Abstract
T cell development proceeds under the influence of a network of transcription factors (TFs). The precise role of Zeb1, a member of this network, remains unclear. Here, we report that Zeb1 expression is induced early during T cell development in CD4−CD8− double-negative (DN) stage 2 (DN2). Zeb1 expression was further increased in the CD4+CD8+ double-positive (DP) stage before decreasing in more mature T cell subsets. We performed an exhaustive characterization of T cells in Cellophane mice that bear Zeb1 hypomorphic mutations. The Zeb1 mutation profoundly affected all thymic subsets, especially DN2 and DP cells. Zeb1 promoted the survival and proliferation of both cell populations in a cell-intrinsic manner. In the periphery of Cellophane mice, the number of conventional T cells was near normal, but invariant NKT cells, NK1.1+ γδ T cells and Ly49+ CD8 T cells were virtually absent. This suggested that Zeb1 regulates the development of unconventional T cell types from DP progenitors. A transcriptomic analysis of WT and Cellophane DP cells revealed that Zeb1 regulated the expression of multiple genes involved in the cell cycle and TCR signaling, which possibly occurred in cooperation with Tcf1 and Heb. Indeed, Cellophane DP cells displayed stronger signaling than WT DP cells upon TCR engagement in terms of the calcium response, phosphorylation events, and expression of early genes. Thus, Zeb1 is a key regulator of the cell cycle and TCR signaling during thymic T cell development. We propose that thymocyte selection is perturbed in Zeb1-mutated mice in a way that does not allow the survival of unconventional T cell subsets.
Keywords: T cell selection, TCR signaling, Zeb1, transcription, development
Subject terms: T-cell receptor, Lymphocyte differentiation
Introduction
T cell development occurs in the thymus and begins in immature thymocytes that are double negative (DN) for CD4 and CD8 expression. The DN population can be subdivided into four subsets, DN1–DN4, depending on the expression of the cell surface molecules CD44 and CD25 (for a review, see1). DN1 cells (CD44+CD25−) are the most immature progenitors and retain the ability to differentiate into non-T cell lineages. In DN2 cells (CD44+CD25+), the expression of RAG1/2 is induced, which promotes the rearrangement of gene segments encoding the TCR-β, TCR-γ, and TCR-δ subunits. In DN3 cells (CD44−CD25+), the T cell antigen receptor (TCR) β-chain associates with the pre-TCR α-chain and CD3 subunits to form the pre-TCR complex; the pre-TCR complex allows β-selection to occur. During β-selection, DN3 cells with productive TCRβ rearrangements receive survival and proliferative signals and mature into the DN4 (CD44−CD25−) stage. DN4 thymocytes then develop into CD4+CD8+ double-positive (DP) cells.2
At the DP stage, a series of events takes place that determines the fate of developing T cells, including rearrangement of the TCR alpha locus, association of the αβ T cell receptor, and subsequent thymic selection. In general, high-affinity interactions between the αβTCR and self-peptide-MHC complexes presented by different thymic cells lead to negative selection and elimination of self-reactive thymocytes, while low-affinity interactions result in positive selection and development of CD4 or CD8 single-positive (SP) T cells.3–5 Despite this general rule, regulatory T cells and invariant NKT cells (iNKT) receive stronger TCR signals than conventional T cells during their development6 as a result of selection by agonist self-antigens. iNKT cells are a subset of innate-like T cells with a single invariant TCRα chain (Vα14-Jα18 in mice) and a limited repertoire of TCRβ chains (Vβ8.2, Vβ7, or Vβ2) that recognize glycolipid antigens bound to CD1d, a nonpolymorphic MHC molecule.7 iNKT cell development includes discrete stages (stages 0–3) that can be discriminated according to CD44 and NK1.1 expression.8 Three functionally distinct iNKT cell subsets have also been identified: iNKT1 cells, which express T-bet and mainly secrete IFN-γ; iNKT2 cells, which express Gata3 and Plzf and secrete IL-4 and IL-13; and iNKT17 cells, which express Rorγt and secrete IL-17. The TCR signal strength during selection governs the development of iNKT cell subsets, with strong signals promoting iNKT2 and iNKT17 development.9,10 A large number of molecules regulate the strength of TCR-derived signaling. TCR signaling strength can also be modulated at the transcriptional level by transcription factors (TFs) such as Sox411 or at the posttranscriptional level by miR-181.12,13 The loss of either Sox4 or miR-181 blocks iNKT cell development. Mechanistically, miR-181a regulates the expression of multiple phosphatases and other proteins to boost TCR signaling as well as cell metabolism.12,13 Interestingly, mice expressing a hypomorphic form of Zap70, a major TCR-proximal kinase, also show impaired developmental maturation of γδ T cells, suggesting that innate-like T cell subsets are particularly dependent on the tight regulation of the strength of TCR signaling for their development.14
A dense network of TFs has been shown to regulate T cell development.15 Early commitment is dependent on Notch signaling,16 which induces the expression of many TFs and maintains their expression throughout T cell development. Among these TFs, the E-protein family factors E2a, Tcf1 (encoded by Tcf7) and Heb (encoded by Tcf12)17 induce the expression of TCR components and balance the survival and proliferation of thymocytes.18 Many other TFs, such as Gata3, Myb, Runx1, and Bcl11b, also cooperate with E proteins at different developmental stages and further establish T cell identity.15,18
The Zeb family of TFs consists of Zeb1 and Zeb2, which are best known for their role in epithelial-to-mesenchymal transition (EMT). EMT programs operate at different stages of embryonic development and are downstream of Wnt, TGF-β, Bmp, Notch, and other signaling pathways.19 Zeb1−/− mice exhibit multiple developmental defects and die at birth.20 Under pathological conditions, activation of EMT programs contributes to fibrosis and cancer metastasis.21 Zeb1 and Zeb2 are highly homologous and are characterized by two clusters of zinc finger domains at the protein extremities. They also contain a homeodomain and a Smad-binding domain and can interact with many other TFs.22 Zeb1 and Zeb2 are also expressed in a tightly regulated manner in the immune system and regulate cell differentiation.23 We and others have previously shown that Zeb2 regulates terminal NK cell24 and effector CD8 T cell differentiation.25,26 Mutated mice expressing a truncated form of Zeb1 without the C-terminal zinc finger clusters at C727 have a small and hypocellular thymus, which is the result of a reduction in early T cell precursors.27 In Cellophane mutant mice, a T → A mutation in the seventh exon of Zeb1 replaces the tyrosine at position 902 with a premature stop codon.28 The resulting mRNA encodes a truncated protein lacking the C-terminal zinc finger domain, which is predicted to be hypomorphic. Cellophane homozygous mice have small, hypocellular thymi with decreased DP thymocytes. However, the mechanism of Zeb1 action during T cell development and its role in the maturation of T cell subsets remain unclear. Here, we show that Cellophane homozygous mice lack several peripheral T cell subsets, including iNKT cells, NK1.1+ γδ T cells, and Ly49-expressing CD8 T cells. This specific defect involving innate-like T cells is caused by the cell-intrinsic role of Zeb1 in T cell development. We show that Zeb1 expression is maximal in the DN2 and DP stages of T cell development. Furthermore, Zeb1 regulates the transition to the SP stage by promoting cell proliferation and survival and repressing the expression of various molecules that modulate the strength of TCR signaling. Therefore, we propose that Zeb1 is a key regulator of thymocyte selection that is essential for the development and survival of innate-like T cell subsets undergoing agonist-type selection.
Results
Zeb1 is highly expressed in the DN2 and DP stages of T cell development
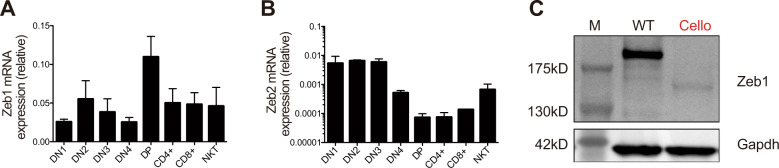
To study the role of Zeb1 in T cell development, we sorted thymocyte subsets and measured Zeb1 transcript levels by semiquantitative (Q) RT-PCR (reverse transcription-polymerase chain reaction). As shown in Fig. 1a, Zeb1 transcript levels were low in DN1, began to increase in the DN2 stage and were maximal in DP thymocytes. Zeb1 expression then decreased as T cells underwent selection and matured into either conventional T cells or iNKT cells. Interestingly, the expression of Zeb2 was somewhat similar to that of Zeb1; high expression of Zeb2 was observed in early thymic progenitors (DN1–DN4), and the lowest expression of Zeb2 was observed in DP cells (Fig. 1b). This pattern of expression was corroborated by data from the ImmGen consortium29 (Fig. S1A). Thus, as was observed in memory T cells,30 Zeb1 and Zeb2 show similar patterns of expression in thymocytes. We used the ImmGen web browser to search for coregulated genes across different immune subsets. The E-protein Heb (encoded by Tcf12) was among the top 3 genes found to be coregulated with Zeb1 (Fig. S1B).29 Heb is well known for its important roles throughout T cell development,31 especially in the DP stage,32 which further indicates that Zeb1 is a potential regulator of the DP developmental stage. We also analyzed the expression of Zeb1 protein in total thymocytes (80% of which consist of DPs). Zeb1 was strongly expressed in WT thymocytes but not in Cellophane thymocytes. Mutant mice expressed only reduced quantities of a truncated form of Zeb1 (Fig. 1c).
Fig. 1.
Zeb1 expression in WT and Cellophane thymocytes. a, b RT-PCR analysis of RNA from sorted thymocyte subsets isolated from C57BL/6 mice, as indicated. The results are presented relative to the expression of the control gene Gapdh. c WB analysis of Zeb1 expression in total thymocytes from WT and Cellophane mice, as indicated. Data are representative of three independent experiments with three to six mice (a, b) or three independent experiments with three mice (c)
Impaired development of both conventional and unconventional T cells in Cellophane mice
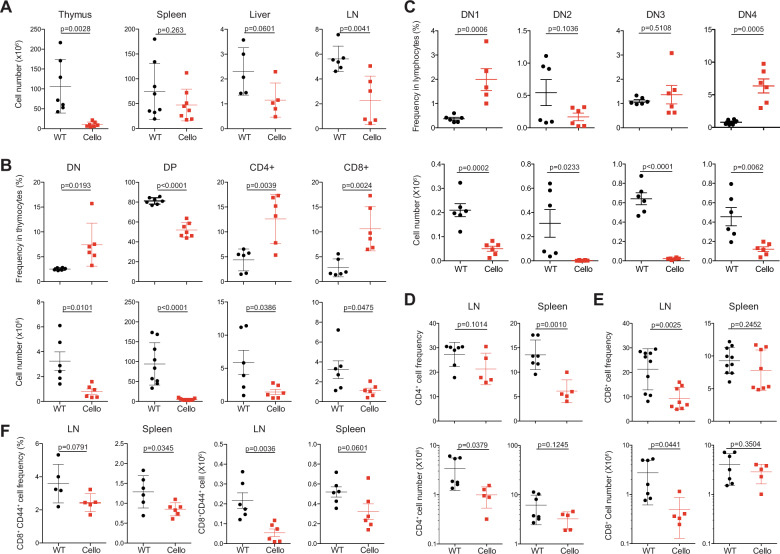
To define the impact of the Cellophane mutation on T cell development, we analyzed the T cell composition in the thymus, spleen, and lymph nodes (LNs) of Cellophane mice. The T cell numbers in the spleen and liver were normal, while the number of lymphocytes was reduced in LNs (Fig. 2a). As shown by previous findings,28 we also observed a strong decrease in the cell numbers in the thymus in Zeb1-mutated mice (Fig. 2a). This decrease in the cell number affected all subsets defined by CD4, CD8, CD44, and CD25 expression (Fig. 2b, c). The CD4+CD8+ DP thymocytes and DN2 populations also decreased in number within Cellophane thymocytes (Fig. 2b, c). In the LN and spleen, the percentages of CD4 T cells and CD8 T cells were reduced (Fig. 2d, e), and the proportion of memory-phenotype CD44+ T cells among total CD8 T cells was decreased by nearly 30% (Fig. 2f).
Fig. 2.
Cellularity and proportions of T cell subsets in Cellophane mice. a Total number of cells from the thymus, spleen, liver, and LN. b Percentages and absolute numbers of the indicated T cell subsets (DN, DP, CD4 SP, and CD8 SP) in the thymus. c Percentages and absolute numbers of the indicated DN subsets defined by CD44 and CD25 expression in the thymus. Percentages and absolute numbers of (d) CD4 T cells and (e) CD8 T cells. (f) Percentages and absolute numbers of CD8+ CD44+ memory T cells in LN and spleen. Each dot represents an individual mouse. Data were pooled from 7 to 8 mice in four experiments (a–e) or three to six mice in two experiments (f). The statistical analysis was performed using an unpaired Student’s t test
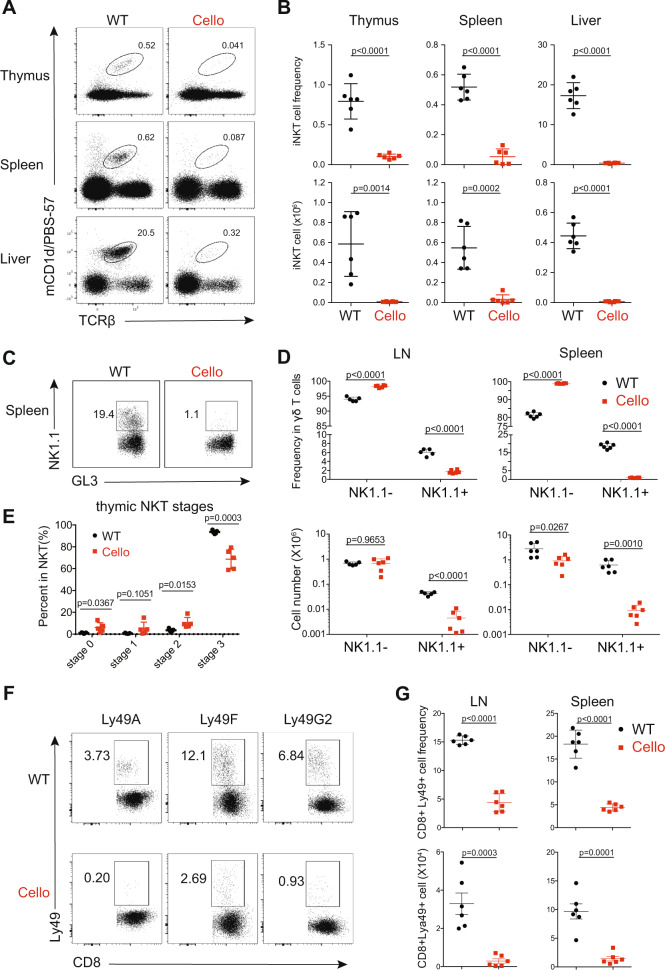
We then investigated the development of unconventional T cell subsets. We observed a drastic decrease in the proportion and in the number of iNKT cells as well as NK1.1+ γδ T cells in Cellophane mice compared with those in littermate controls (Fig. 3a–d). This decrease affected all organs in Cellophane mice (Fig. 3a–d). iNKT cells were mainly affected at stage 3 (Fig. 3e). To complete our analysis, we also studied memory-phenotype Ly49+ CD8 T cells, which are thought to arise “naturally” in the thymus without antigenic exposure.33 All Ly49+ CD8 T cell populations were dramatically reduced in LN and spleen from Cellophane mice in terms of both proportions and numbers, irrespective of the inhibitory Ly49 receptor type that was analyzed (Ly49A, Ly49F, or Ly49G2) (Fig. 3f, g).
Fig. 3.
Cellophane mice have decreased numbers of thymic DP cells and lack iNKT cells, NK1.1+ γδ T cells and Ly49+ CD8 cells. a Flow cytometry analysis of CD1d-tet+ iNKT cells (black gate) from thymus, spleen, and liver in wild-type (WT) and Cellophane (Cello) homozygous mice as indicated. b Percentages and absolute numbers of iNKT cells (mCD1d/PBS-57+ TCRβ+) in the thymus, spleen, and liver. c Flow cytometry analysis of NK1.1− and NK1.1+ γδ T cells (black square gates) from the spleen of WT and Cellophane mice. d Percentages within total γδ T cells and absolute numbers of NK1.1− and NK1.1+ γδ T cells in spleen and LN. e Percentage within total thymic iNKT cells of cells from stages 0–3, as defined by CD24, CD44, and NK1.1 expression. f Flow cytometry analysis of CD8+ CD44+ Ly49A+/Ly49F+/Ly49G2+ T cells (black square gates) from LN and spleen from WT and Cellophane mice. g Percentage within CD8+CD44+ T cells and absolute number of CD8+ CD44+ Ly49+ T cells in spleen and LN. Each dot represents an individual mouse. Data are representative of two to four independent experiments with six mice in total in each panel. The statistical analysis was performed using an unpaired Student’s t test
Altogether, these data confirm the important role of Zeb1 in early T cell development. We also demonstrated the essential and specific role of Zeb1 in the development of peripheral T cell subsets expressing NK cell markers, such as iNKT cells, NK1.1+ γδ T cells and Ly49+ CD8 T cells.
Intrinsic role of Zeb1 in thymic progenitors and T cell development
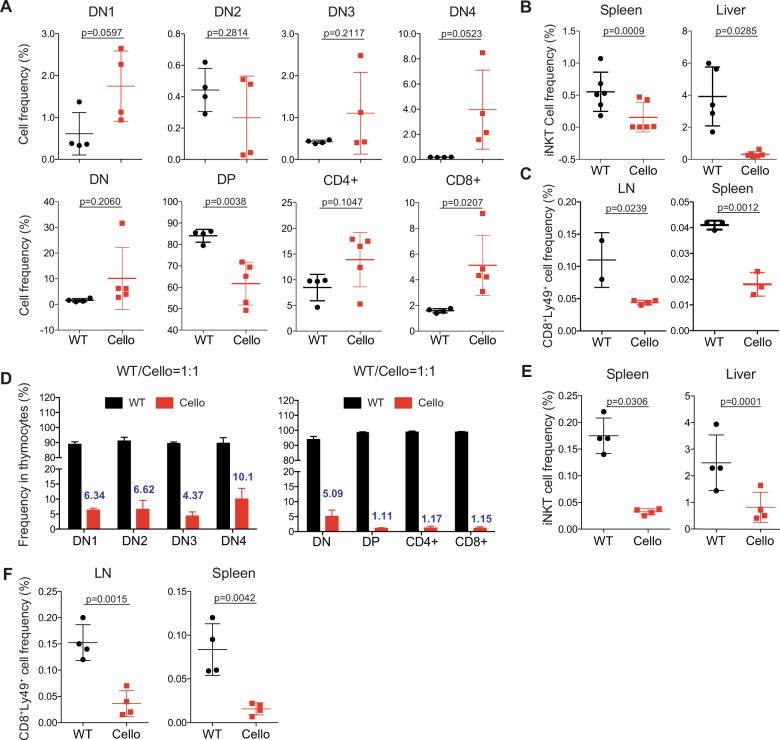
Zeb1 is also required for the development and expression of nonhematopoietic tissues and cell types.20 To test whether Zeb1 played an intrinsic role in T cell development, we generated chimeric mice by transplanting sublethally irradiated Ly5a (CD45.1) mice with BM from Cellophane (CD45.2) or “WT” Ly5a x C57BL/6 (CD45.1/2) mice. In the thymus of chimeric mice, the proportions of DN2 and DP cells were dramatically decreased in Cellophane BM-transplanted mice compared with those in WT BM-transplanted mice, while the proportions of other cell populations defined by CD4 and CD8 were increased (Fig. 4a). The iNKT and Ly49+ T cell subsets were also dramatically decreased in the peripheral organs of Cellophane → Ly5a BM chimeric mice compared with those of WT → Ly5a chimeric mice, indicating that Zeb1 intrinsically regulated T cell development (Fig. 4b, c). The number of cells in all thymic T cell subsets and the numbers of peripheral iNKT cells and Ly49+ T cells were decreased in Cellophane → Ly5a BM chimeric mice compared with those in WT → Ly5a chimera mice (Fig. S2). NK1.1+ γδ T cells were not analyzed because many of them are derived from fetal precursors34 and were not reconstituted in BM chimera mice. To further test the role of Zeb1 in the environment of developing T cells, we also generated different BM chimeric mice using WT and Cellophane mice as both BM recipients and donors (WT → WT, WT → Cellophane, Cellophane → WT, and Cellophane → Cellophane). As shown in Fig. S3A, B, the proportions and numbers of the DN and DP cells subsets were determined by the genotype of the BM donor rather than that of the recipient. Similar conclusions could be reached upon examination of the proportion and number of iNKT cells in the thymus and the liver (Fig. S3C).
Fig. 4.
The role of Zeb1 in T cell development is cell-intrinsic. a Flow cytometry analysis of the indicated thymocyte subsets from BM chimeras. Frequencies are shown for each subset. Recipients were Ly5a mice (CD45.1), and the donor BM was obtained from either Ly5a x C57BL/6 (CD45.1/2) or Cellophane (CD45.2) mice. b iNKT cell and c CD8+ Ly49+ T cell frequencies in the spleen, liver, or LN from the same BM chimera described in a. (d) DN1-4, DP, CD4+, and CD8+ cell reconstitution in the thymus of competitive BM chimeras. Recipients were Ly5a mice (CD45.1), and the donor BM was a 1:1 mix of cells from Ly5a x C57BL/6 (CD45.1/2) and Cellophane (CD45.2) mice. Eight weeks after BM transplantation, the cells were analyzed by flow cytometry. The number above the red bar shows the percentage of the indicated cells originating from Cellophane BM. e, f Frequencies of iNKT cells (e) and Ly49+ cells within CD8+ CD44+ T cells in spleen, liver or LN from chimera BM described in d. Each dot represents an individual mouse. The results show the pooled data from two to three independent experiments for a total of two to six mice per group in each panel. Statistical analyses were performed using an unpaired Student’s t test
We then generated mixed BM chimeras by transplanting lethally irradiated Ly5a mice with a 1:1 mixture of BM from Cellophane and Ly5a x C57BL/6 (WT) mice.
Cellophane T cell progenitors showed poor competitive fitness in BM chimeric mice (Fig. 4d). Indeed, the percentage of cells originating from the Cellophane BM progenitors was already low in the DN stage and further decreased during the transition between the DN and DP stages (Fig. 4d). In the periphery of mixed BM chimeric mice, we found that the proportions of iNKT and Ly49+ CD8 T cells were greatly reduced among Cellophane T cells compared with those among WT lymphocytes (Fig. 4e, f), thus revealing that the role of Zeb1 in T cell development is cell-intrinsic and is not due to a defective stromal environment. Of note, we also analyzed the reconstitution of myeloid cells as a control. In the spleen, on average, 20% of macrophages, 25% of dendritic cells, and 28% of neutrophils originated from Cellophane mice (Fig. S3D), suggesting that Zeb1 regulated the development of all hematopoietic subsets, perhaps by regulating multipotent progenitors. However, the most important effects were observed for thymocytes and peripheral T cell subsets expressing NK cell markers.
Reduced survival and proliferation of Cellophane DN2 and DP cells
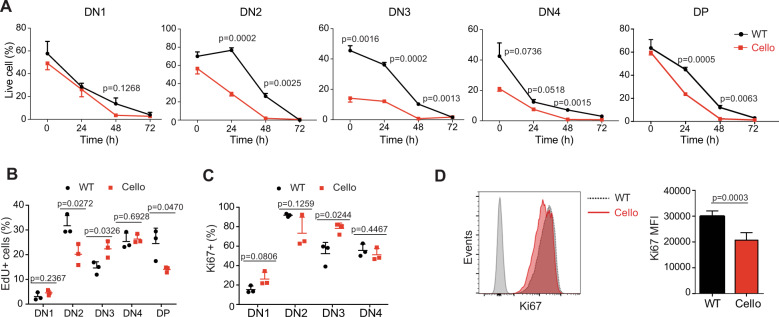
The decreased cellularity of the Cellophane thymi could be due to the reduced proliferation or increased apoptosis of thymocytes. To address this point, we first compared the survival of WT and Cellophane thymocytes during ex vivo culture. We found that Cellophane thymocytes in DN2, DN3, and DN4 showed reduced ex vivo viability compared with their WT counterparts (Fig. 5a). Moreover, after 24 or 48 h in culture, Cellophane DN2, DN3, DN4, and DP cells also showed reduced viability compared with control cells (Fig. 5a).
Fig. 5.
Cellophane DP cells show reduced survival and proliferation. a Percentages of annexin-V-negative cells (live cells) among wild-type and Cellophane thymocytes cultured for 0–72 h. Each graph shows a different subset, as indicated. b EdU incorporation in wild-type and Cellophane thymocytes after a 12-h in vivo pulse of 0.2 mg EdU. c Frequencies of Ki67+ cells in DN1-4 cell subsets from WT and Cellophane mice. d Ki67 nuclear staining in DP thymocytes from WT and Cellophane mice. Bar graphs (right panel) show the mean ± SD fluorescence intensity (MFI). Each dot represents an individual mouse. Data are pooled from three independent experiments with three mice in each group (a–d). Statistical analysis was performed using an unpaired Student’s t test
Next, we compared the in vivo proliferation of WT and Cellophane thymocytes, as measured by EdU incorporation. Cellophane DN2 and DP cells showed less proliferation than their WT counterparts, while Cellophane cells proliferated more than WT DN3 cells (Fig. 5b). Ki67 staining corroborated the data we obtained by using EdU incorporation (Fig. 5c, d). As all DP cells were Ki67 positive, we only reported the changes in the mean fluorescence intensity (MFI) (Fig. 5d).
Thus, the Cellophane mutation affects both the survival and proliferation of developing DN2 and DP thymocytes, which could account for the decreased number of such cells in the Cellophane thymi.
Zeb1 modulates TCR signaling strength
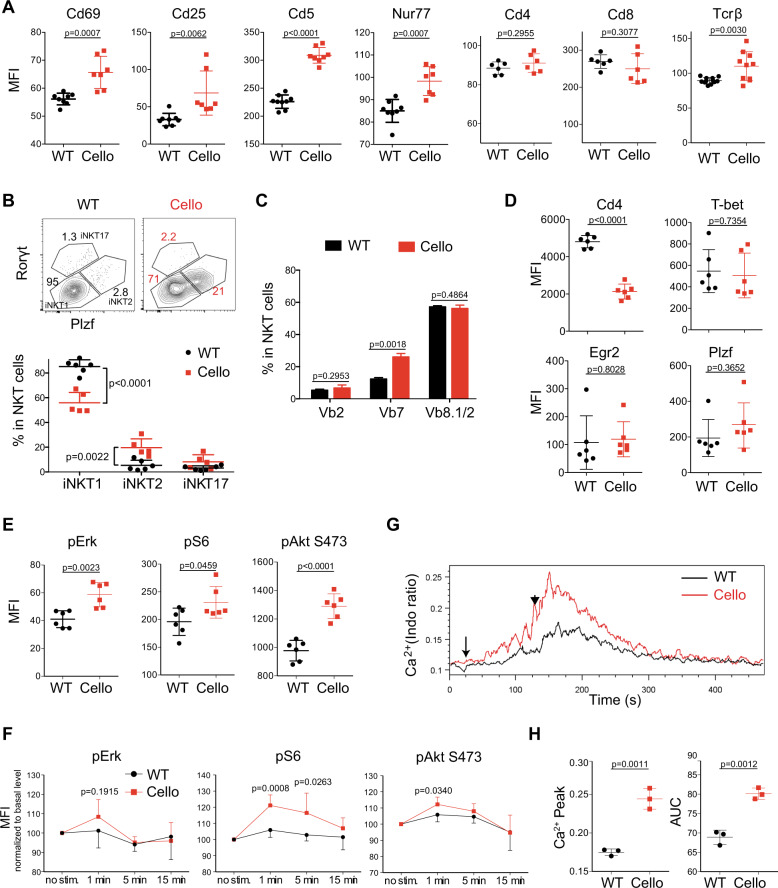
To gain insight into the mechanism of Zeb1 function, we focused on DP cells, as they expressed the highest level of Zeb1 among thymocytes (Fig. 1a). We first compared the expression of membrane proteins involved in thymocyte selection in Cellophane versus WT DP cells by flow cytometry. Cellophane DP cells expressed higher levels of Cd69, Cd25, and Cd5 than WT thymocytes (Fig. 6a). Nur77 is an early response gene expressed in T cells within hours after TCR stimulation. We observed an increase in the intracellular expression of Nur77, which was more highly correlated with Tcrβ levels in Cellophane cells than in WT DP thymocytes (Fig. 6a). Notably, similar levels of Cd4 and Cd8 were detected (Fig. 6a).
Fig. 6.
Cellophane DP thymocytes display increased TCR signaling. a Flow cytometry analysis of Cd69, Cd25, Cd5, Nur77, Cd4, Cd8, and Tcrβ expression (mean fluorescence intensity) in DP thymocytes of wild-type and Cellophane mice. b Left panels: Representative flow cytometry analysis of iNKT1 (Plzf lo, Rorγt-, T-bet+), iNKT2 (Plzf hi, Rorγt-, T-bet-), and iNKT17 (Plzf int, Rorγt+, T-bet-) cell subsets in wild-type and Cellophane mice (as defined by the gating strategy shown in the upper panel). Right panel: graphs of the percentages of iNKT cell subsets within total iNKT cells. c The TCRVβ repertoire of thymic iNKT cells was analyzed by flow cytometry. Bar graphs show the mean ± SD frequencies of individual Vβ chains within iNKT cells. d Expression of Cd4, T-bet, Egr2, and Plzf in iNKT cells as measured by flow cytometry (mean fluorescence intensity). e Phosphorylation levels of Erk, S6, and Akt (S473) in resting DP thymocytes from WT and Cellophane mice. f Phosphorylation levels of Erk, S6, and Akt (Ser473) in DP cells stimulated with anti-CD3 antibody for the indicated times. The results are normalized to the nonstimulated condition (i.e., basal level is 100%) for each group. g Representative histogram overlay showing the Ca2+ flux in thymic DP cells from wild-type or Cellophane mice stimulated with anti-CD3 antibodies, as measured by flow cytometry. Thymocytes were activated following incubation with biotinylated anti-CD3 (arrow) followed by cross-linking with streptavidin (arrowhead). h Dot plots showing the AUC (area under the curve) and Ca2+ peak. Statistical analysis was performed using an unpaired Student’s t test. Each dot represents an individual mouse. Data are pooled from three experiments with a total of eight mice in each group (a), three experiments with six mice (b), three mice (c), two experiments with six mice (d–f) or three experiments with three mice (g, h)
Next, we focused our attention on thymic iNKT cell subsets. The mouse thymi is known to contain at least three iNKT subsets, iNKT1, iNKT2, and iNKT17, which are thought to play distinct roles in the immune response.35 iNKT1 cells comprise mainly stage 3 iNKT cells. TCR signaling strength governs the development of iNKT cell subsets in the thymus, in which high signaling strength is necessary for iNKT2 and iNKT17 development.9,10 We examined iNKT cell subsets by staining for Plzf and Rorγt36 in Cellophane mice and WT mice. The results in Fig. 6b show a significant increase in the proportions of iNKT2 and iNKT17 cells and a decrease in the proportion of iNKT1 cells in Cellophane mice compared with those in control mice. These data indicated an increase in the TCR signaling from DP progenitors of iNKT cells in Cellophane mice. The change in iNKT subsets was associated with subtle changes in the TCR repertoire, as assessed by measuring the frequencies of Vβ8-, Vβ7-, and Vβ2-positive cells among iNKT cells of each genotype. We observed a twofold increase in Vβ7 in Cellophane mice (Fig. 6c). This could reflect the increase in iNKT2 cells, as a previous study showed that Vβ7 was more often associated with iNKT2 cells.9 Thymic Cellophane iNKT cells expressed normal levels of the TFs T-bet and Egr2 but strongly reduced levels of Cd4 (Fig. 6d). Since CD4 is known to sustain TCR signaling strength,5 the selection of CD4 low iNKT cells in Cellophane mice could reflect the adaptation to overt TCR signaling in Cellophane DP cells.
We then specifically analyzed TCR signaling in developing thymocytes. We started by measuring the phosphorylation (p) levels of a series of signaling proteins involved in TCR-mediated activation, either at the steady-state in freshly isolated thymocytes or following TCR engagement by cross-linking with anti-CD3 antibodies. To minimize the experimental variation, we used a barcoding strategy that allowed stimulation and then staining of WT and Cellophane thymocytes simultaneously (see “Materials and methods”). The results in Fig. 6e, f show increases in pAkt (Ser473), ribosomal protein pS6, and, to a lesser extent, pErk either at the steady-state or following TCR engagement in Cellophane DP cells compared with WT DP cells. Thus, the MAPK and PI3K/Akt pathways are more active in thymocytes undergoing selection in Cellophane mice compared with those in control mice. To complement this analysis, we also assessed the calcium response of DP cells of both genotypes in response to TCR engagement. As shown in Fig. 6g, h, this response was stronger for Cellophane DP cells than control DP cells in terms of both intensity (peak) and duration (area under the curve, AUC). Thus, Zeb1 modulates the signaling strength downstream of the engaged TCR at the DP stage, and Cellophane DP cells show increased TCR signaling, which may increase negative selection and therefore account for defective T cell development in Cellophane mice. We also measured pre-TCR signaling in DN cells following stimulation with anti-CD3 antibodies. This analysis did not reveal any difference between WT and Cellophane DN cells (data not shown).
Zeb1 broadly shapes transcription during the DP → SP transition to promote proliferation and repress TCR signaling
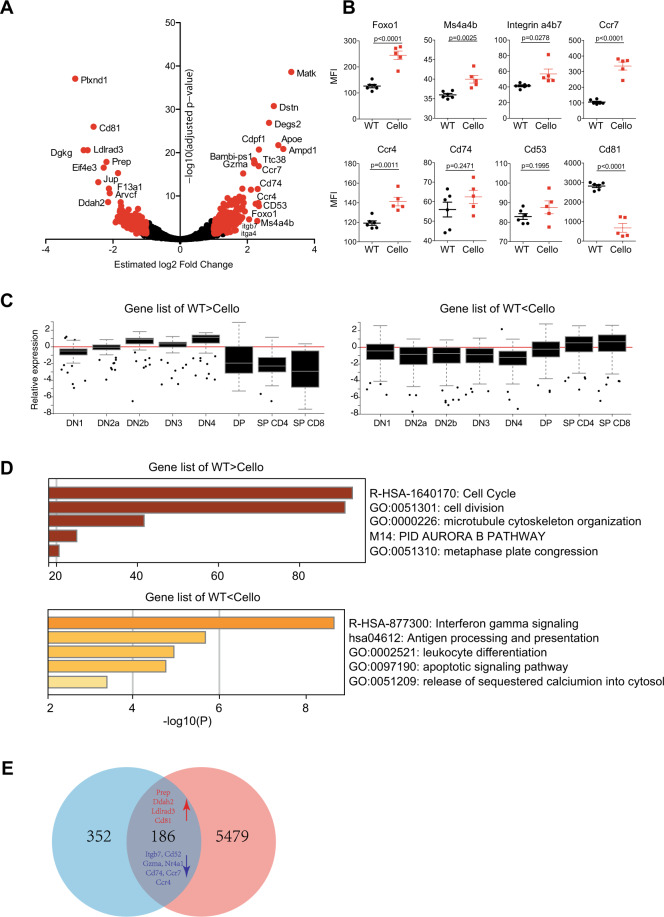
To further uncover the mechanisms of Zeb1 function during T cell development, we performed RNA-seq to compare the WT and Cellophane DP transcriptomes. We found 538 differentially expressed genes (DEGs, p value < 0.05, log2-fold change > 1). A total of 204 genes were increased, and 334 were downregulated in Cellophane DP cells compared with WT DP cells. These data reveal that Zeb1 broadly shapes the genetic program of developing thymocytes (Fig. 7a and Table S1). For some of the DEGs identified, antibodies were available, and we were thus able to confirm the higher expression of Foxo1, Ms4a4b, Itgb7, Ccr7, and Ccr4 in Cellophane cells compared with that in control DP cells; in contrast, we observed lower Cd81 expression in Cellophane DP cells than in WT cells (Fig. 7b).
Fig. 7.
RNA-seq analysis of Zeb1-regulated genes. a Volcano plot representing the −log10 adjusted p value as a function of the estimated log2-fold-change. Significant genes were selected using the following thresholds: an adjusted p value lower than 0.05 and an absolute value of the log2-fold-change greater than 1. For significant genes (plotted in red), a selection of the gene names is displayed. The results are representative of the biological replicates in each group. b Flow cytometry analysis of several genes identified by RNA-seq as either upregulated (Foxo1, Ms4a4b, integrin α4:β7, Ccr7, Ccr4, Cd74, and Cd53) or downregulated (Cd81) in Cellophane mice. Data are pooled from two independent experiments with a total of six mice. c Expression level of genes in two selected lists (200 genes in each list) that were either upregulated (WT < Cello) or downregulated (WT > Cello) in different αβ T cell subsets in Cellophane mice with a fold change > 2 and a p value < 0.05, as indicated. Graphs are adapted from the ImmGen website. d Pathway analysis of DEGs in a using Metascape. The selected terms are shown among the most significant terms. e Venn diagram showing the overlap between DEGs in a and genes located within 100 kb of OCRs containing Zeb1 motifs for which chromatin accessibility changed during T cell development (the corresponding clusters are visualized in Fig. S4B). The genes upregulated in WT mice are shown in red, while the genes that were downregulated are shown in blue
Next, we queried the ImmGen database to retrieve the expression profile of the Zeb1-regulated gene set (induced or repressed) across all thymocyte subsets. Interestingly, genes downregulated in Cellophane DP cells (i.e., normally induced by Zeb1) correspond to genes that are normally expressed at high levels in early T cell progenitors and at low levels in mature T cells (Fig. 7c). Their expression level normally drops during the DP to SP transition, which is when Zeb1 is highly expressed. Genes upregulated in Cellophane DP cells correspond to genes that show a reciprocal pattern of expression (Fig. 7c). This pattern of expression also correlates with cell proliferation and TCR responsiveness in thymocytes. Indeed, irrespective of the mouse genotype, SP T cells are much more responsive to TCR signaling-mediated calcium responses than DP cells but also do not cycle as much as DP cells (data not shown). This suggests that Zeb1 promotes cell proliferation and represses TCR signaling specifically at the DP stage, presumably to ensure proper selection.
A functional annotation analysis of DEGs identified by our RNA-seq analysis using “Metascape”37 highlighted the cell cycle as the most downregulated biological process in Cellophane DP cells compared with control DP cells (Fig. 7d and Table S2), confirming the findings in Fig. 3. Pathways linked to Ifnγ (and also type I-Ifn; Table S2), antigen presentation, leukocyte differentiation, and apoptosis were significantly associated with genes upregulated in Cellophane DP cells compared with those in WT cells (Fig. 7d). Of note, a modest but significant enrichment of genes involved in the calcium response was also associated with these upregulated genes, corroborating the data shown in Fig. 6. To further annotate this dataset, we performed individual PubMed searches to look for connections between genes upregulated in Cellophane DP cells compared with control DP cells and “T cell activation”, “TCR signaling”, or “T cell development”. Interestingly, this analysis showed that more than 25% of the genes in the list played a known role in T cell activation or TCR signaling, and 10% played a role in T cell development, as defined using loss-of-function mouse strains (Table S3). Moreover, we used the STRING database of physical and functional protein interactions38 to further annotate genes that were up- or downregulated in Cellophane DP cells compared with control DP cells. In particular, we used the PubMed module that searches for the enrichment of gene lists in articles in PubMed. This unbiased analysis showed that genes upregulated in Cellophane DP cells were significantly enriched for genes involved in negative selection39 or T cell maturation regulated by Bcl11b40 (Table S4), which corroborated our manual PubMed searches.
Chromatin regions remodeled at the DP stage contain Zeb1 binding motifs
Next, we wanted to determine whether Zeb1 could regulate chromatin remodeling at the DP stage of T cell development. For this, we took advantage of a recently published large-scale analysis of chromatin accessibility and gene expression across 86 immune cell subsets, including subsets representing T cell developmental stages in the thymus.41 In this study, in silico predictions identified Zeb1 as one of the few TFs whose expression was correlated with modifications of chromatin accessibility during thymic T cell development and for which the corresponding chromatin regions contained sites predicted to be bound by Zeb1. Other TFs in this category included Gata3, Tcf7, Lef1, Tcf12, and Zfp740 (Fig. S4A), especially Tcf7 and Tcf12, whose roles in T cell development have been well established.32 We retrieved the open chromatin regions (OCRs) for which Zeb1 motifs were discovered in this study and whose accessibility changed during T cell development (see the corresponding clusters in Fig. S4B) and compared the list of corresponding genes with the DEGs between Cellophane and WT DP cells identified in our own study. We found an important overlap between both lists that included many of the genes previously highlighted in our analysis (Fig. 7e, p value = 1.839413e−46). Altogether, these data suggest that Zeb1 is a direct transcriptional regulator of T cell development that is especially involved in the DP to SP transition that promotes cell proliferation and ensures proper selection.
Discussion
Here, we demonstrated that Zeb1 is essential for the transition through the DN2 and DP stages of T cell development as well as for the differentiation of iNKT cells, NK1.1+ γδ T cells and Ly49+ CD8 T cells. Mechanistically, Zeb1 regulates the expression of a number of genes that are notably involved in cell proliferation or in TCR signaling at the DP stage. In Cellophane mice, these events may perturb thymic development and selection in a way that does not allow the production of the NK1.1+ and Ly49+ T cell subsets.
Zeb1 expression was found to increase at the DN2 stage and to be maximal at the DP stage of T cell development. Accordingly, we found decreases in the frequencies of DN2 and DP thymocytes in Cellophane mice. This could be due to the cell-intrinsic role of Zeb1 in DN2 and DP proliferation. A number of genes involved in the cell cycle were differentially expressed between WT and Cellophane DP cells, as revealed by our RNA-seq analysis. There is also a strong link in the literature between Zeb1 and cell proliferation in cancer. In particular, Zeb1 interacts with many TFs involved in the regulation of cell growth, such as Smad TFs, which are downstream of several growth factor pathways.22 Moreover, Zeb1 is known to repress cyclin-dependent kinases during EMT.21 However, the levels of CDkn2c and CDkn3 were decreased by Zeb1 in DP thymocytes, suggesting the different roles of Zeb1 in epithelial versus lymphoid cells. The decreased proliferation of DN2 and DP cells is expected to have important consequences for overall thymic output. Indeed, we found decreased numbers of peripheral T cells in Cellophane mice. However, this defect was much more pronounced for iNKT cells, NK1.1+ γδ T cells, and Ly49+ CD8 T cells. In particular, iNKT cells were virtually absent from the periphery. The altered development of T cells was associated with increased TCR signaling at the DP stage, which was verified by increased basal levels of Cd5 and Nur77 and increased mTOR activity and calcium flux upon CD3 engagement. iNKT cells are known to receive stronger TCR signals than conventional T cells during their development.6 Thus, increased TCR signaling in Cellophane DP cells could trigger cell death via negative selection of iNKT precursors. The increased negative selection of iNKT cells has already been shown to occur in mice in which transgenic TCR-β chains confer high affinity for self-lipid/CD1d complexes when they are randomly paired with Vα14-Jα18 rearrangements.42 Thus, increased negative selection could impair the development of iNKT cells in Cellophane mice and perhaps that of other T cell subsets expressing NK cell markers. Indeed, strong TCR-mediated signals are also important for γδ T cell development.43,44 In particular, NK1.1+ γδ T cells have an oligoclonal TCR repertoire and accumulate in mouse models of decreased TCR signaling,45 suggesting that this subset of γδ cells can also be negatively selected. The ontogeny of Ly49+ CD8 T cells is not very well known, but our data suggest that their development and selection could share common mechanisms with those of NK1.1+ T cells. How does Zeb1 regulate TCR signaling strength? Cellophane DP thymocytes expressed higher levels of Tcrβ than control DP thymocytes, and this could certainly lead to increased TCR signaling. Moreover, the RNA-seq analysis we performed suggested that there were multiple connections between Zeb1 and signaling transduction through the TCR. For example, multiple members of the GTPases of the IMmunity-Associated Proteins (GIMAP) family (GIMAP5 and GIMAP8) were upregulated in Cellophane DP cells. Interestingly, Gimap5 enhances calcium influx following TCR stimulation.46 Several members of the Ms4a family of receptors, which have four transmembrane domains, are also upregulated in Cellophane DP cells and could reinforce TCR signaling. Indeed, the transduction of signaling by Ms4a4b in naive T cells can heighten their sensitivity to antigens through a process that could involve association with costimulatory molecules.47 Several phosphatases and kinases are also deregulated in Cellophane DP cells and could perhaps alter TCR signaling. In particular, the expression of Pyk2 (encoded by Ptk2b), Rasgrp4, Rasl1, or Rasa3 could all contribute to increased TCR signaling via calcium flux or the MAPK pathway.
A series of TFs were also deregulated in Cellophane DP cells, which showed notable upregulation of JunB, Jun, Atf6, Foxo1, Stat4, or Irf7/9. JunB and Jun are essential components of AP-1 TFs and are typically activated downstream of TCR stimulation. Similar to Nur77, they could represent surrogate markers of increased TCR signaling in Cellophane DP. The derepression of Foxo1 could in part account for altered T cell development, as it regulates Ccr7, CD62L, and S1pr1 via Klf2.48 The lack of control by Klf2 could perturb and perhaps even accelerate the normal migration of developing thymocytes in the medullary region where negative selection occurs. Foxo1 deletion in thymocytes was reported to decrease the number of DP thymocytes, and Foxo1-deficient peripheral T cells seem to be refractory to TCR stimulation through unknown mechanisms.49 Moreover, upregulation of Foxo1 in Cellophane DP cells could in part explain the inverse changes in cell proliferation observed in these cells compared with those in control DP cells, since Foxo TFs are known to promote stem cell quiescence50 and clearly contribute to the regulation of cell division, survival, and metabolism in T cells.51 A recent study showed that the transcriptional repressor Gfi1 is important in maintaining Foxo1 expression at low levels in DP thymocytes.52 In the absence of Gfi1, premature expression of genes normally expressed in mature T cells and accelerated maturation of DP cells into SP thymocytes occurred, which was largely attributable to Foxo1 derepression. Zeb1 and Gfi1 could therefore cooperate to repress Foxo1.
There are many similarities in the phenotypes of Tcf12-deficient53 and Cellophane mutant mice, particularly in terms of the susceptibility of DP cells to cell death and the impaired development of iNKT cells. Moreover, microarray data from the ImmGen consortium suggest that Tcf12 and Zeb1 are strongly coregulated, and ATAC-seq data predict that they control chromatin accessibility during thymic T cell development together with Gata3, Tcf1, Lef1, and Zfp740 (Fig. S4 and ref. 41). Altogether, these data suggest a strong functional link between Zeb1, Heb (encoded by Tcf12), and perhaps Tcf1, which acts in coordination with Heb.32 The fact that Zeb1 is known to bind tandem E-box motifs suggests that there is possibly competition between Zeb members and E proteins for binding of those genes regulated by tandem E-boxes. Such competition has been previously established in the context of the CD4 enhancer, which is repressed by Zeb1, through competition with Heb for E-box binding.54 Moreover, Zfh-1 and Daughterless, the Drosophila homologs of Zeb1 and Tcf12, are also known to compete for the same genomic sites.55 The deletion of Tcf12 and Tcf1 in thymocytes results in the opposite phenotype as the Cellophane Zeb1 mutation in terms of DP proliferation.32 This suggests that Heb and Zeb1 could have partially antagonistic activities in the regulation of genes bearing tandem E-box elements. Competition between Zeb1 and E proteins has already been suggested to play a role in the control of GATA3 expression in human CD4 T cells.56 Cellophane mice express a truncated form of Zeb1 that is expressed at lower levels than WT Zeb1. As the phenotype of these mice is milder than that of Zeb1−/− mice, we assumed that the Cellophane mutation was hypomorphic. However, we cannot rule out that Cellophane Zeb1 may retain some DNA-binding capability and therefore act as a dominant negative molecule by preventing the binding of E-box proteins. Further work will be needed to precisely map the interactions between Zeb TFs and E proteins. The regulatory network may also include inhibitors of the differentiation genes Id2 and Id3, which are TFs that bind and inactivate E proteins, thereby regulating their function. Moreover, a deficiency in Id3 has the same impact on NK1.1+ γδ T cells as a deficiency in TCR signaling,45 which indicates a links between both factors.
The Zeb1 genomic region is frequently deleted in cutaneous T cell lymphomas (CTCLs).57 Such deletions are often associated with genetic mutations in components of the TCR signaling machinery (recurrent alterations in Card11, Plcg1, Lat, Rac2, Prkcq, CD28, and genes that encode calcium channel subunits). This observation, together with our own data showing the role of Zeb1 in repressing TCR signaling, suggests that Zeb1 deletion could promote lymphomagenesis by releasing the normal constraints on TCR signaling. Of note, a previous study proposed the essential role of IL-15 in CTCL development and showed that IL-15 expression was suppressed in patients with CTCL due to promoter hypermethylation and the failure of Zeb1 to gain access to and repress the IL-15 regulatory region.58 However, IL-15 expression was not detected in developing thymocytes in our RNA-seq analysis, excluding the possibility that the IL-15 pathway could play a role in the mechanism of action of Zeb1 in T cell development. However, we found that the transcriptional responses to different cytokines, such as interferons or IL-6, were increased in Cellophane DP cells (Table S2). Zeb1 may therefore normally repress the responses to these cytokines, which presumably occurs to ensure proper selection. TGF-β is a known regulator of iNKT cell development59 that promotes early differentiation and prevents the apoptosis of developing iNKT cells. A recent study showed that Zeb1 expression was induced by TGF-β in conventional CD8 T cells stimulated through the TCR and was essential for memory T cell survival and function.30 Although we failed to detect any effect of recombinant TGF-β on Zeb1 expression in thymocytes (data not shown), it would be interesting to address this question in vivo using appropriate genetic models.
In summary, Zeb1 is an essential member of the TF network that regulates T cell development and selection in the DN2 and DP stages. Furthermore, we have also shown that Zeb1 facilitates the development of iNKT cells and other T cell subsets expressing NK cell markers by regulating the cell cycle and TCR signaling in developing thymocytes.
Materials and methods
Mice
Mice that were 8–24 weeks old were used. Wild-type C57BL/6 mice were purchased from Charles River Laboratories (L’Arbresle). Cellophane mice were previously described,28 and their littermates were used as controls. This study was carried out in accordance with the French recommendations in the Guide for the Ethical Evaluation of Experiments Using Laboratory Animals and the European guidelines 86/609/CEE. All experimental studies were approved by the local bioethics committee CECCAPP. Mice were bred at the Plateau de Biologie Expérimentale de la Souris (ENS, Lyon).
Bone marrow chimeric mice
8- to 10-week-old Ly5a mice or Ly5a x C57BL/6 mice were anesthetized with ketamine/xylazine before irradiation at a dose of 9 Gray with an X-ray irradiator XRAD-320. After irradiation, they were intravenously injected with 2–5 × 106 cells collected from either wild-type or mutant murine bone marrow or a mix of both (as indicated in the figures). Immune cell reconstitution was analyzed 8 weeks post BM injection.
Flow cytometry
Single-cell suspensions of thymus, spleen, and liver were used for flow cytometry. Cell viability was measured using annexin-V (BD Biosciences)/live-dead fixable (eBiosciences) stain. Intracellular staining for TFs was performed using a Foxp3 kit (eBioscience). Lyse/Fix and PermIII buffers (BD Biosciences) were used for intracellular staining of phosphorylated proteins. Flow cytometry was carried out on a FACS Canto, a FACS LSRII, or a FACS Fortessa (Becton-Dickinson). Data were analyzed using FlowJo (Treestar). Antibodies were purchased from eBioscience, BD Biosciences, R&D Systems, Beckman Coulter, Miltenyi, or Biolegend. We used the following antibodies: anti-mouse CD3 (clone 145-2C11), anti-mouse CD4 (clone GK1.5), anti-mouse CD8 (clone 53-6.7), anti-mouse TCRβ (clone H57-597), anti-mouse CD69 (clone H1.2F3), anti-mouse TCRγδ (clone GL3), anti-mouse NK1.1 (clone PK136), anti-mouse CD24 (clone M1/69), anti-mouse CD44 (clone IM7), anti-mouse CD27 (clone LG.7F9), anti-mouse TCRVβ2 (clone B20.6), anti-mouse TCRVβ7 (clone TR310), anti-mouse TCRVβ8.1/8.2 (clone KJ16-133), anti-mouse Ly49A (clone A1), anti-mouse Ly49E/F (clone REA218), anti-mouse Ly49G2 (clone 4D11), anti-mouse CD45.1 (clone A20), anti-mouse CD45.2 (clone 104), anti-mouse Nur77 (clone 12.14), anti-mouse Ccr7 (clone 4B12), anti-mouse CD5 (clone 53-7.3), anti-mouse CD81 (clone Eat-2), anti-mouse CD53 (clone OX79), anti-mouse Lpam-1 (clone DATK-32), anti-mouse Foxo1 (clone C29H4), anti-mouse Ms4a4b (clone 444008), anti-mouse CD74 (clone In1/CD74), anti-mouse T-bet (clone 4B10), anti-mouse Egr2 (clone erongr2), anti-mouse Plzf (clone Mags.21F7), anti-mouse Rorγt (clone AFKJS-9), anti-mouse pErk (clone 20A), anti-mouse pAkt (Ser473) (clone M89-61), and anti-mouse pS6 (clone D57.2.2E). For the staining of iNKT cells, phycoerythrin (PE)-conjugated PBS-57 loaded on mouse CD1d tetramers (mCD1d/PBS-57) was obtained from the Tetramer Core Facility of the National Institute of Health.
Measurement of in vivo cell proliferation and ex vivo survival
Mice were given one intraperitoneal injection of 0.2 mg EdU (BD Bioscience). Twelve hours after EdU injection, the mice were sacrificed, and the organs were harvested. Cells derived from the thymus were stained with the antibodies specific for the cell surface antigens described above. After fixation and permeabilization, cells were stained with FITC anti-EdU antibody and 7-AAD (BD Pharmingen) according to the manufacturer’s instructions. EdU incorporation into different cell populations was measured by flow cytometry.
For the measurement of cell viability, we stained the thymocyte suspensions with 7-AAD and antibodies against annexin-V (BD Biosciences) and other surface markers, such as CD4, CD8, CD69, TCRβ, CD25, and CD44, either ex vivo or 24, 48, or 72 h after in vitro culture in complete medium.
Cell sorting and RNA preparation
Lymphocytes were obtained from the thymus. Immune cell populations, including DN1-4, DP, SP CD4+ and CD8+, and iNKT cells, were stained in combination with antibodies against the cell-specific markers CD4, CD8, CD69, TCRβ, CD25, CD44, and mCD1d/PBS-57 and were subsequently sorted into different subsets using a FACSAria Cell Sorter (Becton-Dickinson, San Jose, USA). The purity of the sorted cell populations was over 98%, as validated by flow cytometry. The sorted cells were lysed using TRIzol reagent (Invitrogen) or RLT buffer from the RNeasy Micro kit (Qiagen), and RNA was extracted according to the manufacturer’s instructions.
Quantitative RT-PCR
We used a high capacity RNA-to-cDNA kit (Applied Biosystems, Carlsbad, USA) or iScript cDNA synthesis kit (Bio-Rad) to generate cDNA for RT-PCR. PCR was carried out with a SYBR Green-based kit (FastStart Universal SYBR Green Master, Roche, Basel, Switzerland) or SensiFast SYBR No-ROX kit (Bioline) on a StepOne plus instrument (Applied Biosystems, Carlsbad, USA) or a LightCycler 480 system (Roche). Primers were designed using software from Roche. We used the following primers for mouse QPCR: Zeb1 forward primer, 5′-GCCAGCAGTCATGATGAAAA-3′; Zeb1 reverse primer, 5′-TATCACAATACGGGCAGGTG-3′; Zeb2 forward primer, 5′-CCAGAGGAAACAAGGATTTCAG-3′; Zeb2 reverse primer, 5′-AGGCCTGACATG
TAGTCTTGTG-3′; Gapdh forward primer, 5′-GCATGGCCTTCCGTGTTC-3′; Gapdh reverse primer, 5′- TGTCATCATACTTGGCAGGTTTCT-3′. The relative expression of Zeb1 and Zeb2 were normalized to Gapdh expression.
Western blotting
Cells were lysed in NP40 lysis buffer (20 mM Tris, HCl pH 7.4; 150 mM NaCl; 2 mM EDTA; 1% NP40) containing protease inhibitors for 30 min on ice. The supernatant was collected following 10 min of centrifugation at 12,000 g at 4 °C, and the protein concentration was quantified by a μBCA quantification kit (Thermo Fisher Scientific). Fifty micrograms of total cellular protein from the thymus was incubated for 5 min at 95 °C. Protein samples were separated by electrophoresis using Novex 4–12% Tris-Glycine gels (Life Technologies) for 1 h at 120 V. The proteins were then transferred to a PVDF membrane (Bio-Rad). After blocking with PBS containing 0.1% Tween and 5% milk for 1 h, the membranes were probed with the following primary antibodies: anti-Gapdh (Cell Signaling Technology, 2118) and anti-Zeb1 (Cell Signaling Technology, 3396; the antibody was raised against a peptide with Asp846, the Cellophane mutation truncating the protein after Tyr902) overnight at 4 °C. Membranes were washed three times with PBS containing 0.1% Tween, and secondary antibodies were added for incubation for one hour at RT. Anti-rabbit and anti-mouse HRP conjugate secondary antibodies were provided by Jackson ImmunoResearch. Proteins were revealed with a Chemiluminescence Western Lightening Plus kit (Perkin-Elmer).
RNA-seq analysis
Thymic suspensions were stained in combination with anti-CD3, anti-CD4, anti-CD8, anti-CD69, and anti-TCRβ and subsequently sorted into different subsets using a FACSAria Cell Sorter (Becton-Dickinson, San Jose, USA). The purity of the sorted cell populations was over 98%, as measured by flow cytometry. RNA libraries were prepared as previously described.60 Briefly, total RNA was purified from 5 × 104 sorted thymocytes using the Direct-Zol RNA microprep kit (Zymo Research) according to the manufacturer’s instructions and was quantified using the QuantiFluor RNA system (Promega). One microliter of 10 µM oligo-dT primer and 1 µl of 10 mM dNTP mix were added to 0.15 ng of total RNA in a final volume of 2.3 µl. Oligo-dT cells were hybridized for 3 min at 72 °C, and reverse transcription (11 cycles) was performed. PCR preamplification was then conducted using 16 cycles. The cDNA was purified with Ampure XP beads (Beckman Coulter), and the cDNA quality was checked with D5000 screen tape and analyzed on a Tape Station 4200 (Agilent). Three nanograms of cDNA was tagmented using a NextEra XT DNA sample preparation kit (Illumina). The tagged fragments were further amplified and purified with Ampure XP beads (Beckman Coulter). The tagged library quality was checked with D1000 screen tape and analyzed on a Tape Station 4200 (Agilent). Sequencing was performed with the GenomEast platform by a member of the “France Génomique” consortium (ANR-10-INBS- 0009) on an Illumina HiSeq 4000 sequencing instrument (read length of 1×50 nt).
Measurements of TCR signaling
Calcium response
WT and Cellophane thymocytes were first barcoded with anti-CD45 coupled with different fluorochromes and then stained at RT with fluorescent anti-CD4, anti-CD8, anti-CD69, anti-TCRβ, anti-CD25, and anti-CD44 antibodies, followed by staining with Indo-1 (1 µM, Life Technologies) at a concentration of 1 × 107 cells/ml for 30 min at 37 °C. Following two washes at 4 °C, the cells were resuspended in RPMI medium (0.2% BSA and 25 mM HEPES) and were incubated at 37 °C for 5–10 min prior to acquisition. The samples were acquired on an LSRII (BD) as follows: 15 s of baseline acquisition, addition of anti-CD3 biotin (2C11, 10 µg/ml), acquisition for 1 min 30 s, addition of streptavidin (Life Technologies, 10 µg/ml), and acquisition for another 3–5 min.
Phosphorylation events
Different samples corresponding to different mice were barcoded by labeling them with a series of anti-CD45 antibodies coupled with different fluorochromes. For phospho-flow staining, 3 × 106 mixed thymocytes were stained using biotinylated CD3 (2C11, 5 µg/ml) and other surface markers for 15 min, followed by streptavidin (Life Technologies, 10 µg/ml) stimulation and fixation by the addition of 10 volumes of Lyse/Fix at the indicated time points. The levels of pErk, pS6, or pAkt were normalized according to the MFI, which was detected in the nonstimulated condition (regarded as 100%) for each mouse.
In silico analyses
The functional annotations of DEGs were performed using Metascape37 or STRING38 using the default parameters. In addition, we used several functionalities of the ImmGen database browser29 to generate some of the figures included in the supplementary information.
Statistical analysis
Statistical analyses were performed using Prism 5 (GraphPad Software). Two-tailed unpaired t-tests, paired t-tests, and ANOVA with Bonferroni correction were used as indicated. We used the hypergeometric test and the Benjamini–Hochberg p value correction algorithm to calculate if the enrichment of the overlap between the gene lists was statistically significant.
Supplementary information
Acknowledgements
The authors acknowledge the contribution of the SFR Biosciences facilities (UMS3444/CNRS, ENSL, UCBL, and US8/INSERM), particularly the Plateau de Biologie Expérimentale de la Souris and the flow cytometry facility. We thank Bruce Beutler for sharing the Cellophane mutant mice. We also thank Andrew Griffiths and Kiyoto Kurima for discussions regarding Twirler mutant mice and Fotini Gounari and Christophe Benoist for providing RNA-seq/ChIP-seq data on T cell development. The TW lab is supported by the Agence Nationale de la Recherche (ANR GAMBLER to TW and ANR JC BaNK to AM) and the Institut National du Cancer and receives institutional grants from the Institut National de la Santé et de la Recherche Médicale (INSERM), Centre National de la Recherche Scientifique (CNRS), Université Claude Bernard Lyon and ENS de Lyon, and the Joint Research Institute for Science and Society (JORISS). JZ is the recipient of a fellowship from the China Scholarship Council (CSC). RS and YGH were funded by an FRM grant (AJE20161236686) to YGH.
Author contributions
JZ, AB, MW, DL, DEC, ALM, AR, and AM performed the experiments. RS and QM performed the in silico analyses. JC, LG, and YGH provided reagents and conceptual insight and helped write the paper. TW wrote the paper and supervised the work.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version of this article (10.1038/s41423-020-0459-y) contains supplementary material.
References
- 1.Shah DK, Zúñiga-Pflücker JC. An overview of the intrathymic intricacies of T cell development. J. Immunol. 2014;192:4017–4023. doi: 10.4049/jimmunol.1302259. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurd N, Robey EA. T-cell selection in the thymus: a spatial and temporal perspective. Immunol. Rev. 2016;271:114–126. doi: 10.1111/imr.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogquist KA, Jameson SC. The self-obsession of T cells: how TCR signaling thresholds affect fate “decisions” and effector function. Nat. Immunol. 2014;15:815–823. doi: 10.1038/ni.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gascoigne NRJ, Rybakin V, Acuto O, Brzostek J. TCR signal strength and T cell development. Annu. Rev. Cell Dev. Biol. 2016;32:327–348. doi: 10.1146/annurev-cellbio-111315-125324. [DOI] [PubMed] [Google Scholar]
- 6.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat. Immunol. 2015;16:1114–1123. doi: 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- 8.Kronenberg M, Kinjo Y. Innate-like recognition of microbes by invariant natural killer T cells. Curr. Opin. Immunol. 2009;21:6. doi: 10.1016/j.coi.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuttle KD, et al. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat. Commun. 2018;9:2650. doi: 10.1038/s41467-018-05026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao M, et al. Altered thymic differentiation and modulation of arthritis by invariant NKT cells expressing mutant ZAP70. Nat. Commun. 2018;9:2627. doi: 10.1038/s41467-018-05095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malhotra N, et al. SOX4 controls invariant NKT cell differentiation by tuning TCR signaling. J. Exp. Med. 2018;215:2887–2900. doi: 10.1084/jem.20172021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziętara N, et al. Critical role for miR-181a/b-1 in agonist selection of invariant natural killer T cells. Proc. Natl Acad. Sci. USA. 2013;110:7407–7412. doi: 10.1073/pnas.1221984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henao-Mejia J, et al. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity. 2013;38:984–997. doi: 10.1016/j.immuni.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wencker M, et al. Innate-like T cells straddle innate and adaptive immunity by altering antigen-receptor responsiveness. Nat. Immunol. 2014;15:80–87. doi: 10.1038/ni.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo W, Taniuchi I. Transcriptional regulation of early T-cell development in the thymus. Eur. J. Immunol. 2016;46:531–538. doi: 10.1002/eji.201545821. [DOI] [PubMed] [Google Scholar]
- 16.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev. Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 17.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 18.Hosokawa H, Rothenberg EV. Cytokines, transcription factors, and the initiation of T-cell development. Cold Spring Harb. Perspect. Biol. 2018;10:a028621. doi: 10.1101/cshperspect.a028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gheldof A, Hulpiau P, van Roy F, De Craene B, Berx G. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell Mol. Life Sci. CMLS. 2012;69:2527–2541. doi: 10.1007/s00018-012-0935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takagi T, Moribe H, Kondoh H. Higashi Y. DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Dev. Camb. Engl. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 21.Caramel J, Ligier M, Puisieux A. Pleiotropic roles for ZEB1 in cancer. Cancer Res. 2018;78:30–35. doi: 10.1158/0008-5472.CAN-17-2476. [DOI] [PubMed] [Google Scholar]
- 22.Conidi A, et al. Few Smad proteins and many Smad-interacting proteins yield multiple functions and action modes in TGFβ/BMP signaling in vivo. Cytokine Growth Factor Rev. 2011;22:287–300. doi: 10.1016/j.cytogfr.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Scott CL, Omilusik KD. ZEBs: novel players in immune cell development and function. Trends Immunol. 2019;40:431–446. doi: 10.1016/j.it.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 24.van Helden MJ, et al. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J. Exp. Med. 2015;212:2015–2025. doi: 10.1084/jem.20150809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez CX, et al. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 2015;212:2041–2056. doi: 10.1084/jem.20150186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Omilusik KD, et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 2015;212:2027–2039. doi: 10.1084/jem.20150194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higashi Y, et al. Impairment of T cell development in deltaEF1 mutant mice. J. Exp. Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnold CN, et al. A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity. Proc. Natl Acad. Sci. 2012;109:12286–12293. doi: 10.1073/pnas.1209134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heng TSP, Painter MW, Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 30.Guan T, et al. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8+ T cell fates. J. Exp. Med. 2018;215:1153–1168. doi: 10.1084/jem.20171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones ME, Zhuang Y. Stage-specific functions of E-proteins at the β-selection and T-cell receptor checkpoints during thymocyte development. Immunol. Res. 2011;49:202–215. doi: 10.1007/s12026-010-8182-x. [DOI] [PubMed] [Google Scholar]
- 32.Emmanuel AO, et al. TCF-1 and HEB cooperate to establish the epigenetic and transcription profiles of CD4+CD8+ thymocytes. Nat. Immunol. 2018;19:1366–1378. doi: 10.1038/s41590-018-0254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahim MMA, et al. Ly49 receptors: innate and adaptive immune paradigms. Front. Immunol. 2014;5:145. doi: 10.3389/fimmu.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grigoriadou K, Boucontet L, Pereira P. Most IL-4-producing gamma delta thymocytes of adult mice originate from fetal precursors. J. Immunol. 2003;171:2413–2420. doi: 10.4049/jimmunol.171.5.2413. [DOI] [PubMed] [Google Scholar]
- 35.Gapin L. iNKT cell autoreactivity: what is “self” and how is it recognized? Nat. Rev. Immunol. 2010;10:272–277. doi: 10.1038/nri2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuttle KD, Gapin L. Characterization of thymic development of natural killer T cell subsets by multiparameter flow cytometry. Methods Mol. Biol. Clifton NJ. 2018;1799:121–133. doi: 10.1007/978-1-4939-7896-0_11. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liston A, et al. Impairment of organ-specific T cell negative selection by diabetes susceptibility genes: genomic analysis by mRNA profiling. Genome Biol. 2007;8:R12. doi: 10.1186/gb-2007-8-1-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kastner P, et al. Bcl11b represses a mature T-cell gene expression program in immature CD4(+)CD8(+) thymocytes. Eur. J. Immunol. 2010;40:2143–2154. doi: 10.1002/eji.200940258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida H, et al. The cis-regulatory atlas of the mouse immune system. Cell. 2019;176:897–912.e20. doi: 10.1016/j.cell.2018.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bedel R, et al. Effective functional maturation of invariant natural killer T cells is constrained by negative selection and T-cell antigen receptor affinity. Proc. Natl Acad. Sci. 2014;111:E119–E128. doi: 10.1073/pnas.1320777110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayes SM, Love PE. Strength of signal: a fundamental mechanism for cell fate specification. Immunol. Rev. 2006;209:170–175. doi: 10.1111/j.0105-2896.2006.00356.x. [DOI] [PubMed] [Google Scholar]
- 44.Haks MC, et al. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 45.Alonzo ES, et al. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J. Immunol. 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ilangumaran S, et al. Loss of GIMAP5 (GTPase of immunity-associated nucleotide binding protein 5) impairs calcium signaling in rat T lymphocytes. Mol. Immunol. 2009;46:1256–1259. doi: 10.1016/j.molimm.2008.09.031. [DOI] [PubMed] [Google Scholar]
- 47.Howie D, et al. MS4A4B is a GITR-associated membrane adapter, expressed by regulatory T cells, which modulates T cell activation. J. Immunol. 2009;183:4197–4204. doi: 10.4049/jimmunol.0901070. [DOI] [PubMed] [Google Scholar]
- 48.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 49.Gubbels Bupp MR, et al. T cells require Foxo1 to populate the peripheral lymphoid organs. Eur. J. Immunol. 2009;39:2991–2999. doi: 10.1002/eji.200939427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, Bhatia R. Molecular pathways: stem cell quiescence. Clin. Cancer Res J. Am. Assoc. Cancer Res. 2011;17:4936–4941. doi: 10.1158/1078-0432.CCR-10-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hedrick SM, Michelini RH, Doedens AL, Goldrath AW, Stone EL. FOXO transcription factors throughout T cell biology. Nat. Publ. Group. 2012;12:649–662.. doi: 10.1038/nri3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi LZ, et al. Gfi1-Foxo1 axis controls the fidelity of effector gene expression and developmental maturation of thymocytes. Proc. Natl Acad. Sci. USA. 2017;114:E67–E74.. doi: 10.1073/pnas.1617669114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D’Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat. Immunol. 2010;11:240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brabletz T, et al. Negative regulation of CD4 expression in T cells by the transcriptional repressor ZEB. Int. Immunol. 1999;11:1701–1708. doi: 10.1093/intimm/11.10.1701. [DOI] [PubMed] [Google Scholar]
- 55.Postigo AA, Ward E, Skeath JB, Dean DC. zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol. Cell Biol. 1999;19:7255–7263. doi: 10.1128/mcb.19.10.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grégoire JM, Roméo PH. T-cell expression of the human GATA-3 gene is regulated by a non-lineage-specific silencer. J. Biol. Chem. 1999;274:6567–6578. doi: 10.1074/jbc.274.10.6567. [DOI] [PubMed] [Google Scholar]
- 57.Wang L, et al. Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat. Genet. 2015;47:1426–1434. doi: 10.1038/ng.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mishra A, et al. Mechanism, consequences, and therapeutic targeting of abnormal IL15 signaling in cutaneous T-cell lymphoma. Cancer Discov. 2016;6:986–1005. doi: 10.1158/2159-8290.CD-15-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doisne J-M, et al. iNKT cell development is orchestrated by different branches of TGF- signaling. J. Exp. Med. 2009;206:1365–1378. doi: 10.1084/jem.20090127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.