Keywords: acute kidney injury, chronic kidney disease, lactate, metabolism, lactate dehydrogenase
Abstract
Cellular metabolic rates in the kidney are critical for maintaining normal renal function. In a hypoxic milieu, cells rely on glycolysis to meet energy needs, resulting in the generation of pyruvate and NADH. In the absence of oxidative phosphorylation, the continuation of glycolysis is dependent on the regeneration of NAD+ from NADH accompanied by the fermentation of pyruvate to lactate. This reaction is catalyzed by lactate dehydrogenase (LDH) isoform A (LDHA), whereas LDH isoform B (LDHB) catalyzes the opposite reaction. LDH is widely used as a potential injury marker as it is released from damaged cells into the urine and serum; however, the precise isoform-specific cellular localization of the enzyme along the nephron has not been characterized. By combining immunohistochemistry results and single-cell RNA-sequencing data on healthy mouse kidneys, we identified that LDHA is primarily expressed in proximal segments, whereas LDHB is expressed in the distal parts of the nephron. In vitro experiments in mouse and human renal proximal tubule cells showed an increase in LDHA following hypoxia with no change in LDHB. Using immunofluorescence, we observed that the overall expression of both LDHA and LDHB proteins decreased following renal ischemia-reperfusion injury as well as in the adenine-diet-induced model of chronic kidney disease. Single-nucleus RNA-sequencing analyses of kidneys following ischemia-reperfusion injury revealed a significant decline in the number of cells expressing detectable levels of Ldha and Ldhb; however, cells that were positive showed increased average expression postinjury, which subsided during the recovery phase. These data provide information on the cell-specific expression of LDHA and LDHB in the normal kidney as well as following acute and chronic kidney disease.
NEW & NOTEWORTHY Cellular release of lactate dehydrogenase (LDH) is being used as an injury marker; however, the exact localization of LDH within the nephron remains unclear. We show that LDH isoform A is expressed proximally, whereas isoform B is expressed distally. Both subunit expressions were significantly altered in models of acute kidney injury and chronic kidney disease. Our study provides new insights into basal and postinjury renal lactate metabolism.
INTRODUCTION
The kidney exhibits the highest resting energy metabolic rate in the human body (1) and has the second highest oxygen consumption and mitochondrial content, superseded only by the heart (2, 3). Such extensive energy production machinery is critical in maintaining normal renal function, which requires active transport and reabsorption of solutes, amino acids, sugars, and other essential elements back into the blood.
Generation of ATP, a cellular energy carrier, occurs by the intracellular breakdown of simple sugars into pyruvate via glycolysis and by subsequent oxidation of pyruvate, fatty acids, and amino acids by the mitochondria using the tricarboxylic acid cycle and oxidative phosphorylation (OXPHOS). Oxidative metabolism is roughly 15 times more efficient in the production of ATP compared with glycolysis. Hence, it is the preferred mechanism for energy production in the kidney. Cells with large numbers of mitochondria such as proximal tubule cells (PTCs) require substantial quantities of oxygen. They are predisposed to injury from hypoxic conditions as would occur during ischemia-reperfusion injury (IRI) in the kidney (4). Renal IRI induces metabolomic changes within the kidney including activation of glycolysis and fatty acid metabolism as well as mitochondrial dysfunction and impaired ATP production (5). Under hypoxic conditions, when OXPHOS is impaired, cells are forced to rely on glycolysis to meet their energy needs. This is a highly inefficient way to produce ATP and results in the accumulation of pyruvate and NADH. Regeneration of NAD+ from NADH is essential for the continuation of glycolysis, and, in the absence of OXPHOS, this process is accomplished via the fermentation of pyruvate to lactate catalyzed by the enzyme lactate dehydrogenase (LDH) (6–8).
LDH is a tetramer encoded by Ldha and Ldhb genes. The Ldha gene encodes the LDH A isoform (LDHA), which converts pyruvate to lactate and NADH to NAD+, whereas the Ldhb gene encodes for LDH isoform B (LDHB), which catalyzes the opposite reaction (Fig. 1A). The LDHA-to-LDHB subunit ratio in the LDH tetramer determines the overall direction of the reaction. The LDH-5 enzyme, consisting of four LDHA subunits, has a high affinity for pyruvate to lactate conversion, whereas the LDH-1 enzyme, consisting entirely of LDHB subunits, primarily converts lactate to pyruvate. LDH complexes that have both subunits present would display activity in between LDH-1 and LDH-5. Ldhc is a testis-specific gene and will not be discussed in this work (9).
Figure 1.
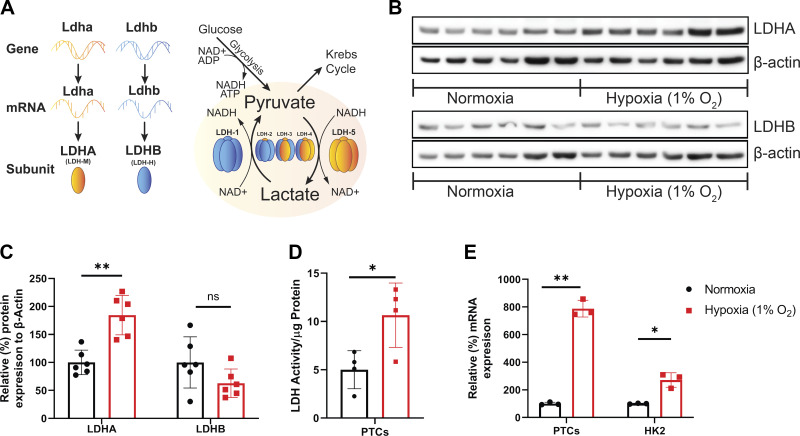
Response of lactate dehydrogenase (LDH) isoform A (LDHA) and LDH isoform B (LDHB) to hypoxia in vitro. A: schematic representation of the LDH complex subunit composition and the reaction governed by its LDHA and LDHB distribution. LDH-1 consists of four LDHB subunits and converts lactate to pyruvate, whereas LDH-5 consists of four LDHA subunits and catalyzes the opposite reaction. B: Western blots of protein lysates from mouse primary proximal tubular cells (PTCs) that were subjected to 24-h hypoxia (1% O2) and probed for LDHA and LDHB. Each lane represents PTCs from an individual mouse kidney (n = 6 animals). C: densitometry results normalized to β-actin (n = 6 animals). D: LDH activity was measured in cells subjected to hypoxia and was normalized to the total amount of protein detected by the bicinchoninic acid (BCA) assay (n = 4 samples). E: the percent change in mRNA levels of Ldha was analyzed in PTCs and HK-2 cells that were subjected to hypoxia and normalized to β-actin mRNA expression (n = 3 samples). *P < 0.05; **P < 0.001. One-way ANOVA was used to obtain P values.
LDH plays a critical role in the aerobic glycolytic switch (Warburg effect) that is seen in many cancers and is being investigated as a potential therapeutic target (10). Immunologists have been interested in LDH as a potential activator of M1 macrophages and its effect on inflammation and cancer (11). It is highly resistant to protein degradation and persists after initial release from damaged cells and tissues, making it useful as an indicator of cell death and injury. For example, after renal injury, LDH is released into the urine and serum (12), which correlates with other markers for renal injury (13). It is notable that renal tubule cells also undergo glycolytic switch postischemic injury during their dedifferentiation and repair process, causing cells to rely more on glycolysis for their energy needs (14). LDH is crucial for recovery from renal injury and the maintenance of a healthy NAD+-to-NADH ratio (15). De novo synthesis and recycling of NAD+ is inhibited in renal IRI (16), which also might contribute to the dependence of epithelial cells on LDH to continue glycolysis.
Previous studies have confirmed the presence of LDH in the kidney (17, 18) and showed that LDH expression is altered in postischemic injury (13, 19); however, these studies did not investigate LDH isoform-specific localization. In this work, we tested the hypothesis that LDHA and LDHB are differentially localized within the nephron and that their expression patterns are altered in response to acute and chronic kidney disease (CKD).
MATERIALS AND METHODS
Animals
C57BL/6J male mice (Jackson Laboratories) aged 10–12 wk were used for the study. Bilateral renal IRI was performed as previously described (20). Briefly, ketamine-xylazine was used to anesthetize mice. Using a dorsal approach, both renal pedicles were cross clamped for 20 min. Kidney ischemia was confirmed by renal color change. Body temperature was maintained at 37°C throughout the procedure. Kidneys were inspected for color change within 1 min after clamp removal to ensure uniform reperfusion. Sham surgeries were performed as a control. To induce CKD, 8- to 12-wk-old DBA/2J female mice (Jackson Laboratories) were placed on an adenine diet (0.2% adenine, 0.6% Ca, and 0.9% phosphate) for 16 wk (diet manufactured by Envigo, Madison, WI). Control mice received 0.6% Ca and 0.9% phosphate without adenine supplementation. Mice were harvested at the end of 16 wk to assess kidney function and collect tissues. All procedures involving mice were performed per National Institutes of Health guidelines regarding the care and use of live animals and were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham.
Cell Culture
Primary PTC cultures were generated from the kidneys of mice as previously described (21–23). Briefly, kidneys were decapsulated, minced, and filtered through a 70-μm cell strainer over a 50-mL conical tube with media (Renal Epithelial Cell Growth Medium, PromoCell). The medium containing the tubules was centrifuged and plated on collagen-coated culture plates and then incubated for 72 h at 37°C in 5% CO2. Each experiment used PTCs generated from a single animal and cells were not passaged. Cells were examined for the expression of γ-glutamyltranspeptidase as a marker for PTCs, as previously described (22). An established and well-characterized human renal proximal tubular epithelial cell line, HK-2, was purchased from the American Type Culture Collection (Manassas, VA). Cell cultures were incubated under hypoxic conditions using an anaerobic chamber equilibrated with 1% O2, 5% CO2, and 94% N2 at 37°C. Control cells were incubated at 37°C under 95% air and 5% CO2. Cells were immediately harvested upon opening the chamber and processed for Western blot or RNA analyses as previously described (21).
Western Blot Analysis
Cells were harvested by scraping in radioimmunoprecipitation assay buffer (50 mM Tris, 1% Nonidet P-40, 0.25% deoxycholic acid, 150 mM NaCl, 1 mM EDTA, 1 mM sodium orthavanadate, and 1 mM sodium fluoride) with protease inhibitor (Sigma-Aldrich, St. Louis, MO). Protein concentration was measured by the bicinchoninic acid (BCA) assay. Total protein (15 µg) was resolved with 12% Tris-glycine SDS-PAGE and transferred to a PVDF membrane (Millipore). Membranes were blocked with 5% nonfat dry milk in PBS with Tween 20 for 1 h and then incubated with the appropriate antibodies followed by peroxidase-conjugated goat anti-rabbit IgG antibody (Jackson ImmunoResearch Laboratories, 1:10,000). Horseradish peroxidase activity was detected using the enhanced chemiluminescence KwikQuant detection system (Kindle Biosciences, Greenwich, CT). The membrane was stripped and probed with anti-β-actin antibody (Sigma-Aldrich, 1:5,000) to confirm loading and transfer. Densitometry analysis was performed, and the results were normalized to β-actin expression using Image Studio Lite (LI-COR Biosciences).
Real-Time Quantitative PCR
Ldha mRNA expression analysis was performed as previously described (20, 24). Briefly, total RNA was isolated from tissue culture plates using TRIzol (Thermo Fisher Scientific, Waltham, MA) and quantified using RT-PCR using SYBR Green Master Mix (Thermo Fisher, Waltham, MA). The reaction was done in triplicate to ensure accurate reading, and melting curves were inspected. Relative mRNA expression was quantified using the ΔΔCt method (where Ct is threshold cycle) and normalized to β-actin mRNA as an internal control. The following primers were used: mouse Ldha, forward 5′- TGTGGCAGACTTGGCTGAGA-3′ and reverse 5′- CTGAGGAAGACATCCTCATTGATTC-3′; and human Ldha, forward 5′- AGGCTACACATCCTGGGCTAT-3′ and reverse 5′- CCCAAAATGCAAGGAACACTA-3′.
LDH Activity Assay
LDH enzymatic activity in fresh cell lysates was determined as previously described (25) with minor modifications. Briefly, LDH activity was measured by the change in absorbance at 340 nm in 0.1 M PBS buffer with 0.25% Triton X-100 (BP151, Fisher BioReagents) at pH 7.4 at 37°C with 10 mM pyruvate and 0.3 mM NADH (Fluka, St. Louis, MO).
Immunohistochemistry
Kidneys were preserved by in vivo cardiac perfusion with PBS (pH 7.4) containing 6,000 U/L of Na-heparin followed by periodate-lysine-2% paraformaldehyde, cut transversely into several 2- to 3-mm-thick slices, and then immersed for 24–30 h at 4°C in the same fixative. Kidney samples from each animal were embedded in polyester wax using polyethylene glycol 400 distearate (Polysciences, Warrington, PA) with 10% 1-hexadecanol, and 2-μm-thick sections were cut and mounted on gelatin-coated glass slides.
Immunolocalization was accomplished using standard procedures as previously described (26–28). Briefly, sections were dewaxed in ethanol, rehydrated, heated in Trilogy (Cell Marque, Rocklin, CA) to 88°C for 30 min and then to 96°C for 30 min, cooled for 30 min, and rinsed in PBS. Endogenous peroxidase activity was blocked by incubating sections in 3% H2O2 in distilled water for 45 min. Sections were blocked for 15 min with Serum-Free Protein Block (DakoCytomation) and then incubated at 4°C overnight with primary antibody. Sections were washed in PBS, incubated for 30 min with polymer-linked, peroxidase-conjugated goat anti-rabbit IgG (MACH2, Biocare Medical, Concord, CA), washed again with PBS, and then exposed to diaminobenzidine for 5 min. Sections were washed in distilled water, dehydrated with xylene, mounted, and then observed by light microscopy.
Immunofluorescence
Harvesting, processing, and staining procedures were done following previously published procedures (20). Briefly, mouse kidney slices were fixed in 10% formalin solution overnight and then switched to 70% ethanol to be later embedded in paraffin. Five-micrometer-thick sections were cut, deparaffinized with xylene, and then rehydrated in graded ethanol and ultimately water. Trilogy solution (Cell Marque) was used as an antigen retrieval in which tissues were incubated for 20 min at 98°C and then cooled to room temperature. Sections were then incubated with LDHA or LDHB antibody overnight at 4°C. Incubation of anti-rabbit Alexa Fluor 594 (30 min at room temperature, 1:100–1:200 dilution, Thermo Fisher) and fluorescein-linked lotus lectin (FL-1321, Vector Laboratories, Burlingame, CA) was subsequently done after washes. As a control, unrelated appropriate IgG antibodies (Santa Cruz Biotechnology, Houston, TX) were used. Images were captured by a Leica DMi8 microscope (Leica Microsystems) using Image LAS X (Leica Microsystems). Fluorescence signal quantification was done using LAS X analysis software using at least 15 high-resolution images per sample per each isoform. Average positive percent area and intensity outputs from the software were used for the analysis.
Antibodies
Antibodies to LDHA (Cat. No. NBP1-48336, Novus Biologicals, Centennial, CO) and LDHB (Cat. No. 19988-1-AP, Proteintech, Rosemont, IL) were used for immunohistochemistry and immunofluorescence. For Western blot analysis, we used LDHA-specific antibody (Cat. No. 19987-1-AP, Proteintech) instead. We confirmed the specificity of these antibodies to recombinant protein of LDHA (Cat. No. NBP1-40407, Novus Biologicals) and LDHB (Cat. No. NBP1-45281, Novus Biologicals) using Western blot analysis (Supplemental Fig. S1B; all Supplemental material is available at https://doi.org/10.6084/m9.figshare.14055275).
RNA Sequencing
In this study, we used one set of single-cell and two sets of single-nucleus RNA-sequencing data from previously published work (29–31). To investigate Ldha and Ldhb transcripts in healthy mouse kidneys, we used the Kidney Cell Explorer tool (https://cello.shinyapps.io/kidneycellexplorer/) developed for the single-cell RNA-sequencing dataset (GSE129798) (31). This dataset consisted of two male and two female adult C57BL/6J mice. To investigate differential transcript expression of Ldha and Ldhb post-IRI, we used single-nucleus RNA-sequencing datasets: GSE151167 (15 min of bilateral IRI, n = 1 per group) (30) and GSE139107 (10 min of bilateral IRI, n = 4 per group) (29). Analyses were carried out using packages created for the R statistical analysis environment (version 3.6). The primary package used was Seurat (v3.2.1) (32) and associated dependencies. To compare the experimental conditions, the datasets were integrated based on common sources of variation (33) as implemented in the Seurat package. For GSE151167, clusters were determined using the FindClusters function with a resolution constant of 0.2. The dimensional reduction was performed using uniform manifold approximation and projection. For GSE139107, provided metadata were used to determine clustering to minimize variation with the author’s study.
Statistics
Statistical analyses were performed using Graphpad Prism (version 8.4). Specific statistical tests are indicated in the figures and include one-way ANOVA with a post hoc test. Data are presented as means ± SE. Values of P < 0.05 were considered statistically significant.
RESULTS
In Vitro Analysis of LDHA and LDHB
Western blots of lysates from primary mouse PTCs were probed with LDHA and LDHB isoform-specific antibodies and revealed a significant increase in proximal tubule LDHA expression 24 h after 1% hypoxia (Fig. 1B). LDHB levels were not significantly altered by hypoxia exposure. The expression of both isoforms was quantified by densitometry (Fig. 1C). Exposure to hypoxia resulted in a significant increase in overall LDH activity in fresh PTC protein lysates (Fig. 1D). To confirm the response in human cells, we subjected HK-2 cells to identical hypoxic conditions. Ldha mRNA levels were significantly increased following hypoxia in mouse PTCs and human HK-2 cells (Fig. 1E). These results demonstrate that both LDHA and LDHB are present in renal PTCs and that LDHA protein, mRNA, and LDH activity are upregulated in cells subjected to hypoxic conditions.
Immunolocalization of LDHA in a Healthy Kidney
Both LDHA and LDHB subunits are present in most cells as these proteins are essential for basal metabolism. The antibodies used were specific to LDHA and LDHB subunits and could not differentiate between LDH complex isoforms (LDH-1−LDH-5). If both LDHA and LDHB subunits were present within the complex, we would have observed labeling from both antibodies. It is difficult to determine the exact ratio of LDH isoforms using subunit-specific antibodies as it relies on the assumption that antibodies produce an equal signal to both subunits. However, we compared the distribution of each subunit throughout the nephron. We found that LDHA subunit was detected at higher levels proximally, whereas LDHB was expressed at higher levels distally. This indicates that proximal tubules are more skewed toward the production of lactate whereas distal segments are skewed toward its utilization.
Specific labeling with anti-LDHA antibodies was observed in the cortex and outer stripe of the outer medulla (OSOM) with a clear decrease in label in the inner stripe of the outer medulla (ISOM) and inner medulla (IM) (Fig. 2A). Proximal convoluted tubule (PCT) cells displayed substantial uniform cytoplasmic LDHA staining with a stronger signal in the brush border and strongest at the base of the brush border (Fig. 2C). Proximal straight tubule cells (PSTs) showed similar cytoplasmic LDHA expression as PCT cells; however, they showed weaker, yet still significant, expression in the brush border (Fig. 2E). Control sections did not reveal any nonspecific staining in the brush border in the absence of primary antibody, suggesting that such staining was specific (Supplemental Fig. S1A). LDHA staining retained similar intensity throughout the entire length of the PST starting at the cortex and ending at the ISOM. The thick ascending limb (TAL) displayed no significant LDHA staining (Fig. 2G). Distal convoluted tubule (DCT) cells displayed nominal LDHA staining; however, in distant segments of the DCT, some of the cells showed a strong LDHA label, which can be attributed to the incorporation of connecting tubule (CNT) cells into the DCT (Fig. 2C). CNT cells had a strong LDHA label, which was intensified at the apical side of the cell. Collecting duct (CD) cells were weakly labeled except for cortical intercalated cells, which showed a much stronger label with increased intensity at the apical side of the cell (Fig. 2E). Positively labeled intercalated cells were localized in the cortex and OSOM, but as the CD advanced toward the IM, the signal disappeared (Fig. 2I).
Figure 2.
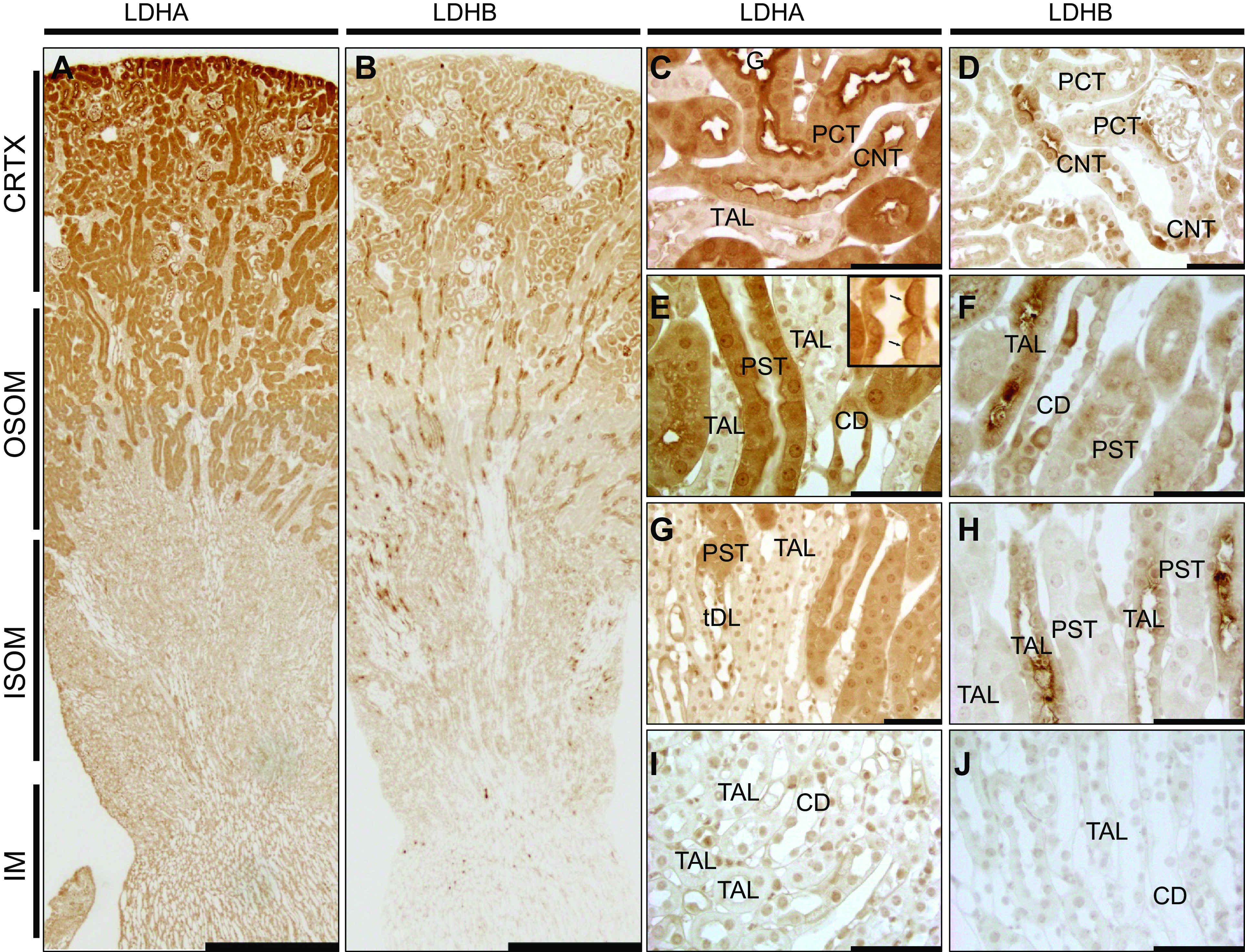
Lactate dehydrogenase (LDH) isoform A (LDHA) and LDH isoform B (LDHB) localization in the healthy mouse kidney. Immunohistochemistry for LDHA and LDHB expression in the normal mouse kidney is shown. A and B: low-power micrographs (scale bars = 500 µm) of LDHA and LDHB immunolabel, respectively. LDHA was strongest in the cortex (CRTX) and outer stripe of the outer medulla (OSOM) but was weak in the inner stripe of the outer medulla (ISOM) and inner medulla (IM). LDHB label was present in the CRTX but was strongest in medullary rays going through the ISOM. C−J: higher-power micrographs (scale bars = 50 µm) of LDHA and LDHB showing adjacent images for the CRTX (C and D), OSOM (E−H), and IM (I and J). The arrows in the inset of E points to an intercalated cell. Micrographs are representative of n = 3 animals per group. CD, collecting duct; CNT, connecting tubule; PCT, proximal convoluted tubule; PST, proximal straight tubule cell; TAL, thick ascending limb.
Immunolocalization of LDHB in the Normal Kidney
LDHB label was present in the cortex; however, unlike LDHA, it was especially concentrated in medullary rays extending deep into the ISOM (Fig. 2B). PCT LDHB expression was similarly intense between the cell body and brush border (Fig. 2D). The PST displayed weak cytoplasmic and even weaker brush border staining (Fig. 2F). LDHB expression was gradually weakened as PST progressed from the cortex to the outer medulla (Fig. 2H). TAL LDHB label was observed throughout the entire cell body with a very strong concentration on the apical side of the cell; however, the intensity of staining was lost in the IM (Fig. 2J). The DCT showed a weak uniform LDHB label compared with PCT cells (Fig. 2D). Intercalated cells in the late DCT/CNT showed a strong LDHB signal with the concentration on the apical membrane (Fig. 2D). Principal cells of the CD showed a significant yet weak label of LDHB, however, some intercalated cells stained more strongly for both isoforms with increased intensity at the apical side of the cell. Positively labeled intercalated cells were seen in the cortex and OSOM, but as the CD advanced toward the IM, the signal disappeared (Fig. 2J).
Transcript Expression of Ldha and Ldhb in the Normal Mouse Kidney
To investigate transcript levels of both LDH isoforms in cells of specific tubular segments in a healthy state, we used the Kidney Cell Explorer tool, which was developed by Ransick et al. (31) for their single-cell RNA-sequencing dataset (GSE129798). Both male and female mice were present in these data with 32 clusters identified (Fig. 3A). Ldha gene transcripts were notable in the proximal segments and in the loop of Henle as well as in principal cells (Fig. 3, B and D). In contrast, Ldhb transcripts were maximal in the distal cell types in the nephron, including the distal tubule and CD cells, consistent with the immunostaining results (Fig. 3, C and D). Expression proportion data highlighted the average expression findings (Fig. 3E).
Figure 3.
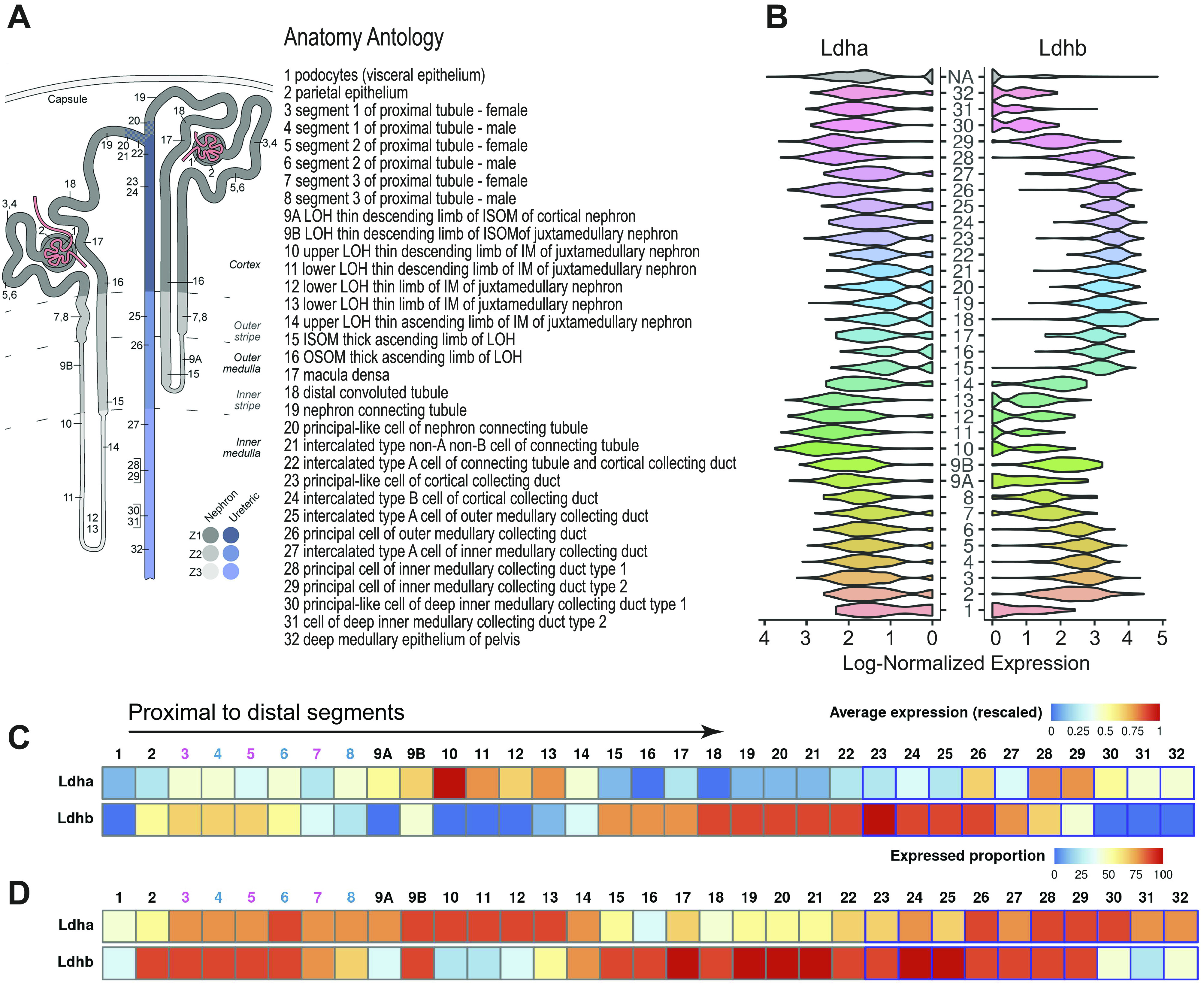
Lactate dehydrogenase (LDH) isoform A and isoform B (Ldha and Ldhb, respectively) mRNA expression in a healthy mouse nephron. Using publicly available mouse renal single-cell RNA-sequencing data [GSE129798, Ransick et al. (30)] and the Kidney Cell Explorer tool (https://cello.shinyapps.io/kidneycellexplorer/), we investigated Ldha and Ldhb transcript expression throughout nephron segment-specific clusters. A: multiple cell populations were identified in this dataset, which is shown in the schematic along with the description of each population ID (NA indicates unidentified cells). The distinction between female (4, 6, and 8) and male (3, 5, and 7) segments (S1−S3) of proximal tubule cells is also shown. B: log-normalized expression plots illustrating the Ldha and Ldhb transcript distribution throughout the nephron. Ldha transcript expression was strongest in proximal segments and the loop of Henle, whereas Ldhb expression was present in proximal segments but was strongest in the distal tubule and collecting duct segments. C: average transcript expression in nephron segments of both isoforms. D: proportion of cells expressing the gene highlighting the differential distribution.
Differential Expression of LDHA and LDHB Protein in Acute Kidney Injury
To examine the specific changes of LDH isoform protein expression after renal injury, we conducted immunofluorescence experiments on kidney sections from mice that underwent renal IRI and were harvested at days 1, 3, 7, 14, and 28 post-IRI. All samples were stained and imaged at constant exposure to allow consistency with the quantification. LDHA label was strongly present in PSTs, which are more susceptible to IRI (Fig. 4A). At day 3 postinjury, obvious signs of tubular injury and necrosis characterized by dispersed lotus lectin staining were observed (Fig. 4C). LDHA expression in the cortex decreased postinjury at day 3. As PST cells proceeded to repair and recover their normal appearance postinjury, LDHA levels were restored by day 28 (Fig. 4E); however, some of the tubules stopped showing any significant label of LDHA altogether, resulting in a checkered pattern. This pattern was progressively more prominent at day 28 postinjury (Fig. 4, K and M). We confirmed that tubular cells that lack LDHA showed normal levels of LDHB by examining the same tubules using serial sections (Fig. 4, L and N). The percent area positive for LDHA expression significantly decreased postinjury but partially recovered on day 28 (Fig. 4G). Despite the decreased percentage of LDHA-positive area post-IRI, the overall intensity of the label divided by the percent area did not change significantly, indicating a potential compensatory mechanism (Fig. 4H).
Figure 4.
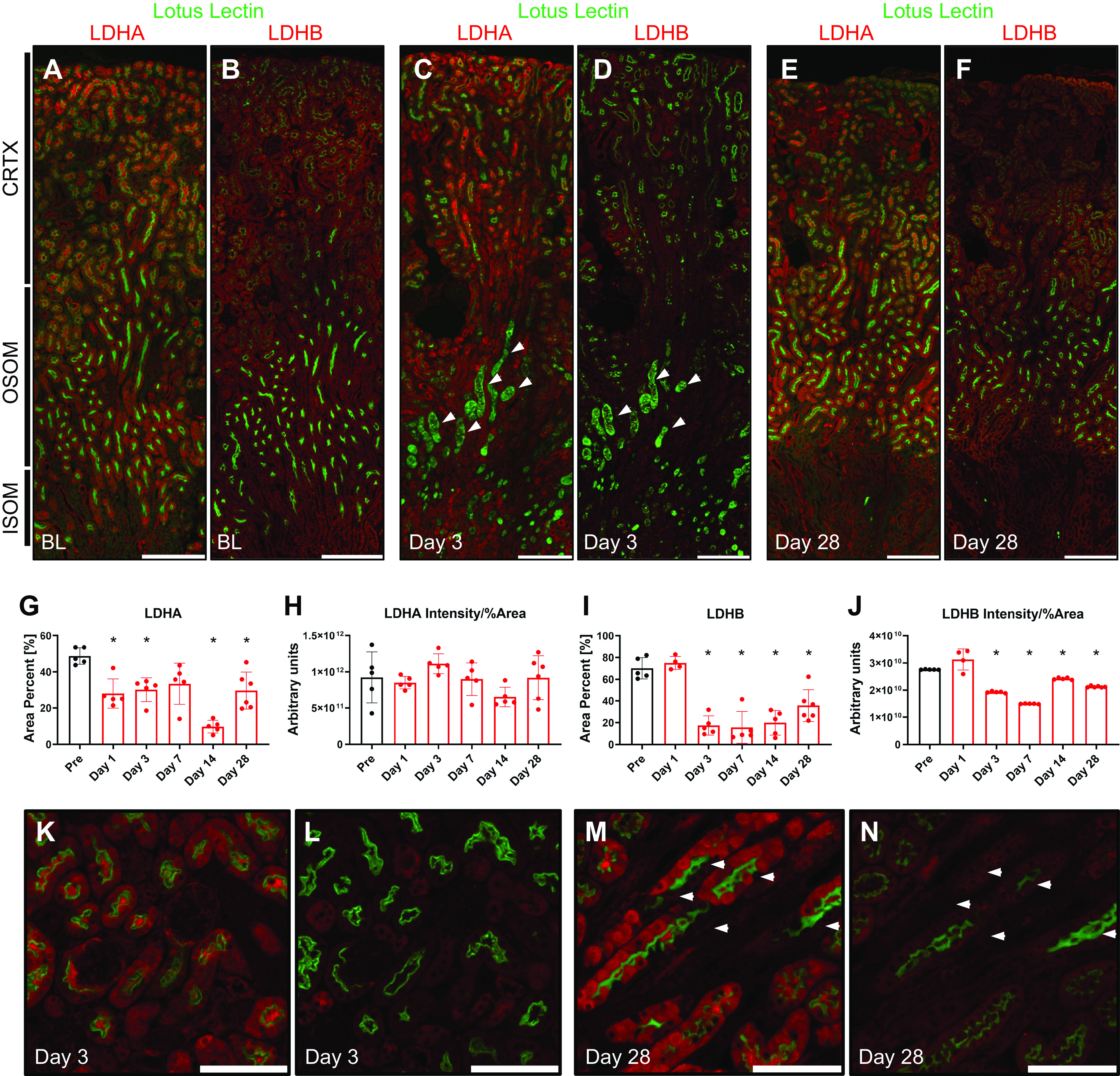
Lactate dehydrogenase (LDH) isoform A (LDHA) and LDH isoform B (LDHB) expression in the mouse kidney after ischemia-reperfusion injury. Immunofluorescence was performed for LDHA and LDHB on consecutive sections of kidneys after ischemia-reperfusion injury. Lotus lectin was used to identify the brush border of proximal tubules. A−F: low-power micrographs of baseline (A and B), day 3 (C and D), and day 28 (E and F) mouse kidney sections were stained for LDHA and LDHB, respectively. The arrows in C and D highlight the damaged proximal straight tubule. LDHA expression was then measured using LAX software to calculate the percent area (G) and total intensity/percent area (H). Similar calculations were done for LDHB (I and J). K−N: high-power micrographs of LDHA and LDHB were done on consecutive sections of the same area at day 3 (K and L) and day 28 (M and N) postischemia. The arrows in M and N point at the same cells in serial sections highlighting the expression of LDHB but not LDHA. Scale bars = 400 µm in for A−F and 200 µm in K−N. n = 5 animals per group (4 per LDHB day 1). *P < 0.05 using one-way ANOVA. CRTX, cortex; ISOM, inner stripe of the outer medulla; OSOM, outer stripe of the outer medulla.
LDHB expression did not alter at day 1 postinjury but appeared to be decreased on day 3 in the entire section (Fig. 4D). Distal tubules and connecting segments showed LDHB expression, whereas levels in proximal tubules remained downregulated (Fig. 4L). By day 28, the overall expression of LDHB appeared to remain slightly decreased; however, the pattern of expression was similar to the uninjured kidney (Fig. 4, F and N). The LDHB-positive area slightly increased on day 1 but significantly decreased on day 3 and failed to recover to nominal levels by day 28 postinjury (Fig. 4I). When total intensity divided by the percentage of the LDHB-positive area was taken into account, it also showed a slight but significant decrease by day 28 (Fig. 4J). Significant LDHA and LDHB labeling was detected in the tubular lumen postinjury, suggesting the potential release of LDH into the lumen or nonspecific staining. Sections used for immunohistochemistry were perfusion fixed, whereas immunofluorescence sections were not. We believe that this intense staining of the tubular lumen could be nonspecific staining of cellular debris from the cells damaged during sample preparation. In noninjured kidneys, immunofluorescent staining was observed in the lumen whereas no such staining was observed in perfusion-fixed sections in which lumens were open and clean of debris.
Proximal tubules, selected by positive lotus lectin labeling, were analyzed for LDHA and LDHB expression. LDHA expression in these tubules decreased significantly postinjury at day 1 and showed a recovery trend yet remained slightly below preinjury levels even after 28 days (Supplemental Fig. S1, C and D). LDHB expression in these tubules remained the same on day 1 but decreased by day 3, showing a recovery trend that, similarly to LDHA, remained below preinjury levels by day 28 postinjury (Supplemental Fig. S1, C and D).
Cell-Specific Differential Transcript Expression of Ldha and Ldhb in Acute Kidney Injury
To validate immunofluorescent findings and obtain higher resolution cell and segment-specific expression of Ldha and Ldhb, we used two publicly available single-nucleus RNA-sequencing datasets from mouse kidneys following IRI. We first evaluated single-nucleus RNA-sequencing data published by Legouis et al. (30) (GSE151167) for clusters containing cells expressing tubule segment-specific genes and Ldha and Ldhb expression. The percentage of cells expressing Ldha transcripts initially showed an increase at 64 h but decreased in most compartments of the nephron by 96-h postinjury (Fig. 5A) except for intercalated cells. The percentage of cells that expressed detectable levels of Ldha was significantly smaller than the percentage of cells that expressed Ldhb. The percentage of Ldhb-positive cells decreased at 64-h postinjury and remained low by 28 days (Fig. 5B). To investigate this further, we evaluated the percentage of cells that were exclusively Ldha positive, exclusively Ldhb positive, Ldha and Ldhb positive, or Ldha and Ldhb negative in the noninjured and injured state (Fig. 5C). The proportion of cells expressing detectable levels of either Ldha or Ldhb decreased from 47% in the noninjured state to 19% at 96-h postinjury. The percentage of positive cells expressing either isoform did not fully recover to the healthy state and remained low at 28 days postinjury, indicating that fewer cells remained expressing Ldha or Ldhb post-IRI. The percentage of cells that had detectable levels of both isoforms was smaller than the percentage of cells that expressed Ldha or Ldhb isoform only. Since a significant percentage of cells in these data did not show detectable levels of Ldha and Ldhb, we used violin plots to visualize isoform expression between the groups using subsets of Ldha-positive (Fig. 5D) and Ldhb-positive (Fig. 5E) cells. Both Ldha and Ldhb gene transcripts were increased post-IRI, peaking at 96 h, and remained elevated by day 28.
Figure 5.
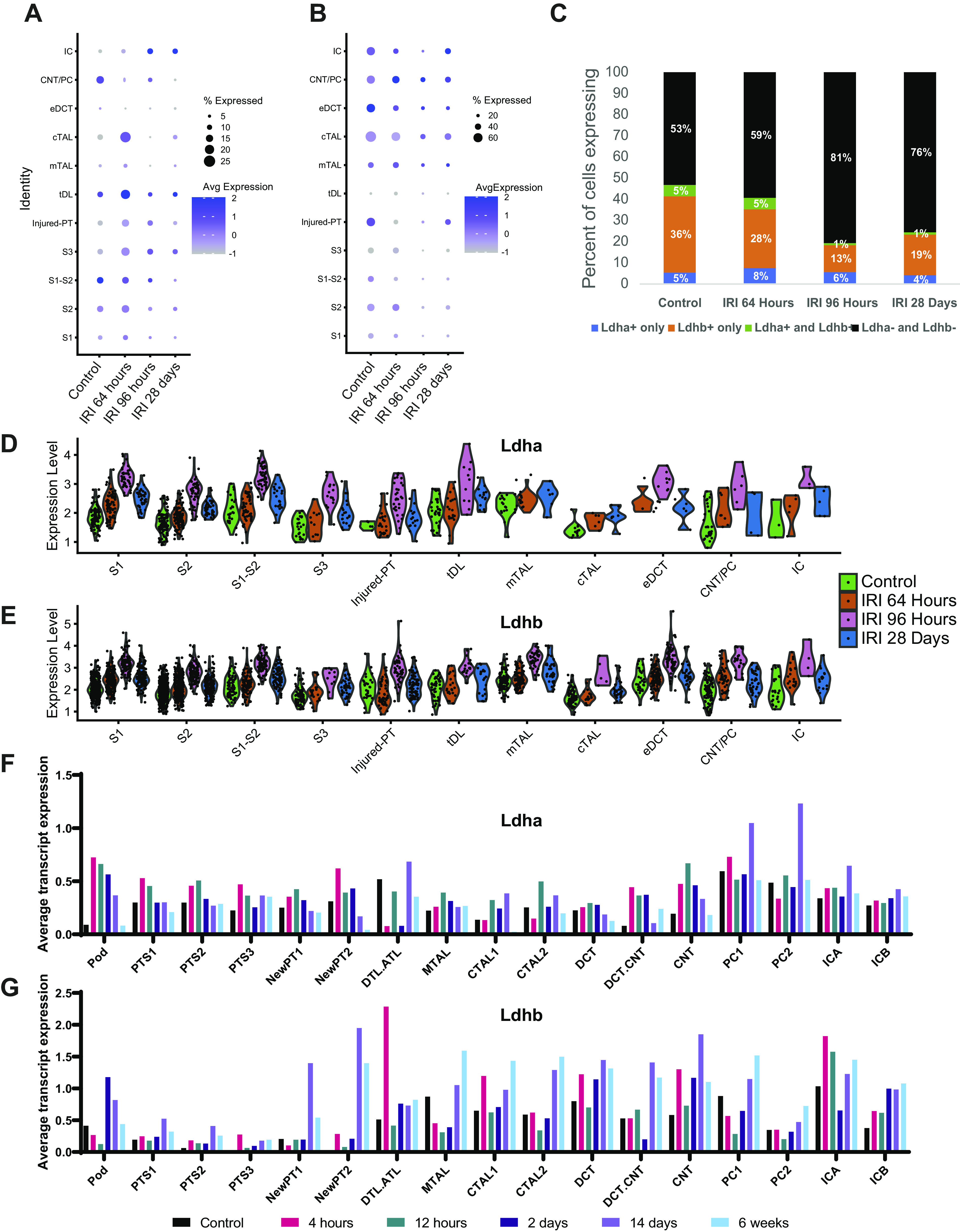
Lactate dehydrogenase (LDH) isoform A and isoform B (Ldha and Ldhb, respectively) gene expression changes in mouse kidneys after ischemic acute kidney injury. Single-nucleus RNA-sequencing data [GSE151167, Legouis et al. (30)], containing one animal per group, was used in A−E. Dot charts of Ldha (A) and Ldhb (B) expression in identified clusters corresponding to specific tubular segments compared with control, 64 h, 96 h, and 28 days postischemic injury. C: comparison of the percent abundance between Ldha-positive, Ldhb-positive, Ldha and Ldhb-positive, and Ldha and Ldhb-negative cells. The Ldha-positive subset was analyzed for Ldha expression (D) and the Ldhb-positive subset was analyzed for Ldhb expression (E). The single-nucleus RNA-sequencing data [GSE139107, Kirita et al. (29)], containing four animals per group, was used in F and G. Metadata were used to assign cell clusters. Average transcript expression of Ldha (F) and Ldhb (G) for control, 4 h, 12 h, 2 days, 14 days, and 6- wk post-ischemia-reperfusion (IRI).
To ensure the accuracy of our findings, we used an additional single-nucleus RNA-sequencing dataset containing a larger amount of cells and animals per group as well as earlier time points published by Kirita et al. (29) (GSE139107). Provided metadata was used to ensure a consistent clustering assignment with the original study. Expression of Ldha transcripts was upregulated even 4 h after IRI, especially in proximal tubules and connecting segments, and then returned to preinjury levels by 6 wk (Fig. 5F). In contrast, Ldhb transcripts did not increase in proximal tubules until 2 days postinjury, increased at 4 h in the connecting segment, and were elevated at 6 wk in both segments (Fig. 5G). These data highlight that both isoform and cell/segment specificity in the response of Ldha and Ldhb to injury.
Expression of LDHA and LDHB in a Model of CKD
To investigate LDHA and LDHB expression in CKD, we used the adenine-induced nephropathy model. Immunofluorescence revealed an apparent decrease of lotus lectin-positive tubules, which indicated a diminished number of functioning tubules with a healthy brush border. LDHA expression was decreased globally (Fig. 6, A and C). LDHB expression was also nominal in the tubules that were intact and followed a similar decrease in overall expression as LDHA (Fig. 6, B and D). No tubule-specific differences were observed.
Figure 6.
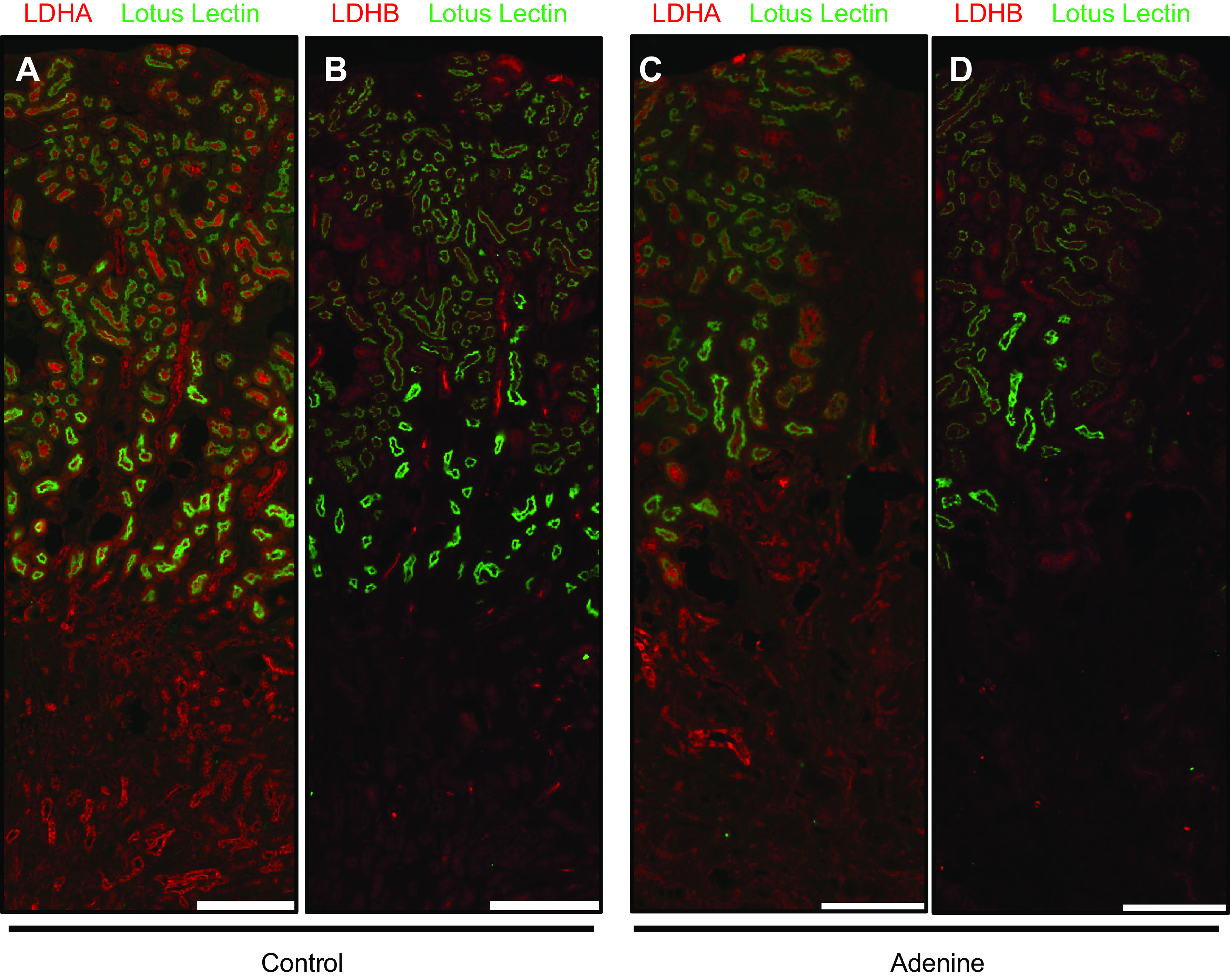
Lactate dehydrogenase (LDH) isoform A (LDHA) and LDH isoform B (LDHB) expression in chronic kidney disease. Immunofluorescence was performed for LDHA and LDHB expression on consecutive sections using kidneys from mice on an adenine diet. Lotus lectin was colocalized to identify the brush border of proximal tubules. Control kidneys are shown in A and B; kidneys with chronic kidney disease are shown in C and D. Scale bars = 400 µm. Representative images from n = 5/group are shown. Parts of the graphical abstract were created with BioRender.com.
DISCUSSION
Our study provides evidence that LDH isoforms (LDHA and LDHB) are differentially expressed throughout the nephron and are subject to spatial- and time-dependent responses to ischemic injury in the kidney. In vitro, using isoform-specific antibodies, we showed that LDH is expressed in both human and mouse PTCs. LDHA expression increased significantly during hypoxia, yet LDHB remained largely unchanged. Our study also demonstrates the differential in vivo expression of LDH isoforms, both at the level of RNA transcript and protein, throughout the normal nephron and their response to IRI in the mouse kidney. Finally, we show that both LDHA and LDHB expression is decreased globally in a model of CKD.
Several previous studies have shown a significant increase in the renal pyruvate-to-lactate ratio postinjury (34) as well as LDH release during hypoxic injury (13, 35); however, these studies did not investigate native LDH expression or distinguish between isoforms. Here, we show that mouse proximal tubules express detectable levels of both isoforms and that LDHA mRNA, protein, and LDH activity are significantly upregulated 24 h after hypoxia. Basal levels of LDHA protein in human HK-2 cells were very high, and we were unable to detect changes in LDH activity or protein levels (data not shown). However, LDHA mRNA levels increased more than fivefold after hypoxia. This can be attributed to the fact that HK-2 cells are an immortalized cell line and may have altered basal metabolic rates. Our findings of increased expression of LDHA, but not LDHB, after 24-h hypoxia in primary PTCs are reinforced by single-nucleus RNA-sequencing data. Single-nucleus RNA-sequencing data showed that while Ldha transcripts are elevated even after 4-h post-IRI in proximal tubules, Ldhb transcripts are only upregulated by day 2, which highlights the differential response of both isoforms to the injury.
LDH is released from cells upon injury, and increased LDH levels in serum and urine have been suggested as a potential marker of renal injury (13). Both LDHA and LDHB contribute to LDH enzymatic activity, which is usually measured by the conversion of NAD+ to NADH. Distinguishing between LDHA and LDHB localization patterns provides better insight into the specific damaged nephron segments during injury. Our results showed that LDHA is mostly expressed in the PCT and PST, tubules that are highly sensitive to ischemic injury; however, LDHB is more highly expressed in distal nephron segments in the TAL, DCT, CNT, and CD. In addition to being an end product of glycolysis, lactate also plays a role in inflammation, energy regulation, wound healing, epigenetics (histone lactylation) (36), and ischemic tissue injury (37). The lactate shuttle discovered by Brooks (38) in brain astrocytes may play a role in the kidney. Our observation that LDHA, the lactate-producing isoform, being expressed more in proximal segments of the nephron when LDHB, the lactate-utilizing isoform, is expressed in distal segments suggests potential disparate roles for LDH and lactate in the kidney. LDH potentially could be supplying lactate as a nutrient to the more distal areas of the nephron where glucose and oxygen are not as readily available, analogous to the lactate shuttle, or producing lactate as a signaling mechanism to establish communication within the nephron.
Renal IRI, a major underlying cause of acute kidney injury (AKI), is characterized by an initial decrease of blood flow followed by its subsequent restoration. Currently, there is an unmet need for approaches to circumvent injury and to develop therapies to prevent IRI and subsequent AKI (8). Dysregulation of energy metabolism is a potential driver of AKI and decreased LDH levels have been reported in models of renal IRI (13). Our results correlate with such previously reported data showing that overall cortical LDH protein levels decrease following AKI. However, we found LDHA and LDHB initially decreased and showed a trend toward recovery. However, LDHA decreased on day 1, whereas the decrease in LDHB was delayed at day 3 postinjury. We also observed a change in the localization of LDHB expression at days 3−7 with an increase in distal tubule expression, which then returned to basal levels at days 14 and 28. This suggests that LDHA and LDHB are differentially regulated throughout the nephron during injury even though the overall LDH levels are decreased.
Prior studies have demonstrated the accumulation of lactate in the kidney in models of AKI; however, those studies, which were limited to the whole kidney, did not consider expression in kidney substructures or cell types. Differential expression of LDH isoforms in the healthy kidney suggests potential variations in lactate and pyruvate metabolism in the kidney following recovery from the injury. Although it is known that LDH is regulated by hypoxia-inducible factor-1α, the molecular mechanisms underlying its differential expression in nephron segments following IRI is not known. We observed that after injury, renal tubules return to their normal appearance, yet some cells show almost no LDHA expression but strong LDHB expression. We speculate that this lack of LDHA expression may be due to altered cellular metabolism that occurs in the cells during repair and regeneration. Dualistic expression of renal LDH isoforms suggests a potential presence of a lactate shuttle in the kidney, similar to the lactate shuffle observed in astrocytes and neurons (39, 40). LDHA produces lactate proximally whereas LDHB converts it to pyruvate distally, which may be satisfying the energy demands of distal nephron segments. Such differential lactate metabolism within a nephron might allude to the role of lactate as a signaling metabolite within the kidney.
The results of our study were complemented by single-cell RNA-sequencing and single-nucleus RNA-sequencing data allowing exploration of isoform-specific expression in cells that belong to specific nephron segments identified by highly specific tubular markers. Single-nucleus RNA-sequencing analysis revealed that fewer cells express Ldha or Ldhb transcripts after IRI, which correlates well with the immunostaining results showing an overall decrease in the percentage of the area in expression. Despite that, cells that are positive for a specific LDH isoform express it at a higher level, which might explain why the overall intensity/percent area is not diminished to a similar extent. An increase in the abundance of cells that express only Ldhb, and not Ldha, transcripts might also explain our findings that some PST cells stop expressing detectable levels of LDHA protein postinjury, whereas LDHB protein is observed in the same cells.
A previously published study by Wu et al. (41) compared single-cell and single-nucleus RNA-sequencing techniques in kidneys. This study concluded that single-nucleus RNA sequencing had advantages with consistency with transcript expression of most genes and was superior to single-cell RNA sequencing, particularly in fibrotic kidneys. During our analysis of two independent data sets of single-nucleus RNA sequencing, we noticed that only a small percentage of cells expressed detectable transcripts levels of Ldha and Ldhb, in contrast to the wide expression of both isoforms in single-cell RNA-sequencing data used to analyze LDH expression in normal kidneys. The rapid increase in Ldha mRNA transcript expression in proximal tubules highlights the initial glycolytic switch after injury. As tubular cells recover, the global decline in protein expression of both LDH isoforms suggests a long-lasting change in renal cellular metabolism as it remained significantly altered even 28 days after injury. This study provides support that AKI and CKD result in persistent effects on lactate metabolism and could potentially limit the kidney’s ability to engage the glycolytic switch required for cellular proliferation and recovery after injury.
Rapid loss of kidney function during AKI frequently transitions to progressive CKD. We investigated whether there are longitudinal effects of the injury on LDH isoform expression. Despite the changes observed in AKI, neither LDHA nor LDHB showed a compensatory mechanism in the adenine-induced model of CKD, suggesting that changes in LDH expression were due to an acute response to the injury. These results suggest that LDHA and LDHB are important response factors in the immediate ischemic cellular injury response and their expression is not a consequence of decreased renal function. Despite the decreasing number of viable proximal tubules in both AKI and CKD, the remaining PTCs showed decreased levels of both LDHA and LDHB compared with preinjury levels. This suggests a potential switch of the cellular metabolic profile after injury. The precise mechanism for this change remains unclear.
Overall, this study describes the nominal expression of LDHA and LDHB isoforms throughout the kidney at both protein and gene transcript levels. We investigated the differential renal response of both isoforms to ischemic AKI showing the time- and location-sensitive response. These findings are novel as they highlight that both LDH isoforms play an important role in the renal response to injury as an innate metabolic enzyme. Changes of expression between isoforms suggest a significant role for lactate and pyruvate metabolism in the kidney.
GRANTS
This work was supported by grants from National Institutes of Health Grant R01DK59600 and U54ES030246 (to A.A.), T32DK007545 (to L.M.B.), and K08HL140294 (to A.Z.), the Univeristy of Alabama at Birmingham-University of California-San Diego O’Brien Center for Acute Kidney Injury Research Grant P30DK079337 (to A.A.), a Merit Award from the Department of Veterans Affairs Grant 1I01BX004047 (to A.A.), and a Predoctoral Fellowship Award from the American Heart Association Grant 20PRE35200054 (to G.O.).
DISCLOSURES
A.A. serves as a consultant for Dynamed and is on the advisory boards of Goldilocks Therapeutics, Reata, Akebia, Alpha Young, and Angion for work outside the scope of this manuscript. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
AUTHOR CONTRIBUTIONS
G.O. and A.A. conceived and designed research; G.O., D.S., and J.W.V. performed experiments; G.O., A.M.T., L.M.B., D.S., J.F.G., A.Z., J.W.V., and A.A. analyzed data; G.O., L.M.B., A.Z., and A.A. interpreted results of experiments; G.O. prepared figures; G.O., J.F.G., and A.Z. drafted manuscript; G.O., A.M.T., L.M.B., D.S., J.F.G., A.Z., J.W.V., and A.A. edited and revised manuscript; G.O., A.M.T., D.S., J.F.G., A.Z., J.W.V., and A.A. approved final version of manuscript.
ACKNOWLEDGMENTS
We appreciate the technical assistance of Yanlin Jiang in conducting animal surgeries.
REFERENCES
- 1.Wang Z, Ying Z, Bosy-Westphal A, Zhang J, Schautz B, Later W, Heymsfield SB, Muller MJ. Specific metabolic rates of major organs and tissues across adulthood: evaluation by mechanistic model of resting energy expenditure. Am J Clin Nutr 92: 1369–1377, 2010. doi: 10.3945/ajcn.2010.29885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Connor PM. Renal oxygen delivery: matching delivery to metabolic demand. Clin Exp Pharmacol Physiol 33: 961–967, 2006. doi: 10.1111/j.1440-1681.2006.04475.x. [DOI] [PubMed] [Google Scholar]
- 3.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134: 112–123, 2008. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venkatachalam MA, Bernard DB, Donohoe JF, Levinsky NG. Ischemic damage and repair in the rat proximal tubule: differences among the S1, S2, and S3 segments. Kidney Int 14: 31–49, 1978. doi: 10.1038/ki.1978.87. [DOI] [PubMed] [Google Scholar]
- 5.Huang H, van Dullemen LFA, Akhtar MZ, Faro ML, Yu Z, Valli A, Dona A, Thezenas ML, Charles PD, Fischer R, Kaisar M, Leuvenink HGD, Ploeg RJ, Kessler BM. Proteo-metabolomics reveals compensation between ischemic and non-injured contralateral kidneys after reperfusion. Sci Rep 8: 8539, 2018. doi: 10.1038/s41598-018-26804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basile DP. The endothelial cell in ischemic acute kidney injury: implications for acute and chronic function. Kidney Int 72: 151–156, 2007. doi: 10.1038/sj.ki.5002312. [DOI] [PubMed] [Google Scholar]
- 7.Kalogeris T, Baines CP, Krenz M, Korthuis RJ. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol 298: 229–317, 2012. doi: 10.1016/B978-0-12-394309-5.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kezic A, Stajic N, Thaiss F. innate immune response in kidney ischemia/reperfusion injury: potential target for therapy. J Immunol Res 2017: 6305439, 2017. doi: 10.1155/2017/6305439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valvona CJ, Fillmore HL, Nunn PB, Pilkington GJ. The regulation and function of lactate dehydrogenase A: therapeutic potential in brain tumor. Brain Pathol 26: 3–17, 2016. doi: 10.1111/bpa.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9: 425–434, 2006[Erratum inCancer Cell10: 172, 2006]. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 11.Sriram R, Nguyen J, Santos JD, Nguyen L, Sun J, Vigneron S, Van Criekinge M, Kurhanewicz J, MacKenzie JD. Molecular detection of inflammation in cell models using hyperpolarized 13C-pyruvate. Theranostics 8: 3400–3407, 2018. doi: 10.7150/thno.24322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girgis H, Masui O, White NM, Scorilas A, Rotondo F, Seivwright A, Gabril M, Filter ER, Girgis AH, Bjarnason GA, Jewett MA, Evans A, Al-Haddad S, Siu KM, Yousef GM. Lactate dehydrogenase A is a potential prognostic marker in clear cell renal cell carcinoma. Mol Cancer 13: 101, 2014. doi: 10.1186/1476-4598-13-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zager RA, Johnson AC, Becker K. Renal cortical lactate dehydrogenase: a useful, accurate, quantitative marker of in vivo tubular injury and acute renal failure. PLoS One 8: e66776, 2013. doi: 10.1371/journal.pone.0066776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 15: 243–256, 2014. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershberger KA, Martin AS, Hirschey MD. Role of NAD+ and mitochondrial sirtuins in cardiac and renal diseases. Nat Rev Nephrol 13: 213–225, 2017. doi: 10.1038/nrneph.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poyan Mehr A, Tran MT, Ralto KM, Leaf DE, Washco V, Messmer J, Lerner A, Kher A, Kim SH, Khoury CC, Herzig SJ, Trovato ME, Simon-Tillaux N, Lynch MR, Thadhani RI, Clish CB, Khabbaz KR, Rhee EP, Waikar SS, Berg AH, Parikh SM. De novo NAD+ biosynthetic impairment in acute kidney injury in humans. Nat Med 24: 1351–1359, 2018. doi: 10.1038/s41591-018-0138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobsen NO. The histochemical localization of lactic dehydrogenase isoenzymes in the rat nephron by means of an improved polyvinyl alcohol method. Histochemie 20: 250–265, 1969. doi: 10.1007/BF00306013. [DOI] [PubMed] [Google Scholar]
- 18.Smith CH, Kissane JM. Distribution of forms of lactic dehydrogenase within the developing rat kidney. Dev Biol 8: 151–164, 1963. doi: 10.1016/0012-1606(63)90039-3. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen PM, Laustsen C, Bertelsen LB, Qi H, Mikkelsen E, Kristensen ML, Norregaard R, Stodkilde-Jørgensen H. In situ lactate dehydrogenase activity: a novel renal cortical imaging biomarker of tubular injury? Am J Physiol Renal Physiol 312: F465–F473, 2017. doi: 10.1152/ajprenal.00561.2015. [DOI] [PubMed] [Google Scholar]
- 20.Zarjou A, Black LM, Bolisetty S, Traylor AM, Bowhay SA, Zhang MZ, Harris RC, Agarwal A. Dynamic signature of lymphangiogenesis during acute kidney injury and chronic kidney disease. Lab Invest 99: 1376–1388, 2019. doi: 10.1038/s41374-019-0259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolisetty S, Traylor AM, Kim J, Joseph R, Ricart K, Landar A, Agarwal A. Heme oxygenase-1 inhibits renal tubular macroautophagy in acute kidney injury. J Am Soc Nephrol 21: 1702–1712, 2010. doi: 10.1681/ASN.2010030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kie JH, Kapturczak MH, Traylor A, Agarwal A, Hill-Kapturczak N. Heme oxygenase-1 deficiency promotes epithelial-mesenchymal transition and renal fibrosis. J Am Soc Nephrol 19: 1681–1691, 2008. doi: 10.1681/ASN.2007101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taub M, Sato G. Growth of functional primary cultures of kidney epithelial cells in defined medium. J Cell Physiol 105: 369–378, 1980. doi: 10.1002/jcp.1041050220. [DOI] [PubMed] [Google Scholar]
- 24.Bolisetty S, Zarjou A, Hull TD, Traylor AM, Perianayagam A, Joseph R, Kamal AI, Arosio P, Soares MP, Jeney V, Balla J, George JF, Agarwal A. Macrophage and epithelial cell H-ferritin expression regulates renal inflammation. Kidney Int 88: 95–108, 2015. doi: 10.1038/ki.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renner K, Amberger A, Konwalinka G, Kofler R, Gnaiger E. Changes of mitochondrial respiration, mitochondrial content and cell size after induction of apoptosis in leukemia cells. Biochim Biophys Acta 1642: 115–123, 2003. doi: 10.1016/s0167-4889(03)00105-8. [DOI] [PubMed] [Google Scholar]
- 26.Handlogten ME, Osis G, Lee HW, Romero MF, Verlander JW, Weiner ID. NBCe1 expression is required for normal renal ammonia metabolism. Am J Physiol Renal Physiol 309: F658–F666, 2015. doi: 10.1152/ajprenal.00219.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HW, Osis G, Handlogten ME, Verlander JW, Weiner ID. Proximal tubule glutamine synthetase expression is necessary for the normal response to dietary protein restriction. Am J Physiol Renal Physiol 313: F116–F125, 2017. doi: 10.1152/ajprenal.00048.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osis G, Webster KL, Harris AN, Lee HW, Chen C, Fang L, Romero MF, Khattri RB, Merritt ME, Verlander JW, Weiner ID. Regulation of renal NaDC1 expression and citrate excretion by NBCe1-A. Am J Physiol Renal Physiol 317: F489–F501, 2019. doi: 10.1152/ajprenal.00015.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirita Y, Wu H, Uchimura K, Wilson PC, Humphreys BD. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc Natl Acad Sci USA 117: 15874–15883, 2020. doi: 10.1073/pnas.2005477117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Legouis D, Ricksten SE, Faivre A, Verissimo T, Gariani K, Verney C, Galichon P, Berchtold L, Feraille E, Fernandez M, Placier S, Koppitch K, Hertig A, Martin PY, Naesens M, Pugin J, McMahon AP, Cippa PE, de Seigneux S. Altered proximal tubular cell glucose metabolism during acute kidney injury is associated with mortality. Nat Metab 2: 732–743, 2020[Erratum inNat Metab2: 989, 2020]. doi: 10.1038/s42255-020-0238-1. [DOI] [PubMed] [Google Scholar]
- 31.Ransick A, Lindstrom NO, Liu J, Zhu Q, Guo JJ, Alvarado GF, Kim AD, Black HG, Kim J, McMahon AP. Single-cell profiling reveals sex, lineage, and regional diversity in the mouse kidney. Dev Cell 51: 399–413.e7, 2019. doi: 10.1016/j.devcel.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21, 2019. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420, 2018. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zager RA, Johnson AC, Becker K. Renal cortical pyruvate depletion during AKI. J Am Soc Nephrol 25: 998–1012, 2014. doi: 10.1681/ASN.2013070791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breggia AC, Himmelfarb J. Primary mouse renal tubular epithelial cells have variable injury tolerance to ischemic and chemical mediators of oxidative stress. Oxid Med Cell Longev 1: 33–38, 2008. doi: 10.4161/oxim.1.1.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, Ding J, Czyz D, Hu R, Ye Z, He M, Zheng YG, Shuman HA, Dai L, Ren B, Roeder RG, Becker L, Zhao Y. Metabolic regulation of gene expression by histone lactylation. Nature 574: 575–580, 2019. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun S, Li H, Chen J, Qian Q. Lactic acid: no longer an inert and end-product of glycolysis. Physiology (Bethesda) 32: 453–463, 2017. doi: 10.1152/physiol.00016.2017. [DOI] [PubMed] [Google Scholar]
- 38.Brooks GA. The science and translation of lactate shuttle theory. Cell Metab 27: 757–785, 2018. doi: 10.1016/j.cmet.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Muraleedharan R, Gawali MV, Tiwari D, Sukumaran A, Oatman N, Anderson J, Nardini D, Bhuiyan MAN, Tkac I, Ward AL, Kundu M, Waclaw R, Chow LM, Gross C, Rao R, Schirmeier S, Dasgupta B. AMPK-regulated astrocytic lactate shuttle plays a non-cell-autonomous role in neuronal survival. Cell Rep 32: 108092, 2020. doi: 10.1016/j.celrep.2020.108092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pellerin L, Pellegri G, Bittar PG, Charnay Y, Bouras C, Martin JL, Stella N, Magistretti PJ. Evidence supporting the existence of an activity-dependent astrocyte-neuron lactate shuttle. Dev Neurosci 20: 291–299, 1998. doi: 10.1159/000017324. [DOI] [PubMed] [Google Scholar]
- 41.Wu H, Kirita Y, Donnelly EL, Humphreys BD. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol 30: 23–32, 2019. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]