Abstract
Deep learning (DL) is a relatively newer subdomain of machine learning (ML) with incredible potential for certain applications in the medical field. Given recent advances in its use in neuro-oncology, its role in diagnosing, prognosticating, and managing the care of cancer patients has been the subject of many research studies. The gamut of studies has shown that the landscape of algorithmic methods is constantly improving with each iteration from its inception. With the increase in the availability of high-quality data, more training sets will allow for higher fidelity models. However, logistical and ethical concerns over a prospective trial comparing prognostic abilities of DL and physicians severely limit the ability of this technology to be widely adopted. One of the medical tenets is judgment, a facet of medical decision making in DL that is often missing because of its inherent nature as a “black box.” A natural distrust for newer technology, combined with a lack of autonomy that is normally expected in our current medical practices, is just one of several important limitations in implementation. In our review, we will first define and outline the different types of artificial intelligence (AI) as well as the role of AI in the current advances of clinical medicine. We briefly highlight several of the salient studies using different methods of DL in the realm of neuroradiology and summarize the key findings and challenges faced when using this nascent technology, particularly ethical challenges that could be faced by users of DL.
Keywords: Deep learning, Glioma prognostication, Machine learning, Neuro-oncology

INTRODUCTION
Amidst contemporary shifts in the global “smart-tech” arena, there has been a parallel shift in the dynamics of technological integration in health care and its rapidly growing role in supplementing traditional methods of patient care.[36] An integral aspect of this global movement is the marriage of artificial intelligence (AI) with vast databases as a tool to assist physicians to not only diagnose but also subsequently manage the ever-growing burden of disease.
The realm of AI contains within itself machine learning (ML), as well as a further subset known as deep learning (DL). ML takes use of statistical models to enable these algorithms to improve as more data are introduced to them [Figure 1].[28] DL is also a process by which computers glean patterns from past experiences and large amounts of data to generate algorithms designed to make increasingly accurate predictions about future events.[3,5] What further specializes it from ML is the multilayered neural networks used to train on vast amounts of data. Neural networks much like the brain use nodes of information (neurons) that are arranged in layers. Each neuron uses data from the training set (or previous neuronal connections) and calculates a new piece of datum using weightage and bias factors before communicating the new piece to neuron(s) in the next layer, eventually generating an outcome.[39]
Figure 1:
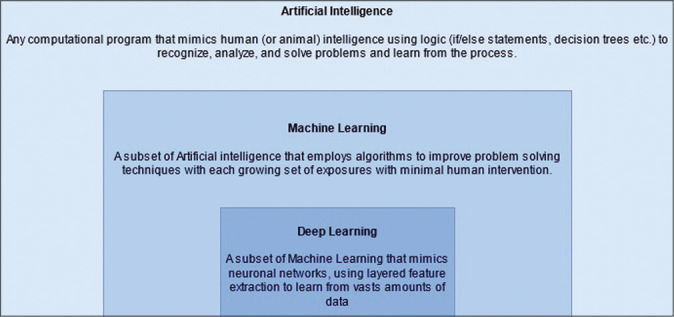
The subsets of artificial intelligence.
DL algorithms may then function when provided sets of parameters to evaluate a plethora of aspects of clinical importance in medicine, including screening, diagnosis, and treatment efficacy. Each algorithm must first be assessed by the relevant performance metrics of the extracted patterns when extrapolating beyond the model dataset.[39] In practice, however, the presence of unknown confounders and imprecise measurements endows a random component to our estimates that varies from individual to individual and population to population.[5] As a result, the diversity of a training dataset will enable the DL algorithm to learn “better,” which, in turn, would allow for greater and more valid generalizability of predicted patterns on a wider range of populations.[5,39]
Consequently, this generalizability requires a revolution in data mining to generate large volumes of data from which this subset of AI technology can learn. This is vital to overcome the challenges of data sparsity, multicollinearity, and overfitting, and therefore, to effectively minimize internal bias in the model developed.[38]
Based on a subtle balance between use and limitations, DL promises to play a crucial role in healthcare, allowing us to find a niche between generalized practice guidelines and personalized patient-centric care. These algorithms have already shown a high level of diagnostic performance based on imaging data for conditions including diabetic retinopathy, skin cancer, and pneumonia.[36] Salim et al., for example, have recently commented on the rates of diagnostic capabilities of AI (92% area under the receiver operating curve [AUROC] for three different commercially available computer-aided detection [CADe] algorithms) in assessments of screening mammograms, with one algorithm demonstrating comparable sensitivity (81.9%) to those of first-reader (77.4%) and second-reader (80.1%) radiologists given that the AI and radiologists’ readings were equally probable to rule in the disease (at the same specificity).[30] Other applications include but are not limited to the ability to predict genomic features such as clinically relevant mutations, to predict and build models of regulatory and enhancing elements in the DNA replication and transcription, and to predict patterns of disease based on population statistics such as infectious disease epidemics and pandemics.[38]
Amidst these enhancements in health care across disciplines, the realm of prediction and management in oncology stands to benefit greatly from such technologies.[26] In 2014, Louis et al. published an update on behalf of the International Society of Neuropathology stating that non-tissue-based information (e.g. clinical and radiological information), could be of clear utility to aid in reaching a diagnosis. This support from an international body has led neuroradiology to play an increasingly important role in the potential treatment of neuro-oncological patients, with promises of AI-enhanced readings further elevating the importance of neuroradiology.[24] As such, our paper, as summarized in [Figure 2], aims to assess the state of DL algorithms in their efficacy and precision of pre-, peri-, and post-operative decision-making in neuro-oncology, using gliomas as an oncologic model.
Figure 2:
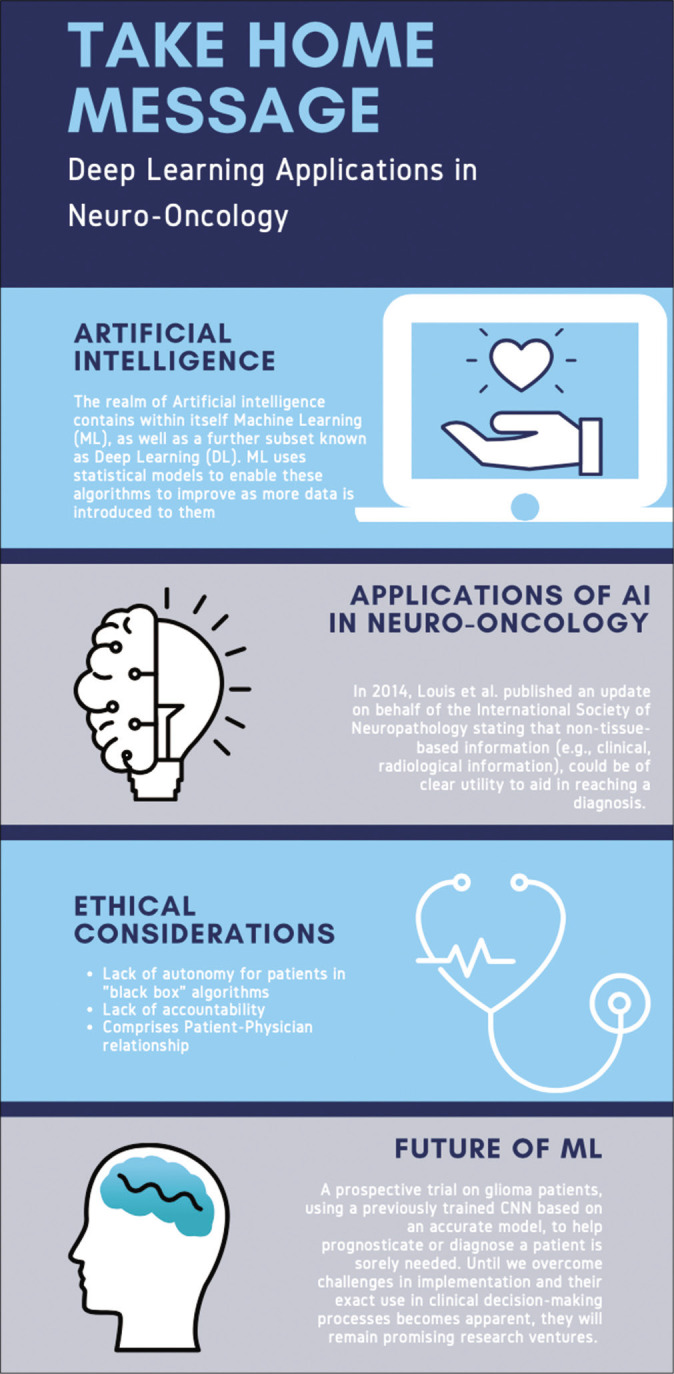
The take-home message.
DIAGNOSIS, GRADING, AND CLASSIFICATION OF GLIOMAS
The clinical course of a neoplasm is dependent on the efficacy of several stages of medical intervention including the recognition of clinical manifestations, initial screening of the lesion (employing imaging or laboratory testing), the decision to observe or treat, postscreening histopathological assessment/gold standard confirmation, and predicting morbidity and mortality outcomes. ML can be used to aid this paradigm of medical intervention at several of these levels [Figure 3].[28]
Figure 3:
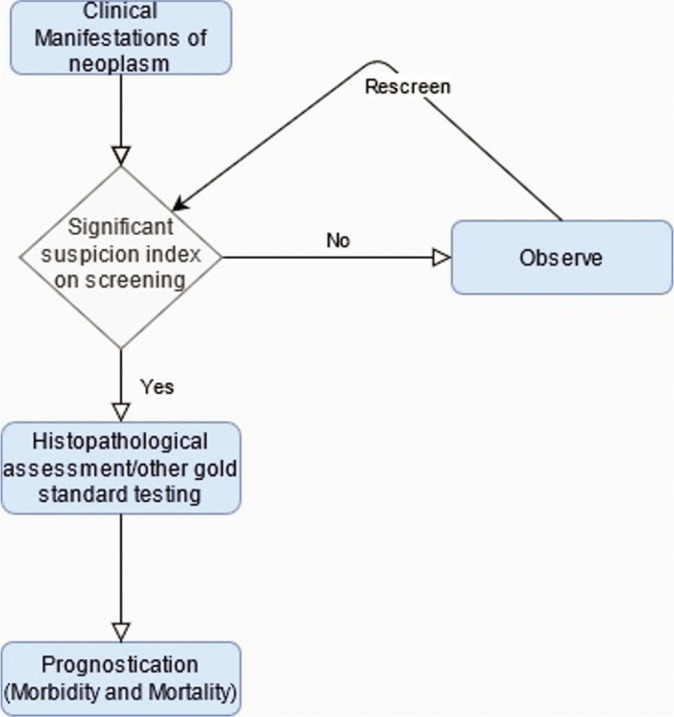
Levels of entry for deep learning applications in neuro-oncology.
As mentioned before, the strength of ML as a tool to evaluate radiological screening independently or augmentatively is yielding promising results, with this excitement not only limited to mammography but also in the realms of lung cancer, prostate cancer, and brain metastases with the use of CADe and computer-aided diagnoses techniques.[2,7,35] Prospectively, ML may be able to correlate radiological imaging detailing the extent of disease with genomic data of the lesion such as mutations and gene expressions and histopathological findings to establish an accurate diagnosis in an emerging field known as imaging genomics.[4]
The promise of ML in the field of oncology in addition to the growing recognition of an integrated approach to neuropathology lends optimism to the treatment of a subset of neuro-oncological entities known as gliomas. These tumors are difficult to render an exact judgment on, due to similarity of cellular structures among lower grade lesions; with estimations varying due to inter-reader variability depending on the professional experience of histopathologists and radiologists, introducing an element of subjectivity.[37] Although the simple grading of these tumors is not a principle bottleneck in their treatment paradigm, some difficulties remain in the presence of standardization efforts, such as the use of objective classification criteria, the most prevalent of which is the WHO classification.[34] Furthermore, although histopathological and molecular analysis remains the gold standard for the diagnosis of gliomas, the very notion of a biopsy raises a few concerns. There are many inherent risks with this method of diagnosis, with inevitable errors due to inadequate sampling and time inefficiency.[43] Thus, the need for elimination of subjectivity and improvement of sensitivity and timeliness of histopathological diagnosis as well as the growing logistical feasibility of ML imparts a growing confidence in the use of such technology in the diagnosis of gliomas.
Broadly, ML techniques can be divided into those that learn to predict outcomes based on input-output paired training, termed supervised learning, or those that find patterns within input data itself without preset outcomes, termed unsupervised learning. Supervised learning itself can be achieved through several techniques such as support vector matrixes (SVMs) which aim to delineate a function that separates two sets of data.[33]
Previously, studies have demonstrated that gliomas can be classified according to their clinical grade using linear SVMs which were trained on descriptive features such as amount of mass effect of blood supply. The limitation of these studies, such as one by Li et al., is that these quantitative features were estimated by domain experts and the definition of these features was limited to expert opinion and hence not reproducible.[23] Two other studies subsequently used other parametric features to improve accuracy; Devos et al. used MR intensities with spectroscopy using linear discriminant analysis, linear and nonlinear least squares SVMs (AUROC of 0.94 in distinguishing low-grade vs. high-grade gliomas),[11] whereas Rajendran made use of association rule mining (specifically pruned association rule), a technique in ML that aims to predict certain characteristics of a set based on other characteristics, to categorize CT scan brain images as simply either normal, benign, or malignant with a 96% sensitivity and 93% accuracy.[29]
It stands to reason from this discussion that the beginnings of ML in neuropathology must be the generation of a dataset from which newly developed AI can learn. For gliomas, radiological evidence in addition to matched clinical, genetic, and pathological data collections reside in the Cancer Imaging Archive and Genomics Data Commons Data Portal respectively, as a joint effort between the National Cancer Institute and the National Human Genome Research Institute from 2006 onward.[13] Since 2012, ML and DL techniques applied to carefully selected data subsets from these archives have already generated numerous algorithms related to tumor segmentation (delineating the location and extension of a lesion from surrounding tissue) under the brain tumor segmentation challenge.
At present, algorithms generated by ML have shown promise in accurate recognition of characteristics used to define gliomas. Noninvasive neuroimaging tools for glioma grading using a multitude of quantitative parameters obtained from advanced magnetic resonance imaging (MRI) techniques, such as dynamic contrast-enhanced MRI, arterial spin labeling, and diffusion-weighted imaging, are currently being utilized in many ML/DL models.[40] Skogen et al. were able to discriminate high-grade gliomas from low-grade gliomas using MRI texture analysis with a fine texture scale resulting in a sensitivity and specificity of 93% and 81%, out of a total of 95 glioma patients, but fared worse at differentiating within those grades.[32]
The emerging field of radiogenomics breaks the restrictions of applying AI technology to segmentation only. Advancement in our understanding of key molecular differences in tumor cells in comparison to normal cells and how these differences drive changes in the microenvironment of the lesions now lends itself to radiological patterns that can more accurately identify a tumor. For example, certain glioma varieties exhibit differences in isocitrate dehydrogenase (IDH), a key enzyme in the Krebs cycle. Gliomas that potentiate the activity of certain genotypes can drive the production of 2-hydroxyglutaric acid, vascular endothelial growth factor, and hypoxia-induced factor-1a. The activity of these molecules may, in turn, produce characteristic radiological findings such as alterations in histogram analysis.[21]
In fact, the current literature supports the idea of using radiological imaging to predict the presence of such biomolecular differences. Specifically in the context of gliomas, studies have been conducted on IDH mutations, O6-methylguanine methyltransferase (MGMT) hypermethylation, epidermal growth factor receptors (EGFRs), and 1p/19q chromosomal codeletions.
Zhang et al. were able to generate a predictive model for IDH genotypes (of which genotypes 1 and 2 predict good response to therapy) within high-grade glioma lesions based on clinical data and radiological features obtained through conventional MRI with an AUROC of 0.9231 in a validation cohort.[41] The radiological features considered included anatomical location, texture, and shape, among others. An analogous non-AI-assisted study conducted by Zhou et al. was also able to generate a similar algorithm to predict IDH1 mutation (AUROC 0.86 ± 0.01), 1p/19q codeletion status (conferring increased survival) (AUROC of 0.96 ± 0.01), and histological grade (AUROC 0.86 ± 0.01).[42] In contrast, a convolution neural network (CNN), a form of DL especially suited to image recognition, developed by Chang et al. demonstrated a high degree of accuracy in predicting IDH genotypes from MRI features in Grade II–IV gliomas in a validation test (AUROC 0.95).[8] Chang et al. also developed a neural network predictive of several factors, of which its accuracy in predicting IDH1 mutation status was 94%.[9]
Similarly to IDH, MGMT hypermethylation is an important identifying marker due that also happens to predict higher response to treatment with combination temozolomide and radiation.[17] Korfiatis et al. evaluated three residual deep neural network architectures in their ability to predict the MGMT methylation status. The three 18, 34, and 50 layered architectures were able to achieve an accuracy of 76.75% (±20.67%), 80.72% (±13.61%), and 94.90% (±3.92%), respectively, lending credibility to higher-order processing providing better results.[22] Similarly, Han and Kamdar obtained a 67% accuracy on their validation set exploring MGMT methylation status from MRI scans obtained from glioblastoma multiforme (GBM) (Stage IV glioma) patients in the TCIA/ TCGA archives.[16] Chang et al. developed a neural network from data for 259 patients from the TCIA/TCGA archives, showing 83% accuracy in predicting methylation status.[9]
Other studies have also shown the ability of ML to detect 1p19q chromosomal codeletion (recognized as predictor of chemotherapeutic response in gliomas), and EGFR amplification status, another avenue for targeted therapy. The aforementioned neural network developed by Chang et al. manages to predict 1p19q codeletion status at an accuracy of 92%.[9] A multiscale CNN developed by Akkus et al. similarly predicts the codeletion at an 87.7% accuracy.[1] Kickingereder et al. set out to use ML techniques to identify a myriad of molecular characteristics including MGMT hypermethylation and hallmark copy number variations. Of these, they managed to develop a model that predicted EGFR with a 63% accuracy.[21]
With such exciting advancements, comparable outputs to traditional methods of algorithmic generation, and a yet unmet ceiling for improvement, it is thought that in the near future, with the increasing availability of high-quality data, CNNs will be the mainstay in both diagnosing and prognosticating cancer, using widely available imaging modalities.
PROGNOSIS PREDICTION
While ML can be useful in predicting prognosis based just on imaging characteristics before surgery, we have more information after surgical resection of the tumor that can be incorporated in making extensive ML algorithms. Along with imaging characteristics, the cumulative biopsy and clinical data such as tumor markers can help generate ML algorithms for estimating prognosis, complications, and other outcomes. It can also overcome challenges such as the correct recognition of tumor progression versus pseudoprogression, a transient MRI finding mimicking progression with eventual improvement. One such study aiming to delineate the two conducted by Jang et al. developed a CNN model with an AUROC of 83% that was trained on 59 individuals and tested on 19 (Seoul National University Hospital dataset).[19] Repeat model development by Jang et al. on the Korean Radiation Oncology Data (n = 182) yielded a model with an AUROC of 86%.[20] The necessity to develop a second model shall be discussed in the section on ML and challenges.
Conventionally, prognosticating for GBM involves accounting for a multitude of independent risk factors impacting overall survival (OS) such as male gender, age >60 years), poor preoperative Karnofsky scores of <70, Caucasian ethnicity, advanced tumor with partial resection, and surgery without adjuvant chemoradiation.[35] Using several of these factors, Gittleman et al. developed an independently validated nomogram with a concordance index (equivalent to AUROC for the censored data used in the study) of 0.657.[14] Notably, however, this study was conducted on a Caucasian majority sample and utilizes only MGMT hypermethylation as a prognostic factor of the molecular characteristics seen in gliomas.
A study by Nie et al. demonstrated that through a 3D CNN, they were able to train a support vector machine to predict a long or short OS time. Their experimental results were able to achieve 89.9% accuracy through their methods.[27] They were able to feed single-channel (T1 MRI) multi-channel (fMRI and DTI) data through a binary classifier (e.g. SVM) to produce a framework for high-level brain tumor prognosis. The study elucidates the incredible potential application that a neural network possesses in prognosticating neurooncological patients based on just imaging alone. Similarly, a study by Sanghani et al. was able to produce comparable results on GBM patients based on routinely acquired MR images alone. MR image derived texture features, tumor shape, and volumetric features, and patient age was obtained for 163 patients, which after put through a similar SVM, was capable of producing 2-class and 3-class OS group prediction accuracy of 98.7% and 88.95%, respectively.[31]
Another study by Zhou et al. utilized nonquantitative spatial-correlated features from MRI defined tumor subregions (termed habitats) in developing a computational framework. This framework was able to use intratumoral grouping and spatial mapping to identify GBM tumor subregions and yield habitat-based features. After separating data sets into those that underwent resection with GBM (32 patients), and those that did not (22 patients), they were able to achieve 87.50% and 86.36% accuracies for survival group prediction, respectively.[44]
The role of computational networks in prognosticating overall long- and short-term survival is one that has been evaluated through dozens of studies in low- to mid-grade tumors, yielding similar results when applied to high-grade tumors as well.[14,19,20,27,31,35,44] These networks are capable of functioning on routinely acquired data, as demonstrated by Sanghani et al., but also provide considerable information to both the health-care provider and the patient. Eventually, these systems are thought to be able to augment a neurooncologist’s decision-making ability, however, it would not be long before they are potentially able to surpass human performance in terms of prognostication.
CHALLENGES AND LIMITATIONS
Ethical considerations
ML, like any early adopted technology, is not without its potential drawbacks in terms of ethical dilemmas.[10] ML applied in other fields has already shown some biases, with the problem stemming from the fact that algorithms may mirror human biases. These biases may be introduced into a medical setting where patients receive notably different treatment regimes leading to an unoptimized approach and potential harm.
There is an incredibly urgent need for ethical guidelines for medical practitioners to use these advancing AI and machine learning technologies. Time-constrained consultants are expected to understand the inner workings of these insular programs, as without knowing the data sets they are built on, the technology can progress into a black box leading to ethically gray outcomes. The lack of transparency inherent with these systems may lead to physicians doubt either the algorithm, or themselves, when their decision is conflicting with one made by a machine.
It is also important to recall that the foundation of the current medical system in many nations was based on a patient-doctor relationship, not a patient-health-care system relationship. Many doctors (and patients) feel uneasy at the growing rate at which we are adopting these machines into our health-care decisions. One study showed that over 63% of adults in the United Kingdom felt uncomfortable with AI replacing the conventional decision-making process.[12] Their lack of acceptance of AI is, to some degree, warranted. In many fields, such as engineering, AI and ML represent efficiency and will play an important role in the future. However, one of the basic ethical tenets of medicine is autonomy, which requires physicians to have a deeper understanding of the clinical validation of ML in studies. Health-care professionals operate on their own basis of expertise and clinical acumen. When their decision-making skills are infringed upon, by blindly accepting a prognosis, diagnosis, or route of treatment provided by an AI or algorithm, they could be seen as violating their Hippocratic oath: primum non nocere (“first, do no harm”).[10] It is this lack of transparency and explainability that is vital in the role of medicine that limits its use when compared to its role in business or engineering. Autonomy and nonmaleficence are ethical principles grounded in “consequentialist” ethical theory, which holds that the moral quality of an action depends on its consequences. If the consequences of choosing an AI favored diagnosis over a physician diagnosis, whether for a high- or low-grade tumor, have the possibility (however minute as shown by the accurate results of ML) of resulting in an increased mortality – should we still use this technology?[12]
Finally, the laws and rules of ethics are a worldwide debate but enforced by national laws. Ethical laws on the potential of AI and ML in medicine have been hotly contentious but are virtually absent in low- and middle-income countries.[15] This is not based on a lack of acceptance of AI/ML, as most health-care professionals are in favor of their use (despite ethical dilemmas), rather it is contingent on lack of infrastructure, high cost, and lack of training in the region.[18]
Another reason for the impediment in widespread adoption, and perhaps the most important one, is a lack of accountability.[12] Allocation and grounds for liability for adverse events related to the ML tools used with actual patients need to be clarified by a national/international ethical body. There is currently no conceivable method to predict how these tools, along with other ML models in medicine, will expose hospitals, physicians, and patients to liability.
Other logistical drawbacks to the use of AI in medicine include the scale of computational power required to develop useful algorithms and prioritization of resource allocation toward more pragmatic algorithms to maximize their effects.
Specifically in the field of radiological AI interpretations, a major drawback in considering findings and pattern recognition in any image modality is a lack of consensus on which features would add most value to a preoperative diagnosis, whether from a histogram parameters or image texture attributes.[6] Attribute selection is a concept that aims to remedy this challenge. It revolves around choosing a division with the chosen least number of variables providing the highest accuracy, alongside the most informative attributes. The rigorous selection process searches through possible combinations of attributes to find a particular subset that can reliably predict outcomes, resulting in an algorithm that is characterized by the method used to define the predictive value of each subset of attributes (feature evaluator) and the method determining the search over the attributes (search method).[40]
A prospective trial on glioma patients, using a previously trained CNN based on an accurate model, to help prognosticate or diagnose a patient is sorely needed. Until, we overcome challenges in implementation and their exact use in clinical decision-making processes becomes apparent, they will remain promising research ventures.
CONCLUSION
The use of AI to augment the clinical judgment and practice of a physician is an exciting prospect, specifically in the field of neuro-oncology where neuroradiology and neuroradiogenomics are prime candidates for the use of the algorithmic learning in order. In a noninvasive manner, these tools can predict the presence of several factors that can be helpful in the diagnosis and prognostication of gliomas. Current literature already demonstrates a high sensitivity and accuracy of individual learning techniques in determining genetic markers and radiological features. The challenge lies in creating algorithms that are applicable on much larger scale, with greater amounts of learning and practice sets for AI learning techniques to be tested on, while still remaining logistically feasible to be run within the strict timeframes within which health-care systems operate. In addition, there are ethical considerations to be made regarding the use of AI within the clinical realm and their impact on a physician’s decision-making.
Footnotes
How to cite this article: Khan AA, Ibad H, Ahmed KS, Hoodbhoy Z, Shamim SM. Deep learning applications in neuro-oncology. Surg Neurol Int 2021;12:435.
Contributor Information
Adnan A. Khan, Email: adnan@khan.health.
Hamza Ibad, Email: hamzaibad5@gmail.com.
Kaleem Sohail Ahmed, Email: kaleem.s.ahmed@gmail.com.
Zahra Hoodbhoy, Email: zahra.hoodbhoy@aku.edu.
Shahzad M. Shamim, Email: shahzad.shamim@aku.edu.
Declaration of patient consent
Patient’s consent not required as there are no patients in this study.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Akkus Z, Ali I, Sedlář J, Agrawal JP, Parney IF, Giannini C, et al. Predicting deletion of chromosomal arms 1p/19q in low-grade gliomas from MR images using machine intelligence. J Digit Imaging. 2017;30:469–76. doi: 10.1007/s10278-017-9984-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrosini RD, Wang P, O’Dell WG. Computer-aided detection of metastatic brain tumors using automated three-dimensional template matching. J Magn Reson Imaging. 2010;31:85–93. doi: 10.1002/jmri.22009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baştanlar Y, Özuysal M. Introduction to machine learning. Methods Mol Biol. 2014;1107:105–28. doi: 10.1007/978-1-62703-748-8_7. [DOI] [PubMed] [Google Scholar]
- 4.Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin. 2019;69:127–57. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bzdok D, Krzywinski M, Altman N. Points of significance: Machine learning: A primer. Nat Methods. 2017;14:1119–20. doi: 10.1038/nmeth.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caruana R, Caruana R, Freitag D. Proceedings of the elevator International Conference Machine Learning. 1994. Greedy attribute selection; pp. 28–36. [Google Scholar]
- 7.Chan HP, Hadjiiski L, Zhou C, Sahiner B. Computer-aided diagnosis of lung cancer and pulmonary embolism in computed tomography-a review. Acad Radiol. 2008;15:535–55. doi: 10.1016/j.acra.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, et al. Residual convolutional neural network for the determination of IDH status in low-and high-grade gliomas from mr imaging. Clin Cancer Res. 2018;24:1073–81. doi: 10.1158/1078-0432.CCR-17-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol. 2018;39:1201–7. doi: 10.3174/ajnr.A5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Char DS, Shah NH, Magnus D. Implementing machine learning in health care-addressing ethical challenges. N Engl J Med. 2018;378:981–3. doi: 10.1056/NEJMp1714229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devos A, Simonetti AW, van der Graaf M, Lukas L, Suykens JA, Vanhamme L, et al. The use of multivariate MR imaging intensities versus metabolic data from MR spectroscopic imaging for brain tumour classification. J Magn Reson. 2005;173:218–28. doi: 10.1016/j.jmr.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Fenech M, Strukelj N, Buston O. Ethical Social and Political Challenges of Artificial Intelligence in Health. 2018.
- 13.Freymann J. TCGA Glioma Phenotype Research Group-The Cancer Imaging Archive (TCIA) Public Access-Cancer Imaging Archive Wiki. 2021. Available from: https://www.wiki.cancerimagingarchive.net/display/public/tcga+glioma+phenotype+research+group [Last accessed on 2021 Apr 25]
- 14.Gittleman H, Cioffi G, Chunduru P, Molinaro AM, Berger MS, Sloan AE, et al. An independently validated nomogram for isocitrate dehydrogenase-wild-type glioblastoma patient survival. Neurooncol Adv. 2019;1:vdz007. doi: 10.1093/noajnl/vdz007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J, Li B. The application of medical artificial intelligence technology in rural areas of developing countries. Health Equity. 2018;2:174–81. doi: 10.1089/heq.2018.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han L, Kamdar MR. MRI to MGMT: Predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Pac Symp Biocomput. 2018;23:331–42. [PMC free article] [PubMed] [Google Scholar]
- 17.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 18.Hoodbhoy Z, Hasan B, Siddiqui K. Does artificial intelligence have any role in healthcare in low resource settings? J Med Artif Intell. 2019;2:13. [Google Scholar]
- 19.Jang BS, Jeon SH, Kim IH, Kim IA. Prediction of pseudoprogression versus progression using machine learning algorithm in glioblastoma. Sci Rep. 2018;8:12516. doi: 10.1038/s41598-018-31007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang BS, Park AJ, Jeon SH, Kim IH, Lim DH, Park SH, et al. Machine learning model to predict pseudoprogression versus progression in glioblastoma using mri: A multi-institutional study (KROG 18-07) Cancers (Basel) 2020;12:2706. doi: 10.3390/cancers12092706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kickingereder P, Bonekamp D, Nowosielski M, Kratz A, Sill M, Burth S, et al. Radiogenomics of glioblastoma: Machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology. 2016;281:907–18. doi: 10.1148/radiol.2016161382. [DOI] [PubMed] [Google Scholar]
- 22.Korfiatis P, Kline TL, Lachance DH, Parney IF, Buckner JC, Erickson BJ. Residual deep convolutional neural network predicts MGMT methylation status. J Digit Imaging. 2017;30:622–8. doi: 10.1007/s10278-017-0009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li GZ, Yang J, Ye CZ, Geng DY. Degree prediction of malignancy in brain glioma using support vector machines. Comput Biol Med. 2006;36:313–25. doi: 10.1016/j.compbiomed.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Louis DN, Perry A, Guido R, von Deimling A, FigarellaBranger D, Webster K, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 25.Machine Bias-ProPublica. 2021. Available from: https://www.propublica.org/article/machine-bias-risk-assessments-incriminal-sentencing [Last accessed on 2021 Apr 25]
- 26.Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20:e262–73. doi: 10.1016/S1470-2045(19)30149-4. [DOI] [PubMed] [Google Scholar]
- 27.Nie D, Zhang H, Adeli E, Liu L, Shen D. Vol. 9901. Berlin, Germany: Springer; 2016. 3D deep learning for multi-modal imaging-guided survival time prediction of brain tumor patients, Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) pp. 212–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panch T, Szolovits P, Atun R. Artificial intelligence, machine learning and health systems. J Glob Health. 2018;8:020303. doi: 10.7189/jogh.08.020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajendran P, Madheswaran M. An improved image mining technique for brain tumour classification using efficient classifier. Int J Comput Sci Inf Secur. 2010;6:1–10. [Google Scholar]
- 30.Salim M, Wåhlin E, Dembrower K, Azavedo E, Foukakis T, Liu Y, et al. External evaluation of 3 commercial artificial intelligence algorithms for independent assessment of screening mammograms. JAMA Oncol. 2020;6:1581–8. doi: 10.1001/jamaoncol.2020.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanghani P, Ang BT, King NK, Ren H. Overall survival prediction in glioblastoma multiforme patients from volumetric, shape and texture features using machine learning. Surg Oncol. 2018;27:709–14. doi: 10.1016/j.suronc.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Skogen K, Schulz A, Dormagen JB, Ganeshan B, Helseth E, Server A. Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol. 2016;85:824–9. doi: 10.1016/j.ejrad.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Sotoudeh H, Shafaat O, Bernstock JD, Brooks MD, Elsayed GA, Chen JA, et al. Artificial intelligence in the management of glioma: Era of personalized medicine. Front Oncol. 2019;9:768. doi: 10.3389/fonc.2019.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valdebenito J, Medina F. Machine learning approaches to study glioblastoma: A review of the last decade of applications. Cancer Rep. 2019;2:e1226. doi: 10.1002/cnr2.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Hu G, Quan X. Analysis of the factors affecting the prognosis of glioma patients. Open Med. 2019;14:331–5. doi: 10.1515/med-2019-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson DS, Krutzinna J, Bruce IN, Griffiths CE, McInnes IB, Barnes MR, et al. Clinical applications of machine learning algorithms: Beyond the black box. BMJ. 2019;364:l886. doi: 10.1136/bmj.l886. [DOI] [PubMed] [Google Scholar]
- 37.Whittle IR. The dilemma of low grade glioma. J Neurol Neurosurg Psychiatry. 2004;75(Suppl 2):ii31–6. doi: 10.1136/jnnp.2004.040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu C, Jackson SA. Machine learning and complex biological data. Genome Biol. 2019;20:76. doi: 10.1186/s13059-019-1689-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang S, Zhu F, Ling X, Liu Q, Zhao P. Intelligent health care: Applications of deep learning in computational medicine. Front Genet. 2021;12:607471. doi: 10.3389/fgene.2021.607471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zacharaki EI, Kanas VG, Davatzikos C. Investigating machine learning techniques for MRI-based classification of brain neoplasms. Int J Comput Assist Radiol Surg. 2011;6:821–8. doi: 10.1007/s11548-011-0559-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang B, Chang K, Ramkissoon S, Tanguturi S, Bi WL, Reardon DA, et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol. 2017;19:109–17. doi: 10.1093/neuonc/now121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou H, Vallières M, Bai HX, Su C, Tang H, Oldridge D, et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol. 2017;19:862–70. doi: 10.1093/neuonc/now256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou M, Scott J, Chaudhury B, Hall L, Goldgof D, Yeom KW, et al. Radiomics in brain tumor: Image assessment, quantitative feature descriptors, and machine-learning approaches. Am J Neuroradiol. 2018;39:208–16. doi: 10.3174/ajnr.A5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou M, Chaudhury B, Hall LO, Goldgof DB, Gillies RJ, Gatenby RA. Identifying spatial imaging biomarkers of glioblastoma multiforme for survival group prediction. J Magn Reson Imaging. 2017;46:115–23. doi: 10.1002/jmri.25497. [DOI] [PubMed] [Google Scholar]


