Abstract
The cdc25A gene encodes a tyrosine phosphatase which activates cyclin-dependent kinase activity in the G1 phase of the cell cycle. cdc25A RNA levels are induced from 3 to 6 h after serum induction of serum-starved NIH 3T3 cells, suggesting that the cdc25A gene is a delayed-early gene. Analysis of cdc25A promoter constructs showed that the cdc25A promoter is sufficient for serum induction. Surprisingly for a gene expressed in early to mid-G1, serum induction of the promoter requires an E2F site at position −62 in the promoter. Deletion or point mutation of the E2F site resulted in activation of expression in serum-starved cells and no further induction by serum treatment. E2F factors were found to bind to the cdc25A E2F site along with the retinoblastoma protein (Rb) family members p130 and p107. A shift in mobility of the E2F-p107 complex in extracts of cells induced for 6 h correlated with induction of cdc25A expression. These results suggest that serum induction of cdc25A expression is mediated by inactivation of p107 or p130, both of which repress transcription when bound to the promoter through E2F.
Cell growth is a process of progression of cells through the cell cycle, which contains four defined phases: G1, S, G2, and M (reviewed in reference 52). Mammalian cell cycle progression has often been studied by synchronizing cells in a quiescent state, termed G0, at the beginning of the G1 phase. Serum-starved, quiescent cells reenter the cell cycle after serum treatment and enter S phase after about 12 h depending on the cell type. This process of progression from G0 to S phase is marked by specific changes in gene expression. The first genes to be induced, cellular immediate-early genes, are activated rapidly by serum stimulation, with a peak of gene expression within 30 to 60 min (25). Immediate-early genes are typified by the c-fos and c-jun proto-oncogenes, which are themselves transcription factors. A second class of genes, delayed-early response genes, are induced after 3 to 6 h of serum induction and, in contrast to immediate-early genes, require new protein synthesis for induction of expression (36). There is a third class of genes that are activated late in G1 and that seem to herald the G1-to-S transition. Many of these genes are regulated by the E2F transcription factor and retinoblastoma protein (Rb) family members (13).
The connection, if any, among the different classes of serum-inducible genes has yet to be established. Since some immediate-early genes are transcription factors, it is possible that they serve to activate subsequent gene expression of the delayed-early or late G1 genes. To begin to investigate this possibility, we have studied the regulation of a delayed-early gene, cdc25A, known to be important for cell cycle progression.
cdc25 was originally identified in the fission yeast, Schizosaccharomyces pombe, as a phosphatase that controls cdc2/cyclin B kinase activity and activates mitosis (reviewed in reference 12). cdc2 must be dephosphorylated by cdc25 at a conserved tyrosine residue, tyrosine 15, in order to be activated (40). Mammalian cells have three cdc25 homologues, cdc25A, -B, and -C (16, 42, 47). These phosphatases can dephosphorylate cyclin-dependent kinase (cdk)-related kinases in vitro and induce their activity (16, 19, 23, 27, 51). cdc25A can also activate cdc2 kinase activity by competitively blocking its binding to the p21 cdk inhibitor (48).
The three mammalian cdc25 family members are differentially expressed during the cell cycle following serum stimulation. cdc25A is induced in mid-G1, similar to other delayed-early genes (32). cdc25B is expressed later, during S phase, while cdc25C is expressed in G2 phase (32, 34, 47). Besides undergoing transcriptional induction, cdc25A is also activated by cdk2/cyclin E phosphorylation in late G1 and by cyclin B in vitro (16, 26). Immunodepletion of cdc25A in rat cells by microinjection blocked cell entry into S phase, demonstrating that cdc25A is critical for G1 phase progression (16, 26, 32). cdc25A is also a candidate oncogene since it can cooperate with ras in oncogenic transformation of primary rat fibroblasts (18).
We have investigated the mechanism of serum induction of cdc25A in order to understand how delayed-early genes are activated and because cdc25A is an important gene for G1 phase progression. It was previously determined that the proto-oncogene and immediate-early gene c-myc can induce cdc25A expression and that there are myc binding sites in the cdc25A gene (17). It was not determined, however, whether c-myc or the myc binding sites were required for G1 phase induction of cdc25A. We have found that the myc sites in the cdc25A gene are not required for serum induction. Instead we found that cdc25A expression is regulated by an E2F site and Rb family members.
MATERIALS AND METHODS
Cell culture and cell cycle analysis.
NIH 3T3 cells were grown in Dulbecco’s modified Eagle medium (DMEM) with 10% newborn calf serum (NCS). For experiments performed to isolate RNA and determine the cell cycle, NIH 3T3 cells were plated at a density of 5 × 105 per 10-cm-diameter dish, grown for 24 h, starved in DMEM plus 0.2% NCS for 48 h, and then induced with 20% NCS for various periods of time. For cell cycle analysis, the serum-starved or -induced cells were trypsinized, centrifuged, resuspended in 0.6 ml of phosphate-buffered saline, and fixed by addition of 10 ml of methanol. The fixed cells were centrifuged and resuspended in 2 ml of a 60-μg/ml solution of propidium iodide in phosphate-buffered saline containing 20 μg of RNase A/ml. The stained cells were analyzed with a FACSCalibur flow cytometer (Becton Dickinson), and the distribution of cells at each stage of cell cycle was determined by using the CELLQuest and ModFit LT programs (Becton Dickinson).
RNase protection assay.
Total RNA were prepared from serum-starved or -induced NIH 3T3 cells by using Trizol reagent (Gibco/BRL). For RNase protection, an antisense cdc25A RNA probe was generated from a mouse cDNA clone containing the 3′ end of the gene (IMAGE consortium clone no. 441394). The plasmid was digested with PvuII and transcribed with T3 RNA polymerase, yielding a 1,100-nucleotide probe and a 420-nucleotide protected fragment. Total RNA (10 μg) from each time point was hybridized with 2 × 105 cpm of probe at 49°C overnight and digested with RNase A and RNase T1 at 30°C for 1 h as described elsewhere (49). The products were analyzed on a 4% polyacrylamide–7 M urea gel.
The cyclin E probe was made from a mouse cyclin E cDNA in pBSK (24). It was digested with PvuII and transcribed with T7 polymerase to generate a 550-nucleotide probe and a 470-nucleotide protected fragment.
Acidic ribosomal phosphoprotein P0 (ARPP-P0) RNA was measured as a loading control, using a full-length mouse cDNA in pBSK (29). The plasmid was digested with StuI and transcribed with T3 polymerase to yield a 230-nucleotide probe and a 150-nucleotide protected fragment.
Plasmid construction.
The human cdc25A promoter constructs (pNPGL3 with and without genomic fragments) were generously provided by David Beach (17). All of the 5′ deletion constructs were prepared by PCR with pNPGL3 as a template and appropriate primers, with a XhoI site being added at the 5′ end of the cdc25A promoter deletions. For the pCF series, various regions of the cdc25A promoter were generated as XhoI-to-BglII fragments and cloned upstream of position −53 of the c-fos promoter in p0-FlucGL3 (8). For pNP-E2F−, the E2F site was replaced by a BamHI site in pNPGL3. The wild-type cdc25A E2F site sequence at position −62, TTTGGCGC, was changed to TGGATCCC by using PCR primers. The identities of all constructs were confirmed by sequencing. The internal control plasmid pRL-SV40P was generated by placing the simian virus 40 (SV40) promoter from pGL3-Promoter (Promega) into pRL-TK (Promega) such that the SV40 promoter was driving the Renilla luciferase gene.
Transfection and dual luciferase assay.
NIH 3T3 cells were plated at a density of 2 × 105 per 6-cm-diameter culture dish. After 24 h, the cells were transfected by the calcium phosphate-DNA coprecipitation method as described elsewhere (49). Firefly luciferase reporter constructs (2 μg of the pNP series and 4 μg of the pCF series) were transfected together with 1 μg of the Renilla luciferase reporter plasmid pRL-SV40P as an internal control. Expression vectors (3 μg) for wild-type or mutant E1A (57), E2F-1, or a control vector, pcDNA3, were cotransfected with the reporter genes. The E2F-1 expression vector was kindly provided by Srikumar Chellapan. Herring sperm DNA was used to bring the total amount of DNA to 10 μg per dish. The cells were incubated with the transfection cocktail for 16 h and then grown in DMEM with 0.2% NCS for 36 h for serum starvation. The cells were then stimulated with DMEM containing 20% NCS for 13 h or as indicated in the figures. Cells were lysed in 200 μl of passive lysis buffer (Promega), and 5 μl of lysate was assayed for firefly and Renilla luciferase activity by using a dual luciferase kit as described by the manufacturer (Promega), except that one-fourth of the volume was used. The firefly luciferase activities were normalized to the Renilla luciferase activities to compensate for variability in transfection efficiencies. All experiments were performed with duplicate plates of cells for each time point.
Gel mobility shift assay.
Cell extracts were prepared from four semiconfluent 15-cm-diameter plates of NIH 3T3 cells as described elsewhere (9). Gel mobility shift assays utilized 32P-end-labelled double-stranded oligonucleotide probes, 5′-CCGCT CGAGG GATTC CGTTT GGCGC CAACTA-3′ and 5′-GGAAGATCTAGTTGGCGCCAAACGGAATCC-3′, spanning positions −70 to −49 of the human cdc25A promoter. Cell extracts (7 μg) were incubated with 1 ng of probe for 30 min at room temperature in 10-μl reaction mixtures containing 50 μg of sonicated herring sperm DNA/ml, 20 mM HEPES (pH 7.6), 4% glycerol, 2.5 mM MgCl2, 40 mM KCl, 0.1 mM EGTA, and 0.5 mM dithiothreitol. For competition assays, a 40-fold excess of unlabelled specific or nonspecific serum response element oligonucleotides was used. The samples were subsequently separated by electrophoresis at 4°C in a 4% polyacrylamide gel in a solution consisting of 22 mM Tris, 22 mM boric acid, and 0.6 mM EDTA.
Supershift assays were performed in an identical manner except that the cell extracts were preincubated with antibodies (1 μl) at 4°C for 30 min. The anti-p130 (sc-317) and anti-DP-1 (sc-610) sera were from Santa Cruz Biotechnology. Monoclonal antibodies to p107 (SD15) and Rb (21C9) were previously described (14, 55).
RESULTS
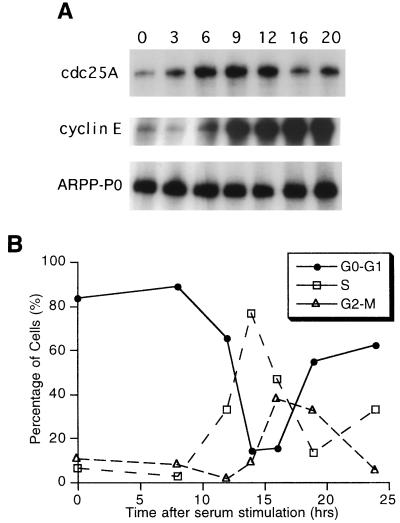
cdc25A mRNA was previously found to be induced by serum in rat NRK cells (32). We chose to examine serum induction of cdc25A expression in NIH 3T3 cells since serum-induced expression of immediate-early and delayed-early genes has been well studied in this cell line. We measured cdc25A mRNA levels by RNase protection assay. cdc25A expression showed an increase as early as 3 h after the onset of serum stimulation, with maximal induction by 6 h (Fig. 1A). The level decreased by 16 h. The level of a control mRNA of ARPP-P0 was unchanged by serum stimulation (Fig. 1A). The time course of the increase of cdc25A expression in NIH 3T3 cells is similar, though more gradual, than that of the serum-induced increase of cdc25A expression in rat NRK cells (32). We compared cdc25A induction to that of cyclin E, which is regulated by E2F and Rb family members (4, 20, 45). Induction of cdc25A mRNA occurred earlier than induction of cyclin E mRNA, which started to increase at 6 h but was not maximal until 9 h after the onset of serum induction (Fig. 1A). Other E2F-regulated genes, such as that encoding dihydrofolate reductase, are activated even later in G1 or S phase (29).
FIG. 1.
(A) Serum induction of cdc25A mRNA in NIH 3T3 cells. NIH 3T3 cells were serum starved for 48 h and induced with serum for the time periods (in hours) indicated above the lanes. Total RNA was isolated and analyzed by RNase protection for cdc25A, cyclin E, and ARPP-P0 as described in Materials and Methods. The areas of separate gels with specifically protected bands are shown. (B) The cell cycle phase of serum-starved and -induced NIH 3T3 cells was determined by propidium iodide staining and flow cytometry.
To correlate cdc25A expression with cell cycle progression, we analyzed the percentage of cells in each phase of the cell cycle by flow cytometry. The serum-stimulated NIH 3T3 cells began to enter S phase after 12 h (Fig. 1B). This indicates that cdc25A expression is induced in early to mid-G1.
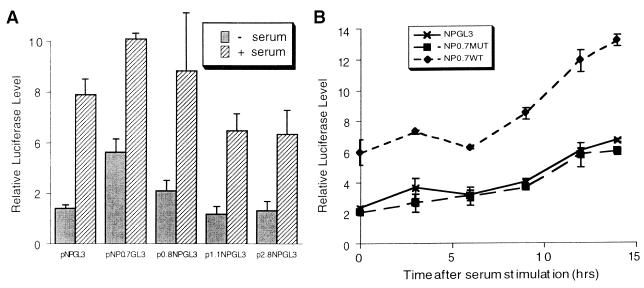
To determine which sequence elements of the cdc25A promoter are required for serum induction, we transfected into NIH 3T3 cells cdc25A promoter constructs driving a luciferase reporter gene. A construct, pNPGL3, containing positions −450 to +126 of the cdc25A gene was induced over threefold (Fig. 2A). Since c-myc, itself an immediate-early gene, was found to activate the cdc25A gene through myc/max binding sites in intronic regions of the gene, we tested several constructs containing genomic fragments of the cdc25A gene in pNPGL3 (17). The 0.7- and 2.8-kb fragments contain myc-max binding sites, while the 0.8-kb fragment is upstream of the promoter region (17). We found that all of the constructs were induced similarly to pNPGL3, although the levels of expression differed (Fig. 2A). The level of expression of pNP0.7GL3, which contains a myc binding site, was significantly higher in serum-starved cells, but the subsequent fold serum induction was lower. These results suggest that the myc-max binding sites are not required for serum induction of the cdc25A gene. Similarly, it was found that the myc-max binding sites are not required for transforming growth factor beta (TGF-β) repression of cdc25A expression (31).
FIG. 2.
Serum induction of cdc25A promoter constructs. (A) NIH 3T3 cells were transfected with the indicated cdc25A promoter-luciferase constructs containing nucleotides −450 to +126 of the cdc25A promoter, pNPGL3, and additional genomic fragments of the cdc25A gene (17). An internal control plasmid, pRL-SV40P, expressing Renilla luciferase was cotransfected. The transfected cells were serum starved (−serum) and then induced with 20% serum for 13 h (+ serum). Firefly luciferase activities were measured and normalized to the internal control Renilla luciferase activity. Shown are the averages of data from three experiments and the standard errors of the means (SEM; error bars). (B) NIH 3T3 cells were transfected with the indicated reporter genes and pRL-SV40P, serum starved, and then induced with serum for the indicated periods. The results are the average normalized luciferase activities of three experiments ± the standard errors of the means. NP0.7WT = pNP0.7GL3; NP0.7MUT contains a double point mutation in the myc binding site in NP0.7WT.
We tested the time course of serum induction of the pNPGL3 reporter gene to determine whether it correlated with induction of endogenous cdc25A mRNA. Induction of expression was gradual, reaching a peak at 14 h after serum stimulation (Fig. 2B). A modest induction, however, was apparent as early as 6 h. Since c-myc is an immediate-early gene, and since overexpression of c-myc can activate the cdc25A gene through a myc site in the 0.7-kb cdc25A genomic fragment (17), we tested whether this region could cause an earlier induction of the reporter gene. While this fragment in NP0.7WT (= pNP0.7GL3) caused a higher level of basal expression, the time course of induction was similar to that of NPGL3 (Fig. 2B). The level of induction of NP0.7WT (2.2-fold) was actually lower than the fold induction of NPGL3 (2.9-fold) in these experiments. Mutation of the myc binding site in the 0.7-kb fragment in NP0.7MUT (17) reduced the level of expression to that of NPGL3 (Fig. 2B). These results suggest that the myc binding site, CACGTG, contributes to expression but does not significantly affect the level or timing of induction. Besides myc family members, this E box element can bind several constitutive transcriptional activators, such as TFE3, TFEB, and USF, which may cause the higher level of expression (1, 7, 22).
Induction of the luciferase reporter gene was somewhat slower than induction of the endogenous message. There may be a lag between the induction of transcription and the accumulation of luciferase protein. Nevertheless, we cannot rule out the possibility that other mechanisms besides those reproduced by the pNPGL3 reporter contribute to serum induction of cdc25A expression. These could involve sequence elements outside the tested regions, chromatin or locus-specific regulation, or posttranscriptional regulation. While the myc binding site in pNP0.7 did not affect the kinetics of induction, it is still possible that this and other myc binding sites in other parts of the cdc25A gene together contribute to the regulation of the kinetics of induction of chromosomal (rather than transfected) c-myc genes.
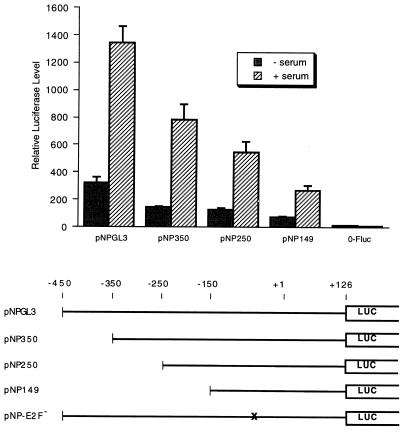
We sequenced the region of the human cdc25A promoter in pNPGL3 (Fig. 3). Sequence elements for CAAT box binding factors, SP1 and E2F, among others, were identified by using the MatInspector program (46). To determine regions of the promoter required for serum induction, we made 5′ deletions in pNPGL3 (Fig. 4). We found that all of the constructs, with deletions down to position −149 in the promoter, were still inducible by serum, although the levels of expression were reduced. This suggests that sequence elements between positions −149 and +126 of the cdc25A gene are sufficient for serum induction. Elements upstream of position −149 increased the level of expression but were not required for induction. As an internal control, we included an SV40 promoter-Renilla luciferase reporter, for which the Renilla luciferase activity can be measured separately from the firefly luciferase activity. Expression from this promoter was not significantly induced, and all points were normalized to this internal control to correct for differences in transfection efficiency. An SV40 promoter construct driving the firefly luciferase gene was also not induced by serum (data not shown). In addition, we tested a c-fos minimal promoter reporter, p0-Fluc, which was not induced (Fig. 4).
FIG. 3.
Sequence of the cdc25A promoter. The sequence of the human cdc25A promoter in pNPGL3 was determined. Position +1 indicates the 5′ end of the full-length cDNA (16). Two potential E2F sites are boxed, CAAT boxes are underlined, and a potential SP1 site has a dashed underline. The start codon for cdc25A is at position +460.
FIG. 4.
Serum induction of cdc25A promoter deletions. The constructs indicated at the bottom of the figure were transfected into NIH 3T3 cells with pRL-SV40P as an internal control and analyzed for serum induction as described in the legend to Fig. 2A. A minimal c-fos promoter plasmid, p0-Fluc (see Fig. 5), was tested as a control. Shown are the averages of data from three experiments and the standard errors of the means (error bars). − serum, no serum induction; + serum, serum induction performed.
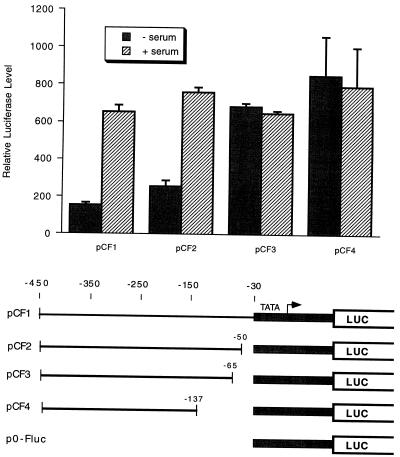
To further narrow the region required for serum induction, we made heterologous constructs with nucleotides −450 to −30 of the cdc25A gene fused to the c-fos minimal promoter of p0-Fluc. Deletions from the 3′ end of the cdc25A region were then made (Fig. 5). The heterologous construct pCF1 was induced by serum similarly to pNPGL3. Deletion to position −50 had no effect, while deletion to nucleotide −65 or −137 resulted in activation of the promoter in serum-starved cells, with no further increase upon serum stimulation (Fig. 5). These results suggest that deletion of a repressor element between positions −50 and −65 activates the promoter and that serum induction functions by inactivating the repressor. This segment of the cdc25A promoter contains a putative E2F site.
FIG. 5.
Serum induction of heterologous-promoter constructs. Regions of the cdc25A promoter were fused upstream of a c-fos minimal promoter construct as indicated at the bottom of the figure (LUC, luciferase gene). The constructs were transfected into NIH 3T3 cells with pRL-SV40P and either not serum induced (− serum) or serum induced (+ serum) as described in the legend to Fig. 2A. Shown are the averages of data from three experiments and the standard errors of the means (error bars). The c-fos minimal promoter alone, p0-Fluc, was not inducible (Fig. 4).
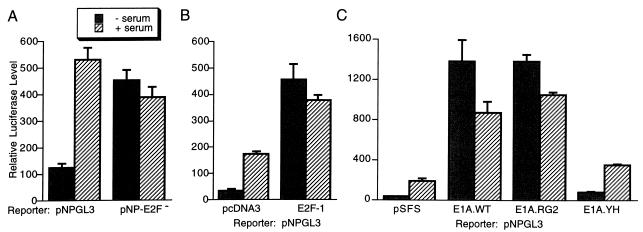
The E2F site at position −62 of the cdc25A promoter, TTTGGCGC, is identical to E2F sites in the dihydrofolate reductase, DNA polymerase α, and N-myc genes (44). To test whether this E2F element is involved in serum induction of the cdc25A promoter, we mutated the E2F site (pNP-E2F− [Fig. 4]) and tested for serum induction. As with the heterologous-promoter constructs, mutation of the E2F site resulted in constitutive activation of the reporter gene (Fig. 6A). We also found that mutation of the E2F site in the context of pNP0.7GL3 (which contains a myc binding site) resulted in high-level expression in serum-starved cells (data not shown). Overexpression of E2F-1 also activated the cdc25A promoter further, suggesting that it contains a functional E2F site (Fig. 6B). An additional putative E2F site at position −3 of the cdc25A promoter was deleted in the pCF series of constructs such that it is not required for serum induction or repression of the promoter (Fig. 5). Since it is present in the pNP-E2F− construct, the E2F site at nucleotide −3 is also not sufficient for repression of the reporter gene in serum-starved cells, and we have not studied it further.
FIG. 6.
E2F regulation of the cdc25A promoter. (A) The cdc25A E2F site at position −62 was mutated as diagrammed in Fig. 4 (pNP-E2F−) and tested for serum induction in NIH 3T3 cells. (B) The cdc25A reporter construct pNPGL3 was transfected with an E2F-1 expression vector or vector pcDNA3 and tested for serum-induced expression. (C) Adenovirus E1A expression vectors were transfected with pNPGL3 and pRL-SV40P. Wild-type E1A (E1A.WT) and mutants were transfected. A plasmid with a frameshift of E1A, pSFS, which does not express E1A served as a control vector. The E1A.RG2 mutant binds Rb family proteins but not p300 or CBP (57). The E1A YH47/928 mutant (E1A.YH) binds p300 and CBP but not Rb family proteins (57). Shown are the averages of data from three experiments and the standard errors of the means (error bars). − serum, no serum induction; + serum, serum induction performed.
E2F family members are regulated by Rb family proteins. Rb-related proteins (Rb, p107, and p130) either suppress E2F activity or repress the activity of transcription factors binding nearby in the promoter (reviewed in reference 13). Phosphorylation of Rb proteins during G1 progression leads to their inactivation and, hence, the activation of promoters with E2F sites (13). To test whether Rb family members are required for the low level of expression of the cdc25A reporter in serum-starved cells, we transfected an adenovirus E1A expression vector. E1A binds and neutralizes Rb family proteins (13). E1A strongly activated pNPGL3, and this activation was not further elevated by serum induction (Fig. 6C). Since E1A binds several cellular proteins, we used E1A point mutants to correlate its binding with specific proteins to its activation of the cdc25A reporter gene. A point mutation, RG2, that strongly reduces binding to p300 and CREB binding protein (CBP) (57) had no effect on activation of pNPGL3. In contrast, a double point mutation, YH47/928, that abolishes Rb, p107, and p130 binding (57) no longer activated the reporter (Fig. 6C). These results suggest that Rb family members are required to repress cdc25A expression in serum-starved cells.
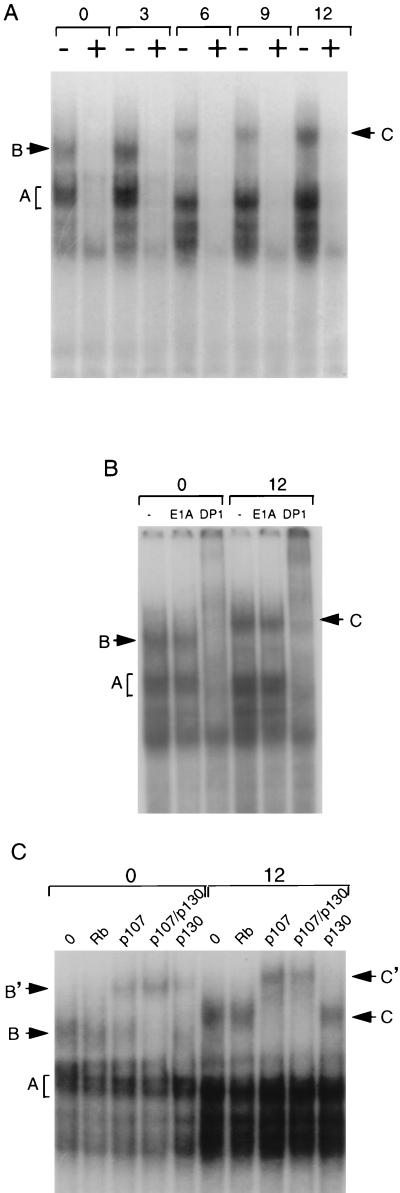
We examined binding of nuclear proteins to the putative E2F site by gel mobility shift assays. A double-stranded oligonucleotide spanning the site was used as a probe. Nuclear extracts were made from serum-starved and -stimulated NIH 3T3 cells. With each extract, several complexes were observed that were specifically competed by excess unlabelled probe (Fig. 7A). To confirm that these complexes contain E2F-related proteins, we performed a gel mobility shift assay with anti-DP1 serum. DP1 is a common heterodimer partner for E2F family members (13). Anti-DP1 serum disrupted the binding of complexes A, B, and C, while control antiserum (anti-E1A) had no effect (Fig. 7B).
FIG. 7.
Binding of E2F and Rb family proteins to the cdc25A E2F site. (A) Extracts were prepared from NIH 3T3 cells that were serum starved and then induced with serum for the time periods (in hours) indicated above the lanes. Extracts were incubated with 32P-labelled cdc25A E2F site oligonucleotide with a 40-fold excess of unlabelled probe (+) or nonspecific competitor (−). (B) Extracts from serum-starved (0) or 12-h serum-stimulated (12) cells were incubated with the E2F site probe, as described for panel A, without antiserum (−), with a nonspecific antiserum (anti-E1A), or with anti-DP1. (C) Extracts from serum-starved (0) or 12-h serum-stimulated (12) cells were incubated with antiserum to Rb, p107, or p130 or with a combination of anti-p107 and anti-p130 sera as indicated. Complexes A, B, and C indicate E2F containing the complexes discussed in the text. B’ and C’ indicate complexes supershifted by antiserum.
By comparison to previous data on E2F, we determined that it is likely that complex A consists of E2F family members complexed with DP-1 while complexes B and C are complexes of Rb family member bound to E2F-DP-1 (29, 54). We observed a shift from complex B to C at between 3 and 6 h of serum induction (Fig. 7A). There was also a shift of complex A from a doublet to a single band between 3 and 6 h. These shifts correlated with increased expression of the cdc25A gene (Fig. 1A). The change in complex A could be due to the presence of different E2F family proteins or differential binding to Rb family members. To determine which Rb family members are present in complexes B and C, we used specific antisera to supershift the complexes. At the 0-h point, complex B was partially supershifted by either anti-p107 or anti-p130 serum. A combination of both antisera completely supershifted the complex (Fig. 7C). At 12 h of serum induction, antiserum to p107 completely supershifted complex C while anti-p130 serum had no effect. Antiserum to Rb had no effect at either time point. From these results, we concluded that E2F is complexed with p107 and p130 in nuclear extracts of serum-starved cells while it is complexed with only p107 in nuclear extracts of serum-induced cells. The shift from complex B to C is likely due to the association of the E2F-p107 complexes with cdk’s (6, 11, 53). Among the E2F family members E2F-1 to -5, the p107 protein binds preferentially to E2F-4 (3, 9, 21, 50, 56), suggesting that complex C contains E2F-4.
DISCUSSION
We have found that serum induction of cdc25A expression is regulated by an E2F site in the cdc25A promoter. Mutation of the cdc25A E2F site resulted in derepression of a cdc25A reporter gene and constitutive expression in serum-starved and -induced cells. These results suggest that serum induction of cdc25A expression is mediated by inactivation of repressors acting via the E2F site. This type of regulation by E2F sites has been well characterized in a number of genes, including cdc2, B-myb, and E2F-1 (10, 28, 33, 35, 43), and is due to repression by Rb family members binding to E2F factors (reviewed in reference 13). Rb family members can repress transcriptional activation either by E2F or by adjacent factors binding to the promoter. In the former case, E2F function is required. Since a site mutation in E2F results in activation of the cdc25A reporter, it appears that direct transcriptional activation by E2F is not required and that Rb family proteins repress the activity of other transcriptional activators binding to the cdc25A promoter.
Overexpression of E2F-1 activated the cdc25A reporter, further suggesting that the latter contains a functional E2F site. Overexpression of E2F-1 may have activated the promoter by bringing a potent transcriptional activation domain to the promoter or by titrating out Rb family repressors. E1A can bind and titrate out Rb family members (13, 57), and transfection of E1A with the cdc25A promoter activates the reporter. In contrast, point mutations in E1A that specifically reduce Rb binding abolish the activation of the cdc25A reporter gene. These results support the conclusion that cdc25A is regulated by E2F and Rb family members.
We found that E2F-related factors bound to the cdc25A E2F site and that there was a shift in the mobilities of E2F complexes from 3 to 6 h after serum induction. This shift correlates well with induction of cdc25A expression and suggests that changes in the E2F complex could control cdc25A expression.
We initially investigated cdc25A as an example of a delayed-early gene since it is expressed in early to mid-G1 phase after serum induction of quiescent cells (Fig. 1) (32). Serum induction of cdc25A expression was apparent by 3 h after the onset of stimulation and peaked by 6 h in NIH 3T3 cells. This was about 3 h before the peak for cyclin E, a known E2F-regulated gene (4, 20, 45). It is thus surprising that cdc25A expression is regulated by E2F factors, which have generally been associated with late-G1- and S-phase gene expression (reviewed in reference 13). The peak of induction of the cdc25A promoter reporter was delayed compared to the induction of endogenous cdc25A mRNA. This partially reflects a lag in accumulation of luciferase protein but leaves open the possibility of additional mechanisms for early induction of cdc25A expression that are not reproduced by the reporter gene. The inclusion of myc binding sites from genomic fragments of the cdc25A gene (17) did not significantly affect the timing or level of induction, suggesting that the myc site is not required for serum induction of the promoter. In addition, it was recently found that cdc25A induction was not significantly affected in myc−/− null fibroblasts (5). Thus, while the myc binding sites may contribute to regulation of cdc25A in other contexts, they do not appear to mediate activation of the promoter by myc in serum-stimulated cells. In fact, since mutation of the E2F site results in derepression and constitutive expression of the cdc25A reporter gene, it appears that other activating factors binding to the promoter are constitutively active but are sensitive to E2F repression.
Since the E2F site mediates repression, activation early in G1 must overcome that repression. The correlation of changes in E2F complex formation with cdc25A expression further suggests that there is direct regulation via factors complexed with E2F. An alternative model, however, is that factors binding elsewhere in the cdc25A gene overcome repression by E2F-associated factors early in G1. While this work was under review, Iavarone and Massagué (30) also identified the E2F site in the cdc25A promoter as being critical for TGF-β repression of cdc25A expression. Since TGF-β causes cell cycle arrest, together these results suggest that control of E2F can either activate or repress expression of cdc25A in response to growth-activating or -inhibitory signals, respectively.
In analyzing factors binding to the cdc25A E2F site, we found a complex B that contains E2F and either p130 or p107 in extracts from serum-starved cells and a complex C that contains E2F and p107 in serum-stimulated cells (Fig. 7). The pattern of complexes observed is similar to that observed by Hurford et al. (29) in extracts of serum-induced mouse embryo fibroblasts (MEFs) except that the shift of the E2F complex in MEFs did not occur until 14 h after serum stimulation. The slower time course likely occurs because MEFs take several hours longer than NIH 3T3 cells to enter S phase. In contrast to our results, Smith et al. (54) found only p130 in E2F complexes from serum-starved REF52 cells. They then observed a loss of the p130 complex followed by gradual accumulation of a p107-E2F complex as cells entered S phase. While we observed a loss of the p130 complex, the p107 complex was present in serum-starved and -induced cells. These differences may be due to the use of different cell types and, possibly, to different mechanisms or extents of quiescence.
Since we find p107-E2F complexes in serum-starved and -stimulated cells, the shift in migration of the complex after 6 h must reflect a change in modifications or complexing proteins. Cyclins A and E have been found in complexes with p107- or p130-E2F such that binding of cyclin-cdk factors could account for the shift of the complex (6, 9, 11, 53). Cyclin D1-cdk4 complexes are induced earlier in G1 phase than cyclin E, and hence they are good candidates for complexing proteins (38). In fact, cyclin D1-cdk4 has been found to phosphorylate p107 and p130 and to stably complex with Rb and p107 (2, 15, 39, 58). It will be interesting to determine whether cyclin D1 becomes associated with E2F and whether this association or phosphorylation of p107 or p130 is required for derepression by the E2F site.
Another difference in our E2F gel mobility shift assays from those of others is in complex A (Fig. 7). This complex appears to contain E2F without complexing factors. We observed a generally constant amount of E2F binding, while others have found an increase following serum stimulation (29, 54). We also observed a conversion of a doublet to a single band. This may reflect a change in E2F family members. Leone et al. (37) found E2F-4 and E2F-5 complexes in serum-starved cells and a stimulation of E2F-3 complex formation 14 h after serum stimulation of REF52 cells. Determination of the exact nature of the E2F complexes in NIH 3T3 cells and the importance of the changes will require further investigation.
Regulation by E2F and Rb family members is complex because of the multiple members of each family and the multiple cyclin-cdk kinases that can phosphorylate them (13, 41). We have shown here that an E2F site can mediate repression of the cdc25A promoter in serum-starved cells and that this repression can be relieved as early as 6 h after the onset of serum stimulation. It will be interesting to determine how the E2F system is regulated at this point of the cell cycle.
ACKNOWLEDGMENTS
We thank David Beach, Elizabeth Moran, and Srikumar Chellapan for cdc25A, E1A, and E2F-1 plasmids, respectively. We especially thank David Cobrinik for antisera and critical comments.
This work was supported by grant CA 50329 from the National Cancer Institute to R.P.
REFERENCES
- 1.Beckmann H, Su L K, Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990;4:167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- 2.Beijersbergen R L, Carlee L, Kerkhoven R M, Bernards R. Regulation of the retinoblastoma protein-related p107 by G1 cyclin complexes. Genes Dev. 1995;9:1340–1353. doi: 10.1101/gad.9.11.1340. [DOI] [PubMed] [Google Scholar]
- 3.Beijersbergen R L, Kerkhoven R M, Zhu L, Carlee L, Voorhoeve P M, Bernards R. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 1994;8:2680–2690. doi: 10.1101/gad.8.22.2680. [DOI] [PubMed] [Google Scholar]
- 4.Botz J, Zerfass-Thome K, Spitkovsky D, Delius H, Vogt B, Eilers M, Hatzigeorgiou A, Jansen-Dürr P. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol Cell Biol. 1996;16:3401–3409. doi: 10.1128/mcb.16.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush A, Mateyak M, Dugan K, Obaya A, Adachi S, Sedivy J, Cole M. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes Dev. 1998;12:3797–3802. doi: 10.1101/gad.12.24.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao L, Faha B, Dembski M, Tsai L H, Harlow E, Dyson N. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature. 1992;355:176–179. doi: 10.1038/355176a0. [DOI] [PubMed] [Google Scholar]
- 7.Carr C S, Sharp P A. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol Cell Biol. 1990;10:4384–4388. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke N, Arenzana N, Hai T, Minden A, Prywes R. Epidermal growth factor induction of the c-jun promoter by a Rac pathway. Mol Cell Biol. 1998;18:1065–1073. doi: 10.1128/mcb.18.2.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobrinik D, Whyte P, Peeper D S, Jacks T, Weinberg R A. Cell cycle-specific association of E2F with the p130 E1A-binding protein. Genes Dev. 1993;7:2392–2404. doi: 10.1101/gad.7.12a.2392. [DOI] [PubMed] [Google Scholar]
- 10.Dalton S. Cell cycle regulation of the human cdc2 gene. EMBO J. 1992;11:1797–1804. doi: 10.1002/j.1460-2075.1992.tb05231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devoto S H, Mudryj M, Pines J, Hunter T, Nevins J R. A cyclin A-protein kinase complex possesses sequence-specific DNA binding activity: p33cdk2 is a component of the E2F-cyclin A complex. Cell. 1992;68:167–176. doi: 10.1016/0092-8674(92)90215-x. [DOI] [PubMed] [Google Scholar]
- 12.Draetta G, Eckstein J. Cdc25 protein phosphatases in cell proliferation. Biochim Biophys Acta. 1997;1332:M53–M63. doi: 10.1016/s0304-419x(96)00049-2. [DOI] [PubMed] [Google Scholar]
- 13.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 14.Dyson N, Dembski M, Fattaey A, Ngwu C, Ewen M, Helin K. Analysis of p107-associated proteins: p107 associates with a form of E2F that differs from pRB-associated E2F-1. J Virol. 1993;67:7641–7647. doi: 10.1128/jvi.67.12.7641-7647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 16.Galaktionov K, Beach D. Specific activation of cdc25 tyrosine phosphatases by B-type cyclins: evidence for multiple roles of mitotic cyclins. Cell. 1991;67:1181–1194. doi: 10.1016/0092-8674(91)90294-9. [DOI] [PubMed] [Google Scholar]
- 17.Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 18.Galaktionov K, Lee A K, Eckstein J, Draetta G, Meckler J, Loda M, Beach D. CDC25 phosphatases as potential human oncogenes. Science. 1995;269:1575–1577. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 19.Gautier J, Solomon M J, Booher R N, Bazan J F, Kirschner M W. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- 20.Geng Y, Eaton E N, Picon M, Roberts J M, Lundberg A S, Gifford A, Sardet C, Weinberg R A. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- 21.Ginsberg D, Vairo G, Chittenden T, Xiao Z X, Xu G, Wydner K L, DeCaprio J A, Lawrence J B, Livingston D M. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 1994;8:2665–2679. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- 22.Gregor P D, Sawadogo M, Roeder R G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990;4:1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- 23.Gu Y, Rosenblatt J, Morgan D O. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992;11:3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrera R E, Sah V P, Williams B O, Mäkelä T P, Weinberg R A, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herschman H R. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann I, Draetta G, Karsenti E. Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E dependent phosphorylation at the G1/S transition. EMBO J. 1994;13:4302–4310. doi: 10.1002/j.1460-2075.1994.tb06750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda R, Ohba Y, Nagata A, Okayama H, Yasuda H. Dephosphorylation of human p34cdc2 kinase on both Thr-14 and Tyr-15 by human cdc25B phosphatase. FEBS Lett. 1993;318:331–334. doi: 10.1016/0014-5793(93)80540-b. [DOI] [PubMed] [Google Scholar]
- 28.Hsiao K M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 29.Hurford R K, Jr, Cobrinik D, Lee M H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 30.Iavarone A, Massagué J. E2F and histone deacetylase mediate transforming growth factor β repression of cdc25A during keratinocyte cell cycle arrest. Mol Cell Biol. 1999;19:916–922. doi: 10.1128/mcb.19.1.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iavarone A, Massagué J. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-β in cells lacking the CDK inhibitor p15. Nature. 1997;387:417–422. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- 32.Jinno S, Suto K, Nagata A, Igarashi M, Kanaoka Y, Nojima H, Okayama H. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J. 1994;13:1549–1556. doi: 10.1002/j.1460-2075.1994.tb06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 34.Kakizuka A, Sebastian B, Borgmeyer U, Hermans-Borgmeyer I, Bolado J, Hunter T, Hoekstra M F, Evans R M. A mouse cdc25 homolog is differentially and developmentally expressed. Genes Dev. 1992;6:578–590. doi: 10.1101/gad.6.4.578. [DOI] [PubMed] [Google Scholar]
- 35.Lam E W, Watson R J. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanahan A, Williams J B, Sanders L K, Nathans D. Growth factor-induced delayed early response genes. Mol Cell Biol. 1992;12:3919–3929. doi: 10.1128/mcb.12.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J-Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayol X, Garriga J, Grana X. Cell cycle-dependent phosphorylation of the retinoblastoma-related protein p130. Oncogene. 1995;11:801–808. [PubMed] [Google Scholar]
- 40.Millar J B, McGowan C H, Lenaers G, Jones R, Russell P. p80cdc25 mitotic inducer is the tyrosine phosphatase that activates p34cdc2 kinase in fission yeast. EMBO J. 1991;10:4301–4309. doi: 10.1002/j.1460-2075.1991.tb05008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 42.Nagata A, Igarashi M, Jinno S, Suto K, Okayama H. An additional homolog of the fission yeast cdc25+ gene occurs in humans and is highly expressed in some cancer cells. New Biol. 1991;3:959–968. [PubMed] [Google Scholar]
- 43.Neuman E, Flemington E K, Sellers W R, Kaelin W G., Jr Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 45.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadhu K, Reed S I, Richardson H, Russell P. Human homolog of fission yeast cdc25 mitotic inducer is predominantly expressed in G2. Proc Natl Acad Sci USA. 1990;87:5139–5143. doi: 10.1073/pnas.87.13.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saha P, Eichbaum Q, Silberman E D, Mayer B J, Dutta A. p21CIP1 and Cdc25A: competition between an inhibitor and an activator of cyclin-dependent kinases. Mol Cell Biol. 1997;17:4338–4345. doi: 10.1128/mcb.17.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 50.Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg R A. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc Natl Acad Sci USA. 1995;92:2403–2407. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sebastian B, Kakizuka A, Hunter T. Cdc25M2 activation of cyclin-dependent kinases by dephosphorylation of threonine-14 and tyrosine-15. Proc Natl Acad Sci USA. 1993;90:3521–3524. doi: 10.1073/pnas.90.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 53.Shirodkar S, Ewen M, DeCaprio J A, Morgan J, Livingston D M, Chittenden T. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992;68:157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 54.Smith E J, Leone G, DeGregori J, Jakoi L, Nevins J R. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol Cell Biol. 1996;16:6965–6976. doi: 10.1128/mcb.16.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Templeton D J, Park S H, Lanier L, Weinberg R A. Nonfunctional mutants of the retinoblastoma protein are characterized by defects in phosphorylation, viral oncoprotein association, and nuclear tethering. Proc Natl Acad Sci USA. 1991;88:3033–3037. doi: 10.1073/pnas.88.8.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vairo G, Livingston D M, Ginsberg D. Functional interaction between E2F-4 and p130: evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 57.Wang H-G H, Rikitake Y, Carter M C, Yaciuk P, Abraham S E, Zerler B, Moran E. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao Z X, Ginsberg D, Ewen M, Livingston D M. Regulation of the retinoblastoma protein-related protein p107 by G1 cyclin-associated kinases. Proc Natl Acad Sci USA. 1996;93:4633–4637. doi: 10.1073/pnas.93.10.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]