ABSTRACT
In this study, we sought to determine whether an in vivo assay for studying antibiotic mechanisms of action could provide insight into the activity of compounds that may inhibit multiple targets. Thus, we conducted an activity screen of 31 structural analogs of rhodanine-containing pan-assay interference compounds (PAINS). We identified nine active molecules against Escherichia coli and classified them according to their in vivo mechanisms of action. The mechanisms of action of PAINS are generally difficult to identify due to their promiscuity. However, we leveraged bacterial cytological profiling, a fluorescence microscopy technique, to study these complex mechanisms. Ultimately, we found that although some of our molecules promiscuously inhibit multiple cellular pathways, a few molecules specifically inhibit DNA replication despite structural similarity to related PAINS. A genetic analysis of resistant mutants revealed thymidylate kinase (essential for DNA synthesis) as an intracellular target of some of these rhodanine-containing antibiotics. This finding was supported by in vitro activity assays, as well as experiments utilizing a thymidylate kinase overexpression system. The analog that demonstrated the half-maximal inhibitory concentration in vitro and MIC in vivo displayed the greatest specificity for inhibition of the DNA replication pathway, despite containing a rhodamine moiety. Although it is thought that PAINS cannot be developed as antibiotics, this work showcases novel inhibitors of E. coli thymidylate kinase. Moreover, perhaps more importantly, this work highlights the utility of bacterial cytological profiling for studying the in vivo specificity of antibiotics and demonstrates that bacterial cytological profiling can identify multiple pathways that are inhibited by an individual molecule.
IMPORTANCE We demonstrate that bacterial cytological profiling is a powerful tool for directing antibiotic discovery efforts because it can be used to determine the specificity of an antibiotic’s in vivo mechanism of action. By assaying analogs of PAINS, molecules that are notoriously intractable and nonspecific, we (surprisingly) identify molecules with specific activity against E. coli thymidylate kinase. This suggests that structural modifications to PAINS can confer stronger inhibition by targeting a specific cellular pathway. While in vitro inhibition assays are susceptible to false-positive results (especially from PAINS), bacterial cytological profiling provides the resolution to identify molecules with specific in vivo activity.
KEYWORDS: E. coli, PAINS, SAR, antibiotic, bacterial cytological profiling, mechanism
INTRODUCTION
Due to the increasing emergence of multidrug-resistant (MDR) bacteria, there is a desperate need for new antibiotics to effectively treat persistent infections (1–3). In 2015, ceftazidime-avibactam-resistant Klebsiella pneumoniae was identified within months of the antibiotic’s approval for use in the clinic (4). In addition, as the rate at which pathogens are developing resistance to clinically administered antibiotics continues to increase, many companies once actively involved in antibiotic discovery are abandoning these efforts (5–11). Consequently, the speed at which antibiotics are achieving clinical status is not fast enough to keep pace with evolving resistance (9–11). For this reason, the continued research and development of potential drug candidates is critical for combatting one of the greatest threats to global health today.
Vast libraries of natural products and/or synthetic molecules are frequently screened in order to identify those with antibacterial activity (12). These screens often yield a number of active molecules; however, potent inhibitors are rarely pursued if their mechanisms of action (MOA) are difficult to classify. Consequently, many of these molecules do not make it to the development stage of antibiotic research (13). Pan-assay interference compounds (PAINS) constitute a structurally diverse class of molecules that contain highly reactive functional groups (such as toxoflavin, isothiazolone, curcumin, hydroxyphenol hydrazine, phenol-sulfonamide, or rhodanine) that frequently interfere with the integrity of biological assays and yield false-positive results (14). Their tendency to react with or bind promiscuously to many different targets makes PAINS notoriously difficult to study and, for this reason, they are often discarded from antibiotic investigations (15, 16). In fact, filters have been developed for removing PAINS from compound libraries prior to screening (17). Consequently, many PAINS remain unstudied.
Previously, we developed bacterial cytological profiling (BCP) as a method for rapidly pinpointing the cellular pathway(s) targeted by an antibiotic (18, 19). BCP, which has been used to ascertain the MOA of a variety of compounds active against Gram-negative and Gram-positive bacteria, is predicated on the different cytological changes that occur in cells exposed to antibiotics with various mechanisms (18–22). For example, inhibitors that strongly block protein synthesis (i.e., tetracycline) cause the DNA to assume a toroidal structure, while inhibitors of transcription (i.e., rifampin) cause the DNA to decondense (see Fig. S1 in the supplemental material). Inhibitors of cell wall biogenesis induce cell shape defects and cause lysis, while antibiotics affecting DNA replication (i.e., ciprofloxacin) yield elongated cells that contain a single, centrally located nucleoid. We previously demonstrated that the inhibition of multiple pathways within a single cell results in a combination of distinguishable phenotypes (22, 23). Thus, BCP enables us to study mechanistically complex molecules, which would otherwise present an insurmountable challenge due to their multiple cellular targets or general reactivity. When used for analyses of structure-activity relationships (SAR), BCP facilitates the identification of molecular analogs that have a greater specificity for a single target pathway.
Here, we conducted an antibiotic screen and identified a chemical series of PAINS-related molecules with antibacterial activity against an Escherichia coli ΔtolC strain. With the understanding that PAINS have little direct clinical relevance, we pursued these molecules in order to validate BCP as an approach for studying mechanistically complex antibiotics. Using BCP, we discovered that some PAINS-related molecules inhibit multiple pathways in vivo, but other structural analogs exhibited high specificity for the DNA synthesis pathway. In vivo and in vitro experiments revealed that some of these molecules specifically inhibit thymidylate kinase (TMPK), an essential enzyme that catalyzes the synthesis of dTDP from dTMP (24). Conserved in many bacterial species, TMPK is a promising target for new antibiotics (25). Thus, our study identifies potentially powerful cell biology tools: molecules with specific activity against TMPK in an isolated E. coli system, as well as a novel application of BCP for studying families of antibiotics that have long been considered too much of a “pain” to study.
RESULTS AND DISCUSSION
Characterizing the cellular pathways inhibited by two PAINS using BCP and viability studies.
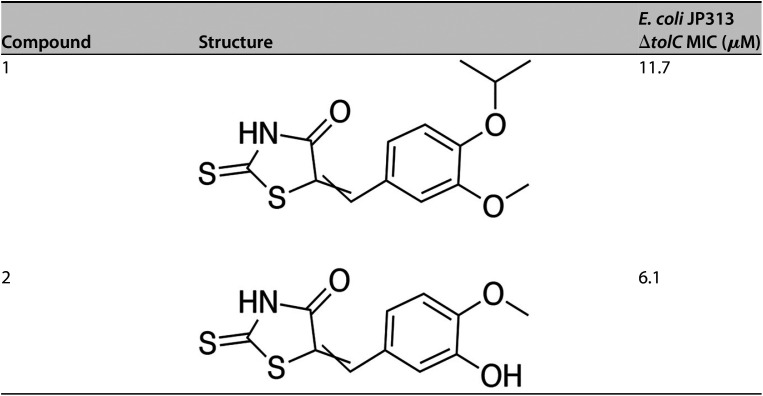
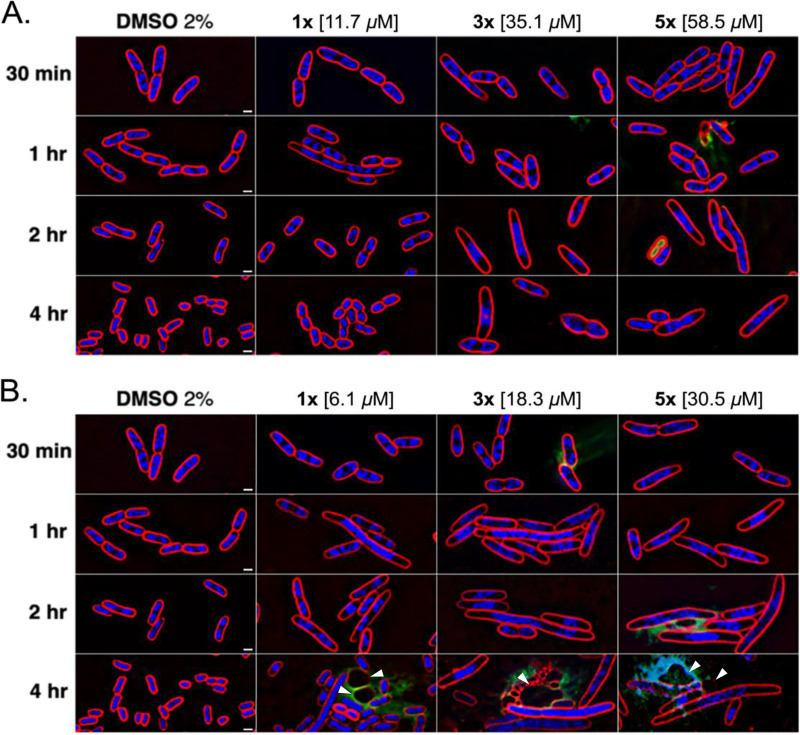
A library of 1,798 synthetic small molecules was screened to identify those that inhibit the growth of the E. coli ΔtolC strain. In conducting this screen, we identified two active compounds that shared a similar backbone structure containing a rhodanine moiety, suggesting that they might be PAINS. These molecules, designated compound 1 and compound 2, are shown in Table 1. The MICs of compounds 1 and 2 against the E. coli ΔtolC strain were determined to be 11.7 and 6.1 μM, respectively. To rapidly identify the pathway(s) inhibited by these compounds, we performed BCP. E. coli ΔtolC cells were treated with each compound at concentrations equivalent to 1×, 3×, and 5× MIC for durations of 30 min, 1, 2, and 4 h. After treatment, the cells were stained with fluorescent dyes and imaged using high-resolution fluorescence microscopy (Fig. 1). Cells treated with compound 1 at 3× and 5× MIC for 2 h or more were elongated and contained a single, centrally located nucleoid (Fig. 1A), resembling cells treated with known DNA replication inhibitors (see Fig. S1) (19–22). At these concentrations, some cells also appeared to be swollen or lysed, typical of cells treated with cell wall inhibitors. Compound 2 also induced DNA replication defects after 1 h of treatment at all tested concentrations, and a severe degree of cell lysis and spheroplast formation were observed after 4 h of treatment (Fig. 1B, arrows). Thus, consistent with the knowledge that PAINS often target multiple pathways, compounds 1 and 2 inhibited two distinct metabolic processes, DNA replication and cell wall biogenesis.
TABLE 1.
Structures and MICs of PAINS compounds 1 and 2
FIG 1.
Cytological profiles of the E. coli JP313 ΔtolC strain treated with two PAINS. Cells were exposed to compound 1 (A) or compound 2 (B) at 1×, 3×, and 5× MIC for 30 min, 1 h, 2 h, and 4 h. Images were taken after staining the cells with FM4-64 (red), DAPI (blue), and SYTOX-green (green). Arrowheads mark spheroplasts indicative of inhibited cell wall biogenesis. Scale bars, 1 μm.
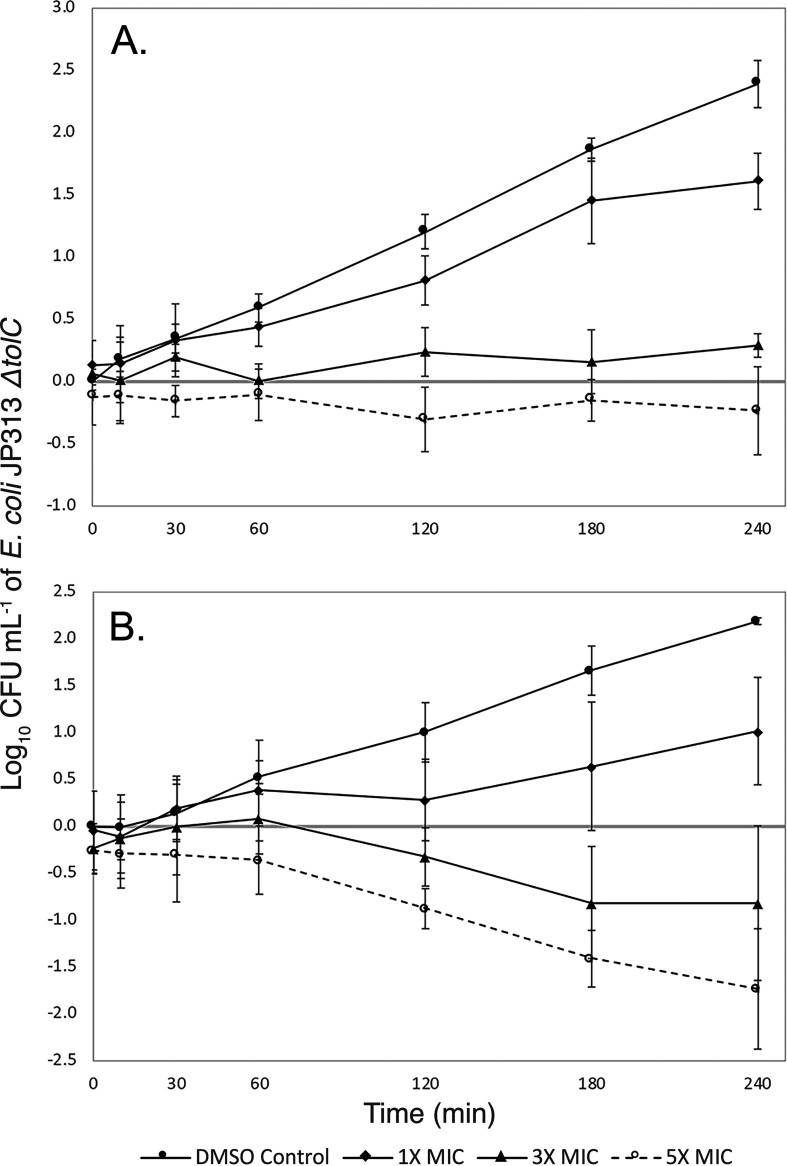
Our viability experiments further characterized the antibacterial effects of compounds 1 and 2 against E. coli ΔtolC. Compound 1 appeared to be largely bacteriostatic, for even when cells were treated at 5× MIC, viability never decreased considerably (Fig. 2A). Compound 2, on the other hand, caused more than a 10-fold average reduction in the number of viable E. coli ΔtolC cells after treatment at 5× MIC (Fig. 2B). These data are consistent with our BCP observations because, while both compounds inhibited DNA replication, compound 2 caused more significant cell lysis and appeared to inhibit cell wall biogenesis to a much greater degree (Fig. 1B, arrows).
FIG 2.
Viability of the E. coli JP313 ΔtolC strain treated with two PAINS. Cells were treated with various concentrations of compound 1 (A) or compound 2 (B). CFU/ml was measured at 30 min, 1 h, 2 h, 3 h, and 4 h. Standard errors were calculated from three independent trials.
Identifying TMPK as a cellular target by isolating resistant mutations and performing in silico docking.
Though we suspected that compounds 1 and 2 inhibited the DNA replication pathway, we did not know which protein within the pathway was inhibited. To identify a potential enzymatic target, we passaged E. coli ΔtolC serially in the presence of increasing concentrations of both compounds and obtained one resistant mutant for each compound. The MIC of compound 1 against its corresponding resistant mutant (mutant 1) was six times higher than the MIC against the E. coli ΔtolC parent strain, and mutant 1 was also resistant (∼2.8-fold) to compound 2 (Table 2). Mutant 2 (from compound 2) was roughly 10-fold resistant to both compounds. Whole-genome sequencing revealed that both resistant mutants contained missense mutations in the tmk gene, which codes for TMPK (see Fig. S2). In mutant 1, proline replaced glutamine at residue 45, and in mutant 2, threonine replaced alanine at residue 69. Mutant 1 contained an additional mutation in the dksA gene, and mutant 2 contained an additional mutation in the yjeA gene. However, because tmk was the only gene that was mutated in both strains and because cross-resistance was conferred, we suspected that these mutated tmk alleles were likely responsible for conferring some resistance to the compounds. Therefore, we moved the tmk allele from the more resistant mutant into a clean strain background via phage P1 transduction. In isolation, the tmk(A69T) allele conferred 6-fold resistance to compound 1 and 2.4-fold resistance to compound 2 (Table 2). These results suggest that TMPK is a cellular target of both compounds in vivo.
TABLE 2.
MICs of compounds 1 and 2 against resistant mutants
| Strain | MIC (μM) |
|
|---|---|---|
| Compound 1 | Compound 2 | |
| E. coli JP313 ΔtolC | 11.7 | 6.1 |
| E. coli JP313 ΔtolC mutant 1 | 75.0 | 17.2 |
| E. coli JP313 ΔtolC mutant 2 | >100 | 81.0 |
| E. coli JP313 ΔtolC tmk(A69T) | 68.8 | 14.8 |
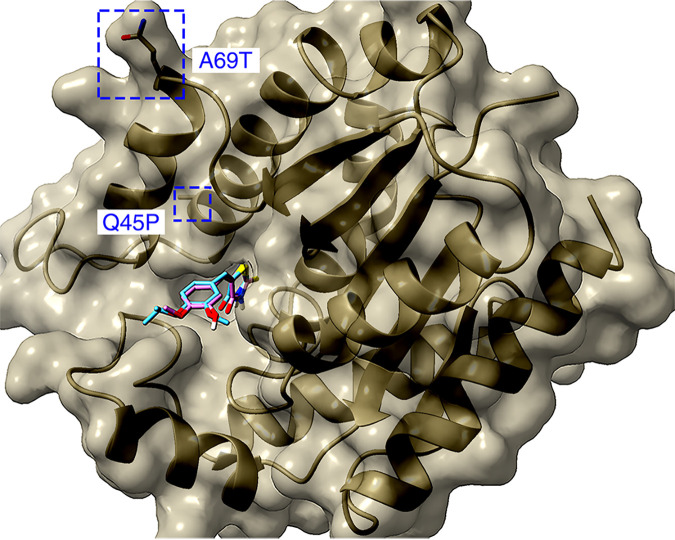
In order to probe the potential molecular mechanism of our compounds, we used computational modeling to dock compounds 1 and 2 to TMPK (Fig. 3). This analysis revealed binding sites for compounds 1 and 2 that overlap considerably within the TMPK active site. The high-probability binding orientation of our inhibitors closely mimicked that of the substrate (dTMP) and known analog inhibitors (26, 27). It is likely that by binding at this location, compounds 1 and 2 compete with dTMP binding and block the enzyme’s catalytic activity. Neither of the resistant mutations that we selected was located in the TMPK active site, and it remains unclear how these mutations confer resistance to compounds 1 and 2.
FIG 3.
Compounds 1 and 2 docked to the active site of TMPK. Compound 1 (cyan) and compound 2 (magenta) are shown overlapping. Mutated residues conferring resistance are labeled in blue. Heteroatoms are colored such that sulfur is yellow, oxygen is red, and nitrogen is blue.
Screening analogs of compounds 1 and 2.
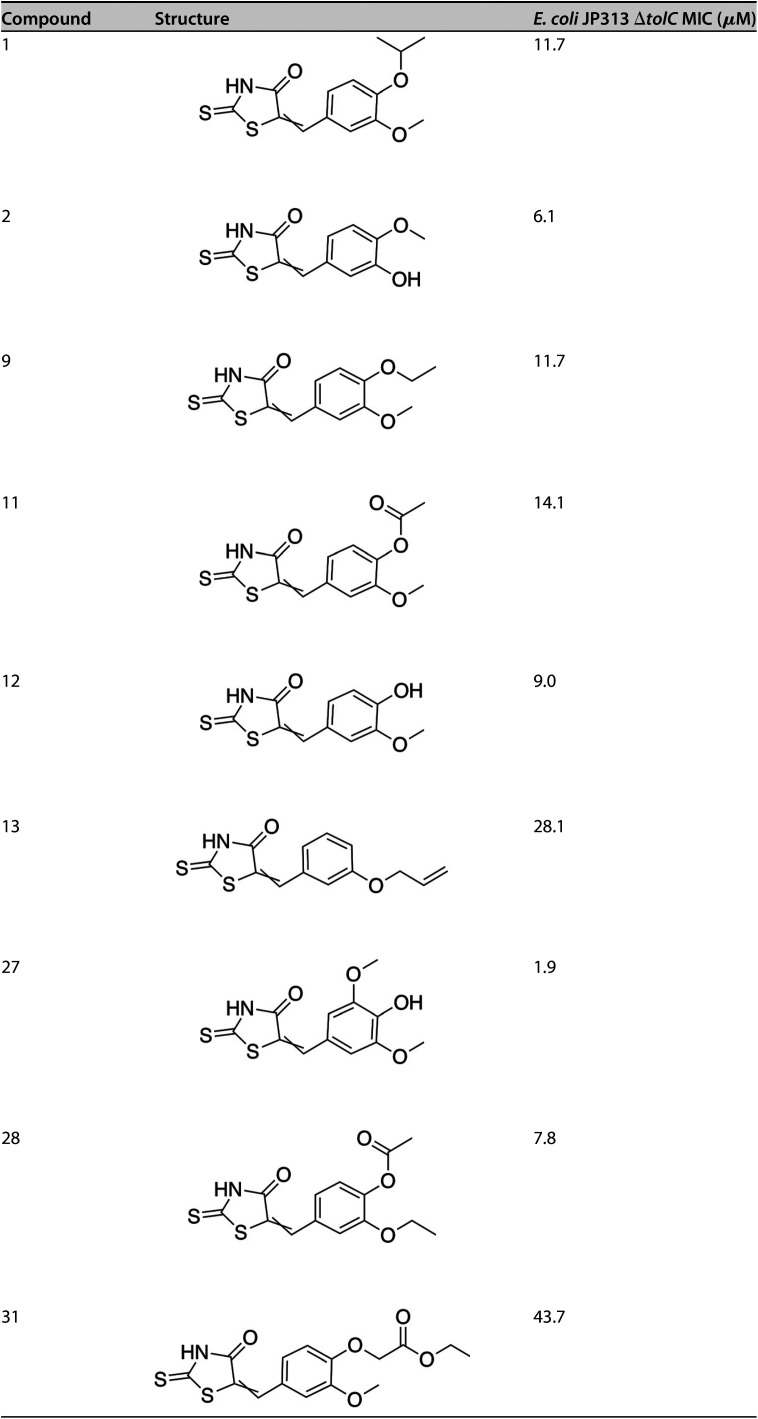
In order to identify specific inhibitors of TMPK, we screened 29 structural analogs of compounds 1 and 2 for their antibacterial activity and MOA against the E. coli ΔtolC mutant (see Fig. S3). Of these analogs, seven showed antibacterial activity with MICs less than 50 μM (Table 3; see also Table S1 in the supplemental material). Compound 27 was the most potent, with an MIC of 1.9 μM. BCP was used to determine whether these analogs inhibit DNA replication specifically or have other mechanisms of action (Fig. 4). Many of the compounds induced filamentation and chromosomal replication defects, indicating that they inhibit DNA replication (Fig. 4), but most of these molecules also inhibited other pathways, such as cell wall biogenesis, which was evidenced by cell lysis. Compound 13 induced membrane permeability and cell lysis with no sign of inhibited DNA replication. This compound had a relatively high MIC compared to most other active molecules. In addition to nonspecific inhibitors, we identified three molecules (compounds 9, 11, and 27) that selectively inhibited DNA replication. The molecule with perhaps the most promise as a TMPK inhibitor was compound 27, since it displayed the lowest MIC and a clear cytological profile indicating high specificity for the DNA replication pathway.
TABLE 3.
Structures and MICs of PAINS analogs
FIG 4.
Cytological profiles of the E. coli JP313 ΔtolC strain treated with PAINS analogs. E. coli JP313 ΔtolC cells were treated with 2% DMSO as a control. Analogs were administered at 5× MIC (concentrations shown on the left), and images were taken after 30 min and 2 h. Scale bars, 1 μm.
Testing mutant and plasmid-overexpressing strains for resistance to analogs.
To provide evidence that TMPK is the molecular target of some of our PAINS analogs, we tested the activity of these molecules against the tmk(A69T) mutant strain. This mutant exhibited some degree of resistance to all of the analogs (Table 4). The MIC of compound 27 was most drastically increased (10-fold) in the E. coli strain containing tmk(A69T), further suggesting that this analog specifically inhibits thymidylate kinase in vivo. Conversely, those analogs that appeared not to inhibit DNA replication as strongly since compound 27 showed only a modest increase in MIC against the tmk(A69T) strain. For example, the MICs of compounds 13 and 31 were only ∼2-fold higher against this strain compared to the strain with wild-type tmk.
TABLE 4.
MICs of PAINS analogs against E. coli ΔtolC tmk(A69T) and plasmid-overexpressing strains, as well as IC50 values for the inhibition of E. coli TMPK in vitro
| Compounda | Allele-conferred resistance (μM) |
Plasmid overexpression (μM) |
In vitro inhibition (μM) |
|||||
|---|---|---|---|---|---|---|---|---|
|
E. coli
ΔtolC tmk + |
E. coli ΔtolC tmk(A69T) |
Conferred fold resistance |
Vector control |
Plasmid+ tmk |
Plasmid+ tmk(Q45P) |
Plasmid+ tmk(A69T) |
E. coli TMPK IC50 | |
| 1 | 11.7 | 68.8 | 5.9 | 15.5 | 62 | 62 | 62 | 0.39 |
| 2 | 6.1 | 14.8 | 2.4 | 7.8 | 31 | 62 | 62 | 2.50 |
| 8a | >100 | 1.20 | ||||||
| 9 | 11.7 | 43.8 | 3.7 | 15.5 | 62 | 62 | 62 | 0.41 |
| 11 | 14.1 | 37.5 | 2.7 | 15.5 | 62 | >62 | >62 | 0.48 |
| 12 | 9.0 | 20.3 | 2.3 | 7.8 | 31 | >62 | >62 | 0.50 |
| 13 | 28.1 | 56.3 | 2.0 | 31.3 | 62.5 | 62.5 | 62.5 | 1.00 |
| 14* | >100 | 4.20 | ||||||
| 16* | >100 | 5.60 | ||||||
| 27 | 1.9 | 18.8 | 10 | 3.9 | 15.5 | 15.5 | 15.5 | 0.25 |
| 28 | 7.8 | 19.4 | 2.5 | 7.8 | 31.3 | 64 | 64 | 0.47 |
| 31 | 43.7 | 93.8 | 2.1 | 31.3 | 125 | 125 | 125 | 0.68 |
*, inactive in vivo.
To provide additional evidence that thymidylate kinase might be the target of this family of compounds, we tested whether TMPK overexpression conferred resistance. To accomplish this, we cloned the wild-type tmk gene, as well as the tmk(A69T) and tmk(Q45P) resistant alleles, on multicopy number plasmids and measured the MICs of all nine of our active compounds against the E. coli ΔtolC mutant transformed with these plasmids. Overexpression of the wild-type or mutant tmk genes resulted in at least a 4-fold increase in the MICs of all compounds with BCP phenotypes characteristic of DNA replication inhibitors (Table 4). In the cases of compounds 2, 12, and 28, an even greater increase in MIC (up to 8-fold) was observed when either tmk(A69T) or tmk(Q45P) was overexpressed. In contrast, the MIC of compound 13, which had no obvious effect on DNA replication in vivo, was increased only 2-fold upon overexpression of tmk alleles.
PAINS analogs inhibit thymidylate kinase in vitro.
Because our genetic and cell biology studies suggested that thymidylate kinase is a target of this family of molecules, we determined whether or not these compounds inhibited the purified E. coli thymidylate kinase in vitro. We obtained 50% half-maximal concentrations (IC50; indicating 50% inhibition of purified TMPK activity in vitro) for all nine active compounds and also for three compounds with no activity against the E. coli ΔtolC mutant (compounds 8, 14, and 16) (Table 4). The inactive compounds studied were chosen on the basis of their structural diversity. The IC50 values for all 12 compounds ranged from 250 nM (compound 27) to 5.6 μM (compound 16) (Table 4). As might be expected, the three compounds for which MICs could not be measured had some of the highest IC50 values and yielded poor dose-response curves (see Fig. S4). Compound 27, which is the most potent and specific inhibitor in vivo, displayed the lowest IC50. Despite this, our data seemed to demonstrate only a weak positive correlation between compound MIC and IC50. This correlation was primarily affected by the unexpectedly high IC50 value associated with compound 2, one of the strongest inhibitors in vivo. The r2 value for the linear correlation between IC50 values and MICs was 0.001; however, the removal of compound 2 yielded a significantly improved value of 0.536 (see Fig. S5A and B). In addition, an analysis of the relationship between IC50 values and MICs using logistic regression yielded a pseudo r2 value of 0.246 with the inclusion of compound 2 but achieved perfect separation (pseudo r2 value of 1.0) when compound 2 was excluded (see Fig. S5C and D). We suspect that compound 2 disrupts the correlation between IC50 values and MICs because it inhibits non-TMPK targets with greater potency than the other molecules. Of the compounds that appeared based on BCP to inhibit cell wall biogenesis, compound 2 was the most potent. In addition, the tmk(A69T) mutation conferred only 2.4-fold resistance against compound 2, whereas mutant 2 which contained this mutation and one additional mutation in yjeA was >13-fold resistant. Together, these data suggest that TMPK is likely not the primary target of compound 2 and therefore that compound 2 should not be expected to contribute to the correlation between MICs and IC50 values against purified TMPK. That being said, inconsistencies between MICs and IC50 values could potentially be explained by other factors. For instance, the cell envelope is a significant barrier to antibiotic infiltration, and different structural features of our antibiotic analogs could affect their ability to enter the cell (28). In addition, some of these molecules might behave as PAINS and yield misleading data when studied using in vitro assays such as this one.
Here, we present a collection of genetic, cell biological, and biochemical data that demonstrate that many of the molecules within a particular rhodanine-containing chemical series inhibit cellular TMPK. Although some of these molecules have additional targets in vivo, three compounds appeared to specifically target TMPK. Compound 27, stands out among this group because, in addition to its specificity (eliciting clear DNA replication defects in vivo), it had the most potent IC50 against the purified protein in vitro and inhibited the growth of the E. coli ΔtolC mutant with the lowest MIC. It may be the case that PAINS cannot be developed as antibiotics, for even if compounds with specific antibacterial targets can be developed, any given compound may have toxic targets in the human host. This study nonetheless highlights the utility of combining an in vivo MOA assay with medicinal chemistry to study the antibacterial mechanisms of molecules.
MATERIALS AND METHODS
Construction of the E. coli JP313 ΔtolC mutant.
The ΔtolC mutation is derived from strain PB3 and was introduced into strain JP313 by P1 transduction (29, 30). JP313 was transduced to tetracycline resistance with a lysate of strain CAG18475 (metC162::Tn10), and the methionine requirement of the transductants was confirmed. This strain was then transduced to prototrophy with a lysate of PB3, and these transductants were screened on MacConkey agar for the presence of the ΔtolC mutation. Transductants that acquired ΔtolC were unable to grow on this medium owing to their sensitivity to bile salts such as deoxycholate, and these arose at about 6%, in keeping with previous linkage data (29, 31). PB3 and CAG18475 were obtained from the Coli Genetic Stock Center at Yale University.
Synthetic compound library screen.
A synthetic small molecule library consisting of ∼1,800 compounds was obtained from the ChemBridge EXPRESS-pick library stock collection and screened in 96-well plates at 100 μg/ml in Luria-Bertani (LB) medium to identify compounds active against the E. coli ΔtolC mutant.
Determining MICs.
MICs were determined using the broth microdilution method performed in triplicate. All compounds used in this study were purchased from ChemBridge and solubilized in dimethyl sulfoxide (DMSO). Concentrated stocks of each compound were prepared at 20 mM and stored at −80°C. A 1:100 dilution of an overnight culture of E. coli JP313 ΔtolC mutant was prepared in LB liquid medium and grown at 30°C to an optical density at 600 nm (OD600) between 0.2 and 0.4. With the exception of a medium control column, 1 μl of cells diluted to an OD600 of 0.05 was added to a 96-well plate containing 2-fold serial dilutions of eight different starting concentrations of each compound in 100 μl of LB medium. An initial cell density count was determined using a plate reader prior to incubation at 30°C for 24 h while shaking. After 24 h, the optical density was determined, and OD600 readings were corrected by subtracting the initial reading at T0 from the final OD600 reading at T24. Readings of 0.5 and below constituted inhibition.
Bacterial cytological profiling.
High-resolution fluorescence microscopy and bacterial cytological profiling (BCP) were performed as previously described by Nonejuie et al. (19). Briefly, overnight cultures of the E. coli JP313 ΔtolC mutant were diluted 1:100 in LB medium, followed by incubation with rolling at 30°C until they reached an OD600 between 0.15 and 0.17. Next, 300 μl of cells was treated with compounds prepared to the desired test concentration and incubated while rolling at 30°C. Microscopy images were taken after 30 min, 1 h, 2 h, and/or 4 h. Prior to applying the treated culture to an agarose pad for imaging, the cells were stained with FM4-64, DAPI (4′,6′-diamidino-2-phenylindole), and SYTOX-green as previously described (19).
Cell viability.
Viability assays were performed in triplicate for the E. coli JP313 ΔtolC strain treated with compounds 1 and 2. Cells were plated for colony counting at the same time points used for BCP imaging. The plates were incubated at room temperature overnight, then individual colonies were counted. The number of CFU per milliliter was calculated for each treatment condition. These values were then normalized using the T0 measurement of the untreated cells and subsequently log10 transformed. The resulting viability measurements were averaged across three independent trials and plotted with standard errors.
Resistant mutant selection.
Mutants resistant to compounds 1 and 2 were obtained via serial passaging. A 1:500 dilution of an overnight culture of the E. coli JP313 ΔtolC strain was prepared in 6 ml of LB medium and incubated at 30°C with rolling until the cells reached an OD600 of 0.2 to 0.25. In a small glass culture tube, 1 μl of cells was added to 1 ml of LB medium containing compound at a concentration of 0.5× MIC. The cultures were incubated at 30°C for 1 day while rolling. If growth occurred at the starting concentration, the culture was diluted 1,000-fold using LB medium containing a slightly higher concentration of compound. Serial passaging was performed until the highest concentration of each compound was reached, at which point colonies were selected and purified from plates containing the compound. Only one mutant colony from each independent culture was selected for sequencing.
Genomic DNA extraction and quantification.
Genomic DNA was extracted using a protocol adapted for use with a Qiagen DNeasy blood and tissue kit. The gDNA concentration was quantified using a Thermo Scientific NanoDrop One Microvolume UV-Vis spectrophotometer. gDNA was stored at −20°C.
Genome sequencing, assembly, and variant analyses.
The genome sequences of two E. coli JP313 ΔtolC mutants were generated using the Illumina MiSeq sequencing platform with a V2 500 paired-end read kit at the La Jolla Institute for Allergy and Immunology Research, San Diego, CA. All sequence processing and analyses were performed in Geneious Prime. Two FastQ files containing forward and reverse sequence read lists were generated for each genome. The files were imported into Geneious and simultaneously paired (input of expected insert size of 500 bp), resulting in a single paired read list. The sequences were trimmed using the BBDuk v38.37 plugin. BBDuk identifies and trims adapters based on presets for Illumina adapters and paired read overhangs, trims the end of sequences based on quality (Q score), and discards short reads and their associated pair mate. Adapter sequences trimmed using presets for Illumina adapters were trimmed from the right end of the sequence with the Kmer length set at 27 with a maximum substitution allotment of 1. A minimum overlap value of 25 was set for adapter trimming based on paired read overhangs. Low-quality sequences were trimmed on both ends with the minimum quality score set at 30, and reads shorter than 30 bp were discarded. After trimming, the paired-end reads were merged into a single read with BBMerge. The resulting merged and unmerged sequences were mapped to the previously sequenced and annotated genome of the E. coli JP313 ΔtolC parent strain using the Geneious assembler set to medium sensitivity and to find structural variants, short insertions, and deletions of any size. Further variation analysis was performed with the resulting alignment using the Geneious SNP finder, set to use the bacterial genetic code and the following parameters—a minimum variant frequency of 0.25, a maximum variant P value of 6, and a minimum strand-bias P value of 5 when exceeding 65% bias—and to calculate an approximate P value for identified variants. Visual inspection of these resulting data led to the identification of mutations present in each genome.
Primer design and PCR.
Thymidylate kinase DNA templates (∼1,099 bp) were amplified from E. coli JP313 ΔtolC strains using a Q5 high-fidelity DNA PCR (New England Biolabs). The primers used for these reactions were EM029-TMKF1 (5′-ATGGCAAACTTTCTCGTG-3′) and EM030-TMKR1 (5′-GGTGTAGTAATCGGGATG-3′). Each PCR mixture (50 μl) contained 100 ng of template genomic DNA, 500 pmol of each primer, and 200 μM deoxynucleoside triphosphates. The thermocycling conditions were 30 s of initial denaturation at 98°C, followed by 30 cycles of denaturation (98°C, 10 s), annealing (61°C, 15 s), and extension (72°C, 2 min). A final extension was performed at 72°C for 5 min. PCR products were purified with the oligonucleotide cleanup protocol, as described in the Monarch PCR & DNA cleanup kit (5 μg) user manual (NEB, T1030). Purified PCR products were sequenced using Sanger methods by Eton Biosciences and trimmed for quality prior to analysis.
E. coli genome manipulation by P1 virulent transduction.
P1 transductions of the tmk(A69T) allele were carried out according to the protocol described by Miller and Miller (32). The tmk(A69T) mutation was backcrossed into the E. coli JP313 ΔtolC strain by cotransduction with the closely linked fhuE gene. A P1vir lysate of strain JW1088-5 (ΔfhuE764::kan) was used to transduce the original tmk(A69T) mutant to kanamycin resistance, and transductants were screened for retention of resistance to compound 2. A P1vir lysate of one such transductant was then used to transduce the E. coli JP313 ΔtolC strain to kanamycin resistance. Colonies that were also resistant to compound 2, having acquired the tmk(A69T) mutation by cotransduction, represented the desired backcross. JW1088-5 was obtained from the Coli Genetic Stock Center at Yale University.
TMK expression in the E. coli JP313 ΔtolC mutant.
Three full-length E. coli K-12 MG1655 thymidylate kinase alleles (wild type, Q45P, and A69T) were amplified and cloned into the plasmid expression vector pRSFDuet-1 (GenScript). Expression is under the control of the T7 lac promoter. Plasmids were transformed into the E. coli JP313 ΔtolC strain to give the following strains: JP313 ΔtolC-pRSFDuet, JP313 ΔtolC-pRSFDuet-tmk+, JP313 ΔtolC-pRSFDuet-tmk-134, and JP313 ΔtolC-pRSFDuet-tmk-205.
These strains were grown in LB medium in the presence of kanamycin (30 μg/ml) to an OD600 of 0.1 to 0.2 and then treated with the compounds. The assay was performed with eight different concentrations of each compound, serially diluted 2-fold, in 96-well plates in 100 μl of LB medium with kanamycin at 30 μg/ml. Cell densities were determined using a plate reader before and after a 24-h incubation at 30°C with shaking. Final OD600 values were determined by subtracting the values from the initial reading at T0. MICs were determined as the lowest concentration of compound resulting in an OD600 of 0.5 or below, indicating growth inhibition.
Molecular docking.
The protein docking was performed using Autodock Vina (v1.1.2). The crystal structure of E. coli thymidylate kinase (TMPK) was obtained in complex with P1-(5′-adenosyl)-P5-(5′-thymidyl)pentaphosphate and P1-(5′-adenosyl)P5-[5′-(3′-azido-3′-deoxythymidine)] pentaphosphate from the Protein Data Bank (PDB 4TMK) (26). Water molecules, cofactors, and the originally docked ligand were removed, while polar hydrogens and partial charges were added using Autodock Tools (v1.5.6), which is available as a part of MGLtools v1.5.6 through The Scripps Research Institute (http://mgltools.scripps.edu/downloads). Docking was carried out according to the standard procedures for Autodock Vina, and the resulting docking configuration was viewed and illustrated using Chimera X (https://www.rbvi.ucsf.edu/chimerax/) (33).
E. coli TMPK IC50 determination.
Inhibition of E. coli TMPK catalysis was measured in the direction of ATP synthesis using an ATPlite luminescence assay (Perkin-Elmer, catalog no. 6016739) in white 384-well plates (Corning catalog no. 3825). The assay buffer consisted of 50 mM HEPES (pH 8.0), 25 mM sodium acetate, 10 mM MgCl2, 5 mM dithiothreitol, 0.01% Triton X-100, and 0.5 mM EDTA. The dTDP and ADP concentrations were 65 and 10 mM, respectively. The TMPK concentration was 400 pM. Test compounds stock solutions were prepared at 10 μM in DMSO. Serial 2-fold dilutions were prepared in buffer supplemented with DMSO such that the final DMSO concentration in the assay was constant at 0.33% and the highest compound concentration tested was 33 μM. A replicate plate was prepared with no TMPK to serve as the 100% inhibition control and to allow correction for signal suppression by the test compounds, according to the method described by Shapiro et al. (34). The assay volume during the 1-h TMPK reaction was 10 μl. The reactions were quenched by the addition of 5 μl of ATPlite reagent. Background luminescence was measured in wells containing only buffer and ATPlite reagent. Luminescence was measured, after 5 min of incubation in the dark, using a Pherastar FS plate reader (BMG Labtech), with 1-s integration/well.
The IC50 values were calculated as follows. The average luminescence of the background wells was subtracted from the luminescence of all the other wells. The correction for signal suppression was made using the data from the plate with no TMPK. The average uninhibited control luminescence (MAX) was measured in the wells containing TMPK but no inhibitor. The average fully inhibited control (MIN) was measured in the wells containing no inhibitor or TMPK. The percent inhibition at each inhibitor concentration was calculated for each compound using the following equation: % inhibition = 100[1 – (X – MIN)/(MAX – MIN)], where X is the measurement in the well with TMPK and the specific concentration of inhibitor. The IC50 was calculated by nonlinear regression from the % inhibition data using the Excel add-on XLfit (ID Business Solutions, Ltd., UK), equation 205, in which the minimum and maximum percent inhibitions are fixed at 0 and 100, respectively, as follows: % inhibition = 100[I]n/(IC50n + [I]n), where [I] is the inhibitor concentration and n is the Hill slope.
ACKNOWLEDGMENTS
These studies were supported by the National Institutes of Health (RO1AI117712).
K.P. and J.P. have an equity interest in Linnaeus Bioscience, Inc., and receive consulting income from the company. The terms of this arrangement have been reviewed and approved by the University of California—San Diego in accordance with its conflict-of-interest policies.
E.T.M., J.F.N., J.S., E.E., A.B.S., H.T., and A.I.D. conducted experiments and analyzed data. The manuscript was written by E.T.M., J.F.N., A.I.D., K.P., and J.P. All authors have read and approved the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Joe Pogliano, Email: jpogliano@ucsd.edu.
William W. Metcalf, University of Illinois at Urbana Champaign
REFERENCES
- 1.Wencewicz TA. 2019. Crossroads of antibiotic resistance and biosynthesis. J Mol Biol 431:3370–3399. 10.1016/j.jmb.2019.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown ED, Wright GD. 2016. Antibacterial drug discovery in the resistance era. Nature 529:336–343. 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 3.Pogue JM, Kaye KS, Cohen DA, Marchaim D. 2015. Appropriate antimicrobial therapy in the era of multidrug-resistant human pathogens. Clin Microbiol Infect 21:302–312. 10.1016/j.cmi.2014.12.025. [DOI] [PubMed] [Google Scholar]
- 4.Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, Gregson A. 2015. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59:6605–6607. 10.1128/AAC.01165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dougherty TJ, Pucci MJ. 2014. Antibiotic discovery and development. Springer, New York, NY. 10.1007/978-1-4614-1400-1. [DOI] [Google Scholar]
- 6.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. P T 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 7.Livermore DM, Blaser M, Carrs O, Cassell G, Fishman N, Guidos R, Levy S, Powers J, Norrby R, Tillotson G, Davies R, Projan S, Dawson M, Monnet D, Keogh-Brown M, Hand K, Garner S, Findlay D, Morel C, Wise R, Bax R, Burke F, Chopra I, Czaplewski L, Finch R, Livermore D, Piddock LJV, White T, on behalf of the British Society for Antimicrobial Chemotherapy Working Party on The Urgent Need: Regenerating Antibacterial Drug Development. 2011. Discovery research: the scientific challenge of finding new antibiotics. J Antimicrob Chemother 66:1941–1944. 10.1093/jac/dkr262. [DOI] [PubMed] [Google Scholar]
- 8.Lewis K. 2013. Platforms for antibiotic discovery. Nat Rev Drug Discov 12:371–387. 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 9.Hede K. 2014. Antibiotic resistance: an infectious arms race. Nature 509:S2–S3. 10.1038/509S2a. [DOI] [PubMed] [Google Scholar]
- 10.Frieri M, Kumar K, Boutin A. 2017. Antibiotic resistance. J Infect Public Health 10:369–378. 10.1016/j.jiph.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 11.da Cunha R, Fonseca LP, Calado CRC. 2019. Antibiotic discovery: where have we come from, where do we go? Antibiotics 8:45. 10.3390/antibiotics8020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newman D. 2017. Screening and identification of novel biologically active natural compounds. F1000Res 6:783. 10.12688/f1000research.11221.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes JP, Rees S, Kalindjian SB, Philpott KL. 2011. Principles of early drug discovery. Br J Pharmacol 162:1239–1249. 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baell J, Walters MA. 2014. Chemistry: chemical con artists foil drug discovery. Nature News 513:481–483. 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 15.Tomasic T, Peterlin Masic L. 2012. Rhodanine as a scaffold in drug discovery: a critical review of its biological activities and mechanisms of target modulation. Expert Opin Drug Discov 7:549–560. 10.1517/17460441.2012.688743. [DOI] [PubMed] [Google Scholar]
- 16.Baell JB, Nissink JWM. 2018. Seven year itch: pan-assay interference compounds (PAINS) in 2017—utility and limitations. ACS Chem Biol 13:36–44. 10.1021/acschembio.7b00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baell JB, Holloway GA. 2010. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 53:2719–2740. 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 18.Lamsa A, Liu WT, Dorrestein PC, Pogliano K. 2012. The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Mol Microbiol 84:486–500. 10.1111/j.1365-2958.2012.08038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nonejuie P, Burkart M, Pogliano K, Pogliano J. 2013. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc Natl Acad Sci U S A 110:16169–16174. 10.1073/pnas.1311066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Htoo HH, Brumage L, Chaikeeratisak V, Tsunemoto H, Sugie J, Tribuddharat C, Pogliano J, Nonejuie P. 2019. Bacterial cytological profiling as a tool to study mechanisms of action of antibiotics that are active against Acinetobacter baumannii. Antimicrob Agents Chemother 63:e02310-18. 10.1128/AAC.02310-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamsa A, Lopez-Garrido J, Quach D, Riley EP, Pogliano J, Pogliano K. 2016. Rapid inhibition profiling in Bacillus subtilis to identify the mechanism of action of new antimicrobials. ACS Chem Biol 11:2222–2231. 10.1021/acschembio.5b01050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nonejuie P, Trial RM, Newton GL, Lamsa A, Ranmali Perera V, Aguilar J, Liu WT, Dorrestein PC, Pogliano J, Pogliano K. 2016. Application of bacterial cytological profiling to crude natural product extracts reveals the antibacterial arsenal of Bacillus subtilis. J Antibiot (Tokyo) 69:353–361. 10.1038/ja.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters CE, Lamsa A, Liu RB, Quach D, Sugie J, Brumage L, Pogliano J, Lopez-Garrido J, Pogliano K. 2018. Rapid inhibition profiling identifies a keystone target in the nucleotide biosynthesis pathway. ACS Chem Biol 13:3251–3258. 10.1021/acschembio.8b00273. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Botella G, Breen JN, Duffy JE, Dumas J, Geng B, Gowers IK, Green OM, Guler S, Hentemann MF, Hernandez-Juan FA, Joseph-McCarthy D, Kawatkar S, Larsen NA, Lazari O, Loch JT, Macritchie JA, McKenzie AR, Newman JV, Olivier NB, Otterson LG, Owens AP, Read J, Sheppard DW, Keating TA. 2012. Discovery of selective and potent inhibitors of gram-positive bacterial thymidylate kinase (TMK). J Med Chem 55:10010–10021. 10.1021/jm3011806. [DOI] [PubMed] [Google Scholar]
- 25.Keating TA, Newman JV, Olivier NB, Otterson LG, Andrews B, Boriack-Sjodin PA, Breen JN, Doig P, Dumas J, Gangl E, Green OM, Guler SY, Hentemann MF, Joseph-McCarthy D, Kawatkar S, Kutschke A, Loch JT, McKenzie AR, Pradeepan S, Prasad S, Martínez-Botella G. 2012. In vivo validation of thymidylate kinase (TMK) with a rationally designed, selective antibacterial compound. ACS Chem Biol 7:1866–1872. 10.1021/cb300316n. [DOI] [PubMed] [Google Scholar]
- 26.Lavie A, Ostermann N, Brundiers R, Goody RS, Reinstein J, Konrad M, Schlichting I. 1998. Structural basis for efficient phosphorylation of 3′-azidothymidine monophosphate by Escherichia coli thymidylate kinase. Proc Natl Acad Sci U S A 95:14045–14050. 10.1073/pnas.95.24.14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haouz A, Vanheusden V, Munier-Lehmann H, Froeyen M, Herdewijn P, Van Calenbergh S, Delarue M. 2003. Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase new insights into the phosphoryl transfer mechanism. J Biol Chem 278:4963–4971. 10.1074/jbc.M209630200. [DOI] [PubMed] [Google Scholar]
- 28.Zgurskaya HI, Lopez CA, Gnanakaran S. 2015. Permeability barrier of gram-negative cell envelopes and approaches to bypass it. ACS Infect Dis 1:512–522. 10.1021/acsinfecdis.5b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitney EN. 1971. The tolC locus in Escherichia coli K-12. Genetics 67:39–53. 10.1093/genetics/67.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. 1995. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83:1171–1181. 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 31.Morona R, Reeves P. 1981. Molecular cloning of the tolC locus of Escherichia coli K-12 with the use of transposon Tn10. Mol Gen Genet 184:430–433. 10.1007/BF00352517. [DOI] [PubMed] [Google Scholar]
- 32.Miller JH, Miller JB. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 33.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. 2018. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27:14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shapiro AB, Walkup GK, Keating TA. 2009. Correction for interference by test samples in high-throughput assays. J Biomol Screen 14:1008–1016. 10.1177/1087057109341768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4 and Table S1. Download JB.00105-21-s0001.pdf, PDF file, 3.6 MB (3.6MB, pdf)