Abstract
Early diagnosis and timely management of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) are the keys to preventing the spread of the epidemic and controlling new infection clues. Therefore, strengthening the surveillance of the epidemic and timely screening and confirming SARS-CoV-2 infection is the primary task. In this work, we first proposed the idea of activating CRISPR-Cas12a activity using double-stranded DNA amplified by a three-dimensional (3D) DNA walker. We applied it to the design of an electrochemiluminescent (ECL) biosensor to detect the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) gene. We first activated the cleavage activity of CRISPR-Cas12a by amplifying the target DNA into a segment of double-stranded DNA through the amplification effect of a 3D DNA walker. At the same time, we designed an MXene based ECL material: PEI-Ru@Ti3C2@AuNPs, and constructed an ECL biosensor to detect the RdRp gene based on this ECL material as a framework. Activated CRISPR-Cas12a cleaves the single-stranded DNA on the surface of this sensor and causes the ferrocene modified at one end of the DNA to move away from the electrode surface, increasing the ECL signal. The extent of the change in electrochemiluminescence reflects the concentration of the gene to be measured. Using this system, we detected the SARS-CoV-2 RdRp gene with a detection limit of 12.8 aM. This strategy contributes to the rapid and convenient detection of SARS-CoV-2-associated nucleic acids and promotes the clinical application of ECL biosensors based on CRISPR-Cas12a and novel composite materials.
Keywords: SARS-CoV-2, RdRp gene, CRISPR-Cas12a, ECL, Mxene, PEI-Ru@Ti3C2@AuNPs
Graphical abstract
1. Introduction
SARS-CoV-2 is a positive single-stranded RNA virus that coexists with zoonotic animals and is a coronavirus. The disease caused by it was officially named Coronavirus Disease 2019 (COVID-19). High-throughput sequencing showed that the pathogenic virus is a new type of severe acute respiratory syndrome (SARS) -coronavirus (SARS-CoV-2) [1]. The source of Covid-19 infection is mainly SARS-CoV-2 infected patients, including asymptomatic infections. Respiratory droplets and contact transmission are the main transmission routes. The transmission routes such as aerosols and digestive tract have yet to be clarified. SARS-CoV-2 infected patients usually present with pneumonia-like symptoms (fever, dry cough, and dyspnea) and gastrointestinal symptoms such as diarrhea, followed by severe acute respiratory infections [2]. Some cases may have acute respiratory distress with severe respiratory symptoms, complications, and even death. At present, there is no specific treatment for SARS-CoV-2 infected patients. Early diagnosis and timely management are the keys to preventing the spread of the epidemic and controlling new infection clues [3]. Therefore, strengthening the surveillance of the epidemic and timely screening and confirming SARS-CoV-2 infection is the primary task. Reverse transcription-polymerase chain reaction (RT-PCR) to detect viral nucleic acid is considered the gold standard for diagnosing SARS-CoV-2 [4]. However, this method has higher requirements for experimental conditions, facilities, personnel, many influencing factors, operation steps, and long detection time [1]. More importantly, although the specificity of nucleic acid detection is high, due to various reasons, the false-negative rate is high, which limits the detection of nucleic acid. There is no doubt that the COVID-19's rapid and accurate identification can significantly help control the emerging pandemic.
Biological protein machinery is ubiquitous in biological systems and performs various physiological functions, including mechanical drive, intracellular transport, and signal transduction [5]. Inspired by these biological machines, researchers have tried to create various artificial molecular machines and motors that can use controlled molecular-level motion to perform specific tasks. Relying on the predictability, specificity, and versatility of Watson-Crick base coordination theory, researchers have designed and synthesized various mechanical devices made of DNA molecules, including DNA Walker, DNA tweezers, DNA motors, and DNA robots [6]. DNA Walker, which can be precisely controlled on the micron or nano DNA tracks, has shown great potential in biosensor analysis [7]. At present, the emergence of three-dimensional (3D) DNA orbits has brought new perspectives to DNA walkers' research [8,9]. Compared with the restricted one-dimensional DNA track or two-dimensional DNA origami such as the DNA Walker track, the 3D DNA track composed of micro- or nano-particles has a larger specific surface area [6]. So the three-dimensional DNA track has a more vital ability to concentrate and expand. For example, Ellington and his colleagues first developed a 3D DNA Walker on the surface of particles using the principle of catalytic hairpin self-assembly (CHA) [10]. In this way, similar DNA Walkers based on CHA principles have appeared one after another. Besides, Yin and his colleagues reasonably designed an entropy-driven 3D DNA walking sensor [11]. Different from the DNA Walker based on the CHA principle, the entropy-driven DNA Walker uses a single-stranded linear DNA molecular sequence, thereby avoiding the use of complex pseudoknots, the secondary structure of the kiss ring, or high background signals in the DNA molecules. A simple DNA hybridization reaction activates the enzyme-free three-dimensional DNA Walker. Various signal amplification methods for protein detection have been proposed.
Compared with previous biosensors constructed based on isothermal amplification strategy [12,13], this 3D-DNA Walker can be based on a highly localized entropy-driven reaction, and the chain substitution reaction triggered by the walking arm walking will be strictly limited to the Au particle surface, and no chain substitution reaction will occur across the particles. Thus the highly localized motion of a single molecule walking arm on the surface of a single nanoparticle will generate enough nucleic acid duplexes to allow a more complete DNA walker amplification reaction on the surface of that nanoparticle.
As we all know, the polymerase chain reaction (PCR) is one of the most widely used amplification techniques and is very important for biological research. Instead, it is limited by labor-intensive work, precise temperature control, and potential differences between batches. In contrast, CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is a relatively new nucleic acid detection method. Since the application of the modified CRISPR/Cas9 system for gene editing of mammalian genomes, the CRISPR/Cas system has taken center stage in biotechnology [2]. A large number of researchers have worked to develop a variety of nucleic acid amplification systems by exploiting the side-cleavage (Trans) activity of CRISPR/Cas nucleases such as CRISPR/Cas12a, CRISPR/Cas12b, and CRISPR/Cas13a [14]. This technology is now considered an innovative approach for next-generation diagnostics and has been identified as having the potential to significantly impact the development of biosensors by providing a faster and more accurate ultra-sensitive nucleic acid detection method [15]. The CRISPR system consists of a guide RNA (gRNA) and a Cas12a protein, and the gRNA is able to direct Cas to recognize and cleave RNA or DNA molecules with specific sequences [16,17]. Among them, Cas12a protein will continue to cut non-target DNA indiscriminately when it explicitly cuts the target DNA. Using this feature, DNA signal molecules with signal markers are added to the whole system, which will cause a change in signal once the DNA with signal is cut, and this technology combined with isothermal amplification technology can detect the nucleic acid to be detected with high specificity and sensitivity [16].
Electrochemiluminescence (ECL) technology has been active in the research of disease marker analysis. ECL not only has the advantages of ultra-sensitive detection and wide range of detection by traditional electrochemical methods, but also has the characteristics of popularized analytical tools and good signal stability [[18], [19], [20], [21]]. In recent years, ECL has been widely used as a powerful analytical technique for the detection of biomolecules in clinical, environmental, and industrial fields. It has the advantages of high sensitivity, low cost, simple operation, and easy miniaturization and intelligence to meet the development needs of clinical treatment [3]. However, traditional electrochemiluminescence-based biosensors are not perfect and need new breakthroughs due to the disadvantages of electrode interface potential resistance that affects the binding efficiency of enzyme molecules, complex interface environment that increases the probability of non-specific adsorption, easy diffusion of luminescent reagents in homogeneous solutions, poor luminescence stability, inability to modulate ECL spectra and reversible modulation, etc. [16,22] Therefore, this solution is not perfect and needs new breakthroughs. The continuous discovery of graphene derivatives and graphene-like 2D nanomaterials provides new development opportunities to develop solid-phase material-based luminescent electrochemical biosensors [23]. The reason is that 2D nanomaterials have the following advantageous properties: a) The electronic properties are significantly enhanced because the electrons are confined within the 2D planar structure, which makes 2D nanomaterials ideal electron transport carriers for optoelectronic sensors. b) The strong covalent bonds within the lamellae and the thickness of the atomic layers make 2D nanomaterials exhibit excellent mechanical strength, flexibility and light transmission. Among them, Ti3C2Tx (MXene), as a new 2D lamellar material, possesses high intrinsic photoelectric conversion effect, while the abundant functional groups on its surface enable efficient modification and good homogeneous dispersion, which makes it highly promising as an ECL material for the construction of various ECL sensors [16].
Considering that the amplification product of DNA track-based DNA Walker contains DNA double-stranded, we pioneered the combination of DNA walker and CRISPR/Cas12a amplification by using the amplification product double-stranded DNA as a linkage, in order to achieve highly sensitive detection of novel coronaviruses. This double-stranded DNA can further activate CRISPR/Cas 12a, which can indiscriminately cut the single-stranded DNA on the surface of the electrode, thus causing a change in the ECL signal, which can reflect the concentration of RdRp gene to be tested. Using this detection strategy, gene detection at 12.8 aM can be achieved. Therefore, the present method has potential application in the clinical screening of SARS-CoV-2 gene.
2. Experimental section
2.1. Materials and reagent
Gold chloride trihydrate (HAuCl4·3H2O), sodium citrate, sodium borodeuteride (NaBH4) were obtained from Aladdin Biochemical Technology Co. Ltd. (Shanghai, China). Ti3C2 dispersion liquid was obtained from Jiangsu XFNANO Materials Tech, Co. Ltd. (Nanjing, China). Tris (4,4′-dicarboxylic acid-2,2′-bipyridyl) ruthenium (II) dicbloride (Ru(dcbpy)3Cl2) was obtained from Suna Tech Inc. (Suzhou, China). Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) and 6-mercaptohexanol (MCH) were gained from Sigma-Aldrich (St Louis, MO, USA). The purified DNA sequences (Table S1) were synthesized by Genscript Bio-technology Co. Ltd. (Nanjing, China).
2.2. Preparation of the PEI-Ru@Ti3C2@AuNPs-DNA7
The PEI-Ru@Ti3C2@AuNPs-DNA7 probe was successfully synthesized with slight modification by referring to our previous literature [16]. 10 mg of Ru(dcbpy)3 2+ and EDC (200 mM)/NHS (50 mM) were simultaneously dissolved in 5 mL of PBS (0.1 M, pH = 7.4) with continuous stirring for 2 h. The Ti3C2 suspension and PEI (1%, w/v) were mixed well and added to the PBS (0.1 M, pH = 7.4) of Ru(dcbpy)3 2+ for 1 h. Subsequently, 600 μL of HAuCl4 (0.01 M) and 600 μL of fresh NaBH4 (0.01 M) were added to the above solution and stirred for 1 h. To remove excess impurities, the prepared PEI-Ru@Ti3C2@AuNPs were centrifuged and washed three times and redispersed in 1 mL of PBS (0.1 M, pH = 7.4). To prepare PEI-Ru@Ti3C2@AuNPs-DNA7 probes, we added 200 μL of DNA7 (10 μM) to 1 mL of Ru@Ti3C2@AuNPs and stored in a refrigerator at 4 °C for overnight. Finally, 100 μL of MCH (100 μM) was added to reduce non-specific adsorption. In this way, PEI-Ru@Ti3C2@AuNPs-DNA7 probes were successfully prepared.
2.3. Preparation of 3D DNA walker
Gold nanoparticles (AuNP) with a diameter of 13 nm were prepared as follows: after heating 200 mL of HAuCl4 solution (0.01%) to 100 °C, 5.0 mL of trisodium citrate (1%) was rapidly added to the boiling solution under continuous stirring with a rotor. The reaction mixture was then continued to stir at 100 °C for 1 h until the color of the solution turned deep red and then stored at 4 °C.
The DNA1, DNA2 and DNA3 (20 μM each) solutions were first mixed and heated to 95 °C for 10 min, and then the nucleic acid mix was cooled naturally to room temperature for approximately 4 h. Next, 40 μL of the nucleic acid mix (20 μM DNA4 and 20 μM DNA1/DNA2/DNA3 complex with a volume ratio of 20:1) was incubated with 5 mM TCEP for 15 min and then mixed with AuNPs to a total volume of 6 μL. The above solutions were then mixed with 40 μL phosphate buffer (pH 7.5, 100 mM) and 180 μL ddH2O to make a final volume of 400 μL and incubated for 15 min at room temperature. An equal volume of 400 μL NaCl solution (2.0 M) was added to the above solution in ten drops and shaken vigorously, and then incubation was continued for 2 h at room temperature. Afterward, the excess thiol modified ligonucleotides were completely removed by centrifugation at 12 000 rpm for 15 min at 4 °C and the precipitated AuNPs were resuspended in 400 μL of 1 × PBS containing 0.01% Tween-20. Next, the above DNA-functionalized AuNP solution was mixed with 12 μL of MCH (100 M) and incubated for 1 h, thereby sealing the excess sites on the surface of the gold nanoparticles. Then 1 × PBS containing 0.01% Tween-20 was added and washed three times by centrifugation at 12 000 rpm for 15 min at 4 °C. The DNA-functionalized AuNPs were then resuspended in 400 μL of 1 × PBS containing 0.01% Tween-20.
2.4. 3D-DNA walker and CRISPR-Cas12a assay
Different concentration of RdRp gene incubated with 100 pM DNA-AuNP (DNA1/DNA2/DNA3 complex and DNA4), 5 nM DNA5 and 320 nM DNA6 for 60 min to generate DNA2/DNA6 duplex (contains Prototype Spacer Adjacent Mass (PAM) Sequence 5′-TTTA-3′). Next, 50 μL of the above reaction solution was added into the cleavage buffer (20 mM tris HCl, 100 mM KCl, 5 mM MgCl2, 5% glycerol, and 1 mM DTT) containing 100 nM CRISPR-Cas12a/gRNA and incubated for 30 min to remove the excess free DNA from the solution by the shearing effect of CRISPR/Cas12a. For ECL assay, the DNA7 modified PEI-Ru@Ti3C2@AuNPs biosensor was immersed in the above prepared mixed solution for 20 min. The ECL signal was captured by ECL-6B which is a gift from the State Key Laboratory of Analytical Chemistry for Life Sciences, Nanjing University, and the scanning range was from 0 to −1.25 V. Three-electrode system was used for testing, with a modified 3 mm GCE as the working electrode, platinum wire electrode as the counter electrode and Ag/AgCl as the reference electrode.
3. Results and discussion
3.1. The working principle
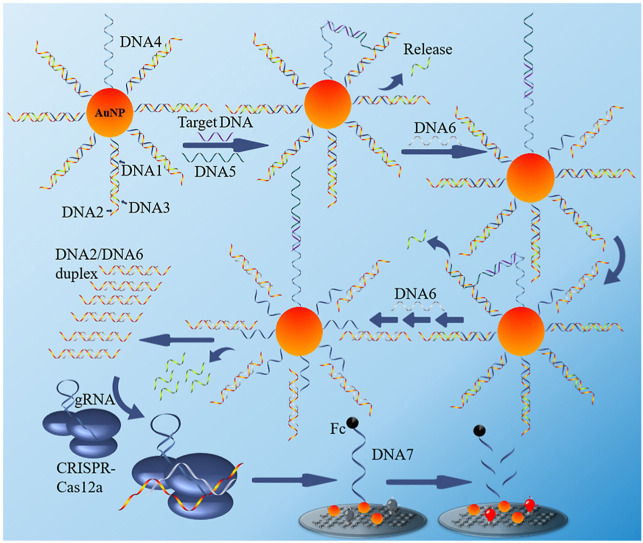
As shown in Scheme 1 , The system we designed contains two parts: a 3D nano-machine and a CRISPR/Cas12a-based nucleic acid amplification ECL sensor. In the first part, our nano-machine mainly contains a DNA-modified AuNP (DNA-AuNP) a walking leg (DNA5), and a fuel (DNA6). The DNA-AuNP is a conjugate with a three-stranded substrate complex (DNA1/DNA2/DNA3) and affinity ligand (DNA4) on a single AuNP. The detailed sequence information for the nano-machine is shown in Table S1. DNA1 was modified with a thiol at 5′ end to modified on the AuNP surface and hybridized with part of DNA2, which co-hybridizes to DNA3. This forms a sandwich structure with a toehold at the 5′ end of DNA2. The recognition sequences for target DNA (RdRp gene) are designed to embed in DNA4 and DNA5, respectively. The binding target DNA and DNA5 to DNA4 bring DNA5 into proximity to the AuNP surface, leading to tethered to the AuNP to form a walkable leg with the capability to perform highly effective intramolecular hybridization. The entropy-driven catalytic reaction occurs as follows: DNA5 interacts with DNA2 via toehold and displaces DNA3 from DNA1/DNA2/DNA3 complex via toehold-mediated strand displacement to expose a new toehold on DNA2. DNA6 hybridizes to the new toehold and triggers branch migration to form a DNA2/DNA6 duplex (contains PAM Sequence 5′-TTTA-3′), resulting in the complete disassembly of the DNA1/DNA2/DNA3 complex. The interaction of DNA2 with DNA5 liberates DNA4 to move along the DNA-AuNP track to react with a toehold on another DNA1/DNA2/DNA3 complex, triggering autonomous surface-bound DNA1/DNA2/DNA3 disassembly the formation of more DNA2/DNA6 duplex. Following this pattern of continued operation, more and more DNA2/DNA6 duplex forms and leaves the AuNP surface.
Scheme 1.
Schematic illustration of the 3D-DNA walker and CRISPR Cas 12a based ECL biosensor for the detection of SARS-COV-2 RdRp gene.
The second part of the system mainly consists of the electrochemiluminescent sensor and CRISPR/Cas12a. We ingeniously designed sequences in the DNA2/DNA6 duplex that can activate CRISPR/Cas12a activity. When the first part of the DNA walker reaction is complete, the remaining gold nanoparticles/DNA complexes in the above solution are separated from the solution by centrifugation, and CRISPR/Cas12a is added to the solution and incubated for an hour to remove the excess free DNA from the solution by the shearing effect of CRISPR/Cas12a. Before cleavage, the electrode detects a low ECL signal caused by the system because the other end of the single-stranded DNA modified on the electrode surface is modified with ferrocene (Fc), which quenches the ECL signal on the electrode surface. In contrast, when activated CRISPR/Cas12a is present in the system, CRISPR/Cas12a cuts off the single-stranded DNA, causing the ferrocene to move away from the electrode surface, resulting in an increase in the ECL signal. Therefore, the intensity of the ECL signal can reflect the concentration of the RdRp gene in the system to be tested.
3.2. Feasibility assay
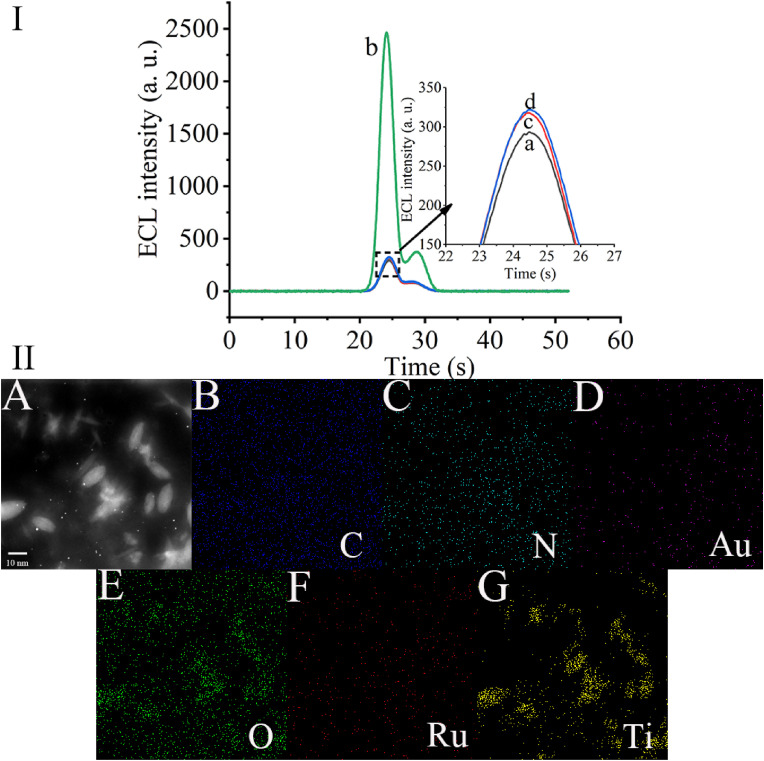
We first performed the feasibility analysis of this system (Fig. 1 I). When no nucleic acid to be tested was present in the system, the system showed an extremely low ECL signal (curve a) because it could not trigger the amplification system to produce DNA2/DNA6 duplex, so it could not further activate the cleavage activity of CRISPR-Cas 12a. When 1000 aM of the nucleic acid to be tested was added to the system, a large amount of DNA2/DNA6 duplex was generated and successfully activated the cleavage activity of CRISPR-Cas 12a through the amplification effect of the DNA walker on the surface of the gold electrode, which in turn led to an increase in the ECL signal (curve b). When no gold nanoparticle-modified DNA walker was present in the system, the same low ECL signal was generated because the DNA walker reaction could not be triggered and no raw DNA2/DNA6 duplex could be produced to trigger the cleavage activity of CRISPR-Cas 12a (curve c). When CRISPR-Cas 12a was not added to the system, although the nucleic acid to be tested was able to trigger the DNA walker reaction and produce the DNA2/DNA6 duplex, CRISPR-Cas 12a was not present in the system and therefore did not cleave DNA7 on the electrode surface, and thus, only a low ECL signal was obtained (curve d). All these data corroborate the mechanism of this experiment and prove the feasibility of our designed system.
Fig. 1.
(I) Feasibility assay: (a) DNA7/PEI-Ru@Ti3C2@AuNPs modified electrode, (b) 1000 aM target DNA tested by using this system (with 100 pM DNA modified AuNP and 50 nM CRISPR-Cas12a/gRNA), (c) DNA7/PEI-Ru@Ti3C2@AuNPs modified electrode treated with 1000 aM target DNA and 50 nM CRISPR-Cas12a/gRNA (without DNA modified AuNP), and (d) DNA7/PEI-Ru@Ti3C2@AuNPs modified electrode treated with 1000 aM target DNA and 100 pM DNA modified AuNP (without CRISPR-Cas12a/gRNA). (II) SEM image of PEI-Ru@Ti3C2@AuNPs composites. (B–G) Elemental distribution mapping spectra of PEI-Ru@Ti3C2@AuNPs composites, C, N, Au, O, Ru, and Ti.
3.3. SEM characterization of PEI-Ru@Ti3C2@AuNPs
The PEI-Ru@Ti3C2@AuNPs composites were characterized by EDX (Energy dispersive X-ray spectroscopy) mapping as shown in Fig. 1(II). Fig. 1(II)A shows the SEM image of the composites, and it can be seen that the major elements, C (Fig. 1(II)B), N (Fig. 1(II)C), Au (Fig. 1(II)D), O (Fig. 1(II)E), Ru (Fig. 1(II)F), and Ti (Fig. 1(II)G), are uniformly dispersed in the composites. It shows that we successfully synthesized PEI-Ru@Ti3C2@AuNPs composites.
3.4. The mechanism of the ECL biosensor
The ECL enhancement mechanism of the ternary system (Ru(bpy)3 2+/PEI/Ti3C2) can be summarized as follows: (1) the co-reactants are modified PEI modified on MXene; (2) PEI is directly oxidized to PEI+ under applied voltage and then directly deprotonated to PEI• radicals (PEI•); (3) PEI• reduced Ti3C2/Ru(bpy)3 2+ to form Ti3C2/Ru(bpy)3 2+, which in turn reacts with PEI•+ in the oxidized state to form Ti3C2/Ru(bpy)3 2+*. Eventually, Ti3C2/Ru(bpy)3 2+* is finally converted to Ru(bpy)3 2+ and the ECL light signal is generated.
3.5. Characterization of electrode modification process
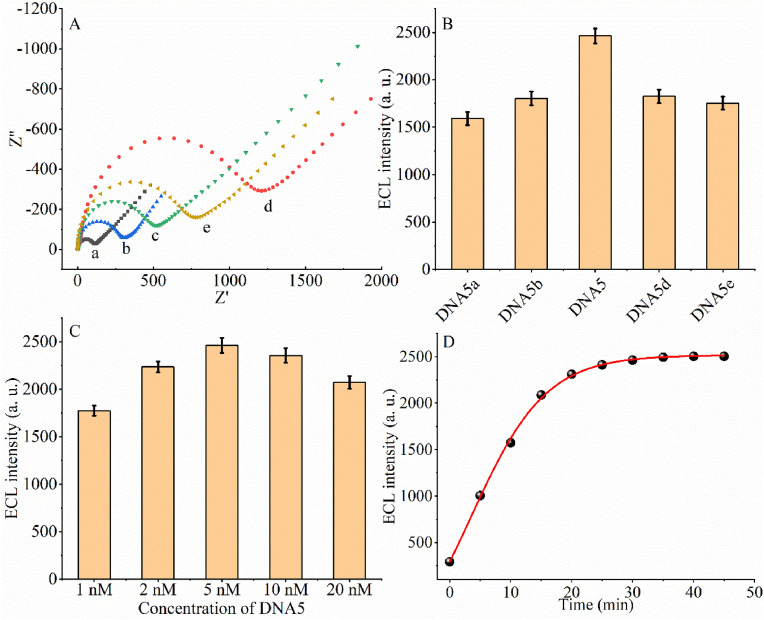
Electrochemical impedance spectroscopy (EIS), an effective method to probe the interfacial properties of electrode surface modifications, was also used to characterize the interfacial changes during the preparation of DNA walker and CRISPR-Cas12a based biosensors, and the results are shown in Fig. 2 A. The impedance plot of the bare gold electrode shows that the native impedance semicircle diameter is less than 200 (curve a), which indicates that the bare gold electrode has excellent conductivity and low impedance. When PEI-Ru@Ti3C2@AuNPs were modified on the electrode surface and became a uniform film, the electrode impedance increased significantly (curve b), which is mainly due to the dense PEI-Ru@Ti3C2@AuNPs film hindering the electron transfer, thus leading to a more significant impedance of the sensor. When DNA7 was assembled on the electrode surface through Au–S bonding, the impedance of the biosensor increased significantly (curve c). This indicates that DNA7 further hinders the electron transfer. When MCH was modified on the AuNP surface, the impedance value increased further (curve d), indicating that MCH sealed the electrode surface even further. When the prepared biosensor was incubated with the DNA2/DNA6 duplex activated CRISPR Cas12a/gRNA complex, the impedance value of the biosensor decreased significantly (curve e), indicating that DNA7 was cleaved by the CRISPR Cas12a/gRNA complex, resulting in an increased electron transfer rate. Therefore, this data suggests the successful assembly of the target sensor.
Fig. 2.
Optimization of the conditions: (A) EIS of the biosensor at different stages of modification: (a) bare GCE; (b) PEI-Ru@Ti3C2@AuNPs film modified GCE. (c) DNA7 and PEI-Ru@Ti3C2@AuNPs film modified GCE (d) MCH, DNA7, and PEI-Ru@Ti3C2@AuNPs film modified GCE. (e) DNA2/DNA6 duplex activated CRISPR Cas12a/gRNA complex treated DNA7 and PEI-Ru@Ti3C2@AuNPs film modified GCE. (B) DNA5 length optimization experiments. (C) DNA5 concentration optimization experiments. (D) CRISPR-Cas12a/gRNA complexes cleavage time optimization experiments.
3.6. Optimization of analytical conditions
Since DNA5 is the key to the DNA walker reaction, we optimized the length and concentration of DNA5. Firstly, for length optimization, different lengths of DNA5 (DNA5a-DNA5e) were added to the reaction system (Fig. 2B), which was prepared as follows: DNA-AuNP (DNA1/DNA2/DNA3 complex and DNA4), DNA5, DNA6 and SARS-COV-2 RdRp gene at concentrations of 100 pM, 5 nM, 320 nM and 1 fM, respectively. After incubation at room temperature for 60 min and then incubated with the CRISPR system for 20 min at 37 °C, ECL signal values were recorded. From Fig. 2B we see that the signal value is the largest when employing DNA5 for the DNA walker reaction, so we chose DNA5 for the next experiments. In turn, to investigate the effect of DNA5 concentration (Fig. 2C), ECL signals were measured using different concentrations of DNA5 (1, 2, 5, 10, and 20 nM). The concentrations of DNA-AuNP (assembled with DNA1/DNA2/DNA3 complex and DNA4), DNA6, and SARS-COV-2 RdRp gene were 100 pM, 320 nM, and 1 pM, respectively. After incubation for 20 min at 37 °C, the ECL signal values were recorded by incubating with the CRISPR system. As can be seen in Fig. 2C, the ECL signal value reaches a maximum when the concentration of DNA5 is 5 nM, which may be because DNA5 reaches 5 nM to maximize the DNA4/target DNA/DNA5 complex formed. And when the concentration of DNA5 is higher than 5 nM, it may lead to more free target DNA/DNA5 duplex formation, which affects the formation of DNA4/target DNA/DNA5 complex and thus the 3D-DNA walker-based amplification reaction.
Next, we optimized the time of cleavage of single-stranded nucleic acids on the electrode surface by activated CRISPR-Cas12a. The CRISPR-Cas12a/gRNA complex was activated by 1 nM DNA2/DNA6 duplex after treating the electrode with modified DNA7 (Fig. 2D). We recorded the ECL signal values at different time points. We found that the signal reached a maximum at 20 min after DNA2/DNA6 duplex activation of CRISPR-Cas12a treatment of the ECL sensor. It indicates that the DNA7 on the electrode surface is completely cleaved at that time point. Therefore, we next chose 20 min as the time of CRISPR-Cas12a treatment of the electrode.
3.7. Detection of SARS-COV-2 RdRp gene with the 3D DNA walker and CRISPR-Cas12 biosensor
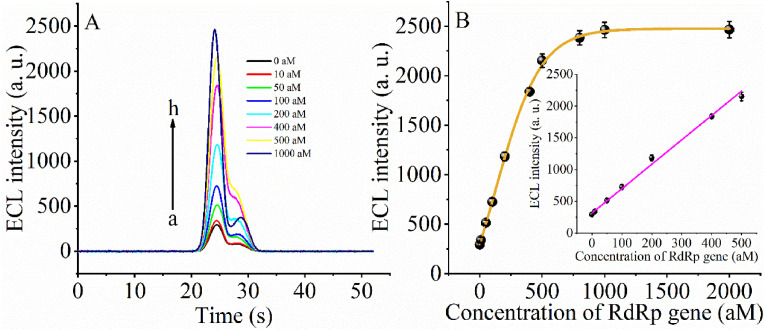
Under the optimized conditions, we used the ECL biosensor system to detect the SARS-COV-2 RdRp gene. Fig. 3 A depicts the increase in ECL signal intensity as the SARS-COV-2 RdRp concentration increased from 10 aM to 500 aM. The reason may be due to the fact that the RdRp gene triggers a 3D walker reaction on the gold electrode surface and generates a DNA2/DNA6 duplex. This duplex further activates the cleavage activity of CRISPR-Cas12a. The activated activity of CRISPR-Cas12a cleaves the DNA7 on the electrode surface, causing an increase in ECL signal. Fig. 3B insert also depicts an excellent linear relationship between the change in ECL signal intensity and the concentration of the RdRP gene, i.e., ECL = 3.84163 × C(RdRp)+314.72, R2 = 0.9952. The detection limit was 12.8 aM. In addition, we also did a comparison data between the gene to be tested and the previously reported SARS-COV-2 gene (Table 1 ). We can see that our detection system has a more comprehensive linear range and lower detection limit compared to conventional sensors.
Fig. 3.
(A) Time-based ECL spectra for the different concentration of SARS-CoV-2 RdRp gene: curve a to curve h indicate the concentration of target DNA from 0 aM to 1000 aM. (B) The relationship between the ECL intensity and the concentration of SARS-CoV-2 RdRp gene. Insert shown the linear part of the relationship between ECL intensity and the concentration of SARS-CoV-2 RdRp gene.
Table 1.
Comparison of different methods for SARS-COV-2.
| Method | Target | LOD | Reference |
|---|---|---|---|
| RT-qPCR | RdRp gene | 1.26 aM | [24] |
| E gene | 1.73 aM | ||
| RT-qPCR | ORF1b/N | 4.15 aM | [25] |
| Colorimetric | N gene | 43 nM | [26] |
| Real-time optomagnetic | RdRp gene | 0.4 fM | [27] |
| Dual-functional plasmonic | RdRp gene | 0.22 pM | [28] |
| Electrochemical | ORF1ab gene | 0.33 aM | [29] |
| Reverse-transcription recombinase-aided amplification assay | N gene | 3.32 aM | [30] |
| ECL | RdRp gene | 2.67 fM | [3] |
| ECL | RdRP gene | 12.8 aM | This work |
3.8. Specificity and stability of the strategy
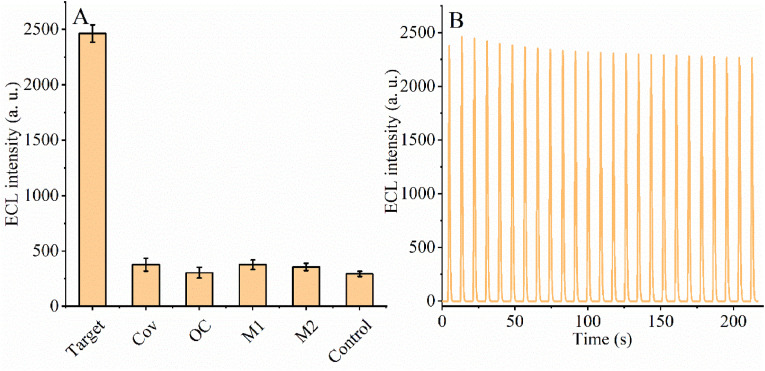
Specificity and stability are essential metrics for evaluating CRISPR-Cas12a and 3D DNA walker-based biosensors for RdRp gene detection. Therefore, next, we tested various nucleic acids to assess the specificity and stability of our constructed biosensing system (Fig. 4 A). We selected a blank solution (no target DNA), SARS-COV RdRp gene (COV, 1 fM), ORF1ab-COVID (OC, 1 fM) DNA1 with random mutation sites (M1, 1 fM), and DNA2 with random mutation sites (M2, 1 fM) as controls. As shown in Fig. 4A, there was no significant difference in the electrochemiluminescence signal after using the blank solution, SARS-COV RdRp gene, DNA1 with random mutation sites and DNA2 with random mutation sites acting on this assay system. In contrast, the electrochemiluminescence signal had a greater degree of variation after acting the SARS-CoV-2 RdRp gene (1 fM) on the CRISPR-Cas 12a-based assay system. The reason for this experimental phenomenon may be that DNA containing the gene mutation site cannot promote the 3D DNA walker reaction on the surface of gold nanoparticles, and therefore does not activate the shearing activity of CRISPR-Cas12a. The results show that our constructed CRISPR-12a-based and 3D DNA walker has good selectivity for the RdRp gene of SARS-COV-2.
Fig. 4.
(A) The specificity of ECL biosensor for different DNA; (B) The stability of ECL biosensor.
Stability is also an important performance indicator for the success of CRISPR-Cas12a and 3D DNA walker-based biosensor construction. We measured the stability of the electrochemiluminescence signal, and the results are shown in Fig. 4B. It can be seen from the figure that the electrochemiluminescence signal of this biosensor has good stability (RSD = 4.21%) despite performing twenty scans three times. Thus, this indicates that the constructed CRISPR-Cas12a and 3D DNA walker-based biosensor is expected to be used for the detection of SARS-COV-2 RdRp gene.
3.9. Real sample analysis in pharyngeal swabs
The detection capability of biosensors in real samples is a key metric to assess the potential clinical applications of the sensors. To examine the potential applications of ECL biosensors in clinical medicine. We performed recovery experiments by detecting the signal response of different concentrations of RdRp gene added in pharyngeal swabs according to the standard addition method. As described in Table S2, the experimental recoveries ranged from 98.97% to 102.51%, and the RSD values ranged from 3.2% to 4.57%. It indicates that the novel ECL biosensor has a good signal response for RdRp and can be applied to detect RdRp gene in real samples.
4. Conclusions
Unlike most 1D or 2D DNA nanomachines, our 3D DNA walker is built on a 3D DNA AuNP track. Combining all DNA components on a single AuNP ensures space for DNA walking. The corresponding increase in the effective local concentration of DNA probes also accelerates the entropy-based drive reaction, enabling the 3D DNA walker to perform the task of rapid release of double-stranded DNA (the activated nucleic acid sequence of CRISPR-12a). The concept of integrating multiple functional nucleic acids on a single nanoparticle and releasing CRISPR-12a triggered nucleic acid sequences also makes it easier to scale up the functionality of isothermal amplification and CRISPR-12a based detection strategies, making it a highly sensitive and fast nanosensor system for novel coronavirus target nucleic acids. More importantly, the 3D DNA walker technology can accurately distinguish between the RdRp genes of SARS-CoV and SARS-CoV-2. In the context of the SARS-CoV-2 outbreak, this biosensor can provide a reliable and easy-to-implement diagnostic platform to improve the diagnostic accuracy of clinical tests and reduce the pressure of traditional PCR assays. Thus, our design of the 3D DNA walker and CRISPR-12a assay system opens up new concepts and strategies for the design of biosensing systems that may lead to the development of new DNA nanodevices, nucleic acid diagnostics, and biosensors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (21705061), the Jiangsu Provincial Key Medical Discipline (Laboratory) (ZDXKA2016017), and the Innovation Capacity Development Plan of Jiangsu Province (BM2018023).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2021.122868.
Credit author statement
Kai Zhang: Conceptualization, Drafting the manuscript, Methodology, Zhenqiang Fan: acquisition of data, Analysis of data, Yue Huang: Methodology. Yuedi Ding: Analysis of data. Minhao Xie: Conceptualization, Resources
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ji T., Liu Z., Wang G., Guo X., Akbar khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Detection of COVID-19: a review of the current literature and future perspectives Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nouri R., Tang Z., Dong M., Liu T., Kshirsagar A., Guan W. CRISPR-based detection of SARS-CoV-2: a review from sample to result Biosens. Bioelectron. 2021;178:113012. doi: 10.1016/j.bios.2021.113012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Z., Yao B., Ding Y., Zhao J., Xie M., Zhang K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds Biosens. Bioelectron. 2021;178:113015. doi: 10.1016/j.bios.2021.113015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelmann I., Alidjinou E.K., Ogiez J., Pagneux Q., Miloudi S., Benhalima I., Ouafi M., Sane F., Hober D., Roussel A., Cambillau C., Devos D., Boukherroub R., Szunerits S. Preanalytical issues and cycle threshold values in SARS-CoV-2 real-time RT-PCR testing: should test results include these? ACS Omega. 2021;6(10):6528–6536. doi: 10.1021/acsomega.1c00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mason S.D., Tang Y., Li Y., Xie X., Li F. Emerging bioanalytical applications of DNA walkers. Trac. Trends Anal. Chem. 2018;107:212–221. doi: 10.1016/j.trac.2018.08.015. [DOI] [Google Scholar]
- 6.Yang X., Tang Y., Mason S.D., Chen J., Li F. Enzyme-powered three-dimensional DNA nanomachine for DNA walking, payload release. and Biosensing ACS Nano. 2016;10(2):2324–2330. doi: 10.1021/acsnano.5b07102. [DOI] [PubMed] [Google Scholar]
- 7.Chen J., Luo Z., Sun C., Huang Z., Zhou C., Yin S., Duan Y., Li Y. Research progress of DNA walker and its recent applications in biosensor TrAC. Trends Anal. Chem. 2019;120:115626. doi: 10.1016/j.trac.2019.115626. [DOI] [Google Scholar]
- 8.Li W., Wang L., Jiang W. A catalytic assembled enzyme-free three-dimensional DNA walker and its sensing application. Chem. Commun. 2017;53(40):5527–5530. doi: 10.1039/C7CC02306E. [DOI] [PubMed] [Google Scholar]
- 9.Peng H., Li X.-F., Zhang H., Le X.C. A microRNA-initiated DNAzyme motor operating in living cells Nat. Commun. Now. 2017;8:14378. doi: 10.1038/ncomms14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung C., Allen P.B., Ellington A.D. A stochastic DNA walker that traverses a microparticle surface. Nat. Nanotechnol. 2016;11(2):157–163. doi: 10.1038/nnano.2015.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang C.-P., Ma P.-Q., Liu H., Guo X., Yin B.-C., Ye B.-C. Rational engineering of a dynamic, entropy-driven DNA nanomachine for intracellular MicroRNA imaging. Angew. Chem. Int. Ed. 2017;56(31):9077–9081. doi: 10.1002/anie.201704147. [DOI] [PubMed] [Google Scholar]
- 12.Zhang K., Huang W., Li H., Xie M., Wang J. Ultrasensitive detection of hERG potassium channel in single-cell with photocleavable and entropy-driven reactions by using an electrochemical biosensor Biosens. Bioelectron. 2019;132:310–318. doi: 10.1016/j.bios.2019.02.065. [DOI] [PubMed] [Google Scholar]
- 13.Zhang K., Wang K., Zhu X., Xie M. Ultrasensitive fluorescence detection of transcription factors based on kisscomplex formation and the T7 RNA polymerase amplification method. Chem. Commun. 2017;53(43):5846–5849. doi: 10.1039/C7CC02231J. [DOI] [PubMed] [Google Scholar]
- 14.Feng W., Newbigging A.M., Tao J., Cao Y., Peng H., Le C., Wu J., Pang B., Li J., Tyrrell D.L., Zhang H., Le X.C. CRISPR technology incorporating amplification strategies: molecular assays for nucleic acids, proteins, and small molecules. Chem. Sci. 2021;12(13):4683–4698. doi: 10.1039/D0SC06973F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou T., Huang R., Huang M., Shen J., Shan Y., Xing D. CRISPR/Cas13a powered portable electrochemiluminescence chip for ultrasensitive and specific MiRNA detection. Adv. Sci. 2020;7(13):1903661. doi: 10.1002/advs.201903661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang K., Fan Z., Yao B., Ding Y., Zhao J., Xie M., Pan J. Exploring the trans-cleavage activity of CRISPR-Cas12a for the development of a Mxene based electrochemiluminescence biosensor for the detection of Siglec-5 Biosens. Bioelectron. 2021;178:113019. doi: 10.1016/j.bios.2021.113019. [DOI] [PubMed] [Google Scholar]
- 17.Zeng R., Wang W., Chen M., Wan Q., Wang C., Knopp D., Tang D. CRISPR-Cas12a-driven MXene-PEDOT:PSS piezoresistive wireless biosensor. Nanomater. Energy. 2021;82:105711. doi: 10.1016/j.nanoen.2020.105711. [DOI] [Google Scholar]
- 18.Fan Z., Lin Z., Wang Z., Wang J., Xie M., Zhao J., Zhang K., Huang W. Dual-wavelength electrochemiluminescence ratiometric biosensor for NF-κB p50 detection with dimethylthiodiaminoterephthalate fluorophore and self-assembled DNA tetrahedron nanostructures probe. ACS Appl. Mater. Interfaces. 2020;12(10):11409–11418. doi: 10.1021/acsami.0c01243. [DOI] [PubMed] [Google Scholar]
- 19.Liu W., Chen A., Li S., Peng K., Chai Y., Yuan R. Perylene derivative/luminol nanocomposite as a strong electrochemiluminescence emitter for construction of an ultrasensitive MicroRNA biosensor. Anal. Chem. 2019;91(2):1516–1523. doi: 10.1021/acs.analchem.8b04638. [DOI] [PubMed] [Google Scholar]
- 20.Qi H., Li M., Dong M., Ruan S., Gao Q., Zhang C. Electrogenerated chemiluminescence peptide-based biosensor for the determination of prostate-specific antigen based on target-induced cleavage of peptide. Anal. Chem. 2014;86(3):1372–1379. doi: 10.1021/ac402991r. [DOI] [PubMed] [Google Scholar]
- 21.Wu F., Zhou Y., Zhang H., Yuan R., Chai Y. Electrochemiluminescence peptide-based biosensor with hetero-nanostructures as coreaction accelerator for the ultrasensitive determination of tryptase. Anal. Chem. 2018;90(3):2263–2270. doi: 10.1021/acs.analchem.7b04631. [DOI] [PubMed] [Google Scholar]
- 22.Zhang K., Fan Z., Yao B., Zhang T., Ding Y., Zhu S., Xie M. Entropy-driven electrochemiluminescence ultra-sensitive detection strategy of NF-κB p50 as the regulator of cytokine storm Biosens. Bioelectron. 2021;176:112942. doi: 10.1016/j.bios.2020.112942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J.-H., Feng Y.-G., Wang A.-J., Mei L.-P., Luo X., Feng J.-J. A signal-on photoelectrochemical aptasensor for chloramphenicol assay based on 3D self-supporting AgI/Ag/BiOI Z-scheme heterojunction arrays Biosens. Bioelectron. 2021;181:113158. doi: 10.1016/j.bios.2021.113158. [DOI] [PubMed] [Google Scholar]
- 24.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):23. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14(6):7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence Biosens. Bioelectron. 2020;165:112356. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- 28.Qiu G., Gai Z., Tao Y., Schmitt J., KullakUblick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H., Liu F., Xie W., Zhou T., OuYang J., Jin L., Li H., Zhao C., Zhang L., Wei J., Zhang Y., Li C. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu T., Ge Y., Zhao K., Zhu X., Chen Y., Wu B., Zhu F., Zhu B., Cui L. A reverse-transcription recombinase-aided amplification assay for the rapid detection of N gene of severe acute respiratory syndrome coronavirus 2(SARS-CoV-2) Virology. 2020;549:1–4. doi: 10.1016/j.virol.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.