Abstract
Frameshift mutations occur when the coding region of a gene is altered by addition or deletion of a number of base pairs that is not a multiple of three. The occurrence of a deletion versus an insertion type of frameshift depends on the nature of the transient intermediate structure formed during DNA synthesis. Extrahelical bases on the template strand give rise to deletions, whereas extrahelical bases on the strand being synthesized produce insertions. We previously used reversion of a +1 frameshift mutation to analyze the role of the mismatch repair (MMR) machinery in correcting −1 frameshift intermediates within a defined region of the yeast LYS2 gene. In this study, we have used reversion of a −1 frameshift mutation within the same region of LYS2 to analyze the role of the MMR machinery in the correction of frameshift intermediates that give rise to insertion events. We found that insertion and deletion events occur at similar rates but that the reversion spectra are very different in both the wild-type and MMR-defective backgrounds. In addition, analysis of the +1 spectra revealed novel roles for Msh3p and Msh6p in removing specific types of frameshift intermediates.
The addition or removal of one or more base pairs in the coding region of a gene generates a frameshift mutation if the number of inserted or deleted base pairs is not a multiple of three. Frameshift mutations, when encountered by a translating ribosome, result in the incorporation of variant amino acids specified by an alternative reading frame. Stop codons in the alternative reading frame usually result in truncation of the protein as well. Unlike the majority of base substitutions, frameshift mutations almost invariably destroy or drastically alter the function of a protein. Because of their deleterious nature, it is particularly important for cells to recognize and remove frameshift intermediates.
The frequency of frameshift events has been shown to increase in regions of repeated base composition such as mono-, di-, or trinucleotide repeats (13, 29). This presumably occurs because DNA polymerase has a higher propensity to “slip” at repetitive sequences during DNA synthesis (39). DNA polymerase slippage occurs when the nascent strand (the strand being synthesized) and the template strand transiently dissociate and then reanneal incorrectly, resulting in the presence of one or more extrahelical nucleotides in either the nascent or the template strand. A failure to repair the resulting loop before the next round of DNA replication will result in a deletion if the unpaired base(s) is on the template strand or an addition if the unpaired base(s) is on the nascent strand. In addition to DNA polymerase slippage events at tandem repeats, slippage events between noncontiguous direct repeats have been proposed to account for the occurrence of large deletion and duplication events (30, 33, 36, 41).
Three processes affect the rate of mutational events: (i) the frequency of incorporation of incorrect nucleotides by DNA polymerase, (ii) the efficiency with which incorporation errors are corrected by the exonucleolytic proofreading activity of DNA polymerase, and (iii) the efficiency with which the mismatch repair (MMR) system removes replication errors that escape the proofreading activity of DNA polymerases. The best-understood MMR system is that of the bacterium Escherichia coli. The E. coli MMR system contains three dedicated Mut proteins (MutS, MutL, and MutH), mutations in any one of which result in a strong mutator phenotype (for a review, see reference 23). The MutS protein binds preferentially to mismatched DNA substrates as a homodimer, and the MutH protein nicks the unmethylated strand of a nearby, hemimethylated GATC site. The creation of nicks on the unmethylated strand by the MutH protein marks the newly replicated strand for subsequent removal and resynthesis. The MutL protein interacts with MutS and is important in the activation of the endonuclease activity of MutH. In yeast, six MutS (Msh1p to Msh6p) and four MutL (Pms1p and Mlh1p to Mlh3p) homologs have been identified but no MutH homologs have been found (for reviews, see references 5 and 17). The signal for the biased removal of newly synthesized DNA in eukaryotes is unclear but may involve strand nicks and/or a direct interaction of the MMR machinery with the replication machinery (14, 44).
The major players in the correction of mismatches generated during nuclear DNA replication in yeast are the MutS homologs Msh2p, Msh3p, and Msh6p and the MutL homologs Pms1p and Mlh1p (15, 22, 26). Biochemical experiments have shown that Msh2p can heterodimerize with either Msh6p or Msh3p and that the heterodimeric Msh2p-Msh3p and Msh2p-Msh6p complexes have different binding specifies (1, 2, 9, 12, 21, 25). Msh2p-Msh6p, for example, recognizes a G/T mismatch but not a +CA or +(CA)5 loop, while the converse is true for the Msh2p-Msh3p heterodimer (1, 2). Distinct roles for the two Msh2p-containing complexes also have been identified in vivo (8, 22, 35, 37), where both are thought to work with a Pms1p-Mlh1p heterodimer (26, 27). Based on available genetic data, the yeast Msh2p-Msh6p heterodimer appears to recognize both base substitutions and small insertion or deletion mismatches, while the Msh2p-Msh3p heterodimer appears to recognize only insertion or deletion mismatches (15, 22). Recent in vivo data also have implicated an Mlh1p-Mlh3p complex in the repair of specific types of frameshift intermediates (7). The remaining members of the yeast MutL and MutS family either do not function in nuclear mismatch repair or have no known function (11, 28, 31).
We previously examined the in vivo specificities for the MMR machinery in the removal of −1 frameshift intermediates within a defined 150-bp region of the yeast LYS2 locus (8). In this report, we describe a system for examining +1 frameshift events within the same region of the LYS2 locus. This system has been used to examine the roles of individual MMR proteins in the recognition and correction of +1 frameshift intermediates. These data reveal distinct differences between the −1 and +1 frameshift spectra and suggest novel roles for individual MMR components in the removal of frameshift intermediates.
MATERIALS AND METHODS
Media and growth conditions.
Yeast strains were grown nonselectively in YEP medium (1% yeast extract, 2% Bacto peptone [with 2.5% agar for plates]) supplemented with either 2% dextrose (YEPD) or 2% glycerol–2% ethanol (YEPGE). Synthetic complete (SC) medium (34) containing 2% dextrose and lacking the appropriate amino acid was used for selective growth. LB medium (1% yeast extract, 0.5% Bacto Tryptone, 1% NaCl [with 1.5% agar for plates]) supplemented with ampicillin (100 μg/ml), as appropriate, was used for growth of Escherichia coli strains. Yeast and bacterial strains were grown at 30 and 37°C, respectively.
Strain constructions.
An assay system that specifically detects +1 frameshift intermediates was derived by deleting nucleotide (nt) 746 of the LYS2 gene (nucleotides are numbered beginning at the upstream XbaI site) and concurrently removing two potential stop codons present in the −1 reading frame relative to the normal reading frame. This was accomplished in two steps by using in vitro site-directed mutagenesis (Stratagene Chameleon Double-Stranded, Site-Directed Mutagenesis Kit). First, the two potential stop codons were changed to sense codons (TAG to TCG and TGA to CGA at positions 767 and 781, respectively) by using the mutagenesis primer 5′-gcatcatttCgtggactttgcttCgaatttggatacc (altered base pairs are in uppercase). This manipulation also created a BstBI restriction site (underlined). Next, mutagenesis primer 5′-ccaagatttcaaatt*gacgagCtcaagcatc was used to delete the A at position 746 (asterisk) and change nt 753 from a T to a C (uppercase), creating a SacI restriction site (underlined). The resulting plasmid, pSR585, was used to introduce the lys2ΔA746 allele into SJR195 (MATα ade2-101oc his3Δ200 ura3ΔNco) by two-step allele replacement (32), thus creating strain SJR922.
All repair-defective strains were isogenic derivatives of SJR922 derived by transformation. msh2Δ, msh3Δ, msh6Δ, pms1Δ, and mlh1Δ strains were constructed as described by Greene and Jinks-Robertson (8). The rad1Δ::hisG-URA3-hisG allele was introduced by transformation of SJR922 with SalI/EcoRI-digested pR1.6 (obtained from L. Prakash).
Mutation rates and spectra.
Rates of reversion to lysine prototrophy were determined by the method of the median (20) by using data from 12 to 24 cultures of each strain. For the rate measurements, 5 ml of YEPGE medium was inoculated with single colonies from YEPD plates and the cultures were incubated for 2 days on a roller drum. Cells were harvested by centrifugation, washed with sterile H2O, and resuspended in 1 ml of H2O. Aliquots (100 μl) of appropriate dilutions were plated onto SC-Lys to identify Lys+ revertants and on YEPD to determine viable cell numbers. Lys+ colonies were counted 2 days after selective plating.
To isolate independent Lys+ revertants for DNA sequence analysis, YEPGE cultures were grown as described above and plated on SC-Lys. To ensure independence, only one revertant from each culture was purified for subsequent molecular analysis. Manual or automated DNA sequence analysis of PCR-amplified genomic fragments was performed as described previously (4, 8) by using primer 5′-CGCAACAATGGTTACTCT.
RESULTS
Creation of a +1 frameshift assay system.
The 4.2-kb LYS2 locus has been widely used in genetic assays to study both the reversion rates and spectra of spontaneous mutations in wild-type and mutant yeast strains (8, 40, 41, 43). Previously, Greene and Jinks-Robertson (8) described a −1 frameshift assay system based on reversion of a +4 frameshift allele (lys2ΔBgl; the BglII site is at nt 763) at the LYS2 locus. The 150-bp reversion window for the lys2ΔBgl allele was defined as the region of the LYS2 gene in which a compensatory frameshift mutation must occur in order to restore a functional Lys2 protein. For the current study, we constructed a yeast system to specifically study the correction of +1 frameshift intermediates, which correspond to slippage events that place the extrahelical base on the newly synthesized strand (the nascent strand) rather than on the template strand. This system is based on the reversion of a −1 frameshift allele (lys2ΔA746) and was designed so that the +1 reversion events would be in essentially the same 150-bp reversion window as the previously characterized −1 frameshift events.
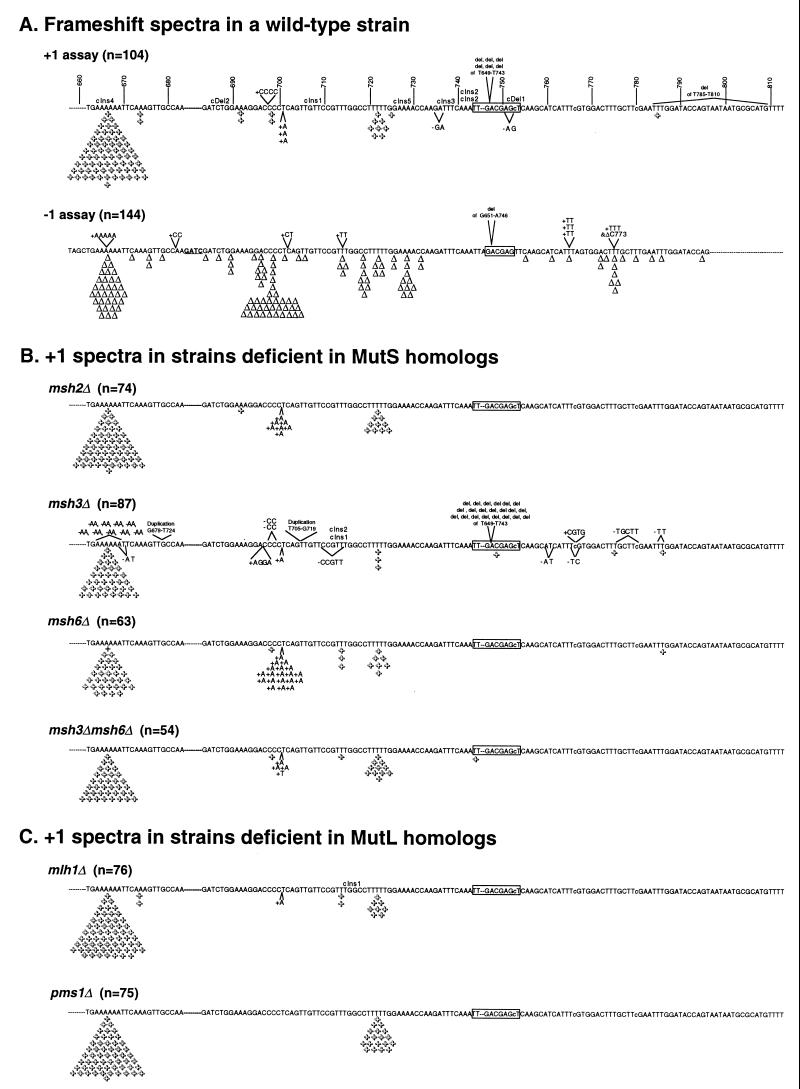
The lys2ΔA746 allele was constructed by deleting nt 746 from the LYS2 coding sequence. Deletion of this nucleotide results in an approximately 80-bp upstream region where a compensatory +1 frameshift can occur but only a 20-bp downstream region. In order to extend the downstream reversion region and to make it roughly coincide with that of the lys2ΔBgl allele, two nonsense codons were changed to sense codons in the relevant reading frame. The resulting lys2ΔA746 reversion window differs from the lys2ΔBgl reversion window in several ways (Fig. 1A). First, the lys2ΔA746 reversion window lacks 5 bp and contains an additional 18 bp relative to the 5′ and 3′ ends, respectively, of the lys2ΔBgl reversion window. At least 14 of the additional 18 bp present in the lys2ΔA746 reversion window are not essential for Lys2p function (see below; Fig. 1A). Second, the lys2ΔA746 allele lacks the internal 4-bp duplication present in the lys2ΔBgl allele. Finally, the lys2ΔA746 allele is missing bp 746 and contains three base pair substitutions (T753C, A767C, and T781C) that are not present in the lys2ΔBgl allele.
FIG. 1.
Sequence spectra of +1 frameshift events in wild-type and MMR-defective strains. The sequences of the entire +1 and −1 assay reversion windows are shown; nucleotides are numbered from the XbaI site upstream of the LYS2 gene. Nt 746 was deleted, and 3 nt were changed (A767C, T781C, and T753C, lowercase letters; see Materials and Methods) to create the +1 assay strain. The −1 assay strain was created by filling in a BglII site (added nucleotides are underlined and in boldface; 8). Dashes denote sequences that are present in the −1 assay system but are absent in the +1 reversion window. The −1 mutation spectrum was adopted from Greene and Jinks-Robertson (8). The locations of single base pair insertions (+) and deletions (Δ) are indicated. The locations of deletion events that occurred between two perfect direct repeats are indicated by the abbreviation del. One copy of the 10-bp direct repeat at the endpoints of the 94-bp deletion is boxed; the second 10-bp direct repeat lies 5′ of the reversion window and is not shown. The deletion extending from T785 to T810 has 4-bp direct repeats (TTTG) at its ends; the G of the second repeat is outside of the reversion window. cIns and cDel denote the locations of complex insertion and deletion events, respectively. The complex events in the lys2ΔA746 reversion spectrum in wild-type cells were as follows (underlining indicates the positions of base substitutions; asterisks indicate the positions of the inserted or deleted bases; base changes are in uppercase): cIns1, t*gttccgtttggc changed to tAgttccgtttgTc; cIns2, tttc*aaa changed to tttAAaaa; cIns3, ga*tttcaaattg changed to gaAtttAaaaAtg; cIns4, aaaaaa*ttc changed to aaaaaaAAtc; cIns5, tttttgg*aaa changed to tttttAgAaaa; cDel1, gagctcaag changed to ga*TCc*ag; cDel2, tctggaaa changed to tTt**aaa. The complex events in the msh3Δ spectrum were as follows: cIns1, ccg*******tttggcc changed to ccgCATAAGGtttgTcc; cIns2, ccggttt*gcc changed to ccTgtttTTcc.
Reversion of the lys2ΔA746 allele in a wild-type strain.
The simplest way for a lys2ΔA746 strain to acquire a Lys+ phenotype is to restore the correct open reading frame of the encoded protein by the addition of a single base pair. We will therefore refer to a strain containing the lys2ΔA746 allele as a +1 assay strain. The rate of spontaneous Lys+ revertants in the wild-type +1 assay strain was 1.4 × 10−9, a rate very similar to that previously reported for a −1 assay strain containing the lysΔBgl allele (2.8 × 10−9; reference 8). To determine the molecular nature of the lys2ΔA746 revertants, 104 independent reversion events were sequenced (the spectrum is presented in Fig. 1A). As expected, 83% (86 of 104) of the reversion events were single base pair insertions and the majority of these (67 of 86 [78%]) were in the longest mononucleotide run in the reversion window, a run of six adenines (six-A run beginning at nt 664) near the 5′ end of the window. The second largest mononucleotide run (five-T run beginning at nt 720) present in the reversion window contained 9% (8 of 86) of the single base pair insertions. Single base pair insertions occurred infrequently (≤2% of the total events analyzed) in runs of four C, A, or T nucleotides. In addition to the single base pair insertions, there were two 2-bp deletions, one 4-bp insertion, one deletion of 26 bp, six 94-bp deletions, and eight complex events (a complex event is defined here as an insertion or deletion event that is accompanied by a base substitution). It should be noted that in the engineering of the +1 assay strain, we increased the size of a naturally occurring direct repeat from 6 to 10 bp (see the boxed sequences in Fig. 1A). The six 94-bp deletions recovered in the +1 assay strain occurred between the 10-bp repeats. The remaining 26-bp deletion occurred between 4-bp direct repeats. For each type of event, the total reversion rate and the percentage of the relevant event were used to calculate the rate of each type of reversion event. These rates are given in Table 1.
TABLE 1.
Reversion of the lys2ΔA746 frameshift allele
| Genotype | Total rate (109) | Fold increase in rate of specific type of frameshift event relative to rate in wild-type straina
|
|||||
|---|---|---|---|---|---|---|---|
| +1 events in 6-A run | +A next to 4-C run | +1 events in 5-T run | 2-bp deletions | Large deletions | Other | ||
| Wild type | 1.4 | 1 (67/104) | 1 (3/104) | 1 (8/104) | 1 (2/104) | 1 (7/104) | 1 (17/104) |
| msh2 | 220 | 190 (56/74) | 470 (7/74) | 280 (10/74) | NA (0/74) | NA (0/74) | NA (1/74) |
| msh3 | 1.4 | 0.64 (36/87) | NA (1/87) | 0.38 (3/87) | 8.5 (15/87) | 3.4 (21/87) | 0.8 (11/87) |
| msh6 | 15 | 8.5 (32/63) | 110 (19/63) | 15 (7/63) | NA (0/63) | NA (0/63) | 5.3 (5/63) |
| msh3 msh6 | 220 | 160 (36/54) | 310 (3/54) | 390 (11/54) | NA (0/54) | NA (0/54) | 73 (4/54) |
| mlh1 | 520 | 480 (62/76) | NA (1/76) | 510 (8/76) | NA (0/76) | NA (0/76) | 150 (5/76) |
| pms1 | 360 | 300 (56/76) | NA (0/75) | 800 (19/75) | NA (0/75) | NA (0/75) | NA (0/75) |
The values in parentheses following the fold increase for each class correspond to the proportion of the sequenced events that were of that particular class. The proportion of a given class was multiplied by the total mutation rate to obtain the rate of mutations in that class. The fold increase for each frameshift class in a given MMR-deficient strain was obtained by dividing the rate in the MMR-deficient strain by that in wild-type control strain. The “other” class includes all events that did not fall into one of the specified classes; some of these events are simple +1 insertions. NA, not applicable because one or no events of the class in question was obtained.
Reversion of the lys2ΔA746 allele in strains defective in MutS homologs.
Three yeast MutS homologs (Msh2p, Msh3p, and Msh6p) have been shown to play a role in the correction of spontaneous mitotic frameshift intermediates in both vertebrate and invertebrate organisms (for reviews, see references 5 and 45). The effects of the individual disruption of MSH2, MSH3, or MSH6 on both the rate (Table 1) and spectra (Fig. 1B) of +1 frameshift mutations in the lys2ΔA746 reversion window were analyzed. As expected, elimination of Msh2p or concurrent disruption of its heterodimeric partners Msh3p and Msh6p resulted in a strong mutator phenotype, with the rate of Lys+ revertants increased 150-fold over that observed in a wild-type strain. Disruption of MSH6 alone resulted in a relatively modest 11-fold increase in the mutation rate, while disruption of MSH3 alone did not cause a detectable mutator phenotype.
The lys2ΔA746 reversion window was sequenced in independent Lys+ revertants to uncover the mutations responsible for restoring Lys2p function. The reversion spectra were very similar in the msh2Δ and msh3Δ msh6Δ strains (Fig. 1B), with all of the events analyzed being insertions of single base pairs. The vast majority of reversion events were found to occur in the six-A mononucleotide run (56 of 74 [76%] for msh2Δ and 36 of 54 [67%] for msh3Δ msh6Δ). The msh2Δ and msh3Δ msh6Δ strains also contained two additional insertion hot spots: one within the five-T mononucleotide run and a second immediately 3′ of the four-C mononucleotide run. The insertion hot spot after the four-C tract showed a very strong preference for the addition of a single adenine. For the msh2Δ and msh3Δ msh6Δ strains, the rate increase in each of the individual hot spots (the six-A run, the five-T run, and the +A insertion) was similar to the overall increase in the reversion rate (Table 1).
Although there was no detectable elevation in the rate of lys2ΔA746 reversion in the msh3Δ strain, the +1 mutational spectrum differed from the wild-type spectrum in several ways. First, in the msh3Δ strain, the 94-bp deletion accounted for 24% (21 of 87) of the total number of reversion events, whereas this deletion accounted for only 6% (6 of 104) of the events in the wild-type background (Table 1). The proportion of 94-bp deletions observed in the wild-type versus the msh3Δ mutant strain is significantly different (P < 0.01 by contingency χ2) and corresponds to a 3.4-fold increase in the rate of the 94-bp deletion in the msh3Δ strain relative to the wild-type strain. A second, very striking difference between the reversion spectra in the wild-type and msh3Δ +1 assay strains was the prominence in the msh3Δ strain of 2-bp deletions. The 2-bp deletions accounted for 17% (15 of 87) of the total number of reversion events in the msh3Δ strain but only 2% (2 of 104) of the events in the wild-type background. This corresponds to an 8.5-fold increase in the rate of 2-bp deletions in the msh3Δ strain. The rate of neither the 94-bp deletion nor the 2-bp deletions was elevated in any of the other MMR-deficient strains analyzed.
In the msh6Δ strain, the percentages of lys2ΔA746 reversion events occurring in the six-A and five-T tracts were similar to those in the wild-type strain (Table 1). Most of the remaining reversion events resulted from the addition of a single adenine immediately 3′ of the four-C run. This hot spot accounted for 30% (19 of 63) of the events in the msh6Δ strain but only 3% (3 of 86) of the total +1 events in a wild-type strain, corresponding to a 110-fold increase in the rate of this novel insertion.
Reversion of the lys2ΔA746 allele in strains defective in MutL homologs.
Disruption of PMS1 or MLH1 resulted in a 250- or 350-fold increase, respectively, in the spontaneous reversion rate of the lys2ΔA746 allele (Table 1). This is similar to the lys2ΔBgl reversion rates obtained when these genes were disrupted in the −1 assay strain (8). In both the pms1Δ and mlh1Δ +1 mutation spectra, the reversion events occurred almost exclusively in the six-A and five-T tracts; the +A hot spot adjacent to the four-C run was not prominent in these spectra (Fig. 1C). Although similar, the pms1Δ and mlh1Δ spectra are statistically significantly different if the distributions of events among the six-A run, the five-T run, and all other classes are compared (P < 0.05 by χ2 contingency test). It should be noted that a subtle difference between the pms1Δ and mlh1Δ spectra also was detected by the −1 assay system (8).
DISCUSSION
We previously described a frameshift detection assay based on reversion of the +4 frameshift allele lys2ΔBgl (8). Reversion of this allele specifically detects compensatory mutational events that alter the number of base pairs within a defined reversion window by 3N − 1, where N is the number of base pairs. The majority of frameshift events detected by this system are single base pair deletions derived from slippage events that place an extrahelical base on the template strand during DNA synthesis. Because most reversion events of the lys2ΔBgl allele arise through the deletion of a single base pair, we will refer to this system as the −1 assay system. In the current study, we have developed a similar +1 assay system based on reversion of the −1 frameshift allele lys2ΔA746. An important feature of the −1 and +1 frameshift assay systems is that the mutational events are constrained to occur within a common, approximately 150-bp region of the LYS2 locus. This feature allows direct comparison of the −1 spectra generated previously (8) with the +1 frameshift spectra generated in this study. In both studies, we have focused on the removal of frameshift intermediates by the yeast MMR machinery and have generated frameshift spectra in wild-type strains and in strains defective in individual MMR proteins. Because proofreading is still operative in the MMR-defective strains, the events that are proportionally elevated in MMR mutants correspond to mutational intermediates that are processed primarily via the MMR system. Although we assume below that most frameshift intermediates are generated during DNA replication, we note that such intermediates may also arise during DNA repair processes. It is not known whether the MMR machinery corrects errors generated during DNA repair.
+1 versus −1 frameshift events in wild-type strains.
The rates at which mutation events occurred in the −1 and +1 assay systems were similar in a wild-type background: 2.8 × 10−9 reported previously for the −1 assay strain (8) versus 1.4 × 10−9 for the +1 assay strain in this study. This twofold difference in wild-type rates is due to a slight variability in experimental procedures, as plating of both strains in parallel yields indistinguishable rates (10). Although the mutation rates are similar, the spectra obtained with the +1 and −1 assays are very different (Fig. 1A). For example, single base pair insertion or deletion events represent 94% of the −1 spectrum but only 83% of the +1 spectrum (P < 0.01 by χ2 contingency test). Also, events generally are much less clustered in the −1 spectrum than in the +1 spectrum.
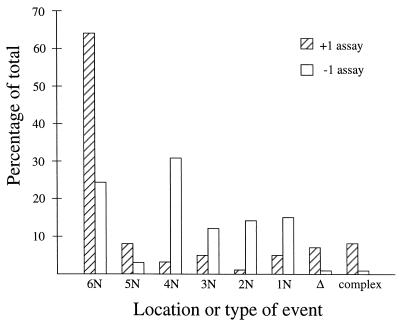
A useful way to classify the mutational events is by type according to whether they occur in a noniterated sequence (1N sequence) or in repeated sequences of defined lengths (2N to 6N runs). Additional classifications include large deletion events and complex events. Figure 2 graphically compares the proportions of the different types of events obtained with the +1 and −1 assay systems. Our previous analyses with the −1 system indicated that runs of ≥4N are hot spots for frameshift events (8). Relative to the −1 assay system, there was a significant decrease in compensatory frameshift events in mononucleotide runs of <4N and in noniterated sequences in the +1 assay system (P < 0.001 by χ2 contingency test; complex reversion events and deletions between direct repeats were omitted from this analysis). Mutations in short runs and noniterated sequences have been proposed to occur through a dislocation mechanism in which a misincorporation is followed by a misalignment that restores base pairing of the 3′ end of the nascent strand with the template (19). A decrease in these types of events in the +1 assay system relative to the −1 assay system suggests that the misaligned intermediate either is repaired more efficiently in this system or does not lead to insertion events as frequently as deletion events.
FIG. 2.
Distributions of frameshift mutations in the +1 versus −1 assay systems in wild-type strains. Events were classified by location in noniterated sequences (1N) or in tandem repeated sequences (2N to 6N). Large deletions (Δ; >3 bp) and complex events were considered separate classes. The percentage of total events in each class is indicated.
The distribution of reversion events among the 4N (single four-C and four-A runs), 5N (a single five-T run), and 6N (a single six-A run) runs also differed between the two systems. In the +1 assay system, an increase in the number of reversion events was observed at both the six-A (threefold) and five-T (twofold) tracts compared to the number of events occurring at these sites in the −1 system. With the 4N tracts, the reverse was true, with 20-fold fewer reversion events being observed in the +1 assay system than in the −1 system (see below for further discussion). In addition to the simple insertions or deletions, six complex insertions and two complex deletions were observed in the +1 assay strain (8 of 104 [7.7%]) whereas only a single complex insertion (1 of 144 [0.7%]) was observed in the −1 system. This represents an 11-fold increase in complex events in the +1 system relative to the −1 assay system. Finally, there was a 10-fold increase in large deletion events in the +1 assay system (7 of 104 [6.7%]) relative to the −1 assay system (1 of 144 [0.7%]). In the −1 assay system, the deletions were flanked by a 6-bp direct repeat that was enlarged to a 10-bp direct repeat during the engineering of the +1 assay system (Fig. 1A). Six of the seven large deletions observed in the +1 system occurred between the 10-bp repeats; the remaining deletion occurred between 4-bp direct repeats. Because the occurrence of deletions between direct repeats is known to increase as a function of repeat size (30, 33, 36, 41), the increase in repeat size likely accounts for the observed differences in the rates of large deletions in the +1 and −1 assay systems.
Perhaps the most notable feature of the distributions of +1 versus −1 events is the large deficit in +1 events occurring in the two 4N runs (3 of 104 [2.9%]) relative to −1 events in these runs (44 of 144 [31%]). The 10-fold difference in the rates of +1 versus −1 events in the 4N runs can be accounted for if one assumes either (i) that DNA polymerase generates 10 times more −1 frameshift intermediates than +1 intermediates, (ii) that exonucleolytic proofreading of extrahelical bases on the nascent strand is 10-fold more efficient than that of extrahelical bases on the template strand, or (iii) that the removal of +1 frameshift intermediates by the MMR system is 10-fold more efficient than removal of −1 intermediates. It also is possible that the overall difference in the rates of +1 versus −1 frameshifts observed in this system reflects differences at more than one step. We think it unlikely that the disparity originates entirely in the polymerization reaction, as such large differences are not evident in in vitro systems (18). The fact that the large disparity is evident in MMR-deficient strains as well (Fig. 1 and reference 8) argues that the MMR system removes +1 and −1 frameshift intermediates with similar efficiencies. Thus, the most likely source of the observed disparity is more efficient proofreading of extrahelical bases on the nascent strand than on the template strand.
Based on bacterial studies, Streisinger et al. (39) proposed that a loop on the nascent strand might be more efficiently removed by proofreading than a loop on the template strand because of its accessibility to the 3′ exonuclease editing activity of DNA polymerase. In vitro evidence for this phenomenon has been obtained by Kroutil et al. (18). They demonstrated that base pair additions are proofread by the 3′ exonuclease activity of T7 DNA polymerase more efficiently than are deletions in AT tracts of 5, 6, or 7 bp, although the opposite appeared to be true for a smaller AT tract of 4 bp. It should be noted that the vast majority of events in our assays occurred in the four-C tract rather than in the four-A tract, so sequence context (run composition and/or flanking sequences), in addition to run length, clearly impacts frameshift spectra. In addition to the bias reported here, a general bias for deletion versus insertion types of frameshift events has been reported in studies of dinucleotide repeat stability in MMR-defective yeast (35, 37, 38), an observation that can be explained in the context of a potential proofreading bias. Although our data indicate that the bias does not originate with the MMR system, it must be demonstrated that the bias does not originate with polymerization in order to conclude that proofreading is the source of the bias. It should be possible to address the inherent error rate of the yeast DNA polymerases by examining frameshift rates and spectra in strains that are simultaneously proofreading defective and MMR defective.
+1 frameshift events in MMR-defective strains.
The reversion rates of MMR-defective strains, with the exception of those of the msh6Δ and msh3Δ strains, were similar to the rates obtained in the −1 assay strain (8). Disruption of MSH6 resulted in an 11-fold increase in the mutation rate in the +1 assay system but only a 1.6-fold increase in the −1 assay strain. The reverse was true of an msh3Δ strain, where a 3.8-fold increase was seen with the −1 assay system, but no increase was detectable with the +1 assay. With the −1 assay system, more than 90% of the frameshift events in the MMR-defective strains with strong mutator phenotypes were in the six-A and four-C runs.
Mutation rates at three +1 hot spots were elevated more than 100-fold in an msh2 strain: the six-A run, the five-T run, and the insertion of a single adenine (+A) immediately 3′ of the four-C run. Although homopolymer runs have been shown previously to be hot spots for frameshift events in MMR-defective strains (8, 22, 35, 43), the +A adjacent to the four-C run is unique. In 37 of 38 cases, where an insertion occurred next to the four-C run, the inserted base was an adenine. Elimination of Msh6p resulted in a more-than-100-fold increase in the occurrence of the unique A insertion, but this hot spot was not evident in strains lacking Msh3p. The presence of the +A mutational hot spot in strains where Msh2p and Msh3p were present (i.e., in an msh6Δ strain) demonstrates either that the Msh2p-Msh3p complex cannot correct this type of mistake or that the repair process is very inefficient. It should be noted that by using the −1 assay strain, we previously identified an msh6-specific hot spot in a three-T run. Together, these data suggest novel functions for the Msh2p-Msh6p complex in resolving both +1 and −1 frameshift intermediates. We were not able to determine if either Mlh1p or Pms1p was required along with Msh6p and Msh2p to correct the novel +A frameshift intermediate due to the large reversion rate increases at the six-A and five-T tracts in these mutant strains. In addition to the unique +A mutation in the +1 assay system, the msh6 strain exhibited 9-fold and 15-fold increases in the rate of reversion events at the six-A and five-T tracts, respectively, relative to the wild-type strain. In an msh3 mutant strain, these rate increases were not observed, again supporting the idea that Msh3p and Msh6p are both capable of recognizing similar types of frameshift intermediates but have distinct specificities and/or efficiencies in vivo. Although our data do not address the basis of these differences, recent in vitro data obtained by using human Msh6p have suggested that the sequence context flanking a frameshift intermediate may influence the manner in which the MMR machinery corrects DNA mismatches (21).
The repeated recovery of revertants containing the addition of a single adenine after the four-C tract suggests that this insertion event is templated from another location in the genome (29). Visual analysis of sequences surrounding this site has identified two possible regions that might serve as templates. The first site is 5 bp in length (5′-CATCA) and is located within the reversion window, approximately 55 bp downstream of the +A hot spot. The +A mutation converts an imperfect direct repeat (C-TCA; the dash corresponds to the position of the +A hot spot) to a perfect direct repeat (5′-CATCA). The second site (5′-TGATGGGTGTC; underlined bases are not present at the +A hot spot) is an imperfect inverted repeat of sequences surrounding the +A hotspot (5′-GACCCC*TCA; the asterisk corresponds to the site of the adenine insertion) and is located approximately 500 bp downstream of the +A hotspot. Mispairing with either of these sites could result in the templated insertion of the adenine immediately 3′ of the four-C run. Further studies involving site-directed mutagenesis of the candidate template sequences are needed in order to determine whether either of these sites indeed is relevant to the +A hot spot. In addition to a templating mechanism to explain the +A hot spot, the repeated insertion of an A could also be due to a dislocation (i.e., misincorporation followed by slippage) type of mechanism. Given the flanking sequences, however, the dislocation mechanism would involve either tandem misincorporations or mispairing at the 3′ end following slippage.
In the msh3Δ strain, we observed a threefold increase in the occurrence of 94-bp deletions with endpoints in 10-bp direct repeats. The elevated rate of this deletion specifically in the msh3Δ strain suggests that Msh3p participates in the removal of the large looped structure formed by slipped mispairing between the 10-bp direct repeats. Tran et al. (42) previously identified a role for Msh3p in resolving DNA loops of 7 bp or less, but the resolution of larger DNA loops did not appear to be dependent on Msh3p. In their study, however, it was necessary to use a pol3-t mutant in order to detect deletion events of greater than 1 bp and use of the mutant polymerase may have impacted the results. Genetic data indicate that Msh2p and Rad1p are involved together in the repair of 26-nt loops formed during meiotic recombination (16), in the removal of nonhomologous DNA tails formed during mitotic recombination (6), and in the recognition of 12-nt loops formed during mitotic recombination (24). It also has been demonstrated that yeast MMR proteins interact physically with components of the nucleotide excision repair machinery (3). We therefore examined the role of Rad1p in the removal of intermediates for the 94-bp deletion in our +1 assay system. We observed a 3.7-fold increase in the rate of the 94-bp deletion in a rad1 mutant (10), an increase that is similar to the 3.4-fold increase observed in the msh3 mutant. Our data thus are consistent with the involvement of an Msh3p/Rad1p-containing complex in the removal of large loops leading to the formation of deletions. Whether a similar complex is involved in the removal of large loops leading to the formation of duplications is not known, although this could be examined by using the lys2ΔBgl −1 assay system in a rad27 mutant (40).
In addition to the significant increase in the rate of the 94-bp deletion, a 10-fold increase in rate of 2-bp deletions was observed specifically in the msh3 mutant (the 2-bp deletions were not elevated in a rad1 mutant; 10). Msh3p has been shown to be involved in the removal of −2 frameshift intermediates in poly(GT) tracts, with Msh6p appearing to play a relatively minor role (35). Because of the overall reversion rate increase observed in the msh6 mutant, it is not possible to determine whether Msh6p similarly is involved in the removal of 2-nt loops in our assay system. Although 2-bp insertions can be detected in the −1 assay system, very few +2 events have been seen and there is no evidence that they are elevated in MMR-defective strains (8).
The +1 assay system developed in this study extensively overlaps the −1 assay system described previously (8), thus allowing direct comparison of +1 versus −1 frameshift spectra for a defined region of the LYS2 locus. Our analyses of wild-type strains have demonstrated a clear difference in the +1 and −1 mutational spectra, which correspond to slippage events generating extrahelical bases on the nascent and template strands, respectively, during DNA synthesis. Analyses of MMR-defective strains have revealed a role for Msh3p in the recognition and correction of large DNA loops, as well as a novel role for Msh6p in the removal of a unique +A intermediate. We suggest that the nature and/or location of the initial slippage event, on either the template or the nascent DNA strand, plays an important role in determining how a cell will repair the error.
ACKNOWLEDGMENTS
We gratefully acknowledge the contributions of C. Greene and N. Morey in constructing and generously donating strain SJR922 for use in this study. We thank K. Hill, G. F. Crouse, and members of the S.J.-R. lab for critically reading the manuscript.
This work was funded by a grant from the National Science Foundation.
REFERENCES
- 1.Acharya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol Cell Biol. 1996;17:2436–2447. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertrand P, Tishkoff D X, Filosi N, Dasgupta R, Kolodner R D. Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc Natl Acad Sci USA. 1998;95:14278–14283. doi: 10.1073/pnas.95.24.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, Jinks-Robertson S. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol Cell Biol. 1998;18:6525–6537. doi: 10.1128/mcb.18.11.6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crouse G F. Mismatch repair systems in Saccharomyces cerevisiae. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. 1. DNA repair in prokaryotes and lower eukaryotes. Totowa, N.J: Humana Press; 1998. pp. 411–448. [Google Scholar]
- 6.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 7.Flores-Rozas H, Kolodner R D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greene C N, Jinks-Robertson S. Frameshift intermediates in homopolymer runs are removed efficiently by yeast mismatch repair proteins. Mol Cell Biol. 1997;17:2844–2850. doi: 10.1128/mcb.17.5.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habraken Y, Sung P, Prakash L, Prakash S. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr Biol. 1996;6:1185–1187. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- 10.Harfe, B. D., and S. Jinks-Robertson. Unpublished data.
- 11.Hollingsworth N M, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- 12.Iaccarino I, Palombo F, Drummond J, Totty N F, Hsuan J J, Modrich P, Jiricny J. MSH6, a Saccharomyces cerevisiae protein that binds to mismatches as a heterodimer with MSH2. Curr Biol. 1996;6:484–486. doi: 10.1016/s0960-9822(02)00516-x. [DOI] [PubMed] [Google Scholar]
- 13.Jinks-Robertson S, Greene C, Chen W. Genetic instabilities in yeast. In: Wells R D, Warren S T, editors. Genetic instabilities and hereditary neurological diseases. San Diego, Calif: Academic Press; 1998. pp. 485–507. [Google Scholar]
- 14.Johnson R E, Kivvali G K, Guzder S N, Amin N S, Holm C, Habraken Y, Sung P, Prakash L, Prakash S. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J Biol Chem. 1996;271:27987–27990. doi: 10.1074/jbc.271.45.27987. [DOI] [PubMed] [Google Scholar]
- 15.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 16.Kirkpatrick D T, Petes T D. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature. 1997;387:929–931. doi: 10.1038/43225. [DOI] [PubMed] [Google Scholar]
- 17.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 18.Kroutil L C, Register K, Bebenek K, Kunkel T A. Exonucleolytic proofreading during replication of repetitive DNA. Biochemistry. 1996;35:1046–1053. doi: 10.1021/bi952178h. [DOI] [PubMed] [Google Scholar]
- 19.Kunkel T A, Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- 20.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 21.Macpherson P, Humbert O, Karran P. Frameshift mismatch recognition by the human MutSα complex. Mutat Res. 1998;408:55–66. doi: 10.1016/s0921-8777(98)00017-2. [DOI] [PubMed] [Google Scholar]
- 22.Marsischky G T, Filosi N, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 23.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson, A., M. Hendrix, S. Jinks-Robertson, and G. F. Crouse. Unpublished data.
- 25.Palomba F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMUTSβ, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 26.Prolla T A, Christie D-M, Liskay R M. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prolla T A, Pang Q, Alani E, Kolodner R D, Liskay R M. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science. 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 28.Reenan R A G, Kolodner R D. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochrondrial and nuclear functions. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ripley L S. Frameshift mutation: determinants of specificity. Annu Rev Genet. 1990;24:189–213. doi: 10.1146/annurev.ge.24.120190.001201. [DOI] [PubMed] [Google Scholar]
- 30.Ripley L S, Clark A, deBoer J G. Spectrum of spontaneous frameshift mutations: sequences of bacteriophage T4 rII gene frameshifts. J Mol Biol. 1986;191:601–613. doi: 10.1016/0022-2836(86)90448-1. [DOI] [PubMed] [Google Scholar]
- 31.Ross-Macdonald P, Roeder G S. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994;79:1069–1080. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 32.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 33.Schaaper R M, Dunn R L. Spontaneous mutation in the Escherichia coli lacI gene. Genetics. 1991;129:317–326. doi: 10.1093/genetics/129.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 35.Sia E A, Kokoska R J, Dominska M, Greenwell P, Petes T D. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer B S, Westlye J. Deletion formation in bacteriophage T4. J Mol Biol. 1988;202:233–243. doi: 10.1016/0022-2836(88)90454-8. [DOI] [PubMed] [Google Scholar]
- 37.Strand M, Earley M C, Crouse G F, Petes T D. Mutations in the MSH3 gene preferentially lead to deletions within tracts of simple repetitive DNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:10418–10421. doi: 10.1073/pnas.92.22.10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strand M, Prolla T A, Liskay R M, Petes T D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 39.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 41.Tran H T, Degtyareva N P, Koloteva N N, Sugino A, Masumoto H, Gordenin D A, Resnick M A. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran H T, Gordenin D A, Resnick M A. The prevention of repeat-associated deletions in Saccharomyces cerevisiae by mismatch repair depends on size and origin of deletions. Genetics. 1996;143:1579–1587. doi: 10.1093/genetics/143.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran H T, Keen J D, Kricker M, Resnick M A, Gordenin D A. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 45.Umar A, Kunkel T A. DNA-replication fidelity, mismatch repair and genome instability in cancer cells. Eur J Biochem. 1996;238:297–307. doi: 10.1111/j.1432-1033.1996.0297z.x. [DOI] [PubMed] [Google Scholar]