Abstract
In each of six gastric cancer patients, repetitive extragenic palindromic PCR DNA fingerprints of 18 single colonies of Helicobacter pylori from the gastric antrum, corpus, and cardia were identical and matched that of the parental isolate. In three additional gastric cancer patients, 17 of 18 single-colony DNA fingerprints were identical to each other and to the DNA fingerprint of the corresponding parental isolate.
The large degree of genetic heterogeneity among Helicobacter pylori strains allows individual clinical H. pylori isolates to be reliably distinguishable from one another through the use of DNA fingerprinting methods (1, 7). Repetitive extragenic palindromic (REP) sequences are well-characterized repetitive DNA elements occurring in various bacterial species, including H. pylori (4, 11). PCR amplification of genomic DNA located between these repetitive sequences within the H. pylori genome (REP-PCR) has been shown to be useful to generate stable and reproducible DNA fingerprints despite the large degree of genetic heterogeneity among H. pylori strains (4). cagA indicates the presence of a so-called pathogenicity island which harbors other potential virulence determinants (3).
From nine patients with intestinal-type gastric adenocarcinoma (four men and five women; age range, 24 to 90 years; mean age, 59 years) evaluated at the University Hospital San Juan de Dios, Bogota, Colombia, gastric biopsy specimens from cardia, corpus, and antrum were obtained. H. pylori isolates from all gastric biopsy specimens were recovered under standard conditions and identified by Gram-stain morphology and biochemical testing. Six single colonies from each H. pylori isolate were obtained, giving a total of 18 single colonies per patient. Genomic DNA extraction and generation of REP-PCR DNA fingerprints were performed as described previously (4). cagA was detected by PCR amplification of a 297-bp region (nucleotides 1751 to 2048) and a ∼1.4-kb DNA fragment further downstream in the cagA gene as described previously (6).
REP-PCR DNA fingerprints were generated for a total of 162 single colonies of H. pylori obtained from three different gastric sites and were compared with that of the parental isolate. Each patient harbored an H. pylori strain with a unique DNA fingerprint. DNA fingerprints from the H. pylori parent isolate and the corresponding 18 single colonies obtained from six of nine patients demonstrated identical DNA band profiles (Fig. 1). In one patient, the H. pylori DNA fingerprints of 17 of the 18 single colonies matched the parental isolate, and the remaining single-colony fingerprint was highly similar (Fig. 2). In two patients, two single-colony band patterns were highly similar but not identical to the parental isolate DNA fingerprint, with one single colony being very different from the parental isolate DNA fingerprint. Both the 297-bp and the ∼1.4-kb PCR products for the cagA gene were detected in all H. pylori isolates and the respective single colonies, indicating the ubiquitous presence of the cagA gene in this population (data not shown).
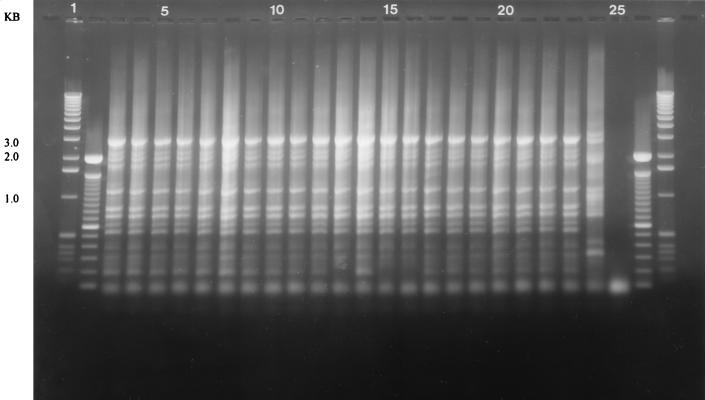
FIG. 1.
DNA fingerprints of parental isolate and single colonies from a single patient showing identical fingerprints from parental and single-colony isolates. Lanes: 1 and 27, 1-kb molecular mass standards; 2 and 26, 100-bp ladder; 3, antral parental isolate; 4 to 9, single colonies from antrum; 10, corpus parental isolate; 11 to 16, single colonies from corpus; 17, cardia parental isolate; 18 to 23, single colonies from cardia; 24, ATCC 49503 reference strain fingerprint; 25, negative control.
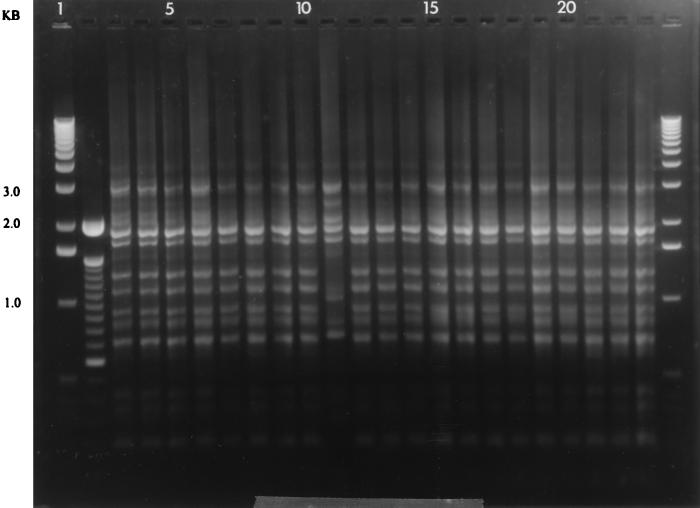
FIG. 2.
DNA fingerprints of parental isolate and single colonies from a single patient showing identical single-colony fingerprints except one single colony from the corpus. Lanes: 1 and 24, 1-kb molecular mass standards; 2, 100-bp ladder; 4, parental isolate from antrum; 5 to 9, single colonies from antrum; 10, corpus parental isolate; 11 to 16, single colonies from corpus; 17, cardia parental isolate; 18 to 23, single colonies from cardia.
Our data suggest that gastric cancer patients from a country with a high prevalence of H. pylori infection appear to be infected with a genetically predominant strain throughout the stomach. Jorgensen et al. utilized randomly amplified polymorphic DNA (RAPD)-PCR and found that the majority of 106 patients were infected with a predominant strain, but multiple patients harbored more than one strain (5). However, this study was limited by the fact that no single colonies were investigated. Similarly, Taylor et al. detected identical DNA patterns produced by restriction enzyme analysis, RAPD-PCR, and PCR-restriction fragment length polymorphism, suggesting infection with a single strain in 56% of patients. However, this analysis was also not extended to single colonies (9).
With respect to H. pylori isolates obtained from different gastroduodenal sites, our data are in agreement with a study by Prewett et al. (8), who identified a single H. pylori strain in biopsy specimens from the antrum, corpus, and duodenum of 13 of 15 patients. Similarly, a study by Cellini et al. included antrum and corpus biopsy specimens from 32 H. pylori-infected patients and identified a single predominant strain infection in 27 patients (2).
The present study is unique in that H. pylori from the cardia region in addition to that from the gastric antrum and corpus was cultured, and in that six single colonies were subcultured from the parental isolate of each gastric site. Our finding of a homogeneous H. pylori population throughout the stomach may be explained by the following possibilities. (i) An individual patient may be exposed and colonized by one unique strain. (ii) A patient may have been infected with multiple strains, with one strain becoming dominant over time. Or (iii) the presence of a particular strain may inhibit or prevent colonization by different strains upon sequential exposure. In our study, all single colonies of H. pylori and the respective parental isolates possessed the cagA gene, which is considered a marker for the presence of the so-called pathogenicity island. The presence of two different regions of the cagA gene further supports the homogeneity in these isolates. This is in contrast to the observation by van der Ende et al. that three of eight family members from whom multiple single colonies were obtained contained both cagA-positive and cagA-negative H. pylori strains (10).
From our data, we conclude that REP-PCR can be used to generate reproducible and stable DNA fingerprints of all H. pylori strains we examined. Individual strains (single colonies) of the H. pylori population from individual patients almost always yield DNA fingerprints identical or highly similar to that of the parent isolate. H. pylori-positive individuals are infected with a single major genotype. If a single colony is selected from an individual patient for molecular analysis, it is possible but unlikely to select a single colony not representative of the H. pylori population infecting that individual host.
Acknowledgments
We thank Susan M. Small for her meticulous technical assistance in the culture and genetic characterization of these H. pylori isolates.
The research was supported by the NIH National Cancer Institute grant R01CA67469.
REFERENCES
- 1.Akyopyants N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cellini L, Allocati N, Di Campeli E, Masuli M, Di Bartolomeo S, Danielli B. Helicobacter pylori isolated from stomach corpus and antrum: comparison of DNA patterns. J Infect. 1996;32:219–221. doi: 10.1016/s0163-4453(96)80022-3. [DOI] [PubMed] [Google Scholar]
- 3.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Go M F, Chan K Y, Versalovic J, Koeuth T, Graham D Y, Lupski J R. Cluster analysis of Helicobacter pylori genomic DNA fingerprints suggests gastroduodenal disease-specific associations. Scand J Gastroenterol. 1995;30:640–646. doi: 10.3109/00365529509096306. [DOI] [PubMed] [Google Scholar]
- 5.Jorgensen M, Daskalopoulos G, Waburton V, Mitchel H M, Hazell S L. Multiple strain colonization and metronidazole resistance in Helicobacter pylori-infected patients: identification from sequential and multiple biopsy specimens. J Infect Dis. 1996;174:631–635. doi: 10.1093/infdis/174.3.631. [DOI] [PubMed] [Google Scholar]
- 6.Miehlke S, Kibler K, Kim J G, Figura N, Small S M, Graham D Y, Go M F. Allelic variation in Helicobacter pylori cagA gene from Korea compared to the United States. Am J Gastroenterol. 1996;91:1322–1325. [PubMed] [Google Scholar]
- 7.Owen R J, Bickley J, Costas M, Morgan D R. Genomic variation in Helicobacter pylori: application for identification of strains. Scand J Gastroenterol. 1991;181:43–50. doi: 10.3109/00365529109093207. [DOI] [PubMed] [Google Scholar]
- 8.Prewett E J, Bickley J, Owen R J, Pounder R E. DNA patterns of Helicobacter pylori isolated from the antrum, body and duodenum. Gastroenterology. 1992;102:829–833. doi: 10.1016/0016-5085(92)90165-u. [DOI] [PubMed] [Google Scholar]
- 9.Taylor N S, Fox J G, Akopyants N S, Berg D E, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter F M, Correa P. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol. 1995;33:918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Ende A, Rauws E A J, Feller M, Mulder C J J, Tytgat G N J, Dankert J. Heterogeneous Helicobacter pylori isolates from members of a family with a history of peptic ulcer disease. Gastroenterology. 1996;111:638–647. doi: 10.1053/gast.1996.v111.pm8780568. [DOI] [PubMed] [Google Scholar]
- 11.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]