Abstract
Background and objectives: One of the most important causes of urinary tract infections (UTI) is Escherichia coli. The infection is mainly due to the uropathogenic strain (UPEC), which has key virulence factors, including hemolysis. In this study, we evaluated the frequency of hlyA, hlyB, hlyC and hlyD genes in UPEC strains isolated from clinical samples from Shiraz city, Iran.
Materials and methods: 130 urine samples with suspected UTI were collected from Shiraz medical centers and cultured on blood agar and EMB media. Colonies were then characterized by biochemical methods. The genomes were extracted and the presence of hemolysis genes was detected by polymerase chain reaction (PCR) using hly gene specific primers and 16S rRNA. Drug resistance was assessed by using 10 antibiotic disks in the disk diffusion method, according to CLSI criteria.
Results: Out of the 130 collected UTI samples, 100 were identified as UPECs. Within isolates, the hlyD gene had the highest frequency – 95% – and hlyC had the lowest, with 23%. The frequencies of hlyA and hlyB genes were calculated as 50% and 43%, respectively. The rates of antibiotic resistance to Azithromycin, Ampicillin, Cefotaxime, Nalidixic Acid, Tetracycline, Trimethoprim-Sulfamethoxazole, Cefepime, Aztreonam, Gentamicin, and Nitrofurantoin were 95%, 86%, 68%, 66%, 65%, 64%, 51%, 46%, 44%, 14%, respectively. 98% of these isolates belonged to the MDR group.
Conclusion: This study shows diversity of hemolysis virulence factor in UPECs and unique UPEC drug resistance that would indicate a high antibiotic use in the general population.
Keywords: antibiotic resistance, hly gene, uropathogenic Escherichia coli, urinary tract infection, 16S rRNA
Zusammenfassung
Hintergrund und Zielsetzung: Eine der wichtigsten Ursachen für Harnweginfektionen (HWI) ist Escherichia coli. Die Infektion ist hauptsächlich auf den uropathogenen Stamm (UPEC) zurückzuführen, der wichtige Virulenzfaktoren einschließlich Hämolyse aufweist. In dieser Studie evaluieren wir die Häufigkeit der hlyA, hlyB, hlyC und hlyD Gene in UPEC Stämmen, die aus klinischen Proben in Shiraz isoliert wurden.
Material und Methode:130 HWI-verdächtige Urinproben wurden auf Blut Agar und EMB Medien kultiviert. Die Kolonien wurden dann mit biochemischen Methoden charakterisiert. Die Genome wurden extrahiert und das Vorhandensein von Hämolyse Genen wurde durch Polymerasekettenreaktion (PCR), hly Gene spezifischer Primer und 16S rRNA nachgewiesen. Die Arzneimittelresistenz wurde anhand von 10 Antibiotikaplättchen im Diffusionsverfahren gemäß den CLSI Kriterien bewertet.
Ergebnisse: Von den 130 HWI Proben wurden 100 als UPECs identifiziert. Innerhalb der Isolate hatte das hlyD Gen die höchste Häufigkeit von 95% und das hlyC die niedrigste mit 23%. Die Häufigkeit der hlyA und hlyB Genen wurde als 50% bzw. 43% berechnet. Die Raten der Antibiotikaresistenz gegen Azithromycin, Ampicillin, Cefotaxim, Nalidixinsäure, Tetracycline, Trimethoprim-Sulfamethoxazole, Cefepime, Aztreonam, Gentamicin und Nitrofurantoin betrugen 95%, 86%, 68%, 66%, 65%, 64%, 51%, 46%, 44% bzw. 14%. 98% dieser Isolation gehörten der MDR-Gruppe an.
Schlussfolgerungen: Die Studie zeigt die Vielfalt des Hämolysevirulenzfaktors in UPECs und die UPEC-Arzneimittelresistenz, die auf die häufige Antibiotikaanweneung in der Bevölkerung hinweist.
Introduction
Urinary tract infection (UTI) is one of the most prevalent bacterial infectious diseases, and is an important factor in the mortality of infants and the elderly [1]. Generally, UTIs are more common among females than males, mainly because of the proximity of the urinary tract and the anus in women. While the male urinary tract is longer than that of in the female and far from the anus, the prostatic fluids in males are also antimicrobially active [2]. Infections of the urinary tract are consistent with a variety of clinical symptoms, affecting the upper urinary tract (kidneys and ureter) and the lower urinary tract (bladder and urethra). It can also appear as symptomatic or non-symptomatic cystitis, pyelonephritis, and in severe cases urosepsis [3]. Escherichia coli, from the Enterobacteriaceae family, is one of the most common pathogens of the digestive system in human and domestic animals. As a commensal bacterium, it shows a strong association with its host, and it is well known that it causes diseases harshlyas a pote pathoge [4].
Among different strains of E. coli, only a few can cause urinary tract infections: these that are known as uropathogenic Escherichia coli (UPEC) [5]. To effectively reside in the host's urinary tract, uropathogenic E. coli strains express a set of virulence factors, including flagella to move the bacteria, adhesions in order to attach to the tissues and resist the flow of urine [6]. Also, a variety of toxins enable the bacteria to alter the host’s immune responses and escape them [7].
One of those toxins is the pore-forming toxin hemolysin, which belongs to the RTX family. As a result, there is a strong connection between increasing urinary tract infection and hlyA gene expression in UPEC strains [8].
UTIs have a huge impact on the economy and public health, and alter the quality of life of those who are affected [9].
Antimicrobial resistance is an important problem in global health care. After the advent of penicillin, which was a major breakthrough in antibacterial treatment, different bacteria have developed strong resistance to antibiotics. Bacteria acquire resistance and can transfer it to other species [10]. Increased use and misuse of antimicrobial agents are among the factors that have increased drug resistance. Furthermore, constant travel between countries plays a significant role in the multidrug-resistance (MDR) of many species [11]. MDR strains are increasing worldwide, due to the spread of genes located on mobile genetic elements such as plasmids, integrons, and transposons. The combination of these genes with chromosomally encoded resistance genes often leads to the development of bacterial resistance to the primary classes of antimicrobials [1], [4].
A few studies exist on virulence genes and antibiotic resistance among the UPEC strains causing UTI in Iran. Based on these, this study evaluated the association of UPEC isolates from Shiraz city, Iran, and determined the prevalence of hemolysis genes as well as their correlation with antibiotic resistance.
Materials and methods
In this descriptive cross-sectional study, a total of 130 urine samples suspected of UTI were obtained from Taghizadegan, Farhangiyan, and Farzanegan laboratories in Shiraz city over a period of six months from February to July, 2019.
Standard bacteriological and biochemical tests were done to identify and isolate the E. coli strains. The urine samples were cultured on eosin methylene blue (EMB) agar and blood agar and incubated at 37°C for 24 hours. The colonies with metallic-green color were selected for further phenotypic identification tests, including TSI, Citrate, SIM, Urease, MR-VP, LD and OD. Finally, the confirmed E. coli isolates were suspended in skim-milk media to be preserved for further experiments.
Antimicrobial susceptibility testing
The Kerby-Bauer disk diffusion method was carried out on Muller Hinton agar medium to determine the antibiotic susceptibility of the E. coli isolates to cefotaxime (CTX) (30 µg), nalidixic acid (NA) (30 µg), cefepime (FEP) (30 µg), tetracycline (TET) (30 µg), azithromycin (AZM) (15 µg), nitrofurantoin (FM) (300 µg), gentamicin (GM) (10 µg), aztreonam (AZ) (30 µg), ampicillin (AMP) (10 µg) and trimethoprim-sulfamethoxazole (SXT) (1.25/ 23.75 µg), according to the Clinical Laboratory Standard Institute Criteria [12]. The E. coli PTCC 1338 strain was used as quality control.
DNA extraction and detection of virulence factor
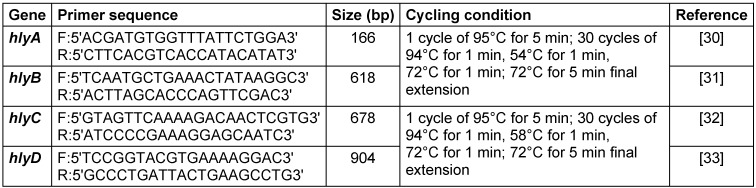
The genomic DNA and the hemolysis virulence genes were extracted from 100 UPEC isolates using a DNA extraction kit (CinnaGen Co., Iran). Extracted DNA was kept in skim milk between –50° C and –70° C until it was required for other tests. DNA was amplified using a thermal cycler (Eppendorf, Germany) in a total volume of 25 µL. The liquid mixture consisted of 4 µL DNA template, 12.5 µL Master Mix (2X), 2.5 µL of specific primers (10 pmol) and 6 µL distilled water. A standard polymerase chain reaction (PCR) was performed according to the manufacturer’s instructions (see Table 1 (Tab. 1)). The PCR products were analyzed by electrophoresis on 2% agarose gel along with the 100-bp DNA ladder as a marker. Gels were stained with ethidium bromide and detection of the amplified DNA was conducted using a UV transilluminator (Unico, China).
Table 1. Primers and PCR cycling conditions.
Statistical analysis
The statistical analysis was done using SPSS software version 20 (SPSS Inc., Chicago, IL, USA). The chi-squared test was used to assess the correlation between variables. A p-value <0.05 was considered significant.
Results
Frequency of virulence genes
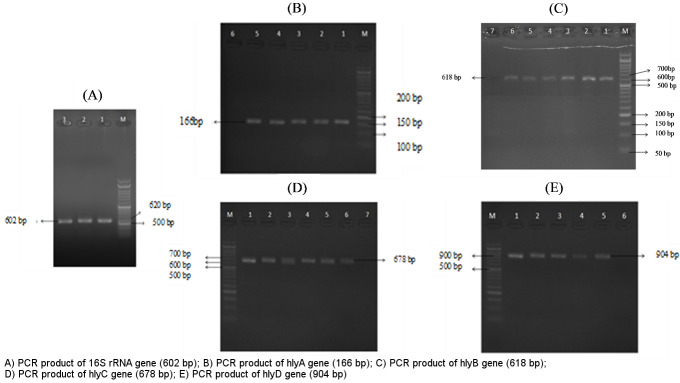
Approximately 100 (90.80%) E. coli were confirmed out of the 130 isolates using differential and biochemical tests and the PCR of specific 16S rRNA. They were proven by the appearance of a 620 bp band on 2% agarose gel.
Analysis of hemolysis genes showed that among 100 UPEC isolates, hlyD was the most frequent virulence gene, detected in 95% of the isolates, followed by hlyA with 50%, hlyB with 43%, and hlyC with 23% (Figure 1 (Fig. 1)). The PCR products were evaluated on 2% agarose gel.
Figure 1. Gel electrophoresis of PCR products.
In the phenotypic study of the hly genes, wide genetic diversity was observed and only 19% of the isolates had all the genes of the hly operon at the same time, which indicates that the isolates may have undergone a series of mutations.
The present study also evaluated the likelihood of the simultaneous presence of a gene pair. The chi-squared test indicates a statistically significant correlation between the simultaneous presence of hlyC and hlyD genes in study subjects (p<0.05).
Antibiotic susceptibility among UPEC isolates
In this study, the examined isolates showed more than 50% resistance to 70% of the tested antibiotics.
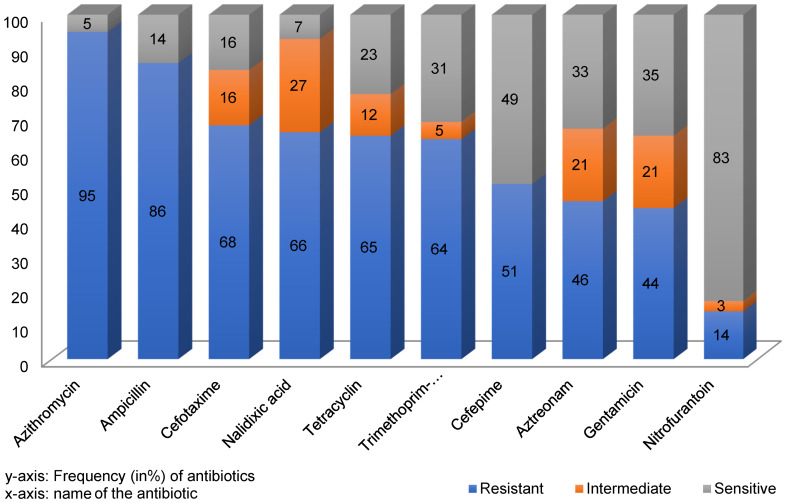
The results of the antibiotic susceptibility test showed that Azithromycin (95%) and Ampicillin (86%) had the highest resistance rates. The isolates had the lowest resistance to Gentamicin (44%) and Nitrofurantoin (14%). The prevalence of resistance against Cefotaxime, Nalidixic Acid, Tetracycline, Trimethoprim-Sulfamethoxazole, Cefepime, and Aztreonam was 68%, 66%, 65%, 64%, 51%, and 46%, respectively (Figure 2 (Fig. 2)).
Figure 2. Frequency of antibiotic resistance or susceptibility.
Based on the one sample T test, the study population became resistant to other antibiotics except Azteronam, gentamicin and nitrofurantine antibiotics and a significant relationship was formed between resistance and microbial population.
According to the chi-square test, in the study population, there is a significant relationship between resistance and the use of more than one antibiotic. The simultaneous use of these antibiotics or replacement of one for the others had no impact on the treatment result.
This study also shows some unique antibiotic resistance patterns in the studied population. For instance, eight strains demonstrated resistance to all antibiotics. Thus, this finding highlights the importance of knowing the patient’s specific infection, awareness of the diversity of antibiotic resistance and therefore the fact that the medication might not be effective against some infections yet very successful against others.
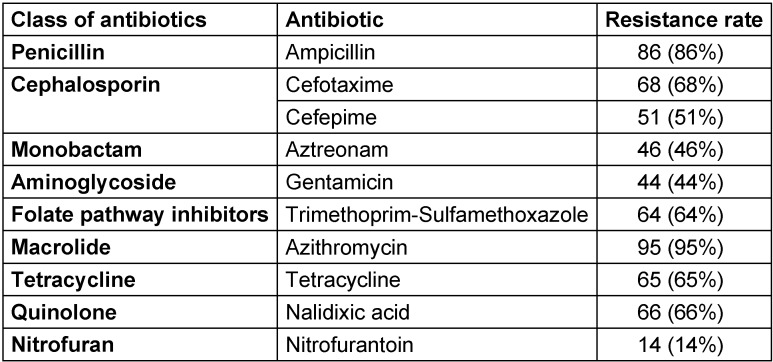
As shown in Table 2 (Tab. 2), 10 antibiotics from 9 classes of antibiotics were used and 98% of the isolates were verified to be multidrug-resistant (MDR).
Table 2. Frequency of antibiotic resistance to different antibiotic classes.
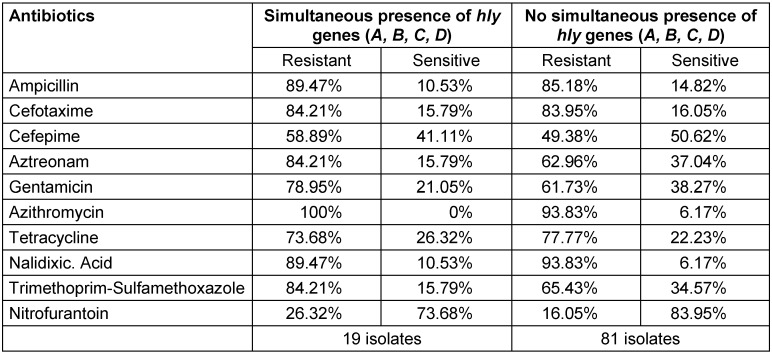
In the case of hemolysis genes, although the studied operons were important factors in the invasion of bacteria, our results (Table 3 (Tab. 3)) indicate no significant correlation between the presence or absence of these genes and antibiotic resistance.
Table 3. Comparison of the presence or absence of hly genes with antibiotic resistance.
Discussion
In humans, after respiratory tract infections, urinary tract infections are the second most important bacterial infection. In many cases, infection recurrence has been reported, which makes the treatment very difficult. UTI can spread and damage the parenchyma of the kidneys, which leads to kidney failure. Within the UTIs, different clinical steps and complications, such as the stages of diagnosis, management, side effects, and in some cases severity that can cause death, impose a heavy burden on society and health care systems. In addition to these, with increasing resistance to important antimicrobial agents, new pathogenic strains will emerge. These factors can lead to more expensive, resource-consuming ways of controlling these infections. Some studies showed that severe UTIs are due to the existence of a variety of pathogens. Therefore, studies targeting pathogens are of particular importance and might play a crucial role in the production and development of effective treatment or vaccines against such infections. Studying the pathogenic factors involved in UTIs highlights the role of uropathogenic Escherichia coli as one of the main bacteria causing UTIs. We may be able to provide solutions to this problem by studying patient-specific antibiotic resistance [2].
In the current study, 98% of the UPEC isolates were multidrug-resistant. This finding was consistent with those reported by Spindola et al. [13] and Rehman et al. [14], but differed from reports by Navidinia et al. (26%) [15].
The specific evaluations in Mostafavi’s study showed that resistance to cefotaxime and tetracycline were very high, with 100% and 80%, respectively, followed by 20/24% resistance to gentamycin, 0% to nitrofurantoin, and a 32/60% frequency of hlyA gene, so the reducing process of resistance to these antibiotics is similar to the results of this study [16].
According to studies by Navidinia [15] and Shabani [17] on UTI patients, the lowest resistances were to gentamicin and nitrofurantoin and the highest were to ampicillin and azithromycin, which is consistent with our study.
Comparison with other study populations indicates statistically different significantly results. For instance, studies performed on Indian [18] or Turkish subjects [19] showed the highest resistance was found to gentamicin and nitrofurantoin, while our study population demonstrated the lowest resistance to these two antibiotics. These comparisons emphasize the importance of geographical status and antibiotic regimens in different medical and health facilities.
In another study, Karami et al. [20] evaluated 205 Swedish one-year-olds and showed that the lowest resistance rates were to gentamicin and nitrofurantoin. Karimi et al. concluded that there was a significant increase in the resistance of uropathogenic Escherichia coli to antibiotics, which may be due to the transmission of resistance genes between strains of bacteria over the years. In their study population, they showed a 22% presence of the hlyA gene, which is different from our study population.
The simultaneous presence of invasive agents and antibiotic resistance could be involved in the pathogenicity, but the current study did not detect a significant correlation between these two factors in our subjects.
Comparing our results with other reports, a 19% presence of the hly gene was compatible with the research of Tarchouna et al. [21] and Staji et al. [22] on Tunisian subjects, but interestingly different from a study population from the northern part of Iran reported by Raespour et al. [23] (60%).
Paniagua-Cotreras et al. [24] concluded that the hlyA gene was present in 15.4% of the isolates, and most of these strains were resistant to ampicillin and cefotaxime, the results of which were quite similar to those obtained in this study. In contrast, Moustafa et al. [25] found 11% hlyA and 10% hlyB, 10% hlyC, and 10% hlyD genes in 85 stool samples from people with intestinal inflammation at San Diego University in the USA, which contradicts the results of this study.
In Romania, Cristea et al. [26] reported 47.52% antibiotic resistance to ampicillin and 41.16% to tetracycline. Those authors also found the frequency of hlyA and hlyD genes to be 12.45% and 44.34%, respectively, which did not agree with the results of our study population. On the other hand, similar to their study, this study showed that hlyD has a higher frequency than hlyA. Furthermore, in the Cristea et al. study, the MDR value was 35.19%, which is much lower than the MDR in our subjects, i.e., 98%.
Studies done by Ali et al. [27] and Ghosh et al. [28] in 2019 indicated that the frequencies of the hlyA gene in Pakistan and the hly gene in India were 12% and 10%, respectively, which were different from this study’s results. The resistance rate to cefotaxime, gentamycin, and nitrofurantoin were 100%, 50/57%, and 12/50% respectively, according to Ghosh et al. [28].
In Iraq, Mohammad et al. [29] reported a 100% frequency of the hlyA gene, which was much higher than the rate of that gene in the current study.
In our research, we could not find any similar study on Iranian subjects evaluating the four hly genes in UPEC isolates, so we cannot compare our study results with previous ones in similar populations.
Conclusions
Assessment of UPEC isolates in present study showed that out of 100 Escherichia coli isolates from patients suspected of UTI, 19% harbored the four evaluated hly genes. 81% of the samples had a hemolysin operon mutation, so that part of the hemolysin structure was lost due to the loss of stated genes. Nevertheless, there is no influence on the etiology of the disease, indicating the existence of other pathogens in UPECs. As the activity of hemolysin could cause destruction of kidney cells and nephropathogenicity, the observed 19% frequency of these four genes in UTI patients should raise the alarm and encourage molecular analysis of these genes in UTI patients to prevent very adverse outcomes, such as kidney failure in UTI patients.
Evaluation of the results for antibiotic resistance suggests that UTI strains become highly resistant to azithromycin and ampicillin, so patients and physicians should be much more careful in the usage or prescription of these antibiotics in UTIs. In UTIs, alternative antibiotics such as nitrofurantoin and gentamicin might be effective substitutes. It is essential to note that given the high resistance to important antibiotics and the occurrence of 98% multidrug-resistant strains reported here and elsewhere, it is even more important to focus on the existence of resistance genes and Escherichia coli’s proclivity for acquiring them through horizontal gene transfer (HGT).
Overall, the noted differences in antibiotic resistance and abundance of hemolysin genes may be due to the influence of routine treatment regimens in each population and the effect of environmental factors and the different pathogenic strains in different regions.
Notes
Competing interests
The authors declare that they have no competing interests. This article is an excerpt from a student’s thesis with code 1602922953320951397119700.
No human samples were used in this article by any of the authors directly.
We received the anonymous samples randomly from the laboratory.
Funding
Funds were provided by the student Ms. Heliyaneh Moeinizadeh.
Acknowledgement
We are highly grateful to the Cell Biotechnology Saba Arna knowledge-based company, Shiraz, Iran, for their support in this study.
This article is part of the MSc thesis by Ms. Heliyaneh Moeinizadeh under the supervision of Dr. M. Shaheli.
References
- 1.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015 May;13(5):269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kudinha T. The pathogenesis of Escherichia coli urinary tract infection. In: Samie A, editor. Escherichia coli-Recent Advances on Physiology, Pathogenesis and Biotechnological Applications. London: IntechOpen; 2017. pp. 45–61. [DOI] [Google Scholar]
- 3.Schwab S, Jobin K, Kurts C. Urinary tract infection: recent insight into the evolutionary arms race between uropathogenic Escherichia coli and our immune system. Nephrol Dial Transplant. 2017 Dec;32(12):1977–1983. doi: 10.1093/ndt/gfx022. [DOI] [PubMed] [Google Scholar]
- 4.Allocati N, Masulli M, Alexeyev MF, Di Ilio C. Escherichia coli in Europe: an overview. Int J Environ Res Public Health. 2013 Nov;10(12):6235–6254. doi: 10.3390/ijerph10126235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arabi S, Tohidi F, Naderi S. The common fimbarie genotyping in Uropathogenic Escherichia coli. Ann Biol Res. 2012;3:4951–4954. [Google Scholar]
- 6.Bower JM, Eto DS, Mulvey MA. Covert operations of uropathogenic Escherichia coli within the urinary tract. Traffic. 2005 Jan;6(1):18–31. doi: 10.1111/j.1600-0854.2004.00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhakal BK, Kulesus RR, Mulvey MA. Mechanisms and consequences of bladder cell invasion by uropathogenic Escherichia coli. Eur J Clin Invest. 2008 Oct;38 Suppl 2:2–11. doi: 10.1111/j.1365-2362.2008.01986.x. [DOI] [PubMed] [Google Scholar]
- 8.Ristow LC, Welch RA. Hemolysin of uropathogenic Escherichia coli: A cloak or a dagger? Biochim Biophys Acta. 2016 Mar;1858(3):538–545. doi: 10.1016/j.bbamem.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Kostakioti M, Hultgren SJ, Hadjifrangiskou M. Molecular blueprint of uropathogenic Escherichia coli virulence provides clues toward the development of anti-virulence therapeutics. Virulence. 2012 Nov;3(7):592–594. doi: 10.4161/viru.22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Baum H, Marre R. Antimicrobial resistance of Escherichia coli and therapeutic implications. Int J Med Microbiol. 2005 Oct;295(6-7):503–511. doi: 10.1016/j.ijmm.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004 Feb;2(2):123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 12.Humphries RM, Kircher S, Ferrell A, Krause KM, Malherbe R, Hsiung A, Burnham CA. The Continued Value of Disk Diffusion for Assessing Antimicrobial Susceptibility in Clinical Laboratories: Report from the Clinical and Laboratory Standards Institute Methods Development and Standardization Working Group. J Clin Microbiol. 2018 Aug;56(8) doi: 10.1128/JCM.00437-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spindola MG, Cunha MPV, Moreno LZ, Amigo CR, Silva APS, Parra BM, Poor AP, de Oliveira CH, Perez BP, Knöbl T, Moreno AM. Genetic diversity, virulence genotype and antimicrobial resistance of uropathogenic Escherichia coli (UPEC) isolated from sows. Vet Q. 2018 Dec;38(1):79–87. doi: 10.1080/01652176.2018.1519321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rehman MU, Zhang H, Iqbal MK, Mehmood K, Huang S, Nabi F, Luo H, Lan Y, Li J. Antibiotic resistance, serogroups, virulence genes, and phylogenetic groups of isolated from yaks with diarrhea in Qinghai Plateau, China. Gut Pathog. 2017;9:24. doi: 10.1186/s13099-017-0174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navidinia M, Najar PS, Fallah F, Bakhshi B, Adabian S, Alimehr S, Gholinejad Z. Distribution of the Pathogenicity Islands markers (PAIs) in uropathogenic E. coli isolated from children in Mofid children hospital. Arch Pediatr Infect Dis. 2013;1(2):75–79. doi: 10.5812/pedinfect.9083. [DOI] [Google Scholar]
- 16.Shokouhi Mostafavi SK, Najar-Peerayeh S, Mohabbati Mobarez A, Kardoust Parizi M. Serogroup distribution, diversity of exotoxin gene profiles, and phylogenetic grouping of CTX-M-1- producing uropathogenic Escherichia coli. Comp Immunol Microbiol Infect Dis. 2019 Aug;65:148–153. doi: 10.1016/j.cimid.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Shabani A, Amini A, Ebrahimzadeh Namvar A. Frequency of Necrotizing Factor Type 1 and Hemolysin Genes Among Escherichiacoli Strains Isolated from Hospitalized Patients of Rouhani Hospital in Babol, IRAN in 2017: A Short Report. J Rafsanjan Univ Med Sci. 2018;17(7):681–688. [Google Scholar]
- 18.Harwalkar A, Gupta S, Rao A, Srinivasa H. Lower prevalence of hlyD, papC and cnf-1 genes in ciprofloxacin-resistant uropathogenic Escherichia coli than their susceptible counterparts isolated from southern India. J Infect Public Health. 2014 Sep-Oct;7(5):413–419. doi: 10.1016/j.jiph.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Düzgün AÖ, Okumuş F, Saral A, Çiçek AÇ, Cinemre S. Determination of antibiotic resistance genes and virulence factors in Escherichia coli isolated from Turkish patients with urinary tract infection. Rev Soc Bras Med Trop. 2019 Jun;52:e20180499. doi: 10.1590/0037-8682-0499-2018. [DOI] [PubMed] [Google Scholar]
- 20.Karami N, Wold AE, Adlerberth I. Antibiotic resistance is linked to carriage of papC and iutA virulence genes and phylogenetic group D background in commensal and uropathogenic Escherichia coli from infants and young children. Eur J Clin Microbiol Infect Dis. 2017 Apr;36(4):721–729. doi: 10.1007/s10096-016-2854-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int J Infect Dis. 2013 Jun;17(6):e450–e453. doi: 10.1016/j.ijid.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 22.Staji H, Rassouli M, Jourablou S. Comparative virulotyping and phylogenomics of Escherichiacoli isolates from urine samples of men and women suffering urinary tract infections. Iran J Basic Med Sci. 2019;22(2):211–214. doi: 10.22038/ijbms.2018.28360.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raeispour M, Ranjbar R. Antibiotic resistance, virulence factors and genotyping of Uropathogenic strains. Antimicrob Resist Infect Control. 2018;7:118. doi: 10.1186/s13756-018-0411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paniagua-Contreras GL, Monroy-Pérez E, Rodríguez-Moctezuma JR, Domínguez-Trejo P, Vaca-Paniagua F, Vaca S. Virulence factors, antibiotic resistance phenotypes and O-serogroups of Escherichia coli strains isolated from community-acquired urinary tract infection patients in Mexico. J Microbiol Immunol Infect. 2017 Aug;50(4):478–485. doi: 10.1016/j.jmii.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Moustafa A, Li W, Anderson EL, Wong EHM, Dulai PS, Sandborn WJ, Biggs W, Yooseph S, Jones MB, Venter JC, Nelson KE, Chang JT, Telenti A, Boland BS. Genetic risk, dysbiosis, and treatment stratification using host genome and gut microbiome in inflammatory bowel disease. Clin Transl Gastroenterol. 2018 Jan;9(1):e132. doi: 10.1038/ctg.2017.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cristea VC, Gheorghe I, Czobor Barbu I, Popa LI, Ispas B, Grigore GA, Bucatariu I, Popa GL, Angelescu MC, Velican A, Marutescu L, Popa M, Chifiriuc MC, Popa IM. Snapshot of Phylogenetic Groups, Virulence, and Resistance Markers in Uropathogenic Strains Isolated from Outpatients with Urinary Tract Infections in Bucharest, Romania. Biomed Res Int. 2019;2019:5712371. doi: 10.1155/2019/5712371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ali I, Rafaque Z, Ahmed I, Tariq F, Graham SE, Salzman E, Foxman B, Dasti JI. Phylogeny, sequence-typing and virulence profile of uropathogenic Escherichia coli (UPEC) strains from Pakistan. BMC Infect Dis. 2019 Jul;19(1):620. doi: 10.1186/s12879-019-4258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh A, Mukherjee M. Incidence of multidrug resistance, pathogenicity island markers, and pathoadaptive FimH mutations in uropathogenic Escherichia coli isolated from asymptomatic hospitalized patients. Folia Microbiol (Praha) 2019 Jul;64(4):587–600. doi: 10.1007/s12223-019-00685-4. [DOI] [PubMed] [Google Scholar]
- 29.Mohammed GJ, Abdul-Razaq MS. Grouping and Revelation the significant Virulence genes of Escherichiacoli isolated from Patients with Urinary Tract Infections. Res J Pharm Technol. 2018;11(12):5483–5489. doi: 10.5958/0974-360X.2018.00999.X. [DOI] [Google Scholar]
- 30.Emara N, Saad S, El-shatter M. Molecular characteristics of E. coli contaminating from meat products. Benha Vet Med J. 2016;31(1):60–63. doi: 10.21608/bvmj.2016.31220. [DOI] [Google Scholar]
- 31.Cleven BE, Palka-Santini M, Gielen J, Meembor S, Krönke M, Krut O. Identification and characterization of bacterial pathogens causing bloodstream infections by DNA microarray. J Clin Microbiol. 2006 Jul;44(7):2389–2397. doi: 10.1128/JCM.02291-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgos YK, Pries K, Pestana de Castro AF, Beutin L. Characterization of the alpha-haemolysin determinant from the human enteropathogenic Escherichia coli O26 plasmid pEO5. FEMS Microbiol Lett. 2009 Mar;292(2):194–202. doi: 10.1111/j.1574-6968.2009.01496.x. [DOI] [PubMed] [Google Scholar]
- 33.Navidinia M, Peerayeh SN, Fallah F, Bakhshi B, Sajadinia RS. Phylogenetic grouping and pathotypic comparison of urine and fecal Escherichia coli isolates from children with urinary tract infection. Braz J Microbiol. 2014;45(2):509–514. doi: 10.1590/s1517-83822014000200019. [DOI] [PMC free article] [PubMed] [Google Scholar]