Abstract
Locomotor problems are among one of the main concerns in the current poultry industry, causing major economic losses and affecting animal welfare. The most common bone anomalies in the femur are dyschondroplasia, femoral head separation (FHS), and bacterial chondronecrosis with osteomyelitis (BCO), also known as femoral head necrosis (FHN). The present study aimed to identify differentially expressed (DE) genes in the articular cartilage (AC) of normal and FHS-affected broilers by RNA-Seq analysis. In the transcriptome analysis, 12,169 genes were expressed in the femur AC. Of those, 107 genes were DE (FDR < 0.05) between normal and affected chickens, of which 9 were downregulated and 98 were upregulated in the affected broilers. In the gene-set enrichment analysis using the DE genes, 79 biological processes (BP) were identified and were grouped into 12 superclusters. The main BP found were involved in the response to biotic stimulus, gas transport, cellular activation, carbohydrate-derived catabolism, multi-organism regulation, immune system, muscle contraction, multi-organism process, cytolysis, leukocytes and cell adhesion. In this study, the first transcriptome analysis of the broilers femur articular cartilage was performed, and a set of candidate genes (AvBD1, AvBD2, ANK1, EPX, ADA, RHAG) that could trigger changes in the broiler´s femoral growth plate was identified. Moreover, these results could be helpful to better understand FHN in chickens and possibly in humans.
Subject terms: Animal breeding, Functional genomics
Introduction
In the last decades, an intense selection has been performed for greater feed efficiency and faster growth of broiler chickens1. However, a significant increase in locomotor problems has been detected, causing a negative impact on welfare, feed efficiency, performance and other characteristics2,3, and consequently, economic losses2. These losses are due to increase in mortality, reduction in feed conversion and weight gain caused directly or indirectly by skeletal problems4. Currently, bone disorders are still considered one of the main concerns for the poultry industry5. Among the locomotor problems, bacterial chondronecrosis with osteomyelitis (BCO) is the most common cause of claudication, affecting approximately 1.5% of chickens slaughtered at 42 days of age in the United States, as well as an important cause of mortality in broilers6. This condition, also known as femoral head necrosis (FHN)7, is one of the most important disturbances in the locomotor system in commercial chickens worldwide8. Besides its importance in poultry production, there are few studies on this pathology, especially related to its genetics and molecular mechanisms5,7,9–12.
The separation of the articular cartilage (AC) from the growth plate (GP), known as proximal femoral head separation (FHS) is a risk factor for infection that may cause BCO in broilers13,14. This can occur because the BCO pathogenesis seems to be initiated by damage of the poorly mineralized chondrocyte (cartilage cells) columns in the epiphyseal and physeal growth plates of the leg bones, followed by colonization of the osteochondral clefts by opportunistic bacteria6,7.
Genetics play a considerable role in the skeleton development, where genetic selection and gene mutations influence the development of the skeletal system2. DNA mutations in different genes are involved with different skeletal clinical phenotypes15. There is a controversy in the literature regarding BCO and FHN terminology. Some authors consider BCO and FHN as the same pathology16,17, while others consider them different pathologies9,18,19. Usually, when there is separation of the AC and necrosis, this condition is called FHN, while when there is evidence of bacterial infection, it is referred to as BCO17. Previous studies evaluating the chicken femoral growth plate found that the genes RUNX2, SPARC11, ADIPOQ, PRRX1, ANGPTL5, ANGPTL7, GFRA2, SFRP5, COL14A1, ABI3BP, COL8A1, SLC30A1020, bFGF5, LEPR21, LRP1B, COL28A1, PTHrP, PERP1, FAM180A and CHST112 were associated with the FHN. According to the Chicken Quantitative Trait Loci (QTL) Database (Chicken QTLdb) (https://www.animalgenome.org/cgi-bin/QTLdb/GG/summary), some QTL for morphometric traits, mineral composition, and tibia and femur resistance were mapped to several chicken chromosomes, indicating important regions related to bone development22.
Recently, differences in the cartilage morphology and metabolism were observed when normal and FHN-affected broilers were compared (Liu et al. 2021). However, there are no studies investigating the molecular and genetic pathways involved with FHS in broilers articular cartilage, and its etiology is still unknown in chickens22,23. The functional analysis of genes is important to elucidate their contribution to the development of locomotor problems in chickens. Therefore, this study aimed to identify differentially expressed (DE) genes in the femoral head articular cartilage between healthy and FHS-affected broilers using RNA-Seq analysis.
Results
RNA sequencing and differential expression analysis
The sequencing of the femoral head articular cartilage samples generated around 190 million (2 × 100 bp) reads. An average of 26.82 million paired-end reads was obtained per sample, remaining about 23.7 million after the QC. Approximately, 94.6% of the reads were mapped against the chicken reference genome (GRCg6a) available at Ensembl 95. The percentage of reads mapped per sample ranged from 93.45 to 95.2% and were similar between the normal and FHS-affected group (Supplementary File 1: Table S1). The MDS plot showed the separation between the affected and control samples evaluated in the current study (Supplementary File 2: Fig S1).
A total of 12,169 genes were expressed in the femoral articular cartilage, of which 107 genes were differentially expressed (FDR < 0.05) between the two groups (Supplementary File 1: Table S2). From those, 91 were annotated and 16 were uncharacterized (Table 1). Out of the 107 DE genes, 98 (91.6%) were upregulated and 9 (8.4%) were downregulated in the FHS-affected compared with the healthy control group (Supplementary File 1: Table S2). Hierarchical clustering analysis of the 107 DE genes showed different expression patterns between FHS-affected and control sample groups and homogeneity among samples from each group (Supplementary File 2: Fig S2).
Table 1.
Characterization of the articular cartilage transcriptome showing the total number of expressed genes and the differentially expressed ones, according to Ensembl 95 gene type information.
| Gene type | Expressed genes | DE |
|---|---|---|
| lncRNA | 409 | 5 |
| miRNA | 18 | 0 |
| IG_V_gene | 3 | 0 |
| Pseudogenes | 154 | 2 |
| snRNA | 10 | 0 |
| rRNA | 2 | 1 |
| Misc_RNA | 1 | 0 |
| Mt_rRNA | 2 | 0 |
| Protein-coding genes | 10,496 | 83 |
| Uncharacterized-protein genes | 1,074 | 16 |
| Total | 12,169 | 107 |
Considering the top 9 DE genes, those related to collagen (COL13A1), myosin (MYH15), phosphatases (PSPH) and transferases (GGT1) were downregulated, while genes involved with response to microorganisms and immune system were upregulated in the FHS-affected group (Table 2).
Table 2.
Top 9 downregulated and upregulated differentially expressed genes in the articular cartilage between normal and FHS-affected broilers.
| Ensembl Gene ID | Gene symbol | Gene description | Log2FC |
|---|---|---|---|
| ENSGALG00000039489 | RF00002 | 5.8S ribosomal RNA | − 2.23 |
| ENSGALG00000010490 | DPYSL4 | dihydropyrimidinase like 4 | − 2.00 |
| ENSGALG00000015358 | MYH15 | Gallus gallus myosin, heavy chain 15 | − 1.92 |
| ENSGALG00000006565 | GGT1 | gamma-glutamyltransferase 1 | − 1.50 |
| ENSGALG00000038225 | SEMA3E | semaphorin 3E | − 1.44 |
| ENSGALG00000002397 | PSPH | phosphoserine phosphatase | − 1.42 |
| ENSGALG00000004286 | COL13A1 | collagen type XIII alpha 1 chain | − 1.29 |
| ENSGALG00000043671 | − 1.23 | ||
| ENSGALG00000014686 | FBN2 | fibrillin 2 | − 1.08 |
| ENSGALG00000016669 | AvBD2 | Gallus gallus avian beta-defensin 2 | 3.66 |
| ENSGALG00000019696 | CATHL2 | Gallus gallus cathelicidin antimicrobial peptide | 3.69 |
| ENSGALG00000028273 | HBE1 | Gallus gallus hemoglobin subunit epsilon 1 | 3.73 |
| ENSGALG00000024272 | S100A9 | Gallus gallus S100 calcium binding protein A9 | 4.04 |
| ENSGALG00000006572 | TNNT3 | troponin T3 | 4.04 |
| ENSGALG00000023953 | C4BPA | Gallus gallus complement component 4 binding protein | 4.05 |
| ENSGALG00000002907 | MYL1 | myosin, light chain 1 | 4.17 |
| ENSGALG00000043254 | EPX | eosinophil peroxidase | 4.26 |
| ENSGALG00000014463 | ACTN2 | Gallus gallus actinin alpha 2 | 4.33 |
qPCR validation
In the qPCR analysis, six out of the 11 analyzed genes were DE between the FHS-affected and the normal group (Table 3). Moreover, the same expression profile between both approaches was observed for all evaluated genes, except for FBN2 (Table 3), confirming the RNA-Seq results obtained in the current study.
Table 3.
Relative expression (Log2FC) and statistical significance between normal and FHS-affected broilers obtained with qPCR and comparison with RNA-Seq results for 11 candidate genes.
| Gene | qPCR | p-value | RNA-Seq | FDR |
|---|---|---|---|---|
| AvBD1 | 3.21 | 0.005 | 3.55 | 1.23E-07 |
| AvBD2 | 3.65 | 0.006 | 3.32 | 0.0006 |
| FBN2 | 0.52 | 0.227 | − 1.08 | 0.037 |
| ANK1 | 2.45 | 0.014 | 2.12 | 0.023 |
| RHAG | 3.04 | 0.006 | 2.44 | 0.010 |
| CSF3R | 0.86 | 0.356 | 2.28 | 0.003 |
| EPX | 3.48 | 0.007 | 4.27 | 3.23E-09 |
| ADA | 2.14 | 0.003 | 1.72 | 0.008 |
| COL13A1 | − 0.22 | 0.562 | − 1.30 | 0.038 |
| S100A9 | 2.51 | 0.093 | 4.04 | 1.84E-06 |
| PSPH | − 1.13 | 0.218 | − 1.43 | 0.012 |
Gene ontology and gene network analyses
In the gene ontology (GO) analysis, DE genes with defined biological functions were grouped into 79 functional clusters according to their most relevant biological processes (BP) identified through the GO enrichment analysis (Supplementary File 1: Table S3). These BP were summarized in 12 superclusters (Fig. 1). The main BP identified were related to immune system processes, response to biotic stimulus, striated muscle tissue development, carbohydrate derivative catabolism, gas transport and cell activation.
Figure 1.
Superclusters of biological processes enriched for up- and downregulated genes in the articular cartilage related to FHS using REVIGO104. Different colors show different superclusters, and the size of each box is determined by the uniqueness of the categories.
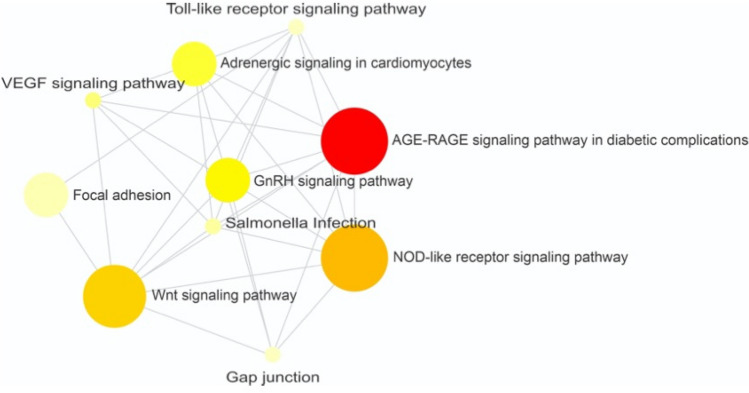
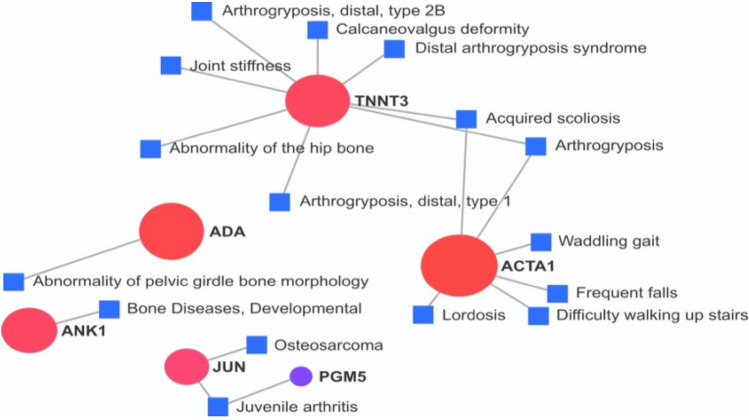
To verify the known interactions among the DE genes aiming to improve the knowledge about them, an analysis at the NetworkAnalyst platform was performed with the chicken database. An enriched network was generated, where genes were linked to pathways, such as Wnt signaling pathway (PLCB2, CCND3, JUN, RAC2), GnRH signaling pathway (PLCB2, MAPK12, JUN), adrenergic signaling in cardiomyocytes (PLCB2, MYH15, MAPK12), VEGF signaling pathway (MAPK12, RAC2), bacterial infection (MAPK12, JUN), focal adhesion (CCND3, JUN, RAC2), gap junction (TUBB1, PLCB2) and toll-like receptor signaling pathway (MAPK12, JUN) (Fig. 2, Supplementary File 1: Table S4). Furthermore, some DE genes were associated with bone-related disease problems when compared with a human database (Fig. 3). The JUN and PGM5 genes were related to osteosarcoma, ACTA1 with waddling gait and difficulty to walk, ADA with abnormality of pelvic girdle bone morphology, TNNT3 with joint stiffness, arthrogryposis distal type 1, ulnar deviation of the wrist, ulnar deviation of the fingers, abnormality of the hip bone and metatarsus varus, and ANK1 with bone development (Fig. 3).
Figure 2.
Gene network of differentially expressed genes-enrichment analysis using KEGG with chicken information. Circles represent metabolic pathways and connecting lines represent interactions between them, according to the active NetworkAnalyst prediction method. Nodes are colored according to their over-representation analysis (ORA) p-value. Darker colors are the smallest p-values. The node size is related to the number of genes enriched in each pathway.
Figure 3.
Gene network of differentially expressed genes constructed with the human gene-disease associations database (DisGeNET). In the figure, the genes previously associated with locomotor diseases in humans are shown. Circles represent the DE genes, the squares represent the diseases and connecting lines represent the association between the genes and bone/locomotor diseases, according to the active prediction method of NetworkAnalyst. The size of the circles was related to all conditions associated to the gene, however, for better visualization, we have shown only those related to bone/locomotor diseases.
Discussion
Studies related to bone integrity problems, such as FHS and FHN or BCO are scarce in chicken models7,11,12,24,25. Most of the bone tissue studies reported to date are in humans and rodents22,26–28. Furthermore, there are no studies evaluating the transcriptional profile of femoral articular cartilage from chickens and its impacts in bone-related problems. Therefore, in the present study, a global gene expression profile of the femoral head articular cartilage of normal and FHS-affected broilers at 35 days of age was reported for the first time.
In the cartilage transcriptome, approximately 91.5% of the DE genes were upregulated while 8.5% were downregulated in the FHS-affected compared to the normal group (Supplementary File 1: Table S2). The expression profile of the AC tissue differed from the femoral growth plate reported in a previous study, in which approximately 49% of the genes were upregulated and 51% downregulated in the FHS-affected group12. In the femoral growth plate, several biological processes identified were related to angiogenesis, blood coagulation, cell adhesion, bone development and lipid metabolism, with several genes downregulated in the FHS-affected group12. Here, in the AC tissue, 79 BP were found (Supplementary File 1: Table S3) and, after REVIGO clusterization, the genes that draw the most attention due to their function were those related to response to biotic stimulus, immune response, cytolysis, striated muscle development, carbohydrate derivative catabolism and cell activation processes. Several BP were similar between the growth plate12 and the AC. However, in the AC transcriptome, a higher proportion of the genes had an increased level of expression in the affected animals than in the GP transcriptome. In our study, 107 genes were identified as DE in the AC and 28 of those were also found by Peixoto et al.12 in the bone GP. Moreover, some genes previously described as potentially involved with FHS, such as interferon alfa-6 inducible protein (IFI6), adenosine deaminase (ADA), cathelicidin-3 (CATH3), avian beta defensin 1 (AvBD1), avian beta defensin 2 (AvBD2), ankirine 1 (ANK 1), leukocyte cell-derived chemotaxin 2 (LECT2) and collagen type XII alpha chain 1 (COL13A1)5,11,12,14,20 were also DE in the current study. Among the DE genes, some of them (AvBD1, AvBD2, ANK1, RHAG, ADA and EPX) were confirmed by qPCR, which demonstrates the reliability of the results found with the RNA-Seq approach (Table 3).
Here, only 9 DE genes were downregulated in the cartilage of broilers with FHS, in which the COL13A1 can be highlighted. This gene shows a wide tissue distribution and occurs at the cellular junctions and cell–matrix interaction sites in epithelial, mesenchymal, and neural tissues. It is a component of focal adhesion in cultured fibroblasts29 interacting with the collagen-binding integrin α1β1, suggesting its involvement in multiple cell–matrix interactions30,31. Therefore, the downregulation of COL13A1 and possibly other collagen genes could reduce the synthesis of the extracellular matrix, facilitating the separation of the cartilage and bone tissues, favoring the occurrence of proximal FHS. Considering all downregulated DE genes, the BP involved were mainly related to muscle development processes (muscle cell differentiation, contraction, development, striated muscle cell development and tissue development) (Supplementary File 1: Table S3) and could be associated to the cytoskeleton, since the lower expression of these genes can lead to malformation in the femur cartilage structure, which could contribute to the separation of the articular cartilage from the growth plate.
Through the evaluation of the upregulated genes, most of the BP was involved with the recruitment of immune cells to enhance the adaptive immune response, blood circulation, angiogenesis, circulatory system development and cellular adhesion. The CCND3, CDK6, JUN, ADD2, ANK1, RHAG, EPB42, SLC4A1, STOM, CAMP, SERPINB10, LYG2, CATHL3, AvBD1, AvBD2, S100A9, CSF3R, RAC2, FGL2, PTPRC, LYVE1 and ITGAB2 genes (Supplementary File 1: Table S3, Fig. 1) can be highlighted due to their involvement in direct antimicrobial activities and immunomodulatory responses32,33. It is important to highlight that, since the information regarding FHN is scarce in chickens, it is hard to prospect BP and pathways involved with this condition. Moreover, as we had a relatively small number of DE genes, the non-corrected p-values were used for enrichment analysis. The main BP identified in the AC from DE genes between FHS-affected and normal broilers are discussed below.
Genes related to immune response
Several studies have reported bacterial infection as one of the causes of the BCO. However, there is a controversy in the literature regarding the bacterial infections in the FHS and whether it is related or not to the cause of this condition9,16–19. The genes MAPK12 and JUN were related to the bacterial infection and to the toll-like receptor signaling pathway BP. The later BP plays a key role in the innate immune system. The upregulation of those genes can be a consequence of the FHS, through the recognition of structurally conserved molecules derived from microbes that breached physical barriers, and are recognized by the toll-like receptors, activating the immune response34.
Immune biological processes had 18 genes present in DAVID and REVIGO (Supplementary File 1: Table S3, Fig. 1). The gene expression profile observed in this study showed a global activation of the immune system (Fig. 1). Among the enriched genes, ANK1, AvBD1, ACTN2, ADA, C7, CATH2, CCND3, CSF3R, EDN2, JUN, TF, RHAG, S100A9, SERPINB10, SSTR2, AvBD2, EPB42, LECT2, LYG2, PTPRC and STOM were upregulated in the FHS-affected group. The AvBD1, AvBD2, AvBD7, CATHL2, CATHB1, LECT2, SERPINB10 and S100A9 genes were enriched in the host immune response BP. Mainly AvBD1, AvBD2 and CATHL2 are key components of the innate immune system35,36. LECT2 (Leukocyte cell-derived chemotaxin 2) encodes a multifunctional protein characteristically similar to cytokines that improve protective immunity in bacterial sepsis37.
The BP of defense response to other organisms was enriched with RSFR, LYG2, AvBD1, STOM, CATH2, AvBD2 and SERPINB10 genes. The identification of this BP indicates a probable presence of pathogenic microorganisms in the AC tissue. It has been suggested that BCO can be initiated by a mechanical micro fracturing of the growth plate, followed by colonization of osteochondrotic clefts by different opportunistic bacteria circulating in the blood25. Moreover, one of the causes of BCO is bacterial translocation from the intestinal tract and their proliferation in bone fissures6, and Staphylococcus aureus was found to be the most frequent bacteria associated to osteomyelitis38,39.
The host-defense peptides (HDPs) are a group of small molecules that have direct antimicrobial activities and immunomodulatory properties that are responsible for the recruitment of immune cells to enhance the adaptive immune response32,33. Its antimicrobial activity aims to eliminate bacteria, enveloped viruses, fungi and protozoa by binding the cell and producing pores that lead to cell leakage and lysis, while the immunomodulatory properties help boosting the adaptive immunity through chemotaxis of lymphocytes40. In avian species, three classes of HDP are described: avian beta-defensins (AvBDs), cathelicidins (CATHs), and liver-expressed antimicrobial peptide 2 (LEAP-2)41,42.
The AvBDs group comprises 14 genes, which encode proteins that are different in their chemical structure, mainly amino acid sequence and composition43. In this study, the beta-defensins AvBD1 and AvBD2 were upregulated in the FHS-affected broilers, confirming the possible role of these genes in controlling the FHS development and, in consequence, interrupting the FHS progression towards BCO.
The ANK1 gene encodes a protein related to the binding of the structural constituent of the cytoskeleton, protein that aids in the attachment of other proteins in the membrane to the actin-spectrin cytoskeleton44,45. According to Hall et al.46, ANK1 has an adaptive function as a membrane adapter protein, making connections between the cell membrane proteins and the spectrin-actin cytoskeleton, resulting in cell migration. Ankirin-1 has a role in supporting cell movement after damage. As the ankirin-1 can affect the structure of the actin filament and the cellular motility, it is possible that increased levels of ankirin-1 may inhibit the organization of the actin filament by increasing the binding of the spectrin-actin or, alternatively, the ankyrin-1 could act modulating the signaling pathways of actin remodeling46. Moreover, the gene ANK1 is co-regulated by p53, which is involved in a variety of cellular functions, including cell-cycle arrest, DNA repair, and apoptosis46,47. The upregulation of ANK1 can be related to the FHS and BCO since its high expression can alter the structure of actin cytoskeleton affecting the structural integrity of the femur articular cartilage, contributing to the occurrence of proximal FHS. On the other hand, ANK1 expression is related to cellular damage, so it could also be a consequence, since after the damage process from the FHS is initiated, the upregulation of ANK1 can act as a sign of trying to combat the progression of this condition.
The inflammation is a vital component of the host defenses, but on the other hand, excessive inflammation can cause tissue damage48. The adenosine deaminase (ADA) is an enzyme that acts as an endogenous regulator of the adaptive immune response, playing an important role on T-lymphocytes proliferation and differentiation49. Furthermore, adenosine regulates cell metabolism and triggers a variety of physiological effects in cell proliferation50. The ADA gene acts as a sensor and provides information to the immune system about tissue damage, protecting the host cells from excessive tissue injury associated with strong inflammation51. The upregulation of ADA could downregulate the activation of lymphocytes during inflammation, and also play a regulatory role on neutrophils in immune responses50,52. Extracellular adenosine signaling has been shown to play a role in inflammation during hypoxia and ischemia–reperfusion injuries, usually resulting in vascular leakage, accumulation of inflammatory cells, and elevated cytokine levels in serum. Moreover, just as hypoxia can induce inflammation, inflamed tissues often become severely hypoxic53. The ADA upregulation can be considered a consequence of the proximal FHS, since its high expression is related to immune responses, trying to combat inflammatory process already installed. Therefore, it gives rise to the hypothesis that through the increased number of bacteria in the tissue, there is an upregulation of the ADA gene, aiming to fight and eliminate the bacteria that are causing damage.
The EPX gene is activated during an immune response, releasing proteins and other components in the area of injury or inflammation that have a toxic effect on severely damaged cells or infecting pathogens. One of these proteins is called eosinophil peroxidase, that are extremely cytotoxic to bacteria54,55, parasites56,57, eukaryotic cells58 and neoplastic cells59,60. The upregulation of the EPX may be a consequence of FHS, since a possible infection could pressure the bone structure, impairing the blood supply to the affected area, developing necrosis. Furthermore, EPX could regulate the inflammatory process to control the infection.
Chemokines are a group of chemoattractant cytokines released by tissues in the beginning of infection. They are usually produced by different cell types in response to bacterial products and other pathogens. Besides the promotion of immune cells chemotaxis to the site of infection, they regulate a variety of biological processes related to cellular activation, differentiation and survival61, such as those found in our study (Supplementary File 1: Table S3). In mice, it has been shown that in the presence of bacterial infection, there is an increase of inflammatory cytokines, which can lead to osteocyte apoptosis and consequently osteonecrosis62. In our study, two chemokines (CCL26 and CCR5) were DE in the chicken articular cartilage. The CCL26 has a bactericidal activity verified against pathogens Streptococcus pneumoniae, Staphylococcus aureus, Nontypeable Haemophilus influenzae, and Pseudomonas aeruginosa63, while CCR5 has already been identified as biomarker for osteonecrosis of the femoral head in human plasma64.
Our results showed several biological processes and genes related to immune response, indicating that the overexpression of these genes is activating the immune system to fight against the progression of FHS, evidencing the presence of an inflammatory process, even at the early stages of FHS.
Bone-related bioprocesses
The results of the NetworkAnalyst platform indicates associations between DE genes and Wnt signaling pathway, GnRH signaling pathway, Adrenergic signaling in cardiomyocytes, VEGF signaling pathway, Bacterial infection, Focal adhesion, Gap junction, Toll-like receptor signaling pathway, AGE-RAGE signaling pathway in diabetic complications and NOD-like receptor signaling pathway (Fig. 2). The Wnt signaling is an ancient and evolutionarily conserved pathway responsible for the regulation of crucial aspects of cell fate determination, cell migration, cell polarity, neural patterning and organogenesis during embryonic development65, which are important for bone development. Most of the genes enriched in this BP also appeared in focal adhesion and Toll-like receptors. Some of these processes have already been described by Peixoto et al.12 and could be intrinsically correlated with FHS.
Durairaj et al.66 suggested that FHS could be a metabolic problem, related to fat metabolism disorders, facilitating an unbalanced growth in the articular-epiphyseal complex that leads to its separation under shear stress. They observed that the blood parameters such as cholesterol, triglycerides, and low-density lipoproteins were slightly increased in FHS-affected chickens. Despite the physiological differences between humans and chickens, the appearance of the GnRH signaling pathway, AGE-RAGE signaling pathway in diabetic complication and adrenergic signaling in cardiomyocytes, indicates that chickens may have a similar physiology, needing more studies to better elucidate these pathways.
The genes MAPK12 and RAC2 were connected to the VEGF signaling pathway. This pathway is crucial to the vascular development stages and processes, like vasculogenesis, angiogenesis and lymphangiogenesis, which are essential for specification, morphogenesis, differentiation, and homeostasis of vessels during development and in the adulthood67. The involvement of VEGF signaling pathway in the FHN and BCO in chickens has already been observed11,12,20,24 and could affect the cells regeneration and maintenance68. Furthermore, the MAPK12 gene is located in a QTL for bone mineral density in humans69, while the activity of RAC2 gene has been observed in the osteocalastogenesis70,71, involved in the development of tibial dyschondroplasia in chickens72 and osteoarthritis in humans73.
Moreover, the DE genes identified in the current study were investigated for associations with locomotor problems using the human curated information of DisGeNET database from the NetworkAnalyst 3.0 (Fig. 3). This analysis showed that the ADA, ANK1, JUN, ACTA1, TNNT3 and ACTA1 genes were also related to human locomotor problems, evidencing a similar pattern in chickens and humans. Although the knowledge of the chicken transcriptomic profile is increasing, the functional annotation of its genome remains incomplete. In this way, human databases are still needed to infer pathway information in the chicken74. Therefore, more studies are needed to better understand the role of these genes in the development of locomotor problems in chickens.
Response to biotic stimulus, gas transport, cell activation and cytolysis
Biomechanical continuous local stress and impaired blood flow to the epiphyseal-physical cartilage are some of the factors that favor the pathogenesis of osteochondrosis, reported in several animal species75–80. The FHS has been associated with the growing phase and a large number of DE genes in this study were involved in BP response to biotic stimulus and regulation of multi-organism processes, which are relevant to the animal locomotor system development. The DE genes associated to FHS in these BP are ADA, AvBD1, AvBD2, C7, CCND3, CSF3R, EDN2, EPB42, GGT1, JUN, LECT2, LYG2, PTPRC, RAC2, RHAG, RSFR, S100A9, SELP, SERPINB10, STOM.
One of the main BP enriched in the current study was gas transport and cell migration (Fig. 1). The RHAG is one of the genes of the Rh gene family81. This gene is usually expressed in tissues that produce blood cells, but it is also expressed in heart cells and those related to the gas transferring system from the lungs to organelles within cells82,83. The elevated expression of genes related to the gas transfer system can indicate a more pronounced O2 reduction or CO2 enhancement. The imbalance between the O2 supply and CO2 removal of the gas transferring elements has already been associated to hypoxia or hypercapnia, which could lead to the damage of the heart cells metabolism84,85. The upregulation of RHAG can be related to the proximal FHS development, since in consequence of the pressure in the bone structure, caused by inflammation, the blood supply is reduced, causing hypoxia due to the lack of oxygen. RHAG upregulation can also be related to a consequence of FHS since its upregulation leads to increased oxygenation of the affected tissue. This gene was also enriched in several others BP, such as those related to immune response.
The apoptosis is probably involved in the FHS in broilers, since it is a physiological mechanism crucial in the development and tissue homeostasis. In our study, this BP was not enriched in the DAVID database, but some genes associated with apoptosis were DE (ADA, JUN, IFI6). The gene IFI6, also known as ISG12, has an important role in apoptosis regulation20,86. In humans, this gene encodes a hydrophobic protein that acts in intracellular signaling87,88, but in birds, it does not have its function fully established. Furthermore, the ISGs family is known to generate cellular and physiological diversity and it is associated with antiviral, anti-tumor and immunomodulatory activity mechanisms89. In our results, the gene IFI6 was DE and co-located with the gene STEAP4. These genes are expressed at the same site (cell or tissue), and their functions are related to regulation of cellular metabolism during osteoblast differentiation and regulation of apoptosis90. The upregulation of the IFI6 can be related to a causal factor, stimulating an excessive apoptosis at the articular cartilage, turning the animal more susceptible to FHS.
Extracellular matrix
The carbohydrate derivate catabolism was one of the superclusters observed, which contained the carbohydrate derivate catabolism and glycosaminoglycan catabolic BP (Fig. 1, Supplementary File 1: Table S3). The extracellular matrix (ECM) is a structurally stable component that is located under the epithelium and surrounds connective tissue cells91. Due to its structure, ECM is responsible for providing support and resistance to tissues and organs throughout the body, and acts in biochemical processes related to tissue morphogenesis, differentiation and homeostasis92. In addition, in the ECM there are molecules, like glycosaminoglycan, responsible for cell modulation, such as adhesion, migration, proliferation, differentiation and cell survival of the tissue93.
The glycosaminoglycans are fundamental components fulfilling various ECM biological functions. They are highly polar and can also contribute to permeability properties, connective tissue structure and as a guide to enzymes and growth factors in both the matrix and cell surface94. The DE genes enriched in the glycosaminoglycan and aminoglycan BP were upregulated in FHS-affected broilers (ADA, SERPINB10, AvBD1, STOM, JUN, RHAG, KEL, TF, EDN2, EPB42) (Supplementary File 1: Table S3). Here, important upregulated genes are ADA, RHAG and JUN, which participate in the glycosaminoglycan (GAGs) and aminoglycan metabolic processes involved in the ECM metabolism. The RHAG, ADA and AvBD1 differential expression pattern between healthy and FHS-affected group were also confirmed by qPCR (Table 3).
Genes grouped in these previous BP were upregulated in the FHS-affected broilers, indicating that the body tries to fix the damage through remodeling. Altogether, the results indicate that the upregulation of the genes could be a consequence of the damage by the FHS, where the upregulation of those genes is an attempt to diminish the injury, since glycosaminoglycans mediate various receptor-ligand interactions on the cell surface and, as a result, play an important role in development, as well as in lesion repair94.
In this study, response to biotic stimulus, immune response and cell activation processes were BP highly represented. FHS may cause important physiological implications to the broiler’s development, which leads to more severe disorders.
There are some studies conducted with chicken bone tissue evaluating locomotor problems, but just one has recently been performed with cartilage tissue95. Both tissues are important to the development of those problems. Therefore, the knowledge of the relation between bone and cartilage tissues with these disorders is essential to provide alternative strategies to counteract these complex production problems. The identification of young broilers with vulnerable femoral joint can help genetic selection to reduce this anomaly. The BCO pathology does not show clinical signs at early stages, only at late stages or after necropsy when the diagnostic is possible14. The use of infrared thermography (IRT) was suggested as a technique to detect lesions attributed to BCO96. The IRT consists of a noninvasive technique that measures infrared radiation from an object and can be a useful tool to evaluate clinical health. Although there are options to confirm the diagnosis of this condition, there are still limitations. The functional analyses of the DE genes help to elucidate their involvement in the development of FHS. These results contribute to a better understanding of the FHN in chickens and possibly other femur disorders in humans.
In summary, the first transcriptome of the femoral articular cartilage was generated, and biological processes and genes involved with femur head separation in rapid growth chickens were identified. Some genes such as AvBD1, AvBD2, ANK1, RHAG, ADA and EPX were firstly associated to FHS in broilers, indicating that the disruption in the articular cartilage could favor the development of this condition. These results might help the development of strategies to reduce the manifestation of this disorder in poultry, improving welfare and reducing economic losses.
Material and methods
Ethics statement
All of the experimental procedures were conducted in conformity with the guidelines of the Ethics Committee for Animal Use (CEUA) from the Embrapa Swine and Poultry National Research Center, with approval protocol number 012/2012, in agreement with the rules of the National Council of Animal Experimentation Control (CONCEA) to ensure compliance with international guidelines for animal welfare.
Animals and sample collection
A total of 29 Cobb500 commercial male broilers from a poultry farm, located in Concórdia/SC, Brazil, was used in this study. Broilers were housed according to the standard practices, raised with free access to both feed and water. To reduce environmental effects, the broilers used in this study were sampled from the same flock, in a darkhouse system managed by a high standard producer. At the farm, the animals were selected based on the absence or presence of lameness and split into two groups: 14 normal and 15 chickens showing lameness as described by Peixoto et al.12. At the necropsy, the animals were evaluated for the presence or absence of FHS according to Wideman and Prisby7 and Paludo et al.11. Broilers showing separation between femoral GP an AC were included in the FHS-affected group and broilers with good adhesion of the AC and GP were considered normal and were included in the control group, as in Peixoto et al.12. From those, eight samples of AC, four normal (average weight 2,401 g ± 31.19) and four FHS-affected (average weight 2,406 ± 148.45 g) were randomly chosen for RNA-Seq, collected in liquid nitrogen and stored at − 80 °C. None of the chosen broilers were visually affected by BCO.
Total RNA extraction, library preparation and sequencing
For total RNA extraction, eight samples of femoral articular cartilage (4 from each group) were homogenized in liquid nitrogen and 100 mg of tissue was added in 1 mL of TRIzol reagent. Then, 200 μL of chloroform were added, the tubes were homogenized for 15 s and incubated at room temperature for five minutes, centrifuged at 16,000 × g at 4 °C for 15 min. The aqueous phase was separated into a new microtube, mixed with 70% ethanol. This solution was added to a Qiagen RNeasy silica column (Qiagen, Hilden, NRW, Germany), and the RNA extraction followed the standard protocol of Qiagen RNeasy kit (Qiagen, Hilden, NRW, Germany), according to the manufacturer´s instructions. Total RNA was quantified using the BioDrop spectrophotometer (Biodrop, Cambridge, UK) and samples with OD260:OD280 ratio greater than 1.9 were considered pure. The RNA integrity was confirmed in a 1% agarose gel electrophoresed for 90 min with limit of 90 V in 1X TBE buffer and in Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Samples with RNA integrity number (RIN) greater than 8.0 were used for RNA libraries preparation.
Approximately 2 μg of total RNA was submitted to library preparation using the TruSeq Stranded mRNA Library Prep Kit (Illumina, Inc., San Diego CA, USA), according to the manufacturer's recommendations. The size of the libraries was confirmed in Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Libraries were quantified with qPCR using primers with Illumina adapters and then sequenced in an Illumina HiSeq2500 (Illumina Inc., San Diego, CA, USA) at the Center for Functional Genomics at ESALQ, University of São Paulo, Piracicaba—SP, Brazil, using HiSeq SBS Kit (Illumina Inc., San Diego, CA, USA) following a paired-end (2 × 100 bp) protocol. All samples were sequenced in the same lane.
Quality control, mapping and differential expression analyses
The reads quality control (QC) was performed with Trimmomatic v. 0.3897 to remove short reads (< 70 bp), low-quality reads (QPhred < 20) and adapter sequences. Mapping was performed with STAR 2.798 using the chicken reference genome (Gallus gallus, assembly GRCg6a) available at the Ensembl 95 database (www.ensembl.org). The reads were counted in exon regions using HTSeq v. 0.11.299 and the EdgeR100 implemented in R language101 was used to identify DE genes between the normal and affected groups. Genes with false discovery rate (FDR) < 0.05 were considered DE, after correcting for the Benjamini-Hochberg (BH) multiple-tests102. Genes were considered upregulated and downregulated according to the positives and negatives log2 fold-change (Log2FC), respectively, in the affected compared to the normal broilers. The multidimensional scaling (MDS) plot was created using the normalized read counts for each sample using the plotMDS function in edgeR100. Based on DE genes, a heatmap was generated to check the consistence between samples using the heatmap.2 function from gplots in R101. The FASTQ files sequenced in this study were deposited in the SRA database, with BioProject number PRJNA350521.
qPCR validation
To confirm the results obtained in the RNA-Seq analysis, a quantitative PCR analysis (qPCR) was performed using the same eight AC samples from the normal and FHS-affected animals. Total RNA was extracted as described above. The cDNA synthesis was performed according to the recommendations of the SuperScript III, First-Strand Synthesis Supermix protocol (Invitrogen, Carlsbad, CA, USA). For validation, 11 DE genes in the RNA-Seq analysis were chosen based on FDR, Log2FC and their function as candidates to be involved with FHN: avian beta-defensin 1 (AvBD1), avian beta-defensin 2 (AvBD2), fibrillin 2 (FBN2), ankyrin 1 (ANK-1), phosphoserine phosphatase (PSPH), eosinophil peroxidase (EPX), adenosine deaminase (ADA), collagen type XIII alpha 1 chain (COL13A1), Rh associated glycoprotein (RHAG) and S100 calcium-binding protein A9 (S100A9). Primers were designed using the NCBI Primer-BLAST tool103 (Table 4) and the quality was evaluated and confirmed with the Netprimer program (http://www.premierbiosoft.com/NetPrimer). For the relative quantification analyses, reactions were prepared using 1X GoTaq qPCR Master Mix (Promega, Madison, WI, USA), with BRYT Green Dye and CRX as reference dye, 0.13 μM of forward and reverse primers, 2 μL cDNA at 1:10 dilution and ultrapure water (Nuclease Free Water, Qiagen) to complete a 15 μL reaction. The reactions were performed in duplicate and submitted to the QuantStudio 6 Real-Time PCR equipment (Applied Biosystems, Foster City, CA, USA) with an initial cycling of 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for one minute, with melting curve of 95 °C for 15 s, 60 °C for 1 min and 95 °C for 15 s.
Table 4.
Primers used for the qPCR analysis of the target candidate genes for FHS in the femur articular cartilage of broilers.
| Gene | Ensembl ID | Primer sequences (5'-3') |
|---|---|---|
| AvBD1 | ENSGALG00000022815 | F: CAGGATCCTCCCAGGCTCTA |
| avian beta-defensin 1 | R: GATGAGAGTGAGGGAAGGGC | |
| AvBD2 | ENSGALG00000016669 | F: TTCTCCAGGGTTGTCTTCGC |
| avian beta-defensin 2 | R: TGCATTCCAAGGCCATTTGC | |
| FBN2 | ENSGALG00000014686 | F: TGCATCGATAGCCTGAAGGG |
| fibrillin 2 | R: CTAATTCACACCGCTCACATGG | |
| ANK1 | ENSG00000029534 | F: CCACCATCCCACCATTCAGT |
| Ankyrin 1 | R: ACGGTCACAAACTCCAGCAT | |
| PSPH | ENSGALG00000002397 | F:CAGGAATACGGGAGCTGGTG |
| phosphoserine phosphatase | R: CCCAGAGACCAGGAAGACCT | |
| EPX | ENSGALG00000043254 | F:AAAGGAGGTGGCATTGACCC |
| eosinophil peroxidase | R: GCCACGCTGCATGTTAAGAG | |
| ADA | ENSGALG00000004170 | F:TTCGGCAAGAAAAGAGGGGT |
| adenosine deaminase | R: GTGTTTGGTAGCTGACGTGC | |
| COL13A1 | ENSGALG00000004286 | F:CCAAGCAAGGACTAGACACTCA |
| collagen type XIII alpha 1 chain | R: ACCCTTCATGCCATGTCTTCC | |
| CSF3R | ENSGALG00000002112 | F: TCATCCGGGACAGCATTGAG |
| colony stimulating factor 3 receptor | R: TGTAGAGGGGGTACACCGAG | |
| RHAG | ENSGALG00000016684 | F:TCTGGAGATCACGGCCTTTG |
| Rh associated glycoprotein | R:GCTCCAATATCTGTGGCCTGA | |
| S100A9 | ENSGALG00000024272 | F:GGGGACAAAGACACCCTGAC |
| S100 calcium binding protein A9 | R:TTCACGTGCTTCAGGTAGTTGG |
The Ct means of each sample were obtained to perform the relative expression ratio analysis104. For the data normalization, the geometric mean of the Ct values from the reference genes RPL5 (Ribosomal Protein L5) and RPLP1 (Ribosomal Protein Lateral Stalk Subunit P1) were used. These genes were chosen based on their stability evaluation in chicken femoral articular cartilage105. The Relative Expression Software Tool (REST)104 was used to perform the relative quantification and the statistical test, using the non-parametric Pair Wise Fixed Reallocation Randomization Test106. Genes with p-values ≤ 0.05 were considered DE.
Functional annotation
To investigate the role of DE genes in known metabolic pathways, the list of all DE genes was analyzed with the Functional Annotation Clustering (FAC) implemented in the DAVID database (http://david.abcc.ncifcrf.gov/)107,108, considering a p-value of < 0.10 as significant. Subsequently, the biological processes were clustered in the REVIGO109. An enrichment analysis was also performed using the chicken genome in the NetworkAnalyst110, where new biological functions were obtained with the protein–protein interactions (PPI). Moreover, a gene-disease association network was created with the DE genes using human curated information of DisGeNET database available in the NetworkAnalyst110, considering a p-value of < 0.05 as significant.
Supplementary Information
Acknowledgements
This study was financed by Project # 01.11.07.002.04.03 from the Brazilian Agricultural Research Corporation (EMBRAPA). The authors are grateful to A. L. Tessmann for technical assistance. LTF is supported by a visiting specialist fellowship from the National Council of Scientific and Technological Development (CNPq) Grant # 380282/2021-6, Brazil. LLC and MCL are recipient of a productivity fellowship from CNPq. We thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil—Finance Code 001 for the free access to the journals used in the literature review.
Author contributions
J.O.P., A.M.G.I., M.C.L. conceived and designed the experiment. J.O.P., A.M.G.I., M.C.L., L.L.C., M.E.C., L.M.H., D.E.P.M., I.R.S. performed the experiment. L.M.H., A.M.G.I., J.O.P., M.C.L., M.E.C., P.F.G., R.H.H. performed data analysis and curation. L.M.H., A.M.G.I., J.O.P., M.C.L., L.T.F. prepared the original draft manuscript. J.O.P. and M.C.L. were responsible for funding acquisition. J.O.P., A.M.G.I., M.C.L. supervised the research. All authors have read and approved the final manuscript.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. The fastq files used in this study were submitted to SRA database and will be available as BioProject number PRJNA350521.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-97306-3.
References
- 1.Zuidhof MJ, Schneider BL, Carney VL, Korver DR, Robinson FE. Growth, efficiency, and yield of commercial broilers from 1957, 1978, and 20051. Poult. Sci. 2014;93:2970–2982. doi: 10.3382/ps.2014-04291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook ME. Skeletal deformities and their causes: Introduction. Poult. Sci. 2000;79:982–984. doi: 10.1093/ps/79.7.982. [DOI] [PubMed] [Google Scholar]
- 3.Havenstein GB, Ferket PR, Qureshi MA. Carcass composition and yield of 1957 versus 2001 broilers when fed representative 1957 and 2001 broiler diets. Poult. Sci. 2003;82:1509–1518. doi: 10.1093/ps/82.10.1509. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan TW. Skeletal problems in poultry: Estimated annual cost and descriptions. Poult. Sci. 1994;73:879–882. doi: 10.3382/ps.0730879. [DOI] [PubMed] [Google Scholar]
- 5.Li PF, Zhou ZL, Shi CY, Hou JF. Downregulation of basic fibroblast growth factor is associated with femoral head necrosis in broilers. Poult. Sci. 2015;94:1052–1059. doi: 10.3382/ps/pev071. [DOI] [PubMed] [Google Scholar]
- 6.Wideman RF, Prisby RD. Bone circulatory disturbances in the development of spontaneous bacterial chondronecrosis with osteomyelitis: A translational model for the pathogenesis of femoral head necrosis. Front. Endocrinol. (Lausanne) 2013;3:1–14. doi: 10.3389/fendo.2012.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wideman RF, et al. A wire-flooring model 1 for inducing lameness in broilers: Evaluation of probiotics as a prophylactic treatment. Poult. Sci. 2012;91:870–883. doi: 10.3382/ps.2011-01907. [DOI] [PubMed] [Google Scholar]
- 8.McNamee PT, Smyth JA. Bacterial chondronecrosis with osteomyelitis ('femoral head necrosis’) of broiler chickens: A review. Avian Pathol. 2000;29:477–495. doi: 10.1080/030794500750047243. [DOI] [PubMed] [Google Scholar]
- 9.Almeida Paz ICL, et al. Seguimiento del desarrollo de lesiones por degeneración en pollos de engorde. Int. J. Morphol. 2009;27:571–575. doi: 10.4067/S0717-95022009000200042. [DOI] [Google Scholar]
- 10.Olkowski AA, et al. Biochemical and physiological weaknesses associated with the pathogenesis of femoral bone degeneration in broiler chickens. Avian Pathol. 2011;40:639–650. doi: 10.1080/03079457.2011.626017. [DOI] [PubMed] [Google Scholar]
- 11.Paludo E, et al. The involvement of RUNX2 and SPARC genes in the bacterial chondronecrosis with osteomyelitis in broilers. Animal. 2017;11:1063–1070. doi: 10.1017/S1751731116002433. [DOI] [PubMed] [Google Scholar]
- 12.Peixoto JO, et al. Proximal femoral head transcriptome reveals novel candidate genes related to epiphysiolysis in broiler chickens. BMC Genom. 2019;20:1031. doi: 10.1186/s12864-019-6411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riddell C, King MW, Gunasekera KR. Pathology of the skeleton and tendons of broiler chickens reared to roaster weights II. Normal chickens. Avian Dis. 1983;27:980–991. doi: 10.2307/1590199. [DOI] [PubMed] [Google Scholar]
- 14.Packialakshmi B, Rath NC, Huff WE, Huff GR. Poultry femoral head separation and necrosis: A review. Avian Dis. 2015;59:349–354. doi: 10.1637/11082-040715-Review.1. [DOI] [PubMed] [Google Scholar]
- 15.Geister KA, Camper SA. Advances in skeletal dysplasia genetics. Annu. Rev. Genomics Hum. Genet. 2015;16:199–227. doi: 10.1146/annurev-genom-090314-045904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorp BH, et al. Proximal femoral degeneration in growing broiler fowl. Avian Pathol. 1993;22:325–342. doi: 10.1080/03079459308418924. [DOI] [PubMed] [Google Scholar]
- 17.Wilson FD, Stayer P, Pace LW, Hoerr FJ, Magee DL. Disarticulation-associated femoral head separation in clinically normal broilers: Histologic documentation of underlying and predisposing cartilage abnormalities. Avian Dis. 2019;63:495. doi: 10.1637/19-00090.1. [DOI] [PubMed] [Google Scholar]
- 18.Santili C, et al. Southwickʼs head-shaft angles: Normal standards and abnormal values observed in obesity and in patients with epiphysiolysis. J. Pediatr. Orthop. B. 2004;13:244–247. doi: 10.1097/01.bpb.0000111042.46580.68. [DOI] [PubMed] [Google Scholar]
- 19.Mestriner MB, et al. Radiographic evaluation in epiphysiolysis: Possible predictors of bilaterality. Acta Ortop. Bras. 2012;20:203–206. doi: 10.1590/S1413-78522012000400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petry B, et al. New genes involved in the Bacterial Chondronecrosis with Osteomyelitis in commercial broilers. Livest. Sci. 2018;208:33–39. doi: 10.1016/j.livsci.2017.12.003. [DOI] [Google Scholar]
- 21.Marchesi, J.A.P, Ibelli, A.M.G., Paludo, E., Tavernari, F.C., Peixoto, J.O., Ledur, M.C. Expressão do gene receptor da leptina (LEPR) em frangos de corte normais e afetados pela necrose da cabeça do fêmur. In: 11ª Jornada de Iniciação Científica (JINC), 2017, Concórdia. p 43–44. Available online: http://www.cnpsa.embrapa.br/11jinc/docs/anais2017.pdf (2017).
- 22.Johnsson M, Jonsson KB, Andersson L, Jensen P, Wright D. Genetic regulation of bone metabolism in the chicken: Similarities and differences to mammalian systems. PLOS Genet. 2015;11:e1005250. doi: 10.1371/journal.pgen.1005250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faveri JC, et al. Quantitative trait loci for morphometric and mineral composition traits of the tibia bone in a broiler × layer cross. Animal. 2019;13:1563–1569. doi: 10.1017/S175173111800335X. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, et al. Evaluation of housekeeping genes for normalizing real-time quantitative PCR assays in pig skeletal muscle at multiple developmental stages. Gene. 2015;565:235–241. doi: 10.1016/j.gene.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 25.Wideman RF, Blankenship J, Pevzner IY, Turner BJ. Efficacy of 25-OH vitamin D3 prophylactic administration for reducing lameness in broilers grown on wire flooring. Poult. Sci. 2015;94:1821–1827. doi: 10.3382/ps/pev160. [DOI] [PubMed] [Google Scholar]
- 26.Simpson AH, Murray IR. Osteoporotic fracture models. Curr. Osteoporos. Rep. 2015;13:9–15. doi: 10.1007/s11914-014-0246-8. [DOI] [PubMed] [Google Scholar]
- 27.Abubakar AA, Noordin MM, Azmi TI, Kaka U, Loqman MY. The use of rats and mice as animal models in ex vivo bone growth and development studies. Bone Joint Res. 2016;5:610–618. doi: 10.1302/2046-3758.512.BJR-2016-0102.R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabriele Sommer, N., Hahn, D., Okutan, B., Marek, R. & Weinberg, A.-M. Animal models in orthopedic research: The proper animal model to answer fundamental questions on bone healing depending on pathology and implant material. in Animal Models in Medicine and Biology, IntechOpen 10.5772/intechopen.89137 (2019).
- 29.Hägg P, et al. Type XIII collagen: A novel cell adhesion component present in a range of cell-matrix adhesions and in the intercalated discs between cardiac muscle cells. Matrix Biol. 2001;19:727–742. doi: 10.1016/S0945-053X(00)00119-0. [DOI] [PubMed] [Google Scholar]
- 30.Nykvist P, et al. Distinct recognition of collagen subtypes by α1β1 and α2β1. integrins α1β1 mediates cell adhesion to type XIII collagen. J. Biol. Chem. 2000;275:8255–8261. doi: 10.1074/jbc.275.11.8255. [DOI] [PubMed] [Google Scholar]
- 31.Tu H, et al. The type XIII collagen ectodomain is a 150-nm rod and capable of binding to fibronectin, nidogen-2, perlecan, and heparin. J. Biol. Chem. 2002;277:23092–23099. doi: 10.1074/jbc.M107583200. [DOI] [PubMed] [Google Scholar]
- 32.van Dijk A, Veldhuizen EJA, Haagsman HP. Avian defensins. Vet. Immunol. Immunopathol. 2008;124:1–18. doi: 10.1016/j.vetimm.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang G. Database-guided discovery of potent peptides to combat HIV-1 or superbugs. Pharmaceuticals. 2013;6:728–758. doi: 10.3390/ph6060728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahla RS, Reddy MC, Prasad DVR, Himanshu Kumar H. Sweeten PAMPs: Role of Sugar Complexed PAMPs in Innate Immunity and Vaccine Biology. Front. Immunol. 2013;4:248. doi: 10.3389/fimmu.2013.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harwig SSL, et al. Gallinacins: Cysteine-rich antimicrobial peptides of chicken leukocytes. FEBS Lett. 1994;342:281–285. doi: 10.1016/0014-5793(94)80517-2. [DOI] [PubMed] [Google Scholar]
- 36.Cheng CY, et al. Annotation of differential gene expression in small yellow follicles of a broiler-type strain of Taiwan country chickens in response to acute heat stress. PLoS One. 2015;10:e0143418. doi: 10.1371/journal.pone.0143418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu XJ, et al. LECT2 protects mice against bacterial sepsis by activating macrophages via the CD209a receptor. J. Exp. Med. 2013;210:5–13. doi: 10.1084/jem.20121466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berendt T, Byren I. Bone and joint infection. Clin. Med. J. R. Coll. Phys. Lond. 2004;4:510–518. doi: 10.7861/clinmedicine.4-6-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright JA, Nair SP. Interaction of staphylococci with bone. Int. J. Med. Microbiol. 2010;300:193–204. doi: 10.1016/j.ijmm.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L, Weiss TM, Lehrer RI, Huang HW. Crystallization of antimicrobial pores in membranes: Magainin and protegrin. Biophys. J. 2000;79:2002–2009. doi: 10.1016/S0006-3495(00)76448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuperus T, Coorens M, van Dijk A, Haagsman HP. Avian host defense peptides. Dev. Comp. Immunol. 2013;41:352–369. doi: 10.1016/j.dci.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 42.Zhang G, Sunkara L. Avian antimicrobial host defense peptides: From biology to therapeutic applications. Pharmaceuticals. 2014;7:220–247. doi: 10.3390/ph7030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klüver E, Adermann K, Schulz A. Synthesis and structure–activity relationship of β-defensins, multi-functional peptides of the immune system. J. Pept. Sci. 2006;12:243–257. doi: 10.1002/psc.749. [DOI] [PubMed] [Google Scholar]
- 44.Rubtsov AM, Lopina OD. Ankyrins. FEBS Lett. 2000;482:1–5. doi: 10.1016/S0014-5793(00)01924-4. [DOI] [PubMed] [Google Scholar]
- 45.Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J. Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall AG, et al. The cytoskeleton adaptor protein ankyrin-1 is upregulated by p53 following DNA damage and alters cell migration. Cell Death Dis. 2016;7:e2184. doi: 10.1038/cddis.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 48.Sitkovsky MV, et al. Physiological control of immune response and inflammatory tissue damage by hypoxia-inducible factors and adenosine A2A receptors. Annu. Rev. Immunol. 2004;22:657–682. doi: 10.1146/annurev.immunol.22.012703.104731. [DOI] [PubMed] [Google Scholar]
- 49.Aran JM, Colomer D, Matutes E, Vives-Corrons JL, Franco R. Presence of adenosine deaminase on the surface of mononuclear blood cells: Immunochemical localization using light and electron microscopy. J. Histochem. Cytochem. 1991;39:1001–1008. doi: 10.1177/39.8.1856451. [DOI] [PubMed] [Google Scholar]
- 50.Melanie DD, Katherine MC, et al. Adenosine desamination sustains dendritic cell activation in inflmmation. J. Immunol. 2007;179:1884–1892. doi: 10.4049/jimmunol.179.3.1884. [DOI] [PubMed] [Google Scholar]
- 51.Kumar V, Sharma A. Adenosine: An endogenous modulator of innate immune system with therapeutic potential. Eur. J. Pharmacol. 2009;616:7–15. doi: 10.1016/j.ejphar.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 52.Mills JH, Alabanza LM, Mahamed DA, Bynoe MS. Extracellular adenosine signaling induces CX3CL1 expression in the brain to promote experimental autoimmune encephalomyelitis. J. Neuroinflammation. 2012;9:683. doi: 10.1186/1742-2094-9-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N. Engl. J. Med. 2011;364:656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnston RB, Baehner RL. Chronic granulomatous disease: correlation between pathogenesis and clinical findings. Pediatrics. 1971;48:730–739. [PubMed] [Google Scholar]
- 55.Mickenberg ID, Root RK, Wolff SM. Bactericidal and metabolic properties of human eosinophils. Blood. 1972;39:67–80. doi: 10.1182/blood.V39.1.67.67. [DOI] [PubMed] [Google Scholar]
- 56.Auriault C, Capron M, Capron A. Activation of rat and human eosinophils by soluble factor(s) released by Schistosoma mansoni schistosomula. Cell. Immunol. 1982;66:59–69. doi: 10.1016/0008-8749(82)90157-5. [DOI] [PubMed] [Google Scholar]
- 57.Locksley RM, Wilson CB, Klebanoff SJ. Role for endogenous and acquired peroxidase in the toxoplasmacidal activity of murine and human mononuclear phagocytes. J. Clin. Invest. 1982;69:1099–1111. doi: 10.1172/JCI110545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gleich GJ, Adolphson CR. The eosinophilic leukocyte: Structure and function. Adv. Immunol. 1986;39:177–253. doi: 10.1016/S0065-2776(08)60351-X. [DOI] [PubMed] [Google Scholar]
- 59.Jong EC, Klebanoff SJ. Eosinophil-mediated mammalian tumor cell cytotoxicity: Role of the peroxidase system. J. Immunol. 1980;124:1949–1953. [PubMed] [Google Scholar]
- 60.Nathan CF, Klebanoff SJ. Augmentation of spontaneous macrophage-mediated cytolysis by eosinophil peroxidase. J. Exp. Med. 1982;155:1291–1308. doi: 10.1084/jem.155.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soni B, Singh S. Cytokine milieu in infectious disease: A sword or a Boon? J. Interferon Cytokine Res. 2020;40:24–32. doi: 10.1089/jir.2019.0089. [DOI] [PubMed] [Google Scholar]
- 62.Morita M, et al. Elevation of pro-inflammatory cytokine levels following anti-resorptive drug treatment is required for osteonecrosis development in infectious osteomyelitis. Sci. Rep. 2017;7:46322. doi: 10.1038/srep46322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gela A, et al. Eotaxin-3 (CCL26) exerts innate host defense activities that are modulated by mast cell proteases. Allergy Eur. J. Allergy Clin. Immunol. 2015;70:161–170. doi: 10.1111/all.12542. [DOI] [PubMed] [Google Scholar]
- 64.Li T, et al. Discovery and validation an eight-biomarker serum gene signature for the diagnosis of steroid-induced osteonecrosis of the femoral head. Bone. 2019;122:199–208. doi: 10.1016/j.bone.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Habas R, Dawid IB. Dishevelled and Wnt signaling: Is the nucleus the final frontier? J. Biol. 2005;4:2. doi: 10.1186/jbiol22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Durairaj V, Okimoto R, Rasaputra K, Clark FD, Rath NC. Histopathology and serum clinical chemistry evaluation of broilers with femoral head separation disorder. Avian Dis. 2009;53:21–25. doi: 10.1637/8367-051908-Reg.1. [DOI] [PubMed] [Google Scholar]
- 67.Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat. Rev. Mol. Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- 68.Hato T, Tabata M, Oike Y. The role of angiopoietin-like proteins in angiogenesis and metabolism. Trends Cardiovasc. Med. 2008;18:6–14. doi: 10.1016/j.tcm.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 69.Morris JA, et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019;51:258–266. doi: 10.1038/s41588-018-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y, et al. Identifying the relative contributions of Rac1 and Rac2 to osteoclastogenesis. J. Bone Miner. Res. 2007;23:260–270. doi: 10.1359/jbmr.071013. [DOI] [PubMed] [Google Scholar]
- 71.Itokowa T, et al. Osteoclasts lacking Rac2 have defective chemotaxis and resorptive activity. Calcif. Tissue Int. 2011;88:75–86. doi: 10.1007/s00223-010-9435-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jahejo AR, et al. Transcriptome-based screening of intracellular pathways and angiogenesis related genes at different stages of thiram induced tibial lesions in broiler chickens. BMC Genom. 2020;21:50. doi: 10.1186/s12864-020-6456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, X. et al. Identification of potential diagnostic gene biomarkers in patients with osteoarthritis. Sci. Rep, 10(1), 13591 (2020). [DOI] [PMC free article] [PubMed]
- 74.Burt, D. W. Emergence of the chicken as a model organism: Implications for agriculture and biology. Poultry Science86 (7), 1460–1471 (2007). [DOI] [PubMed]
- 75.Trueta J, Amato VP. The vascular contribution to osteogenesis III. Changes in the growth cartilage caused by experimentally induced Ischaemia. J. Bone Jt. Surg. 1960;42:571–587. doi: 10.1302/0301-620X.42B3.571. [DOI] [PubMed] [Google Scholar]
- 76.Riddell C. Studies on the pathogenesis of tibial dyschondroplasia in chickens. I. Production of a similar defect by surgical interference. Avian Dis. 1975;19:483. doi: 10.2307/1589073. [DOI] [PubMed] [Google Scholar]
- 77.Boss JH, Misselevich I. Osteonecrosis of the femoral head of laboratory animals: the lessons learned from a comparative study of osteonecrosis in man and experimental animals. Vet. Pathol. 2003;40:345–354. doi: 10.1354/vp.40-4-345. [DOI] [PubMed] [Google Scholar]
- 78.Ytrehus B, et al. Vascularisation and osteochondrosis of the epiphyseal growth cartilage of the distal femur in pigs—Development with age, growth rate, weight and joint shape. Bone. 2004;34:454–465. doi: 10.1016/j.bone.2003.07.011. [DOI] [PubMed] [Google Scholar]
- 79.Ytrehus B, Ekman S, Carlson CS, Teige J, Reinholt FP. Focal changes in blood supply during normal epiphyseal growth are central in the pathogenesis of osteochondrosis in pigs. Bone. 2004;35:1294–1306. doi: 10.1016/j.bone.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 80.Ytrehus B, Carlson CS, Ekman S. Etiology and pathogenesis of osteochondrosis. Vet. Pathol. 2007;44:429–448. doi: 10.1354/vp.44-4-429. [DOI] [PubMed] [Google Scholar]
- 81.Kitano T, Satou M, Saitou N. Evolution of two Rh blood group-related genes of the amphioxus species Branchiostoma floridae. Genes Genet. Syst. 2010;85:121–127. doi: 10.1266/ggs.85.121. [DOI] [PubMed] [Google Scholar]
- 82.Scheele CW, et al. Ascites and venous carbon dioxide tensions in juvenile chickens of highly selected genotypes and native strains. Worlds. Poult. Sci. J. 2005;61:113–129. doi: 10.1079/WPS200447. [DOI] [Google Scholar]
- 83.Navarro P, Visscher PM, Chatziplis D, Koerhuis ANM, Haley CS. Segregation analysis of blood oxygen saturation in broilers suggests a major gene influence on ascites. Br. Poult. Sci. 2006;47:671–684. doi: 10.1080/00071660601077931. [DOI] [PubMed] [Google Scholar]
- 84.Enkvetchakul B, Bottje W, Anthony N, Moore R, Huff W. Compromised antioxidant status associated with ascites in broilers. Poult. Sci. 1993;72:2272–2280. doi: 10.3382/ps.0722272. [DOI] [PubMed] [Google Scholar]
- 85.Daneshyar M, Kermanshahi H, Golian A. The effects of turmeric supplementation on antioxidant status, blood gas indices and mortality in broiler chickens with T 3 -induced ascites. Br. Poult. Sci. 2012;53:379–385. doi: 10.1080/00071668.2012.702340. [DOI] [PubMed] [Google Scholar]
- 86.GENECARDS. IFI6 Gene - GeneCards | IFI6 Protein | IFI6 Antibody. Accessed 31 Aug 2021. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=IFI6. (2019).
- 87.Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 88.Sato M, Taniguchi T, Tanaka N. The interferon system and interferon regulatory factor transcription factors - Studies from gene knockout mice. Cytokine Growth Factor Rev. 2001;12:133–142. doi: 10.1016/S1359-6101(00)00032-0. [DOI] [PubMed] [Google Scholar]
- 89.Parker, N. & Porter, A. C. G. Identification of a novel gene family that includes the interferon-inducible human genes 6-16 and ISG12. BMC Genom.5, 8 (2004). [DOI] [PMC free article] [PubMed]
- 90.Zhou J, Ye S, Fujiwara T, Manolagas SC, Zhao H. Steap4 plays a critical role in osteoclastogenesis in vitro by regulating cellular iron/reactive oxygen species (ROS) levels and cAMP response element-binding protein (CREB) activation. J. Biol. Chem. 2013;288:30064–30074. doi: 10.1074/jbc.M113.478750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hay, E. D. Extracellular Matrix. J. Cell Biol.91(3), 205s–223s . 10.1083/jcb.91.3.205s (1981). [DOI] [PMC free article] [PubMed]
- 92.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J. Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Daley WP, Peters SB, Larsen M. Extracellular matrix dynamics in development and regenerative medicine. J. Cell Sci. 2008;121:255–264. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 94.Cechowska-Pasko M. Decrease in the glycosaminoglycan content in the skin of diabetic rats. The role of IGF-I, IGF-binding proteins and proteolytic activity. Mol. Cell. Biochem. 1996;154:1–8. doi: 10.1007/BF00248454. [DOI] [PubMed] [Google Scholar]
- 95.Liu K, Wang K, Wang L, Zhou Z. Changes of lipid and bone metabolism in broilers with spontaneous femoral head necrosis. Poult. Sci. 2021;100:100808. doi: 10.1016/j.psj.2020.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weimer SL, et al. The utility of infrared thermography for evaluating lameness attributable to bacterial chondronecrosis with osteomyelitis. Poult. Sci. 2019;98:1575–1588. doi: 10.3382/ps/pey538. [DOI] [PubMed] [Google Scholar]
- 97.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria https://www.R-project.org/ (2020).
- 102.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 103.Ye J, et al. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hul, L. M. et al. Reference genes for proximal femoral epiphysiolysis expression studies in broilers cartilage. PLoS One, 15(8), e0238189 (2020). [DOI] [PMC free article] [PubMed]
- 106.Manly, B.F.J. Randomization, Bootstrap and Monte Carlo Methods in Biology (3rd ed.). Chapman and Hall/CRC (2007).
- 107.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 108.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Supek F, Bošnjak M, Kunca S, Muc S. Summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou G, et al. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. The fastq files used in this study were submitted to SRA database and will be available as BioProject number PRJNA350521.