Abstract
MHR3, a homolog of the retinoid orphan receptor (ROR), is a transcription factor in the nuclear hormone receptor family that is induced by 20-hydroxyecdysone (20E) in the epidermis of the tobacco hornworm, Manduca sexta. Its 2.7-kb 5′ flanking region was found to contain four putative ecdysone receptor response elements (EcREs) and a monomeric (GGGTCA) nuclear receptor binding site. Activation of this promoter fused to a chloramphenicol acetyltransferase (CAT) reporter by 2 μg of 20E per ml in Manduca GV1 cells was similar to that of endogenous MHR3, with detectable CAT by 3 h. When the ecdysone receptor B1 (EcR-B1) and Ultraspiracle 1 (USP-1) were expressed at high levels under the control of a constitutive promoter, CAT levels after a 3-h exposure to 20E increased two- to sixfold. In contrast, high expression of EcR-B1 and USP-2 caused little increase in CAT levels in response to 20E. Moreover, expression of USP-2 prevented activation by EcR-B1–USP-1. Deletion experiments showed that the upstream region, including the three most proximal putative EcREs, was responsible for most of the 20E activation, with the EcRE3 at −671 and the adjacent GGGTCA being most critical. The EcRE1 at −342 was necessary but not sufficient for the activational response but was the only one of the three putative EcREs to bind the EcR-B1–USP-1 complex in gel mobility shift assays and was responsible for the silencing action of EcR-B1–USP-1 in the absence of hormone. EcRE2 and EcRE3 each specifically bound other protein(s) in the cell extract, but not EcR and USP, and so are not EcREs in this cellular context. When cell extracts were used, the EcR-B1–USP-2 heterodimer showed no binding to EcRE1, and the presence of excess USP-2 prevented the binding of EcR-B1–USP-1 to this element. In contrast, in vitro-transcribed-translated USP-1 and USP-2 both formed heterodimeric complexes with EcR-B1 that bound ponasterone A with the same Kd (7 × 10−10 M) and bound to both EcRE1 and heat shock protein 27 EcRE. Thus, factors present in the cell extract appear to modulate the differential actions of the two USP isoforms.
Insect molting and metamorphosis are controlled by ecdysteroids and juvenile hormone (JH), with the ecdysteroids initiating and orchestrating the molt and with JH preventing metamorphosis (46). The ecdysteroids act through a heterodimeric receptor complex consisting of the ecdysone receptor (EcR) and Ultraspiracle (USP), the insect homolog of the retinoid X receptor (RXR) in vertebrates (see reference 7 for a review), to initiate a cascade of transcription factors that serve to inactivate and/or activate tissue-specific genes as well as to activate directly a few structural genes. In Drosophila melanogaster, some of the transcription factors are activated immediately by 20-hydroxyecdysone (20E), such as E74B, an ETS oncogene homolog (4, 29, 51), and E75A, the homolog of the vertebrate EAR1 (48), whereas others, such as DHR3, the homolog of the vertebrate retinoid orphan receptor (ROR) (13), appear later and require protein synthesis for their full expression (22, 31). Relatively little is known about the details of the regulation of these later factors. Ectopic expression of E74B can partially repress the appearance of the ecdysteroid-induced transcripts of the E78B and DHR3 genes, both delayed-early genes, indicating that E74B is likely involved in the timing of expression of these genes (11).
In the larval epidermis of the tobacco hornworm, Manduca sexta, a similar response to 20E is seen. E75A RNA appears first during the molt and within 30 min of exposure to 20E in vitro (58), whereas MHR3 (the DHR3 homolog) RNA is not seen until later in the molt or after about 3 h of exposure to 20E in vitro and requires protein synthesis for its full expression (40). During the intermolt, EcR-B1 is the predominant EcR isoform in the epidermis, with both EcR-B1 and EcR-A RNAs appearing during the rise in ecdysteroid titer for the molt in a complex pattern that is partly regulated by the presence or absence of JH (20, 21, 26). Unlike in Drosophila, which apparently has only one isoform of USP (18, 39), Manduca has two isoforms, with a switch from USP-1 to USP-2 occurring in the epidermis during the larval and pupal molts due to the rising titer of ecdysteroid (2, 21, 27). In mosquitoes (28) and the tick Amblyoma americanum (14), both isoforms of USP or RXR, respectively, heterodimerize with the single EcR isoform so far isolated and bind ponasterone A. Also, they both form functional complexes that bind the heat shock protein 27 (hsp27) EcRE. Yet, whether or not they function interchangeably in the regulation of particular genes is not known. Therefore, it was of interest to determine which EcR-USP isoform combination is involved in the up-regulation of MHR3.
To answer this question, we studied the regulation of the MHR3 promoter by using transient transfection assays in M. sexta GV1 cells, which are responsive to 20E (35, 36) and in which the amounts of the various EcR and USP isoforms could be manipulated. In this paper, we show that the sequences responsible for 20E-induced activity of this promoter are located within the first 1 kb of the transcription initiation site. Cotransfection of baculovirus-derived expression vectors of EcR-B1 and USP-1 with the MHR3 promoter caused increased transcription of the reporter that depended on the presence of at least two of the putative ecdysone response elements (EcREs), but the EcR-B1–USP-1 complex bound only to the most proximal EcRE. In contrast, expression of USP-2 blocked binding of EcR-B1–USP-1 to the proximal EcRE and thus its activation of MHR3 expression. Thus, the two isoforms can play different roles in the cells, even though both form functional heterodimers which can bind to ecdysteroid and to the usual EcREs in an in vitro system.
MATERIALS AND METHODS
Cells and hormones.
The Manduca GV1 cell line was maintained in modified Grace’s medium supplemented with 10% bovine fetal serum and exposed to 20E as previously described (35).
20E was purchased from Rohto Pharmaceutical Co., Osaka, Japan, or was a gift of Takeshi Matsumoto (99% pure [32a]; Daicel Chemical Co., Tokyo, Japan). A stock solution was prepared in ethanol, and the concentration was determined spectrophotometrically (ɛ240 = 12,677 in ethanol for 20E [33]).
Sequencing, S1 nuclease protection, and primer extension assays.
A genomic DNA clone containing the N terminus of MHR3, its 5′ untranslated region (UTR), and 6.4 kb of its promoter (Fig. 1) was obtained by S. R. Palli in this laboratory (41). The 5′ UTR and 2.8 kb of the promoter were sequenced in both directions by using either the dideoxy chain termination reaction (47) with the enzyme Sequenase, version 2.0 (U.S. Biochemical, Cleveland, Ohio), or the Dye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, Calif.) and automated sequencing. The sequence data were edited with the DNASIS DNA analysis software (Hitachi Software Engineering) and GCG (Wisconsin package, version 9.1; Genetics Computer Group, Madison, Wis.).
FIG. 1.
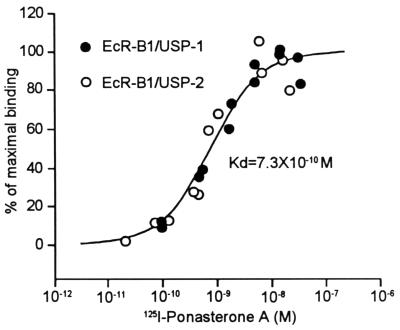
Binding of in vitro-transcribed-translated EcR-B1 and USP-1 or USP-2 to ponasterone A. The nonspecific binding was determined by addition of 10 μM muristerone A and subtracted from the total binding, and then the percentage of maximal binding was obtained as described in Materials and Methods. Kd was calculated from all of the data shown.
S1 nuclease protection assays were done with a kit according to the Ambion manual (Austin, Tex.). The uniform labeled antisense probe is indicated in Fig. 4. The primer extension assays were done as described previously (3).
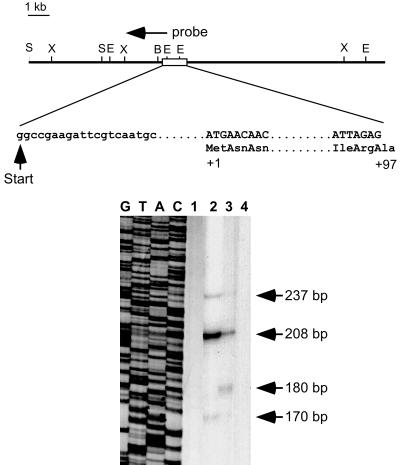
FIG. 4.
(Top) Endonuclease restriction map of a cloned genomic DNA fragment of the MHR3 gene. B, BamHI; E, EcoRI; S, SalI; and X, XbaI. The transcription start site (+1) and the DNA (capital letters) and amino acid (titlecase) sequences bordering the first intron are indicated. Sequence data for the MHR3 upstream region are deposited in GenBank (AF086951). (Bottom) S1 nuclease protection assay for MHR3 mRNA. The antisense labeled probe is as indicated in the map. Lanes: 1, yeast RNA; 2, total RNA from GV1 cells treated with 2 μg of 20E per ml for 6 h; 3, total RNA from day 2 4th instar larval epidermis treated with 2 μg of 20E per ml for 21 h; 4, total RNA from GV1 cells. A DNA sequence reaction (lanes G, T, A, and C) was used to indicate the precise sizes of the S1 nuclease-protected fragments.
Plasmid construction and deletion analysis.
The 6.4-kb promoter and 241 bp of the 5′ UTR were ligated to bacterial chloramphenicol acetyltransferase (CAT) that had been excised from pCAT basic (Promega, Madison, Wis.). Exonuclease III deletion was conducted from the 5′ end of the promoter toward the 3′ end by using the ExoIII deletion kit (Stratagene, La Jolla, Calif.). Series deletion clones were then selected and sequenced to ensure that the sequence matched the genomic DNA.
For the in vitro transcription-translation studies of binding of ponasterone A, the entire 2.7-kb Manduca EcR-B1 cDNA (12) was cloned into BamHI (5′) and KpnI (3′) sites of the cMX vector. These sites were derived from the artificial adapter used for cloning the cDNA library. Both cDNA ends have the same adapters, but directional cloning into the cMX plasmid vector was possible because the BamHI site on the 3′ end of EcR-B1 cDNA was defective. The USP-1a cDNA (27) was in Bluescript SK−, in EcoRI sites 5′ to the T3 promoter. The USP-2-specific cDNA (27) was fused to the common cDNA of USP-1 through an endogenous MluI site and cloned into BamHI (5′) and EcoRI (3′) sites of pcMX. In all three pcMX plasmids, the 5′ ends of the cDNAs were ligated to the T7 promoter; linearization on the 3′ end was possible with NotI.
Expression vectors using the promoter of the immediate-early gene of the Autographa californica baculovirus (AcMNPV) (43) were constructed as follows. The parental plasmid pIE1hr/PA was a gift from Paul D. Friesen (Robert M. Bock Laboratory, Madison, Wis.). For pIE1hr/EcR-B1, the entire EcR-B1 cDNA was excised from the cMX vector with BamHI and NotI and inserted into the pIE1hr/PA vector 3′ of the transcription initiation site. For pIE1hr/USP-1 and pIE1hr/USP-2, the coding region and 1.3 kb of the 3′ UTR were removed from the USP cMX vector by using BamHI and NotI and inserted into the pIE1hr/PA vector.
Binding of EcR-B1–USP-1 and EcR-B1–USP-2 to 125I-ponasterone A.
M. sexta EcR-B1, USP-1, and USP-2 were synthesized in the TNT Coupled Reticulocyte Lysate system (Promega). Equal volumes of EcR-B1 and either USP1 or USP2 were mixed and incubated with 125I-ponasterone A according to the method of Cherbas et al. (8) at room temperature for 1 h and then spotted on a GF/C filter and washed three times at 4°C. Filters were counted on a Packard model B5002 gamma counter. Concentrations of free probe were determined by counting a portion of the mixture. The nonspecific binding was measured at each concentration of 125I-ponasterone A in the presence of a large excess (10 μM) of muristerone A and subtracted from the total binding to give the specific binding.
Each set of data was fitted with a Sigma Plot program to the first-order binding curve y = m · x/(Kd + x), where y is the specific binding and x is the free ligand, to obtain the maximal binding, m, and the dissociation constant, Kd. To plot the two sets of binding data together, each set was normalized to the percentage of maximal binding, m, at the plateau level. The two sets of normalized data were then plotted together, and Kd was calculated with the equation y = 100 · x/(Kd + x), where y is the percentage of maximal binding and x is the concentration of free ligand. The smooth curve was generated by using the calculated Kd value.
RNA and protein analyses.
Total RNA was extracted from the GV1 cells by the modified method of Chomczynski and Sacchi (9) as previously described (20). Briefly, after the first precipitation with isopropanol, the RNAs were dissolved in TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]) followed by ethanol precipitation. The RNA concentrations were determined spectrophotometrically (10).
For dot blot hybridization analysis, 5 μg of total RNA was denatured and applied to a Duralon UV membrane (27). cRNA probes for USP-1 and USP-2 (26) were synthesized by using RNA polymerase and [α-32P]UTP (3,000 Ci/mmol; Amersham, Arlington Heights, Ill.) as described by a Promega manual. Hybridization and washing conditions at high stringency were as described previously (27). The hybridized filters were analyzed by the Molecular Imager System, model GS-363 (Bio-Rad, Hercules, Calif.). Each blot contained standard RNA from day 2 5th instar, W0, W1, and W3 epidermis for normalization of different blots.
Cellular proteins were extracted in a mixture of 250 mM Tris-HCl (pH 8.0), 0.3% Triton X-100, 1 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonylfluoride (PMSF) by three cycles of freeze-thaw followed by centrifugation (12,000 × g, 5 min). The protein concentration was determined with the bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.). Fifteen micrograms of total proteins was separated by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis and then transferred onto Protran nitrocellulose (Schleicher & Schuell, Keene, N.H.). The filters were blocked overnight in phosphate-buffered saline (150 mM NaCl, 2 mM NaH2PO4, 7 mM Na2HPO4) (PBS) containing 0.1% Tween 20 (PBST) containing 5% bovine serum albumin and 5% dried nonfat milk. They were then incubated with the USP monoclonal antibody (MAb) AB11 (1:2,000) (30) and the EcR-B1-specific MAb 6B7 (1:1,000) (26) in PBST for 2 h, followed by incubation for 1 h in 1:1,500 goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, Pa.). Detection was performed by chemiluminescence (Renaissance; DuPont NEN, Boston, Mass.) by using the Molecular Imager System, model GS-363 (Bio-Rad).
Transient transfection assays.
All transfections were conducted as described previously (34) with 5 μg of total plasmid DNA per ml and 15 μg of Lipofectin reagent in 1 ml of transfection medium. An hsp70–β-galactosidase plasmid, pXH70ZT, was cotransfected in all experiments for normalization of transfection efficiency; this plasmid shows a modest constitutive activity in transfected GV1 cells (34). Cells were treated with 2 μg of 20E per ml 45 h after the initial transfection. Both β-galactosidase activity and CAT protein assays were performed as previously described (34).
The relative CAT protein level was calculated by normalizing CAT protein with the β-galactosidase activity from the same sample. The fold increased CAT was determined by comparing the CAT level in a 20E-treated cell extract with that of untreated cells.
GMSA.
The pIE1hr expression vectors of EcR-B1, USP-1, USP-2, and E75A were transfected into GV1 cells in different combinations, and the whole-cell extract of the cellular proteins (about 107 cells) was prepared 45 h after transfection according to the method described in reference 34. Cell pellets were lysed in 200 μl of EcR buffer (60 mM KCl, 25 mM HEPES [pH 7.5], 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 1 μg of antipain per ml), sonicated at 4°C for 30 s, and centrifuged at 12,000 × g at 4°C for 10 min. The supernatant was collected as 40-μl aliquots and stored at −70°C. As a control, an extract of GV1 cells that was transfected with the empty plasmid pIE1hr/PA was used. All of the extracts were frozen only once at −70°C until use. In vitro-transcribed-translated EcR-B1, USP-1, and USP-2 were prepared with the TNT Coupled Reticulocyte Lysate system (Promega) for the gel mobility shift assay (GMSA).
The oligonucleotides used as probes (EcRE1, 5′-CGGGGTCAATGAACCGGT-3′; EcRE2, 5′-GAACGTTGATTGCGCAAA-3′; EcRE3, 5′-TATCGTTGACAACTCATT-3′; monomeric receptor element [MRE], 5′-TTTCCGGGGTCAACGACC-3′; hsp27 response element, 5′-AAGTGCATTGAACCCTTG-3′) were synthesized by Gibco BRL (Gaithersburg, Md.). The probes were labeled as described previously (19). One strand of the oligonucleotide was 32P labeled at the 5′ end with T4 polynucleotide kinase and [γ-32P]ATP, 10-fold molar excess of the cold complementary strand was added, and the mixture was heated at 95°C for 2 min, followed by a slow cooling to the room temperature. The double-stranded labeled probes were purified by Sephadex G-50 columns. The cold nucleotides used for competitors were prepared as follows: equimolar amounts of both strands of oligonucleotides were heated at 95°C for 2 min and then annealed by cooling slowly to the room temperature in TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]).
Five micrograms of cell extract or 1 μl of reticulocyte lysate for each of the tested translation products in 19 μl of buffer containing 1 μg of poly(dI-dC)(dI-dC) (Sigma, St. Louis, Mo.), 12 mM HEPES (pH 7.9), 60 mM KCl, 65 mM NaCl, 7.5 mM MgCl2, 6.6 mM EDTA, 1.2 mM DTT, and 12% glycerol was preincubated on ice for 5 to 10 min, and then 50 fmol of labeled probe and competitor DNA were added. The reaction mixture was incubated for 20 min at 0°C. When 1 μl of the EcR-B1 or USP MAb supernatant (as used for immunoblotting above) was added, the mixture was incubated on ice for another 1.5 h. The mixture was then run on a 4% polyacrylamide gel containing 2.5% glycerol in 1× TBE (45 mM Tris-borate, 1 mM EDTA), which had been prerun for 3 to 4 h in 1× TBE. The gels were dried and then analyzed by the Molecular Imager System, model GS-363 (Bio-Rad).
Nucleotide sequence accession number.
The sequence of the first 2.6 kb of the MHR3 promoter and 898 bp of the 5′ UTR of the 4.5-kb transcript has been deposited in GenBank under accession no. AF086951.
RESULTS
Effects of EcR-B1 and USP isoforms on 20E induction of MHR3 RNA.
In Manduca larval epidermis, MHR3 RNA appears during a molt as the ecdysteroid titer reaches its peak and requires 20E-induced protein synthesis for its full expression (40). Both EcR-B1 and USP-1 mRNA are predominant at the onset of the molt, but USP-1 mRNA then declines and USP-2 mRNA increases with the rising ecdysteroid titer (21, 26, 27). Therefore, it was of interest to determine whether the induction of MHR3 was dependent on a particular isoform of USP.
(i) Functionality of EcR-B1–USP-1 and EcR-B1–USP-2 heterodimers.
To determine whether the two heterodimers differed in their binding of ligand, we tested the binding of the potent ecdysteroid ponasterone A to in vitro-transcribed-translated products of EcR-B1 and either USP-1 or USP-2. As seen in Fig. 1, both EcR-B1–USP-1 and EcR-B1–USP-2 heterodimers bound ponasterone A and showed an indistinguishable binding affinity with a Kd of 7.3 × 10−10 M.
These in vitro-transcribed-translated products of EcR-B1 and either USP-1 or USP-2 also bound to the hsp27 EcRE in gel mobility shift assays (n = 2; data not shown). When tested alone, no binding was seen with the translated EcR-B1, USP-1, or USP-2. Thus, both USP-1 and USP-2 form functional heterodimers with EcR-B1.
(ii) Effects of constitutive expression of EcR-B1 and USP isoforms in GV1 cells.
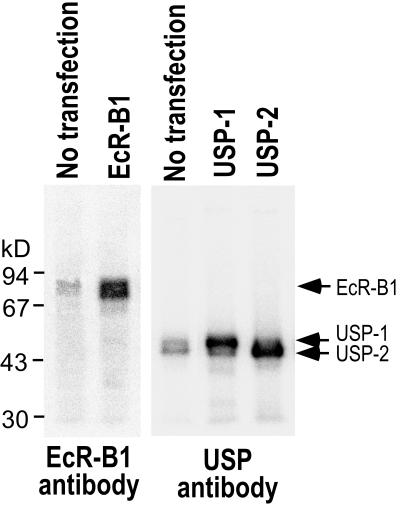
MHR3 RNA is induced in the Manduca GV1 cell line by 2 μg of 20E per ml, with a delayed time course similar to that seen in the epidermis (trace amounts at 3 h, maximal amounts at 6 h) (35). To examine the roles of EcR-B1 and the USP isoforms in 20E activation of MHR3 expression, we used the Autographa californica baculovirus immediate-early gene 1 promoter to express these various proteins constitutively in transfected GV1 cells. Immunoblotting with both the common USP MAb and the EcR-B1-specific MAb showed that nontransfected GV1 cells contain trace amounts of EcR-B1 and USP (Fig. 2). Forty-five hours after transfection of pIE1hr–EcR-B1, pIE1hr–USP-1, or pIE1hr–USP-2, high levels of EcR-B1 and the USPs were observed (Fig. 2). The intensity of the bands as determined using the Multi-Analyst software for the phosphorimager showed that, compared to the level in nontransfected cells, the EcR-B1 level was about 25 times higher. The two expressed USP proteins were of the expected sizes for USP-1 and USP-2 (55 and 46 kDa, respectively) (2, 27).
FIG. 2.
Expression of EcR-B1 and USPs in GV1 cells. Immunoblotting of cell extracts with EcR-B1-specific and USP-common antibodies. The leftmost lane in each blot contains extract from nontransfected cells. The right lanes in each blot contain extracts transfected with the pIE1hr expression vectors for EcR-B1 or USP-1 or USP-2 as designated. Fifteen micrograms of soluble protein was applied.
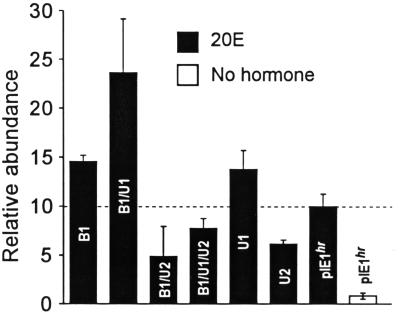
Figure 3 shows that when both EcR-B1 and USP-1 were constitutively expressed at high levels, the level of MHR3 RNA expression after exposure to 2 μg of 20E per ml for 6 h was 2.4-fold higher than that in cells containing only the pIEhr vector and about 29-fold higher than that in vector-containing cells in the absence of 20E. Transfection of either pIEhr–EcR-B1 or pIEhr–USP-1 alone caused about a 1.4-fold increase, presumably interacting with endogenous pools of the other member of the pair. In contrast, constitutively high expression of pIEhr–USP-2 alone suppressed the normal induction of MHR3 RNA by 20E by about 40% (Fig. 3). These increased levels of USP-2 also suppressed the higher MHR3 response to 20E seen in the presence of increased EcR-B1 or EcR-B1–USP-1. These results suggest that combined EcR-B1–USP-1 is critical to the induction of MHR3 and that USP-2 is inhibitory to this expression.
FIG. 3.
The induction of MHR3 mRNA in GV1 cells in 6 h by 2 μg of 20E per ml in the presence of excess EcR-B1, USP-1, USP-2, or various combinations thereof under the control of the pIE1hr constitutive promoter. The empty pIE1hr was transfected as a control. Cells were cultured in the presence and absence of 20E hormone. Relative abundance refers to MHR3 RNA induced by 20E in 6 h in cells which contained the empty pIE1hr expression vector, designated as 10. Means ± standard deviations (error bars) are shown (n = 3 to 5).
Organization of the MHR3 promoter.
To explore this phenomenon in more detail, we isolated a genomic DNA fragment containing 6.4 kb of the 5′ flanking region, exon I, and part of intron I of the MHR3 gene from the Manduca genomic DNA library (41). Figure 4 shows the organization of the MHR3 promoter and exon I, which includes 1 kb of 5′ UTR and the first 97 amino acids of the N-terminal A/B domain of the MHR3 protein followed by intron I, which is greater than 10 kb. This intron begins at predicted amino acid 97 (Ala) of the previously sequenced MHR3 cDNA (40). The transcription start site was located by using S1 nuclease mapping (Fig. 4) and primer extension (data not shown). The 237-bp S1 nuclease-protected band was chosen as the full transcript because it matched with the results from primer extension, although the 208-bp band was present in larger amounts in the S1 nuclease protection assays (Fig. 4). Furthermore, a TATA box is 21 bp upstream of the presumed transcription start site, and a cap site consensus sequence (5) was found at +22 downstream of the transcription start site. No CAAT box was found in the upstream region. The nucleotide sequence at the 3′ end of the 993-bp 5′ UTR overlapped with the 5′ UTR of the published cDNA sequence of MHR3 (40). Therefore, this transcript represents the 4.5-kb MHR3 mRNA, which corresponds to the larger isoform of the two MHR3 mRNAs (40). The transcription start site for the 3.8-kb MHR3 mRNA has not yet been determined.
There are four sequences that are similar to the EcRE consensus sequence (7) within the first 2.7 kb upstream of the 5′ transcription start site, which we designate as putative EcREs: EcRE1, GGGGTCAATGAACCg (−342 to −328; boldface uppercase letters indicate matches with the consensus sequence; the italicized letter represents the one nucleotide separating the palindromic pair); EcRE2, AcGTTGATTGcgCaa (−534 to −520); EcRE3, tcGTTGACaACtCaT (−671 to −657); and EcRE4, ctGGTCATTGtCaCT (reverse orientation at −1930 to −1911). A nuclear receptor monomeric binding sequence (37) (MRE), tttccgGGGTCAacgacc, was present near EcRE3 (−656 to −638).
Ecdysteroid-inducible activity of the MHR3 promoter.
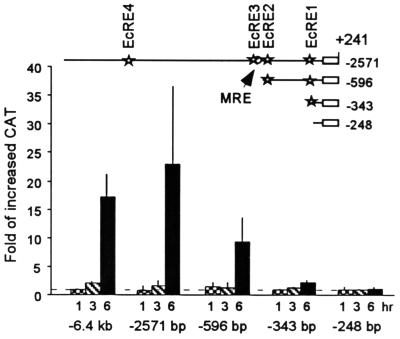
The entire 6.4 kb of the 5′ flanking region of the MHR3 promoter was ligated to a bacterial CAT cDNA as a reporter, and the construct was transiently transfected into the Manduca GV1 cells. Addition of 2 μg of 20E per ml to these cells 45 h later caused the appearance of CAT protein by 3 h, indicating an activation of the MHR3 promoter (Fig. 5 [6.4-kb construct]). By 6 h, the CAT level had increased over 17-fold (Fig. 5). Deletion of the distal 3.7 kb of the 5′ region had no significant effect on the 20E responsiveness of the promoter (Fig. 5 [−2571 construct]). In contrast, the promoter was not activated by 20E when only 248 bp of the proximal promoter sequence remained (Fig. 5 [−248 construct]). This result is consistent with the fact that all EcRE-like sequences are located more distally in the promoter. Removal of the upstream sequence, including EcREs 3 and 4 (Fig. 5 [−596 construct]), caused at least a 2.5-fold (range, 2.5- to 5-fold) reduction in 20E-induced activity, and further truncation removing EcRE2 nearly abolished the 20E induction. Thus, EcRE1 is not sufficient for 20E induction of MHR3.
FIG. 5.
Transient transfection assays of MHR3 promoter activity as determined by the increase in CAT protein after exposure to 2 μg of 20E per ml for 1 ( ), 3 (▧), or 6 (■) h over the uninduced level at 0 h (set at 1). The deletion constructs tested are shown in the map above. Stars, putative EcRE; open circle, GGGTCA sequence. Means ± standard deviations (error bars) are shown (n = 3).
Roles of EcR-B1 and USP isoforms in activation of the MHR3 promoter.
For assays of promoter activity in the presence of the expressed EcR-USP isoforms, we chose a 3-h 20E treatment, since at that time both endogenous MHR3 RNA (35) and MHR3 promoter-driven CAT (Fig. 5) are just beginning to increase, and only relatively small changes in the endogenous EcR and USP mRNA levels are seen (35). Each set of experiments was repeated at least three times with the same transfected batch of cells. In different sets of transfected cells, the levels of 20E-induced promoter activity were variable (the n values given in the text below refer to different transfections). However, the differences among the various deletion constructs within a batch were reproducible, so that only a representative set of experiments is shown.
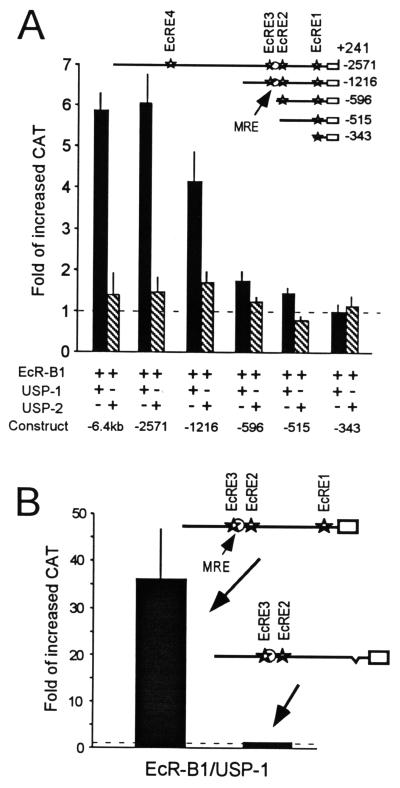
When the pIE1hr–EcR-B1 and pIE1hr–USP-1 cDNAs were cotransfected and expressed with either the −6.4-kb or −2,571-bp MHR3 promoter constructs, the addition of 20E for 3 h caused a sixfold increase in CAT activity over that of the transfected cells not treated with 20E (Fig. 6A). In contrast, when both pIE1hr–EcR-B1 and pIE1hr–USP-2 were present, there was no significant increase in CAT activity after a 3-h exposure to 20E. For the −2.6-kb construct in the absence of hormone, the presence of the excess EcR-B1–USP-1 suppressed basal levels of CAT activity seen with the construct alone to 60% ± 9% (n = 4 different transfections), whereas EcR-B1–USP-2 showed relatively little suppression (85% ± 20%; n = 3). Such suppression in the presence of the EcR complex in the absence of 20E is typical (6). In similar experiments with the −2.6-kb promoter construct and cotransfection of only pIE1hr–EcR-B1, the increase in CAT activity after 3 h of exposure to 20E was similar to that seen with the cotransfected pIE1hr–EcR-B1–USP-1 (92% ± 2%; n = 3). In contrast, after cotransfection of either pIE1hr–USP-1 or pIE1hr–USP-2 alone, there was no increase in CAT activity after 3 h of exposure to 20E (n = 2 for USP-1 and n = 1 transfection set for USP-2). Therefore, increased levels of EcR-B1 are necessary for the increased transcription from the MHR3 promoter elicited by 20E after 3 h. This effect of increased EcR-B1, however, is not seen when increased levels of USP-2 are also present.
FIG. 6.
(A) Effects of increased levels of EcR-B1 and either USP-1 or USP-2 on 20E induction of MHR3 promoter activity. The pIE1hr expression vectors for EcR-B1 and USP-1 or USP-2 were cotransfected with the MHR3 promoter. Solid bars, EcR-B1–USP-1; hatched bars, EcR-B1–USP-2. (B) Reduction of 20E-induced promoter activity by the deletion of EcRE1. EcRE1 was deleted from the −1,216-bp promoter, and then either this or the parent construct was cotransfected with the pIE1hr expression vectors for EcR-B1 and USP-1. Transfection assays for MHR3 promoter activity were done 48 h after the cotransfection and 3 h after exposure to 2 μg of 20E per ml as described in Materials and Methods. Constructs are shown in the maps above the figures. Stars, putative EcRE; open circle, GGGTCA sequence (MRE). Means ± standard deviations (error bars) are shown (n = 3). Fold increased CAT was determined as in Fig. 5.
Removal of upstream sequence including EcRE4 at −1928 only slightly reduced the response in the presence of increased EcR-B1–USP-1 (Fig. 6A [construct −1216]). In contrast, further truncation removing EcRE3 and the adjacent MRE abolished nearly all of the induction (Fig. 6A [construct −596]), indicating a role for this upstream region in the 20E activation of this promoter. Removal of EcRE2 leaving only the EcRE1 site abolished all stimulation above the control level (Fig. 6A [construct −343]). Interestingly though, this EcRE1 was sufficient for the suppressive effect of the nonliganded EcR-B1–USP-1 (67% for one transfection set). Thus, EcRE1 (a perfect match to the EcRE consensus sequence [6]) by itself was insufficient for the increased responsiveness of this promoter in the presence of large amounts of the EcR-B1–USP-1 complex, but was able to mediate suppression.
The GMSAs of DNA binding (see Fig. 8) discussed below showed that only EcRE1 binds the EcR-B1–USP-1 complex. To determine whether EcRE1 was necessary for 20E activation, we deleted this EcRE and 80 bp 3′ (−342 to −247, by using SmaI and BamHI) from the −1,216-bp promoter, leaving all other sequence intact so that EcRE2, EcRE3, and the monomeric binding site were all present. As seen in Fig. 6B, this modified promoter lost its 20E-inducible activity. This deletion also abolished the suppressive effect of EcR-B1–USP-1 in the absence of ligand (1.4 ± 0.3 for the deleted construct versus 0.7 ± 0.1 for the intact one in three replicates of one transfection set). Thus, EcRE1 is necessary but not sufficient for the induction of MHR3 by 20E and also is important for the suppressive effect of the EcR-USP heterodimer in the absence of hormone.
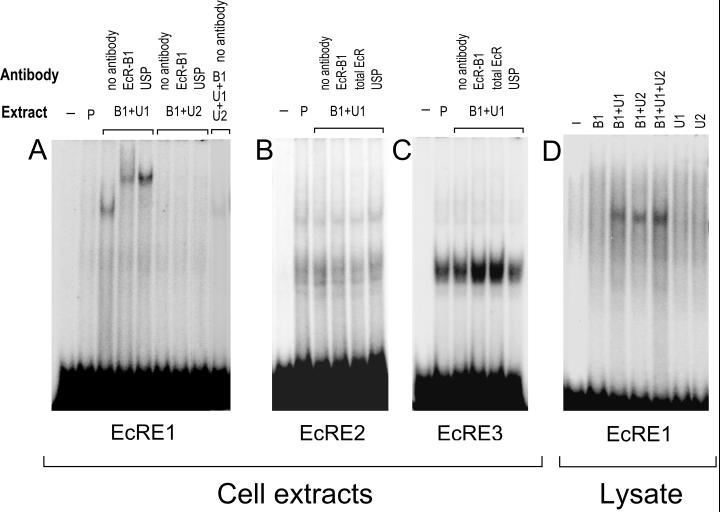
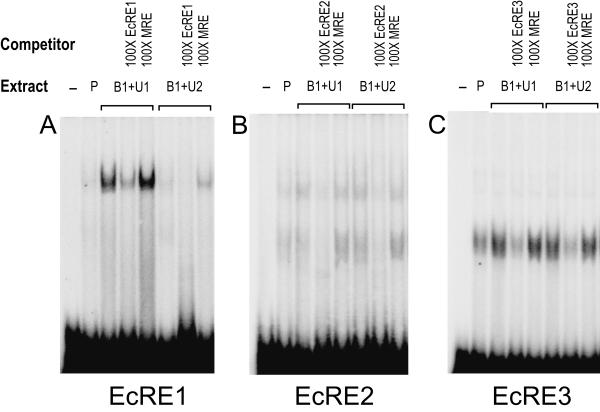
FIG. 8.
GMSAs using the EcRE1 (A), EcRE2 (B), and EcRE3 (C) oligonucleotides and lysate (D) (see Materials and Methods). Cotransfections were as follows: Extract P, empty pIE1hr/PA; Extract B1+U1, pIE1hr–EcR-B1 and pIE1hr–USP-1; Extract B1+U2, pIE1hr-EcR-B1 and pIE1hr-USP-2. Five micrograms of the extracts was used. The antibodies used are indicated above the gels.
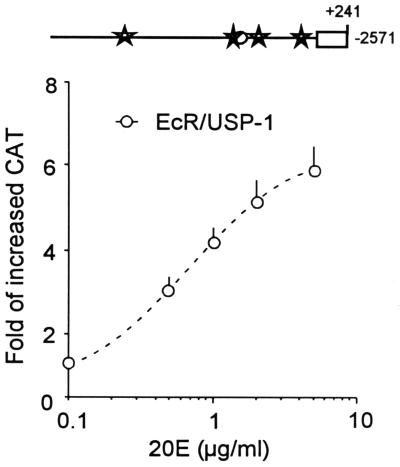
Activation of the −2.6-kb MHR3 promoter in the presence of increased levels of EcR-B1 and USP-1 was dependent on the concentration of 20E added. The 50% effective concentration was about 0.7 μg of 20E per ml (Fig. 7) which is slightly less than that seen for endogenous MHR3 mRNA (1 μg/ml) (34). This increased sensitivity of the MHR3 promoter to 20E in the presence of large amounts of EcR-B1 and USP-1 suggests that the promoter selectively responds to the EcR-B1–USP-1–20E complex.
FIG. 7.
Concentration-response curves for the −2,571-bp MHR3 promoter construct cotransfected with pIE1hr–EcR-B1 and pIE1hr–USP-1. Forty-five hours later, cells were treated with the given concentration of 20E for 6 h, and then the CAT level was assessed as in Fig. 5. Means ± standard deviations (error bars) are shown (n = 3).
Binding of EcR-USP to the putative EcREs.
To examine whether or not the putative EcREs in the MHR3 promoter region bind the EcR-USP complex, we used oligonucleotides corresponding to the sequences of EcREs 1, 2, and 3 (see Materials and Methods) for GMSAs. Figure 8A shows that EcRE1 strongly bound to protein(s) in the cell extracts containing EcR-B1–USP-1. This binding was specific, because it could be severely reduced by addition of 100-fold excess EcRE1, but not by 100-fold excess of an oligonucleotide to the putative monomeric binding site (MRE) (Fig. 9A). The bands were further shifted by the addition of the antibodies against EcR-B1 or USP (Fig. 8A), indicating that the complex contained both EcR and USP.
FIG. 9.
Competition GMSAs using 100× excess probe DNA used in Fig. 8 (EcRE1 [A], EcRE2 [B], and EcRE3 [C]) and the combination of the extracts, which show that all of the major shifted bands are probe specific. As a nonspecific competitor, 100× MRE DNA (see Materials and Methods) was used. Five micrograms of the extracts was used. See the legend to Fig. 8 for definitions of the abbreviations.
When USP-1 was replaced by USP-2 in the cell extracts, binding was undetectable (Fig. 8A and 9A). Also, there was no binding of EcR-B1–USP-2 in the cell extract to the hsp27 EcRE (n = 3 [data not shown]). Importantly, when both USP-1 and USP-2 were overexpressed together, only traces of the complex could be seen (Fig. 8A and 9A), indicating that USP-2 somehow interfered with the binding of EcR-B1–USP-1 to EcRE1. In none of these GMSAs did the presence of 2 μg of 20E per ml have any effect on the results obtained (data not shown). Thus, USP-2 did not mediate EcR binding to EcRE1 and in fact inhibited the binding of the EcR-B1–USP-1 complex to this element. Importantly, however, translation products of in vitro-transcribed EcR-B1 and either USP-1 or USP-2 heterodimerized and bound to EcRE1, whereas EcR-B1, USP-1, or USP-2 alone showed no binding (Fig. 8D). Therefore, some factor(s) in the cell may interact with USP-2 and prevent its binding as a heterodimer with EcR-B1 to EcRE1.
When either EcRE2 or EcRE3 was used as a probe in the GMSA, both the cell extracts containing pIE1hr-expressed EcR-B1 and USP-1 and the control extracts containing only endogenous levels of these proteins (Fig. 2) showed one major broad band and one or two (respectively) minor bands of higher molecular weight (Fig. 8B and C). The same bands were seen in cells transfected with EcR-B1 and USP-2 (Fig. 9B and C). No differences were seen when 20E was present. For EcRE2, where binding was not as strong as for EcRE3, the major band could be resolved into at least two components. These bands were specific, because they were nearly eliminated when 100-fold excess unlabeled probe was present, but not when a 100-fold excess of the MRE was present (Fig. 9B and C). The addition of EcR-B1 or USP antibodies (Fig. 8B and C) or the EcR-common (26) antibody (data not shown) had no effect on the size of the shifted bands, indicating that neither EcR nor USP is involved in these complexes. These findings indicate that neither EcRE2 nor EcRE3 binds the EcR-B1–USP-1 or -2 heterodimer, and therefore they are not EcREs in this context. Apparently, these elements bind proteins present in the cell extract which may be important in the activating complex.
DISCUSSION
These studies have shown that both isoforms of Manduca USP form heterodimers with Manduca EcR-B1 which bind ponasterone A with equal affinity and which bind to both the hsp27 EcRE and a natural EcRE (EcRE1) found in the Manduca MHR3 promoter when tested as in vitro translation products. In contrast, activation of the MHR3 promoter in the Manduca GV1 cell line was found to require the EcR-B1–USP-1 complex, EcRE1, and an upstream region between −596 and −1216 which contains an EcRE-like consensus sequence adjacent to a putative monomeric nuclear hormone binding site, GGGTCA. Only EcRE1 binds the EcR-B1–USP-1 complex in cell extracts, whereas the two EcRE-like sequences upstream bind other components of the cell extract. Surprisingly, USP-2 did not promote binding of EcR-B1 to EcRE1 in the cell extracts, but instead large amounts of USP-2 inhibited binding of EcR-B1–USP-1 and hence prevented 20E activation of the MHR3 promoter.
Both EcR and USP are present in the Manduca GV1 cell line, and addition of 20E causes an initial increase in EcR and later a down-regulation of both EcR and USP (35). Analysis of isoform-specific RNAs indicates that EcR-B1 and USP-2 RNAs increase during the first 6 h, whereas USP-1 RNA decreases (n = 3 to 6 [data not shown]). Expression of high levels of EcR-B1 and USP-1 in the cells under the control of the baculovirus promoter caused a 2.4-fold-increased induction of the endogenous MHR3 by 20E in 6 h, whereas either alone had only a slight effect. High expression of USP-2 in the cells inhibited this enhanced induction caused by the excess EcR-B1–USP-1 and also the normal induction mediated by the endogenous EcR-B1–USP-1 in the cells. Thus, it is clear that EcR-B1–USP-1 is the heterodimeric pair that binds 20E and activates MHR3 in both the GV1 cells (35) and the epidermis (40), which is consistent with the finding that both EcR-B1 and USP-1 are present in the epidermal cells when MHR3 RNA first appears (2, 26, 27, 46a). The earlier induction of increased levels of EcR-B1 in both the GV1 cells (33a, 35) and the epidermis (26) before the increase in MHR3 suggests that the increase in EcR-B1 may be one of the 20E-induced proteins necessary for full expression of MHR3 (40). The inhibitory effect of high USP-2 suggests that its high expression at the time of head capsule slippage and the peak of the ecdysteroid titer (2, 21, 27) may be one of the factors contributing to the down-regulation of MHR3 at this time.
The 20E-induced activation of the transfected MHR3 promoter was similar to that of the endogenous 4.5-kb MHR3 RNA (35), with increasing amounts of CAT protein beginning after 3 h of exposure. Most important for this activation were the first 924 bp of the promoter which contain three EcRE-like sequences and a nuclear hormone receptor monomeric binding sequence, GGGTCA, just 6 bp downstream from the distal EcRE3. However, only the most proximal, EcRE1 (−342 to −328), bound the EcR-B1–USP-1 heterodimer. This element perfectly matches the EcRE consensus sequence (6) and differs by 4 of the 15 bp (1 in the 5′ half and 3 in the 3′ half) from the perfect palindromic EcRE that was recently found to have the highest DNA binding affinity and activation-inducing activity in Drosophila SL-2 cells (52). Although EcRE1 was found to be necessary for 20E activation of MHR3 expression, it was not sufficient. Truncation experiments showed that upstream sequences which included EcRE3 located at −671 bp (and the closely associated monomeric binding site) were also critical for activation of the promoter. Both this and EcRE2 at −520 bind a protein(s) in the GV1 cell extract, but none of the complexes involves EcR and USP, as indicated by the absence of binding of the specific antibodies to the complex. The proteins that bind to both EcRE2 and EcRE3 apparently are present in the GV1 cells before exposure to 20E, since the binding pattern is the same, irrespective of whether or not 20E is present. Therefore, these two elements are not acting as EcREs in the GV1 cellular context and will now be referred to as “EcRE-like”. A likely explanation for these findings is that the EcRE1 binds the 20E–EcR-B1–USP-1 complex, and EcRE-like sequences 2 and 3 bind other transcription factors present in the cell extract. Interactions among these proteins at the different sites and their associated coactivators (25) then presumably cause the necessary DNA bending in order to activate the transcription complex.
As was originally shown by Cherbas et al. (6) for the ecdysone receptor complex and by many for the nuclear hormone receptors that interact with the RXR (25, 37), the receptor dimer binds to its response element in the absence of hormone and reduces the basal transcription (usually referred to as silencing). Importantly, even though EcRE1 alone could not support activation by increased EcR-B1–USP-1, the silencing effect in the absence of 20E was still seen when only EcRE1 was present. Therefore, EcRE1 allows the assembly of the EcR-B1–USP-1 heterodimer on the MHR3 promoter and the suppression of its basal transcription, even though factors interacting further upstream are necessary for the observed activation by hormone.
The EcR-B1–USP-2 heterodimer binds ponasterone A, an active ecdysteroid, and binds to both the hsp27 EcRE and EcRE1 in an in vitro system indistinguishably from the EcR-B1–USP-1 heterodimer. The novel finding is that the USP-2 does not do so in the GV1 cell extract and hence does not function with EcR-B1 either to silence the MHR3 promoter in the absence of hormone or to activate it in the presence of 20E. The only other insect in which two USP isoforms have been identified is the mosquito Aedes aegypti (28). Both of these isoforms in conjunction with the mosquito EcR as in vitro translation products bind ponasterone A and to the hsp27 EcRE, albeit with slightly different affinities (28). Only one isoform, Aedes USP-b, has been tested and shown to activate via this element in mammalian CV-1 cells (54). The other insect EcR-USP complexes that have been tested in a cellular context (1, 6, 7, 42, 50, 52, 56, 57) show binding and activation on hsp27 and other functional EcREs, although relative affinities and levels of activation may vary with the cells used. Apparently there is a factor in the GV1 cells that interacts with USP-2 that either prevents its heterodimerization with EcR-B1 or its ability after heterodimerization to mediate binding to EcREs. The ability of highly constitutively expressed USP-2 to prevent EcR-B1–USP-1 binding to EcRE1, and hence its activation of the MHR3 promoter, suggests that USP-2 is likely competing with USP-1 for heterodimerization with EcR-B1 in the cells. It also may be competing for the coactivators that are necessary for EcR-B1–USP-1 to activate the MHR3 promoter when 20E is present, since coactivators primarily interact with regions in the ligand binding domains (25), which are the same in both USP isoforms.
In vertebrates, many factors are now known to interact with nuclear hormone receptors to mediate their interactions with other transcription factors and with the transcription initiation complex, the best known of which are coactivators and corepressors (25). The corepressors are present in the absence of the hormonal ligand and are responsible for transcriptional silencing. Binding of hormone to the receptor causes a conformational change leading to loss of the corepressor and binding of the coactivator. These molecules mainly bind to certain areas in the hinge (D) or the ligand binding (E) domain, but there is also an activation domain (AF1) in the N-terminal A/B domain (37, 55). Interactions between the A/B domain and the D domain of RXRa and RXRb (the vertebrate counterparts of USP) have been shown to be necessary for cell-specific activation (16). Also, recent studies have shown that the steroid receptor coactivator 1 (SRC-1) has several domains, some of which interact with the N-terminal AF1 domain and are necessary for transactivation (38), and that different SRC-1s may either enhance or inhibit interactions between the N and C termini of the androgen receptor that are necessary for its transcriptional activity (24). In addition to the AF1 domain, there are other motifs in the N-terminal domain that can modulate DNA binding and dimerization (15) or inhibit transcriptional activation (23).
The reason for lack of DNA binding by the EcR-B1–USP-2 complex in the cellular context must lie with the region missing in the N-terminal A/B domain of USP-2 compared to that of USP-1. Both of the Manduca USPs are identical in the C-terminal 51 amino acids of the A/B domain (27), the last 13 of which are 100% conserved among all insect USPs (28). The latter region is all that is necessary for Drosophila USP to heterodimerize with the human thyroid hormone receptor β (TRβ) and activate the human apolipoprotein A-II promoter (17). USP-2 has only 8 amino acids upstream of this common region, whereas USP-1 has 61. In the unique portion of USP-1 is a stretch of 22 amino acids containing 9 (41%) serines and threonines which are putative phosphorylation sites. When a similar 19-amino-acid stretch of high serine/threonine content was deleted from the A/B region of TRβ1, the hormone-dependent activation response was abolished in mammalian COS-1 cells (55). In this case, however, truncation of the entire N terminus, including this domain, increased rather than decreased the DNA binding affinity of TRβ1 to several different TREs. In the Chironomus tentans epithelial cell line, the presence of 20E increases the phosphorylation of USP, likely in the highly conserved protein kinase C sites in the DNA binding domain and near the C terminus of the ligand binding domain (45), but also possibly in the A/B domain. Also, phosphorylation of both Manduca USP isoforms occurs in the prothoracic glands (49), indicating that the common phosphorylation sites are used, but not precluding the phosphorylation of the A/B domain sites in USP-1 as well. Possibly phosphorylation of the latter sites in USP-1 is necessary for high-affinity binding of the EcR-USP heterodimer to and activation of the EcRE.
These studies have clearly shown that 20E is able to activate the MHR3 promoter only when bound to the EcR-B1-USP-1 heterodimer, with this binding and activation prevented by the presence of high levels of USP-2. This is the first example of the discriminated usage of USP isoforms in the cellular context and demonstrates the power of changing EcR-USP isoforms in the context of orchestrating a transcription factor cascade. Similar selective usage of RXR, retinoic acid receptor (RAR), and ROR isoforms have been reported in vertebrate systems. The cellular retinol-binding protein type II promoter can be activated only by 9-cis-retinoic acid in NIH 3T3 cells in combination with RXRα and not with RXRβ, but both isoforms activate the same reporter in P19 embryonal carcinoma cells (16). Giguére et al. (13) showed that the isoform-specific N-terminal domains of ROR determined its binding to different monomeric response elements, and RORα-1 was the effective isoform for activation of the apolipoprotein A-I gene (53). Also, RARγ was preferred in the activation of the uncoupling protein gene promoter (44). How the interacting factors with the different N-terminal domains mediate these changes in DNA binding and transactivation is a challenging question for the future.
ACKNOWLEDGMENTS
We thank Paul Friesen for the gift of the pIE1hr/PA expression vector, Fotis Kafatos for the gift of the USP antibody, and James Truman and Peter Cherbas for a critical reading of the manuscript.
The work was supported by a grant from NIH AI12459. Que Lan was supported by a fellowship from the American Cancer Society, and Kiyoshi Hiruma was supported by a grant from the USDA (97-35302-4433).
REFERENCES
- 1.Antoniewski C, Mugat B, Delbac F, Lepesant J-A. Direct repeats bind the EcR/USP receptor and mediate ecdysteroid responses in Drosophila melanogaster. Mol Cell Biol. 1996;16:2977–2986. doi: 10.1128/mcb.16.6.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asahina M, Jindra M, Riddiford L M. Developmental expression of Ultraspiracle proteins in the epidermis of the tobacco hornworm, Manduca sexta, during larval life and the onset of metamorphosis. Dev Genes Evol. 1997;20:381–388. doi: 10.1007/s004270050127. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, vol. 1, section 4.8. New York, N.Y: John Wiley & Sons; 1994. [Google Scholar]
- 4.Burtis K C, Thummel C S, Jones W C, Karim F D, Hogness D S. The Drosophila E74 early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell. 1990;61:85–90. doi: 10.1016/0092-8674(90)90217-3. [DOI] [PubMed] [Google Scholar]
- 5.Cherbas L, Cherbas P. The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem Mol Biol. 1993;23:81–90. doi: 10.1016/0965-1748(93)90085-7. [DOI] [PubMed] [Google Scholar]
- 6.Cherbas L, Lee K, Cherbas P. Identification of ecdysone response elements by analysis of the DrosophilaEip28/29 gene. Genes Dev. 1991;5:120–131. doi: 10.1101/gad.5.1.120. [DOI] [PubMed] [Google Scholar]
- 7.Cherbas P, Cherbas L. Molecular aspects of ecdysteroid hormone action. In: Gilbert L I, Tata J R, Atkinson B G, editors. Metamorphosis: postembryonic reprogramming of gene expression in amphibian and insect cells. San Diego, Calif: Academic Press; 1996. pp. 175–221. [Google Scholar]
- 8.Cherbas P, Cherbas L, Lee S-S, Nakanishi K. 26-[125I]iodoponasterone A is a potent ecdysone and a sensitive radioligand for ecdysone receptors. Proc Natl Acad Sci USA. 1988;85:2096–2100. doi: 10.1073/pnas.85.7.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski O, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Davis L G, Dibner M D, Battey J F. Basic methods in molecular biology. Amsterdam, The Netherlands: Elsevier; 1986. [Google Scholar]
- 11.Fletcher J C, D’Avino P P, Thummel C S. A steroid-triggered switch in E74 transcription factor isoforms regulates the timing of secondary-response gene expression. Proc Natl Acad Sci USA. 1997;94:4582–4586. doi: 10.1073/pnas.94.9.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara H, Jindra M, Newitt R, Palli S R, Hiruma K, Riddiford L M. Cloning of an ecdysone receptor homolog from Manduca sextaand the developmental profile of its mRNA in wings. Insect Biochem Mol Biol. 1995;25:845–856. doi: 10.1016/0965-1748(95)00023-o. [DOI] [PubMed] [Google Scholar]
- 13.Giguère V, Tini M, Flock G, Ong E, Evans R M, Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of RORa, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 14.Guo X, Harmon M A, Jin X, Laudet V, Mangelsdorf D J, Palmer M J. Isolation of two functional retinoid X receptor subtypes from the ixodid tick, Amblyoma americanum(L.) Mol Cell Endocrinol. 1998;139:45–60. doi: 10.1016/s0303-7207(98)00073-2. [DOI] [PubMed] [Google Scholar]
- 15.Hadzic E, Habeos I, Raaka B M, Samuels H H. A novel multifunctional motif in the amino-terminal A/B domain of T3R alpha modulates DNA binding and receptor dimerization. J Biol Chem. 1998;273:10270–10278. doi: 10.1074/jbc.273.17.10270. [DOI] [PubMed] [Google Scholar]
- 16.Hallenbeck P L, Minucci S, Lippoldt P, Phyillaier M, Horn V, Ozato K, Nikodem V M. Differential 9-cis-retinoic acid-dependent transcriptional activation by murine retinoid X receptor alpha (RXR alpha) and RXR beta—role of cell type and RXR domains. J Biol Chem. 1996;271:10503–10507. doi: 10.1074/jbc.271.18.10503. [DOI] [PubMed] [Google Scholar]
- 17.Hatzivassiliou E, Cardot P, Zannis V I, Mitsialis S A. Ultraspiracle, a Drosophilaretinoic X receptor α homologue, can mobilize the human thyroid hormone receptor to transactivate a human promoter. Biochemistry. 1997;36:9221–9231. doi: 10.1021/bi963145k. [DOI] [PubMed] [Google Scholar]
- 18.Henrich V C, Szekely A A, Kim S J, Brown N E, Antoniewski C, Hayden M A, Lepesant J-A, Gilbert L I. Expression and function of the ultraspiracle (usp) gene during development of Drosophila melanogaster. Dev Biol. 1994;165:38–52. doi: 10.1006/dbio.1994.1232. [DOI] [PubMed] [Google Scholar]
- 19.Hiruma K, Carter M S, Riddiford L M. Characterization of the dopa decarboxylase gene of Manduca sextaand its suppression by 20-hydroxyecdysone. Dev Biol. 1995;169:195–209. doi: 10.1006/dbio.1995.1137. [DOI] [PubMed] [Google Scholar]
- 20.Hiruma K, Böcking D, Lafont R, Riddiford L M. Action of different ecdysteroids on the regulation of mRNAs for the ecdysone receptor, MHR3, dopa decarboxylase, and a larval cuticle protein in the larval epidermis of the tobacco hornworm, Manduca sexta. Gen Comp Endocrinol. 1997;107:84–97. doi: 10.1006/gcen.1997.6901. [DOI] [PubMed] [Google Scholar]
- 21.Hiruma K, Shinoda T, Malone F, Riddiford L M. Juvenile hormone modulates 20-hydroxyecdysone-inducible ecdysone receptor and ultraspiracle gene expression in the tobacco hornworm, Manduca sexta. Dev Genes Evol. 1999;209:18–30. doi: 10.1007/s004270050223. [DOI] [PubMed] [Google Scholar]
- 22.Horner M A, Chen T, Thummel C S. Ecdysteroid regulation and DNA binding properties of Drosophilanuclear hormone receptor superfamily members. Dev Biol. 1995;168:490–502. doi: 10.1006/dbio.1995.1097. [DOI] [PubMed] [Google Scholar]
- 23.Hovland A R, Powell R L, Takimoto G S, Tung L, Horwitz K B. An N-terminal inhibitory function, IF, suppresses transcription by the A-isoform but not the B-isoform of human progesterone receptors. J Biol Chem. 1998;273:5455–5460. doi: 10.1074/jbc.273.10.5455. [DOI] [PubMed] [Google Scholar]
- 24.Ikonen T, Palvimo J P, Jänne O A. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 25.Jenster G. Coactivators and corepressors as mediators of nuclear receptor function: an update. Mol Cell Endocrinol. 1998;143:1–7. doi: 10.1016/s0303-7207(98)00145-2. [DOI] [PubMed] [Google Scholar]
- 26.Jindra M, Malone F, Hiruma K, Riddiford L M. Developmental profiles and ecdysteroid regulation of the mRNAs for two ecdysone receptor isoforms in the epidermis and wings of the tobacco hornworm, Manduca sexta. Dev Biol. 1996;180:258–272. doi: 10.1006/dbio.1996.0299. [DOI] [PubMed] [Google Scholar]
- 27.Jindra M, Huang J-Y, Malone F, Asahina M, Riddiford L M. Identification and mRNA developmental profiles of two Ultraspiracle isoforms in the epidermis and wings of Manduca sexta. Insect Mol Biol. 1997;6:41–53. doi: 10.1046/j.1365-2583.1997.00153.x. [DOI] [PubMed] [Google Scholar]
- 28.Kapitskaya M, Wang S, Cress D E, Dhadialla T S, Raikhel A S. The mosquito ultraspiracle homologue, a partner of ecdysteroid receptor heterodimer: cloning and characterization of isoforms expressed during vitellogenesis. Mol Cell Endocrinol. 1996;121:119–132. doi: 10.1016/0303-7207(96)03847-6. [DOI] [PubMed] [Google Scholar]
- 29.Karim F D, Thummel C S. Temporal coordination of regulatory gene expression by the steroid hormone ecdysone. EMBO J. 1992;11:4083–4093. doi: 10.1002/j.1460-2075.1992.tb05501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khoury Christianson A N, King D L, Hatzivassiliou E, Casa J E, Hallenbeck P L, Nikodem V M, Mitsalis S A, Kafatos F C. DNA binding and heteromerization of the Drosophilatranscription factor chorion factor 1/ultraspiracle. Proc Natl Acad Sci USA. 1992;89:11503–11507. doi: 10.1073/pnas.89.23.11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koelle M R, Segraves W A, Hogness D S. DHR3: a Drosophilasteroid receptor homolog. Proc Natl Acad Sci USA. 1992;89:6167–6171. doi: 10.1073/pnas.89.13.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koelle M R, Talbot W S, Segraves W A, Bender M T, Cherbas P, Hogness D S. The Drosophila EcRgene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- 32a.Lafont, R. Personal communication.
- 33.Lafont R, Wilson I D. The ecdysone handbook. 1992. Chromatographic Society, Nottingham, England. [Google Scholar]
- 33a.Lan, Q. Unpublished observations.
- 34.Lan Q, Riddiford L M. DNA transfection in the ecdysteroid-responsive GV1 cell line from the tobacco hornworm, Manduca sexta. In Vitro Cell Dev Biol. 1997;33:615–621. doi: 10.1007/s11626-997-0111-5. [DOI] [PubMed] [Google Scholar]
- 35.Lan Q, Wu Z-N, Riddiford L M. Regulation of the ecdysone receptor, USP, E75, and MHR3 genes by 20-hydroxyecdysone in the GV1 cell line of the tobacco hornworm, Manduca sexta. Insect Mol Biol. 1997;6:3–10. doi: 10.1046/j.1365-2583.1997.00151.x. [DOI] [PubMed] [Google Scholar]
- 36.Lynn D E, Oberlander H. The effect of cytoskeletal disrupting agents on the morphological response of a cloned Manduca sextacell line to 20-hydroxyecdysone. Wilhelm Roux Arch. 1981;190:150–156. doi: 10.1007/BF00867801. [DOI] [PubMed] [Google Scholar]
- 37.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 38.Onate S A, Boonyaratanakornkit V, Spencer T E, Tsai S Y, Tsai M-J, Edwards D P, O’Malley B W. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 39.Oro A E, McKeown M, Evans R M. The Drosophila retinoid X receptor homolog ultraspiraclefunctions in both female reproduction and eye morphogenesis. Development. 1992;115:449–462. doi: 10.1242/dev.115.2.449. [DOI] [PubMed] [Google Scholar]
- 40.Palli S R, Hiruma K, Riddiford L M. An ecdysteroid-inducible Manduca gene similar to the DrosophilaDHR3 gene, a member of the steroid hormone receptor superfamily. Dev Biol. 1992;150:306–318. doi: 10.1016/0012-1606(92)90244-b. [DOI] [PubMed] [Google Scholar]
- 41.Palli S R, Riddiford L M, Hiruma K. Juvenile hormone and “retinoic acid” receptors in Manducaepidermis. Insect Biochem. 1991;21:7–15. [Google Scholar]
- 42.Perera S C, Palli S R, Ladd T R, Krell P J, Retnakaran A. The ultraspiracle gene of the spruce budworm, Choristoneura fumiferana: cloning of cDNA and developmental expression of mRNA. Dev Genet. 1998;22:169–179. doi: 10.1002/(SICI)1520-6408(1998)22:2<169::AID-DVG6>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 43.Pullen S S, Friesen P D. The CAGT motif functions as an initiator element during early transcription of the baculovirus transregulator ie-1. J Virol. 1995;69:3575–3583. doi: 10.1128/jvi.69.6.3575-3583.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabelo R, Reyes C, Schifman A, Silva J E. A complex retinoic acid response element in the uncoupling protein gene defines a novel role for retinoids in thermogenesis. Endocrinology. 1996;137:3488–3496. doi: 10.1210/endo.137.8.8754778. [DOI] [PubMed] [Google Scholar]
- 45.Rauch P, Grebe M, Elke C, Spindler K-D, Spindler-Barth M. Ecdysteroid receptor and ultraspiracle from Chironomus tentans(Insecta) are phosphoproteins and are regulated differently by molting hormone. Insect Biochem Mol Biol. 1998;28:265–275. doi: 10.1016/s0965-1748(98)00026-5. [DOI] [PubMed] [Google Scholar]
- 46.Riddiford L M. Molecular aspects of juvenile hormone action in insect metamorphosis. In: Gilbert L I, Tata J R, Atkinson B G, editors. Metamorphosis: postembryonic reprogramming of gene expression in amphibian and insect cells. San Diego, Calif: Academic Press; 1996. pp. 223–251. [Google Scholar]
- 46a.Riddiford, L. M., M. Asahina, and R. Langelan. Unpublished observations.
- 47.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segraves W A, Hogness D S. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophilaencodes two new members of the steroid receptor superfamily. Genes Dev. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- 49.Song Q, Gilbert L I. Alteration in ultraspiracle (USP) content and phosphorylation state accompany feedback regulation of ecdysone synthesis in the insect prothoracic gland. Insect Biochem Mol Biol. 1998;28:849–860. doi: 10.1016/s0965-1748(98)00075-7. [DOI] [PubMed] [Google Scholar]
- 50.Swevers L, Cherbas L, Cherbas P, Iatrou K. Bombyx EcR (BmEcR) and BombyxUSP (BmCF1) combine to form a functional ecdysone receptor. Insect Biochem Mol Biol. 1996;26:217–221. doi: 10.1016/0965-1748(95)00097-6. [DOI] [PubMed] [Google Scholar]
- 51.Thummel C S, Burtis K C, Hogness D S. Spatial and temporal patterns of E74 transcription during Drosophiladevelopment. Cell. 1990;61:101–111. doi: 10.1016/0092-8674(90)90218-4. [DOI] [PubMed] [Google Scholar]
- 52.Vögtli M, Elke C, Imhof M O, Lezzi M. High level transactivation by the ecdysone receptor complex at the core recognition motif. Nucleic Acids Res. 1998;26:2407–2414. doi: 10.1093/nar/26.10.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vu-Dac N, Gervois P, Grotzinger T, DeVos P, Schoonjans K, Fruchart J C, Auwerx J, Mariani J, Tedgui A, Staels B. Transcription regulation of apolipoprotein AI gene expression by the nuclear receptor RORα. J Biol Chem. 1997;272:22401–22404. doi: 10.1074/jbc.272.36.22401. [DOI] [PubMed] [Google Scholar]
- 54.Wang S-F, Miura K, Miksicek R J, Segraves W A, Raikhel A S. DNA binding and transactivation characteristics of the mosquito ecdysone receptor-Ultraspiraclecomplex. J Biol Chem. 1998;273:27531–27540. doi: 10.1074/jbc.273.42.27531. [DOI] [PubMed] [Google Scholar]
- 55.Wilkinson J R, Towle H C. Identification and characterization of the AF-1 transactivation domain of thyroid hormone receptor β1. J Biol Chem. 1997;272:23824–23832. doi: 10.1074/jbc.272.38.23824. [DOI] [PubMed] [Google Scholar]
- 56.Yao T-P, Forman B M, Jiang Z, Cherbas L, Chen J-D, McKeown M, Cherbas P, Evans R M. Functional ecdysone receptor is the product of EcR and Ultraspiraclegenes. Nature. 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 57.Yao T-P, Segraves W A, Oro A E, McKeown M, Evans R M. Drosophilaultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- 58.Zhou B, Hiruma K, Jindra M, Shinoda T, Malone F, Segraves W A, Riddiford L M. Regulation of the transcription factor E75 by 20-hydroxyecdysone and juvenile hormone in the epidermis of the tobacco hornworm, Manduca sexta, during larval molting and metamorphosis. Dev Biol. 1998;193:127–138. doi: 10.1006/dbio.1997.8798. [DOI] [PubMed] [Google Scholar]