Abstract
Aims/hypothesis
ZBED6 (zinc finger, BED-type containing 6) is known to regulate muscle mass by suppression of Igf2 gene transcription. In insulin-producing cell lines, ZBED6 maintains proliferative capacity at the expense of differentiation and beta cell function. The aim was to study the impact of Zbed6 knockout on beta cell function and glucose tolerance in C57BL/6 mice.
Methods
Beta cell area and proliferation were determined in Zbed6 knockout mice using immunohistochemical analysis. Muscle and fat distribution were assessed using micro-computed tomography. Islet gene expression was assessed by RNA sequencing. Effects of a high-fat diet were analysed by glucose tolerance and insulin tolerance tests. ZBED6 was overexpressed in EndoC-βH1 cells and human islet cells using an adenoviral vector. Beta cell cell-cycle analysis, insulin release and mitochondrial function were studied in vitro using propidium iodide staining and flow cytometry, ELISA, the Seahorse technique, and the fluorescent probes JC-1 and MitoSox.
Results
Islets from Zbed6 knockout mice showed lowered expression of the cell cycle gene Pttg1, decreased beta cell proliferation and decreased beta cell area, which occurred independently from ZBED6 effects on Igf2 gene expression. Zbed6 knockout mice, but not wild-type mice, developed glucose intolerance when given a high-fat diet. The high-fat diet Zbed6 knockout islets displayed upregulated expression of oxidative phosphorylation genes and genes associated with beta cell differentiation. In vitro, ZBED6 overexpression resulted in increased EndoC-βH1 cell proliferation and a reduced glucose-stimulated insulin release in human islets. ZBED6 also reduced mitochondrial JC-1 J-aggregate formation, mitochondrial oxygen consumption rates (OCR) and mitochondrial reactive oxygen species (ROS) production, both at basal and palmitate + high glucose-stimulated conditions. ZBED6-induced inhibition of OCR was not rescued by IGF2 addition. ZBED6 reduced levels of the mitochondrial regulator PPAR-γ related coactivator 1 protein (PRC) and bound its promoter/enhancer region. Knockdown of PRC resulted in a lowered OCR.
Conclusions/interpretation
It is concluded that ZBED6 is required for normal beta cell replication and also limits excessive beta cell mitochondrial activation in response to an increased functional demand. ZBED6 may act, at least in part, by restricting PRC-mediated mitochondrial activation/ROS production, which may lead to protection against beta cell dysfunction and glucose intolerance in vivo.
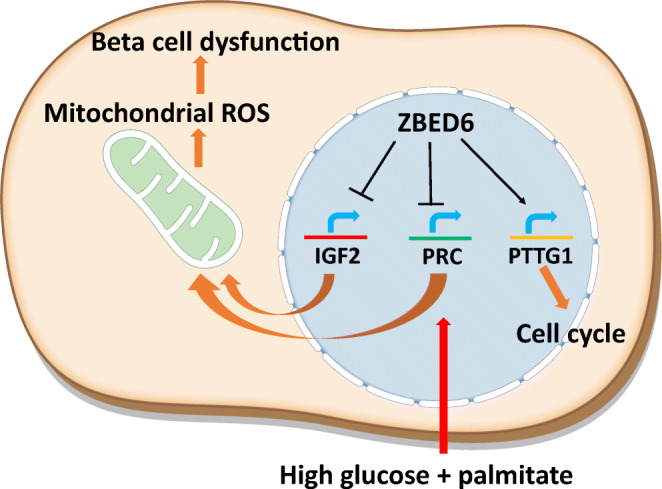
Graphical abstract
Supplementary Information
The online version contains peer-reviewed but unedited supplementary material available at 10.1007/s00125-021-05517-0.
Keywords: Beta cell proliferation, Glucose intolerance, High-fat diet, IGF2, Insulin release, PPAR-gamma related coactivator 1 protein, Pttg1, ZBED6
Introduction
ZBED6 (zinc finger, BED-type containing 6) is a transcriptional modulator that is unique to placental mammals and has evolved from a DNA transposon that integrated in an ancestor of mammals more than 200 million years ago [1]. The fact that the DNA-binding domain of ZBED6 was found to show 100% sequence identity among 25 placental mammals implies that it serves an essential function. ZBED6 acts as a repressor of Igf2 expression in multiple tissues through the binding of the GCTCG motif located in an intron of Igf2 [1, 2]. The ZBED6-mediated lowering of Igf2 expression results not only in decreased muscle mass, but also affects the size of organs such as the liver, kidney and heart [3]. ZBED6 is also expressed in insulin-producing beta cells, where it has been shown to modulate reactive oxygen species (ROS) production, insulin production, proliferation and cell death [4]. In vitro, ZBED6 seems to maintain the capacity of beta cells to proliferate at the expense of specialised function, negatively affecting cell-to-cell interactions [5], stimulus-secretion coupling involving cAMP and Ca2+ signalling events, and neuronal/beta cell differentiation pathways [6]. Thus, it is possible that ZBED6 controls the balance between proliferation and function/differentiation of beta cells, thereby preventing either process from unwanted domination. Indeed, in early stages of the pathogenesis of type 2 diabetes, beta cells are often hyperactivated in response to hyperglycaemia, hyperlipidaemia or inflammatory signals [7], leading to hyperinsulinaemia [8]. Later, as peripheral insulin resistance develops and the insulin requirement augments further, the beta cell mass fails to adapt by proliferation [9], and instead de- or transdifferentiate [10], or even undergo apoptosis [11], leading to a worsened glucose homeostasis. This may suggest that initial activation traps the beta cells in a non-proliferative state, which does not allow expansion of the beta cell mass under conditions of continued and intensified stimulation, subsequently causing beta cell damage [12]. Therefore, ZBED6 may be a factor that modulates the balance between proliferation and function. Consequently, in this study we aimed to evaluate the effects of the transcriptional modulator ZBED6 on beta cell proliferation and function in vivo and in vitro.
Methods
Animals and high-fat diet treatment
The Zbed6−/− and Igf2pA/mG models were generated by homologous recombination in mouse Nnt+/+ C57BL/6 embryonic stem cells as previously described [3], without affecting the host gene Zc3h11a [13]. The two transgenic mouse models were both maintained on a C57BL/6 background and bred to generate Zbed6+/+ Igf2G/G (wild-type [WT] G), Zbed6−/− Igf2G/G (knockout [KO] G), Zbed6+/+ Igf2pA/mG (WT G/A) and Zbed6−/− Igf2pA/mG (KO G/A) mice. The G to A mutation in the ZBED6 binding site in the Igf2 intronic CpG island abolishes ZBED6 binding and was present on the paternal chromosome, as Igf2 is only paternally expressed.
At 5 weeks of age, Zbed6 KO G and WT G mice (both female and male) were divided into two groups with 16 mice in each. One group was given a control diet (CD) and the other was given a high-fat diet (HFD). The HFD (D12492, Research Diets, USA) contained 60% energy from fat, whereas the CD (D12450B, Research Diets) contained only 10% energy from fat.
Ethics permission and animal welfare
All animals were group-housed with free access to food and water in the pathogen-free facilities of Uppsala University and Karolinska Institute. All procedures described in this study were approved by the Uppsala Ethical Committee on Animal Research (#C63/15 and #C143/15) and the Stockholm Ethical Committee (#N38/15), following the rules and regulations of the Swedish Animal Welfare Agency, and were in compliance with the European Communities Council Directive of 22 September 2010 (2010/63/EU). All efforts were made to minimise animal suffering and to reduce the number of animals used.
Blood glucose tolerance test
The mice, after having fasted for approximately 8 h, were given a single i.p. dose of 2.5 g/kg body weight of 30% wt/vol. d-glucose. Blood was drawn from the tail and measured with Freestyle Mini System (Abbott, TheraSense, USA). Blood glucose was determined prior to injection and then at 10, 30, 60 and 120 min after injection.
Insulin sensitivity test
Mice were given an i.p. injection (1.6 U/kg body weight) of the insulin analogue Actrapid (Novo Nordisk, Bagsværd, Denmark). The animals had free access to food before the insulin injection and were transferred to new cages without food during the measurements.
Micro-computed tomography
After 10 weeks on a CD or HFD (15–17 weeks of age) the mice were scanned by micro-computed tomography (micro-CT) (Bruker, USA) directly after euthanasia. The scanning protocol was set using voltage 50 kV, current 500 μA and exposure time 60 ms. We scanned at 36 μm with filter Al 0.5 mm, rotation step 0.7, and two connected scans with a total duration time of 3 min 30 s. The amount of abdominal fat was analysed by reconstructing the whole scan in NRecon. In CTAn the volume of interest was determined by the start and end points of the lumber vertebrae. The amount of fat and muscle between vertebrae L6–L4 was analysed.
Immunohistochemistry
Pancreatic sections, 5 μm thick, were immunostained as previously described [4]. Sections were incubated with primary antibodies (guinea pig anti-human insulin, [1:3000; Reactionlab, USA], anti-glucagon and anti-Ki-67 [1:250, Abcam, UK]) overnight at 4°C. The secondary antibodies (highly cross-adsorbed Alexa Fluor 633 goat anti-guinea pig [1:500; Life Technologies, USA], highly cross-adsorbed Alexa Fluor 555 goat anti-rabbit [1:500; Life Technologies], Alexa Fluor 647 donkey anti-rabbit [1:200, Jackson ImmunoResearch Laboratories, UK]) were added for 1 h. The total pancreas area for each section was measured using ImageJ and a mean for all sections for each mouse, expressed in arbitrary units, was calculated. Beta cell (and alpha cell) mass was semi-quantified similarly and expressed as a ratio to the total pancreas area obtained for the same section. Number of Ki-67-positive beta and alpha cells were counted and expressed as ratio to total number of beta and alpha cells in the same section. At least 30 islets per pancreas, located at three different regions of the pancreas, were analysed.
Quantitative real-time PCR analysis
Pancreatic islets from WT G and Zbed6 KO G C57BL/6 mice were prepared by collagenase digestion and cultured overnight as previously described [14]. RNA was purified from islet cells using the Qiagen RNeasy kit (Qiagen, Hilden, Germany), and cDNA was synthesised using avian myeloblastosis virus (AMV) reverse transcriptase (Bio-Rad, Hercules, CA, USA). Real-time q-PCR was performed using the Roche Light Cycler System and the FastStart DNA Master DNA SYBR Green I kit (Roche Diagnostics, Mannheim, Germany). Values were normalised to the relative amounts of GAPDH cDNA. Primer sequences can be found in electronic supplementary material (ESM Table 1).
Whole transcriptome analysis
Strand-specific mRNA sequencing libraries were generated using the QuantSeq 3′ mRNA-seq library prep kit (Lexogen, Austria). 100 ng of total RNA was poly-A selected using magnetic beads. The libraries were sequenced as 50 bp single reads using an Illumina HiSeq instrument (Illumina, USA). Sequence reads were mapped to the reference mouse genome (mm10) using STAR 2.5.1b. HTSeq-0.6.1 package was used to generate read counts and edgeR package was used to analyse differentially expressed genes. The gene expression was normalised by trimmed mean of M-values method [15] and the abundance of gene expression was calculated as count-per-million reads. For gene ontology analysis, the differentially expressed genes were analysed using the ClusterProfiler R package [16].
Mitochondrial ROS and membrane potential analysis
Mycoplasma free βTC-6 (RRID: CVCL_0605) and MIN6 (RRID: CVCL_0431) cells were cultured as previously described [4, 6]. Cells were cultivated in 24-well plates (Falcon, USA) to a density about 5 × 105 cells/well. In experiments, cells were treated with sodium palmitate (0.5 mmol/l in 0.5% fatty acid free BSA) + 20 mmol/l glucose. Human EndoC-βH1 (RRID: CVCL_L909) cells were cultured as previously described [17].
Cells were labelled with 1 μmol/l MitoSOX (Thermo Fisher, USA) for 60 min. Cells were then trypsinised and red fluorescence (FL2) was measured using a flow cytometer. For the analysis of mitochondrial membrane potential, cells were labelled with 5 μmol/l of the fluorescent probe JC-1 (Thermo Fisher), which is activated in the mitochondrial matrix when the membrane is hyperpolarised [18], for 30 min. The ratio between the 585 and 530 nm signals was calculated to monitor changes in mitochondrial membrane potential.
Immunoblotting
Immunoblotting was performed as previously described [4]. The following primary antibodies were used: mouse anti-PRC (1:400; Santa Cruz Biotechnology, USA), rabbit anti-human phospho-PGC1α (S571) (1:200; R & D systems, USA), and mouse anti-α-tubulin (TU-02) (1:500, Santa Cruz). Fluorescent anti-mouse/rabbit secondary antibodies were from Abcam (1:15,000). The bound antibodies were visualised with a LI-COR Odyssey Fc system (LI-COR Biosciences, Lincoln, USA).
Adenoviral-mediated ZBED6 overexpression
We used an adenoviral vector expressing human ZBED6 protein (untagged) behind the cytomegalovirus (CMV) promoter from Applied Biological Materials (Abm, Canada). An adenoviral vector without any protein expression was used as a control virus. Virus stocks were purified by caesium chloride density-gradient centrifugation (L-80 ultracentrifuge; Beckman Coulter, Fullerton, CA, USA) and semi-quantified by titration. For the adenoviral-mediated transduction, EndoC-βH1 cells were incubated at 37°C for 1 h in a volume of 0.4 ml RPMI-1640 supplemented with 2% FCS and adenoviral vectors at equal concentrations. Two and 3 days after transfection with viral particles at a concentration of 10 and 50 multiplicity of infection (MOI), cells were fixed in ethanol (−20°C) and then stained for 20 min in 5 μg/ml propidium iodide. Cell cycle phases were analysed by flow cytometry.
Adenoviral vectors (10 and 50 MOI) were also used for transduction of human islets. Permission to obtain pancreatic islet tissue from the Nordic Network for Clinical Islet Transplantation was reviewed and approved by the local ethics committee (Regionala etikprövningsnämnden, Uppsala) in Uppsala, Sweden. Human islets were cultured free-floating in Sterilin dishes in CMRL 1066 medium (ICN Biomedicals, Costa Mesa, CA, USA) containing 5.6 mmol/l glucose, 10% FCSand 2 mmol/l l-glutamine for 1–5 days and then dispersed by trypsin treatment for 7–8 min. Dispersed cells were then transduced as given above. Three days after the transduction procedure islet cells were incubated for 60 min in 1.7 mmol/l glucose and an additional 60 min in 17 mmol/l glucose. Insulin release was determined using an ELISA (Mercodia, Sweden).
Oxygen consumption rates
Oxygen consumption rates (OCR) and extracellular acidification rate (ECAR), which usually represents glycolytic rates, in EndoC-βH1 cells were measured by Extracellular Flux Analyzer XFe96 (Seahorse Bioscience, USA) as previously described [19]. EndoC-βH1 cells were transduced with adenoviral vectors, and after a 48 h culture period, assayed for OCR and ECAR in 5.6 mmol/l glucose, combination of palmitate (1.5 mmol/l with 2% BSA) and high glucose (20 mmol/l), or mouse IGF2 100 ng/ml (R & D Systems) for 1 h.
Statistical analysis
SigmaStat 4.0 (SYSTAT, USA) was used for all statistical computations. Results are given as means ± SEM. The differences between two groups were determined by Student’s paired t test. For multiple comparisons repeated measurements two-way ANOVA followed by Student Newman Keuls post hoc test was used.
Results
Knockout of Zbed6, but not knockin of Igf2, results in reduced beta cell area
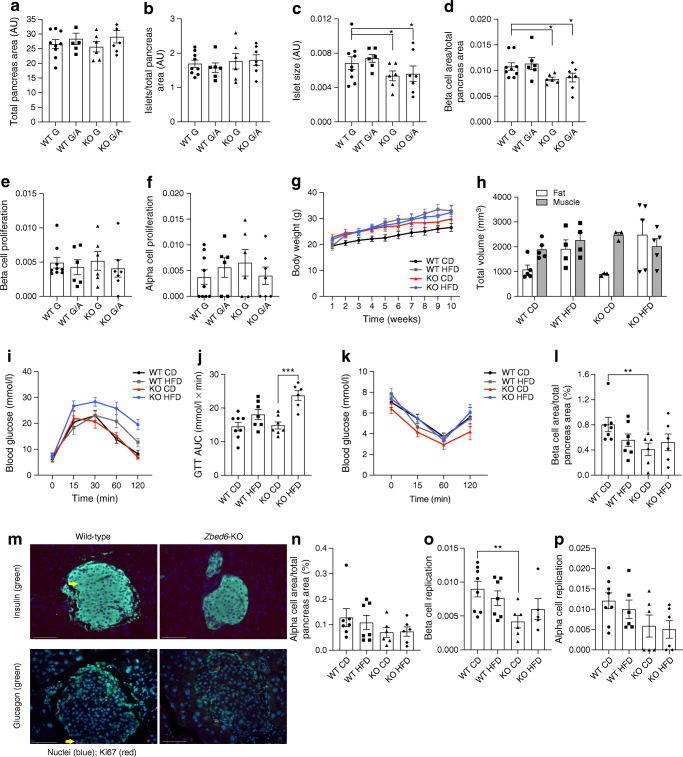
The Zbed6 KO mice were generated using the C57BL/6 embryonic stem cell line Bruce4 [20], which are Nnt+/Nnt+ as assessed by RNA sequencing (RNA-Seq) analysis of the Nnt transcripts (ESM Fig. 1). The general characteristics of the Zbed6 KO and Igf2 knockin mice have been recently characterised [3]. To specifically study insulin-producing beta cells, we euthanised Zbed6+/+ Igf2G/G (WT G), Zbed6−/− Igf2G/G (KO G), Zbed6+/+ Igf2pA/mG (WT G/A) and Zbed6−/− Igf2pA/mG (KO G/A) mice when 25–28 weeks of age. WT G mice express ZBED6 and low levels of IGF2, the KO G mice lack ZBED6 and express high IGF2 levels, the WT G/A mice do not lack ZBED6 but express high IGF2 levels, and the KO G/A mice lack ZBED6 and express high IGF2 levels [3]. Presently, the total pancreas area and the number of islets per total pancreas area were not affected by ZBED6 or IGF2 (Fig. 1a,b). However, the mean islet size, as well as the beta cell area per total pancreas area, were reduced in both KO G and KO G/A mice (Fig. 1c,d). Thus, the reduced beta cell area occurred in response to Zbed6 KO and independently from Igf2 expression. Despite this lowered beta cell area, neither alpha- nor beta cell proliferation were decreased by Zbed6 KO or Igf2 knockin (Fig. 1e,f).
Fig. 1.
Islet size and beta cell area are reduced in Zbed6 KO G and Zbed6 KO G/A mice and Zbed6 KO promotes HFD-induced glucose intolerance. WT G, KO G, WT G/A and KO G/A mice were killed when 25–28 weeks of age and analysed for (a) total pancreas area, (b) islet number, (c) islet size, (d) beta cell area, (e) beta cell proliferation and (f) alpha cell proliferation. Results are means ± SEM for 6–9 mice per group. *p < 0.05 vs corresponding WT mice. (g) Weight curve of WT G and KO G mice given a CD or an HFD from 6 to 15 weeks of age; 7–8 mice per group were analysed. (h) Micro-CT was performed on the mice shortly after euthanasia and fat and muscle mass was quantified; 3–5 mice per group were analysed. (i) GTT of WT G and KO G mice treated with HFD or CD. Mice were fasted for 8 h and injected intraperitoneally with glucose (2.5 g/kg). Blood glucose levels were analysed at the time points given. Results are means ± SEM for 6–8 mice in each group. (j) Data from (i) were recalculated to AUC. *** p < 0.001 vs KO mice given a CD. (k) Insulin sensitivity test of WT G and KO G mice treated for 9 weeks with an HFD. Insulin (1.6 U/kg Actrapid) was injected i.p. and blood glucose was analysed at the time points given in the figure. Results are means ± SEM for 6–8 mice in each group. (l) Beta cell area was assessed by morphometric analysis. ** p < 0.01 vs WT CD (n; = 6–8 mice). (m) Representative immunostainings showing morphology of beta cells (insulin, green), alpha cells (glucagon, green) and Ki-67 positive cells (red). Arrows point to two Ki-67 positive beta/alpha cells. White scale bars, 100 μm. (n) Alpha cell area was assessed by morphometric analysis (n = 6–8 mice). Beta cell (o) and alpha cell (p) replication was assessed by Ki-67 immunostaining. ** p < 0.01 vs WT CD mice (n = 6–8 mice)
Zbed6 KO promotes glucose intolerance in HFD-fed mice
Both WT G and KO G mice, 5–7 weeks old, were given a control diet (WT CD and KO CD, respectively) or an HFD (WT HFD and KO HFD, respectively) for 10 weeks. These mice are anticipated to present a mild beta cell phenotype [21]. The HFD increased the weight of WT G mice, but not of KO G mice (Fig. 1g). The weight of KO G mice was higher than that of WT G mice at start of the HFD, which is in line with a previous report stating that muscle and internal organ mass is increased in KO mice [3]. Indeed, using micro-CT we observed an augmented muscle mass of KO CD as compared with WT CD at the level corresponding to the lumbar vertebrae 4 to 6 (L4-L6) (Fig. 1h). In HFD mice, however, this effect was abolished (Fig. 1h).
As in a previous study [3], a GTT showed that KO CD mice responded similarly to the glucose injection as WT CD mice (Fig. 1i). The GTT of WT CD and WT HFD mice were similar. The glucose levels of KO HFD mice were, however, augmented when compared with WT HFD mice (Fig. 1i). Furthermore, calculations of the AUC showed that KO worsened the glucose tolerance in HFD mice (Fig. 1j). We also performed an ITT and observed no differences between the different groups (Fig. 1k), suggesting that neither the 10-week HFD, nor the Zbed6 KO, affected peripheral insulin sensitivity.
The beta cell area was decreased in KO CD mice as compared with WT CD mice (Fig. 1l), which is in line with the findings with older KO G mice (25–28 weeks) (Fig. 1d). A representative picture of WT G and KO G islets shows that the morphology and size of islet cells was not affected (Fig. 1m). This was paralleled by a lowered beta cell replication rate in the younger (15–17 weeks) KO CD mice (Fig. 1o). This lowering effect by Zbed6 KO was not observed in HFD-treated mice. There was a non-significant trend to similar effects of Zbed6 KO on alpha cells (Fig. 1n and p).
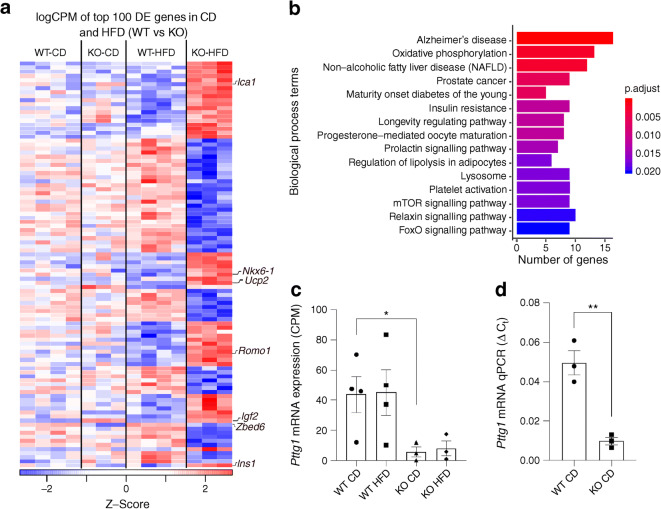
Zbed6 KO reduces Pttg1 and increases levels of mRNAs coding for mitochondrial oxidative phosphorylation proteins when exposed to an HFD
RNA-Seq analysis of Zbed6-KO G and WT G islets, under CD conditions, revealed up- and downregulation of only six and two genes, respectively (False discovery rate [FDR] < 0.05, Fig. 2a). In contrast, during HFD treatment there was upregulation of 377 genes and downregulation of 409 genes (Fig. 2a, ESM Table 2). Among the downregulated genes we observed that the cell cycle gene Pttg1 was reduced by almost 90% in the KO islets (ESM Table 2 and Fig. 2c), which was confirmed using RT-qPCR (Fig. 2d). Chromatin immunoprecipitation sequencing (ChIP-seq) using the mouse MIN6 beta cell line indicated a binding site of ZBED6 in the Pttg1 gene (ESM Fig. 2). In addition to Pttg1, we also observed a lowered expression of the cell cycle protein cyclin C (Table 1 and ESM Table 2). Other noteworthy upregulated genes were Ins1/2, Pcsk2 and Glut2/Slc2a2, all essential for insulin production (ESM Fig. 3 and Table 1). Beta cell transcription factors, Nkx6-1, FoxO1 and Pax6 [22], were increased in KO islets (Fig. 2a and Table 1), concurring with previous studies [4, 6]. Also in line with previous reports [5], E-cadherin was downregulated, and N-cadherin and the cAMP-generating enzyme Adcy9 were upregulated (Table 1). The most important beta cell autoantigens in type 1 diabetes (Ins1/2, Slc30a8, Ica1, Gad1, Ptprn2) were expressed at higher levels in Zbed6-KO islets. Among the upregulated genes in the HFD conditions we observed also Igf2, which confirms that ZBED6 acts as a repressor of Igf2 expression in islet cells (Table 1).
Fig. 2.
Zbed6 KO results in decreased islet expression of Pttg1 and increased expression of oxidative phosphorylation genes when given an HFD. Islets from 3–4 mice in each group were isolated and analysed by RNA-Seq. (a) Heat map of the top 100 differentially expressed genes (DEs) in WT HFD vs KO HFD islets, colour scale represents their expression across groups. (b) Gene ontology analysis of the upregulated differentially expressed genes in KO HFD. (c, d) Pttg1 mRNA expression, as analysed by RNA-Seq (counts per min [CPM]) (c) and qPCR (d)
Table 1.
Genes affected by Zbed6 knockout in mice. Validations from in vitro beta cell studies were also included
| Cellular process | Gene symbol | Gene name | Comment/Reference |
|---|---|---|---|
| Insulin production | Ins1 | Insulin 1 | Upregulated [4, 6] |
| Ins2 | Insulin 2 | Upregulated [4, 6] | |
| Pcsk2 | Proprotein convertase subtilisin/kexin type 2 | Upregulated | |
| Slc2a2 | Solute carrier family 2 (facilitated glucose transporter), member 2 | Upregulated | |
| Transcription factors | Nkx6-1 | NK6 homeobox 1 | Upregulated [6] |
| Foxo1 | Forkhead box O1 | Upregulated | |
| Pax6 | Paired box gene 6 | Upregulated [6] | |
| Cell cycle | Pttg1 | Pituitary tumour-transforming gene 1 | Downregulated |
| Ccnc | Cyclin C | Downregulated | |
| Beta cell autoantigens | Ins1/2 | Insulin 1/Insulin 2 | Upregulated |
| Slc30a2 | Solute carrier family 30 (zinc transporter), member 8 | Upregulated | |
| Ica1 | Islet cell autoantigen 1 | Upregulated | |
| Gad1 | Glutamic acid decarboxylase 1 | Upregulated | |
| Ptprn2 | Protein tyrosine phosphatase, receptor type, N polypeptide 2 | Upregulated | |
| Cell–cell contacts | Cdh1 | Cadherin 1 (E-cadherin) | Downregulated [5] |
| Cdh2 | Cadherin 2 (N-cadherin) | Upregulated [5] | |
| cAMP-signalling | Adcy9 | Adenylate cyclase 9 | Upregulated [6] |
| Mitochondrial ROS | Romo1 | Reactive oxygen species modulator 1 | Upregulated |
| Ucp2 | Uncoupling protein 2 (mitochondrial, proton carrier) | Upregulated | |
| Miscellaneous: | Igf2 | Insulin-like growth factor 2 | Upregulated |
| Zbed6 | Zinc finger, BED-type containing 6 | Downregulated |
Gene ontology enrichment (GOE) analysis of the KO differentially expressed genes in HFD conditions showed clustering (FDR < 0.05) in the category oxidative phosphorylation among the upregulated genes (Fig. 2b). The 13 genes belonging to this category are shown in ESM Table 3. This was paralleled by increased expression of uncoupling protein 2 (Ucp2) and reactive oxygen species modulator 1 (Romo1) (Table 1), two proteins involved in mitochondrial ROS production and protection from ROS [23, 24].
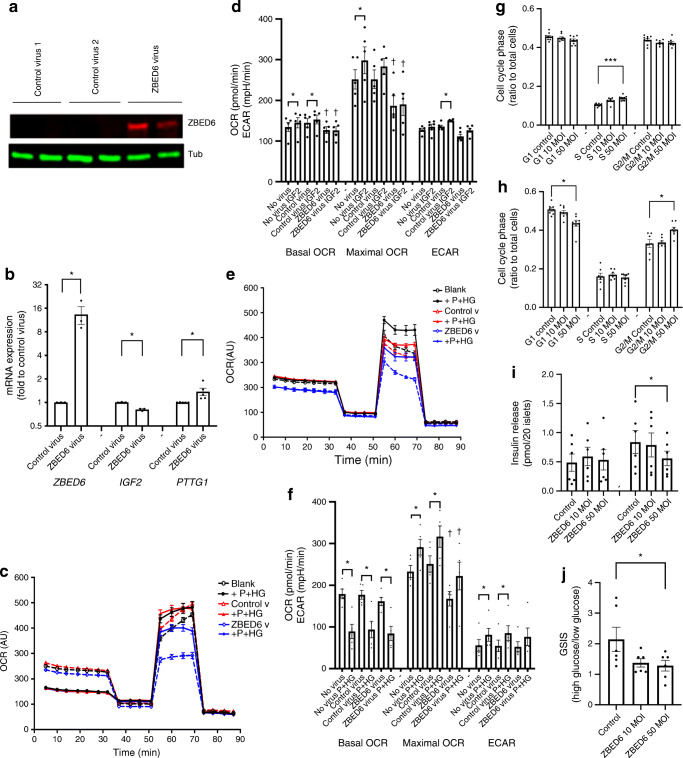
ZBED6 decreases EndoC-βH1 cell basal and maximal OCR
We next determined EndoC-βH1 cell OCR at basal (no addition) and carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP)-stimulated (maximal OCR) conditions 2 days after ZBED6 adenoviral transduction (Fig. 3a). ZBED6 adenoviral particles increased ZBED6 protein levels in EndoC-βH1 beta cells without signs of cytotoxicity (Fig. 3a, results not shown) and generated an approximate tenfold increase in ZBED6 mRNA expression (Fig. 3b). ZBED6 overexpression was associated with a lowered IGF2 mRNA level and an increased PTTG1 mRNA level (Fig. 3b). ZBED6 overexpression resulted in lower basal and maximal OCR as compared with control virus exposed cells (Fig. 3c,d). IGF2 addition for 1 h stimulated both basal and maximal OCR in control cells, but not in ZBED6 cells. ECAR was unaffected by ZBED6, and was stimulated by IGF2 in control cells, but not in ZBED6 cells (Fig. 3d). Thus, ZBED6 restricts respiration, but not via repression of the IGF2 gene.
Fig. 3.
ZBED6 overexpression reduces mitochondrial respiration and glucose-induced insulin release. (a) EndoC-βH1 cells were transduced with control (no protein expression) or ZBED6 adenoviral particles and analysed after 2 days by immunoblotting for ZBED6 protein levels. An adenoviral vector without transgene protein expression was used as control virus 1. Control virus 2 was a SIRT1-expressing adenoviral vector. The immunoblot is representative for three observations. α-Tubulin (Tub) was used as a control. (b) EndoC-βH1 cells were transduced with control (no protein expression) or ZBED6 adenoviral particles and analysed after two days for ZBED6, IGF2 and PTTG1 mRNA expression levels. Results are normalised to β-actin mRNA and expressed as a ratio to control virus. *p < 0.05 vs control virus for 3–4 independent experiments. (c, d) Control and ZBED6 overexpressing EndoC-βH1 cells were cultured for 48 h and then pre-incubated at 5.6 mmol/l glucose for 1 h prior to measuring mitochondrial respiration with or without 100 ng/ml IGF2 using the Seahorse technique. Basal OCR, maximal OCR (after FCCP addition) and ECAR were calculated. (c) Representative OCR recordings in which each line represents 6–10 replicates, and (d) the results from five independent experiments as means ± SEM. *p < 0.05 vs corresponding no IGF2 treatment cells. †p < 0.05 vs corresponding control virus cells. (e, f) Control and ZBED6 overexpressing EndoC-βH1 cells were incubated in the absence and presence of palmitate (1.5 mmol/l in 2% BSA) and high glucose (20 mmol/l) for 1 h during measurement of mitochondrial respiration. Results are mean ± SEM of five independent experiments. *p < 0.05 vs corresponding no treatment cells. † denotes p < 0.05 vs corresponding control virus cells. (g) EndoC-βH1 cells were transduced with ZBED6 adenoviral vectors at 10 and 50 MOI and then cell cycle phases were analysed by flow cytometry 2 days later. Results are mean ± SEM of five independent experiments. ***p < 0.001 vs corresponding control cells. (h) EndoC-βH1 cells were transduced with ZBED6 adenoviral vectors at 10 and 50 MOI and then analysed by flow cytometry 3 days later. Results are mean ± SEM of five independent experiments. *p < 0.05 vs corresponding control cells. (i) Human islets were dispersed by trypsin treatment and then transduced with ZBED6 adenoviral vectors at 10 and 50 MOI. Three days later cells were exposed to 1.7 and 17 mmol/l glucose and the insulin release was analysed by ELISA. Results are mean ± SEM of six independent experiments using islets from two human donors. *p < 0.05 vs corresponding control cells. (j) Insulin release data from (i) were used to calculate the glucose-stimulated insulin release expressed as ratio 17 mmol/l (high) glucose to 1.7 mmol/l (low) glucose. *p < 0.05 vs corresponding control cells
We also analysed effects of ZBED6 overexpression in cells stimulated by palmitate (1.5 mmol/l) and high glucose (20 mmol/l) for 1 h. Addition of palmitate and high glucose increased ECAR and reduced basal OCR (Fig. 3e,f), indicating that EndoC-βH1 cells have a high capacity to upregulate glycolytic ATP production in response to a high glucose concentration (Warburg effect). However, in the presence of FCCP, maximal respiration was increased by palmitate and high glucose in control cells (Fig. 3f), suggesting that palmitate and high glucose can further stimulate respiration at conditions of low mitochondrial ATP. ZBED6 promoted a lowered maximal OCR with and without palmitate and high glucose (Fig. 3f).
ZBED6 overexpression also affected EndoC-βH1 cell proliferation as assessed by analysis of the distribution of cells in the different cell cycle phases. We observed that the fraction of cells in the S phase was increased 2 days after transduction at the concentration of 50 MOI (Fig. 3g). Three days after transfection, the fraction of cells in G1 and G2/M were decreased and increased, respectively, by ZBED6 (Fig. 3h), suggesting that ZBED6 promotes increased proliferation.
To analyse whether ZBED6 affects glucose-induced insulin release we transduced dispersed human islets with ZBED6 adenoviral vectors. After 3 days, the higher concentration of the vector (50 MOI) resulted in a lowered insulin release at 17 mmol/l glucose (Fig. 3i). This resulted in a lowered glucose-stimulated insulin release ratio (Fig. 3j).
ZBED6 reduces mitochondrial membrane potential and mitochondrial ROS production
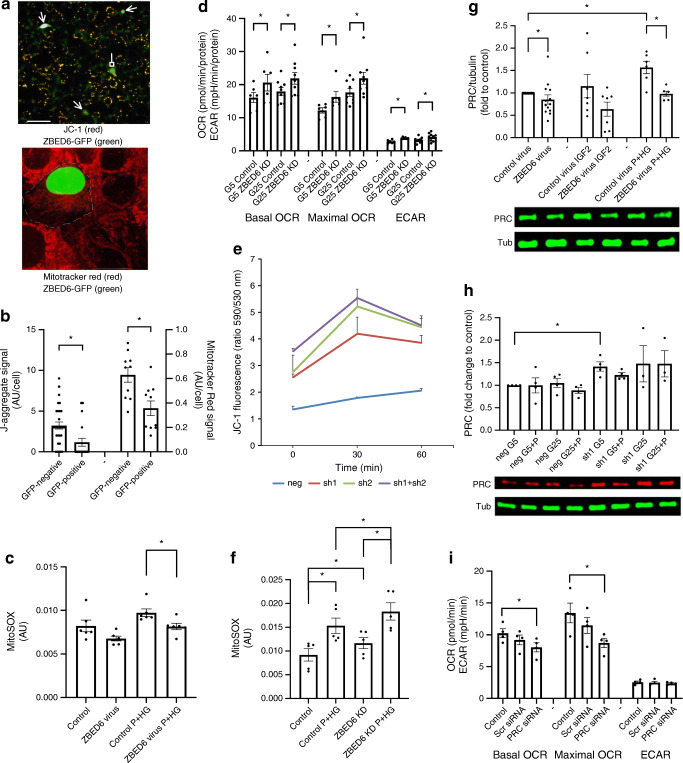
We next transfected EndoC-βH1 cells with a ZBED6-GFP expression vector and immunostained for respiratory chain protein ATPase inhibitory factor 1 (ATPIF1), a mitochondrial marker protein, which allows visualisation of mitochondrial density and morphology. ZBED6-GFP-positive cells displayed mitochondria with similar morphology and number as those of non-GFP-positive cells 2 days after transfection (ESM Fig. 4). However, when cells were stained with JC-1, which emits red J-aggregate light when the inner membranes of mitochondria are polarised, GFP-positive cells showed fewer J-aggregate positive mitochondrial structures than GFP-negative cells (Fig. 4a,b). Also, MitoTracker Red uptake into mitochondria of GFP-positive cells was lower than that of GFP-negative cells (Fig. 4a,b). This supports the hypothesis that ZBED6 lowers mitochondrial activity, as MitoTracker Red is not efficiently taken up by inactive mitochondria. Mitochondrial ROS production, assessed using the MitoSOX probe, was lower in ZBED6-overexpressing EndoC-βH1 cells cultured with palmitate and high glucose (Fig. 4c).
Fig. 4.
ZBED6 overexpression in EndoC-βH1 cells results in lowered J-aggregate fluorescence, MitoTracker Red uptake and mitochondrial ROS production. Downregulation of ZBED6 results in increased OCR, ECAR, J-aggregate fluorescence and mitochondrial ROS production. ZBED6 overexpression reduces expression level of PRC, and ZBED6 downregulation increases expression level of PRC. (a) A fluorescence microscopic image with JC-1 staining (3 μmol/l for 10 min, red colour) of EndoC-βH1 cells showing three ZBED6-GFP-positive cells (green colour, arrows) with no active mitochondria, one ZBED6-GFP-positive cell with active mitochondria (green, block head). Scale bar, 10 μm. Confocal image showing MitoTracker Red staining (10 μmol/l for 10 min, red colour) of a ZBED6-GFP expressing cell (green nucleus) surrounded by non-transfected cells. The dotted line shows the probable cell membrane position of the ZBED6-GFP-positive cell. Scale bar, 50 μm. (b) Left axis: quantification of J-aggregate positive mitochondrial structures in GFP-positive and GFP-negative cells. Results are means from 21 (GFP-negative) and 29 (GFP-positive) cells obtained from six randomly photographed culture plate areas. Right axis: quantification of Mitotracker Red signal in GFP-positive and GFP-negative cells. Results are means from ten randomly photographed GFP-positive cells with an adjacent GFP-negative cell. (c) MitoSOX fluorescence in EndoC-βH1 control and ZBED6 overexpressing cells quantified by flow cytometry. Cells were cultured for 24 h at standard conditions (5 mmol/l glucose, control) or supplemented with 0.5 mmol/l palmitate + 20 mmol/l glucose (P+HG), and then labelled for 40 min with 1 μmol/l of the MitoSOX probe. Results are means ± SEM for six independent experiments. *p < 0.05. (d) Control and Zbed6 knockdown (ZBED6 KD) MIN6 cells were cultured for 24 h in 5 mmol/l (G5) or 25 mmol/l (G25) glucose and then pre-incubated at 5.6 mmol/l glucose for 1 h prior to measuring mitochondrial respiration at 5 mmol/l (G5) or 25 mmol/l (G25) glucose using the Seahorse technique. Basal OCR, maximal OCR (after FCCP addition) and ECAR were calculated. Results are from six independent experiments. *p < 0.05 vs corresponding control cells. (e) βTC-6 cells treated with scrambled shRNA lentiviral particles (negative control; neg) or with two different anti-Zbed6 shRNA lentiviral particles (sh1 and sh2) were labelled with 3 μmol/l JC-1 for 10 min and then treated with 0.5 mmol/l palmitate +25 mmol/l glucose (P+HG) for 0, 30 or 60 min. J-aggregate fluorescence was obtained by flow cytometry analysis and by calculation of the FL2/FL1 (590/530 nm) ratio. Results are means ± SEM for three experiments. (f) MitoSOX fluorescence in MIN6 control and ZBED6 KD cells quantified by flow cytometry. Cells were cultured for 24 h under standard conditions (25 mmol/l glucose, control) or supplemented with 0.5 mmol/l palmitate + 25 mmol/l glucose (P+HG), and then labelled for 40 min at 1 μmol/l MitoSOX probe. Results are means ± SEM for five independent experiments. *p < 0.05. (g−i) ZBED6 overexpression reduces and ZBED6 downregulation increases expression level of PRC. (g) Control and ZBED6 overexpressing EndoC-βH1 cells were cultured for 48 h and then treated with or without mouse IGF2 (100 ng/ml) or the combination palmitate (1.5 mmol/l in 2% BSA) and high glucose (20 mmol/l) for 3 h. The expression level of PRC was measured by immunoblotting analysis. Results are normalised to α-tubulin (Tub) expression level and are expressed as means ± SEM for 6–13 independent experiments. *p < 0.05. (h) MIN6 cells expressing scramble shRNA (neg) or anti-Zbed6 shRNA (sh1) were analysed for PRC expression using immunoblot analysis. Cells were cultured for 24 h in the presence of 5 mmol/l glucose (G5), 5 mmol/l glucose + 0.5 mmol/l palmitate (G5+P), 25 mmol/l glucose (G25) or 25 mmol/l glucose + 0.5 mmol/l palmitate (G25+P). Results are from 3–4 independent experiments. (i) Knockdown of PRC results in reduced basal and maximal OCR. EndoC-βH1 cells were transfected with 50 nmol/l siRNA targeting to PRC and then were analysed for OCR using the Seahorse technique. Results are from four independent experiments. *p < 0.05 vs control
Zbed6 knockdown results in increased OCR, J-aggregate formation and mitochondrial ROS production
Zbed6 was knocked down in MIN6 cells by using anti-Zbed6 short hairpin RNA (shRNA) lentiviral particles [4–6]. As control cells we used scramble shRNA lentiviral-transduced cells (neg) and found that Zbed6 knockdown (sh1) resulted in higher basal and maximal OCR (Fig. 4d). Also, ECAR was higher in sh1 cells than in neg cells (Fig. 4d). The increased OCR of sh1 cells was observed at both 5 mmol/l and 25 mmol/l glucose.
Silencing of Zbed6 was also performed in insulin-producing βTC-6 cells [4], and in this case the Zbed6 knockdown sh1, sh2 and sh1+sh2 cells displayed increased JC-1 J-aggregate fluorescence both at basal conditions and after 30 and 60 min of stimulation with palmitate and high glucose (Fig. 4e).
The MIN6 cells were also used for analysis of mitochondrial ROS production using the MitoSOX probe. The MIN6 sh1 cells produced more mitochondrial ROS, both at basal and palmitate stimulated conditions, than neg cells (Fig. 4f).
ZBED6 reduces the expression of the mitochondrial regulator PRC
As ZBED6-restricted mitochondrial activity is not restored by exogenous IGF2, we looked for other ZBED6 targets. ZBED6 binds with moderate strength to the PPAR coactivator-1 (PRC) promoter region in MIN6 cells, but very weakly in C2C12 myotube cells [25] (ESM Fig. 2). PRC, together with PGC-1α and PGC-1β, belongs to the PPAR coactivator family and are known activators of mitochondrial protein transcription [26]. In ZBED6-overexpressing EndoC-βH1 cells, PRC levels were lowered at basal conditions and after culture for 24 h in the presence of palmitate and high glucose (Fig. 4g,h). In MIN6 sh1 cells (Zbed6-knockdown), PRC was increased at basal conditions (Fig. 4h), suggesting that ZBED6 controls PRC levels in beta cells.
Knockdown of PRC results in lowered basal and maximal OCR
We next performed RNA interference (RNAi) to downregulate PRC protein levels in EndoC-βH1 cells. In a small interfering RNA (siRNA) titration experiment, we observed after 2 and 3 days an approximate 40–50% lowering of the PRC protein (ESM Fig. 5). In such PRC knockdown cells, basal and maximal OCR were decreased when comparing with scrambled siRNA treatment (Fig. 4i). PRC knockdown did not affect ECAR, suggesting that ZBED6 suppresses mitochondrial function by reducing PRC levels.
Discussion
ZBED6 is a transcriptional regulator that represses postnatal IGF2 expression [3]. However, we report here an unaltered beta cell area in IGF2 overexpressing WT G/A mice. It is therefore likely that ZBED6-dependent IGF2 expression is without physiological consequence for beta cells, and that the observed ZBED6-induced effects were instead due to other growth-controlling genes. One such gene, which contains ZBED6 binding sites in its promoter/enhancer region and which is downregulated in Zbed6-KO mice, is Pttg1. The Pttg1 gene product modulates G1/S cell phase transition [27], functions as a securin during chromosome separation [28] and is required for beta cell development and proliferation [29]. It is therefore possible that ZBED6, by enhancing expression of this and other growth-controlling genes, maintains normal beta cell replication, ensuring an adequate beta cell mass for the long-term preservation of glucose tolerance. Interestingly, ZBED6 expression appears to be reduced in human beta cells with increasing age [30].
The lowered beta cell mass of Zbed6-KO mice did not promote glucose intolerance when given an CD, but as the mice were of the Nnt+/Nnt+ genotype, a milder beta cell phenotype can be expected [21]. Therefore, it is not surprising that the lowered beta cell mass, without HFD treatment, did not provoke glucose intolerance. However, in Zbed6-KO (Nnt+/Nnt+) mice fed an HFD, glucose intolerance developed, suggesting that the combination of a reduced beta cell mass and a diet overload is necessary to cause impaired beta cell function.
A clue to why Zbed6-KO promotes impaired glucose tolerance in HFD-treated mice may be that the expression of islet oxidative phosphorylation genes was increased in vivo. Interestingly, Zbed6 knockdown increased and ZBED6 overexpression decreased OCR in vitro, clearly supporting the in vivo results. Mitochondrial respiration is regulated by members of the PPAR coactivator family PRC, PGC-1α and PGC-1β [26]. Human islets express high levels of PRC (6.8 reads per kilobase million [RPKM]), intermediate levels of PGC-1α (2.6 RPKM) and low levels of PGC-1β (0.3 RPKM) [31], and ZBED6 binds to the PRC promoter in beta cells [6]. In line with this, PRC protein levels were increased by Zbed6-KO and decreased by ZBED6 overexpression, and PRC knockdown resulted in reduced OCR. This suggests that PRC is an important mitochondrial regulator in human beta cells, and that ZBED6 may control expression of this PPAR coactivator via a direct interaction with the PRC gene promoter/enhancer region. PRC has, to our knowledge, not been investigated in beta cells. Additionally, in other cell types PRC is rather poorly characterised, but it has been proposed that PRC acts rapidly upon extracellular signals, for example growth factor stimulation, to increase mitochondrial biogenesis [26].
There seems to exist a correlation between maximal OCR, inner membrane hyperpolarisation and mitochondrial ROS production, as all three variables were increased by the nutrients palmitate and high glucose. Indeed, it is known that mitochondria of primary beta cells respond to nutrients with stimulation of the Krebs cycle and the respiratory chain [32]. Furthermore, in vitro addition of metabolites, ADP and uncouplers results in superoxide and hydrogen peroxide leakage from the respiratory complexes, and this occurs via chemical reduction of electron leaking sites and a strong protonmotive force [33–35]. Excess nutrients also evoke an increased functional demand (higher need for insulin production), which consumes ATP and increases ADP levels [35], further stimulating the respiratory chain. Moreover, our results demonstrate that mitochondria are activated by the PRC gene, a process controlled by ZBED6. Thus, it seems that overnutrition predisposes for augmented beta cell ROS production, and in the case when ZBED6 does not restrict mitochondrial activation by dampening PRC gene expression, potentially harmful levels of ROS may be generated. High mitochondrial ROS levels cause beta cell dysfunction [36, 37], and the present ROS increase, in response to nutrients and Zbed6-KO, could therefore explain, at least in part, why Zbed6-KO mice become glucose intolerant when given an HFD.
Type 2 diabetes is associated with beta cell loss of identity, dedifferentiation or beta to alpha cell transdifferentiation [10, 38], and can in later stages often result in loss of transcription factors such as MafA, Nkx6-1 and Pdx1 [10]. Interestingly, in this study we observed the opposite, namely increased expression of beta cell-specific genes during conditions of glucose intolerance. In addition, important beta cell autoantigens in type 1 diabetes were also increased. Thus, Zbed6-KO in functionally challenged mice promoted a beta cell hyper-differentiated and hyper-immunogenic state that mediates not only a higher insulin production, but also might facilitate inflammatory/autoimmune reactions. It is not clear why beta cells of glucose-intolerant Zbed6-KO mice become hyper-differentiated rather than dedifferentiated, but it could be envisaged that the islet cells were in an early stage of metabolic stress and still had not started the process of trans-/dedifferentiation [38].
A limitation of this study is that a global knockout was used, making it harder to pinpoint whether ZBED6 affects beta cells directly or via other organs/cell types. Another limitation is that the in vitro techniques were not easily adapted for the in vivo situation. Nevertheless, ZBED6 appears to perform an important function in balancing differentiation vs proliferation. This could promote a more sustainable beta cell phenotype in which mitochondrial ROS production is restrained. The role of ZBED6 in the pathophysiology of type 2 diabetes remains to be clarified, but it is tempting to speculate that ZBED6 maintains beta cell survival and proliferation at the cost of decreased insulin production, and that this is beneficial during long-term high-fat overfeeding.
Supplementary Information
(PDF 1126 kb)
(XLSX 645 kb)
(XLSX 14 kb)
Acknowledgements
Some of the data of this article were presented as an abstract at the 55th EASD Annual Meeting in 2019.
Authors’ relationships and activities
The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Abbreviations
- CD
Control diet
- CT
Computed tomography
- ECAR
Extracellular acidification rate
- ES
Embryonic stem
- FCCP
Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone
- FDR
False discovery rate
- HFD
High-fat diet
- KO
Knockout
- MOI
multiplicity of infection
- OCR
Oxygen consumption rate
- PRC
PPAR-gamma related coactivator 1 protein
- RNA-Seq
RNA sequencing
- ROS
Reactive oxygen species
- RPKM
Reads per kilobase million
- shRNA
Short hairpin RNA
- siRNA
Small interfering RNA
- WT
Wild-type
- ZBED6
Zinc finger, BED-type containing 6
Contribution statement
All authors participated in generating and analysing data and revising or drafting the article. All authors have approved the final version to be published. LA and NW are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Funding
Open access funding provided by Uppsala University. This work was supported by the Swedish Diabetes Foundation (DIA-2020-540), the family Ernfors Fund, Barndiabetesfonden, EXODIAB and Vetenskapsrådet (to LA) and Knut and Alice Wallenberg Foundation (to LA).
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author upon reasonable request.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xuan Wang and Shady Younis contributed equally
Contributor Information
Leif Andersson, Email: leif.andersson@imbim.uu.se.
Nils Welsh, Email: nils.welsh@mcb.uu.se.
References
- 1.Markljung E, Jiang L, Jaffe JD, et al. ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 2009;7:e1000256. doi: 10.1371/journal.pbio.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Younis S, Naboulsi R, Wang X, et al. The importance of the ZBED6-IGF2 axis for metabolic regulation in mouse myoblast cells. FASEB J. 2020;34:10250–10266. doi: 10.1096/fj.201901321R. [DOI] [PubMed] [Google Scholar]
- 3.Younis S, Schonke M, Massart J, et al. The ZBED6-IGF2 axis has a major effect on growth of skeletal muscle and internal organs in placental mammals. Proc Natl Acad Sci U S A. 2018;115:E2048–E2057. doi: 10.1073/pnas.1719278115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Jiang L, Wallerman O, et al. Transcription factor ZBED6 affects gene expression, proliferation, and cell death in pancreatic beta cells. Proc Natl Acad Sci U S A. 2013;110:15997–16002. doi: 10.1073/pnas.1303625110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Xie B, Qi Y, et al. Knock-down of ZBED6 in insulin-producing cells promotes N-cadherin junctions between beta-cells and neural crest stem cells in vitro. Sci Rep. 2016;6:19006. doi: 10.1038/srep19006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Jiang L, Wallerman O, et al. ZBED6 negatively regulates insulin production, neuronal differentiation, and cell aggregation in MIN6 cells. FASEB J. 2019;33:88–100. doi: 10.1096/fj.201600835R. [DOI] [PubMed] [Google Scholar]
- 7.Erion K, Corkey BE. Beta-cell failure or beta-cell abuse? Front Endocrinol (Lausanne) 2018;9:532. doi: 10.3389/fendo.2018.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trico D, Natali A, Arslanian S, Mari A, Ferrannini E (2018) Identification, pathophysiology, and clinical implications of primary insulin hypersecretion in nondiabetic adults and adolescents. JCI Insight 3. 10.1172/jci.insight.124912 [DOI] [PMC free article] [PubMed]
- 9.Tiwari S, Roel C, Tanwir M, et al. Definition of a Skp2-c-Myc pathway to expand human beta-cells. Sci Rep. 2016;6:28461. doi: 10.1038/srep28461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter CS, Stein RW. Evidence for loss in identity, de-differentiation, and trans-differentiation of islet beta-cells in type 2 diabetes. Front Genet. 2017;8:35. doi: 10.3389/fgene.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathis D, Vence L, Benoist C. Beta-cell death during progression to diabetes. Nature. 2001;414:792–798. doi: 10.1038/414792a. [DOI] [PubMed] [Google Scholar]
- 12.Halban PA, Polonsky KS, Bowden DW, et al. Beta-cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. Diabetes Care. 2014;37:1751–1758. doi: 10.2337/dc14-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Younis S, Kamel W, Falkeborn T, et al. Multiple nuclear-replicating viruses require the stress-induced protein ZC3H11A for efficient growth. Proc Natl Acad Sci U S A. 2018;115:E3808–E3816. doi: 10.1073/pnas.1722333115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersson A. Isolated mouse pancreatic islets in culture: effects of serum and different culture media on the insulin production of the islets. Diabetologia. 1978;14:397–404. doi: 10.1007/BF01228134. [DOI] [PubMed] [Google Scholar]
- 15.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu G, Wang L, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravassard P, Hazhouz Y, Pechberty S, et al. A genetically engineered human pancreatic beta cell line exhibiting glucose-inducible insulin secretion. J Clin Invest. 2011;121:3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbu A, Welsh N, Saldeen J. Cytokine-induced apoptosis and necrosis are preceded by disruption of the mitochondrial membrane potential (Deltapsi(m)) in pancreatic RINm5F cells: prevention by Bcl-2. Mol Cell Endocrinol. 2002;190:75–82. doi: 10.1016/S0303-7207(02)00009-6. [DOI] [PubMed] [Google Scholar]
- 19.Cen J, Sargsyan E, Bergsten P. Fatty acids stimulate insulin secretion from human pancreatic islets at fasting glucose concentrations via mitochondria-dependent and -independent mechanisms. Nutr Metab. 2016;13:59. doi: 10.1186/s12986-016-0119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes ED, Qu YY, Genik SJ, et al. Genetic variation in C57BL/6 ES cell lines and genetic instability in the Bruce4 C57BL/6 ES cell line. Mamm Genome. 2007;18:549–558. doi: 10.1007/s00335-007-9054-0. [DOI] [PubMed] [Google Scholar]
- 21.Attané C, Peyot M-L, Lussier R, et al. Differential insulin secretion of high-fat diet-fed C57BL/6NN and C57BL/6NJ mice: implications of mixed genetic background in metabolic studies. PLoS One. 2016;11:e0159165. doi: 10.1371/journal.pone.0159165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernardo AS, Hay CW, Docherty K. Pancreatic transcription factors and their role in the birth, life and survival of the pancreatic beta cell. Mol Cell Endocrinol. 2008;294:1–9. doi: 10.1016/j.mce.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Li N, Karaca M, Maechler P. Upregulation of UCP2 in beta-cells confers partial protection against both oxidative stress and glucotoxicity. Redox Biol. 2017;13:541–549. doi: 10.1016/j.redox.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung YM, Kim JS, Yoo YD. A novel protein, Romo1, induces ROS production in the mitochondria. Biochem Biophys Res Commun. 2006;347:649–655. doi: 10.1016/j.bbrc.2006.06.140. [DOI] [PubMed] [Google Scholar]
- 25.Jiang L, Wallerman O, Younis S, et al. ZBED6 modulates the transcription of myogenic genes in mouse myoblast cells. PLoS One. 2014;9:e94187. doi: 10.1371/journal.pone.0094187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villena JA. New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. FEBS J. 2015;282:647–672. doi: 10.1111/febs.13175. [DOI] [PubMed] [Google Scholar]
- 27.Tong Y, Tan Y, Zhou C, Melmed S. Pituitary tumor transforming gene interacts with Sp1 to modulate G1/S cell phase transition. Oncogene. 2007;26:5596–5605. doi: 10.1038/sj.onc.1210339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salehi F, Kovacs K, Scheithauer BW, Lloyd RV, Cusimano M. Pituitary tumor-transforming gene in endocrine and other neoplasms: a review and update. Endocr Relat Cancer. 2008;15:721–743. doi: 10.1677/ERC-08-0012. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Moro E, Kovacs K, Yu R, Melmed S. Pituitary tumor transforming gene-null male mice exhibit impaired pancreatic beta cell proliferation and diabetes. Proc Natl Acad Sci U S A. 2003;100:3428–3432. doi: 10.1073/pnas.0638052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seiron P, Stenwall A, Hedin A, et al. Transcriptional analysis of islets of Langerhans from organ donors of different ages. PLoSOne. 2021;3:e0247888. doi: 10.1371/journal.pone.0247888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fred RG, Kappe C, Ameur A, et al. Role of the AMP kinase in cytokine-induced human EndoC-betaH1 cell death. Mol Cell Endocrinol. 2015;414:53–63. doi: 10.1016/j.mce.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Welsh M. Claes Hellerström and Cartesian diver microrespirometry. Ups J Med Sci. 2016;121:77–80. doi: 10.3109/03009734.2015.1109012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong HS, Dighe PA, Mezera V, Monternier PA, Brand MD. Production of superoxide and hydrogen peroxide from specific mitochondrial sites under different bioenergetic conditions. J Biol Chem. 2017;292:16804–16809. doi: 10.1074/jbc.R117.789271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HJ, Kim JH, Noh S, et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J Proteome Res. 2011;10:722–731. doi: 10.1021/pr100892r. [DOI] [PubMed] [Google Scholar]
- 35.Gorlach A, Dimova EY, Petry A, et al. Reactive oxygen species, nutrition, hypoxia and diseases: problems solved? Redox Biol. 2015;6:372–385. doi: 10.1016/j.redox.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlsson C, Borg LA, Welsh N. Sodium palmitate induces partial mitochondrial uncoupling and reactive oxygen species in rat pancreatic islets in vitro. Endocrinology. 1999;140:3422–3428. doi: 10.1210/endo.140.8.6908. [DOI] [PubMed] [Google Scholar]
- 37.Weiss H, Wester-Rosenloef L, Koch C, et al. The mitochondrial Atp8 mutation induces mitochondrial ROS generation, secretory dysfunction, and beta-cell mass adaptation in conplastic B6-mtFVB mice. Endocrinology. 2012;153:4666–4676. doi: 10.1210/en.2012-1296. [DOI] [PubMed] [Google Scholar]
- 38.Mezza T, Cinti F, Cefalo CMA, Pontecorvi A, Kulkarni RN, Giaccari A. Beta-cell fate in human insulin resistance and type 2 diabetes: a perspective on islet plasticity. Diabetes. 2019;68:1121–1129. doi: 10.2337/db18-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1126 kb)
(XLSX 645 kb)
(XLSX 14 kb)
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author upon reasonable request.