Abstract
A constitutive DNase I-hypersensitive site 5′ of the chicken β-globin locus, termed 5′HS4 or cHS4, has been shown to insulate a promoter from the effect of an upstream enhancer and to reduce position effects on mini-white expression in Drosophila cells; on the basis of these findings, it has been designated a chromatin insulator. We have examined the effect of the cHS4 insulator in a system that assays both the level of gene expression and the rate of transcriptional silencing. Because transgenes flanked by insulator elements are shielded from position effects in Drosophila cells, we tested the ability of cHS4 to protect transgenes from position effects in mammalian cells. Flanking of an expression vector with the cHS4 insulator in a colony assay did not increase the number of G418-resistant colonies. Using lox/cre-based recombinase-mediated cassette exchange to control integration position, we studied the effect of cHS4 on the silencing of an integrated β-geo reporter at three genomic sites in K562 erythroleukemia cells. In this assay, enhancers act to suppress silencing but do not increase expression levels. While cHS4 blocked enhancement at each integration site, the strength of the effect varied from site to site. Furthermore, at some sites, cHS4 inhibited the enhancer effect either when placed between the enhancer and the promoter or when placed upstream of the enhancer. These results suggest that the activity of cHS4 is not dominant in all contexts and is unlikely to prevent silencing at all genomic integration sites.
It has been hypothesized that the genomes of higher eukaryotes are organized in domains similar in chromatin structure and that “boundary” or “insulator” elements may form borders that separate active and inactive chromatin (9, 10, 12, 20, 22). These elements would ensure that transcriptionally active genes or loci are sequestered from those that are inactive or that the influence of regulatory structures in a locus is restricted from spreading to neighboring regions. Such boundaries may be formed by attachment to the nuclear matrix or the formation of higher-order chromatin structures; no clear evidence for either mechanism is available, although recent observations suggest that insulator proteins may localize to the nuclear periphery (21). Thus, while it is believed that insulators are important for the functional organization of complex genomes in higher eukaryotes, their structure and mechanism of action are not well understood.
While the ability to block the effect of an enhancer at ectopic sites of chromosomal integration in gene transfer experiments is an effect attributed to insulators, the relationship of this effect to the function of these elements in their native contexts remains uncertain. For example, the insulators scs and scs′ located downstream of hsp70 genes in the Drosophila 87A7 heat shock locus block repressive and activating chromosomal position effects from random sites of integration in Drosophila DNA (25, 26). On the basis of their ability to establish domains of independent gene activity, these elements were identified as insulators. Establishment of domains of independent activity and blockage of activation by enhancers are effects consistent with a simple model of chromatin insulation. However, there is no direct evidence that these effects are characteristics shared by all insulators, related to their normal function, or apparent in every chromosomal context. In fact, it is not clear if most gene domains have discrete boundaries or if these boundaries create discrete regions of transition in chromatin structure in vivo.
A region of constitutive DNase I hypersensitivity located near the upstream boundary of the chicken β-globin locus exhibits characteristics and effects similar to those of other chromatin insulators (7, 8, 12). Chromatin upstream of this site is condensed. Downstream, the chromatin is open, histone H4 is more abundant in the acetylated state, and genes are transcriptionally active (34). In human K562 erythroleukemia cells, chicken 5′HS4 (cHS4) had enhancer-blocking activity: in a colony assay, it effectively inhibited the stimulatory effect of an enhancer (5′HS2 of the mouse β-globin locus control region [LCR]) when placed between the enhancer and a promoter. In addition, when cHS4 flanked the white minigene in transgenic Drosophila DNA, it protected white expression from position effects (7). Much of the insulator activity was mapped to a 250-bp G-C-rich core that contains CpG sites that are hypomethylated in erythroid and nonerythroid tissues (8). These CpG “islands” are not associated with significant promoter activity. The core contains canonical binding motifs for the transcriptional activator SP-1 and for the yeast α-2 repressor, but it is not known if these proteins (or related ones) are important for the function of cHS4. By itself, cHS4 has shown little stimulatory or repressive effect on gene expression, and its function in its native context has not been established. One possible function of the cHS4 element is to restrict upstream propagation of the active β-globin domain, but it has also been speculated that cHS4 may protect the β-globin locus from the repressive effect of neighboring heterochromatin and so maintain expression of downstream β-globin genes (8). If cHS4 has an autonomous ability to counteract silencing of gene expression by flanking repressive chromatin at sites of ectopic chromosomal integration, it could be an extremely useful tool in the design of replacement gene therapy vectors (35).
In earlier studies, we showed that in transgenic mice and cells lines, 5′HS2 and other enhancers have little effect on the level of gene expression in cells that actively express a gene but act to increase the proportion of cells that express a gene and to maintain expression over time (39–41). These results are consistent with all evidence on enhancer action, since most assays are not able to distinguish effects on transcription rate from effects on the recruitment of templates to an active state. They suggest that enhancers function primarily to ensure that a gene is established and maintained in an active state in the appropriate lineage and have little or no direct effect on the rate of transcription.
Here we describe studies on the cHS4 insulator that were designed to test its ability to block enhancer activity under conditions that control for position effects. Using a modification of the recombinase mediated cassette exchange (RMCE) system (2), we studied the effect of cHS4 on 5′HS2 enhancer activity at three genomic sites in K562 cells. cHS4 blocks this effect of the enhancer, but surprisingly, at some sites, it also reduced the enhancer effect when placed on the distal flank of the enhancer. The function of the cHS4 element in the β-globin locus remains uncertain, but we believe these results suggest that its function as an insulator in the locus may be more complex than suggested by a domain model of genomic regulation.
MATERIALS AND METHODS
Plasmid construction. (i) L1L2 plasmids.
Plasmid pL1HYGL2 (2, 44) was digested with BamHI, and a synthetic polylinker was inserted in place of the HYG sequences to create pL1L2. Parent vector pL1CMV-HyTKL2 was constructed by digesting a plasmid containing the cytomegalovirus (CMV) promoter driving expression of the fusion protein HyTK with XhoI (kindly provided by P. Greenberg, Fred Hutchinson Cancer Research Center) and ligating the ends to a BamHI linker. A 3.1-kb CMV-HyTk BamHI fragment was subsequently inserted into the BamHI site of the pL1L2 polylinker (schematically shown in Fig. 1). Plasmid pL1γ β-geoL2 was made by digesting plasmid γ β-geo (described in reference 41) with Asp718, ligating it to a BglII linker, and purifying a 4.3-kb BglII γ β-geo fragment. The γ β-geo BglII fragment was inserted into the BamHI site of the pL1L2 polylinker. pL1HS2γ β-geoL2 was constructed by combining a 1.0-kb BglII/SmaI fragment from 5′HS2 of the human β-globin LCR, a 4.3-kb γ β-geo fragment containing a blunt 5′ end (digested initially with Asp718 and ends subsequently filled in with Klenow fragment) and a SalI 3′ end, and a BamHI/SalI pL1L2 DNA fragment in a three-way ligation reaction (pL1-βgeoL2 plasmids are shown in Fig. 1).
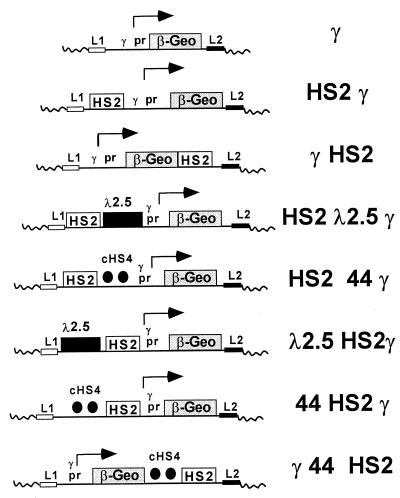
FIG. 1.
β-geo reporter constructs containing the cHS4 boundary element. All contain the β-geo reporter driven by the human γ-globin promoter (pr) and are flanked by lox elements L1 and L2 for site-specific integration with the RMCE system. HS2 is a strong erythroid enhancer derived from the human β-globin LCR and was used in the original characterization of cHS4. Two copies of a 1.2-kb fragment containing cHS4 were placed in various orientations with respect to the promoter and enhancer, as indicated in the diagrams. In addition, a 2.5-kb DNA fragment from bacteriophage λ was used as a control for enhancer-promoter spacing.
Control plasmids containing bacteriophage λ DNA were created by inserting a 2.5-kb HindIII/SmaI λ DNA fragment upstream from HS2 and between the HS2 enhancer and the γ promoter in pL1HS2γ β-geoL2. The latter was accomplished by converting the λ DNA HindIII end to an Asp718 end after KpnI linker ligation and combining it in a four-way ligation reaction with a 1.0-kb BglII/SmaI HS2 fragment, a 4.3-kb Asp718/SalI γ β-geo fragment, and a BamHI/SalI pL1L2 fragment. pL1λ2.5HS2γ β-geo was constructed in two steps. First, the modified 2.5-kb SmaI/Asp718 λ DNA fragment described above was inserted upstream from γ β-geo to form an intermediate plasmid, pL1λ2.5γ β-geoL2. Second, the intermediate pL1λ2.5γ β-geoL2 was digested with Asp718, the ends were filled in with Klenow fragment, and a 1.0-kb HS2 BglII/SmaI fragment modified to have blunt ends was inserted between the λ DNA and the γ-globin promoter to create pL1λ2.5HS2γ β-geoL2.
(ii) cHS4 plasmids.
Plasmids containing the boundary element from cHS4 were derived from pJC13-1, kindly provided by Jay Chung. pJC13-1 was altered by removal of the neo gene (digested with BamHI and self-ligated after neo gene removal) and the LCR sequences (digested with EcoRI and self-ligated). Two copies of the original 1.2-kb SacI/SspI cHS4 fragment were placed upstream of γ β-geo by performing a three-way ligation. This ligation included a 2.4-kb Asp718/SalI fragment which had two copies of cHS4 from pJC13-1, a 4.3-kb Asp718/BglII γ β-geo fragment, and a SalI/BamHI pL1L2 fragment and generated pL144γ β-geoL2. Two copies of cHS4 were placed downstream of γ β-geo by ligating a SalI linker to the BamHI end of a 2.4-kb BamHI/SalI two-copy cHS4 fragment and inserting this fragment into a SalI site located downstream from β-geo to make pL1γ β-geo44L2. A construct with the γ β-geo cassette flanked by cHS4 was created by replacing a 1.3-kb ClaI fragment from pL1γ β-geo44L2 containing γ promoter and upstream β-geo sequences with a 3.8-kb ClaI fragment from pL144γ β-geoL2. Insertion of the 3.8-kb ClaI replacement fragment added two copies of cHS4 to this construct, positioned upstream from the β-geo and γ promoter sequences to produce pL144γ β-geo44L2.
Two copies of the cHS4 boundary element were placed upstream of 5′HS2 in pL1HS2γ β-geoL2 and between the HS2 enhancer and the γ-globin promoter to make 44HS2γ β-geo and HS244γ β-geo, respectively. First, the cHS4 DNA fragment was made by digesting the modified pJC-1 plasmid with SalI, filling the ends with Klenow fragment, and digesting it with Asp718 to produce a two-copy cHS4 fragment with Asp718 and blunt ends. This cHS4 fragment was combined with a 1.0-kb 5′HS2 fragment with SmaI/BglII ends, a fragment containing the γ promoter and β-geo with Asp718 and SalI ends, and the pL1L2 vector with BamHI and SalI ends in a four-way ligation to produce pL1HS244γ β-geo. pL144HS2γ β-geoL2 was made by ligating the 1.0-kb HS2 enhancer element with blunt ends to an Asp718 site located upstream of the γ-globin promoter in pL144γ β-geo L2. The Asp718 site was first modified by the Klenow fragment to create blunt ends for the ligation. Proper content and orientation of the promoter, enhancer, and insulator elements in these RMCE constructs were confirmed by restriction digests and by DNA sequencing.
Site-specific recombination plasmids for the single loxP expression trap method.
Plasmids were constructed to perform a second cre recombinase-based method for generating site-specific recombinants (see Fig. 7A). The parent plasmid (loxPHS2γ-loxPβgeo) was constructed in two steps. In the first, a three-way ligation was performed with a 1.0-kb HS2 enhancer element (BglII and SmaI ends), the Aγ promoter fragment (blunted Asp718 end and HindIII end), and the pBS246loxP plasmid (kindly provided by Brian Sauer) (HindIII and BamHI ends). In the second step, the loxP-HS2γ-loxP intermediate fragment with NotI ends was ligated to the βgeo backbone (described in reference 41) after the γ-βeo plasmid was first digested with Asp718 and HindIII, the ends were blunted with Klenow fragment, and the fragment was ligated to a NotI linker. Target replacement plasmids were also constructed. These included γ-loxP, HS2γ-loxP, HS2 44 γ-loxP, and HS2-spacer-γ-loxP. γ-loxP was made by performing a three-way ligation with loxP (HindIII and EcoRI ends), the Aγ promoter (HindIII and Asp718 ends), and Bluescript (pKS+; Stratagene) (EcoRI and Asp718 ends) fragments. HS2γ-loxP was made by ligating HS2 (SmaI and BglII ends) and the Aγ promoter (HindIII and blunted Asp718 ends) to a Bluescript plasmid containing a single loxP site with HindIII and BamHI ends. HS244-γ-loxP was generated in two steps. In the first, a three-way ligation involving the Bluescript-loxP vector (HindIII and BamHI ends), the Aγ promoter (with HindIII and blunted Asp718 ends), and two copies of cHS4 (with EcoRV and BamHI ends) was performed. In the second step, this intermediate vector with BamHI and blunted NotI ends was ligated to a 1.0-kb 5′HS2 fragment with BglII and SmaI ends. Finally, a construct with a plasmid spacer between the 5′HS2 enhancer and γ promoter was created by ligating the Aγ-loxP plasmid with BamHI and SmaI ends to the 1.0-kb HS2 enhancer element (BglII and SmaI ends) to make HS2-spacer-γ-loxP.
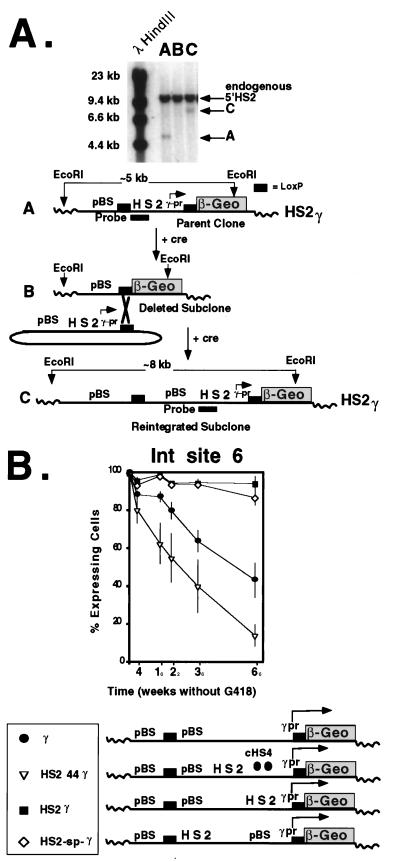
FIG. 7.
Effect of plasmid spacer DNA on expression after site-specific recombination. (A) A single-copy clone carrying the parent plasmid, loxP-HS2γ-loxPβ-Geo was isolated in K562 cells (A). After electroporation with a cre recombinase expression plasmid, subclones with deletions and carrying expressionless β-geo were isolated on the basis of sensitivity to G418 (B). After cotransfection with cre recombinase and the circular target plasmid, site-specific recombinants carrying the reintegrated target plasmid were selected in G418 (C). Site-specific recombination was confirmed by Southern blotting. After digestion of genomic DNA with EcoRI, hybridization to the HS2 probe generated a 5-kb fragment in the parent clone (lane A, band A), no hybridization signal in the deletion-containing subclone (lane B), and an 8-kb fragment in the reintegrated subclone (lane C, band C). (B) Effect of the 3.0-kb Bluescript DNA spacer on silencing. At this site, the spacer DNA had no effect on silencing when positioned between the 5′HS2 enhancer and the γ-globin promoter (pr). cHS4 effectively blocked the enhancer effect at this integration (Int) site.
Cell culture and transfection.
Conditions for growth and electroporation of K562 cells were as previously described (41). To select for RMCE events in clones containing a single copy of pL1CMVHyTKL2, 5 × 106 cells were electroporated with 10 μg of cre expression plasmid pBS185 (Life Technologies, Gaithersburg, Md.) and 100 μg of the target vector. At 48 h after electroporation, cells were plated by limiting dilution on 96-well microtiter plates in the presence of G418 (1 mg/ml) and ganciclovir (4 μg/ml).
To select for recombination events using the expression trap method, cells containing the single-copy promoterless β-geo target were electroporated with the cre expression plasmid and the replacement vector as described above and plated by limiting dilution on 96-well microtiter plates in the presence of G418 (1 mg/ml).
Confirmation of RMCE.
After 2 weeks in culture, subclones resistant to G418 and ganciclovir were expanded, genomic DNA was prepared, and Southern blotting was performed to confirm site-specific integration. This was done by documenting the presence of junction fragments whose sizes were predicted from junction fragment sizes in the HyTK parental clones (see Fig. 4). The expected junction fragment sizes after RMCE for each of the three integration sites are listed in Table 1. Digestion of genomic DNA with PstI generated downstream junction fragments in all constructs. PstI digestion generated upstream junction fragments in all constructs except those which contained the cHS4 element, due to the presence of a PstI restriction site in cHS4. To confirm site-specific recombination in the constructs containing cHS4, genomic DNA was digested with HindIII and the presence of a 5′ junction fragment of the correct size was verified by hybridization to a cHS4 probe (for construct 44HS2γ) or an HS2 probe (for construct HS244γ) (see Fig. 4C, right blots). In this manner, subclones containing single intact copies of each construct depicted in Fig. 1 were generated by RMCE at each of the three integration sites. Three or more subclones were isolated for each construct (with the sole exception of 44HS2γ at site 15, where a single subclone was isolated) and were used in subsequent analyses of gene expression.
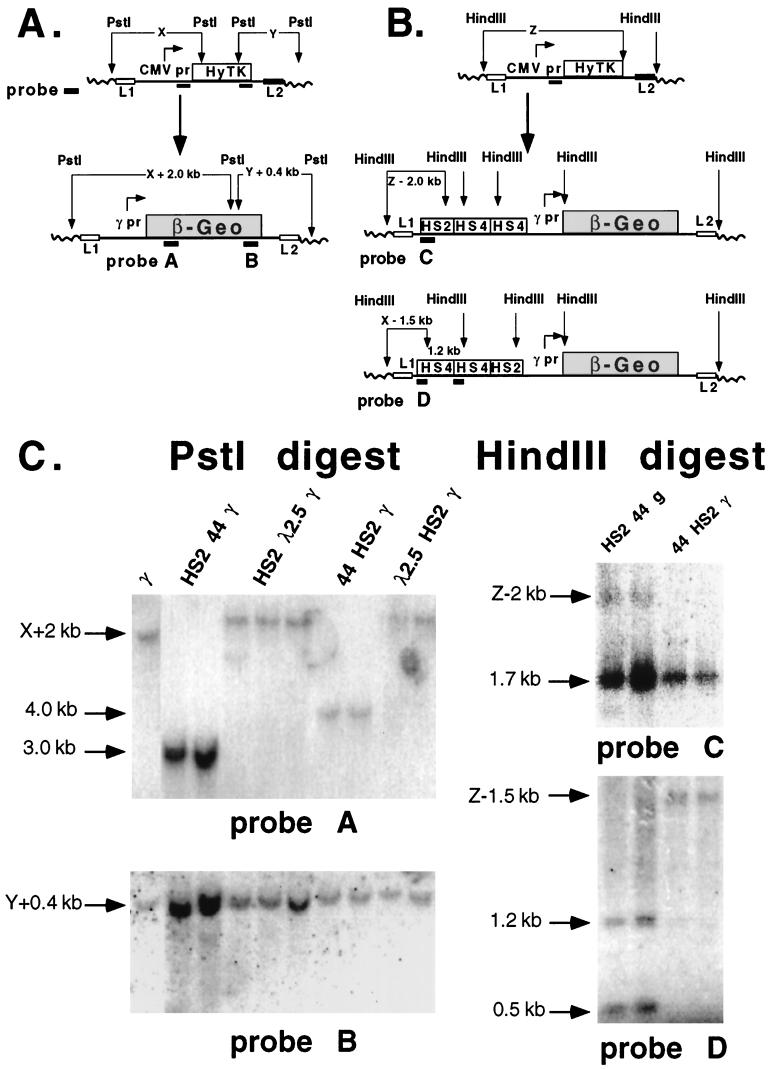
FIG. 4.
Southern blot analysis to confirm site-specific integration by RMCE. (A) Integration of a single copy of the parental CMV-HyTK plasmid was confirmed by the presence of unique junction fragments. Genomic DNA was digested with PstI and hybridized, first to an upstream CMV promoter (pr) fragment probe and subsequently to a downstream HyTK fragment probe. Genomic junction fragment lengths designated X (for the upstream junction fragment) and Y (for the downstream fragment) were used to predict the sizes of upstream (X + 2 kb) and downstream (Y + 0.4 kb) junction fragments produced by correctly integrated β-geo plasmids after hybridization to β-geo probes A and B (Table 1). This strategy is not effective when cHS4 is present at the junction, because cHS4 contains a PstI site. For analysis of cHS4 integration, the strategy in panel B was used. (B) Generation of new upstream HindIII junction fragments by integration of constructs containing cHS4. The size of upstream junction fragment X was determined by digestion of genomic DNAs from HyTK parental clones with HindIII and hybridization to a CMV promoter fragment probe. After exchange of the cassettes by RMCE, genomic DNA was digested with HindIII. The predicted size of the upstream β-geo junction fragment for HS2 44 γ β-geo was X − 2 kb when hybridized to HS2 enhancer probe C; for 44HS2γ β-geo, the junction fragment was X − 1.5 kb when hybridized to cHS4 probe D (Table 1). (C) Southern blot of representative subclones illustrating RMCE. Following RMCE at integration site 30, genomic DNAs from representative subclones were digested with PstI. For the lower left blot, DNA was hybridized to β-geo probe B to generate a unique 4.7-kb downstream junction fragment (Y + 0.4 kb) in all constructs. For the upper left blot, hybridization to lacZ probe A (which recognizes β-geo) generated a unique junction fragment (X + 2 kb) in all constructs (except those containing the cHS4 element, due to the presence of a PstI restriction site in the cHS4 insulator). Upstream PstI junction fragment sizes for the integration site 30 subclones ranged from 9 kb for γ β-geo to 12.5 kb for H λ2.5 γ β-geo, as listed in Table 1. The lacZ probe hybridized to internal fragments that were 3.0 and 4.0 kb in length from HS2 44 γ β-geo and 44 HS2 γ β-geo, respectively. For the upper and lower right blots, to confirm site-specific recombination in subclones containing the cHS4 insulator, DNA was digested with HindIII. For HS2 44 γ β-geo, hybridization to 5′HS2 probe C generated a 1.7-kb endogenous genomic HS2 fragment and a 3.6-kb (X − 2 kb) upstream junction fragment (upper right). For 44 HS2 γ β-geo, hybridization to cHS4 probe D to generated a 1.2-kb internal fragment and a 3.1-kb (X − 1.5 kb) upstream junction fragment (lower right). Subclones with the HS2 44 γ β-geo construct had 0.5- and 1.2-kb internal fragments after hybridization to cHS4 probe D (lower right).
TABLE 1.
Hybridization fragment sizes for β-geo constructs after RMCE in K562 cells
| K562 clone and construct | Junction fragment size (kb) after PstI digestion
|
Upstream junction fragment size(s) (kb) after HindIII digestion
|
||
|---|---|---|---|---|
| Upstream | Downstream | HS4 probe | HS2 probe | |
| 15 | ||||
| γ | 6.0 | 4.8 | ||
| HS2 γ | 7.0 | 4.8 | 2.9 | |
| HS2 λ2.5 γ | 9.5 | 4.8 | ||
| HS2 44 γ | 4.8 | 2.9 | ||
| λ2.5 HS2 γ | 9.5 | 4.8 | ||
| 44 HS2 γ | 4.8 | 2.4, 1.2 | ||
| 19 | ||||
| γ | 4.2 | 4.4 | ||
| HS2 γ | 5.2 | 4.4 | 1.8 | |
| HS2 λ2.5 γ | 7.7 | 4.4 | ||
| HS2 44 γ | 4.4 | 1.8 | ||
| λ2.5 HS2 γ | 7.7 | 4.4 | ||
| 44 HS2 γ | 4.4 | 1.3, 1.2 | ||
| 30 | ||||
| γ | 9 | 4.7 | ||
| HS2 γ | 10 | 4.7 | 3.6 | |
| HS2 λ2.5 γ | 12.5 | 4.7 | ||
| HS2 44 γ | 4.7 | 3.6 | ||
| λ2.5 HS2 γ | 12.5 | 4.7 | ||
| 44 HS2 γ | 4.7 | 3.1, 1.2 | ||
Confirmation of site-specific recombination with the expression trap.
After generation of single-copy integrants containing the loxP-HS2γ-loxPβ-geo parent vector, a promoterless β-geo intermediate was made by electroporation of the parental cells with the cre expression vector. Following electroporation, single cells were cloned by limiting dilution, split into equivalent aliquots, and expanded in the presence and absence of G418. Clones sensitive to G418 were analyzed by Southern blotting to confirm excision of the enhancer-promoter element. After confirmation of excision, the promoterless β-geo target clone was electroporated with the cre expression vector and the replacement plasmid. Clones resistant to G418 were expanded, genomic DNA was prepared, and Southern blotting was performed. Site-specific recombination was confirmed by digestion of genomic DNA with EcoRI and hybridization to a 5′HS2 enhancer probe (see Fig. 7).
Determination of β-gal activity.
The methylumbelliferyl-β-glucuronide (MUG) assay was performed on lysates from cells containing the RMCE constructs. The cellular lysates were placed in 96-well plates and assayed on a Dynatech fluorimeter as described previously (40). Cells were passaged in the presence of G418 to ensure that all of them expressed β-geo. Fluorescence of cells from each subclone was measured in triplicate, and mean β-galactosidase (β-gal) activity was determined after correction for the protein content of the lysates. MUG assays were repeated two times, and mean activity relative to that of a reference sample and the standard error of the mean (SEM) were calculated.
FACS-gal assays.
Fluorescence-activated cell sorter (FACS)-Gal analysis was performed as described previously (40, 41). Cells passaged in the absence of G418 selection were assayed at 2- to 4-week intervals for β-gal activity. Cells were incubated on ice for 4 h in the presence of fluorescein di-β-d-galactopyranoside before analysis to ensure that cells with any β-gal activity were scored as positive. Each subclone found to have a correctly integrated RMCE construct was divided into two aliquots that were separately subjected to FACS-Gal analysis to determine the rate of transcriptional silencing. This rate was determined for each construct by calculating the average fraction of β-gal-expressing cells at 2- to 4-week intervals up to 40 weeks after removal of G418 selection. In order to maintain a uniform rate of cell division, all subclones were expanded to a density of 5 × 105 to 8 × 105 cells/ml and split 1:40 in medium containing calf serum from a single lot. Average values were calculated from at least three different subclones for each construct, and the SEM was determined for each time interval.
Colony assay.
Mid-log-phase K562 cells (5 × 106) were transfected by electroporation as described previously. Linearized DNA (10 μg) was mixed with supercoiled human growth hormone expression plasmid DNA (2.5 μg) as a control for transfection efficiency and electroporated into K562 cells. To generate G418-resistant colonies, the transfected cells were diluted 1:2 (for 44γ β-geo, 44γ44β-geo, γ β-geo44, and γ β-geo constructs) or 1:10 (for HS2γ β-geo and 44HS2γ β-geo constructs) in medium containing 1-mg/ml G418 and plated on 96-well culture plates 2 days after transfection. The G418-resistant colonies were counted 2 to 3 weeks after selection and corrected for transfection efficiency. The experiment was performed three times in duplicate to generate average values and the SEM.
RESULTS
Colony assays suggest that cHS4 does not protect against position effects.
It has been proposed that cHS4 may counteract the repressive effect of chromatin and thereby promote stability of expression at randomly selected sites of integration. We elected to first test this hypothesis in a conventional colony assay. The colony assay measures the ability of a transfected plasmid to integrate into chromatin and express a drug resistance marker with sufficient stability to produce a colony of drug-resistant cells; it was used to characterize cHS4 by Chung et al. (7, 8). In the colony assay, enhancers appear to act by increasing the number of sites at which transcriptional activity can occur after integration. Constructs may be more efficient at initiating and maintaining expression within a region of inactive chromatin when they contain an enhancer. However, there is considerable evidence that they do not affect the rate of transcription in active integrated constructs (28, 39, 40).
If it is assumed that most random integration sites are repressive for expression and cHS4 acts to counteract repressive chromatin and permit expression of insulated genes, then a construct flanked by cHS4 should produce increased numbers of G418-resistant colonies by blocking the repressive influence of flanking chromatin. To test this, we transfected K562 cells with linearized β-geo plasmid DNA, plated transfected cells into medium with G418, and determined the number of G418-resistant colonies after correcting for transfection efficiency. The β-geo reporter is a fusion protein with β-gal and neomycin phosphotransferase activities (41) and thus can be utilized in a colony assay. In each construct, β-geo expression was driven by the human Aγ-globin promoter.
The β-geo constructs used in the colony assay are depicted in Fig. 1, and the results of the colony assay are presented in Fig. 2. First, we observed a significant increase in colonies by addition of the 5′HS2 erythroid enhancer to the β-geo plasmid, similar to what we and others have observed in the past (7, 28, 40). We then tested the effect of the cHS4 insulator in the colony assay. We found that placing cHS4 upstream of the HS2 enhancer (which, in multicopy head-to-tail arrays, would cause the enhancer-promoter sequences to be flanked by cHS4) had little effect on the number of G418-resistant colonies, as observed previously by Chung et al. (7) (Fig. 2). To avoid an enhancer-blocking effect that may antagonize the ability of the 5′HS2 enhancer to establish expression in the colony assay, we also tested the effect of cHS4 in the absence of an enhancer. Two copies of cHS4 were placed upstream or downstream of β-geo, and the construct was also flanked by cHS4 at both ends (Fig. 2B). Placing the cHS4 boundary element upstream or downstream of the γ-globin promoter did not increase the number of colonies relative to that obtained with the promoter alone, and flanking this plasmid with cHS4 elements actually decreased the number of colonies by more than 50% (Fig. 2B). The reduction in colony number after flanking of a plasmid construct has been observed previously (8).
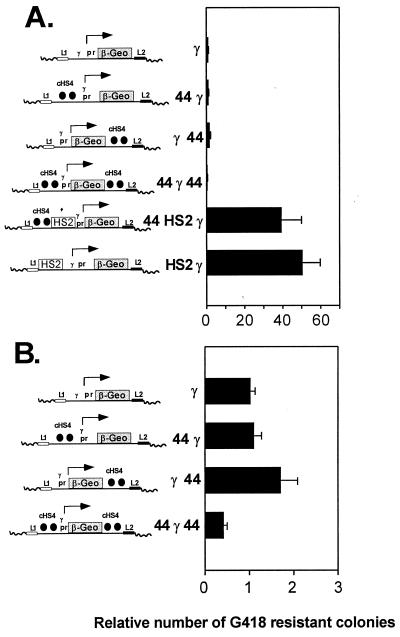
FIG. 2.
Effect of the cHS4 boundary element on expression when flanking the β-geo reporter in a colony assay. Linearized plasmid DNA was transfected in K562 erythroleukemia cells. The cells were plated in medium with G418, and 2 weeks after transfection, G418-resistant colonies were counted. The number of colonies was corrected for the efficiency of transfection with a human growth hormone control plasmid and normalized to the number of G418-resistant colonies obtained with the γ β-geo plasmid. The averages from three transfections performed in duplicate were determined, and the SEM was calculated. (A) Effect of cHS4 on constructs carrying the 5′HS2 enhancer. The constructs (shown on the y axis) include controls with the γ-globin promoter (pr) alone and with an upstream HS2 enhancer driving β-geo expression. Two copies of a 1.2-kb fragment from cHS4 were positioned upstream of the promoter (44 γ), upstream of the enhancer-promoter combination (44 HS2 γ), downstream of β-geo (γ 44), and at both ends of the γ β-geo plasmid (44 γ 44). (B) Effect of cHS4 on expression of constructs lacking an enhancer. cHS4 significantly affects colony number only when placed at each end of the construct (44 γ 44), and in this case it causes a decline in colony number.
Taken together, these results may indicate that cHS4 does not protect against position effects that reduce the number of colonies and may even repress expression. However, this view is at odds with previous work that established the cHS4 element as an insulator that blocks repressive chromatin (8, 31). Moreover, we did not observe a negative effect of cHS4 in the colony assay when the insulator was placed on only one side of the β-geo reporter (Fig. 2B). Thus, it is possible that in the colony assay, the enhancer-blocking activity of cHS4 shields expression from the stimulatory effect of neighboring genomic elements: if most random integration sites in the colony assay are actually permissive for expression, then flanking cHS4 elements may actually reduce the number of colonies by blocking activating effects of an endogenous enhancer near the integration site. Unfortunately, the colony assay is not capable of distinguishing these possibilities, as it does not control for the effect of integration position. We therefore developed a novel system to study cHS4 in a manner that overcomes this limitation of the colony assay.
A system to control integration position in cultured cells.
To further investigate the effect of cHS4 on gene expression, we have used a system that permits detailed analysis of stable gene expression. As a reporter, we use β-geo, a fusion protein with β-gal and neomycin phosphotransferase activities. In addition to acting as a selectable marker (conferring resistance to G418), β-geo expression can be measured with a variety of assays for β-gal. We used conversion of MUG to assess the quantity of β-geo expression, and this level is a reflection of the transcription rate. The FACS-Gal assay (29) is a flow cytometric assay that gives a direct and extremely sensitive measurement of the proportion of cells within a population that express β-gal; cells containing only a few β-gal molecules are scored positive (15), but under these conditions, the assay does not conveniently quantitate β-gal expression. With these two assays, we can assess the amount of transcription from an expressing gene and also measure the proportion of cells in a population that are actively expressing the reporter (40, 41).
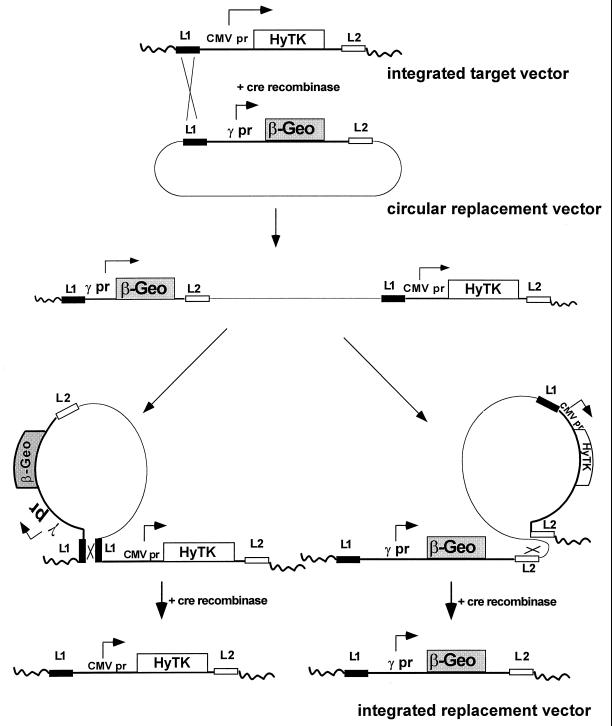
We have further refined these analyses by testing different cis elements at the same integration site by using an adaptation of a novel strategy termed RMCE. RMCE, recently described by Bouhassira et al. (2), makes use of the cre recombinase system and two LoxP recombination target sites designated L1 and L2 (16, 37, 38) (Fig. 3). These sites differ by a single base pair in the spacer region of the LoxP target, which renders L1 and L2 incompatible for recombination. The cre recombinase facilitates exchange of the integrated cassette with a circular plasmid replacement cassette flanked by L1 and L2. The process is initiated by a recombination event between the integrated target vector and the circular plasmid replacement vector, which creates an intermediate that includes the target and replacement plasmids (Fig. 3). The exchange process is completed by a second recombination event (between either two L1 elements or two L2 elements) that reduces the two-plasmid intermediate to either the original integrated vector or the desired replacement. Recombination events that generate the replacement may be selected on the basis of resistance: the original cassette expresses HyTK, which confers sensitivity to ganciclovir, and the replacement cassette expresses β-geo, which gives G418 resistance.
FIG. 3.
Site-specific integration of β-geo constructs by RMCE. The method relies on the use of variant lox elements (L1 and L2) that can recombine with other lox elements of the same type but not with each other (i.e., L1 × L1 or L2 × L2 but not L1 × L2). The target vector contains an expression cassette in which the CMV promoter (pr) drives expression of HyTK, a fusion protein with hygromycin phosphotransferase and thymidine kinase activities; the cassette is flanked by L1 and L2 elements. K562 clones carrying single copies of the L1HyTKL2 plasmid are cotransfected with a cre recombinase expression plasmid and a circular replacement plasmid containing the γ-globin promoter driving β-geo expression that is flanked by lox sites L1 and L2. In the example shown, an initial recombination between identical lox targets (L1) integrates the replacement plasmid, generating an intermediate containing both the replacement and HyTK vectors. This intermediate is resolved by a second recombination, leaving either the original HyTK vector, as shown on the lower left (L1 × L1 recombination), or the replacement β-geo vector, as shown on the lower right (L2 × L2 recombination). The desired replacement was selected by growth in G418 (selection for β-geo expression) and ganciclovir (selection against HyTK expression) and confirmed by Southern blotting.
We used RMCE to introduce a series of plasmids into three unique integration sites in K562 erythroleukemia cells; the replacement cassettes are shown in Fig. 1. The integration sites were generated by isolating single-copy integrants containing the HyTK reporter driven by the CMV enhancer-promoter, flanked by the loxP recombination targets, L1 and L2. Our earlier work supports the view that strong transcriptional control elements are more likely to establish expression in relatively repressive integration sites: when these elements are removed, expression is silenced at greatly increased rates (40). We chose the CMV enhancer-promoter because it is a strong element that is likely to be active at repressive sites, so that randomly selected clones would be more likely to represent these repressive sites. Use of repressive sites allows us to assess the silencing effect of neighboring chromatin and facilitates the study of cHS4 in such a setting.
In our constructs, β-geo expression is driven by the human Aγ-globin promoter, with the erythroid enhancer 5′HS2 (from the human β-globin LCR) placed upstream of the promoter. The γ-globin promoter, 5′HS2 enhancer, and K562 cell line were used in the original description of the HS4 insulator. Two copies of the 1.2-kb 5′HS4 boundary element, as tested in studies performed by Chung et al. (7, 8), were placed either between the 5′HS2 enhancer and the γ-globin promoter or upstream of 5′HS2. To simulate the effect of distance on enhancer activity caused by insertion of cHS4 sequences between the promoter and enhancer, a 2.5-kb DNA control fragment containing bacteriophage λ DNA was placed between the enhancer and promoter. Subclones that had completed RMCE were selected on the basis of G418 and ganciclovir resistance, and site-specific recombination was confirmed by Southern blotting (Fig. 4). The junction fragment sizes expected after RMCE for each of the three integration sites are listed in Table 1.
Neither the 5′HS2 enhancer nor cHS4 affects expression level.
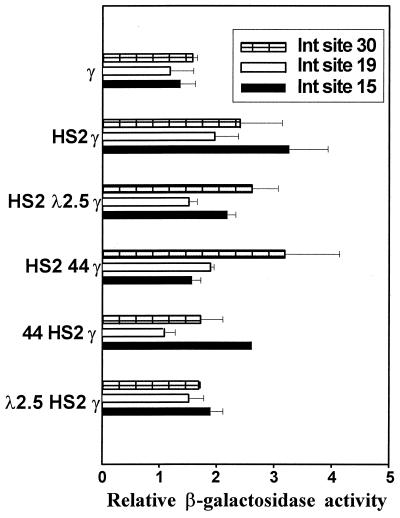
The level of β-geo expression and the rate at which expression was silenced were measured in separate assays. The β-gal expression level was measured by MUG conversion in cells maintained in G418, thereby eliminating the possibility that cells not expressing β-geo might be included in the analysis. To address the possibility that G418 selection excludes cells that have low expression levels and thus die in the presence of G418, we have measured expression levels by MUG conversion in cells passaged in the absence of positive selection (40). We accounted for the fraction of cells not expressing β-gal by performing the FACS-gal assay, corrected for the percentage of expressing cells, and determined β-gal activity per expressing cell. There was no difference in measurements of expression obtained by the two methods (data not shown). Consistent with our previous studies of 5′HS2 (40, 41), the addition of 5′HS2 to the γ-globin promoter resulted in only a slight or no increase in the level of β-gal expression (Fig. 5). 5′HS2 is a well-characterized enhancer, but this result is consistent with earlier studies by us and others (13, 28, 39–41). We have shown that enhancers have little effect on expression level but act to ensure the establishment and maintenance of a domain permissive for stable gene expression (39–41). The absence of a significant stimulatory effect on expression level by the upstream 5′HS2 enhancer does not mean that it has no effect, as the measurement of reporter gene silencing shows (see below). The boundary element from cHS4 had little or no effect on expression level when positioned either upstream or downstream of the enhancer, and neither did the bacteriophage λ spacer DNA. Thus, the enhancer-blocking activity of cHS4 does not appear to be mediated by effects on transcription rate.
FIG. 5.
Level of β-geo expression by constructs integrated by RMCE into three unique genomic sites. K562 cells carrying β-geo constructs after RMCE were expanded in G418 to ensure that all of the cells were expressing β-geo. The β-gal activity in cell lysates was determined by conversion of MUG. Error bars represent the SEM. MUG conversion values were normalized to the β-gal activity level of a reference clone in which β-geo is driven by the γ-globin promoter. For each β-geo construct at each integration site, three or more subclones were generated, and the values shown are averages from the three subclones. Three separate determinations were performed on each subclone in duplicate.
The 5′HS2 enhancer suppresses silencing of the reporter.
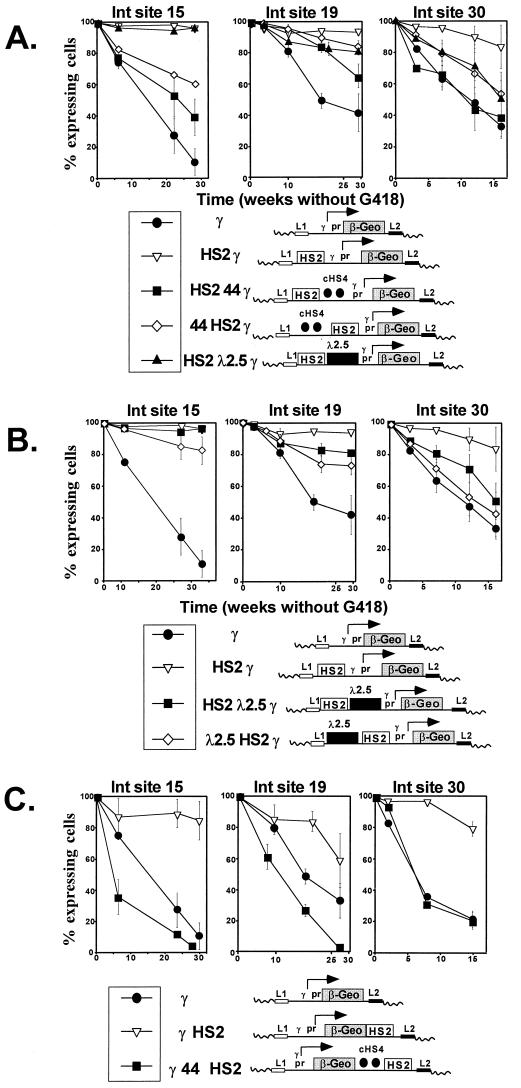
Silencing of expression was measured with the FACS-Gal assay. When cells expressing β-geo are removed from G418 selection, expression is no longer necessary for survival, but the FACS-Gal assay can be used to monitor the proportion of cells that continue to express β-geo. We have previously shown that each integration site silences reporter gene expression over time at a characteristic rate and that enhancers counteract this silencing. To ensure that all cells were initially expressing β-geo, subclones were expanded in G418; after removal of G418 selection, cells were subjected to the FACS-Gal assay at intervals to determine the percentage of cells that continued to actively express β-geo. To assess the effect of clonal variation on the silencing rate, six single-cell subclones were derived from one clone (γ-βgeo at site 30) by limiting dilution and passaged in the absence of G418. There was no significant variation in the rate of silencing among the single-cell subclones (data not shown). Subclones carrying each of the constructs shown in Fig. 1 were passaged separately in duplicate, and FACS-Gal assays were performed at standardized time points. The rate of silencing varied widely among the three integration sites, suggesting that repressive effects caused by the position of integration also varied from site to site (Fig. 6A to C). For example, the elapsed time necessary to observe a 70% decline in the percentage of cells expressing β-gal for the β-geo plasmid driven by the γ-globin promoter was 22 weeks at site 15, more than 30 weeks at site 19, and 16 weeks at site 30. This variation in rates of silencing from the RMCE sites was comparable to that observed with similar constructs integrated into K562 cells we have described previously (40). Thus, the RMCE sites appear to be subject to a variegating effect on gene expression that has been observed after stable integration in cell lines and in transgenic mice (14, 36, 39, 40). At each of the sites, the addition of the 5′HS2 enhancer had a profound effect on silencing, significantly slowing the rate of silencing compared to that of a similar construct lacking the enhancer (Fig. 6A, compare open triangles to closed circles in the three graphs). This observation is completely consistent with our earlier studies of enhancer action (40, 41) and with the hypothesis that enhancer elements stimulate expression by maintaining a domain that is permissive for transcription rather than affecting the rate of transcription.
FIG. 6.
Silencing of β-geo expression at the three integration (Int) sites. A comparison of the rates at which the various constructs are silenced is shown. At time zero, cells were removed from G418 selection, and thereafter, they were expanded continuously without G418 selection. At intervals, the individual subclones were analyzed in the FACS-Gal assay to determine the percentage of cells expressing β-gal. The y axis shows the percentage of cells expressing β-gal, and the x axis shows the time in weeks for which cells were expanded without G418 selection. The percentage of expressing cells was determined by creating a gate in the histograms that separated expressing from nonexpressing cells, based on the distribution of cells from positive and negative controls, and calculating the fraction of expressing cells. Each determination represents the average percentage of cells expressing β-gal from three or more subclones, and the time course of silencing was studied at least twice in each clone. Error bars represent the SEM. (A) The 5′HS2 globin enhancer suppresses silencing of β-geo, and cHS4 blocks this effect. At each of the three integration sites, interposition of cHS4 between the enhancer and the promoter (pr) causes silencing to proceed at a more rapid rate than it does when only the enhancer is present. This acceleration of silencing also occurs when cHS4 is placed upstream of the enhancer at sites 15 and 30 but not at site 19. (B) The effect of the 2.5-kb λ DNA spacer on silencing. At sites 15 and 19, the presence of the spacer had little or no effect, but at site 30, it mimicked the effect of cHS4. (C) The 5′HS2 enhancer suppresses silencing from a distance of 4 kb (downstream of β-geo), and cHS4 antagonizes this effect.
The cHS4 boundary element blocks 5′HS2 enhancer activity.
Based on previous studies by Chung et al. (7, 8), we anticipated that the cHS4 boundary element, when positioned between the promoter and enhancer, would increase silencing of β-gal expression by blocking the ability of the enhancer to maintain expression. Complete abrogation of the enhancer effect by cHS4 should cause the rate of silencing to be indistinguishable from that obtained with the promoter alone. As expected, cHS4 did block the effect of the HS2 enhancer in the silencing assay, but the magnitude of its activity varied among the integration sites. At integration site 30 (Fig. 6A, right graph), cHS4 completely abolished the enhancer effect. At sites 15 and 19 (Fig. 6A, center and left graphs), the interposition of cHS4 between the enhancer and promoter attenuated rather than blocked the effect of the HS2 enhancer. While these results confirm that cHS4 has enhancer-blocking activity, they also suggest that sequences neighboring the three integration sites exerted some influence on expression. It is possible that the variation in insulator activity we observed might be due in part to the influence of noninsulated downstream sequences. However, these results may also indicate that the cHS4 insulator does not exert absolute dominance at all genomic integration sites.
cHS4 enhancer blocking activity is bidirectional.
It has generally been supposed that an insulator blocks transmission of an enhancer effect when placed between an enhancer and a promoter but that an insulator placed upstream of the enhancer would have no effect. To determine if cHS4 had a bidirectional repressive effect on the maintenance of expression, we placed it upstream of the HS2 enhancer at the three integration sites. Surprisingly, in this configuration, cHS4 exhibited significant silencing activity at two of the three sites. At sites 15 and 30 (Fig. 6A, left and right graphs), cHS4 was nearly as effective at blocking enhancer activity when upstream of the enhancer as it was when placed between the enhancer and the promoter. However, at integration site 19, cHS4 had little effect when placed upstream of the enhancer (Fig. 6A, center graph). Again, this suggests that the activity of the cHS4 element is influenced by the site of integration.
If cHS4 insulates a promoter from the influence of repressive flanking chromatin, it might be expected to counteract silencing when placed on the flank of a construct, but in fact, its effect appears to be the opposite, increasing silencing of the reporter. To further test the possibility that cHS4 has bidirectional silencing activity, we attempted to integrate cassettes in which β-geo, driven only by the γ-globin promoter, was flanked on one or both sides by cHS4. In order to identify reintegration events, β-geo must be expressed so that a subclone is selected. We recovered subclones in which the cassette contained either no cHS4 or cHS4 at the 3′ end. However, we did not recover any clones when the cassette contained cHS4 at each end or (significantly) cHS4 at the 5′ end adjoining the promoter (data not shown). While it is not possible to say with certainty why no such subclones were obtained, the result is consistent with the interpretation that the cassettes were integrated but not expressed. While it is not certain if the repressive effects described above and the results of a colony assay indicate that flanking cHS4 elements repress expression, these results do indicate that cHS4 elements do not promote stable expression in every context.
Effect of increased distance on the enhancer effect.
We attempted to control for the effect of altering the spacing between the promoter and enhancer in the constructs containing cHS4 by testing constructs in which a 2.5-kb DNA fragment of bacteriophage λ was placed either between the promoter and the enhancer or upstream of the enhancer. While the λ DNA had no effect on expression level in these configurations (Fig. 5), it did increase silencing at integration site 30, where cHS4 also had significant silencing activity (Fig. 6B, right graph). No significant silencing activity was associated with λ DNA at integration site 15, and λ DNA had a modest repressive effect at site 19 (Fig. 6B, left and center graphs). These findings call into question the notion of functionally inert DNA; the context-dependent silencing activity of the spacer may be due to its prokaryotic origin. While the silencing activity of the spacer, to some extent, impaired our ability to judge the effect of distance from the promoter on enhancer activity, we observed that 5′HS2 significantly reduced the rate of silencing when placed downstream of β-geo, even though the promoter and enhancer were separated by a distance of 4.0 kb (Fig. 6C, compare γ [circles] and γ HS2 [triangles]). When cHS4 was placed between downstream 5′HS2 and the reporter gene (γ 44 HS2), it blocked the enhancer effect (Fig. 6C, squares).
To exclude the possibility that the repressive effect of λ could be a general phenomenon of DNA elements utilized in this system, we tested the effect of Bluescript (Stratagene, Inc.) plasmid DNA at a fourth integration site. At this site, recombinase-mediated site-specific recombination was accomplished through an expression trap method shown schematically in Fig. 7A. We made a parental construct containing the 5′HS2 enhancer upstream of the γ-globin promoter driving β-geo; the enhancer and promoter sequences were flanked by loxP targets. Following cre expression, a promoterless intermediate was created. A second plasmid containing the new transcriptional control elements was then introduced with a cre expression plasmid; site-specific recombination was screened on the basis of restored β-geo expression and confirmed by Southern blotting. As at the three RMCE sites, the effect of the 5′HS2 enhancer at this site was to significantly slow the rate at which expression of the reporter was silenced (Fig. 7B). At this site, the enhancer effect was effectively blocked by cHS4. When the 3.0-kb plasmid spacer was located between the enhancer and the promoter, it had no effect on silencing. This observation argues against the possibility that the repressive effects of cHS4 we observed were due to nonspecific effects of random DNA after insertion at the genomic sites.
DISCUSSION
By using the RMCE system to control for the effect of integration position and β-geo to assay effects on silencing of gene expression, we confirmed that the cHS4 insulator has significant enhancer-blocking activity. We also showed that it blocks the enhancer effect by disrupting the ability of an enhancer to maintain expression rather than by an effect on transcription rate. However, we also found that cHS4 causes more rapid silencing of a linked β-geo reporter, whether it is placed between the enhancer and the promoter or distal to the enhancer. Furthermore, the intensity of its effect varies from site to site, suggesting that it is itself susceptible to position effects. When positioned at the borders of transcription units that either had or lacked an enhancer, cHS4 did not increase the number of G418-resistant colonies in a colony assay. In our experiments, the three integration sites were randomly selected, and there is no reason to suppose that they are unusual. Our results strongly suggest that cHS4 will not shelter randomly integrated genes in mammalian cells from repressive chromatin in all contexts, although it may do so under some circumstances (42).
These observations are not in conflict with those made in previous investigations of cHS4, but they do reveal more complex effects. RMCE allows direct comparison of the effects of cHS4 when it is placed in different configurations with respect to an enhancer or other regulatory elements at the same genomic site. The colony assay assesses the ability of cis-regulatory elements to counteract repressive chromosomal position effects that silence drug resistance reporter genes, i.e., to establish expression and maintain it long enough to score a colony for drug resistance. Colonies derived from such an experiment can be analyzed with the β-geo assay, which permits observation of reporter silencing and separate measurement of the expression level. We previously showed that enhancers are potent antagonists of variegating position effect in the colony assay and so act to increase the number of colonies without acting directly to increase the transcription rate (40, 41). Chung and colleagues demonstrated that cHS4 blocked the stimulatory effect of the 5′HS2 enhancer in a colony assay (7). We have extended these observations and found that, unlike transcriptional enhancers, cHS4 does not suppress position effects in a colony assay. Recently, Chung and his colleagues showed a similar reduction in the number of G418-resistant colonies by flanking a neomycin resistance reporter, driven by the γ-globin promoter and 5′HS2 enhancer, with a single copy of cHS4 (8). We also noted that cHS4 does not suppress a variegating position effect (36) on a globin-lacZ transgene in mice (16a). Taken together, the results of these studies suggest that cHS4 does not universally block the effects of repressive chromatin that borders integration sites in a colony assay. It is possible that cHS4 blocks repressive chromatin in some sites; however, our data argue that the insulator does not exert absolute dominance at all sites of integration.
It is possible that apparent repression by cHS4 is created by insulation of the expression cassette from endogenous genomic enhancers. In the colony assay, this would reduce the number of colonies by insulating the cassette from stimulatory effects of flanking chromatin required for expression and, at the RMCE sites, insulate the cassette from an endogenous enhancer upstream of each of the three integration sites. This would account for both the apparent bidirectional silencing activity of cHS4 and the difficulty in generating subclones carrying the β-geo cassette flanked by cHS4. Nevertheless, this seems unlikely. First, it would require that each of the three integration sites have such a strong enhancer nearby but that this endogenous enhancer not be sufficient to maintain reporter expression. Second, it would require that 5′HS2 have a much stronger effect than the endogenous enhancer (since it maintains expression much more effectively) but that when the endogenous enhancer was blocked by placing cHS4 on the flank of a construct, 5′HS2 would be unable to maintain expression. A possibly simpler explanation for our results is that cHS4 has a local effect that suppresses the activity of enhancer and promoter elements on either side of it. However, we cannot exclude the possibility that an endogenous activator and the 5′HS2 enhancer interact in an additive manner to stabilize expression and that this interaction is effectively blocked by the cHS4 insulator, accounting for its partial effect on expression stability. Taken together, our results suggest either that cHS4 itself has a repressive effect or that it can block the activating effect of flanking chromatin and so enhance silencing of the flanked construct. Either action could cause the silencing of expression associated with cHS4 we observed.
Our observations are not inconsistent with the idea that cHS4 functions as an insulator; the bidirectional silencing effect may or may not bear any relation to its function in the native context of the chicken β-globin domain. If, as Chung et al. have speculated (7, 8), cHS4 functions to separate chromatin domains near the chicken β-globin locus, it is possible that this activity causes a silencing effect in our assay. While insulators have been identified and defined by an effect of unidirectional silencing of gene expression dependent upon enhancer activation, these assays may produce only a partial understanding of their function. The characteristics of some other known insulators suggest more complex functions.
Perhaps the best-studied insulator is that found in the Drosophila gypsy retrotransposon, where binding sites for suppressor of hairy wing [su(Hw)] have silencing and enhancer-blocking activities (3, 4, 19). The silencing activity is modified by trans-acting factors to confer directionality on its enhancer-blocking effects. The unidirectional silencing is mediated by protein-protein interactions involving the su(Hw) protein and its partners (17–19). Mutations in the modifier protein encoded by mod(mdg4) cause these sites to lose their polarity and act as bidirectional silencers of transcription and also become enhancers of position effect variegation. The gypsy insulator may not recapitulate these effects at every site of integration, and there is evidence from mod(mdg4) mutants that selected promoters are spared from its repressive effect (4, 21). Thus, similar to our observations of cHS4, the activity of the su(Hw) insulator is influenced by its position of integration. Moreover, under certain conditions, it too has bidirectional silencing activity.
The Drosophila bithorax complex contains several chromatin insulators that may facilitate the expression of homeotic selector proteins in a developmentally regulated pattern. The best studied is the Fab-7 insulator, positioned between cis-regulatory regions iab-6 and iab-7 of the Abdominal-B gene in the bithorax complex (23, 46). These elements control expression in posterior parasegments 11 and 12 in the developing embryo. Fab-7 was shown to silence linked reporters in transgenic fly strains and has enhancer-blocking activity associated with binding by polycomb (Pc) group proteins. Recently, mapping studies have demonstrated separate Pc response element and boundary element sequences located near the Fab-7 boundary. Thus, the Fab-7 boundary has overlapping affinities for repressive Pc proteins and activating trithorax group proteins (24, 27), which may account for the observation that the Fab-7 boundary functions to block ectopic activation and ectopic silencing when tested in the context of the bithorax complex. These complexes of stimulatory and repressive proteins may form structures that maintain chromatin in a stable state that is initiated by events early during embryogenesis (5, 30). Whether conditioned to be active or inactive, Fab-7 maintains the state in a manner that is stable and heritable.
Thus, the silencing activity associated with insulators may well be linked to their function. This activity is subject to modification by trans-acting factors that may confer directionality or specificity of silencing, or by the transcriptional state of a locus initiated by prior events, to maintain a state of repression (or activation) that is stable through cell divisions and even meiosis (5, 6, 32). It is also influenced by its context. These interactions may account for the variation in silencing and enhancer-blocking activity we observed for cHS4 among the three integration sites studied.
Chromatin insulators such as Fab-7, and an enhancer-blocking element located between the human T-cell receptor α and δ genes, may function to form boundaries between transcriptionally active and inactive domains in the locus (45). However, the function of 5′HS4 in the chicken β-globin locus is not immediately apparent. In erythroid cells, it is located in chromatin that changes from condensed to open, although it is not known if it is required for formation or maintenance of this transition (34). Its position at that locus suggests that it may isolate neighboring loci and associated chromatin domains from the influence of β-globin genes; our results suggest that its function is more likely to block nonspecific activation by globin enhancers than to block the spread of repressive heterochromatin from upstream sites. In the murine and human β-globin loci, 5′HS5 has been thought to act as a boundary element by virtue of its analogous location and properties. However, deletion of mouse 5′HS5 by homologous recombination did not significantly alter the expression of downstream globin genes, nor did its deletion alter the open chromatin structure of the locus, suggesting that other structures in the locus are sufficient for initiation and maintenance of a transcriptionally active state (1, 33).
In erythroid tissues, globin genes are clearly not targets for repression by cHS4. cHS4 may (like the Fab-7 insulator in the bithorax complex) act to propagate either an active or an inactive transcriptional state established during earlier stages of development, or its primary action may be to prevent activation of upstream genes by the β-globin domain. It is clear that cHS4 creates a chromatin structure that affects neighboring elements; the nature and function of this structure remain to be understood.
ACKNOWLEDGMENTS
We thank Mark Groudine and Gary Felsenfeld for helpful discussions.
This work was supported by grants from the NIH to D.I.K.M. and M.C.W. D.I.K.M. is a Scholar of the Leukemia Society of America.
REFERENCES
- 1.Bender M A, Reik A, Close J, Telling A, Epner E, Fiering S, Hardison R, Groudine M. Description and targeted deletion of 5′HS5 and 6 of the mouse β-globin locus control region. Blood. 1998;92:4394–4403. [PubMed] [Google Scholar]
- 2.Bouhassira E E, Westerman K, Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood. 1997;90:3332–3344. [PubMed] [Google Scholar]
- 3.Cai H, Levine M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 4.Cai H N, Levine M. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 1997;16:1732–1741. doi: 10.1093/emboj/16.7.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 6.Chan C-S, Rastelli L, Pirotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung J, Whitely M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 8.Chung J H, Bell A C, Felsenfeld G. Characterization of the chicken β-globin insulator. Proc Natl Acad Sci USA. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corces V G. Chromatin insulators: keeping enhancers under control. Nature. 1995;376:462–463. doi: 10.1038/376462a0. [DOI] [PubMed] [Google Scholar]
- 10.Eissenberg J C, Elgin S C. Boundary functions in the control of gene expression. Trends Genet. 1991;7:335–340. doi: 10.1016/0168-9525(91)90424-o. [DOI] [PubMed] [Google Scholar]
- 11.Enver T, Li Q, Gale K, Hu M, May G, Karlinsey J, Jimenez G, Papayannopoulou T, Costantini F. Analysis of the developmental and transcriptional potentiation functions of 5′HS2 of the murine β-globin locus control region in transgenic mice. Dev Biol. 1994;65:574–584. doi: 10.1006/dbio.1994.1277. [DOI] [PubMed] [Google Scholar]
- 12.Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 13.Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V. Chromatin structure and gene expression. Proc Natl Acad Sci USA. 1996;93:9384–9388. doi: 10.1073/pnas.93.18.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kiousses D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- 15.Fiering S N, Roederer M, Nolan G P, Micklem D R, Parks D R, Herzenberg L A. Improved FACS-Gal: flow cytometric analysis and sorting of viable eukaryotic cells expressing reporter gene constructs. Cytometry. 1991;12:291–301. doi: 10.1002/cyto.990120402. [DOI] [PubMed] [Google Scholar]
- 16.Fukushige S, Sauer B. Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc Natl Acad Sci USA. 1992;89:7905–7909. doi: 10.1073/pnas.89.17.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Garrick, D., and E. Whitelaw. Unpublished data.
- 17.Gdula D A, Corces V G. Characterization of functional domains of the su(Hw) protein that mediate the silencing effect of mod(mdg4) mutations. Genetics. 1997;145:153–161. doi: 10.1093/genetics/145.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gdula D A, Gerasimova T I, Corces V G. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci USA. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerasimova T I, Gdula D A, Gerasimov D V, Siimonova O, Corces V G. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 20.Gerasimova T I, Corces V G. Boundary and insulator elements in chromosomes. Curr Opin Gene Dev. 1996;6:185–192. doi: 10.1016/s0959-437x(96)80049-9. [DOI] [PubMed] [Google Scholar]
- 21.Gerasimova T I, Corces V G. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell. 1998;92:511–521. doi: 10.1016/s0092-8674(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 22.Geyer P K. The role of insulator elements in defining domains of gene expression. Curr Opin Gene Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- 23.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Gene Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 24.Hagstrom K, Muller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 26.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. In situ dissection of the Fab-7 region of the bithroax complex into a chromatin domain boundary and a Polycomb-response element. Development. 1997;124:1809–1820. doi: 10.1242/dev.124.9.1809. [DOI] [PubMed] [Google Scholar]
- 28.Moon A M, Ley T J. Functional properties of the β-globin locus control region in K562 cells. Blood. 1991;77:2272–2284. [PubMed] [Google Scholar]
- 29.Nolan G P, Fiering S, Nicolas F F, Herzenberg L A. Fluorescence-activated cell analysis and sorting of viable mammalian cells based on β-d-galactosidase activity after transduction of Escherichia coli lacZ. Proc Natl Acad Sci USA. 1988;85:2603–2607. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paro R. Propagating memory of transcriptional states. Trends Genet. 1995;11:295–297. doi: 10.1016/s0168-9525(00)89081-2. [DOI] [PubMed] [Google Scholar]
- 31.Pikaart M J, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirrotta V, Rastelli L. White gene expression, repressive chromatin domains and homeotic gene regulation in Drosophila. Bioessays. 1994;16:549–556. doi: 10.1002/bies.950160808. [DOI] [PubMed] [Google Scholar]
- 33.Reik A, Telling A, Zitnik G, Epner E, Groudine M. The locus control region is necessary for gene expression in the human β-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reitman M, Felsenfeld G. Developmental regulation of topoisomerase II sites and DNase I-hypersensitive sites in the chicken β-globin locus. Mol Cell Biol. 1990;10:2774–2786. doi: 10.1128/mcb.10.6.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivella S, Sadelain M. Genetic treatment of severe hemoglobinopathies: the combat against transgene variegation and transgene silencing. Semin Hematol. 1998;35:112–125. [PubMed] [Google Scholar]
- 36.Robertson G, Garrick D, Wu W, Kearns M, Martin D, Whitelaw E. Position-dependent variegation of globin transgene expression in mice. Proc Natl Acad Sci USA. 1995;92:5371–5375. doi: 10.1073/pnas.92.12.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauer B. Manipulation of transgenes by site-specific recombination: use of cre recombinase. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 38.Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 39.Sutherland H G E, Martin D I K, Whitelaw E. A globin enhancer acts by increasing the proportion of erythrocytes expressing a linked transgene. Mol Cell Biol. 1997;17:1607–1614. doi: 10.1128/mcb.17.3.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walters M C, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin D I K. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev. 1996;10:185–196. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 41.Walters M C, Fiering S, Eidemiller J, Magis W, Groudine M, Martin D I K. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci USA. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, DeMayo F J, Tsai S, O’Malley B W. Ligand-inducible and liver-specific target gene expression in transgenic mice. Nat Biotechnol. 1997;15:239–243. doi: 10.1038/nbt0397-239. [DOI] [PubMed] [Google Scholar]
- 43.Weintraub H. Formation of stable transcriptional complexes as assayed by analysis of individual templates. Proc Natl Acad Sci USA. 1988;92:5819–5823. doi: 10.1073/pnas.85.16.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westerman K A, Leboulch P. Reversible immortalization of mammalian cells mediated by retroviral transfer and site-specific recombination. Proc Natl Acad Sci USA. 1996;93:8971. doi: 10.1073/pnas.93.17.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong X-P, Krangel M S. An enhancer-blocking element between α and δ gene segments within the human T cell receptor α/δ locus. Proc Natl Acad Sci USA. 1997;94:5219–5224. doi: 10.1073/pnas.94.10.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 1996;10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]