approach relies on reported clinical new cases over the course of the pandemic, and is thus subject to the limitations of clinical testing criteria and capacity (Fig. 2). This early lack of testing is especially evident given that SARS-CoV-2 was detectable in wastewater when there were only 2 reported cases in the sewershed, which is certainly underreported and would decrease the robustness of the model (Fig. 1A). However, our finding of a narrow-peaked viral shedding function was confirmed when repeating the modeling with data after August 15, 2020, when clinical testing capacity was well established in Massachusetts, lending more confidence to this result. Third, the exact percentage of asymptomatic infections in the sewershed remains unknown. We here incorporated z to reflect an estimate of the true number of infections into our model and analyzed when z equals 6 or 24× the clinically reported cases based on serological reports (Havers et al., 2020). We recognize that z may be a function of disease spread and clinical testing capacity.
Fourth, the wastewater viral concentrations reported were not corrected for viral RNA degradation in sewage lines and during sample storage at 4 °C or losses during experimental procedures. In this study, some samples were stored for up to 2.5 months at 4 °C before being reprocessed with Method II. We compared the SARS-CoV-2 RNA concentrations measured by Method I and Method II. As shown in Fig. S9A, the general magnitude and trends of SARS-CoV-2 concentrations were similar between the two methods, and samples detected positive in Method I were also positive in Method II. These results indicated that SARS-CoV-2 is not highly degraded during storage, which is consistent with previous work reporting that SARS-CoV-2 copy numbers were stable at 4 °C for one to three months (Hokajärvi et al., 2021). We also compared PMMoV concentrations in January, February, and March wastewater samples, which were stored at 4 °C for 1 to 97 d before processed in April. No significant difference was found between those samples by month (Fig. S9B). Together, these results suggested that SARS-CoV-2 and PMMoV were not highly degraded after storage at 4 °C for up to three months. However, further experiments using live SARS-CoV-2 viral stocks of known concentrations to quantify the degradation rate and recovery efficiency from wastewater samples would improve our model inference.
This work demonstrates the power of longitudinal wastewater surveillance in tracking the emergence and spread of an infectious disease in a population, in advance of clinical case reporting – as well as its potential to shed light on infection characteristics that may be challenging to capture clinically. It also highlights a key question for future investigation. To what degree can our collective insights from SARS-CoV-2 wastewater surveillance be applied to other infectious diseases? Whether or not they generalize will depend on how excreted pathogen (or other infection biomarkers) signals relate to the course of each individual disease. This relationship may affect the sensitivity and accuracy of wastewater surveillance efforts, as well as determine whether wastewater signals correlate best with new cases or cumulative cases, if at all. Finally, this work highlights the importance of addressing batch effects in wastewater-based disease monitoring, where rapid availability of data is critical to enable real-time public health responses to disease dynamics. As we collectively explore the limitations and future directions of wastewater surveillance, its application in communities at this time can provide important guidelines for responding to the current or future public health crisis.
5. Conclusions
In this study, we used longitudinal wastewater surveillance to track the appearance and spread of SARS-CoV-2 in the Greater Boston area during the early phase of the pandemic, and demonstrated the application of temporal wastewater data in inferring viral shedding dynamics. Our results showed:
-
•
SARS-CoV-2 was first detected in Massachusetts wastewater on March 3, 2020, when there were only two clinically confirmed cases in the state.
-
•
Viral concentrations in wastewater correlate with newly diagnosed COVID-19 cases, with trends in wastewater occurring 4–10 days earlier than in clinical data.
-
•
The viral shedding function inferred from wastewater data shows an early burst of high viral shedding for infected individuals, suggesting that wastewater data may be more specific to newly infected cases.
Data availability
The data presented in the figures, and the code for the analysis, and data that support the other findings of this study are available from the corresponding author on reasonable request. The code used for the analysis will be released as soon as the paper is published.
CRediT authorship contribution statement
Fuqing Wu: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft, Visualization. Amy Xiao: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft, Visualization. Jianbo Zhang: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft. Katya Moniz: Conceptualization, Writing – original draft, Project administration. Noriko Endo: Investigation, Writing – review & editing. Federica Armas: Validation, Writing – review & editing. Richard Bonneau: Resources, Writing – review & editing. Megan A. Brown: Writing – review & editing. Mary Bushman: Writing – review & editing. Peter R. Chai: Writing – review & editing. Claire Duvallet: Writing – review & editing. Timothy B. Erickson: Writing – review & editing. Katelyn Foppe: Investigation, Writing – review & editing. Newsha Ghaeli: Writing – review & editing. Xiaoqiong Gu: Methodology, Validation, Writing – review & editing. William P. Hanage: Writing – review & editing, Supervision. Katherine H. Huang: Visualization, Writing – review & editing. Wei Lin Lee: Validation, Writing – review & editing. Mariana Matus: Funding acquisition, Resources. Kyle A. McElroy: Investigation, Writing – review & editing. Jonathan Nagler: Resources. Steven F. Rhode: Resources, Writing – review & editing. Mauricio Santillana: Resources, Writing – review & editing. Joshua A. Tucker: Resources. Stefan Wuertz: Writing – review & editing. Shijie Zhao: Writing – review & editing. Janelle Thompson: Conceptualization, Funding acquisition, Writing – review & editing, Supervision. Eric J. Alm: Conceptualization, Methodology, Funding acquisition, Formal analysis, Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
MM and NG are cofounders of Biobot Analytics. EJA is advisor to Biobot Analytics. CD, KAM, KF, and NE are employees at Biobot Analytics, and all these authors hold shares in the company. PRC and TBE have a financial interest in Biobot Analytics, a company engaged in the collection and analysis of wastewater to develop epidemiological data. PRC and TBE's interests were reviewed and are managed by Brigham and Women's Hospital and Mass General Brigham in accordance with their conflict-of-interest policies.
Acknowledgment
We thank the management and sampling team at the Massachusetts wastewater treatment facility who worked with us in providing the samples for analysis, in particular Conor Donovan, Jim Fitzgerald, Louis Logan, Nicole Mangano, Keith Stocks, Sean Winter, and David Wu. We thank Lisa Wong (MWRA) for providing flow data, and Stephen Estes-Smargiassi (MWRA) and Betsy Reilley (MWRA) for helpful discussion. We thank Penny Chisholm (MIT) and Allison Coe (MIT) for access to equipment and other supplies, Mathilde Poyet (MIT) and Shandrina Burns (MIT) for logistical support, Andrew Tang (Broad Institute) for assistance with Fig. 2, Nicholas Santos-Powell for thoughtful comments on the manuscript, Kyle Bibby (University of Notre Dame) for suggestions on PMMoV, and Karina Gin (National University of Singapore) and Lee Ching Ng (National Environmental Agency, Singapore) for helpful discussion and for sharing the Amicon protocol. We also thank Dr. Victor M. Corman (Charité Universitätsmedizin, Germany) for sharing data of viral concentrations in patients' stool samples, and the SafeGraph team for providing free access to their data for COVID-19-related research. Finally, we express our deep gratitude to all healthcare professionals and first-line responders who have been caring for patients with COVID-19.
This work was supported by the Center for Microbiome Informatics and Therapeutics and Intra-CREATE Thematic Grant (Cities) grant NRF2019-THE001-0003a to JT and EJA; National Institute on Drug Abuse of the National Institutes of Health award numbers K23DA044874; and R44DA051106 to MM and PRC, Hans and Mavis Psychosocial Foundation funding, and e-ink corporation funding to PRC; funding from the Morris-Singer Foundation and NIH award R01AI106786 to WPH; funds from the Massachusetts Consortium on Pathogen Readiness and China Evergrande Group to TBE, PRC, MM, and EJA; funding from the Ministry of Education - Singapore and National Research Foundation through an RCE award to Singapore Centre for Environmental Life Sciences Engineering (SCELSE) to SW and JT; and a National Institute of General Medical Sciences of the National Institutes of Health award, number R01GM130668, to MS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding institutions.
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.150121.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerswald H., Yann S., Dul S., In S., Dussart P., Martin N.J., Karlsson E.A., Garcia-Rivera J.A. Assessment of inactivation procedures for SARS-CoV-2. J. Gen. Virol. 2021;102 doi: 10.1099/jgv.0.001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Polito Administration Announces New Initiatives to Stop Spread of COVID-19 | Mass.gov [WWW Document], n.d. URL https://www.mass.gov/news/baker-polito-administration-announces-new-initiatives-to-stop-spread-of-covid-19 (accessed 6.9.21).
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., Qu Y., Li F., Lv Q., Wang W., Xue J., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao Linna, Liu P., Zhao Li, Ye F., Wang H., Zhou W., Zhu N., Zhen W., Yu H., Zhang X., Guo L., Chen L., Wang C., Wang Y., Wang X., Xiao Y., Sun Q., Liu H., Zhu F., Ma C., Yan L., Yang M., Han J., Xu W., Tan W., Peng X., Jin Q., Wu G., Qin C. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Bar-Or I., Weil M., Indenbaum V., Bucris E., Bar-Ilan D., Elul M., Levi N., Aguvaev I., Cohen Z., Shirazi R., Erster O., Sela-Brown A., Sofer D., Mor O., Mendelson E., Zuckerman N.S. Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci. Total Environ. 2021;789 doi: 10.1016/j.scitotenv.2021.148002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benefield A.E., Skrip L.A., Clement A., Althouse R.A., Chang S., Althouse B.M. SARS-CoV-2 viral load peaks prior to symptom onset: a systematic review and individual-pooled analysis of coronavirus viral load from 66 studies. medRxiv. 2020 doi: 10.1101/2020.09.28.20202028. [DOI] [Google Scholar]
- CDC Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes [WWW Document]. Centers for Disease Control and Prevention. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html (accessed 6.22.20)
- Centers for Disease Control and Prevention FluView: National, Regional, and State Level Outpatient Illness and Viral Surveillance [WWW Document]. Centers for Disease Control and Prevention. 2020. https://gis.cdc.gov/grasp/fluview/fluportaldashboard.html (accessed 9.16.20)
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C., Yang J., Ye G., Cheng Z. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Cholankeril G., Podboy A., Aivaliotis V.I., Tarlow B., Pham E.A., Spencer S.P., Kim D., Hsing A., Ahmed A. High prevalence of concurrent gastrointestinal manifestations in patients with severe acute respiratory syndrome coronavirus 2: early experience from California. Gastroenterology. 2020;159:775–777. doi: 10.1053/j.gastro.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citymapper Mobility Index [WWW Document] 2020. https://citymapper.com/cmi (accessed 9.16.20)
- CrowdTangle Team . CrowdTangle; 2020. Facebook, Menlo Park, California, United States. [Google Scholar]
- Dane B., Brusca-Augello G., Kim D., Katz D.S. Unexpected findings of coronavirus disease (COVID-19) at the lung bases on abdominopelvic CT. Am. J. Roentgenol. 2020;215:603–606. doi: 10.2214/AJR.20.23240. [DOI] [PubMed] [Google Scholar]
- Daughton C.G. Monitoring wastewater for assessing community health: sewage chemical-information mining (SCIM) Sci. Total Environ. 2018;619–620:748–764. doi: 10.1016/j.scitotenv.2017.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Department of Public Health Mass.gov [WWW Document] 2020. https://www.mass.gov/orgs/department-of-public-health (accessed 9.16.20)
- Ferretti L., Wymant C., Kendall M., Zhao L., Nurtay A., Abeler-Dörner L., Parker M., Bonsall D., Fraser C. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368 doi: 10.1126/science.abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuschi C., Pu H., Negri M., Colwell R., Chen J. Wastewater-based epidemiology for managing the COVID-19 pandemic. ACS EST Water. 2021;1:1352–1362. doi: 10.1021/acsestwater.1c00050. [DOI] [PubMed] [Google Scholar]
- Gandhi M., Yokoe D.S., Havlir D.V. Asymptomatic transmission, the Achilles’ heel of current strategies to control Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMe2009758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Yu., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang Jin-lin, Liang Z., Peng Y., Wei L., Liu Y., Hu Ya.-hua., Peng P., Wang Jian-ming, Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza H., Rizk N.M., Gad M.A., Hamza I.A. Pepper mild mottle virus in wastewater in Egypt: a potential indicator of wastewater pollution and the efficiency of the treatment process. Arch. Virol. 2019;164:2707–2713. doi: 10.1007/s00705-019-04383-x. [DOI] [PubMed] [Google Scholar]
- Han M.S., Seong M.-W., Kim N., Shin S., Cho S.I., Park H., Kim T.S., Park S.S., Choi E.H. Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19, Seoul, South Korea. Emerg. Infect. Dis. 2020;26:2497–2499. doi: 10.3201/eid2610.202449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havers F.P., Reed C., Lim T., Montgomery J.M., Klena J.D., Hall A.J., Fry A.M., Cannon D.L., Chiang C.-F., Gibbons A., Krapiunaya I., Morales-Betoulle M., Roguski K., Rasheed M.A.U., Freeman B., Lester S., Mills L., Carroll D.S., Owen S.M., Johnson J.A., Semenova V., Blackmore C., Blog D., Chai S.J., Dunn A., Hand J., Jain S., Lindquist S., Lynfield R., Pritchard S., Sokol T., Sosa L., Turabelidze G., Watkins S.M., Wiesman J., Williams R.W., Yendell S., Schiffer J., Thornburg N.J. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, march 23-may 12, 2020. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.4130. [DOI] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hoffmann T., Alsing J. Faecal shedding models for SARS-CoV-2 RNA amongst hospitalised patients and implications for wastewater-based epidemiology. medRxiv. 2021 doi: 10.1101/2021.03.16.21253603. [DOI] [Google Scholar]
- Hokajärvi A.-M., Rytkönen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.-M., Kankaanpää A., Gunnar T., Al-Hello H., Blomqvist S., Miettinen I.T., Savolainen-Kopra C., Pitkänen T. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh G.T., Rong L. Modeling the dynamics of virus shedding into the saliva of Epstein-Barr virus positive individuals. J. Theor. Biol. 2012;310:105–114. doi: 10.1016/j.jtbi.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Information on the Outbreak of Coronavirus Disease 2019 (COVID-19) | Mass.gov [WWW Document], n.d. URL https://www.mass.gov/resource/information-on-the-outbreak-of-coronavirus-disease-2019-covid-19 (accessed 9.16.20).
- Johns Hopkins University Center for Systems Science and Engineering COVID-19 map - Johns Hopkins coronavirus resource center [WWW Document] 2020. https://coronavirus.jhu.edu/map.html (accessed 9.11.20)
- Jones D.L., Baluja M.Q., Graham D.W., Corbishley A., McDonald J.E., Malham S.K., Hillary L.S., Connor T.R., Gaze W.H., Moura I.B., Wilcox M.H., Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaashoek J., Santillana M. Social Science Research Network; Rochester, NY: 2020. COVID-19 Positive Cases, Evidence on the Time Evolution of the Epidemic or an Indicator of Local Testing Capabilities? A Case Study in the United States (SSRN Scholarly Paper No. ID 3574849) [DOI] [Google Scholar]
- Kitajima M., Sassi H.P., Torrey J.R. Pepper mild mottle virus as a water quality indicator. npj Clean Water. 2018;1:1–9. doi: 10.1038/s41545-018-0019-5. [DOI] [Google Scholar]
- Kocamemi B.A., Kurt H., Hacioglu S., Yarali C., Saatci A.M., Pakdemirli B. First data-set on SARS-CoV-2 detection for Istanbul Wastewaters in Turkey. medRxiv. 2020 doi: 10.1101/2020.05.03.20089417. [DOI] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020 doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de R. Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020 doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F.S., Nguyen A.T., Link N.B., Molina M., Davis J.T., Chinazzi M., Xiong X., Vespignani A., Lipsitch M., Santillana M. Estimating the cumulative incidence of COVID-19 in the United States using influenza surveillance, virologic testing, and mortality data: four complementary approaches. PLoS Comput. Biol. 2021;17 doi: 10.1371/journal.pcbi.1008994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020;54:3733–3735. doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- Massachusetts Department of Transportation MBTA Gated Station validations by line 2018–20 [WWW Document] 2020. https://massdot.app.box.com/s/xockmt85gk2o997dcowkp24bozbslgaz (accessed 9.16.20)
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Milbrath M.O., Spicknall I.H., Zelner J.L., Moe C.L., Eisenberg J.N.S. Heterogeneity in norovirus shedding duration affects community risk. Epidemiol. Infect. 2013;141:1572–1584. doi: 10.1017/S0950268813000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orive G., Lertxundi U., Barcelo D. Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Sci. Total Environ. 2020;732 doi: 10.1016/j.scitotenv.2020.139298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino B., Touret F., Gilles M., de Lamballerie X., Charrel R.N. Heat inactivation of different types of SARS-CoV-2 samples: what protocols for biosafety, molecular detection and serological diagnostics? Viruses. 2020;12 doi: 10.3390/v12070735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K., Symonds E.M., Sinigalliano C., Stewart J., Breitbart M. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol. 2009;75:7261–7267. doi: 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SafeGraph The Impact of Coronavirus (COVID-19) on Foot Traffic. [WWW Document]. SafeGraph. 2020. https://safegraph.com/dashboard/ (accessed 9.16.20)
- Shirasaki N., Matsushita T., Matsui Y., Koriki S. Suitability of pepper mild mottle virus as a human enteric virus surrogate for assessing the efficacy of thermal or free-chlorine disinfection processes by using infectivity assays and enhanced viability PCR. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116409. [DOI] [PubMed] [Google Scholar]
- Siegel A., Chang P.J., Jarou Z.J., Paushter D.M., Harmath C.B., Arevalo J.B., Dachman A. Lung base findings of coronavirus disease (COVID-19) on abdominal CT in patients with predominant gastrointestinal symptoms. Am. J. Roentgenol. 2020;215:607–609. doi: 10.2214/AJR.20.23232. [DOI] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tausczik Y.R., Pennebaker J.W. The psychological meaning of words: LIWC and computerized text analysis methods. J. Lang. Soc. Psychol. 2010;29:24–54. doi: 10.1177/0261927X09351676. [DOI] [Google Scholar]
- U.S. Outpatient Influenza-like Illness Surveillance Network (ILINet): Overall Percentage of Visits for ILI | CDC [WWW Document], n.d. URL https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/04102020/ilinet-regions.html (accessed 9.11.20).
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Lien C., Liu S., Selveraj P. Effective heat inactivation of SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.04.29.20085498. 2020.04.29.20085498. [DOI] [Google Scholar]
- Wei W.E. Presymptomatic transmission of SARS-CoV-2 — Singapore, January 23–March 16, 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69 doi: 10.15585/mmwr.mm6914e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;1–10 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Chai P., Thompson J., Alm E. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. The Lancet Gastroenterology & Hepatology. 2020 doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., McElroy K.A., Rhode S.F., Matus M., Wuertz S., Thompson J., Alm E.J. Wastewater surveillance of SARS-CoV-2 across 40 U.S. states from february to june 2020. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Eurosurveillance. 2020;25:2000776. doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W.H., Run J.-Q., Wei C.L., Soh S.W.L., Hibberd M.L., Liu E.T., Rohwer F., Ruan Y. RNA viral Community in Human Feces: prevalence of plant pathogenic viruses. PLoS Biol. 2005;4 doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., Chen W., Wang Q., Zhang D., Liu Y., Gong R., Ma Z., Lu S., Xiao Y., Gu Y., Zhang J., Yao H., Xu K., Lu X., Wei G., Zhou J., Fang Q., Cai H., Qiu Y., Sheng J., Chen Y., Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The data presented in the figures, and the code for the analysis, and data that support the other findings of this study are available from the corresponding author on reasonable request. The code used for the analysis will be released as soon as the paper is published.