Abstract
Listeria monocytogenes are Gram-positive, facultatively anaerobic, non-spore-forming bacteria that easily adapt to changing environmental conditions. The ability to grow at a wide range of temperatures, pH, and salinity determines the presence of the pathogen in water, sewage, soil, decaying vegetation, and animal feed. L. monocytogenes is an etiological factor of listeriosis, especially dangerous for the elderly, pregnant women, and newborns. The major source of L. monocytogenes for humans is food, including fresh and smoked products. Its high prevalence in food is associated with bacterial adaptation to the food processing environment (FPE). Since the number of listeriosis cases has been progressively increasing an efficient eradication of the pathogen from the FPE is crucial. Understanding the mechanisms of bacterial adaptation to environmental stress will significantly contribute to developing novel, effective methods of controlling L. monocytogenes in the food industry.
Keywords: Listeria monocytogenes, stress factors, adaptive response, disinfectants, food processing environment
Introduction
Listeria monocytogenes are Gram-positive, rod-shaped, non-spore-forming bacteria, widespread in the environment, i.e., water, soil, sewage, decaying vegetation, silage, and animals (O'Neil and Marquis, 2006; Gandhi and Chikindas, 2007). These bacteria tolerate low water activity (≥ 0.9) and are able to survive in a wide range of temperature (0–45°C), pH (4.3–9.6), and salinity (up to 10% NaCl; Renier et al., 2011). L. monocytogenes is a causative agent of human listeriosis, especially dangerous for the elderly, pregnant women, newborns, and immunocompromised patients. The major source of the pathogen for humans is food, including meat, fish (raw and smoked), mold cheese, unpasteurized milk, ready-to-eat (RTE) food, and fresh produce (Gandhi and Chikindas, 2007). Despite the low annual incidence of listeriosis ranging from 0.1 to 10 cases per million people, the mortality rate is high and may exceed 15–20% (World Health Organization (WHO), 2018a). To date, many listeriosis outbreaks have been noted worldwide, including the deadliest outbreak in United States history (1985 – soft cheese; Beckers et al., 1987), the deadliest outbreak in Canada (2008 – deli meat; Birk-Urovitz, 2011), and the deadliest outbreak ever in South Africa (2017–2018, deli meat; Smith et al., 2019). Table 1 summarizes data on the most important epidemics of listeriosis. Despite the development of therapies and biocontrol measures, L. monocytogenes remains a serious threat to food safety, as evidenced by the outbreak in South Africa. Environmental stress affects the morphology, pathogenicity, gene expression, and antimicrobial resistance (AMR) of L. monocytogenes (Matereke and Okoh, 2020). Understanding the adaptive mechanisms of the pathogen can facilitate the planning of disinfection strategies (more effective and less expensive methods) in a food processing environment (FPE), thus ensuring food safety.
Table 1.
The most important epidemics of listeriosis.
| Year | Country | Food | Number of cases | Number of deaths | Number of miscarriages | References | |
|---|---|---|---|---|---|---|---|
| United States | 1985 | Los Angeles | Mexican-style cheese | 142 | 41 | 0 | Beckers et al., 1987 |
| 1998–1999 | 11 states | Hot dog | 108 | 14 | 4 | Centers of Disease Control and Prevention (CDC), 1999 | |
| 2000 | 10 states | Deli turkey meat | 29 | 4 | 3 | Centers of Disease Control and Prevention (CDC), 2000 | |
| 2002 | Eight states | Fresh and frozen, Ready-to-eat turkey, and chicken | 46 | 7 | 3 | Centers of Disease Control and Prevention (CDC), 2002 | |
| 2010 | Texas | Pre-cut celery | 10 | 5 | 0 | Gaul et al., 2012 | |
| 2011 | 28 states | Cantaloupe | 147 | 33 | 1 | Centers of Disease Control and Prevention (CDC), 2012a | |
| 2012 | 13 states | Ricotta salata Cheese | 22 | 4 | 1 | Centers of Disease Control and Prevention (CDC), 2012b | |
| 2013 | Five states | Crave brothers farmstead cheeses | 6 | 1 | 1 | Centers of Disease Control and Prevention (CDC), 2012c | |
| 2010–2015 | Four states | Blue bell ice cream | 10 | 3 | 0 | Centers of Disease Control and Prevention (CDC), 2015a | |
| 2015 | 10 states | Soft cheeses distributed by Karoun dairies | 30 | 3 | 1 | Centers of Disease Control and Prevention (CDC), 2015b | |
| 2014–2015 | 12 states | Caramel apples | 35 | 7 | 1 | Centers of Disease Control and Prevention (CDC), 2015c | |
| 2016 | Two states | Raw milk | 2 | 1 | 0 | Centers of Disease Control and Prevention (CDC), 2016a | |
| 2016 | Four states | Frozen vegetables | 9 | 3 | 0 | Centers of Disease Control and Prevention (CDC), 2016b | |
| 2016 | Nine states | Packaged salads | 19 | 1 | 0 | Centers of Disease Control and Prevention (CDC), 2016c | |
| 2017 | Four states | Soft raw milk cheese Made by vulto creamery |
8 | 2 | 0 | Centers of Disease Control and Prevention (CDC), 2017 | |
| 2018 | Two states | Deli ham | 4 | 1 | 0 | Centers of Disease Control and Prevention (CDC), 2018 | |
| 2019 | Five states | Deli-sliced meats and cheeses | 10 | 1 | 0 | Centers of Disease Control and Prevention (CDC), 2019a | |
| 2019 | Four states | Pork products | 4 | 0 | 0 | Centers of Disease Control and Prevention (CDC), 2019b | |
| 2019 | Five states | Hard-boiled eggs | 8 | 1 | 0 | Centers of Disease Control and Prevention (CDC), 2020a | |
| 2020 | 17 states | Enoki mushrooms | 36 | 4 | 2 | Centers of Disease Control and Prevention (CDC), 2020b | |
| 2020 | Four states | Deli meats | 12 | 1 | 0 | Centers of Disease Control and Prevention (CDC), 2021a | |
| 2021 | Four states | Queso Fresco | 11 | 1 | 0 | Centers of Disease Control and Prevention (CDC), 2021b | |
| Europe | 2003 | The Swindon area of the United Kingdom | Prepacked sandwiches from a retail outlet | 5 | 0 | 0 | Dawson et al., 2006 |
| 2005 | Switzerland | Soft cheese (known as “tomme”) | 10 | 3 | 2 | Bille et al., 2006 | |
| 2015–2018 | Austria, Denmark, Finland, Sweden, and the United Kingdom | Frozen corn | 41 | 6 | 0 | European Centre for Disease Prevention and Control (ECDC), 2018a | |
| 2017–2019 | Netherlands, Belgium | Ready-to-eat meat products | 21 | 3 | 1 | European Centre for Disease Prevention and Control (ECDC), 2019a | |
| Sience 2015 | Denmark, Germany, and France | Ready-to-eat salmon products | 12 | 4 | 0 | European Centre for Disease Prevention and Control (ECDC), 2018b | |
| 2014–2019 | Denmark, Estonia, Finland, France, and Sweden | Cold-smoked fish products | 22 | 5 | 0 | European Centre for Disease Prevention and Control (ECDC), 2019b | |
| 2019 | Spain | Chilled roasted pork meat product | 222 | 3 | 6 | World Health Organization (WHO), 2018b | |
| Australia | 2018 | Australia | Rockmelons | 20 | 7 | 1 | World Health Organization (WHO), 2018c |
| Africa | 2017–2018 | Republic of South Africa | Ready-to-eat processed meat products | 1,024 | 200 | – | Smith et al., 2019 |
The review aims to summarize knowledge about the stress response of L. monocytogenes in FPE. A deeper insight into the stress response of L. monocytogenes may help implement well-designed food preservation and disinfection protocols, reducing the risk of the bacteria in the food and FPE and ensuring consumer safety.
Stress Factors in the Food Processing Environment
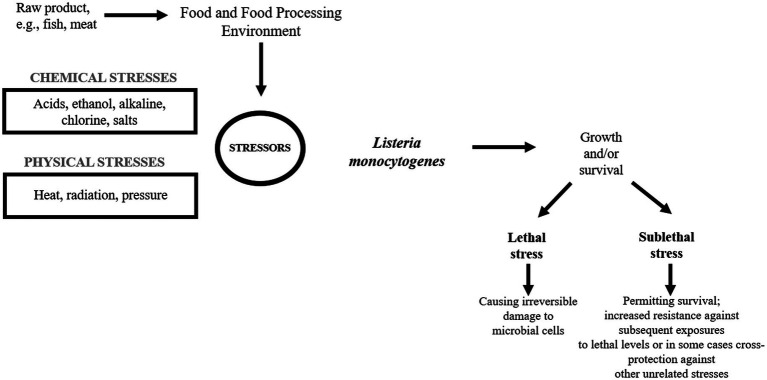
Listeria monocytogenes is a serious problem in the food industry as the pathogen easily contaminates a variety of food products leading to food recall (Miladi et al., 2017). In FPE, L. monocytogenes encounters many adverse stress factors, e.g., low/high temperature, low pH, high salinity, and the concentration of chemicals, which may be sublethal or lethal (Figure 1).
Figure 1.
Listeria monocytogenes in the food processing environment.
Environmental stress tolerance and virulence may be considered as overlapping aspects of the biology of L. monocytogenes (O’Byrne and Karatzas, 2008). Without a strong stress response, the pathogen would not survive and persist in the FPE and withstand the passage through the human gastrointestinal tract (NicAogain and O’Byrne, 2016). One of the major stress response mechanisms is associated with alternative sigma factors σB, σC, σH, and σL, of which σB plays the pivotal role (Lungu et al., 2009). In L. monocytogenes, these factors control over 300 genes, including stress-associated and virulence genes. σB helps L. monocytogenes survive under acid, osmotic, oxidative, and other stress conditions (Figure 2). The activity of σB and hence the general stress response activation in L. monocytogenes, is regulated by a complex structure known as the stressosome (NicAogain and O’Byrne, 2016). Analyses of the proteome and transcriptome of L. monocytogenes revealed many proteins activated in response to stress. Table 2 presents an overview of L. monocytogenes main stress-response and virulence genes and proteins.
Figure 2.
The stress response in L. monocytogenes.
Table 2.
Listeria monocytogenes stress-response and virulence genes and proteins.
| Stress | System | Gene | Protein |
|---|---|---|---|
| Acid | Glutamate decarboxylase system (GAD) | gadA (formerly gadB, gadD, gadD3, or lmo2434) | Glutamate decarboxylase |
| gadCB (formerly gadD2T2 or lmo2363–lmo2362) | Glutamate decarboxylase and a glutamate-γ-aminobutyric acid (GABA) antiporter | ||
| Osmotic | Glycine betaine transport system | betL | Glycine betaine transporter (BetL) |
| Glycine betaine ABC transport system | three-gene operon (gbuABC) |
Gbu | |
| Carnitine ATP-binding cassette (ABC) transport system | Four-gene operon (opuCABCD) | OpuC | |
| Cold | – | cspD | Cold-shock protein D |
| ltrC | Low-temperature requirement C protein | ||
| Heat | – | grpE | Heat-shock protein GrpE |
| Oxidative | – | lmo0669 | Oxidoreductase |
| lmo1433 | Glutathione reductase | ||
| Other genes involved in the stress response | – | clpB | ClpB, similar to endopeptidase Clp ATP-binding chain B; transcribed in an operon with lmo2205 |
| – | clpP | ClpP, an ATP-dependent Clp protease proteolytic subunit |
|
| – | clpX | ClpX, an ATP-dependent Clp protease ATP-binding subunit |
|
| – | lmo1138 | Similar to ATP-dependent Clp protease proteolytic component | |
| – | lmo1601 | General-stress protein; transcribed in an operon with lmo1602, which encodes an unknown protein |
|
| – | lstR | Lineage-specific thermal regulator; transcribed in an operon with sigC | |
| – | rsbVWX | Regulators of σB activity, RsbV, RsbW, and RsbX | |
| Bile-exclusion system | bilE (formerly lmo1421-lmo1422, or opuBAB) | Two-gene operon (bilEAB) encodes a bile-exclusion system; responsible for bile resistance | |
| Bile tolerance | bsh | Bile salt hydrolase | |
| clpC operon | comprises ctsR, mcsA, mcsB, and clpC, | CtsR, McsA, and McsB, which both regulate CtsR activity, and the ClpC endopeptidase and chaperone |
σB is responsible for the transcription of genes involved in the stress response. In turn, PrfA (transcriptional activator of virulence genes) controls the expression of virulence genes. There is an extensive cross-talk system between σB and PrfA ensuring the optimal expression of genes required in extrahost environments (including suppression of genes that reduce efficiency) and in intrahost environments (Gaballa et al., 2019). The stress conditions encountered during food production and the adaptive response of L. monocytogenes affect the virulence of the bacilli and thus pose a much higher risk to consumers. The knowledge on the adaptive mechanisms of L. monocytogenes to the changing environmental conditions will help design appropriate methods of production process control and disinfection (mainly lethal effect). As a result, we expect to limit the acquisition of resistance and prevent the increase of virulence of L. monocytogenes.
The presented review focuses on the stress factors encountered by L. monocytogenes in FPE and the stress response mechanisms.
Salt
One of the main preservatives in FPE is salt contributing to osmotic stress. Salt is frequently used in the food industry as a preservative and antibacterial agent, especially in RTE meat and fish products (Desmond, 2006). In addition, salt is widely used as a texture and flavor improver (Ruusunen and Puolanne, 2005). Salt disturbs homeostasis between the internal and external environment of the bacterial cells. All changes of the osmolarity induce osmotic stress resulting in hypotonia (swelling of the cell) or hypertonia (shrinking of the cell; Sleator and Hill, 2002). The change of osmolarity may also affect biophysical properties (surface tension), decrease cell membrane fluidity, and destroy proteins and DNA. Moreover, salt can contribute to bacterial cell desiccation, making it vulnerable to the action of free radicals, which results in cell death. Reversely, osmotic stress may lead to cross-resistance and help bacteria survive in the next stage of food processing (Burgess et al., 2016).
Listeria monocytogenes can withstand up to 10% of NaCl in the environment (Chihib et al., 2003). Many proteins play role in salt stress response, including osmolyte transporters, cell wall modifying proteins, regulatory proteins, and general stress response proteins (Figure 3). The response to increased osmolality in L. monocytogenes is two-step. First, K+ and glutamate are collected, and then the accumulation of compatible solutes/osmolytes occurs. Compatible solutes are small organic molecules that function as osmoprotectants and are indispensable for cell turgor maintenance. In response to elevated osmolarity, bacterial cell accumulates very high levels of compatible solutes (restoring the cell turgor without the influence on the cytoplasmic functions). Additionally, compatible solutes stabilize the structure and functions of enzymes under stress conditions (Matereke and Okoh, 2020). A number of osmolytes have been identified in L. monocytogenes, e.g., glycine betaine, carnitine, proline, acetylcarnitine, gammabutyrobetaine, and dimethylsulfoniopropionate. Since these bacteria are unable to synthesize osmolytes de novo they must be transported from the environment. L-carnitine is commonly found in raw meat, whereas vegetables (sugar beet and spinach) and cornflakes are sources of glycine betaine (Chan et al., 2007).
Figure 3.
Transport of compatible solutes in response to osmotic stress in L. monocytogenes (CtsR – negative transcription regulator; ClpP, ClpC – ATP-dependent Clp protease; HtrA – serine protease; Csp – cold shock proteins; Hfq – the RNA binding protein; Ctc – 50S ribosomal protein L25; ProBA – proline synthesis enzyme system; Kdp – operon response regulator; Gbu – glycine betaine; OpuC – ATP-dependent carnitine transporter; ATP – adenosine triphosphate; ADP – Adenosine 5'-diphosphate; and BetL – glycine betaine transporter).
The analysis of L. monocytogenes genome revealed three σB–dependent transporters of compatible solutes, i.e., betaine symporter (BetL), Gbu, and OpuC (Figure 3; Bucur et al., 2018). Glycine betaine accumulation in the cytosol may occur without impairing protein structure and enzyme-substrate interaction (Angelidis and Smith, 2003). There are two transporters of glycine betaine, i.e., ATP-dependent porter II (Gbu) and Na+- glycine BetL. BetL, encoded by betL gene, is activated immediately upon stress exposure but provides long-term protection only under low salt stress. In turn, Gbu, encoded by gbuABC operon, plays the predominant role in long-term adaptation, especially at higher concentrations of salt (Mendum and Smith, 2002). OpuC, encoded by opuCABCD operon, is an ATP-dependent carnitine transporter activated in response to osmotic and cold stress (Figure 3; Bucur et al., 2018). In the absence of osmoprotectants, the osmotic stress response mechanism stress relies on Ctc protein. Ctc most likely binds to the 5S RNA sub-principle (Duché et al., 2002).
An important role in osmotolerance plays also general stress response proteins, including serine protease HtrA (degradation of improperly folded proteins), two-component regulatory system LisRK, ClpC (ATPase), CLpP (protease), and RelA (synthesis of guanosine pentaphosphate [(p)ppGpp]), Ctc. Researchers have suggested that RelA is involved in osmotic stress response independent of compatible solutes accumulation (Burall et al., 2012).
In σB regulon, scientists have described eight genes encoding putative osmotic stress-associated proteins, i.e., LMRG 01658, LMRG 00208, LMRG 00211-00212, hfq, and dtpT in L. monocytogenes. However, the role of these genes has not been elucidated yet (Liu et al., 2019). Ribeiro et al. (2014) have suggested that under salt stress, upregulation of these genes enables the increased synthesis of exopolysaccharides. The Hfq protein binds sRNA during intracellular growth. The mutants devoid of this protein were not able to respond properly to osmotic and ethanol stress (Christiansen et al., 2006). The DtpT protein plays important role in the transport of di- and tripeptides involved in osmotic stress protection (Wouters et al., 2005). In turn, Kdp is a transcriptional response regulator enabling quick reaction to stress via K+ import. Also, trehalose was reported to be linked to osmotolerance, and the treA gene, encoding phosphotrehalase was detected in L. monocytogenes (Ells and Truelstrup Hansen, 2011). Researchers described proteins modifying cell membrane, i.e., putative peptidoglycan-associated protein (lmo2085 gene) and putative UDP-glucose phosphorylase (lmo1078 gene) as osmotic stress proteins (Burgess et al., 2016).
The main mechanism of the osmotic stress response in L. monocytogenes involves osmoprotectants. In their absence, Ctc protein provides protection. The contribution of the remaining proteins presented is not fully understood. Food manufacturers should pay attention to the rightness of using salt as a preservative in the elimination of microorganisms from food, especially L. monocytogenes, due to the advanced mechanisms of adaptation to high salt concentration.
In recent years, there has been a demand for low-salt products, which forces searching for new technologies to control microbiological hazards in low-salt products. The currently used alternative to salt (especially in meat products) is the treatment with high isostatic pressures, activated plasma, pulsed UV light, or active packaging (Fraqueza et al., 2021). In our opinion, an attractive solution would be natural substances with antimicrobial activity (e.g., essential oils and plant extracts).
Temperature
Another factor ensuring food safety is temperature, both high and low. Heat treatment (pasteurization and sterilization) is one of the most common methods of assuring food safety and microbial elimination. However, too low temperature of such a process might not effectively eliminate the pathogen, and too high temperature may affect the organoleptic properties of food. In turn, low temperatures (e.g., cold stores) aim to limit the growth of undesirable microorganisms in food.
Listeria monocytogenes is able to survive and grow in a wide temperature range, i.e., 0–45°C (Arioli et al., 2019). Depending on the physiological state of the cell, inactivation of L. monocytogenes ranges from 10 min to 12 s at 55–65°C (Smelt and Brul, 2014).
Reaction to elevated temperature is associated with increased expression of heat shock proteins (Hsp). Hsp stabilize proteins and prevent improper folding and aggregation of proteins (Figure 4). In response to heat stress, L. monocytogenes increase the production of these proteins, thereby stimulating the repair of thermally denatured proteins. Listeria monocytogenes possess III classes of heat shock-associated genes. Genes of class I (groE, dnaK, dnaJ, groEL, and groES) are overexpressed during the accumulation of denatured proteins in the cytosol and act as intracellular chaperones. Class III genes encode ATP-dependent proteins of caseinolytic activity (ClpB, ClpC, ClpP, and ClpE; Bucur et al., 2018). These proteins cooperate to maintain the proper protein structure at high temperatures. The proteins GroEL and GroES (regulation of basic cellular processes) and DnaK, and DnaJ (stabilization of the conformation of unfolded proteins) are the main chaperones protecting L. monocytogenes against heat stress. Also, ClpE and ClpC (caseinolytic activity) are indispensable during the replication at elevated temperatures and nutrients deficiency. During heat stress, these proteins participate in the regulation of proteolysis and proper protein folding. Genes of class I and III are negatively regulated by HrcA and CtsR, respectively, whereas class II genes are under positive control of σB (Nair et al., 2000). GroE regulates HrcA activity. Under optimal growth conditions, the interaction of HrcA with GroE inhibits transcription of class I genes. At elevated temperature, unfolded proteins bind GroE contributing to HrcA inactivation and enabling binding of RNA-s32 polymerase with promoters and gene expression.
Figure 4.
Regulation of heat shock response of L. monocytogenes (ROS – reactive oxygen species; Msc – tyrosine kinase; McsA – zinger finger protein; HtrA – serine protease; ClpC – ATP-dependent Clp protease; HrcA – Heat-inducible transcription repressor; CtsR – negative transcription regulator; Fri – ferritin-like protein; and RNAP – RNA polymerase enzyme).
At 37°C, CtsR is stabilized by McsA, which results in class III gene repression. At increased temperature, McsB modifies CtsR conformation preventing its binding with gene promoters. As a result, RNA-s32 polymerase binds with promoters leading to gene expression, and CtsR is degraded (Figure 4; Kocaman and Sarimehmetoğlu, 2016). ClpC expression is negatively controlled at the transcription level, either directly or indirectly, by PrfA (Ripio et al., 1998).
Heat shock proteins, i.e., DnaK, ClpC ATPase, ClpP serine prosthesis, and Ctc protein also participate in the osmotic stress response. Thus, proteins involved in the reaction to osmotic and heat stress interact with each other (Figure 5).
Figure 5.
Interaction of various stress response mechanisms in L. monocytogenes.
During food processing, microbes experience low temperatures. Cold stress response in bacteria is three-step, i.e., cold shock, acclimatization, and adaptation (Kocaman and Sarimehmetoğlu, 2016). At 10°C, amino-acid starvation, oxidative stress, synthesis of misfolded proteins, reconstruction of a cell wall, changes of metabolism, and induction of global regulatory reactions occur (Lianou and Sofos, 2007). A fast decrease below optimal temperature, termed cold shock, triggers the synthesis of cold shock proteins (Csp). Csp are small proteins (65–70 aa) of highly conserved structure, expressed at a low level at optimal temperature (37°C; Schmid et al., 2009). Csp function as a molecular chaperone, enabling replication, transcription, and translation at low temperatures (Lee et al., 2012). Csps bind to a single-stranded nucleic acid via ribonucleoprotein binding motifs RNP1 and RNP2, thereby stabilizing nucleic acid conformation and preventing its degradation (Barria et al., 2013). CspA, CspB, and CspD contribute to adaptation to a varying extent (Schmid et al., 2009). Schmid et al. (2009) have shown that L. monocytogenes ∆cspA and ∆cspD mutants significantly worse responded to cold stress. Stress conditions also induce cold acclimatization proteins (Cap; Figure 6; Barria et al., 2013). The main mechanism of response to cold stress in L. monocytogenes relies on the action of both, Csp and Cap proteins. There are also other mechanisms involved in the cold stress response, supporting Csp and Cap action. Another cold stress response mechanism involves osmolytes and cryoprotectants (Angelidis and Smith, 2003). A key role as a cryoprotectant is attributed to branched-chain fatty acids of the cell membrane, ensuring membrane integrity at low temperatures (Becker et al., 2000). During cold shock, σB confers growth phase-dependent adaptation and assures efficient accumulation of cryoprotectants such as betaine and carnitine. Eight Rsb proteins modulate σB binding to the primary regulator through protein-protein interaction and phosphates transfer (Liu et al., 2019).
Figure 6.
Cold stress response of L. monocytogenes (OpuC – ATP-dependent carnitine transporter; Gbu – glycine betaine; ROS – reactive oxygen species; Csp – cold-shock protein; Cap – cold acclimatization protein; and Flp – ferritin-like proteins).
High temperature is a frequently used factor for the elimination of microorganisms in the food industry. To prevent loss of organoleptic properties, producers shorten the thermal processing of food. Unfortunately, such an approach may generate sub-lethal stress leading to the resistance acquisition in microorganisms. Listeria monocytogenes has developed several adaptation mechanisms to both high and low temperatures. Therefore, food producers should consider combining the thermal treatment with other methods, e.g., chemicals or natural preservatives (e.g., bacteriocins). The combination of methods can provide a lethal effect on microorganisms (safety for the consumer) without affecting the properties of the food.
pH
Acid stress is one of the most common stresses faced by foodborne pathogens. Low pH increases H+ concentration, decreasing an internal pH (pHi) and inhibiting microbial growth. The ability to withstand acid stress determines bacterial survival in acid food products (Ryan et al., 2008). Bacteria need to restore homeostasis inside the cell via a passive mechanism (an increase of cytoplasm buffer capacity) or an active one (using a proton pump) to survive (Ryan et al., 2008). In food processing plants, L. monocytogenes is exposed to many acids (benzoic, salicylic, lactic, and propionic acid), commonly used as food preservatives and disinfectants. Other acid challenges, i.e., gastric juice and bile salts, the pathogen encounters in the human food tract (Begley et al., 2002).
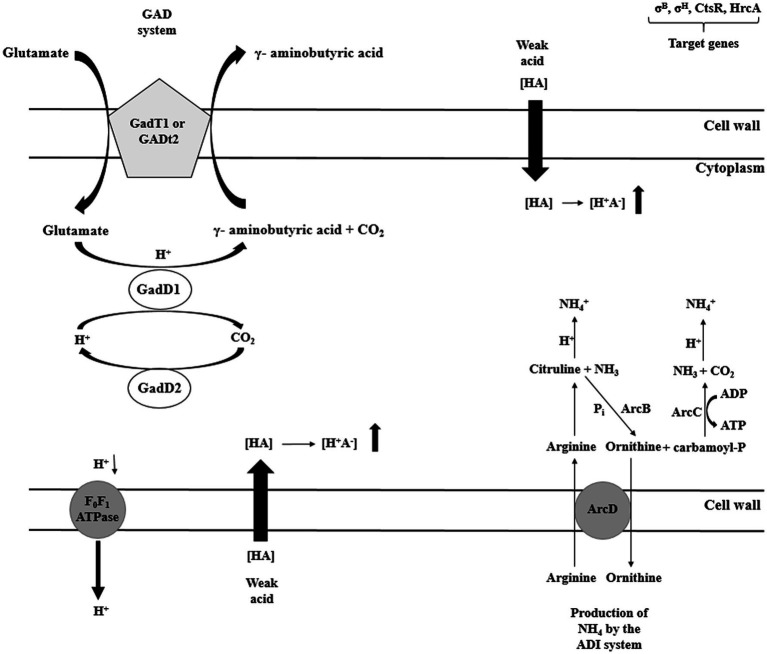
There are four potential mechanisms responsible for the homeostasis maintenance in L. monocytogenes, i.e., acid tolerance response (ATR), glutamate decarboxylase (GAD), arginine deiminase (ADI), and F1F0-ATPase (Cotter et al., 2005). The major role of ATR mechanism is the cell protection against the lethal effect of stress following short exposure to mild acids (Koutsoumanis et al., 2003). The GAD system affects survival in food products of low pH (juices, yogurts, salads, and mayonnaise). The GAD system compromises five genes. Three genes (gadD1, gadD2, and gadD3) encode decarboxylases and two genes encode (gadT1, gadT2) antiporters (Melo et al., 2015). The GAD system converts extracellular glutamate to γ-aminobutyrate acid (GABA), elevating pHi (Werbrouck et al., 2009). Subsequently, GABA is exchanged for glutamate at the cell membrane by GadT2 antiporter. As a consequence, an intracellular proton is consumed, contributing to alkalization of the environment and pH homeostasis. Researchers have shown that strains with low GAD activity are susceptible to gastric juice (Ryan et al., 2009). Extremely acid pH activates another system: the ADI pathway (Soares and Knuckley, 2016). ADI converts, imported from the external environment, arginine to ornithine, CO2, ammonia, and ATP. Three enzymes participate in the arginine conversion, i.e., ADI, carbamoyltransferase, and carbamate kinase, encoded by arcA, arcB, and arcC, respectively. Ornithine is then transported from the cell, in an energy-independent manner, via membrane-bound antiporter, encoded by arcD gene. In turn, ammonia, produced as a by-product, reacts with intracellular protons generating NH4+, thereby increasing cytoplasmic pH and protecting the cell from the acid environment (Figure 7; Matereke and Okoh, 2020). ATP produced during arginine conversion may be used by F1F0-ATPase. This multisubunit enzyme generates a proton gradient, enabling H+ expulsion and homeostasis restoration (Ryan et al., 2009). The enzyme consists of two distinct domains: the membrane domain F0, which functions as a channel for protons translocation, and the cytoplasmic domain F1 catalyzing ATP synthesis and hydrolysis (Figure 7; Smith et al., 2013). The three discussed systems, i.e., ATR, GAD, and F1F0-ATPase, act at the same time (Figure 5). Interactions between them ensure survival and adaptation to acid stress.
Figure 7.
Selected mechanisms of acid stress response of L. monocytogenes (GAD – glutamate decarboxylase system; ADI – arginine deiminase system; HrcA – Heat-inducible transcription repressor; CtsR – negative transcription regulator; ArcA – arginine deiminase; ArcB – catabolic ornithine carbamoyltransferase; ArcC – carbamate kinase; ArcD – membrane-bound arginine-ornithine antiporter; and F0F1 –ATPase – proton efflux membrane ATPase).
In the food production environment, L. monocytogenes may also face sublethal alkaline stress associated with the use of detergents and disinfectants. The adaptation to such conditions involves increased production of acids and induction of transporters and enzymes responsible for protons retention and cell surface modifications (Soni et al., 2011). Usually, in response to high pH, the general stress proteins production, ATP generation (via ATP synthase conjugation with H+), and increased expression and activity of the monovalent cation/proton antiporters are observed (Giotis et al., 2010).
Many foodborne pathogens, including L. monocytogenes, develop adaptation to low pH and cross-resistance to other stresses upon exposure to acid stress. This cross-protection phenomenon may have severe consequences for food safety, as L. monocytogenes can survive various treatments, such as heat stress, cold stress, and osmotic stress. It is necessary to search for new disinfection methods in the food industry, limiting acids and alkalis use. An attractive solution could be substances of natural origin (herbs extracts and essential oils) or bacteriocins (environmentally friendly and safe for the consumer). Lately, increasing attention is being paid to essential oils as natural additives to extend the shelf life of food products (Tongnuanchan and Benjakul, 2014). Work (Shan et al., 2009; Efenberger-Szmechtyk et al., 2021) on natural preservatives is ongoing, and the results are promising. It is also necessary to understand the impact of such new applications on the survival of L. monocytogenes.
Nutrients
The availability of ingredients in the FPE varies. Availability is low in post-disinfection areas. In turn, during food processing, there may be different types of waste, such as blood, fruit juices, etc. The availability of nutrients influences microbial biofilm formation, undesirable in the food industry. Listeria monocytogenes is able to use various compounds in the form of carbon, and/or energy sources to thrive in potential niches also within the food industry (Tapia et al., 2020). Thus, L. monocytogenes can survive and multiply in a variety of environments with different availability of nutrients. The pathogen possesses transport systems for various sugars allowing adaption to changing environmental conditions.
The genome of L. monocytogenes contains a significant number of ABC transporters phosphoenolpyruvate: sugar phosphotransferase (PTS) systems, most of which have not yet been fully characterized (Stoll and Goebel, 2010; Deutscher et al., 2014). Additionally, L. monocytogenes can utilize many carbon sources to produce biofilms on food processing surfaces (Tapia et al., 2020). In laboratory conditions, L. monocytogenes can grow in commonly used bacteriological media. Increased growth rates occur in the presence of fermentable sugars. However, nutritional requirements of L. monocytogenes are strain-dependent and reflect the ability of the pathogen to survive in a wide range of environmental conditions (Lungu et al., 2009). Premaratne et al. (1991) have shown that L. monocytogenes requires glucose, glutamine, leucine, isoleucine, arginine, methionine, valine, cysteine, riboflavin, biotin, and thiamine for growth. On the other hand, fructose, mannose, cellobiose, trehalose, maltose, glycerol, glucosamine, and N-acetylglucosamine support its growth in the absence of glucose. Tapia et al. (2020) have observed that the utilization of lactose by L. monocytogenes triggers a strong σB-dependent stress response, which may have implications for L. monocytogenes in the food chain. Tapia et al. (2020) have demonstrated the potential role of lactose in the activation of the σB-dependent stress response, which may have implications for L. monocytogenes in the food chain. They have observed that the pathogen in the presence of lactose, compared to other carbon sources tested, increased virulence, biofilm production potential, and resistance to heat and acids (Tapia et al., 2020). Thus, L. monocytogenes ability to utilize lactose may increase its survival in food processing (dairy), resulting in a higher risk of reinfection. Collectively, the ability to use alternative energy sources, e.g., carbohydrate polymers, proteins, nucleic acids, and lipids, determines the survival and growth of the pathogen in the human gastrointestinal tract (Lungu et al., 2009).
Zhou et al. (2012) have found that limited access to nutrients increased the biofilm formation ability of L. monocytogenes. Also, the presence of food by-products (meat juice, pork serum, or fat) in the processing environment stimulated biofilm formation (Van Houdt and Michiels, 2010). Listeria monocytogenes in the biofilm are resistant to disinfection methods. Therefore, efficient cleaning and disinfection procedures are of great importance.
Reactive Oxygen Species
Disinfectants, commonly used in the food industry, trigger in bacteria oxidative stress. Reactive oxygen species (ROS), such as hydrogen peroxide or hydroxyl radicals, can be produced as by-products of metabolism or during reduction of the respiratory chain or other stress factors (Bowman et al., 2010; Burgess et al., 2016). In order to withstand exposure to oxidizing agents, e.g., sodium hypochlorite, cells must activate mechanisms to repair damages of proteins, the cytoplasmic membrane, and nucleic acids. Listeria monocytogenes under oxidative stress activates a survival strategy that includes the expression of sigB, cold and heat shock proteins, proteases (ClpC, ClpP, and GroEL), and ROS detoxification systems (Manso et al., 2020). ROS detoxification systems include superoxide dismutase (Sod), catalase (Kat), and alkylhydroperoxidase (AhpCF; Bucur et al., 2018). Strains without Kat and Sod activity showed increased sensitivity to oxidative stress and were low virulent (Archambaud et al., 2006; Azizoglu and Kathariou, 2010). Also, ferritin plays an essential role in the oxidative stress response of L. monocytogenes (Dussurget et al., 2005). Rea et al. (2005) have reported that L. monocytogenes ∆perR mutants (peroxide regulon repressor) displayed elevated sensitivity to hydrogen peroxide and limited growth.
Manso et al. (2020) have shown that oxidative stress is temperature-dependent. The lower temperature increased oxidative stress response. Both stressful conditions caused similar damage to nucleic acids and cell membranes (Manso et al., 2020). Most of the disinfectants used in the food industry are applied at low temperatures, favoring the acquisition of resistance by L. monocytogenes. Therefore, food manufacturers should consider the type of active substances in disinfectants and the disinfection process temperature.
Disinfectants
One method to maintain food and consumer safety is the use of appropriate cleaning and disinfection procedures. Chemical preparations are most common. An important problem is the acquisition of resistance to disinfectants by L. monocytogenes due to the residues of chemical substances in sub-lethal concentrations after disinfection. The most commonly used disinfectants for L. monocytogenes are quaternary ammonium compounds (QACs) and chlorine-based biocides (Minarovicova et al., 2018; Aryal and Muriana, 2019). Listeria monocytogenes strains resistant to disinfectants contribute to sporadic cases as well as outbreaks of listeriosis. The antimicrobial effectiveness of biocides may be affected by the presence of organic pollutants. Also, the biofilm structure protects L. monocytogenes against the action of disinfectants (Duze et al., 2021). The main cause of resistance of L. monocytogenes to antimicrobial agents is horizontal gene transfer (HGT) of mobile genetic elements such as plasmids and transposons carrying resistant genes. Another mechanism relies on the activation of efflux pump systems. To date, six genes (mdrL, lde, cassette bcrABC, qacH, emrE, and emrC), located on mobile genetic elements, related to the efflux system, have been identified in L. monocytogenes in response to disinfectants action. Table 3 presents genes conferring biocide tolerance. Two major efflux pump genes, mdrL and lde, are present in nearly all L. monocytogenes serotypes (Haubert et al., 2019). The MDRL pump detoxifies macrolide, cefotaxime, heavy metals, and ethidium bromide (EtBr). In turn, the Lde pump detoxifies fluoroquinolones, antibiotics, and intercalating dyes such as EtBr and acridine orange (Mata et al., 2000). Jiang et al. (2019) confirmed the role of MdrL in disinfectants tolerance in L. monocytogenes (in the survival of biocidal stress in food). Other efflux pump genes responsible for the increased tolerance to QAC are emrE and emrC genes, located on the LGI1 genomic mobile island and pLMST6 plasmid, respectively (Kovacevic et al., 2016; Kremer et al., 2017). Kropac et al. (2019) showed that plasmid pLMST6 increased tolerance only to QAC-based disinfectants. However, as plasmids aid L. monocytogenes to survive in FPE, they may help in biocide tolerance. Kremer et al. (2017) have shown a link between isolates with plasmid pLMST6, pump efflux, emrC gene, and reduced antibiotic susceptibility. In turn, Noll et al. (2020) have studied the impact of adaptation L. monocytogenes against antimicrobials. Listeria monocytogenes cells after exposure to benzalkonium chloride showed a 2-fold increase in MIC compared to the disinfectant treatment and reduced sensitivity to antibiotics (ceftriaxone, gentamicin, linezolid, tetracycline, and a combination of trimethoprim and sulfamethoxazole). The tolerance to disinfectants does not always induce cross-resistance to antibiotics. Roedel et al. (2019) have revealed that tolerance to biocides did not induce AMR. Antibiotic resistance in L. monocytogenes may severely influence public health due to poor treatment outcomes and sequelae treatment failure. The frequent use of disinfectants in the food processing industry can lead to selective pressure and promote the absorption of plasmides, e.g., containing brcABC from other L. monocytogenes strains (Hingston et al., 2019).
Table 3.
Genes involved in biocide tolerance in L. monocytogenes.
| The source of the isolation L. monocytogenes | Biocide and exposure time | Presence/expression of genes | Reference |
|---|---|---|---|
| L. monocytogenes 08–5,578 [clinical strain responsible for the Canadian deli meat listeriosis outbreak (2008)] | Benzalkonium chloride – 16 h | emrELm | Kovacevic et al., 2016 |
| Isolates from product contact and non-product contact surfaces | Benzalkonium chloride – 24 h | Transposon Tn6188 | Ortiz et al., 2016 |
| L. monocytogenes H7550 (strain implicated in the 1998–1999 multistate outbreak involving contaminated hot dogs) | Benzalkonium chloride – 48 h | bcrABC | Elhanafi et al., 2010 |
| Isolates from nine Norwegian meat- and salmon processing plants | Benzalkonium chloride – 48 h | bcrABC | Møretrø et al., 2017 |
| L. monocytogenes strains from Switzerland and Finland | Benzalkonium chloride – 48 h | bcrABC, emrE | Meier et al., 2017 |
| Isolates from food and food processing plants | Benzalkonium chloride – 7 days | mdrL | Yu et al., 2018 |
| Isolates from foods and food processing plants | Benzalkonium chloride, Cadmium & arsenic – 48 h | CadA 1, CadA2 | Ratani et al., 2012 |
| Isolates from food and food-processing environments in southern Brazil | Benzalkonium chloride & Cadmium chloride – 48 h | mdrL, lde | Haubert et al., 2019 |
| Isolates from a dairy cattle farm | Benzalkonium chloride, Virkon, H2O2, and sodium Hypochlorite – 12-48 h | qacED1 | Mohammed et al., 2017 |
| Isolates from food production facilities | Benzalkonium chloride and Cetylpyridinium chloride – 72 h | Pdu, Cob-cbi, and Eut | Fox et al., 2011 |
It is relevant to control the used disinfectants to ensure the effective elimination of microorganisms from the food industry. In our opinion, periodical change of the active substances would have a beneficial impact. Such an action would limit the acquisition of resistance traits among microorganisms. Food manufacturers should monitor the effectiveness of agents and avoid extensive storage of open packages of disinfectants.
Bacteriocins
Researchers have been constantly searching for new methods allowing microbes elimination from the FPE. These methods should be cheap, environmentally friendly, and, above all, safe for consumers. One alternative is the use of bacteriocins. Bacteriocins are antimicrobial peptides produced extensively by lactic acid bacteria. These molecules are mainly active against Gram-positive bacteria, including L. monocytogenes (Chikindas et al., 2018). Bacteriocins are natural and safe food preservatives (Silva et al., 2018). Nisin is the class I bacteriocin, approved as a food preservative. Listeria monocytogenes resistance to nisin is associated with changes in the cytoplasmic membrane. Ming and Daeschel (1993) have demonstrated an increased proportion of straight-chain fatty acids and a decreased branched-chain fatty acids ratio. In turn, Wu et al. (2018) have reported that L. monocytogenes bacilli exposed to a high nisin concentration had lowered expression of genes dltA and dltB, indicating that D-alanine residue is not linked to bacteriocin resistance. Scientists emphasized the role of regulation of the expression of two cell wall synthesis-associated genes: lmo2714 (membrane-anchored peptidoglycan protein) and lmo2522 (for cell wall-binding protein with possible effects on nisin tolerance; Wu et al., 2018). Other genes/operons that affect nisin resistance include lmo2229 (Collins et al., 2012), telA (Collins et al., 2010a), mprF (Thedieck et al., 2006), anrAB (Collins et al., 2010b), and dltABCD (Abachin et al., 2002). Many class II bacteriocins with antilisterial activity, including pediocin, sacacin P, leukocin, enterococin, creaticine Y105, garvicin, and linocin M18 have been also isolated and characterized (Ríos Colombo et al., 2018). Natural resistance of L. monocytogenes ranges from 1 to 8%, depending on bacteriocin and tested strain (Macwana and Muriana, 2012).
Listeria monocytogenes response to bacteriocins involves different mechanisms, i.e., changes in cell structures and the participation of relevant proteins which work together.
Stress Adaptation
Listeria monocytogenes can regulate and fine-tune gene expression to adapt to diverse stress conditions encountered during foodborne transmission (Orsi et al., 2021). The mechanism allowing L. monocytogenes to respond to stress conditions is the cross-talk between σB and PrfA.
Application of sublethal doses of stress factors in the FPE may increase AMR or stimulate the resistance to other stress conditions (cross-resistance) and higher doses of the same stress (stress adaptation; Faezi-Ghasemi and Kazemi 2015). Two-component signal transduction systems (TCS) enable bacteria to detect and adapt to a variety of stressors. The prototype modular TCS consists of a periplasmic sensor histidine kinase (HK) and a cognate cytoplasmic response regulator (RR; Gao and Stock, 2009). The action of TCS relies on the phosphorylation of proteins. In a direct phosphotransfer system, the HK sensor domain detects the stimulus leading to ATP-dependent autophosphorylation at a specific histidine (His) residue catalyzed by the kinase domain. Cognate RR then catalyzes the transfer of a phosphoryl group to its own aspartate (Asp) residue in the regulatory domain. Phosphorylation of the RR regulatory domain leads to activation of the effector domain and ultimately to an appropriate response to a specific stimulus (Casino et al., 2010).
Mobile genetic elements play a crucial role in the environmental adaptation and stability of L. monocytogenes. Such elements are common among L. monocytogenes isolates from FPEs (Hurley et al., 2019) and may contain genes mediating tolerance to heat shock (Pöntinen et al., 2017), salt and acid stress (Hingston et al., 2019; Naditz et al., 2019), and biocides (Meier et al., 2017). Also, plasmids are relevant for rapid adaptation to changing environmental conditions. The frequency of plasmids in L. monocytogenes is up to 79% (Elhanafi et al., 2010). The majority of data concerns plasmids containing genes conditioning resistance to disinfectants (Elhanafi et al., 2010; Rakic-Martinez et al., 2011), heavy metals (Rakic-Martinez et al., 2011), increased acid tolerance, and sensitivity to cold (Hingston et al., 2017), reduced tolerance to salt stress (Zhang et al., 2018), and antibiotics (Hadorn et al., 1993). Plasmids were more frequently isolated from food strains than from clinical strains (McLauchlin et al., 1997; Hingston et al., 2019). More, plasmids were more common among recurrent L. monocytogenes strains (75%) than among sporadic strains (35%) isolated from food or food processing plants (Harvey and Gilmour, 2001). These observations suggest that genes found in L. monocytogenes plasmids may be helpful for the pathogen’s survival in such environments (Hingston et al., 2019). A phenomenon found in the food industry is also HGT. The phenomenon is responsible for features critical to the evolution, such as antibiotic resistance or increased virulence. HGT ensures increased survival of L. monocytogenes strains exposed to stress factors related to FPE, which prevents their elimination, especially in places that are difficult to clean (Palma et al., 2020).
Cross-Resistance
Cross-resistance is an essential aspect in the food industry, which often applies many stressors to assure food safety and food quality (Bergholz et al., 2012). Bergholz et al. (2012) have found that pre-exposure to osmotic stress enabled bacterial growth at low temperatures, whereas cold shock-induced cross-protection against salt stress. More, L. monocytogenes subjected to acidic conditions displayed increased resistance to H2O2, heat shock, ethanol, and oxidative and osmotic stress (Lou and Yousef, 1997; Ferreira et al., 2003). Adaptation to acid conditions may also increase L. monocytogenes resistance to nisin. Since the sublethal concentration of acids is frequently applied during food processing, cross-resistance of the acid-adapted cell is of great importance (Mastronicolis et al., 2010). Alonso-Hermando et al. (2009) have revealed that sodium hypochlorite induced AMR of L. monocytogenes. In turn, Faezi-Ghasemi and Kazemi (2015) have demonstrated increased susceptibility of L. monocytogenes to penicillin, ampicillin, gentamycin, tetracycline, trimethoprim/sulfamethoxazol, and rifampicin after stress exposure [7% NaCl, pH = 5.0, and ethanol (5% w/v)]. The researchers claimed that an acid environment leads to an influx of protons and anions to the cell cytoplasm, disturbing its metabolic functions and inducing the damage of proteins, nucleic acids, and cell membranes (Faezi-Ghasemi and Kazemi 2015). The knowledge of the cross-resistance phenomenon may help in the appropriate designing of food processes, limiting the risk for public health.
Conclusion
In the presented review, we summarize the current knowledge about the adaptation of L. monocytogenes to stress factors in food processing plants. Although we know a lot about the stress response of L. monocytogenes, the pathogen is still present in the FPE and is a threat to the consumer. Mobile genetic elements determine the acquisition of new resistance traits to physical and chemical disinfection methods among L. monocytogenes strains. Food producers should not underestimate the risk of microorganisms present in the processing environment, especially L. monocytogenes. Therefore, high sanitary regimes and food processing control are indispensable for the reduction of foodborne listeriosis. Understanding the mechanisms of adaptation to environmental stress would significantly develop new, efficient, and cost-effective methods of controlling L. monocytogenes in the food industry. The knowledge of L. monocytogenes biology and constant control of the pathogen in FPE are critical to ensure food production safety.
Author Contributions
NW-K and KS: conceptualization and project administration. KG-B and EW-Z: formal analysis. NW-K and JK: writing – original draft preparation and visualization. KS, KG-B, and EW-Z: writing – review and editing. KS and EG-K: supervision. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This review was created during the research work carried out as part of a grant for participants of doctoral studies at the Faculty of Pharmacy of Collegium Medicum in Bydgoszcz (no. MN-SDF-4/WF/2019).
Footnotes
Funding. The article is financed with a fund from the contest for doctoral students under the “Excellence Initiative Research University” program in Nicolaus Copernicus University in Toruń.
References
- Abachin E., Poyart C., Pellegrini E., Milohanic E., Fiedler F., Berche P., et al. (2002). Formation of D-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43, 1–14. 10.1046/j.1365-2958.2002.02723.x, PMID: [DOI] [PubMed] [Google Scholar]
- Alonso-Hermando A., Capita R., Prieto M., Alonso-Calleja C. (2009). Adaptation and cross-adaptation of Listeria monocytogenes and Salmonella enterica to poultry decontaminations. J. Microbiol. 47, 142–146. 10.1007/s12275-008-0237-5, PMID: [DOI] [PubMed] [Google Scholar]
- Angelidis A. S., Smith G. M. (2003). Three transporters mediate uptake of glycine betaine and carnitine by Listeria monocytogenes in response to hyperosmotic stress. Appl. Environ. Microbiol. 69, 1013–1022. 10.1128/AEM.69.2.1013-1022.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambaud C., Nahori M. A., Pizarro-Cerda J., Cossart P., Dussurget O. (2006). Control of Listeria superoxide dismutase by phosphorylation. J. Biol. Chem. 281, 31812–31822. 10.1074/jbc.M606249200, PMID: [DOI] [PubMed] [Google Scholar]
- Arioli S., Montanari C., Magnani M., Tabanelli G., Patrignani F., Lanciotti R., et al. (2019). Modelling of Listeria monocytogenes Scott A after a mild heat treatment in the presence of thymol and carvacrol: effects on culturability and viability. J. Food Eng. 240, 73–82. 10.1016/j.jfoodeng.2018.07.014 [DOI] [Google Scholar]
- Aryal M., Muriana P. M. (2019). Efficacy of commercial sanitizers used in food processing facilities for inactivation of Listeria monocytogenes, E. coli O157:H7, and Salmonella biofilms. Foods 8:639. 10.3390/foods8120639, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizoglu R. O., Kathariou S. (2010). Temperature dependent requirement for catalase in aerobic growth of Listeria monocytogenes F2365. Appl. Environ. Microbiol. 76, 6998–7003. 10.1128/AEM.01223-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barria C., Malecki M., Arraiano C. M. (2013). Bacterial adaptation to cold. Microbiology 159, 2437–2443. 10.1099/mic.0.052209-0, PMID: [DOI] [PubMed] [Google Scholar]
- Becker L. A., Evans S. N., Hutkins R. W., Benson A. K. (2000). Role of σB in adaptation of Listeria monocytogenes to growth at low temperature. J. Bacteriol. 182, 7083–7087. 10.1128/JB.182.24.7083-7087.2000, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers H. J., Soentoro P. S. S., Delfgou-van Asch E. H. M. (1987). The occurrence of Listeria monocytogenes in soft cheeses and raw milk and its resistance to heat. Int. J. Food Microbiol. 4, 249–256. 10.1016/0168-1605(87)90041-9 [DOI] [Google Scholar]
- Begley M., Gahan C. G., Hill C. (2002). Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 68, 6005–6012. 10.1128/AEM.68.12.6005-6012.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergholz T. M., Bowen B., Wiedmann M., Boor K. J. (2012). Listeria monocytogenes shows temperature-dependent and independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl. Environ. Microbiol. 78, 2602–2612. 10.1128/AEM.07658-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bille J., Blanc D. S., Schmid H., Boubaker K., Baumgartner A., Siegrist H. H., et al. (2006). Outbreak of human listeriosis associated with Tomme cheese in Northwest Switzerland, 2005. Euro Surveill. 11, 91–93. PMID: [PubMed] [Google Scholar]
- Birk-Urovitz E. (2011). The 2008 Canadian Listeriosis outbreak: a result of knowledge ignored. McMaster University Medical Journal 8, 65–67. [Google Scholar]
- Bowman J. P., Lee Chang K. J., Pinfold T., Ross T. (2010). Transcriptomic and phenotypic responses of Listeria monocytogenes strains possessing different growth efficiencies under acidic conditions. Appl. Environ. Microbiol. 76, 4836–4850. 10.1128/AEM.00315-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucur F. I., Grigore-Gurgu L., Crauwels P., Riedel C. U., Nicolau A. I. (2018). Resistance of Listeria monocytogenes to stress conditions encountered in food and food processing environments. Front. Microbiol. 9:2700. 10.3389/fmicb.2018.02700, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burall L. S., Laksanalamai P., Datta A. R. (2012). Listeria monocytogenes mutants with altered growth phenotypes at refrigeration temperature and high salt concentrations. Appl. Environ. Microbiol. 78, 1265–1272. 10.1128/AEM.06576-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess C. M., Gianotti A., Gruzdev N., Holah J., Knøchel S., Lehner A., et al. (2016). The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. Int. J. Food Microbiol. 16, 37–53. 10.1016/j.ijfoodmicro.2015.12.014, PMID: [DOI] [PubMed] [Google Scholar]
- Casino P., Rubio V., Marina A. (2010). The mechanism of signal transduction by two-component systems. Curr. Opin. Struct. Biol. 20, 763–771. 10.1016/j.sbi.2010.09.010, PMID: [DOI] [PubMed] [Google Scholar]
- Centers of Disease Control and Prevention (CDC) (1999). Update: multistate outbreak of listeriosis–United States, 1998–1999. MMWR Morb. Mortal. Wkly Rep. 47:1117., PMID: [PubMed] [Google Scholar]
- Centers of Disease Control and Prevention (CDC) (2000). Multistate outbreak of listeriosis–United States, 2000. MMWR Morb. Mortal. Wkly Rep. 49:1129, PMID: [PubMed] [Google Scholar]
- Centers of Disease Control and Prevention (CDC) (2002). Public health dispatch: outbreak of listeriosis—Northeastern United States, 2002. MMWR Morb. Mortal. Wkly Rep. Available at: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5142a3.html (Accessed April 1, 2021). 51, 950–951. [PubMed] [Google Scholar]
- Centers of Disease Control and Prevention (CDC) (2012a). Timeline of events: multistate outbreak of listeriosis linked to whole cantaloupes from Jensen Farms, Colorado. Available at: https://www.cdc.gov/listeria/outbreaks/cantaloupes-jensen-farms/timeline.html (Accessed April 12, 2021).
- Centers of Disease Control and Prevention (CDC) (2012c). Multistate outbreak of Listeriosis linked to crave brothers farmstead cheeses (Final Update). Available at: https://www.cdc.gov/listeria/outbreaks/cheese-07-13/index.html (Accessed April 1, 2021).
- Centers of Disease Control and Prevention (CDC) (2015b). Multistate outbreak of Listeriosis linked to soft cheeses distributed by Karoun Dairies, Inc. (Final Update). Available at: https://www.cdc.gov/listeria/outbreaks/soft-cheeses-09-15/index.html (Accessed March 30, 2021).
- Centers of Disease Control and Prevention (CDC) (2015c). Multistate outbreak of listeriosis linked to commercially produced, prepackaged caramel apples made From Bidart Bros. Apples (Final Update); Centers for Disease Control and Prevention: Atlanta, GA, USA, 2015. Available at: https://www.cdc.gov/listeria/outbreaks/caramel-apples-12-14/index.html (Accessed March 25, 2021).
- Centers of Disease Control and Prevention (CDC) (2016a). Multistate outbreak of listeriosis linked to raw milk produced by miller’s organic farm in Pennsylvania (Final Update). Available at: https://www.cdc.gov/listeria/outbreaks/raw-milk-03-16/index.html (Accessed March 30, 2021).
- Centers of Disease Control and Prevention (CDC) (2016b). Multistate Outbreak of Listeriosis Linked to Frozen Vegetables (Final Update); Centers for Disease Control and Prevention: Atlanta, GA, USA, 2016. Available at: https://www.cdc.gov/listeria/outbreaks/frozen-vegetables-05-16/index.html (Accessed March 30, 2021).
- Centers of Disease Control and Prevention (CDC) (2016c). Multistate Outbreak of Listeriosis Linked to Packaged Salads Produced at Springfield, Ohio Dole Processing Facility (Final Update); Centers for Disease Control and Prevention: Atlanta, GA, USA, 2016. Available at: https://www.cdc.gov/listeria/outbreaks/bagged-salads-01-16/index.html (Accessed March 30, 2021).
- Centers of Disease Control and Prevention (CDC) (2017). Multistate Outbreak of Listeriosis Linked to Soft Raw Milk Cheese Made by Vulto Creamery (Final Update); Centers for Disease Control and Prevention: Atlanta, GA, USA, 2017. Available at: https://www.cdc.gov/listeria/outbreaks/soft-cheese-03-17/index.html (Accessed April 1, 2021).
- Centers of Disease Control and Prevention (CDC) (2018). Outbreak of Listeria Infections Linked to Deli Ham (Final Update); Centers for Disease Control and Prevention: Atlanta, GA, USA, 2018. Available at: https://www.cdc.gov/listeria/outbreaks/countryham-10-18/index.html (Accessed April 1, 2021).
- Centers of Disease Control and Prevention (CDC) (2019a). Outbreak of Listeria Infections Linked to Deli-Sliced Meats and Cheeses; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019. Available at: https://www.cdc.gov/listeria/outbreaks/deliproducts-04-19/index.html (Accessed April 1, 2021).
- Centers of Disease Control and Prevention (CDC) (2019b). Outbreak of Listeria Infections Linked to Pork Products. Available at: https://www.cdc.gov/listeria/outbreaks/porkproducts-11-18/index.html (Accessed April 1, 2021).
- Centers of Disease Control and Prevention (CDC) (2020a). Outbreak of Listeria infections linked to hard-boiled eggs. Available at: https://www.cdc.gov/listeria/outbreaks/eggs-12-19/index.html (Accessed April 1, 2021).
- Centers of Disease Control and Prevention (CDC) (2020b). Outbreak of Listeria infections linked to enoki mushrooms. Available at: https://www.cdc.gov/listeria/outbreaks/enoki-mushrooms-03-20/index.html (Accessed April 1, 2021).
- Centers of Disease Control and Prevention (CDC) (2021a). Outbreak of Listeria Infections Linked to Deli Meats. Available at: https://www.cdc.gov/listeria/outbreaks/delimeat-10-20/index.html (Accessed April 1, 2021).
- Centers of Disease Control and Prevention (CDC) (2021b). Listeria Outbreak Linked to Queso Fresco Made by El Abuelito Cheese Inc. Available at: https://www.cdc.gov/listeria/outbreaks/hispanic-soft-cheese-02-21/index.html (Accessed April 1, 2021).
- Centers of Disease Control and Prevention (CDC) (2012b). Multistate outbreak of listeriosis linked to imported frescolina marte brand ricotta salata cheese (Final Update); Centers for Disease Control and Prevention: Atlanta, GA, USA. Available at: https://www.cdc.gov/listeria/outbreaks/cheese-09-12/index.html (Accessed April 1, 2021).
- Centers of Disease Control and Prevention (CDC) (2015a). Multistate outbreak of listeriosis linked to blue bell creameries products (Final Update); Centers for Disease Control and Prevention: Atlanta, GA, USA. Available at: https://www.cdc.gov/listeria/outbreaks/ice-cream-03-15/index.html (Accessed March 30, 2021).
- Chan Y. C., Raengpradub S., Boor K. J., Wiedmann M. (2007). Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl. Environ. Microbiol. 73, 6484–6498. 10.1128/AEM.00897-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihib N.-E., da Silva M. R., Delattre G., Laroche M., Federighi M. (2003). Different cellular fatty acid pattern behaviours of two strains of Listeria monocytogenes Scott A and CNL 895807 under different temperature and salinity conditions. FEMS Microbiol. Lett. 218, 155–160. 10.1111/j.1574-6968.2003.tb11512.x, PMID: [DOI] [PubMed] [Google Scholar]
- Chikindas M. L., Weeks R., Drider D., Chistyakov V. A., Dicks L. M. T. (2018). Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 49, 23–28. 10.1016/j.copbio.2017.07.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J. K., Nielsen J. S., Ebersbach T., Valentin-Hansen P., Sogaard-Andersen L., Kallipolitis B. H. (2006). Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 12, 1383–1396. 10.1261/rna.49706, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B., Curtis N., Cotter P. D., Hill C., Ross R. P. (2010b). The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various β-lactam antibiotics. Antimicrob. Agents Chemother. 54, 4416–4423. 10.1128/AAC.00503-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B., Guinane C. M., Cotter P. D., Hill C., Ross P. R. (2012). Assessing the contributions of the lias histidine kinase to the innate resistance of Listeria monocytogenes to nisin, cephalosporins, and disinfectants. Appl. Environ. Microbiol. 78, 2923–2929. 10.1128/AEM.07402-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B., Joyce S., Hill C., Cotter P. D., Ross R. P. (2010a). TelA contributes to the innate resistance of Listeria monocytogenes to nisin and other cell wallacting antibiotics. Antimicrob. Agents Chemother. 54, 4658–4663. 10.1128/AAC.00290-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P. D., Ryan S., Gahan C. G., Hill C. (2005). Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71, 2832–2839. 10.1128/AEM.71.6.2832-2839.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S. J., Evans M. R. W., Willby D., Bardwell J., Chamberlain N., Lewis D. A. (2006). Listeria outbreak associated with sandwich consumption from a hospital retail shop. Euro Surveill. 11, 89–91. PMID: [PubMed] [Google Scholar]
- Desmond E. (2006). Reducing salt: a challenge for the meat industry. Meat Sci. 74, 188–196. 10.1016/j.meatsci.2006.04.014, PMID: [DOI] [PubMed] [Google Scholar]
- Deutscher J., Moussan Désirée Aké F., Zebre A., Nguyen Cao T., Kentache T., Mai Ma Pham Q., et al. (2014). “Carbohydrate utilization by Listeria monocytogenes and its influence on virulence gene expression,” in Listeria monocytogenes: Food sources, prevalence and management strategies. ed. Edmund C. Hambrick. (Hauppauge, New York, USA: Nova Science Publishers; ), 49–76. [Google Scholar]
- Duché O., Trémoulet F., Glaser P., Labadie J. (2002). Salt stress proteins induced in Listeria monocytogenes. Appl. Environ. Microbiol. 68, 1491–1498. 10.1128/AEM.68.4.1491-1498.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussurget O., Dumas E., Archambaud C., Chafsey I., Chambon C., Hébraud M., et al. (2005). Listeria monocytogenes ferritin protects against multiple stresses and is required for virulence. FEMS Microbiol. Lett. 250, 253–261. 10.1016/j.femsle.2005.07.015, PMID: [DOI] [PubMed] [Google Scholar]
- Duze S. T., Marimani M., Patel M. (2021). Tolerance of Listeria monocytogenes to biocides used in food processing environments. Food Microbiol. 97:103758. 10.1016/j.fm.2021.103758, PMID: [DOI] [PubMed] [Google Scholar]
- Efenberger-Szmechtyk M., Gałązka-Czarnecka I., Otlewska A., Czyżowska A., Nowak A. (2021). Aronia melanocarpa (Michx.) Elliot, Chaenomeles superba Lindl. And Cornus mas L. leaf extracts as natural preservatives for pork meat products. Molecules 26:3009. 10.3390/molecules26103009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhanafi D., Dutta V., Kathariou S. (2010). Genetic characterization of plasmid associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998-1999 outbreak. Appl. Environ. Microbiol. 76, 8231–8238. 10.1128/AEM.02056-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ells T. C., Truelstrup Hansen L. (2011). Increased thermal and osmotic stress resistance in Listeria monocytogenes 568 grown in the presence of trehalose due to inactivation of the phosphotrehalase-encoding gene treA. Appl. Environ. Microbiol. 77, 6841–6851. 10.1128/AEM.00757-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control (ECDC) (2018a). Epidemiological update: multi-country outbreak of Listeria monocytogenes serogroup IVb, multi-locus sequence type 6 infections. Available at: https://www.ecdc.europa.eu/en/news-events/epidemiological-update-multi-country-outbreak-listeria-monocytogenes-serogroup-ivb (Accessed March 20, 2021).
- European Centre for Disease Prevention and Control (ECDC) (2018b). Multi-country outbreak of Listeria monocytogenes infections linked to consumption of salmon products. Available at: https://www.ecdc.europa.eu/en/news-events/multi-country-outbreak-listeria-monocytogenes-infections-linked-consumption-salmon (Accessed March 20, 2021).
- European Centre for Disease Prevention and Control (ECDC) (2019a). Rapid outbreak assessment: Multi-country outbreak of Listeria monocytogenes sequence type 6 infections linked to ready-to-eat meat products. Available at: https://www.ecdc.europa.eu/en/publications-data/rapid-outbreak-assessment-multi-country-outbreak-listeria-monocytogenes-sequence (Accessed March 20, 2021).
- European Centre for Disease Prevention and Control (ECDC) (2019b). Multi-country outbreak of Listeria monocytogenes clonal complex 8 infections linked to consumption of cold-smoked fish products. Available at: https://www.ecdc.europa.eu/en/publications-data/multi-country-outbreak-listeria-monocytogenes-fish-products (Accessed March 20, 2021).
- Faezi-Ghasemi M., Kazemi S. (2015). Effect of sub-lethal environmental stresses on the cell survival and antibacterial susceptibility of Listeria monocytogenes PTCC1297. Zahedan J. Res. Med. Sci. 17, 1–6. [Google Scholar]
- Ferreira A., Sue D., O’Byrne C. P., Boor K. J. (2003). Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69, 2692–2698. 10.1128/AEM.69.5.2692-2698.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox E. M., Leonard N., Jordan K. (2011). Physiological and transcriptional characterization of persistent and nonpersistent Listeria monocytogenes isolates. Appl. Environ. Microbiol. 77, 6559–6569. 10.1128/AEM.05529-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraqueza M. J., Laranjo M., Elias M., Patarata L. (2021). Microbiological hazards associated with salt and nitrite reduction in cured meat products: control strategies based on antimicrobial effect of natural ingredients and protective microbiota. Curr. Opin. Food Sci. 38, 32–39. 10.1016/j.cofs.2020.10.027 [DOI] [Google Scholar]
- Gaballa A., Guariglia-Oropeza V., Wiedmann M., Boor K. J. (2019). Cross talk between SigB and PrfA in Listeria monocytogenes facilitates transitions between extra- and intracellular environments. Microbiol. Mol. Biol. Rev. 83, e00034–e00019. 10.1128/MMBR.00034-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M., Chikindas L. M. (2007). Listeria: A foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 113, 1–15. 10.1016/j.ijfoodmicro.2006.07.008, PMID: [DOI] [PubMed] [Google Scholar]
- Gao R., Stock A. M. (2009). Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63, 133–154. 10.1146/annurev.micro.091208.073214, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaul L. K., Farag N. H., Shim T., Kingsley M. A., Silk B. J., Hyytia-Trees E. (2012). Hospital-acquired listeriosis outbreak caused by contaminated diced celery—Texas, 2010. Clin. Infect. Dis. 56, 20–26. 10.1093/cid/cis817, PMID: [DOI] [PubMed] [Google Scholar]
- Giotis E. S., Muthaiyan A., Natesan S., Wilkinson B. J., Blair I. S., McDowell D. A. (2010). Transcriptome analysis of alkali shock and alkali adaptation in Listeria monocytogenes 10403S. Foodborne Pathog. Dis. 7, 1147–1157. 10.1089/fpd.2009.0501, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadorn K., Hächler H., Schaffner A., Kayser F. H. (1993). Genetic characterization of plasmid-encoded multiple antibiotic resistance in a strain of Listeria monocytogenes causing endocarditis. Eur. J. Hum. Genet. 12, 928–937. 10.1007/BF01992167, PMID: [DOI] [PubMed] [Google Scholar]
- Harvey J., Gilmour A. (2001). Characterization of recurrent and sporadic Listeria monocytogenes isolates from raw milk and nondairy foods by pulsed-field gel electrophoresis, monocin typing, plasmid profiling, and cadmium and antibiotic resistance determination. Appl. Environ. Microbiol. 67, 840–847. 10.1128/AEM.67.2.840-847.2001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubert L., Zehetmeyr M. L., da Silva W. P. (2019). Resistance to benzalkonium chloride and cadmium chloride in Listeria monocytogenes isolates from food and foodprocessing environments in southern Brazil. Can. J. Microbiol. 65, 429–435. 10.1139/cjm-2018-0618, PMID: [DOI] [PubMed] [Google Scholar]
- Hingston P., Brenner T., Hansen L. T., Wan S. (2019). Comparative analysis of Listeria monocytogenes plasmids and expression levels of plasmid-encoded genes during growth under salt and acid stress conditions. Toxins 11:426. 10.3390/toxins11070426, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingston P., Chen J., Dhillon B. K., Laing C., Bertelli C., Gannon V., et al. (2017). Genotypes associated with Listeria monocytogenes isolates displaying impaired or enhanced tolerances to cold, salt, acid, or desiccation stress. Front. Microbiol. 8:369. 10.3389/fmicb.2017.00369, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley D., Luque-Sastre L., Parker C. T., Huynh S., Eshwar A. K., Nguyen S. V., et al. (2019). Whole-genome sequencing-based characterization of 100 Listeria monocytogenes isolates collected from food processing environments over a four-year period. mSphere 4, e00252–e00219. 10.1128/mSphere.00252-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Yu T., Xu Y., Wang H., Korkeala H., Shi L. (2019). MdrL, a major facilitator superfamily efflux pump of Listeria monocytogenes involved in tolerance to benzalkonium chloride. Appl. Microbiol. Biotechnol. 103, 1339–1350. 10.1007/s00253-018-9551-y, PMID: [DOI] [PubMed] [Google Scholar]
- Kocaman N., Sarimehmetoğlu B. (2016). Stress responses of Listeria monocytogenes. Ankara Üniv Vet Fak Derg. 63, 421–427. [Google Scholar]
- Koutsoumanis K. P., Kendall P. A., Sofos J. N. (2003). Effect of food processing-related stresses on acid tolerance of Listeria monocytogenes. Appl. Environ. Microbiol. 69, 7514–7516. 10.1128/AEM.69.12.7514-7516.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic J., Ziegler J., Walecka-Zacharska E., Reimer A., Kitts D. D., Gilmour M. W. (2016). Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 82, 939–953. 10.1128/AEM.03741-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer P. H. C., Lees J. A., Koopmans M. M., Ferwerda B., Arends A. W. M., Feller M. M., et al. (2017). Benzalkonium tolerance genes and outcome in Listeria monocytogenes meningitis. Clin. Microbiol. Infect. 23, 265.e1–265.e7. 10.1016/j.cmi.2016.12.008e1-265.e7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropac A. C., Eshwar A. K., Stephan R., Tasara T. (2019). New insights on the role of the pLMST6 plasmid in Listeria monocytogenes biocide tolerance and virulence. Front. Microbiol. 10:1538. 10.3389/fmicb.2019.01538, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Jeong K. W., Kim Y. M. (2012). Purification and structural characterization of cold shock protein from Listeria monocytogenes. Bull. Kor. Chem. Soc. 33, 2508–2512. 10.5012/bkcs.2012.33.8.2508 [DOI] [Google Scholar]
- Lianou A., Sofos J. N. (2007). A review of the incidence and transmission of Listeria monocytogenes in ready-to-eat products in retail and food service environments. J. Food Prot. 70, 2172–2198. 10.4315/0362-028X-70.9.2172, PMID: [DOI] [PubMed] [Google Scholar]
- Liu Y., Orsi R. H., Gaballa A., Wiedmann M., Boor K. J., Guariglia-Oropeza V. (2019). Systematic review of the Listeria monocytogenes σB regulon supports a role in stress response, virulence and metabolism. Future Microbiol. 14, 801–828. 10.2217/fmb-2019-0072, PMID: [DOI] [PubMed] [Google Scholar]
- Lou Y., Yousef A. E. (1997). Adaptation to sub-lethal environmental stresses protects Listeria monocytogenes against lethal preservation factors. Appl. Environ. Microbiol. 63, 1252–1255. 10.1128/AEM.63.4.1252-1255.1997, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lungu B., Ricke S. C., Johnson M. G. (2009). Growth, survival, proliferation and pathogenesis of Listeria monocytogenes under low oxygen or anaerobic conditions. Anaerobe 15, 7–17. 10.1016/j.anaerobe.2008.08.001, PMID: [DOI] [PubMed] [Google Scholar]
- Macwana S., Muriana P. M. (2012). Spontaneous bacteriocin resistance in Listeria monocytogenes as a susceptibility screen for identifying different mechanisms of resistance and modes of action by bacteriocins of lactic acid bacteria. J. Microbiol. Methods 88, 7–13. 10.1016/j.mimet.2011.09.009, PMID: [DOI] [PubMed] [Google Scholar]
- Manso B., Melero B., Stessl B., Jaime I., Wagner M., Rovira J., et al. (2020). The response to oxidative stress in Listeria monocytogenes is temperature dependent. Microorganisms 8:521. 10.3390/microorganisms8040521, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronicolis S. K., Berberi A., Diakogiannis I., Petrova E., Kiaki I., Baltzi T., et al. (2010). Alteration of the phospho- or neutral lipid content and fatty acid composition in Listeria monocytogenes due to acid adaptation mechanisms for hydrochloric, acetic and lactic acids at pH 5.5 or benzoic acid at neutral pH. Antonie Van Leeuwenhoek 98, 307–316. 10.1007/s10482-010-9439-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata M. T., Baquero F., P’erez-Díaz J. C. (2000). A multidrug efflux transporter in Listeria monocytogenes. FEMS Microbiol. Lett. 15, 185–188. 10.1016/S0378-1097(00)00199-3, PMID: [DOI] [PubMed] [Google Scholar]
- Matereke L. T., Okoh A. I. (2020). Listeria monocytogenes virulence, antimicrobial resistance and environmental persistence: a review. Pathogens 9:528. 10.3390/pathogens9070528, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlin J., Hampton M. D. D., Shah S., Threlfall E. J. J., Wieneke A. A. A., Curtis G. D. W. D. W. (1997). Subtyping of Listeria monocytogenes on the basis of plasmid profiles and arsenic and cadmium susceptibility. J. Appl. Microbiol. 83, 381–388. 10.1046/j.1365-2672.1997.00238.x, PMID: [DOI] [PubMed] [Google Scholar]
- Meier A. B., Guldimann C., Markkula A., Pöntinen A., Korkeala H., Tasara T. (2017). Comparative phenotypic and genotypic analysis of Swiss and Finnish Listeria monocytogenes isolates with respect to benzalkonium chloride resistance. Front. Microbiol. 8:397. 10.3389/fmicb.2017.00397, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J., Andrew P. W., Faleiro M. L. (2015). Listeria monocytogenes in cheese and the dairy environment remains a food safety challenge: the role of stress responses. Food Res. Int. 67, 75–90. 10.1016/j.foodres.2014.10.031 [DOI] [Google Scholar]
- Mendum M. L., Smith L. T. (2002). Characterization of glycine betaine porter I from Listeria monocytogenes and its roles in salt and chill tolerance. Appl. Environ. Microbiol. 68, 813–819. 10.1128/AEM.68.2.813-819.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miladi H., Elabed H., Slama R. B., Rhim A., Bakhrouf A. (2017). Molecular analysis of the role of osmolyte transporters opuCA and betL in Listeria monocytogenes after cold and freezing stress. Arch. Microbiol. 199, 259–265. 10.1007/s00203-016-1300-y, PMID: [DOI] [PubMed] [Google Scholar]
- Minarovicova J., Veghova A., Mikulasova M., Chovanova R., Soltys K., Drahovska H., et al. (2018). Benzalkonium chloride tolerance of Listeria monocytogenes strains isolated from a meat processing facility is related to presence of plasmidborne bcrABC cassette. Int. J. Gen. Mol. Microbiol. 111, 1913–1923. 10.1007/s10482-018-1082-0, PMID: [DOI] [PubMed] [Google Scholar]
- Ming X., Daeschel M. A. (1993). Nisin resistance of foodborne bacteria and the specific resistance responses of Listeria monocytogenes Scott A. J. Food Prot. 56, 944–948. 10.4315/0362-028X-56.11.944, PMID: [DOI] [PubMed] [Google Scholar]
- Mohammed A. N., Abdel S., Abdel A. (2017). Research article ecological study on Listeria monocytogenes and the extent of its resistance to different disinfectants in dairy farm for improving animal health. Asian J. Anim. Vet. Adv. 12, 302–310. 10.3923/ajava.2017.302.310 [DOI] [Google Scholar]
- Møretrø T., Schirmer B. C. T., Heir E., Fagerlund A., Hjemli P., Langsrud S. (2017). Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry. Int. J. Food Microbiol. 241, 215–224. 10.1016/j.ijfoodmicro.2016.10.025, PMID: [DOI] [PubMed] [Google Scholar]
- Naditz A. L., Dzieciol M., Wagner M., Schmitz-Esser S. (2019). Plasmids contribute to food processing environment–associated stress survival in three Listeria monocytogenes ST121, ST8, and ST5 strains. Int. J. Food Microbiol. 299, 39–46. 10.1016/j.ijfoodmicro.2019.03.016, PMID: [DOI] [PubMed] [Google Scholar]
- Nair S., Milohanic E., Berche P. (2000). ClpC ATPase is required for cell adhesion and invasion of Listeria monocytogenes. Infect. Immun. 68, 7061–7068. 10.1128/IAI.68.12.7061-7068.2000, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- NicAogain K., O’Byrne C. P. (2016). The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front. Microbiol. 7:1865. 10.3389/fmicb.2016.01865, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Trunzer K., Vondran A., Vincze S., Dieckmann R., Al Dahouk S., et al. (2020). Benzalkonium chloride induces a VBNC state in Listeria monocytogenes. Microorganisms 8:184. 10.3390/microorganisms8020184, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Byrne C. P., Karatzas K. A. (2008). The role of sigmaB (σB) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv. Appl. Microbiol. 65, 115–140. 10.1016/S0065-2164(08)00605-9, PMID: [DOI] [PubMed] [Google Scholar]
- O'Neil S. H., Marquis H. (2006). Listeria monocytogenes flagella are used for motility, not as adhesins to increase host cell invasion. Infect. Immun. 74, 6675–6681. 10.1128/IAI.00886-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi R. H., Chaturongakul S., Oliver H. F., Ponnala L., Gaballa A., Wiedmann M. (2021). Alternative factors regulate overlapping as well as distinct stress response and metabolic functions in Listeria monocytogenes under stationary phase stress condition. Pathogens 10:411. 10.3390/pathogens10040411, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz S., López-Alonso V., Rodríguez P., Martínez-Suárez J. V. (2016). The connection between persistent, disinfectant-resistant Listeria monocytogenes strains from two geographically separate Iberian pork processing plants: evidence from comparative genome analysis. Appl. Environ. Microbiol. 82, 308–317. 10.1128/AEM.02824-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma F., Brauge T., Radomski N., Mallet L., Felten A., Mistou M. Y., et al. (2020). Dynamics of mobile genetic elements of Listeria monocytogenes persisting in ready-to-eat seafood processing plants in France. BMC Genomics 21:130. 10.1186/s12864-020-6544-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöntinen A., Aalto-Araneda M., Lindström M., Korkeala H. (2017). Heat resistance mediated by pLM58 plasmid-borne ClpL in Listeria monocytogenes. mSphere 2, e00364–e00317. 10.1128/mSphere.00364-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premaratne R. J., Lin W., Johnson E. A. (1991). Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57, 3046–3048. 10.1128/aem.57.10.3046-3048.1991, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic-Martinez M., Drevets D. A., Dutta V., Katic V., Kathariou S. (2011). Listeria monocytogenes strains selected on ciprofloxacin or the disinfectant benzalkonium chloride exhibit reduced susceptibility to ciprofloxacin, gentamicin, benzalkonium chloride, and other toxic compounds. Appl. Environ. Microbiol. 77, 8714–8721. 10.1128/AEM.05941-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratani S. S., Siletzky R. M., Dutta V., Yildirim S., Osborne J. A., Lin W., et al. (2012). Heavy metal and disinfectant resistance of Listeria monocytogenes from foods and food processing plants. Appl. Environ. Microbiol. 78, 6938–6945. 10.1128/AEM.01553-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea R., Hill C., Gahan C. G. (2005). Listeria monocytogenes PerR mutants display a small-colony phenotype, increased sensitivity to hydrogen peroxide, and significantly reduced murine virulence. Appl. Environ. Microbiol. 71, 8314–8322. 10.1128/AEM.71.12.8314-8322.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier S., Hebraud M., Desvaux M. (2011). Molecular biology of surface colonization by Listeria monocytogenes: an additional facet of an opportunistic gram-positive foodborne pathogen. Environ. Microbiol. 13, 835–850. 10.1111/j.1462-2920.2010.02378.x, PMID: [DOI] [PubMed] [Google Scholar]
- Ribeiro V. B., Mujahid S., Orsi R. H., Bergholz T. M., Wiedmann M., Boor K. J., et al. (2014). Contributions of σB and PrfA to Listeria monocytogenes salt stress under food relevant conditions. Int. J. Food Microbiol. 177, 98–108. 10.1016/j.ijfoodmicro.2014.02.018, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos Colombo N. S., Chalón M. C., Navarro S. A., Bellomio A. (2018). Pediocin-like bacteriocins: new perspectives on mechanism of action and immunity. Curr. Genet. 64, 345–351. 10.1007/s00294-017-0757-9, PMID: [DOI] [PubMed] [Google Scholar]
- Ripio M. T., Vaèzquez-Boland J.-A., Vega Y., Nair S., Berche P. (1998). Evidence for expressional crosstalk between the central virulence regulator PrfA and the stress response mediator ClpC in Listeria monocytogenes. FEMS Microbiol. Lett. 158, 45–50. 10.1111/j.1574-6968.1998.tb12798.x, PMID: [DOI] [PubMed] [Google Scholar]
- Roedel A., Dieckmann R., Brendebach H., Hammerl J. A., Kleta S., Noll M., et al. (2019). Biocide-tolerant Listeria monocytogenes isolates from German food production plants do not show cross-resistance to clinically relevant antibiotics. Appl. Environ. Microbiol. 85, e01253–e01219. 10.1128/AEM.01253-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruusunen M., Puolanne E. (2005). Reducing sodium intake from meat products. Meat Sci. 70, 531–541. 10.1016/j.meatsci.2004.07.016, PMID: [DOI] [PubMed] [Google Scholar]
- Ryan S., Begley M., Gahan C. G. M., Hill C. (2009). Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11, 432–445. 10.1111/j.1462-2920.2008.01782.x, PMID: [DOI] [PubMed] [Google Scholar]
- Ryan S., Hill C., Gahan C. G. (2008). Acid stress responses in Listeria monocytogenes. Adv. Appl. Microbiol. 65, 67–91. 10.1016/S0065-2164(08)00603-5, PMID: [DOI] [PubMed] [Google Scholar]
- Schmid B., Klumpp J., Raimann E., Loessner M. J., Stephan R., Tasara T. (2009). Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl. Environ. Microbiol. 75, 1621–1627. 10.1128/AEM.02154-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B., Cai Y.-Z., Brooks J. D., Corke H. (2009). Antibacterial and antioxidant effects of five spice and herb extracts as natural preservatives of raw pork. J. Sci. Food Agric. 89, 1879–1885. 10.1002/jsfa.3667 [DOI] [Google Scholar]
- Silva C. C. G., Silva S. P. M., Ribeiro S. C. (2018). Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 9:594. 10.3389/fmicb.2018.00594, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleator R. D., Hill C. (2002). Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26, 49–71. 10.1111/j.1574-6976.2002.tb00598.x, PMID: [DOI] [PubMed] [Google Scholar]
- Smelt J. P. P. M., Brul S. (2014). Thermal inactivation of microorganisms. Crit. Rev. Food Sci. Nutr. 54, 1371–1385. 10.1080/10408398.2011.637645, PMID: [DOI] [PubMed] [Google Scholar]
- Smith J. L., Liu Y., Paoli G. C. (2013). How does Listeria monocytogenes combat acid conditions? Can. J. Microbiol. 59, 141–152. 10.1139/cjm-2012-0392, PMID: [DOI] [PubMed] [Google Scholar]
- Smith A. M., Tau N. P., Smouse S. L., Allam M., Ismail A., Ramalwa N. R., et al. (2019). Outbreak of Listeria monocytogenes in South Africa, 2017-2018: laboratory activities and experiences associated with whole-genome sequencing analysis of isolates. Foodborne Pathog. Dis. 16, 524–530. 10.1089/fpd.2018.2586, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares C. A., Knuckley B. (2016). Mechanistic studies of the agmatine deiminase from Listeria monocytogenes. Biochem. J. 473, 1553–1561. 10.1042/BCJ20160221, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni K. A., Nannapaneni R., Tasara T. (2011). The contribution of transcriptomic and proteomic analysis in elucidating stress adaptation responses of Listeria monocytogenes. Foodborne Pathog. Dis. 8, 842–852. 10.1089/fpd.2010.0746, PMID: [DOI] [PubMed] [Google Scholar]
- Stoll R., Goebel W. (2010). The major PEP-phosphotransferase systems (PTSs) for glucose, mannose and cellobiose of Listeria monocytogenes, and their significance for extra- and intracellular growth. Microbiology 156, 1069–1083. 10.1099/mic.0.034934-0, PMID: [DOI] [PubMed] [Google Scholar]
- Tapia N. C., Dorey A. L., Gahan C. G. M., den Besten H. M. W., O'Byrne C. P., Abee T. (2020). Different carbon sources result in differential activation of sigma B and stress resistance in Listeria monocytogenes. Int. J. Food Microbiol. 320:108504. 10.1016/j.ijfoodmicro.2019.108504, PMID: [DOI] [PubMed] [Google Scholar]
- Thedieck K., Hain T., Mohamed W., Tindall B. J., Nimtz M., Chakraborty T., et al. (2006). The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 62, 1325–1339. 10.1111/j.1365-2958.2006.05452, PMID: [DOI] [PubMed] [Google Scholar]
- Tongnuanchan P., Benjakul S. (2014). Essential oils: extraction, bioactivities, and their uses for food preservation. J. Food Sci. 79, 1231–1249. 10.1111/1750-3841.12492, PMID: [DOI] [PubMed] [Google Scholar]
- Van Houdt R., Michiels C. W. (2010). Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 109, 1117–1131. 10.1111/j.1365-2672.2010.04756.x, PMID: [DOI] [PubMed] [Google Scholar]
- Werbrouck H., Vermeulen A., Van Coillie E., Messens W., Herman L., Devlieghere F., et al. (2009). Influence of acid stress on survival, expression of virulence genes and invasion capacity into Caco-2 cells of Listeria monocytogenes strains of different origins. Int. J. Food Microbiol. 134, 140–146. 10.1016/j.ijfoodmicro.2009.03.022, PMID: [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) . (2018a). Listeriosis. Available at: https://www.who.int/news-room/fact-sheets/detail/listeriosis (Accessed April 17, 2021).
- World Health Organization (WHO) (2018b). Listeriosis– Spain. Available at: https://www.who.int/csr/don/16-september-2019-listeriosis-spain/en/ (Accessed April 17, 2021).
- World Health Organization (WHO) . (2018c). Listeriosis—Australia. Available at: https://www.who.int/csr/don/09-april-2018-listeriosis-australia/en/ (Accessed April 17, 2021).
- Wouters J. A., Hain T., Darji A., Hüfner E., Wemekamp-Kamphuis H., Chakraborty T., et al. (2005). Identification and characterization of di- and tripeptide transporter DtpT of Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 71, 5771–5778. 10.1128/AEM.71.10.5771-5778.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Yu P.-L., Wheeler D., Flint S. (2018). Transcriptomic study on persistence and survival of Listeria monocytogenes following lethal treatment with nisin. J. Glob. Antimicrob. Resist. 15, 25–31. 10.1016/j.jgar.2018.06.003, PMID: [DOI] [PubMed] [Google Scholar]
- Yu T., Jiang X., Zhang Y., Ji S., Gao W., Shi L. (2018). Effect of benzalkonium chloride adaptation on sensitivity to antimicrobial agents and tolerance to environmental stresses in Listeria monocytogenes. Front. Microbiol. 9:2906. 10.3389/fmicb.2018.02906, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Hu Y., Zhou C., Yang Z., Wu L., Zhu M., et al. (2018). Stress resistance, motility and biofilm formation mediated by a 25kb plasmid pLMSZ08 in Listeria monocytogenes. Food Control 94, 345–352. 10.1016/j.foodcont.2018.07.002 [DOI] [Google Scholar]
- Zhou Q., Feng X., Zhang Q., Feng F., Yin X., Shang J., et al. (2012). Carbon catabolite control is important for Listeria monocytogenes biofilm formation in response to nutrient availability. Curr. Microbiol. 65, 35–43. 10.1007/s00284-012-0125-4, PMID: [DOI] [PubMed] [Google Scholar]