Abstract
The tumor suppressor p16INK4a inhibits cyclin-dependent kinases 4 and 6. This activates the retinoblastoma protein (pRB) and, through incompletely understood events, arrests the cell division cycle. To permit biochemical analysis of the arrest, we generated U2-OS osteogenic sarcoma cell clones in which p16 transcription could be induced. In these clones, binding of p16 to cdk4 and cdk6 abrogated binding of cyclin D1, p27KIP1, and p21WAF1/CIP1. Concomitantly, the total cellular level of p21 increased severalfold via a posttranscriptional mechanism. Most cyclin E-cdk2 complexes associated with p21 and became inactive, expression of cyclin A was curtailed, and DNA synthesis was strongly inhibited. Induction of p21 alone, in a sibling clone, to the level observed during p16 induction substantially reproduced these effects. Overexpression of either cyclin E or A prevented p16 from mediating arrest. We then extended these studies to HCT 116 colorectal carcinoma cells and a p21-null clone derived by homologous recombination. In the parental cells, p16 expression also augmented total cellular and cdk2-bound p21. Moreover, p16 strongly inhibited DNA synthesis in the parental cells but not in the p21-null derivative. These findings indicate that p21-mediated inhibition of cdk2 contributes to the cell cycle arrest imposed by p16 and is a potential point of cooperation between the p16/pRB and p14ARF/p53 tumor suppressor pathways.
p16INK4a/CDKN2A/MTS1 is a specific inhibitor of the closely related cyclin-dependent kinases (cdk’s) 4 and 6 (96, 109). Mutations at the INK4a locus segregate with tumor formation in familial melanoma pedigrees, and the locus is within a small region of chromosome 9 that is deleted in multiple human tumor cell lines (45, 46, 73). The locus encodes a second protein, p14ARF (ARF), from an alternative reading frame (19, 63, 81, 100). ARF is unrelated in sequence to p16 but, surprisingly, can also inhibit cell cycle progression (56, 81). Although p16 is preferentially inactivated by point mutation or promoter methylation in a number of tumors (35, 80), recent data suggest that ARF is a tumor suppressor protein in its own right, potentiating effects of p53 (14, 47, 78, 111). Thus, the INK4a locus regulates the two most pervasive tumor suppressor pathways.
p16 is a potent mediator of G1 cell cycle arrest in tissue culture cells (52, 58, 67, 95). The only known effector of the arrest is the retinoblastoma protein (pRB), consistent with evidence that a major role of cdk4/6 is to phosphorylate and inactivate pRB (98). Prominent among pRB’s properties is that it binds E2F transcription factors and represses the activity of promoters with the corresponding response elements (20, 39, 106). These elements are found in several genes required for cell replication, including the cdk2 partners cyclins E and A, cdk1 (cdc2), ribonucleotide reductase, and DNA polymerase alpha (15). Overexpression of a dominant-negative form of DP-1, the major heterodimeric partner of E2Fs, can inhibit G1 progression in mammalian cell lines (107). However, substantial cell replication occurs in Drosophila melanogaster embryos with mutations in E2F or DP-1 after apparent exhaustion of the maternally derived proteins (88). Hence, it is unclear whether pRB-mediated repression of E2F-responsive genes is sufficient to account for the ability of pRB and p16 to rapidly impose a cell cycle arrest.
There is increasing evidence that inhibition of cdk2 complexes may be sufficient and even necessary for effective cell cycle inhibition in many settings. Inhibitors of the Cdk-interacting protein (CIP)/Clk-inhibitin, protein (KIP) class, although capable of binding many cdk’s, appear to primarily act through inhibition of cdk2 (8, 11, 12, 60, 99). Efficient G1 arrest in response to DNA damage is brought about by the ARF/p53 pathway and requires induction of p21WAF1/CIP1/sdi1/CDKN1 transcription (10, 16, 22). Inhibition of the cell cycle by glucocorticoids also appears to involve p21 induction and can occur in pRB-deficient cells (87). p27KIP1 contributes to cell cycle arrests mediated by transforming growth factor β and contact inhibition (76). Null alleles of p27 in mice and the structurally related inhibitor dacapo in Drosophila melanogaster enhance cell cycling in diverse tissues (17, 27, 50, 54, 72).
Overexpression studies suggest that dysregulation of cyclin E can bypass normal G1 controls. Forced expression of cyclin E can induce DNA synthesis in most postmitotic cells in the Drosophila embryo (51). In mammalian cells, ectopic expression of cyclin E-cdk2 can rescue an arrest mediated by repression of c-Myc-responsive genes (7). Overexpression of cyclin E has recently been found to rescue an arrest mediated by dominant-negative DP-1, a phosphorylation-deficient mutant of pRB, or p16 in rodent fibroblasts (2, 57). This occurs without reversing pRB hyperphosphorylation or its association with E2F transcription factors. Although overexpression studies must be interpreted cautiously, p16-mediated activation of pRB and sequestration of E2F appear to be insufficient to maintain a cell cycle arrest in these settings if cyclin E-cdk2 is ectopically hyperactivated. On the other hand, these studies have not addressed whether cell cycle inhibition by p16 normally involves and/or requires inhibition of endogenous cdk2. Studies of p15INK4b, a cdk inhibitor structurally related to p16, indicate that its binding to cdk4 and cdk6 results in redistribution of p27 and p21 to cdk2, potentially contributing to cell cycle arrest (83).
To further explore the mechanism of action of p16, we performed a biochemical analysis of events that occur during p16-mediated cell cycle inhibition. We then manipulated these events through targeted genetic experiments. We generated clones of the human osteogenic sarcoma cell line U2-OS which could be induced to express p16. U2-OS is the cell line in which the p16’s function has been most extensively studied (52, 58, 59, 67). p16 is not expressed endogenously in these cells, and its exogenous expression strongly inhibits DNA synthesis. This inhibition appears to be specific, because it is abrogated by coexpression of cdk4 or cdk6 or by introducing tumor-associated mutations into p16 (52, 58, 67).
Using such clones, we found that cell cycle inhibition by p16 is associated with a posttranscriptional induction of p21 and a strong inhibition of cyclin E-cdk2 kinase activity. In a sibling clone, induction of p21 alone to the level induced by p16 was sufficient to significantly inhibit cyclin E-cdk2 activity and DNA synthesis. We then extended our studies to the HCT 116 human colorectal carcinoma cell line and a p21-null clone derived by two rounds of homologous recombination (104). p16 expression in the parental HCT 116 cells also caused an increase in total cellular and cdk2-bound p21. Moreover, p16 inhibited DNA synthesis in these cells much more effectively than in the p21-null derivative. These results indicate that p21-mediated inhibition of cdk2 is a key means by which p16 achieves cell cycle arrest and a common element of the ARF/p53 and p16/pRB response pathways.
MATERIALS AND METHODS
Cell culture and generation of inducible cell lines.
Stable p16-inducible clones were generated by transfection. A U2-OS osteogenic sarcoma cell clone (U24) containing an integrated pUHD15 vector (31), which encodes a tetracycline (TET)-repressible transcriptional activator, was kindly provided by Liang Zhu (Albert Einstein College of Medicine) (43). The p16 cDNA was cloned into the pUHD10-3 vector (31) and cotransfected into U24 with a puromycin resistance-encoding plasmid. Puromycin-resistant colonies were maintained in the presence of TET. There were no gross differences in the numbers or sizes of puromycin-resistant colonies obtained with the p16 construct and those obtained with the empty target vector. This suggests that in the presence of TET, the level of expression of p16 was sufficiently low in most transfected clones that their growth was not inhibited; it also indicates that there was little selection pressure for lesions in the p16 response pathway.
We randomly picked 34 well-separated clones obtained by transfection of approximately 5 × 105 cells with the p16 construct. We also picked two vector-transfected control clones and pooled the rest as OSvec.pool. p16-inducible clones were identified by immunofluorescence, using the JC6 monoclonal antibody provided by Jim Koh (University of Vermont Cancer Center) (24). Four clones showed levels of staining distinctly above background in the absence of TET and were amplified for further analysis. p21-inducible cells were generated in a similar fashion, except that an anti-p21 antibody (WAF1; Oncogene Research) was used to screen for expression.
Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) containing 10% fetal bovine serum (Life Technologies), penicillin (100,000 U/liter), and streptomycin sulfate (100,000 U/liter). TET was added to the culture medium at 1 to 2 μg/ml to suppress expression of the inducible proteins. To allow induction, cells were trypsinized, washed with DMEM, pelleted at 300 × g for 5 min, and plated in DMEM without or with appropriate concentrations of TET. Cells were generally maintained at no more than 50 to 70% confluence. At greater cell densities, U2-OS cells display moderate contact inhibition of growth, associated with increased p21 levels (70a).
Parental and p21−/− HCT 116 cells were obtained from Bert Vogelstein (Howard Hughes Medical Institute, The Johns Hopkins Oncology Center) and maintained in McCoy’s 5A medium supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin. The p21−/− cells were periodically treated with neomycin and puromycin to prevent outgrowth of potential residual parental cells.
Cell cycle synchronization.
Cells were synchronized with lovastatin (Merck Research Laboratories, West Point, Pa.) by the following method (33, 49, 108). Twenty-four to 36 h after seeding of cells, fresh medium containing 40 μM lovastatin was added, and the cells were then grown for 40 h with or without TET. At the end of 40 h (0 h time point; see Results), the cells were released from lovastatin-induced G1 arrest by addition of fresh medium containing 4 mM mevalonic acid (Sigma). Cells were harvested at various times and processed either for biochemical analysis or for cell cycle position analysis by flow cytometry.
Flow cytometry.
For flow cytometry, cells were harvested by trypsinization, centrifuged at 300 × g for 5 min, and resuspended in 100 μl of phosphate-buffered saline (PBS). The cells were fixed by dropwise addition of 1 ml of 100% ethanol and stained for 30 min at 37°C with a solution consisting of 0.001% propidium iodide and 250 μg of RNase A/ml. Total cellular DNA content was determined on a Becton-Dickinson flow cytometer, using ModFit and SynchWizard software. Small debris (less than about one-quarter of the diameter of a cell) and cell aggregates were gated out of the analysis, and each profile was compiled from approximately 5,000 gated events.
Analysis of CD20-stained cells was performed as described previously (103).
Tritiated-thymidine incorporation.
Cell proliferation was also measured by [3H]thymidine uptake as follows. Cells were pulse-labeled with 2.0 μCi of [3H]thymidine/ml for 45 min prior to being harvested. After two washes with PBS, the cells were trypsinized and retrieved by centrifugation as described above. The cell pellet was resuspended in 0.3 N NaOH for 30 min on ice prior to the addition of 20% cold trichloroacetic acid (TCA). The TCA precipitate was filtered through glass fiber filters (Whatman GF/C) by the use of a benchtop apparatus (Millipore model 1225). The filter was washed with 5 ml of cold 10% TCA and then with 5 ml of 95% ethanol. After the filter was air dried, the radioactivity associated with it was counted by the use of a fluor scintillant (Beckman model LS6000IC counter).
Preparation of cell extracts.
Subconfluent cells on 10-cm2 plates were washed twice with PBS and lysed by addition of 0.8 ml of cold Ela lysis buffer (38) with protease inhibitors (50 mM HEPES [pH 7.0], 250 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA, 0.5 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, and 0.1 mM sodium vanadate). Lysed cells were scraped off the plates, incubated on ice for 15 min, and subjected to centrifugation at 14,000 × g for 15 min at 4°C. The protein contents of the supernatants were assayed by the Bradford method and confirmed by Coomassie blue staining following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Abs.
All antibodies (Abs) were from Santa Cruz Biotechnology unless otherwise indicated. For immunoblotting of p16, we used a mouse monoclonal Ab, JC6, raised against human p16, at a 1:4 dilution (24). Immunoprecipitation (IP) of p16 was performed with a polyclonal rabbit anti-p16 Ab from PharMingen (San Diego, Calif.), raised against full-length human recombinant glutathione S-transferase (GST) fusion protein, at 2 μl/200 μg of total cell extract. For immunoblotting of p21, an affinity-purified mouse monoclonal Ab, WAF1 (Oncogene Research Products, Cambridge, Mass.), raised against full-length recombinant human p21 WAF1, was used at a 1:200 dilution (with overnight incubation). p21 was immunoprecipitated with a rabbit polyclonal Ab (C19) at 0.5 μg/200 μg of total cell extract. Immunoblotting and IP of cdk2 was performed with a rabbit polyclonal Ab (M2) at a 1:500 dilution. A mouse monoclonal anti-cdk2 Ab (D12) was used to confirm the levels of cdk2 immunoprecipitated with the rabbit Ab. D12 was raised against a peptide corresponding to amino acids (aa) 1 to 290 of full-length human cdk2 and was used at a 1:100 dilution. For both IP and immunoblotting of cdk4, C22, a rabbit polyclonal Ab raised against a peptide corresponding to aa 282 to 303 C at the carboxy terminus of mouse cdk4, was used at a 1:100 dilution. The anti-cdk6 Ab used for both IP and immunoblotting (1:500 dilution) was a rabbit polyclonal Ab raised against the carboxy-terminal peptide. A mouse monoclonal immunoglobulin G1 (IgG1) Ab (HD11) raised against recombinant human cyclin D1 was used for both IP and immunoblotting of cyclin D. We used two cyclin E antibodies. A rabbit polyclonal Ab (C19) raised against a peptide corresponding to aa 377 to 395 of human cyclin E was used for IP of cyclin E from extracts (1.5 μg/200 μg of total cell extract). A mouse monoclonal IgG2b Ab raised against recombinant human cyclin E protein (HE12) was used at a 1:100 dilution for immunoblotting of the protein in immunoprecipitates. The 12CA5 Ab, directed against the influenza virus hemagglutinin epitope, was used as a control in some precipitations. For IP as well as immunoblotting of pRB, a rabbit polyclonal Ab (C15) raised against a peptide corresponding to aa 914 to 928 at the carboxy terminus of human pRB (110 kDa) was used at a 1:1,000 dilution. The anti-rabbit and anti-mouse secondary antibodies (Amersham) were used at a 1:5,000 dilution.
Immunoblotting and IP.
Cell extracts were normalized for protein concentration and resolved by SDS-PAGE. The proteins were transferred to polyvinylidene difluoride membranes (Immobilon P or Hybond P; Amersham Life Science) either for 2 h or overnight. The membrane was blocked in 5% nonfat milk in TBS-Tween (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1% Tween 20) for 1 h. The membrane was then washed for 1 h in TBS-Tween with three changes prior to incubation with the primary Ab for 1 h or overnight. After a second 1-h wash in TBS-Tween, the membrane was incubated with the appropriate secondary Ab for 1 h. The membrane was washed in TBS-Tween for 1 h, and the bound antigen was detected by enhanced chemiluminescence (ECL; Amersham). For IP, cell extracts were precleared with a 20-μl packed volume of protein G-agarose beads (Life Technologies) for 1 h at 4°C. The precleared extracts were incubated for 2 h with fresh, washed protein G-agarose and an appropriate Ab in a total volume of 300 μl of ELB. After IP, the beads were washed three times with ELB, heated at 90°C for 3 min with 2× sample buffer (100 mM Tris-HCl [pH 6.8], 200 mM DTT, 4% SDS, 0.2% bromophenol blue, and 20% glycerol), and the supernatant proteins were resolved by SDS-PAGE. Immunoblotting of the precipitated antigen was performed by enhanced chemiluminescence as described above. All images were scanned with a PhosphorImager (Molecular Dynamics Storm).
IP-kinase assays. (i) Histone H1 as substrate.
Precleared cell extracts were subjected to IP with the appropriate antibody (anti-cyclin E or anti-cdk2) as described above. After the immunoprecipitation, the beads were washed three times with ELB and once with 1× kinase assay buffer (25 mM HEPES buffer [pH 7.4], 1 mM phenylmethylsulfonyl fluoride, 0.1 mM sodium vanadate, 10 μg of aprotinin/ml, and 10 μg of leupeptin/ml) and then incubated with a kinase reaction mix (5 μg of histone H1 [Sigma], 0.1 mM ATP, and 0.2 μCi of [γ-32P]ATP in a total volume of 10 μl of 1× kinase buffer) for 30 min at 30°C with shaking. The reaction was stopped by addition of 2× sample buffer to the suspension; the reaction mixture was then heated at 90°C for 3 min, and the products were resolved by SDS-PAGE (10% gels).
(ii) GST-pRB as substrate.
GST-tagged carboxy-terminal fragment of pRB was prepared as described elsewhere (68). The cdk4 kinase assay method followed was as described elsewhere with slight modifications (53). Cell extracts were precleared with a 20-μl packed volume of protein G-agarose for 30 min at 4°C. The precleared extracts were immunoprecipitated with anti-cdk4 antibody for 2 h at 4°C in a total volume of 300 μl of ELB buffer. The immunoprecipitated complexes were washed three times with 300 μl of ELB buffer and once with pRB kinase buffer (50 mM HEPES [pH 7.5] containing 10 mm MgCl2 and 5 mM MnCl2). The beads were resuspended in kinase reaction mix (0.125 mM EGTA, 10 μg of bovine serum albumin/ml, 2.5 μM cold ATP, 5 μCi of [32P]ATP, and 2 μg of GST-pRB/reaction) and incubated for 30 min at 30°C. The reaction was quenched by addition of 2× sample buffer; then the reaction mixture was heated for 3 min at 90°C, and the products were resolved by SDS-PAGE (10% gels). The signal intensities of the bands were quantified by using a PhosphorImager fitted with ImageQuant version 1.1 software.
Northern blotting.
Total RNA was isolated from cultured cells by using Trizol reagent (Life Technologies), and 20 μg of each preparation was subjected to electrophoresis in a 1% agarose gel containing formaldehyde (91). RNA was blotted onto a nylon membrane and detected with 32P-labeled probes generated from the entire 2.1-kb p21WAF1/CIP1 cDNA. As a loading control, the same blot was rehybridized with an RNase P probe (a gift from Robert P. Ricciardi [University of Pennsylvania Dental School]).
Metabolic labeling of cells and IP of p21.
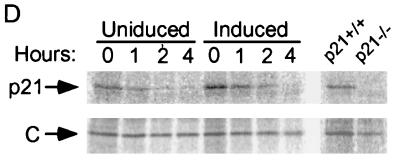
Cells were preincubated for 30 min (37°C; 5% CO2 environment) in methionine-deficient DMEM containing 10% dialyzed FCS (Life Technologies). Fresh medium containing 25 μCi of [35S]methionine-[35S]cysteine (>1,000 Ci/mmol) (Translabel; Amersham)/ml was added for either 1 h or 30 min. The cells were washed two times with PBS and once with complete DMEM containing 10% FCS and 100 μg of unlabeled l-methionine andl-cysteine per ml and then incubated in this medium. After another 1, 2, and 4 h (or 30 min, 1 h, and 2 h for cells exposed to the label for 30 min), extracts were prepared as described above. Protein content was normalized by the Bradford reaction and confirmed by Coomassie blue staining of proteins separated by SDS-PAGE. We found that this also effectively normalized incorporation of the radiolabel in extracts prepared from the pulse phase, with or without p16 induction. p21 was immunoprecipitated as described above, using the C19 antibody (Santa Cruz). Proteins were resolved by SDS-PAGE (12% gels), and the gels were fixed for 30 min in a 50% methanol–10% acetic acid solution, dried, and applied to a PhosphorImager. Parental and p21-null HCT 116 cells were labeled and processed in the same way to confirm the identity of the p21 band. Other bands in the lanes derived from cells subjected to p16-induction typically exhibited either threefold-higher intensity than in immunoprecipitates from cells without p16 induction (data not shown), suggesting co-IP with p21, or similar intensity, suggesting cross-reactivity of the antibody with unrelated cellular proteins. One of the latter is shown in Fig. 2D, as a loading control.
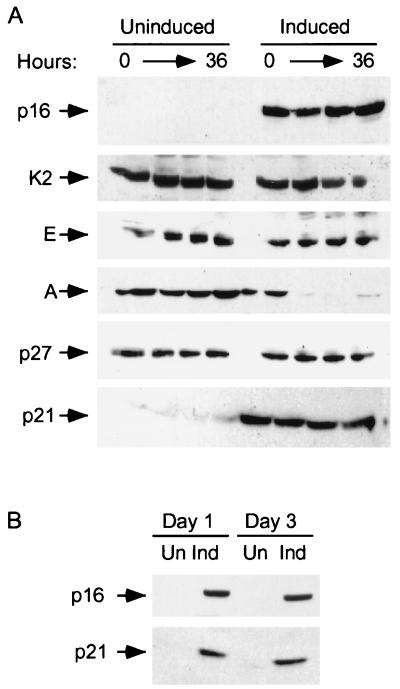
FIG. 2.
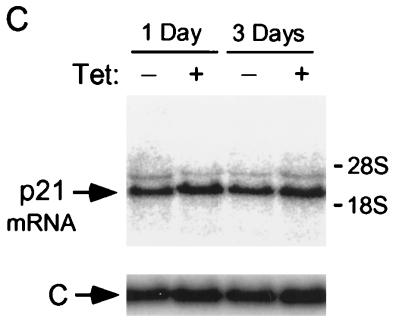
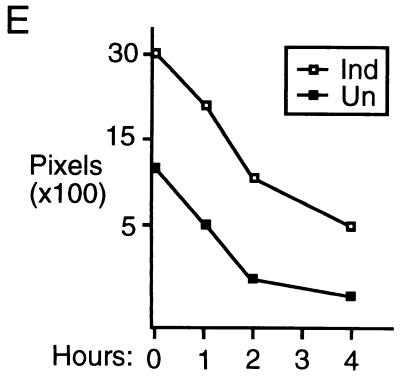
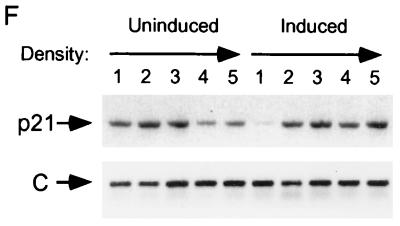
Induction of p21 by p16. (A) Lovastatin-synchronized cells. Extracts were prepared from cells synchronized with lovastatin and rescued with mevalonic acid, in parallel with those cells described in the legend to Fig. 1D, and normalized for protein content. The designated proteins (p16, cdk2 [K2], cyclin E [E], cyclin A [A], p27, and p21) were detected by immunoblotting. (B) Unsynchronized cells. Extracts (normalized for protein content) were prepared from cells initially in log-phase growth, then plated with (Ind) or without (Un) p16 induction for 1 or 3 days. p16 and p21 were detected by immunoblotting. (C) p21 mRNA levels are not changed by p16 induction. RNA was prepared from cells that were or were not subjected to p16 induction for 1 and 3 days, and Northern blotting was performed with the p21 cDNA as a probe. The band labeled p21 mRNA was not present in RNA prepared from p21-null HCT 116 cells (data not shown) (see below). As a control for loading and transfer, the blot was rehybridized with a probe to RNase P (labeled “C”). Similar results were obtained in three other experiments (data not shown). (D and E) Increased metabolic labeling of p21 following p16 induction. Cells that were or were not subjected to p16 induction for 3 days were pulse-labeled for 1 hour with [35S]methionine-[35S]cysteine, and extracts were prepared at the end of this period and at 1, 2, and 4 h after addition of an excess of unlabeled amino acids. (D) The extracts were normalized for protein content (we confirmed that this also resulted in normalization for incorporation of label in the pulse phases), and p21 was immunoprecipitated and resolved on an SDS-polyacrylamide gel. A relatively stable protein that appears to cross-react with the antibody (labeled “C”) is shown as a loading control. HCT 116 p21+/+ and p21−/− cells (see below) were treated identically to confirm that the band shown corresponds to p21. This band also was not seen in control IPs with nonspecific rabbit IgG and comigrated exactly with p21 detected in the same gels by immunoblotting (data not shown). Similar results were obtained in two other experiments (data not shown). (E) The p21 band intensity was quantitated and graphed (pixels [× 100], on a semilog scale; y axis) versus time (hours; x axis). Open squares represent results from cells with p16 induction (Ind), and closed squares represent results from uninduced (Un) cells. (F) Increased sedimentation of p21 mRNA in polyribosome gradients following p16 induction. Cytoplasmic lysates were prepared from cells that were (right) or were not (left) subjected to 3 days of p16 induction and were fractionated by sucrose gradient density sedimentation. Preliminary Northern blotting studies established that p21 mRNA was largely confined to the bottom half of each gradient (e.g., in contrast to the 36B4 mRNA [data not shown]). For finer fractionation, RNA isolated from the top half of each gradient was pooled as fraction 1 and that from the bottom half of the gradient was further subdivided into four fractions (2 to 5), in order of increasing density. A single major PCR product, of the predicted size, was obtained from each fraction, employing p21- and glyceraldehyde 6-phosphate dehydrogenase (“C”)-specific primers, respectively. Titrations showed that the PCRs were substrate limited under these conditions (data not shown). Similar results were obtained in a second experiment (data not shown).
Transient transfection.
Cells (2 × 105) plated in 35-mm-diameter dishes were transiently transfected for 6 h by using 2 μl of Lipofectin reagent (Life Technologies/Gibco BRL) and 2 to 4 μg of DNA diluted in 100 μl of reduced-serum medium (OPTIMEM; Gibco BRL) per the manufacturer’s instructions. HCT 116 p21−/− cells are more transfectable than the parental p21+/+ cells; somewhat less Lipofectin was required to achieve similar fractions of transfected cells and similar levels of protein expression with the same amounts of DNA.
Adenoviruses.
A recombinant adenovirus expressing p16 (Ad5CMV/p16, or Ad5p16) was generated by established protocols (32). Briefly, p16 cDNA (provided by David Beach, Cold Spring Harbor Laboratories) was cloned into shuttle vector pAD/CMV (Vector Core, Institute for Human Gene Therapy, University of Pennsylvania, Philadelphia) and cotransfected with E3-deleted human adenovirus dl70001 into 293 cells, using calcium phosphate precipitation. Positive plaques were identified by Southern blotting, repurified three times, and propagated in 293 cells. Subsequently, the recombinant virus was produced on a large scale and purified twice by cesium chloride density gradient centrifugation (93, 94). Preparation of Ad5CMV/lacZ (Ad5lacZ) has been described previously (94).
HCT 116 cells were infected at 10% confluence 2 days after being replated. The cells were washed once with PBS and incubated with ca. 20 PFU (determined on 293 cells) of virus/cell in serum-free McCoy’s 5A medium for 3 h. The medium was removed, the cells were washed once with PBS, and growth medium was added. Cell extracts were prepared extracts 24 h after infection.
BrdU labeling and immunofluorescence.
Bromodeoxyuridine (BrdU) labeling was performed as described previously (24) with the following modifications. For labeling of U2-OS cells, BrdU was added in aliquots 24 to 32 h after TET withdrawal. For labeling of HCT 116 cells, BrdU was added in aliquots 24 to 48 h after the end of the transfection period. Immunofluorescence staining was performed as described in reference 24, except that an incubation time of 1 h was routinely used. Nuclear DNA was stained, with bisbenzimide (1 μg/ml) (Hoechst 33258; Sigma) being included in the final wash. The results were scored by direct microscopic examination of randomly chosen medium-power fields by an observor blinded to the treatment conditions. For photography, images of each fluorescence wavelength were independently and quantitatively captured in greyscale using a cooled charge-coupled device camera (Sensys model KF 1400) mounted to a Leica DM IRB inverted microscope and controlled by IP Lab Spectrum software (Scanalytics, Fairfax, Va.). These images were recombined and pseudocolored by using the Multiprobe software (Scanalytics). Each field presented was treated with the same refinements of contrast and color.
Polyribosome preparation and RT-PCR.
OSp16.1 cells were lysed in a solution containing 10 mM Tris (pH 8.0), 30 mM MgCl2, 150 mM KCl, 1 mM DTT, 1% Nonidet P-40, and 20 mM ribonucleoside-vanadate complexes/ml (23). Nuclei and cell debris were removed by centrifugation at 10,000 × g for 10 min. Equal amounts of total RNA from the cytoplasmic extracts (estimated from samples prepared by Trizol extraction) in 1.5-ml volumes were layered onto continuous 10 to 50% sucrose gradients in a solution containing 20 mM Tris (pH 8.0), 50 mM KCl, and 5 mM MgCl2. Centrifugation was carried out at 28,000 rpm for 4 h in an SW41 rotor. Preliminary experiments employing Northern blotting had demonstrated that the p21 mRNA was largely confined to the lower half of a gradient. After the run, the upper halves of all gradient were therefore pooled as fraction 1 and the lower halves was separated into four fractions (2 to 5). tRNA (50 μg; Gibco BRL) was added to each of the fractions as a carrier to facilitate RNA recovery. RNA in the fractions was precipitated with 2.5 volumes of 100% ethanol and with 3 M sodium acetate (10% of the total volume), treated with RNase-free DNase (Gibco BRL), and extracted with Trizol. Equal portions (by volume) were reverse transcribed, using 10 U of avian myeloblastosis virus reverse transcriptase (Promega), 40 U of RNasin, 70 ng of random primer, and 0.4 mM each dATP, dCTP, dGTP and dTTP in a total volume of 20 μl. Reverse transcription (RT) was conducted in a model 9600 thermal cycler (Perkin-Elmer) for 1 h at 42°C. An appropriate volume of the RT reaction product was used in PCR reaction mixtures containing 10 mM Tris (pH 8.3), 50 mM KCl, 1.45 mM MgCl2, 0.4 mM each deoxynucleoside triphosphate, 0.8 U of Taq DNA polymerase (Boehringer Mannheim), and 125 ng each of the 5′ and 3′ primers in a total volume of 20 μl. The WAF1/p21 primers used were as follows: 5′-AGGATCCATGTCAGAACCGGCTGG-3′ and 5′-CAGGATCCTGTGGGCGGATTAGGGCT-3′ (37). A three-step cycling PCR program was used, consisting of 40 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 1 min, with a final 5-min strand extension at 72°C. Glyceraldehyde 3-phosphate dehydrogenase used as internal standard was amplified for 24 cycles with the following primers: 5′-TGGTATCGTGGAAGGACTCATGAC-3′ and 5′-ATGCCAGTGAGCTTCCCGTTCAGC-3′ (73).
RESULTS
p16 disrupts cyclin D1-cdk4 binding.
We generated U2-OS cell clones in which p16 expression is regulated by the “TET-off” transcription system (31). Induction of p16 and its effects on DNA synthesis over various time courses in these clones will be presented in detail in the future (14a). In a representative clone, OSp16.1, TET withdrawal yielded an eightfold induction of p16 at 24 h, to a level moderately higher than that in SaOS osteogenic sarcoma cells (Table 1). DNA synthesis was inhibited by 75% during this period, as determined by monitoring tritiated-thymidine incorporation or by flow cytometry. By day 3 following TET removal, the p16 level was 25-fold above the baseline and DNA synthesis was inhibited by more than 90% compared to that of uninduced cells (Table 1). Strong inhibition of DNA synthesis was observed on TET withdrawal in three other clones with inducible p16 expression, but not in two control clones transfected with the empty vector (data not shown). Within 24 h of p16 induction in OSp16.1, most pRB shifted from a slowly migrating form to a more rapidly migrating form on SDS-PAGE, suggesting that it was predominantly hypophosphorylated (data not shown) (see references 26, 62, 92, and 102). Thereafter there was little change in the ratio of the faster-migrating and slower-migrating pRB forms, but the overall levels of the protein declined, as reported by others (92).
TABLE 1.
Inhibition of [3H]thymidine incorporation by induction of p16 in OSp16.1
| Day | Fold p16 induction | % Inhibitiona |
|---|---|---|
| 1 | 8 | 75 ± 9 |
| 2 | 18 | 85 ± 7 |
| 3 | 25 | 92 ± 2 |
Values, normalized to percent [3H]thymidine incorporation in uninduced cells, are means ± standard deviations of data from three determinations.
cdk4 (Fig. 1A) and cdk6 (data not shown) activities, assayed by IP and in vitro phosphorylation of a GST-pRB fusion protein (gstRB), were largely inhibited by p16 induction. This coincided with p16 binding and with the loss of cdk4 from cyclin D1 complexes (Fig. 1B). Similarly, most cdk6 was lost from cyclin D1 immunoprecipitates and most cyclin D1 was lost from cdk6 immunoprecipitates at days 1 and 3 of p16 induction (data not shown). These results differ from those reported for p15INK4b, in which evidence for trimeric p15-cyclin D-cdk complexes was obtained (82). Whether this can be explained by differences in the cellular background or by intrinsic differences between the two inhibitors remains to be determined.
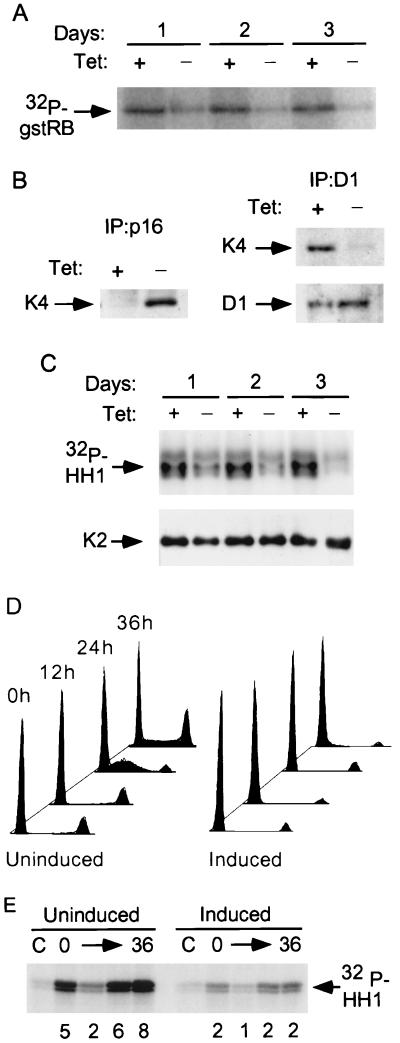
FIG. 1.
p16 mediates inhibition of both cdk4 and cdk2 in OSp16.1 cells. (A) Inhibition of cdk4 kinase activity. Extracts were prepared from cells cultured with (+) or without (−) TET (1 μg/ml) for 1 to 3 days and were normalized for protein content. cdk4 was immunoprecipitated, and its kinase activity was assayed using gstRB as a substrate. (B) (Left panel) Binding of p16 to cdk4. Extracts were prepared from cells cultured with or without TET for 3 days and were normalized for protein content. p16 complexes were immunoprecipitated and immunoblotted for cdk4 (K4). (Right panel) Inhibition of cyclin D1 binding to cdk4 on p16 induction. Extracts were prepared from cells that were or were not subjected to p16 induction for 3 days. Cyclin D1 complexes were immunoprecipitated (D1). These samples were normalized for cyclin D1 content by immunoblotting (below) and immunoblotted for cdk4 (K4). (C) Inhibition of cyclin E-associated kinase activity. Extracts were prepared from cells that were or were not subjected to p16 induction for 1 to 3 days. Cyclin E complexes were immunoprecipitated and normalized for cdk2 content by immunoblotting (K2) (shown below). Kinase activities were then assayed using histone H1 (HH1) as a substrate. (D) p16 induction prevents S phase entry following cell cycle synchronization with lovastatin and rescue with mevalonic acid. OSp16.1 cells were treated with lovastatin for 40 h in the presence (Uninduced) and absence (Induced) of TET. At the end of this period, a portion of the cells was harvested and assayed for DNA content by flow cytometry (0 h; x axis, DNA content; y axis, cell number, normalized to each G1 peak). The lovastatin block was rescued in the remaining cells by addition of mevalonic acid for another 36 h. Cells were removed at 12-h intervals for analysis. The S phase fraction at 24 h was 43% in the uninduced cells versus 5% in the induced cells. (E) Inhibition of cyclin E-associated kinase activity by p16 induction in synchronized cells. Extracts were prepared from cells treated as described for panel D and normalized for protein content. Cyclin E complexes were immunoprecipitated, and their kinase activities were assayed using histone H1 (HH1) as a substrate. C, control (employing nonspecific rabbit IgG). The relative signal intensity is given below each lane.
p16 induction yields an inhibition of cdk2 complex activity.
We asked whether cdk2 activity was affected by the p16-mediated arrest. We prepared extracts from cells which were or were not subjected to 1 to 3 days of p16 induction and normalized them for protein content. We immunoprecipitated cdk2 and cyclin E complexes and assayed their kinase activity in vitro, using histone H1 as a substrate. cdk2- and cyclin E-associated kinase activities were inhibited in all lysates of cells subjected to p16 induction (data not shown). To distinguish a decrease in the specific activity of the complexes from a decrease in either their abundance or the ability of the antibodies to precipitate them, we assayed kinase activity in the immunoprecipitates after normalizing for cdk2 content. p16 induction resulted in a marked decrease in the specific activity of cdk2 complexes (data not shown) and cyclin E complexes (Fig. 1C).
Because p16 induction causes a cell cycle arrest in late G1, we asked whether cell cycle position alone could account for the observed decrease in the specific activity of cdk2 complexes. We induced p16 during an arrest mediated by the cholesterol biosynthesis inhibitor lovastatin. This drug has been shown to effectively synchronize U2-OS cells in early G1 phase and is thought to interfere with critical membrane signaling events (49, 79, 108). Following lovastatin treatment, we restored cell cycling by addition of the downstream metabolite mevalonic acid and assayed cell cycle position by flow cytometry. Lovastatin caused a strong, predominantly G1 arrest in a pool of vector-transfected control clones (data not shown) and in OSp16.1 (Fig. 1D). Mevalonic acid treatment restored cell cycling in about half the control cells within 24 h, in the presence or absence of TET (data not shown). About half of the OSp16.1 cells also resumed cycling in the presence of TET, but entry into S phase was blocked by p16 induction in the absence of TET (Fig. 1D).
We prepared lysates from OSp16.1 cells at successive 12-h intervals following mevalonic acid rescue, with or without p16 induction, and normalized these samples for protein content. Total cdk2 activity (data not shown) and cyclin E-associated kinase activity were largely inhibited by p16 induction (Fig. 1E). Note that this inhibition occurred even at the 0-h time point of mevalonic acid addition, a point at which the two cultures have similar flow cytometry profiles. Thus, inhibition of cyclin E-associated kinase activity mediated by p16 was observed in a comparison of cells synchronously arrested in early G1 and synchronously released from this block.
Induction of p21 by p16.
A variety of experiments have demonstrated that p16 does not associate with cdk2 (52, 57, 96), and we were unable to detect such an association in OSp16.1 cells (data not shown). We asked whether the inhibition of cyclin E-associated kinase activity mediated by p16 could be explained by a decrease in expression of cyclin E or cdk2 in the synchronized cells. Cyclin D1 and cdk4/6 levels were not affected by the treatments (data not shown). Cyclin E expression was also unaffected, and cdk2 levels dropped only slightly in the cultures subjected to p16 induction (Fig. 2A and data not shown). Expression of cyclin A was strongly inhibited by p16 induction, although this effect was less pronounced in unsynchronized cells (data not shown). The decrease in cyclin A levels following addition of mevalonic acid illustrates the point that the culture in which p16 was induced was arrested at 0 h by the effects of lovastatin but at later points by the effects of p16.
We asked whether the observed decrease in cdk2-associated kinase activity could be accounted for by a change in the total cellular level of p27 or p21. p27 expression was not affected, but p21 levels were more than eightfold higher in the cells subjected to p16 induction (Fig. 2A). Note that like the decrease in cyclin E-associated kinase activity, induction of p21 occurred by the 0-h time point. The two cultures have similar cell cycle profiles at this point (Fig. 1D), with the vast majority of cells being in G1, before the normal arrest point mediated by p16. In fact, synchronization of these cells in early G1 by lovastatin treatment results in a slight decrease in p21 levels compared to those of untreated cells, accentuating p16’s effect (data not shown) (see below). Furthermore, passage through late G1 did not result in a prominent increase in the p21 level. We conclude that p16-mediated induction of p21 is not simply a secondary effect of arresting cells in G1.
We then asked whether induction of p21 is seen in cultures under standard passage conditions. Figure 2B shows that p21 was induced within 1 day of p16 induction in unsynchronized cells (passage of U2-OS cells causes a moderate, transient synchronization in G1 phase [data not shown], but we refer to them as unsynchronized for simplicity). Over a series of more than 10 experiments with unsynchronized cells, we found that p21 was typically induced three- to fourfold after 3 days of p16 induction (Fig. 2B [see also Fig. 4A below] and data not shown). Induction of p21 was also seen on TET withdrawal in three other clones with inducible p16 expression, but not in two control clones (data not shown). We also analyzed p21 expression by immunohistochemistry. There was a greater degree of p21 staining in cells subjected to p16 induction but no detectable change in the subcellular distribution, which remained preferentially nuclear (data not shown).
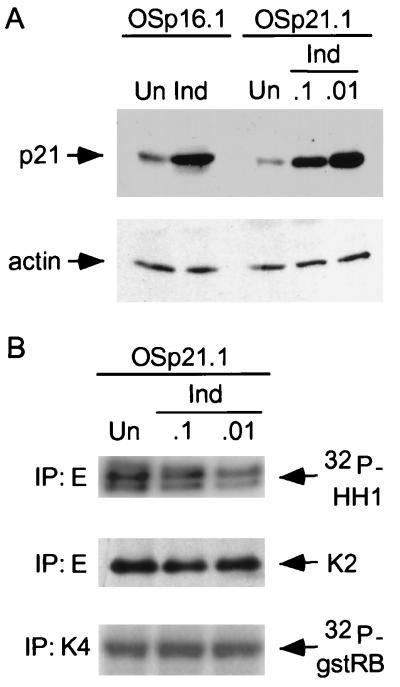
FIG. 4.
Inhibition of cyclin E-associated kinase activity by induction of p21 alone to levels induced by P16. (A) OSp21.1 cells were either maintained in medium with suppressive levels of TET (1.5 μg/ml; uninduced [Un]) or treated with concentrations of TET (0.1 and 0.01 μg/ml; Ind) chosen to induce p21 to a level (right) similar to that observed in OSp16.1 cells upon full induction of p16 (left). p21 levels in extracts normalized for protein content were determined by immunoblotting. Protein content was documented by immunoblotting for actin. (B) Inhibition of cyclin E-associated kinase activity by p21 induction in OSp21.1 cells. Extracts were prepared from OSp21.1 cells maintained for 3 days in media containing different concentrations of TET, as described for panel A. Cyclin E was immunoprecipitated. The complexes were normalized for cdk2 (K2) content and assayed for kinase activity with histone H1 as a substrate. cdk4 (K4) activity was also assayed, in a like manner, using gstRB as a substrate (normalization data are not shown). Quantitation of these signals revealed that cyclin E-associated kinase activity was inhibited in OSp21.1 cells by 40 and 75% in 0.1 and 0.01 μg of TET/ml, respectively. Since p21 levels under these conditions were, respectively, slightly lower and higher than that observed with p16 induction, we estimate that 50% inhibition of cyclin E-associated kinase activity can be expected for a comparable level on p21 induction alone. This compares with 75% inhibition of cyclin E-associated kinase activity seen with p16 induction (Fig. 1C).
We performed initial studies to characterize the level at which p21 expression is regulated. We prepared RNA from OSp16.1 cells maintained in the presence or absence of TET for 1 and 3 days. In Northern hybridizations using the p21 cDNA as a probe, a single major species, of the expected size (2.1 kb), was present at similar levels in all samples (Fig. 2C). Metabolic pulse-labeling with [35S]methionine-[35S]cysteine showed that the half-life of p21 remained about 1 h in the presence or absence of p16 induction for 3 days (Fig. 2D and E). In contrast, threefold more label was incorporated into p21 during the pulse-labeling phase in the setting of p16 induction than in the controls. This ratio was unaffected by further reducing the pulse duration from 1 h to 30 min (3.3- and 2.8-fold in two independent experiments) (data not shown). The increased metabolic labeling of p21 accounts well for the three- to fourfold increase in the steady-state level of the protein detected by immunoblotting following p16 induction and points to regulation of p21 expression at the level of translation. Most translation appears to be controlled at the initiation step, correlating with an increased number of ribosomes per mRNA (64). Consistent with this, we observed increased sedimentation of p21 mRNA in polyribosome gradients on p16 induction (Fig. 2F). Further work will be required to elucidate more fully the mechanism of regulation.
Inhibition of cdk2 complex activity by p21.
We examined the physical association of p27 and p21 with cdk’s by IP followed by immunoblotting. We noted an increase in p27 associated with cdk2 after 1 day of p16 induction, but this increase was lost by day 3 (Fig. 3A). Association of p21 with cdk2 was also increased by day 1 and had increased further by day 3. The displacement of cyclin D1 from cdk4/6 complexes mediated by p16 binding (Fig. 1B) would be expected to prevent the binding of CIP/KIP inhibitors, since they require cyclins for efficient binding to cdk’s (12, 90). Consistent with this, we found that binding of p21 to cdk4 (Fig. 3A) and cdk6 (data not shown) was disrupted by induction of p16 at both the 1-day and 3-day time points. Thus, binding of p16 to cdk4/6 prevents the binding of p21, even as p21 levels increase.
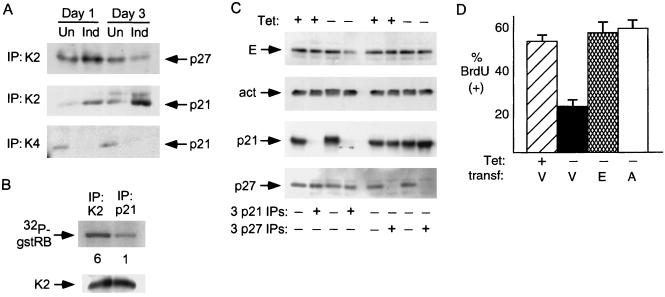
FIG. 3.
Inhibition of cdk2 by p21 in cells with p16 induction. (A) Increased association of p21 with cdk2 and decreased association with cdk4 following p16 induction. Extracts were prepared from cells cultured with or without p16 induction for 1 and 3 days and were normalized for protein content. cdk2 (K2) or cdk4 (K4) complexes were immunoprecipitated and probed for coprecipitated p21 or p27 by immunoblotting. Un, uninduced; Ind, induced. (B) Inhibition of cdk2 kinase activity in p21-bound complexes. Extracts were prepared from cells with p16 induction for 3 days, and p21 and cdk2 were immunoprecipitated. The immunoprecipitates were normalized for cdk2 protein level and assayed for cdk2 activity with gstRB as a substrate. Quantitation of the relative band intensity is provided below each lane. (C) Immunodepletion of cyclin E bound to p21 following 3 days of p16 induction. Extracts were prepared from cells with (+) or without (−) TET for 3 days and subjected to three successive immunoprecipitations with antibodies directed against p21 or p27. Cyclin E, actin (act; a negative control), p21, and p27 levels in the starting extracts and in the supernatant following the third IP were assessed by immunoblotting. Clearing of a major fraction of cyclin E from the supernatant occurred only following p21 IP, and only in extracts prepared from cells with p16 induction in this and two other experiments (data not shown). Parallel IPs with a negative-control antiserum (anti-hemagglutinin [12CA5]) had no effect on the level of any of these antigens (data not shown). (D) Overexpression of cyclin E or A can prevent the arrest mediated by p16. OSp16.1 cells were cotransfected with a CMV vector expressing β-gal together with an empty expression vector (V) or one expressing cyclin E (E) or cyclin A (A). A portion of the vector-transfected (transf.) cells was replated in the presence of TET as a positive control for BrdU incorporation. p16 was induced in the remaining cells by replating in the absence of TET for 24 h. The cells were then incubated with BrdU for 8 h, fixed, and stained for β-gal and BrdU. One hundred β-gal-positive cells per condition were scored by an observer blinded to the treatment groups, and the results confirmed by a second, similarly blinded observer. The mean percentage of BrdU-positive cells from three experiments is depicted. Error bars indicate standard deviations.
cdk complexes associated with p21 can retain catalytic activity, typically at low inhibitor stoichiometries (11, 53, 110). We therefore asked whether p21-bound cdk2 complexes were indeed inhibited. We immunoprecipitated cdk2 and p21 complexes and normalized these precipitates for cdk2 content. The p21-bound complexes retained far less catalytic activity for histone H1 or gstRB (Fig. 3B and data not shown).
We next asked whether p21 or p27 complexes bound a major fraction of cyclin E complexes in extracts prepared from cells that were or were not subjected to 3 days of p16 induction. We performed successive immunodepletions with antibodies directed against p27, p21, or a negative-control antigen and measured cyclin E in the remaining supernatants by immunoblotting. We observed the loss of the majority of cyclin E from the supernatant only after immunodepletion with anti-p21 antibodies and induction of p16 (Fig. 3C). These results indicate that the majority of cyclin E in the extract from cells subjected to p16 induction is bound to p21. Since p21 cannot bind efficiently to free cyclins (12, 90) and cdk2 is the only major cdk partner for cyclin E (18, 55, 75, 101), we infer that most of the cyclin E-cdk2 complexes in these extracts are bound by p21. These results do not exclude the possibility that p27 binds a significant fraction of cyclin E complexes in the initial stages of the arrest.
We asked whether overexpression of cyclin E or A could prevent a p16-mediated arrest of these cells. We transfected OSp16.1 cells with an empty cytomegalovirus (CMV) expression vector or one expressing cyclin E or A, induced p16 for 24 h, and assayed DNA synthesis by measuring BrdU incorporation. Successfully transfected cells were identified by cotransfection with a vector expressing β-galactosidase (β-gal) (see Fig. 6A for examples of the technique). Expression of either cyclin effectively prevented p16 from inhibiting DNA synthesis (Fig. 3D). Immunoblotting demonstrated about a twofold increase in cyclin levels in the respective transfected cultures (data not shown). Immunofluorescence staining demonstrated that about 25% of the cells were successfully transfected (data not shown). We therefore estimate that cyclins E and A were expressed at about eight times the endogenous level in the successfully transfected cells.
FIG. 6.
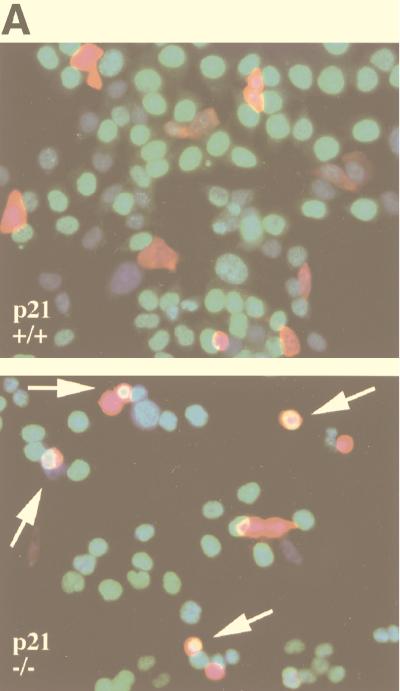
p16 expression strongly inhibits S phase entry in p21+/+ cells but not in p21−/− cells. (A and B) BrdU incorporation. Parental (p21+/+) and p21−/− HCT 116 cells were cotransfected with a constant amount of a CMV vector expressing β-gal and either the same empty vector or one expressing p16 or cdk2DN. BrdU was added in several additions 24 to 48 h after transfection, and the cells were fixed and stained for β-gal and BrdU. (A) Quantitative fluorescent micrographs were obtained from a single field of p21+/+ cells (top) or p21−/− cells (bottom) transfected with the p16 expression vector. β-Gal is stained red, BrdU is stained green (giving a yellow tinge when combined with red), and nuclear DNA (detected with bisbenzimide) is stained blue. (B) One hundred β-gal-positive cells per transfection condition were scored through direct microscopic examination of random medium-power fields by an observer blinded to the treatment groups. The results were confirmed by a second, similarly blinded observer. The mean percentage of BrdU-positive cells from three experiments is depicted. Error bars indicate standard deviations. (C) Flow cytometry. The respective cells were cotransfected with a constant amount of a CMV vector expressing CD20 and either the same empty vector or one expressing p16. Single, CD20-positive (green fluorescent) cells were identified by flow cytometry 36 h after transfection and analyzed for DNA content. One hundred thousand events were collected for each transfection condition and subjected to identical gating parameters. This analysis yielded a similar number of cells from each transfection condition: 3,812 (p21+/+; vector), 2,810 (p21+/+; p16), 2,718 (p21−/−; vector), 3,503 (p21−/−; p16), and 49 (mock transfected; data not shown). The S phase fraction is highlighted with cross-hatching and quantitated above each profile. Comparable results were obtained in a second experiment (data not shown).
Induction of p21 to the level attained during p16 induction is sufficient to inhibit cdk2 complex activity and DNA synthesis.
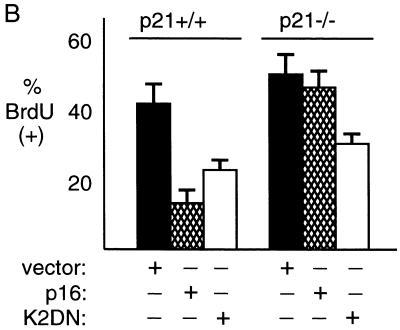
We then asked whether the level of p21 induction observed was alone sufficient to inhibit cyclin E kinase activity and/or DNA synthesis. From the same parental cell used to derive OSp16.1, we generated clones in which p21 expression was inducible. We identified levels of TET that permitted induction of p21 in one such clone (OSp21.1) to levels bracketing that achieved by induction of p16 (0.1 and 0.01 μg of Tet/ml, respectively) (Fig. 4A). Each of these levels of p21 yielded a significant inhibition of the specific activity of cyclin E complexes (Fig. 4B). By interpolation, we estimate that p21 induction alone to the level attained during p16 induction is sufficient to account for more than half of the decrease in cyclin E-associated kinase activity observed in the latter setting (50% versus 75% inhibition of kinase activity, respectively) (see the legend to Fig. 4). cdk4 kinase activity was not detectably affected by p21 induction at these levels in OSp21.1 (Fig. 4B), but cyclin A expression was moderately inhibited (data not shown). p21 induction alone at these levels also inhibited DNA synthesis by 40 and 75%, respectively, compared to that in uninduced cells (Table 2) (versus >90% with p16 [Table 1]). We conclude that induction of p21 alone to the level observed in p16-inhibited cells is sufficient to significantly inhibit cyclin E-cdk2 complex activity, cyclin A expression, and DNA synthesis, but it does not quantitatively account for p16’s effects (see Discussion).
TABLE 2.
Inhibition of [3H]thymidine incorporation by induction of p21 in OSp21.1
| TET concn (μg/ml) | % Inhibition of [3H]thymidine incorporationa
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| 1.0 | 0 | 0 |
| 0.1 | 35 ± 14 | 44 ± 4 |
| 0.01 | 75 ± 4 | 76 ± 6 |
Values, normalized to percent [3H]thymidine incorporation in uninduced cells, are means ± standard deviations of data from three determinations.
p21 is required for efficient p16-mediated cell cycle arrest in HCT 116 cells.
We sought to determine whether the effects of p16 on p21 can be observed in other cell lines and whether p21 is required for p16 to efficiently arrest the cell cycle. We therefore extended our studies to HCT 116 colorectal carcinoma cells. A p21-null derivative of these cells was generated by two rounds of targeted homologous recombination (104). Colorectal carcinoma cell lines nearly uniformly express full-length, apparently intact pRB (3, 42). Although p16 mutations are uncommon in these cell lines, most show hypermethylation of the p16 promoter, and in one study about 40% of primary colon carcinomas also showed this modification, consistent with a possible role for p16 in suppression of these tumors (6, 41). In HCT 116 cells, the promoter of one p16 allele is hypermethylated, the other allele bears a frameshift mutation, and functional p16 is not expressed (71).
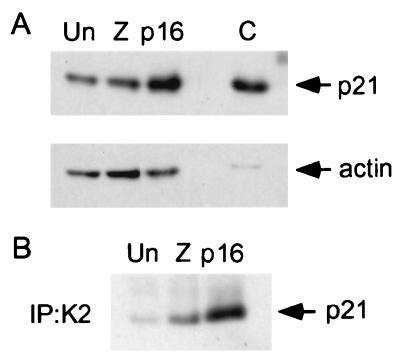
We first asked whether p16 expression in the parental cells is associated with an increase in total cellular and/or cdk2-bound p21. We expressed p16 for 24 h, using a replication-defective adenovirus (Ad5p16), and compared p21 levels in these cells to those in cells that were either mock infected or infected with a control adenovirus expressing β-gal (Ad5lacZ). Immunofluorescence staining of cells exposed to similar viral multiplicities of infection (determined on 293 cells) demonstrated that p16 expression occurred in about 25% of the Ad5p16-infected cells and that β-gal expression occurred in about 40% of the Ad5lacZ-infected cells (data not shown). (Use of higher multiplicities of infection resulted in nonspecific toxicity and was therefore avoided.) We observed induction of p21 and increased binding of p21 to cdk2 in the Ad5p16-infected cells compared to the two controls (Fig. 5). When the fraction of p16-expressing cells is taken into account, induction of p21 mediated by p16 in the HCT 116 cells appears to be somewhat stronger than that in U2-OS cells.
FIG. 5.
Increase in total cellular and cdk2-associated p21 levels mediated by p16 expression in HCT 116 cells. (A) Induction of p21 by p16 in adenovirus-infected cells. Parental HCT 116 cells (p21+/+) were mock infected (Un), infected with Ad5lacZ (Z), or infected with Ad5p16 for 24 h. Extracts were prepared and normalized for protein content. p21 was detected by immunoblotting. The same extracts were probed for actin by immunoblotting (below) as a control to assess protein content. An extract prepared from OSp21.1 cells maintained for 3 days in the absence of TET served as a positive control (C). (B) p16 mediates an increase in association of p21 with cdk2 (K2) in adenovirus-infected cells. cdk2 complexes were immunoprecipitated from the extracts described above, and associated p21 was detected by immunoblotting.
We then assayed the ability of p16 to inhibit DNA synthesis in the parental and p21-null cells. To facilitate comparison of the effects of p16 expression in these cells with those of both negative and positive controls, we employed cotransfection of test plasmids in the presence of a constant amount of marker plasmid, as described above (Fig. 3D). We monitored DNA synthesis by performing serial additions of BrdU at between 24 and 48 h after transfection. Transfection of the p16 expression vector strongly inhibited DNA synthesis in the parental cells, relative to transfection with the empty vector, but was ineffective in the p21-null derivative (Fig. 6A and B). A similar result was observed in a comparison of Ad5p16- and Ad5lacZ-infected cells, although the level of BrdU incorporation was moderately lower in the virus-infected cells than in the transfected cells (data not shown). Immunofluorescence staining demonstrated that expression of the β-gal marker (Fig. 6A) and p16 (data not shown) in the culture of cells with wild-type p21 was similar to that in the p21-null cell cultures. We then asked whether the p21-null cells were nonspecifically insensitive to cdk inhibition by transfecting a vector expressing a dominant-negative mutant of cdk2 (cdk2DN) (103). Although the p21-null cells showed a trend toward more BrdU incorporation at baseline than the parental cells, cdk2DN showed the same percentage of inhibition of DNA synthesis in the two cultures, in marked contrast to p16 (Fig. 6B).
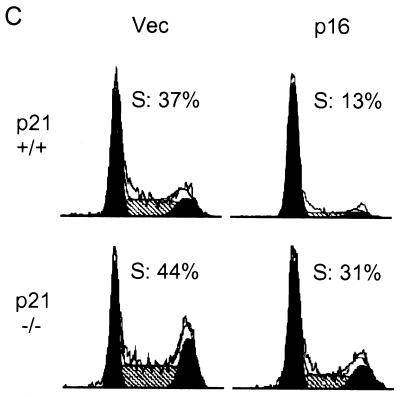
We then sought to confirm by flow cytometry that the effects of p16 represented an inhibition of cell cycle progression rather than an effect on DNA repair synthesis or on BrdU uptake. We cotransfected cells with a vector expressing a cell surface marker (CD20) and used flow cytometry to focus the analysis on successfully transfected cells. Equivalent fractions of cells for each set of transfection conditions were identified as successfully transfected (see the legend to Fig. 6C), and histograms of anti-CD20 fluorescence intensity and direct staining for p16 expression in parallel transfections showed that the levels of expression of the transfected proteins were similar in the two cell types (data not shown). The flow cytometry results showed that p16 expression inhibited S phase entry by 65% in the parental cells but by only 29% in the p21-null derivative (Fig. 6C). The large difference between the responses to p16 of the two cell types was maintained across all levels of CD20 staining intensity (data not shown). We conclude that in the absence of the p21 gene, p16 inhibits DNA synthesis in HCT 116 cells much less efficiently than in its presence.
DISCUSSION
We have investigated the mechanism of cell cycle inhibition by the tumor suppressor p16. In U2-OS osteogenic sarcoma cells, p16 induction causes a marked reduction in the specific activity of cyclin E-cdk2 complexes. Binding of p16 to cdk4 and cdk6 blocks the binding of cyclin D1, p21, and p27 to these cdk’s. Concomitantly, the total cellular level of p21 increases severalfold. The majority of cyclin E complexes become bound by p21, and these complexes retain little catalytic activity. Induction of p21 alone can substantially reproduce this effect and is sufficient to significantly inhibit DNA synthesis without detectably affecting cdk4 activity. p21 induction and its binding to cdk2 are also observed following p16 expression in HCT 116 colorectal carcinoma cells. Targeted deletion of the p21 genes in these cells severely compromises p16’s ability to arrest the cell cycle.
Cyclin A expression is moderately inhibited by induction of p16 or p21 alone. This suggests that p21 induction can also account, at least in part, for the effect of p16 on cyclin A. We found that overexpression of cyclin A, as with cyclin E, can prevent p16 from arresting the cells. Thus, our results are consistent with the following model: cyclin E kinase activity stimulates the expression of cyclin A, which then initiates DNA synthesis or cooperates with cyclin E to do so. This model remains speculative, however, because this study did not address whether cyclin A expression is directly regulated by cyclin E kinase activity and because, when overexpressed, these cyclins may gain functional capabilities not present at normal expression levels.
Induction of p21 by p16.
The induction of p21 (and inhibition of cdk2) does not appear to be simply secondary to cell synchronization at the p16 arrest point in late G1 (58), because it does not occur in cells lacking p16 during synchronous transit of these cells through late G1 or during an early G1 arrest imposed by lovastatin. If p16 is expressed during the lovastatin block, then p21 is induced, without marked changes in the cell cycle profile. p21 levels also do not change significantly when the lovastatin block is relieved and the cells become arrested by effects of p16. Thus, the induction of p21 appears to be a specific consequence of p16 induction rather than simply a result of arresting cells at a point at which p21 levels are normally high. On the other hand, it would not be surprising if induction of p21 by p16 were dependent in part on the cell cycle position; for example, we do not know yet whether expression of p16 can cause an increase in p21 levels or an inhibition of cdk2 within S or G2/M phase.
p21 induction in response to p16 was not observed in studies of a U2-OS cell clone which were published while this work was under review (65). The reason for this discrepancy is not clear. We have observed that in dense U2-OS cell cultures, p21 levels can increase as cells not subjected to p16 induction approach confluence, counterbalancing to some extent the induction of p21 due to p16. Our studies were routinely performed with cell cultures that were less than 50% confluent.
Our initial studies of the mechanism of p21 induction by p16 in U2-OS cells suggest an increase in translation of the p21 mRNA. Most known means of p21 regulation occur at the mRNA level (21, 22, 74). Evidence for translational control of p21 has been found previously during serum stimulation of quiescent normal breast epithelial cells and during a second phase of p21 induction in irradiated breast carcinoma cells that follows the initial increase in its mRNA (34). cdk4, p27, and the yeast cyclin CLN3 are three other proteins for which translational regulation is thought to play a significant role in influencing entry into the cell cycle (25, 40, 69, 77). However, we did not detect changes in p27 or cdk4 levels in our experiments, and there is no evidence at this point for a common mechanism regulating their translation. p21 induction by p16 is, likewise, not obviously accounted for known effects of pRB or p53, but further work will be required to define the mechanism in detail.
Inhibition of cdk2 through CIP/KIP redistribution and induction of p21.
The effect of induction of p21 on cdk2 complexes is likely augmented by p16-mediated displacement of cyclin D1 from cdk4 and cdk6. This prevents binding of p21 and shunts preexisting and newly synthesized protein to other binding partners, including cdk2. Because the half-life of p21 in U2-OS cells is only an hour, redistribution of preexisting p21 is likely to have a minimal impact. We also observed a transient increase in p27 levels associated with cdk2 during p16 induction, even though total cellular levels of p27 remained constant. These results are consistent with prior studies with keratinocytes that revealed a redistribution of CIP/KIP inhibitors from cdk4/6 to cdk2 on induction of p15 (82, 83) and with two recent, independent studies demonstrating redistribution of p21 and/or p27 on induction of p16 for 24 h in U2-OS cells (43, 65). These findings confirm the recent predictions, derived from the X-ray crystal structure of p16 bound to cdk6, that p16 binding likely antagonizes the binding of cyclin D and the CIP/KIP inhibitors (89). p16-mediated shunting of p27 and p21 to cdk2 may account for the residual gap between the effects of p21 induction alone and p16’s ability to inhibit cyclin E kinase activity (e.g., 50% versus ca. 75%, respectively, at day 3 in the U2-OS clones) and DNA synthesis (e.g., 50% versus ca. 90%, respectively).
An exact accounting of the relative contributions of p21 induction and CIP/KIP redistribution to p16’s effects is not possible with these data, because the latter and/or other effects of p16 may accentuate (or, conceivably, mute) the effects of the former assayed in isolation. However, our results suggest that redistribution of p21 and p27 is likely markedly potentiated by the increase in the total cellular level of p21. p21 induction begins within the first 24 h of p16 expression in both U2-OS cells and HCT 116 cells, and it continues, at least in U2-OS cells, over the next 2 days. By day 3, the majority of cyclin E in the cells is associated with p21. Moreover, the absence of p21 at between 24 and 48 h of p16 expression in HCT 116 cells abrogates efficient cell cycle arrest.
Our finding, and that of others (43, 65), that p16 induction results in inhibition of cyclin E complex activity differs from that of an earlier study (59). One potential explanation for this difference is that in our hands, the monoclonal antibody used in the latter study did not efficiently immunoprecipitate CIP/KIP-bound cyclin E complexes (data not shown). Under conditions in which the antibody was limiting, we found that it did not detect CIP/KIP-mediated inhibition of cyclin E kinase activity.
Our observations suggest that p16-mediated inhibition of cdk4/6 per se may not be sufficient to impose a cell cycle arrest. Support for this notion comes from recent results of Jiang et al. (43), who found that a dominant-negative mutant of cdk4 (cdk4DN) could effectively inhibit endogenous cyclin D1-associated kinase activity without imposing an arrest. It will also be interesting to examine whether expression of cdk4DN yields induction of p21. In this setting, cdk2 activity is unaffected and pRB remains in a slow-migrating, apparently hyperphosphorylated form (43). Thus, inhibition of cdk4 and cdk6 appears to be dissociated from hypophosphorylation of pRB or cell cycle arrest. If p21 is not induced in this setting, then one (or both) of these latter features is likely required to achieve p21 induction, in addition to inhibition of cdk4 and cdk6. Alternatively, p21 may be induced by inhibition of cdk4 and cdk6 per se, and the additional cyclin D-cdk4DN complexes may sequester p21, preventing it from binding and inhibiting cdk2.
In considering the time course of p16’s effects, we note that there is currently no known physiologic setting in which p16 is acutely induced. Expression of the protein appears to increase gradually and persist at high levels during cellular senescence (1, 36, 84, 86, 113). Although little is yet known about p16 regulation during tumorigenesis, high-level p16 expression is found in some tumors, presumably reflecting sustained expression (4, 14a, 28, 30). We find that the cell cycle arrest mediated by induction of p16 for 24 h in U2-OS cells is reversible if TET is restored and p16 levels are allowed to return to baseline (14a). In contrast, following sustained induction of p16 for 3 days, a significant fraction of cells are no longer able to resume growth during the ensuing 12 days, long after the return of p16 expression to the baseline. This latter state is accompanied by features of cellular senescence (14a, 48, 66, 102). Thus, the events occurring within the first 24 h of p16 induction, including the conversion of most of the pRB to a hypophosphorylated form and redistribution of preexisting CIP/KIP inhibitors, appear to be insufficient to mediate a durable cell cycle inhibition. This observation suggests the importance of delineating subsequent elements of the response to p16, such as the increasing inhibition of cdk2 activity described in this study.
p21 and tumor suppression.
If p21 is a key mediator of p16’s effects, then why is p21 not also a major tumor suppressor? This question has, in fact, been posed since the identification of p21 as an effector for p53 (10, 16, 22). A variety of recent data suggest that p21 does have tumor suppressor properties. p21 mutations are found at a low frequency in several human tumors (61), and a p21 mutation demonstrated to specifically abrogate its binding to cdk’s was identified in a primary breast tumor (5). The human papillomavirus E7 transforming protein interacts with p21, disrupting its interaction with cdk’s (29, 44). Keratinocytes from p21-null mice are more highly tumorigenic than their wild-type counterparts following expression of an activated ras allele (70), and high-intensity Raf signaling is opposed by p21 (97). Finally, mice doubly nullizygous for p21 and pRB are more tumor prone than those that are null for pRB alone (9). The latter finding also reinforces evidence that p21 does not act solely through its effects on pRB.
Several possible explanations for why p21 gene alterations are not found more frequently in tumors have been put forward. p21 does not appear to be required for most forms of p53-mediated apoptosis (10), and it may protect against apoptosis in myocytes (105). Therefore, the loss of p21 in some neoplastic cells may avoid a G1 arrest but yield apoptosis instead. p21 has several properties that may be useful to tumor cells, potentially negating the growth advantage of a loss of the protein. These include the abilities to interact with proliferating cell nuclear antigen and DNA methyltransferase and to promote assembly of cyclin-cdk complexes (11, 13, 53, 109). Our results and those of others suggest that p27 may, in some instances, act redundantly with p21 in mediating cell cycle arrest (43, 82, 83). Finally, in some tumors, a loss of E2F-dependent transcription and/or other effects of pRB activation might eventually be sufficient to cause cell cycle arrest, without the need for CIP/KIP-mediated inhibition of cdk2. Our data suggest that p21 is not absolutely essential for p16 to mediate cell cycle inhibition, because some inhibition was reproducibly observed in the p21-null HCTl16 cells, albeit at a much lower efficiency than in the parental cells.
Potential interactions of the p16 response pathway.
The identification of p21 as an effector for p16 and cdk2 as a target of its actions delineates a new branch of the p16 response pathway. Therefore, other influences on p21 and cdk2 activities may affect p16’s ability to mediate cell cycle arrest. An example is presented by the ARF/p53 pathway. p16 and ARF are often coexpressed in tumors, tumor cell lines, and senescing cells, even though transcription of the two proteins is not always coordinately regulated (47, 85, 112, 113). Our results indicate that p21-mediated inhibition of cdk2 is a common element of the ARF/p53 and p16/pRB response pathways and, thus, a potential point of cooperation between them.
ACKNOWLEDGMENTS
We are grateful to Youngsup Byun, who contributed to this project during undergraduate studies at Harvard University, and to Ed Harlow, in whose laboratory this project was initiated. We thank Jim Koh for antibodies and Liang Zhu for sharing data prior to publication.
W.S.E.-D. is an Assistant Investigator of the Howard Hughes Medical Institute. This work was supported in part by the following grants to G.H.E.: a pilot project grant from the University of Pennsylvania Comprehensive Cancer Center, an American Cancer Society institutional research grant administered through this Center, and grant 7-K08-CA61412 from the National Cancer Institute (NCI). We also acknowledge use of facilities of the Penn Digestive Disease Center, supported by National Institutes of Health grant P30 DK50306, and the Comprehensive Cancer Center, supported by grants from the NCI and the Markey Charitable Trust.
REFERENCES
- 1.Alcorta D A, Xiong Y, Phelps D, Hannon G, Beach D, Barret J C. Involvement of the cyclin-dependent kinase inhibitor p16INK4a in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci USA. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alevizopoulos K, Vlach J, Hennecke S, Amati B. Cyclin E and c-myc promote cell proliferation in the presence of p16INK4a and of hypophosphorylated retinoblastoma family proteins. EMBO J. 1997;16:5322–5333. doi: 10.1093/emboj/16.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali A A, Marcus J N, Harvey J P, Roll R, Hodgson C P, Wildrick D M, Chakraborty A, Boman B M. RB1 protein in normal and malignant human colorectal tissue and colon cancer cell lines. FASEB J. 1993;7:931–937. doi: 10.1096/fasebj.7.10.8344490. [DOI] [PubMed] [Google Scholar]
- 4.Asamoto M, Hori T, Baba-Toriyama H, Sano M, Takahashi S, Tsuda H, Shirai T. p16 gene overexpression in mouse bladder carcinomas. Cancer Lett. 1998;127:9–13. doi: 10.1016/s0304-3835(97)00447-3. [DOI] [PubMed] [Google Scholar]
- 5.Balbin M, Hannon G J, Pendas A M, Ferrando A A, Vizoso F, Fueyo A, Lopez-Otin C. Functional analysis of a p21WAF1/CIP1/SDI1 mutant (Arg94-Trp) identified in a human breast carcinoma. J Biol Chem. 1996;271:15782–15786. doi: 10.1074/jbc.271.26.15782. [DOI] [PubMed] [Google Scholar]
- 6.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J-P. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 7.Berns K, Hijmans E M, Bernards R. Repression of c-Myc responsive genes in cycling cells causes G1 arrest through reduction of cyclin E/CDK2 kinase activity. Oncogene. 1997;15:1347–1356. doi: 10.1038/sj.onc.1201280. [DOI] [PubMed] [Google Scholar]
- 8.Blain S W, Montalvo E, Massague J. Differential interaction of the cyclin-dependent kinase (Cdk) inhibitor p27Kip1 with cyclin A-Cdk2 and cyclin D2-Cdk4. J Biol Chem. 1997;272:25863–25872. doi: 10.1074/jbc.272.41.25863. [DOI] [PubMed] [Google Scholar]
- 9.Brugarolas J, Bronson R T, Jacks T. p21 is a critical CDK2 regulator essential for proliferation control in Rb-deficient cells. J Cell Biol. 1998;141:503–514. doi: 10.1083/jcb.141.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Jackson P, Kirschner M W, Dutta A. Separate domains of p21 involved in the inhibition of cdk kinase and PCNA. Nature. 1995;374:386–388. doi: 10.1038/374386a0. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuang L S, Ian H I, Koh T W, Ng H H, Xu G, Li B F. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 14.Clurman B E, Groudine M. The CDKN2A tumor-suppressor locus—a tale of two proteins. N Engl J Med. 1998;338:910–912. doi: 10.1056/NEJM199803263381312. [DOI] [PubMed] [Google Scholar]
- 14a.Dai, C. Y., and G. H. Enders. Unpublished data.
- 15.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21Cip1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 17.de Nooij J C, Letendre M A, Hariharan I K. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell. 1996;87:1237–1247. doi: 10.1016/s0092-8674(00)81819-x. [DOI] [PubMed] [Google Scholar]
- 18.Dulic V, Lees E, Reed S I. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 19.Duro D, Bernard O, Della Valle V, Berger R, Larsen C J. A new type of p16INK4/MTS1 gene transcript expressed in B-cell malignancies. Oncogene. 1995;11:21–29. [PubMed] [Google Scholar]
- 20.Dyson N. pRB, p107, and the regulation of the E2F transcription factor. J Cell Sci Suppl. 1994;18:81–87. doi: 10.1242/jcs.1994.supplement_18.12. [DOI] [PubMed] [Google Scholar]
- 21.El-Deiry W S. p21/p53, cellular growth control, and genomic integrity. Curr Top Microbiol Immunol. 1998;227:121–137. doi: 10.1007/978-3-642-71941-7_6. [DOI] [PubMed] [Google Scholar]
- 22.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 23.Enders G H, Ganem D, Varmus H E. 5′-Terminal sequences influence the segregation of ground squirrel hepatitis virus RNAs into polyribosomes and viral core particles. J Virol. 1987;61:35–41. doi: 10.1128/jvi.61.1.35-41.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enders G H, Koh J, Missero C, Rustgi A K, Harlow E. p16 inhibition of transformed and primary squamous epithelial cells. Oncogene. 1996;12:1239–1245. [PubMed] [Google Scholar]
- 25.Ewen M E, Oliver C J, Sluss H K, Miller S J, Peeper D S. p53-dependent repression of CDK4 translation in TGF-β-induced G1 cell-cycle arrest. Genes Dev. 1995;9:204–217. doi: 10.1101/gad.9.2.204. [DOI] [PubMed] [Google Scholar]
- 26.Fahraeus R, Paramio J M, Ball K L, Lain S, Lane D P. Inhibition of pRb phosphorylation and cell-cycle progression by a 20-residue peptide derived from p16CDKN2/INK4A. Curr Biol. 1996;6:84–91. doi: 10.1016/s0960-9822(02)00425-6. [DOI] [PubMed] [Google Scholar]
- 27.Fero M L, Rivkin M, Tasch M, Porter P, Carow C E, Firpo E, Polyak K, Tsai L H, Broudy V, Perlmutter R M, Kaushansky K, Roberts J M. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27Kip1-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 28.Foster S A, Wong D J, Barrett M T, Galloway D A. Inactivation of p16 in human mammary epithelial cells by CpG island methylation. Mol Cell Biol. 1998;18:1793–1801. doi: 10.1128/mcb.18.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funk J O, Waga S, Harry J B, Espling E, Stillman B, Galloway D A. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geradts J, Wilson P A. High frequency of aberrant p16INK4A expression in human breast carcinoma. Am J Pathol. 1996;149:15–20. [PMC free article] [PubMed] [Google Scholar]
- 31.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham F L, Prevec L. Methods for construction of adenovirus vectors. Mol Biotechnol. 1995;3:207–220. doi: 10.1007/BF02789331. [DOI] [PubMed] [Google Scholar]
- 33.Gray-Bablin J, Rao S, Keyomarsi K. Lovastatin induction of cyclin-dependent kinase inhibitors in human breast cells occurs in a cell cycle-independent fashion. Cancer Res. 1997;57:604–609. [PubMed] [Google Scholar]
- 34.Gudas J, Nguyen H, Li T, Hill D, Cowan K H. Effects of cell cycle, wild-type p53 and DNA damage on p21CIP1/Waf1 expression in human breast epithelial cells. Oncogene. 1995;11:253–261. [PubMed] [Google Scholar]
- 35.Haber D. Splicing into senescence: the curious case of p16 and p19ARF. Cell. 1997;91:555–558. doi: 10.1016/s0092-8674(00)80441-9. [DOI] [PubMed] [Google Scholar]
- 36.Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harada N, Gansauge S, Gansauge F, Gause H, Shimoyama S, Imaizumi T, Mattfeld T, Schoenberg M, Beger H. Nuclear accumulation of p53 correlates significantly with clinical features and inversely with the expression of the cyclin-dependent kinase inhibitor p21WAF1/CIP1 in pancreatic cancer. Br J Cancer. 1997;76:299–305. doi: 10.1038/bjc.1997.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 39.Helin K, Harlow E. The retinoblastoma protein as a transcriptional repressor. Trends Cell Biol. 1993;3:43–46. doi: 10.1016/0962-8924(93)90150-y. [DOI] [PubMed] [Google Scholar]
- 40.Hengst L, Reed S I. Translation control of p27KIP1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- 41.Herman J G, Merlo A, Lapidus R G, Issa J-P, Davidson N E, Sidransky D, Baylin S B. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 42.Horowitz J M, Park S H, Bogenmann E, Cheng J C, Yandell D W, Kaye F J, Minna J D, Dryja T P, Weinberg R A. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc Natl Acad Sci USA. 1990;87:2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang H, Chou H S, Zhu L. Requirement of cyclin E-Cdk2 inhibition in p16INK4a-mediated growth suppression. Mol Cell Biol. 1998;18:5284–5290. doi: 10.1128/mcb.18.9.5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones D L, Alani R M, Munger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamb A, Shattuck-Eidens D, Eeles R, Liu Q, Gruis N A, Ding W, Hussey C, Tran T, Miki Y, Weaver-Feldhaus J, McClure M, Aitken J F, Anderson D E, Bergman W, Frants R, Goldgar D E, Green A, MacLennan R, Martin N G, Meyer L J, Youl P, Zone J J, Scolnick M H, Cannon-Albright L A. Analysis of the p16 gene (cdkN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994;8:22–26. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- 46.Kamb A, Gruis N A, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian S V, Stockert E, Day R S, Johnson B E, Skolnick M H. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 47.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashburn R A, Grosveld G, Sherr C J. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–660. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 48.Kato D, Miyazawa K, Ruas M, Starborg M, Wada I, Oka T, Sakai T, Peters G, Hara E. Features of replicative senescence induced by direct addition of antennapedia-p16INK4A fusion protein to human diploid fibroblasts. FEBS Lett. 1998;427:203–208. doi: 10.1016/s0014-5793(98)00426-8. [DOI] [PubMed] [Google Scholar]
- 49.Keyomarsi K, Sandoval L, Band V, Pardee A. Synchronization of tumor and normal cells from G1 to multiple cell cycles by lovastatin. Cancer Res. 1991;51:3602–3609. [PubMed] [Google Scholar]
- 50.Kiyokawa H, Kineman R D, Manova-Todorova K O, Soares V C, Hoffman E S, Ono M, Khanam D, Hayday A C, Frohman L A, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27Kip1. Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 51.Knoblich J, Sauer K, Jones L, Richardson H, Saint R, Lehner C. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 52.Koh J, Enders G H, Dynlacht B D, Harlow E. Tumor-derived p16 alleles encoding proteins defective in cell cycle inhibition. Nature. 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 53.LaBaer J, Garret M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. New functional activities for the p21 family of cdk inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 54.Lane M E, Sauer K, Wallace K, Jan Y N, Lehner C F, Vaessin H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- 55.Lees E, Faha B, Dulic V, Reed S I, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- 56.Liggett W H, Jr, Sewell D A, Rocco J, Ahrendt S A, Koch W, Sidransky D. p16 and p16 beta are potent growth suppressors of head and neck squamous carcinoma cells in vitro. Cancer Res. 1996;56:4119–4123. [PubMed] [Google Scholar]
- 57.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin K, Reed S I, Bartek J. Cyclin E-induced S phase without activation of the pRB/E2F pathway. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 58.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent inhibition by the tumor-suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 59.Lundberg A S, Weinberg R A. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo Y, Hurwitz J, Massague J. Cell-cycle inhibition by independent CDK and PCNA binding domains in p21Cip1. Nature. 1995;375:159–161. doi: 10.1038/375159a0. [DOI] [PubMed] [Google Scholar]
- 61.Malkowicz S B, Tomaszewski J E, Linnenbach A J, Cangiano T A, Maruta Y, McGarvey T W. Novel p21WAF1/CIP1 mutations in superficial and invasive transitional cell carcinomas. Oncogene. 1996;13:1831–1837. [PubMed] [Google Scholar]
- 62.Mann D J, Jones N C. E2F-1 but not E2F-4 can overcome p16-induced G1 cell cycle arrest. Curr Biol. 1996;6:474–483. doi: 10.1016/s0960-9822(02)00515-8. [DOI] [PubMed] [Google Scholar]
- 63.Mao L, Merlo A, Bedi G, Shapiro G I, Edwards C D, Rollins B J, Sidransky D. A novel p16INK4A transcript. Cancer Res. 1995;55:2995–2997. [PubMed] [Google Scholar]
- 64.Mathews M, Sonnenburg N, Hershey J. Origins and targets of translational control. In: Hershey J, Mathews M, Sonnenburg N, editors. Translational control. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 1–29. [Google Scholar]
- 65.McConnell B B, Gregory F J, Stott F J, Hara E, Peters G. Induced expression of p16INK4a inhibits both CDK4- and CDK2-associated kinase activity by reassortment of cyclin-CDK-inhibitor complexes. Mol Cell Biol. 1999;19:1981–1989. doi: 10.1128/mcb.19.3.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McConnell B B, Starborg M, Brookes S, Peters G. Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr Biol. 1998;8:351–354. doi: 10.1016/s0960-9822(98)70137-x. [DOI] [PubMed] [Google Scholar]
- 67.Medema R H, Herrera R E, Lam F, Weinberg R A. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc Natl Acad Sci USA. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Millard S S, Yan J S, Nguyen H, Pagano M, Kiyokawa H, Koff A. Enhanced ribosomal association of p27Kip1 mRNA is a mechanism contributing to accumulation during growth arrest. J Biol Chem. 1997;272:7093–7098. doi: 10.1074/jbc.272.11.7093. [DOI] [PubMed] [Google Scholar]
- 70.Missero C, Di Cunto F, Kiyokawa H, Koff A, Dotto G P. The absence of p21Cip1/WAF1 alters keratinocyte growth and differentiation. Genes Dev. 1996;10:3065–3075. doi: 10.1101/gad.10.23.3065. [DOI] [PubMed] [Google Scholar]
- 70a.Mitra, J., and G. H. Enders. Unpublished data.
- 71.Myohanen S K, Baylin S B, Herman J G. Hypermethylation can selectively silence individual p16ink4A alleles in neoplasia. Cancer Res. 1998;58:591–593. [PubMed] [Google Scholar]
- 72.Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh D Y. Mice lacking p27Kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 73.Nobori T, Miura K, Wu D J, Lois A, Takabayashi K, Carson D A. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- 74.Noda A, Ning Y, Venable S F, Pereira-Smith O M, Smith J R. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;211:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 75.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Polyak K, Lee M-H, Erdjument-Bromage H, Koff A, Roberts J M, Tempst P, Massague J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 77.Polymenis M, Schmidt E V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 79.Poon R Y C, Hunter T. Dephosphorylation of Cdk2 Thr-160 by the cyclin-dependent kinase-interacting phosphatase KAP in the absence of cyclin. Science. 1995;270:90–93. doi: 10.1126/science.270.5233.90. [DOI] [PubMed] [Google Scholar]
- 80.Quelle D E, Cheng M, Ashmun R A, Sherr C J. Cancer-associated mutations at the INK4a locus cancel cell cycle arrest by p16INK4a but not by the alternative reading frame protein p19ARF. Proc Natl Acad Sci USA. 1997;94:669–673. doi: 10.1073/pnas.94.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 82.Reynisdottir I, Massague J. The subcellular locations of p15Ink4b and p27Kip1 coordinate their inhibitory interactions with cdk4 and cdk2. Genes Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- 83.Reynisdottir I, Polyak K, Iavarone A, Massague J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-β. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 84.Reznikoff C A, Yeager T R, Belair C D, Savelia E, Puthenveetil J A, Stadler W M. Elevated p16 at senescence and loss of p16 at immortalization in human papillomovirus 16 E6, but not E7, transformed human uroepithelial cells. Cancer Res. 1996;56:2886–2890. [PubMed] [Google Scholar]
- 85.Robertson K D, Jones P A. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rogan E M, Bryan T M, Hukku B, Maclean K, Chang A C-M, Moy E L, Englezou A, Warneford S G, Dalla-Pozza L, Reddel R R. Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li-Fraumeni syndrome fibroblasts. Mol Cell Biol. 1995;15:4745–4753. doi: 10.1128/mcb.15.9.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rogatsky I, Trowbridge J M, Garabedian M J. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol Cell Biol. 1997;17:3181–3193. doi: 10.1128/mcb.17.6.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Royzman I, Whittaker A J, Orr-Weaver T L. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev. 1997;11:1999–2011. doi: 10.1101/gad.11.15.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Russo A, Tong L, Lee J-O, Jeffrey P, Pavletich N. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 90.Russo A A, Jeffrey P D, Patten A K, Massague J, Pavletich N P. Crystal structure of the p27Kip1 cyclin dependent kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 91.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 92.Sandig V, Brand K, Herwig S, Lukas J, Bartek J, Strauss M. Adenovirally transferred p16INK4/CDKN2 and p53 cooperate to induce apoptotic tumor cell death. Nat Med. 1997;3:313–319. doi: 10.1038/nm0397-313. [DOI] [PubMed] [Google Scholar]
- 93.Satyamoorthy K, Nesbit M, Hsu M-Y, Herlyn M. Utility of adenoviruses as gene expression modules in melanoma. In: Maio M, editor. Immunology of human melanoma. Amsterdam, The Netherlands: IOS Press; 1996. pp. 71–77. [Google Scholar]
- 94.Satyamoorthy K, Soballe P W, Soans F, Herlyn M. Adenovirus infection enhances killing of melanoma cells by a mitotoxin. Cancer Res. 1997;57:1873–1876. [PubMed] [Google Scholar]
- 95.Serrano M, Gomez-Lahoz E, DePinho R A, Beach D, Bar-Sagi D. Inhibition of ras-induced proliferation and cellular transformation by p16INK4. Science. 1995;267:249–252. doi: 10.1126/science.7809631. [DOI] [PubMed] [Google Scholar]
- 96.Serrano M, Hannon G J, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 97.Sewing A, Wiseman B, Lloyd A C, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 99.Soos T J, Kiyokawa H, Yan J S, Rubin M S, Giordano A, DeBlasio A, Bottega S, Wong B, Mendelsohn J, Koff A. Formation of p27-CDK complexes during the human mitotic cell cycle. Cell Growth Differ. 1996;7:135–146. [PubMed] [Google Scholar]
- 100.Stone S, Jiang P, Dayananth P, Tavtigian S V, Katcher H, Parry D, Peters G, Kamb A. Complex structure and regulation of the P16 (MTS1) locus. Cancer Res. 1995;55:2988–2994. [PubMed] [Google Scholar]
- 101.Tsai L-H, Lees E, Faha B, Harlow E, Riabowol K. The cdk2 kinase is required for the G1-to-S transition in mammalian cells. Oncogene. 1993;8:1593–1602. [PubMed] [Google Scholar]
- 102.Uhrbom L, Nister M, Westermark B. Induction of senescence in human malignant glioma cells by p16INK4A. Oncogene. 1997;15:505–514. doi: 10.1038/sj.onc.1201227. [DOI] [PubMed] [Google Scholar]
- 103.van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 104.Waldman T, Kinzler K W, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 105.Wang J, Walsh K. Resistance to apoptosis conferred by Cdk inhibitors during myocyte differentiation. Science. 1996;273:359–361. doi: 10.1126/science.273.5273.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 107.Wu C-L, Classon M, Dyson N, Harlow E. Expression of dominant-negative mutant DP-1 blocks cell cycle progression in G1. Mol Cell Biol. 1996;16:3698–3706. doi: 10.1128/mcb.16.7.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiao Z-H, Ginsberg D, Ewen M, Livingston D M. Regulation of the retinoblastoma protein-related protein p107 by G1 cyclin-associated kinases. Proc Natl Acad Sci USA. 1996;93:4633–4637. doi: 10.1073/pnas.93.10.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xiong Y, Zhang H, Beach D. Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev. 1993;7:1572–1583. doi: 10.1101/gad.7.8.1572. [DOI] [PubMed] [Google Scholar]
- 110.Zhang H, Hannon G J, Beach D. p21-containing cyclin kinases exist in both active and inactive states. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- 111.Zhang Y, Xiong Y, Yarbrough W G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 112.Zindy F, Eischen C, Randle D, Kamijo T, Cleveland J, Sherr C, Roussel M. Myc signalling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zindy F, Quelle D E, Roussel M F, Sherr C J. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]