SUMMARY
Human brain organoids represent remarkable platforms for recapitulating features of human brain development and diseases. Existing organoid models do not resolve fine brain subregions, such as different nuclei in the hypothalamus. We report the generation of arcuate organoids (ARCOs) from human induced pluripotent stem cells (iPSCs) to model the development of the human hypothalamic arcuate nucleus. Single-cell RNA sequencing of ARCOs revealed significant molecular heterogeneity underlying different arcuate cell types, and machine learning-aided analysis based on the neonatal human hypothalamus single-nucleus transcriptome further showed a human arcuate nucleus molecular signature. We also explored ARCOs generated from Prader-Willi syndrome (PWS) patient iPSCs. These organoids exhibit aberrant differentiation and transcriptomic dysregulation similar to postnatal hypothalamus of PWS patients, indicative of cellular differentiation deficits and exacerbated inflammatory responses. Thus, patient iPSC-derived ARCOs represent a promising experimental model for investigating nucleus specific features and disease relevant mechanisms during early human arcuate development.
Graphical Abstract
eTOC
Current brain organoid models do not resolve fine subregions of the human brain. Ming and colleagues established a method to generate hypothalamic arcuate organoids from human induced pluripotent stem cells, which exhibit neuronal subtype diversity and molecular signatures of human arcuate nucleus and can be used to model Prader-Willi syndrome.
INTRODUCTION
Three-dimensional (3D) organoids have emerged as an experimental model system for recapitulating complex in vivo physiological phenomena ranging from investigating human brain development to predicting pharmacogenomic interactions in cancer not detected using 2D assays, to modeling patient responses to drug treatment and resistance to therapy (Clevers, 2016; Lancaster and Knoblich, 2014). In particular, brain region-specific organoids generated from human induced pluripotent stem cells (iPSCs) have provided an unprecedented window to investigate the development of distinct human brain regions and generation of diverse cell types, including the neocortex, median ganglionic eminence, midbrain, and thalamus, as well as to model various brain disorders (Arlotta and Pasca, 2019; Marton and Pasca, 2020; Qian et al., 2016; Qian et al., 2019; Xiang et al., 2019). However, current organoid protocols lack the resolution to recapitulate cell type identities and molecular features of finer brain structures with subregional identity, such as different areas of the cortex or distinct nuclei in the hypothalamus, an evolutionarily conserved brain region that maintains systemic homeostasis and mediates critical physiological functions (Puelles and Rubenstein, 2015; Sternson, 2013).
Among many nuclei in the hypothalamus, the arcuate nucleus (ARC) is an essential structure that integrates circulating signals of hunger and satiety reflecting energy stores and nutrient availability (Andermann and Lowell, 2017). ARC, a small brain region, exhibits tremendous neuronal subtype diversity (Campbell et al., 2017; Huisman et al., 2019). Previous studies have generated 2D hypothalamic-like neurons and 3D hypothalamic organoids from human or mouse iPSCs (Merkle et al., 2015; Qian et al., 2016; Rajamani et al., 2018; Wang et al., 2016; Wataya et al., 2008). However, no methods are currently available to generate hypothalamus nucleus-specific organoids. Most studies of ARC development have used mouse models (Bluet-Pajot et al., 2001; Hill et al., 2008), whereas little is known about human ARC development (Zhou et al., 2020). One disease linked to ARC dysfunction is Prader-Willi Syndrome (PWS), a neurodevelopmental disorder characterized by impaired satiety and severe childhood obesity, and social and learning deficits (Angulo et al., 2015; Mercer et al., 2013). Although clinical abnormalities in PWS have been reported in fetal and early childhood development (Miller et al., 2006; Miller et al., 2011; Xu et al., 2017), little is known about the pathophysiology of PWS in the developing human hypothalamus, in part due to the lack of nucleus-specific in vitro models for this genetic disorder.
Here we developed a protocol to generate arcuate nucleus-like organoids (ARCOs) from human iPSCs that recapitulate diverse cell types of the ARC based on known markers. As a reference, we performed single-nucleus RNA-seq of the neonatal human hypothalamus to characterize the detailed molecular heterogeneity underlying specific subpopulations of hypothalamic cells, and identified a putative human molecular ARC signature at the single-cell level through a transfer learning approach using published mouse hypothalamus single-cell transcriptomes (Campbell et al., 2017; Kim et al., 2019; Mickelsen et al., 2019; Moffitt et al., 2018; Romanov et al., 2020; Wen et al., 2020). We showed that ARCOs exhibit a high similarity to this human molecular ARC signature. In addition, we generated ARCOs from PWS patient iPSCs and revealed that they exhibit aberrant differentiation patterns during development, defective leptin responses, and convergent global transcriptomic dysregulation with published PWS patient hypothalamus datasets (Bochukova et al., 2018; Falaleeva et al., 2015) suggestive of impaired cellular processes and exacerbated inflammatory responses. Together, ARCOs derived from human iPSCs can recapitulate distinct molecular signatures of neurotypical human ARC and patient-derived ARCOs represent an amenable system for modeling and identifying cellular and molecular deficits in ARC-associated brain disorders.
RESULTS
Generation and Characterization of ARCOs from Human iPSCs
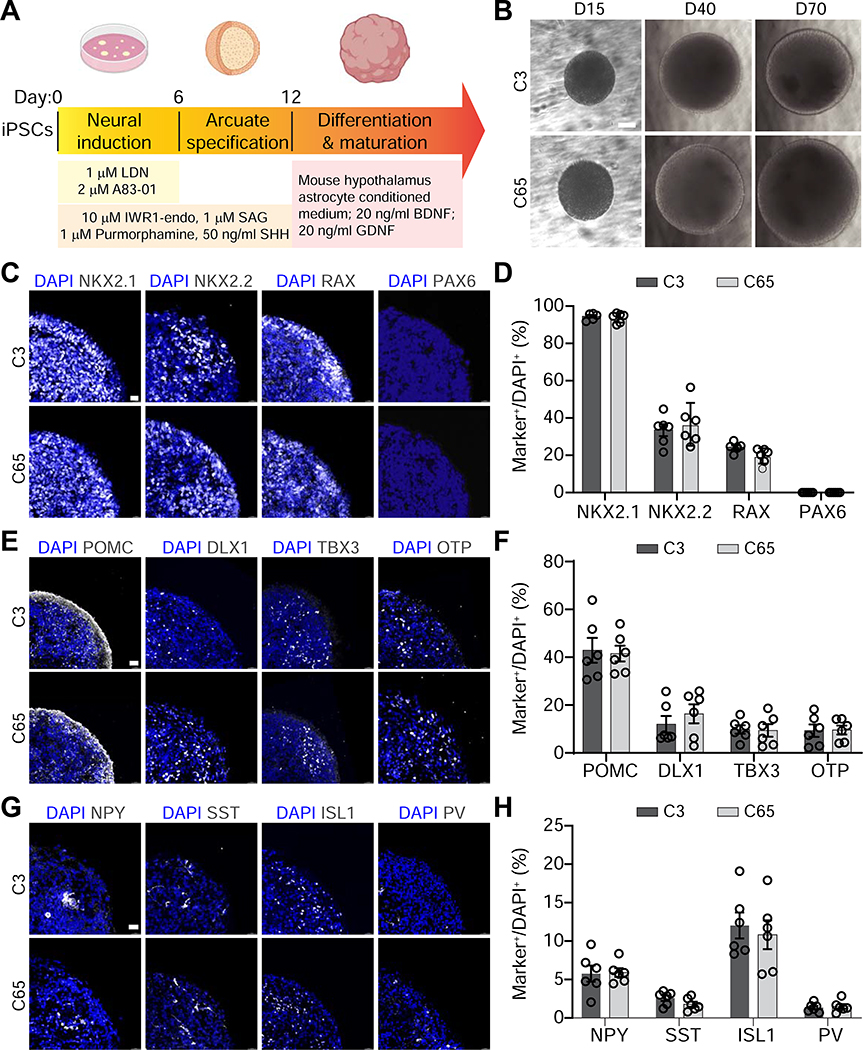
In the developing brain, the hypothalamus is generated from the diencephalon, which is located in the caudal region of the forebrain and is comprised of multiple discrete nuclei, including the arcuate nucleus (ARC), lateral hypothalamus (LHA), ventromedial hypothalamus (VMH), and suprachiasmatic nucleus (SCN) (Figure S1A). Based on developmental patterns of morphogen expression and the requirement of Sonic Hedgehog (Shh) signaling activation and Wnt inhibition during hypothalamic ARC development (Blaess et al., 2014; Corman et al., 2018; Kapsimali et al., 2004), we developed a protocol to generate ARCOs from human iPSCs by an extended induction of hypothalamic patterning using combined Shh activation (with recombinant SHH protein, SAG and Purmorphamine) and Wnt signaling inhibition (with IWR-1-endo) starting at the neural ectoderm specification stage by dual SMAD inhibition (A83–01 and LDN-193189), followed by maturation of hypothalamic neurons using hypothalamus astrocyte conditioned medium and trophic factors (Figure 1A). ARCOs derived from two independent control iPSC lines C3 and C65 (Figures S1B–D and Table S1) maintain a spherical morphology and relatively small size for prolonged periods up to 100 days in vitro (DIV), facilitating a permissive environment for diffusion of nutrients and oxygen with low numbers of cleaved caspase-3+ apoptotic cells (C3 iPSC line: 0.044 ± 0.023%, C65 iPSC line: 0.087 ± 0.033%; n = 3; Figure 1B).
Figure 1. Generation and Characterization of Arcuate Organoids from Human iPSCs.
(A) Schematic describing the protocol for generating arcuate organoids (ARCOs) from human iPSCs.
(B) Sample bright-field images of ARCOs at 15, 40 and 70 days in vitro (DIV). Scale bar, 300 μm.
(C-H) Sample confocal images of immunostaining for NKX2.1, NKX2.2, RAX and PAX6 in ARCOs at 15 DIV (C, scale bar, 15 μm), for POMC, DLX1, TBX3 and OTP at 40 DIV (E, scale bar, 30 μm), for NPY, SST, ISL1 and PV at 40 DIV (G, scale bar, 30 μm), and quantifications (D, F, H). Values represent mean ± SEM with individual data points plotted (n = 6 organoids per iPSC line).
We first examined early patterning of cellular identities in ARCOs by performing immunostaining at 15 DIV. We found expression of hypothalamic progenitor markers NKX2.1, NKX2.2 and RAX (Blackshaw et al., 2010; Romanov et al., 2020), in conjunction with a lack of expression of the forebrain neural progenitor marker PAX6 (Figures 1C–D and Table S2). We further identified top marker genes expressed during ARC development based on published single-cell transcriptomic data (Huisman et al., 2019) (Figure S1E) and our own characterization of the mouse ARC (Ma et al., 2021), including the transcription factors OTP, DLX, and TBX3 (Figure S1F), and the canonical ARC marker pro-opiomelanocortin (POMC). We found robust expression of these markers in ARCOs at 40 DIV (Figures 1E–F). ARCOs also expressed other markers that are known to be expressed in diverse hypothalamic neurons, such as NPY, SST, ISL1, and PV (Figures 1G–H). Together, these data indicate that our protocol leads to the generation of a diverse ARC-like neuronal population. Quantitative analysis showed consistent differentiation and temporal progression of iPSC lines derived from two independent subjects (Figures 1C–H).
Modeling ARC-Specific Cell Type Diversity and Molecular Signatures by ARCOs
To systematically investigate the cell-type diversity and molecular signatures of ARCO subpopulations, we performed single-cell transcriptomic analysis of ARCOs from C3 and C65 iPSC lines at two developmental timepoints each (20 DIV: 8,146 cells; 40 DIV: 4,369 cells) (Figure 2A). A total of 14 broad clusters were obtained from ARCO samples, which were further categorized into multiple cell types, including different populations of neural progenitor cells (NPCs), intermediate progenitor cells (IPCs), and neurons (Figures 2A–B). These cell types can be reproducibly generated in ARCOs from two different control iPSC lines at different developmental timepoints (Figure S2A). While ARCOs contained large populations of hypothalamic neural progenitor cells expressing NESTIN and SOX2, and immature neurons expressing DCX and SYT1, strong expression of hypothalamic ARC markers, such as POMC, OTP, DLX1, and TBX3, was found in specific populations (Figures 2C and S2B).
Figure 2. Single-Cell RNA-Seq Analyses of ARCOs.
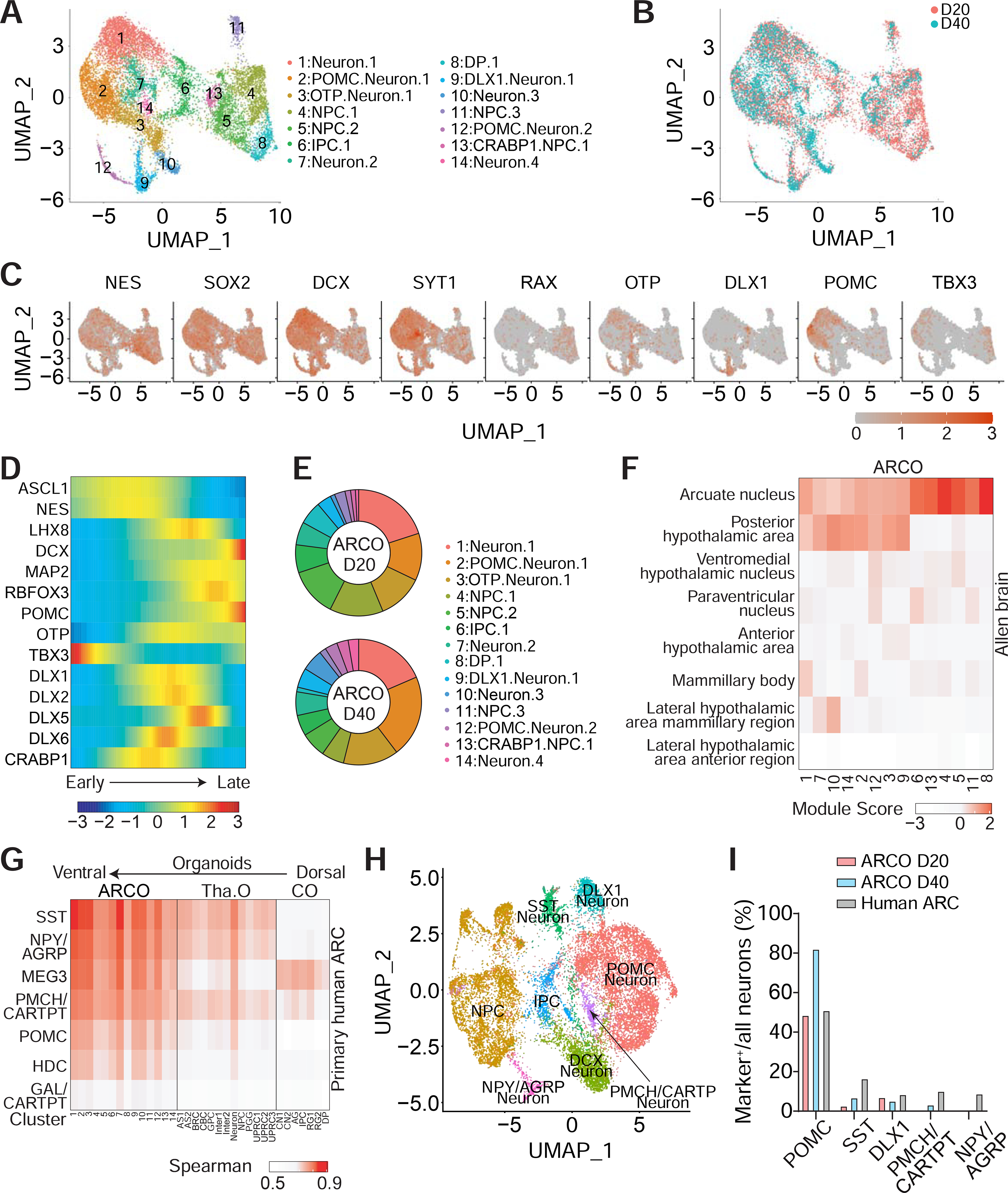
(A-C) Sample UMAP plots of single-cell RNA-seq analysis of ARCOs at 20 and 40 DIV, colored and labeled by cluster (A) or by timepoint (B), and representative feature plots of genes expressed in ARCOs (C).
(D-E) Pseudotime trajectory analysis of selected genes in ARCOs (E) and proportion of cellular subtypes in ARCOs at 20 and 40 DIV (E).
(F) Heatmap showing average module scores across all ARCO clusters for each hypothalamic nuclei-specific gene list compiled from the published Allen Brain Adult Human database (Hawrylycz et al., 2012; Jones et al., 2009; Shen et al., 2012; Sunkin et al., 2013), plotted as the column Z-score per ARCO cluster for visualization. See Methods section for detailed explanation of the module score.
(G) Transcriptomic comparison of the predicted human ARC neuronal subtypes with different cell clusters from ARCOs, thalamus organoids (Tha. O) (Xiang et al., 2019) and neocortical organoids (CO) (Qian et al., 2020).
(H-I) Integrated analysis of single-cell RNA-seq of ARCOs and predicted neonatal human ARC populations. Shown are UMAP of different clusters of the integrated dataset (H) and quantification of different neuronal subtypes as percentages of total neurons from ARCOs at 20 or 40 DIV and predicted human neonatal ARC populations (I).
To investigate developmental changes in molecular trajectories in ARCOs, we performed an inferred pseudotime analysis across ARCOs at 20 and 40 DIV using Monocle (Qiu et al., 2017a; Qiu et al., 2017b; Trapnell et al., 2014), which revealed subpopulation-specific marker expression at different inferred states. For example, neural progenitor markers, such as NESTIN, and ASCL1, were expressed during early stages, whereas ARC-enriched markers POMC and OTP were mainly expressed later, together with the immature neuronal marker DCX and mature neuronal markers MAP2 and RBFOX3/NEUN (Figures 2D and S2C). We found the presence of similar molecular states in both 20 DIV and 40 DIV and the proportion of these states changed over time (Figure 2E).
Next, we compared the general transcriptomic signatures of ARCOs to different subregions of the human hypothalamus using the published subregion-specific Allen Brain Adult Human database (Hawrylycz et al., 2012; Jones et al., 2009; Shen et al., 2012; Sunkin et al., 2013). Indeed, we found that the majority of subpopulations in ARCOs most closely resemble the human ARC compared to other hypothalamic nuclei, such as the VMH, mamillary body and LHA (Figure 2F). Therefore, ARCOs appear to recapitulate not only the cell type diversity, but also the molecular properties of the human hypothalamic ARC.
Identification of ARC-Specific Cell Type Heterogeneity in ARCOs via Machine Learning-Aided Modeling
Currently there is no reference single-cell transcriptome dataset for either embryonic or adult human ARC. To further confirm the brain subregion-specific molecular signatures of human ARCOs, we performed single-nucleus RNA-seq analysis of the neonatal human hypothalamus to generate a reference dataset. The human neonatal hypothalamus exhibits an adult nucleus-like appearance (Koutcherov et al., 2002) and its smaller size makes it feasible to sequence enough cells to capture cells within ARC, an anatomically small region that is difficult to precisely dissect. We generated a total of 70,628 single-nuclei transcriptomes across 6 human samples (Table S1), capturing diverse cellular populations, including neurons (SYT+, SNAP25+), ependymal cells (VIM+, SOX2+), tanycytes (RAX+,FRZB+), oligodendrocyte lineage cells (MOG+, MBP+, MOBP+, OLIG2+), oligodendrocyte precursor cells/NG2 cells (PDGFRA+, FYN+), endothelial cells (CLDN5+, CD34+), and astrocytes (GFAP+, AQP4+, APOE+) (Figures S2D–E). To identify the putative ARC transcriptomic profiles in the human hypothalamus, we applied a machine learning approach based on the recently published Seurat transfer learning workflow (Stuart et al., 2019). We used a published mouse adult ARC dataset (Campbell et al., 2017) as a reference together with published datasets of other mouse hypothalamic regions (Kim et al., 2019; Mickelsen et al., 2019; Moffitt et al., 2018; Romanov et al., 2020; Wen et al., 2020) as negative controls to predict ARC populations in the human hypothalamus at the single-cell level (Figure S2D). In conjunction with supervised analysis of known marker genes and published annotations with reference to the mouse ARC, we obtained a population of predicted ARC cells in our human neonatal hypothalamus dataset comprising 3,563 cells and multiple cellular subpopulations (Figures S2F–G). Characterization of this putative ARC population revealed a high degree of conservation between specific ARC subpopulations in both human and mouse hypothalamus based on the expression of ARC marker genes POMC, TBX3, DLX1 and OTP (Figure S2H). As a validation, we found that the neuropeptide expression patterns in our predicted human ARC population matched previous immunostaining data in the human ARC, including populations expressing NPY, AGRP, CARTPT, CCK and GAL, as well as similar co-expression patterns in the macaque ARC, such as high co-expression of AGRP and NPY, and minimal co-localization of CARTPT and POMC (Figures S2I–J) (Skrapits et al., 2015; True et al., 2017). We then compared individual clusters of cells from the ARCO dataset with the putative human ARC cells in the neonatal human hypothalamus. We found that ARCOs exhibit significantly higher correlation to the human ARC cells at the single-cell level as compared to thalamus organoids (Xiang et al., 2019) and forebrain cortical organoids (Qian et al., 2020) derived from human iPSCs (Figure 2G).
For a direct comparison, we integrated single-cell RNA-seq datasets from ARCOs and predicted human neonatal ARC. We found a largely overlapped distribution of cells for these two samples, suggesting similarity in cell type composition and molecular signatures (Figure S2K). In contrast, similar integration of datasets from ARCO and embryonic mouse ARC (Huisman et al., 2019), fetal (Zhong et al., 2018) and adult (Hodge et al., 2019) human cortex showed less overlap (Figures S2L–N). We further performed cluster analysis of integrated datasets for ARCOs and predicted human ARC neurons (Figure 2H). Quantification of the presence of different neuronal subtypes showed higher percentages of POMC+ neurons and lower percentages of SST+ and NPY+ neurons in ARCOs compared to the human neonatal ARC (Figure 2I). Together, these analyses showed that ARCOs exhibit similar cell type diversity and molecular signatures compared to the human ARC.
PWS-Derived ARCOs Exhibit Aberrant Differentiation and Leptin Responses
PWS is a neurodevelopmental disorder caused by a lack of expression of paternally inherited genes located in the 15q11.2-q13 chromosome region and the major symptoms are attributed to hypothalamic dysfunction (Kalsner and Chamberlain, 2015). To investigate potential cellular and molecular deficits in the ARC of PWS patients at early neurodevelopmental stages, we generated and characterized two iPSC lines each from two PWS patients with two different translocation breakpoints, one between exon 2 and 3 (“major deletion”) and the other between exon 17 and 18 (“minor deletion”) on the chromosome 15q11.2-q13 Prader-Willi SNRPN locus (Figures 3A and S3A–C). qRT-PCR of specific exonic regions revealed a respective loss of gene expression in major and minor deletion ACROs derived from the two patients, indicating that these cell lines can recapitulate disease-specific genetic features of PWS (Figure S3D).
Figure 3. Aberrant Differentiation and Leptin Responses of ARCOs Derived from PWS Patient iPSCs.
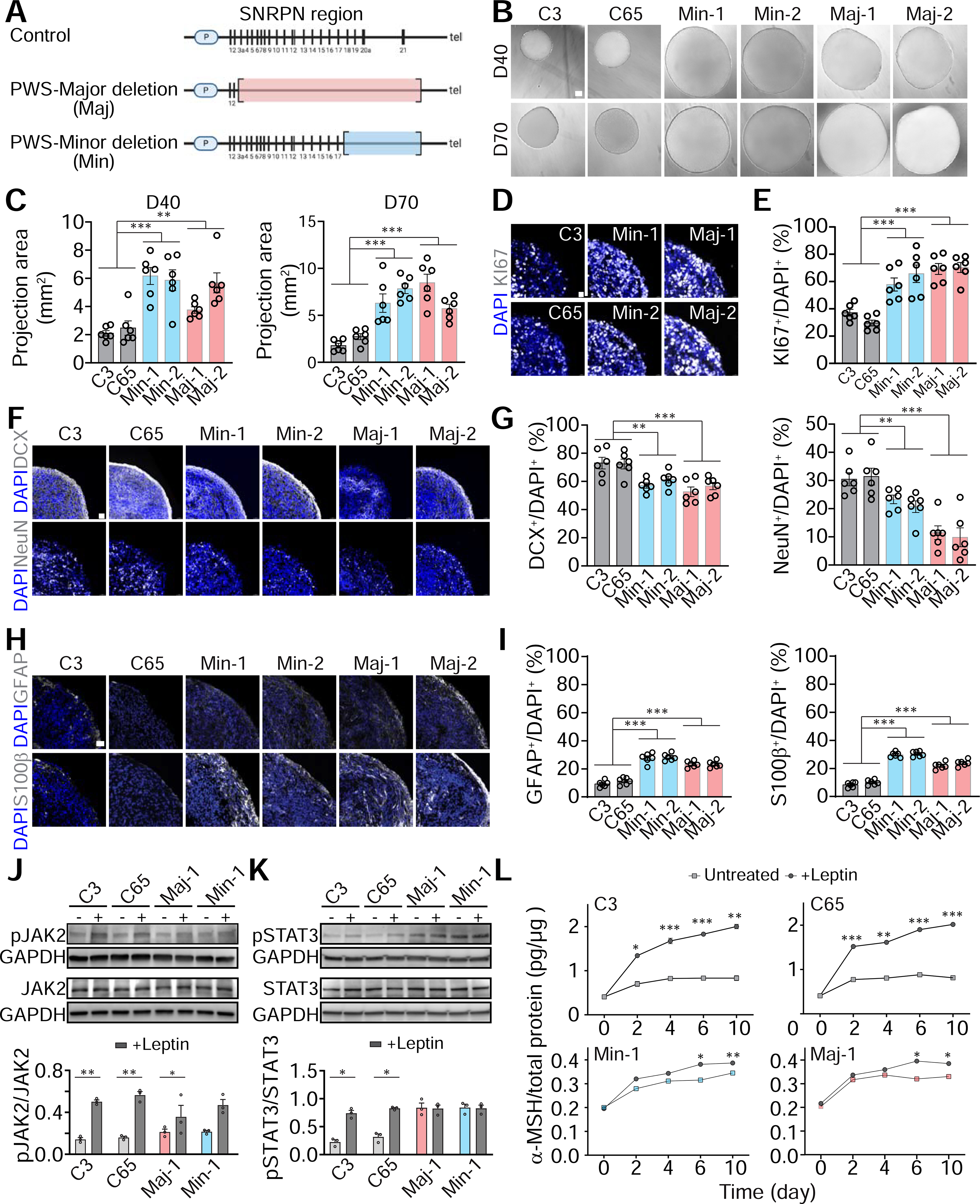
(A) Schematic map of the human paternal chromosome region 15q11-q13 comparison between control, PWS major and minor deletion iPSC lines.
(B-C) Sample bright-field images of ARCOs generated from control and PWS iPSC lines at 40 and 70 DIV (B, scale bar, 300 μm) and quantification of projected area (mm2; C). Values represent mean ± SEM with individual data points plotted (n = 6 organoids per iPSC line; **p < 0.01; ***p < 0.001; One-way ANOVA)
(D-I) Sample confocal images of immunostaining for KI67 in ARCOs from control and PWS iPSC lines at 15 DIV (D, scale bar, 15 μm), for DCX and NeuN at 40 DIV (F, scale bar, 30 μm), for GFAP and s100β at 100 DIV (H, scale bar, 20 μm), and quantifications (E, G, I). Values represent mean ± SEM with individual data points plotted (n = 6 organoids per iPSC line; **p < 0.01; ***p < 0.001; One-way ANOVA).
(J-K) Leptin-induced activation of JAK2 (J) and STAT3 (K) signaling pathway. ARCOs at 60 DIV were treated with Leptin (2 μg/ml) and lysates were subject to Western blot analysis 1 hr later. Shown are sample Western blot images (top panel) and quantification (bottom panel). Values represent mean ± SEM (n = 3; *p < 0.05; ***p < 0.01; Paired t test).
(L) Leptin-induced release of MCH. ARCOs at 60 DIV were treated with Leptin (2 μg/ml) every two days and conditioned medium was subjected to ELSA analysis of MCH. Values represent mean ± SEM (n = 3; *p < 0.05; **p < 0.01; ***p < 0.001; Paired t test).
Morphologically, ARCOs derived from iPSCs of both PWS patients were significantly larger in size compared to control ARCOs (Figures 3B–C). Immunostaining showed that PWS ARCOs contained a significantly higher percentage of KI67+ cells at 15 DIV (Figures 3D–E), but no differences in the expression of NKX2.1 and RAX, suggesting normal patterning of PWS ARCOs (Figures S3E–G). We further found reduced percentages of cells expressing DCX and NeuN (Figures 3F–G and S3H–J), with an increased percentage of cells expressing NESTIN (Figures S3K–M) at both 40 and 70 DIV. We confirmed this result with single-cell RNA-seq analysis of PWS ARCOs at both 20 and 40 DIV, which showed an increased percentage of neural progenitors and a decreased percentage of neurons (Figures S3N–P). This analysis also showed a decreased percentage of POMC+ neurons in PWS ARCOs, but no change in the percentage of SST+ neurons (Figure S3P). We further examined gliogenesis at 100 DIV and found increased percentages of GFAP+ or s100β+ astrocytes in PWS ARCOs (Figures 3H–I). Notably, an increase in s100β+ astrocytes was also observed in postmortem hypothalamus samples of PWS patients compared to control subjects (Bochukova et al., 2018). Together, these results suggest that PWS ARCOs exhibit marked neural proliferation and differentiation deficits compared to control ARCOs.
Given the differentiation deficit in PWS ARCOs, we examined its functional consequence on properties of ARCOs. We plated ARCOs at 60 DIV on a multi-electrode array (MEA) for electrophysiological analysis (Figure S3Q). Two weeks after plating, we observed active neuronal firing for both control and PWS ARCOs, and, furthermore, a decreased frequency of neuronal firing for both PWS major deletion and minor deletion ARCOs (Figures S3R–S). One functional hallmark of ARC is activation of POMC+ neurons in response to leptin, an anorexigenic neuropeptide, with the release of melanocyte stimulating hormone (MSH) (Cowley et al., 2001). Upon leptin treatment, we found elevated levels of phosphorylated forms of JAK2 and STAT3 in control ARCOs at 60 DIV, which were significantly attenuated in PWS ARCOs (Figures 3J–K), suggesting defective leptin signaling (Figures 3J–K). Furthermore, ELISA analysis of conditioned medium of ARCOs treated with leptin showed increased MSH levels in control ARCOs, but much lower levels in PWS ARCOs (Figures 3L). Together, these results showed functional deficits of PWS ARCOs.
Recapitulation of Disease Molecular Signatures in Human PWS Hypothalamus by PWS Patient-derived ARCOs
Next, we performed bulk RNA-seq of ARCOs generated from 2 control, 2 PWS minor deletion, and 2 PWS major deletion iPSC lines at early (20 DIV) and late (100 DIV) stages of maturation to compare transcriptional changes between control and PWS ARCOs in more detail. Principal component and correlation analyses showed clustering of biological replicates within different cell lines across both timepoints (Figures S4A–B and S4E–F). Comparison of control and PWS ARCOs revealed 2,501 (minor) and 5,031 (major) downregulated genes and 2,407 (minor) and 5,194 (major) upregulated genes at 20 DIV (Figures S4C–D and Table S3), as well as 5,444 (minor) and 7,816 (major) downregulated genes and 4,781 (minor) and 7,439 (major) upregulated genes at 100 DIV (Figures S4G–H and Table S4), respectively, indicating the presence of widespread transcriptional dysregulation in PWS ARCOs.
Given that the PWS major deletion ARCO iPSC lines were derived from a PWS patient with a classical clinical presentation, we first examined in more detail downregulated genes in PWS major deletion ARCOs compared to controls at 20 and 100 DIV. We found a decrease in the expression of many genes involved in nervous system development, such as ADCYAP1, CALM2 and RBFOX1/NEUN, further supporting that PWS ARCOs exhibit aberrant development (Figure 4A). In addition, there was a downregulation of synapse-related genes, including SYNJ1 and SYN2 (Figure 4A). Synapse dysfunction has been linked to cognitive defects in a variety of neurodevelopmental disorders and PWS patients are well known to exhibit cognitive defects throughout early childhood development (Dimitropoulos et al., 2019). More detailed gene ontology (GO) analysis of downregulated genes showed enrichment for terms related to regulation of neuron differentiation, axon development, regulation of synaptic plasticity and ion transport (Figure 4B).
Figure 4. Convergent Dysregulated Transcriptomic Signatures in PWS ARCOs and PWS Patient Hypothalamus.
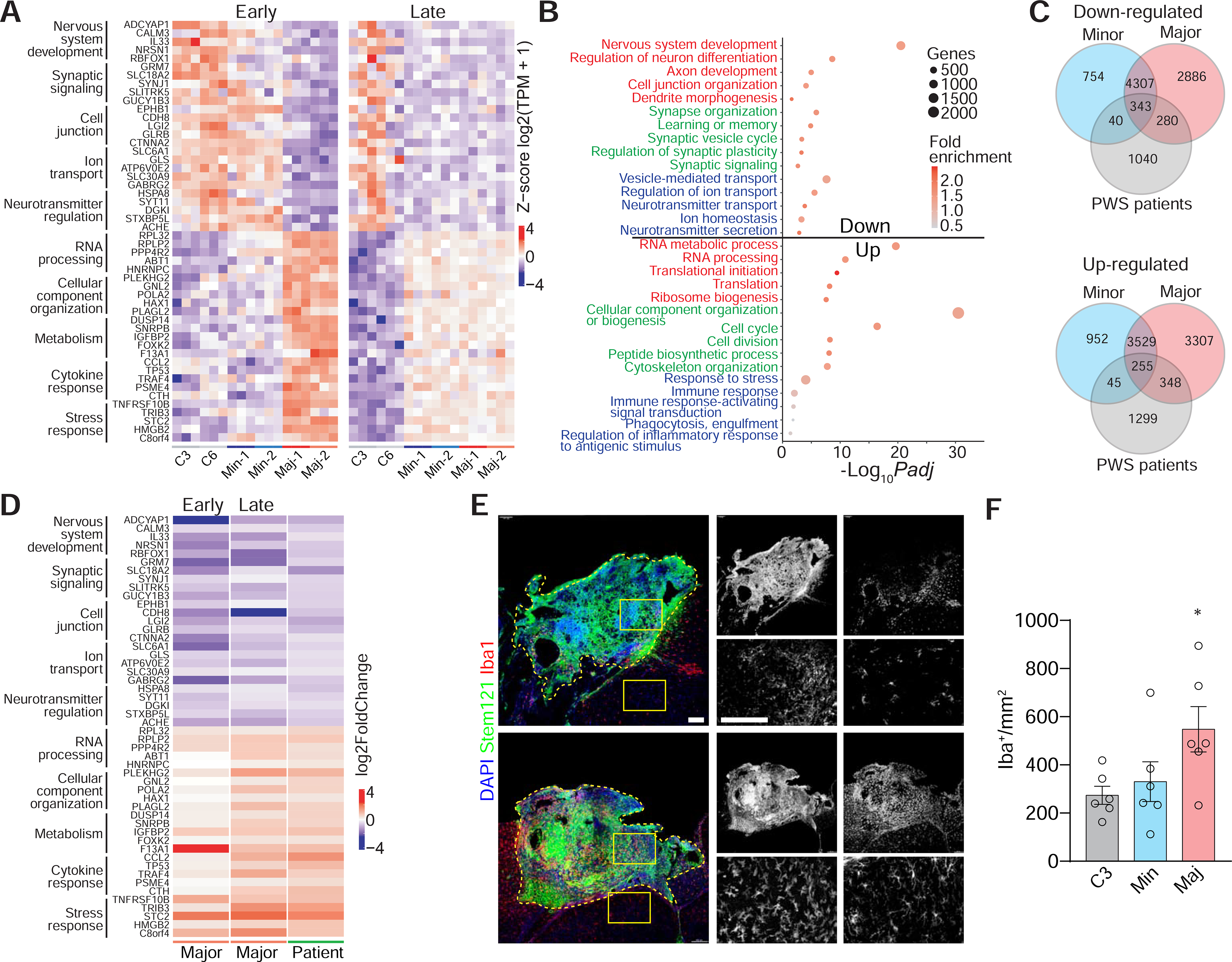
(A) Heatmap of selected downregulated and upregulated genes comparing PWS minor and major deletion iPSC-derived ARCOs with control ARCOs at early (20 DIV) and late (100 DIV) stages. Genes related to different biological functions are grouped and labeled. Values are shown for each biological replicate as the row Z-score per gene of Log2(TPM+1)-transformed values.
(B) Dot plot of selected enriched gene ontology (GO) terms for downregulated and upregulated genes comparing PWS major deletion iPSC-derived ARCOs with control ARCOs. Differentially expressed genes at both 20 and 100 DIV were combined.
(C) Venn diagrams comparing the overlap of downregulated and upregulated genes in PWS minor and major deletion iPSC-derived ARCOs and PWS patient hypothalamus (Bochukova et al., 2018). Differentially expressed genes in ARCOs at both 20 and 100 DIV were combined. The overlap of downregulated genes between PWS major deletion ARCOs and PWS patient hypothalamus is significant (p = 7.26e-07; Fisher’s exact test).
(D) Comparison of selected downregulated and upregulated gene fold-changes in PWS major deletion ARCOs at early (20 DIV) and later (100 DIV) stages and PWS patient hypothalamus. Genes related to different biological functions are grouped and labeled.
(E-F) Sample confocal images of immunostaining for microglia marker IBA1 and human cell marker STEM121 in transplanted ARCOs derived from control and PWS major deletion iPSC lines (E) and quantification (F). Note increased densities of IBA1+ microglia both within (box 1) and near (box 2) the transplanted ARCO derived from PWS major deletion iPSCs compared to the ARCO derived from the control iPSCs. Scale bars, 100 μm. Values represent mean ± SEM with individual data points plotted (n = 6 organoids per iPSC line; *p < 0.05; Student’s t test).
On the other hand, closer examination of upregulated genes in PWS major deletion ARCOs compared to control ARCOs revealed an increase in genes involved in RNA processing, such as RPLP2 and ABT1 (Figure 4A), consistent with findings of abnormal post-transcriptional processing in a mouse model for PWS (Doe et al., 2009; Garfield et al., 2016). Upregulation of genes linked to metabolism, such as IGFBP2 and DUSP14, is also consistent with studies reporting abnormal metabolic processes in children with PWS, such as heightened insulin sensitivity (Haqq et al., 2011) and glucose homeostasis alterations (Fintini et al., 2016). Interestingly, we observed an upregulation of cytokine and stress response genes, such as CCL2, TRAF4 and TNFRSF10B (Figure 4A). In one study, analysis of inflammatory cytokines present in the plasma of 23 PWS patients identified CCL2 as one of the four cytokines enriched in the patients’ plasma compared to controls (Butler et al., 2015), while another study reported that a pro-inflammatory phenotype is associated with behavioral traits in children with PWS (Krefft et al., 2020). Together, GO analysis of upregulated genes revealed enrichment for terms related to translational initiation, ribosome biogenesis, peptide biosynthetic process, as well as immune responses (Figure 4B).
Comparison of the dysregulated gene list in minor and major deletion PWS ARCOs revealed an overlap of 4,650 and 3,784 of downregulated and upregulated genes, respectively (Figure 4C), with dysregulated genes enriched for GO processes including the oxidation-reduction process, response to nutrient levels, and chromatin organization (Figure S4I).
To examine whether PWS ARCOs can recapitulate certain features of transcriptional dysregulation in the human PWS brain, we compared transcriptional changes in PWS ARCOs with a published PWS patient hypothalamus RNA-seq dataset with 4 patients and 4 controls (Bochukova et al., 2018) (Figure 4C). There was an overlap of 623 downregulated and 603 upregulated genes between the PWS major deletion ARCOs and PWS patient hypothalamus when compared to their respective controls, indicating that transcriptomic differences reported postnatally in PWS patients may be initiated during early neurodevelopment (Figure 4C). Additionally, a number of these common differentially expressed genes showed comparable or higher fold-changes in PWS major deletion ARCOs compared to PWS patient hypothalamus (Figure 4D), suggesting a convergent dysregulation in cellular processes, such as those related to nervous system development, which may begin during embryonic development. We also found many more dysregulated genes in PWS ARCOs compared to PWS patient hypothalamus (Figure 4C).
In addition to RNA processing, translation and extracellular matrix organization, GO analysis of common upregulated genes in PWS major deletion ARCOs and PWS patient hypothalamus showed enrichment for terms related to response to cytokines and stress response (Figure S4J). These results raise the possibility that cells in PWS ARCOs may signal to promote infiltration of immune cells and microglia during systemic inflammation, similar to the chronic low-grade inflammation found in the hypothalamus of obese patients (Le Thuc et al., 2017). To directly test this possibility, we transplanted PWS major and minor deletion ARCOs and control ARCOs into adult mouse brains to provide exposure to an extrinsic immune system. Interestingly, we found a higher number of IBA1+ microglia, with activated morphology, infiltrating transplanted major deletion PWS ARCOs compared to minor deletion PWS and control ARCOs, consistent with an exacerbated inflammatory response signature in major deletion PWS ARCOs (Figures 4E–F).
To further examine gene expression dysregulation similarities between PWS ARCOs and patient hypothalamus, we compared a previously published microarray dataset with 2 PWS patients and 3 controls (Falaleeva et al., 2015). We found an overlap of 1,867 downregulated genes and 158 upregulated genes for major deletion PWS ARCOs and patient hypothalamus (Figure S4K).
DISCUSSION
Despite significant advances in the organoid field, there are currently no reported organoid protocols that model fine brain regions with a subregional identity. A remarkable feature of the hypothalamus is the complex cellular composition and tremendous neuronal diversity within individual nuclei, which have essential physiological functions and have been associated with various disorders when dysregulated. Here, we developed a protocol for generating ARCOs from human iPSCs that share the expression of markers observed in the mouse ARC. Single-cell RNA profiling of ARCOs and neonatal human hypothalamus aided by a machine learning approach further reveals a predicted human ARC signature in ARCOs. Using ARCOs generated from multiple iPSC lines derived from PWS patients, we found aberrant differentiation patterns and functional properties in PWS ARCOs. Transcriptomic comparison of PWS ARCOs and published PWS patient hypothalamus datasets showed convergent signatures of transcriptional dysregulation related to downregulation of neural development and synaptic function in conjunction with upregulation of RNA processing and inflammatory responses. Our study provides a human organoid platform for investigating molecular and cellular processes during ARC development and potential mechanisms underlying disorders associated with hypothalamic ARC dysfunction.
Modeling Hypothalamic ARC Development Using Brain Organoids
Brain organoids have recently emerged as a promising in vitro model to study fetal human brain development (Qian et al., 2020). Our protocol relies on similar underlying principles of morphogen expression patterns as previous findings reporting the generation of hypothalamic neurons from human iPSCs, wherein 2D hypothalamic-like neurons were obtained through early simultaneous activation of Shh and inhibition of Wnt signaling to promote a high percentage of hypothalamic progenitors (Rajamani et al., 2018). Another more recent protocol generates a functional hypothalamic-pituitary unit using human iPSCs, which gives rise to both anterior pituitary and hypothalamic neurons via culture conditions with both SAG to induce Shh activation and bone morphogenetic protein 4 (BMP4) (Kasai et al., 2020). Using a precisely controlled targeted differentiation protocol to induce prolonged ventralization of progenitor fate, we generated and characterized ARC-specific hypothalamic organoids across multiple time points. Our protocol generates consistent organoids reliably across different iPSC lines. Compared to forebrain organoids, ARCOs are much smaller, with no apparent necrosis in the core even at 100 DIV. ARCOs showed robust expression of hypothalamic progenitor markers and transcription factors enriched in diverse ARC neuronal populations, suggesting that cellular compositions within fine brain structures with a subregional identity can be recapitulated with specific patterning factors. Unlike the laminarly organized cortical layers of the cortex, ARC as a nucleus does not exhibit obvious cytoarchitecture organization with a mixed distribution of different neuronal subtypes in vivo in mice (Ma et al., 2021). Similarly, we did not observe obvious cytoarchitectural organization or a distinct pattern of distribution of different neuronal subtypes in ARCOs.
We compared unbiased single-nucleus profiling of the human neonatal hypothalamus across multiple subjects and examined the transcriptome of distinct cellular populations within the human hypothalamus to serve as a reference dataset for organoid comparisons. We identified an ARC molecular signature from the human hypothalamus based on mouse hypothalamic ARC and previously identified markers, which was partially validated with published immunostaining data from the human and macaque ARC (Skrapits et al., 2015; True et al., 2017). We further used this information to validate human ARC-specific signatures in our ARCOs and quantitatively compared the composition of different neural subtypes. Here we provide an example of how our unbiased label transfer approach can be used to resolve transcriptomic heterogeneity in brain regions for systematic molecular characterization of cellular subpopulations, as well as identification of candidate marker genes for downstream validation. This approach may also be applied to organoid protocols for other fine brain structures wherein both cross-species and cross-platform datasets can be applied in tandem to identify less well characterized organ subregions.
Modeling Developmental Deficits in Prader-Willi Syndrome with ARCOs
ARCOs contain neuronal diversity and key molecular features of the human ARC and can be efficiently maintained in culture for prolonged periods of time, offering the opportunity to model hypothalamus-associated developmental brain disorders. PWS is a complex genetic disorder linked to hypothalamic dysfunction that causes obesity and intellectual disability. Given that PWS is directly linked to hypothalamus ARC function, current cellular models such as 2D cortical neurons (Soeda et al., 2019) may not adequately recapitulate the diverse cellular features of PWS pathogenesis compared to our platform. Although some of the PWS-like phenotypes can be mimicked in PWS mouse models wherein the function of one or more PWS genes is disrupted (Carias and Wevrick, 2019), much less is known about cellular and molecular mechanisms underpinning PWS in human hypothalamus. As the ARC represents only one of the many nuclei of the human hypothalamus, a brain region making up only approximately 0.4% of the whole brain, it is challenging to specifically assess the changes in ARC from post-mortem samples. Brain organoids provide an opportunity to temporally observe phenotypic characteristics of PWS in vitro, with the option of adding external perturbations such as therapeutic compounds to determine changes in cellular functions. The development of protocols modeling fine brain structures, such as ARCOs, adds to the current 3D organoid tool box for investigating cell type diversity and molecular signatures in subregions of the brain in health and disease.
In our study, we modeled PWS using ARCOs derived from different PWS patients to investigate how cellular, molecular and functional phenotypes in vitro may be associated with clinical manifestations of the disease. We showed that ARCOs derived from patients with PWS maintain disease and patient-specific gene signatures and exhibit marked deficits in differentiation during early development, raising the possibility of altered neurodevelopmental progression prenatally. This is consistent with studies that have found an increase in s100β+ astrocytes (Bochukova et al., 2018), decreased grey matter in the hypothalamus of children with PWS, as well as hypotonia in PWS infants and reduced fetal movement in utero in PWS patients (Miller et al., 2006; Miller et al., 2011; Xu et al., 2017). We further showed functional deficits using PWS ARCOs, including reduced neuronal firing and defective responses to leptin treatment in activation of downstream signaling and release of MCH.
Implications of Potential Impairments in Fetal Development in Prader-Willi Syndrome
Our finding of an overlap of dysregulated gene expression between the PWS major deletion ARCO and PWS patient hypothalamus suggests that disease signatures at the transcriptional level may be conserved to some degree from embryonic to postnatal development, highlighting the importance of developmental models of genetic disorders in unraveling the etiologies of such diseases. Specifically, common genes that show transcriptional dysregulation between PWS major deletion ARCOs and PWS patient hypothalamus suggest that there may be perturbations to multiple cellular functions. For example, downregulation of many genes implicated in nervous system development is consistent with the cellular phenotype of aberrant differentiation in patient-specific organoids as well as developmental delays reported in PWS patients (Festen et al., 2007), whereas decreased expression of synapse-linked genes may be indicative of synaptic dysregulation, which could be associated with compromised intellectual abilities. Furthermore, downregulation of multiple genes related to axonal development is consistent with findings in mouse models of PWS that show aberrant axonal projections (Maillard et al., 2016; Miller et al., 2009; Pagliardini et al., 2005), and may contribute to the cognitive and somatosensory deficits presented in PWS patients. Upregulation of extracellular matrix organization genes in both PWS ARCOs and PWS patient hypothalamus suggests that there may be increased signaling of PWS ARC cells to the adjacent vasculature for the recruitment of inflammatory cells, given that the extracellular matrix is known to be important in modulating immune cell behavior in tissues which are inflamed (Sorokin, 2010). Indeed, inflammatory cytokines, such as CCL2, which play a key role in modulating the recruitment of immune cells to infection sites, were also upregulated. Consistent with these results, we showed that PWS major deletion patient-derived ARCOs exhibit an increased number of infiltrating microglia when transplanted into the mouse brain. Notably, our study reveals similar trends to those reported in a recently published study showing that the transcriptomic signature in the hypothalamus of PWS patients is characterized by neuronal loss, which could be indicative of early deficits in differentiation, and upregulation of neuroinflammatory processes (Bochukova et al., 2018), which implicates these biological processes as potential contributors to PWS disease pathogenesis.
Limitations of the Study
A limited number of iPSC lines, especially PWS donors, were included in the current study. Future studies using a larger cohort with more iPSC lines are needed for better modeling of PWS. While our study demonstrates that dysregulated transcriptomic signatures are conserved between ARCOs in vitro and PWS human hypothalamus, a detailed analysis of PWS patient hypothalamus samples is necessary to verify the disease phenotypes in vivo. There are also caveats of using brain organoids that limit the interpretation of results. For example, since brain organoids are more similar to the developing fetal brain than the mature postnatal brain, there may exist important differences in PWS disease pathophysiology between immature and mature neural cells, given that technical barriers have largely precluded the investigation of PWS disease phenotypes in the fetal brain. The lack of certain cell types, such as immune cells, in brain organoid cultures also indicates that regulation of inflammatory processes important to PWS disease pathogenesis in vivo may not be well represented. Extending studies of disease modeling in ARCOs to include examining functional interactions with other cell types and/or different brain region-specific organoids would be important in providing mechanistic insights into disease pathogenesis. Despite our findings that cellular functions are disrupted in PWS ARCOs and that the associated transcriptional changes are similar in PWS patient hypothalamus, it is unclear if these phenotypic deficits have functional consequences in vivo. It remains poorly understood as to what the contributing factors to clinical symptoms in PWS patients are, be it dysregulation of processes such as impaired prenatal neurogenesis, excessive extracellular matrix remodeling, circulating cytokines leading to chronic inflammation of the hypothalamic ARC, or a combination of other factors. Future studies in PWS patients will be important to shed light on the correlation between in vitro phenotypes and clinical symptoms.
In summary, our study provides a scalable, efficient and robust protocol to generate ARC-specific organoids, which can be used to model early hypothalamic ARC developmental processes and related brain diseases. Validation of nuclei-specific markers in ARCOs shows that distinct fine brain regions with tremendous neuronal diversity and subregional identities can be modeled through careful adjustment of the timing of patterning and combinations of induction factors used in culture. Our study further provides a comparative analysis of a single-cell human hypothalamus dataset and a comprehensive view of transcriptome signatures in distinct brain nuclear structures, providing a resource for exploring conserved and divergent signatures between the human and mouse hypothalamus. Finally, modeling early ARC development using this protocol would also provide access to phenotypes during the early stages of hypothalamus-associated diseases that precede the onset of clinical symptoms, which could be useful for development of diagnostic biomarkers and testing new therapeutics.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Guo-li Ming (gming@pennmedicine.upenn.edu).
Material Availability
All unique/stable reagents and biological material generated in this study are available from the Lead Contact, Dr. Guo-li Ming (gming@pennmedicine.upenn.edu), with a completed Materials Transfer Agreement.
Data and Code Availability
The RNA-seq data reported in this study is deposited in GEO: GSE164101 (human hypothalamus) and GSE164102 (human arcuate organoids). the published article includes all data generated during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Induced Pluripotent Stem Cells and Human Brain Tissue
Human iPSC lines used in the current study were either previously generated and fully characterized (Chiang et al., 2011; Wen et al., 2014; Yoon et al., 2014) or generated and characterized in this study (Table S1). C3 and C65 iPSCs were generated from fibroblasts of normal adult subjects (Kim et al., 2021). PWS iPSCs for the minor (Schüle et al., 2005) and major (Kuslich et al., 1999) deletion were generated from previously reported fibroblasts obtained from individuals with PWS from the Coriell Institute Cell Repository (minor deletion: GM21890, major deletion: GM21889). Generation of iPSC lines followed institutional IRB and ISCRO guidelines and was approved by Johns Hopkins University School of Medicine. Karyotyping analysis by standard G-banding technique was carried out by the Cytogenetics Core Facility at the Johns Hopkins Hospital or Cell Line Genetics. Results were interpreted by clinical laboratory specialists of the Cytogenetics Core or Cell Line Genetics. De-identified human brain tissues were obtained from the Brain and Tissue Repository of the NIH NeuroBioBank following institutional IRB approved by University of Pennsylvania Perelman School of Medicine (Table S1).
Animals
All animal procedures used in this study were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Animals were housed at a maximum of five per cage with a 14-hr light/10-hr dark cycle with food and water ad libitum. Female 4–8-week-old athymic nude (NU/J) mice (The Jackson Laboratory) were used for transplantation experiments. P3 CD-1 mice pups (Charles River) were used for isolation of hypothalamic astrocytes to prepare astrocyte-conditioned medium.
METHOD DETAILS
Maintenance of Human iPSCs
All studies involving human iPSCs were performed under approved protocols of the University of Pennsylvania. All human iPSC lines were confirmed to have a normal karyotype. For organoid generation, iPSCs were cultured on mouse embryonic fibroblast feeder (MEF) cells in stem cell medium consisting of DMEM:F12 supplemented with 20% KnockOut Serum Replacement, 1X MEM-NEAAs, 1X GlutaMAX, 1X Penicillin-Streptomycin, 1X 2-mercaptoethanol, and 10 ng/mL bFGF in a 5% CO2, 37°C, 90% relative humidity incubator as previously described (Qian et al., 2018). Culture medium was replaced every day. Human iPSCs were passaged every week onto a new tissue-treated culture plate coated with 0.1% gelatin for 2 hr and pre-seeded with γ-irradiated CF1 MEF cells one day in advance. Colonies were detached by washing with DPBS and treating with 1 mg/mL Collagenase Type IV for 30–60 min. Detached colonies were washed 3 times with 5 mL DMEM:F12 and dissociated into small clusters by trituration with a P1000 pipette.
Generation of ARCOs
Human iPSC colonies were detached with Collagenase Type IV 7 days after passage and washed with fresh stem cell medium in a 15 mL conical tube. On Day 0, detached iPSC colonies were transferred to an ultra-low attachment 6-well plate (Corning Costar), containing induction medium I consisting of DMEM:F12 and neurobasal medium in a 1:1 ratio, plus 1 μM LDN, 2 μM A83–01, 10 μM IWR1-endo, 1 μM SAG, 1 μM purmorphamine, and 50 ng/mL recombinant Sonic Hedgehog. Organoids were placed on a CO2 resistant orbital shaker (ThermoFisher) starting from Day 3. On Day 6, organoids were transferred to induction medium II (same as the induction medium I but excluding 1 μM LDN, and 2 μM A83–01). From Day 12, organoids were transferred to differentiation medium consisting of neurobasal medium conditioned by mouse (CD1) hypothalamus astrocytes, 20 ng/ml BDNF, 20 ng/ml GDNF, 1X B27 Supplement, 0.5 μM Dibutyryl-cAMP, 0.2 mM Ascorbic Acid, 1X GlutaMAX, and 1X NEAA. Organoids were maintained with media changes every other day. To obtain the conditioned medium, astrocytes isolated from P3 CD-1 mouse hypothalamus were grown on 75-cm culture flask until confluency as previously described (Song et al., 2002). Every 24 hours, astrocyte conditioned medium was collected and replaced with fresh medium. Astrocyte conditioned medium was sterile filtered and stored at −80 °C until use.
ARCO Immunohistochemistry and Microscopy
Organoids were placed directly in 4% methanol-free formaldehyde (Polysciences) diluted in DPBS (Thermo Fisher Scientific) overnight at 4°C on a tube rotator (Boekel). After fixation, ARCOs were washed in DPBS and cryoprotected by overnight incubation in 30% sucrose (Sigma-Aldrich) in DPBS at 4°C. ARCOs were placed in a plastic cryomold (Electron Microscopy Sciences) and snap frozen in tissue freezing medium (General Data) on dry ice. Frozen tissue was stored at −80°C until processing. Serial tissue sections (20 μm for organoids) were sliced using a cryostat (Leica, CM3050S), and melted onto charged slides (Thermo Fisher Scientific). Slides were dried at room temperature and stored at −20°C until ready for immunohistology.
For immunostaining, cryosectioned slides were washed with PBS before permeabilization with 0.5% Triton-X in PBS for 1 hr (Jacob et al., 2020a). Tissues were incubated with blocking medium consisting of 10% donkey serum in PBS with 0.05% Triton-X (PBST) for 30 min. Primary antibodies diluted in blocking solution were applied to the sections overnight at 4°C. The primary antibodies used and their dilution are summarized in the Key Resources Table. After washing with PBST for a minimum of 5 times, secondary antibodies diluted in blocking solution were applied to the sections for 1–4 hr at room temperature or overnight at 4°C. Sections were washed with PBST for a minimum of 5 times before mounting in mounting solution (Vector laboratories), cover-slipped, and sealed with nail polish. Secondary antibodies were: AlexaFluor 488, 546, 594, or 647 - conjugated donkey antibodies (Invitrogen) used at 1:500 dilution. Images were captured by a confocal microscope (Zeiss LSM 800). Sample images were prepared in Imaris (Bitplane) and Photoshop (Adobe) software.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-AFP | Dako | Cat#: A0008; RRID: AB_2650473 |
| Rabbit anti-CC3 | Cell Signaling Technology | Cat#: 9661; RRID: AB_2341188 |
| Goat anti-DCX | Santa Cruz | Cat#: sc-8066; RRID: AB_2088494 |
| Rabbit anti-DLX1 | (Lee et al., 2018) | N/A |
| Rabbit anti-GFAP | Dako Agilent | Cat#: Z033429-2 |
| Rabbit anti-GAPDH (14C10) | Cell Signaling Technology | Cat#: 2118S; RRID: AB_561053 |
| Rabbit anti-IBA1 | Wako | Cat#: 019-19741; RRID: AB_839504 |
| Mouse anti-ISL1 | DSHB | Cat#: 40.3A4; RRID: AB_528313 |
| Rabbit anti-Jak2 (D2E12) XP | Cell Signaling Technology | Cat#: 3230S; RRID: AB_2128522 |
| Mouse anti-KI67 | BD | Cat#: 550609; RRID: AB_393778 |
| Rabbit anti-KLF4 | Santa Cruz | Cat#: sc-20691; RRID: AB_669567 |
| Mouse anti-NESTIN | Stemcell Technologies | Cat#: 60091; RRID: AB_2650581 |
| Rabbit anti-NKX2.1 | Santa Cruz | Cat#: sc-13040; RRID: AB_793532 |
| Mouse anti-NKX2.2 | DSHB | Cat# 74.5A5; RRID:AB_531794 |
| Mouse anti-NEUN | Millipore | Cat#: MAB377B; RRID: AB_177621 |
| Rabbit anti-NPY | Millipore | Cat#: ab9608; RRID: AB_2153720 |
| Mouse anti-OCT4 | Santa Cruz | Cat#: sc-5279; RRID: AB_628051 |
| Guinea Pig anti-OTP | Clontech | Cat#: M195 |
| Mouse anti-PAX6 | BD | Cat#: 561664; RRID: AB_10895587 |
| Rabbit anti-POMC | Phoenix Pharmaceuticals | Cat#: H-029-30; RRID: AB_2307442 |
| Rabbit anti-PV | Abcam | Cat#: ab11427; RRID: AB_298032 |
| Rabbit anti-Phospho-Jak2 (Tyr1007/1008) (C80C3) | Cell Signaling Technology | Cat#: 3776S; RRID: AB_2617123 |
| Rabbit anti-Phospho-Stat-3 (tyr705) | Cell Signaling Technology | Cat#: 9131S; RRID: AB_331586 |
| Mouse anti-RAX | Santa Cruz | Cat#: sc-271889; RRID: AB_10708730 |
| Rabbit anti-SMA | Abcam | Cat#: AB5694; RRID: AB_2223021 |
| Goat anti-SOX2 | Santa Cruz | Cat#: sc-17320; RRID: AB_2286684 |
| Mouse anti-SSEA4 | Millipore | Cat#: MAB4304; RRID: AB_177629 |
| Rabbit anti-SST | Santa Cruz | Cat#: sc-13099 RRID: AB_2195930 |
| Mouse anti-STEM121 | Takara | Cat#: Y40410; RRID: AB_2801314 |
| Mouse anti-S100B (CL2720) | Sigma-Aldrich | Cat#: AMAb91038; RRID: AB_2665776 |
| Mouse anti-Stat3 (124H6) | Cell Signaling Technology | Cat#: 9139S; RRID: AB_331757 |
| Goat anti-Tbx3 | Santa Cruz | Cat#: sc-17871; RRID: AB_661666 |
| Mouse anti-TRA-1-60 | Millipore | Cat#: MAB4360; RRID: AB_2119183 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 2-Mercaptoethanol | Thermo Fisher Scientific | 21985023 |
| A83-01 | Stemcell Technologies | 72024 |
| Advantage 2 PCR Kit | Takara Bio | 639206 |
| Advantage UltraPure PCR deoxynucleotide mix (10mM each dNTP) | Takara Bio | 639125 |
| AMPure XP beads | Beckman Coulter | A63880 |
| Ascorbic acid | Sigma-Aldrich | A0278 |
| B-27 Supplement (50X), minus vitamin A | Thermo Fisher Scientific | 12587010 |
| bFGF | Peprotech | 100-18B |
| Bovine serum albumin (BSA) | Sigma-Aldrich | B6917; CAS# 904846-8 |
| Bio-Rad Protein Assay Dye Reagent Concentrate | Bio-Rad | 500-0006 |
| Collagenase Type IV | Invitrogen | 17104019 |
| complete™, Mini, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich | 11836170001 |
| DAPI | Thermo Fisher Scientific | D1306 |
| Dibutyryl cyclic AMP | Sigma-Aldrich | D0627 |
| DL-Dithiothreitol solution | Millipore Sigma | 43816 |
| Donkey serum | Millipore | S30-100ML |
| Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM:F12) | Thermo Fisher Scientific | 11320033 |
| Dulbecco’s phosphate-buffered saline (DPBS), calcium, magnesium | Thermo Fisher Scientific | 14040133 |
| Dulbecco’s phosphate-buffered saline (DPBS), no calcium, no magnesium | Thermo Fisher Scientific | 14190144 |
| Durapore® Membrane Filter, 0.22 μm | Millipore | GVWP04700 |
| EZ PREP Nuclei Isolation Kit | Sigma-Aldrich | NUC101-1KT |
| EZ-Tn5 Transposase | Lucigen | TNP92110 |
| GlutaMAX supplement | Gibco | 35050061 |
| IWR-1 Endo | Cayman Chemical | 13659-25 |
| KAPA HiFi Hotstart Readymix | Emsco/Fisher | KK2601 |
| KnockOut Serum Replacement | Thermo Fisher Scientific | 10828028 |
| LDN-193189 | Stemcell Technologies | 72147 |
| Recombinant Human Leptin | Peprotech | 300-27 |
| MEM Non-Essential Amino Acids Solution (100X) | Thermo Fisher Scientific | 11140050 |
| MgCl2 (1M) | Thermo Fisher Scientific | AM9530G |
| N-2 Supplement (100X) | Thermo Fisher Scientific | 17502048 |
| Neurobasal medium | Thermo Fisher Scientific | 21103049 |
| Nuclease-Free Water (not DEPC-Treated) | Thermo Fisher Scientific | AM9937 |
| Formaldehyde, 16%, methanol free, Ultra Pure | Polysciences | 18814-10 |
| Penicillin-Streptomycin (5,000 U/mL) | Thermo Fisher Scientific | 15070063 |
| Polyethylene glycol (PEG) solution, 40% | Sigma-Aldrich | P1458 |
| Purmorphamine | Tocris | 455110 |
| PhosSTOP™ | Sigma-Aldrich | 4906845001 |
| Recombinant Human/Murine/Rat BDNF | Peprotech | 450-02 |
| Recombinant Human GDNF | Peprotech | 450-10 |
| Recombinant Human SHH | BioLegend | 753506 |
| RNA Clean & Concentrator 5 | Zymo Research | R1013 |
| RNase Inhibitor, Murine | New England Biolabs | M0314S |
| SAG | Cayman Chemicals | 11914-1 |
| SDS (10% w/v) | Fisher Scientific | 50-751-7490 |
| SMARTScribe Reverse Transcriptase | Takara | 639537 |
| Sodium hydroxide solution (1 N) | Sigma-Aldrich | 1091371000 |
| Sterile saline solution injection | Midwest Veterinary Supply | 193.74500.3 |
| Sucrose | Sigma-Aldrich | S0389 |
| SuperSignal™ West Femto Maximum Sensitivity Substrate | Thermo Fisher Scientific | 34096 |
| TFM Tissue Freezing Medium | General Data | TFM-5 |
| Tris (1 M), pH 8.0, RNase-free | Thermo Fisher Scientific | AM9855G |
| Triton X-100 | Sigma-Aldrich | T9284 |
| TRIzol reagent | Thermo Fisher Scientific | 15596026 |
| Trypan blue stain, 0.4% | Thermo Fisher Scientific | T10282 |
| TWEEN 20 | Sigma-Aldrich | P1379 |
| VECTASHIELD Vibrance Antifade Mounting Medium | Vector Laboratories | H170010 |
| Critical Commercial Assays | ||
| BrainPhys™ Neuronal Medium N2-A & SM1 Kit | STEMCELL Technologies | 05793 |
| CytoView MEA 12 | Axion Biosystems | M768-GL1-30Pt200 |
| ELISA Kit for Alpha-Melanocyte Stimulating Hormone (aMSH) | Cloud-Clone Corp | CEA239Hu |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Q33231 |
| KAPA Library Quantification Kit for Illumina NGS | Kapa Biosystems | KK4835 |
| Neural Tissue Dissociation Kit | Miltenyi Biotech | 130-092-628 |
| NextSeq High Output v2 75 Cycles | Illumina | TG-160-2005 |
| Nextera XT Library Prep Kit | Illumina | FC-131-1024 |
| Deposited Data | ||
| Bulk and single-cell RNA-sequencing of ARCOs | This paper | GEO: GSE164102 |
| Single-nucleus RNA-sequencing of human neonatal hypothalamus | This paper | GEO: GSE164101 |
| Experimental models: Cell lines | ||
| Human C3-1 iPSC line | (Wen et al., 2014) | DOI: 10.1038/nature13716 |
| Human C65 iPSC line | This study | |
| PWS Major Deletion line | This study | (Kuslich et al., 1999) |
| PWS Minor Deletion line | This study | (Schüle et al., 2005) |
| Software and Algorithms | ||
| Adobe Illustrator CC | Adobe | https://www.adobe.com/products/illustrator.html; RRID:SCR_010279 |
| Adobe Photoshop CC | Adobe | https://www.adobe.com/products/photoshop.html; RRID:SCR_014199 |
| bcl2fastq v2.17.1.14 | Illumina | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html; RRID:SCR_015058 |
| DESeq2 v1.24.0 | (Love et al., 2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html; RRID:SCR_015687 |
| Drop-seq tools v.2.1.0 | (Saunders et al., 2018) | https://github.com/broadinstitute/Drop-seq; RRID:SCR_018142 |
| EnhancedVolcano v1.7.8 | GitHub | https://github.com/kevinblighe/EnhancedVolcano; RRID:SCR_018931 |
| GenomicFeatures v1.36.4 | (Lawrence et al., 2013) | http://bioconductor.org/packages/release/bioc/html/GenomicFeatures.html; RRID:SCR_016960 |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| Harmonizome | (Rouillard et al., 2016) | http://amp.pharm.mssm.edu/Harmonizome/; RRID:SCR_016176 |
| Human genome release 28 (GRCh38.p12) | GENCODE | https://www.gencodegenes.org/human/release_28.html |
| Imaris | Bitplane | http://www.bitplane.com/imaris; RRID:SCR_007370 |
| Monocle v2.14.0 | (Qiu et al., 2017a; Qiu et al., 2017b; Trapnell et al., 2014) | http://cole-trapnell-lab.github.io/monocle-release/docs/; RRID:SCR_016339 |
| PANTHER v15.0 | (Mi et al., 2019) | http://www.pantherdb.org/; RRID:SCR_004869 |
| pathwayPCA v1.4.0 | (Odom et al., 2019) | https://www.bioconductor.org/packages/release/bioc/html/pathwayPCA.html |
| R Project v3.6.0 | Open source | https://www.r-project.org/; RRID:SCR_001905 |
| RStudio 1.2.1335 | Open source | https://www.rstudio.com/; RRID:SCR_000432 |
| scater v1.12.2 | (McCarthy et al., 2017) | https://bioconductor.org/packages/release/bioc/html/scater.html; RRID:SCR_015954 |
| Seurat v3.1.5 | (Stuart et al., 2019) | https://github.com/satijalab/seurat; RRID:SCR_007322 |
| STAR v2.5.2a | (Dobin et al., 2013) | https://github.com/alexdobin/STAR; RRID:SCR_015899 |
| Trimmomatic v0.32 | (Bolger et al., 2014) | http://www.usadellab.org/cms/index.php?page=trimmomatic; RRID:SCR_011848 |
| VennDiagram v1.6.20 | (Chen and Boutros, 2011) | https://cran.r-project.org/web/packages/VennDiagram/; RRID:SCR_002414 |
| Zen Blue | Carl Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen-lite.html; RRID:SCR_013672 |
| Experimental models: Organisms/Strains | ||
| CD-1 Mus musculus | Charles River | CRL:022; RRID: IMSR_CRL:022 |
| NU/J Mus musculus, female | The Jackson Laboratory | Cat# 002019; RRID: IMSR_JAX:002019 |
| Oligonucleotides | ||
| CDS Primer sequence: 5′-AAGCAGTGGTATCAACGCAGAGTACT30VN-3′ | IDT | N/A |
| TSO Primer sequence: 5′-AAGCAGTGGTATCAACGCAGAGTACATrGrGrG-3′ | IDT | N/A |
| LS PCR Primer sequence: 5′-AAGCAGTGGTATCAACGCAGAGT-3′ | IDT | N/A |
| Others | ||
| 23G blunt needle | Neta Scientific | B23-100 |
| 6-well ultra-low attachment culture plate | Fisher Scientific | 07200601 |
| Bioanalyzer 2100 | Agilent | G2939BA |
| C&B Metabond Quick adhesive cement system | Benco Dental | 1681-343 |
| Cell counting slides | Thermo Fisher Scientific | C10228 |
| Charged microscope slides | Fisher Scientific | 22-035813 |
| Countess II Automated Cell Counter | Thermo Fisher Scientific | AMQAX1000 |
| Disposable pellet pestle | Fisher | 12-141-368 |
| Fine Forceps - Curved/Serrated | Fine Science Tools | 11065-07 |
| Forma Steri-Cult CO2 Incubator | Thermo Fisher Scientific | 3310 |
| Gelfoam Sponge | Pfizer | 031508 |
| Hydrophobic barrier PAP pen | Vector Laboratories | H-4000 |
| In-VitroCell ES Direct Heat CO2 Incubator | Nuaire | NU-5710 |
| MACS SmartStrainer, 70 μM | Miltenyi Biotech | 130-098-462 |
| Maxi-Cure Super Glue | Bob Smith Industries | BSI-113 |
| MaxQ CO2 Plus Shaker | Thermo Fisher Scientific | 88881102 |
| Microfluidic chips for drop-seq | FlowJEM | Custom order |
| Micromotor drill | Stoelting | 51449 |
| NanoDrop 2000 | Thermo Fisher Scientific | ND-2000 |
| NextSeq550 | Illumina | SY-415-1002 |
| PCR tube strips | Emsco/Fisher | AB0490 |
| Pellet pestle cordless motor | Fisher | 12-141-361 |
| Plastic cryomold | Electron Microscopy Sciences | 62534-10 |
| Qubit 3 Fluorimeter | Thermo Fisher Scientific | Q33216 |
| Tube rotator | Boekel | UX-51202-07 |
| Round Bottom Polystyrene Test Tube, with Cell Strainer Snap Cap | Corning | 352235 |
| T100 Thermal Cycler | Bio-rad | 1861096EDU |
| TB Syringe (26 G × 3/8 in, 1 ml) | BD Biosciences | 309625 |
| Tissue homogenizer | Sigma | D8938 |
| Vannas Spring Scissors - Curved/3mm Cutting Edge | Fine Science Tools | 15000-10 |
Single-Cell and Single-Nucleus RNA-seq Library Preparation and Sequencing
ARCOs were dissociated using a papain-based neural tissue dissociation kit (Miltenyi Biotech). All samples were washed three times by centrifugation at 200 g for 5 min and resuspension in 10 mL of calcium-free, magnesium-free DPBS (Thermo Fisher Scientific). Cells were strained through a 70 μm SmartStrainer (Miltenyi Biotech), analyzed for viability by trypan blue staining (Thermo Fisher Scientific), and counted using an automated cell counter (Thermo Fisher Scientific). Samples had a viability of > 80% and were diluted to a final concentration of 100 viable cells/μL in DPBS with 0.01% BSA (w/v).
Frozen human hypothalamus pieces were dissociated using Nuclei EZ lysis buffer (Sigma) as previously described (Habib et al., 2017; Jacob et al., 2020b). Briefly, tissue samples were minced into small pieces and homogenized using a tissue homogenizer (Sigma) in 2 mL of ice-cold Nuclei EZ lysis buffer (Sigma) (20–25 times with pestle A, 20–25 times with pestle B) and incubated on ice in 2.5 mL of Nuclei EZ lysis buffer. Nuclei were collected by centrifugation at 500 g for 5 min and resuspended in 4 mL of Nuclei EZ lysis buffer. The suspension was incubated on ice for 5 min. Then, nuclei were collected by centrifugation at 500 g for 5 min at 4°C and resuspended in 4 mL of Nuclei Suspension Buffer (NSB; 1x PBS, 0.01% BSA (Sigma), 0.1% RNAse inhibitor (New England Biolabs)). Finally, nuclei were collected by centrifugation at 500 g for 5 min, resuspended in 1 mL of NSB, and filtered through a 35 μm cell strainer (Corning). Nuclei were counted using an automated cell counter (Thermo Fisher Scientific) and diluted to a final concentration of 100 nuclei/μL in NSB.
Single cell droplet encapsulation and library preparation was performed using the drop-seq method with minor modifications (Macosko et al., 2015b). Microfluidic chips were obtained from FlowJEM with plasma bonding and aquapel treatment using the same design as previously published (Macosko et al., 2015a). Each droplet co-encapsulation run was performed with 50% additional reagent volume to account for syringe and tubing dead volume as well as occasional microfluidic channel clogging and re-starts. Each sample was loaded onto 2–4 droplet generation runs, and cell suspensions and droplets were kept on ice prior to droplet encapsulation and droplet breakage. In the cDNA PCR amplification stage, between 4,000–8,000 beads were combined in a single PCR tube reaction and purified individually using 0.6x AMPure XP beads (Beckman Coulter). cDNA across multiple runs of the same sample were pooled together and used as input for tagmentation, where the tagment reaction time was increased to 8 min. Final samples were purified twice with 0.6x AMPure XP beads (Beckman Coulter). Sequencing library fragment sizes were quantified by bioanalyzer (Agilent), and concentrations were quantified by qPCR (KAPA). Samples were pooled, loaded at 2.2 pM, and sequenced on a NextSeq 550 (Illumina) with a 20 bp Read 1 and 64 bp Read 2. The custom Read 1 primer was spiked into the usual Illumina sequencing primer well (#20) at the manufacturer’s recommended concentration.
Single-Cell and Single-Nucleus RNA-seq Bioinformatics Analyses
Data Processing
Raw sequencing data was demultiplexed with bcl2fastq2 (Illumina) with adaptor trimming turned off. Additional processing was performed using drop-seq tools v.2.1.0 (Saunders et al., 2018) with GRCh38 as the reference genome, and GEBCODE v.28 GTF as the annotation file. Seurat v3.1.5 was used to analyze all single-cell and single-nucleus data (Stuart et al., 2019).
ARCO Single-cell RNA-seq Analysis
For control, PWS minor deletion, and PWS major deletion ARCO single-cell RNA-seq data from 20 DIV and 40 DIV (two iPSC cell line clones per genotype), single cells with > 300 unique genes detected and < 5% mitochondrial counts were retained for further analysis. Data were normalized and percent mitochondrial counts regressed out using the SCTransform function in Seurat. PCA and UMAP dimensionality reduction were performed using the first 30 empirically selected PCs with standard pipelines. Control ARCO 20 DIV and 40 DIV data were integrated across time points to remove batch effects using the standard SCTransform integration workflow in Seurat prior to clustering analysis. Cluster markers were identified using the FindAllMarkers function in Seurat. ARCO control line data were also integrated across C3 and C65 iPSC lines using the standard SCTransform integration workflow in Seurat to demonstrate reproducibility between the control cell lines. Control, PWS minor deletion, and PWS major deletion ARCO data from 20 DIV and 40 DIV were integrated using similar pipelines. Pseudotime trajectory analysis for ARCO 20 DIV and 40 DIV integrated data was performed using Monocle v2.14.0 (Qiu et al., 2017a; Qiu et al., 2017b; Trapnell et al., 2014).
To compare transcriptomic signatures identified in the ARCO 20 DIV and 40 DIV integrated data to adult human hypothalamic nuclei transcriptomic signatures, data from the Allen Brain Adult Human database (Hawrylycz et al., 2012; Jones et al., 2009; Shen et al., 2012; Sunkin et al., 2013) were downloaded as upregulated gene sets in gene matrix transposed (.gmt) format from the Harmonizome database (Rouillard et al., 2016). These gene sets were read using the read gmt function in pathwayPCA v1.4.0 (Odom et al., 2019). The union of gene sets between each left-right hemisphere identified from dissected hypothalamic nuclei populations were taken for further analysis. To infer the average expression of each hypothalamic nuclei-specific gene list per single cell in the ARCO dataset compared to random control genes, the module score for each gene list was calculated using the AddModuleScore function in Seurat as per detailed methods outlined in (Stuart et al., 2019; Tirosh et al., 2016). Average module scores per ARCO cluster for selected hypothalamic nuclei were plotted as the column Z-score per ARCO cluster for visualization.
Neonatal Human Hypothalamus Single-nucleus RNA-seq Analysis
Neonatal human hypothalamus single-nucleus RNA-seq data, single cells from each patient sample (Table S1) with > 400 unique genes detected and < 5% mitochondrial counts were retained for further analysis. Data were normalized and percent mitochondrial counts regressed out using the SCTransform function in Seurat. Clustering and UMAP/tSNE dimensionality reduction were performed using the first 30 empirically selected PCs with standard pipelines prior to clustering analysis. Cluster markers were identified using the FindAllMarkers function in Seurat.
Transfer Learning Workflow
To identify the putative ARC transcriptomic signature in the neonatal human hypothalamus, we adapted the recently published Seurat transfer learning approach (Stuart et al., 2019). We first compiled an atlas of published adult mouse single-cell RNA-seq datasets with known spatial hypothalamic nuclei specificity. We defined an ARC population of Arcuate-ME cells from “Chow”-fed mice (Campbell et al., 2017) with > 400 unique genes detected and < 5% mitochondrial counts for further analysis. We next compiled a hypothalamus-wide non-ARC population of cells consisting of five single-cell RNA-seq datasets to capture as much of the remaining adult mouse hypothalamus as possible. We selected cells with > 400 unique genes detected and < 5% mitochondrial counts from the hypothalamic preoptic region (Moffitt et al., 2018), “Control” cells from the ventromedial hypothalamus (Kim et al., 2019), cells from the suprachiasmatic nucleus (Wen et al., 2020), and “P23” cells from the whole hypothalamus (Romanov et al., 2020). Additionally, we selected cells with > 400 unique genes detected and < 40% mitochondrial counts from the lateral hypothalamic area of “P30” male and female mice (Mickelsen et al., 2019). In total, we retained 7,395 ARC cells and 56,081 non-ARC cells. The non-ARC population was randomly downsampled to the size of the ARC population, to achieve an ARC population and non-ARC population with 7,395 cells each. These two populations were used as reference data for a transfer learning workflow, where mouse ARC cells and non-ARC cells were used to classify putative ARC and non-ARC cells in the neonatal human hypothalamus dataset in the space of mouse and human orthologous genes. Pairwise correspondences or “anchors” between individual cells in the mouse ARC-non-ARC reference dataset and human hypothalamus query dataset were first defined using the FindTransferAnchors function in Seurat, followed by binary classification of the query human hypothalamus cells as ARC or non-ARC based on the ARC-non-ARC reference mouse dataset using the TransferData function in Seurat. Human cells with a predicted ARC score ≥ 0.95 were defined as ARC cells, while remaining cells were defined as non-ARC cells. We further identified ARC neurons and non-ARC neurons within the neuronal clusters of the human hypothalamus dataset, together with remaining cells in the hypothalamus. In summary, we idenfitied 3,563 cells as predicted ARC neurons and next subclustered this population using methods similar to the original human hypothalamus dataset to identify transcritptional heterogeneity within the predicted ARC neurons. PCA and tSNE dimensionality reduction were performed using the first 30 empirically selected PCs with standard pipelines. Cluster markers were identified using the FindAllMarkers function in Seurat. These markers were subsequently validated using known immunostaining expression patterns from human and macaque ARC (Skrapits et al., 2015; True et al., 2017).
Comparison of Predicted Human ARC to Brain Region-specific Organoids
To compare transcriptomic signatures from ARCOs, thalamus organoids (Xiang et al., 2019), and forebrain cortical organoids (Qian et al., 2020) to the predicted ARC neuron population from the neonatal human hypothalalmus dataset, we removed mature neuron clusters from the thalamus organoid dataset and forebrain cortical organoid dataset, and performed correlation analysis using remaining organoid clusters and all populations defined in the predicted ARC neuron dataset. Normalized expression values for all organoid clusters and predicted ARC neuron clusters were averaged across all single cells per cluster and correlation analysis performed by calculating Spearman correlation coefficients in the space of shared genes across the whole transcriptome of all datasets.
Single-cell RNA-seq Dataset Integration and Quantification of Different Cell Types
To explore cell type differences between ARCOs and the predicted human ARC dataset, control ARCO 20 DIV and 40 DIV data were integrated with the predicted human ARC dataset using the standard SCTransform integration workflow in Seurat prior to clustering analysis. Different neuronal subtypes identified within the integrated dataset were quantified as a percentage of total neurons from the ARCO or predicted human ARC populations. Integration of control ARCO with embryonic mouse ARC (Huisman et al., 2019), fetal human cortex (Zhong et al., 2018), and adult human cortex (Hodge et al., 2019) were performed with similar pipelines.
Western Blot Analysis
To detect Leptin-induced changes in the JAK-STAT signaling pathway, ARCOs at 60 DIV were treated with 2 μg/ml Leptin for 1 hr. After treatment, ARCOs were washed three times with cold 1× PBS and lysed with 100 μl of RIPA buffer (Sigma-Aldrich) with protease inhibitor (Sigma-Aldrich) and phosphatase inhibitor (Sigma-Aldrich). Lysates were centrifuged for 30 min at 13,000 × g and protein contents in the supernatant were quantified using Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad). Protein samples were prepared by mixing 5× SDS sample buffer (250 mM Tris·HCl, pH 6.8, 10% SDS, 0.2% bromophenoblue, 50% glycerol, 4% 2-mercaptoethanol) with 80 μg of cell lysates and separated using SDS/PAGE 10% gels. After completion of electrophoresis, proteins were transferred onto PVDF membrane (Millipore). Membranes were blocked by 3% BSA in TBS-T (50 mM Tris·HCl, pH 7.4, 150 mM NaCl, and 0.05 % Tween-20) for 1 hr and then incubated with primary antibody (1:500) overnight at 4 °C. The next day membranes were incubated in secondary antibodies (1: 2000) overnight at 4 °C. The chemiluminescence was detected via SuperSignal™ West Femto Maximum Sensitivity Substrate (34096, Thermo Fisher Scientific). Images were captured by Amersham imager 600 (version 1.2.0, GE Healthcare). Membranes were stripped and processed for detection for GAPDH to ensure equal loading of samples. Intensities of the bands of the correct molecular weights were quantified and were first normalized to that of GAPDH and then calculated to obtain the ratio of phosphorylated levels over the total levels of JAK2 or STAT3 under different conditions.
ELISA Analysis
To determine levels of secreted MSH, ARCOs were treated with 2 μg/ml Leptin at 60, 62, 64 and 66 DIV. Conditioned medium was collected before treatment and after treatment on day 2, 4, 6, and 10. Alpha-melanocyte stimulating hormone ELISA kit (Cloud-Clone, CEA239Hu) was applied according to the manufacturer’s instructions.
MEA Analysis
To measure neuronal firing activity, ARCOs at 60 DIV were plated onto 12-well microelectrode array (MEA) plates (Axion Biosystems) precoated with Matrigel and maintained in BrainPhys neuronal culture media (Stemcell Technologies). Data of spontaneous neuronal firing were collected for 120 s in 37°C at 2 weeks after plating using the default neural activity settings from the manufacturer (Axion Biosystems Maestro Axis software version 2.4.2.13) (Wang et al., 2019). Mean firing rate was measured by counting spikes from active electrodes in each well per minute. Spike and burst detection were further analyzed and plotted by Neural Metric Tool version 2.2.3.
RNA Isolation, Bulk RNA-Seq Library Preparation, and Sequencing
To minimize variability due to sampling and processing, each biological replicate consisted of 3 organoids and the replicates for all experimental conditions were processed in parallel for RNA-extraction, library preparation and sequencing as previously described (Berg et al., 2019; Weng et al., 2017). At the desired experimental endpoints, organoids were homogenized in TRIzol (Thermo Fisher Scientific) using a disposable pestle and handheld mortar and stored at −80 °C until processing. RNA clean-up was performed using the RNA Clean & Concentrator kit (Zymo Research) after TRIzol phase separation according to the manufacturer’s protocol. RNA concentration and quality were assessed using a Nanodrop 2000 (Thermo Fisher Scientific).
Library preparation was performed as previously described with some minor modifications (Weng et al., 2017). About 300 ng of RNA in 3.2 μL was combined with 0.25 μL RNase inhibitor (New England Biolabs) and 1 μL CDS primer (5’-AAGCAGTGGTATCAACGCAGAGTACT30VN-3’) in an 8-well PCR tube strip (Emsco/Fisher), heated to 70 °C for 2 min, and immediately placed on ice. 5.55 μL RT mix, containing 2 μL of 5X SMARTScribe RT buffer (Takara), 0.5 μL of 100 mM DTT (Millipore Sigma), 0.3 μL of 200 mM MgCl2 (Thermo Fisher Scientific), 1 μL of 10 mM dNTPs (Takara), 1 μL of 10 μM TSO primer (5’-AAGCAGTGGTATCAACGCAGAGTACATrGrGrG-3’), 0.25 μL of RNase inhibitor (New England Biolabs), and 0.5 μL SMARTScribe reverse transcriptase (Takara) was added to the reaction. RT was performed under the following conditions: 42 °C for 90 min, 10 cycles of 50 °C for 2 min and 42 °C for 2 min, 70 °C for 15 min, and 4 °C indefinitely. For cDNA amplification, 2 μL of the RT reaction was combined with 2.5 μL of 10X Advantage 2 buffer (Takara), 2.5 μL of 2.5 mM dNTPs (Takara), 0.25 μL of 10 μM IS PCR primer (5’-AAGCAGTGGTATCAACGCAGAGT-3’), 17.25 μL nuclease free water (ThermoFisher), and 0.5 μL Advantage 2 DNA Polymerase (Takara). Thermocycling conditions were as follows: 94 °C for 3 min, 8 cycles of 94 °C for 15 s, 65 °C for 30 s, and 68 °C for 6 min, 72 °C for 10 min, and 4 °C indefinitely. Amplified cDNA was purified using 0.8X AMPure XP beads (Beckman Coulter), eluted in 15 μL nuclease-free water, and quantified using Qubit dsDNA HS assay kit (Thermo Fisher Scientific). cDNA was fragmented by combining 100 pg cDNA in 1 μL nuclease free water, 2X TD buffer (20 mM Tris, pH 8.0; Thermo Fisher Scientific), 10 mM MgCl2, and 16% PEG 8000 (Sigma), and 0.5 μL EZ-Tn5 (Lucigen). The mixture was heated to 55 °C for 12 min, and the reaction was terminated upon the addition of 1.25 μL of 0.2% SDS (Fisher) and incubated at room temperature for 10 min. Fragments were amplified by adding 16.75 μL nuclease free water (Thermo Fisher Scientific), 1 μL of 10 mM Nextera i7 primer, 1 μL of 10 mM Nextera i5 primer, and 25 μL KAPA HiFi hotstart readymix (EMSCO/FISHER). Thermocycling conditions were as follows: 72 °C for 5 min, 95 °C for 1 min, 14 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, 72 °C for 1 min, and 4 °C indefinitely. DNA was purified twice with 0.8X AMPure XP beads (Beckman Coulter) and eluted in 10 μL of 10 mM Tris, pH 8 (Thermo Fisher Scientific). Samples were quantified by qPCR (KAPA) and pooled at equal molar amounts. Final sequencing library fragment sizes were quantified by bioanalyzer (Agilent) with an average size of ~420 bp, and concentrations were determined by qPCR (KAPA). Samples were loaded at concentrations of 2.7 pM and sequenced on a NextSeq 550 (Illumina) using 1 X 72 bp reads to an average depth of 10 million reads per sample.
Bulk RNA-Seq Bioinformatics Analyses
ARCO raw sequencing data were demultiplexed with bcl2fastq2 v2.17.1.14 (Illumina) with adapter trimming using Trimmomatic v0.32 software (Bolger et al., 2014). Alignment was performed using STAR v2.5.2a (Dobin et al., 2013). GENCODE human genome release 28 (GRCh38.p12) was used for alignment. Multimapping and chimeric alignments were discarded, and uniquely mapped reads were quantified at the exon level and summarized to gene counts using STAR -- quantMode GeneCounts. All further analyses were performed in R v3.6.0. Transcript lengths were retrieved from GTF annotation files using GenomicFeatures v1.36.4 (Lawrence et al., 2013) and raw counts were converted to units of transcripts per million (TPM) using the calculateTPM function in scater v1.12.2 (McCarthy et al., 2017) to obtain TPM values for human transcripts.
Differential gene expression analyses for ARCOs among controls, PWS minor deletion, and PWS major deletion iPSC lines at 20 and 100 DIV were performed using DESeq2 v1.24.0 (Love et al., 2014). Variance stabilizing transformation (VST) of raw counts was performed prior to whole-transcriptome principal component analysis (PCA) using DESeq2 v1.24.0. Upregulated and downregulated genes for comparisons were filtered by adjusted p-value < 0.05. Volcano plots for comparisons were plotted using EnhancedVolcano v1.7.8. Shared and unique signatures for upregulated and downregulated genes between datasets were visualized using VennDiagram v1.6.20 (Chen and Boutros, 2011). Upregulated and downregulated gene lists from the respective comparisons were used for gene ontology (GO) (Ashburner et al., 2000) enrichment analysis using PANTHER v15.0 (Mi et al., 2019). For each gene list, a list of terms for GO biological process complete, GO molecular function complete, and GO cellular component complete were filtered by false discovery rate (FDR) < 0.05. Selected genes from notable GO terms were log2(TPM + 1) normalized and converted to row Z-scores per gene for visualization. To systematically explore variability in differentiation across iPSC clones and patient donors, iPSC cell line clonal identity and patient control, PWS minor genotype, and PWS major genotype donor identity were compared across replicates and datasets for 20 and 100 DIV timepoints, respectively, by calculating Spearman correlation coefficients in the space of shared genes across the whole transcriptome of all datasets.
For PWS human hypothalamus comparison analysis, we downloaded a list of upregulated and downregulated differentially expressed genes (DEGs) in the hypothalamus of PWS patients compared to controls from two published gene expression datasets. Four PWS patients compared to four control patients profiled by bulk RNA-seq with DEGs defined by Benjamini-Hochberg false discovery rate [FDR] < 0.25 (Bochukova et al., 2018) and two PWS patients compared to three control patients profiled by gene expression microarray with DEGs defined by p ≤ 0.05 (Falaleeva et al., 2015) were used for further analysis. Shared and unique signatures for upregulated and downregulated genes for the respective comparisons were visualized using VennDiagram v1.6.20, and statistical significance of gene overlaps tested using Fisher’s exact test.
Transplantation of ARCOs into the Adult Mouse Brain
Transplantation of ARCOs was performed using a method described for organoid transplantation into adult immunodeficient mice (Jacob et al., 2020b). Mice were induced into anesthesia with 5% and maintained with 2% isoflurane in oxygen. An approximately 1 mm2 craniotomy above the right cerebral cortex at the intersection between the sagittal and lambdoid sutures was performed using a micromotor drill (Stoelting). After removing the meninges, the underlying brain tissue was aspirated using a 23G blunt needle (Neta Scientific) to create a 1 mm3 cavity. Bleeding was controlled using Gelfoam (Pfizer) and sterile saline. Between 3–5 ARCOs were transferred into the cavity and sealed by 3-mm coverslip and super glue (Bob Smith Industries), followed by application of dental cement (Benco Dental) onto the surrounding skull area. Each transplanted ARCO was approximately 1 mm in diameter and consisted of roughly 1 million cells as determined by automated cell counting (Countess II, Thermo Fisher Scientific) of similarly sized ARCOs. Animals were weighed twice a week and euthanized immediately after weight loss and/or the onset of neurological symptoms. Anesthesia by intraperitoneal injection of a lethal dose of ketamine, xylazine and acepromazine cocktail was performed. At 2 months after transplantation, mice were transcardially perfused with cold 0.1 M phosphate buffer followed by cold 4% formaldehyde in 0.1 M phosphate buffer (pH 7.4). Brains were carefully removed from the skull and fixed in 4% formaldehyde at 4°C overnight, washed with DPBS, and cryoprotected in 30% sucrose (w/v) overnight at 4°C. Brains were placed in plastic cryomolds and snap frozen in tissue freezing medium (General Data) on dry ice. Frozen brains were stored at −80°C until processing. Serial tissue sections (40 μm for the mouse brain) were sliced using a cryostat (Leica, CM3050S), and melted onto charged slides (Thermo Fisher Scientific). Slides were dried at room temperature and stored at −20°C until ready for immunohistology.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical tests and sample sizes are included in the Figure Legends and text. All data are shown as mean ± SEM as stated in the Figure Legends. In all cases, the p values are represented as follows: ***p < 0.001, **p < 0.01, *p < 0.05, and not statistically significant (n.s.) when p > 0.05. In all cases, the stated “n” value is individual organoids with multiple independent images used to obtain data points for each or batches of experiments. No statistical methods were used to predetermine sample sizes. For all quantifications of immunohistology, the samples being compared were processed and imaged in parallel for confocal microscopy. Expression of all markers were quantified over DAPI+ cells in individual organoid sections and the counts were used to calculate the percentage of marker positive cells, and represented as one data point in the plot. For organoid transplantation quantifications, STEM121 immunostaining was used to identify the graft site and boundary for quantification of IBA1+ cells within the organoid. Quantifications were performed using the cell counter function in Imaris (Bitplane). Statistical analysis was performed using GraphPad Prism (GraphPad Software Inc).
Supplementary Material
HIGHLIGHTS.
A protocol for arcuate organoids (ARCOs) to model a discrete brain subregion
A single-cell transcriptome atlas of human neonatal hypothalamus
Similar molecular signatures between ARCOs and human arcuate nucleus
Differentiation and functional deficits in Prader-Willi syndrome patient ARCOs
ACKNOWLEDGEMENTS
We thank members of Ming and Song laboratories for comments and suggestions, B. Temsamrit and E. LaNoce for technical support, J. Schnoll for lab coordination, and Jae W. Lee for Dlx1 antibodies. Schematics were created using illustrations from https://www.biorender.com. This work was supported by grants from National Institutes of Health (R35NS097370 and RF1MH123979 to G-l.M., and R35NS116843 to H.S.), and from The Foundation for Prader-Willi Research (FPWR; to G-l.M.).
Footnotes
DECLARE OF INTEREST
G-l.M. is on the advisory board of Cell Stem Cell. The authors declare no other competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andermann ML, and Lowell BB (2017). Toward a Wiring Diagram Understanding of Appetite Control. Neuron 95, 757–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo MA, Butler MG, and Cataletto ME (2015). Prader-Willi syndrome: a review of clinical, genetic, and endocrine findings. J Endocrinol Invest 38, 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, and Pasca SP (2019). Cell diversity in the human cerebral cortex: from the embryo to brain organoids. Curr Opin Neurobiol 56, 194–198. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg DA, Su Y, Jimenez-Cyrus D, Patel A, Huang N, Morizet D, Lee S, Shah R, Ringeling FR, Jain R, et al. (2019). A Common Embryonic Origin of Stem Cells Drives Developmental and Adult Neurogenesis. Cell 177, 654–668 e615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S, Scholpp S, Placzek M, Ingraham H, Simerly R, and Shimogori T (2010). Molecular pathways controlling development of thalamus and hypothalamus: from neural specification to circuit formation. J Neurosci 30, 14925–14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaess S, Szabó N, Haddad-Tóvolli R, Zhou X, and Álvarez-Bolado G (2014). Sonic hedgehog signaling in the development of the mouse hypothalamus. Front Neuroanat 8, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluet-Pajot MT, Tolle V, Zizzari P, Robert C, Hammond C, Mitchell V, Beauvillain JC, Viollet C, Epelbaum J, and Kordon C (2001). Growth hormone secretagogues and hypothalamic networks. Endocrine 14, 1–8. [DOI] [PubMed] [Google Scholar]
- Bochukova EG, Lawler K, Croizier S, Keogh JM, Patel N, Strohbehn G, Lo KK, Humphrey J, Hokken-Koelega A, Damen L, et al. (2018). A Transcriptomic Signature of the Hypothalamic Response to Fasting and BDNF Deficiency in Prader-Willi Syndrome. Cell Rep 22, 3401–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Hossain W, Sulsona C, Driscoll DJ, and Manzardo AM (2015). Increased plasma chemokine levels in children with Prader-Willi syndrome. Am J Med Genet A 167a, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, Goldman M, Verstegen AM, Resch JM, McCarroll SA, et al. (2017). A molecular census of arcuate hypothalamus and median eminence cell types. Nat Neurosci 20, 484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carias KV, and Wevrick R (2019). Preclinical Testing in Translational Animal Models of Prader-Willi Syndrome: Overview and Gap Analysis. Mol Ther Methods Clin Dev 13, 344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, and Boutros PC (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, Song H, and Ming GL (2011). Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry 16, 358–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H (2016). Modeling Development and Disease with Organoids. Cell 165, 1586–1597. [DOI] [PubMed] [Google Scholar]
- Corman TS, Bergendahl SE, and Epstein DJ (2018). Distinct temporal requirements for Sonic hedgehog signaling in development of the tuberal hypothalamus. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley MA, Smart JL, Rubinstein M, Cerdan MG, Diano S, Horvath TL, Cone RD, and Low MJ (2001). Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411, 480–484. [DOI] [PubMed] [Google Scholar]
- Dimitropoulos A, Zyga O, and Russ SW (2019). Early Social Cognitive Ability in Preschoolers with Prader-Willi Syndrome and Autism Spectrum Disorder. J Autism Dev Disord 49, 4441–4454. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CM, Relkovic D, Garfield AS, Dalley JW, Theobald DE, Humby T, Wilkinson LS, and Isles AR (2009). Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour. Hum Mol Genet 18, 2140–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaleeva M, Surface J, Shen M, de la Grange P, and Stamm S (2015). SNORD116 and SNORD115 change expression of multiple genes and modify each other’s activity. Gene 572, 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festen DA, Wevers M, de Weerd AW, van den Bossche RA, Duivenvoorden HJ, Otten BJ, Wit JM, and Hokken-Koelega AC (2007). Psychomotor development in infants with Prader-Willi syndrome and associations with sleep-related breathing disorders. Pediatr Res 62, 221–224. [DOI] [PubMed] [Google Scholar]
- Fintini D, Grugni G, Bocchini S, Brufani C, Di Candia S, Corrias A, Delvecchio M, Salvatoni A, Ragusa L, Greggio N, et al. (2016). Disorders of glucose metabolism in Prader-Willi syndrome: Results of a multicenter Italian cohort study. Nutr Metab Cardiovasc Dis 26, 842847. [DOI] [PubMed] [Google Scholar]
- Garfield AS, Davies JR, Burke LK, Furby HV, Wilkinson LS, Heisler LK, and Isles AR (2016). Increased alternate splicing of Htr2c in a mouse model for Prader-Willi syndrome leads disruption of 5HT(2C) receptor mediated appetite. Mol Brain 9, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, et al. (2017). Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14, 955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqq AM, Muehlbauer MJ, Newgard CB, Grambow S, and Freemark M (2011). The metabolic phenotype of Prader-Willi syndrome (PWS) in childhood: heightened insulin sensitivity relative to body mass index. J Clin Endocrinol Metab 96, E225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JW, Elmquist JK, and Elias CF (2008). Hypothalamic pathways linking energy balance and reproduction. Am J Physiol Endocrinol Metab 294, E827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman C, Cho H, Brock O, Lim SJ, Youn SM, Park Y, Kim S, Lee SK, Delogu A, and Lee JW (2019). Single cell transcriptome analysis of developing arcuate nucleus neurons uncovers their key developmental regulators. Nat Commun 10, 3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Pather SR, Huang WK, Zhang F, Wong SZH, Zhou H, Cubitt B, Fan W, Chen CZ, Xu M, et al. (2020a). Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 27, 937–950 e939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, Thokala R, Sheikh S, Saxena D, Prokop S, et al. (2020b). A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity. Cell 180, 188–204.e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Overly CC, and Sunkin SM (2009). The Allen Brain Atlas: 5 years and beyond. Nat Rev Neurosci 10, 821–828. [DOI] [PubMed] [Google Scholar]
- Kalsner L, and Chamberlain SJ (2015). Prader-Willi, Angelman, and 15q11-q13 Duplication Syndromes. Pediatr Clin North Am 62, 587–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapsimali M, Caneparo L, Houart C, and Wilson SW (2004). Inhibition of Wnt/Axin/beta-catenin pathway activity promotes ventral CNS midline tissue to adopt hypothalamic rather than floorplate identity. Development 131, 5923–5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai T, Suga H, Sakakibara M, Ozone C, Matsumoto R, Kano M, Mitsumoto K, Ogawa K, Kodani Y, Nagasaki H, et al. (2020). Hypothalamic Contribution to Pituitary Functions Is Recapitulated In Vitro Using 3D-Cultured Human iPS Cells. Cell Rep 30, 18–24 e15. [DOI] [PubMed] [Google Scholar]
- Kim DW, Yao Z, Graybuck LT, Kim TK, Nguyen TN, Smith KA, Fong O, Yi L, Koulena N, Pierson N, et al. (2019). Multimodal Analysis of Cell Types in a Hypothalamic Node Controlling Social Behavior. Cell 179, 713–728.e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NS, Wen Z, Liu J, Zhou Y, Guo Z, Xu C, Lin YT, Yoon KJ, Park J, Cho M, et al. (2021). Pharmacological rescue in patient iPSC and mouse models with a rare DISC1 mutation. Nature communications 12, 1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutcherov Y, Mai JK, Ashwell KW, and Paxinos G (2002). Organization of human hypothalamus in fetal development. J Comp Neurol 446, 301–324. [DOI] [PubMed] [Google Scholar]
- Krefft M, Frydecka D, Zalsman G, Krzystek-Korpacka M, Smigiel R, Gebura K, Bogunia-Kubik K, and Misiak B (2020). A pro-inflammatory phenotype is associated with behavioural traits in children with Prader-Willi syndrome. Eur Child Adolesc Psychiatry, Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuslich CD, Kobori JA, Mohapatra G, Gregorio-King C, and Donlon TA (1999). Prader-Willi syndrome is caused by disruption of the SNRPN gene. Am J Hum Genet 64, 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, and Knoblich JA (2014). Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125. [DOI] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, and Carey VJ (2013). Software for computing and annotating genomic ranges. PLoS Comput Biol 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thuc O, Stobbe K, Cansell C, Nahon JL, Blondeau N, and Rovère C (2017). Hypothalamic Inflammation and Energy Balance Disruptions: Spotlight on Chemokines. Front Endocrinol (Lausanne) 8, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Wong SZH, Lee B, Ming G. l., and Song H (2021). Decoding neuronal composition and ontology of individual hypothalamic nuclei. Neuroin, Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015a). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015b). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard J, Park S, Croizier S, Vanacker C, Cook JH, Prevot V, Tauber M, and Bouret SG (2016). Loss of Magel2 impairs the development of hypothalamic Anorexigenic circuits. Hum Mol Genet 25, 3208–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton RM, and Pasca SP (2020). Organoid and Assembloid Technologies for Investigating Cellular Crosstalk in Human Brain Development and Disease. Trends Cell Biol 30, 133–143. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun AT, and Wills QF (2017). Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer RE, Michaelson SD, Chee MJ, Atallah TA, Wevrick R, and Colmers WF (2013). Magel2 is required for leptin-mediated depolarization of POMC neurons in the hypothalamic arcuate nucleus in mice. PLoS Genet 9, e1003207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Maroof A, Wataya T, Sasai Y, Studer L, Eggan K, and Schier AF (2015). Generation of neuropeptidergic hypothalamic neurons from human pluripotent stem cells. Development 142, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, and Thomas PD (2019). PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47, D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelsen LE, Bolisetty M, Chimileski BR, Fujita A, Beltrami EJ, Costanzo JT, Naparstek JR, Robson P, and Jackson AC (2019). Single-cell transcriptomic analysis of the lateral hypothalamic area reveals molecularly distinct populations of inhibitory and excitatory neurons. Nat Neurosci 22, 642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, Kranzler J, Liu Y, Schmalfuss I, Theriaque DW, Shuster JJ, Hatfield A, Mueller OT, Goldstone AP, Sahoo T, et al. (2006). Neurocognitive findings in Prader-Willi syndrome and early-onset morbid obesity. J Pediatr 149, 192–198. [DOI] [PubMed] [Google Scholar]
- Miller JL, Lynn CH, Driscoll DC, Goldstone AP, Gold JA, Kimonis V, Dykens E, Butler MG, Shuster JJ, and Driscoll DJ (2011). Nutritional phases in Prader-Willi syndrome. Am J Med Genet A 155a, 1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NL, Wevrick R, and Mellon PL (2009). Necdin, a Prader-Willi syndrome candidate gene, regulates gonadotropin-releasing hormone neurons during development. Hum Mol Genet 18, 248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, et al. (2018). Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom GJ, Ban Y, Liu L, Sun X, Pico AR, Zhang B, Wang L, and Chen X (2019). pathwayPCA: an R package for integrative pathway analysis with modern PCA methodology and gene selection. bioRxiv, 615435. [Google Scholar]
- Pagliardini S, Ren J, Wevrick R, and Greer JJ (2005). Developmental abnormalities of neuronal structure and function in prenatal mice lacking the prader-willi syndrome gene necdin. Am J Pathol 167, 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L, and Rubenstein JL (2015). A new scenario of hypothalamic organization: rationale of new hypotheses introduced in the updated prosomeric model. Front Neuroanat 9, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Jacob F, Song MM, Nguyen HN, Song H, and Ming GL (2018). Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat Protoc 13, 565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. (2016). Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165, 1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Song H, and Ming GL (2019). Brain organoids: advances, applications and challenges. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Su Y, Adam CD, Deutschmann AU, Pather SR, Goldberg EM, Su K, Li S, Lu L, Jacob F, et al. (2020). Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell 26, 766–781 e769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C (2017a). Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017b). Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamani U, Gross AR, Hjelm BE, Sequeira A, Vawter MP, Tang J, Gangalapudi V, Wang Y, Andres AM, Gottlieb RA, et al. (2018). Super-Obese Patient-Derived iPSC Hypothalamic Neurons Exhibit Obesogenic Signatures and Hormone Responses. Cell Stem Cell 22, 698–712 e699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov RA, Tretiakov EO, Kastriti ME, Zupancic M, Häring M, Korchynska S, Popadin K, Benevento M, Rebernik P, Lallemend F, et al. (2020). Molecular design of hypothalamus development. Nature 582, 246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, and Ma’ayan A (2016). The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüle B, Albalwi M, Northrop E, Francis DI, Rowell M, Slater HR, Gardner RJ, and Francke U (2005). Molecular breakpoint cloning and gene expression studies of a novel translocation t(4;15)(q27;q11.2) associated with Prader-Willi syndrome. BMC Med Genet 6, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EH, Overly CC, and Jones AR (2012). The Allen Human Brain Atlas: comprehensive gene expression mapping of the human brain. Trends Neurosci 35, 711–714. [DOI] [PubMed] [Google Scholar]
- Skrapits K, Borsay B, Herczeg L, Ciofi P, Liposits Z, and Hrabovszky E (2015). Neuropeptide co-expression in hypothalamic kisspeptin neurons of laboratory animals and the human. Front Neurosci 9, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda S, Saito R, Fujita N, Fukuta K, and Taniura H (2019). Neuronal differentiation defects in induced pluripotent stem cells derived from a Prader-Willi syndrome patient. Neurosci Lett 703, 162–167. [DOI] [PubMed] [Google Scholar]
- Song H, Stevens CF, and Gage FH (2002). Astroglia induce neurogenesis from adult neural stem cells. Nature 417, 39–44. [DOI] [PubMed] [Google Scholar]
- Sorokin L (2010). The impact of the extracellular matrix on inflammation. Nat Rev Immunol 10, 712–723. [DOI] [PubMed] [Google Scholar]
- Sternson SM (2013). Hypothalamic survival circuits: blueprints for purposive behaviors. Neuron 77, 810–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL, Thompson CL, Hawrylycz M, and Dang C (2013). Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res 41, D996–d1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True C, Takahashi D, Kirigiti M, Lindsley SR, Moctezuma C, Arik A, Smith MS, Kievit P, and Grove KL (2017). Arcuate nucleus neuropeptide coexpression and connections to gonadotrophin-releasing hormone neurones in the female rhesus macaque. J Neuroendocrinol 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Egli D, and Leibel RL (2016). Efficient Generation of Hypothalamic Neurons from Human Pluripotent Stem Cells. Curr Protoc Hum Genet 90, 21.25.21–21.25.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ye F, Wen Z, Guo Z, Yu C, Huang WK, Rojas Ringeling F, Su Y, Zheng W, Zhou G, et al. (2019). Structural interaction between DISC1 and ATF4 underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders. Mol Psychiatry, Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wataya T, Ando S, Muguruma K, Ikeda H, Watanabe K, Eiraku M, Kawada M, Takahashi J, Hashimoto N, and Sasai Y (2008). Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proc Natl Acad Sci U S A 105, 11796–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S, Ma D, Zhao M, Xie L, Wu Q, Gou L, Zhu C, Fan Y, Wang H, and Yan J (2020). Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nat Neurosci 23, 456–467. [DOI] [PubMed] [Google Scholar]
- Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim NS, Yoon KJ, Shin J, Zhang C, et al. (2014). Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515, 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng YL, An R, Cassin J, Joseph J, Mi R, Wang C, Zhong C, Jin SG, Pfeifer GP, Bellacosa A, et al. (2017). An Intrinsic Epigenetic Barrier for Functional Axon Regeneration. Neuron 94, 337–346 e336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, Kang YJ, Zhong M, Liu X, Patra P, et al. (2019). hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell 24, 487–497.e487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Zhang Y, von Deneen KM, Zhu H, and Gao JH (2017). Brain structural alterations in obese children with and without Prader-Willi Syndrome. Hum Brain Mapp 38, 4228–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, Makri G, Nauen D, Shin JH, Park Y, et al. (2014). Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 15, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Sun L, Pan N, et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. [DOI] [PubMed] [Google Scholar]
- Zhou X, Zhong S, Peng H, Liu J, Ding W, Sun L, Ma Q, Liu Z, Chen R, Wu Q, et al. (2020). Cellular and molecular properties of neural progenitors in the developing mammalian hypothalamus. Nat Commun 11, 4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data reported in this study is deposited in GEO: GSE164101 (human hypothalamus) and GSE164102 (human arcuate organoids). the published article includes all data generated during this study.