Abstract
Pancreatic cancer is a highly malignant cancer of the pancreas with a very poor prognosis. Methylation of histone lysine residues is essential for regulating cancer physiology and pathophysiology, mediated by a set of methyltransferases (KMTs) and demethylases (KDMs). This study surveyed the expression of methylation regulators functioning at lysine 9 of histone 3 (H3K9) in pancreatic lesions and explored the underlying mechanisms. We analyzed KDM1A and KDM3A expression in clinical samples by immunohistochemical staining and searching the TCGA PAAD program and GEO datasets. Next, we identified the variation in tumor growth in vitro and in vivo after knockdown of KDM1A or KDM3A and explored the downstream regulators of KDM1A and KDM3A via RNA-seq, and gain- and loss-of-function assays. Eleven H3K9 methylation regulators were highly expressed in pancreatic cancer, and only KDM1A and KDM3A expression positively correlated with the clinicopathological characteristics in pancreatic cancer. High expression of KDM1A or KDM3A positively correlated with pathological grade, lymphatic metastasis, invasion, and clinical stage. Kaplan–Meier analysis indicated that a higher level of KDM1A or KDM3A led to a shorter survival period. Knockdown of KDM1A or KDM3A led to markedly impaired tumor growth in vitro and in vivo. Mechanistically, CCNA2, a cell cycle-associated gene was partially responsible for KDM1A knockdown-mediated effect and CDK6, also a cell cycle-associated gene was partially responsible for KDM3A knockdown-mediated effect on pancreatic cancer cells. Our study demonstrates that KDM1A and KDM3A are highly expressed in pancreatic cancer and are intimately correlated with clinicopathological factors and prognosis. The mechanism of action of KDM1A or KDM3A was both linked to the regulation of cell cycle-associated genes, such as CCNA2 or CDK6, respectively, by an H3K9-dependent pathway.
Keywords: Pancreatic cancer, methylation, KDM1A, KDM3A, cell cycle
Impact statement
H3K9 methylation plays an essential role in the biology of pancreatic cancer (PC), while the clinicopathological significances and the underlying downstream of H3K9 regulators remains largely unexplored. Our work was designed and performed to solve this issue through systematical analysis of the expression of methyltransferases (KMTs) and demethylases (KDMs) functioning at H3K9 methylation in PC, and then unveiling the key downstream of KDM1A and KDM3A by using RNA-seq, and gain- and loss-of-function assays. Our data firmly demonstrated that KDM1A and KDM3A were the main dysregulated H3K9 regulators in PC, determining the tumor growth in vitro and in vivo and regulating the expression of many cell cycle-associated genes (such as CCNA2 and CDK6). We further demonstrated that KDM1A or KDM3A-mediated epigenetic regulation of these genes was relied on an H3K9-dependent pathway.
Introduction
Pancreatic cancer (PC) is a highly aggressive cancer of the pancreas and was the seventh leading cause of death in 2018 worldwide. 1 In China, there were an estimated 90,100 new diagnoses and 79,400 deaths in 2015. 2 Despite the pronounced advances achieved in cancer diagnosis and treatment in recent decades, the five-year overall survival rate of patients with PC is still less than 30%, and the median survival time is only three to six months for metastatic patients.3,4 Resectability or borderline resectability is only possible for a small proportion of patients (approximately 20%). 5 Chemotherapy-based first-line treatment is recommended for borderline resectable disease, metastatic disease, and recurrent disease. 6 However, the overall efficiency of these treatments remains less satisfactory for patients with PC than for patients with other cancers. Thus, intensive exploration of the underlying disease markers and targeting molecules is essential for early diagnosis and targeted therapy for PC.
Methylation of histone lysine residues is a vital mechanism for genomic stability and cell fate maintenance, influencing numerous biological processes. 7 Histone methylation is dynamically regulated via the addition of monomethyl, dimethyl, or trimethyl groups to lysine residues by methyltransferases (KMTs) and, conversely, the removal of these groups by specific demethylases (KDMs), which possess various catalytic mechanisms. 8 In this regard, the biological and physiological significance of histone methylation in tumorigenesis and therapeutic response is attracting tremendous attention. For instance, EZH2, an active enzyme with methylation activity at the lysine residue at position 27 of histone 3 (H3K27), has exhibited high expression in various cancers, such as breast, bladder, and pancreatic cancer, and correlates with poor prognosis.9,10 It is vital for cell proliferation, and forced expression of it confers a replication advantage in noncancerous cells. 10 In PC, ANLN-mediated upregulation of EZH2 is responsible for cell proliferation, migration, and tumor growth in vivo through the miR-218-5p/LASP1 signaling axis. 11 The protumorigenic function of NSD1-mediated H3K36 methylation was also found in solid tumors. 12 Thus, elucidating the clinicopathological significance of methylation regulators and the potential mechanism is essential for the treatment of PC.
Apart from methylation at H3K27 and H3K36, methylation at other lysine residues, including H3K4, H3K9, H3K79, and H4K20, is also involved in cancer. We focused our attention on the KMTs and KDMs regulating H3K9 methylation in PC. Several H3K9 methylation regulators have been demonstrated to be associated with the prognosis of solid cancers, and certain underlying mechanisms have been unveiled. In breast cancer, G9a, a KMT with enzymatic activity for H3K9 methylation, exhibited a significant prognostic value along with its negatively regulated genes. 13 Enhanced G9a was associated with silencing of two suppressor genes, DSC3 and MASPIN, while inhibiting G9a resulted in downregulation of H3K9 dimethylation and promoted the expression of the two genes. 14 KDM1A, also known as LSD1, is highly expressed in squamous cell carcinoma and closely correlated with tumor size, pathological grade, and prognosis. 15 Knockdown of it attenuated cell proliferation and tumor growth via various mechanisms.15,16 In PC, an initial study suggested that KDM1A was upregulated in cancerous tissues compared with corresponding adjacent tissues, while the lack of normal tissues and limited samples impaired the significance. 16 In addition, KDM3A was also found to be deregulated in cancer, and enhanced expression of KDM3A predicted poor prognosis in colorectal cancer patients. 17 Loss of function assays indicated that KDM3A positively regulated migration and invasion by influencing EMT and matrix metalloproteinases. Thus, a systematical understanding of the regulators of H3K9 methylation in PC remains to be explored.
In this work, we systematically surveyed the expression of H3K9 methylation regulators in PC tissues compared to normal tissues, pericancerous tissues, and benign lesion tissues based on our clinical samples and incorporating data from the TCGA PAAD program and GEO datasets. The knockdown of KDM1A or KDM3A significantly impaired tumor growth in vitro and in vivo by depressing the expression of cell cycle-associated genes, such as CCNA2 or CDK6, respectively, by an H3K9-dependent pathway. Thus, we found that two H3K9 methylation regulators, KDM1A and KDM3A determined the growth of PC through regulating cell cycle-associated genes.
Materials and methods
Analysis of TCGA datasets
TCGA datasets of patients with PC (PAAD program) were abstracted and analyzed using the UALCAN online tool (www.ualcan.path.uab.edu). A total of 174 patients with PC were involved. We searched and compared the mRNA of 16 H3K9 methylation regulators in PC and paired normal tissues. We further compared the expression of these genes stratified by pathological grade. Coexpression analysis between two genes was also performed by UALCAN. Kaplan–Meier analysis of TCGA data was conducted with the OncoLnc online tool (www.oncolnc.org).
Analysis of GEO datasets
Two datasets were used, GEO16515 and GEO15471, to compare the expression of H3K9 methylation regulators. The expression levels of genes were downloaded from the Oncomine database (www.oncomine.org) and expressed by the log2 median-centered ratio. GEO16515 comprises 36 tumor samples and 16 normal samples sequenced with the Affymetrix Human Genome U133 Plus 2.0 Array. GEO15471 includes 36 pancreatic ductal adenocarcinoma samples and paired normal tissues sequenced with the same platform. We also downloaded data from the GEO database (www.ncbi.nlm.nih.gov/geo) and selected potential genes using a cutoff value of P = 0.001.
Clinical case selection and cell lines
The present retrospective study was approved by the Ethics Committee for Human Research, Central South University, and was conducted according to the approved guidelines and the principles of the Declaration of Helsinki. A total of 106 PC tissues, 35 pericancerous tissues, 55 benign lesion tissues (20 tissues of chronic pancreatitis, 20 tissues of adenoma, and 15 tissues of intraepithelial neoplasia), and 13 normal pancreatic tissues were obtained at Second Xiangya Hospital. Follow-up of the patients was performed by letters and phone calls.
Thirty-five pericancerous tissues were collected from patients with PC. Fifty-five benign lesion tissues were collected from 29 males and 26 females. Normal pancreatic tissues were obtained from surrounding healthy areas of the adenomas.
The PC cell lines Panc-1, BxPC-3, and CFPAC-1 were purchased from ZSBIO, China, which also performs cell line STR genotyping. Panc-1 and BxPC-3 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS), while CFPAC-1 cells were maintained in RPMI-1640 medium supplemented with 10% FBS and placed in a humidified incubator with 5% CO2 at 37°C.
Immunohistochemistry
Antibodies against KDM1A and KDM3A were provided by Dako Corporation Company (Carpentaria, CA, USA). The EnVisionTM Detextion Kit was provided by the same company. EnVision immunohistochemical staining of KDM1A and KDM3A was conducted following the operation guideline. Briefly, the sample sections were prepared from paraffin-embedded samples. To deparaffinization, the sections were heated, followed by incubation with 3% H2O2 for proper time. Epitope retrieval was performed by heating in sodium citrate buffer (10 mM sodium citrate and 0.05% Tween 20, pH 6.0) at 96°C for 30 min. For antigen-antibody reaction, samples were incubated with rabbit anti-human KDM1A and KDM3A primary antibodies (1:100 dilution) for 2 h. The sections were incubated with 20–50 µl Solution A (ChemMate™ EnVison+/HRP) for 30 min followed by DAB staining and hematoxylin counterstaining. The samples were dehydrated, soaked in xylene, mounted with neutral balsam, and further fastened by enamel. 18 About 500 cells from 10 random selected fields were evaluated per section by two pathologists independently. Cases with ≥25% positive cells were considered positive, whereas the other cases were considered negative. For samples from animals, antibodies against KDM1A, KDM3A, CCNA2, CDK6, and Ki67 were purchased from Abclonal, China and IHC was performed using ZSGB-BIO IHC kit.
Colony formation assay
The PC cells (Panc-1 and CFPAC-1) were seeded in 6-well plates at 5000 cells per well and cultured for eight days. Then, wash the cells with PBS two times, fix with 4% paraformaldehyde, and stain with crystal violet staining solution (Beyotime, China). After airing, images of colonies were captured by a digital camera.
Subcutaneous xenograft model
BALB/c nude mice received human care in compliance with the guidelines implemented at the Second Xiangya Hospital. The study was performed according to the international, national, and institutional rules considering animal experiments and biodiversity rights. In brief, 4 × 106 Panc-1 cells transfected with KDM1A or KDM3A shRNA or scramble were subcutaneously injected into the right dorsal of six-week-old male nude mice (n = 5). The animals were carefully nursed for four weeks. Then, the animals were sacrificed, and tumors were collected and measured.
Western blot analysis
PC cells were lysed in RIPA buffer added by protease and phosphatase inhibitors (TargetMol, USA) and incubated on ice. After centrifugation, the supernatant was harvested. The denatured proteins were electrophoresed and electrotransfer onto PVDF. After blocking with 3% BSA, the membranes were incubated with diluted primary antibody overnight at 4°C. After washed by TBST, diluted secondary antibody was added and incubated for 1 h at room temperature. The bands were detexted via an enhanced chemiluminescence system (Life Tec, USA). Analysis of the bands was performed using ImageJ software (Version 11). The primary antibodies involved in this work included the following: KDM1A, KDM3A, CCNA2, CDK6, and GAPDH. Primary and secondary antibodies were purchased from Abclonal, China.
Quantitative real-time PCR
The RNA of tissues and cells was extracted using standard methods, as previously described. 19 Briefly, TRIzol reagent was added to the samples, followed by lysis for 10 min, centrifugation at 12,000g for 15 min, and then reduction with isopropanol. The RNA purity and concentration were tested using a Nanodrop 2000 spectrophotometer (Thermo Fisher, USA). cDNA was synthesized using a high-capacity cDNA reverse transcription kit (Life Tec, USA) according to the manufacturer’s guidelines. The primers were listed as follows: KDM1A, 5′- CCAGAGATATTACTGCCGAGTT-3′ (forward) and 5′- GCTTTCCTTGTGTTTCAGCTAA-3′ (reverse); KDM3A, 5′- AAGGTGTGTGTGGAATTTGATG-3′ (forward) and 5′- AAAATGCTCTCCTTAGAAGGCT-3′ (reverse). CCNA2 promoter, AGTAGTTCAAGGTGCCATC (forward), TTCAGTATCTCCGATTCTCC (reverse); CDK6 promoter, CCACCTCTGCTACCTACA (forward), TCGCTTGTCGTCTCCTTA (reverse). 2× Universal SYBR Green Fast qPCR mix (Abclonal, China) was used for qRT–PCR in a LightCycler 96 system (Roche, America).
EdU assay
PC cells were planted in 24-well plates at 2 × 105 per well and cultured for two days. Then, the cells were incubated with EdU for 2 h at 37°C, and then rinsed three times with PBS. After permeabilization with 0.25% Triton X-100 (Sigma-Aldrich, USA), the cells were treated by Click Additive Solution, washed by PBS, stained with DAPI. The signal was captured using a fluorescence microscope (Olympus Inc., USA).
siRNA transfection or plasmid transfection
For siRNA transfection, the PC cells were planted in 6-well plates and cultured overnight. Then, siRNAs or negative controls were transfected using Lipofectamine™ RNAiMAX transfection reagent (Invitrogen, USA), while CCNA2 or CDK6 plasmid was transfected using Lipo8000™ reagent (Beyotime, China). The targeted sequences of KDM1A were GCTCGACAGTTACAAAGTT and for KDM3A were GAAGGCTTCTTAACACCAA. After incubation for two days, the cells were treated as indicated.
Lentivirus-mediated shRNA knockdown of gene expression
pHBLV-lentiviral shRNA vectors targeting human KDM1A and KDM3A gene and pHBLV control vector were purchased from Hanbio., Tec, China and the targeted sequences were commercially preserved. Panc-1 cells were transfected by the lentiviral particles at a multiplicity of infection (MOI) of 30 followed by puromycin selection for two weeks. The clones stably transfected with pHBLV-lentiviral shRNA was referred to as shRNA cells, whereas the cells stably transfected with pHBLV control vector as scrambled cells. The knockdown of KDM1A or KDM3A was evaluated by qRT-PCR and Western blot analysis.
Flow cytometry assay
Apoptosis rate was determined by flow cytometry assay. The Annexin V/PI Apoptosis Detextion Kit and Cell Cycle Analysis Kit were purchased from Genview, China and used according to the manufacturer’s instructions. In brief, PC cells after treatment as indicated were harvested, washed with PBS, resuspended in 1× binding buffer, and added with the Annexin V and PI solution or stained with PI solution after fixed by 70% alcohol. Then, the cells were harvested and detexted by BD FACSARIA II flow cytometry (Becton Dickinson, USA).
RNA-sequencing
A total amount of 1 µg RNA per sample was used as input material for the RNA sample preparations. The sequencing libraries were generated using the NEBNext® UltraTM RNA Library Prep Kit. After cluster generation, the library preparations were sequenced using an Illumina Novaseq platform and 150 bp paired-end reads were generated. The RNA-sequence procedure and data analysis were performed by Novogene, China.
Chromatin immunoprecipitation
ChIP assay was performed by a ChIP kit (Abcam, USA) and followed by the manufacturer’s instructions. In brief, cells were planted in 6-cm dish and transfected by siRNA or negative control. After two days’ culture, the cells were harvested and fixed in formaldehyde for 10 min. The cell pellets were sonicated, the residue was removed by centrifugation, and the primary antibody (ChIP grade) was added and incubated overnight at 4°C. After treated by proteinase K, the DNA was abstracted. The expression of promoters in DNA after ChIP or input was analyzed by qRT–PCR using Universal SYBR Green Fast qPCR mix (Abclonal, China). The primers are listed in qRT–PCR section.
Statistical analysis
All the data were analyzed by SPSS 17.0 (Version 17.0) or GraphPad (Version 6). The correlations between expression of KDM1A and KDM3A and between their expression and clinicopathological characteristics were analyzed using the χ2 test or Fisher’s exact test, when appropriate. 20 The overall survival of patients with PC was calculated according to Kaplan–Meier univariate survival analysis and log-rank tests. Multivariate analysis was performed with a Cox proportional hazards model, and a 95% confidence interval was calculated. Two-tailed Student’s t test was used to assess significant differences between two groups. For three or more groups, one-way ANOVA was used. P < 0.05 was considered statistically significant.
Results
Evaluating the expression of H3K9 regulators in PC
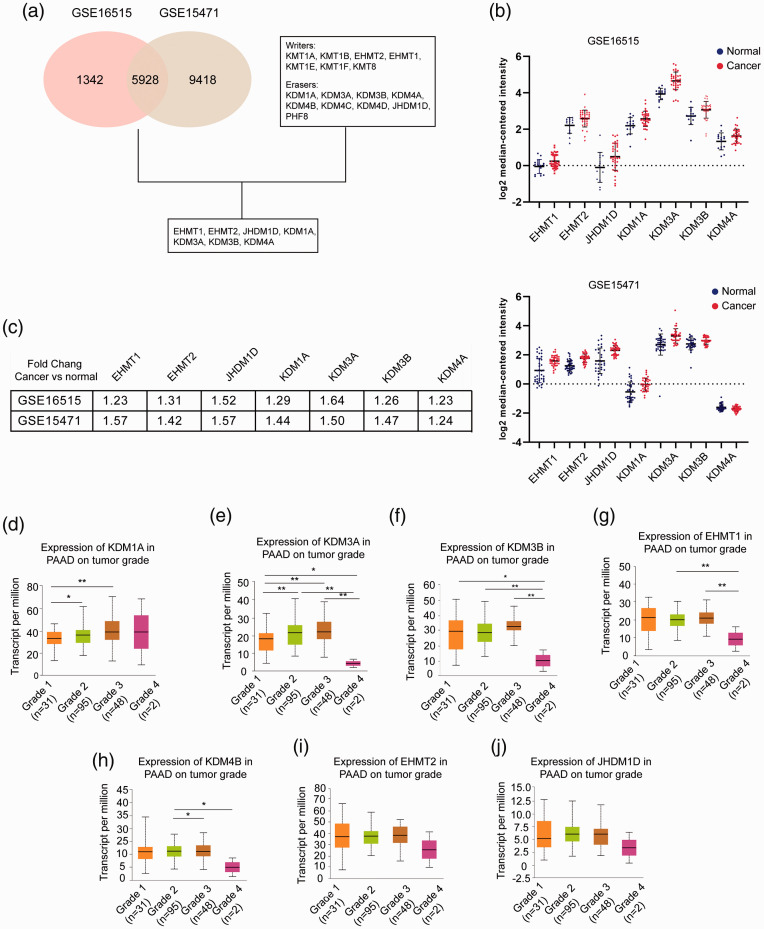
To systematically evaluate the expression of H3K9 regulators in PC, we analyzed two datasets, GSE16515 and GSE15471. A cutoff value of P < 0.001 was applied to select the candidate genes with significant differences between cancerous and normal tissues, resulting in 7270 and 15,346 genes, respectively. Among them, 5928 overlapping genes were identified. H3K9 was regulated by a series of lysine KMTs and KDMs; among them, KMT1A, KMT1B, EHMT1, EHMT2, KMT1E, KMT1F, and KMT8 served as writers, while KDM1A, KDM3A, KDM3B, KDM4A, KDM4B, KDM4C, KDM4D, JHDM1D, and PHF8 served as erasers. After correlating these data with data from the GEO datasets, we found that seven genes were significantly dysregulated in PC, including EHMT1, EHMT2, JHDM1D, KDM1A, KDM3A, KDM3B, and KDM4A (Figure 1(a)). Figure 1(b) shows the expression of these genes in GSE16515 (upper) and GSE15471 (lower) by dot plots. Compared to their expression in normal tissues, these genes were upregulated in PC, and the fold changes are presented in Figure 1(c). The data from the TCGA PAAD program were used to survey the potentially pathological significance of these genes. KDM1A and KDM3A were closely associated with pathological grade, and increased expression of KDM1A or KDM3A was found in grade 2 and grade 3 tumors (Figure 1(d) and (e)). Although the expression of KDM1A and KDM3A in grade 4 tumors was not elevated or was otherwise downregulated compared with the respective expression levels in grade 3 tumors, the limited number of samples in the grade 4 group greatly attenuated the statistical significance. Figure 1(f) shows that KDM3B expression in grade 1, grade 2, and grade 3 tumors was upregulated compared with that in grade 4 tumors, while the variability among the comparisons between other grades hindered a definitive negative correlation of KDM3B with pathological features in PC. A similar result was also observed in the association of EHMT1 with pathological grade (Figure 1(g)). In addition, a weak correlation of KDM4B with pathological grade was found and is presented in Figure 1(h). EHMT2 and JHDM1D showed no significance between various pathological grades (Figure 1(i) and (j)). Collectively, we found that the H3K9 regulators KDM1A and KDM3A were significantly upregulated in PC and correlated with pathological grade.
Figure 1.
Expression of H3K9 regulators in pancreatic cancer. (a) GSE16515 and GSE15471 data were derived from GEO datasets (www.ncbi.nlm.nih.gov/gds/) and analyzed by GEO2R. The cutoff value was 0.001. Writers are lysine KTMs that function at the H3K9 site, and erasers are lysine KDMs that function at the H3K9 site. (b) Scatter plots of the expression of EHMT1, EHMT2, JHDM1D, KDM1A, KDM3A, KDM3B, and KDM4A in GSE16515 (upper) and GSE15471 (lower). Blue indicates normal tissues, and red indicates cancerous tissues. The expression value for each gene was downloaded from the Oncomine database (www.oncomine.org) and is presented as the log2 median-centered ratio. (c) Summary of the fold changes of the genes in GSE16515 and GSE15471. (d–j) mRNA expression of the genes stratified by pathological grade in pancreatic cancer. Data were derived from the TCGA PAAD program. A total of 176 patients were involved, including 31 patients with grade 1 tumors, 95 patients with grade 2 tumors, 48 patients with grade 3 tumors, and 2 patients with grade 4 tumors. The mRNA level is presented as transcripts per million. *P < 0.05, **P < 0.01, ***P < 0.001. (A color version of this figure is available in the online journal.)
High expression of KDM1A and KDM3A in PC
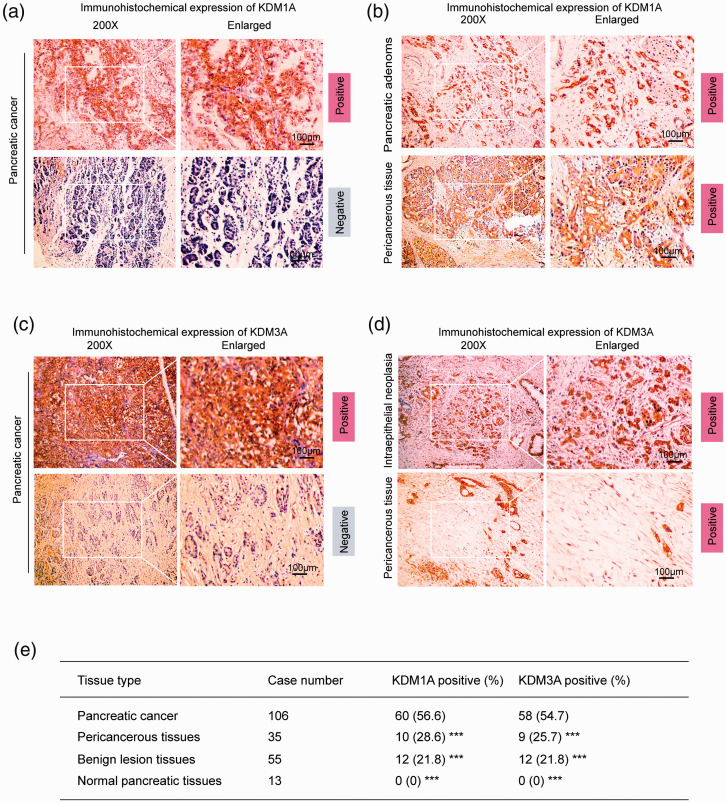
We confirmed that either KDM1A or KDM3A was upregulated in pancreatic cancer cell lines compared to normal pancreatic duct cells (Supplementary Figure 1(a)). To analyze the expression of KDM1A and KDM3A in PC and compare it to the expression in nonmalignant tissues or normal tissues, we performed IHC assays on surgical samples. A total of 209 samples were involved in the analysis, including 106 PC tissues, 35 pericancerous tissues, 55 precursor lesion tissues (chronic pancreatitis, adenoma, and intraepithelial neoplasia), and 13 normal tissues. In cancerous tissues, the positive expression rates for KDM1A and KDM3A were 56.6% and 54.7%, respectively (Figure 2(a) and (c)). In precursor lesion tissues, the positive expression rate for both KDM1A and KDM3A was 21.8% (Figure 2(b) upper and (d) upper). A total of 28.6% and 25.7% of pericancerous tissues were positive for KDM1A and KDM3A, respectively (Figure 2(b) lower and (d) lower). All normal tissues showed negative staining for KDM1A and KDM3A. Figure 1(e) summarizes the positive rates of KDM1A and KDM3A expression in PC tissues, pericancerous tissues, precursor lesion tissues, and normal tissues. Specifically, the positive rates of KDM1A expression in chronic pancreatitis, adenoma, and intraepithelial neoplasia were 15.0% (3/20), 25.0% (5/20), and 26.7% (4/15), respectively. Similarly, KDM3A was also poorly expressed in these tissues (the positive expression rates were 10.0%, 25.0%, and 33.3%, respectively). Thus, our results showed that PC tissues had upregulated expression of KDM1A and KDM3A, suggesting the potential clinicopathological significance of these genes.
Figure 2.
High expression of KDM1A and KDM3A in pancreatic cancer. (a) Immunohistochemical staining of KDM1A in pancreatic cancer tissues (n = 106). The upper panel shows positive staining for KDM1A, and the lower panel shows negative staining for KDM1A. (b) Immunohistochemical staining of KDM1A in benign pancreatic lesions (n = 55) and pericancerous tissues (n = 35). Benign lesions included chronic pancreatitis (n = 20), adenoma (n = 20), and intraepithelial neoplasia (n = 15). The results showed positive staining of KDM1A in all tissues. (c) Immunohistochemical expression of KDM3A in cancerous tissues (n = 106). The upper image indicates positive expression of KDM3A, and the lower image indicates negative expression. (d) Immunohistochemical expression of KDM3A in benign lesions (n = 55) and pericancerous tissues (n = 35). Benign lesions included chronic pancreatitis (n = 20), adenoma (n = 20), and intraepithelial neoplasia (n = 15). The results showed positive staining in all tissues. (e) Summary of positive staining of KDM1A and KDM3A in pancreatic cancer tissues, pericancerous tissues, benign lesion tissues, and normal tissues. ***P < 0.001, compared with pancreatic cancer tissues. Scale bars correspond to 100 μm. (A color version of this figure is available in the online journal.)
Expression of KDM1A and KDM3A was associated with clinicopathological characteristics in PC
To evaluate the association of KDM1A and KDM3A expression with clinicopathological characteristics in PC, we compared the rates of positive KDM1A and KDM3A expression with various factors after stratification of the samples into subgroups. As presented in Table 1, the expression of KDM1A and KDM3A was coincidently associated with pathological grade, lymphatic metastasis, invasion, and TNM stage. However, age, sex, and tumor size were not significantly associated with the expression of KDM1A or KDM3A. Furthermore, of the cases with positive expression of KDM1A, 42 cases positively expressed KDM3A; conversely, 46 cases with negative expression of KDM1A exhibited negative expression of KDM3A. In addition, we used the χ2 test to determine the relationship between KDM1A and KDM3A and showed a close association between these genes (χ2=13.033, P = 0.009).
Table 1.
Correlations of KDM1A and KDM3A protein expression with clinicopathological characteristics in pancreatic cancer.
| CPC | Case no. | KDM1A |
KDM3A |
|||||
|---|---|---|---|---|---|---|---|---|
| Pos no. (%) | χ2 | P value | Pos no. (%) | χ2 | P value | |||
| Age (year) | ||||||||
| ≤45 years | 22 | 15 (68.4) | 1.515 | 0.218 | 14 (63.6) | 0.891 | 0.345 | |
| >45 years | 84 | 45 (53.6) | 44 (52.4) | |||||
| Sex | ||||||||
| Male | 61 | 35 (57.4) | 0.035 | 0.852 | 32 (52.5) | 0.296 | 0.587 | |
| Female | 45 | 25 (55.6) | 26 (57.8) | |||||
| Differentiation | ||||||||
| Well | 38 | 14 (36.8) | 14.659 | 0.001 | 15 (39.5) | 9.613 | 0.008 | |
| Moderately | 35 | 19 (54.3) | 18 (51.4) | |||||
| Poorly | 33 | 27 (81.8) | 25 (75.8) | |||||
| Tumor size | ||||||||
| ≤3 cm | 13 | 6 (46.3) | 1.149 | 0.563 | 6 (46.2) | 1.341 | 0.512 | |
| 3–5 cm | 68 | 38 (55.9) | 36 (52.9) | |||||
| >5 cm | 25 | 16 (64.0) | 17 (68.0) | |||||
| Lymphatic metastasis | ||||||||
| No | 77 | 36 (46.8) | 11.118 | 0.001 | 35 (45.5) | 9.745 | 0.002 | |
| Yes | 29 | 24 (82.8) | 23 (79.3) | |||||
| Invasion | ||||||||
| No | 42 | 18 (42.9) | 5.351 | 0.021 | 9 (21.4) | 12.838 | <0.001 | |
| Yes | 64 | 42 (65.6) | 31 (48.4) | |||||
| TNM stage | ||||||||
| I + II | 53 | 24 (45.3) | 5.530 | 0.016 | 29 (54.7) | 12.336 | <0.001 | |
| III + IV | 53 | 36 (67.9) | 37 (69.8) | |||||
CPC: clinicopathological characteristics; No.: number; Pos: positive.
Expression of KDM1A and KDM3A was correlated with the prognosis of PC
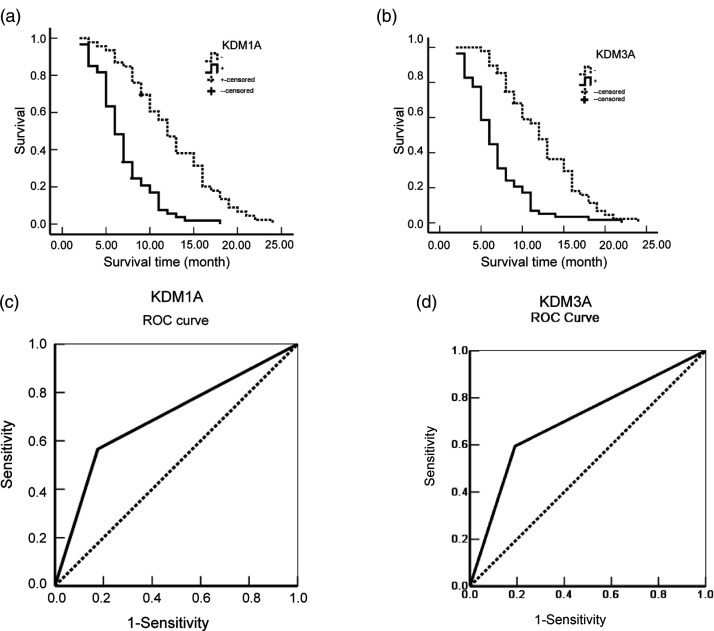
To survey whether the expression of KDM1A and KDM3A was a prognostic factor for PC, we collected the survival data of each patient. During the one-year follow-up, 29 patients survived, and 77 patients died. As summarized in Table 2, it was suggested that the average overall survival time of PC patients was intimately correlated with pathological grade, tumor size, lymphatic metastasis, invasion, and TNM stage. Furthermore, after stratification by expression of KDM1A or KDM3A, positive expression indicated a significantly shorter survival time than that seen with negative expression of either (6.95 vs. 12.52 for KDM1A; 6.91 vs. 12.45 for KDM3A) and correlated with the average overall survival time (Figure 3(a) and (b)). The Cox multivariate analysis showed that, as expected, clinicopathological factors, including tumor size >5 cm, poor pathological grade, lymphatic metastasis, invasion, and high clinical stage were positively associated with mortality (Table 3). Furthermore, positive expression of KDM1A and KDM3A was significantly correlated with high mortality in patients with PC. To evaluate the efficiency of KDM1A or KDM3A expression in the differentiation of PC from benign lesions, an AUC was calculated, and the result was 0.695 (95% CI: 0.616–0.774) for KDM1A and 0.685 (95% CI: 0.606–0.765) for KDM3A (Figure 3(c) and (d)).
Table 2.
Correlations of clinicopathological characteristics and KDM1A and KDM3A expression with mean survival in patients with pancreatic cancer.
| Group | Case no. (n) | Mean survival (month) | Chi-square | P value |
|---|---|---|---|---|
| Sex | ||||
| Male | 61 | 9.98 (2–24) | 1.656 | 0.198 |
| Female | 45 | 8.61 (2–21) | ||
| Age (year) | ||||
| ≤45 | 22 | 8.18 (3–19) | 2.144 | 0.143 |
| >45 | 84 | 9.73 (2–24) | ||
| Differentiation | ||||
| Well | 38 | 11.27 (3–24) | 17.786 | <0.001 |
| Moderately | 35 | 9.74 (3–1) | ||
| Poorly | 33 | 6.86 (2–14) | ||
| Tumor size | ||||
| ≤3 cm | 13 | 13.46 (5–21) | 7.504 | 0.023 |
| 3–5 cm | 68 | 9.34 (2–22) | ||
| >5 cm | 25 | 7.40 (3–24) | ||
| TNM stage | ||||
| I | 11 | 16.46 (11–24) | 80.807 | <0.001 |
| II | 42 | 11.37 (2–22) | ||
| III | 37 | 7.14 (2–17) | ||
| IV | 16 | 4.56 (2–8) | ||
| Lymph node metastasis | ||||
| No | 77 | 10.64 (2–24) | 27.120 | <0.001 |
| Yes | 29 | 6.35 (2–12) | ||
| Invasion | ||||
| No | 42 | 13.33 (5–24) | 46.949 | <0.001 |
| Yes | 54 | 6.35 (2–17) | ||
| KDM1A | ||||
| − | 46 | 12.52 (2–18) | 36.204 | <0.001 |
| + | 60 | 6.95 (3–24) | ||
| KDM3A | ||||
| − | 48 | 12.45 (5–24) | 30.982 | <0.001 |
| + | 58 | 6.91 (5–30) | ||
No.: number.
Figure 3.
KDM1A and KDM3A correlated with the prognosis of pancreatic cancer patients. (a) Kaplan–Meier plots of the overall survival period in patients with KDM1A-positive and KDM1A-negative cancers. The solid line indicates positive expression, and the dotted line indicates negative expression. (b) Kaplan–Meier plots of the overall survival period in patients with KDM3A-positive and KDM3A-negative cancers. The solid line indicates positive expression, and the dotted line indicates negative expression. (c) ROC curves were produced to assess the predictive ability of KDM1A expression in pancreatic cancer. (d) ROC curves were produced to assess the predictive ability of KDM3A expression in pancreatic cancer.
Table 3.
Multivariate Cox regression analysis of survival rate in patients with pancreatic cancer.
| 95%CI |
||||||||
|---|---|---|---|---|---|---|---|---|
| Groups | Factors | B | SE | Wald | P | RR | Lower | Upper |
| Differentiated degree | Well/moderately/poorly | 0.371 | 0.152 | 5.957 | 0.015 | 1.449 | 1.076 | 1.952 |
| Tumor size | ≤3 cm/3–5 cm/>3 cm | 0.252 | 0.203 | 1.541 | 0.214 | 1.287 | 0.864 | 1.915 |
| Lymph node | ||||||||
| metastasis | No/yes | 0.885 | 0.300 | 8.703 | 0.003 | 2.423 | 1.346 | 4.362 |
| Invasion | No/yes | 0.993 | 0.341 | 8.480 | 0.004 | 2.699 | 1.384 | 5.266 |
| TNM stage | I/II/III/IV | 0.829 | 0.244 | 11.543 | 0.001 | 2.291 | 1.420 | 3.696 |
| KDM1A | −/+ | 1.242 | 0.348 | 12.738 | <0.001 | 3.463 | 1.751 | 6.849 |
| KDM3A | −/+ | 0.811 | 0.295 | 7.558 | 0.006 | 2.250 | 1.262 | 4.012 |
B: regression coefficients; SE: standard error; RR: relative risk.
Downregulation of KDM1A or KDM3A significantly impaired tumor growth in vitro and in vivo through cell cycle arrest
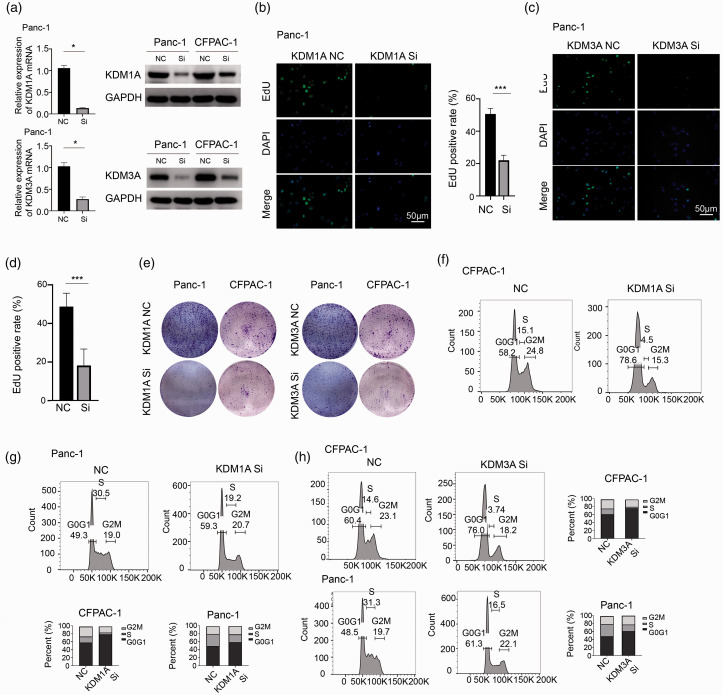
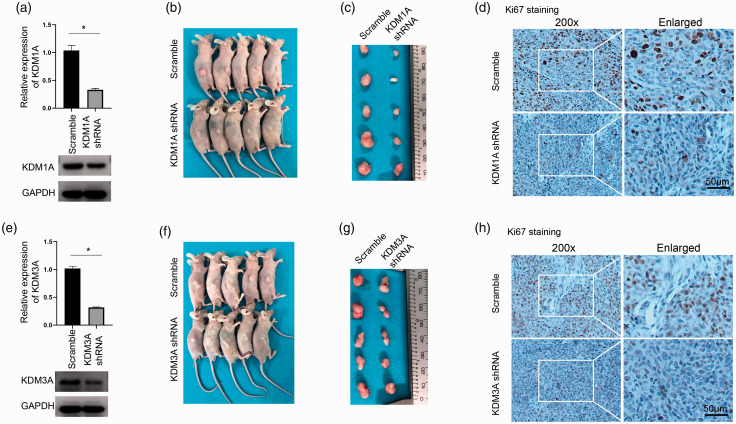
To explore the function of KDM1A and KDM3A in PC, we used siRNA to knockdown KDM1A or KDM3A, verified by mRNA and protein examination (Figure 4(a)). The EdU assay showed that either downregulation of KDM1A or KDM3A significantly inhibited cell proliferation in Panc-1 cells (Figure 4(b) to (d)). Then, colony formation assay further demonstrated a markedly impaired cell growth in Panc-1 and CFPAC-1 cells after KDM1A or KDM3A knockdown (Figure 4(e)). Cell cycle analysis was conducted by flow cytometry and results showed that downregulation of KDM1A or KDM3A would similarly lead to cell cycle arrest by G0G1 stage in CFPAC-1 and Panc-1 cells (Figure 4(f) to (h)). Furthermore, we used shRNA to knockdown KDM1A or KDM3A and the stably transfected cells were subcutaneously injected into the nude mice. The knockdown efficiency of three distinct shRNAs on KDM1A or KDM3A was verified by Western blot and the biological influence of them was confirmed by colony formation assay in Panc-1 cells (Supplementary Figure 1(b) and (c)). After a period of four weeks, mice were sacrificed and the tumors were collected. Either knockdown of KDM1A or KDM3A led to significantly impaired tumor growth in vivo compared to scrambled group (Figure 5(a) to (c) and (e) to (g)). The Ki-67 staining showed that knockdown of KDM1A or KDM3A decreased the expression of Ki-67 (Figure 5(d) and (h)), consistently demonstrating both KDM1A and KDM3A are essential for tumor growth in vivo. Thus, these data showed that knockdown of KDM1A or KDM3A significantly inhibited tumor growth in vitro and in vivo through leading to cell cycle arrest in PC.
Figure 4.
Downregulation of KDM1A or KDM3A significantly impaired tumor growth in vitro through cell cycle arrest. (a) siRNA was used to inhibit KDM1A or KDM3A expression in Panc-1 and CFPAC-1 cells and was verified by qRT–PCR (left) and Western blotting (right), n = 3. (b) The EdU assay showed that the knockdown of KDM1A significantly reduces cell proliferation in Panc-1 cells, n = 6. (c–d) The EdU assay showed that the knockdown of KDM3A significantly reduces cell proliferation in Panc-1 cells, n = 6. (e) The colony formation assay was performed to detext the ability of colony formation upon KDM1A or KDM3A knockdown. Either the knockdown of KDM1A or KDM3A reduced colony formation in Panc-1 and CFPAC-1 cells. (f–g) Cell cycle analysis was performed by flow cytometry. KDM1A siRNA led to cell arrest in G0G1 in CFPAC-1 and Panc-1 cells. (h) Knockdown of KDM3A also led to cell arrest in G0G1 in BxPC-3 and CFPAC-1. NC: negative control; Si: siRNA; ***P < 0.001. Scale bars correspond to 50 μm. (A color version of this figure is available in the online journal.)
Figure 5.
Downregulation of KDM1A or KDM3A significantly impaired tumor growth in vivo. (a) KDM1A shRNA successfully downregulated the mRNA (upper) and protein (lower) expression of KDM1A in Panc-1 cell, n = 3. (b) Panc-1 cells with stable transfection of KDM1A shRNA or control were subcutaneously injected into nude mice. After one month, the mice were sacrificed. The knockdown of KDM1A markedly inhibited tumor group in vivo. (c) The tumors were collected and the sizes of tumors in KDM1A shRNA group were much smaller than the scrambled group. (d) IHC staining of Ki67 in samples from both groups. (e) KDM3A shRNA successfully downregulated the mRNA (upper) and protein (lower) expression of KDM3A in Panc-1 cell, n = 3. (f) Panc-1 cells with transfection of KDM3A shRNA or control were subcutaneously injected into nude mice. The knockdown of KDM3A markedly inhibited tumor group in vivo. (g) The sizes of tumor were evidently smaller than the control group. (h) IHC staining of Ki67 in KDM3A knockdown or control group. ***P < 0.001; Scramble, scrambled control. Scale bars correspond to 50 μm. (A color version of this figure is available in the online journal.)
CCNA2 was partially responsible for KDM1A knockdown-mediated effect on PC cells
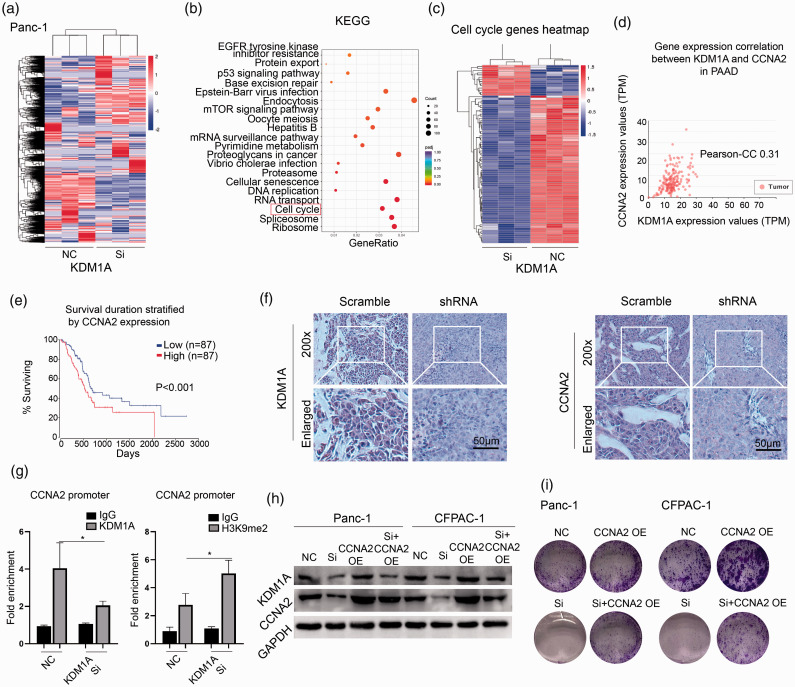
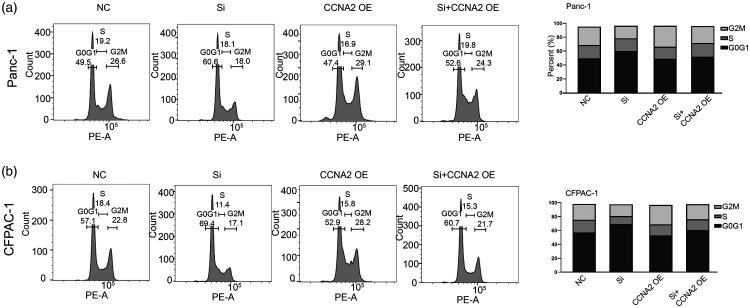
To delineate the underlying mechanism of KDM1A knockdown-mediated effect, we analyzed global gene expression variations by RNA-seq in Panc-1 cell—after KDM1A knockdown (Figure 6(a)). KEGG signaling analysis showed that KDM1A knockdown led to significant enrichment of cell cycle-associated signaling, which suggested the potential role of it in regulating cell cycle (Figure 6(b)). Indeed, knockdown of KDM1A markedly dysregulated a series of cell cycle-associated genes in Panc-1 cell (Figure 6(c)). We focused on CCNA2, an essential regulator of cell cycle control and was depressed by 2.03-fold upon KDM1A knockdown. We used TCGA PAAD data to explore the association of KDM1A and CCNA2 in PC, and found that the expression of KDM1A was significantly correlated with that of CCNA2 (Figure 6(d)). Furthermore, high expression of CCNA2 suggested a poor prognosis in PC (Figure 6(e)). A marked inhibition of CCNA2 was observed in in vivo model when knockdown of KDM1A (Figure 6(f)). Furthermore, ChIP-qPCR assay showed that knockdown of KDM1A decreased the enrichment of CCNA2 promoter by KDM1A antibody while increased it by H3K9me2 antibody (Figure 6(g)), indicating that KDM1A negatively regulated the expression of CCNA2 by an H3K9-dependent pathway. Knockdown of KDM1A impaired the protein expression of CCNA2 in Panc-1 and CFPAC-1 cells, while transfection of CCNA2 plasmid rescued its expression (Figure 6(h)). The colony formation assay indicated that CCNA2 overexpression improves the ability of colony formation by partially reversing KDM1A knockdown-mediated growth arrest in Panc-1 and CFPAC-1 cells (Figure 6(i)). Cell cycle analysis further demonstrated that CCNA2 overexpression could rescue the KDM1A knockdown-mediated cell cycle arrest in Panc-1 and CFPAC-1 cells (Figure 7(a) and (b)). These data unveiled that CCNA2 was partially responsible for KDM1A knockdown-mediated effect on PC cells.
Figure 6.
CCNA2 was partially responsible for KDM1A knockdown-mediated effect on PC cells. (a) RNA-seq was performed to compare whole-gene expression variations between the KDM1A siRNA and control group in Panc-1 cells. (b) The genes of significantly differentiated expression were enriched by KEGG signal pathway. The red box indicated the pathway of cell cycle. (c) The heatmap showed the differentiated genes associated with cell cycle between siRNA and control group. (d) The Pearson-CC was 0.31 between the expression of KDM1A and CCNA2 in PAAD datasets. (e) High expression of CCNA2 suggested a poor prognosis in patients with PAAD, P < 0.001. (f) KDM1A shRNA successfully downregulated the expression of KDM1A and CCNA2 in vivo compared to scrambled group. (g) ChIP assay using KDM1A and H3K9me2 antibody was performed and the enrichment of CCNA2 promoter was examined by qPCR, n = 3. (h) Knockdown of KDM1A depressed the expression of KDM1A and CCNA2, while overexpression of CCNA2 rescued the expression of CCNA2 in Panc-1 and CFPAC-1 cells. (i) Knockdown of KDM1A reduced colony formation in Panc-1 and CFPAC-1 cells and overexpression of CCNA2 partially rescued the ability of colony formation. NC: negative control; Si: siRNA; OE: overexpression. Scale bars correspond to 50 μm. (A color version of this figure is available in the online journal.)
Figure 7.
(a–b) Overexpression of CCNA2 partially rescued the cell cycle arrest in G0G1 upon KDM1A knockdown in Panc-1 and CFPAC-1 cells.
CDK6 was partially responsible for KDM3A knockdown-mediated effect on PC cells
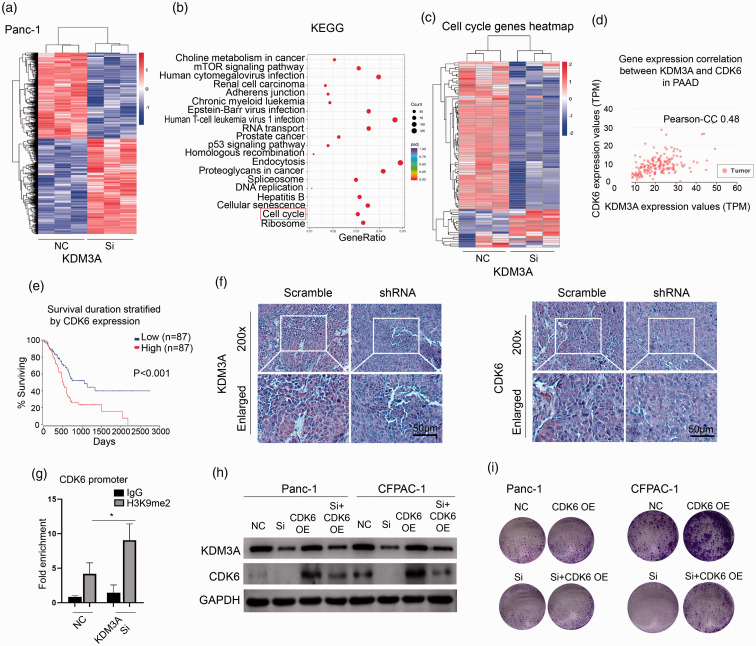
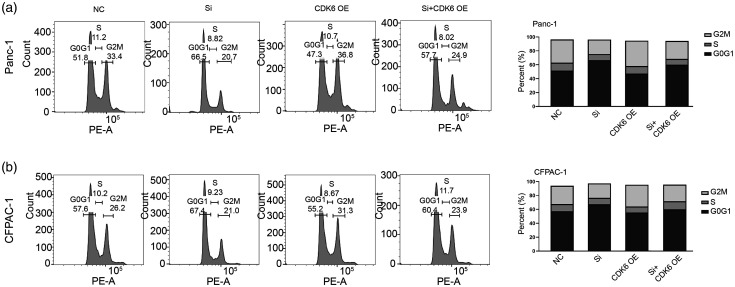
To explore the mechanism of KDM3A knockdown-mediated effect, we used RNA-seq to determine the global gene expression variations in Panc-1 cell—after KDM3A knockdown (Figure 8(a)). KEGG signaling analysis showed that KDM3A knockdown led to significant enrichment of cell cycle-associated signaling (Figure 8(b)) and a series of cell cycle-regulated genes were detexted (Figure 8(c)). Among them, CDK6 was significantly depressed by 2.20-fold upon KDM3A knockdown. As shown in Figure 8(d) and (e), the expression of KDM3A was significantly associated with CDK6 and high expression of CDK6 indicated a poor prognosis in PC. In vivo model, knockdown of KDM3A led to remarkable inhibition of CDK6 (Figure 8(f)). Furthermore, ChIP-qPCR assay showed that knockdown of KDM3A increased the enrichment of CDK6 promoter by H3K9me2 antibody (Figure 8(g)), suggesting that KDM3A regulated the expression of CDK6 by an H3K9-dependent pathway. Knockdown of KDM3A indeed decreased the expression of CDK6 and transfection of CDK6 plasmid could rescue its expression in Panc-1 and CFPAC-1 cells (Figure 8(h)). Functionally, CDK6 overexpression could rescue the KDM3A knockdown-mediated impairment of colony formation and cell cycle arrest in PC cells (Figures 8(i) and 9(a) and (b)). Thus, these data firmly demonstrated that CDK6 was partially responsible for KDM3A knockdown-mediated effect on PC cells.
Figure 8.
CDK6 was partially responsible for KDM3A knockdown-mediated effect on PC cells. (a) Compare whole-gene expression variations between the KDM3A siRNA and control group in Panc-1 cells by RNA-seq. (b) The genes of significantly differentiated expression were enriched by KEGG signal pathway. The red box indicated the pathway of cell cycle. (c) The heatmap showed the differentiated genes associated with cell cycle between siRNA and control group. (d) The Pearson-CC was 0.48 between the expression of KDM3A and CDK6 in PAAD datasets. (e) High expression of CDK6 was associated with a poor prognosis in patients with PAAD, P<0.001. (f) KDM3A shRNA successfully downregulated the expression of KDM3A and CDK6 in vivo compared to scrambled group. (g) ChIP assay using H3K9me2 antibody was performed and the enrichment of CDK6 promoter was examined by qPCR, n = 3. (h) Knockdown of KDM3A depressed the expression of KDM3A and CDK6, while overexpression of CDK6 rescued the expression of CDK6 in Panc-1 and CFPAC-1 cells. (i) Downregulation of KDM3A reduced colony formation in Panc-1 and CFPAC-1 cells and overexpression of CDK6 partially rescued the effect. NC: negative control; Si: siRNA; OE: overexpression. Scale bars correspond to 50 μm. (A color version of this figure is available in the online journal.)
Figure 9.
(a–b) Overexpression of CDK6 partially rescued the cell cycle arrest in G0G1 upon KDM3A knockdown in Panc-1 and CFPAC-1 cells.
Discussion
In our analysis, we found that seven H3K9-regulating genes, including EHMT1, EHMT2, JHDM1D, KDM1A, KDM3A, KDM3B, and KDM4A, were dysregulated in PC tissues compared with normal tissues. Our IHC staining of KDM1A and KDM3A verified that the expression of KDM1A and KDM3A in PC was significantly higher than that in pancreatic benign lesions, pericancerous tissues, and normal tissues. Positive expression of KDM1A and KDM3A was negatively correlated with pathological grade, lymphatic metastasis, invasion, and TNM stage. These data emphasized the clinicopathological significance of KDM1A and KDM3A in PC and suggested its potential role in tumorigenesis in PC.
Histone lysine methylation is an essential bioprocess responsible for the posttranslational modification of numerous genes. The most characterized histone lysine methylation sites include H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20, which are summarized in detail in an excellent review. 8 Methylation of lysine residues is accomplished by a series of KMTs, and most lysine methylation is currently verified as reversible, which is mediated by KDMs.21,22 G9a is the most intensively studied H3K KTM and is responsible for catalyzing H3K9 monomethylation and demethylation. 23 G9a-like proteins (GLPs) interact with G9a, forming a heterodimeric complex and marking transcriptional silencing at the targeted site. 24 In various cancers, studies have demonstrated that high expression of G9a is critical for the promotion of tumorigenesis and serves as a predictor of poor prognosis.25–27 Thus, elucidating the major dysregulated regulators of histone lysine methylation and exploring the underlying mechanisms are essential for understanding the development of cancer. We systematically explored the expression of H3K9 regulators in PC and concluded that KDM1A and KDM3A were upregulated and were correlating with clinicopathological features and prognosis.
KDM1A was the first identified lysine-specific KDM responsible for removing monomethyl and dimethyl groups at H3K4 and H3K9 sites.21,28 Previous reports have demonstrated that KDM1A is highly expressed in hematological cancers, sarcomas, breast cancers, colorectal cancers, and prostate cancers. 29 Although an initial study suggested that KDM1A was overexpressed in PC tissues compared to paired adjacent tissues, lack of comparison to normal tissues and lack of association with clinicopathological features attenuated the meaningfulness of the results. Impairment of KDM1A inhibits cancer growth and migration and the self-renewal of cancer stem cells. 30 Mechanistically, KDM1A regulates the activity of β-catenin, maintains the stability of HIF1α, and induces the ER stress response.16,28,30 Furthermore, GSK3β stabilized KDM1A protein by reducing its ubiquitination in glioblastoma. 31 We found that knockdown of KDM1A markedly impaired tumor growth in vitro and in vivo, and the expression of KDM1A was correlated with that of CCNA2 and overexpression of CCNA2 partially rescued of KDM1A knockdown-mediated impairment of colony formation and cell cycle arrest in PC cells. Although our evidence demonstrated the direct linkage between KDM1A and CCNA2 promoter, a detailed study to understand how KDM1A regulating the expression of CCNA2 is still needed.
KDM3A was demonstrated to specifically regulate gene expression via demethylase activity at promoters. 32 In cancers, emerging evidence suggests that dysregulation of KDM3A is found in breast, colorectal, prostate and brain cancers and Ewing sarcoma.33–36 Knockdown of KDM3A led to improved cell cycle arrest, apoptosis, and depressed migration in cancers. 37 Forced expression of KDM3A promoted the growth of primary tumors and, interestingly, induced more lung and liver metastases, which was attributed to an enhanced stemness of PC cells, in an animal model. Furthermore, recent studies suggested that KDM3A conferred tumorigenicity via its methylation of nonhistone lysines. 38 For instance, KDM3A was shown to interact with PGC-1α, a vital protein regulating the biogenesis of mitochondria under normal conditions, and rescue PGC-1α activity by enhancing its monomethylation in hypoxic conditions, contributing to tumor growth in vivo. 36 In our study, CDK6 was involved in the underlying mechanism of KDM3A in PC by an H3K9-dependent pathway, which was partially responsible for KDM3A knockdown-mediated effect on PC cells, while the thorough mechanism underlying KDM3A-regulated CDK6 remains elusive.
In sum, our research found that KDM1A and KDM3A were highly expressed in PC compared to benign lesions, pericancerous tissues, and normal tissues, among other H3K9 regulators. Both genes were intimately associated with clinicopathological features and prognosis in patients with PC. Mechanically, we demonstrated that CCNA2 was partially responsible for KDM1A knockdown-mediated effect and CDK6 for KDM3A knockdown-mediated effect in PC. However, a thorough understanding of KDM1A or KDM3A-mediated epigenetic regulation of cell cycle-associated genes will be needed that might involve a coordinated interplay among DNA methylation, histone modifications, or even nucleosome structure.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702211023473 for KDM1A and KDM3A promote tumor growth by upregulating cell cycle-associated genes in pancreatic cancer by Xuyang Hou, Qiuguo Li, Leping Yang, Zhulin Yang, Jun He, Qinglong Li and Daming Li in Experimental Biology and Medicine
Footnotes
AUTHORS’ CONTRIBUTIONS: XH and QL performed most experiments; LY, ZY, and JH assisted; QL provided the funds; DL designed and supervised the project and wrote the manuscript.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ETHICAL APPROVAL: This study was approved by the Ethics Committee for Human Research, Central South University, and was conducted according to the approved guidelines and the principles of the Declaration of Helsinki.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Hunan Provincial Natural Science Foundation (2020JJ4790).
ORCID iD: Daming Li https://orcid.org/0000-0002-5109-4655
Supplemental material: Supplemental material for this article is available online.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66:115–32 [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin 2006; 56:106–30 [DOI] [PubMed] [Google Scholar]
- 4.Katz MH, Wang H, Fleming JB, Sun CC, Hwang RF, Wolff RA, Varadhachary G, Abbruzzese JL, Crane CH, Krishnan S, Vauthey J, Abdalla E, Lee J, Pisters P, Evans D. Long-term survival after multidisciplinary management of resected pancreatic adenocarcinoma. Ann Surg Oncol 2009; 16:836–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khorana AA, Mangu PB, Berlin J, Engebretson A, Hong TS, Maitra A, Mohile SG, Mumber M, Schulick R, Shapiro M. Potentially curable pancreatic cancer: American Society of Clinical Oncology Clinical Practice guideline. J Clin Oncol 2016; 34:2541–56 [DOI] [PubMed] [Google Scholar]
- 6.Sohal DP, Kennedy EB, Khorana A, Copur MS, Crane CH, Garrido-Laguna I, Krishnamurthi S, Moravek C, O’Reilly EM, Philip PA, Ramanathan R, Ruggiero J, Shah M, Urba S, Uronis H, Lau M, Laheru D. Metastatic pancreatic cancer: ASCO clinical practice guideline update. J Clin Oncol 2018; 36:2545–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 2012; 48:491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morera L, Lübbert M, Jung M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenet 2016; 8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin M, Chinnaiyan A. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419:624–9 [DOI] [PubMed] [Google Scholar]
- 10.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB‐E2F pathway, essential for proliferation and amplified in cancer. EMBO J 2003; 22:5323–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang A, Dai H, Gong Y, Zhang C, Shu J, Luo Y, Jiang Y, Liu W, Bie P. ANLN-induced EZH2 upregulation promotes pancreatic cancer progression by mediating miR-218-5p/LASP1 signaling axis. J Exp Clin Cancer Res 2019; 38:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bianco-Miotto T, Chiam K, Buchanan G, Jindal S, Day TK, Thomas M, Pickering MA, O'loughlin MA, Ryan NK, Raymond WA, Horvath L, Kench J, Stricker P, Marshall V, Sutherland R, Henshall S, Gerald W, Scher H, Risbridger G, Clements J, Butler L, Tilley W, Horsfall D, Ricciardelli C. Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol Biomarkers Prev 2010; 19:2611–22 [DOI] [PubMed] [Google Scholar]
- 13.Casciello F, Al-Ejeh F, Kelly G, Brennan DJ, Ngiow SF, Young A, Stoll T, Windloch K, Hill MM, Smyth MJ, Gannon F, Lee J. G9a drives hypoxia-mediated gene repression for breast cancer cell survival and tumorigenesis. Proc Natl Acad Sci U S A 2017; 114:7077–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wozniak R, Klimecki W, Lau S, Feinstein Y, Futscher BW. 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene 2007; 26:77–90 [DOI] [PubMed] [Google Scholar]
- 15.Yuan C, Li Z, Qi B, Zhang W, Cheng J, Wang Y. High expression of the histone demethylase LSD1 associates with cancer cell proliferation and unfavorable prognosis in tongue cancer. J Oral Pathol Med 2015; 44:159–65 [DOI] [PubMed] [Google Scholar]
- 16.Qin Y, Zhu W, Xu W, Zhang B, Shi S, Ji S, Liu J, Long J, Liu C, Liu L, Xu J, Yu X. LSD1 sustains pancreatic cancer growth via maintaining HIF1α-dependent glycolytic process. Cancer Lett 2014; 347:225–32 [DOI] [PubMed] [Google Scholar]
- 17.Wang HY, Long QY, Tang SB, Xiao Q, Gao C, Zhao QY, Li QL, Ye M, Zhang L, Li LY, Ye M, Zhang L, Li LY, Wu M. Histone demethylase KDM3A is required for enhancer activation of hippo target genes in colorectal cancer. Nucleic Acids Res 2019; 47:2349–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimaldi F, Muser D, Beltrami CA, Machin P, Morelli A, Pizzolitto S, Talmasson G, Marciello F, Colao AA, Monaco R, Monaco G, Faggiano A. Partitioning of bronchopulmonary carcinoids in two different prognostic categories by ki-67 score. Front Endocrinol 2011; 2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong F, Liu X, Zhou Y, Hou X, He J, Li Q, Miao X, Yang L. Downregulation of METTL14 increases apoptosis and autophagy induced by cisplatin in pancreatic cancer cells. Int J Biochem Cell Biol 2020; 122:105731. [DOI] [PubMed] [Google Scholar]
- 20.Xiong L, Yang Z, Yang L, Liu J, Miao X. Expressive levels of MUC1 and MUC5AC and their clinicopathologic significances in the benign and malignant lesions of gallbladder. J Surg Oncol 2012; 105:97–103 [DOI] [PubMed] [Google Scholar]
- 21.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119:941–53 [DOI] [PubMed] [Google Scholar]
- 22.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006; 439:811–6 [DOI] [PubMed] [Google Scholar]
- 23.Casciello F, Windloch K, Gannon F, Lee JS. Functional role of G9a histone methyltransferase in cancer. Front Immunol 2015; 6:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001; 410:116–20 [DOI] [PubMed] [Google Scholar]
- 25.Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, Shiah SG, Chen PS, Jeng YM, Cheng TY, Lai TC, Chang JS, Jan YH, Chien MH, Yang CJ, Huang MS, Hsiao M, Kuo ML. H3k9 Histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res 2010; 70:7830–40 [DOI] [PubMed] [Google Scholar]
- 26.Huang J, Dorsey J, Chuikov S, Zhang X, Jenuwein T, Reinberg D, Berger SL. G9a and glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 2010; 285:9636–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong X, Chen X, Guan X, Zhang H, Ma Y, Zhang S, Wang E, Zhang L, Han Y. Overexpression of G9a and MCM7 in oesophageal squamous cell carcinoma is associated with poor prognosis. Histopathology 2015; 66:192–200 [DOI] [PubMed] [Google Scholar]
- 28.Pishas KI, Drenberg CD, Taslim C, Theisen ER, Johnson KM, Saund RS, Pop IL, Crompton BD, Lawlor ER, Tirode F, Mora J, Delattre O, Beckerle M, Callen D, Sharma S, Lessnick S. Therapeutic targeting of KDM1A/LSD1 in Ewing sarcoma with SP-2509 engages the endoplasmic reticulum stress response. Mol Cancer Ther 2018; 17:1902–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theisen ER, Pishas KI, Saund RS, Lessnick SL. Therapeutic opportunities in Ewing sarcoma: EWS-FLI inhibition via LSD1 targeting. Oncotarget 2016; 7:17616–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang M, Chen C, Geng J, Han D, Wang T, Xie T, Wang L, Wang Y, Wang C, Lei Z, Chu X. Targeting KDM1A attenuates wnt/β-catenin signaling pathway to eliminate sorafenib-resistant stem-like cells in hepatocellular carcinoma. Cancer Lett 2017; 398:12–21 [DOI] [PubMed] [Google Scholar]
- 31.Zhou A, Lin K, Zhang S, Chen Y, Zhang N, Xue J, Wang Z, Aldape KD, Xie K, Woodgett JR, Huang S. Nuclear GSK3β promotes tumorigenesis by phosphorylating KDM1A and inducing its deubiquitylation by USP22. Nat Cell Biol 2016; 18:954–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 2007; 450:119–23 [DOI] [PubMed] [Google Scholar]
- 33.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 2006; 125:483–95 [DOI] [PubMed] [Google Scholar]
- 34.Ramadoss S, Guo G, Wang CY. Lysine demethylase KDM3A regulates breast cancer cell invasion and apoptosis by targeting histone and the non-histone protein p53. Oncogene 2017; 36:47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sechler M, Parrish JK, Birks DK, Jedlicka P. The histone demethylase KDM3A, and its downstream target MCAM, promote Ewing sarcoma cell migration and metastasis. Oncogene 2017; 36:4150–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian X, Li X, Shi Z, Bai X, Xia Y, Zheng Y, Xu D, Chen F, You Y, Fang J, Hu Z, Zhou Q, Lu Z. KDM3A senses oxygen availability to regulate PGC-1α-mediated mitochondrial biogenesis. Mol Cell 2019; 76:885–95.e7 [DOI] [PubMed] [Google Scholar]
- 37.Dandawate P, Ghosh C, Palaniyandi K, Paul S, Rawal S, Pradhan R, Sayed AAA, Choudhury S, Standing D, Subramaniam D, Padhye S, Gunewardena S, Thomas S, Neil M, Tawfik O, Welch D, Jensen R, Maliski S, Weir S, Iwakuma T, Anant S, Dhar A. The histone demethylase KDM3A, increased in human pancreatic tumors, regulates expression of DCLK1 and promotes tumorigenesis in mice. Gastroenterology 2019; 157:1646–59.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamamoto R, Saloura V, Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer 2015; 15:110–24 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702211023473 for KDM1A and KDM3A promote tumor growth by upregulating cell cycle-associated genes in pancreatic cancer by Xuyang Hou, Qiuguo Li, Leping Yang, Zhulin Yang, Jun He, Qinglong Li and Daming Li in Experimental Biology and Medicine