Abstract
Homeoproteins are known to participate in development and cell type specification. The homeoproteins CCAAT displacement protein (CDP) and special AT-rich sequence binding protein 1 (SATB1) have been shown to bind to nuclear matrix-associated regions and to act as repressors of many cellular genes. Moreover, binding of SATB1 to the mouse mammary tumor virus (MMTV) promoter region dramatically affects the tissue-specific transcription of this retrovirus. Because protein-protein interactions are a common means of regulating homeoprotein function, we tested whether SATB1 and CDP interact in vivo and in vitro. SATB1 interacted with CDP through its DNA-binding domain, as demonstrated by glutathione S-transferase (GST) pull-down assays. GST pull-down assays also showed that CDP associated with SATB1 through three of its four DNA-binding domains (CR1, CR2, and the homeodomain). SATB1-specific antisera, but not preimmune sera, precipitated CDP from nuclear extracts, and CDP-specific antisera precipitated SATB1 from the same extracts. Far-Western blotting detected interaction of SATB1 and CDP in several different tissue extracts. Association of purified SATB1 and CDP in vitro resulted in the inability of each protein to bind to DNA in gel retardation assays. CDP overexpression in cultured T cells led to a loss of detectable SATB1 binding to the MMTV promoter region, as measured by gel shift experiments. CDP overexpression also elevated MMTV long terminal repeat reporter gene activity in transient-transfection assays, a result consistent with neutralization of the SATB1 repressor function in T cells. SATB1 is very abundant in certain tissues, particularly thymus, whereas CDP is relatively ubiquitous, except in certain terminally differentiated cell types. Because of the tissue and cell type distribution of SATB1 and CDP, we propose that the SATB1-to-CDP ratio in different tissues is a novel mechanism for homeoproteins to control gene expression and differentiation in mammals.
The tissue-specific regulation of transcription is critical to the development and function of all higher eukaryotic organisms. Tissue-specific transcription is controlled by the presence of factors, such as MyoD and NF-κB (29, 55), that bind DNA to activate or suppress the expression of a subset of genes in particular cell types. Because eukaryotic viruses contain relatively few genes, these viruses must rely heavily on processes operative in the cells they infect. Thus, viruses have offered numerous insights into cellular mechanisms that control transcription.
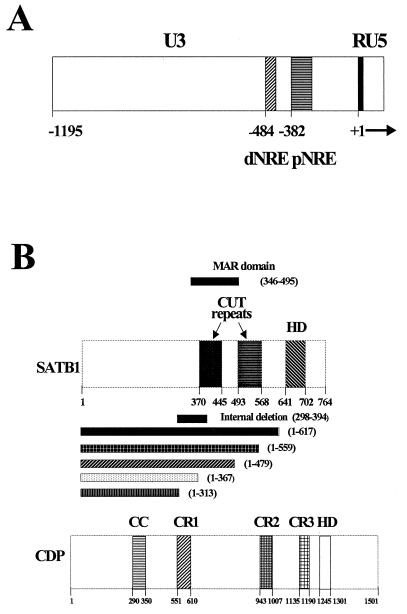
Members of our group previously have shown that negative regulation is an important component of the tissue-specific expression of mouse mammary tumor virus (MMTV), a retrovirus that is transmitted through the milk of infected mothers to offspring (9, 32). Milk-borne MMTV must replicate in gut-associated B and T cells prior to transmission to target cells in the mammary gland (6, 9, 17, 22, 48). High-level transcription and replication of MMTV in mammary tissue result in overexpression of cellular oncogenes near viral integration sites (42, 44). MMTV variants that cause leukemia, rather than mammary cancer, are expressed at much higher levels in T cells than are mammotropic MMTVs (9, 22, 48). Loss of transcriptional suppression by leukemogenic MMTV variants is due to deletion of negative regulatory elements (NREs), including a matrix-associated region (MAR) within the promoter-proximal NRE in the MMTV long terminal repeat (LTR) (22) (Fig. 1A). The protein complexes that bind to this MMTV MAR include NRE-binding protein (NBP) and upper binding protein (UBP) (9). The NBP and UBP complexes recently were shown to contain the homeodomain proteins special AT-rich sequence binding protein 1 (SATB1) and CCAAT displacement protein (CDP), respectively (5, 12, 13, 41). Mutation of the promoter-proximal SATB1 binding site dramatically increased MMTV LTR reporter gene expression in lymphoid tissues of transgenic mice and destabilized the binding of SATB1 to the MMTV LTR in vitro (32).
FIG. 1.
(A) The MMTV LTR, divided into U3, R, and U5 regions. The U3 region contains the transcription regulatory signals for the standard MMTV promoter that allows RNA initiation at the U3-R border. The approximate positions of the promoter-proximal (pNRE) and promoter-distal (dNRE) negative elements, as determined by reporter gene expression in transient-transfection assays, are shown. The probes used in this study include a 120-bp fragment that spans the pNRE and an oligonucleotide probe that includes an inverted repeat at the 3′ end of the 120-bp probe (see Materials and Methods). (B) Domain structures of SATB1 and CDP. The cut domains CR1, CR2, and CR3 and the homeodomain (HD) in CDP all can bind DNA independently, whereas the coiled-coil (CC) domain cannot (2, 18). An internal MAR domain, including the CR domains, contains the DNA-binding domain of SATB1 (40). Numbers indicate amino acid positions. CDP (the human protein) is known in other species as Cut (Drosophila), Clox (dogs), and Cux (mice) (1, 8, 53). In this paper, we use CDP to refer to proteins from all species.
In addition to their role in MMTV transcriptional regulation, SATB1 and CDP appear to control expression of many cellular genes. Binding sites for SATB1 and CDP are found in the MARs associated with the promoters or enhancers of at least two other genes, those coding for CD8α and immunoglobulin heavy chain, that show tissue-specific expression in T- or B-lymphoid cells (4, 54a). Binding of SATB1 and CDP to regulatory elements has been associated with transcriptional repression of numerous genes expressed in differentiated cells (4, 15, 21, 24, 32, 52, 53). Therefore, SATB1-mediated suppression of RNA synthesis from the MMTV LTR appears to be a good model for studies of transcriptional repression in differentiated tissues.
SATB1 is expressed predominantly in thymus but also in brain and several other organs (12, 13, 32). CDP expression is ubiquitous except in terminally differentiated cell types (5, 32, 41). MMTV transcription is highest in lactating mammary gland, a tissue that lacks SATB1 and CDP NRE-binding activity, whereas MMTV expression is diminished or undetectable in other tissues (32, 48). SATB1 and CDP are homeoproteins, and homeoprotein interaction with other proteins affects cell fate determination in Drosophila and mammals (45, 47, 57, 58). Therefore, we hypothesized that SATB1 could associate with CDP and that this association might affect the ability of these proteins to act as transcriptional suppressors. In this paper, we show that SATB1 and CDP interact and that this interaction eliminates the DNA-binding ability of both proteins.
MATERIALS AND METHODS
Cell culture and transient transfections.
The growth of Jurkat human T cells, LBB.A mouse B cells, and CCL-64 mink lung cells has been described previously (9, 32). Cells were passaged the day prior to transfection to achieve 95% confluence on the following day. SV40-CDP plasmid or plasmid lacking CDP sequences (pMT2) (30 μg) (30) and 5 μg of pBAG (46) were transfected transiently into Jurkat T cells. Cells were transfected with an electroporator (BTX, San Diego, Calif.) at 1,700 μF and 126 V in a 0.2-cm cuvette at a concentration of 2 × 106 cells/200 μl in RPMI medium containing 10% fetal bovine serum. Each electroporation was placed into a 100-mm-diameter tissue culture dish and incubated for 48 h prior to preparation of nuclear extracts for Western blotting and gel shift experiments. In a second set of experiments, very similar conditions were used, except the plasmid amounts were different, as follows: 15 μg of SV40-CDP or pMT2 vector, 20 μg of an MMTV LTR luciferase plasmid (pLC-LUC) (9), and 5 μg of pBAG. Transfections were normalized for DNA uptake by monitoring the β-galactosidase activity of the pBAG reporter plasmid (46) as previously described (9). A portion of the transfected cells was frozen and thawed three times, and the cell debris was removed by centrifugation at 10,000 × g for 10 min at 4°C.
Transfections also were performed in Jurkat cells by using SuperFect (Qiagen, Inc., Valencia, Calif.) according to the manufacturer’s instructions. Samples included 0.1 μg of pRL-TK (Promega Corporation, Madison, Wis.), 1 μg of pLC-LUC, and either 0.5 or 1 μg of SV40-CDP. All transfections were adjusted to a total of 2 μg of DNA with pMT2 control vector. Transfected cells were incubated for 48 h at 37°C and assayed for sea pansy (Renilla reniformis) luciferase and firefly luciferase by using a Dual Luciferase Reporter Assay System (Promega). Firefly luciferase activity from the MMTV LTR (pLC-LUC) was normalized for DNA uptake by using Renilla luciferase activity.
Nuclear extract preparation and gel shift assays.
Cell line and tissue nuclear extracts were prepared according to the method of Dignam et al. (14) with modifications as described by Liu et al. (32). The 120-bp proximal NRE probe from the C3H MMTV LTR (position +813 to +930 on the C3Hvx sequence of Brandt-Carlson et al. [10]) was obtained by PCR and cloned into pCRII (Invitrogen, San Diego, Calif.) as described by Liu et al. (32). The plasmid was digested with EcoRI, purified on polyacrylamide gels as described by Maxam and Gilbert (35), and end labeled with Sequenase (Amersham, Arlington Heights, Ill.). The 22-bp proximal NRE probe was made by the annealing of two oligonucleotides, 5′ gggGACTAATAGAACATTATTC 3′ and 5′ cccGAATAATGTTCTATTAGTC 3′, spanning the imperfect inverted repeat at the 3′ end of the 120-bp LTR sequence, followed by end labeling with Sequenase. Nucleotides in lowercase letters were added to facilitate labeling. The NF-κB probe was prepared by annealing the oligonucleotides 5′ AATTCAGGGGAATTCCCCTAAGCTTGAGCT 3′ and 5′ CAAGCTTAGGGGAATTCCCCTG 3′. Gel shift assays were performed essentially as described by Bramblett et al. (9) and modified by Liu et al. (32). Anti-SATB1 and anti-CDP polyclonal sera against recombinant proteins were prepared in rabbits as described previously (32). Antibody ablation assays were performed as described for gel shift assays, except that antibody (2 μl of 10-fold-diluted antiserum) was added to the reaction mixtures and incubated on ice for 10 min before addition of labeled probe.
Plasmid constructions.
Various glutathione S-transferase (GST) fusions of CDP have been described previously (2). Deletions of the full-length SATB1 cDNA in the pmAT vector (40) were constructed by digestion with Bal31 nuclease for various periods of time following linearization with HindIII. After repair of ends and digestion with MluI, appropriate DNA fragments were purified and truncated SATB1 cDNAs were substituted into the full-length pmAT vector that had been digested with MluI and EcoRV. The internal DNA-binding domain of SATB1 was removed by digestion with BspEI and MluI; the ends were repaired and ligated, yielding an in-frame deletion of 291 bp. Sequences corresponding to both C-terminal and internal SATB1 deletions in pmAT were digested with AvaI and HindIII, treated with Klenow enzyme, and ligated with pGEX-2T (Pharmacia, Uppsala, Sweden) that had been digested with EcoRI and treated with Klenow enzyme. All constructs were confirmed by sequencing.
Preparation of recombinant proteins.
Recombinant proteins were cleaved with thrombin (26) and purified according to standard methods (51). Briefly, bacterial cultures were induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h followed by pelleting and resuspension of cells in lysis buffer (1× phosphate-buffered saline [137 mM NaCl, 2 mM KCl, 8 mM Na2HPO4, and 1.7 mM KH2PO4], 1% Triton X-100, 100 mM EDTA, 10 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.32 μg of pepstatin A per ml, 10 μg of leupeptin per ml, and 2 μg of aprotinin per ml). Cells were lysed with an ultrasonic processor (model 501; PGC Scientific, Gaithersburg, Md.). The lysed cell suspension was centrifuged at 10,000 rpm for 5 min at 4°C in a Sorvall SS-34 rotor to remove insoluble material. The supernatant was mixed with a 50% slurry of glutathione-agarose beads (1 ml) (Sigma Chemical Co., St. Louis, Mo.), and the mixture was incubated at room temperature for 15 min on a rotating wheel. The suspension was centrifuged at 1,800 rpm (600 × g) in an IEC Centra-7R rotor for 1 min at 4°C. The beads were washed twice with wash buffer (1× phosphate-buffered saline, 1% Triton X-100, 10 mM DTT, 1 mM PMSF, 0.32 μg of pepstatin A per ml, 10 μg of leupeptin per ml, and 2 μg of aprotinin per ml), followed by one wash with cleavage buffer I (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10 mM DTT, 1 mM PMSF, 0.32 μg of pepstatin A per ml, 10 μg of leupeptin per ml, and 2 μg of aprotinin per ml). Cleavage buffer II (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 2.5 mM CaCl2) (1 ml) was used to resuspend the beads. Beads were collected by centrifugation at 10,000 rpm for 20 s (Fisher microcentrifuge) and then resuspended in 0.8 ml of cleavage buffer II. Thrombin (Sigma) (20 U) was added to the slurry, which was then incubated at room temperature for 1 h. The beads were collected by centrifugation at 10,000 rpm for 20 s and washed with cleavage buffer I (1 ml). An aliquot of the supernatant containing the purified protein was analyzed on a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel.
Affinity chromatography.
GST pull-down assays were performed essentially according to the method of Melcher and Johnston (36), with modifications. Briefly, the GST fusion protein was bound to glutathione-agarose beads (Sigma), and the bead-bound protein then was used to test for binding to in vitro-translated SATB1 or CDP. 35S-labeled proteins were prepared with a TNT-coupled in vitro transcription-translation system (Promega). Resin (20 μl) bearing equal amounts of either GST or the fusion proteins (ca. 5 μg) was incubated with 10 μl of labeled proteins in 300 μl of buffer D (14) containing 5 μg of bovine serum albumin for 2 h at 4°C on a rotating wheel. The resin was washed thrice in 1 ml of buffer D containing 0.5% Nonidet P-40 and then resuspended in SDS-polyacrylamide gel electrophoresis buffer (32). Samples were boiled for 5 min and then centrifuged at 10,000 × g for 30 s to remove the resin, and the supernatant was analyzed on SDS–10% polyacrylamide gels prior to autoradiography.
Immunoprecipitations and far-Western analysis.
Immunoprecipitation conditions were described previously (32). Immunoprecipitations also were performed in the presence of ethidium bromide as described by Lai and Herr (25). Far-Western blotting was performed essentially as described by Oliner et al. (43), with modifications. Briefly, proteins were separated on SDS–8% polyacrylamide gels, transferred to nitrocellulose membranes, and incubated for 1 h at room temperature in buffer D containing 5% nonfat dry milk. The blot then was incubated with 35S-labeled in vitro-translated protein in buffer D for 2 h at room temperature. The blot was washed thrice with 40 ml of buffer D containing 1% Nonidet P-40 for 30 min prior to autoradiography.
RESULTS
SATB1 and CDP homeoprotein interaction in GST pull-down experiments.
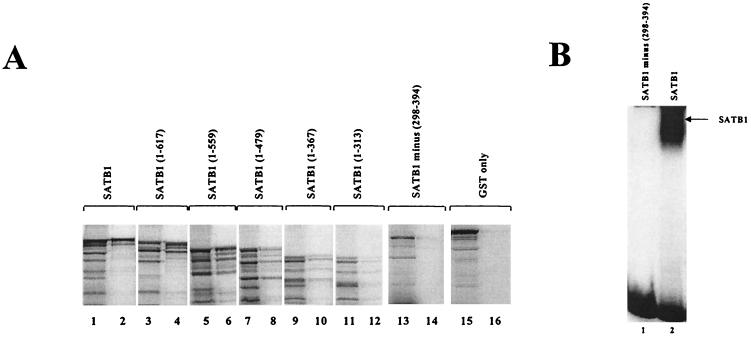
To test for SATB1-CDP association, a GST-CDP fusion protein containing the C-terminal 100 kDa of CDP [also called CDP (CR2-Cterm)] (30) (Fig. 1B) was bound to a column containing glutathione beads and then incubated with [35S]methionine-labeled SATB1 obtained by in vitro translation (36). Although translation yielded some truncated SATB1 proteins (Fig. 2A, lane 1), the full-length protein showed the highest level of GST-CDP binding (Fig. 2A, lane 2) and little binding to GST alone (Fig. 2A, lanes 15 and 16). Thus, the C-terminal two-thirds of CDP appears to be capable of specific SATB1 association, and it contains the cut domains (cut repeat 2 [CR2] and CR3) and the homeodomain that have been shown to bind DNA independently (2, 18). Subsequently, C-terminally truncated forms of SATB1 were translated in vitro for GST-CDP binding studies (Fig. 2A, lanes 3 to 12). SATB1-CDP association diminished with SATB1 mutants that lacked the cut domains and the homeodomain (Fig. 2A, lanes 7 to 12).
FIG. 2.
SATB1 and CDP interaction in vitro. (A) Mapping of the SATB1 region necessary for CDP association. GST-CDP (CR2-Cterm) was immobilized on glutathione-agarose beads and incubated with 35S-labeled in vitro-translated full-length SATB1 (lanes 1 and 2), SATB1 mutants with C-terminal truncations (lanes 3 to 12), an internal-deletion mutant (lanes 13 and 14), or GST alone (lanes 15 and 16). After affinity chromatography, bound proteins were resolved on SDS–10% polyacrylamide gels (even-numbered lanes) and compared with input labeled protein (odd-numbered lanes). (B) Lack of DNA binding of internally deleted SATB1. Wild-type SATB1 and an internal-deletion mutant were purified and used for gel shift assays with a labeled 120-bp proximal NRE probe prior to electrophoresis on a 4% nondenaturing polyacrylamide gel. The 120-bp NRE probe contains two binding sites for CDP and two binding sites for SATB1 (59). Numbers indicate amino acid positions.
An internal-deletion mutant of SATB1 was used to determine whether a SATB1 DNA-binding domain was essential for the interaction with CDP (Fig. 2A, lanes 13 and 14). No CDP interaction was detected with the internal-deletion mutant, suggesting that the amino acids between positions 298 and 394 are essential for SATB1-CDP association. Similar to results previously reported for SATB1 binding to the MAR of the immunoglobulin intronic enhancer (40), this internal-deletion mutant does not bind to DNA from the MMTV proximal NRE (Fig. 2B). SATB1 mutants with C-terminal truncations up to position 479 bound strongly to the MMTV NRE, but mutants with more N-terminal truncations did not (data not shown). These data suggest that the SATB1 DNA-binding domain is responsible for association with CDP.
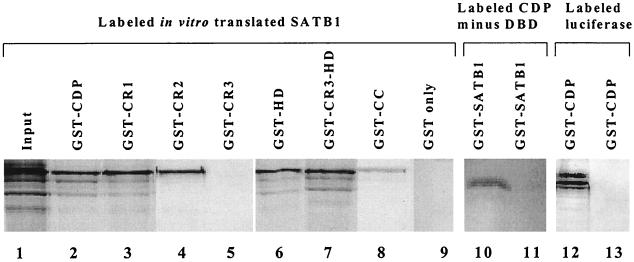
To determine the region of CDP necessary for association with SATB1, various truncated forms of CDP fused to GST were bound to glutathione beads and incubated with radiolabeled SATB1 (Fig. 3). These experiments showed that SATB1 bound to CR1, CR2, and the homeodomain (Fig. 3, lanes 3, 4, and 6), but not to CR3 (lane 5). The N-terminal region containing the coiled-coil domain (lane 8) and a spacer region between CR1 and CR2 (lane 11) bound poorly to labeled SATB1. As expected, SATB1 bound to a CDP fusion protein containing both CR3 and the homeodomain (Fig. 3, lane 7). In vitro-translated luciferase did not bind to GST-CDP (Fig. 3, lane 13). With the exception of CR3, it appears that each of the CDP DNA-binding domains is sufficient for binding to full-length SATB1. Together, these experiments indicate that CDP and SATB1 interact in vitro via their DNA-binding domains in the absence of DNA.
FIG. 3.
Binding of different CDP domains to SATB1. GST-CDP (CR2-Cterm), GST-CR1, GST-CR2, GST-CR3, GST-HD, GST-CR3 plus HD, GST-coiled-coil (GST-CC), or GST only (11) (lanes 2 to 9) were incubated with in vitro-translated labeled SATB1 (lane 1), as described for Fig. 2A. As controls, labeled in vitro-translated CDP containing the spacer region between CR1 and CR2 (lanes 10 and 11) or firefly luciferase (lanes 12 and 13) also was incubated with a GST–full-length SATB1 fusion (lane 11) or a GST-CDP (CR2-Cterm) fusion (lane 13). Lanes 1, 10, and 12 show the input radioactively labeled protein.
Detection of SATB1-CDP association by immunoprecipitation and far-Western analysis.
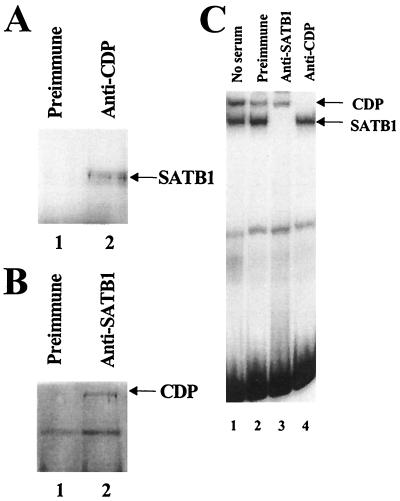
We also determined whether SATB1 and CDP were capable of association in vivo. Members of our group previously showed that Jurkat T cells have high levels of SATB1 and moderate levels of CDP (32). By using preimmune serum (Fig. 4A, lane 1) or polyclonal antibody directed against CDP (lane 2), whole-cell extracts of Jurkat cells were immunoprecipitated, and the immunoprecipitates were subjected to denaturing polyacrylamide electrophoresis and Western blotting. After incubation of blots with anti-SATB1, a 100-kDa band (consistent with the molecular weight of SATB1) was detected by Western analysis in immunoprecipitates with anti-CDP serum but not preimmune serum (Fig. 4A, lanes 1 and 2). Reciprocally, immunoprecipitates with anti-SATB1 serum but not preimmune serum contained a 180-kDa protein that could be detected with anti-CDP serum (Fig. 4B, lane 2). In addition, we performed this experiment in the presence of 200 or 400 μg of ethidium bromide per ml to eliminate the possibility that the CDP-SATB1 interaction was the result of contaminating DNA. As expected, the presence of ethidium bromide did not abolish the association of CDP with SATB1 (data not shown). Immunoprecipitation results also are not due to cross-reactivity of the antisera, since anti-SATB1 serum specifically abolished SATB1-DNA complexes and anti-CDP serum specifically abolished CDP-DNA complexes in gel shift assays (Fig. 4C, lanes 3 and 4).
FIG. 4.
SATB1 and CDP interact in vivo. (A) Coimmunoprecipitation of SATB1 with CDP-specific antibody. Jurkat T-cell lysate was incubated with an insoluble Staphylococcus aureus suspension to eliminate nonspecific binding prior to being immunoprecipitated with preimmune (lane 1) or anti-CDP (lane 2) serum. Immunoprecipitates were resolved on an SDS–8% polyacrylamide gel, transferred to nitrocellulose, and incubated with SATB1-specific serum, followed by development as suggested by the instructions with the Amersham ECL kit. (B) Coimmunoprecipitation of CDP with SATB1-specific antibody. Western blotting was performed as described for panel A, except that immunoprecipitations were performed with preimmune (lane 1) or anti-SATB1 (lane 2) serum prior to incubation of nitrocellulose transfers with anti-CDP. (C) Specificity of anti-SATB1 and anti-CDP sera. Mink lung cell nuclear extracts were incubated with no serum (lane 1), preimmune serum (lane 2), anti-SATB1 serum (lane 3), or anti-CDP serum (lane 4), followed by a gel shift assay with the 120-bp MMTV promoter-proximal NRE probe (32) (Fig. 1A). Reactions were analyzed on a 4% nondenaturing polyacrylamide gel. The detection of both SATB1 and CDP DNA-binding activities in mink lung extracts may indicate that certain modified forms of these proteins do not interact.
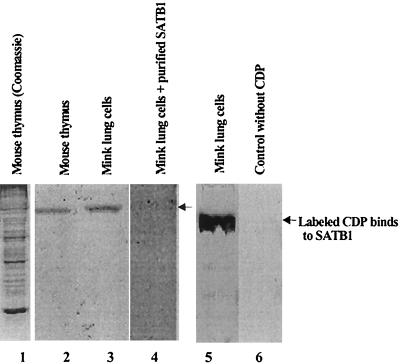
Further evidence for the association of SATB1 and CDP was obtained by far-Western analysis (43). Crude nuclear extracts from mouse thymus and mink lung cells previously were shown to have both CDP and SATB1 (32); extracted proteins were separated by SDS-polyacrylamide gel electrophoresis, blotted onto nitrocellulose membranes, and incubated with radiolabeled CDP. Labeled CDP bound to a 100-kDa protein in both mouse and mink cell nuclear extracts (Fig. 5, lanes 2, 3, and 5); CDP binding could be competed by a molar excess of unlabeled purified SATB1 (lane 4) and was dependent upon the presence of labeled CDP in the reaction (compare lanes 5 and 6). Together, these experiments indicated that CDP and SATB1 from different mammalian species form a complex in vivo and in vitro.
FIG. 5.
Far-Western blotting to detect SATB1-CDP association. Crude nuclear lysates from murine thymus (lanes 1 and 2) and mink lung cells (lanes 3 to 6) were prepared as described previously (32), resolved on SDS–8% polyacrylamide gels, and stained with Coomassie brilliant blue (lane 1) or electrotransferred to nitrocellulose (lanes 2 to 6). 35S-labeled in vitro-translated CDP was incubated in the absence of competitor protein (lanes 2, 3, and 5) or in the presence of purified recombinant SATB1 (lane 4). Mink lung cell proteins on the membrane also were incubated with an in vitro translation reaction mixture containing [35S]methionine, but no DNA template (lane 6). Lanes 1 to 4 are derived from a single gel; lanes 5 and 6 are from a second gel. The arrows show the positions of SATB1 protein detected by interaction with labeled CDP.
SATB1-CDP association abolishes the ability of both proteins to bind to DNA.
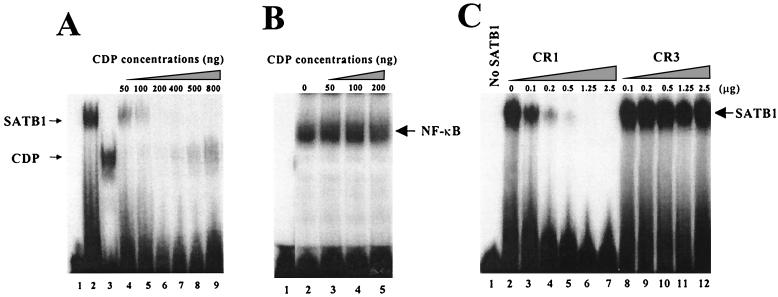
Because these data indicated that SATB1 and CDP interact via their DNA-binding domains (Fig. 2 and 3), we performed experiments to determine if this interaction affected the DNA-binding ability of these proteins. SATB1 and CDP purified from bacterial lysates were used in titration experiments to determine if formation of protein-protein complexes would prevent DNA binding. When either protein alone was added to a labeled probe with both SATB1 and CDP binding sites, distinct protein-DNA complexes were detected (Fig. 6A, lanes 2 and 3). However, addition of CDP to a constant amount of SATB1 prior to probe addition resulted in the loss of SATB1 DNA-binding ability (Fig. 6, lanes 4 to 9), and the ability of CDP to bind to DNA was not recovered until CDP was in molar excess (lane 9). Moreover, these data indicate that SATB1 and CDP do not form heteromeric complexes that bind to DNA. Control experiments indicated that CDP did not interfere with the ability of NF-κB to bind to DNA (Fig. 6B). These experiments support the idea that SATB1 associates with CDP to prevent DNA binding to the MMTV LTR.
FIG. 6.
Interaction of SATB1 and CDP interferes with the DNA-binding ability of both proteins. (A) Inhibition of DNA binding after SATB1-CDP association. N-terminally truncated SATB1 and CDP were purified from recombinant fusion proteins and used for gel shift assays with the 22-bp proximal NRE probe (32) prior to electrophoresis on 4% nondenaturing polyacrylamide gels. SATB1 (500 ng, or 5.6 pmol) (lane 2) was mixed with 50 to 800 ng (0.7 to 11 pmol) (lanes 4 to 9) of CDP before addition of the labeled probe. Lane 1 shows results achieved with no added protein, whereas lane 3 shows results for 50 ng of CDP alone. The estimated molecular masses of SATB1 and CDP were 90 and 70 kDa, respectively. The 22-bp NRE probe appears to have a single binding site for CDP and for SATB1 (59). A 12-h exposure of the autoradiogram is shown. (B) NF-κB DNA binding is unaffected by CDP. LBB.A B-cell (23) nuclear extracts (4 μg) were incubated with no CDP (lane 2) or 50 to 200 ng (0.7 to 2.8 pmol) of recombinant purified CDP (lanes 3 to 5) prior to addition of the labeled NF-κB probe from the interleukin-2 receptor promoter (32) and analysis as described above. (C) The CR1, but not CR3, domain of CDP interferes with SATB1 binding to DNA. Increasing amounts of purified CR1 (9 to 225 pmol) (lanes 2 to 7) or CR3 (5.6 to 140 pmol) (lanes 8 to 12) were added to 2.5 μg (28 pmol) of purified SATB1 and incubated with the 120-bp MMTV proximal NRE probe (32) prior to analysis on a native polyacrylamide gel. Lane 1 contains no recombinant protein. Because the individual CDP cut domains do not bind well to the 120-bp probe relative to SATB1 or CDP containing the homeodomain [CDP(CR2-Cterm)], binding of CR1 and CR3 alone is not detectable under these conditions (not shown). The calculated molecular masses of the purified CR1 and CR3 proteins were approximately 11 and 18 kDa, respectively. A 2-h exposure of the autoradiogram is shown.
Previous experiments indicated that SATB1 and CDP associate through specific DNA-binding domains to prevent binding to the MMTV NRE (Fig. 2, 3, and 6A). These results also indicated that the CR1 domain, but not the CR3 domain, of CDP bound to SATB1 in GST pull-down experiments. Thus, our data suggested that CDP CR1, but not CR3, would prevent SATB1 from binding to DNA. Gel retardation experiments with the 120-bp proximal NRE probe showed that CDP CR1 inhibited binding of SATB1 to the MMTV NRE but the CR3 domain of CDP did not (Fig. 6C). Based on the calculated molecular weights of SATB1 and CDP CR1, one or two CR1 molecules are sufficient to prevent binding of an SATB1 molecule to the NRE. These results support our prediction that association of SATB1 with specific CDP domains (Fig. 3) prevents SATB1 from binding to DNA.
Overexpression of CDP in transfection assays.
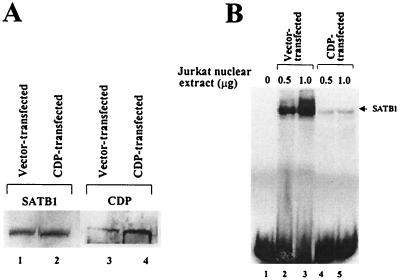
If SATB1 and CDP associate in vivo and this association abolishes their ability to bind to DNA, then overexpression of CDP relative to SATB1 should alter the DNA-binding activity observed in nuclear extracts. Jurkat T cells that contain a large amount of SATB1 DNA-binding activity relative to that of CDP (32) were transfected transiently with a CDP expression vector prior to preparation of nuclear extracts. Western blots of the extracts verified that CDP transfection increased CDP expression fourfold in Jurkat cells, whereas the total amount of SATB1 was unchanged (Fig. 7A). Gel shift assays with the same extracts showed that CDP overexpression greatly diminished the level of SATB1 binding to the MMTV proximal NRE sites relative to that observed from vector control-transfected cells, yet no CDP binding was detectable, despite the presence of CDP binding sites in the NRE (Fig. 7B). Together, these results indicated that the level of SATB1 binding to the MMTV LTR decreased when the intracellular level of CDP increased in T cells. These data support the idea that formation of CDP-SATB1 complexes via their DNA-binding domains prevents binding of both proteins to an MMTV regulatory region that contains a MAR and is involved in the tissue-specific regulation of transcription (32).
FIG. 7.
CDP overexpression leads to reduced SATB1 DNA binding. (A) Overexpression of CDP determined by Western blotting. Nuclear extracts were prepared 48 h following transfection and used for Western analysis with anti-SATB1 or -CDP serum. Equal amounts of extract were loaded in each lane, and the labeled bands were quantitated by densitometry. (B) CDP overexpression prevents SATB1 DNA binding to the MMTV LTR. The same nuclear extracts as used for panel A were used for gel shift assays with the labeled 120-bp proximal NRE probe (32). The upper band observed in lanes 2 and 3 does not contain CDP, as determined with anti-CDP serum in gel shift assays (not shown). This band may represent binding of two molecules of SATB1 to the 120-bp NRE probe.
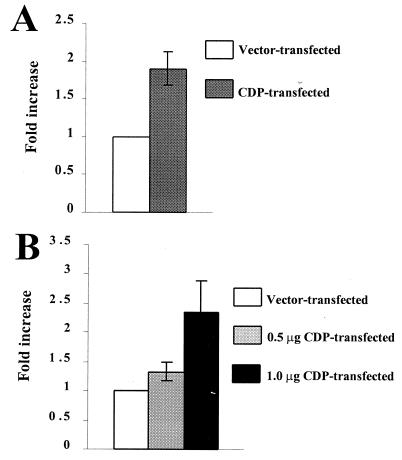
If the CDP-SATB1 interaction results in a loss of SATB1 DNA-binding activity for the MMTV NRE, then CDP overexpression in a cell line that shows SATB1 DNA-binding activity will titrate out the SATB1 DNA-binding and repressor functions. Thus, CDP overexpression in Jurkat T cells that express high levels of SATB1 should elevate expression of an MMTV LTR reporter gene construct (pLC-LUC). Jurkat cells were cotransfected with a CDP expression plasmid or a CDP-negative control vector and pLC-LUC by electroporation. A β-galactosidase expression plasmid also was included to normalize for DNA uptake (Fig. 8A). Compared to that in cells transfected with the CDP-negative vector, CDP overexpression in Jurkat cells increased MMTV LTR-directed luciferase activity approximately twofold. To ensure that this increase was not due to the method of DNA introduction or the expression plasmid used to control for DNA uptake, we also overexpressed CDP in Jurkat cells by using the SuperFect method (Fig. 8B). Again, this experiment showed that CDP overexpression resulted in a two- to threefold increase in luciferase expression and that this increase was proportional to the amount of CDP transfected. Therefore, these experimental results are consistent with the idea that association of SATB1 and CDP results in the mutual loss of their DNA-binding activities and that this loss of activity leads to inactivation of the SATB1 repressor function in T cells.
FIG. 8.
Overexpression of CDP in Jurkat cells elevates expression from the MMTV LTR. (A) Overexpression of CDP in Jurkat cells by electroporation. Transfections of the CDP expression vector (15 μg) were performed in triplicate, and results are expressed as an average increase over the value determined from the average of three determinations of the CDP-negative control vector (assigned a value of 1). Error bar indicates the standard deviation from the mean. (B) Overexpression of CDP in Jurkat cells by using the SuperFect transfection method. Values were calculated as described for panel A. Addition of more DNA (from any source) in the transfection assays resulted in lower reporter gene activity.
DISCUSSION
Interaction of SATB1 and CDP.
Previously, it was shown that SATB1 and CDP bind to NREs located upstream of the MMTV promoter (9, 32). These NREs were localized to a region that is deleted in MMTV variants that cause T-cell lymphomas rather than mammary tumors (3, 22, 27, 37). The promoter-proximal NRE appears to act as a MAR, and deletion of either the promoter-proximal or promoter-distal NREs elevated reporter gene expression from the MMTV LTR in tissues of transgenic mice other than the mammary gland (48). Further studies with transgenic mice showed that a substitution mutation in the proximal NRE destabilized SATB1 binding and elevated reporter gene expression in nonmammary tissues (32). Together, these experiments indicated that SATB1 binding to the proximal NRE is critical for transcriptional suppression of MMTV in nonmammary tissues. Loss of MMTV transcriptional suppression in T cells may lead to high levels of viral transcription and replication, resulting in the onset of T-cell lymphomas (9, 22).
Experiments presented here suggest that the availability of SATB1 for DNA binding to the MMTV LTR in different cell types is affected by binding to other factors, such as the homeoprotein CDP. GST pull-down assays showed that the DNA-binding domain of SATB1 is needed for interaction with CDP (Fig. 2). Similarly, three of the four DNA-binding domains of CDP (CR1, CR2, and the homeodomain) formed complexes with SATB1 (Fig. 3). Because the DNA-binding domains of these two proteins interact, perhaps it is not surprising that SATB1 and CDP association results in the failure of either protein to bind to the MMTV promoter (Fig. 6A). In agreement with the idea that SATB1-CDP interaction leads to a loss of DNA-binding ability, CR1, but not CR3, association with SATB1 abolished detectable binding of SATB1 to the MMTV proximal NRE (Fig. 6C). CR3 is the only known DNA-binding domain of CDP that does not associate with SATB1. Although the nature of the molecular interaction between CDP and SATB1 is unknown, the failure of the CDP CR3 domain to bind to SATB1 may be due to the presence of a predicted helical region found in CR1, CR2, and the homeodomain and the absence of this region in CR3 (as determined by using the GeneStream Align program).
Consequences of SATB1 association with CDP.
What are the functional consequences of SATB1-CDP interaction? Since association of SATB1 with CDP resulted in a loss of the DNA-binding activity of both proteins in vitro in gel retardation assays, we transfected a CDP expression vector into human Jurkat T cells that have a large amount of SATB1 relative to CDP (32). Such experiments showed that CDP overexpression resulted in decreased SATB1 binding to the MMTV proximal NRE without altering the amount of SATB1 present in nuclear extracts (Fig. 7). Thus, in vivo association of CDP and SATB1 appears to affect binding of these homeoproteins to DNA, and transcription of a variety of genes, including MMTV, may be altered by CDP-SATB1 interaction.
CDP or SATB1 overexpression in tissue culture cells leads to some cytotoxic effects (24, 30), and these effects may obscure the results of reporter gene assays. However, we were able to overexpress CDP approximately fourfold in Jurkat T cells (Fig. 7). Such experiments clearly showed that CDP overexpression in Jurkat cells resulted in a loss of SATB1 binding to the MMTV proximal NRE. This loss of NRE binding allowed approximately two- to threefold increases in reporter gene expression from the MMTV LTR in two types of transient-transfection assays (Fig. 8). Such small effects are typical of nuclear matrix-binding factors in transfection experiments (7, 16). Previous experiments showed that mutation of a SATB1-binding site in the MMTV NRE region leads to low-level effects on MMTV reporter gene expression in transient-transfection assays (9, 32). However, the same mutation leads to a dramatic elevation of MMTV expression in transgenic mice, particularly in lymphoid tissues relative to other tissue types (32).
SATB1 and CDP binding sites are located within the MARs of several genes, including those coding for CD8α and immunoglobulin heavy chain, that are expressed in lymphoid cells (4, 49, 54a). Both SATB1 and CDP have been implicated in the repression of genes expressed in highly differentiated cell types (4, 15, 21, 24, 32, 52, 53). Binding sites for SATB1 and CDP also are located within a MAR in the MMTV NRE, a region that is deleted in MMTV strains that exhibit high expression in T cells, leading to leukemia rather than mammary tumors (9, 22, 32).
How is the association between SATB1 and CDP different from interactions between other homeodomain-containing proteins? Clearly, homeoproteins have shown associations with numerous proteins, including members of the same homeoprotein family, members of different homeoprotein families, and nonhomeodomain proteins (58). An example of homeoprotein interactions between members of different homeoprotein families is the interaction of Hox and Pbx proteins (34), whereas association of Oct-1 and VP16 represent interactions between nonhomeoproteins (11). SATB1-CDP interactions appear to be associations between members of the same homeoprotein family, i.e., those containing CR domains. Unlike many homeoprotein interactions, however, the SATB1-CDP association can involve regions outside the homeodomains, most notably the CRs. SATB1 and CDP are unusual proteins in which the homeodomains are not required for DNA binding in vitro, although the contribution of the homeodomain to DNA binding appears to be greater for CDP than for SATB1 (2, 12). Similar to SATB1-CDP interaction, the association of murine Mlx and Dlx homeoproteins results in inactivation of their DNA-binding activity (58). However, the association involves the homeodomains of each protein and results in the loss of the Mlx repressor activity and the Dlx activator function (58). Thus, the CDP-SATB1 interaction is unique because it involves protein-protein associations outside the homeodomain and because these associations potentially would inactivate the repressor activity of both proteins.
Model for tissue-specific regulation by SATB1 and CDP.
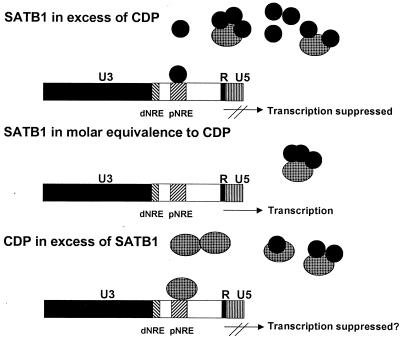
The homeodomain proteins SATB1 and CDP appear to regulate the expression of MMTV and other genes in highly differentiated cell types (32; also this study). We propose a model where the ratio of SATB1 to CDP determines whether gene transcription is suppressed, perhaps through modulation of protein factor binding to transcriptional control regions and MARs (Fig. 9). In this model, certain tissues, such as thymus, would have SATB1 in excess of CDP. If the DNA-binding ability of SATB1 is abolished by interaction of a single molecule of SATB1 with each of the CDP DNA-binding domains (CR1, CR2, and the homeodomain) (Fig. 6), then SATB1 repression would be extinguished in most tissues, except those that have very high SATB1 levels (e.g., T cells). Binding of available CDP to SATB1 would abolish the DNA-binding activity of both proteins, although any excess SATB1 would act as a repressor for the MMTV promoter. This model is supported by our transfection experiments in Jurkat T cells, in which we were able to titrate out the repressor activity of SATB1 by CDP overexpression. In other cell types, CDP may exceed the available SATB1. Preliminary data suggest that CDP also may act as a repressor of MMTV transcription (17a). If CDP is a repressor of MMTV expression, a high CDP/SATB1 ratio would suppress MMTV transcription. Alternatively, other cell types may have nearly equimolar amounts of SATB1 and CDP, and these cells may have little repressor activity for the MMTV promoter from either CDP or SATB1. B cells may represent such a cell type, since they are relatively permissive for MMTV expression (23, 50).
FIG. 9.
Model for SATB1-CDP interaction in different mouse tissues. The large cylinders represent CDP protein, whereas the smaller, dark cylinders represent SATB1. CDP may be inactivated for DNA binding by association with a single molecule of SATB1. Alternatively, binding of SATB1 to one of three possible sites within CR1, CR2, or the homeodomain may alter the specificity of CDP binding to different promoters. The large box represents the MMTV LTR that is divided into U3, R, and U5 domains. Transcription is initiated at the U3-R border of the 5′ LTR within the MMTV provirus. Upstream of the transcription initiation site are two regions termed the promoter-proximal and promoter-distal NREs (pNRE and dNRE, respectively). For simplicity, the binding of a single SATB1 or CDP molecule to the pNRE or dNRE is shown, although at least two binding sites each for SATB1 and CDP are located within the pNRE (59).
Members of our group have shown previously that various mouse tissues express CDP and SATB1 DNA-binding activity to different extents (32) and that the level of MMTV expression in these tissues differs from intermediate to undetectable (20, 48). Although there is not a perfect correlation between the level of SATB1 and CDP DNA-binding activity for the MMTV promoter and levels of MMTV transcription in different tissues, it is likely that tissue-specific levels of MMTV RNA represent the interplay between the absence of functional transcription suppressors and the presence of transcriptional activators. For example, lactating mammary gland is known to express the highest levels of MMTV RNA, and neither SATB1 nor CDP DNA-binding activity for the MMTV NRE is detectable in this tissue (32). However, MMTV transcription in lactating mammary tissue also appears to be regulated by positive factors that bind the mammary gland enhancer in the LTR (19, 28, 38, 39, 56).
Different SATB1-to-CDP ratios also may alter the activity or DNA sequence recognition by CDP. Because the CDP CRs and homeodomain each can bind DNA independently and with unique specificity, binding of SATB1 to a single CDP-binding domain may change CDP recognition of DNA target sequences (2, 18) or alter its interaction with other proteins, such as CDP alternatively spliced product or retinoblastoma-related factors (31, 54). Independent interactions of SATB1 with different CDP domains are indicated by experiments showing that CR1, but not CR3, neutralize SATB1 DNA binding (Fig. 6C). Thus, the ability of CDP and SATB1 to act as repressors depends upon the relative interaction of these homeoproteins and the requirement of these proteins for gene regulation in different tissues. Because CDP and its homologues each can serve in Drosophila sensory organ development (33), these data have broad implications for organismal development as well as for the cell-type-specific expression of many genes (including c-myc and c-mos, as well as those coding for globin, CD8, immunoglobulin, histones, and Ncam) regulated by SATB1 or CDP (4, 15, 21, 24, 49, 52, 53).
ACKNOWLEDGMENTS
We thank Henry Bose, Paul Gottlieb, Phil Tucker, Jim Bull, and Susan Ross for useful discussions and T. Kohwi-Shigematsu and H. Bose for reagents. We also thank Mary Lozano for help with transfection experiments.
This work was supported by NIH grants CA34780 to J.P.D. and DK01977 and HL49196 to E.J.N.
REFERENCES
- 1.Andres V, Nadal-Ginard B, Mahdavi V. Clox, a mammalian homeobox gene related to Drosophila cut, encodes DNA-binding regulatory proteins differentially expressed during development. Development. 1992;116:321–334. doi: 10.1242/dev.116.2.321. [DOI] [PubMed] [Google Scholar]
- 2.Aufiero B, Neufeld E J, Orkin S H. Sequence-specific DNA binding of individual cut repeats of the human CCAAT displacement/cut homeodomain protein. Proc Natl Acad Sci USA. 1994;91:7757–7761. doi: 10.1073/pnas.91.16.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball J K, Diggelmann H, Dekaban G A, Grossi G F, Semmler R, Waight P A, Fletcher R F. Alterations in the U3 region of the long terminal repeat of an infectious thymotropic type B retrovirus. J Virol. 1988;62:2985–2993. doi: 10.1128/jvi.62.8.2985-2993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banan M, Rojas I C, Lee W H, King H L, Harriss J V, Kobayashi R, Webb C F, Gottlieb P D. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8α gene. J Biol Chem. 1997;272:18440–18452. doi: 10.1074/jbc.272.29.18440. [DOI] [PubMed] [Google Scholar]
- 5.Barberis A, Superti-Furga G, Busslinger M. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell. 1987;50:347–359. doi: 10.1016/0092-8674(87)90489-2. [DOI] [PubMed] [Google Scholar]
- 6.Beutner U, Kraus E, Kitamura D, Rajewsky K, Huber B T. B cells are essential for murine mammary tumor virus transmission, but not for presentation of endogenous superantigens. J Exp Med. 1994;179:1457–1466. doi: 10.1084/jem.179.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 8.Bodmer R, Barbel S, Sheperd S, Jack J W, Jan L Y, Jan Y N. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell. 1987;51:293–307. doi: 10.1016/0092-8674(87)90156-5. [DOI] [PubMed] [Google Scholar]
- 9.Bramblett D, Hsu C-L L, Lozano M, Earnest K, Fabritius C, Dudley J. A redundant nuclear protein binding site contributes to negative regulation of the mouse mammary tumor virus long terminal repeat. J Virol. 1995;69:7868–7876. doi: 10.1128/jvi.69.12.7868-7876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandt-Carlson C, Butel J S, Wheeler D. Phylogenetic and structural analyses of MMTV LTR ORF sequences of exogenous and endogenous origins. Virology. 1993;193:171–185. doi: 10.1006/viro.1993.1113. [DOI] [PubMed] [Google Scholar]
- 11.Cleary M A, Stern S, Tanaka M, Herr W. Differential positive control by Oct-1 and Oct-2: activation of a transcriptionally silent motif through Oct-1 and VP16 corecruitment. Genes Dev. 1993;7:72–83. doi: 10.1101/gad.7.1.72. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson L A, Dickinson C D, Kohwi-Shigematsu T. An atypical homeodomain in SATB1 promotes specific recognition of the key structural element in a matrix attachment region. J Biol Chem. 1997;272:11463–11470. doi: 10.1074/jbc.272.17.11463. [DOI] [PubMed] [Google Scholar]
- 13.Dickinson L A, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 14.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 15.Dufort D, Nepveu A. The human Cut homeodomain protein represses transcription from the c-myc promoter. Mol Cell Biol. 1994;14:4251–4257. doi: 10.1128/mcb.14.6.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forrester W C, van Genderen C, Jenuwein T, Grosschedl R. Dependence of enhancer-mediated transcription of the immunoglobulin μ gene on nuclear matrix attachment regions. Science. 1994;265:1221–1225. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- 17.Golovkina T V, Chervonsky A, Dudley J P, Ross S R. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell. 1992;69:637–645. doi: 10.1016/0092-8674(92)90227-4. [DOI] [PubMed] [Google Scholar]
- 17a.Gregg, K., and J. Dudley. Unpublished data.
- 18.Harada R, Dufort D, Denis-Larose C, Nepveu A. Conserved cut repeats in the human cut homeodomain protein function as DNA binding domains. J Biol Chem. 1994;269:2062–2067. [PubMed] [Google Scholar]
- 19.Haraguchi S, Good R A, Engelman R W, Greene S, Day N K. Prolactin, epidermal growth factor or transforming growth factor-α activate a mammary cell-specific enhancer in mouse mammary tumor virus-long terminal repeat. Mol Cell Endocrinol. 1997;129:145–155. doi: 10.1016/s0303-7207(97)04053-7. [DOI] [PubMed] [Google Scholar]
- 20.Henrard D, Ross S R. Endogenous mouse mammary tumor virus is expressed in several organs in addition to the lactating mammary gland. J Virol. 1988;62:3046–3049. doi: 10.1128/jvi.62.8.3046-3049.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgy N A, Tarnasky H A, Valarche I, Nepveu A, van der Hoorn F A. Cux/CDP homeodomain protein binds to an enhancer in the rat c-mos locus and represses its activity. Biochim Biophys Acta. 1997;1351:313–324. doi: 10.1016/s0167-4781(96)00221-7. [DOI] [PubMed] [Google Scholar]
- 22.Hsu C-L L, Fabritius C, Dudley J. Mouse mammary tumor virus proviruses in T-cell lymphomas lack a negative regulatory element in the long terminal repeat. J Virol. 1988;62:4644–4652. doi: 10.1128/jvi.62.12.4644-4652.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King L B, Corley R B. Lipopolysaccharide and dexamethasone induce mouse mammary tumor proviral gene expression and differentiation in B lymphocytes through distinct regulatory pathways. Mol Cell Biol. 1990;10:4211–4220. doi: 10.1128/mcb.10.8.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohwi-Shigematsu T, Maass K, Bode J. A thymocyte factor SATB1 suppresses transcription of stably integrated matrix-attachment region-linked reporter genes. Biochemistry. 1997;36:12005–12010. doi: 10.1021/bi971444j. [DOI] [PubMed] [Google Scholar]
- 25.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavallie E R, McCoy J M, Smith D B, Riggs P. Enzymatic and chemical cleavage of fusion proteins. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 16.4.5–16.4.17. [DOI] [PubMed] [Google Scholar]
- 27.Lee W T, Prakash O, Klein D, Sarkar N H. Structural alterations in the long terminal repeat of an acquired mouse mammary tumor virus provirus in a T-cell leukemia of DBA/2 mice. Virology. 1987;159:39–48. doi: 10.1016/0042-6822(87)90345-x. [DOI] [PubMed] [Google Scholar]
- 28.Lefebvre P, Berard D S, Cordingley M G, Hager G L. Two regions of the mouse mammary tumor virus long terminal repeat regulate the activity of its promoter in mammary cell lines. Mol Cell Biol. 1991;11:2529–2537. doi: 10.1128/mcb.11.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenardo M J, Baltimore D. NF-κB: a pleiotropic mediator of inducible and tissue-specific gene control. Cell. 1989;58:227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- 30.Lievens P M, Donady J J, Tufarelli C, Neufeld E J. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J Biol Chem. 1995;270:12745–12750. doi: 10.1074/jbc.270.21.12745. [DOI] [PubMed] [Google Scholar]
- 31.Lievens P M, Tufarelli C, Donady J J, Stagg A, Neufeld E J. CASP, a novel, highly conserved alternative-splicing product of the CDP/cut/cux gene, lacks cut-repeat and homeo DNA-binding domains, and interacts with full-length CDP in vitro. Gene. 1997;197:73–81. doi: 10.1016/s0378-1119(97)00243-6. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross S R, Dudley J P. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol Cell Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludlow C, Choy R, Blochlinger K. Functional analysis of Drosophila and mammalian cut proteins in flies. Dev Biol. 1996;178:149–159. doi: 10.1006/dbio.1996.0205. [DOI] [PubMed] [Google Scholar]
- 34.Mann R S, Chan S K. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 35.Maxam A M, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melcher K, Johnston S A. GAL4 interacts with TATA-binding protein and coactivators. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michalides R, Wagenaar E. Site-specific rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in murine T-cell leukemias. Virology. 1986;154:76–84. doi: 10.1016/0042-6822(86)90431-9. [DOI] [PubMed] [Google Scholar]
- 38.Mink S, Härtig E, Jennewein P, Doppler W, Cato A C B. A mammary cell-specific enhancer in mouse mammary tumor virus DNA is composed of multiple regulatory elements including binding sites for CTF/NFI and a novel transcription factor, mammary cell-activating factor. Mol Cell Biol. 1992;12:4906–4918. doi: 10.1128/mcb.12.11.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mok E, Golovkina T V, Ross S R. A mouse mammary tumor virus mammary gland enhancer confers tissue-specific but not lactation-dependent expression in transgenic mice. J Virol. 1992;66:7529–7532. doi: 10.1128/jvi.66.12.7529-7532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakagomi K, Kohwi Y, Dickinson L A, Kohwi-Shigematsu T. A novel DNA-binding motif in the nuclear matrix attachment DNA-binding protein SATB1. Mol Cell Biol. 1994;14:1852–1860. doi: 10.1128/mcb.14.3.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neufeld E J, Skalnik D G, Lievens P M, Orkin S H. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet. 1992;1:50–55. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 42.Nusse R, Varmus H E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 43.Oliner J D, Andresen J M, Hansen S K, Zhou S, Tjian R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- 44.Peters G, Brookes S, Smith R, Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983;33:369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- 45.Pinsonneault J, Florence B, Vaessin H, McGinnis W. A model for extradenticle function as a switch that changes HOX proteins from repressors to activators. EMBO J. 1997;16:2032–2042. doi: 10.1093/emboj/16.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price J, Turner D, Cepko C. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc Natl Acad Sci USA. 1987;84:156–160. doi: 10.1073/pnas.84.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rieckhof G E, Casares F, Ryoo H D, Abu-Shaar M, Mann R S. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 48.Ross S R, Hsu C-L L, Choi Y, Mok E, Dudley J P. Negative regulation in correct tissue-specific expression of mouse mammary tumor virus in transgenic mice. Mol Cell Biol. 1990;10:5822–5829. doi: 10.1128/mcb.10.11.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheuermann R H, Chen U. A developmental-specific factor binds to suppressor sites flanking the immunoglobulin heavy-chain enhancer. Genes Dev. 1989;3:1255–1266. doi: 10.1101/gad.3.8.1255. [DOI] [PubMed] [Google Scholar]
- 50.Sharma S, King L B, Corley R B. Molecular events during B lymphocyte differentiation. Induction of endogenous mouse mammary tumor proviral envelope transcripts after B cell stimulation. J Immunol. 1988;141:2510–2518. [PubMed] [Google Scholar]
- 51.Smith D B. Purification of glutathione-S-transferase fusion proteins. Methods Mol Cell Biol. 1993;4:220–229. [Google Scholar]
- 52.Superti-Furga G, Barberis A, Schaffner G, Busslinger M. The −117 mutation in Greek HPFH affects the binding of three nuclear factors to the CCAAT region of the γ-globin gene. EMBO J. 1988;7:3099–3107. doi: 10.1002/j.1460-2075.1988.tb03176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valarche I, Tissier-Seta J P, Hirsch M R, Martinez S, Goridis C, Brunet J F. The mouse homeodomain protein Phox2 regulates Ncam promoter activity in concert with Cux/CDP and is a putative determinant of neurotransmitter phenotype. Development. 1993;119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- 54.van Wijnen A J, van Gurp M F, de Ridder M C, Tufarelli C, Last T J, Birnbaum M, Vaughan P S, Giordano A, Krek W, Neufeld E J, Stein J L, Stein G S. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc Natl Acad Sci USA. 1996;93:11516–11521. doi: 10.1073/pnas.93.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54a.Wang Z, Goldstein A, Zong R-T, Lin D, Neufeld E J, Scheuermann R H, Tucker P W. Cux/CDP homeoprotein is a component of NF-μNR and represses the immunoglobulin heavy chain introhic enhancer by antagonizing the Bright transcription activator. Mol Cell Biol. 1999;19:284–295. doi: 10.1128/mcb.19.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 56.Yanagawa S-I, Tanaka H, Ishimoto A. Identification of a novel mammary cell line-specific enhancer element in the long terminal repeat of mouse mammary tumor virus, which interacts with its hormone-responsive element. J Virol. 1991;65:526–531. doi: 10.1128/jvi.65.1.526-531.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zappavigna V, Sartori D, Mavilio F. Specificity of HOX protein function depends on DNA-protein and protein-protein interactions, both mediated by the homeo domain. Genes Dev. 1994;8:732–744. doi: 10.1101/gad.8.6.732. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen M M, Abate-Shen C. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, Q., and J. Dudley. Unpublished data.