Abstract
SARS-CoV-2 nucleocapsid (N) protein undergoes RNA-induced phase separation (LLPS) and sequesters the host key stress granule (SG) proteins, Ras-GTPase-activating protein SH3-domain-binding protein 1 and 2 (G3BP1 and G3BP2) to inhibit SG formation. This will allow viral packaging and propagation in host cells. Based on a genomic-guided meta-analysis, here we identify upstream regulatory elements modulating the expression of G3BP1 and G3BP2 (collectively called G3BP1/2). Using this strategy, we have identified FOXA1, YY1, SYK, E2F-1, and TGFBR2 as activators and SIN3A, SRF, and AKT-1 as repressors of G3BP1/2 genes. Panels of the activators and repressors were then used to identify drugs that change their gene expression signatures. Two drugs, imatinib, and decitabine have been identified as putative modulators of G3BP1/2 genes and their regulators, suggesting their role as COVID-19 mitigation agents. Molecular docking analysis suggests that both drugs bind to G3BP1/2 with a much higher affinity than the SARS-CoV-2 N protein. This study reports imatinib and decitabine as candidate drugs against N protein and G3BP1/2 protein.
Keywords: SARS-CoV-2, Nucleocapsid protein, Stress granule, G3BP1/2, Gene set enrichment, Imatinib, Decitabine
1. Introduction
The recent outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become one of the greatest public health challenges in recent times, leading to unprecedented growth in research efforts to develop vaccines and therapeutics [1]. The novel SARS-CoV-2 is an enveloped, single-stranded, positive-sense ~30 kb RNA virus of the family Coronaviridae [1]. The SARS-CoV-2 membrane contains the spike (S) glycoprotein, a membrane (M) protein, the envelope (E) protein, and nucleocapsid (N) protein. The N protein encapsulates the viral genome and thereby protects it from the host cell environment [2]. In the infected cells, the N protein is produced at high levels and is necessary for virion assembly and replication [3], [4]. Moreover, it enhances the transcription efficiency of sub-genomic viral RNA, thus modulates host cell metabolism [2]. SARS-CoV-2 N protein is a 46 kDa RNA-binding protein, containing an N-terminal RNA-binding domain, and a C-terminal oligomerization domain. These two domains are connected by a linker region comprising a serine/arginine-rich (SR rich) domain (SRD) [5], [6], [7].
Recent studies showed that SARS-CoV-2 N protein undergoes RNA-induced phase separation [8], [9], [10] and this behavior is critical in viral genome packaging and virion assembly. SARS-CoV-2 N protein is involved in the formation of phase-separated RNA-protein granules [11], [12], [13]. Several of these proteins such as Ras-GTPase-activating protein SH3-domain-binding protein 1 and 2 (G3BP1and G3BP2) are comprised of disordered regions and form stress granule (SG) through liquid-liquid phase separation (LLPS) [14], [15]. SGs are normally thought to play an antiviral role and many viruses employ SGs to escape host responses through inhibiting the post-translational modifications [16], excluding the key SG components such as TIA-1 and G3BP [17], [18], and via the formation of stable viral ribonucleoprotein complexes with key SG proteins [19]. G3BP1 and G3BP2 are key nucleators of SG formation [14], [20], and overexpression of these proteins leads to SG formation even in the absence of stress [21].
Very recently, Syed Nabeel-Shah et al. [13] have shown that the SARS-CoV-2 N protein interacts and sequesters G3BP1 and decreases SG formation. In contrast, ectopic expression of G3BP1 attenuates this inhibition in N-expressing cells. Therefore, the upregulation of G3BP1/2 may have a protective effect on SARS-CoV-2 infections by decreasing the viral load.
To get more insights into the regulation of G3BP1/2 expression, we have performed gene set enrichment analysis (GSEA) to find out the upstream regulatory elements modulating the expression of G3BP1/2 genes. Drugs were then identified based on their ability to change the gene expression of the identified regulators of G3BP1/2 genes. Using this strategy, we have identified and evaluated imatinib and decitabine as the two drugs that could be repurposed to ameliorate the outcomes of COVID-19. Further, similar to our previous studies [22], [23], [24], we have utilized the network-based methods to examine the drug-gene interactions between drugs and human targets of SARS-CoV-2 proteins identified through three different proteomics-based studies [11], [12], [13].
The advantage of GSEA is that it provides a framework to investigate the gene expression alterations related to a disease that can be evident at the level of biological pathways or co-regulated gene sets, rather than individual genes. Thus, we checked here the expression changes of individual genes reported in different GEO studies.
2. Methodology
2.1. Gene set enrichment analyses (GSEA) analysis of G3BP1 and G3BP2
In this study, GSEA was carried out to identify the genes linked to G3BP1 and G3BP2 for the significant enrichment of different functional categories. Gene enrichment analysis offers information about the over-representation of a given gene in a particular pathway. GSEA was performed through the Enrichr bioinformatics platform (http://amp.pharm.mssm.edu/Enrichr), which allows the investigation of nearly 200,000 genes from more than 100 gene set libraries [25], [26]. To this end, GSEA was performed using G3BP1/2 genes as baits applied to the genomic databases about the functional, regulatory, and pathophysiological features associated with these genes. Enrichment analysis infers knowledge about gene (s) by comparing it to gene sets that have already been annotated and searching for overlap.
The transcription factors regulating the expression of G3BP1/2 and the kinases co-expressed with G3BP1/2 were identified and considered as regulators of G3BP1/2 genes. Drugs were then identified based on their effects on gene expression signatures of the regulators of G3BP1/2 genes. The screening for enrichment was based on the “combined score” which is a product of the log of the p-value obtained from the Fisher's exact test and the z-score, which is a deviation from the expected rank (i.e., combined score, c = log(p)*z). Next, the validation of the GSEA results was done by using the GEO2R tool of the NCBI and manual curations of the gene expression profiles of the Gene Expression Omnibus (GEO) database. GEO2R tool compares two or more groups of samples to identify the genes that are differentially expressed across experimental conditions. The tool in the background uses the limma package of R language to perform widely used statistical tests to identify the differentially expressed genes.
2.2. Drug-protein interaction network
Recently, many studies have focused on characterizing the host-virus interactome in SARS-CoV-2 infection. We have extracted the protein-protein interactions from three different studies [11], [12], [13] and identified 809 human proteins target of SARS-CoV-2 proteins. Further, the drug-protein interaction network of the predicted drugs with these 809 human proteins was constructed and studied using Cytoscape [27].
2.3. Molecular docking
Using a network-based approach, we have identified imatinib and decitabine as target molecules for G3BP1 and G3BP2 genes. The three-dimensional structures of G3BP1 (PDB Id: 4FCJ) and G3BP2 (PDB Id: 5DRV) along with the N-terminal domain of SARS-CoV-2 N protein (NTD, PDB Id: 6M3M) were retrieved from the protein data bank database [28]. The three-dimensional structures were energy minimized using the Swiss PDB Viewer (SPDBV) tool [29]. For protein-protein docking, the HDOCK tool was used [30]. HDOCK is based on a hybrid algorithm of template-based modeling and ab initio free docking. The mol file of imatinib and decitabine was retrieved from the DrugBank database [31]. The OpenBabel software [32] was used for converting the mol file into a PDB file having 3D structural coordinates of the drug molecules. For protein-ligand docking, AutoDock-Vina [33] was utilized. The site-specific docking was performed with a grid spacing of 1 Å, and exhaustiveness of 8. From the binding affinity of protein-ligand complexes, the best docking pose was generated and analysed using PyMOL (https://pymol.org/2/) and Discovery studio [34].
2.4. Binding free energy calculations
The molecular mechanics/generalized Born surface area (MM/GBSA) analysis was performed to calculate the binding free energy of the docked complexes. In the MM/GBSA calculation, the binding free energy of the protein-ligand complex is calculated using the following equations:
The binding free energy (∆G bind) is separated into different energy terms. The gas-phase interaction energy, ΔEgas represents the summations of internal energies (∆E int), electrostatic energies (∆E ELE), and van der Waal interactions (∆E VDW). The solvation free energy (∆Gsol) is represented by the polar (∆GGB) and non-polar (∆G Surf) energy terms.
2.5. Molecular dynamics simulation
All-atom MD simulations were performed using Gromacs v5.18.3 on the atomic coordinates of G3BP1 and G3BP1-drug complexes. The CHARMM27 force field was utilized and the TIP3P was used as a water model to solvate the systems [35]. The PRODRG server [36] was utilized to generate molecular topologies and coordinate files of drug-like molecules. The G3BP1 systems were placed in the octahedral simulation box with the water molecules filling around the system. The 0.15 M counter ions (Na+ and Cl−) were added to neutralize the system [37]. The energy minimization of the neutralized systems was achieved using the steepest descent and conjugate gradients (50,000 steps for each). NVT (constant volume) and NPT (constant pressure) for 500 ps were then used for the equilibration of the system. Berendsen weak coupling method [38] and Parrinello-Rahman barostat [39] methods were employed to maintain the temperature and pressure of G3BP1 systems at 300 K, and 1 bar, respectively. LINC algorithm was used to constrain the bonds and angles [40]. The van der Waals interactions were calculated by Lennard–Jones (LJ) potential with a cut-off of 0.10 nm. All MD simulation experiments were performed for 100 ns with a time interval of 10 ps. MD analysis of the different structural order parameters was performed using Gromacs utilities and python scripts with MDTraj [41].
2.5.1. Principal component analysis (PCA) and free-energy landscape (FEL) analysis
The principal component analysis (PCA) is performed to extract the information on the motion of the molecules during MD simulation which is associated with biological function. The first top two principal components (PC1and PC2) of PCA were investigated using the essential dynamics (ED) method by g_covar, g_anaeig modules. Next, PC1 and PC2 were selected as reaction coordinates for the calculation of free energy, Gα using:
where k is the Boltzmann constant, T is the simulation temperature. P(qα) represents the probability density function of reaction coordinates (PC1 and PC2). Pmax(q) represents the probability distribution of the most probable state.
3. Results
3.1. GSEA of genomic features associated with G3BP1 and G3BP2
We have used the Enrichr bioinformatics platform [26] to identify human genes involved in regulatory cross-talks and modulating the expression level of G3BP1 and G3BP2 genes, thus potentially affecting SG formation and SARS-CoV-2 infection.
Expression profiling of G3BP1/2 genes revealed ubiquitous patterns of expression across human tissues. G3BP1 is highly expressed in the lung as well as gastrointestinal and respiratory tract tissues. In contrast, G3BP2 is highly expressed in different regions of the brain (Supplementary Fig. S1). G3BP1 showed very low expression in brain and whole blood tissues whereas G3BP2 showed very low expression in whole blood, heart, and liver.
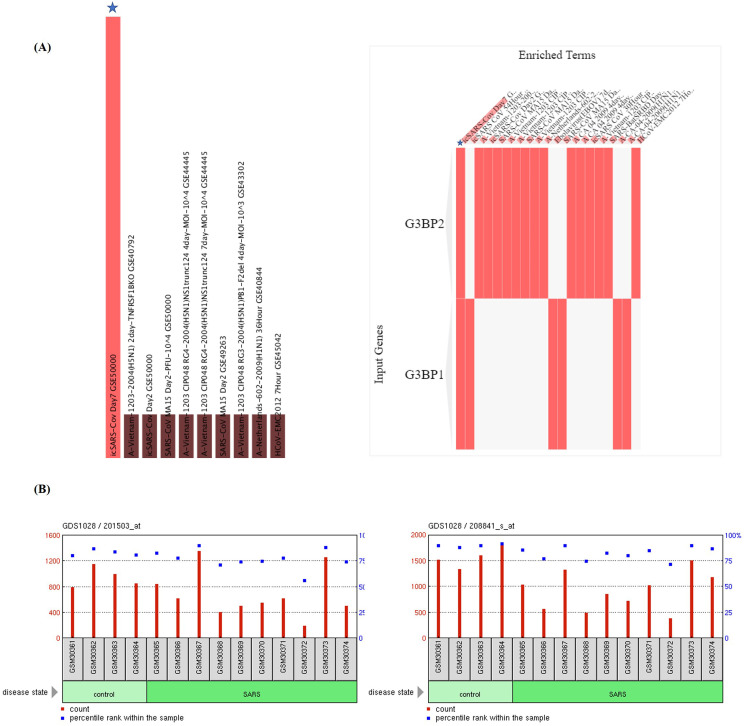
GSEA of the COVID-19 related gene sets revealed that both G3BP1 and G3BP2 belong to SARS-CoV-2 N protein host PPI from the Gordon study [42]. The different GEO records revealed that G3BP2 is a member of SARS-CoV-2 downregulated genes in Vero E6 cells (GSE153940) and it is also downregulated in SARS-CoV infection of Vero E6 cells (GSE30589). G3BP1 is downregulated by cystine and theanine in the IAV-infected mouse spleen gene set (Supplementary Table S1). The virus perturbations data sets among GEO records of downregulated genes in Enrichr identified SARS-Bat SRBD 48Hour (GSE47960) and SARS-Bat SRBD 84Hour (GSE47962) as the most significantly enriched record (Fig. 1A), marked by the downregulation of G3BP1/2 genes in human airway epithelial cells. These findings were substantiated by the decreased expression of G3BP1/2 genes reported in the peripheral blood mononuclear cells (PBMCs) of patients with SARS infection (GSE1739) (Fig. 1B and C) [43].
Fig. 1.
Effects of SARS-CoV-2 challenges on the expression of G3BP1 and G3BP2 genes. (A) GSEA of the virus perturbations from GEO focused on upregulated genes. Star in the figure represents the SARS-CoV-2 infection at 7 days for G3BP1/2 genes. GEO profiles of (B) G3BP1, and (C) G3BP2 expression in peripheral blood mononuclear cells (PBMCs) of patients with SARS.
GSEA identified lung injury as a common human disease perturbation of an enriched record through the downregulation of these genes (Supplementary Fig. S2), consistent with the clinical observations of comorbidity associated with COVID-19. The other significant diseases associated with the downregulation of these genes were pulmonary hypertension, amyotrophic lateral sclerosis, schizophrenia, and rheumatoid arthritis.
Gene Ontology (GO) analyses of G3BP1/2 revealed overlapping records of the biological process as small GTPase mediated signal transduction (GO:0007264) and Ras protein signal transduction (GO:0007265). The common significantly enriched records for the molecular process are mRNA binding (GO:0003729) and RNA binding (GO:0003723), whereas the common significantly enriched record for Jensen compartments is the intracellular ribonucleoprotein complex (Supplementary Fig. S3).
3.2. Identification of the transcription factor binding sites modulating the G3BP1/2 expression
Transcription factors (TFs) control the gene dynamics and expression and we, therefore, looked for TFs regulating G3BP1/2. GSEA of the enriched records of TF binding sites (TFBs) using ChEA 2016 and ENCODE TF ChIP-seq 2015 databases revealed common human TFBs shared by G3BP1/2 genes are FOXA1, CREB1, and SMAD4 according to ChEA 2016 and TAF1, SIN3A, CREB1, and NFYB according to ENCODE TF ChIP-seq 2015 (Supplementary Fig. S4). The GSEA analysis reported the differential regulation of these TFs in the SARS-CoV-2 gene sets available in the Enrichr platform (Supplementary Table S1). FOXA1 is downregulated by SARS-CoV-2 infection in mouse lung (GSE19137) and in lung alveolar A549 cells (GSE147507) while upregulated by SARS-CoV in intestinal organoids (GSE149312). CREB1 has also been downregulated by SARS-CoV-2 in intestinal organoids (GSE149312). TAF1 is downregulated by SARS-CoV-2 infection in lung tissue (GSE147507) and in intestinal organoids (GSE149312). SIN3A has also been shown to be downregulated upon SARS-CoV-2 infection in intestinal organoid (GSE149312) and upregulated by SARS-CoV-2 in A549 cells transduced with ACE2 (GSE147507). In contrast, NFYB is upregulated by SARS-CoV-2 infection in A549 cells (GSE147507). Moreover, the GEO records of SARS infected human peripheral blood mononuclear cells (GSE1739) indicate the mixed responses of FOXA1 and CREB1 expression while the expression of TAF1 and NFYB largely decreased during SARS-CoV infection (Supplementary Table S2).
We have also identified the TFBs in G3BP1/2 using the position weight matrices (PWMs) analysis from TRANSFAC and JASPAR. The common human binding motif site identified in the G3BP1/2 is the transcription factor, Yin Yang 1 (YY1) (Supplementary Fig. S5). YY1 is downregulated by SARS-CoV infection in bronchial epithelial 2B4 cells (GSE17400) and is upregulated by MERS-CoV infection in Calu-3 cells (GSE139516) (Supplemental Table S1). Interestingly, the expression of YY1 showed a mixed response in human PBMC samples infected with SARS-CoV (GSE1739) (Supplementary Table S2).
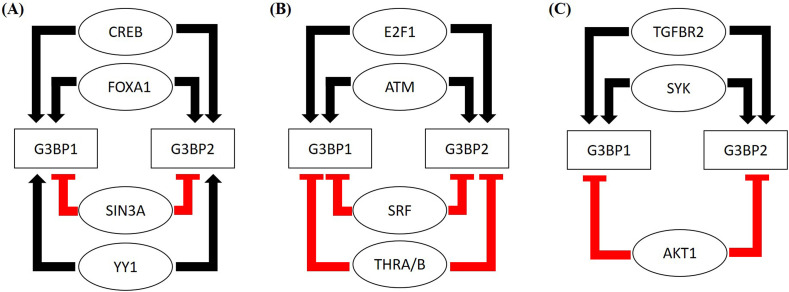
Next, we looked at the GEO database to find out the effects of these TFs on the expression profile of G3BP1/2 genes. The GEO records indicate that FOXA1 overexpression in the human prostate cancer cell line (LNCaP) increased the expression of G3BP1/2 genes, suggesting the activation effects of FOXA1. The expression of G3BP2 decreased due to CREB depletion and heterozygous CREB increased the expression of both G3BP1and G3BP2 genes, suggesting the activator role of CREB (Supplementary Table S2). SIN3A knockdown showed mixed response towards the expression of G3BP1, while SIN3A knockdown increased the expression of G3BP2 (Supplementary Table S2). Similarly, knockdown of transcription factor YY1 decreased the expression of G3BP2 in mouse skeletal muscle and HeLa cells (Supplementary Table S2), while YY1 depletion leads to a mixed response in G3BP1 expression, suggesting its role as an activator of G3BP2 expression while both as an activator and repressor for G3BP1 expression. The summary of these observations is reported in Fig. 2A.
Fig. 2.
Identification of Upstream regulatory elements and their potential mechanisms of affecting the expression of G3BP1 and G3BP2 genes. (A) The summarized figure represents FOXA1, CREB1, and YY1 as potential activators and Sin3A as repressors for the expression of G3BP1/2 genes, (B) overall summary of the effect of TFs on the expression of G3BP1/2 genes. Two TFs, ATM and E2F-1 upregulate whereas, SRF and THRA/B downregulate the G3BP1/2 expression and (C) effect of co-expressed kinases on the regulation of G3BP1/2 expression. TGFBR2 and SYK-1 activate the expression while AKT-1 inhibits the expression of G3BP1/2 genes.
3.3. Identification of TFs as putative modulators of the G3BP1/2 expression
The Enrichr results of the TF perturbations identified SRF (serum response factor) as potential repressors of G3BP1/2 expression and THRA/B mutants (thyroid hormone receptor A and B) as the potential activator. The Enrichr record showed that SRF knockout and THRA/B mutants upregulated the expression of G3BP1/2 genes, indicating their role as a repressor (Supplementary Fig. S6). These findings were further validated by analyzing the gene expression profiles reported in GEO records. Many GEO records showed that SRF depletion or null mutant increased the expression of G3BP1/2 in human prostate cancer cells, mouse liver, hematopoietic stem cells, and neonatal cardiomyocytes (Supplementary Table S2). Both THRA and THRB decreased G3BP1 protein expression and assembly in stress granules after arsenite stimulation [44]. Notably, THRA/B mutants increased the expression of G3BP1/2 expression in the human HepG3 cell line, suggesting the role of the THRA/B gene as possible inhibitors of G3BP1/2 (Supplementary Table S2).
Moreover, SRF is a member of upregulated genes by SARS-CoV-2 infection in Vero E6 cells (GSE153940) while THRA is a member of downregulated genes by SARS-COV-2 infection in Calu3 cells and A549 cells (GSE147507) and is upregulated in intestinal organoids (GSE149312). In contrast, THRB is a member of upregulated gene sets in COVID-19 (Supplemental Table S1). Also, the GEO gene perturbations database focused on down-regulated genes identified ATM (Ataxia-Telangiectasia-Mutated) and E2F-1 (E2F transcription factor 1) as putative activators and repressor of G3BP1/2, respectively (Supplementary Fig. S7). This is further validated by investigating the different GEO records of G3BP1/2 genes. ATM knockdown decreased G3BP1 and G3BP2 expression in HEK293 cells. E2F-1 upregulated the expression of G3BP1/2 genes in mouse, while both knockdown and overexpression of E2F-1 decrease the expression of G3BP1/2 genes (Supplementary Table S2). The summary of these observations is reported in Fig. 2B.
3.4. Identification of kinases as putative modulators of the G3BP1/2 expression
A recent study presents SARS-CoV-2 infection-induced global phosphorylation changes which lead to disruption of kinases and associated pathways [45]. Virus-host interactions could modulate the phosphorylation events by affecting the subcellular localization of host protein or by blocking access of kinases. The ARCHS4 kinases co-expression analysis indicates that kinases like MAPK6, PRKCI, EPHA1, LRRK1, CSNK2A, and NEK4 are co-expressed with both G3BP1 and G3BP2 genes (Supplementary Fig. S8). The kinases including MAPK6, EPHA1, and LRRK1 were found to be upregulated in SARS-CoV-2 infection, whereas CSNK2A and NEK4 were downregulated in SARS-CoV-2 infection. Next, the GSEA of the kinase perturbations from the GEO database focused on downregulated genes identified SYK (spleen tyrosine kinase) and TGFBR2 (TGF-beta receptor type II) as putative activators of the G3BP1/2 expression (Supplementary Fig. S8). Also, the kinase perturbations from the GEO database focused on upregulated genes revealed AKT1 (AKT serine/threonine kinase 1) as a potential repressor of G3BP1/2 genes (Supplementary Fig. S8). Moreover, Enrichr analysis of COVID-19 related gene sets indicates that SYK is a member of up-regulated genes by SARS-CoV-2 in pancreatic organoids. AKT1 is down-regulated in COVID-19 infected bronchoalveolar lavage, whereas TGFBR2 is both up-regulated and down-regulated by SARS-CoV-2 in Vero E6 cells and lung tissue, respectively (Supplementary Table S1).
The analysis of GEO records indicated that G3BP1/2 expression decreased in the presence of SYK deficient human breast epithelial cell line and the presence of SYK inhibitor in human B-cell lymphoma cell lines, indicating the activator role of SYK in G3BP1/2 expression. AKT repression increased the expression of G3BP1/2 genes in the human melanoma cell line and mouse heart, suggesting that it acts as a repressor. Also, TGFBR2 knockdown decreased the expression of G3BP1/2 genes in mouse embryonic palatal tissue and embryonic palatal mesenchymal cells, indicating the activator effect of G3BP1/2 (Supplementary Table S2). A summary of the results is provided in Fig. 2C.
3.5. GSEA identify imatinib and decitabine as potential drugs for mitigating SARS CoV-2 infection
Enrichr analysis of the drug perturbations increasing the expression of G3BP1/2 from the GEO signatures of differentially expressed genes and from GEO 2014 records revealed imatinib and decitabine as the top significantly enriched candidates (Supplementary Fig. S9). The search in GEO data sets suggested that both imatinib and decitabine exert biological activities that would alleviate the SARS-CoV-2 infection. Imatinib seems to both upregulate and downregulate G3BP1 and G3BP2 expression in different GEO records, while decitabine largely increased the expression of G3BP1/2 genes (Supplementary Table S2). Consistent with these findings, the investigation of GEO records indicated that imatinib appears to regulate different modulators of G3BP1/2 and thus affecting the SARS-CoV-2 infection. Imatinib appears to upregulate the TFs including FOXA1, CREB1, and YY1 in human K562 leukemia cell line, chronic myeloid leukemia (CML) cell lines, and in CML patients (Supplementary Table S2). Whereas, imatinib downregulated the expression of G3BP1/2 repressors, SRF, THRA/B, and AKT1 genes. Imatinib also decreased the expression of the repressor, SIN3A in CML patients and leukemia cell lines. The administration of imatinib also appears to increase the expression of ATM and E2F-1 and inhibit the expression of SRF and THRA/B. Moreover, imatinib increased the expression of SYK and TGFBR2 and inhibits AKT-1 in leukemia patients and cell lines (Supplementary Table S2). These results indicate the role of imatinib as a potential drug against COVID-19.
Similar to imatinib, decitabine decreased the expression of activators like CREB1 and YY1, as well as the repressors like SRF, SYK, and THRA/B in human B-lymphoma cell lines (Supplementary Table S2). Decitabine also increased the expression of other activators including ATM, E2F-1, SYK, and TGFBR2, and repressors including SIN3A and AKT-1 in B-lymphoma cell lines (Supplementary Table S2).
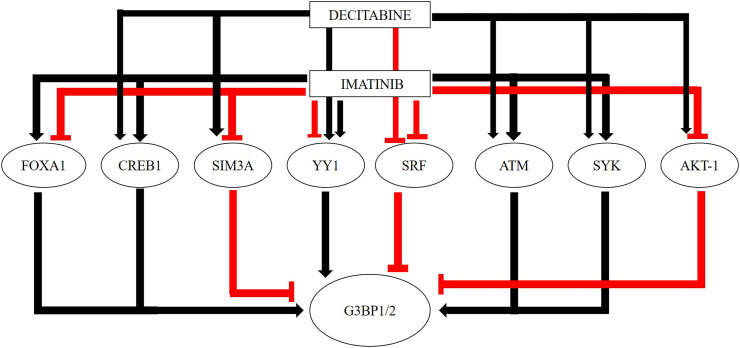
Fig. 3 and Table 1 summarizes the overall effects of imatinib and decitabine on the G3BP1/2 gene and their putative modifiers.
Fig. 3.
Overall summary of the effects of Imatinib and Decitabine on the G3BP1/2 and the potential modifiers of the G3BP1/2 genes.
Table 1.
Summary of the regulators of G3BP1 and G3BP2 and the effect of drugs, Imatinib and Decitabine on the putative G3BP1/2 regulators.
| Regulators of G3BP1/2 | Effect on G3BP1/2 | Imatinib | Decitabine |
|---|---|---|---|
| FOXA1 | Activator | ↑↓ | |
| CREB1 | Activator | ↑ | ↓ |
| SIN3A | Repressor | ↓ | ↑↓ |
| YY1 | Activator/Repressor | ↓↑ | ↓ |
| SRF | Repressor | ↓ | ↓ |
| THRA/B mutants | Repressor | ↓ | ↓ |
| ATM | Activator | ↑ | ↑ |
| E2F-1 | Activator | ↑ | ↑ |
| SYK | Activator | ↓↑ | ↑ |
| AKT-1 | Repressor | ↓ | ↓ |
| TGFBR2 | Activator | ↑ | ↑ |
3.6. Drug-gene interactions network of imatinib and decitabine in SARS-CoV2-human interactome
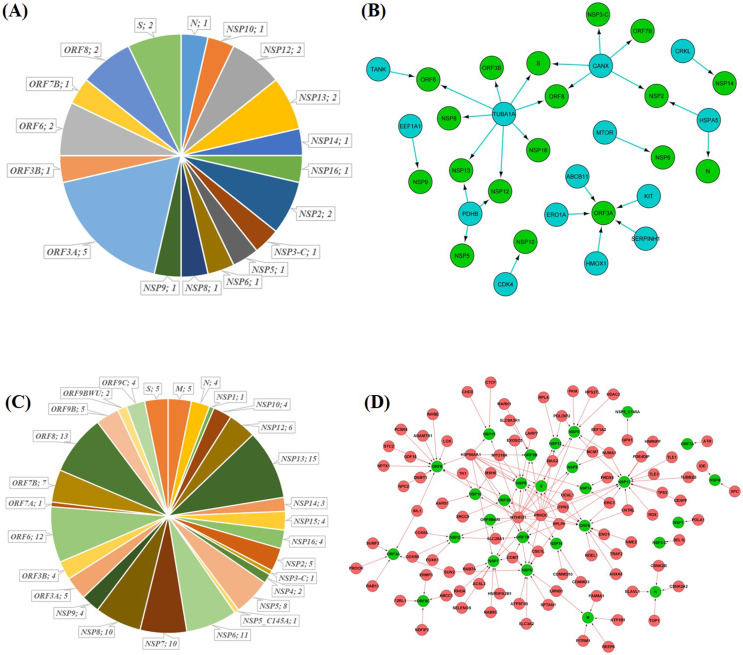
Many different SARS-CoV-2-host interactome studies have identified human proteins that could be considered as important drug targets to inhibit SARS-CoV-2 infection. The three excellent SARS-CoV-2-human interactome studies identified almost 809 high-confidence human proteins interacting with the 27 SARS-CoV-2 proteins [11], [12], [13]. Assuming that the drugs which significantly alter these protein-protein interactions will also inhibit the growth of the virus, we here investigated the drug-protein interactions. We found that out of the total 809 human proteins, imatinib interacts with a total of 14 (i.e., ~1.7%) proteins and potentially interacts with 18 of 27 (i.e., 66%) of SARS-CoV-2 proteins (Fig. 4A). These 14 proteins make at least 1–2 interactions with 18 SARS-CoV-2 proteins, making a total of 28 interactions (Fig. 4B). However, decitabine interacts with 92 (i.e., ~11%) proteins and interacts with 26 of 27 (i.e., 96%) SARS-CoV-2 proteins (Fig. 4C). These 92 proteins make 156 interactions with 26 SARS-CoV-2 proteins with maximum interactions to NSP13 (15), ORF8 (13), ORF 6 (12), NSP6 (11), NSP7, and NSP8 (10) protein (Fig. 4D). Thus, decitabine and imatinib manifest significant interference with the SARS-CoV-2-human interactome. Interestingly, a combination of both decitabine and imatinib interacts with 106 of 809 (13%) human proteins prey of SARS-CoV-2, making a total of 184 interactions with all 27 SARS-CoV-2 proteins (Supplementary Fig. S10).
Fig. 4.
Effects of imatinib and decitabine on the gene-drug interactions network of SARS-CoV-2-human interactome. (A) Imatinib interacts with 14 (i.e. ~1.7%) proteins, making 28 interactions in total, thus potentially interfering with functions of 18 of 27 (i.e. 66%) of SARs-Cov2 viral protein in human cells. (B) Interaction network of imatinib and SARS-CoV-2 viral proteins. The total number of interactions shown was 28. The green color represents viral proteins and the blue color represents human proteins targets of SARS-CoV-2 interacting with imatinib. (C) Out of 809 SARS-CoV-2 human target proteins, decitabine interacts with 92 (i.e. ~11%) proteins, making 156 interactions in total, thus potentially interfering 26 of 27 (i.e. 96%) of SARS-Cov2 viral protein in human cells. (D) Interaction network of the 92 decitabine target human proteins with 26 SARS-CoV-2 proteins. The total number of interactions shown was 156. The green color represents viral proteins and the red color represents decitabine targeted human genes.
3.7. Identification of potential miRNAs involved in the regulation of G3BP1 and G3BP2
Micro-RNAs (miRNAs) are small non-coding RNAs that can bind to 3'UTR of mRNA and inhibit translation or induce mRNAs degradation [46]. Many studies showed that host cellular miRNAs can target both the coding region and 3'UTR of the viral genome to induce an antiviral effect [47], [48]. To identify potential miRNAs that can directly regulate G3BP1 and G3BP2, TargetScan microRNA 2017 and miRTarBase 2017 from the Enrichr platform have been explored. The analysis revealed two common miRNAs, hsa-miR-3925-5p, and hsa-miR-486-5p against the G3BP1/2 genes (Supplementary Fig. S11). A recent study reported the presence of hsa-miR-3925-5p in different COVID-19 isolates [49]. Another study identified hsa-miR-486-5p as a potential antiviral against the influenza A virus [50].
3.8. Molecular docking of imatinib and decitabine to G3BP1/2 and SARS-CoV-2 N protein
Molecular docking studies are largely utilized to explore the binding modes of receptor and ligand molecules and are generally used in drug discovery. Through the molecular docking studies, we here determined the binding of imatinib and decitabine to G3BP1 and G3BP2 genes. The proposed modes of drug binding are presented in Fig. 5, Fig. 6 , and the docking results based on the binding affinity are represented in Table 2 .
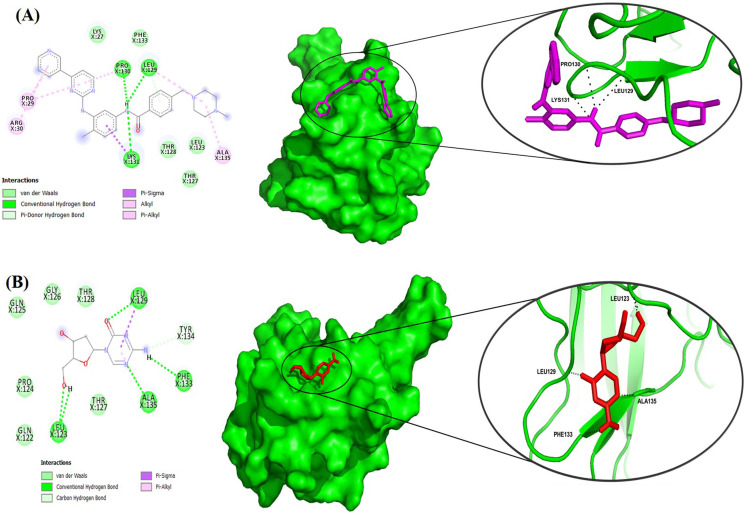
Fig. 5.
Molecular docking interactions of imatinib and decitabine to SARS-CoV-2 NTD. (A) Two-dimensional (2D) representations of NTD-imatinib interactions using Ligplot+ are shown in the left panel. The information about the residues and interactions has been indicated in the figure. The potential binding poses in the 3-D structure of the protein are shown in the right panel. The intermolecular hydrogen bonds are shown as a black dotted line. (B) Similarly, 2D diagrams of NTD-decitabine interactions and binding poses in the 3-D structure are shown in the left and right panels, respectively.
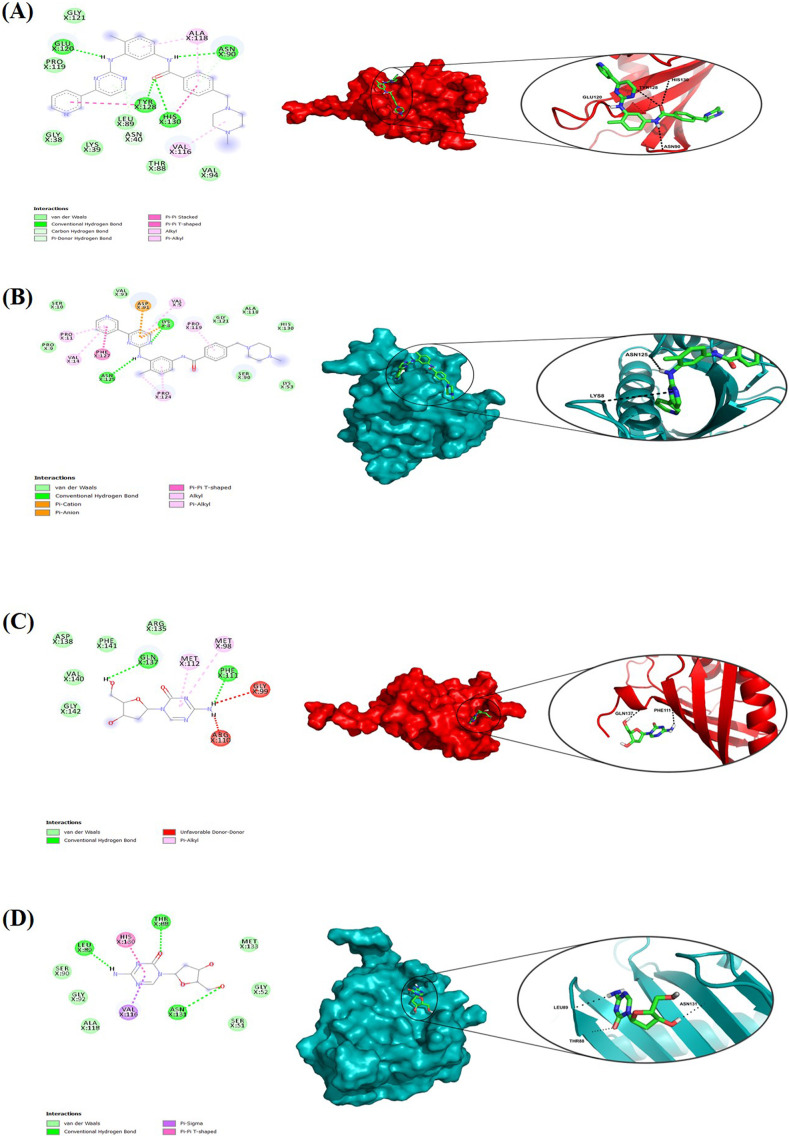
Fig. 6.
Molecular docking interactions of G3BP1 and G3BP2 genes to imatinib and decitabine. Two-dimensional (2D) diagrams and the binding pose of (A) G3BP1- imatinib, (B) G3BP2-imatinib, (C) G3BP1-decitabine, and (D) G3BP2-decitabine interactions using Ligplot+. The information about the residues and interactions has been indicated in the figure. The potential binding poses in the 3-D structure of the protein are shown in the right panel.
Table 2.
Molecular docking results of imatinib and decitabine binding to SARS-CoV-2 NTD and G3BP1/2 genes. The binding affinity has been calculated from three different docking software.
| Protein-drug | Binding energy (kcal/mol) | H-bond | Vander Waal bond | Other bonds |
|---|---|---|---|---|
| NTD-imatinib | −7.2 | Leu129, Pro130, Lys131 | Leu27, Leu123, Thr127, Thr128, Phe133 | Pro29, Arg30, Ala135 |
| NTD-decitabine | −4.7 | Leu123, Leu129, Phe133, Ala135 | Gln122, Pro124, Gln125, Gly126, Thr127, Thr128 | Tyr134 |
| G3BP1-imatinib | −8.7 | Asn90, Glu120, Tyr128, His130 | Gly38, Lys39, Thr88, Leu89, Val94, Pro119, Gly121 | Asn40, Val116, Ala118 |
| G3BP2-imatinib | −7.7 | Lys8, Asn125 | Pro9, Ser10, Lys53, Ser90, Val93, Ala118, Gly121, His130 | Val5, Pro11, Val14, Asp91, Pro119, Pro124, Phe127 |
| G3BP1-decitabine | −4.9 | Phe111, Gln137 | Arg135, Asp138, Val140, Phe141, Gly142 | Met98, Gly99, Arg110, Met112 |
| G3BP2-decitabine | −4.8 | Thr88, Leu89, Asn131 | Ser51, Gly52, Ser90, Gly92, Ala118, Met133 | Val116, His130 |
As G3BP1 and G3BP2 interact directly with the SARS-CoV-2 N protein, we have utilized the HDock web server for protein-protein docking. Docking studies of G3BP1 and G3BP2 with N-protein results in the docking score of −182.53 and −202.03 respectively, indicating the stringent binding affinity with the N-protein (Supporting Fig. S12). Furthermore, the binding study of decitabine and imatinib with G3BP1/2 and N-protein was also performed. To predict the binding pocket in the protein, a ligand-independent binding site identification was done using CASTp Server [51]. For the NTD (PDB ID:6M3M), a binding pocket having a solvent accessible surface area of 3364 Å2 and volume of 3192 Å3 was identified with the residues Lys62-Pro81, Pro123-Ala139, and Pro163-Pro169. These residues are largely found in turn and coil regions, and in β6 strand. The residues in these predicted binding pockets are largely involved in ribonucleotide binding and protein-protein interface. The binding pocket of G3BP1 (PDB ID: 4FCJ) has a solvent-accessible surface area of 145 Å2 and a volume of 104 Å3 which consists of residues Ala84-Val90 in 310 helix. For G3BP2 (PDB ID: 5DRV), the binding pocket has a solvent-accessible surface area of 108 Å2 and a volume of 66 Å3 consists of Thr85-Gly89 (310 helix), Val113-Pro116 (β-strand).
After identification of the putative binding pockets, we performed the molecular docking of the decitabine and imatinib on these binding pockets. Imatinib displayed a binding affinity of −7.2 kcal/mol and forms three hydrogen (H) bonds with NTD at positions Leu129, Pro130, and Lys131 along with five Vander Waals bonds (Fig. 5A). Decitabine binds loosely to NTD with a binding affinity of −4.7 kcal/mol and forms four H-bonds at positions Leu123, Leu129, Phe133, and Ala135 along with six Vander Waal interactions (Fig. 5B). The results revealed that although both drugs bind strongly with the N-protein, imatinib showed a stronger affinity to the NTD compared to decitabine.
Imatinib also showed strong binding to G3BP1 and G3BP2 with −8.7 kcal/mol and −7.7 kcal/mol, respectively. Imatinib forms four H-bond interactions with G3BP1 at positions Asn90, Glu120, Tyr128, and His130, and also forms seven Vander Waals interactions (Fig. 6A). In contrast, imatinib formed two H-bonds at position Lys8 and Asn125, along with eight Vander Waals interactions with G3BP2 (Fig. 6B). However, decitabine binds weakly with G3BP1 and G3BP2 with binding affinities of −4.9 kcal/mol and −4.8 kcal/mol respectively. Interaction studies of decitabine with G3BP1 showed two H-bonds at positions Gln137 and Phe111, along with five Vander Waal interactions (Fig. 6C). With G3BP2, decitabine forms three H-bonds at positions Thr88, Leu89, and Asn131 and six Vander Waals interactions (Fig. 6D). The above results suggested that imatinib binds strongly with G3BP1 and G3BP2 protein compared to decitabine.
3.9. Molecular dynamics simulations analyses
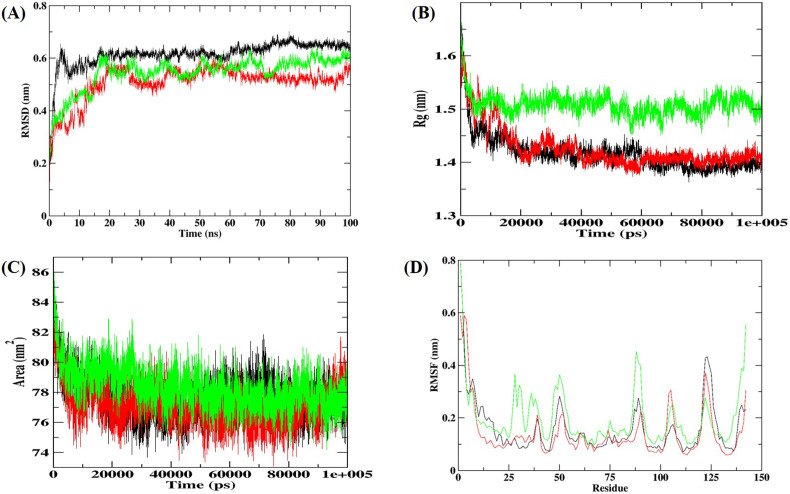
To investigate the conformational stability of the G3BP1-drug complexes, we next performed MD simulation for 100 ns and computed root mean square deviation (RMSD) of the Cα atoms, the radius of gyration (R g), root mean square fluctuations (RMSF), and solvent accessible surface area (SASA) of the protein. The Cα-RMSD provides a global picture of the conformational deviation in structure with respect to the initial structure during the simulation. As can be seen from Fig. 7A, the drug binding induces an increase in conformational stability in the protein with the decrease in Cα-RMSD. The other structural parameters, R g and SASA were calculated to assess the structural integrity and compactness of G3BP1-drug complexes (Fig. 7B and C). The time evolution of R g showed that the imatinib bound G3BP1 complex has increased R g and SASA values, suggesting the potential unfolding effect of imatinib on G3BP1. However, the decitabine bound G3BP1 complex becomes more compact with lesser R g and SASA. Furthermore, RMSF showed the fluctuations of each residue in the unbound and bound G3BP1 (Fig. 7D). For the unbound G3BP1, high fluctuations were significantly found in the C-terminal β-strand. Compared to the unbound G3BP1, the G3BP1-imatinib drug complex increased the fluctuations throughout the protein except at the C-terminal β-strand. The maximum fluctuations were observed for β hairpin residues (85–90) near the predicted drug binding site. However, decitabine binding showed decreased RMSF, indicating significant stabilization of the G3BP1.
Fig. 7.
MD simulation results of G3BP1-drug complexes for 100 ns at 300 K. (A) RMSD of the Cα backbone (B) Rg, (C) SASA, and (D) RMSF of residues. In all the panels the color code represents as: unbound G3BP1 (black), G3BP1-imatinib (green), and G3BP1-decitabine (red).
3.9.1. Principal component analysis (PCA) and Free energy landscape (FEL) analysis
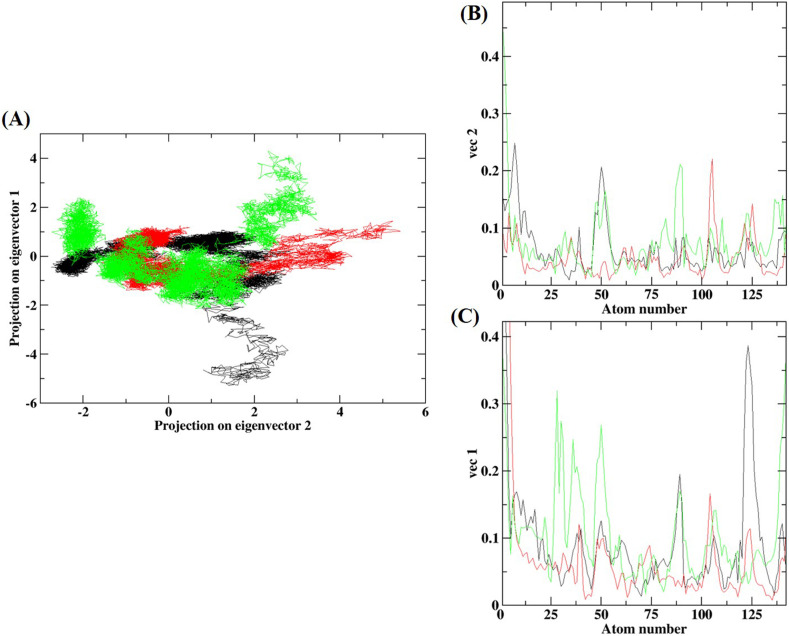
The PCA was studied to determine the important motions of the protein bound to the ligand. The two-dimensional projection of PCA (PC1 and PC2) showed that the G3BP1-drug complexes cover a larger and different conformational subspace with high flexibility as compared to unbound G3BP1 (Fig. 8A). The unbound G3BP1 exhibited lesser stable clusters as compared to G3BP1-drug complexes, particularly to G3BP1-decitabine. G3BP1-decitabine complex showed a more stable cluster with the increased amount of the accessible subspace along PC2. Similarly, the G3BP1-imatinib complex occupied a more stable cluster and wider phase space along PC1. Thus, the G3BP1-imatinib complex has increased fluctuations and might affect the protein motions upon drug binding. These findings were further confirmed through investigating the residue displacements along with PC1 and PC2 for the G3BP1-drug complexes (Fig. 8B and C). The average fluctuation for the G3BP1-decitabine complex was smaller as compared to the G3BP1-imatinib complex.
Fig. 8.
PCA of G3BP1-drug complexes. (A) Projection of the motion of the unbound G3BP1 (black), G3BP1-imatinib (green), and G3BP1-decitabine (red) in phase space along the PC1 and PC2. (C) Average Eigen RMSF values for G3BP1 systems along with PC1 and PC2.
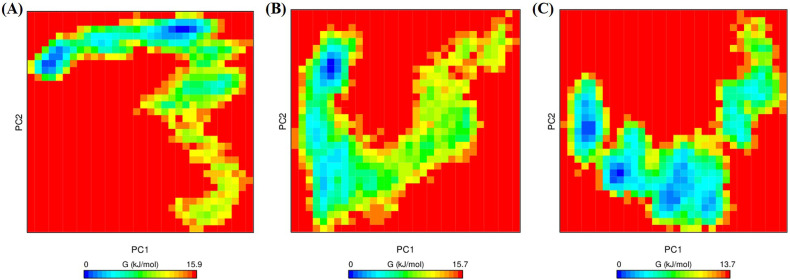
To examine the effects of the drug on the conformational stability of G3BP1, FEL was evaluated as a function of the top two principal components, PC1, and PC2. FEL has been utilized to successfully investigate the conformational redistributions induced by ligand binding [52], [53], [54], [55]. Fig. 9 showed the relative conformational changes of G3BP1 systems where the lower energy conformational states are indicated by the deeper blue color. The FEL plots revealed ΔG values 0 to 15.9, 15.7, and 13.7 kJ/mol (Fig. 9) for unbound G3BP1, G3BP1-decitabine, and G3BP1-imatinib complex, respectively. The FEL of unbound G3BP1 is populated by two different conformational subspaces linked by a low energy barrier (Fig. 9A). G3BP1-decitabine complex showed a significant population shift towards a large, single, and wider energy basin consisting of a single metastable state (Fig. 9B). The G3BP1-imatinib complex is characterized by the presence of different energy basins and a reverse population shift relative to the unbound G3BP1 system (Fig. 9C).
Fig. 9.
The FEL of the simulated G3BP1 and G3BP1-drug complexes. (A) unbound G3BP1, (B) G3BP1-decitabine, and (C) G3BP1-imatinib complex. The color scale represents the free energy value in kcal/mol. The dark blue spot represents the energy minima and energetically favored protein conformations, and the yellow spot indicates the unfavorable high-energy conformation.
Further, the atomic density distribution analysed using densmap indicated that decitabine and imatinib binding increased the partial density area of G3BP1 from 1.97 nm−3 to 3.21 nm−3 and 2.27 nm−3, respectively (Supplementary Fig. S13). The density maps thus revealed that binding of the drug especially imatinib induced a large conformation change to G3BP1. Overall, the MD results revealed that decitabine weakly modulates G3BP1 conformations whereas, imatinib binding significantly changes G3BP1 conformations and thus could significantly regulate the protein's function.
4. Discussion and conclusion
G3BP1 and G3BP2 bind strongly with the N protein of SARS-CoV-2 and SARS-CoV than other human coronaviruses, and the binding to MERS-CoV N protein was the weakest [13]. Recently, SARS-CoV-2 N protein has been shown to undergo phase separation that enhances viral RNA replication and translation and mediates the packaging of viral genome in host cells. The N protein sequester the SG essential protein G3BP1/2 and attenuates SG formation [8]. G3BP1/2 are core components of SGs and promote both the inflammatory and the IFN responses. G3BP1/2 thus could be considered as an important target for the induction of antiviral activities, i.e., SG nucleation and immune regulation. Importantly, we here suggest that targeting the transcriptional regulation of G3BP1/2 may be an attractive treatment approach for mitigating SARS-CoV-2 infection. Our data showed that through inhibition or activation of TFs and kinases involved in G3BP1/2 regulation could be considered as a potential way to tackle SARS-CoV-2 infection.
We here utilized multiple approaches to analyze the gene expression modulators of stress granule core genes, G3BP1 and G3BP2. The GSEA represents a threshold-free overrepresentation analysis strategy to identify the potential regulators of G3BP1/2. We here utilized GSEA to evaluate genome-wide expression profiles to determine whether the identified sets of regulators show statistically significant, collective changes in gene expression of G3BP1/2 that can be correlated with SARS-CoV-2 infection. The identified regulators act as activators and/or repressors of G3BP1 and G3BP2 and provide necessary information to build a model of genomic regulatory interactions observed during SARS-CoV-2 infection. A panel of existing drugs and ligands against these regulatory genes were then identified that could be considered for drug-repurposing to mitigate the outcomes of COVID-19. Two of the most promising candidate drugs, namely imatinib and decitabine alter the gene expression of these regulatory genes and thus could act as antiviral agents. Interestingly, a hypothetical bipartite combination (imatinib and decitabine) indicated more robust effects on the interactome of SARS-CoV-2 target human host genes compared to monotherapies.
Imatinib is a tyrosine kinase inhibitor used to treat chronic myeloid leukemia (CML) and has been reported to inhibit SARS-CoV infection and MERS-CoV [56], [57], [58]. Mechanistic studies suggested that imatinib will be considered as a good therapeutic possibility in the COVID-19 for its demonstrated anti-inflammatory activity, SARS-CoV-2 entry inhibitor, and kinase inhibitory activity. Imatinib has also been shown to induce eIF2α phosphorylation and thus induces SG formation and promotes apoptosis [59]. Moreover, a recent study showed that the kinase inhibitor, nilotinib affects N protein phase separation with viral RNA and disrupts condensates formed by the N protein in mammalian cells [60]. Previous studies have also shown that nilotinib inhibited the proliferation of both SARS-CoV and SARS-CoV-2 [61], [62], [63]. Another study indicated imatinib as an important SARS-CoV-2 protease inhibitor using the Reframe library screen of clinical and near-clinical compounds [64]. Our results thus provide a rationale for evaluating imatinib as an antiviral agent against COVID-19. Interestingly, three potential randomized clinical trials are underway to study the therapeutic efficacy of imatinib against COVID-19, including NCT04357613 (France), NCT04394416 (USA), and EudraCT2020-001236-10 (The Netherlands). In another study, Imatinib will be compared to hydroxychloroquine, Lopinavir/ritonavir, and Baricitinib (NCT04346147).
Decitabine falls under the nucleotide and nucleoside analogs which are broad-spectrum antiviral drugs that inhibit transcription and/or replication of different RNA and DNA viruses by inhibition of DNA methylation [65]. A recent study showed decitabine as the potent inhibitor of RNA-dependent RNA polymerase and thus could be regarded as a drug that would inhibit viral replication and growth [66]. Recently, a randomized double-blind placebo-controlled Phase 2 trial has been initiated to assess the efficacy of decitabine in the treatment of critically ill patients with COVID-ARDS (Clinical Trial ID: NCT04482621, IRB00247544).
In conclusion, our results suggest that by targeting G3BP1/2 and their regulators, the drugs imatinib and decitabine affect the SG formation in host cells and thus could represent a viable approach in future studies of COVID-19.
The following are the supplementary data related to this article.
Supplementary figures
Involvement of Putative modulators of G3BP1 and G3BP2 genes in COVID-19 related gene sets obtained from Enrichr
Differential regulation of G3BP1 and G3BP2 genes and their modulators according to the date retrieved from GEO database.
CRediT authorship contribution statement
Neemat Ali, Kartikay Prasad: Methodology, Data curation, Software. Summya Rashid: Visualization, Investigation. Abdullah F. AlAsmari: Supervision. Metab Alharbi: Software, Validation. Vijay Kumar: Conceptualization, Supervision, Writing, Reviewing and Editing.
Declaration of competing interest
No potential conflict of interest was reported by the authors.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group no. RG-1441-451.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12(2) doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C., Xie J., Ding C., Ma X., Weng J., Gao Y., He H., Jin T. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527(3):618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10(7):1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye Q., West A.M.V., Silletti S., Corbett K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020;29(9):1890–1901. doi: 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savastano A., Ibanez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020;11(1):6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N.L. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020;39(24) doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., Bowman G.R., Hall K.B., Soranno A., Holehouse A.S. 2020. The SARS-CoV-2 Nucleocapsid Protein is Dynamic, Disordered, and Phase Separates With RNA. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y., Wang C., Zhang Y., Zhu Z., Liu Z., Peng C., Zhu T., Liang Q. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med (N Y) 2021;2(1):99–112. doi: 10.1016/j.medj.2020.07.002. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nabeel-Shah Syed, Lee Hyunmin, Ahmed Nujhat, Marcon Edyta, Farhangmehr Shaghayegh, Pu Shuye, Burke Giovanni L., Ashraf Kanwal, Wei Hong, Zhong Guoqing, Tang Hua, Yang Jianyi, Blencowe Benjamin J., Zhang Zhaolei, Greenblatt J.F. 2020. SARS-CoV-2 Nucleocapsid Protein Attenuates Stress Granule Formation and Alters Gene Expression via Direct Interaction With Host mRNAs. bioRxiv. [Google Scholar]
- 14.Yang P., Mathieu C., Kolaitis R.M., Zhang P., Messing J., Yurtsever U., Yang Z., Wu J., Li Y., Pan Q., Yu J., Martin E.W., Mittag T., Kim H.J., Taylor J.P. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020;181(2):325–345. doi: 10.1016/j.cell.2020.03.046. e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillen-Boixet J., Kopach A., Holehouse A.S., Wittmann S., Jahnel M., Schlussler R., Kim K., Trussina I., Wang J., Mateju D., Poser I., Maharana S., Ruer-Gruss M., Richter D., Zhang X., Chang Y.T., Guck J., Honigmann A., Mahamid J., Hyman A.A., Pappu R.V., Alberti S., Franzmann T.M. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020;181(2):346–361. doi: 10.1016/j.cell.2020.03.049. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linero F.N., Thomas M.G., Boccaccio G.L., Scolaro L.A. Junin virus infection impairs stress-granule formation in vero cells treated with arsenite via inhibition of eIF2alpha phosphorylation. J. Gen. Virol. 2011;92(Pt 12):2889–2899. doi: 10.1099/vir.0.033407-0. [DOI] [PubMed] [Google Scholar]
- 17.Emara M.M., Brinton M.A. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 2007;104(21):9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikolic J., Civas A., Lama Z., Lagaudriere-Gesbert C., Blondel D. Rabies virus infection induces the formation of stress granules closely connected to the viral factories. PLoS Pathog. 2016;12(10) doi: 10.1371/journal.ppat.1005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abrahamyan L.G., Chatel-Chaix L., Ajamian L., Milev M.P., Monette A., Clement J.F., Song R., Lehmann M., DesGroseillers L., Laughrea M., Boccaccio G., Mouland A.J. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 2010;123(Pt 3):369–383. doi: 10.1242/jcs.055897. [DOI] [PubMed] [Google Scholar]
- 20.Panas M.D., Kedersha N., Schulte T., Branca R.M., Ivanov P., Anderson P. Phosphorylation of G3BP1-S149 does not influence stress granule assembly. J. Cell Biol. 2019;218(7):2425–2432. doi: 10.1083/jcb.201801214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tourriere H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;160(6):823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Prasad K., Khatoon F., Rashid S., Ali N., AlAsmari A.F., Ahmed M.Z., Alqahtani A.S., Alqahtani M.S., Kumar V. Targeting hub genes and pathways of innate immune response in COVID-19: a network biology perspective. Int. J. Biol. Macromol. 2020;163:1–8. doi: 10.1016/j.ijbiomac.2020.06.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khatoon F., Prasad K., Kumar V. Neurological manifestations of COVID-19: available evidences and a new paradigm. J. Neurovirol. 2020;26(5):619–630. doi: 10.1007/s13365-020-00895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad K., AlOmar S.Y., Alqahtani S.A.M., Malik M.Z., Kumar V. Brain disease network analysis to elucidate the neurological manifestations of COVID-19. Mol. Neurobiol. 2021;58(5):1875–1893. doi: 10.1007/s12035-020-02266-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-pdb viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 30.Yan Y., Tao H., He J., Huang S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020;15(5):1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 31.Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., Chang Z., Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(suppl_1) doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open babel: an open chemical toolbox. J. Cheminformatics. 2011;3(1):1–14. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Systèmes D. Dassault Systèmes Biovia; San Diego, CA, USA: 2016. Biovia, Discovery Studio Modeling Environment. [Google Scholar]
- 35.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 36.SchuÈttelkopf A.W., Van Aalten D.M. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004;60(8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 37.Joung I.S., Cheatham T.E., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112(30):9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berendsen H.J.C., Grigera J.R., Straatsma T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271. [Google Scholar]
- 39.Parrinello M., Rahman A. Crystal structure and pair potentials: a molecular-dynamics study. Phys. Rev. Lett. 1980;45(14):1196–1199. [Google Scholar]
- 40.Berk H., Henk B., B.H.J. C., M F.J.G.E. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18(12):1463–1472. [Google Scholar]
- 41.McGibbon R.T., Beauchamp K.A., Harrigan M.P., Klein C., Swails J.M., Hernandez C.X., Schwantes C.R., Wang L.P., Lane T.J., Pande V.S. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015;109(8):1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reghunathan R., Jayapal M., Hsu L.Y., Chng H.H., Tai D., Leung B.P., Melendez A.J. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 2008;10(10):1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Marrero M.Correa, Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., Batra J., Richards A.L., Stevenson E., Gordon D.E., Rojc A., Obernier K., Fabius J.M., Soucheray M., Miorin L., Moreno E., Koh C., Tran Q.D., Hardy A., Robinot R., Vallet T., Nilsson-Payant B.E., Hernandez-Armenta C., Dunham A., Weigang S., Knerr J., Modak M., Quintero D., Zhou Y., Dugourd A., Valdeolivas A., Patil T., Li Q., Huttenhain R., Cakir M., Muralidharan M., Kim M., Jang G., Tutuncuoglu B., Hiatt J., Guo J.Z., Xu J., Bouhaddou S., Mathy C.J.P., Gaulton A., Manners E.J., Felix E., Shi Y., Goff M., Lim J.K., McBride T., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Wit E.De, Leach A.R., Kortemme T., Shoichet B., Ott M., Saez-Rodriguez J., Mullins R.D., Fischer E.R., Kochs G., Grosse R., Garcia-Sastre A., Vignuzzi M., Johnson J.R., Shokat K.M., Swaney D.L., Beltrao P., Krogan N.J., tenOever B.R. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182(3):685–712. doi: 10.1016/j.cell.2020.06.034. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimakami T., Yamane D., Jangra R.K., Kempf B.J., Spaniel C., Barton D.J., Lemon S.M. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. U. S. A. 2012;109(3):941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piedade D., Azevedo-Pereira J.M. The role of microRNAs in the pathogenesis of herpesvirus infection. Viruses. 2016;8(6) doi: 10.3390/v8060156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo X.K., Zhang Q., Gao L., Li N., Chen X.X., Feng W.H. Increasing expression of microRNA 181 inhibits porcine reproductive and respiratory syndrome virus replication and has implications for controlling virus infection. J. Virol. 2013;87(2):1159–1171. doi: 10.1128/JVI.02386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fulzele S., Sahay B., Yusufu I., Lee T.J., Sharma A., Kolhe R., Isales C.M. COVID-19 virulence in aged patients might be impacted by the host cellular microRNAs abundance/profile. Aging Dis. 2020;11(3):509–522. doi: 10.14336/AD.2020.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng S., Wang J., Wei S., Li C., Zhou K., Hu J., Ye X., Yan J., Liu W., Gao G.F., Fang M., Meng S. Endogenous cellular MicroRNAs mediate antiviral defense against influenza a virus. Mol. Ther.–Nucleic Acids. 2018;10:361–375. doi: 10.1016/j.omtn.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46(W1):W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prakash A., Kumar V., Meena N.K., Hassan M.I., Lynn A.M. Comparative analysis of thermal unfolding simulations of RNA recognition motifs (RRMs) of TAR DNA-binding protein 43 (TDP-43) J. Biomol. Struct. Dyn. 2019;37(1):178–194. doi: 10.1080/07391102.2017.1422026. [DOI] [PubMed] [Google Scholar]
- 53.Kumar V., Pandey P., Idrees D., Prakash A., Lynn A.M. Delineating the effect of mutations on the conformational dynamics of N-terminal domain of TDP-43. Biophys. Chem. 2019;250 doi: 10.1016/j.bpc.2019.106174. [DOI] [PubMed] [Google Scholar]
- 54.Kumar V., Prakash A., Pandey P., Lynn A.M., Hassan M.I. TFE-induced local unfolding and fibrillation of SOD1: bridging the experiment and simulation studies. Biochem. J. 2018;475(10):1701–1719. doi: 10.1042/BCJ20180085. [DOI] [PubMed] [Google Scholar]
- 55.Kumar V., Prakash A., Lynn A.M. Alterations in local stability and dynamics of A4V SOD1 in the presence of trifluoroethanol. Biopolymers. 2018;109(3) doi: 10.1002/bip.23102. [DOI] [PubMed] [Google Scholar]
- 56.Galimberti S., Baldini C., Barate C., Ricci F., Balducci S., Grassi S., Ferro F., Buda G., Benedetti E., Fazzi R., Baglietto L., Lucenteforte E., Di Paolo A., Petrini M. The CoV-2 outbreak: how hematologists could help to fight Covid-19. Pharmacol. Res. 2020;157 doi: 10.1016/j.phrs.2020.104866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sisk J.M., Frieman M.B., Machamer C.E. Coronavirus S protein-induced fusion is blocked prior to hemifusion by abl kinase inhibitors. J. Gen. Virol. 2018;99(5):619–630. doi: 10.1099/jgv.0.001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coleman C.M., Sisk J.M., Mingo R.M., Nelson E.A., White J.M., Frieman M.B. Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. J. Virol. 2016;90(19):8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bordeleau M.E., Robert F., Gerard B., Lindqvist L., Chen S.M., Wendel H.G., Brem B., Greger H., Lowe S.W., Porco J.A., Jr., Pelletier J. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Invest. 2008;118(7):2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jack A., Ferro L.S., Trnka M.J., Wehri E., Nadgir A., Costa K., Schaletzky J., Yildiz A. 2020. SARS CoV-2 Nucleocapsid Protein Forms Condensates With Viral Genomic RNA. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trezza A., Iovinelli D., Santucci A., Prischi F., Spiga O. An integrated drug repurposing strategy for the rapid identification of potential SARS-CoV-2 viral inhibitors. Sci. Rep. 2020;10(1):13866. doi: 10.1038/s41598-020-70863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jr., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58(8):4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gustavo Garcia Arun Sharma, Jr., Ramaiah Arunachalam, Jr., Sen Chandani, Jr., Kohn Donald, Jr., Gomperts Brigitte, Jr., Svendsen Clive N., Jr., Damoiseaux Robert D., Jr., Arumugaswami V., Jr. 2020. Antiviral Drug Screen of Kinase Inhibitors Identifies Cellular Signaling Pathways Critical for SARS-CoV-2 Replication. bioRxiv. [Google Scholar]
- 64.Janes J., Young M.E., Chen E., Rogers N.H., Burgstaller-Muehlbacher S., Hughes L.D., Love M.S., Hull M.V., Kuhen K.L., Woods A.K., Joseph S.B., Petrassi H.M., McNamara C.W., Tremblay M.S., Su A.I., Schultz P.G., Chatterjee A.K. The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Proc. Natl. Acad. Sci. U. S. A. 2018;115(42):10750–10755. doi: 10.1073/pnas.1810137115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ianevski A., Zusinaite E., Kuivanen S., Strand M., Lysvand H., Teppor M., Kakkola L., Paavilainen H., Laajala M., Kallio-Kokko H., Valkonen M., Kantele A., Telling K., Lutsar I., Letjuka P., Metelitsa N., Oksenych V., Bjoras M., Nordbo S.A., Dumpis U., Vitkauskiene A., Ohrmalm C., Bondeson K., Bergqvist A., Aittokallio T., Cox R.J., Evander M., Hukkanen V., Marjomaki V., Julkunen I., Vapalahti O., Tenson T., Merits A., Kainov D. Novel activities of safe-in-human broad-spectrum antiviral agents. Antivir. Res. 2018;154:174–182. doi: 10.1016/j.antiviral.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang A., Tang X., Wu H., Zhang J., Wang W., Wang Z., Song L., Zhai M., Zhao L., Yang H., Ma X., Zhou S., Cai J. 2020. Virtual Screening and Molecular Dynamics on Blockage of Key Drug Targets as Treatment for COVID-19 Caused by SARS-CoV-2. Preprints 2020030239. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Involvement of Putative modulators of G3BP1 and G3BP2 genes in COVID-19 related gene sets obtained from Enrichr
Differential regulation of G3BP1 and G3BP2 genes and their modulators according to the date retrieved from GEO database.