Abstract
Background
Adrenocortical carcinoma (ACC) is an extremely rare malignant tumor with poor prognosis. Existing treatment options have limited effects, and new therapeutic targets urgently need to be discovered. TNFSF13B has been reported to be associated with the prognosis of clear cell renal cell carcinoma, but it has not been studied in ACC.
Methods
TNFSF13B expression was analyzed and compared between ACC tumors and normal tissues by using public datasets from TCGA and GTEx. Kaplan-Meier analysis was employed to evaluate survival, and Cox regression was employed to evaluate clinicopathologic features. The upstream and downstream regulatory mechanisms of TNFSF13B were also analyzed. GSEA was performed to explore the mechanisms of TNFSF13B in ACC. Finally, 14 ACC clinical samples were used to verify the relationships between TNFSF13B expression and disease-free survival (DFS) and overall survival (OS).
Results
TNFSF13B expression was significantly higher in ACC tissues than in normal tissues. The prognosis of ACC patients with high TNFSF13B expression was worse than that of patients with low TNFSF13B expression. High TNFSF13B expression was strongly correlated with poor prognosis, and TNFSF13B was a prognostic factor. TNFSF13B expression is modified by upstream miRNAs, methylation and ubiquitination, and downstream, it interacts with other proteins. GSEA showed that regulation of cholesterol biosynthesis by SREBP and SREBF, downstream signaling events of the B cell receptor (BCR) and activation of gene expression by SREBF and SREBP were significantly enriched in the TNFSF13B high-expression phenotype. Clinical samples confirmed that TNFSF13B expression was significantly associated with DFS but not with OS.
Conclusions
TNFSF13B may be a potential prognostic molecular marker of poor survival in ACC patients, offering a new therapeutic target.
Keywords: Tumor necrosis factor ligand superfamily member 13B (TNFSF13B), adrenocortical carcinoma (ACC), prognosis
Introduction
Adrenocortical carcinoma (ACC) is a rare malignant tumor originating from the adrenal cortex, with an incidence rate of approximately 2.0 per million per year (1,2). The disease can occur at any age, but it has two peak ages of onset in childhood and middle age (3), and the incidence in females is slightly higher than that in males (2,4). The prognosis of the disease is extremely poor, and the overall survival (OS) time of patient is usually short (2,5-7). The complete removal of the primary tumor is one of the most important treatment methods, but local recurrence, infiltration, or distant metastasis often occur after surgery (4,8). Adjuvant therapy, such as radiotherapy or chemotherapy, has been investigated extensively in recent years. However, the exact therapeutic effect is ambiguous among multiple research teams (6,7,9-17). Mitotane is the standard treatment for patients with end-stage ACC, and it is the only drug approved by the Food and Drug Administration (FDA) and by the European Medicines Agency (EMA) for the treatment of metastatic ACC (18). The use of mitotane has a certain curative effect on the survival of patients with ACC, but the effect is limited and side effects occur (5,19-21). Therefore, new therapeutic targets urgently need to be identified. The identification of novel hub genes specific to ACC may improve the effect of targeted therapy and bring significant benefits to patients.
Tumor necrosis factor ligand superfamily member 13B (TNFSF13B), also known as BAFF, is a cytokine that belongs to the tumor necrosis factor (TNF) ligand family encoded by the TNFSF13B gene (22). This cytokine is an effective B cell activator and plays an important role in the proliferation and differentiation of B cells. It is expressed in a variety of cells, including monocytes, macrophages and dendritic cells (23,24). It is a ligand for three TNF receptors named TACI, BCMA and BAFF-R (23,25). In a bioinformatics analysis study, TNFSF13B was identified as a prognostic biomarker for renal clear cell carcinoma (26). That study concluded that high TNFSF13B expression in clear cell renal cell carcinoma leads to poor prognosis. GSEA and TIMER analyses showed that the expression of TNFSF13B was related to immune signaling pathways and lymphocyte infiltration.
However, few studies focusing on the relationship between TNFSF13B and ACC have been reported thus far, and the role of TNFSF13B in ACC remains elusive. Therefore, TNFSF13B expression was analyzed in ACC, and its expression between tumor tissues and normal tissues was compared using public datasets from The Cancer Genome Atlas (TCGA) database and the Genotype-Tissue Expression (GTEx) project. We then analyzed the relationship between TNFSF13B expression and the clinical parameters of ACC patients and assessed the correlation of TNFSF13B expression with the disease-free survival (DFS) and OS of patients. An analysis of the upstream and downstream regulatory mechanisms of TNFSF13B was also implemented. Gene set enrichment analysis (GSEA) was performed to explore the mechanisms of TNFSF13B in ACC. Finally, we used 14 ACC clinical samples of patients recruited from Ruijin Hospital from 2013 to 2020 to verify the relationships between TNFSF13B gene expression and DFS and OS. The purpose of this study was to explore whether TNFSF13B can be used as a prognostic target in ACC patients. The significant diagnostic and prognostic value of TNFSF13B indicated that it may also be used in clinical practice to assist clinical workers in the diagnosis and treatment of ACC patients.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tau-21-232).
Methods
Databases and identification of TNFSF13B differential expression
Gene expression profiles of ACC tissues and normal adrenal glands were downloaded from TCGA (https://portal.gdc.cancer.gov/) and GTEx (https://www.gtexportal.org/home/index.html). EdgeR (http://bioconductor.org/) was utilized to identify differential TNFSF13B expression between normal adrenal tissues and adrenal cortical carcinoma tissues. The difference threshold was |log2FC| ≥1 and P value <0.05. The clinical data of patients, such as age, sex, pathological stage, survival and outcome, were downloaded from the TCGA database.
TNFSF13B expression and prognosis
Based on the median expression of the TNFSF13B gene, the cancer samples were divided into high and low expression groups. The correlations between the expression levels of TNFSF13B and patient DFS and OS were analyzed using the Kaplan-Meier method. Univariate Cox regression analysis was used to select possible prognostic factors.
MiRNA regulation and DNA methylation
Based on the predictions of multiple databases, the upstream miRNA of TNFSF13B might be regulated, and the prediction results of multiple databases were analyzed by VEEN. Through the miRDB, miRTarBase, miRMap, miRanda and TargetScan online prediction websites, miRNAs targeting the TNFSF13B gene were predicted, and the multiple predicted miRNAs were summarized. The miRNAs that were predicted by all five sites were considered intersecting miRNAs. We also analyzed the correlation between TNFSF13B methylation and TNFSF13B expression in ACC.
Query for E3-TNFSF13B interactions in UbiBrowser
UbiBrowser (http://ubibrowser.ncpsb.org/) is an integrated bioinformatics website tool designed to help users explore the predicted and reported E3 substrate interactions and ubiquitination sites of substrate proteins by querying E3 or substrate; it contains 1,295 reported E3-substrate interactions and 8,255 predicted E3-substrate interactions (27). In the network view, the central node is the substrate to be queried, and the surrounding nodes are the predicted E3 ligases. We used UbiBrowser to predict the potential E3 ligases of TNFSF13B.
Protein-protein interaction (PPI) network construction and analysis
STRING (version 11.0, http://string-db.org) is a database of known and predicted PPI networks (27). We used this tool to construct a PPI network and predict potential interactions between TNFSF13B and its targets. Interactions with a cutoff score of 0.4 and above were considered significant.
GSEA
GSEA software (http://software.broadinstitute.org/gsea/downloads.jsp) was used to identify the enrichments for biological functions and signal pathways based on the TCGA clinical data of the ACC patients. According to the expression level of TNFSF13B, the patients were divided into two groups by a median threshold: the high expression group and the low expression group. Enrichment analysis of the GSEA gene set used the C2 set (curated gene sets) in MSigDB (http://software.broadinstitute.org/gsea/msigdb). Finally, we sorted all enriched pathways according to the normalized enrichment score (NES).
Sample collection and quantitative real-time polymerase chain reaction (qPCR)
From 2013 to 2020, 14 ACC specimens were collected from the Department of Urology at Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013). The institutional Committee abandoned the ethical review because it did not affect the treatment strategy. Informed consent to use their tissues for research was obtained from each patient prior to surgery. The samples were stored in liquid nitrogen. Then, total RNA was extracted by the Eastep® Super Total RNA Extraction Kit (Promega, China). A reverse-transcription kit (Promega, China) was used to reverse transcription of total RNA into the first strand of complementary DNA. QPCR was then performed with a Quantstudio™ Dx Real-Time PCR System (Thermo Fisher Scientific, USA). Endogenous gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a reference control. Genes were amplified by the following specific primers: GAPDH forward (5'-GGAGCGAGATCCCTCCAAAAT-3'), GAPDH reverse (5'-GGCTGTTGTCATACTTCTCATGG-3'); TNFSF13B forward (5'-GGGAGCAGTCACGCCTTAC-3'), and TNFSF13B reverse (5'-GATCGGACAGAGGGGCTTT-3'). The experiment was repeated twice, and the average Ct from the values obtained for each reaction was calculated. The fold change of the target genes against that of the reference gene was calculated from the 2−ΔΔCt values.
Statistical analysis
R 4.0.2 (http://www.r-project.org) and GraphPad Prism 8 (https://www.graphpad.com) were used for all data analyses. EdgeR was performed to explore the differential expression levels of TNFSF13B between ACC tissues and normal tissues. Kaplan-Meier survival analysis was performed to estimate the correlations between TNFSF13B expression and DFS and OS. The associations between prognosis and clinical characteristics were evaluated by univariate Cox regression analysis. The mRNA expression 2−ΔΔCt values of clinical samples were converted logarithmically. Then, the patients were divided into a high TNFSF13B expression group and a low TNFSF13B expression group according to the median value. With the follow-up data of patients, the survival function of GraphPad Prism software was used to draw K-M survival curves. Statistical significance was defined as P<0.05.
Results
High expression of TNFSF13B leads to poor prognosis in ACC
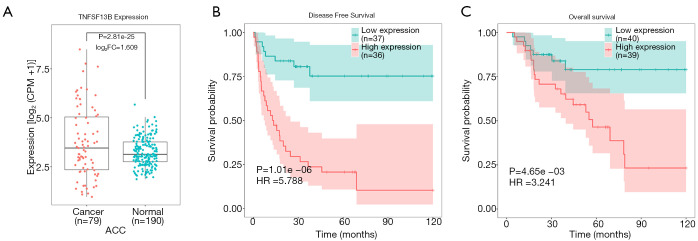
We compared the expression levels of TNFSF13B in 79 ACC tissues and 190 normal tissues. Table S1 lists the expression levels of TNFSF13B in a total of 269 samples. TNFSF13B was expressed at higher levels in ACC tissues than in normal adrenal gland tissues (P<0.0001, Figure 1A). Kaplan-Meier survival curves revealed that the prognosis of ACC patients with high TNFSF13B expression was worse than that of ACC patients with low TNFSF13B expression in terms of both DFS (P<0.0001) and OS (P<0.01) (Figure 1B,C). The univariate Cox regression analysis results showed that high TNFSF13B expression was correlated with poor prognosis [hazard ratio (HR) =1.01, 95% confidence interval: 1–1.01, P<0.01; Table 1]. Moreover, other variables, such as pathologic stage, T stage, M stage and lymph nodes, were also associated with the prognosis of ACC patients. These results indicate that the expression of TNFSF13B is a prognostic factor and that increased TNFSF13B levels are associated with poor DFS and OS.
Figure 1.
High expression of TNFSF13B leads to poor prognosis in ACC. (A). TNFSF13B was more highly expressed in ACC than in normal tissues. (B,C) K-M survival curves revealed that the prognosis of ACC patients with high TNFSF13B expression was worse than that of patients with low TNFSF13B expression in terms of both DFS (B) and OS (C). ACC, adrenocortical carcinoma; CPM, counts per million; DFS, disease-free survival; FC, fold change; HR, hazard ratio; OS, overall survival; TNFSF13B, tumor necrosis factor ligand superfamily member 13B.
Table 1. Univariate Cox regression analysis of the relationship between clinicopathological variables and prognosis.
| Clinicopathological variables | Hazard ratio | 95% CI | P value |
|---|---|---|---|
| TNFSF13B expression | 1.01 | 1–1.01 | 0.002 |
| Age | 1.01 | 0.987–1.04 | 0.379 |
| Gender | 1 | 0.469–2.14 | 0.999 |
| Pathological stage | 2.91 | 1.86–4.56 | 0.000 |
| T stage | 3.38 | 2.11–5.14 | 0.000 |
| N stage | 2.04 | 0.769–5.4 | 0.152 |
| M stage | 6.15 | 2.71–14 | 0.000 |
| Tumor purity | 20.3 | 0.148–2790 | 0.230 |
| Lymph node | 1.07 | 1.01–1.12 | 0.018 |
CI, confidence interval; TNFSF13B, tumor necrosis factor ligand superfamily member 13B.
Upstream and downstream regulation mechanisms
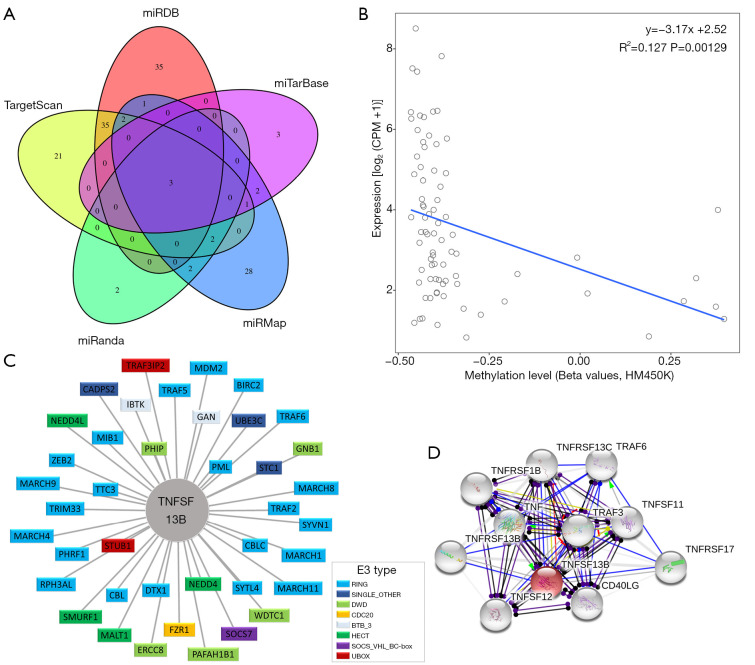
The miRDB, miRTarBase, miRMap, miRanda and TargetScan databases predicted 76, 9, 41, 9, and 64 miRNAs targeting TNFSF13B, among which there were 3 intersecting miRNAs (Figure 2A), namely, hsa-miR-30a-3p, hsa-miR-30d-3p, and hsa-miR-30e-3p. A significant negative correlation was identified between the methylation level of TNFSF13B and the expression level of TNFSF13B (P<0.01) (Figure 2B). To identify the potential E3 ligases of TNFSF13B, we queried TNFSF13B as a substrate in the UbiBrowser web tool. The 4 predicted E3 ligases with middle-confidence interactions and 36 predicted E3 ligases with low-confidence interactions are presented in Table S2. Figure 2C displays them. The protein-protein interaction network is shown in Figure 2D.
Figure 2.
Upstream and downstream regulation mechanisms. (A) The miRDB, miRTarBase, miRMap, miRanda and TargetScan databases predicted 76, 9, 41, 9, and 64 miRNAs targeting TNFSF13B, respectively. The results are shown by Venn Diagram. (B) A significant negative correlation was identified between the methylation level and expression level of TNFSF13B. (C) Network view of the predicted E3-substrate interactions in the UbiBrowser web service. (D) Construction of the protein-protein interaction network between TNFSF13B and its target proteins. Abbreviations of E3 ligases are annotated in Table S2. Abbreviations of E3 type are annotated in Table S3. Abbreviations of protein-protein interaction are annotated in Table S4. CPM, counts per million; HM450K, Illumina 450k methylation arrays; TNFSF13B, tumor necrosis factor ligand superfamily member 13B.
GSEA
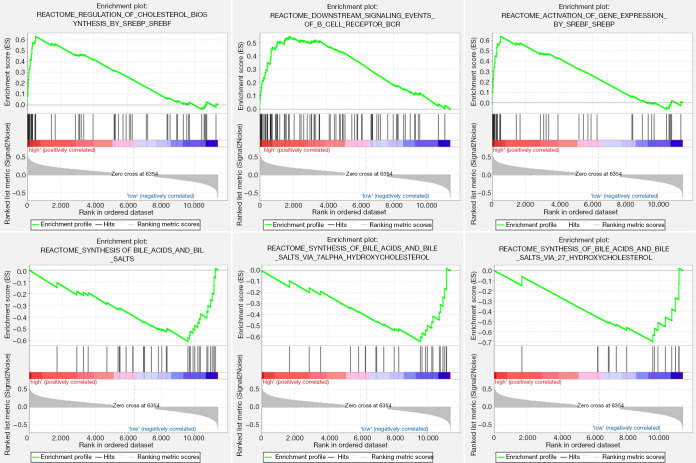
GSEA was performed to identify the potential signaling pathways involved in ACC between the low and high TNFSF13B mRNA expression groups. Figure 3 shows the top 3 pathways in terms of the NES for the high and low TNFSF13B expression groups analyzed by GSEA. The high TNFSF13B group was enriched in the following terms: regulation of cholesterol biosynthesis by SREBP and SREBF, downstream signaling events of the B cell receptor (BCR) and activation of gene expression by SREBF and SREBP. Conversely, the low TNFSF13B group was enriched in the following pathways: synthesis of bile acids and bile salts, synthesis of bile acids and bile salts via 7-alpha-hydroxycholesterol, and synthesis of bile acids and bile salts via 27-hydroxycholesterol.
Figure 3.
Gene set enrichment analysis according to the expression of TNFSF13B in TCGA. BCR, B-cell receptor; SREBF, sterol-regulatory element binding factor; SREBP, sterol-regulatory element binding protein; TCGA, The Cancer Genome Atlas; TNFSF13B, tumor necrosis factor ligand superfamily member 13B.
Validation of clinical samples by qPCR
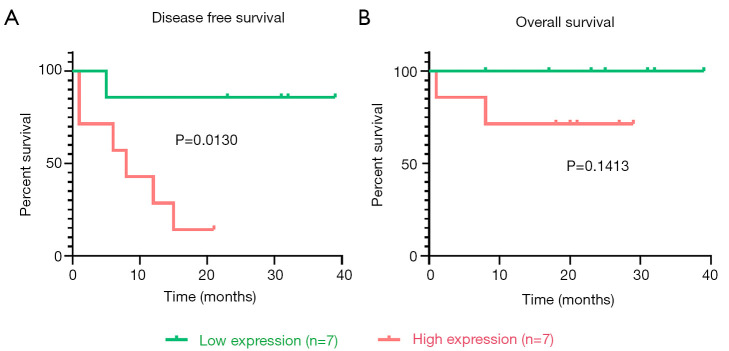
The ACC clinical samples of patients recruited from Ruijin Hospital were used to verify the relationship between TNFSF13B expression and prognosis. Table 2 summarizes the demographic, clinical and pathological characteristics of the 14 patients. The scoring methods of GRAS (28) and mGRAS (29) are described in detail at the bottom of the table. As shown in Figure 4A, Kaplan-Meier survival curves revealed that the prognosis of ACC patients with high TNFSF13B expression was worse than that of patients with low TNFSF13B expression in terms of DFS (P<0.05). However, there was no difference between the two groups in OS (P=0.1413) (Figure 4B).
Table 2. Summary of the demographic, clinical and pathological variables of the 14 patients included in the study.
| Variables | n | % |
|---|---|---|
| Age (year) | ||
| <50 | 5 | 35.71 |
| ≥50 | 9 | 64.29 |
| Gender | ||
| Male | 5 | 35.71 |
| Female | 9 | 64.29 |
| ENSAT (stage) | ||
| I | 1 | 7.14 |
| II | 8 | 57.14 |
| III | 2 | 14.29 |
| IV | 3 | 21.43 |
| Ki67 (%) | ||
| <10 | 4 | 28.57 |
| 10–19 | 4 | 28.57 |
| ≥20 | 6 | 42.86 |
| GRAS▲ | ||
| 0 | 4 | 28.57 |
| 1 | 3 | 21.43 |
| 2 | 4 | 28.57 |
| 3 | 3 | 21.43 |
| mGRAS▼ | ||
| 0–1 | 5 | 35.71 |
| 2–3 | 4 | 28.57 |
| 4–5 | 3 | 21.43 |
| 6–7 | 2 | 14.29 |
| OS (months) | ||
| ≤12 | 3 | 21.43 |
| 13–24 | 5 | 35.71 |
| >24 | 6 | 42.86 |
| DFS (months) | ||
| ≤12 | 6 | 42.86 |
| 13–24 | 4 | 28.57 |
| >24 | 4 | 28.57 |
▲The GRAS score is accumulated by the following four items (28): grade (Weiss score <3 and Ki67 <20% =0; Weiss score ≥3 or Ki67 ≥20% =1), resection status (R0 =0; R1 or R2 =1), age (<50 years =0; ≥50 years =1), and symptoms (absent =0; present =1). ▼The mGRAS score is accumulated by the following five items (29): age (<50 years =0; ≥50 years =1), symptoms (absent =0; present =1), ENSAT (stage I–II =0; stage III =1; stage IV =2), resection status (R0 =0; RX =1; R1 =2; R2 =3), and Ki67 (0–9% =0; 10–19% =1; ≥20% =2). ENSAT, European Network for the Study of Adrenal Tumors; GRAS, Grade, Resection status, Age, and Symptoms of hormone hypersecretion; mGRAS, modified GRAS; OS, overall survival; DFS, disease-free survival.
Figure 4.
Relationship between the mRNA expression of TNFSF13B in ACC clinical samples and prognosis was verified by qPCR. (A). K-M survival curves revealed that the prognosis of ACC patients with high TNFSF13B expression was worse than that of patients with low TNFSF13B expression in terms of DFS. (B) K-M survival curves revealed that there was no difference between the two groups in OS. ACC, adrenocortical carcinoma; DFS, disease-free survival; OS, overall survival; qPCR, quantitative real-time polymerase chain reaction.
Discussion
TNFSF13B is a member of the TNF superfamily. Its coding gene is located on chromosome 13q32-34 (30), and the protein it encodes is involved in many physiological activities of life. As an important B lymphocyte stimulating factor, the normal expression of TNFSF13B is important for the survival, proliferation and differentiation of B cells. TNFSF13B deficiency may lead to a decrease in humoral immune function. Studies have shown that TNFSF13B knockout mice have severe B cell defects (31), and the numbers of B cells in the splenic marginal zone and follicles are significantly reduced, which will cause a sharp drop in serum total immunoglobulin levels (32). In addition, TNFSF13B can effectively promote the apoptosis of tumor cells. TNFSF13B inhibits the growth of human lymphoma cells (U937), prostate cancer cells (PC-3), colon cancer cells (HT-29), cervical cancer cells (HeLa), breast cancer cells (MCF-7) and embryonic kidney cells (A293). In U937 cells, researchers found that TNFSF13B could induce apoptosis by activating caspase-3 to degrade PARP (33).
However, the abnormal expression of TNFSF13B is also closely related to the occurrence and development of certain diseases. First, TNFSF13B is associated with tumorigenesis. In the serum of patients with follicular-type non-Hodgkin's lymphoma, researchers found that the level of TNFSF13B was three times that in normal people (34). In another study, serum levels of TNFSF13B increased in newly diagnosed patients with IgG type multiple myeloma (MM) compared with healthy volunteers, and the concentration of TNFSF13B increased in the late stage with the progression of MM (35). In addition, TNFSF13B is believed to be related to autoimmune diseases such as rheumatoid arthritis, systemic lupus erythematosus and Sjogren’s syndrome. An analysis of biological samples from patients with various autoimmune diseases confirmed that high serum TNFSF13B levels were associated with these diseases (24). Similarly, researchers found that TNFSF13B mRNA transcription levels increased in the brain tissues of experimental autoimmune encephalomyelitis mice (31). Moreover, TNFSF13B may be associated with infectious diseases. Researchers found elevated levels of TNFSF13B in the serum of HIV patients (36). Finally, TNFSF13B has important links with the occurrence and development of diabetes. Diabetes could be prevented in TNFSF13B-deficient NOD mice (37). TNFSF13B blockers inhibit the development of diabetes (38), and TNFSF13B-blocking compounds can delay the onset and reduce the incidence rate of diabetes (37).
At present, ACC is still a serious challenge to human medicine. Due to its low incidence, there are not as many studies on ACC as other tumors, and drugs with definite therapeutic effects are also very limited. For decades, mitotane has been the first choice of adjuvant drug for the treatment of ACC. It is recommended for people with a high risk of recurrence, and it is increasingly used as an adjuvant treatment after surgical resection of ACC. However, mitotane can affect the production of steroids (2). It can cause mitochondrial damage, interfere with the function of the mitochondrial respiratory chain, and induce the morphological fragmentation of the mitochondrial membrane necessary for the activity of the respiratory chain and the production of steroids. In addition, mitotane is a strong inducer of CYP3A4 activity, leading to the inactivation of glucocorticoids (2). Furthermore, mitotane is an inhibitor of sterol-O-acyltransferase 1 (SOAT1) (5), which causes the accumulation of free cholesterol, which is toxic to cells. However, the value of this drug is still a controversial issue, as only a few studies have compared patients in a sufficiently large treatment group with a control group (8). It is worth noting that the use of mitotane is often accompanied by some adverse reactions (5). The most common adverse reactions are neurotoxicity, such as dizziness, headache, drowsiness, and ataxia; liver damage, such as elevated transaminase; and gastrointestinal disorders, such as nausea, vomiting, diarrhea, and upper abdominal discomfort (5). These side effects limit the use of mitotane to a certain extent.
Our study, based on public databases, found that TNFSF13B was highly expressed in ACC, and the expression level of TNFSF13B in ACC was significantly associated with poor prognosis. At the same time, we used a public database to predict factors related to the upstream and downstream mechanisms of TNFSF13B, such as miRNA regulation, methylation, ubiquitination and protein-protein interactions, which provided a direction for further study on the role of TNFSF13B in the occurrence and development of ACC. The abnormal secretion of steroid hormones, such as adrenal cortex hormones and sex hormones, is an important cause of clinical symptoms in patients with adrenal cortical cancer. Cholesterol is the precursor substance of steroid hormone synthesis (39), and its synthesis and decomposition state directly affect the metabolism of steroid hormones. The GSEA results showed that TNFSF13B is significantly related to cholesterol metabolism, which more strongly illustrates the role of TNFSF13B in the occurrence and development of adrenal cortical carcinoma. Based on this, we believe that our research is credible and clinically valuable. Our findings strongly indicate that TNFSF13B may be a potential biomarker or target of ACC, providing a potential new therapeutic target for ACC.
Although this study determined the prognostic value of TNFSF13B in ACC, there are still some limitations. First, the number of clinical samples was too small, and the follow-up time was not long enough. For these two reasons, we failed to verify the difference in OS between the two groups. In future studies, with more cases and longer follow-up times, the important role of the TNFSF13B gene can be confirmed in ACC with poor prognosis. Second, due to the heterogeneity of the data set, there may be some errors in our analysis results. More data sets need to be used to verify our conclusions. Third, the conclusions of our analysis are based on the expression of TNFSF13B mRNA, and further research is needed to evaluate the protein expression and direct mechanism of TNFSF13B.
Conclusions
In our current study, we found that ACC patients with high TNFSF13B expression had a poorer prognosis than those with low TNFSF13B expression. TNFSF13B may be a potential prognostic molecular marker of poor survival in ACC patients, offering a new therapeutic target.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank the public databases for the support of this study. The public data used in this study were all downloaded in September 2020, and we cannot predict the changes that will occur afterwards. We thank WJ Xu for her help in downloading and analyzing the public data. We thank SNAS for the native language correction of this manuscript.
Funding: This study was supported by the National Natural Science Foundation of China (81972494) and Special Research Project of Wise Healthcare of Shanghai Health and Family Planning Commission (2018ZHYL0205).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The public data used in this study were all downloaded in September 2020, and we cannot predict the changes that will occur afterwards. The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013). The Institutional Committee abandoned the ethical review because it did not affect the treatment strategy. Informed consent was obtained from each patient prior to surgery.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tau-21-232
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tau-21-232
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-232). The authors have no conflicts of interest to declare.
References
- 1.Ettaieb M, Kerkhofs T, van Engeland M, et al. Past, Present and Future of Epigenetics in Adrenocortical Carcinoma. Cancers (Basel) 2020;12:1218. 10.3390/cancers12051218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puglisi S, Calabrese A, Basile V, et al. New perspectives for mitotane treatment of adrenocortical carcinoma. Best Pract Res Clin Endocrinol Metab 2020;34:101415. 10.1016/j.beem.2020.101415 [DOI] [PubMed] [Google Scholar]
- 3.Pittaway JFH, Guasti L. Pathobiology and genetics of adrenocortical carcinoma. J Mol Endocrinol 2019;62:R105-19. 10.1530/JME-18-0122 [DOI] [PubMed] [Google Scholar]
- 4.Berruti A, Baudin E, Gelderblom H, et al. Adrenal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2012;23 Suppl 7:vii131-8. 10.1093/annonc/mds231 [DOI] [PubMed] [Google Scholar]
- 5.Creemers SG, Hofland LJ, Korpershoek E, et al. Future directions in the diagnosis and medical treatment of adrenocortical carcinoma. Endocr Relat Cancer 2016;23:R43-69. 10.1530/ERC-15-0452 [DOI] [PubMed] [Google Scholar]
- 6.Else T, Kim AC, Sabolch A, et al. Adrenocortical carcinoma. Endocr Rev 2014;35:282-326. 10.1210/er.2013-1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoff AO, Berruti A. 5th International ACC Symposium: Future and Current Therapeutic Trials in Adrenocortical Carcinoma. Horm Cancer 2016;7:29-35. 10.1007/s12672-015-0241-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fassnacht M, Assie G, Baudin E, et al. Adrenocortical carcinomas and malignant phaeochromocytomas: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2020;31:1476-90. 10.1016/j.annonc.2020.08.2099 [DOI] [PubMed] [Google Scholar]
- 9.Fassnacht M, Hahner S, Polat B, et al. Efficacy of adjuvant radiotherapy of the tumor bed on local recurrence of adrenocortical carcinoma. J Clin Endocrinol Metab 2006;91:4501-4. 10.1210/jc.2006-1007 [DOI] [PubMed] [Google Scholar]
- 10.Gharzai LA, Green MD, Griffith KA, et al. Adjuvant Radiation Improves Recurrence-Free Survival and Overall Survival in Adrenocortical Carcinoma. J Clin Endocrinol Metab 2019;104:3743-50. 10.1210/jc.2019-00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Habra MA, Ejaz S, Feng L, et al. A retrospective cohort analysis of the efficacy of adjuvant radiotherapy after primary surgical resection in patients with adrenocortical carcinoma. J Clin Endocrinol Metab 2013;98:192-7. 10.1210/jc.2012-2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermsen IG, Groenen YE, Dercksen MW, et al. Response to radiation therapy in adrenocortical carcinoma. J Endocrinol Invest 2010;33:712-4. 10.1007/BF03346675 [DOI] [PubMed] [Google Scholar]
- 13.Ho J, Turkbey B, Edgerly M, et al. Role of radiotherapy in adrenocortical carcinoma. Cancer J 2013;19:288-94. 10.1097/PPO.0b013e31829e3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polat B, Fassnacht M, Pfreundner L, et al. Radiotherapy in adrenocortical carcinoma. Cancer 2009;115:2816-23. 10.1002/cncr.24331 [DOI] [PubMed] [Google Scholar]
- 15.Sabolch A, Feng M, Griffith K, et al. Adjuvant and definitive radiotherapy for adrenocortical carcinoma. Int J Radiat Oncol Biol Phys 2011;80:1477-84. 10.1016/j.ijrobp.2010.04.030 [DOI] [PubMed] [Google Scholar]
- 16.Al Asadi A, Hubbs DM, Sweigert PJ, et al. Analysis of adjuvant chemotherapy in patients undergoing curative-intent resection of localized adrenocortical carcinoma. Am J Surg 2021;222:119-25. 10.1016/j.amjsurg.2020.10.038 [DOI] [PubMed] [Google Scholar]
- 17.Varghese J, Habra MA. Update on adrenocortical carcinoma management and future directions. Curr Opin Endocrinol Diabetes Obes 2017;24:208-14. 10.1097/MED.0000000000000332 [DOI] [PubMed] [Google Scholar]
- 18.De Filpo G, Mannelli M, Canu L. Adrenocortical carcinoma: current treatment options. Curr Opin Oncol 2021;33:16-22. 10.1097/CCO.0000000000000695 [DOI] [PubMed] [Google Scholar]
- 19.Berruti A, Grisanti S, Pulzer A, et al. Long-Term Outcomes of Adjuvant Mitotane Therapy in Patients With Radically Resected Adrenocortical Carcinoma. J Clin Endocrinol Metab 2017;102:1358-65. 10.1210/jc.2016-2894 [DOI] [PubMed] [Google Scholar]
- 20.Fassnacht M, Dekkers OM, Else T, et al. European Society of Endocrinology Clinical Practice Guidelines on the management of adrenocortical carcinoma in adults, in collaboration with the European Network for the Study of Adrenal Tumors Eur J Endocrinol 2018;179:G1-G46. [DOI] [PubMed]
- 21.Tang Y, Liu Z, Zou Z, et al. Benefits of Adjuvant Mitotane after Resection of Adrenocortical Carcinoma: A Systematic Review and Meta-Analysis. Biomed Res Int 2018;2018:9362108. 10.1155/2018/9362108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stelmach P, Pütz M, Pollmann R, et al. Alternative splicing of the TNFSF13B (BAFF) pre-mRNA and expression of the BAFFX1 isoform in human immune cells. Gene 2020;760:145021. 10.1016/j.gene.2020.145021 [DOI] [PubMed] [Google Scholar]
- 23.Pieper K, Grimbacher B, Eibel H. B-cell biology and development. J Allergy Clin Immunol 2013;131:959-71. 10.1016/j.jaci.2013.01.046 [DOI] [PubMed] [Google Scholar]
- 24.Mackay F, Ambrose C. The TNF family members BAFF and APRIL: the growing complexity. Cytokine Growth Factor Rev 2003;14:311-24. 10.1016/S1359-6101(03)00023-6 [DOI] [PubMed] [Google Scholar]
- 25.Kampa M, Notas G, Stathopoulos EN, et al. The TNFSF Members APRIL and BAFF and Their Receptors TACI, BCMA, and BAFFR in Oncology, With a Special Focus in Breast Cancer. Front Oncol 2020;10:827. 10.3389/fonc.2020.00827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang M, Lin J, Xing H, et al. Microenvironment-related gene TNFSF13B predicts poor prognosis in kidney renal clear cell carcinoma. PeerJ 2020;8:e9453. 10.7717/peerj.9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Han X, Chen L, et al. TRIM28 is a distinct prognostic biomarker that worsens the tumor immune microenvironment in lung adenocarcinoma. Aging (Albany NY) 2020;12:20308-31. 10.18632/aging.103804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baechle JJ, Marincola Smith P, et al. Cumulative GRAS Score as a Predictor of Survival After Resection for Adrenocortical Carcinoma: Analysis From the U.S. Adrenocortical Carcinoma Database. Ann Surg Oncol 2021. [Epub ahead of print]. doi: . 10.1245/s10434-020-09562-8 [DOI] [PubMed] [Google Scholar]
- 29.Elhassan Y, Altieri B, Berhane S, et al. Modified GRAS Score for Prognostic Classification of Adrenocortical Carcinoma: An ENSAT Multicentre Study. J Endocr Soc 2021;5:A165-6. 10.1210/jendso/bvab048.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider P, MacKay F, Steiner V, et al. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J Exp Med 1999;189:1747-56. 10.1084/jem.189.11.1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou T, Zhang J, Carter R, et al. BLyS and B cell autoimmunity. Curr Dir Autoimmun 2003;6:21-37. 10.1159/000066854 [DOI] [PubMed] [Google Scholar]
- 32.Schiemann B, Gommerman JL, Vora K, et al. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science 2001;293:2111-4. 10.1126/science.1061964 [DOI] [PubMed] [Google Scholar]
- 33.Mukhopadhyay A, Ni J, Zhai Y, et al. Identification and characterization of a novel cytokine, THANK, a TNF homologue that activates apoptosis, nuclear factor-kappaB, and c-Jun NH2-terminal kinase. J Biol Chem 1999;274:15978-81. 10.1074/jbc.274.23.15978 [DOI] [PubMed] [Google Scholar]
- 34.Briones J, Timmerman JM, Hilbert DM, et al. BLyS and BLyS receptor expression in non-Hodgkin's lymphoma. Exp Hematol 2002;30:135-41. 10.1016/S0301-472X(01)00774-3 [DOI] [PubMed] [Google Scholar]
- 35.Bolkun L, Lemancewicz D, Jablonska E, et al. BAFF and APRIL as TNF superfamily molecules and angiogenesis parallel progression of human multiple myeloma. Ann Hematol 2014;93:635-44. 10.1007/s00277-013-1924-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stohl W, Cheema GS, Briggs WS, et al. B lymphocyte stimulator protein-associated increase in circulating autoantibody levels may require CD4+ T cells: lessons from HIV-infected patients. Clin Immunol 2002;104:115-22. 10.1006/clim.2002.5238 [DOI] [PubMed] [Google Scholar]
- 37.Mariño E, Walters SN, Villanueva JE, et al. BAFF regulates activation of self-reactive T cells through B-cell dependent mechanisms and mediates protection in NOD mice. Eur J Immunol 2014;44:983-93. 10.1002/eji.201344186 [DOI] [PubMed] [Google Scholar]
- 38.Mariño E, Villanueva J, Walters S, et al. CD4(+)CD25(+) T-cells control autoimmunity in the absence of B-cells. Diabetes 2009;58:1568-77. 10.2337/db08-1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanukoglu I. Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J Steroid Biochem Mol Biol 1992;43:779-804. 10.1016/0960-0760(92)90307-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as