Main text
We read with great interest the Letter to the Editor entitled ‘“D” Matters in Recombinant AAV packaging’ by Zhang et al.,1 which was recently published in Molecular Therapy. These authors have made major contributions to the adeno-associated virus (AAV) field in the past, and the data presented in their Letter to the Editor are a fine example of their thoughtful analysis of the role of the D-sequence within the AAV inverted terminal repeats (ITRs) in AAV genome packaging. However, in contrast to the critical role that the D-sequence plays in the rescue, replication, and packaging of the single-stranded (ss) wild-type (WT) AAV genomes, based on their data with self-complementary (sc) recombinant AAV (rAAV) vectors containing a reporter gene, Zhang et al.1 concluded that the D-sequence does not matter much in the packaging of scAAV vectors. We wish to further emphasize the following important distinctions between the processing of scAAV vector versus the ss WT-AAV genomes.
-
(1)
Replication of the WT-AAV genome and production of progeny virions during a natural course of infection in the presence of a helper virus may not precisely mimic the rescue, replication, and packaging of the WT-AAV genome from a recombinant plasmid following DNA-mediated transfection.
-
(2)
Rescue, replication, and packaging of the WT-ssAAV genome from a recombinant plasmid, and those of either a ssAAV or a scAAV vector genome, are likely to be different following DNA-mediated transfection in the presence of helper virus genes or in helper virus-infected cells.
Here, we wish to offer an alternative explanation, which may help explain why the role of the D-sequence in the processing of the two genomes is more alike than it would appear, based on our long-term interest in AAV biology in general and in the AAV D-sequence in particular.2, 3, 4, 5, 6, 7, 8, 9 For example, in contrast to the data with a scAAV vector genome containing a reporter gene presented in the letter, we have previously reported that deletions of the D-sequence dramatically reduce the extent of rescue and replication of the WT-AAV genomes from recombinant plasmids (see Figure 4 in Wang et al.2) and that the rescued/replicated genomes fail to undergo encapsidation (see Figure 6 in Wang et al.3) because of failure to generate ssAAV progeny DNA strands (see Figure 7A in Wang et al.3). In subsequent studies, we generated mutations in the D-sequence in which the proximal 5, 10, or 15 nucleotides (nts) to the hairpin structure in the AAV-ITRs were deleted.4 The efficiency of rescue, replication, and packaging of WT-AAV genomes from recombinant plasmids were compared with that from WT-AAV genomes containing the full-length D-sequence (D20). These data documented, once again, that not only low-level rescue/replication occurred in the absence of the D-sequence, but also the inclusion of the proximal 5 nts (D5) significantly restored rescue and replication. It is also evident that, whereas inclusion of 5 nts of the D sequence augmented not only the rescue/replication of the WT-AAV genome from recombinant plasmid (see Figure 2 in Wang et al.4), but also the packaging, albeit at a lower efficiency (see Figure 3 in Wang et al.4).
Interestingly, there was no observable difference among the rescue, replication, and packaging of the AAV genomes containing the proximal 10 nts or the proximal 15 nts compared with those containing the full-length WT-D-sequence.4 Thus, we concluded that, whereas the distal 10 nts are dispensable, the proximal 10 nts of the D-sequence are the minimal requirement for the optimal rescue, replication, and packaging of the WT-ssAAV genomes. Interestingly, the presence of only one D-sequence in the viral genome is necessary and sufficient to mediate successful rescue, replication, and packaging of rAAV vectors.8
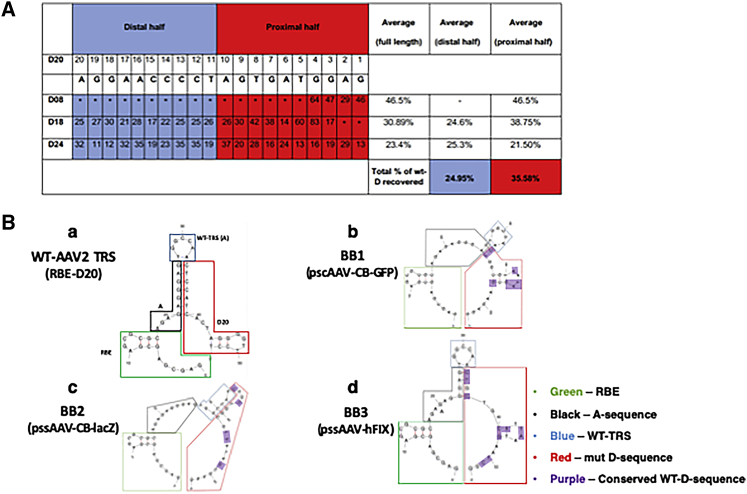
Zhang et al. also utilized a clever strategy in which they used three random nt libraries (D08, D18, and D24) in their studies of scAAV vectors and implied that the D-sequence is not explicitly needed for packaging into capsids. The random library experiment was performed with substitutions along three different truncations of the D-sequence and the first four nts of the A sequence. Their results indicated that the WT-D-sequence was only recovered 25% of the time, indicating a lax requirement of the specific sequence of the D-sequence.
Upon re-evaluation of these results, we noted that, whereas the distal 10 nts of the D-sequence were indeed reconstituted ∼25% of the time, the proximal 10 nts of the D-sequence were recovered ∼36% of the time (Figure 1A). This indicates an increased propensity for the conservation of the proximal 10 nts of the D-sequence over the distal 10 nts, which is consistent with our published studies.4
Figure 1.
Role of proximal half of D-sequence in rescue, replication, and packaging of AAV genomes
(A) Analysis of random sequence library results and recovery of the D-sequence in AAV progeny. Average recovery of the D-sequence represents the total average in the distal half (n = 20) and the proximal half (n = 22). (B) Schematic representations of partially recreated secondary structures of wild-type (wt)-TRS in AAV2 (a), BB1 (b), BB2 (c), and BB3 (d) sequences, containing the full-length A-sequence + wt D-sequence, and the pseudoD-sequences, respectively. Green outline, Rep-binding element (RBE); black, A-sequence; blue, wt-TRS; red, D/pseudoD; purple, represents the sequences with homology to the wt-D-sequence. All DNA-folding analyses were performed using the m-fold software.
Zhang et al.1 also argue that the presence of the “pseudo-D” elements present in the plasmid backbone mediate rescue and replication of the vector backbone, which accounts for the high level of contamination with plasmid backbone during AAV vector production. We have reported that, in the absence of the D-sequence, absolutely no packaging of the plasmid backbone occurs in AAV capsids (see Figure 7B in Wang et al.3). These data further support our contention that the D-sequence is not a lax requirement for packaging. Additionally, the titers of each vector containing these substitutions were reduced by at least 1 log, as compared with those containing the WT-D-sequence, indicating that the presence of the WT-D-sequence dramatically increases the packaging efficiency of AAV vectors. Furthermore, the other constructs were packaged at the same rate as the backbone of the WT-D-sequence, indicating that there is an imperative need for the WT-D-sequence to yield meaningful titers of rAAV vectors.
Since it is well-known that during Rep-mediated resolution the AAV genome, the Rep-complex generates a secondary hairpin structure exposing the terminal resolution site (TRS) for catalysis.10 The stem of this structure is stabilized by partial base-pairing between the A sequence and the first half of the D-sequence (D10).11 We reasoned, therefore, that this partial homology between the A- and the D-sequence, and the secondary structure thus generated, is the ultimate implication of the D-sequence in rescue/replication, which is supported by the minimal need of the proximal D10 sequence and the partial rescue in the D5 construct described above. Thus, rather than the specific sequence of the WT-D being the requirement, it is the consequential secondary structure that the proximal half of the D-sequence, the TRS, and the A-sequence that is the requirement for successful rescue and replication.
We next analyzed the three pseudo-D substitution sequences (BB1, BB2, and BB3) using a DNA-folding software (m-fold) to compare the secondary structure of each and compared with that of the WT-D-sequence. These structures are depicted in Figure 1B. As can be seen, whereas both BB1 and BB3 partially recreated the WT-TRS, BB3 generated a “TRS-like” site in the close vicinity of the WT-TRS.
These partial reconstitutions of secondary structure needed for TRS-exposure and Rep-mediated resolution might be the primary reason for the observed packaging of the transgene and backbone giving these sequences D-like characteristics rather than being an arbitrary replacement. When the D-sequence is either replaced with a sequence that has no partial base-pairing homology to the A sequence or is ablated altogether, there is no rescue/replication or encapsulation. Additionally, when the TRS site is partially reconstituted, as it was in D5, there is partial rescue/replication. In this context, it is noteworthy that “TRS-like” sites also exist in the WT-AAV genome and can and do mediate successful rescue and autonomous replication of the WT-AAV genome.12,13
In summary, we postulate that Rep-mediated resolution at non-canonical sites is the result of secondary structural improvisations at the “TRS-like” sites that have sequence homology to the WT-TRS site that are flanked by homologous sequences that can base-pair with one another, forming the stem-loop needed for Rep-mediated resolution, and that the D-sequence has evolved to be the most biologically efficient iteration of this design with respect to the WT-ITR. Thus, as we have surmised previously, although the precise mechanism of AAV genome encapsidation still remains a puzzle, rather than being a lax requirement, the D-sequence, in addition to Rep proteins, plays a critical role in AAV genome encapsidation.14
References
- 1.Zhang J., Guo P., Xu Y., Mulcrone P.L., Samulski R.J., Xiao W. “D” matters in recombinant AAV DNA packaging. Mol. Ther. 2021;29:1937–1939. doi: 10.1016/j.ymthe.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X.S., Ponnazhagan S., Srivastava A. Rescue and replication signals of the adeno-associated virus 2 genome. J. Mol. Biol. 1995;250:573–580. doi: 10.1006/jmbi.1995.0398. [DOI] [PubMed] [Google Scholar]
- 3.Wang X.S., Ponnazhagan S., Srivastava A. Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: selective encapsidation of viral genomes in progeny virions. J. Virol. 1996;70:1668–1677. doi: 10.1128/jvi.70.3.1668-1677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X.S., Qing K., Ponnazhagan S., Srivastava A. Adeno-associated virus type 2 DNA replication in vivo: mutation analyses of the D sequence in viral inverted terminal repeats. J. Virol. 1997;71:3077–3082. doi: 10.1128/jvi.71.4.3077-3082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qing K., Wang X.S., Kube D.M., Ponnazhagan S., Bajpai A., Srivastava A. Role of tyrosine phosphorylation of a cellular protein in adeno-associated virus 2-mediated transgene expression. Proc. Natl. Acad. Sci. USA. 1997;94:10879–10884. doi: 10.1073/pnas.94.20.10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qing K., Khuntirat B., Mah C., Kube D.M., Wang X.S., Ponnazhagan S., Zhou S., Dwarki V.J., Yoder M.C., Srivastava A. Adeno-associated virus type 2-mediated gene transfer: correlation of tyrosine phosphorylation of the cellular single-stranded D sequence-binding protein with transgene expression in human cells in vitro and murine tissues in vivo. J. Virol. 1998;72:1593–1599. doi: 10.1128/jvi.72.2.1593-1599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong L., Qing K., Si Y., Chen L., Tan M., Srivastava A. Heat-shock treatment-mediated increase in transduction by recombinant adeno-associated virus 2 vectors is independent of the cellular heat-shock protein 90. J. Biol. Chem. 2004;279:12714–12723. doi: 10.1074/jbc.M310548200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling C., Wang Y., Lu Y., Wang L., Jayandharan G.R., Aslanidi G.V., Li B., Cheng B., Ma W., Lentz T. Enhanced transgene expression from recombinant single-stranded D-sequence-substituted adeno-associated virus vectors in human cell lines in vitro and in murine hepatocytes in vivo. J. Virol. 2015;89:952–961. doi: 10.1128/JVI.02581-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon H.-J., Qing K., Ponnazhagan S., Wang X.S., Markusic D.M., Gupte S., Boye S.E., Srivastava A. Adeno-associated virus D-sequence-mediated suppression of expression of a human major histocompatibility class II gene: Implications in the development of adeno-associated virus vectors for modulating humoral immune response. Hum Gene Ther. 2020;31:565–574. doi: 10.1089/hum.2020.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder R.O., Im D.S., Ni T., Xiao X., Samulski R.J., Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J. Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan J.H., Zolotukhin S., Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J. Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X.S., Srivastava A. A novel terminal resolution-like site in the adeno-associated virus type 2 genome. J. Virol. 1997;71:1140–1146. doi: 10.1128/jvi.71.2.1140-1146.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X.S., Srivastava A. Rescue and autonomous replication of adeno-associated virus type 2 genomes containing Rep-binding site mutations in the viral p5 promoter. J. Virol. 1998;72:4811–4818. doi: 10.1128/jvi.72.6.4811-4818.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling C., Wang Y., Lu Y., Wang L., Jayandharan G.R., Aslanidi G.V., Li B., Cheng B., Ma W., Lentz T. The adeno-associated virus genome packaging puzzle. J. Mol. Genet. Med. 2015;9:175. doi: 10.4172/1747-0862.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]