Abstract
Ad libitum high-fat diet (HFD) induces obesity and skeletal muscle metabolic dysfunction. Liver kinase B1 (LKB1) regulates skeletal muscle metabolism by controlling the AMP-activated protein kinase family, but its importance in regulating muscle gene expression and glucose tolerance in obese mice has not been established. The purpose of this study was to determine how the lack of LKB1 in skeletal muscle (KO) affects gene expression and glucose tolerance in HFD-fed, obese mice.
KO and littermate control wild-type (WT) mice were fed a standard diet or HFD for 14 weeks. RNA sequencing, and subsequent analysis were performed to assess mitochondrial content and respiration, inflammatory status, glucose and insulin tolerance, and muscle anabolic signaling.
KO did not affect body weight gain on HFD, but heavily impacted mitochondria-, oxidative stress-, and inflammation-related gene expression. Accordingly, mitochondrial protein content and respiration were suppressed while inflammatory signaling and markers of oxidative stress were elevated in obese KO muscles. KO did not affect glucose or insulin tolerance. However, fasting serum insulin and skeletal muscle insulin signaling were higher in the KO mice. Furthermore, decreased muscle fiber size in skmLKB1-KO mice was associated with increased general protein ubiquitination and increased expression of several ubiquitin ligases, but not muscle ring finger 1 or atrogin-1. Taken together, these data suggest that the lack of LKB1 in skeletal muscle does not exacerbate obesity or insulin resistance in mice on a HFD, despite impaired mitochondrial content and function and elevated inflammatory signaling and oxidative stress.
Keywords: Skeletal muscle, AMPK, Obesity, Mitochondria, Inflammation, Atrophy
1. Introduction
Liver kinase B1 (LKB1), the product of the serine/threonine kinase 11 (STK11) gene, is conserved throughout evolution from worms to mammals [1]. It is a major regulator of carbohydrate, lipid and protein metabolism [2–9], and these effects are exerted primarily through its action on downstream targets of the AMP-activated protein kinase (AMPK) family. In addition to the AMPK α1 and α2 catalytic subunits, the AMPK family of serine/threonine kinases that are regulated by LKB1 includes salt-inducible kinase 1 (SIK1) and 3 (SIK3), sucrose nonfermenting AMPK-related kinase (SNARK), AMPK-related kinase 5 (Ark5), and 6 other protein kinases [10].
AMPK itself is the best-characterized of the LKB1 targets. It has been well-established as a cellular “energy-gauge” that responds to increased AMP/ATP or ADP/ATP ratio by promoting catabolic processes that restore ATP, and by inhibiting ATP-consuming anabolic processes that are not required for the immediate survival of the cell [11,12]. It is a heterotrimeric protein consisting of α, β and γ subunits, each of which consists of different isoforms. Classic activation of AMPK by AMP (or, to an extent, by ADP) involves displacement of ATP on the regulatory γ subunit by AMP. This leads to a conformational change in AMPK that results in increased phosphorylation at Thr172 on the α subunit, as well as allosteric effects that lead to AMPK activation [12]. In many tissues, including skeletal muscle, LKB1 is the primary kinase responsible for activation of AMPK via phosphorylation at Thr172 [3,8,13], although calcium calmodulin-dependent protein kinase kinase 2 (CamKKβ) is also able to do so in some tissues under certain conditions [14].
Ad libitum high-fat diet (HFD) feeding is a well-established model in rodents for the induction of diet-induced obesity (DIO). This accumulation of body fat is associated with the development of insulin resistance and eventually leads to the development of diabetes with increased fasting blood glucose and insulin levels [15–17]. Although many factors likely contribute to peripheral insulin resistance with HFD-induced obesity, lipid-induced ROS production, mitochondrial dysfunction, chronic low-grade inflammation, and a generalized loss of muscle mass all seem to play prominent roles [18–21].
The role LKB1 plays in the adaptation of skeletal muscle to DIO is not completely clear. Since LKB1 regulation of the AMPK family plays well-established roles in increasing glucose uptake, fat oxidation [2,5], and mitochondrial accretion [3,4,22], while attenuating inflammation [23,24] and mediating part of the response to oxidative stress [23,25], it stands to reason that it should beneficially impact skeletal muscle metabolism and function under HFD feeding, including insulin sensitivity. Indeed, multiple studies showed that activation of skeletal muscle AMPK is associated with improved insulin sensitivity under high-fat diet feeding. For instance, daily treatment of mice on a HFD with AICAR, an AMPK-activating drug, largely prevented the development of glucose intolerance and insulin resistance in obesity [26]. Furthermore, a gain-of-function mutation for the AMPK γ3 subunit that led to an overall increase in AMPK activity protected skeletal muscle against HFD-induced insulin resistance [27]. Similarly, global AMPK β2 knockout increased weight gain and impaired glucose tolerance in HFD-induced obesity [28]. Muscle specific deletion of AMPK α1 under HFD increased accumulation of muscle lipid and adipogenic gene expression, along with decreased expression of fat-oxidation genes [29], the last two effects both being potential mechanistic links between obesity and insulin resistance. Muscle-specific deletion of LKB1 likewise increased adipogenic gene expression and increased myofiber lipid content while on a HFD [30].
On the other hand, and contrary to what might be expected based on AMPK’s well-documented effects, muscle-specific AMPK α2 knockout mice gained less weight, less fat, and had improved glucose tolerance and insulin sensitivity compared to WT mice after 14 weeks of HFD feeding [31]. Muscle-specific knockout of NUAK1 and SIK1 (other LKB1-regulated AMPK family members) also improved glucose tolerance and insulin sensitivity [16,32]. Thus, the composite role of skeletal muscle LKB1 remains somewhat obscure, and the effect of skeletal muscle LKB1 deficiency on global gene expression and whole-body glucose and insulin tolerance has not been explored. The purpose of this study, therefore, was to determine the effect of skeletal muscle specific LKB1 deficiency on whole-body glucose and insulin tolerance as well as skeletal muscle gene expression in HFD-induced obese mice.
2. Materials and methods
2.1. Animals
The Institutional Animal Care and Use Committee (IACUC) of Brigham Young University or of the University of California, Los Angeles approved all experimental procedures involving animals, and the Guide For The Care and Use of Laboratory Animals (National Institutes of Health) was followed.
2.1.1. Hybrid Mouse Diversity Panel (HMDP)
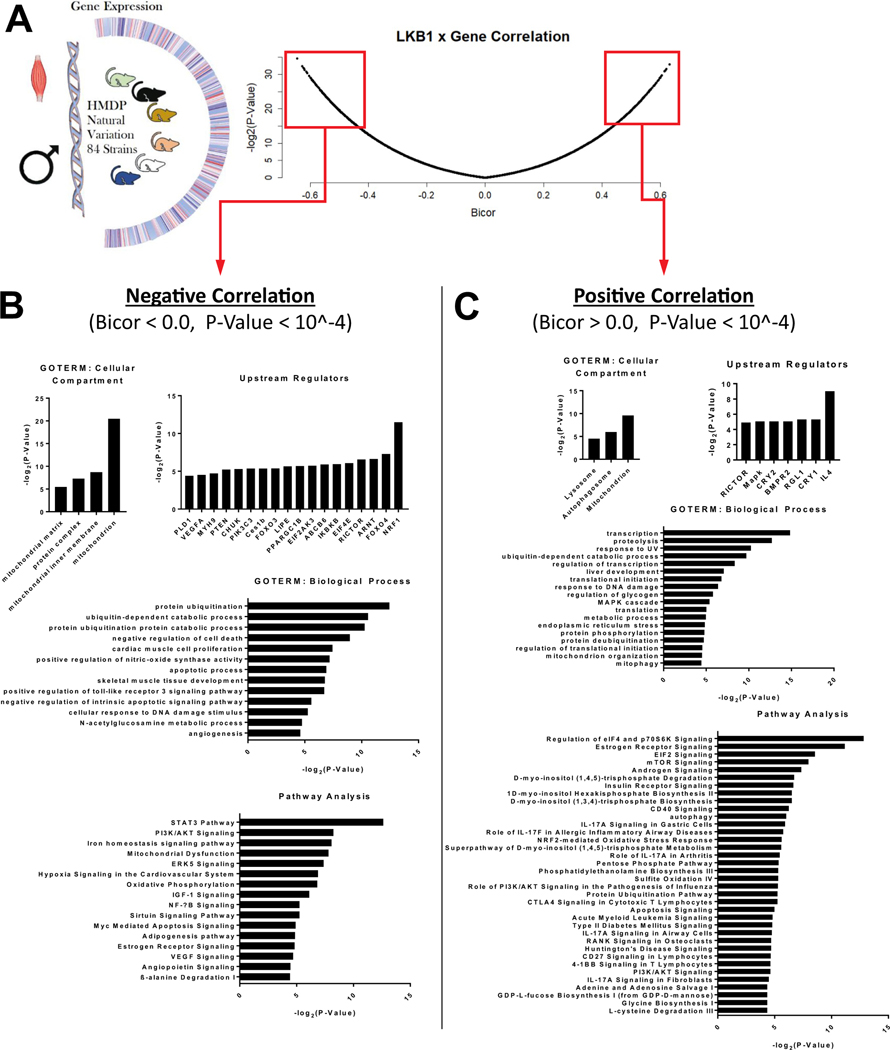
Studies from the Hybrid Mouse Diversity Panel (HMDP) followed the procedure outlined previously [33]. Briefly, mice obtained from The Jackson Laboratory were bred at the University of California, Los Angeles and maintained on a chow diet (Ralston Purina Company, St. Louis, MO, USA) until 8 weeks of age. They were then provided a high fat/high sucrose diet (HF/HS Research Diets D12266B; 8 weeks) with the following composition: 16.8% kcal protein, 51.4% kcal carbohydrate, 31.8% kcal fat. Total RNA from HMDP mouse muscle (228 males) was hybridized to Affymetrix HT_MG-430A arrays and scanned using standard Affymetrix protocols. To reduce the chances of spurious association results, RMA normalization was performed after removing all individual probes with SNPs and all probesets containing 8 or more SNP-containing probes, which resulted in 22,416 remaining probe sets. To determine the accuracy of our microarray data, we tested by qPCR the expression of a dozen genes and found an r = 0.7 between qPCR and microarray. LKB1 gene expression was correlated with all other genes using the Midweight Bicorrelation Method (Bicor) function within the Weighted Gene Correlation Network Analysis Package (WGCNA; Version 1.66) [34] within R (Version 3.5). Genes found to strongly correlate (−log2 (P-Value) > or < 15 and Bicor> or < ± 0.4) with LKB1 either positively or negatively were analyzed using DAVID Functional Annotation (Version 6.8) or Ingenuity Pathway Analysis (IPA, Version 46901286, Qiagen, Hilden, Germany). Data are presented using GraphPad Prism (Version 8.0).
2.1.2. Skeletal muscle specific LKB1 (skmLKB1) knockout mouse
Skeletal muscle specific knockout of floxed LKB1 (KO) was accomplished through Myf6 promoter-driven Cre expression on a FVB background as described previously [4,23]. Our KO mice were bred onto the FVB strain. Compared to C57/Bl6 mice, which are prone to diet-induced obesity, female FVB mice gain less body mass, body fat and have less impairment in glucose tolerance after high-fat diet feeding [35]. Male FVB mice may [36] or may not [37,38] be resistant to HFD-induced obesity. Preliminary experiments in our lab in which female mice were resistant to HFD-induced weight gain were in agreement with this finding and we therefore used only male mice in the current study.-Genotyping was performed by PCR followed by western blotting for LKB1 protein as previously described [3]. All mice were housed in a pathogen-free facility (Life Science Building, Brigham Young University, Provo, UT, USA) at 21–23 °C with a 12:12 h light-dark cycle.
Beginning at 5 weeks of age, wild type (WT) and KO mice were given ad libitum access to either high-fat chow (Envigo, TD.06414; 18.3% protein, 21.4% carbohydrate, 60.3% fat, by kcal) or continued ad libitum access to standard chow (Envigo, TD.8604; 32% protein, 54% carbohydrate, 14% fat, by kcal) for 14 weeks after which the mice were euthanized and tissue collected as described below.
2.1.3. Ambulatory activity monitoring
In-cage ambulatory activity was measured in a subset of individually housed mice for 3 consecutive days in between days 74 and 88 of the feeding period. This was done using an infrared beam activity monitoring system (Columbus Instruments, Columbus, OH, USA). The numbers of beam breaks for every 30-minute period were recorded by computer software.
2.1.4. Intraperitoneal glucose tolerance testing (IPGTT)
On day 90 of the feeding period, and after a 6 h fast, topical anesthetic (EMLA 2.5% lidocaine/prilocaine; Hi-Tech Pharmacal, Amityville, NY, USA) was applied to the tip of the mouse’s tail. 20–30 min later, ∼1 mm was cut from the distal tip of the tail with a scalpel blade. After discarding 3 small drops of blood, blood glucose concentration was measured using a glucometer (Freestyle Lite Blood Glucose Monitoring System, Abbot Laboratories, Abbott Park, IL, USA). The mice were then injected intraperitoneally with freshly-prepared sterile 20% glucose solution [2 mg glucose/g body weight (BW)]. At 15, 30, 60, and 120 min after the injection, blood glucose was measured again from the tail by removing the clot from the first incision.
2.1.5. Intraperitoneal insulin tolerance testing (IPITT)
On day 94 of the feeding period, IPITT was performed as described above for IPGTT except for the mice being fasted for 4 rather than 6 h, and they were injected with insulin (0.5 U/kg BW; Novolin R, Novo Nordisk, Bagsvaerd, DK) rather than glucose.
2.1.6. Tissue collection
Mice were anesthetized with 2–2.5% isoflurane in supplemental oxygen. Serum was collected by withdrawing approximately 0.5 ml of blood from the inferior vena cava and allowing it to coagulate at room temperature for 30 min prior to centrifugation at 4 °C and 1000 g for 10 min. Gastrocnemius, soleus, tibialis anterior (TA) and quadriceps (QUAD) muscles, the retroperitoneal fat pad and heart were harvested and clamp-frozen in liquid nitrogen. Contralateral QUAD muscles were embedded in optimal cutting temperature (OCT) medium and frozen in isopentane at the temperature of liquid nitrogen. Tissues were stored at −90 °C until further analysis.
2.2. Muscle homogenization
QUAD muscles were powdered in liquid nitrogen then ground-glass homogenized in 19 volumes of homogenization buffer (50 mM Tris, pH 7.4; 250 mM mannitol; 50 mM NaF; 5 mM sodium pyrophosphate; 1 mM EDTA; 1 mM EGTA; 1% Triton X-100; 50 mM β-glycerophosphate; 1 mM sodium orthovanadate; 1 mM dithiothrietol; 1 mM benzamidine; 0.1 mM phenylmethanesufonyl fluoride; 5 μg/ml soybean trypsin inhibitor). The homogenate was slowly frozen at −90 °C and thawed three times to ensure rupture of cell and organelle membranes, then centrifuged at 10,000 g for 10 min at 4 °C. Supernatants were analyzed for protein content (DC Protein Assay; Biorad Laboratories, Hercules, CA, USA), and stored at −90 °C until further analysis.
2.3. Western blot and immunodetection
Protein homogenates were diluted in sample loading buffer (125 mM Tris, pH 6.8; 20% glycerol, 4% sodium dodecyl sulfate; 5% β-mercaptoethanol; 0.01% bromophenol blue), then loaded on Tris-HCl gels (Criterion System; Biorad Laboratories, Hercules, CA, USA). After electrophoresis, proteins were transferred to polyvinylidene difluoride (PVDF) or nitrocellulose membranes and stained with Ponceau S (0.1% Ponceau S in 5% acetic acid) and verified for equal protein loading within each lane. A representative ponceau stain is included for reference in the supplemental materials (Fig. S1). Membranes were then washed with tris-buffered saline + tween (TBST), blocked with 5% non-fat dry milk in TBST for 1 h, then rotated overnight at 4 °C in primary antibody (see Table S1) dissolved in 1% bovine serum albumin in TBST. Appropriate HRP- or infrared dye-conjugated secondary antibody diluted 1:20,000 in 2% non-fat dry milk in TBST was then applied for 1 h at room temperature, followed by washing in TBST and application of enhanced chemiluminescent substrate (Immobilon Western Chemiluminescent HRP Substrate; EMD Millipore, Burlington, MA, USA) or direct infrared imaging (Odyssey CLx, LICOR Biosciences, Lincoln, NE, USA). Chemiluminescent signals were detected with autoradiography film. Band intensities were quantified using Gel-Pro Analyzer 6.0 (Media Cybernetics, Inc. Bethesda, MD, USA).
2.4. Insulin radioimmunoassay (RIA)
Fasting serum (6–8 h fast) was diluted 1:50 in PBS. Serum insulin was measured using a mouse insulin RIA kit (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer’s directions.
2.5. Glycogen assay
Powdered QUAD was boiled for 30 min in 10 μl of ice-cold 30% KOH/mg tissue. Samples were then neutralized (pH = 5–7.5) with 10.7 M acetic acid. 0.1 ml of the sample was then transferred to 0.9 ml of amyloglucosidase buffer (1 μg amyloglucosidase/ml in 50 mM acetate buffer, pH 4.7) in a fresh microcentrifuge tube and incubated at 55 °C for 1 h. Samples were centrifuged for 10 min at 13,000 g. 20 μl of supernatant or glucose standard (0–3.2 mM glucose) was added in triplicate to a 96-well plate. 180 μl of reaction buffer (100 mM Tris, pH 8.8; 10.7 mM MgCl2; 100 μM DTT; 500 μM ATP; 400 μM NADP) with or without enzymes (0.37 U/ml glucose-6-phosphate dehydrogenase; 0.8 U/ml hexokinase) was then added to the wells (2 wells per sample each). The plate was incubated for 15 min at room temperature and absorbance was measured at 340 nm. Glycogen concentration was calculated by subtracting the absorbance without enzyme from the absorbance with enzyme, then fitting the difference to the glucose standard curve.
2.6. RNA isolation
Powdered QUAD was homogenized in Trizol (Life Technologies, Carlsbad, CA, USA) in 1.5 ml lock-top microcentrifuge tubes and the Bullet Blender Storm (Next Advance, Troy, NY, USA) using 0.9–2.0 mm stainless steel beads. RNA was then isolated using a Direct-zol RNA miniprep kits (Zymo Research, Irvine, CA, USA) following the manufacturer’s directions. RNA concentration and purity (260:280 ratio ≥ 1.9) was assessed by spectrophotometry.
2.7. RNA sequencing (RNA-seq)
Total RNA samples were checked for integrity by measuring the 28 s/18 s ratio on a TapeStation machine (Agilent Genomics, Santa Clara, CA, USA). All samples had RNA-integrity scores >7. Libraries were constructed using the Truseq RNA Library Prep Kit v2 (illumina, San Diego, CA, USA) and sequenced using 50 bp, single-end reads on two lanes of an Illumina HiSeq 2500 at the University of Utah High Throughput Genomics Core. Quality of the sequenced reads was examined using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Sequence reads were then aligned to the GRCm38 mouse genome reference using Rsubread [39,40], and differentially expressed genes were identified by DESeq2 using default parameters [41]. Genes which had an FDR ≤ 0.05 were retained as “differentially-expressed” between the two phenotypes. Enriched Gene Ontology (GO) categories were determined using the goseq Bioconductor package [42] and transcription factor binding site analysis (TFBS) was performed with Pscan using JASPAR 2016 database and a promoter region of −950 to +50 bp [43].
2.8. Real-time polymerase chain reaction
Synthesis of cDNA was performed from 500 ng RNA using iScript Reverse Transciption Supermix (Bio-Rad, Hercules, CA, USA). The sequences for primers that were used in real-time PCR are listed in Table S2. For the redox genes (NQO1, GCLC, Prx1, Gpx1), the PCR amplification was carried out on a StepOne Plus PCR system (Applied Biosciences, Foster City, CA, USA) using SYBR Green Supermix (2×; Quanta Biosciences, Beverly, MA, USA). For all other genes, the amplification was carried out using SsoFast EvaGreen Supermix (Bio-Rad) according to the manufacturer’s instructions on a CFX96 real-time detection system (Bio-Rad). Fold changes were calculated for all groups versus the control (WT-STD) using the 2-ΔΔCT method. β-actin was used as the internal control except for the myostatin, MuRF1 and atrogin-1 experiments where the internal control was Ppia.
2.9. Malondialdehyde assay
The malondialdehyde (MDA) assay to measure lipid peroxidation was carried out as described previously [44]. Briefly, samples were suspended in 250 μl 0.1% (w/v) trichloroacetic acid (TCA; Sigma-Aldrich, Milwaukee, WI, USA) solution and sonicated briefly (<5 s, 30 kHz). The sonicated solution was centrifuged at 15,000 g for 10 min at 4 °C. Supernatant was collected in a new microcentrifuge tube and mixed with 750 μl 0.5% (w/v) thiobarbituric acid (VWR International, Radnor, PA, USA) diluted in 20% (w/v) TCA. Samples were heated at 95 °C for 20 min and then cooled on ice for 10 min before being centrifuged at 15,000 g for 5 min at 4 °C. Absorbance was measured at 532 nm using a microplate reader. Absorbance values were normalized to protein values obtained from a bicinchoninic acid (BCA, Thermo Fisher Scientific, Waltham, MA, USA) assay and expressed vs. control (WT mouse on STD chow).
2.10. Citrate synthase activity assay
Whole, uncentrifuged homogenates were assayed for citrate synthase activity as described previously [4], with the exception that reaction volumes were scaled down to a final volume of 200 μL to be run in 96-well plates.
2.11. Mitochondrial respiration
Soleus muscles were prepared for mitochondrial respiration as described previously [45] before being transferred to respirometer chambers using the Oroboros O2K oxygraph (Oroboros, Innsbruck, Austria). Electron flow through complex I was supported by glutamate + malate (10 mM and 2 mM, respectively) to determine leak oxygen consumption (GM). Following stabilization, ADP (2.5 mM) was added to determine oxidative phosphorylation capacity (D). Succinate was added for complex I + II electron flow into the Q-junction (S). To determine full electron transport system capacity in cells over oxidative phosphorylation, the chemical uncoupler carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) was added (0.05 μM, followed by 0.025 μM steps until maximal O2 flux was reached). Lastly, residual oxygen consumption was measured by adding antimycin A (2.5 μM) to block complex III action, effectively stopping any electron flow, which provides a baseline rate of respiration. Following the respiration protocol, samples were removed from the chambers and used for further analysis, including protein quantification.
2.12. Measurement of fiber size
10 μm sections from OCT-embedded QUAD muscles were cut onto glass slides using a microtome-cryostat. Sections were fixed with 4% paraformaldehyde for 20 min at room temperature, washed with phosphate-buffered saline (PBS), blocked with 10% normal goat serum (NGS) for 45 min, washed with PBS, blocked with AffiniPure Fab fragment goat anti-mouse IgG (1.3 mg/ml in PBS; 115–007–003; Jackson ImmunoResearch, West Grove, PA, USA) for 30 min, washed with PBS, then incubated at 4 °C overnight in mouse anti-laminin IgG2a primary antibody (1:100 dilution in 10% NGS; D18; Developmental Studies Hybridoma Bank, Iowa City, IA, USA; deposited by Sanes, J.R.). Sections were then washed with PBS, incubated in goat-anti mouse IgG2a 546 secondary antibody (1:500 dilution in 10% NGS; #A21133; Thermo Fisher Scientific, Waltham, MA, USA) and 10 μg/ml DAPI for 45 min, washed with PBS and mounted with a glass coverslip and Fluormount-G (Southern Biotech, Birmingham, AL, USA). 5–7 Images were acquired at 20× magnification using fluorescence microscopy and average fiber area of a total of at least 600 fibers per muscle was measured using Image J software (National Institutes of Health, Bethesda, MD, USA; https://imagej.nih.gov/ij/download.html).
2.13. Statistics
One-way ANOVA, factorial ANOVA or t-testing was performed, as appropriate, using Microsoft Excel or NCSS statistical software. Fisher’s LSD post-hoc analysis as needed to determine group differences when significant (p ≤ .05) interactions were observed in the ANOVA analysis. Values are reported as mean ± SE.
3. Results
3.1. LKB1 gene expression in muscles from HFHS diet-fed mice in 84 different strains of mice correlates with mitochondria-, proteolysis-, inflammation-, insulin signaling, and oxidative stress-related genes
GO-TERM and ingenuity pathway analysis were performed for the RNA expression from skeletal muscles of 84 strains of mice that were fed a HFHS diet for 8 weeks (Fig. 1A). Based on the varying expression level of LKB1 in the skeletal muscle for all 84 strains of mice, LKB1 expression was negatively (Fig. 1B) and positively (Fig. 1C) correlated with various categories of gene expression. Of particular interest was LKB1’s relationship with the following gene categories. LKB1 was negatively correlated with 1) mitochondria-related genes (cellular compartment: mitochondrion, mitochondrial matrix, mitochondrial inner membrane; pathway analysis: mitochondrial dysfunction, oxidative phosphorylation); 2) protein catabolism-related genes (biological process: protein ubiquitination, ubiquitin-dependent catabolic process, protein ubiquitination protein catabolic process); 3) inflammation and oxidative stress-related genes (biological process: positive regulation of toll-like receptor 3 signaling pathway, positive regulation of nitricoxide synthase activity; pathway analysis: STAT3 Pathway, NF-kB signaling); and 4) insulin signaling related genes (pathway analysis: PI3K/AKT signaling). LKB1 was positively correlated with 1) mitochondria-related genes (biological process: mitochondrial organization, mitophagy); 2) protein catabolism-related genes (biological process: proteolysis, protein deubiquitination; pathway analysis: protein ubiquitination pathway); 3) inflammation and oxidative stress-related genes (pathway analysis: NRF2-mediated oxidative stress response and role of IL-17A/F and IL17A signaling); and 4) other insulin signaling related genes (biological process: regulation of glycogen; pathway analysis: regulation of eIF4 and p70S6k signaling, eIF2 signaling, mTOR signaling, insulin receptor signaling, PI3K/AKT signaling).
Fig. 1.
Association of LKB1 with gene expression in HMDP mice. (A) Gene expression was determined for skeletal muscle (gastrocnemius) from 84 strains of male mice from the hybrid mouse diversity panel [HMDP [33,73,74]] after high-fat/high-sugar diet feeding for 8 weeks (N = 4 mice/strain). Genes that were negatively [biweight midcorellation (BICOR) values <0] and positively (BICOR >0) correlated with p-values <.0001 were selected for further analysis. (B) The top cellular compartment and biological process GO-TERMS, upstream regulators and signaling pathways negatively associated with LKB1 gene expression in skeletal muscle. (C) The top cellular compartment and biological process GO-TERMS, upstream regulators and signaling pathways positively associated with LKB1 gene expression in skeletal muscle.
3.2. SkmLKB1 KO does not affect high-fat diet (HFD)-induced obesity
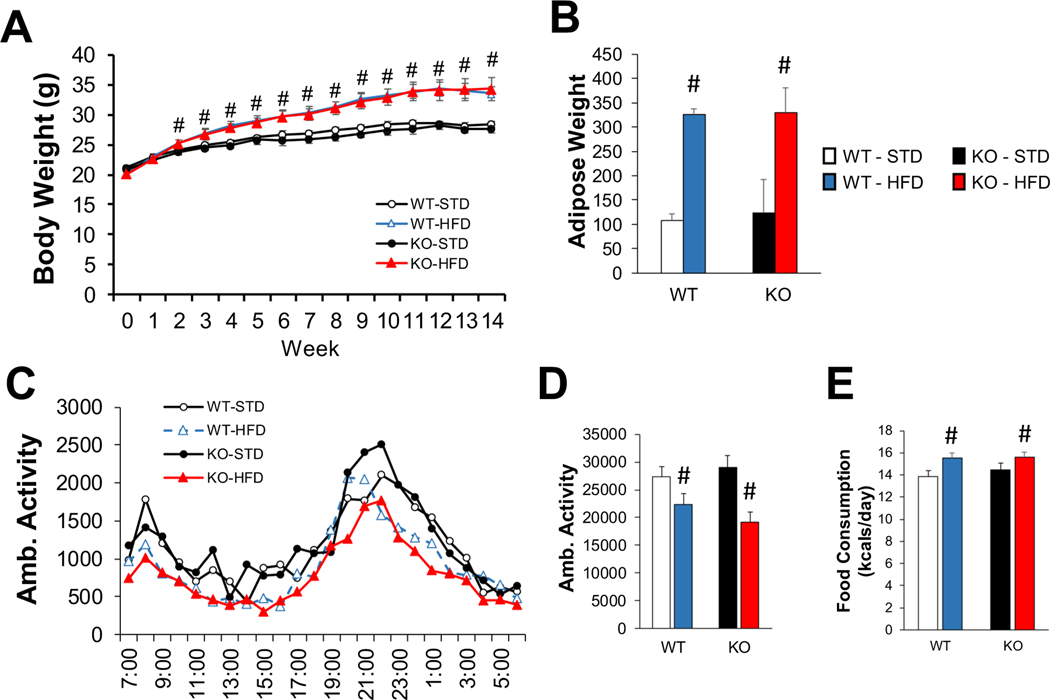
14 weeks of HFD resulted in body weight gain of ∼21% (5.9 g) by the 14th week of feeding compared to standard diet (STD), with no difference in weight between skmLKB1-KO and WT mice at any time-point. (Fig. 2A). Retroperitoneal fat pad weight at 14 weeks was likewise ∼200% higher with HFD vs. STD, with no genotype effect (Fig. 2B). Ambulatory activity decreased (Fig. 2C, D) and caloric consumption increased (Fig. 2E) in a diet-dependent fashion, and these two effects likely combined to drive the HFD induced weight gain.
Fig. 2.
KO does not alter diet-induced obesity. (A) Weekly body weight measurements over the 14-week experimental period (N = 11–15 mice/group). (B) Retroperitoneal fat pad weight at the end of the feeding period. Average hourly (C) and daily (D) in-cage activity was measured over 3 consecutive days between the 74th and 88th day of diet treatment using computer-monitored infrared beam breaks [n = 14 for standard (STD) diet groups, 29 for high fat diet (HFD) groups]. (E) Food consumption was measured over 3 days while the mice were individually housed in activity monitoring cages (n = 15–16 for STD, 30–32 for HFD groups). # = significant (p < .05) main effect of diet. WT = wild-type. KO = skeletal muscle specific LKB1 knockout.
3.3. Skeletal muscle AMPK activity is decreased in skmLKB1-KO mice, but unaffected by 14 weeks of HFD feeding
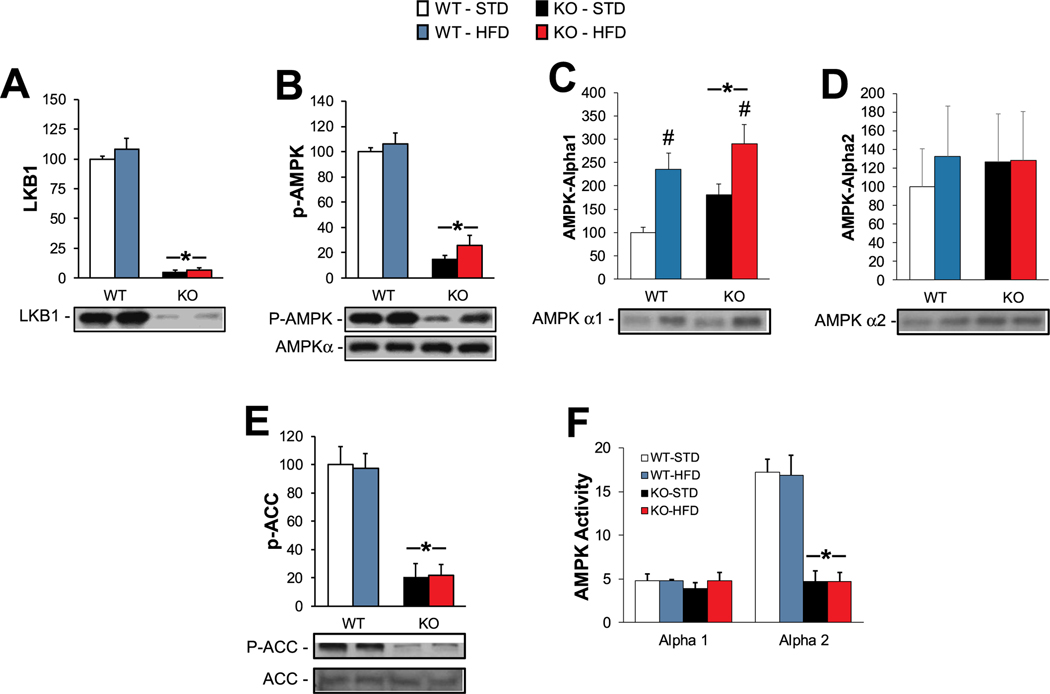
SkmLKB1-KO effectively eliminated LKB1 protein from the QUAD muscles of the KO mice (Fig. 3A). Consistent with LKB1’s role as the primary AMPK kinase in skeletal muscle, AMPK phosphorylation was reduced by ∼85% in KO muscles, while total AMPKα was unaffected by diet or genotype (Fig. 3B). AMPKα1 levels were increased in KO muscles, and were also increased by HFD feeding (Fig. 3C), while AMPKα2 levels were unaffected by genotype and diet (Fig. 3D). However, SAMS peptide-based AMPK activity assay showed AMPK α2 activity was suppressed by KO and unaffected by diet, while AMPKα1 activity was not significantly affected by either diet or genotype (Fig. 3F). The content of acetyl-CoA carboxylase (ACC), a downstream target of AMPK, was not affected by diet or genotype, but its phosphorylation showed a similar pattern as AMPKα2 activity (Fig. 3E).
Fig. 3.
Effects of HFD on LKB1/AMPK content and signaling in muscles from WT and KO mice. Protein content of (A) LKB1, (B) phospho-AMPK (p-AMPK) and total (pan) AMPKα, (C) AMPKα1, and (D) AMPKα2, as determined by western blotting. (E) Phosphorylation of acetyl-CoA carboxylase was measured by western blotting as a marker of in vivo AMPK activity. (F) In vitro AMPK activity was determined for AMPKα1 and α2 subunits that were immunoprecipitated from QUAD homogenates. N = 9–12 / group except for (F) for which N = 6/group. * = significant (p ≤ .05) main effect of KO vs. WT muscles. # = significant main effect of HFD vs. STD.
3.4. SkmLKB1 knockout alters mitochondria-, inflammation- and oxidative stress-related gene expression in muscles from HFD-fed mice
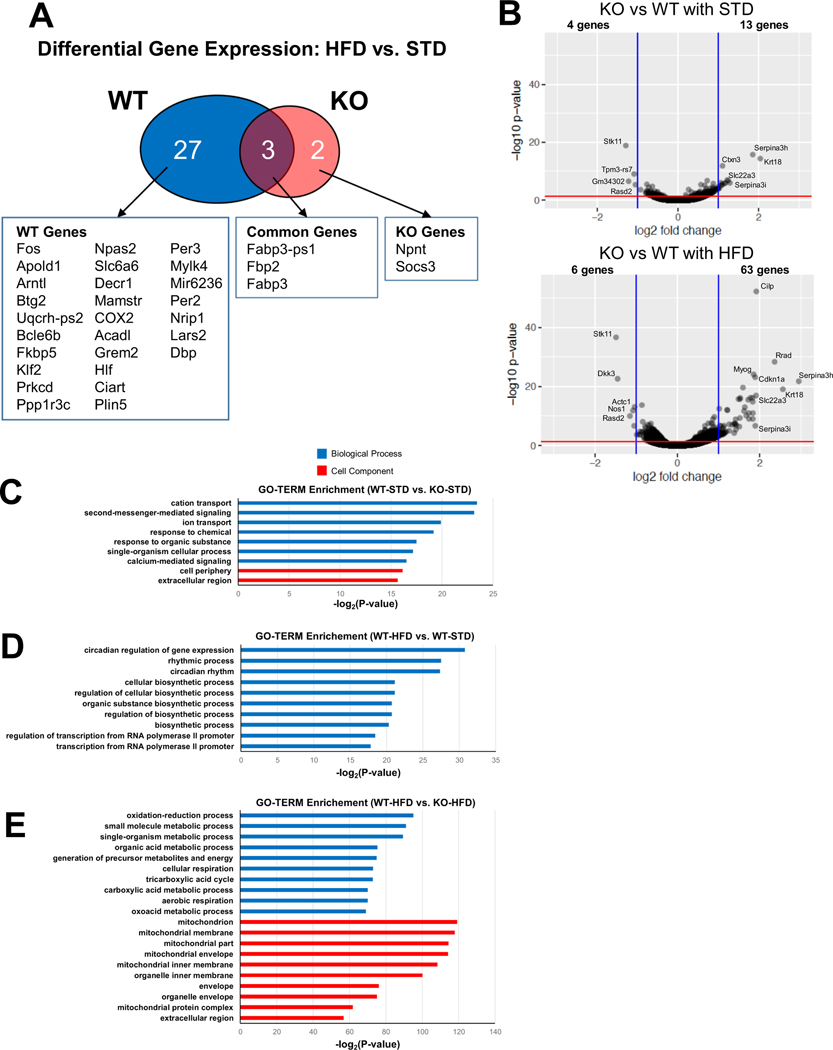
WT and KO mice were fed standard or high-fat chow for 14 weeks after which RNA sequencing was performed on RNA from QUAD muscles. The top 500 genes with the highest variance across all 4 groups (WT-STD, WT-HFD, KO-STD, KO-HFD) were heavily affected by genotype (see heatmap in Fig. S2). Furthermore, while 30 known genes were significantly altered by HFD feeding in WT muscles, only 5 were affected by diet in KO muscles (Fig. 4A, Table S3 and S4). Furthermore, in mice that were fed STD, 17 genes were differentially expressed in KO vs. WT mice (Fig. 4B and Table S5; 4 genes underexpressed and 13 overexpressed). With HFD feeding, the number of differentially expressed genes in KO vs. WT increased to 69 (Fig. 4B and Table S6; 6 genes underexpressed and 63 overexpressed), suggesting that LKB1 is important in the responsiveness of skeletal muscle gene expression to high-fat feeding.
Fig. 4.
RNA sequencing analysis of WT and KO muscles under STD and HFD. (A) Venn diagram showing genes significantly upregulated by HFD feeding in WT and KO muscles. (B) Volcano plots showing the genes significantly reduced or increased by LKB1 knockout under a standard diet (STD) or high fat diet (HFD). (C) Significantly enriched biological process (blue bars) and cell component (red bars) GO TERMs in KO vs. WT mice under STD. (D) Top 10 most-enriched biological process GO TERMs in WT HFD vs. standard diet (STD). No cell component GO TERMS were significantly affected. (E) Top 10 most-enriched biological process and cell component GO TERMs in KO vs. WT muscles under HFD.
To further understand the RNA sequencing data and determine whether general changes in gene expression from skmLKB1-KO muscles under a HFD would be consistent with the gene expression correlations observed in Fig. 1, we also performed GO-TERM analysis on the RNA sequencing data. GO-TERM categories were significantly altered by diet and genotype. In WT-STD vs. KO-STD group, the most enriched GO-TERM categories were mostly membrane-related (e.g. cation transport, second-messenger-mediated signaling, cell periphery; Fig. 4C). Interestingly, the top GO-TERMs enriched by HFD-feeding in muscles from WT mice were circadian rhythm-related (Fig. 4D), while no significant effect of diet on any GO-TERMS was observed in KO muscles (data not shown).
Five-hundred forty-nine GO-TERM categories were significantly different in muscle between WT-HFD vs. KO-HFD groups (data not shown). Consistent with the HMDP data, in HFD-fed mice, mitochondrial-related (e.g. cellular respiration, tricarboxylic acid cycle, mitochondrion, aerobic respiration) and oxidative stress-related (i.e. oxidation-reduction process) GO-TERM categories were among those most highly enriched in WT-HFD vs. KO-HFD muscles (Fig. 4E and Table S7). Inflammation-related GO-TERM categories (e.g. acute inflammatory response, adaptive immune response, innate immune response, macrophage activation) were also significantly affected by genotype (data not shown).
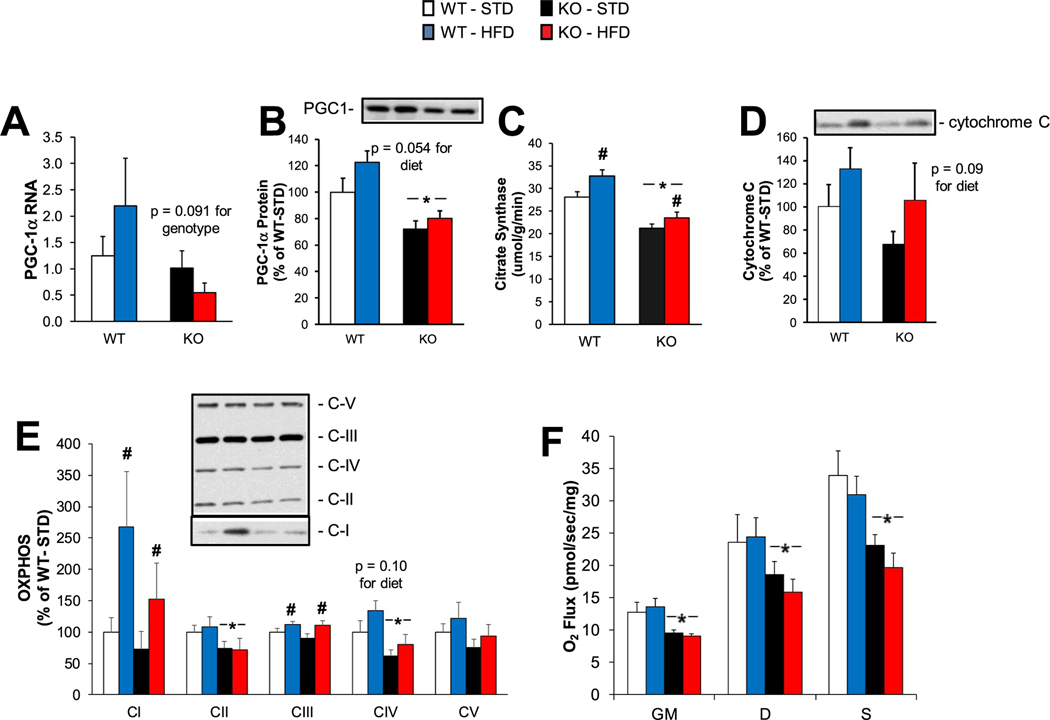
3.5. The lack of LKB1 in muscles from HFD-fed mice results in decreased mitochondrial content and respiration
The HMDP and RNA sequencing data indicated that mitochondrial gene expression in HFD-fed muscles is heavily dependent on LKB1 expression level. Therefore, we examined markers of mitochondrial content and function in skmLKB1-KO muscles. Transcription levels for PGC-1α, a master transcriptional coactivator for mitochondrial gene expression, tended to be lower (Fig. 5A), and the protein level for PGC-1α was significantly lower (Fig. 5B) in KO vs. WT muscles. Citrate synthase activity, reflecting the mitochondrial content in skeletal muscle, was increased by HFD feeding in both KO and WT muscles, but was lower in KO muscles regardless of diet (Fig. 5C). Cytochrome C protein levels were not significantly affected by diet or genotype, though they tended to increase with HFD (Fig. 5D). The protein content for electron transport chain complex I and III proteins increased with HFD, regardless of genotype, while complex II and IV components were decreased in KO muscles, regardless of diet (Fig. 5E). Consistent with lower mitochondrial content, mitochondrial respiration was lower in KO muscles, regardless of diet (Fig. 5F).
Fig. 5.
Effects of LKB1-KO on mitochondrial content and function under a high-fat diet. (A) Gene expression (n = 8/group) and (B) protein content of PGC-1⍺ in QUAD muscles were determined by RT-qPCR and western blotting, respectively. (C) Citrate synthase activity was measured via spectrophotometric assay as a marker of mitochondrial content (n = 8/group). (D) Protein content of Cytochrome C was determined by western blotting (n = 7–9/group). (E) Protein content of electron transport proteins [complex I (CI), complex II (CII), complex III (CIII), complex IV (CIV), and complex V (CV)] were determined by western blotting (n = 9/group). (F) Mitochondrial respiration was measured using the Oroboros O2K oxygraph. Oxygen flux (n = 4–8/group) was determined in permiabilized soleus muscles after stabilization of leak oxygen consumption under glutamate and malate (“GM”); Oxidative phosphorylation capacity was the determined after the addition of ADP (“D”), followed by the addition of succinate (“S”). Data are reported as means ± SEM.* = significant (p < .05) main effect of KO vs. WT muscles. # = significant main effect of HFD vs. STD.
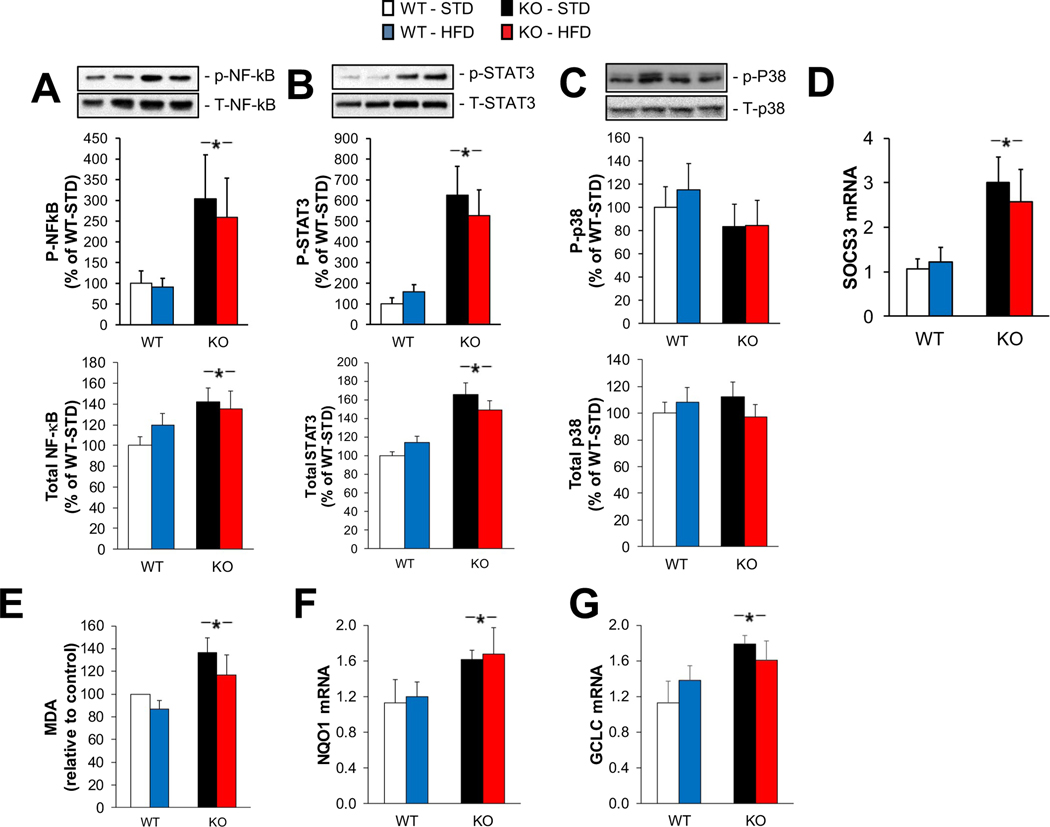
3.6. Markers of inflammation and oxidative stress are elevated in KO muscles
In addition to changes in mitochondrial gene expression, both HMDP and RNA sequencing data indicated that inflammation related genes have a close relationship with LKB1 expression levels under the HFD feeding condition. Therefore, we assessed inflammatory signaling. Phosphorylation of nuclear factor-κB (NFκB; Fig. 6A) and signal transducer of activated T-cells (STAT3; Fig. 6B), but not p38 mitogen-activated protein kinase (p38; Fig. 6C) were elevated in KO muscles compared to WT, but diet had no effect on these inflammatory markers in either genotype. Total protein levels for all three proteins showed the same pattern (Fig. 6A, B, and C). Suppressor of cytokine signaling 3 (SOCS3) gene expression, which is induced by inflammatory signaling, was elevated in KO muscles, with no effect of diet (Fig. 6D).
Fig. 6.
Effects of LKB1-KO on inflammatory signaling and oxidative stress under a HFD. Protein content of phosphorylated (p-) and total NFκB (A), STAT3 (B), and p38 (C) was measured by western blotting (n = 9/group). (D) Gene expression of SOCS3 was determined by RT-qPCR (n = 5/group). * = significant main effect for KO vs. WT; # = significant main effect for HFD vs. STD. (E) Lipid peroxidation was quantified by a spectrophotometric assay of malondialdehyde (MDA). Gene expression of NQO1 (F), and GCLC (G) were determined by RT-PCR. (n = 5/group). * = significant main effect of KO vs. WT.
Indicators of oxidative stress were also elevated in KO muscles. Malondialdehyde (MDA), a lipid peroxidation indicator, increased in KO muscles compared to WT muscles, regardless of diet (Fig. 6E). The transcription levels of the oxidative stress responsive genes NQO1 (Fig. 6F) and GCLC (Fig. 6G) all showed the same change. However, expression of other oxidative stress responsive genes (Prx1 and Gpx1) were not affected by diet or genotype (data not shown).
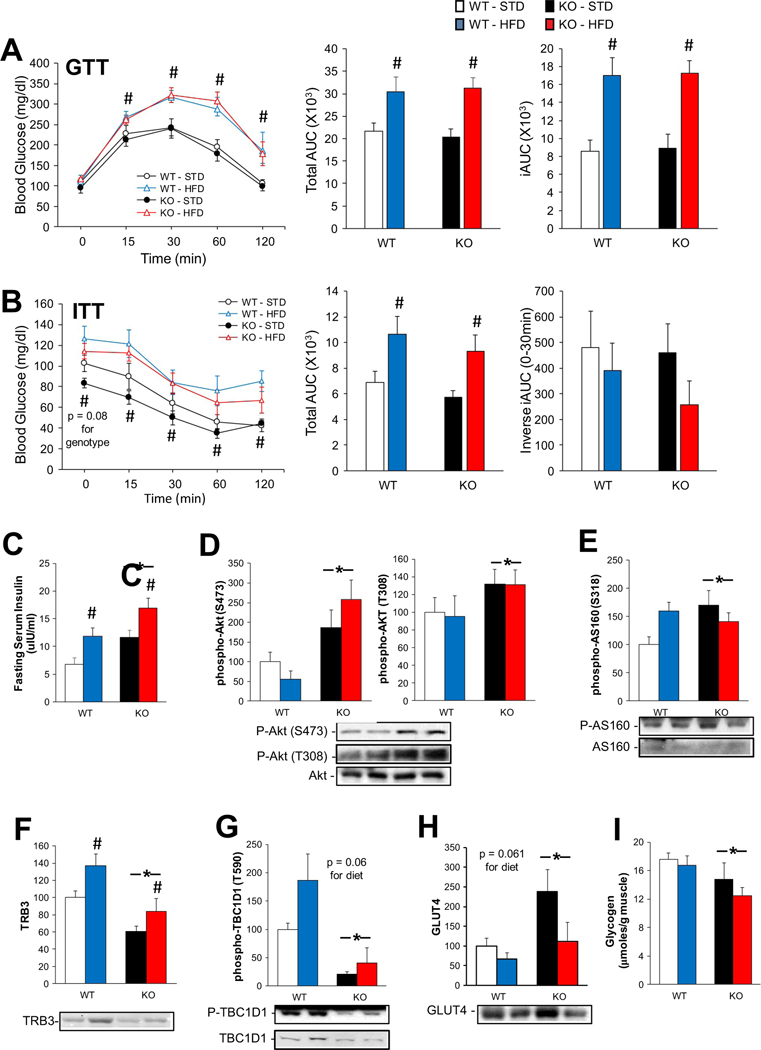
3.7. Glucose tolerance and insulin signaling are disrupted in KO mice under a HFD
WT and skmLKB1-KO mice on HFD had impaired glucose tolerance vs. the STD-fed mice, but genotype did not affect the GTT results (Fig. 7A). ITT showed the same change except for the inverse iAUC (area under the curve) during the first 30 min, which was not affected by diet or genotype (Fig. 7B). Neither diet nor genotype significantly affected the fasting glucose level (Fig. 7A and B), although it tended (p = .08) to be lower in KO vs. WT regardless of diet. The fasting insulin level was increased by HFD and in KO mice (Fig. 7C). Akt phosphorylation (downstream of the insulin receptor) at S473 and T308 was elevated in the KO muscles (Fig. 7D), as was phosphorylation of AS160 at S318, one of the Akt target sites (Fig. 7E). The level of TRB3, an Akt inhibitor, was increased by HFD, and decreased in KO muscles (Fig. 7F). Phosphorylation of TBC1D1 at T590, an AMPK target site, was decreased in the KO muscles, as expected (Fig. 7G). Total TBC1D1 tended (p = .07) to increase with HFD in WT, but not KO muscles (data not shown). HFD feeding tended (p = .061) to decrease muscle GLUT4 levels in both genotypes, and, interestingly, GLUT4 content was elevated in KO muscles (main effect), but this was primarily due to an increase in the STD-fed, but not HFD-fed mice (Fig. 7H). Glycogen levels were slightly lower in KO vs. WT muscles (Fig. 7I).
Fig. 7.
Effects of KO on glucose and insulin tolerance under a HFD. (A) Intraperitoneal glucose tolerance test (GTT); blood glucose levels were determined at the indicated times after an intraperitoneal injection of glucose (2 mg/g BW) in fasted (6 h) WT and KO mice. Total area under the curve (AUC) for 0–120 min of the GTT, and integrated area under the curve (iAUC; area below the curve, but above baseline levels) are shown to the right. (B) Intraperitoneal insulin tolerance test (ITT); blood glucose levels were determined at the indicated times after an intraperitoneal injection of insulin (0.5 U/kg BW) in fasted (4 h) WT and KO mice after 14 weeks of STD or HFD feeding. Total AUC for ITT and the inverse iAUC (area above the curve but below baseline levels) for the 0–30 min time points are shown to the right. (C) Serum insulin concentration as determined by radioimmunoassay after a 6–8 h fast. Content of phosphorylated Akt at Ser473 and Thr308 (D) and of phosphorylated AS160 at Ser318 (E), TRB3 (F), phosphorylated TBC1D1 at Thr590 (G) and GLUT4 (H) in QUAD muscles were measured by western blot (n = 7–9 except for GLUT4 where n = 4–6). (I) Glycogen content of skeletal muscle from fed mice after 14 weeks of STD or HFD feeding (n = 7–8). * = main effect of KO vs. WT; # = main effect of HFD vs. STD.
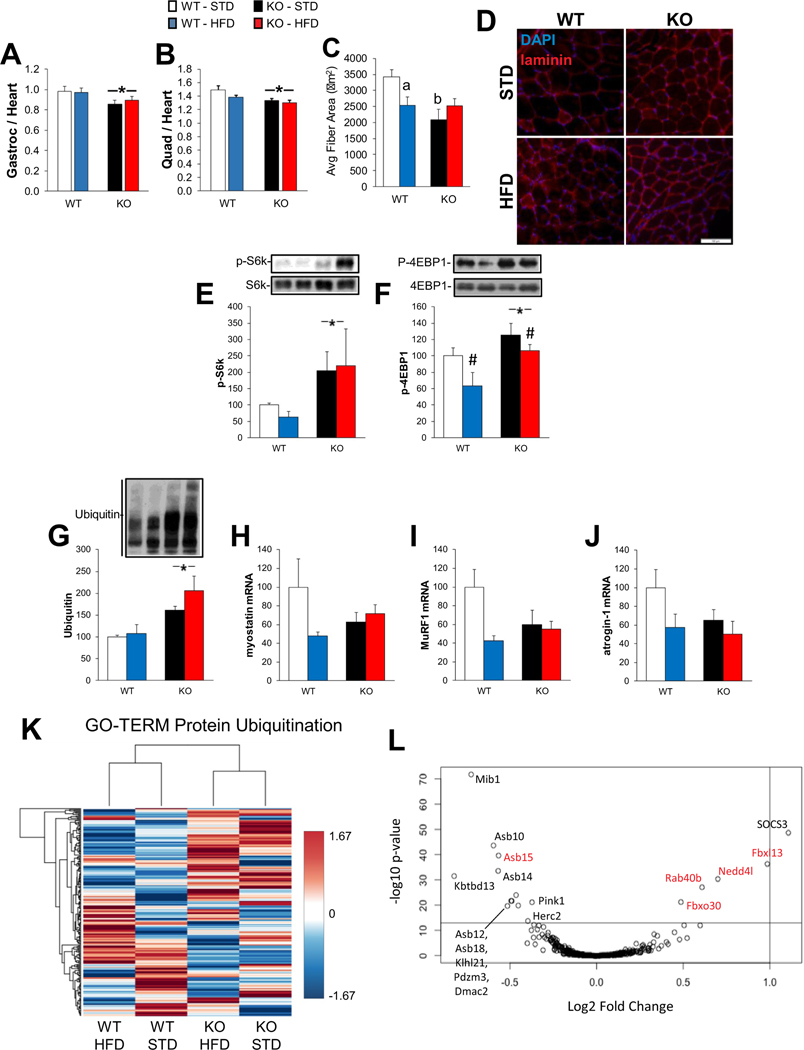
3.8. Muscle mass is decreased, and protein ubiquitination increased in KO mice, despite elevated mTOR anabolic signaling
Gastrocnemius (Fig. 8A) and QUAD (Fig. 8B) weight were both decreased in KO mice, but were not significantly affected by diet. No significant differences were observed for the TA muscle between diets or genotype (data not shown), suggesting that the effect of genotype may be specific to weight-bearing muscles. Average myofiber area was lower in KO vs. WT QUAD muscles under STD (Fig. 8C, D; Fig. S4). HFD resulted in decreased average fiber area in WT, but not KO QUAD (Fig. 8C). No significant differences were observed in WT vs. KO muscle fiber size under HFD. Therefore, knockout of LKB1 seems be able to protect QUAD from HFD induced fiber atrophy. Phosphorylation of ribosomal protein S6 kinase (S6k) and eukaryotic initiation factor 4E binding protein 1 (4EBP1), both readouts for mechanistic target of rapamycin complex 1 (mTORC1) activity, were elevated in KO muscles (Fig. 8E, F). Ubiquitin levels were elevated in KO muscles regardless of diet (Fig. 8G). Expression of myostatin, MuRF1 and atrogin-1 mRNA was not significantly different between genotypes or diet, although a trend was observed in all cases for decreased expression with HFD in WT but not KO muscles, and lower expression in KO-STD vs. WT-STD muscles (Fig. 8H, I, and J). Substantial differences in muscle gene expression between WT and KO mice seem apparent within the “Protein Ubiquitination” GO-TERM category when expressed as a heat-map (Fig. 8K). Expression of several ubiquitin ligase genes (rab40b, nedd4l, fbx13, fbxo30) was increased in KO vs. WT muscles (Fig. 8L). These might contribute to the elevated ubiquitination and decreased muscle mass in KO muscles.
Fig. 8.
Effects of KO and HFD on muscle size and growth-related signaling. (A) Ratio of gastrocnemius (Gastroc) to heart weight (n = 11–12) for wild-type (WT) and skeletal muscle specific LKB1 knockout (KO) mice fed standard diet (STD) or high-fat diet (HFD) for 14 weeks. (B) Ratio of quadricep (QUAD) to heart weight (n = 11–12). (C) Average cross-sectional area of QUAD muscle fibers (n = 5–6). (F) Representative immunofluorescent images showing DAPI (blue) and laminin (red) staining for data in (C). Phosphorylation of S6k at Thr389 (E) and 4E-BP1 at Thr37/46 (F) was measured by western blotting. (G) Ubiquitin content was measured by western blotting (n = 5–6 per group). Gene expression (n = 4–6) for myostatin (H), MuRF1 (I), atrogin-1 (J) were measured by RT-PCR. (K) Heatmap showing gene expression of genes in the “protein ubiquitination” GO-TERM. (L) Volcano plot of gene expression within the ubiquitin GO-TERM with Log2 fold change in KO vs. WT on the X axis, and adjusted p-value on the y-axis. Red genes could explain increased ubiquitination in KO muscles (panel G). * = significant main effect of KO vs. WT; # = significant main effect of HFD vs. STD; a = significant difference between WT-HFD vs. WT-STD; b = significant difference of KO-STD vs. WT-STD.
4. Discussion
The primary purpose of this study was to determine how the lack of skeletal muscle LKB1 alters gene expression in muscles from obese mice. While LKB1 KO substantially affected the expression of many genes regardless of diet and seemed to attenuate high-fat diet induced changes in gene expression, it did not have major effects on the development of obesity, nor did it severely impair insulin or glucose tolerance in obesity.
Previous work using microarray gene expression analysis showed that the greatest gene expression differences in muscles lacking LKB1 were related to energy production and lipid metabolism. This was associated with decreased SIRT1 deacetylase activity toward p53, along with increased HDAC4 content and reduced HDAC4 phosphorylation [2]. We observed similar differences in gene expression in mice fed a HFD, and also observed that the lack of LKB1 appeared to limit adaptation of gene expression to HFD feeding.
LKB1’s effects on gene expression in skeletal muscle are likely mediated by multiple downstream effectors. While LKB1 itself may localize to the nucleus and directly engage in transcriptional regulation by stabilizing transcription factors such as p53 [46], its major effects on transcription are likely mediated through its activation of AMPK family members. For instance, AMPK itself can regulate several transcription factors and coactivators that are key players in the control of the genes affected by LKB1 knockout in the present study, including PPARα and PGC1α [47], CREB [48], FOXO [49], and nuclear factor erythroid 2-related factor (NRF2) [50]. Furthermore, AMPK regulates epigenetic regulators such as histone deacetylase 4 and 5 [51,52] as well as SIRT1 [53], consistent with findings in LKB1-KO muscles [2].
Decreased AMPK activity likely mediates many of the effects of skmLKB1-KO on gene exprerssion. Indeed, global knockout of the AMPKγ3 subunit results in gene expression changes that are very similar to those observed in the present study. The AMPKγ3 subunit is predominantly expressed in skeletal muscle, and accounts for most of the activation of AMPK in exercising muscle [54]. Using both gain of function (AMPKγ3 R225Q transgenic) and loss of function (global AMPKγ3 knockout) genetic models, Nilsson, et al. [55] identified many genes whose expression was regulated conversely by the two models, strongly supporting AMPK’s involvement in their regulation. Upon inspection of their individual expression patterns, the majority of these genes, including specific ones highlighted by those authors related to glycogen synthesis (gbe1, ugp2), p38 MAPK regulation (map2k6, dusp10), lipid metabolism (srebf1, ppargc1a, cd36, oxct1, ces3) as well as nitric oxide synthase (nos1) were similarly regulated in our muscle-specific LKB1 knockout mice (data not shown). This suggests that much of LKB1’s effect on gene expression is mediated through AMPK, although other AMPK family members likely play a role as well.
Multiple models of skeletal muscle LKB1 deficiency have clearly established it as a positive regulator of mitochondrial content and gene expression under a standard diet [3,4,56]. Here, we have extended those observations to muscles from HFD-fed mice. First, we found a positive association between expression of LKB1 and expression of mitochondrial cell component genes across 84 strains of HFD-fed mice. This association was confirmed by RNA sequencing data. GO-TERM analysis showed that 6 of the top 10 most enriched GO-TERM cell component categories in WT-HFD vs. KO-HFD muscles were mitochondria-related. This deficit in mitochondrial gene expression was likely driven in part by the observed decrease in PGC-1α protein content in KO muscles and is consistent with decreased mitochondrial content as evidenced by decreased citrate synthase activity, electron transport chain complex II and IV constituents, and mitochondrial respiration in the KO muscles.
Our data is in general agreement with many previous studies that have demonstrated increased mitochondrial content and/or respiration in skeletal muscle under HFD-induced obesity [38,57–60]. We found that citrate synthase activity and protein content for complex I and III of the electron transport chain increased with HFD feeding. Trends for increased PGC-1α, cytochrome C and ETC complex IV with HFD feeding were also observed. However, the lack of skmLKB1 had no significant effect on these HFD-induced increases in mitochondrial markers. Based on this, we conclude that LKB1 is more important in establishing basal mitochondrial content and function than it is for diet-induced mitochondrial adaptation, and that these basal mitochondrial deficits are maintained under a HFD. In line with the decreased mitochondrial content, previous work has shown that fat oxidation is impaired in LKB1 and AMPK knockout models [5,61]. It would be interesting in future work to determine how substrate-specific respiration is impacted under HFD in the skmLKB1-KO mice.
Diet induced obesity induces a low-level inflammatory response in many tissues of the body, including skeletal muscle, and this has been postulated to contribute to skeletal muscle metabolic dysfunction in obesity [62]. On the other hand, LKB1 and AMPK signaling in muscle generally blocks anti-inflammatory signaling [23,24]. The HMDP pathway analysis indicated that expression of NF-κB signaling pathway components is negatively correlated to LKB1-expression in muscle under HFD. This is consistent with our previous work in contracting skeletal muscle where KO muscle was susceptible to contraction-induced NF-κB activation and inflammation [23]. Several inflammation-related GO-TERM categories were significantly overrepresented in HFD-fed KO vs. WT muscles including “inflammatory response” and “acute inflammatory response” (data not shown). Consistent with this, protein content and phosphorylation for NF-κB and STAT3, both of which are important transcription factors regulating inflammation-related gene expression, were increased in KO vs. WT muscles (Fig. 6). mRNA expression of SOCS3 (an indicator of inflammatory response) was also increased in KO muscles. In this case, the hyper-inflammatory state, however, seems to be mainly a function of genotype, as diet had little effect on any of these markers.
Since inflammation and oxidative stress responses are closely related and synergistic, it is not surprising that indicators of oxidative stress are also elevated in KO muscles, regardless of diet. HMDP analysis identified the NRF2-mediated oxidative stress response pathway as one that is significantly correlated with LKB1 levels. Furthermore, the GO-TERM “oxidation-reduction process” was the cell process most-impacted by LKB1 knockout in muscles from HFD-fed mice. The elevated lipid peroxidation and expression of the oxidative stress-responsive genes NQO1 and GCLC in KO muscles, regardless of diet further illustrate the importance of LKB1 in maintaining proper redox balance in skeletal muscle. However, these indicators of oxidative stress were not impacted by diet in WT or KO muscles.
Insufficient mitochondrial function [63,64], oxidative stress [18], and inflammation and inflammatory signaling [65] are all associated with obesity-related metabolic dysfunction and insulin resistance in skeletal muscle. Therefore, exacerbated HFD-induced insulin resistance might be expected in KO mice vs. WT given the decreased mitochondrial content and elevated markers of inflammation and oxidative stress in their muscles. However, GTT and ITT testing in the present study gave no indication that skmLKB1 knockout induced impairment of glucose or insulin tolerance basally or under HFD, which is consistent with previous work with mice expressing a dominant negative LKB1 construct [56], as well as findings that fasting 2-deoxyglucose clearance and uptake is unaffected in a similar muscle-specific LKB1-KO (mLKB1-KO) mouse model in which LKB1 was knocked out of both cardiac and skeletal muscle [2]. Furthermore, Koh et al. reported that basal insulin sensitivity was increased in mLKB1-KO mice, as measured by hyperinsulinemic-euglycemic clamp. This occurred in conjunction with decreased blood glucose and serum insulin levels, as well as decreased GTT AUC. This improvement in insulin sensitivity was attributed to enhanced Akt function due to decreased expression of TRB3, an Akt inhibitor [9]. Like Koh et al., we also observed decreased TRB3 content and increased Akt phosphorylation in our skmLKB1-KO muscles, but without any indication of improved glucose or insulin tolerance. This is in line with results from global TRB3 knockout mice in which GTT and ITT were unaffected by the lack of TRB3 despite elevated liver Akt phosphorylation [66]. Nonetheless, insignificant trends for lower fasting glucose levels and ITT AUC in our skmLKB1-KO mice might suggest the possibility of mildly improved insulin sensitivity. However, these trends are more likely due to the elevated fasting insulin levels that we observed in skmLKB1-KO mice, which, taken together with the lower glucose levels, suggest that skmLKB1-KO somehow leads to increased insulin release from the pancreatic beta cells. Presumably, since these are skeletal muscle-specific knockouts, the increased insulin might be mediated by release, or cessation of release, of an unknown skeletal muscle endocrine factor that regulates the pancreatic islets. For instance, growth differentiation factor 15 (GDF15) and CX3CL1 are both endocrine factors released from skeletal muscle that have been shown to enhance insulin release from the beta cells [67]. Interestingly, our RNA sequencing data shows that expression of CXCL1 and perhaps GDF15 are elevated in KO muscles (see Fig. S3), suggesting that skeletal muscle LKB1 signaling may be a critical regulator of pancreatic β-cell function. In contrast to our findings, Koh et al. observed lower fasting insulin levels in mLKB1-KO vs. WT mice [9]. While the reason for this discrepancy is unknown, we can speculate that the knockout model is a contributing factor. The lack of cardiac LKB1 in the mLKB1-KO model leads to cardiac failure [6,68], which, in turn, is known to impair insulin release by the pancreas [69]. Thus, early stages of heart failure in the mLKB1-KO mouse may counteract any putative stimulatory effects of skeletal muscle LKB1 deficiency on pancreatic insulin release. This, of course, would need to be substantiated by further study. Taken together with the work of others, our findings indicate that the lack of LKB1 in skeletal muscle does not impair, and may or may not improve, whole body insulin sensitivity basally and under HFD. Further research will be needed to further clarify the mechanism behind this unintuitive finding.
LKB1-AMPK signaling inhibits mTORC1 activity in skeletal muscle [70–72]. Accordingly, S6k and 4EBP1 phosphorylation were both elevated in the KO muscles regardless of diet. This is consistent with previous work from our lab in which phosphorylation of S6k, ribosomal protein S6, and 4E-BP1 was elevated along with increased protein synthesis in skmLKB1-KO muscles [71]. Despite the elevated anabolic signaling through mTORC1, however, the lack of LKB1 resulted in smaller weight-bearing muscles and muscle fibers in STD-fed mice. This was likely due to increased protein degradation since muscle protein ubiquitination was elevated. Strangely, expression of the classic skeletal muscle ubiquitin ligases (MuRF1 and Atrogin-1) tended to be lower in the KO muscles, which would promote decreased ubiquitination. A careful assessment of ubiquitin-related gene expression suggests that the smaller muscle mass may be due to expression of other factors. For instance, RNA sequencing indicated that gene expression for other ubiquitin ligases (Nedd4l, Fbxl13, Rab40b and Fbxo30) was increased in LKB1-KO muscles, while expression of ubiquitin specific peptidase 24 and 47 (USP24 and USP47; deubiquitinating enzymes that are highly expressed in skeletal muscle) was decreased in KO muscles (Fig. S4). Decreased deubiquitination would explain the elevated protein ubiquitin levels, and may contribute, therefore, to the decreased muscle size in the KO mice.
In conclusion, skeletal muscle specific loss of LKB1 in diet-induced obese mice alters expression of many genes regulating mitochondria, inflammation and oxidative stress, but this has no effect on the degree of obesity or glucose/insulin tolerance.
Supplementary Material
Acknowledgments
We are grateful for the kind provision of transgenic “floxed” LKB1 mice and Myf6-Cre mice by R. DePinho (Dana-Farber Cancer Institute, Boston, MA) and M.R. Capecchi (University of Utah, Salt Lake City, UT) respectively, and for the assistance of Marc Matsumura, Michael Harding, Michael Duplisea, Jacob Tsukamoto, Conrad Ashby, Eve Wang, and Stephen Doxey for assistance with the collection of data. Funding for this project came from BYU Mentoring Environment and Graduate Fellowship grants.
Abbreviations:
- 4EBP1
eukaryotic initiation factor 4E binding protein 1
- AMPK
AMP-activated protein kinase
- ARK
AMPK-related kinase
- Ark5
AMPK-related kinase 5
- ASB
ankyrin repeat And SOCS box containing
- AUC
area under the curve
- CamKKβ
calcium calmodulin-dependent protein kinase kinase 2
- DIO
Diet-induced obesity
- Fbx
F box protein
- GO
gene ontology
- GDF15
growth differentiation factor 15
- HFD
high fat diet
- HF/HS
high fat/high sucrose diet
- HMDP
hybrid mouse diversity panel
- IPGTT/GTT
intraperitoneal Glucose Tolerance Testing
- IPITT/ITT
intraperitoneal Insulin Tolerance Testing
- KO
skeletal muscle specific LKB1 knockout mouse
- LKB1
liver kinase B1
- MDA
malondialdehyde
- mTORC1
mechanistic target of rapamycin complex 1
- Nedd4l
neural precursor cell expressed developmentally down-regulated 4 like
- NFκB
nuclear factor-κB
- NUAK1
NUAK family SNF1-like kinase 1
- p38
p38 mitogen-activated protein kinase
- PBS
phosphate-buffered saline
- QUAD
quadriceps
- S6k
ribosomal protein S6 kinase
- SIK
salt-inducible kinase
- SNARK
sucrose nonfermenting AMPK-related kinase
- SOCS3
suppressor of cytokine signaling 3
- STAT3
signal transducer of activated T-cells
- STK11
serine/threonine kinase 11
- STD
standard diet
- TA
tibialis anterior muscle
- TBST
tris-buffered saline + tween
- USP
ubiquitin specific peptidase
- WT
wildtype
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Ting Chen:Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization, Project administration.Jonathon T. Hill:Conceptualization, Methodology, Software, Formal analysis, Data curation, Writing - original draft.Timothy M. Moore:Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing - original draft.Eric C.K. Cheung:Formal analysis, Investigation, Data curation.Zachary E. Olsen:Formal analysis, Investigation, Data curation.Ted B. Piorczynski:Formal analysis, Investigation, Data curation.Tanner D. Marriott:Investigation, Data curation.Jeffery S. Tessem:Methodology, Resources, Formal analysis, Data curation, Supervision.Chase M. Walton:Formal analysis, Investigation, Data curation.Benjamin T. Bikman:Methodology, Resources, Formal analysis, Data curation, Supervision.Jason M. Hansen:Methodology, Resources, Formal analysis, Data curation, Supervision.David M. Thomson:Conceptualization, Methodology, Formal analysis, Investigation, Resources, Data curation, Writing original draft, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbadis.2020.165805.
References
- [1].Sebbagh M, Olschwang S, Santoni MJ, Borg JP, The LKB1 complex-AMPK pathway: the tree that hides the forest, Familial Cancer 10 (2011) 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jeppesen J, Maarbjerg SJ, Jordy AB, Fritzen AM, Pehmoller C, Sylow L, Serup AK, Jessen N, Thorsen K, Prats C, Qvortrup K, Dyck JR, Hunter RW, Sakamoto K, Thomson DM, Schjerling P, Wojtaszewski JF, Richter EA, Kiens B, LKB1 regulates lipid oxidation during exercise independently of AMPK, Diabetes 62 (2013) 1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Thomson DM, Porter BB, Tall JH, Kim HJ, Barrow JR, Winder WW, Skeletal muscle and heart LKB1 deficiency causes decreased voluntary running and reduced muscle mitochondrial marker enzyme expression in mice, Am. J. Physiol. Endocrinol. Metab. 292 (2007) E196–E202. [DOI] [PubMed] [Google Scholar]
- [4].Tanner CB, Madsen SR, Hallowell DM, Goring DM, Moore TM, Hardman SE, Heninger MR, Atwood DR, Thomson DM, Mitochondrial and performance adaptations to exercise training in mice lacking skeletal muscle LKB1, Am. J. Physiol. Endocrinol. Metab. 305 (2013) E1018–E1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Thomson DM, Brown JD, Fillmore N, Condon BM, Kim HJ, Barrow JR, Winder WW, LKB1 and the regulation of malonyl-CoA and fatty acid oxidation in muscle, Am. J. Physiol. Endocrinol. Metab 293 (2007) E1572–E1579. [DOI] [PubMed] [Google Scholar]
- [6].Thomson DM, Hancock CR, Evanson BG, Kenney SG, Malan BB, Mongillo AD, Brown JD, Hepworth S, Fillmore N, Parcell AC, Kooyman DL, Winder WW, Skeletal muscle dysfunction in muscle-specific LKB1 knockout mice, J. Appl. Physiol 108 (2010) (1985) 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Imai K, Inukai K, Ikegami Y, Awata T, Katayama S, LKB1, an upstream AMPK kinase, regulates glucose and lipid metabolism in cultured liver and muscle cells, Biochem. Biophys. Res. Commun 351 (2006) 595–601. [DOI] [PubMed] [Google Scholar]
- [8].Sakamoto K, McCarthy A, Smith D, Green KA, Grahame Hardie D, Ashworth A, Alessi DR, Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction, EMBO J. 24 (2005) 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Koh HJ, Arnolds DE, Fujii N, Tran TT, Rogers MJ, Jessen N, Li Y, Liew CW, Ho RC, Hirshman MF, Kulkarni RN, Kahn CR, Goodyear LJ, Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3, Mol. Cell. Biol 26 (2006) 8217–8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bright NJ, Thornton C, Carling D, The regulation and function of mammalian AMPK-related kinases, Acta Physiol (Oxf) 196 (2009) 15–26. [DOI] [PubMed] [Google Scholar]
- [11].Kahn BB, Alquier T, Carling D, Hardie DG, AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism, Cell Metab. 1 (2005) 15–25. [DOI] [PubMed] [Google Scholar]
- [12].Lin SC, Hardie DG, AMPK: sensing glucose as well as cellular energy status, Cell Metab. 27 (2018) 299–313. [DOI] [PubMed] [Google Scholar]
- [13].Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D, LKB1 is the upstream kinase in the AMP-activated protein kinase cascade, Curr. Biol 13 (2003) 2004–2008. [DOI] [PubMed] [Google Scholar]
- [14].Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG, Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase, Cell Metab. 2 (2005) 9–19. [DOI] [PubMed] [Google Scholar]
- [15].Lemonnier D, Winand J, Furnelle J, Christophe J, Effect of a high-fat diet on obese-hyperglycaemic and non-obese Bar Harbor mice, Diabetologia 7 (1971) 328–333. [DOI] [PubMed] [Google Scholar]
- [16].Inazuka F, Sugiyama N, Tomita M, Abe T, Shioi G, Esumi H, Muscle-specific knock-out of NUAK family SNF1-like kinase 1 (NUAK1) prevents high fat diet-induced glucose intolerance, J. Biol. Chem 287 (2012) 16379–16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Karasawa H, Nagata-Goto S, Takaishi K, Kumagae Y, A novel model of type 2 diabetes mellitus based on obesity induced by high-fat diet in BDF1 mice, Metabolism 58 (2009) 296–303. [DOI] [PubMed] [Google Scholar]
- [18].Di Meo S, Iossa S, Venditti P, Skeletal muscle insulin resistance: role of mitochondria and other ROS sources, J. Endocrinol 233 (2017) R15–R42. [DOI] [PubMed] [Google Scholar]
- [19].Bikman BT, A role for sphingolipids in the pathophysiology of obesity-induced inflammation, Cell. Mol. Life Sci 69 (2012) 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Keane KN, Calton EK, Carlessi R, Hart PH, Newsholme P, The bioenergetics of inflammation: insights into obesity and type 2 diabetes, Eur. J. Clin. Nutr 71 (2017) 904–912. [DOI] [PubMed] [Google Scholar]
- [21].Srikanthan P, Hevener AL, Karlamangla AS, Sarcopenia exacerbates obesity-associated insulin resistance and dysglycemia: findings from the National Health and Nutrition Examination Survey III, PLoS One 5 (2010) e10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kjobsted R, Hingst JR, Fentz J, Foretz M, Sanz MN, Pehmoller C, Shum M, Marette A, Mounier R, Treebak JT, Wojtaszewski JFP, Viollet B, Lantier L, AMPK in skeletal muscle function and metabolism, FASEB J. 32 (2018) 1741–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen T, Moore TM, Ebbert MT, McVey NL, Madsen SR, Hallowell DM, Harris AM, Char RE, Mackay RP, Hancock CR, Hansen JM, Kauwe JS, Thomson DM, Liver kinase B1 inhibits the expression of inflammation-related genes postcontraction in skeletal muscle, J. Appl. Physiol 120 (2016) (1985) 876–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Green CJ, Macrae K, Fogarty S, Hardie DG, Sakamoto K, Hundal HS, Counter-modulation of fatty acid-induced pro-inflammatory nuclear factor kappaB signalling in rat skeletal muscle cells by AMP-activated protein kinase, Biochem. J 435 (2011) 463–474. [DOI] [PubMed] [Google Scholar]
- [25].Morales-Alamo D, Calbet JAL, AMPK signaling in skeletal muscle during exercise: role of reactive oxygen and nitrogen species, Free Radic. Biol. Med 98 (2016) 68–77. [DOI] [PubMed] [Google Scholar]
- [26].Yang Z, Wang X, He Y, Qi L, Yu L, Xue B, Shi H, The full capacity of AICAR to reduce obesity-induced inflammation and insulin resistance requires myeloid SIRT1, PLoS One 7 (2012) e49935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Barnes BR, Marklund S, Steiler TL, Walter M, Hjalm G, Amarger V, Mahlapuu M, Leng Y, Johansson C, Galuska D, Lindgren K, Abrink M, Stapleton D, Zierath JR, Andersson L, The 5’-AMP-activated protein kinase gamma3 isoform has a key role in carbohydrate and lipid metabolism in glycolytic skeletal muscle, J. Biol. Chem 279 (2004) 38441–38447. [DOI] [PubMed] [Google Scholar]
- [28].Steinberg GR, O’Neill HM, Dzamko NL, Galic S, Naim T, Koopman R, Jorgensen SB, Honeyman J, Hewitt K, Chen ZP, Schertzer JD, Scott JW, Koentgen F, Lynch GS, Watt MJ, van Denderen BJ, Campbell DJ, Kemp BE, Whole body deletion of AMP-activated protein kinase {beta}2 reduces muscle AMPK activity and exercise capacity, J. Biol. Chem 285 (2010) 37198–37209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wu W, Xu Z, Zhang L, Liu J, Feng J, Wang X, Shan T, Wang Y, Muscle-specific deletion of Prkaa1 enhances skeletal muscle lipid accumulation in mice fed a high-fat diet, J. Physiol. Biochem 74 (2018) 195–205. [DOI] [PubMed] [Google Scholar]
- [30].Shan T, Zhang P, Bi P, Kuang S, Lkb1 deletion promotes ectopic lipid accumulation in muscle progenitor cells and mature muscles, J. Cell. Physiol 230 (2015) 1033–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen T, Li Z, Zhang Y, Feng F, Wang X, Wang X, Shen QW, Muscle-selective knockout of AMPKalpha2 does not exacerbate diet-induced obesity probably related to altered myokines expression, Biochem. Biophys. Res. Commun 458 (2015) 449–455. [DOI] [PubMed] [Google Scholar]
- [32].Nixon M, Stewart-Fitzgibbon R, Fu J, Akhmedov D, Rajendran K, Mendoza-Rodriguez MG, Rivera-Molina YA, Gibson M, Berglund ED, Justice NJ, Berdeaux R, Skeletal muscle salt inducible kinase 1 promotes insulin resistance in obesity, Mol Metab 5 (2016) 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Moore TM, Zhou Z, Cohn W, Norheim F, Lin AJ, Kalajian N, Strumwasser AR, Cory K, Whitney K, Ho T, Ho T, Lee JL, Rucker DH, Shirihai O, van der Bliek AM, Whitelegge JP, Seldin MM, Lusis AJ, Lee S, Drevon CA, Mahata SK, Turcotte LP, Hevener AL, The impact of exercise on mitochondrial dynamics and the role of Drp1 in exercise performance and training adaptations in skeletal muscle, Mol Metab 21 (2019) 51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis, BMC Bioinformatics 9 (2008) 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Devlin MJ, Robbins A, Cosman MN, Moursi CA, Cloutier AM, Louis L, Van Vliet M, Conlon C, Bouxsein ML, Differential effects of high fat diet and diet-induced obesity on skeletal acquisition in female C57BL/6J vs. FVB/NJ mice, Bone Rep 8 (2018) 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Boudina S, Sena S, Sloan C, Tebbi A, Han YH, O’Neill BT, Cooksey RC, Jones D, Holland WL, McClain DA, Abel ED, Early mitochondrial adaptations in skeletal muscle to diet-induced obesity are strain dependent and determine oxidative stress and energy expenditure but not insulin sensitivity, Endocrinology 153 (2012) 2677–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nascimento-Sales M, Fredo-da-Costa I, Borges Mendes ACB, Melo S, Ravache TT, Gomez TGB, Gaisler-Silva F, Ribeiro MO, Santos AR Jr., Carneiro-Ramos MS, Christoffolete MA, Is the FVB/N mouse strain truly resistant to diet-induced obesity? Physiol. Rep 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Montgomery MK, Hallahan NL, Brown SH, Liu M, Mitchell TW, Cooney GJ, Turner N, Mouse strain-dependent variation in obesity and glucose homeostasis in response to high-fat feeding, Diabetologia 56 (2013) 1129–1139. [DOI] [PubMed] [Google Scholar]
- [39].Liao Y, Smyth GK, Shi W, The subread aligner: fast, accurate and scalable read mapping by seed-and-vote, Nucleic Acids Res. 41 (2013) e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liao Y, Smyth GK, Shi W, The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads, Nucleic Acids Res. 47 (2019) e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol. 15 (2014) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Young MD, Wakefield MJ, Smyth GK, Oshlack A, Gene ontology analysis for RNA-seq: accounting for selection bias, Genome Biol. 11 (2010) R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zambelli F, Pesole G, Pavesi G, Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes, Nucleic Acids Res. 37 (2009) W247–W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ohkawa H, Ohishi N, Yagi K, Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction, Anal. Biochem 95 (1979) 351–358. [DOI] [PubMed] [Google Scholar]
- [45].Dallon BW, Parker BA, Hodson AE, Tippetts TS, Harrison ME, Appiah MMA, Witt JE, Gibbs JL, Gray HM, Sant TM, Bikman BT, Insulin selectively reduces mitochondrial uncoupling in brown adipose tissue in mice, Biochem. J 475 (2018) 561–569. [DOI] [PubMed] [Google Scholar]
- [46].Zeng PY, Berger SL, LKB1 is recruited to the p21/WAF1 promoter by p53 to mediate transcriptional activation, Cancer Res. 66 (2006) 10701–10708. [DOI] [PubMed] [Google Scholar]
- [47].Lee WJ, Kim M, Park HS, Kim HS, Jeon MJ, Oh KS, Koh EH, Won JC, Kim MS, Oh GT, Yoon M, Lee KU, Park JY, AMPK activation increases fatty acid oxidation in skeletal muscle by activating PPARalpha and PGC-1, Biochem. Biophys. Res. Commun 340 (2006) 291–295. [DOI] [PubMed] [Google Scholar]
- [48].Thomson DM, Herway ST, Fillmore N, Kim H, Brown JD, Barrow JR, Winder WW, AMP-activated protein kinase phosphorylates transcription factors of the CREB family, J. Appl. Physiol 104 (2008) (1985) 429–438. [DOI] [PubMed] [Google Scholar]
- [49].Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A, The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor, J. Biol. Chem 282 (2007) 30107–30119. [DOI] [PubMed] [Google Scholar]
- [50].Mo C, Wang L, Zhang J, Numazawa S, Tang H, Tang X, Han X, Li J, Yang M, Wang Z, Wei D, Xiao H, The crosstalk between Nrf2 and AMPK signal pathways is important for the anti-inflammatory effect of berberine in LPS-stimulated macrophages and endotoxin-shocked mice, Antioxid. Redox Signal 20 (2014) 574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].McGee SL, van Denderen BJ, Howlett KF, Mollica J, Schertzer JD, Kemp BE, Hargreaves M, AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5, Diabetes 57 (2008) 860–867. [DOI] [PubMed] [Google Scholar]
- [52].Fritzen AM, Lundsgaard AM, Jeppesen J, Christiansen ML, Bienso R, Dyck JR, Pilegaard H, Kiens B, 5’-AMP activated protein kinase alpha2 controls substrate metabolism during post-exercise recovery via regulation of pyruvate dehydrogenase kinase 4, J. Physiol 593 (2015) 4765–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J, Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle, Cell Metab. 11 (2010) 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Birk JB, Wojtaszewski JF, Predominant alpha2/beta2/gamma3 AMPK activation during exercise in human skeletal muscle, J. Physiol 577 (2006) 1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nilsson EC, Long YC, Martinsson S, Glund S, Garcia-Roves P, Svensson LT, Andersson L, Zierath JR, Mahlapuu M, Opposite transcriptional regulation in skeletal muscle of AMP-activated protein kinase gamma3 R225Q transgenic versus knock-out mice, J. Biol. Chem 281 (2006) 7244–7252. [DOI] [PubMed] [Google Scholar]
- [56].Miura S, Kai Y, Tadaishi M, Tokutake Y, Sakamoto K, Bruce CR, Febbraio MA, Kita K, Chohnan S, Ezaki O, Marked phenotypic differences of endurance performance and exercise-induced oxygen consumption between AMPK and LKB1 deficiency in mouse skeletal muscle: changes occurring in the diaphragm, Am. J. Physiol. Endocrinol. Metab 305 (2013) E213–E229. [DOI] [PubMed] [Google Scholar]
- [57].Turner N, Bruce CR, Beale SM, Hoehn KL, So T, Rolph MS, Cooney GJ, Excess lipid availability increases mitochondrial fatty acid oxidative capacity in muscle: evidence against a role for reduced fatty acid oxidation in lipid-induced insulin resistance in rodents, Diabetes 56 (2007) 2085–2092. [DOI] [PubMed] [Google Scholar]
- [58].Garcia-Roves P, Huss JM, Han DH, Hancock CR, Iglesias-Gutierrez E, Chen M, Holloszy JO, Raising plasma fatty acid concentration induces increased biogenesis of mitochondria in skeletal muscle, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 10709–10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Fillmore N, Jacobs DL, Mills DB, Winder WW, Hancock CR, Chronic AMP-activated protein kinase activation and a high-fat diet have an additive effect on mitochondria in rat skeletal muscle, J. Appl. Physiol 109 (2010) (1985) 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hancock CR, Han DH, Chen M, Terada S, Yasuda T, Wright DC, Holloszy JO, High-fat diets cause insulin resistance despite an increase in muscle mitochondria, Proc. Natl. Acad. Sci. U. S. A 105 (2008) 7815–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].O’Neill HM, Lally JS, Galic S, Thomas M, Azizi PD, Fullerton MD, Smith BK, Pulinilkunnil T, Chen Z, Samaan MC, Jorgensen SB, Dyck JR, Holloway GP, Hawke TJ, van Denderen BJ, Kemp BE, Steinberg GR, AMPK phosphorylation of ACC2 is required for skeletal muscle fatty acid oxidation and insulin sensitivity in mice, Diabetologia 57 (2014) 1693–1702. [DOI] [PubMed] [Google Scholar]
- [62].Wu H, Ballantyne CM, Skeletal muscle inflammation and insulin resistance in obesity, J. Clin. Invest 127 (2017) 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Simoneau JA, Veerkamp JH, Turcotte LP, Kelley DE, Markers of capacity to utilize fatty acids in human skeletal muscle: relation to insulin resistance and obesity and effects of weight loss, FASEB J. 13 (1999) 2051–2060. [DOI] [PubMed] [Google Scholar]
- [64].He J, Watkins S, Kelley DE, Skeletal muscle lipid content and oxidative enzyme activity in relation to muscle fiber type in type 2 diabetes and obesity, Diabetes 50 (2001) 817–823. [DOI] [PubMed] [Google Scholar]
- [65].de Luca C, Olefsky JM, Inflammation and insulin resistance, FEBS Lett. 582 (2008) 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Okamoto H, Latres E, Liu R, Thabet K, Murphy A, Valenzeula D, Yancopoulos GD, Stitt TN, Glass DJ, Sleeman MW, Genetic deletion of Trb3, the mammalian Drosophila tribbles homolog, displays normal hepatic insulin signaling and glucose homeostasis, Diabetes 56 (2007) 1350–1356. [DOI] [PubMed] [Google Scholar]
- [67].Barlow JP, Solomon TP, Do skeletal muscle-secreted factors influence the function of pancreatic beta-cells? Am. J. Physiol. Endocrinol. Metab 314 (2018) E297–E307. [DOI] [PubMed] [Google Scholar]
- [68].Jessen N, Koh HJ, Folmes CD, Wagg C, Fujii N, Lofgren B, Wolf CM, Berul CI, Hirshman MF, Lopaschuk GD, Goodyear LJ, Ablation of LKB1 in the heart leads to energy deprivation and impaired cardiac function, Biochim. Biophys. Acta 1802 (2010) 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tenenbaum A, Fisman EZ, Impaired glucose metabolism in patients with heart failure: pathophysiology and possible treatment strategies, Am. J. Cardiovasc. Drugs 4 (2004) 269–280. [DOI] [PubMed] [Google Scholar]
- [70].Bolster DR, Crozier SJ, Kimball SR, Jefferson LS, AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling, J. Biol. Chem 277 (2002) 23977–23980. [DOI] [PubMed] [Google Scholar]
- [71].Moore TM, Mortensen XM, Ashby CK, Harris AM, Kump KJ, Laird DW, Adams AJ, Bray JK, Chen T, Thomson DM, The effect of caffeine on skeletal muscle anabolic signaling and hypertrophy, Appl Physiol Nutr Metab 42 (2017) 621–629. [DOI] [PubMed] [Google Scholar]
- [72].Thomson DM, Fick CA, Gordon SE, AMPK activation attenuates S6K1, 4E-BP1, and eEF2 signaling responses to high-frequency electrically stimulated skeletal muscle contractions, J. Appl. Physiol 104 (2008) (1985) 625–632. [DOI] [PubMed] [Google Scholar]
- [73].Parks BW, Sallam T, Mehrabian M, Psychogios N, Hui ST, Norheim F, Castellani LW, Rau CD, Pan C, Phun J, Zhou Z, Yang WP, Neuhaus I, Gargalovic PS, Kirchgessner TG, Graham M, Lee R, Tontonoz P, Gerszten RE, Hevener AL, Lusis AJ, Genetic architecture of insulin resistance in the mouse, Cell Metab. 21 (2015) 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Seldin MM, Koplev S, Rajbhandari P, Vergnes L, Rosenberg GM, Meng Y, Pan C, Phuong TMN, Gharakhanian R, Che N, Makinen S, Shih DM, Civelek M, Parks BW, Kim ED, Norheim F, Krishnan K. Chella, Hasin-Brumshtein Y, Mehrabian M, Laakso M, Drevon CA, Koistinen HA, Tontonoz P, Reue K, Cantor RM, Bjorkegren JLM, Lusis AJ, A strategy for discovery of endocrine interactions with application to whole-body metabolism, Cell Metab. 27 (2018) 1138–1155 (e1136). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.