Abstract
Sluggish was identified in a population of third generation mice descended from N-ethyl-N-nitrosourea (ENU3)-mutagenized sires. Macrophages from homozygotes exhibited impaired TNFα production in response to all TLR ligands tested, and displayed impaired type I IFN production in response to TLR7 and 9 stimulations. The phenotype was confined to a critical region on mouse chromosome 18, and then ascribed to a T to A transversion in the acceptor splice site of intron 4 at position 13346 of the Map3k8 gene, resulting in defective splicing. The Map3k8Sluggish mutation does not result in susceptibility to viral infections, but Sluggish mice displayed high susceptibility to group B streptococcus (GBS) infection, with impaired TNFα and type I IFN production in infected macrophages. Our data demonstrate that the encoded protein kinase Tpl2 plays an essential role in cell signaling in the immune response to certain pathogens.
Introduction
Via the TLRs, the innate immune system senses conserved molecules of microbial origin, and triggers innate immune responses that include production of inflammatory cytokines, chemokines and type I IFNs. Most of the TLRs signal through the MyD88 adaptor protein, while TLR3 signals through a MyD88-independent pathway and TLR4 signals through both MyD88-dependent and independent processes [reviewed in (1, 2)].
Cytokines produced in response to TLR signaling, including TNFα and IFNs, are critical for controlling microbial infections. The production of type I IFNs by plasmacytoid dendritic cells (pDCs) in response to the stimulation of TLR7 (which senses ssRNA) and/or TLR9 (which senses CpG DNA) is necessary for the host defense against certain viruses (3), while both TNFα and type I IFNs play important roles in the host response to bacterial infections (4–6). For instance, Tnf−/− and Tnfr1−/− mice are highly susceptible to infection with the intracellular pathogen Listeria monocytogenes (5, 6), and TNF production has been shown to be an important component of the immune response to extracellular group B streptococcus (GBS) infection (7, 8). Mice deficient in type I IFN signaling display impaired immune responses to a variety of bacterial infections including GBS (9), but display increased resistance to L. monocytogenes (10–12).
Tpl2 (tumor progression locus 2)/COT (cancer Osaka thyroid)/MAP3K8 is a serine/threonine protein kinase that is a member of the mitogen-activated protein kinase kinase kinase (MAP3K) family of proteins. In vitro, Tpl2 can activate important immune response factors including the JNK, p38, and ERK protein kinases, as well as NF-κB and NFAT (13–15). Tpl2-deficient mice are resistant to LPS-induced shock, due to low levels of circulating TNFα (16), and Tpl2-deficient macrophages demonstrate impaired ERK activation and TNFα production in response to multiple TLR ligands (17, 18).
We have identified an N-ethyl-N-nitrosourea (ENU)-induced mutation, termed Sluggish, resulting in defective splicing of the Map3k8 encoded Tpl2 transcript. Animals homozygous for the mutation show impaired TNFα secretion in response to all TLR stimuli, while heterozygotes displayed intermediate responses to some TLR ligands. Macrophages isolated from homozygous Map3k8Sluggish/Sluggish mice show impaired type I IFN production specifically in response to TLR7 and 9 stimulations, but are able to control viral infections. In addition, Map3k8Sluggish/Sluggish mice are resistant to mouse cytomegalovirus (MCMV) infection. Although these animals display resistance to infection by L. monocytogenes, they are highly susceptible to GBS infection. Macrophages isolated from Map3k8Sluggish/Sluggish mice exhibit altered type I IFN and TNFα production in response to bacteria. Our results confirm the obligatory role Tpl2 plays in response to TLR signaling, and show that Tpl2 is necessary for type I IFN production downstream of specific signals. Furthermore, we demonstrate that Tpl2 is obligatory for controlling GBS infection.
Materials and methods
Reagents and antibodies
LPS [Salmonella minnesota R959 (Re)], macrophage-activating lipopeptide-2 (Malp2), ODN 2216 (CpG-A) were obtained from Alexis. Unmethylated DNA oligonucleotide (CpG-B) 5’-TCCATGACGTTCCTGATGCT-3’ was synthesized by Integrated DNA Technologies (Coralville, IA). Resiquimod was obtained from 3M. Pam3CSK4 were obtained from EMC Microcollections (Tübingen, Germany). Peptidoglycan was purchased from Fluka. dsRNA poly(I:C) was obtained from Amersham Pharmacia Biotech. Recombinant Mouse IFNγ was obtained from PBL Biomedical Laboratories. DOTAP was obtained from Roche. Antibodies against phosphorylated or total ERK were from Cell Signaling Technology (Beverly, MA). Fluorescent-labeled antibodies against CD11b, and TNFα used in FACS analysis were from eBioscience (San Diego, CA). Antibodies against influenza A virus Hemagglutinin (clone IV.C102), sc-80550 was from Santa Cruz Biotech. Secondary antibody was goat anti-mouse IgG-FITC, sc-2010 (Santa Cruz).
Mice, ENU mutagenesis
C57BL/6J (also referred to as WT), C3H/HeN, MyD88poc/poc, Tlr9CpG1/CpG1, Tlr9CpG3/CpG3, Stat1dom/dom, Tlr7−/− or Tlr7-/Y, Irak4otiose/otiose, and Sluggish/Sluggish mice (MMRRC: 030499-UCD ) were maintained and bred in The Scripps Research Institute vivarium under the supervision of the Department of Animal Resources. All studies involving mice were performed in accordance with the rules of Institutional Animal Care and Use Committee of The Scripps Research Institute. ENU mutagenesis was performed in a C57BL/6J background as described previously (19).
Genetic mapping and position cloning of Sluggish
Sluggish homozygotes were outcrossed to C3H/HeN mice and F1 Hybrids were backcrossed to Sluggish homozygotes. 39 mice were genotyped using microsatellite polymorphisms markers. The mutation was confined upstream of D18mit110 on chromosome 18. Genotyping was carried out by fragment length analysis with fluorescent primers. Genotyping and sequencing were done using the ABI 3100 DNA sequencer.
Peritoneal macrophage response assays
C57BL/6J mice (germline mutants or controls) were injected intraperitoneally with 4% thioglycolate. 3 days later, macrophages were harvested. For the screening of TLR signaling, cells were cultured at a density of 5×104 cells/well (96 well) with varying concentration of TLR agonists as follows: LPS (500 pg/ml), poly(I:C) (5 μg/ml), Lipoprotein Pam3CSK4 (50 ng/ml), resiquimod (100 ng/ml), CpG-B (100 nM), peptidoglycan (2 μg/ml), Malp2 (100 ng/ml) or left unstimulated. Cells were incubated at 37° C for 4h, and culture media were collected for the TNF bioassay or cytokine ELISAs (Ebioscience).
For type I IFN produced by peritoneal macrophages, cells were primed with 10 ng/ml IFNγ for 4 h, and then washed twice with medium; then cells were stimulated with CpG-B (500, 100, 20, 0.1 nM) or resiquimod (100, 20, 4, 0 ng/ml) for another 4 h and media were collected for the TNF and IFN bioassays. Cells were also stimulated with 500, 100, 20, 0 pg/ml LPS and 20, 4, 0.8, 0 μg/ml poly(I:C) for 4 h without priming with IFNγ, and media were collected for the TNF and IFN bioassays.
FACS analysis
Peritoneal macrophages were challenged with Malp2 (100 ng/ml), PGN (2μg/ml) or CpG-B (100 nM) for 4 h in the presence (intracellular staining) or absence (cell surface staining) of 5 μg/ml of Brefeldin A. For intracellular staining, the cells were washed with ice cold FACS Buffer and then incubated with CD11b-APC antibody at 4° C for 30 minutes. The cells were then permeabilised for 20 minutes with BD Fix+Perm solution (BD cytofix/Cytoperm) followed by staining with TNF-PE antibody for 40 minutes. For cell surface staining, cells were washed with ice cold FACS Buffer and then incubated with CD11b-APC and TNF-PE antibody at 4° C for 30 minutes. Cells were acquired by flow cytometry on a FACSCalibur (BD) and data were analyzed using FlowJo software.
Peritoneal macrophages were infected with GFP-tagged MCMV (kindly provided by Christopher Benedict, La Jolla Institute of Allergy and Immunology), GFP-tagged adenoviral vector Ad5.F16-GFP (gift from Glen Nemerow, The Scripps Research Institute), or a mouse-adapted human influenza A (PR8 strain) virus. Cells were collected for FACS analysis after 24 hours for MCMV, 24h for influenza and 72 hours for Ad5.F16-GFP. Medium was collected to determine cytokine production by ELISA or bioassay. Cells were washed twice with PBS, and incubated with trypsin at 37° C for 15 minutes. Cells were collected by centrifugation and resuspended in FACS buffer, and then acquired on a FACSCalibur (BD) and analyzed using FlowJo software.
Biological assays
Type I IFN activity was measured with reference to a recombinant mouse IFNβ standard by using an L-929 cell line transfected with an interferon-sensitive luciferase construct. TNF activity produced by peritoneal macrophages or dendritic cells was determined with reference to a recombinant mouse TNFα standard by using the L-929 cell cytolytic assay (20).
In vivo CpG challenge
Mice were injected intravenously with 2ug CpG-A. Six hours later, blood was collected and serum type IFN was measured by ELISA.
Cytokine measurement
IL-6, IL-12p40, IL-12p70, TNFα, IFNγ concentrations in culture media or sera were measured by ELISA kit (eBioscience). IFNα and IFNβ concentrations in culture media or sera were measured by ELISA (PBL Biomedical Laboratories).
Mouse infection models
MCMV (Smith strain) was prepared as described previously (21). 6 week-old male mice were injected intraperitoneally with 2×105 PFU of MCMV. 36 hours after infection, mice were bled and cytokines in the serum were measured by ELISA.
Bioluminescent bacteria Listeria monocytogenes (strain 10403S; Xenogen) were prepared as described (22). 6 week-old male mice were injected intravenously with 105 CFU of Listeria monocytogenes. Serum cytokines were measured at 24 hours after infection by ELISA or Bioassay.
Type III COH1 strain or type Ib H36B strain of GBS (provided by Giuseppe Teti, University of Messina, Italy) were grown to the early-log phase in Todd-Hewitt broth (BD) and were diluted to the appropriate concentration in PBS. 8 week-old male mice were inoculated intraperitoneally with 106 CFU of the type Ib strain H36B (23). Each experiment included one group of Sluggish mice and wild-type C57BL/6J mice. The groups did not differ in the age or the weight of the animals. Mice were observed for 7 days after infection, but the deaths were never recorded after 4 days. In additional experiments, blood, spleens, and kidneys were harvested 20 hours after GBS infection. Serum cytokine concentrations were measured by ELISA. Bacterial loads in spleen, kidney and blood were determined by colony counts on Todd-Hewitt Agar plates.
Statistical data analysis
Data were analyzed using the GraphPad Prism4® software (GraphPad Software, San Diego, CA). The statistical significance of the differences among groups was determined by Student’s two-tailed test or by one-way ANOVA followed by Dunnett’s test for three or more groups. Survival data were determined by Log-rank (Mantel-Cox) Test. Data were considered significant when P<0.05.
Results
Sluggish macrophages exhibit impaired cytokine production in response to TLR signaling
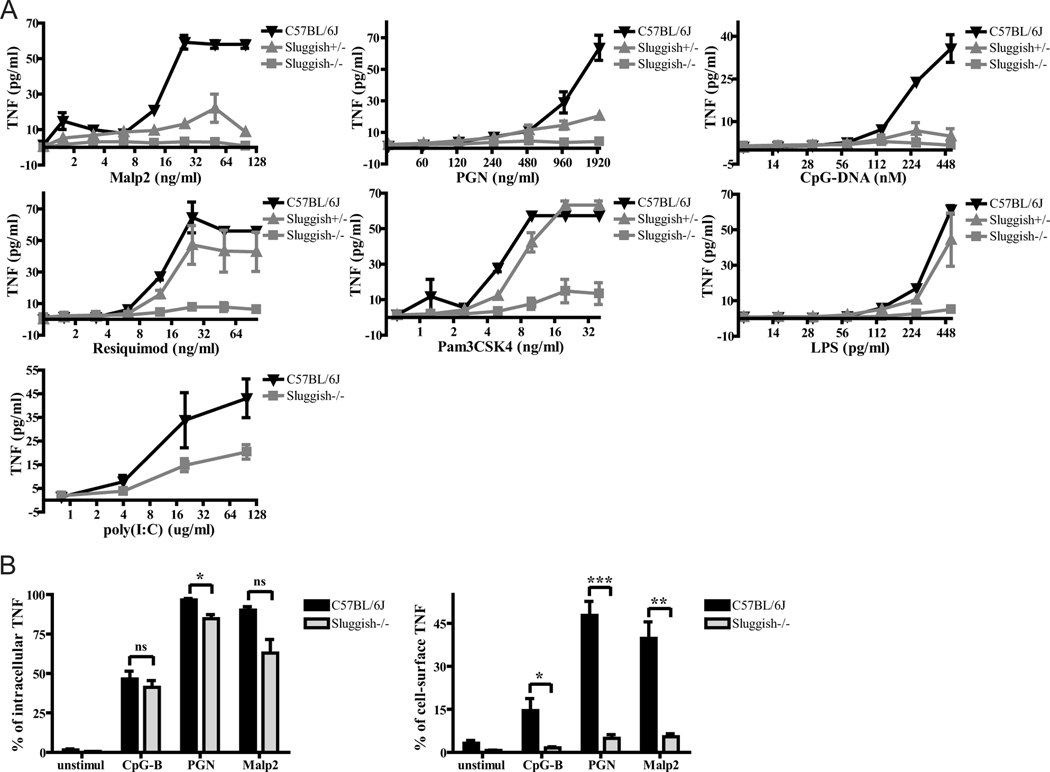
Sluggish was identified as a semidominant mutation in a screen for genetic defects of TLR signaling in peritoneal macrophages derived from third generation C57BL/6J mice homozygous for N-ethyl-N-nitrosourea (ENU)-induced germline mutations (19). Thioglycolate-elicited peritoneal macrophages from Sluggish homozygote mice showed defective responses for all known TLR ligands and secreted little to no TNFα in response to the TLR2/6 ligands peptidoglycan (PGN) and macrophage-activating lipopeptide 2 (Malp2), the TLR9 ligand CpG-B DNA, the TLR4 ligand LPS, the TLR1/2 ligand PAM3CSK4 (a triacyl lipopeptide), the TLR7 ligand resiquimod (a ssRNA mimetic), and the TLR3 ligand poly I:C (a dsRNA mimetic). Heterozygote animals exhibited impaired TNFα responses to some, but not all, TLR ligands (Figure 1A).
Figure 1.
Abnormal cytokine production in Sluggish macrophages in response to TLR signaling. (A) Thioglycolate-elicited peritoneal macrophages from wild-type (C57BL/6J; black triangles), heterozygous Sluggish (gray triangles), and homozygous Sluggish (gray squares) mice were stimulated with various concentrations of TLR agonists as indicated, and after 4 hours of incubation supernatants were tested for TNF bioactivity. n=3 mice per group. (B) In the left panel, peritoneal macrophages of the indicated genotypes were challenged with various TLR ligands in the presence of brefeldin A. After 4 hours hours of stimulation, cells were permeabilized and subjected to intracellular TNFα staining. In the right panel, peritoneal macrophages were challenged with various TLR ligands in the absence of brefeldin A. After 4 hours of stimulation, cells were subjected to cell surface TNFα staining. n=3 mice per group. Data represent mean ± SEM. * = P < 0.05, ** P < 0.01, *** P < 0.005.
In order to identify the stage at which TNFα production is compromised, macrophages from Sluggish homozygotes were tested in an intracellular cytokine staining assay after activation. We noted only a slight, or no, reduction in intracellular TNFα concentration in response to CpG-B DNA, PGN, and Malp2 (Figure 1B, left panel), while macrophages from the appropriate negative controls showed little to no intracellular TNFα (data not shown). While TNF was detected on the surface of wild-type macrophages, it was not detected on the surface of homozygous Sluggish macrophages (Figure 1B, right panel). This strongly suggests that the diminished concentration of soluble TNFα in culture medium conditioned by activated Sluggish macrophages is mainly due to impairment of TNFα secretion rather than impairment of TNFα biosynthesis.
We also examined the early production of other cytokines in peritoneal macrophages stimulated with TLR ligands for 4 hours. The secretion of IL-6, another proinflammatory cytokine, was profoundly impaired in homozygous Sluggish macrophages in response to most TLR ligands (Supplemental Figure 1A). The levels of IL-12p40, which enhances NK and T cell functions, were normal or increased in homozygous Sluggish macrophages in response to TLR signaling (Supplemental Figure 1B). Macrophages from the appropriate negative controls showed no cytokine production (data not shown).
The Sluggish mutation impairs TLR7 and 9-induced type I IFN production in peritoneal macrophages, but does not impair the immune response to viral infections
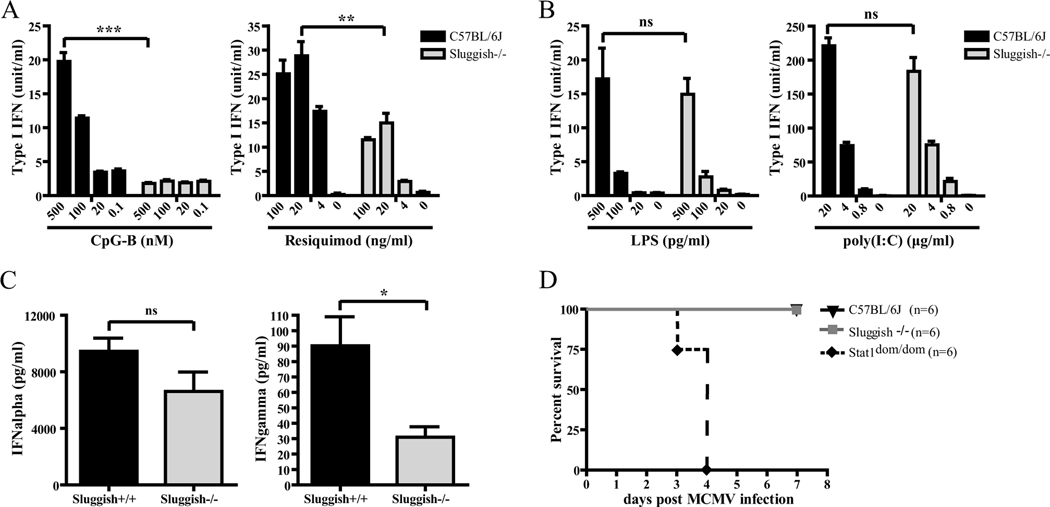
In addition to the induction of genes encoding proinflammatory cytokines, TLR-mediated signaling can induce type I IFN production. TLR3 and 4 primarily elicit IFNβ production through a MyD88-independent pathway (24, 25), while TLR7 and TLR9 signal through MyD88 to induce production of type I IFNs primarily in DCs (3, 23, 26). Following IFNγ-priming, macrophages are able to induce IFNβ production in response to CpG-B (27, 28). In order to examine the effects of the Sluggish mutation on type I IFN production, peritoneal macrophages were pretreated with IFNγ and then challenged with various concentrations of CpG-B or resiquimod, or were exposed directly to LPS or poly(I:C). CpG-B- and resiquimod-induced type I IFN production were significantly reduced in homozygous Sluggish macrophages (Figure 2A), while LPS and poly(I:C)-induced type I IFN production was unaffected (Figure 2B). Macrophages from the appropriate negative controls showed little to no type I IFN production in responses to TLR ligands (data not shown). These results suggest that Tpl2 is important for type I IFN production in peritoneal macrophages specifically in response to MyD88-dependent TLR signals.
Figure 2.
The Sluggish mutation impairs type I IFN production in response to TLR7 and 9 signaling in peritoneal macrophages, but does not affect susceptibility to viral infections. (A) Peritoneal macrophages from the indicated genotypes were primed with IFNγ (10 ng/ml) for 4 hours. Cells were washed with medium twice and then stimulated with various concentrations of CpG-B (500, 100, 20, 0.1 nM left panel) and resiquimod (100, 20, 4, 0 ng/ml right panel) for another 4 hours. The supernatants were collected for a L929-ISRE-Luc-based IFN bioassay. C57BL/6J, n=3; Sluggish−/−, n=5. (B) Peritoneal macrophages from the indicated genotypes were treated with various concentrations of LPS (500, 100, 20, 0 pg/ml left panel) and poly(I:C) (20, 4, 0.8, 0 μg/ml right panel) for 4 hours. C57BL/6J, n=3; Sluggish−/−, n=4. (C) Mice from the indicated genotypes were injected intravenously with CpG-A and DOTAP. Sera were collected and the concentrations of IFNα and IFNγ were determined by ELISA. n=3 mice per group. (D) Homozygous Sluggish mice are not susceptible to MCMV infection. Mice were injected intraperitoneally with 2×105 plaque-forming units (PFU) of MCMV. The survival of these mice was monitored during a 7 day period. C57BL/6J (black triangles), Sluggish−/− (gray squares), Stat1dom/dom (black diamonds). n=6 mice per group. Data represent mean ± SEM. * = P < 0.05, ** P < 0.01, *** P < 0.005.
As pDCs are the primary producers of type I IFN in vivo (29), we wanted to know if the type I IFN defect seen in Sluggish macrophages also appeared in pDCs. We injected mice with CpG-A intravenously together with DOTAP and examined serum concentration of type I IFN. The IFNα production in homozygous Sluggish mice was unaffected. By contrast, IFNγ was decreased in Sluggish mice (Figure 2C), suggesting that IFNγ production in response to TLR signaling is affected by the Sluggish mutation. These results demonstrate that the protein affected by the Sluggish mutation is not critically involved in type I IFN production in response to TLR9 stimulation in pDCs.
To further test whether the Sluggish mutation may affect type I IFN production in vivo, we infected homozygous Sluggish mice with 2×105 PFU of MCMV. Sluggish animals were resistant to MCMV similar to C57BL/6J control mice, while highly susceptible STAT1-deficient mice died within four days (Figure 2D) (30). Furthermore, homozygous Sluggish macrophages infected with either GFP-tagged MCMV, GFP-tagged adenoviral vector, or a mouse-adapted human influenza A (PR8 strain) virus were able to control viral infections (Supplemental Figure 2A) and displayed normal type I IFN production (Supplemental Figure 2B), although TNFα production remained impaired (Supplemental Figure 2C).
Sluggish mice are specifically susceptible to group B streptococci (GBS) infection
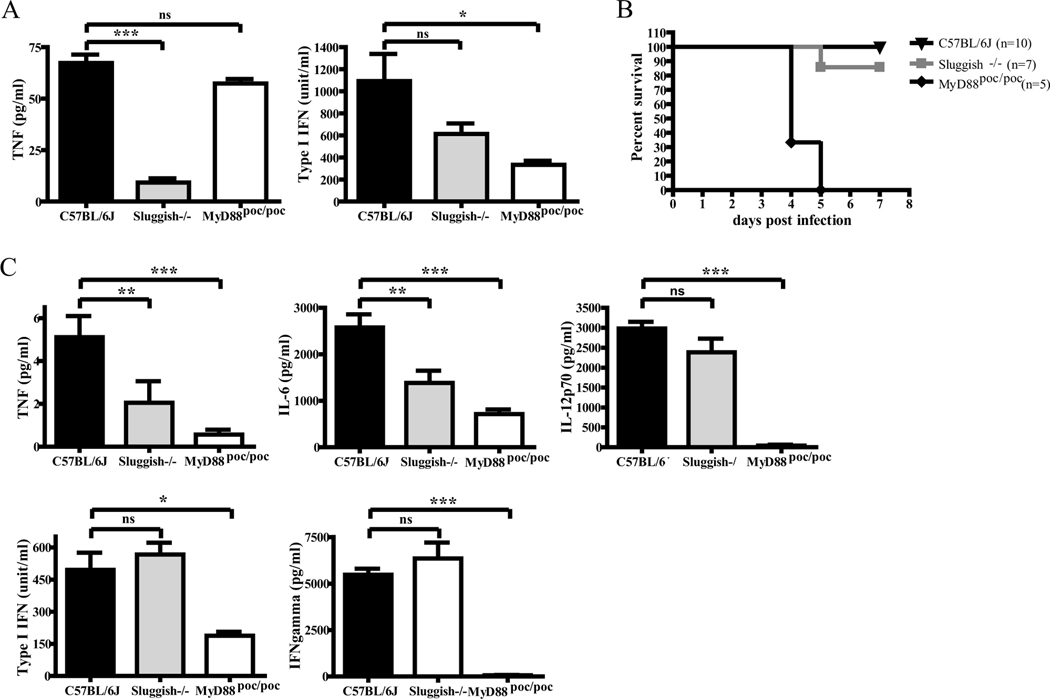
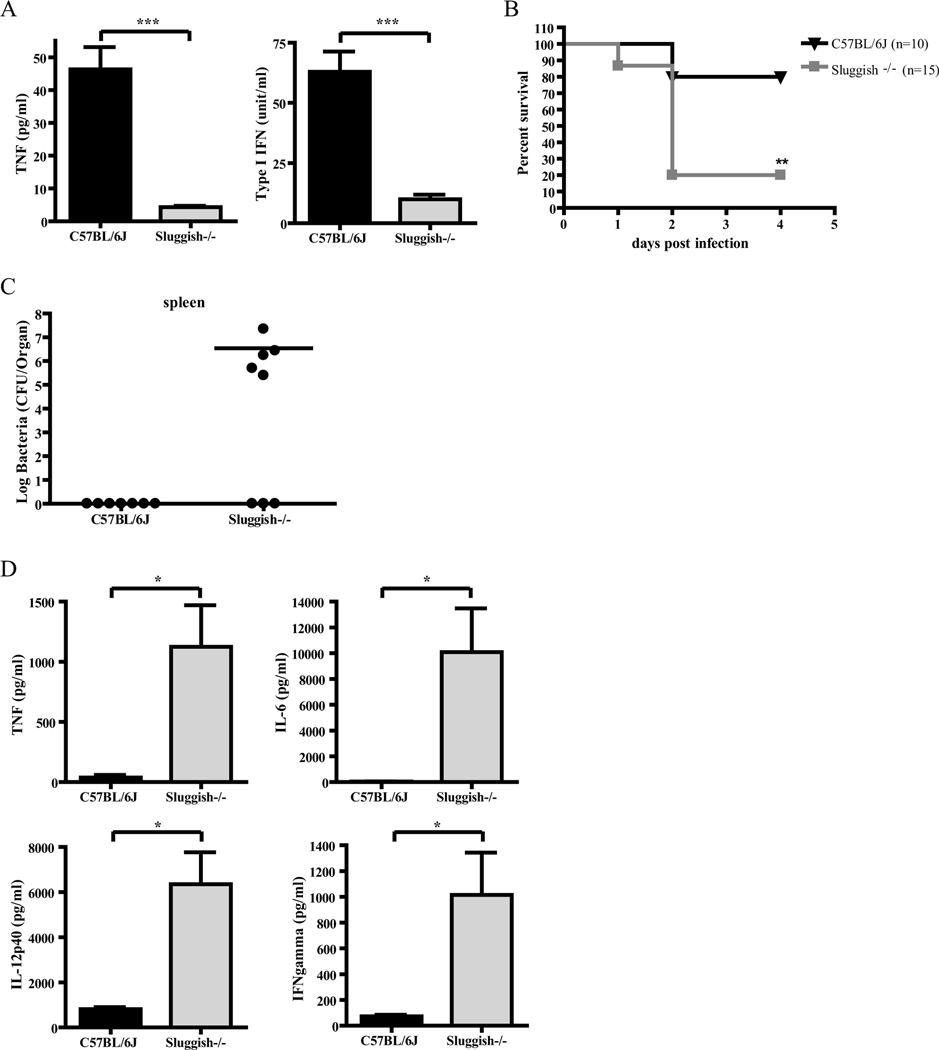
Because macrophages have a central role in innate immunity, we examined the responses of Sluggish animals and macrophages to bacterial infection. Adherent peritoneal macrophages were exposed to L. monocytogenes or live GBS, and at 8 hour post-infection, culture supernatants were collected and subjected to bioassay to determine TNFα and type I IFN production. TNFα production was severely impaired, but type I IFN production was not significantly different, in response to L. monocytogenes infection in Sluggish macrophages, while macrophages isolated from Myd88poc/poc control mice displayed normal TNFα production due to the partial preservation of TLR2/TLR6 signaling (Figure 3A) (28). Despite reductions in TNF and IL-6 production relative to wild-type animals, most homozygous Sluggish mice resisted L. monocytogenes infection. By contrast, Myd88poc/poc animals were highly susceptible to infection by L. monocytogenes and displayed a severe reduction in all serum cytokine levels including IL-12p70, which was unaffected in Sluggish mice (Figure 3B,C). After infection with 1×106 CFU/ml GBS, Sluggish homozygous macrophages displayed severely impaired production of both TNFα and type I IFN (Figure 4A), and most homozygous Sluggish mice died within two days with elevated levels of inflammatory cytokines due to an increased bacterial load in spleen, kidney and blood (Figure 4B-D; data not shown). Surviving Sluggish animals exhibited decreased bacterial loads, which correlated with lower levels of proinflammatory cytokines relative to the susceptible mice.
Figure 3.
Sluggish mice are mostly resistant to Listeria monocytogenes infection. (A) Adherent peritoneal macrophages isolated from the indicated genotypes were exposed to 106 CFU/ml of L. monocytogenes. 8 hours post-infection, culture supernatants were subjected to bioassay to determine TNF and type I IFN production. n=5 per group. (B) Homozygous Sluggish mice were not susceptible to L. monocytogenes infection. Mice were injected intravenously with 105 CFU of L. monocytogenes. The survival of these mice was monitored during a 7 day period. (C) Blood was collected 24 hours after infection, Serum cytokines were measured by ELISA or bioassay. C57BL/6J (black triangles), n=10; Sluggish−/− (gray squares), n=7; Myd88poc/poc (black diamonds), n=5. Data represent mean ± SEM. * = P < 0.05, ** P < 0.01, *** P < 0.005.
Figure 4.
Sluggish mice are specifically susceptible to GBS infection. (A) Adherent peritoneal macrophages isolated from the indicated genotypes were exposed to 1×106 CFU/ml of live GBS strain COH1. 8 hours post-infection, culture supernatants were collected and subjected to TNF bioassay and IFN bioassay. n=5 per group. Data represent mean ± SEM. (B) Survival of adult mice of the indicated genotypes after intraperitoneal challenge with 1×106 CFU of live type Ib GBS strain H36B. C57BL/6J (black triangles), n=10; Sluggish−/− (gray squares), n=15. (C-D) In a similar infection experiment, 20 hours after infection, colony counts were measured in spleen (C), while serum cytokines (D) were determined by ELISA. C57BL/6J, n=7; Sluggish−/−, n=8. Data represent mean ± SEM. * = P < 0.05, ** P < 0.01, *** P < 0.005.
Along with TNFα, type I IFN signaling is critical for host protection against GBS (9, 23). As Tpl2 appears to mediate TLR7 and 9-induced type I IFN production in macrophages, we examined the production of type I IFNs and TNFα in macrophages with mutations in Tlr7 and Tlr9 in response to GBS exposure, as well as macrophages deficient in critical components of the TLR signaling pathway. Macrophages with mutations in Myd88 and Irak4 have profoundly impaired levels of type I IFN and TNFα in response to GBS infection, similar to Sluggish macrophages. In response to GBS exposure, macrophages with mutations in Tlr7 and Tlr9 display intermediate levels of type I IFN, and relatively normal TNFα production (Supplemental Figure 3). These data suggest that multiple TLR signals contribute to GBS sensing and production of important immune factors in response to this pathogen.
Sluggish is a mutation in Tpl2/Map3k8
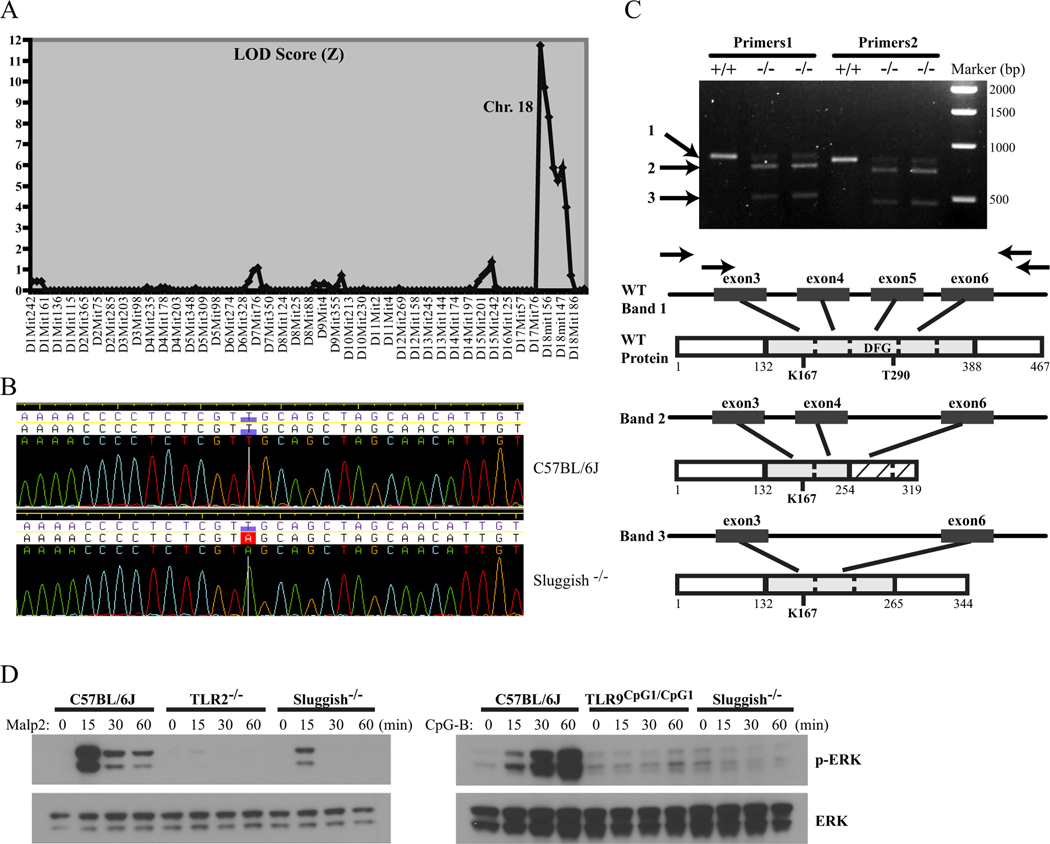
The Sluggish phenotype was mapped by outcrossing the mutant stock to C3H/HeN and backcrossing the F1 hybrids to the mutant stock. F2 mice were scored on the TNFα response of peritoneal macrophages to Malp2 and PGN. On 39 meioses, the mutation was mapped to chromosome 18 (peak LOD score of 11.74; Figure 6A) to an 11.9 Mb critical region upstream of marker D18Mit110 (Figure 6A). The critical region contained a total of 54 annotated genes. Among these was the gene encoding MAP3K8/Tpl2/Cot, a protein kinase previously implicated in TLR signaling (16, 31). Knockout Tpl2 mice and macrophages display a defect in TNFα secretion and pre-TNFα synthesis in response to LPS (16, 32). Moreover, these animals exhibit an impairment of IL-6, but an increase in IL-12, in response to various stimuli (16, 33, 34). These phenotypes paralleled the defects observed in Sluggish mice. Thus, the Map3k8 gene was an excellent candidate for sequencing in Sluggish animals.
The Map3k8 locus was sequenced and found to be modified by a T to A transversion in the acceptor splice site of intron 4 at position 13346 of the Map3k8 gene according to the Genbank genomic region NC_000084 (Figure 5B). Three different splice forms caused by the mutation have been observed, one of which is the wild-type transcript. The splice variants were sequenced, and are shown in Figure 5C. The first splice variant results in skipping the 107-nucleotide exon 5 (out of 8 total exons), causing a frame-shift and insertion of 64 aberrant amino acids (corresponding to positions 255–319) before a premature stop codon, while the second splice variant results in skipping of exons 4 and 5, resulting in an in-frame splice to exon 6 and an internal deletion of 123 amino acids. These splice variants remove exons encoding critical portions of the Tpl2 kinase domain including the activation loop containing the critical threonine 290 residue needed for Tpl2 activation (Figure 5C) (35–37).
Figure 5.
Cloning and sequencing of the Sluggish mutation. (A) Based on 39 meioses, the mutation was mapped to Chromosome 18 with a peak LOD score of 11.74 to an 11.9 Mb critical region upstream of Marker D18Mit110. (B) DNA sequence of the gene coding for MAP3K8/Tpl2 in a C57BL/6J wild-type control (upper panel) and a Sluggish homozygous mouse (lower panel). The mutation is a T to A transversion in the acceptor splice site of intron 4 at position 13346 of the Map3k8 gene. (C) Effect of the Sluggish mutation at the mRNA level. Three different splice forms of Map3k8 have been observed in Sluggish homozygotes; wild-type, one missing exons 4 and 5, and one missing exon 5. Upper panel depicts PCR using two different primer sets. PCR bands are labeled 1, 2, and 3. +/+ and −/− indicates C57BL/6J wild-type controls and Sluggish homozygous samples, respectively. Arrows in the bottom panel indicate the location of primer sets. Also depicted are the wild-type protein and the protein products arising from the aberrantly spliced transcripts. The kinase domain is shaded in gray and subdivided according to exon organization. The conserved ATP-binding lysine (K167), activation loop phosphorylated residue (T290), and conserved activation loop motif DFG are indicated. Frame-shifted amino acids caused by the first aberrantly spliced transcript are depicted by cross-hatching. (D) ERK phosphorylation is impaired in Map3k8Sluggish/Sluggish macrophages. Peritoneal macrophages from the indicated genotypes were stimulated with 100 ng/ml of Malp2 (left panel) or CpG-B (right panel) for the indicated time. Cell lysates were subjected to Western blot with the indicated antibodies.
ERK phosphorylation in response to TLR signaling is absent in homozygous Sluggish macrophages
In the innate immune system, Tpl2 primarily functions to regulate the MEK/ERK pathway downstream of most TLR signaling pathways (16, 18, 33). In order to determine whether the Sluggish mutation abolishes Tpl2 function, we analyzed ERK activation in Map3k8Sluggish/Sluggish (or Map3k8m1Btlr/m1Btlr) macrophages stimulated with the TLR2/6 ligand Malp2 or the TLR9 ligand CpG-B. As shown in Figure 5D, phosphorylation of ERK was observed in C57BL/6J wild-type macrophages in response to Malp2 and CpG-B. However, in Map3k8Sluggish/Sluggish macrophages, both Malp2 and CpG-B-induced phosphorylation of ERK are strongly reduced. These data demonstrate that the Sluggish mutation strongly affects the role of Tpl2 in TLR signaling.
Discussion.
Here, we report the discovery of a splice site mutation in the Map3k8 gene resulting in production of aberrant transcripts that encode proteins lacking critical regions of the Tpl2 kinase domain necessary both for post-translational Tpl2 regulation as well as kinase activity (35, 38). Since low levels of wild-type transcript are produced in Map3k8Sluggish/Sluggish animals, it is possible that small quantities of functional Tpl2 exist in these mice resulting in a hypomorphic phenotype. Alternatively, the aberrant transcripts resulting from the Sluggish mutation may produce proteins with dominant-negative functions. These possibilities may explain the TLR signaling defects also observed in Map3k8Sluggish/+ mice. However, the Map3k8Sluggish/Sluggish mutant shows a strong phenotypic similarity to Map3k8 knockout animals by displaying a lack of ERK activation and the same abnormal cytokine production in response to TLR signaling [reviewed by (31)]. In addition, we observe that Map3k8Sluggish/Sluggish macrophages are able to produce near normal levels of pre-TNFα, but fail to express TNFα on the cell surface. This observation recapitulates the discovery in Tpl2-knockout macrophages that Tpl2-regulated ERK activity is necessary for LPS-induced expression of pre-TNFα on the cell surface, and that ERK activates the TNFα convertase (TACE) needed for TNFα secretion (32). Taken together, these data suggest that Tpl2 function is severely compromised in Map3k8Sluggish/Sluggish mice.
Using the Sluggish allele, we demonstrate that Tpl2 plays a critical role in MyD88-dependent type I IFN production in macrophages. Unlike the general defect seen with TNFα production in response to TLR signaling in Tpl2-deficient mice, Tpl2 appears to be required specifically for type I IFN production from IFNγ-primed macrophages in response to CpG-B and resiquimod, while type I IFN production in response to TLR3 and TLR4 remains intact. Although TLR7 and TLR9-induced type I IFN was affected in Map3k8Sluggish/Sluggish macrophages, we demonstrate that Tpl2 is not necessary for type I IFN production in response to CpG-A in vivo, suggesting that Tpl2 is dispensable for type I IFN production in pDCs (27). As pDCs are important responders and producers of type I IFN in response to some (but not all) viral infections, this may explain why Tpl2-deficient mice and macrophages remain resistant and able to produce appropriate amounts of type I IFN in response to all viral infections tested [(16); this paper]. Concerning the MCMV resistance found in these animals, we have also tested Map3k8Sluggish/Sluggish NK and T cytolytic function, as well as B cell responses, and have found these activities to be completely normal in Map3k8Sluggish/Sluggish mice (data not shown). As NK cell function is important during the innate immune response to MCMV, while T and B cells are required during the adaptive immune response to this virus (1), we suggest that the apparently normal function of these cell types contributes to the MCMV resistance of Map3k8Sluggish/Sluggish animals.
It was recently demonstrated that Tpl2 negatively regulates IFNβ production in bone marrow-derived macrophages (BMDMs) and myeloid dendritic cells in response to both CpG and LPS, while appearing to be necessary for type I IFN production in pDCs in response to CpG (39). These findings apparently contradict our own results that Tpl2 plays a significant role in type I IFN production in response to CpG, but is not important for type I IFN production in response to LPS in macrophages or CpG in vivo. We point out that the type I IFN responses examined in this study occur at a much later time point (24 hours) than the primary responses we have looked at in our own experiments. These discrepancies may also be partially explained by the differences that exist between BMDM and peritoneal macrophages. Previous data suggest that Tpl2-deficient BMDMs and peritoneal macrophages may differ in their response to TLR stimulation (18,33).
Type I IFN signaling is critical for host defense against a variety of pathogenic bacteria, including GBS, pneumococci, and Escherichia coli (9). Recently, it was reported that recognition of GBS RNA by TLR7 in conventional DCs activates a MyD88-dependent pathway leading to type I IFN production that is essential for host response to this disease (23). As we demonstrate that Tpl2 is necessary to produce type I IFNs in macrophages in response to GBS, it is possible that that the impairment of type I IFN responses in Tpl2-deficient macrophages contributes to the high susceptibility of Map3k8Sluggish/Sluggish mice to this pathogen. Conversely, type I IFN signaling has been shown to decrease host resistance against L. monocytogenes (10–12), making it unlikely that the type I IFN defect we observe in Map3k8Sluggish/Sluggish macrophages would negatively impact the host response to this disease. Indeed, a type I IFN defect may contribute to resistance against this pathogen. However, we do not observe any significant differences in the type I IFN response to L. monocytogenes in Sluggish macrophages or animals.
In addition to a type I IFN defect, we demonstrate a profound impairment of TNFα production by Tpl2-deficient mice in response to TLR signaling and suggest that this may be the major cause for GBS susceptibility. TNFα production in response to GBS is important in controlling the infection. Animals with deficiencies in TLR2 or MyD88, or treated with anti-TNF antibodies, were protected against GBS-induced septic shock under certain conditions, but were unable to control the spread of infection (7, 8). In addition, GBS stimulation in human monocytes results in the phosphorylation and activation of various MAPKs, including ERK1/2, which is regulated by Tpl2 kinase activity (16, 18). Inhibition of MAPK activation resulted in reduced TNFα production (40). Finally, TLR9 signaling in macrophages has also been shown to be important for TNFα production in response to GBS (41). This same study demonstrated normal IFN production in TLR9-deficient macrophages in response to GBS infection. Although we were unable to demonstrate that either TLR7 or TLR9 by themselves were essential for TNFα production in response to GBS (Supplemental Figure 1), our data clearly show a lack of TNFα production in Tpl2-deficient macrophages, either in response to TLR signals or to GBS. It is likely that Tpl2 functions downstream of multiple TLRs, including TLR2, TLR7 and TLR9, to induce TNFα production in response to GBS. Thus, lack of appropriate TLR signaling in Tpl2-deficient mice leads to a deficient early response to GBS infection and causes susceptibility. The defect in the primary immune response of Map3k8Sluggish/Sluggish mice to GBS infection results in unrestrained bacterial proliferation, the stimulation of secondary immune responses, production of inflammatory cytokines, and death.
We also demonstrate a reduction in TNFα production in both Map3k8Sluggish/Sluggish macrophages and animals in response to infection with Listeria monocytogenes. The absence of elevated cytokine levels in Map3k8Sluggish/Sluggish mice infected with L. monocytogenes may be due to the resistance of these animals to this pathogen and the failure of secondary immune responses to hide the primary TLR sensing defect. TNFα is an important component of the immune response to this pathogen (5, 6), but Map3k8Sluggish/Sluggish mice appear resistant to this disease. Previous work suggests that only a small amount of TNFα may be sufficient to protect animals against this pathogen, although complete inhibition leads to susceptibility (42). Thus, the resistance of Map3k8Sluggish/Sluggish animals to L. monocytogenes infection may be explained by the presence of residual TNFα levels. We also note that the Myd88poc mutation, which renders mice highly susceptible to L. monocytogenes, allows normal TNFα production in macrophages in responses to this bacterium, suggesting that factors other than TNFα are important in the immune response to L. monocytogenes.
In addition to impairment of type I IFN and TNFα production, Tpl2-deficient mice have significantly reduced levels of IFNγ (type II IFN) in response to CpG stimulation. TNFα and IFNγ play interconnected roles in the host response to GBS. TNFα released from microbe stimulated macrophages has been shown to be important for IFNγ production, and TNFα and IFNγ together activate macrophage killing of intracellular bacteria (41, 43). Moreover, Tpl2 knockout mice exhibit decreased IFNγ production and consequent susceptibility to Toxoplasma gondii infection (34). Although IFNγ levels are markedly increased in GBS-infected animals with unrestrained bacterial infection, it is possible that impaired IFNγ production from specific immune cell types relatively early in the response to bacterial infection may contribute to the GBS-susceptibility of Tpl2-deficient mice.
In summary, we have discovered a novel allele of Map3k8, and have demonstrated a significant role for this kinase in the production of type I IFNs in response to specific stimuli. Furthermore, our data suggest that Tpl2 plays an essential role in the production of multiple factors that are important for the control of GBS infection.
Supplementary Material
Acknowledgements
We would like to thank Xin Du, Elizabeth Hanley, and Christine Domingo for valuable assistance with mouse stocks. We thank Dr. Giuseppe Teti in University of Messini in Italy for providing us with two strains of GBS (COH1 and H36B). We thank Dr. Tamsin Sheen Pointon of San Diego State University for technical assistance in GBS infection.
This work was supported by BAA contract, HHSN272200700038C and NIH grant, 5P01AI070167. PK was supportd by EMBO (European Molecular Biology Organization) and the Swiss National Science Foundation. KB was supported by a fellowship from the German Research Foundation (DFG). ALB was supported by The Irvington Institute Fellowship Program of the Cancer Research Institute.
Footnotes
Abbreviations used in this paper include: BMDM, bone marrow-derived macrophages; DC, dendritic cells; ENU, N-ethyl-N-nitrosourea; GBS, group B streptococcus; Malp2, macrophage-activating lipopeptide-2; MAP3K, mitogen-activated protein kinase kinase kinase; MCMV, mouse cytomegalovirus; MyD88, myeloid differentiation 88; PGN, peptidoglycan; pDC, plasmacytoid dendritic cells; ssRNA, single-stranded RNA; Tpl2, tumor progression locus 2.
References
- 1.Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, and Hoebe K. 2006. Genetic analysis of host resistance: Toll-Like receptor signaling and immunity at large. Annu. Rev. Immunol 24: 353–389. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T. and Akira S. 2007. TLR signaling. Semin. Immunol 19: 24–32. [DOI] [PubMed] [Google Scholar]
- 3.Kaisho T. 2008. Type I interferon production by nucleic acid-stimulated dendritic cells. Front. Biosci 13: 6034–6042. [DOI] [PubMed] [Google Scholar]
- 4.Decker T, Muller M, and Stockinger S. 2005. The yin and yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol 5: 675–687. [DOI] [PubMed] [Google Scholar]
- 5.Pasparakis M, Alexopoulou L, Episkopou V, and Kollias G. 1996. Immune and inflammatory responses in TNFα-deficient mice: A critical requirement for TNFα in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J. Exp. Med 184: 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeffer K, Matsuyama T, K∩┐╜ndig TM, Wakeham A, Kishihara K, Shahinian A, Wiegmann K, Ohashi PS, Kr∩┐╜nke M, and Mak TW 1993. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell 73: 457–467. [DOI] [PubMed] [Google Scholar]
- 7.Teti G, Mancuso G, and Tomasello F. 1993. Cytokine appearance and effects of anti-tumor necrosis factor alpha antibodies in a neonatal rat model of group B streptococcal infection. Infect. Immun 61: 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mancuso G, Midiri A, Beninati C, Biondo C, Galbo R, Akira S, Henneke P, Golenbock D, and Teti G. 2004. Dual role of TLR2 and myeloid differentiation factor 88 in a mouse model of invasive group B streptococcal disease. J. Immunol 172: 6324–6329. [DOI] [PubMed] [Google Scholar]
- 9.Mancuso G, Midiri A, Biondo C, Beninati C, Zummo S, Galbo R, Tomasello F, Gambuzza M, Macri G, Ruggeri A, Leanderson T, and Teti G. 2007. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J. Immunol 178: 3126–3133. [DOI] [PubMed] [Google Scholar]
- 10.Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, and Portnoy DA 2004. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med 200: 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, Miller JF, and Cheng G. 2004. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J. Exp. Med 200: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrero JA, Calderon B, and Unanue ER 2004. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J. Exp. Med 200: 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salmeron A, Ahmad TB, Carlile GW, Pappin D, Narsimhan RP, and Ley SC 1996. Activation of MEK-1 and SEK-1 by Tpl-2 proto-oncoprotein, a novel MAP kinase kinase kinase. EMBO J. 15: 817–826. [PMC free article] [PubMed] [Google Scholar]
- 14.Tsatsanis C, Patriotis C, and Tsichlis PN 1998. Tpl-2 induces IL-2 expression in T-cell lines by triggering multiple signaling pathways that activate NFAT and NF-kappaB. Oncogene 17: 2609–2618. [DOI] [PubMed] [Google Scholar]
- 15.Chiariello M, Marinissen MJ, and Gutkind JS 2000. Multiple mitogen-activated protein kinase signaling pathways connect the cot oncoprotein to the c-jun promoter and to cellular transformation. Mol. Cell. Biol 20: 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumitru CD, Ceci JD, Tsatsanis C, Kontoyiannis D, Stamatakis K, Lin JH, Patriotis C, Jenkins NA, Copeland NG, Kollias G, and Tsichlis PN 2000. TNF-alpha induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 103: 1071–1083. [DOI] [PubMed] [Google Scholar]
- 17.Waterfield MR, Zhang M, Norman LP, and Sun SC 2003. NF-kappaB1/p105 regulates lipopolysaccharide-stimulated MAP kinase signaling by governing the stability and function of the Tpl2 kinase. Mol. Cell 11: 685–694. [DOI] [PubMed] [Google Scholar]
- 18.Banerjee A, Gugasyan R, McMahon M, and Gerondakis S. 2006. Diverse Toll-like receptors utilize Tpl2 to activate extracellular signal-regulated kinase (ERK) in hemopoietic cells. Proc. Natl. Acad. Sci. U. S. A 103: 3274–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO, Goode J, Lin P, Mann N, Mudd S, Crozat K, Sovath S, Han J, and Beutler B. 2003. Identification of Lps2 as a key transducer of MyD88-independent TIR signaling. Nature 424: 743–748. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Z, Georgel P, Du X, Shamel L, Sovath S, Mudd S, Huber M, Kalis C, Keck S, Galanos C, Freudenberg M, and Beutler B. 2005. CD14 is required for MyD88-independent LPS signaling. Nat. Immunol 6: 565–570. [DOI] [PubMed] [Google Scholar]
- 21.Tabeta K, Georgel P, Janssen E, Du X, Hoebe K, Crozat K, Mudd S, Shamel L, Sovath S, Goode J, Alexopoulou L, Flavell RA, and Beutler B. 2004. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. U. S. A 101: 3516–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutschmann S, Hoebe K, Zalevsky J, Du X, Mann N, Dahiyat BI, Steed P, and Beutler B. 2006. PanR1, a dominant negative missense allele of the gene encoding TNF-alpha (Tnf), does not impair lymphoid development. J. Immunol 176: 7525–7532. [DOI] [PubMed] [Google Scholar]
- 23.Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, Akira S, Teti G, and Beninati C. 2009. Bacterial recognition by toll-like receptor 7 in the lysosomes of conventional dendritic cells. Nat. Immunol. in press: [DOI] [PubMed] [Google Scholar]
- 24.Kawai T, Takeuchi O, Fujita T, Inoue J, Muhlradt PF, Sato S, Hoshino K, and Akira S. 2001. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J. Immunol 167: 5887–5894. [DOI] [PubMed] [Google Scholar]
- 25.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, and Hiscott J. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300: 1148–1151. [DOI] [PubMed] [Google Scholar]
- 26.Schroder K, Spille M, Pilz A, Lattin J, Bode KA, Irvine KM, Burrows AD, Ravasi T, Weighardt H, Stacey KJ, Decker T, Hume DA, Dalpke AH, and Sweet MJ 2007. Differential effects of CpG DNA on IFN-beta induction and STAT1 activation in murine macrophages versus dendritic cells: alternatively activated STAT1 negatively regulates TLR signaling in macrophages. J. Immunol 179: 3495–3503. [DOI] [PubMed] [Google Scholar]
- 27.Schmitz F, Heit A, Guggemoos S, Krug A, Mages J, Schiemann M, Adler H, Drexler I, Haas T, Lang R, and Wagner H. 2007. Interferon-regulatory-factor 1 controls Toll-like receptor 9-mediated IFN-beta production in myeloid dendritic cells. Eur. J. Immunol 37: 315–327. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Z, Georgel P, Li C, Choe J, Crozat K, Rutschmann S, Du X, Bigby T, Mudd S, Sovath S, Wilson IA, Olson A, and Beutler B. 2006. Details of Toll-like receptor:adapter interaction revealed by germ-line mutagenesis. Proc. Natl. Acad. Sci. U. S. A 103: 10961–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asselin-Paturel C, Brizard G, Pin JJ, Briere F, and Trinchieri G. 2003. Mouse strain differences in plasmacytoid dendritic cell frequency and function revealed by a novel monoclonal antibody. J. Immunol 171: 6466–6477. [DOI] [PubMed] [Google Scholar]
- 30.Crozat K, Georgel P, Rutschmann S, Mann N, Du X, Hoebe K, and Beutler B. 2006. Analysis of the MCMV resistome by ENU mutagenesis. Mamm. Genome 17: 398–406. [DOI] [PubMed] [Google Scholar]
- 31.Banerjee A. and Gerondakis S. 2007. Coordinating TLR-activated signaling pathways in cells of the immune system. Immunol. Cell Biol 85: 420–424. [DOI] [PubMed] [Google Scholar]
- 32.Rousseau S, Papoutsopoulou M, Symons A, Cook D, Lucocq JM, Prescott AR, O’Garra A, Ley SC, and Cohen P. 2008. TPL2-mediated activation of ERK1 and ERK2 regulates the processing of pre-TNF alpha in LPS-stimulated macrophages. J. Cell. Sci 121: 149–154. [DOI] [PubMed] [Google Scholar]
- 33.Sugimoto K, Ohata M, Miyoshi J, Ishizaki H, Tsuboi N, Masuda A, Yoshikai Y, Takamoto M, Sugane K, Matsuo S, Shimada Y, and Matsuguchi T. 2004. A serine/threonine kinase, Cot/Tpl2, modulates bacterial DNA-induced IL-12 production and Th cell differentiation. J. Clin. Invest 114: 857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watford WT, Hissong BD, Durant LR, Yamane H, Muul LM, Kanno Y, Tato CM, Ramos HL, Berger AE, Mielke L, Pesu M, Solomon B, Frucht DM, Paul WE, Sher A, Jankovic D, Tsichlis PN, and O’Shea JJ 2008. Tpl2 kinase regulates T cell interferon-gamma production and host resistance to Toxoplasma gondii. J. Exp. Med 205: 2803–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho J, Melnick M, Solidakis GP, and Tsichlis PN 2005. Tpl2 (tumor progression locus 2) phosphorylation at Thr290 is induced by lipopolysaccharide via an Ikappa-B Kinase-beta-dependent pathway and is required for Tpl2 activation by external signals. J. Biol. Chem 280: 20442–20448. [DOI] [PubMed] [Google Scholar]
- 36.Luciano BS, Hsu S, Channavajhala PL, Lin LL, and Cuozzo JW 2004. Phosphorylation of threonine 290 in the activation loop of Tpl2/Cot is necessary but not sufficient for kinase activity. J. Biol. Chem 279: 52117–52123. [DOI] [PubMed] [Google Scholar]
- 37.Kornev AP, Haste NM, Taylor SS, and Eyck LF 2006. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. U. S. A 103: 17783–17788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho J. and Tsichlis PN 2005. Phosphorylation at Thr-290 regulates Tpl2 binding to NF-kappaB1/p105 and Tpl2 activation and degradation by lipopolysaccharide. Proc. Natl. Acad. Sci. U. S. A 102: 2350–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaiser F, Cook D, Papoutsopoulou S, Rajsbaum R, Wu X, Yang HT, Grant S, Ricciardi-Castagnoli P, Tsichlis PN, Ley SC, and O’Garra A. 2009. TPL-2 negatively regulates interferon-{beta} production in macrophages and myeloid dendritic cells. J. Exp. Med 206: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mancuso G, Midiri A, Beninati C, Piraino G, Valenti A, Nicocia G, Teti D, Cook J, and Teti G. 2002. Mitogen-activated protein kinases and NF-kappa B are involved in TNF-alpha responses to group B streptococci. J. Immunol 169: 1401–1409. [DOI] [PubMed] [Google Scholar]
- 41.Talati AJ, Kim HJ, Kim YI, Yi AK, and English BK 2008. Role of bacterial DNA in macrophage activation by group B streptococci. Microbes Infect. 10: 1106–1113. [DOI] [PubMed] [Google Scholar]
- 42.Kolls J, Peppel K, Silva M, and Beutler B. 1994. Prolonged and effective blockade of tumor necrosis factor activity through adenovirus-mediated gene transfer. Proc. Natl. Acad. Sci., USA 91: 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodrum KJ, Dierksheide J, and Yoder BJ 1995. Tumor necrosis factor alpha acts as an autocrine second signal with gamma interferon to induce nitric oxide in group B streptococcus-treated macrophages. Infect. Immun 63: 3715–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.