Abstract
We present a live-attenuated RNA hybrid vaccine technology that uses an RNA vaccine delivery vehicle to deliver in vitro-transcribed, full-length, live-attenuated viral genomes to the site of vaccination. This technology allows ready manufacturing in a cell-free environment, regardless of viral attenuation level, and it promises to avoid many safety and manufacturing challenges of traditional live-attenuated vaccines. We demonstrate this technology through development and testing of a live-attenuated RNA hybrid vaccine against Chikungunya virus (CHIKV), comprised of an in vitro-transcribed, highly attenuated CHIKV genome delivered by a highly stable nanostructured lipid carrier (NLC) formulation as an intramuscular injection. We demonstrate that single-dose immunization of immunocompetent C57BL/6 mice results in induction of high CHIKV-neutralizing antibody titers and protection against mortality and footpad swelling after lethal CHIKV challenge.
Keywords: Chikungunya, RNA vaccine, nanostructured lipid carrier, live-attenuated, hybrid vaccine, viral vaccine, virus, arthritis, infection
Graphical abstract
By delivering whole-virus genome RNA with a thermostable RNA delivery nanoparticle, the corresponding author and colleagues present a vaccine technology that combines the benefits of modern RNA vaccine technology with the tried-and-true immunity of traditional live-attenuated vaccines. Their proof-of-concept Chikungunya vaccine is safe and highly effective in a mouse model.
Introduction
Nucleic acid-based vaccines represent attractive alternatives to traditional live-attenuated vaccines due to their ability to be rapidly adapted to new targets and reliably manufactured using pre-developed sequence-independent methods. Recent advances in engineering the structure1 and formulation2 of RNA-based vaccines has led to the advancement of RNA vaccine platforms targeting emerging infectious diseases. Recently, the SARS-CoV-2 pandemic has driven rapid development of RNA vaccines against the coronavirus, including progression through phase I, II, and III clinical trials and culminating in emergency use authorization of multiple RNA-based SARS-CoV-2 vaccines.
RNA vaccine technology may be able to overcome manufacturing and safety challenges typical of traditional live-attenuated vaccines. Manufacture of many attenuated viral vaccines using traditional culture methods can be difficult with a significant failure rate.3,4 The level of viral attenuation in vaccine strains is often high, limiting the rapid replication of virus to high titers. The number of biological substrates allowed for viral culture by regulatory agencies is also highly limited. Even should an excellent culture system exist, high viral titers are often only achieved in adherent cell culture, limiting production capabilities.5 Resulting vaccine product characteristics are often highly variable based on the biological system and culture conditions used,6,7 as are the methods used to analyze the resulting materials.4,8 This results in a high regulatory burden, increased vaccine costs, high failure rates of manufacturing lots, and can lead to severe vaccine shortages.4,9, 10, 11, 12 Safety issues are also inherent in the use of biological culture for vaccine manufacture. Contamination of vaccine materials has resulted from biological culture contamination during manufacture.13,14 Viral source material must also be consistent and regulated, as passage and expansion of live-attenuated viral strains during manufacturing may lead to genetic drift, which may in turn affect vaccine safety and immunogenicity profiles.8,15,16
A hybrid RNA-traditional vaccine approach could harness the strengths of both vaccine types, combining the ease, reliability, and safety inherent in nucleic acid vaccine manufacture with the proven immunogenicity of live-attenuated viral vaccines. Chikungunya virus (CHIKV) is an excellent model system for the testing of such a hybrid live-attenuated RNA hybrid vaccine. CHIKV is an emerging tropical arbovirus transmitted by the mosquito Aedes aegypti that typically results in fever, rash, and debilitating arthralgia and arthritis that can last months to years after infection (reviewed previously17,18), No approved vaccine against CHIKV exists at present. Reactogenicity problems plagued the original traditionally developed, live-attenuated CHIKV vaccine (181/25 strain) derived in the 1980s.19 Despite efficacy demonstrated in phase I and II clinical trials, arthralgia was reported in approximately 8% of 181/25 vaccinees, leading to the halt of 181/25-based vaccine development.20 CHIKV strain 181/25 was also demonstrated to be transmitted by the natural A. aegypti mosquito vector, leading to further concerns about vaccine containment.21 Later studies of the 181/25 viral strain indicated that viral attenuation was due to only two point mutations in the CHIKV envelope protein.22 This led to serious concerns about the genetic stability of the 181/25 vaccine virus strain. Indeed, the noted arthralgia in many vaccinees may be attributable to reversion of the 181/25 virus strain to a fully pathogenic phenotype during or after manufacture, as evidence of such reversion has been observed in experimental 181/25 infection of mice followed by viral sequencing.22
While non-replicating inactivated or virus-like particle (VLP)-based CHIKV vaccines have been described that would overcome such safety concerns,23, 24, 25, 26 VLP-based vaccines often require the use of adjuvants and booster doses,27 while high manufacturing costs often pose a significant challenge to the clinical practicality of such vaccine strategies. Live-replicating CHIKV strains with additional, more stable attenuating mutations and live-replicating chimeric CHIKV vaccines have been created as potential viral vaccines28, 29, 30, 31, 32 and appear to be the most practical candidates for safe and effective single-dose immunization against CHIKV. Manufacture of such live-attenuated CHIKV strains, however, involves all of the manufacturing challenges and safety issues mentioned above. Introduction of such live-attenuated RNA vaccine strains using a hybrid live-attenuated RNA vaccine technology could streamline manufacture of such vaccines, as well as reduce the potential for culture contamination and genetic drift.
In this work we demonstrate the creation of a hybrid live-attenuated RNA vaccine against CHIKV, in which full-length replication-competent attenuated CHIKV genomes are delivered to the site of vaccination using cutting-edge thermostable RNA vaccine delivery technology. This vaccine is an easily manufactured product with no need for biological culture, resulting in a reliable and stable genetic profile ensuring consistent safety and reactogenicity.
Results
Vaccine candidates create VLPs in vitro
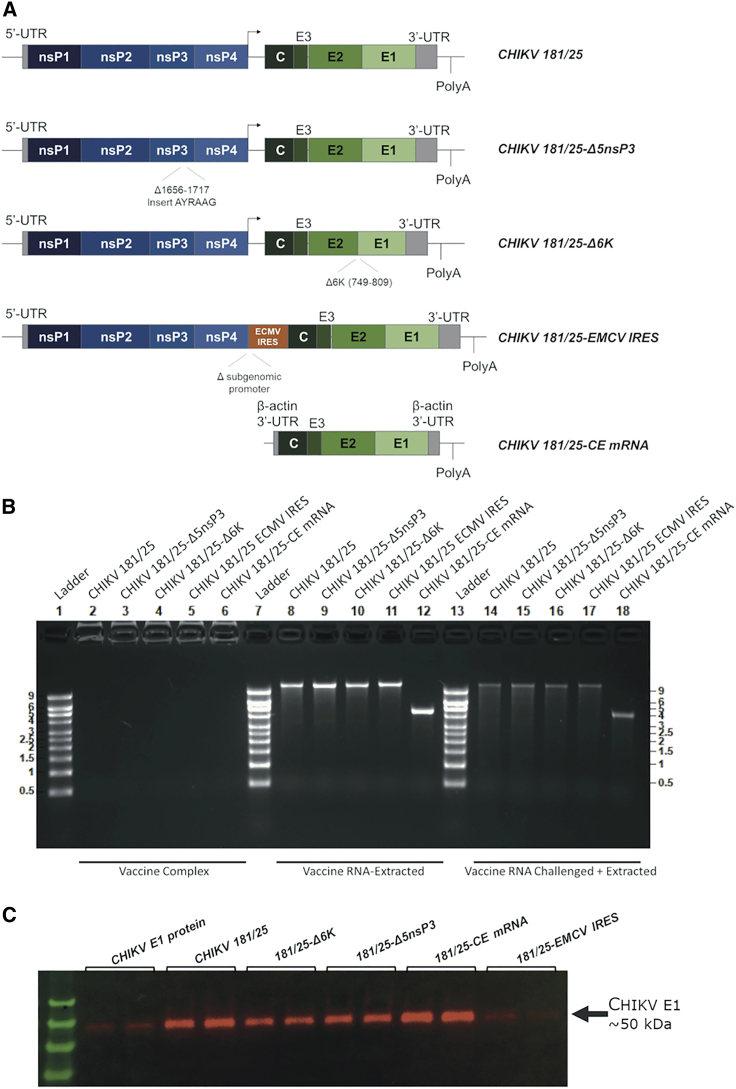
To test the use of whole-genome CHIKV RNA as safe and effective vaccines, we created DNA constructs containing the entire genome of four live-attenuated CHIKV variants (Figure 1A). Construct CHIKV 181/25 contains the full-length 181/25 CHIKV strain sequence. Three further constructs added additional previously described attenuating mutations to the 181/25 sequence in order to achieve genetically stable attenuation and effectively compare whole-genome RNA vaccines to current live-attenuated vaccine candidates, as follows: construct CHIKV 181/25-Δ5nsP3 contains the 181/25 CHIKV strain sequence with a deletion in the P1234 polyprotein of the nsP3 replicase gene, encoding for residues 1,656–1,717.29,30 Construct CHIKV Δ6K contains the 181/25 CHIKV sequence with a deletion in the Δ6K genomic region,29 representing amino acid residues 749–809. Construct CHIKV 181/25-ECMV IRES substitutes an ECMV IRES for the native CHIKV subgenomic promoter, a method previously successfully used to attenuate the virulent La Reunion strain of CHIKV (CHIKV-LR).28,33,34 For comparison with other RNA vaccine technology, we also created construct CHIKV 181/25-CE mRNA, an mRNA-based CHIKV vaccine candidate that expresses the 181/25 strain structural proteins C, E1, and E2 but contains no full-length genomic RNA.
Figure 1.
RNA constructs used as CHIK vaccine candidates
(A) Construct schematics. (B) Agarose gels showing free RNA from each NLC-formulated RNA vaccine candidate, extracted RNA from each NLC-formulated RNA vaccine candidate, and extracted RNA from vaccine candidates after challenge with RNase L. (C) VLPs collected by ultracentrifugation of transfected cell supernates 72 h post-transfection, resuspension of VLP pellets in PBS, bicinchoninic acid (BCA) assay for total protein quantification, and western blot with equal protein loading across samples, alongside purified Chikungunya E protein (~50 kDa).
We created fully functional, capped RNA using each of the DNA constructs as templates using in vitro transcription and capping reactions. We then formulated RNA vaccines with each RNA by complexing with a nanostructured lipid carrier (NLC) for effective delivery into target cells, as described previously.35,36 To verify full and equal loading of RNA onto the nanoparticles, as well as nanoparticle-mediated protection of the RNA from degradation by RNases, we ran a sample of each complexed vaccine, RNA extracted from each vaccine, and RNA extracted from each vaccine after challenge with RNase on an agarose gel (Figure 1B). We saw complete RNA complexing for each RNA vaccine candidate, as indicated by no free RNA present in the vaccine solution. RNA extracted from each vaccine candidate was of the appropriate sizes and showed excellent integrity and equal loading across vaccine candidates. The vaccine nanoparticles also allowed for retention of significant amounts of full-length RNA after challenge with ample RNase to fully degrade non-protected RNA, with protection of vaccine RNA from the action of RNases equal across vaccine candidates.
We tested these RNAs in vitro by transfection into HEK293 cells to validate their ability to induce the production of replication-competent VLPs. Transfected cell supernatants were collected 72 h post-transfection, concentrated, and ultracentrifuged through a 20% sucrose cushion to isolate VLPs produced by cell culture. Western blots were conducted on the resulting isolated VLPs to confirm successful expression of CHIKV proteins from in vitro-transcribed RNAs from each construct (Figure 1B). All four full-genome CHIKV RNAs and the CHIKV structural protein mRNA successfully produced VLPs.
Infectious attenuated virus strains rescued from in vitro transfection of whole-genome RNAs
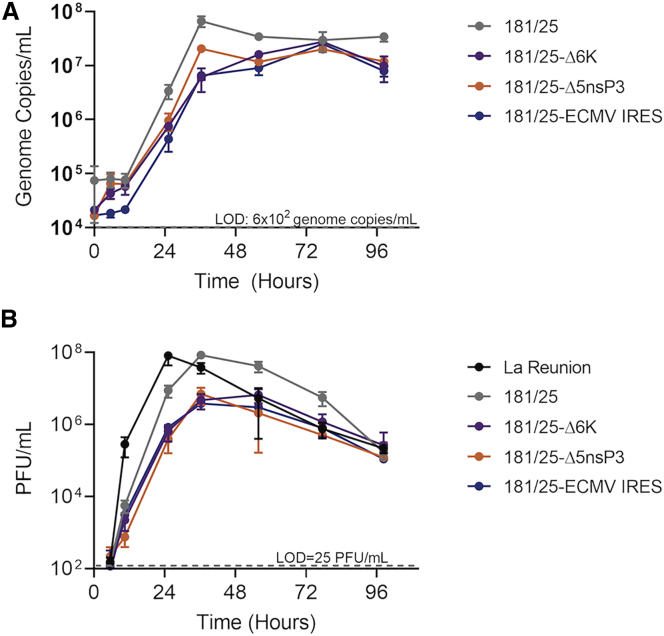
Transfection of Vero cells with the full-length viral genome RNAs allowed for rescue of infectious virus for all four live-attenuated CHIKV virus constructs. After two rounds of passage in cell culture, growth curves for each attenuated viral strain at a fixed multiplicity of infection (MOI) were performed on Vero cells to confirm virus growth and attenuation as measured by both qPCR for viral genomic material and a plaque assay for infectious virus (Figure 2). Similarly, a growth curve for virulent CHIKV-LR was conducted under biosafety level 3 (BSL3) conditions for comparison. The rescued CHIKV 181/25 virus grew to a higher titer (6.6 × 107 genome copies/mL by qPCR and 8.5 × 107 plaque-forming units [PFU]/mL by plaque assay) than those of the more-attenuated CHIKV 181/25-Δ5nsP3, CHIKV 181/25-Δ6K, and CHIKV 181/25-ECMV IRES rescued viral strains (titers of 2.1 × 107, 2.7 × 107, and 2.5 × 107 genome copies/mL by qPCR and 7.0 × 106, 6.7 × 106, and 3.8 × 106 PFU/mL by plaque assay, respectively; p < 0.05 for all). CHIKV-LR replicated to similar titers as CHIKV 181/25 (8.2 × 107 versus 8.5 × 107 PFU/mL, p = 0.93) but reached full titer approximately 12 h sooner. As expected, CHIKV 181/25-CE mRNA VLPs did not allow for rescue of infectious virus.
Figure 2.
Infectious attenuated viral strains rescued from RNA-transfected Vero cells
Rescued virus strains were passaged twice, and the level of attenuation relative to CHIKV-LR was measured by growth curves (MOI of 0.01) on Vero cells. (A and B) Supernate virus content was measured by qRT-PCR of viral genomes (A) or plaque assay (B). Data points represent mean values from biological triplicate samples ± SEM.
Whole-genome RNA vaccines are immunogenic in immunocompetent mice and protect against virulent CHIKV challenge
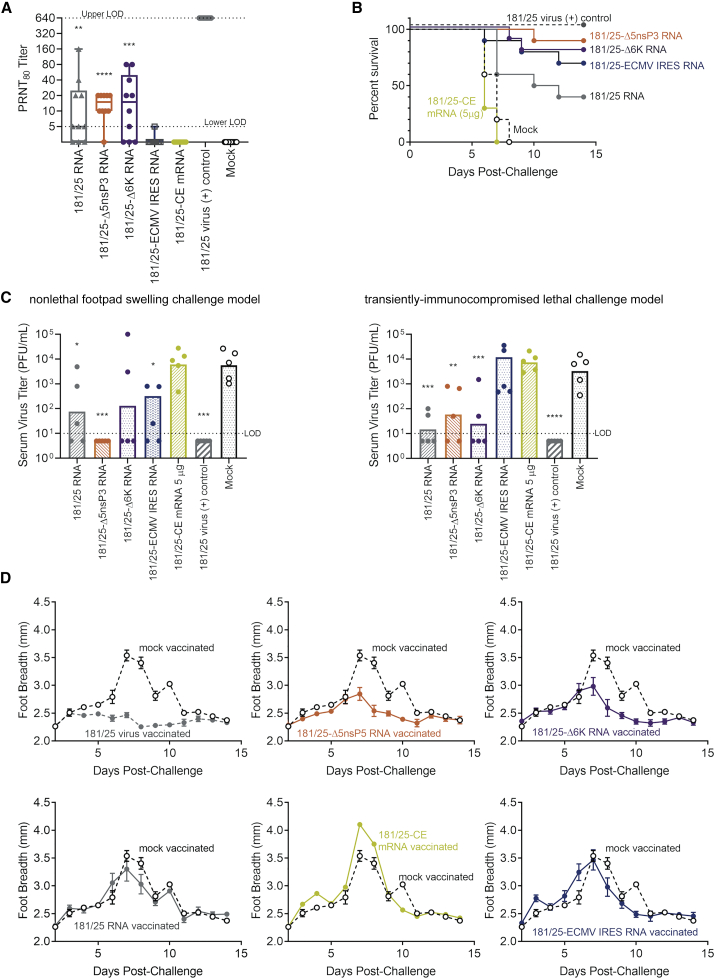
We then tested these RNA vaccine candidates for immunogenicity by injecting groups of immunocompetent C57BL/6 mice with 1 μg (full-length genome RNA and mRNA) or 5 μg (mRNA) of the individual RNA constructs formulated with NLC. While type I interferon (IFN) receptor −/− mice are often used for studies of CHIKV pathogenesis,34,37,38 IFN-competent mice are necessary for studies involving replicating viral vaccines to accurately reflect typical viral replication and immune responses;37 thus, wild-type C57BL/6 mice were used throughout this work. Blood samples were taken from all vaccinated and control mice at 2 days post-vaccination to check for post-vaccination viremia by a plaque assay. Plaque-reduction neutralization titers were measured in serum samples taken at 21 days post-inoculation and compared to control mice vaccinated by footpad injection of 104 PFU of CHIKV-181/25 virus (Figure 3A). The CHIKV-181/25, CHIKV-Δ5nsp3, and CHIKV-Δ6K full-genome RNA vaccines induced significant serum neutralizing antibody titers in vaccinated mice relative to mock-immunized control mouse sera (adjusted p value < 0.005 for each), although these were low relative to plaque reduction neutralization test (PRNT) titers resulting from mouse immunization with CHIKV 181/25 virus (adjusted p value < 0.0001 for all). We detected low levels of post-vaccination viremia in five of the ten 181/25 virally immunized mice, but no viremia in any of the RNA-vaccinated mice (Figure S1). CHIKV 181/25-CE mRNA vaccination did not result in neutralizing antibody titers at either 1- or 5-μg doses.
Figure 3.
Immunogenicity and efficacy screen of CHIKV RNA vaccine candidates
Wild-type C57BL/6 mice (n = 20/group) were immunized with 1 μg (whole-genome) or 5 μg (mRNA) of RNA vaccine candidates complexed with NLC. (A) CHIKV-neutralizing antibody titers 28 days post-vaccination with RNA vaccine candidates or CHIKV 181/25 virus (one-way ANOVA [6 degrees of freedom (DoF), F = 58.5] followed by Dunnett’s multiple comparison test). Mouse groups were then divided in half and challenged either lethally with 103 PFU/mouse of CHIKV-LR after i.p. injection of 2 mg of Mar1 IFNAR-blocking antibody (n = 10/group), or nonlethally challenged with 105 PFU/mouse of CHIKV-LR (n = 10/group) (one-way ANOVA, 6 DoF, F = 14.1 and 7.1, respectively). (B and C) Mouse survival was monitored daily (B), and serum samples were taken from a subset of mice (n = 5) 2 days post-challenge for measurement of viremia (C). (D) For this initial vaccine candidate screening study, footpad breadth alone was measured daily for non-lethally challenged mice (n = 10) to monitor CHIKV-induced arthralgia. Data points represent arithmetic means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 by Dunnett’s multiple comparisons test.
Twenty-eight days post-vaccination, the vaccinated mice from each group were then each split into two groups. A lethal challenge group was used to determine vaccine-induced protection from death, and a nonlethal challenge group was used to examine vaccine-induced protection from footpad swelling, under immunocompetent conditions, as is standard in the field of CHIK vaccine studies.
To create a CHIKV lethal challenge model, we transiently immunocompromised the mice in the lethal challenge group by intraperitoneal (i.p.) injection of 2 mg of MarI IFNAR-blocking monoclonal antibody 18 h prior to challenge with 103 PFU/mouse of virulent CHIKV-LR via footpad injection. While CHIKV-181/25 is nonlethal in immunocompetent C57BL/6 mice,37 CHIKV is known to be type I IFN sensitive,39,40 and temporary inhibition of type I IFN signaling is necessary and sufficient to obtain lethal challenge conditions with CHIKV-LR in otherwise immunocompetent C57BL/6 mice. Survival data are shown in Figure 3B; serum samples were taken from a subset of the challenged mice (n = 5) 2 days after challenge to check for viremia (Figure 3C).
Non-lethal challenge mice for examination of CHIKV-induced footpad swelling did not receive IFNAR-blocking antibody, and they were challenged 28 days post-vaccination by footpad injection of 105 PFU of CHIKV-LR per mouse. Serum samples were taken 2 days after challenge from a subset of the challenged mice (n = 5) to measure viremia (Figure 3C). Footpad breadth was measured daily for each non-lethally challenged mouse for 14 days (Figure 3D). All challenged mice were weighed daily (Figure S2).
Mice vaccinated with CHIKV 181/25 virus showed 100% survival, total suppression of viremia after lethal challenge in the transiently immunocompromised mice, and total suppression of CHIKV-induced footpad swelling in the immunocompetent mice untreated with Mar1 IFNAR-blocking antibody. CHIKV 181/25-CE mRNA inoculation did not result in neutralizing antibody titers at either 1- (data not shown) or 5-μg doses, and the 5-μg dose failed to provide any protection against viremia, death, or footpad swelling relative to unvaccinated mice. CHIKV 181/25-CE mRNA was thus removed from further candidacy. Each whole-genome, live-attenuated RNA vaccine candidate induced partial protection from post-challenge mortality, viremia, and footpad swelling.
The wide range of neutralizing antibody titers induced by any one CHIKV whole-genome RNA vaccine candidate suggested that vaccine dosing was not optimal, leading to launch of the RNA virus in some but not all mice. The full-genome CHIKV 181/25 and CHIKV 181/25Δ5nsP3 RNA vaccine candidates were thus chosen—based on their induction of neutralizing antibody titers and ability to protect mice against viremia, death, and footpad swelling—for further dosing and immunogenicity studies.
Whole-genome RNA vaccine immunogenicity and efficacy is dose-dependent and rivals that of live virus vaccine
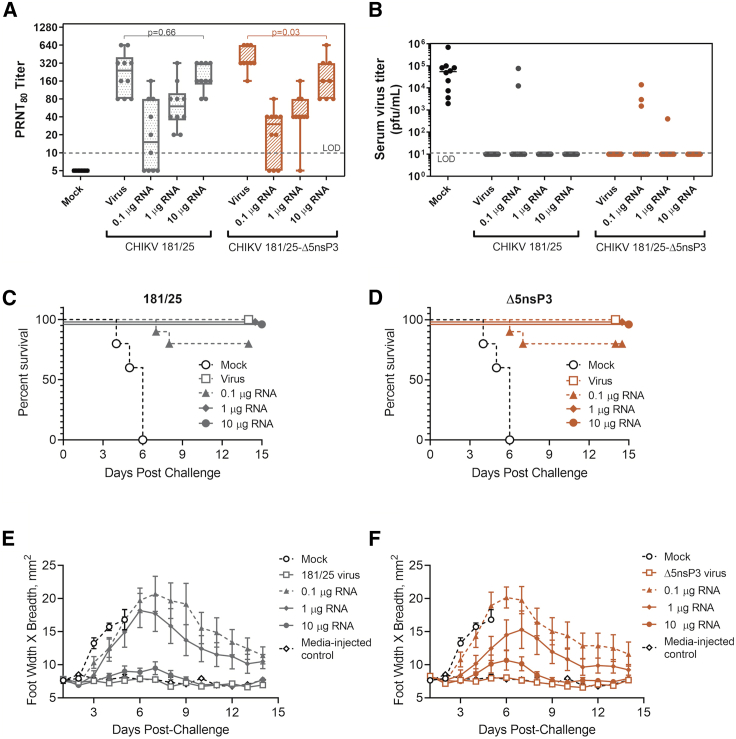
To confirm immunogenicity of and determine suitable dosing for the two lead full-genome RNA CHIK vaccine candidates, we immunized mice with 0.1, 1, or 10 μg of each of the two lead RNA vaccines CHIKV 181/25 and CHIKV 181/25-Δ5nsP3. Vaccination with 104 PFU/mouse of each attenuated virus or plain PBS served as positive and negative vaccination control groups. Serum antibody titers at 28 days post-vaccination were measured by PRNTs (Figure 4A). Each animal group was then injected with 2 mg of murine IFNAR-blocking antibody and lethally challenged with 103 PFU of virulent CHIKV-LR 18 h later. Survival data are displayed in Figure 4B. Footpad area measurements (width × breadth) from the lethally challenged mice were also taken for each mouse, and data for CHIKV 181/25-vaccinated () and CHIKV 181/25-Δ5nsP3-vaccinated () mice are shown in Figures 4C and 4D, respectively. All mice were weighed daily (Figure S3).
Figure 4.
Dosing and efficacy of lead whole-genome CHIKV RNA vaccine candidates
Wild-type C57BL/6 mice (n = 10/group) were immunized i.m. with 0.1, 1, or 10 μg of whole-genome RNA vaccine candidates CHIKV 181/25 or CHIKV 181/25Δ5nsP3. Vaccination with 104 PFU/mouse of each attenuated virus served as positive vaccination control groups. (A) CHIKV-neutralizing antibody titers 28 days post-vaccination. Two-tailed t tests were used on log-normalized PRNT data to compare neutralizing Ab titers induced by the 10-μg RNA vaccine dose with Ab titers induced by the respective live-attenuated viral vaccine. (B) Post-lethal challenge viremia measured by plaque assay (n = 10/group). (C and D) Survival of mice vaccinated with different doses of CHIKV 181/25 and CHIKV 181/25-Δ5nsP3 RNA-based vaccines, respectively, after immunocompromisation and lethal challenge with CHIKV-LR (n = 10/group). (E and F) Mouse footpad swelling as indicated by footpad width × breadth measurements after immunocompromisation and lethal challenge (n = 10/group). Data points represent arithmetic means ± SEM. Two-tailed homoscedastic t tests were used to compare log-normalized mouse serum PRNT titers induced by the 10-μg RNA vaccine doses with those induced by the respective live-attenuated viral vaccine control (DoF = 9).
Both the CHIKV 181/25 and CHIKV 181/25-Δ5nsP3 RNA-based vaccines induced significant neutralizing antibody serum titers at 28 days post-vaccination. Clear dose-dependence was observed for both whole-genome RNA vaccines. Indeed, the highest dose of each whole-genome RNA vaccine (10 μg/mouse) resulted in induction of serum antibody titers not significantly different than antibody titers induced by live viral vaccination (p > 0.05).
Mock-vaccinated mice uniformly died by day 6 post-challenge. Both whole-genome RNA vaccines protected 100% of mice against death at doses of 10 and 1 μg/mouse, and they partially protected mice at the lowest RNA vaccine dose (0.1 μg). Monitoring of CHIKV-induced footpad swelling in this transiently immunocompromised challenge model was highly informative; significant footpad swelling occurred in a dose-dependent manner inversely proportional to vaccine dose. Interestingly, even mice completely protected from mortality by the mid-range 1-μg dose of either full-genome RNA vaccine showed significant footpad swelling, indicating incomplete protection against morbidity. Little to no footpad swelling was seen in virally vaccinated mice, or in mice vaccinated with the highest dose (10 μg RNA) of each RNA vaccine, indicating a high level of protection even upon vaccination with a highly attenuated CHIKV genomic strain.
Discussion
We have demonstrated that an effective CHIKV vaccine can be created by delivering replication-competent attenuated CHIKV genomes to the site of vaccination using cutting-edge RNA vaccine technology. This vaccine technology allowed for the production of replication-competent VLPs in vitro capable of presenting CHIKV epitopes to appropriate immune cells in vivo. In vivo studies demonstrated the ability of this CHIKV hybrid live-attenuated RNA vaccine to induce significant CHIKV-neutralizing antibody titers in immunocompetent mice after a single immunization in a dose-dependent manner. We then used a transiently immunocompromised murine lethal challenge model to demonstrate vaccine-induced protection against CHIKV-mediated morbidity and mortality. The vaccine demonstrated the ability to protect even transiently immunocompromised mice from death, viremia, and footpad swelling after lethal challenge with virulent CHIKV-LR.
Our work also establishes a model for CHIKV lethal challenge in IFN-competent mice. By i.p. injection of IFNAR-blocking antibodies prior to CHIKV-LR virus challenge, wild-type C57BL/6 mice were sufficiently immunocompromised to achieve reliable lethality in unprotected mice. Use of immunocompetent mice with intact innate immune signaling systems is important for live, replicating vaccine efficacy testing to prevent overestimation of vaccine immunogenicity. This model accordingly allows for the progression of normal immune responses to vaccination, while also providing a challenge model for proof of vaccine efficacy beyond footpad swelling measures alone. Indeed, in such transiently immunocompromised mice, footpad swelling in this model appears to be a more sensitive measure of vaccine protection from CHIKV than is footpad swelling in wild-type C57BL/6 mice, a common model of CHIKV protection.
Advantage of live-attenuated RNA hybrid vaccine: delivery
DNA vaccines against CHIKV have previously been created by several scientific teams with a similar goal of harnessing the safety, manufacturability, and reliability of nucleic acid-based vaccines.29,30,41, 42, 43, 44 Indeed, several groups have used DNA to launch full-length replication-competent live attenuated CHIKV strains in a similar bid to harness the advantages of nucleic acid vaccine technology in combination with the proven immunogenicity and reliability of live-attenuated vaccines. In work by Tretyakova et al.,44 “immunization DNA” was used to deliver full-length cDNA of attenuated CHIKV virus genomes to BALB/c mice, and this resulted in the development of CHIKV-neutralizing antibodies and protection of mice against virulent CHIKV challenge. Similarly, Hallengärd et al. administered DNA encoding genomes of the live-attenuated CHIKV strains CHIKV 181/25-Δ5nsP3 and CHIKV 181/25-Δ6K (also used in our work) by electroporation of C57BL/6 mice, resulting in antibody responses and protection against viremia and joint swelling.29 However, all of these DNA-based vaccine platforms require electroporation of vaccine-injected mouse muscle to enable DNA entry into target cells. The ability to administer our live-attenuated RNA hybrid vaccine via a single intramuscular (i.m.) injection is a major improvement to the ease and reliability of vaccine administration, and considerably more adoptable in a clinical setting.
Advantages of live-attenuated RNA hybrid vaccine: safety and genetic stability
Manufacturing for both the RNA and NLC formulation components of these live-attenuated RNA hybrid vaccines is done in cell-free environments, avoiding the potential for biological materials to become contaminated and affect vaccine quality, as has been a rare but serious issue in the manufacture of live-attenuated vaccines. An additional safety benefit is conferred by the nature of the RNA vaccine material, which does not need to be passaged and expanded as do live-attenuated virus strains. As a result of direct translation from a DNA backbone by the relatively low-error T7 RNA polymerase, the CHIKV vaccine RNA has a consistent and easily characterized sequence, unlike the genetically diverse pseudospecies typically found in live-attenuated vaccines against RNA viruses. Even live-attenuated Chikungunya vaccine strains engineered to have a particularly high-fidelity polymerase (fidelity variants), which demonstrated efficacy in mice,45 also showed maintained or even increased accumulation of mutations after passaging in cell culture,45,46 resulting in safety concerns. Increased genetic diversity of live-attenuated Chikungunya vaccines have also been suggested to potentially impair the development of neutralizing antibodies.46 Others have demonstrated that DNA-launched 181/25-derived Chikungunya vaccine virus genomes have a higher level of genetic uniformity than even a minimally passaged 181/25 viral strain, with significantly lower frequency of single-nucleotide polymorphisms, including at the two mutation sites in the 181/25 virus that are responsible for attenuation.47 We would expect similarly reduced genetic diversity in our hybrid live-attenuated RNA vaccine relative to the 181/25 pseudospecies, with any T7 polymerase-introduced mutations randomly assorted across the viral genome rather than due to any selective pressure. Thus, use of in vitro transcription direct from a plasmid likely results in better genetic stability and safety profiles for RNA-delivered CHIKV genomes, free of genetic drift.
Advantage of live-attenuated RNA hybrid vaccine: manufacturability
A major advantage of nucleic acid vaccines is their reliable, sequence-independent manufacturability. Such manufacturing requires little to no specialized equipment not already found in standard good manufacturing practice (GMP) facilities. DNA plasmid manufacture is established GMP technology; in vitro RNA transcription and NLC formulation manufacture are GMP-friendly and easily adapted to new vaccine sequences. We have GMP-ready systems in place for manufacture of such RNA-based vaccines on the scale of tens of thousands of doses per year; similar manufacturing systems could be set up elsewhere including in areas where CHIKV is endemic.
While a number of other CHIKV vaccines are currently in advanced-stage preclinical development or clinical trials,24,30, 31, 32,34 the cost of manufacture, stability, and safety profiles of such vaccines are as yet unclear. The rapid advancement and recent approval under emergency use authorization of nucleic acid-based vaccine technologies against SARS-CoV-2 has established a path for the approval of a first-in-class live-attenuated hybrid RNA vaccine system as proposed herein.
Future work
Future preclinical development of these live-attenuated RNA hybrid CHIKV vaccines should include examination of T cell responses to vaccination. While CHIKV-reactive antibodies are generally considered to be appropriate correlates of protection for CHIKV vaccines,48 T cell responses to our live-attenuated RNA hybrid vaccine platform would of great interest to study as part of future vaccine development efforts. Similarly, this work was conducted in female mice to maximize statistical power to identify significant differences in immunogenicity between vaccine candidates with a limited number of total animals; advanced preclinical testing should include both male and female animals to fully characterize vaccine candidate immunogenicity.
This method of vaccine development may be applied to other positive-stranded RNA viruses, allowing for reliable manufacture of live-attenuated RNA hybrid vaccines of even highly attenuated virus strains. Positive-stranded RNA viruses comprise a broad class of viruses, causing numerous important human pathogens such as SARS, hepatitis C, Coxsackie virus, West Nile virus, polio, and yellow fever, among many others. While this method of vaccine development relies on the existence of a live-attenuated vaccine virus strain, it allows for more straightforward, sequence-independent, cell-free manufacturing compared with traditional live-attenuated vaccine manufacturing methods. Thus, this method may be used to supplement stores of already-existing viral vaccines with manufacturing difficulties, and/or the scale-up and commercialization of otherwise unmanufacturable highly attenuated vaccine strains. Efforts are underway to develop long-term stable storage capabilities for such pre-formulated RNA/NLC vaccines to allow for effective storage and distribution, potentially without the need for a cold-chain.
The yellow fever vaccine is an example of where such a technique could be usefully applied. A live-attenuated vaccine against yellow fever has long been available, but it is notoriously difficult to manufacture. Indeed, difficulties in manufacturing have led to massive shortages in worldwide vaccine supplies,9,49 contributing to the emergence of yellow fever outbreaks throughout Brazil and other endemic countries.50, 51, 52 Our hybrid RNA vaccine technology could potentially allow for the easy manufacture and delivery of YF-17D vaccine virus RNA by standard i.m. injection, bypassing current yellow fever vaccine manufacturing processes and relieving vaccine shortages.
We have demonstrated the use of this hybrid live-attenuated RNA vaccine for a positive-stranded RNA virus as the most straightforward application of this technology. This hybrid live-attenuated RNA vaccine technology could be further adapted to target other classes of viruses with straightforward future development. Negative-strand RNA viruses, for example, could be targeted by addition of a positive-strand gene encoding the appropriate RNA-dependent RNA polymerase to the vaccine RNA encoding the negative-strand RNA genome. Such a gene could successfully be encoded either on a separate mRNA molecule or added to the genome-containing RNA under the control of a separate translation initiation signal.
Conclusions
This work presents a proof of principle that full-genome RNAs can be used to launch live-attenuated viral vaccines, a method we describe as live-attenuated RNA hybrid vaccines. Such hybrid live-attenuated nucleic acid vaccines may be reliably and rapidly manufactured in a cell-free, sequence-independent process that overcomes many of the ongoing production and safety challenges inherent in the manufacture of live-attenuated viral vaccines. As a sequence-independent process, this hybrid live-attenuated/RNA vaccine technology allows for the use of highly attenuated virus strains in vaccines, thereby increasing both the genetic attenuation stability and safety profile of the vaccine.
Materials and methods
Cell and virus culture
Human embryonic kidney cells (293T, ATCC CRL-3216) and African green monkey cells (Vero, ATCC CCL-81) were obtained from the American Type Culture Collection and passaged in antibiotic-free DMEM medium with GlutaMAX (Invitrogen) supplemented with 10% fetal bovine serum. All cell lines were maintained in a humidified incubator at 37°C in a 5% CO2 atmosphere, and prescreened for mycoplasma contamination. CHIKV strains 181/25 and CHIKV-LR (OPY1, passaged five times in Vero cells) were obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch in Galveston, TX, USA, and propagated on Vero cells (MOI of 0.02).
Viral plasmids and cloning
A plasmid containing the full-length CHIKV 181/25 genomic sequence under control of an SP6 promoter was obtained from Kenneth Plante at the World Reference Center for Emerging Viruses and Arboviruses located at the University of Texas Medical Branch in Galveston, TX, USA. We used standard cloning techniques to replace the existing SP6 promoter with a T7 promoter to create the plasmid CHIKV-181/25. Plasmids CHIKV 181/25-Δ5nsP3, CHIKV Δ6K containing CHIKV 181/25 genomes with additional published attenuating deletions,28, 29, 30 and CHIKV 181/25-ECMV IRES33,34 were created from the CHIKV-181/25 plasmid by standard cloning methods. Briefly, gene fragments containing the desired gene edits were synthesized (Integrated DNA Technologies) and cloned into digested, purified CHIKV-181/25 plasmid backbones using InFusion enzyme mix (Clontech) between PpuMI and SfiI (CHIKV 181/25-ECMV IRES), XhoI and SgrAI (CHIKV Δ6K), or PasI and Bpu10I (CHIKV 181/25-Δ5nsP3) restriction enzyme sites. All plasmid sequences were confirmed by Sanger sequencing. Plasmid sequences have been uploaded to GenBank as follows: CHIKV 181/25-CE mRNA, GenBank: MZ671996; CHIKV 181/25, GenBank: MZ671997; CHIKV 181/25-Δ5nsP3, GenBank: MZ671998; CHIKV Δ6K, GenBank: MZ671999; CHIKV 181/25-ECMV IRES, GenBank: MZ672000.
RNA production
Viral genome-containing plasmids were amplified in Top10 cells (Invitrogen) and isolated using QIAGEN Maxiprep kits. Purified plasmids were then linearized with NotI restriction digestion and phenol-chloroform purified. RNA was transcribed in vitro using an in-house optimized protocol using T7 polymerase, RNase inhibitor, and pyrophosphatase enzymes (Aldevron), followed by a DNase incubation (DNase I, Aldevron) and LiCl precipitation. Cap0 structures were added to the RNA by a reaction with vaccinia capping enzyme, guanosine triphosphate (GTP), and S-adenosyl methionine (New England Biolabs). Capped RNA was then precipitated using LiCl and resuspended in nuclease-free water prior to quantification by UV absorbance (NanoDrop 1000) and analysis by agarose gel electrophoresis using Ambion NorthernMax reagents (Invitrogen). All transcribed and capped vaccine RNA was stored at −80°C until use.
RNA vaccine formulation and testing
RNA was complexed with a stable NLC colloidal delivery formulation whose structure and manufacture have previously been described.35 Briefly, a blend of liquid oil (squalene) and solid lipid (Dynasan 114) form a semi-crystalline nanostructure core, stabilized in an aqueous buffer by a hydrophobic sorbitan ester (Span 60), a hydrophilic ethoxylated sorbitan ester (Tween 80), and the cationic lipid DOTAP (N-[1-[2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium chloride), which together allow for long-term colloidal stability. The formulation was prepared as previously described.35
RNA vaccine complexing and characterization
Vaccine RNA was complexed with NLC formulation at a NLC nitrogen/RNA phosphate ratio of 15. Briefly, RNA was diluted in nuclease-free water to twice the desired final vaccine RNA concentration, and dilution of NLC in an aqueous sucrose citrate buffer to a final concentration of 20% sucrose, 10 mM citrate. The diluted RNA and diluted NLC solutions were then combined at a 1:1 ratio and quickly mixed by pipet to form a final 1× RNA concentration complexed in NLC in a 10% sucrose 5 mM citrate isotonic aqueous solution. The resulting vaccine solution was allowed to incubate on ice for 30 min to form stable nanoparticles. For characterization of nanoparticle-loaded RNA, vaccine samples were diluted to a final RNA concentration of 40 ng/μL in nuclease-free water. For verification of full RNA loading on the nanoparticles, vaccine samples containing 200 ng of RNA were mixed 1:1 by volume with glyoxal load dye (Invitrogen), loaded directly on a denatured 1% agarose gel, and run at 120 V for 45 min in NorthernMax Gly running buffer (Invitrogen). Millennium RNA marker (Thermo Fisher Scientific) was included on each gel with markers at 0.5, 1, 1.5, 2, 2.5, 3, 4, 5, 6, and 9 kb. Gels were imaged using ethidium bromide protocol on a ChemiDoc MP imaging system (Bio-Rad). Lack of RNA bands being successfully electrophoresed indicates full complexing of RNA to the nanoparticles. For verification of nanoparticle-loaded RNA integrity, RNA was extracted from the vaccine complexes by addition of 25:24:1 phenol/chloroform/isoamyl alcohol (Invitrogen) 1:1 by volume, vortexing, and centrifuging at 17,000 × g for 15 min. The supernatant for each sample was then mixed 1:1 by volume with glyoxal load dye and incubated at 50°C for 20 min prior to being loaded onto a 1% agarose gel and run as described above. For verification of equal protection of different vaccine RNAs from RNases by the complexes, the diluted vaccine complexes were incubated with RNase A (Thermo Scientific) for 30 min at room temperature at amounts ample to completely degrade un-complexed RNA (ratios of 1:40 RNase/RNA). This treatment was followed by treatment with recombinant Proteinase K (Thermo Scientific) at a ratio of 1:100 RNase A/Proteinase K for 10 min at 55°C. RNA was then extracted from the challenged samples and run on a 1% agarose gel as described above.
Transfection and VLP harvest
We tested the ability of the whole-genome CHIK RNAs to successfully transfect cells when complexed with NLC and induce cellular production of VLPs. HEK293T cells were plated in 12-well plates at a density of 5 × 105 cells/well 24 h prior to transfection. Shortly before transfection, medium was removed from cells and replaced with 450 μL of serum-free Opti-MEM medium (Invitrogen). 500 ng of NLC-complexed RNA was added into each well in a 50-μL volume, and cells were incubated at 37°C and 5% CO2 for 4 h to allow for transfection. After the 4-h incubation, transfection solutions were removed and replaced with 2 mL of DMEM supplemented with 2% fetal bovine serum (FBS). Transfected cell supernatants were collected 72 h post-transfection, concentrated by centrifugation through 30,000-Da molecular weight cutoff (MWCO) Amicon Ultra-15 centrifugal filter tubes (Millipore) at 2,000 × g for 10–15 min, and finally ultracentrifuged through a 20% sucrose in PBS cushion (100,000 × g, 10°C, 2 h) to pellet cellular-produced VLPs. Pelleted VLPs were resuspended in 100 μL of PBS.
Western blotting
Western blots were then conducted on the isolated VLPs. VLP solutions were reduced with NuPage 10× reducing agent and NuPAGE LDS sample buffer (Invitrogen) and denatured by incubation at 95°C for 10 min before loading on duplicate Novex Wedgewell 4%–20% gradient precast PAGE gels and being run at 120 V in NuPAGE 2-(N-morpholino)ethanesulfonic acid (MES) SDS running buffer for 1 h. The gels were then transferred to polyvinylidene fluoride (PVDF) membranes using the Invitrogen iBlot semi-dry transfer system with a 6-min transfer step. The membranes were then blocked overnight in a PBS solution with 5% nonfat dry milk. The blots were then rinsed and incubated for 2 h in a 1:5,000 dilution of anti-CHIK envelope protein antibody in 5% nonfat dry milk. After 3× rinsing in PBS with Tween 20 (PBST), the membranes were incubated in a 1:200 dilution of goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h. After 4× rinsing in PBST, the membranes were developed using West Pico Plus reagents (Thermo Fisher Scientific) and signal was detected on a Bio-Rad Gel Doc XR+ system.
Virus strain growth curves
Infectious CHIKV vaccine virus strains were rescued from full-genome RNAs by 2× passage of VLP-containing supernates from RNA vaccine-transfected HEK293T in Vero cells. CHIKV variant viability and attenuation relative to wild-type CHIKV was measured by infection of Vero cells followed by time course measurements of supernate viral titers by plaque assay (infectious particles) and qPCR (viral genomes). Briefly, medium was removed from 90% confluent monolayers of Vero cells in 12-well tissue culture plates (approximately 1 × 106 cells), and 100 μL of virus solution was added to achieve an MOI of 0.01. After 1 h of adsorption at 37°C and 5% CO2 with gentle rocking every 20 min, the inoculum was removed. One milliliter of DMEM supplemented with 1% FBS was then added. Supernates were harvested from independent biological triplicate wells at the times indicated post-infection and frozen in aliquots for later plaque and qPCR assays.
Plaque assays
For quantification of infectious virus particles in infection supernates, samples were serially diluted in 1:10 dilutions of DMEM supplemented with 1% FBS and 2 mM GlutaMAX. Vero cells were plated 18 h prior to assay at a concentration of 5 × 105 cells/well in six-well tissue culture plates and allowed to form monolayers. Cell monolayers were infected with 200 μL of virus dilution and incubated for 1 h with gentle rocking every 20 min. The virus-containing sample was then removed, and cell monolayers were overlaid with 2 mL of DMEM supplemented with 1% FBS, 2 mM GlutaMAX, and 0.6% melted agar. The plates were cooled until agar solidified and incubated at 37°C, 5% CO2 for approximately 48 h, until plaques appeared. Agar layers were then removed, cells were fixed for 20 min with a formalin solution, and cell layers were stained with 0.1% crystal violet in 20% ethanol to visualize plaques.
Viral genome quantification by quantitative reverse transcriptase PCR
Frozen viral time course supernate samples were thawed and viral genomic RNA was extracted from samples using QIAamp viral RNA mini kits (QIAGEN). Carrier RNA (QIAGEN) was added to each sample to normalize the extraction/reverse transcription process between samples. Total RNA concentrations were normalized between samples to obtain 750 ng of total RNA per random hexamer reverse transcription reaction, conducted using the QuantiTect reverse transcription kit (QIAGEN). qPCR was then conducted on 1 μL of the resulting cDNA, using the following qPCR primers from Lanciotti et al.53 that detect a region of the CHIKV NSP4 gene conserved between all virus strains used in this work: forward, 5′- TCACTCCCTGTTGGACTTGATAGA-3′; reverse, 5′-TTGACGAACAGAGTTAGGAACATACC-3′. A standard curve was formed by serial dilution of NotI-linearized CHIKV 181/25 genomic plasmid of known concentration spanning the entire dynamic range of sample concentrations. This standard curve (genomic plasmid copy number versus CT) was fit with a semi-log line (R2 = 0.993) and used to interpolate absolute CHIKV genome copy numbers in the infection samples. qPCR was performed with technical duplicates of the biological triplicates collected at each time point for each viral variant.
In vivo studies
Female 6- to 8-week old immunocompetent C57BL/6 mice were used for all vaccine immunogenicity studies (The Jackson Laboratory). Only female mice were used to maximize statistical power to detect immunogenicity differences between vaccine variants. Mice were inoculated with full-genome RNA vaccines at doses of 0.1, 1, or 10 μg of RNA complexed with NLC by i.m. injection of 50 μL of vaccine formulation in each rear quadriceps muscle for a total of 100 μL of vaccine per mouse. Mice were inoculated with mRNA vaccine at doses of 1 or 5 μg of RNA complexed with NLC by i.m. injection with the same volume and injection strategy as the full-genome RNA vaccines. Positive vaccination control mice were inoculated by subcutaneous (s.c.) footpad injection of 20 μL containing 104 PFU of CHIKV-181/25 virus. Blood samples were taken at 3, 7, 14, 21, and 27 days post-vaccination to test for viremia (day 3) and to measure the development of CHIKV-neutralizing antibody titers (days 7, 14, 21, and 27). Twenty-eight days post-vaccination mice were challenged with virulent CHIKV-LR (from WRCEVA at UTMB, TVP20521). For lethal challenge, each mouse was injected i.p. with 2 mg of InVivoMAb anti-mouse IFNAR-1 blocking antibody (clone MAR1-5A3, Bio X Cell) in a 300-μL volume 18 h prior to s.c. footpad injection of 80 μL containing 103 PFU/mouse of CHIKV-LR (40 μL/rear footpad). Lethally challenged mice were monitored daily for weight loss and signs of disease.
For non-lethal challenge, each mouse was injected with 105 PFU of CHIKV-LR s.c. into the footpad, and mice were monitored daily for signs of disease, weight loss, and footpad swelling by measurement of footpad width (Figure 3, initial vaccine immunogenicity screen) or footpad width × breadth (Figure 4, detailed lead candidate dosing and efficacy study). Blood samples were taken from all challenged mice 2 and 4 days post-challenge by the retro-orbital route to check for post-challenge viremia. All animal work was carried out in the IDRI Vivarium under ABSL1, ABSL2, or ABSL3 conditions as appropriate under the oversight of the IDRI Institutional Animal Care and Use Committee (IACUC). All challenged mice were monitored daily for weight loss and signs of disease. Mice that lost more than 20% of their pre-challenge weight, or demonstrated lack of mobility, lethargy, or a hunched back that did not resolve, were humanely euthanized by CO2 inhalation. All remaining mice were euthanized at the end of the scheduled study period. All animals were cared for in accordance with the guidelines of the Committee on the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, National Research Council).
PRNT assays
Mouse serum samples were tested for the presence of CHIKV-neutralizing antibody titers by PRNTs. Briefly, mouse sera were diluted 1:10 in DMEM media, then serially diluted further at 1:2 dilutions. Each diluted serum sample was then mixed 1:1 with CHIKV-181/25 virus at a concentration of ∼500 PFU/mL and allowed to incubate for 30 min at room temperature to allow for viral neutralization. 200 μL of the resulting solution was then plated in six-well plates containing 90% confluent Vero cell monolayers and allowed to incubate for 1 h with gentle rocking every 20 min. Similar to the plaque assays described above, the virus-containing sample was then removed, and cell monolayers were overlaid with 2 mL of DMEM supplemented with 1% FBS, 2 mM GlutaMAX, and 0.6% melted agar. The plates were cooled until agar solidified, and incubated at 37°C, 5% CO2 for approximately 48 h, until plaques appeared. Agar layers were then removed; cells were fixed for 20 min with a formamide solution, and cell layers were stained with 0.1% crystal violet in 20% ethanol to visualize plaques. PRNT80 titers were calculated as the mouse serum dilution that resulted in neutralization of ≥80% of the number of CHIKV-181/25 plaques found in control (non-immunized mouse serum) samples.
Statistical analyses
Statistical analysis was performed using GraphPad Prism software. Data distribution and variance were evaluated for normality prior to analyses. All data collected by qPCR, PRNT, or plaque assay were log-normalized prior to analysis. Two-tailed t tests or one-parameter ANOVA analyses followed by Dunnett’s multiple comparison test (α = 0.05) were conducted to determine statistical differences in antibody titers and post-challenge viremia measures between study groups. Test residuals were checked to confirm data normality.
Acknowledgments
Research reported in this work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R43AI127053. 100% percent of total project costs were financed with Federal money. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to sincerely thank Sierra Reed for collecting vaccine complexing data, Raodoh Mohamath for generously providing the purified Chikungunya E1 protein for use as a western blot standard, and Theresa Britschgi for contributing to manuscript revisions.
Author contributions
E.A.V. and N.V.H. conceived experiments. N.V.H. obtained funding for the project. E.A.V. and N.V.H. conceptualized, designed, and did formal analysis of the experiments. Methodology was established by E.A.V., B.G., J.A., and N.V.H. Formal analysis was conducted by E.A.V. and N.V.H., and investigation was by E.A.V., J.F.-S., B.G., and J.A. under the supervision of N.V.H. The original draft of this report was written by E.A.V. and reviewed/edited by E.A.V., J.F.-S., B.G., J.A., and N.V.H.
Declaration of interests
Current affiliations of authors include J.F.-S., the University of Chicago; B.G. and N.V.H., Sana Biotechnology, Inc.; J.A., HDT Bio. N.V.H. is listed as inventor on a patent application describing the NLC formulation. E.A.V. and N.V.H. are listed as inventors on a patent application describing live-attenuated RNA vaccine technology.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.05.018.
Supplemental information
References
- 1.Tavernier G., Andries O., Demeester J., Sanders N.N., De Smedt S.C., Rejman J. mRNA as gene therapeutic: How to control protein expression. J. Control. Release. 2011;150:238–247. doi: 10.1016/j.jconrel.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Midoux P., Pichon C. Lipid-based mRNA vaccine delivery systems. Expert Rev. Vaccines. 2015;14:221–234. doi: 10.1586/14760584.2015.986104. [DOI] [PubMed] [Google Scholar]
- 3.Rodrigues A.F., Soares H.R., Guerreiro M.R., Alves P.M., Coroadinha A.S. Viral vaccines and their manufacturing cell substrates: New trends and designs in modern vaccinology. Biotechnol. J. 2015;10:1329–1344. doi: 10.1002/biot.201400387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plotkin S., Robinson J.M., Cunningham G., Iqbal R., Larsen S. The complexity and cost of vaccine manufacturing—An overview. Vaccine. 2017;35:4064–4071. doi: 10.1016/j.vaccine.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genzel Y., Rödig J., Rapp E., Reichl U. Vaccine production: upstream processing with adherent or suspension cell lines. Methods Mol. Biol. 2014;1104:371–393. doi: 10.1007/978-1-62703-733-4_23. [DOI] [PubMed] [Google Scholar]
- 6.Butler M., Reichl U. Animal cell expression systems. Adv. Biochem. Eng. Biotechnol. 2017 doi: 10.1007/10_2017_31. Published online October 3, 2017. [DOI] [PubMed] [Google Scholar]
- 7.Ng S., Gisonni-Lex L., Azizi A. New approaches for characterization of the genetic stability of vaccine cell lines. Hum. Vaccin. Immunother. 2017;13:1669–1672. doi: 10.1080/21645515.2017.1295191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minor P.D. Live attenuated vaccines: Historical successes and current challenges. Virology. 2015;479–480:379–392. doi: 10.1016/j.virol.2015.03.032. [DOI] [PubMed] [Google Scholar]
- 9.Gershman M.D., Angelo K.M., Ritchey J., Greenberg D.P., Muhammad R.D., Brunette G., Cetron M.S., Sotir M.J. Addressing a yellow fever vaccine shortage—United States, 2016–2017. MMWR Morb. Mortal. Wkly. Rep. 2017;66:457–459. doi: 10.15585/mmwr.mm6617e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulmer J.B., Valley U., Rappuoli R. Vaccine manufacturing: Challenges and solutions. Nat. Biotechnol. 2006;24:1377–1383. doi: 10.1038/nbt1261. [DOI] [PubMed] [Google Scholar]
- 11.Vidor E., Soubeyrand B. Manufacturing DTaP-based combination vaccines: Industrial challenges around essential public health tools. Expert Rev. Vaccines. 2016;15:1575–1582. doi: 10.1080/14760584.2016.1205492. [DOI] [PubMed] [Google Scholar]
- 12.Robbins M.J., Jacobson S.H. Analytics for vaccine economics and pricing: Insights and observations. Expert Rev. Vaccines. 2015;14:605–616. doi: 10.1586/14760584.2015.985662. [DOI] [PubMed] [Google Scholar]
- 13.Pastoret P.P. Human and animal vaccine contaminations. Biologicals. 2010;38:332–334. doi: 10.1016/j.biologicals.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Institute of Medicine (US) Immunization Safety Review Committee . In: Immunization Safety Review: SV40 Contamination of Polio Vaccine and Cancer. Stratton K., Almario D.A., McCormick M.C., editors. National Academies Press; 2002. [PubMed] [Google Scholar]
- 15.Skowronski D.M., Janjua N.Z., De Serres G., Sabaiduc S., Eshaghi A., Dickinson J.A., Fonseca K., Winter A.L., Gubbay J.B., Krajden M. Low 2012-13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS ONE. 2014;9:e92153. doi: 10.1371/journal.pone.0092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kew O. Reaching the last one per cent: Progress and challenges in global polio eradication. Curr. Opin. Virol. 2012;2:188–198. doi: 10.1016/j.coviro.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Weaver S.C., Lecuit M. Chikungunya virus and the global spread of a mosquito-borne disease. N. Engl. J. Med. 2015;372:1231–1239. doi: 10.1056/NEJMra1406035. [DOI] [PubMed] [Google Scholar]
- 18.Goupil B.A., Mores C.N. A review of Chikungunya virus-induced arthralgia: Clinical manifestations, therapeutics, and pathogenesis. Open Rheumatol. J. 2016;10:129–140. doi: 10.2174/1874312901610010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levitt N.H., Ramsburg H.H., Hasty S.E., Repik P.M., Cole F.E., Jr., Lupton H.W. Development of an attenuated strain of chikungunya virus for use in vaccine production. Vaccine. 1986;4:157–162. doi: 10.1016/0264-410x(86)90003-4. [DOI] [PubMed] [Google Scholar]
- 20.Edelman R., Tacket C.O., Wasserman S.S., Bodison S.A., Perry J.G., Mangiafico J.A. Phase II safety and immunogenicity study of live chikungunya virus vaccine TSI-GSD-218. Am. J. Trop. Med. Hyg. 2000;62:681–685. doi: 10.4269/ajtmh.2000.62.681. [DOI] [PubMed] [Google Scholar]
- 21.Turell M.J., Malinoski F.J. Limited potential for mosquito transmission of a live, attenuated chikungunya virus vaccine. Am. J. Trop. Med. Hyg. 1992;47:98–103. doi: 10.4269/ajtmh.1992.47.98. [DOI] [PubMed] [Google Scholar]
- 22.Gorchakov R., Wang E., Leal G., Forrester N.L., Plante K., Rossi S.L., Partidos C.D., Adams A.P., Seymour R.L., Weger J. Attenuation of Chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. J. Virol. 2012;86:6084–6096. doi: 10.1128/JVI.06449-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akahata W., Yang Z.Y., Andersen H., Sun S., Holdaway H.A., Kong W.P., Lewis M.G., Higgs S., Rossmann M.G., Rao S., Nabel G.J. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat. Med. 2010;16:334–338. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang L.J., Dowd K.A., Mendoza F.H., Saunders J.G., Sitar S., Plummer S.H., Yamshchikov G., Sarwar U.N., Hu Z., Enama M.E., VRC 311 Study Team Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: A phase 1 dose-escalation trial. Lancet. 2014;384:2046–2052. doi: 10.1016/S0140-6736(14)61185-5. [DOI] [PubMed] [Google Scholar]
- 25.Metz S.W., Gardner J., Geertsema C., Le T.T., Goh L., Vlak J.M., Suhrbier A., Pijlman G.P. Effective chikungunya virus-like particle vaccine produced in insect cells. PLoS Negl. Trop. Dis. 2013;7:e2124. doi: 10.1371/journal.pntd.0002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saraswat S., Athmaram T.N., Parida M., Agarwal A., Saha A., Dash P.K. Expression and characterization of yeast derived Chikungunya virus like particles (CHIK-VLPs) and its evaluation as a potential vaccine candidate. PLoS Negl. Trop. Dis. 2016;10:e0004782. doi: 10.1371/journal.pntd.0004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cimica V., Galarza J.M. Adjuvant formulations for virus-like particle (VLP) based vaccines. Clin. Immunol. 2017;183:99–108. doi: 10.1016/j.clim.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plante K., Wang E., Partidos C.D., Weger J., Gorchakov R., Tsetsarkin K., Borland E.M., Powers A.M., Seymour R., Stinchcomb D.T. Novel chikungunya vaccine candidate with an IRES-based attenuation and host range alteration mechanism. PLoS Pathog. 2011;7:e1002142. doi: 10.1371/journal.ppat.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallengärd D., Kakoulidou M., Lulla A., Kümmerer B.M., Johansson D.X., Mutso M., Lulla V., Fazakerley J.K., Roques P., Le Grand R. Novel attenuated Chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J. Virol. 2014;88:2858–2866. doi: 10.1128/JVI.03453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roques P., Ljungberg K., Kümmerer B.M., Gosse L., Dereuddre-Bosquet N., Tchitchek N., Hallengärd D., García-Arriaza J., Meinke A., Esteban M. Attenuated and vectored vaccines protect nonhuman primates against Chikungunya virus. JCI Insight. 2017;2:e83527. doi: 10.1172/jci.insight.83527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erasmus J.H., Auguste A.J., Kaelber J.T., Luo H., Rossi S.L., Fenton K., Leal G., Kim D.Y., Chiu W., Wang T. A chikungunya fever vaccine utilizing an insect-specific virus platform. Nat. Med. 2017;23:192–199. doi: 10.1038/nm.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramsauer K., Schwameis M., Firbas C., Müllner M., Putnak R.J., Thomas S.J., Desprès P., Tauber E., Jilma B., Tangy F. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based chikungunya vaccine: A randomised, double-blind, placebo-controlled, active-comparator, first-in-man trial. Lancet Infect. Dis. 2015;15:519–527. doi: 10.1016/S1473-3099(15)70043-5. [DOI] [PubMed] [Google Scholar]
- 33.Roy C.J., Adams A.P., Wang E., Plante K., Gorchakov R., Seymour R.L., Vinet-Oliphant H., Weaver S.C. Chikungunya vaccine candidate is highly attenuated and protects nonhuman primates against telemetrically monitored disease following a single dose. J. Infect. Dis. 2014;209:1891–1899. doi: 10.1093/infdis/jiu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plante K.S., Rossi S.L., Bergren N.A., Seymour R.L., Weaver S.C. Extended preclinical safety, efficacy and stability testing of a live-attenuated Chikungunya vaccine candidate. PLoS Negl. Trop. Dis. 2015;9:e0004007. doi: 10.1371/journal.pntd.0004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erasmus J.H., Khandhar A.P., Guderian J., Granger B., Archer J., Archer M., Gage E., Fuerte-Stone J., Larson E., Lin S. A nanostructured lipid carrier for delivery of a replicating viral RNA provides single, low-dose protection against Zika. Mol. Ther. 2018;26:2507–2522. doi: 10.1016/j.ymthe.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erasmus J.H., Archer J., Fuerte-Stone J., Khandhar A.P., Voigt E., Granger B., Bombardi R.G., Govero J., Tan Q., Durnell L.A. Intramuscular delivery of replicon RNA encoding ZIKV-117 human monoclonal antibody protects against Zika virus infection. Mol. Ther. Methods Clin. Dev. 2020;18:402–414. doi: 10.1016/j.omtm.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haese N.N., Broeckel R.M., Hawman D.W., Heise M.T., Morrison T.E., Streblow D.N. Animal models of Chikungunya virus infection and disease. J. Infect. Dis. 2016;214(suppl 5):S482–S487. doi: 10.1093/infdis/jiw284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan Y.H., Lum F.M., Ng L.F.P. Limitations of current in vivo mouse models for the study of Chikungunya virus pathogenesis. Med. Sci. (Basel) 2015;3:64–77. doi: 10.3390/medsci3030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynaud J.M., Kim D.Y., Atasheva S., Rasalouskaya A., White J.P., Diamond M.S., Weaver S.C., Frolova E.I., Frolov I. IFIT1 differentially interferes with translation and replication of alphavirus genomes and promotes induction of type I interferon. PLoS Pathog. 2015;11:e1004863. doi: 10.1371/journal.ppat.1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Couderc T., Chrétien F., Schilte C., Disson O., Brigitte M., Guivel-Benhassine F., Touret Y., Barau G., Cayet N., Schuffenecker I. A mouse model for Chikungunya: Young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog. 2008;4:e29. doi: 10.1371/journal.ppat.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muthumani K., Block P., Flingai S., Muruganantham N., Chaaithanya I.K., Tingey C., Wise M., Reuschel E.L., Chung C., Muthumani A. Rapid and long-term immunity elicited by DNA-encoded antibody prophylaxis and DNA vaccination against Chikungunya virus. J. Infect. Dis. 2016;214:369–378. doi: 10.1093/infdis/jiw111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muthumani K., Lankaraman K.M., Laddy D.J., Sundaram S.G., Chung C.W., Sako E., Wu L., Khan A., Sardesai N., Kim J.J. Immunogenicity of novel consensus-based DNA vaccines against Chikungunya virus. Vaccine. 2008;26:5128–5134. doi: 10.1016/j.vaccine.2008.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mallilankaraman K., Shedlock D.J., Bao H., Kawalekar O.U., Fagone P., Ramanathan A.A., Ferraro B., Stabenow J., Vijayachari P., Sundaram S.G. A DNA vaccine against chikungunya virus is protective in mice and induces neutralizing antibodies in mice and nonhuman primates. PLoS Negl. Trop. Dis. 2011;5:e928. doi: 10.1371/journal.pntd.0000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tretyakova I., Hearn J., Wang E., Weaver S., Pushko P. DNA vaccine initiates replication of live attenuated chikungunya virus in vitro and elicits protective immune response in mice. J. Infect. Dis. 2014;209:1882–1890. doi: 10.1093/infdis/jiu114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss C.M., Liu H., Riemersma K.K., Ball E.E., Coffey L.L. Engineering a fidelity-variant live-attenuated vaccine for chikungunya virus. NPJ Vaccines. 2020;5:97. doi: 10.1038/s41541-020-00241-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riemersma K.K., Steiner C., Singapuri A., Coffey L.L. Chikungunya virus fidelity variants exhibit differential attenuation and population diversity in cell culture and adult mice. J. Virol. 2019;93:e01606-18. doi: 10.1128/JVI.01606-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hidajat R., Nickols B., Forrester N., Tretyakova I., Weaver S., Pushko P. Next generation sequencing of DNA-launched Chikungunya vaccine virus. Virology. 2016;490:83–90. doi: 10.1016/j.virol.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milligan G.N., Schnierle B.S., McAuley A.J., Beasley D.W.C. Defining a correlate of protection for chikungunya virus vaccines. Vaccine. 2019;37:7427–7436. doi: 10.1016/j.vaccine.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 49.Barrett A.D. Yellow fever in Angola and beyond—The problem of vaccine supply and demand. N. Engl. J. Med. 2016;375:301–303. doi: 10.1056/NEJMp1606997. [DOI] [PubMed] [Google Scholar]
- 50.Goldani L.Z. Yellow fever outbreak in Brazil, 2017. Braz. J. Infect. Dis. 2017;21:123–124. doi: 10.1016/j.bjid.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paules C.I., Fauci A.S. Yellow fever—Once again on the radar screen in the Americas. N. Engl. J. Med. 2017;376:1397–1399. doi: 10.1056/NEJMp1702172. [DOI] [PubMed] [Google Scholar]
- 52.The Lancet Yellow fever: A global reckoning. Lancet. 2016;387:1348. doi: 10.1016/S0140-6736(16)30116-7. [DOI] [PubMed] [Google Scholar]
- 53.Lanciotti R.S., Kosoy O.L., Laven J.J., Panella A.J., Velez J.O., Lambert A.J., Campbell G.L. Chikungunya virus in US travelers returning from India, 2006. Emerg. Infect. Dis. 2007;13:764–767. doi: 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.