Abstract
Diseases caused by Phytophthora pathogens devastate many crops worldwide. During infection, Phytophthora pathogens secrete effectors, which are central molecules for understanding the complex plant–Phytophthora interactions. In this study, we profiled the effector repertoire secreted by Phytophthora sojae into the soybean (Glycine max) apoplast during infection using liquid chromatography–mass spectrometry. A secreted aldose 1-epimerase (AEP1) was shown to induce cell death in Nicotiana benthamiana, as did the other two AEP1s from different Phytophthora species. AEP1 could also trigger immune responses in N. benthamiana, other Solanaceae plants, and Arabidopsis (Arabidopsis thaliana). A glucose dehydrogenase assay revealed AEP1 encodes an active AEP1. The enzyme activity of AEP1 is dispensable for AEP1-triggered cell death and immune responses, while AEP-triggered immune signaling in N. benthamiana requires the central immune regulator BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1. In addition, AEP1 acts as a virulence factor that mediates P. sojae extracellular sugar uptake by mutarotation of extracellular aldose from the α-anomer to the β-anomer. Taken together, these results revealed the function of a microbial apoplastic effector, highlighting the importance of extracellular sugar uptake for Phytophthora infection. To counteract, the key effector for sugar conversion can be recognized by the plant membrane receptor complex to activate plant immunity.
Phytophthora sojae apoplastic effector AEP1 triggers pattern-triggered immunity in nonhost plants and contributes to P. sojae virulence by promoting the uptake of extracellular sugar.
Introduction
In nature, plants are challenged by multiple potential pathogenic microbes. To successfully colonize plants, phytopathogens secrete lots of effectors into plant cells or apoplast. These effectors operate different virulence strategies to interfere with plant immunity, growth, development, and microenvironment (Macho and Zipfel, 2015; Aung et al., 2018; He et al., 2020). To fend off pathogen infection, plants exploit plasma membrane-localized receptors, named pattern recognition receptors (PRRs), to recognize the conserved molecular patterns from microbes (microbe-associate molecular patterns [MAMPs]), and subsequently trigger pattern-triggered immunity (PTI; Schwessinger and Ronald, 2012; Zipfel, 2014 ). In addition, plants contain cytoplasmic receptors, which are named resistant proteins, recognizing pathogen avirulence effectors, and triggering effector-triggered immunity (Eitas and Dangl, 2010; Jizong et al., 2019).
The initial interactions between plants and microbes occur in the apoplast, a space that plays an important role in signal exchange and nutrient uptake (Aung et al., 2018). The plant cell wall provides an important physical barrier to ward off pathogen attacks (Hématy et al., 2009). To establish infection, pathogens penetrate plant cell wall to gain access to nutrients and form small wounds at the penetration sites by secreting a large number of cell wall-degrading enzymes (CWDEs; Kämper et al., 2006). Most CWDEs are carbohydrate enzymes targeted to different components of plant cell wall, including cellulose, pectin, hemicelluloses, and lignin (Kubicek et al., 2014). A broad range of extracellular CWDEs results in the formation of monomeric and small oligomeric components, which are then utilized by pathogens for proliferation and further infection. Nevertheless, the process of how pathogens utilize these products in the apoplast is hardly explored.
In the apoplast, perception of microbe-derived molecules by PRRs allows swift response of plants against microbe challenge. As the trigger of plant immune responses, the currently identified MAMPs are derived from either microbial component, such as flg22, flgII-28 epitopes from bacterial flagellin, or microbial effector proteins (Fliegmann and Felix, 2016; Ranf, 2017). Besides triggering immune responses, like reactive oxygen species (ROS) production, immune marker gene expression, and callose deposition, triggering plant cell death is a common character of the proteinaceous MAMPs (Lloyd et al., 2014; Kettles et al., 2017). Recently, advances have been made on the perception of MAMPs by PRRs and downstream signaling mechanisms (Zhou and Zhang, 2020). The identified PRRs belong to either receptor-like kinases (RLKs) or receptor-like proteins (RLPs; Monaghan and Zipfel, 2012), possessing a ligand-binding ectodomain. PRRs with an extracellular leucine-rich repeat (LRR) domain comprise the largest family of receptors in plants. Upon perception of cognate ligands, LRR-type PRRs form a complex with the coreceptor BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 (BAK1)/SOMATIC EMBRYOGENESIS RECEPTOR KINASE 3 (SERK3). RLK-type PRRs, such as Flagellin-sensing 2, associates with BAK1 and activates signal transduction networks via mutual phosphorylation (Perraki et al., 2018). RLPs resemble RLKs, but lack the intracellular signaling domain. RLP-type PRRs often constitutively associate with an adaptor LRR-RLK, namely, Suppressor of BIR1 (SOBIR1), which was proposed to form bimolecular equivalents of RLKs (Gust and Felix, 2014). RLP-type PRRs must also form a complex with the coreceptor BAK1 to activate downstream immune signaling. This often culminated in a series of immune responses, including ROS production, mitogen-activated protein kinase activation, callose deposition, defense gene expression, and hormone biosynthesis (Boller and Felix, 2009).
Phytophthora sojae is the causal agent of soybean (Glycine max) root and stem rot, leading to serious yield losses every year in soybean-growing countries across the world. During infection, P. sojae secrets, multiple apoplastic effectors, to modulate host cell signaling to favor colonization. For example, P. sojae genome contains multiple genes encode necrosis- and ethylene-inducing peptide-like proteins (NLPs). NLPs are well conserved among different microbial taxa and many are cytotoxic in dicotyledonous plants to facilitate microbial infection (Lenarčič et al., 2017). The glycoside hydrolase, xyloglucanase, is an apoplastic effector of P. sojae that promotes infection by degrading plant cell walls (Ma et al., 2015).
In this study, we profiled the apoplastic proteins secreted by P. sojae during early infection stage and characterized an apoplastic effector aldose 1-epimerase (AEP1). AEP1 encodes an active AEP1. Genetic and biochemical experiments illustrate that AEP1 is a key effector required for P. sojae sugar uptake by mutarotation of extracellular aldose, like glucose. In addition, AEP1 can be recognized by plants and triggers cell death and immune responses.
Results
Phytophthora sojae effector AEP1 was identified from the host apoplast during infection
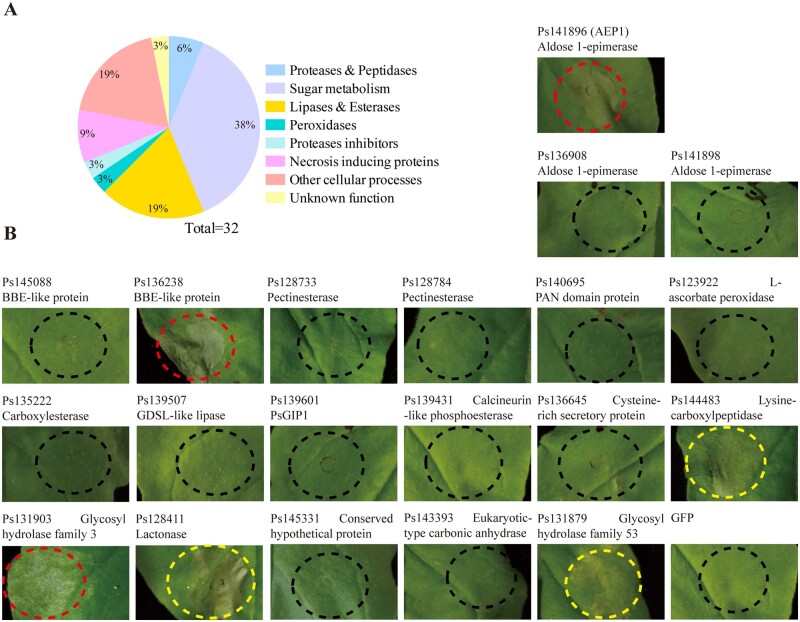
To explore the interactions between Phytophthora and plants in the apoplast space, we inoculated P. sojae zoospores on soybean leaves and collected the apoplastic fluid (AF) from soybean leaves 10 h after inoculation. Gel staining analysis revealed that the protein composition of the AF collected from P. sojae infected leaves are different from those collected from the uninfected control leaves (Supplemental Figure S1B). Infiltration of the AF into Nicotiana benthamiana leaves triggered clear cell death in 2 d (Supplemental Figure S1A). To be noted, the agents in the AF that triggering the plant cell death is sensitive to protease K since the AF after treatment with protease K failed to trigger cell death in N. benthamiana leaves. As a control, the AF from the uninfected soybean leaves could not induce cell death (Supplemental Figure S1A). The AF collected from P. sojae infected or noninfected leaves were then digested by trypsin and analyzed by liquid chromatography–mass spectrometry (LC–MS). Proteins of P. sojae matched by identified peptides were further analyzed whether have a signal peptide. In total, 32 secreted proteins derived from P. sojae were identified and were classified into different groups based on their predicted functions (Figure 1A). Of these, three NLPs and two glycoside hydrolase 12 family proteins were previously reported triggering cell death in multiple plants (Dong et al., 2012; Ma et al., 2015). To investigate whether other P. sojae apoplastic effectors could be perceived by plants, the coding sequences of each effector genes were cloned into PVX and transiently expressed in N. benthamiana leaves. Protein expression was analyzed by western blot (Supplemental Figure S2). For several effectors, including Ps141898, Ps128784, Ps123922, Ps139431, Ps143393, and Ps131879 were not well expressed in N. benthamiana, so we are not sure whether these proteins could be perceived by N. benthamiana. Nevertheless, we constitutively found that Ps141896 triggering strong cell death in N. benthamiana leaves 4 d post agroinfiltration (dpa; Figure 1B). Ps141896 encodes an AEP1 and hereafter was named AEP1. The P. sojae genome also contains another three genes encoding AEP1s, and two other AEP1s, Ps141898, and Ps138908, were also identified in the apoplast fluid (Figure 1B). The unique peptides of AEP1 identified from the apoplast are shown in Supplemental Figure S3.
Figure 1.
AEP1 was identified from apoplast fluid of P. sojae. A, Categories of the proteins isolated from the AF of soybean leaves infected by P. sojae. AF was collected from soybean leaves 10 h after inoculation with P. sojae and assayed by MS. The identified apoplastic effectors were classified based on biological functions. B, Transient expression of apoplastic proteins in N. benthamiana. Red or yellow circles indicate the strong or weak phenotype of cell death. Infiltrated leaves were photographed at 4 dpa.
Apoplastic AEP1 is widely spread among Phytophthora spp
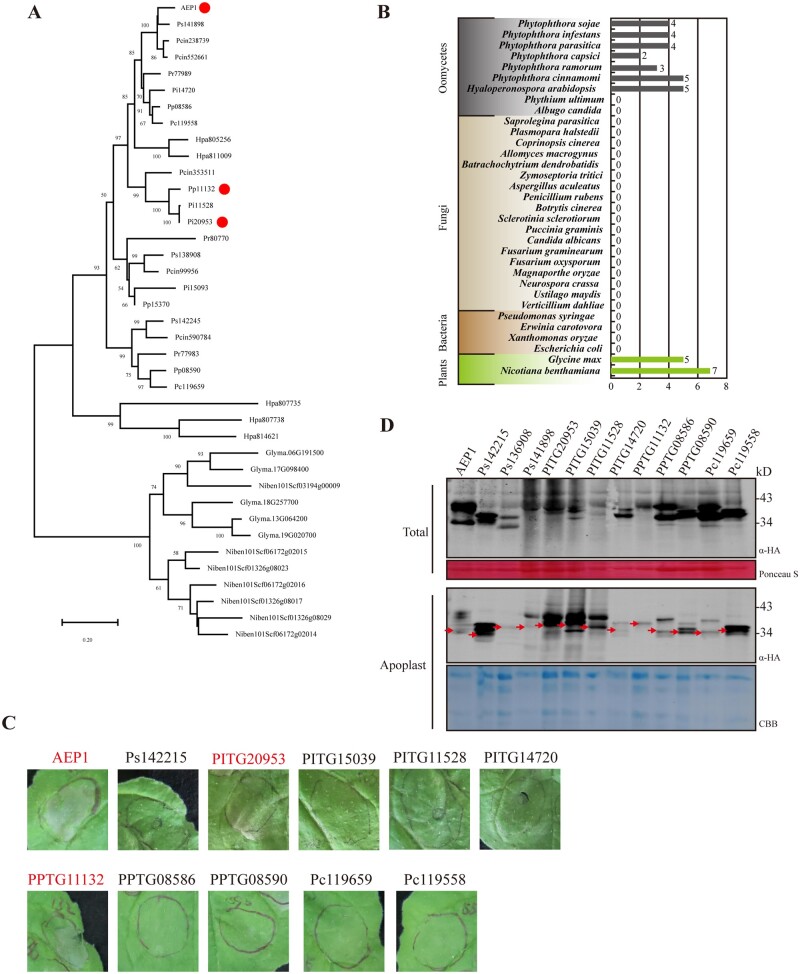
To determine whether the secreted AEP1s represent a type of conserved protein in microbial pathogens, we analyzed the phylogenetic distribution of AEP1in various organisms. AEP1 and the other three paralogs were used to blast 33 genomes from different oomycete, fungal, bacterial, and plant species. The apoplastic AEP1s, which contain a signal peptide were only found in the oomycete and plant species, but not in the fungal or bacterial species we mined (Figure 2B). Apoplastic AEP1s spread among different oomycete pathogens, especially in Phytophthora and Hyaloperonospora species (Figure 2A). Although plants also secret AEP1s, plant AEPs are evolutionarily distant from Phytophthora AEPs (Figure 2A). As AEP1s are conserved among Phytopthora spp., we then determined whether other apoplastic AEPs could be recognized by plants. We cloned the apoplastic AEPs from P. infestans, P. capsici, and P. parasitica transiently expressed in N. benthamiana. The accumulation of AEP1s was monitored in both total extracts and the AFs 36 h after agroinfiltration. Western-blot analysis revealed that all the cloned apoplastic AEP1s were successfully expressed in N. benthamiana, albeit for several samples, even more bands were detected (Figure 2D). Based on this transient expression assay, another two AEPs (PITG20953 and PPTG11132) were found triggering cell death in N. benthamiana, with one from P. infestans which is an adapted pathogen to the solanaceous plants such as potato, and one from P. parasitica which is an adapted pathogen N. benthamiana species (Figure 2C).
Figure 2.
AEP1 is widely spread among Phytophthora spp. A, Phylogenetic analysis of AEP1 and 38 AEP1s containing a signal peptide from the indicated microbial or plant species. AEP1s inducing cell death in N. benthamiana were indicated by red dots. Bootstrap percentage support for each branch is indicated. The scale bar represents 50% weighted sequence divergence. B, The number of AEP1 genes in different microbial or plant species. C, AEP1s containing a signal peptide from Phytophthora species were transiently expressed in N. benthamiana leaves. Infiltrated leaves were photographed 4 dpa. Protein expression assay was repeated more than three times with similar results. Cell death was indicated by red font. D, Western blot analysis of total or apoplastic extractions from N. benthamiana leaves transiently expressing Phytophthora AEP1s. Expected protein bands were indicated by red arrowheads. Ponceau staining or Coomassie brilliant blue (CBB) staining was used to indicate the amount of loading in each sample.
AEP1 targets the apoplast to trigger cell death and immune responses in plant
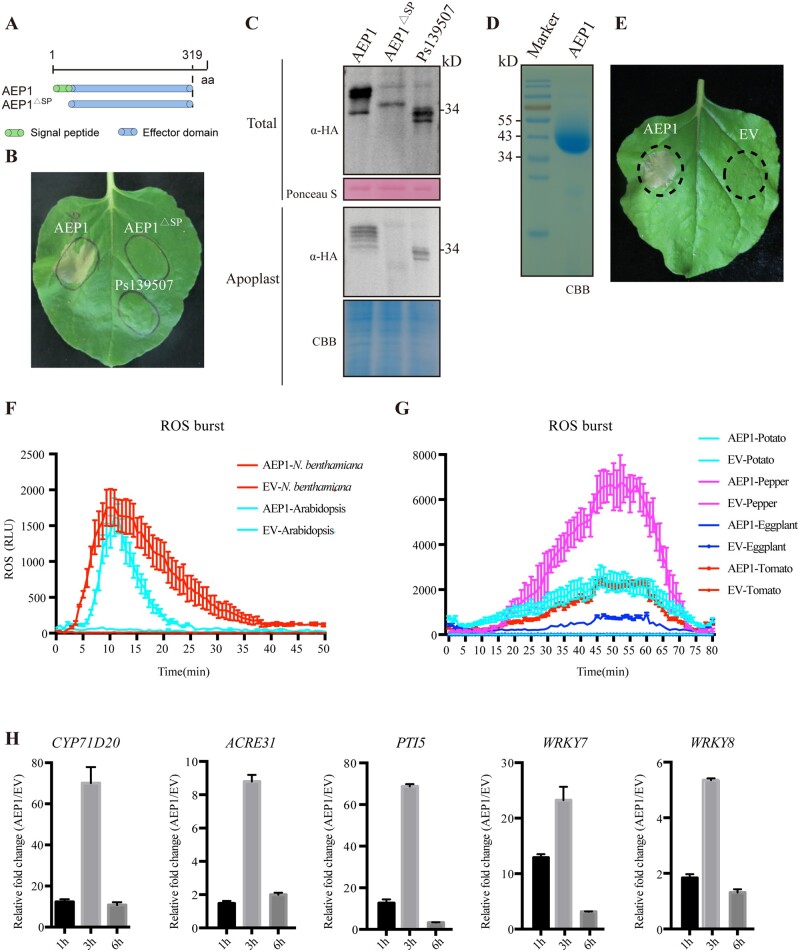
To determine whether AEP1 triggers plant cell death in the apoplast, an AEP1 mutant, AEP1△SP, with deletion of the secretion signal peptide was generated and expressed in N. benthamiana (Figure 3A). In contrast to AEP1, AEP1△SP failed to trigger cell death in N. benthamiana (Figure 3B). Total protein and the AF were collected from the AEP or AEP1△SP expressed N. benthamiana. To be noted, AEP1△SP was successfully expressed in N. benthamiana but could not be detected in the apoplast, as shown in Figure 3C. For the further investigation of the function of AEP1, AEP1 was expressed and purified from Pichia pastoris (Figure 3D). The elutes from the supernatant of P. pastoris transformed with empty vector were named as “EV” and used as control. Again, the purified AEP1 could trigger cell death in N. benthamiana (Figure 3E), while EV failed to trigger cell death. Together, these data demonstrate that only the apoplasic AEP1 could trigger cell death in N. benthamiana.
Figure 3.
AEP1 is secreted to the apoplast triggering cell death and immune responses in plants. A, The schematic of AEP1 and a signal peptide deletion mutant (AEP1△SP). The signal peptide of AEP1 was deleted. Ps139507 was used as a negative control. B, Cell death triggered by AEP1 and the signal peptide mutant AEP1△SP. Both were transiently expressed in N. benthamiana leaves. Cell death was photographed at 4 dpi. C, Detection of AEP1 and signal peptide mutant AEP1△SP in the total protein and the apoplast fluid. Nicotiana benthamiana leaves were collected at 2 dpa and used to isolate total protein or the apoplast fluid. Expression of AEP1 and AEP1△SP was detected by western blot using anti-HA. Ponceau S and CBB staining were used to indicate equal loading in each sample. D, AEP1 was expressed in P. pastoris. CBB staining of the gel to visualize the abundance of purified AEP1. E, Cell death triggered by AEP1. AEP1 (5 µM) and EV were infiltrated into N. benthamiana leaves and photographed at 4 d after infiltration of indicated proteins. F and G, Production of ROS in leaves of N. benthamiana, Arabidopsis, potato, pepper, eggplant, and tomato treated by 1 µM AEP1. EV was used as a negative control. Mean values ± se of three replicates are shown. H, Relative expression of immune marker genes in N. benthamiana leaves triggered by AEP1. Samples were collected 1, 3, and 6 h after infiltration with 200 nM AEP1, and assayed by RT-qPCR. Transcript levels were normalized to EF-1α. Bars represent the mean fold changes of the AEP1-treated leaves relative to EV-treated leaves, which was set as 1. Error bars represent the standard deviation of three biological repeats. Experiments were repeated three times with similar results.
To test whether the perception of AEP1 triggers immune responses, AEP1 was assayed the ability to trigger canonical PTI in N. benthamiana. CYP71D20, ACRE31, PTI5, WRKY7, and WRKY8 are marker genes that are responsive to MAMPs from different microbes in N. benthamiana (Segonzac et al., 2011; Nie et al., 2019). The expression of these marker genes in N. benthamiana was also assayed upon treatment with AEP1. As shown in Figure 3H, the expression of all the five tested marker genes was significantly increased after treatment with AEP1. ROS burst is a potent PTI response triggered by multiple PAMPs (Choi and Klessig, 2016). AEP1-triggered ROS production was examined using a luminol-based assay. As shown in Figure 3F, AEP1 triggers ROS burst in N. benthamiana in a dose-dependent manner (Supplemental Figure S4B). Heat or SDS treatment does not influence AEP1-triggered ROS burst (Supplemental Figure S4A), suggesting that the elicitor activity of AEP1 is probably determined by a derived immunogenic fragment, but not by the tertiary structure. To identify the region required for AEP1-triggered cell death, we employed 2ciq (a hexose-6-phosphate mutarotase, yeast ymr099c) as a template (32.15% identity with AEP1) to build a predicted 3D structure of AEP1 (Supplemental Figures S4, C and D). Multiple loops are exposed on the surface of AEP1, with four loops are relatively longer. Of these, loop 1 is composed of 53 amino acids (AAs), while loops 2, 3, and 4 are composed of 14, 15, and 15 AAs, respectively (Supplemental Figure S5). Based on the predicted structure of AEP1, we constructed four AEP1 mutants (M1, M2, M3, and M4) as shown in the Supplemental Figure S6A, and transiently expressed these mutants in N. benthamiana. The expression of AEP1 and deletion mutants was confirmed by western blot (Supplemental Figure S6C). The mutant M1 and M2, with a deletion of 77 AA or 143 AA at the C-terminus containing loop 4 or loop 3 and loop 4, are still capable to trigger cell death. In contrast, the mutants M3 with a deletion of 195 AA at the C-terminus containing the loops 2, 3, and 4 failed to trigger cell death (Supplemental Figure S6B). In addition, the mutant M4 with a deletion of loop 1 at the N-terminus also failed to trigger cell death (Supplemental Figure S6B). Together, these data showed that both loop 1 and the region between 125th and 177th AAs are indispensable for AEP1-triggered cell death.
Since AEP1 homologs from different Phytophthora species trigger cell death in N. benthamiana, we further determined whether the perception of AEP1 is conserved among different plant species. Leaves of tomato, potato, eggplant, pepper, Arabidopsis, and soybean were treated using the purified AEP1 protein and assayed for ROS burst. AEP1 triggers ROS burst in multiple plant species including tomato, potato, eggplant, pepper, and Arabidopsis, but not in soybean (Figure 3, F and G; Supplemental Figure S6C). In addition, AEP1 could not trigger cell death, or immune marker genes expression in multiple different soybean cultivars (Supplemental Figure S7), demonstrating that soybean failed to respond with AEP1. Taken together, these data illustrated that the AEP1 can be recognized by multiple different plant species to trigger plant immune responses.
AEP1-triggered cell death and immune responses depend on BAK1 but not on SOBIR1
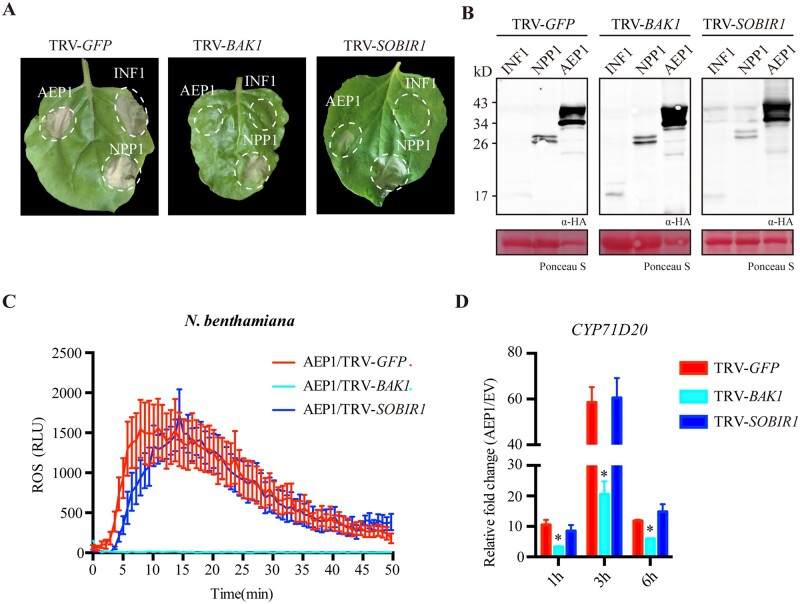
To determine whether the perception of AEP1 requires membrane-localized receptor complex, we assayed AEP1-triggered cell death in N. benthamiana silencing of two central RLK genes, BAK1 and SOBIR1, which mediate multiple proteinous MAMPs recognition. BAK1- or SOBIR1-silenced leaves were agroinfiltrated with AEP1, INF1, and NPP1 expression constructs, and the protein expression was tested by western blot (Figure 4B). AEP1-triggered cell death was abolished in N. benthamiana silencing BAK1, but was not affected in N. benthamiana silencing SOBIR1 (Figure 4A). This also holds true for AEP1-triggered immune responses. AEP1-triggered ROS burst and expression of the marker gene CYP71D20 was significantly reduced in BAK1-silenced N. benthamiana, but not in SOBIR1-silenced plants (Figure 4, C and D). In line with this, AEP1-triggered ROS was almost abolished in Arabidopsis BAK1 mutant (Supplemental Figure S8). Together, these data demonstrate that AEP1-triggered immune signaling requires the central immune sector BAK1.
Figure 4.
BAK1, but not SOBIR1, is required for AEP1-triggered cell death and immune responses. A, Cell death triggered by AEP1, INF1, or NPP1 in TRV: GFP, TRV: BAK1 and TRV-SOBIR1-treated N. benthamiana leaves. AEP1, INF1, or NPP1 were transiently expressed in N. benthamiana leaves by agroinfiltration. Cell death was photographed at 4 dpa. The background of photo was deducted for a better display. B, Western blot of indicated protein expression when transiently expressed in N. benthamiana leaves treated by TRV: GFP, TRV: BAK1 and TRV: SOBIR1. Ponceau S staining was used to indicate equal loading in each sample. C, ROS burst triggered by AEP1 (1 µM) in N. benthamiana leaves treated by TRV: GFP, TRV: BAK1 and TRV: SOBIR1. Mean values ± se of three replicates is shown. D, Relative expression of CYP71D20 in N. benthamiana leaves treated by TRV: GFP, TRV: BAK1 and TRV: SOBIR1. Leaves treated by AEP1 (200 nM) were collected and used to isolate RNA for RT-qPCR. Transcript levels were normalized to EF-1α. Bars represent the mean fold changes of the AEP1-treated leaves relative to EV-treated leaves. The value of EV treated was set as 1. *Significant differences (P < 0.05, t test). Error bars represent the standard deviation of three biological repeats. Experiments were repeated three times with similar results.
Since the adapted pathogens often suppress host PTI by secreted virulence effectors, we then tested whether AEP1-triggered cell death could be suppressed by Phytophthora cytoplasmic effectors. Phytophthora sojae RXLR effectors Avh52, Avh62, Avh109, Avh240, and Avh320 were agroinfiltrated 12 h before AEP1 or INF1 agroinfiltration in N. benthamiana. We found that Avh62, Avh240, and Avh320 could suppress AEP1-triggered cell death in N. benthamiana, while Avh52 and Avh62 failed to do so (Supplemental Figure S9). In contrast, INF1 triggered cell death could be suppressed by Avh52. These results suggest that AEP1- and INF1-induced cell death is activated via different plant signal pathways.
The enzyme activity is not required for AEP1-triggered cell death and PTI
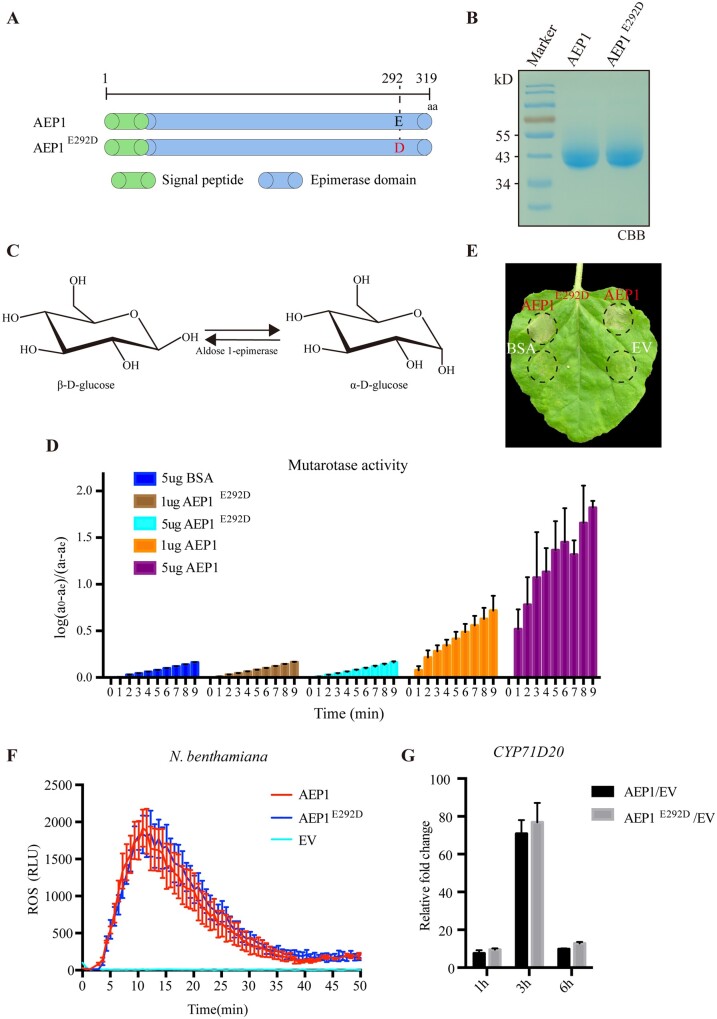
The intracellular AEP1 was previously shown mediating the mutarotation of aldose, like glucose (Figure 5C). To determine whether the extracellular AEP1 AEP1 mediates glucose mutarotation, AEP1 protein was purified from P. pastoris and assayed for the AEP1 activity using the NAD+ and D-glucose dehydrogenase coupled assay. As shown in Figure 5D, AEP1 is capable to convert α-D-glucose to β-D-glucose. Glu-304 is a critical catalytic site for all sugar substrates in Lactococcus lactis (Thoden et al., 2002). Sequence alignment showed that the corresponding catalytic site in P. sojae AEP1 is Glu-292 (Figure 5A) and Glu-292 is conserved among Phytophthora AEP1s (Supplemental Figure S5). We replaced Glu-292 with aspartic acid using site-directed mutagenesis (Figure 5A), and expressed AEP1 and the mutant protein AEP1E292D using P. pastoris (Figure 5B). The enzyme activity assay showed that AEP1E292D completely lost the catalytic activity toward D-glucose (Figure 5D).
Figure 5.
The enzyme activity of AEP1 toward D-glucose. A, Schematic of AEP1 and AEP1 site mutant AEP1E292D. B, CBB staining of the gel to visualize the abundance of purified AEP1 and AEP1E292D mutant. C, The schematic of the AEP1 catalytic reaction. D, The enzyme activity of AEP1 and AEP1E292D mutant. Y-axis indicates the amount of converted β-D-glucose from α-D-glucose with the presence of AEP1 (1 or 5 µg), AEP1E292D mutant (1 or 5 µg) or BSA (5 µg). X-axis indicates the time interval after adding α-D-glucose as substrate into the reaction system. BSA was used as a negative control. Error bars represent the standard deviation of six biological repeats. E, Cell death triggered by AEP1 or AEP1E292D mutant. AEP1 (5 µM), AEP1E292D (5 µM), BSA (5 µM), and EV were infiltrated into N. benthamiana leaves and photographed at 4 d after infiltration of indicated proteins. The background of the photo was deducted for a better display. F, ROS burst in N. benthamiana leaves triggered by 1 µM AEP1 and 1 µM AEP1E292D mutant. Mean values ± se of three replicates are shown. Error bars represent the standard deviation of three biological repeats. G, Relative expression of CYP71D20 in N. benthamiana triggered by AEP1 and AEP1E292D mutant. Nicotiana benthamiana leaves infiltrated with AEP1 (200 nM) and AEP1E292D mutant (200 nM) were collected and assayed by RT-qPCR. Transcript levels were normalized to EF-1α. Bars represent the mean fold changes of the value AEP1- or AEP1E292D mutant-treated leaves relative to EV-treated leaves. The value of EV-treated was set as 1. Error bars represent the standard deviation of three biological repeats. Experiments were repeated three times with similar results.
We further assayed whether the enzyme activity is required for AEP1 mediated immune responses. The AEP1E292D protein could still trigger cell death in N. benthamiana leaves (Figure 5E). Compared with AEP1, AEP1E292D protein triggers similar amount of ROS and expression of CYP71D20 in N. benthamiana (Figure 5, F and G). These results demonstrate that the enzyme activity is not required for AEP1-triggered cell death and immune responses.
AEP1 contributes to virulence of P. sojae by promoting extracellular aldose uptake
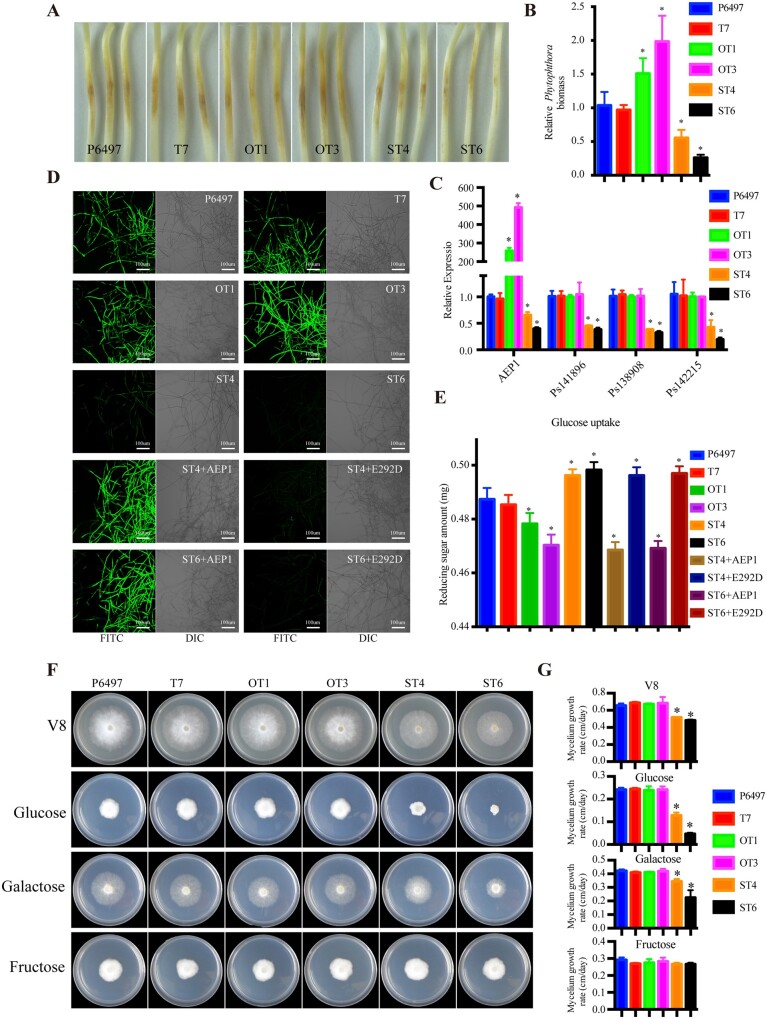
To determine whether AEP1 is implicated in P. sojae development or infection, the expression patterns of AEP1 in different P. sojae development and infection stages were assayed by reverse transcription-quantitative PCR (RT-qPCR; Supplemental Figure S10). AEP1 and the other two paralogs including Ps141898 and Ps138908 were highly upregulated during early infection. To determine the contribution of AEP1 to P. sojae virulence, AEP1 was overexpressed in P. sojae. Overexpression of AEP1 does not significantly affect the morphology and growth of P. sojae (Figure 6F). Two individual transformants, OT1 and OT3, with expression increased by 258- and 492-fold were selected for infection assays (Figure 6C). Zoospores were inoculated onto the etiolated soybean seedlings. The wild-type strain P6497 or the P. sojae transformant T7, with similar AEP1 expression as wild-type strain were used as control. The two overexpressing lines, OT1 and OT3, produced more severe disease symptoms on the etiolated soybean seedlings when compared with the control strains (Figure 6A). Quantitative genomic DNA PCR detected significantly increased P. sojae biomass in the etiolated soybean seedlings inoculated with the two P. sojae overexpessing lines (Figure 6B). To determine whether AEP1 can be recognized by plant as an immunogenic feature during P. sojae infection, we performed infection assays in N. benthamiana using P6497, T7, OT1, and OT3, and tested the triggered immune responses. Zoospores of P. sojae were inoculated on the leaves of N. benthamiana, cell death and H2O2 accumulation were observed at the inoculation sites 2 d postinoculation (dpi; Supplemental Figure S11, A and B). AEP1 overexpression lines OT1 and OT3 elicited stronger responses when compared with wild-type or transformation control (Supplemental Figure S1, A and B1). In addition, OT1 and OT3 also induced higher expression of immune marker gene CYP71D20 (Supplemental Figure S11C). These data illustrate that AEP1 could be recognized by plant and elicit defense against Phytophthora. OT1 and OT3 also induced higher expression of immune marker gene CYP71D20 (Supplemental Figure S11C). To determine whether AEP1 promotes P. sojae infection requires the enzyme activity, enzyme-defect mutant AEP1E292D was also overexpressed in P. sojae strain P6497. Two overexpression lines E292D-1 and E292D-2 were selected for infection assays (Supplemental Figure S10C). In contrast to AEP1, overexpression of AEP1E292D does not enhance P. sojae infection (Supplemental Figure S12, A and B). The growth of these overexpression mutants on medium shows no difference when compared with the wild-type P. sojae strain P6497 (Supplemental Figure S12, D and E). Together, these results demonstrated that AEP1 promotes P. sojae infection by its enzyme activity.
Figure 6.
AEP1 is a virulence factor of P. sojae mediating extracellular aldose uptake. A, Disease symptoms of the etiolated soybean seedlings inoculated with P. sojae wild-type strain P6497 or transformants. Disease symptoms were photographed 2 dpi. B, Relative biomass of P. sojae wild-type strain P6497 and transformants detected in the inoculated etiolated soybean seedlings. Inoculated samples were collected 2 dpi and were used to isolate gDNA for quantitative PCR. The values were calculated by normalizing P. sojae actin to soybean GmCYP2 and the value detected in the P6497-inoculated sample was set as 1. C, Transcript levels of AEP1 in mycelium of P. sojae transformants assayed by RT-qPCR. Phytophthora sojae actin was used as an internal reference. AEP1 expression levels were normalized to that detected in P6497. D, Microscope investigation of the P. sojae wild-type strain P6497 or transformants mycelium taken up 2-NBDG. AEP1 (5 µg) or AEP1E292D (5 µg) were supplemented before inoculation. Bars = 100 µM. E, Amount of rest glucose was measured by reducing sugar quantify Kit. F and G, P6497 wild-type and transformants were cultured on nutrient-rich V8 medium or nutrient-poor Plich medium, in which glucose was substituted with galactose or fructose. Colonies were photographed and measured with growth. *Significant differences (P < 0.05, t test).
To further explore the function of AEP1, we tried to knock AEP1 by CRISPR/Cas9. Six different targets of sgRNA were used for P. sojae transformation (Supplemental Table S2), but no homozygous mutant was identified. Due to the sequence similarity of AEPs, we then generated P. sojae transformants with four AEP1 genes silenced to avoid functional redundancy of AEP1 paralogs (Figure 6C). The RT-qPCR assay shows that the expression of AEP1 and the other three paralogs was significantly reduced in the silencing transformants ST4 and ST6. Strikingly, the growth rate of the silencing mutants ST4 and ST6 was slower than the wild-type P6497 or control transformant T7 when cultured on V8 agar medium (Figure 6, F and G). Since the reported substrate of the intracellular AEP1s are aldoses, such as glucose or galactose, we determined whether silencing of the extracellular AEP1 genes influenced the growth of P. sojae due to reduced ability to uptake aldoses. We replaced the major nutrient source glucose with different aldoses (i.e. glucose or galactose) or ketose (i.e. fructose) in the nutrient-poor Plich medium. The growth of ST4 and ST6 were much slower than the wild-type P6497 and T7 in glucose or galactose but not on fructose medium (Figure 6, F and G), demonstrating that the P. sojae extracellular AEP1s mediate the uptake of aldoses. To test whether AEP1 itself plays a role in the uptake of aldoses, mycelia of P. sojae P6497 and derived transformants were incubated with 20 µM 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl) Amino)-2-Deoxyglucose (2-NBDG), a fluorescent-labeled glucose used to monitor glucose uptake in living cells. The fluorescence signal of the silencing mutants ST4 and ST6 was significantly weaker than the control lines and the overexpression transformants (Figure 6D). Furthermore, supplementing with purified AEP1 protein could rescue the phenotype of ST4 and ST6 in glucose uptake, while the enzyme mutant AEP1E292D failed to do so (Figure 6, D and E). Quantitative measurement of resting glucose after incubating with mycelia further demonstrates that only AEP1, but not the enzyme mutant, could mediate the uptake of glucose in P. sojae (Figure 6E; Supplemental Figure S13). Taken together, these results demonstrate that AEP1 is an essential virulence factor of P. sojae for extracellular sugar uptake during the early infection stage.
Discussion
Effectors are employed as a common weapon in the arsenal of plant pathogens to interrupt host immune system in favor of pathogen propagation. As a counteract, plants developed surveillance system to perceive microbial effectors and activate the innate immunity. As such, effectors are considered as a crucial molecular marker to dissect the activation and suppression of plant immunity. Apoplastic effector proved to be an important player in plant–microbe interactions. In this study, we profiled the apoplastic effectors secreted by P. sojae during the early infection of soybean using LC–MS. AEP1, which encodes an apoplastic AEP1 was identified triggering cell death in N. benthamiana. AEP1 can be recognized by different plant species and trigger immune responses. Apoplastic AEP1s are well conserved among Phytophthora pathogens. Although AEP1 could not be recognized by soybean, the natural host of P. sojae, another two apoplastic AEP1s from N. benthamiana-adapted pathogens P. parasitica and P. infestans are also recognized by N. benthamiana. As such, the apoplastic AEP1 acts as a MAMP. Since AEP1-triggered immunity is resistant to heat and SDS treatments, suggesting that an epitope exposed on the surface of AEP1 protein probably determined the immunogenic activity of AEP1.
Upon pathogen attack, timely activation of plant defense response relies on the effective coordination of microbial perception and downstream signaling events. BAK1 is a central molecular sector that forms complex with PRRs with an extracellular LRR domain upon perception of cognate ligands. Multiple evidence illustrate that the PRR–BAK1 complex is tightly controlled during activation of PTI to maintain the signaling homeostasis. Silencing BAK1 in N. benthamiana could completely destroy the cell death and immune responses triggered by AEP1. AEP1 elicited ROS burst was also severely impaired in Arabidopsis BAK1 null-mutant. These data strongly suggest that AEP1 is perceived by plant through membrane-localized PRR–BAK1 complex. In contrast to other elicitors identified in Phytophthora, such as INF1 and XEG1, AEP1-triggered cell death and immune responses are not dependent on SOBIR1, an adaptor kinase associating with multiple RLP-type PRRs. In addition, we found cell death triggered by INF1 and AEP1 was suppressed by different Phytophthora effectors. These data suggest that AEP1 is recognized by plants via a distinct pathway.
The plant apoplast is a sugar-rich niche, and efficient uptake of sugars is essential for pathogen infection (Naseem et al., 2017). It has been reported that Arabidopsis activates sugar transporters to control extracellular sugars upon recognition of the bacterial PAMPs, and thereby restricting bacterial infection (Yamada et al., 2016). In this study, genetic and biological experiments proved that AEP1 contributes to P. sojae virulence by converting extracellular aldose to the β-anomer, which could be taken up by Phytophthora. Intracellular AEP1 has been extensively studied in the so-called Leloir pathway, which is conserved among most organisms for galactose metabolism (Buttin, 1963; Bouffard et al., 1994). Once β-D-galactose was taken up into the cell, it should be converted to α-anomer by mutarotase before entering the Leloir pathway (Howard and Heinrich, 1965). In contrast, the exact function of the extracellular AEP1, especially in the plant–microbe interface, is hardly explored. In this study, we used D-glucose as the substrate of AEP1, and found that AEP1 promotes the glucose uptake by P. sojae, demonstrating that the extracellular AEP1 AEP1 also mediates the conversion of the α-form aldose to the β-form. Moreover, this process is essential for the sugar uptake by P. sojae, since the AEP1E292D mutant fail to do so. Furthermore, the pathogenicity assays using AEP1 or AEP1E292D overexpressing P. sojae lines demonstrate that modification of plant extracellular aldoses contributes to Phytophthora infection. In nature, the composition of plant extracellular sugar is rather complicated. Although we demonstrate that D-glucose is a substrate of AEP1, there might be other aldoses that can be modulated by AEP1 for efficient sugar uptake of Phytophthora pathogens. Therefore, further analyses of exact sugar profiles exploited by Phytophthora pathogens will promote our how Phytophthora pathogens exploit host nutrient for infection.
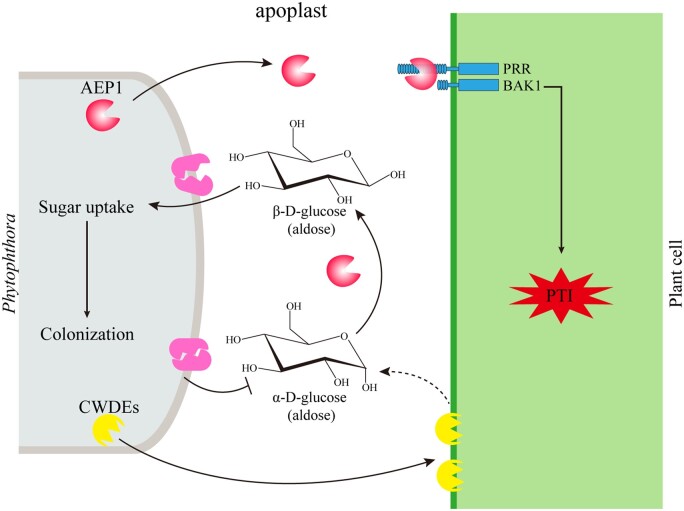
In conclusion, this study proposed a working model of AEP1 (Figure 7). Once the initial interaction with plants occurs in the apoplast, Phytophthora secrets glycoside hydrolases to degrade plant cell wall and convert extracellular sugars for uptake using AEP1, like AEP1. At the same time, plants, such as N. benthamiana, recognize apoplastic AEP1 by plant membrane receptor complex and activate defense signal cascades to mount resistance against Phytophthora.
Figure 7.
Working model for the function of AEP1.
Materials and methods
Phytophthora and plant growth conditions
The P. sojae strain P6497 was routinely grown on V8 juice agar plate at 25°C in the dark. Zoospore suspensions were obtained by repeatedly washing 3-d-old hyphae with sterile distilled water for four times.
Soybean (G. max; Hefeng 47) seeds were grown in vermiculite and maintained at 25°C in the dark for 3 d to harvest the etiolated hypocotyls. Nicotiana benthamiana, Arabidopsis thaliana, tomato (Solanum lycopersicum), potato (Solanum tuberosum L), and eggplant (Solanum megongena L) were grown in a controlled climate chamber at 19°C–22°C with a 14-h photoperiod for 4–6 weeks.
Phylogenetic analysis
The sequence of AEP1 lacking the signal peptide was used for searching AEP1 in microbial and plant species. After correction for redundancy and signal peptide prediction (http://www.cbs.dtudk/services/SignalP/), 38 protein sequences were identified. The mature protein sequences were aligned using the ClustalW2 program and phylogenetic tree was constructed using MEGA5 with maximum likelihood.
Plasmid construction
The coding sequences for AEP1 and other apoplastic effectors were amplified from cDNA of Phytophthora mycelium using primers listed in the Supplemental Table S1. The purified fragments were cloned into the vectors pGR107 or pPic9k, respectively, using the ClonExpress II One Step Cloning Kit (Vazyme Biotech Co. Ltd., Nanjing, China). Each construct was verified by sequencing.
Phytophthora sojae transformation and infection assays
Phytophthora sojae transformants were generated using PEG-mediated protoplast transformation as described previously (Hua et al., 2008). The etiolated hypocotyls of soybean seedlings were inoculated with zoospore suspensions (100 zoospores) of the wild-type or transformants, and maintained at 25°C in the dark. The soybean hypocotyls were photographed 2 d after inoculation and were collected for biomass detection.
For soybean root inoculation, etiolated soybean roots were soaked in zoospores suspensions (100 µL−1) for 30 min and then kept in plastic boxes with high humidity. The inoculated soybean roots were collected at different time points for gene expression analyses.
RNA, DNA extraction, and RT-qPCR
Collected plant leaves or Phytophthora samples were ground in liquid nitrogen and used for RNA or DNA isolation. Genomic DNA was extracted using the genomic DNA kit (TIANGEN Biotech, Beijing, China) following procedures described by the manufacturer. Total RNA was isolated using an RNA kit (Omega Bio-Ten, Norcross, GA, USA) and then used as templates for cDNA synthesis. Reverse transcription was performed using PrimeScript RT reagent Kit (http://www.takara-bio.com). RT-qPCR was performed on an ABI 7500 Fast Real-Time PCR system (Applied Biosystems Inc., Foster City, CA, USA) using ChamQ SYBR Color qPCR Master Mix (Vazyme Biotech Co., Ltd, China) with primers listed in the Supplemental Table S1. Data were analyzed using the 2−ΔΔCT method as described (Wang et al., 2018).
Apoplast fluid isolation
Leaves from 2-week-old soybean seedlings were collected, treated with 0.1% (v/v) Tween-20, and then washed thoroughly using sterile distilled water. Soybean leaves were inoculated with P. sojae zoospores (100 µL−1) and incubated at 25°C for 10 h. The inoculated leaves were soaked with precooling PBS (Phosphate Buffer Saline) buffer and placed in a vacuum pump. Slowly release the vacuum and the PBS buffer was pushed into the leaves apoplast. Leaf surface was dried to remove the remaining PBS buffer and then put into a 10 mL syringe without needle for centrifugation at 1,000g for 30 min at 4°C. The apoplast fluid was collected into a 50 mL centrifuge tubes and was further filtered through 0.25-µm membrane column (Merck Millipore, St. Louis, MO, USA).
The apoplast fluid of N. benthamiana leaves was collected 2 dpa following the abovementioned procedures.
Oxidative burst assay
Leaf discs (Ø 0.5 cm) were collected from 5-week-old N. benthamiana, 4-week-old tomato, potato, eggplant, Arabidopsis, or 2-week-old soybean plants, and then floated in 200 uL sterile water in a 96-well plate overnight. The sterile water was replaced with 200 uL reaction buffer containing luminol/peroxidase (35.4 µg mL−1 luminol, 10 µg mL−1 peroxidase) and different PAMPs (200 nM flg22,1 µM AEP1/1 µM AEP1E292D). Luminescence was measured using the GLOMAX96 microplate luminometer (Promega, Madison, WI, USA).
Agroinfiltration in N. benthamiana
Agrobacterium tumefaciens strain GV3101 was used for both transient expression and VIGS assays in N. benthamiana. Agrobacterium tumefaciens carrying various vectors were grown overnight in LB medium supplemented with Kanamycin (50 mg L−1), Rifampin (50 mg L−1). A. tumefaciens cells were collected by centrifuge for 5 min at 4,000 rpm, washed three times using infiltration buffer (10 mM MgCl, 10 mM MES pH 5.7, 20 nM acetylsyringone) and incubated in the infiltration buffer for 2 h before infiltration. For transient expression assay, A. tumefaciens suspensions were infiltrated into leaves of 4–6-week-old N. benthamiana with appropriate concentrations. Leaves of 4–6-week-old N. benthamiana were used for expression. Two days after infiltration, leaves were harvested for detecting protein accumulation. Cell death was observed 4 d after infiltration. For VIGS assay, 10–12-d-old N. benthamiana seedlings were used. pTRV1 containing Agrobacterium were mixed with pTRV2 containing Agrobacterium in a 1:1 ratio.
Heterologous expression and purification of proteins
Pichia pastoris strain KM71 was used for protein expression. Linearized pPic9k plasmids containing AEP1 or mutant were transformed into KM71 strain under electroporation voltage 1.5 kV. Transformed colonies were verified using PCR and subsequently cultured for 5 d to express proteins. The cultures were centrifuged at 8,000 rpm for 30 min and the supernatant was collected. Proteins in the supernatant were precipitated in Ammonium sulfate (500 g/L) overnight, collected by centrifugation at 8,000 rpm for 1 h and dissolved in sterile distilled water. AEP1 or mutant was purified using AKTA Avant 25 (GE Healthcare, Chicago, IL, USA) in the following flow chart: desalting, Ni-trap, and desalting. HiPrepTM 26/10 Desalting (GE Healthcare) and HisSep Ni-NTA 6FF Chromatography Column (YEASEN, Shanghai, China) were used. The elutes from the supernatant of P. pastoris transformed with EV and followed with the same protein purification procedures with AEP1 or mutant was named as “EV.”
The enzyme activity assay of AEP1 and mutants
The AEP1 activity was detected using the NAD+ and D-glucose dehydrogenase coupled assay. In this assay, the conversion of α-D-glucose to β-D-glucose is coupled with the oxidation of β-D-glucose by β-D-glucose dehydrogenase and reduction of NAD+. The assay buffer contained 0.1 M Tris–HCl buffer (PH 7.5), 3 M NAD+, 10 units of β-D-glucose dehydrogenase and epimerase protein. The reaction was initiated by adding 5 mM freshly dissolved D-glucose and monitored by measuring the increase of the absorbance at 340 nm. The amount of converted D-glucose is indicated by log(a0–ae)/(at–ae). a0, at, and ae are the observed angular rotations at time zero, t, and equilibrium.
Phytophthora sojae sugar uptake assay
The P. sojae strain P6497 and transformants were cultured in 10% (v/v) V8 juice medium at 25°C in the dark for 2 d. Before inoculated with 20 µM 2-NBDG (KGAF017; KeyGEN BioTECH, Nanjing, China), P. sojae mycelia were put into sterile distilled water for half an hour starvation treatment. The mycelia were transferred to distilled water 10 min after incubation with 2-NBDG and then observed by confocal microscope at 540 nm.
The assay for quantitative measure of glucose
The P. sojae strain P6497 and transformants were routinely grown in 10% (v/v) V8 juice at 25°C in the dark for 4 d. Mycelia (10 mg) were incubated with 500 µL glucose solution (100 µg mL−1). The resting glucose was detected by the reducibility using the kit (BC0235; Solarbio LIFE SCIENCES, Beijing, China) following the manufacturer’s instructions.
Statistical analyses
Student’s t test was used for the statistical analysis with excel software. *Significant differences, P < 0.05.
Accession numbers
Sequence data for AEP1, other AEP1s of Phytophthora and apoplastic proteins shown in Figure 1B can be found in Joint Genome Institute (http://ensemblgenomes.org/) with ID shown in Figure 2A, like Ps141896 (AEP1). Sequences of AEP1 from Soybean (G. max) can be found in Phytozome (http://phytozome.jgi.doe.gov/pz/prtal.html) with ID shown in Figure 2A, like Glyma.06G191500. Sequences of AEP1 from N. benthamiana can be found in Solgenomics (http://solgenomics.net) with ID shown in Figure 2A, like Niben101Scf06172g02015.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. AF from P. sojae infected soybean leaves triggers cell death in N. benthamiana.
Supplemental Figure S2. Detection of effector accumulation after transient expression in N. benthamiana by western blot.
Supplemental Figure S3. Unique peptides matching AEP1 identified in the apoplast fluid.
Supplemental Figure S4. AEP1 triggers ROS in N. benthamiana.
Supplemental Figure S5. Sequence (without signal peptide) alignment of secreted AEP1 in different Phytophthora spp.
Supplemental Figure S6. The N-terminal region is required for AEP1-triggered cell death.
Supplemental Figure S7. AEP1 could not trigger cell death or immune responses in soybean.
Supplemental Figure S8. AEP1 could trigger ROS burst in Arabidopsis.
Supplemental Figure S9. AEP1-triggered cell death could be suppressed by P. sojae effectors.
Supplemental Figure S10. AEP1s are highly expressed during infection.
Supplemental Figure S11. Phytophthora sojae AEP1 overexpression lines triggered stronger immune responses on N. benthamiana.
Supplemental Figure S12. AEP1 contributes to P. sojae virulence dependent on enzyme activity.
Supplemental Figure S13. The amount of extracellular D-glucose taken up by Phytophthora strains at different time points.
Supplemental Table S1. List of primers used in this study.
Supplemental Table S2. sgRNA targets for AEP1 knockout in P. sojae.
Supplementary Material
Acknowledgments
We thank Prof. Qing Yang (Dalian University of Technology) for kindly providing the pPic9k vector and KM71 strain for protein heterologous expression.
Funding
This research was supported by the grant to Y.W. from the China National Funds for Innovative Research Groups (grant no. 31721004) and the grants to Y.W. from the China National Funds (grant no. 31872927), from the Natural Science Funds for Distinguished Young Scholars of Jiangsu Province (grant no. BK20190027), and by “the Fundamental Research Funds for the Central Universities” (grant nos. JCQY201904 and KJJQ202002).
Conflict of interest statement. None declared.
Y.W., Y.W., and Y.X. designed the research. Y.X. performed the experiments and analyzed data. Y.Z., Y.S., F.L., and H.W. are involved in experiment performed on the phenotype of Phytophthora mutants. J.Z., B.G., and J.H. are involved in biochemical experiment performed with effector protein. S.M. gave suggestionfor this research. Y.W., Y.W., and Y.X. wrote the manuscript. All authors reviewed and approved the manuscript for publication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Yuanchao Wang (wangyc@njau.edu.cn).
References
- Aung K, Jiang Y, He SY (2018) The role of water in plant–microbe interactions. Plant J 93: 771–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bouffard GG, Rudd KE, Adhya SL (1994) Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J Mol Biol 244: 269–278 [DOI] [PubMed] [Google Scholar]
- Buttin G (1963) [ REGULATORY MECHANISMS IN THE BIOSYNTHESIS OF THE ENZYMES OF GALACTOSE METABOLISM IN ESCHERICHIA COLI K 12. I. THE INDUCED BIOSYNTHESIS OF GALACTOKINASE AND THE SIMULTANEOUS INDUCTION OF THE ENZYMATIC SEQUENCE]. J Mol Biol 7: 164–182 [DOI] [PubMed] [Google Scholar]
- Choi HW, Klessig DF (2016) DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol 16: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Kong G, Qutob D, Yu X, Wang Y (2012) The NLP toxin family in Phytophthora sojae includes rapidly evolving groups that lack necrosis-inducing activity. Mol Plant Microbe Interact 25: 896–909 [DOI] [PubMed] [Google Scholar]
- Eitas TK, Dangl JL (2010) NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 13: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegmann J, Felix G (2016) Immunity: flagellin seen from all sides. Nat Plants 2: 16136. [DOI] [PubMed] [Google Scholar]
- Gust AA, Felix G (2014) Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr Opin Plant Biol 21: 104–111 [DOI] [PubMed] [Google Scholar]
- He Q, McLellan H, Boevink PC, Birch PRJ (2020) All roads lead to susceptibility: the many modes of action of fungal and Oomycete intracellular effectors. Plant Commun 1: 100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K, Cherk C, Somerville S (2009) Host-pathogen warfare at the plant cell wall. Curr Opin Plant Biol 12: 406–413 [DOI] [PubMed] [Google Scholar]
- Howard SM, Heinrich MR (1965) The anomeric specificity of yeast galactokinase. Arch Biochem Biophys 110: 395–400 [DOI] [PubMed] [Google Scholar]
- Hua C, Wang Y, Zheng X, Dou D, Zhang Z, Govers F, Wang Y (2008) A Phytophthora sojae G-protein alpha subunit is involved in chemotaxis to soybean isoflavones. Eukaryot Cell 7: 2133–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jizong W, Meijuan H, Jia J, Qi Z, Han G (2019) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364: eaav5870. [DOI] [PubMed] [Google Scholar]
- Kämper J, Kahmann R, Bölker M, Ma L-J, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Müller O, et al. (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101 [DOI] [PubMed] [Google Scholar]
- Kettles GJ, Bayon C, Canning G, Rudd JJ, Kanyuka K (2017) Apoplastic recognition of multiple candidate effectors from the wheat pathogenZymoseptoria triticiin the nonhost plantNicotiana benthamiana. New Phytol 213: 338–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek CP, Starr TL, Glass NL (2014) Plant cell wall–degrading enzymes and their secretion in plant-pathogenic fungi. Ann Rev Phytopathol 52: 427. [DOI] [PubMed] [Google Scholar]
- Lenarčič T, Albert I, Böhm H, Hodnik V, Pirc K, Zavec AB, Podobnik M, Pahovnik D, Žagar E, Pruitt R (2017) Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 358: 1431. [DOI] [PubMed] [Google Scholar]
- Lloyd SR, Schoonbeek HJ, Trick M, Zipfel C, Ridout CJ (2014) Methods to study PAMP-triggered immunity in Brassica species. Mol Plant Microbe Interact 27: 286–295 [DOI] [PubMed] [Google Scholar]
- Ma Z, Song T, Zhu L, Ye W, Wang Y, Shao Y, Dong S, Zhang Z, Dou D, Zheng X, et al. (2015) A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27: 2057–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C (2015) Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol 23: 14–22 [DOI] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Naseem M, Kunz M, Dandekar T (2017) Plant-pathogen maneuvering over Apoplastic sugars. Trends Plant Sci 22: 740–743 [DOI] [PubMed] [Google Scholar]
- Nie J, Yin Z, Li Z, Wu Y, Huang L (2019) A small cysteine-rich protein from two kingdoms of microbes is recognized as a novel pathogen-associated molecular pattern. New Phytol 222: 995–1011 [DOI] [PubMed] [Google Scholar]
- Perraki A, DeFalco TA, Derbyshire P, Avila J, Sere D, Sklenar J, Qi X, Stransfeld L, Schwessinger B, Kadota Y, et al. (2018) Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 561: 248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S (2017) Sensing of molecular patterns through cell surface immune receptors. Curr Opin Plant Biol 38: 68–77 [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Ronald PC (2012) Plant innate immunity: perception of conserved microbial signatures. Annu Rev Plant Biol 63: 451–482 [DOI] [PubMed] [Google Scholar]
- Segonzac C, Feike D, Gimenez-Ibanez S, Hann DR, Zipfel C, Rathjen JP (2011) Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol 156: 687–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoden JB, Kim J, Raushel FM, Holden HM (2002) Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis. J Biol Chem 277: 45458–45465 [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu Y, Sun Y, Wang H, Wang Y (2018) Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat Commun 9: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Saijo Y, Nakagami H, Takano Y (2016) Regulation of sugar transporter activity for antibacterial defense in Arabidopsis. Science 354: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Zhou JM, Zhang Y (2020) Plant immunity: danger perception and signaling. Cell 181: 978–989 [DOI] [PubMed] [Google Scholar]
- Zipfel C (2014) Plant pattern-recognition receptors. Trends Immunol 35: 345–351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.