Abstract
CRISPR (Clustered regularly interspaced short palindromic repeats)‐based diagnostic technologies have emerged as a promising alternative to accelerate delivery of SARS‐CoV‐2 molecular detection at the point of need. However, efficient translation of CRISPR‐diagnostic technologies to field application is still hampered by dependence on target amplification and by reliance on fluorescence‐based results readout. Herein, an amplification‐free CRISPR/Cas12a‐based diagnostic technology for SARS‐CoV‐2 RNA detection is presented using a smartphone camera for results readout. This method, termed Cellphone‐based amplification‐free system with CRISPR/CAS‐dependent enzymatic (CASCADE) assay, relies on mobile phone imaging of a catalase‐generated gas bubble signal within a microfluidic channel and does not require any external hardware optical attachments. Upon specific detection of a SARS‐CoV‐2 reverse‐transcribed DNA/RNA heteroduplex target (orf1ab) by the ribonucleoprotein complex, the transcleavage collateral activity of the Cas12a protein on a Catalase:ssDNA probe triggers the bubble signal on the system. High analytical sensitivity in signal detection without previous target amplification (down to 50 copies µL−1) is observed in spiked samples, in ≈71 min from sample input to results readout. With the aid of a smartphone vision tool, high accuracy (AUC = 1.0; CI: 0.715 – 1.00) is achieved when the CASCADE system is tested with nasopharyngeal swab samples of PCR‐positive COVID‐19 patients.
Keywords: catalase, CRISPR, mobile phone, SARS‐CoV‐2, viral diagnostics
The CASCADE (Cellphone‐Based Amplification‐Free System with CRISPR/CAS‐Dependent Enzymatic) assay relies on the Cas12‐mediated trans‐cleavage of a Catalase:ssDNA probe, upon recognition of a specific nucleic acid target, such as SARS‐CoV‐2 genomic RNA. This generates a gas bubble signal within a microfluidic channel which is then detected using a smartphone camera and a dedicated smartphone application, without any optical hardware attachment.
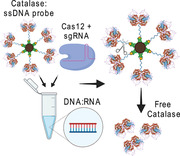
1. Introduction
Since the emergence of the severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) outbreak in December 2019, millions of people have been diagnosed with the infection globally, and the COVID‐19 pandemic has now escalated to become a worldwide crisis.[ 1 , 2 ] Similarly to other emerging viral infectious diseases that have caused epidemic events in recent decades, such as the ones caused by HIV, H1N1, SARS, and ZIKV, the lack of alternative molecular diagnostics technologies has impeded the containment and management of the COVID‐19 epidemic.[ 3 , 4 ] The gold‐standard nucleic acid test (NAT) for COVID‐19 diagnosis, reverse‐transcription quantitative real‐time polymerase chain reaction (RT‐qPCR), has proven to be a robust and reliable assay through detection of specific genomic regions of the SARS‐CoV‐2.[ 5 ] Highly accurate lab‐based RT‐qPCR assays have been quickly developed during the pandemic; however, they still require dedicated bulky instruments, sophisticated protocols, and highly trained personnel. This level of complexity results in high turnaround times, which poses a challenge to the democratization of testing capabilities worldwide.[ 6 , 7 ]
Starting in 2016, various biosensing technologies based on clustered regularly interspaced palindromic repeats (CRISPR)/Cas proteins have emerged as innovative and versatile alternatives to reshape the molecular detection of infectious agents.[ 6 , 8 , 9 ] Different CRISPR‐dependent technologies have been developed for SARS‐CoV‐2 detection,[ 3 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 ] and some have already received the US FDA's emergency use authorization, including DETECTR[ 3 ] and SHERLOCK.[ 11 ] However, common limitations of many CRISPR/Cas‐based diagnostic technologies are the requirement of isothermal amplification[ 8 ] and the dependence on fluorescence signal.[ 18 ] Recent developments aimed at overcoming these drawbacks have included lateral‐flow based results detection,[ 3 , 11 ] a single pot assay with integrated RNA extraction and amplification,[ 11 ] and the use of a smartphone‐attached device for fluorescence results readout in an amplification‐free assay format.[ 14 ]
Herein, we describe an amplification‐free CRISPR/Cas12‐based diagnostic technology for SARS‐CoV‐2 RNA detection using a smartphone camera for results readout, without the need for any external optical hardware smartphone attachments. This technology termed Cellphone‐based amplification‐free system with CRISPR/CAS‐dependent enzymatic (CASCADE) assay works with a catalase‐linked ssDNA probe as a target for the transcleavage collateral activity of Cas12 following specific detection of a reverse‐transcribed SARS‐CoV‐2 DNA/RNA heteroduplex. Results readout relies on the catalase‐mediated gas bubble optical signal generated within a simple, disposable, and inexpensive microfluidic chip that is imaged with a mobile phone camera and without using any optical hardware attachment.
2. Results and Discussion
To first standardize the assay, a single guide RNA (sgRNA) was designed that specifically targets a fragment of the ORF1ab region of the SARS‐Cov‐2 genome. Then, the transcleavage activity of the Cas12 enzyme on non‐related ssDNA fragments upon recognition of different concentrations of the ORF1ab target region by the ribonucleoprotein complex (Cas12+sgRNA) was evaluated (Figure S1, Supporting Information). To standardize the bead‐based assay, we used a FAM‐conjugated biotinylated ssDNA probe immobilized on streptavidin beads as a Cas12 substrate for fluorescence mediated results readout (Figure 1A). Streptavidin magnetic beads were specially chosen for those experiments due to their high capacity for capturing biotinylated molecules. The high loading capacity of the beads increases the test accuracy and provides sensitive detection in low target concentrations.[ 19 ] Incubation of the beads with Cas12:sgRNA ribonucleoprotein complex and the RT‐PCR amplified orf1ab target generated a visible decrease in florescence signal of the beads and noteworthy release of FAM in the supernatant (Figure S2, Supporting Information). Remarkably, a significant amount of FAM was also detected when the reaction was incubated with the reversely transcribed RNA (Figure S2B, Supporting Information).
Figure 1.
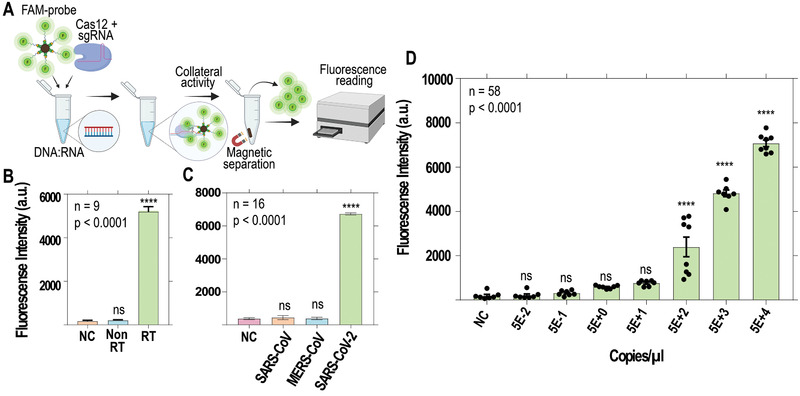
Standardization of the bead‐based SARS‐CoV‐2 detection assay using a fluorescent ssDNA probe for Cas12 activity. A) Samples were reverse transcribed and added to a Cas12 reaction with FAM‐conjugated ssDNA probes immobilized on microbeads. Using a magnetic stand, the supernatant of the samples was separated from magnetic beads and its fluorescence intensity was measured (Ex:488 nm/Em:530 nm). B) A complementary DNA (cDNA) strand is required to drive ssDNA cleavage activity by Cas12; RT = reverse transcribed; NC = negative control; differences were statistically significant (**** p < 0.0001). C) The reaction specifically targets reverse‐transcribed SARS‐CoV‐2 samples, rather than other related Coronaviridae; differences were statistically significant (**** p < 0.0001). D) Ten‐fold serial dilution of RT‐qPCR quantified SARS‐CoV‐2 RNA showing a linear signal output ranging from 50 to 50000 copies µL−1 (R2 = 0.9938). Samples were reverse transcribed before detection.
Cas12‐like enzymes possess DNA‐activated deoxyribonuclease activity and are incapable of stimulating transcleavage of ribonucleic acids;[ 20 ] however, RNA can be detected by Cas12 when a complementary DNA (cDNA) is provided as a heteroduplex (DNA/RNA).[ 21 ] To confirm that our designed orf1ab crRNA is capable of recognizing specifically a DNA/RNA hybrid, SARS‐CoV‐2 genomic RNA was incubated with a reverse transcriptase (RT) enzyme with reduced RNaseH activity and RNAse inhibitor. The fluorescence readings for RT reacted samples was 26‐fold higher compared with no RT samples (Figure 1B). The increased trans‐ssDNA cleavage in the reaction containing the RT enzyme is associated with the stabilizing effect of the nontarget DNA to the Cas12‐crRNA complex in an optimal conformation for collateral activity.[ 20 ] Likewise, we tested if this collateral activity could also be stimulated in the presence of SARS‐CoV and MERS‐CoV genomic RNA (Figure 1C). The incubation of SARS‐CoV‐2 genomic RNA in the reaction released more fluorescence (6723 ± 125 a.u.) compared with RNA from other beta‐coronaviruses, SARS‐CoV (432.5 ± 246 a.u.) and MERS‐CoV (379.5 ± 155.3 a.u.). These results confirm that Cas12 can not only target DNA‐RNA heteroduplexes but also generate a sensitive signal without any cross‐reactivity from other related coronaviruses. This fluorescence‐based assay generated a linear signal output (R2 = 0.9938) when tested with serially diluted SARS‐CoV‐2 target in the range between 50 and 50000 copies µL−1 (Figure 1D).
After confirming the functionality of the used CRISPR‐based signal amplification, we investigated the performance of our newly developed nucleic acid amplification‐free system termed CASCADE (Cellphone‐based amplification‐free system with CRISPR/CAS‐dependent enzymatic) assay in detecting SARS‐CoV‐2. The optical signal in the CASCADE assay is achieved through conjugation of an enzyme (i.e. catalase) with ssDNA targets for detecting the Cas12 transcleavage collateral activity (Figure 2A). The conjugation of 5'phosphorylated and 3'biotinylated ssDNA to primary amines on the surface of catalase was achieved using EDC (1‐ethyl‐3‐(3‐dimethylamino) propyl carbodiimide, hydrochloride) and imidazole (Figure 2B). The successful construction of the catalase‐ssDNA (CD) probe was then confirmed by polyacrylamide gel electrophoresis (Figure 2C), and by Fourier‐transform infrared spectroscopy (FT‐IR) through the presence of characteristic peaks for proteins (amide I at 1600−1800 cm−1, amide II at1470− 1570 cm−1, amide III at 1250−1350 cm−1),[ 22 ] and for the DNA (phosphate backbone at 1090 cm−1, deoxyribose C−O at 1053 cm−1, deoxyribose ring at 899‐890 cm−1) (Figure 2D).[ 23 ]
Figure 2.
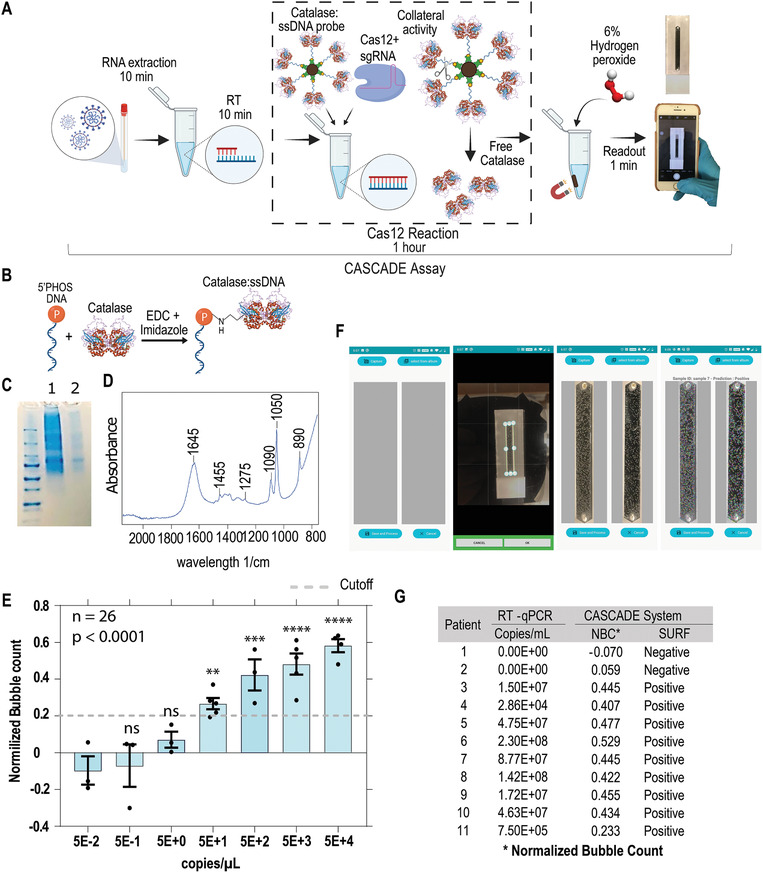
Development of the CASCADE assay for smartphone‐based SARS‐CoV‐2 detection. A) Diagram of the CASCADE system. Viral RNA is extracted from swab samples, reverse transcribed, and added to a Cas12 reaction containing the catalase‐ssDNA (CD) probe. In the presence of a fuel solution, the catalase activity of the probe disproportionates hydrogen peroxide into water and oxygen, then generating a signal output based on oxygen bubbles that can be detected in the microfluidic channel with a smartphone application. B) A phosphorylated and biotinylated ssDNA (45 bases PolyA) was reacted with the catalase enzyme using EDC and imidazole to generate the CD probe. C) SDS‐PAGE analysis of the CD probe after filtration in 50 KDa Amicon Ultra‐15. Lane 1‐ molecular weight marker; Lane 2 – CD probe; Lane 3 – resuspended 60 KDa catalase. D) Fourier‐transform infrared spectroscopy (FT‐IR) analysis of the CD probe after filtration. E) SARS‐CoV‐2 genomic RNA samples were serially diluted and used to standardize on‐chip detection assays using CASCADE; the limit of detection was 5 × 101 copies µL−1 (ANOVA, **** p < 0.0001; Holm–Sidak post hoc test, ** p = 0.0015, *** p = 0.0001, **** p < 0.0001, ns = not significant). The horizontal gray line indicates the normalized bubble count threshold for qualitative assessment of samples based on manual bubble count signal (90% lower confidence interval of the lowest significantly detectable concentration). F) SURF computer vision smartphone application for bubble analysis and sample prediction. G) By measuring SURF keypoints in the cellphone‐based application, the CASCADE assay was able to classify positive and negative samples with 100% accuracy compared with RT‐qPCR results.
The CD probe was then bound to streptavidin beads to separate free catalase from ssDNA conjugated catalase after Cas12 reaction. In previous studies, we have already demonstrated that gas bubbles produced by platinum nanoparticle (PtNPs) based nanoprobes can be used as an optical signal to detect different pathogens with high sensitivity and specificity.[ 24 , 25 ] Here, we have employed the catalase enzyme as a means to generate gas bubbles in the microchip channel, and the results readout was significantly shortened to 1 min due to the faster catalytic activity of catalase to break down hydrogen peroxide. The assay was standardized to provide the most contrasting difference in bubble count between positive and negative samples by adjusting each step involved in the detection protocol (Figure S3, Supporting Information).
The CASCADE assay enables on‐site detection of SARS‐CoV‐2 without the requirement of target nucleic acid amplification in ≈71 min from RNA sample input to signal readout. This new assay requires no instrumentation other than an incubator block, and the whole reaction system comprises simple steps without any complicated technical requirements. The CASCADE system involves sample preparation to generate DNA/RNA heteroduplexes, incubation with Cas12 reaction containing the beads, the magnetic separation of the beads containing catalase, and resuspension with 6% hydrogen peroxide solution to stimulate the production of bubbles. The reaction development takes place in less than 1 min, and the results are displayed as gas bubbles from the disproportionation of hydrogen peroxide into oxygen and water by the ssDNA‐bound catalase enzyme. The images of the on‐chip gas bubbles are analyzed by a smartphone application that classifies the sample as negative or positive outputs (Figure 2A).
To evaluate the analytical sensitivity of the CASCADE assay, we tested serially diluted SARS‐CoV‐2 genomic RNA, along with virus‐free negative control (NC) samples that were included in each run. To quantitatively analyze the sensitivity of the assay, we first relied on gas bubble count as a simple optical signal observed in our test results. The bubble counts of positive samples were normalized based on the bubble counts of the NC samples for each test set (Figure 2E). The normalized bubble count threshold for qualitatively classifying positive and negative samples was 0.20, which was defined as the 90% lower confidence interval in the lowest significantly detectable concentration (5E+1; P = 0.0015) (Figure 2E). The CASCADE assay could detect SARS‐CoV‐2 RNA copies down to 50 copies µL−1, when 10 µL of sample was used in the reverse transcription reaction (Figure 2E). This analytical sensitivity was poorer than the analytical sensitivity of recently reported amplification‐based CRISPR‐diagnostics assays for SARS‐CoV‐2 detection that used RT‐PCR, RT‐LAMP, and RT‐RPA (Table S1, Supporting Information). Nevertheless, the CASCADE assay does not require nucleic acid amplification nor fluorescence‐based readout. We have also shown that the CASCADE system can detect as low as 5 copies per reaction if a previous amplification step is added to the protocol (Supporting Information S4). Moreover, taking into account that most of the nasal swabs have 105 to 109 RNA copies per swab, the sensitivity of the CASCADE assay can be satisfactory for COVID‐19 mass screening purposes.[ 26 ] If we compare with other major state‐of‐the‐art CRISPR‐diagnostics technologies that were similarly developed to simplify sample preprocessing and to generate results with the aid of a smartphone,[ 14 , 27 ] we can conclude that the CASCADE system showed an analytical sensitivity which is comparable to other smartphone‐integrated technologies that rely upon fluorescence detection for results readout.
The studies by Arizti‐Sanz et al.[ 27 ] and Fozouni et al.[ 14 ] reported lower limits of detection of 10 and 100 copies µL−1, respectively, in serially diluted samples of SARS‐CoV‐2 RNA target. However, both studies reported a tradeoff between sensitivity and time for detection of positive fluorescent signal following Cas13 reaction. The lower the viral loads, the higher the times needed for detecting positive signals. In particular, Arizti‐Sanz et al.[ 27 ] reported that 3 h of incubation was needed for a significantly positive result at 10 copies µL−1, even though the general assay time is regarded as being only ≈55 min. Additionally, the performance in true nasopharyngeal swab samples was significantly different, with samples presenting titers <100 copies µL−1 not being detected by the assay.[ 27 ] On the other hand, Fozouni et al.[ 14 ] reported 100% accuracy for 200 copies µL−1 over 30 min of fluorescence measurement on a mobile phone microscope, with accuracy dropping to only 50% at 50 copies µL−1.[ 14 ] In our study, the lower limit of detection of the CASCADE system (50 copies µL−1) was reproducible in 5 of 5 technical replicates. Furthermore, the CASCADE results were 100% concordant with RT‐qPCR in positive COVID‐19 nasopharyngeal swab samples.
In the CASCADE assay, bubble count is just one parameter of the optical signal that can be quantified. However, bubble size, geometry, and distribution may vary for different samples with different viral loads. In our previous studies, we have shown that machine learning methods can be used for qualitatively assessing such optical signals.[ 24 ] Now, we used traditional rule‐based computer vision techniques for developing a smartphone application for automated sample evaluation. The smartphone application enabled us to automatically capture, process, and qualitatively assess microchip images on‐phone (Figure 2F). The use of a Speeded‐Up Robust Features (SURF) based smart vision algorithm reduces immensely the need for manpower and the time to calculate the bubble features and has other advantages over the analysis workflows reported by other major CRISPR‐diagnostics technologies integrated with smartphones.[ 14 , 27 ] Since the image features captured by the SURF application are invariant from different angles, scales, and illumination, a single NC microchip is descriptive enough to be accurately matched against a high number of other test samples. Furthermore, the application does not require much processing power, and any average android‐based phone can be used. Additionally, it has minimized time for computing and matching features, and the measurement is made offline in just 1 min after addition of the H2O2 revealing reagent, with no need to transfer images to a centralized server.
We evaluated the performance of the CASCADE system using PCR‐positive and PCR‐negative nasopharyngeal COVID‐19 swab samples. The smartphone application detects, through SURF, local descriptors on the microchip image of the COVID sample and the NC, and compares them as described in the methods.[ 28 ] The assay achieved 100% negative predictive agreement (NPA) and 100% positive predictive agreement (PPA) using the SURF application, compared with RT‐qPCR as ground truth (Figure 2G). The viral load of the COVID‐19 patient samples used in this study along with the PCR‐based and CASCADE‐based qualitative assessments are reported in Table S2 (Supporting Information).
3. Conclusion
In summary, the CASCADE assay expands on the currently available repertoire of CRISPR‐diagnostics technologies by providing an amplification‐free Cas12‐based detection method with simplified results readout. The simple optical signal in this assay is formed by gas bubbles that can be detected using a smartphone camera and a dedicated smartphone application, without using any optical hardware smartphone attachment. This combination of an amplification‐free assay, visible readout, and cellphone‐based analysis eliminates the requirement of specialized and bulky equipment. Future developments of the system might include integration of multiple sgRNAs for improved sensitivity and elimination of preprocessing steps for simplified and automated on‐chip target extraction and detection. Additionally, we might also explore in future the use of artificial intelligence by generative adversarial networks to construct a synthetic image database covering a high number of realistic microchip images of possible NC samples. This could, in turn, work as the reference image dataset to compare the images resulting from true test samples and then possibly eliminating the need for running NC assays by every new experimenter.
Limitations of our study: We demonstrate in this proof‐of‐concept study that a simple enzymatic reaction, mediated by catalase, can be coupled with the collateral activity of Cas12 to generate a highly sensitive visual signal (gas bubbles) upon target detection, which can be easily integrated with smartphones by using a computer vision tool (SURF). The optical signal was sensitive enough to permit detection of DNA:RNA heteroduplexes (down to 50 target RNA copies µL−1) after a single reverse‐transcription step, with no need for target amplification. Additionally, the results measurement step requires only 1 min for developing the catalase reaction, followed by an uncomplicated image processing on the smartphone. Nevertheless, further developments will be necessary to fully realize the potential of the technology for deployment to the point‐of‐care. One such development could be the elimination of the RNA extraction step, using a method similar to the optimized HUDSON described by Arizti‐Sanz et al.,[ 27 ] which permits nuclease and viral inactivation in the sample. The shelf stabilities of the reagents, in particular of the enzymes Cas12 and Catalase, must also be evaluated in their lyophilized forms for room temperature storage and shipment. Additionally, the thermal stability of the revealing solution, which is based on hydrogen peroxide, must also be taken into consideration. Finally, the time of Cas12 incubation, which is currently 1 h, could be further optimized, potentially with combinations of guide RNAs, such as in Fozouni et al.[ 14 ] Although the need for magnetic bead separation before the catalase reaction development may also bring an additional step that could hamper field deployment, it has been recently demonstrated by another major study with a CRISPR‐diagnostics technology for SARS‐CoV‐2 detection (STOPCovid) that a similar protocol using bead separation is amenable to implementation in low complexity settings.[ 11 ]
4. Experimental Section
Nucleic Acids and Patient Samples
Genomic RNA from SARS‐Related Coronavirus 2, Middle East Respiratory Syndrome Coronavirus (MERS‐CoV), and SARS‐Related Coronavirus were obtained from BEI Resources. The primers for the ORF1ab gene were synthesized based on the Chinese CDC RT‐PCR guideline using ORF1ab as the target sequence. A single guide RNA (sgRNA) targeting ORF1ab was designed using the IDT Alt‐R CRISPR‐Cpf1 crRNA design tool. The modified ssDNA sequences were obtained from Eurofins. A table with the sequences for the ORF1ab, the primers, the crRNA, and the ssDNA can be found in Table S3 (Supporting Information). SARS‐CoV‐2 reactive and nonreactive nasopharyngeal and oropharyngeal specimens were supplied from a certified biorepository providing written informed consent for patient samples (Boca Biolistics; SARS‐CoV‐2 Validation Panel); RNA samples used in the assay were extracted from patient swab samples using QIAamp Viral RNA Mini Kit. More information about the BEI genomic RNA specimens is summarized in Table S4 (Supporting Information) and a list of Boca Biolistics patient samples is found in Table S5 (Supporting Information). The studies on system development and evaluations using COVID‐19 patient samples reported in this manuscript were reviewed and approved by the Brigham and Women's Hospital institutional review board (IRB#: 2020P001065). The viral loads of the samples were quantified using a real‐time RT‐PCR (RT‐qPCR) assay at the laboratory of Drs. Jonathan Li and Daniel Kuritzkes at the Division of Infectious Diseases at Brigham and Women's Hospital.[ 29 ]
Generation of the Fluorescent ssDNA‐Beads Complex
Fluorescence assays were performed with the assistance of bead‐based probes composed of 5'FAM and 3'Biotin‐TEG modified ssDNA and streptavidin‐coated beads. The ssDNA was reacted with the magnetic beads (Invitrogen Dynabeads M‐280 Streptavidin) by resuspending 4 μL of 100 × 10−6 m biotinylated ssDNA solution in 500 μL of beads, then beads were washed to remove the excess of unbound ssDNA.
Fabrication and Characterization of the Catalase‐ssDNA (CD) Probe
Native catalase from Aspergillus niger was conjugated to an ssDNA linker by modifying the 5’ phosphate group of the oligonucleotide. Initially, 7 mg of catalase enzyme (Sigma Aldrich, 21 926) was dissolved in 100 µL of 0.1 m imidazole (Goldbio, I‐901‐25). Phosphorylated ssDNA (Eurofins) was resuspended in 12 µg µL−1 concentration. Then, 2.5 mg of 1‐ethyl‐3‐[3‐dimethylaminopropyl] carbodiimide hydrochloride (EDC) (Thermo Fisher Scientific, 22 980) was dissolved in 7.5 µL of the ssDNA solution, followed by the immediate addition of 5 µL catalase solution. After vortexing, another 20 µL of catalase solution was added and mixed thoroughly. This solution was incubated at 37 °C for 1.5 h. The incubated catalase‐ssDNA probe was then filtered with 1× Tris EDTA (Thermo Fisher Scientific, BP1338‐1) through a 50 KDa Amicon Ultra‐15 Centrifugal Filter Unit (EMD Millipore, UFC905024), for further concentration of the probe. After the filtration, the concentrate probe was analyzed with FT‐IR (Cary 630 ATR‐FTIR). The final product was used immediately to bind to streptavidin‐coupled magnetic beads (Invitrogen Dynabeads M‐280 Streptavidin) or stored in the freezer. The CD probe complexes were conjugated to the beads according to the protocol suggested by Invitrogen, in a 1:80 volumetric ratio of catalase‐ssDNA probe to beads.
Reverse Transcription Reactions and Cas12 Assays
Reverse transcription reactions were conducted using Invitrogen SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, 1 809 005) following manufacturer's guide and using 1 µL of 2 × 10−6 m ORF1ab reverse primer and 1–10 µL of the input RNA sample in low retention tubes. Cas12 assays were performed by incubating 200 × 10−9 m of EnGen Lba Cas12a (Cpf1) with 300 × 10−9 m of sgRNA in 1× NEBuffer 2.1 for 5 min at room temperature. The cDNA was added to the reaction, and then the activated complexes were incubated with either CD or fluorescent probes and incubated at 37 °C for 60 min.
Fluorescent Nucleic Acid Detection
After the Cas12 reaction, samples were placed on a magnetic rack, and supernatants were used to measure the fluorescence generated from the cleavage of ssDNA by collateral activity. Samples were read in a fluorescence plate reader (Tecan infinite 200 Pro). The excitation wavelength was set to 488 nm, the emission wavelength to 530 nm, and the gain to 195.
On‐Chip Viral Nucleic Acid Detection by CASCADE Assay
For the CASCADE assay, samples were placed on a magnetic rack, the supernatants were removed, and beads were resuspended in 6% hydrogen peroxide and 10% glycerol solution (Fuel). Samples were added in a bubble visualization device, and pictures were taken within 1 min. Images were taken using either iPhone 7 or iPhone X.
Microchip Fabrication
Poly(methyl methacrylate) (PMMA) sheets of 3.175 mm thickness (McMaster‐Carr, 8560K239) were initially cut using the VLS 2.30 CO2 laser cutter (Universal Laser Systems Inc.), based on a design generated for the devices in CorelDraw X7 graphical editor. The channel dimensions inside the device are L: 40 mm; W: 5 mm; H: 0.8 mm. Then, the double‐sided adhesive (DSA) sheets (3M, 82 603: 80 µm) were cut using the same design. PMMA and DSA sheets, as well as glass microslides (VWR, 48311–703), were cleaned with hydrogen peroxide, then with water, and lastly with ethanol. The final output is assembled by stacking the parts on top of each other.
Data Analysis and Bubble Visualization
GraphPad Prism software was used to plot the data and to perform statistical analysis. Normality was checked with Shapiro–Wilk normality test and groups were analyzed by one‐way ANOVA with Holm–Sidak's multiple comparisons. NIH ImageJ application (https://imagej.nih.gov/ij/index.html) was used in assisting with counting and analyzing bubble formations. The following formula was applied to provide the normalized Bubble count:
| (1) |
Development of a computer vision applicationAn android application was developed using OpenCV library for image data acquisition, processing, and prediction of viral load. Images are analyzed exploiting Speeded Up Robust Features (SURF) keypoints,[ 28 ] which uses Hessian corner detector algorithm with box filters to find these points of interest. Firstly, it finds distinctive features in both the NC and in the test pictures, which are unvarying in terms of scale, rotation, and illumination toward different viewpoints. Next, these features are matched between the pictures. If the control has more normalized key points than the tested sample, the sample is then classified as positive; otherwise, it is labeled as negative.
| (2) |
The android applications were developed with the OpenCV (ver. 4.5.0) and SmartCropper (ver. 2.1.3) libraries on Android Studio.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Supporting Information
Acknowledgements
The genomic RNA from SARS‐CoV, SARS‐CoV‐2, and MERS‐CoV were deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH. Research reported in this publication was partially supported by the National Institute of Health under award numbers 4U54HL119145‐08 (RADx‐Rad), R01AI118502, R01AI138800, and R61AI140489, and Brigham and Women's Hospital through Innovation Evergreen Fund. L.G.P. was supported through the CAPES/Harvard Junior Visiting Professor/Researcher Program and is currently a recipient of a research fellowship from CNPq‐Brazil. F.S. was supported through the CAPES/Print Program and is the recipient of a Ph.D. scholarship from Fapesb.
Silva F. S. R., Erdogmus E., Shokr A., Kandula H., Thirumalaraju P., Kanakasabapathy M. K., Hardie J. M., Pacheco L. G. C., Li J. Z., Kuritzkes D. R., Shafiee H., SARS‐CoV‐2 RNA Detection by a Cellphone‐Based Amplification‐Free System with CRISPR/CAS‐Dependent Enzymatic (CASCADE) Assay. Adv. Mater. Technol. 2021, 6, 2100602. 10.1002/admt.202100602
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Atzrodt C. L., Maknojia I., McCarthy R. D. P., Oldfield T. M., Po J., Ta K. T. L., Stepp H. E., Clements T. P., FEBS J. 2020, 287, 3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization, COVID‐19 Weekly Epidemiological Update Vol. 38, 2021. [Google Scholar]
- 3. Broughton J. P., Deng X., Yu G., Fasching C. L., Servellita V., Singh J., Miao X., Streithorst J. A., Granados A., Sotomayor‐Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C. Y., Guevara H., Wadford D. A., Chen J. S., Chiu C. Y., Nat. Biotechnol. 2020, 38, 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reperant L. A., Osterhaus A. D. M. E., Vaccine 2017, 35, 4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Feng W., Newbigging A. M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D. B., Cui M., Tao J., Tyrrell D. L., Zhang X. E., Zhang H., Le X. C., Anal. Chem. 2020, 92, 10196. [DOI] [PubMed] [Google Scholar]
- 6. Javalkote V. S., Kancharla N., Bhadra B., Shukla M., Soni B., Sapre A., Goodin M., Bandyopadhyay A., Dasgupta S., Methods 2020, 10.1016/j.ymeth.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Batista A. C., Pacheco L. G. C., J. Microbiol. Methods 2018, 152, 98. [DOI] [PubMed] [Google Scholar]
- 8. Li Y., Li S., Wang J., Liu G., Trends Biotechnol. 2019, 37, 730. [DOI] [PubMed] [Google Scholar]
- 9. Rahimi H., Salehiabar M., Barsbay M., Ghaffarlou M., Kavetskyy T., Sharafi A., Davaran S., Chauhan S. C., Danafar H., Kaboli S., Nosrati H., Yallapu M. M., Conde J., ACS Sens. 2021, 6, 1430. [DOI] [PubMed] [Google Scholar]
- 10. Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y., Chen Y., Feng G., Gu Q., Wang R. R., Zhou Q., Li W., Cell Discov. 2020, 6, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joung J., Ladha A., Saito M., Kim N.‐G., Woolley A. E., Segel M., Barretto R. P. J., Ranu A., Macrae R. K., Faure G., Ioannidi E. I., Krajeski R. N., Bruneau R., Huang M.‐L. W., Yu X. G., Li J. Z., Walker B. D., Hung D. T., Greninger A. L., Jerome K. R., Gootenberg J. S., Abudayyeh O. O., Zhang F., N. Engl. J. Med. 2020, 383, 1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park J. S., Hsieh K., Chen L., Kaushik A., Trick A. Y., Wang T., Adv. Sci. 2021, 8, 2003564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., Wu J., Liao Y., Gou X., Li Y., Wang X., Su H., Gu B., Wang J., Xu T., PLoS Pathog. 2020, 16, e1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fozouni P., Son S., Díaz de León Derby M., Knott G. J., Gray C. N., D'Ambrosio M. V., Zhao C., Switz N. A., Kumar G. R., Stephens S. I., Boehm D., Tsou C. L., Shu J., Bhuiya A., Armstrong M., Harris A. R., Chen P. Y., Osterloh J. M., Meyer‐Franke A., Joehnk B., Walcott K., Sil A., Langelier C., Pollard K. S., Crawford E. D., Puschnik A. S., Phelps M., Kistler A., DeRisi J. L., Doudna J. A., Fletcher D. A., Ott M., Cell 2021, 184, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ning B., Yu T., Zhang S., Huang Z., Tian D., Lin Z., Niu A., Golden N., Hensley K., Threeton B., Lyon C. J., Yin X.‐M., Roy C. J., Saba N. S., Rappaport J., Wei Q., Hu T. Y., Sci. Adv. 2020, 7, eabe3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ding X., Yin K., Li Z., Lalla R., Ballesteros E., Sfeir M., Liu C., Nat. Commun. 2020, 10.1101/2020.03.19.998724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramachandran A., Huyke D. A., Sharma E., Sahoo M. K., Huang C., Banaei N., Pinsky B. A., Santiago J. G., Proc. Natl. Acad. Sci. USA 2020, 117, 29518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gootenberg J. S., Abudayyeh O. O., Kellner M. J., Joung J., Collins J. J., Zhang F., Science (80‐.). 2018, 360, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Borlido L., Azevedo A. M., Roque A. C. A., Aires‐Barros M. R., Biotechnol. Adv. 2013, 31, 1374. [DOI] [PubMed] [Google Scholar]
- 20. Chen J. S., Ma E., Harrington L. B., Da Costa M., Tian X., Palefsky J. M., Doudna J. A., Science (80‐.). 2018, 360, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nguyen L. T., Smith B. M., Jain P. K., Nat. Commun. 2020, 11, 4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ji Y., Yang X., Ji Z., Zhu L., Ma N., Chen D., Jia X., Tang J., Cao Y., ACS Omega 2020, 5, 8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mello M. L. S., Vidal B. C., PLoS One 2012, 7, e43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shokr A., Pacheco L. G. C., Thirumalaraju P., Kanakasabapathy M. K., Gandhi J., Kartik D., Silva F. S. R., Erdogmus E., Kandula H., Luo S., Yu X. G., Chung R. T., Li J. Z., Kuritzkes D. R., Shafiee H., ACS Nano 2021, 15, 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Draz M. S., Vasan A., Muthupandian A., Kanakasabapathy M. K., Thirumalaraju P., Sreeram A., Krishnakumar S., Yogesh V., Lin W., Yu X. G., Chung R. T., Shafiee H., Sci. Adv. 2020, 6, eabd5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weissleder R., Lee H., Ko J., Pittet M. J., Sci. Transl. Med. 2020, 12, eabc1931. [DOI] [PubMed] [Google Scholar]
- 27. Arizti‐Sanz J., Freije C. A., Stanton A. C., Petros B. A., Boehm C. K., Siddiqui S., Shaw B. M., Adams G., Kosoko‐Thoroddsen T. S. F., Kemball M. E., Uwanibe J. N., Ajogbasile F. V., Eromon P. E., Gross R., Wronka L., Caviness K., Hensley L. E., Bergman N. H., MacInnis B. L., Happi C. T., Lemieux J. E., Sabeti P. C., Myhrvold C., Nat. Commun. 2020, 11, 5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bay H., Tuytelaars T., Van Gool L., in Lecture Notes in Computer Science (Including Subseries Lectre Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), 2006, pp. 404–417. [Google Scholar]
- 29. Fajnzylber J., Regan J., Coxen K., Corry H., Wong C., Rosenthal A., Worrall D., Giguel F., Piechocka‐Trocha A., Atyeo C., Fischinger S., Chan A., Flaherty K. T., Hall K., Dougan M., Ryan E. T., Gillespie E., Chishti R., Li Y., Jilg N., Hanidziar D., Baron R. M., Baden L., Tsibris A. M., Armstrong K. A., Kuritzkes D. R., Alter G., Walker B. D., Yu X., Li J. Z., Abayneh B. A. B., Allen P., Antille D., Balazs A., Bals J., Barbash M., Bartsch Y., Boucau J., Boyce S., Braley J., Branch K., Broderick K., Carney J., Chevalier J., Choudhary M. C., Chowdhury N., Cordwell T., Daley G., Davidson S., Desjardins M., Donahue L., Drew D., Einkauf K., Elizabeth S., Elliman A., Etemad B., Fallon J., Fedirko L., Finn K., Flannery J., Forde P., Garcia‐Broncano P., Gettings E., Golan D., Goodman K., Griffin A., Grimmel S., Grinke K., Hartana C. A., Healy M., Heller H., Henault D., Holland G., Jiang C., Jordan H., Kaplonek P., Karlson E. W., Karpell M., Kayitesi C., Lam E. C., LaValle V., Lefteri K., Lian X., Lichterfeld M., Lingwood D., Liu H., Liu J., Lopez K., Lu Y., Luthern S., Ly N. L., MacGowan M., Magispoc K., Marchewka J., Martino B., McNamara R., Michell A., Millstrom I., Miranda N., Nambu C., Nelson S., Noone M., Novack L., Callaghan C. O', Ommerborn C., Osborn M., Pacheco L. C., Phan N., Pillai S., Porto F. A., Rassadkina Y., Reissis A., Ruzicka F., Seiger K., Selleck K., Sessa L., Sharpe A., Sharr C., Shin S., Singh N., Slaughenhaupt S., Sheppard K. S., Sun W., Sun X., (Lizzie) Suschana E., Talabi O., Ticheli H., Weiss S. T., Wilson V., Zhu A., Nat. Commun. 2020, 11, 5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


