ABSTRACT
Zymomonas mobilis has emerged as a promising candidate for production of high-value bioproducts from plant biomass. However, a major limitation in equipping Z. mobilis with novel pathways to achieve this goal is restriction of heterologous DNA. Here, we characterized the contribution of several defense systems of Z. mobilis strain ZM4 to impeding heterologous gene transfer from an Escherichia coli donor. Bioinformatic analysis revealed that Z. mobilis ZM4 encodes a previously described mrr-like type IV restriction modification (RM) system, a type I-F CRISPR system, a chromosomal type I RM system (hsdMSc), and a previously uncharacterized type I RM system, located on an endogenous plasmid (hsdRMSp). The DNA recognition motif of HsdRMSp was identified by comparing the methylated DNA sequence pattern of mutants lacking one or both of the hsdMSc and hsdRMSp systems to that of the parent strain. The conjugation efficiency of synthetic plasmids containing single or combinations of the HsdMSc and HsdRMSp recognition sites indicated that both systems are active and decrease uptake of foreign DNA. In contrast, deletions of mrr and cas3 led to no detectable improvement in conjugation efficiency for the exogenous DNA tested. Thus, the suite of markerless restriction-negative strains that we constructed and the knowledge of this new restriction system and its DNA recognition motif provide the necessary platform to flexibly engineer the next generation of Z. mobilis strains for synthesis of valuable products.
IMPORTANCEZymomonas mobilis is equipped with a number of traits that make it a desirable platform organism for metabolic engineering to produce valuable bioproducts. Engineering strains equipped with synthetic pathways for biosynthesis of new molecules requires integration of foreign genes. In this study, we developed an all-purpose strain, devoid of known host restriction systems and free of any antibiotic resistance markers, which dramatically improves the uptake efficiency of heterologous DNA into Z. mobilis ZM4. We also confirmed the role of a previously known restriction system as well as identifying a previously unknown type I RM system on an endogenous plasmid. Elimination of the barriers to DNA uptake as shown here will allow facile genetic engineering of Z. mobilis.
KEYWORDS: Zymomonas mobilis ZM4, restriction modification system, genome defense, conjugation efficiency of foreign genes, type I restriction enzymes
INTRODUCTION
Zymomonas mobilis has several metabolic attributes that are advantageous for engineering strains to produce biofuels and other valuable commodities from lignocellulosic biomass on an industrial scale (1–4). However, genetic engineering of this organism has been challenging in part due to the presumed restriction of foreign DNA. To optimize the metabolism of Z. mobilis and unlock its full potential for industrial production of compounds of interest, introduction of foreign DNA needs to be more reliable and efficient. Recently, we developed a markerless genetic approach to add or remove genes from Z. mobilis strain ZM4 (5). Here, we used this method to delete genes encoding Z. mobilis restriction systems to test their effects on improving uptake of foreign DNA.
Bacteria have evolved several types of defense systems as barriers to invasion by foreign DNA. These activities usually comprise one or more of the four types of restriction modification (RM) systems or multiple types of CRISPR-Cas systems. The diversity of restriction systems in bacterial species and the challenges encountered in circumventing these systems to facilitate genetic engineering have been well documented (6–15). The type I RM systems are the most complex because the DNA sequence specificity determinant (HsdS), the DNA methyltransferase (HsdM), and the endonuclease (HsdR) are each encoded by separate genes and function as an oligomer. HsdS recognizes the methylation status of a specific bipartite DNA sequence motif. If the DNA motif is hemimethylated, then HsdM in complex with HsdS methylates the unmethylated DNA strand. If the DNA motif is unmethylated or incorrectly methylated, then HsdR cleaves the DNA, usually at a distance from the recognition sequence (6, 16). Type II RM systems also have methyltransferase and endonuclease activities but are quite diverse in their subunit composition and cleave DNA at or near their recognition sequence (12). Nevertheless, as with type I enzymes, base methylation of the target site results in protection from DNA cleavage by the cognate type II endonuclease. Type III RM systems consist of two proteins with endonuclease and DNA methylation activities, but these enzymes usually recognize short asymmetric sequences in an inverted repeat orientation (17, 18). Type IV RM systems contain only endonucleases, and restriction activity is directed against methylated invading DNA (19). These type IV enzymes are suggested to be promiscuous for their target sequence (20). CRISPR-Cas systems are quite diverse and composed of several proteins that defend against invading bacteriophages or plasmids (13, 14).
Because restriction of DNA hinders development of genetic systems in bacteria, several approaches have been exploited to evade RM systems. In some cases, propagating plasmid DNA in a methylation-deficient Escherichia coli strain (21) prior to transformation decreases restriction. An approach to evade type I RM systems is the electroporation of DNA mixed with a type I RM system inhibitor protein, OCR (6, 21, 22). This protein mimics B-form DNA, and binding to a type I restriction enzyme prevents the enzyme from binding to target DNA (23). Genetic engineering to remove type I RM target sequences from heterologous DNA of interest has also been deployed (8, 24). Expression of organism-specific methyltransferases in an E. coli plasmid-propagating strain to methylate heterologous DNA before introduction into relevant bacteria also shows promise (25–27). However, these targeted approaches sometimes do not mitigate restriction from other RM systems; methyltransferases may not express well in E. coli, or the approach may require prior knowledge of the host’s restriction systems to devise an effective strategy. Thus, these approaches may not confer immunity against all RM systems. When gene deletion technologies exist for a strain, an alternative approach is to delete genes involved in restriction.
In the case of Z. mobilis ZM4, the understanding of restriction systems is incomplete, making it challenging to develop an effective strategy to evade all defense systems of this organism. REBASE, a restriction enzyme database (28), provides one resource for the prediction of endogenous restriction enzymes in Z. mobilis ZM4. Independent disruptions of a gene predicted to encode the specificity determinant of a previously annotated type I RM system (hsdSc; ZMO1933) and a gene predicted to encode a type IV RM system (mrr; ZMO0028) (22, 25) resulted in small increases in DNA uptake efficiencies for exogenous plasmids, supporting the predicted roles of mrr and hsdSc in DNA restriction. A CRISPR-Cas system of Z. mobilis ZM4 was also recently characterized (7, 14).
In this study, we extend these earlier findings of Z. mobilis ZM4 RM systems to show that elimination of the previously annotated type I and IV RM systems is not sufficient to improve uptake of foreign DNA, indicating the presence of other restriction systems. Using a bioinformatic approach and high-throughput single-molecule real-time whole-genome methylome sequencing, we identified an additional type I RM system encoded on a native plasmid of Z. mobilis ZM4. This analysis also suggests that the previously annotated chromosomal type I RM system (HsdMSc) appears to have a nuclease domain fused to the HsdM subunit (29, 30). By creation of a series of strains with deletions of one or more restriction system genes, we found that removal of both the chromosomal type I RM system and the plasmid type I RM system was needed to maximally increase the uptake efficiency of foreign DNA. The availability of a suite of markerless strains with different combinations of RM systems eliminated provides more genetically tractable strains for metabolic engineering endeavors.
RESULTS
Bioinformatic predictions of RM systems of Z. mobilis ZM4.
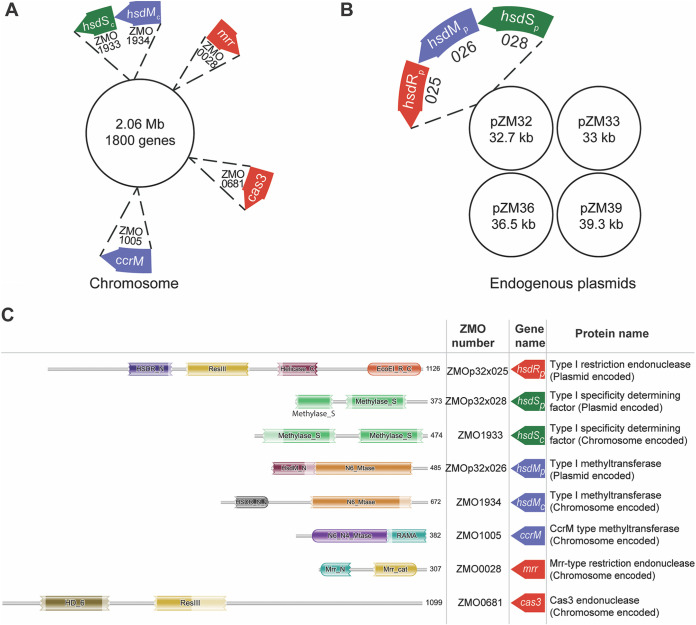
To develop strains with improved uptake efficiency of foreign DNA, we used bioinformatic predictions to identify genes encoding restriction systems in Z. mobilis ZM4 (31) to target for deletion (Fig. 1). As expected, the previously described ZMO0028 (mrr) (25) contains a domain typical of the Mrr superfamily of type IV restriction endonucleases, and ZMO0681 (cas3) (7, 14) contains a domain expected for type I-F CRISPR-Cas systems (13). ZMO1005 contains domains conserved in DNA methyltransferases and is 60% similar with the beta-group methyltransferase, CcrM of Caulobacter sp. (32). The presence of conserved motif IV and motif I of CcrM (33) in ZMO1005 indicates that ZMO1005 is a CcrM-like methyltransferase that is a housekeeping methyltransferase and is not typically associated with RM systems. The previously described ZMO1933 (hsdS) (25) has a domain belonging to the RM TypeI_S_TRD-CR-like superfamily, consistent with its encoding the specificity-determining factor of a type I RM system (30, 34). The neighboring gene ZMO1934 has a domain representative of the N6-methyltransferase superfamily, consistent with its proposed function as the methyltransferase component of a type I RM system along with specificity factor ZMO1933 (34). However, no protein with all of the domains expected for the HsdR subunit of a type I RM system was detected in our bioinformatic search. Rather, ZMO1934 had a conserved domain found in the N terminus of type I restriction endonuclease (HsdR_N), indicating a possible N-terminal fusion of a HsdR with HsdM. Such fusion proteins, which lack the motor and helicase domains of HsdR, have sometimes been reclassified to the quite diverse group of type II RM systems (6, 29).
FIG 1.
Gene location and sequence features of Z. mobilis ZM4 restriction systems. (A and B) Schematic representation showing the location of restriction system genes in the chromosome (A) and plasmids (B). Genes encoding endonucleases are shown in red, those for methyltransferases are in blue, and those for specificity-determining factors are in green. (C) Summary of RM system sequence features and matches with the Pfam database. The region of peptide sequence matching domains of Pfam clans are represented graphically. The protein superfamily clan of the domains for HsdRp identified by the phmmer search tool, CDD domain search tool, and UniProt is cl36022, that for HsdSc is cl38903, those for HsdSp are cl35887 and cl38903, those for HsdMc are cl29110 and cl37510, those for HsdMp are cl37510 and cl13579, those for CcrM are c17173 and cl16759, those for Mrr is cl34341, and that for Cas3 is cl28317. In HsdMc, an N-terminal domain of HsdR that belongs to protein superfamily clan cl29110 is fused with HsdM. The jagged edge of a domain at the N or C terminus indicates that the sequence did not extend to the first or last position in the HMM database, respectively.
We also found a previously unannotated type I RM system encoded on the native plasmid ZMOp32. Three adjacent genes (zmop32x025 [hsdRp], zmop32x026 [hsdMp], and zmop32x028 [hsdSp]) were identified as having conserved domains of the HsdR superfamily, the i6-methyltransferase superfamily, and the RM TypeI_S_TRD-CR-like superfamily, respectively, suggesting that they encode three subunits of a type I RM system in Z. mobilis ZM4. The plasmid-encoded type I RM system appears to be distinct from the chromosomal system, since there is only 34% and 28% amino acid sequence identity between the two HsdSs and the two HsdM protein sequences, respectively. To differentiate between the subunits of the chromosome-encoded type I RM system and the plasmid-encoded type I RM system in Z. mobilis ZM4, we refer to the genes with a subscript “c” for the chromosomal type I RM system (hsdMSc) and “p” for the plasmid type I RM system (hsdRMSp).
Phylogenetic analysis of type I RM systems in Z. mobilis strains.
Since two type I RM systems were found in Z. mobilis ZM4 (31), we asked whether other Z. mobilis strains have the same systems. Using a local tBLASTn function, we searched the genome sequence of 16 Z. mobilis strains for sequences homologous to the HsdRp, HsdMp, HsdSp, HsdMc, and HsdSc (Fig. 2). Two strains (ER79ag and ATC31823) contain the same complement of HsdMSc and HsdRMSp RM systems as strain ZM4, since proteins 100% identical to HsdRMSp and HsdMSc were identified from their genome sequences. Two other strains (DSM12497 and DSM12494) encode proteins identical to HsdRMp, but the ortholog of HsdSc shares only 41% identity to Z. mobilis ZM4. The remaining strains appear to lack orthologs to either RM system, indicating that neither system is part of the core Z. mobilis genome.
FIG 2.
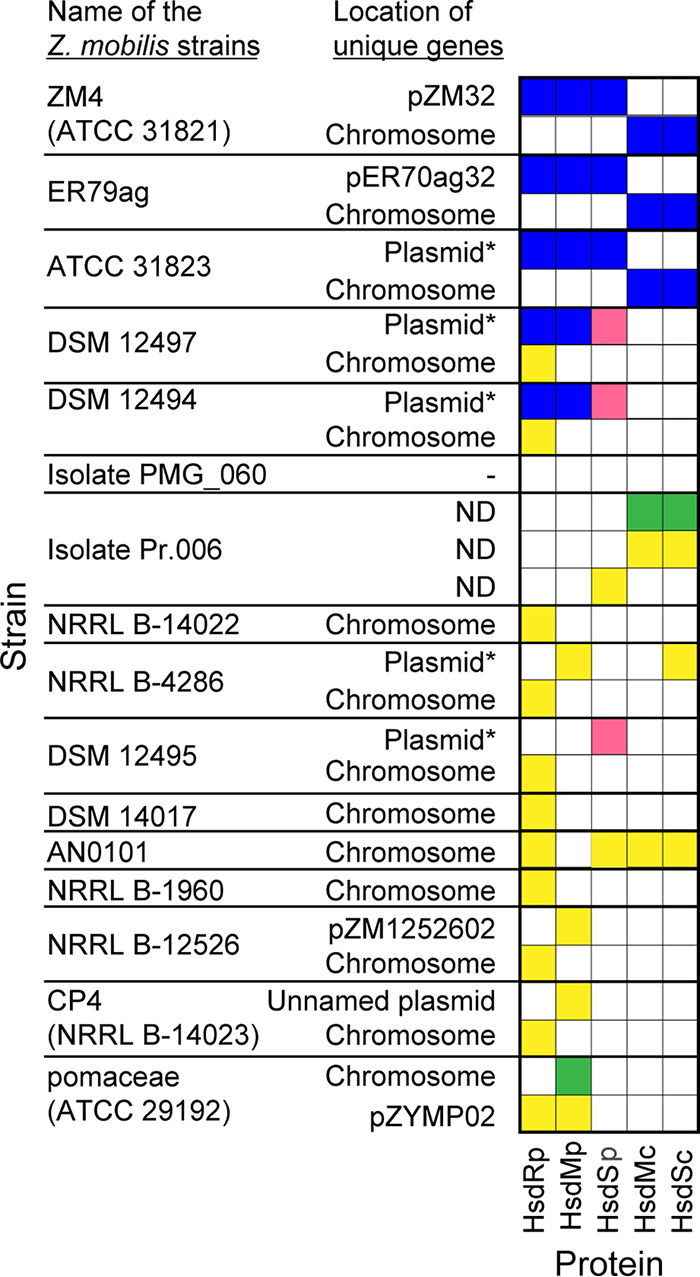
Amino acid sequence identity of Z. mobilis ZM4 HsdRp, HsdMp, HsdSp, HsdMc, and HsdSc across 16 unique Z. mobilis strains. Blue, red, green, and yellow boxes indicate >96%, 41 to 50%, 31 to 40%, and 20 to 30% identity, respectively, to the indicated Z. mobilis ZM4 genes from the tBLASTn result. White boxes indicate that no homolog to the Z. mobilis ZM4 gene was detected in the tBLASTn result. If the genome was not complete, contigs greater than 2 Mb are indicated as chromosomal and the contigs smaller than 50 kb are indicated as plasmid* unless already specified as plasmid in the assembly. The sequence assembly of Z. mobilis isolate Pr.006 contained more than 700 contigs of 10 to 20 kb, and the location of matching regions was not determined (ND).
Methylome sequencing to identify type I RM system target sequences in Z. mobilis ZM4.
To identify the target sequences for the HsdMSc and HsdRMSp RM systems encoded on the chromosome and native plasmid pZM32, respectively, we compared the genomic methylation pattern of the parent strain to mutants lacking one or both Hsd systems. We reasoned that this would allow us to confirm the recognition sequence of HsdMSc and identify the recognition sequence for HsdRMSp. Z. mobilis ZM4 strains lacking the specificity subunits of either the chromosomal (ΔhsdSc) or the chromosomal and plasmid Hsd systems (ΔhsdSc ΔhsdSp), which should eliminate both methylation and restriction activities, were constructed. Using single-molecule real-time DNA sequencing, we identified all methylated base modifications in the genome of the parent Z. mobilis ZM4 strain and compared this pattern with the methylome pattern of the mutant strains lacking one or both of the predicted Hsd systems. The results of the parent strain indicated methylation of adenine to N6-methyladenine at three different target sequences: 5′CAGN4CTG, 5′GAAGN7TCC, and 5′GANTC, where the underline represents adenine methylation (Table 1). The ΔhsdSc strain lacked adenine methylation of the bipartite sequence 5′ CAGN4CTG, confirming this sequence as the HsdSc target site. The ΔhsdSc hsdSp strain additionally lacked adenine methylation of 5′GAAGN7TCC, indicating that the latter sequence is the HsdSp target site. These results also support the bioinformatic analysis that the two Hsd systems are genetically distinct and recognize distinct DNA sequences. GANTC is typically recognized by CcrM, which is presumably the case here as well.
TABLE 1.
Methylome sequencing data
| Genotype of strain | Methylated recognition sequence motifa | Total no. of methylated sequence motifs detectedb | % of motifs methylated in whole genomec |
|---|---|---|---|
| Wild type | CAGN4CTG | 938 | 100 |
| GAAGN7TCC | 325 | 100 | |
| GANTC | 8,599 | 99.24 | |
| hsdSc | CAGN4CTG | 0 | 0 |
| GAAGN7TCC | 325 | 100 | |
| GANTC | 8,470 | 97.78 | |
| hsdSc hsdSp | CAGN4CTG | 0 | 0 |
| GAAGN7TCC | 0 | 0 | |
| GANTC | 8,541 | 98.64 |
Underlining indicates adenine methylation.
Total number of methylated sequence motifs detected by PacBio SMRT DNA sequencing of genomic DNA (chromosomal and plasmids).
Percentage of sequence motifs present in the genome of indicated strain which were methylated.
Establishing the functional relevance of the type I RM systems.
To determine if these systems impact Z. mobilis DNA uptake, we compared the conjugation efficiency of plasmids engineered to contain no, one, or two HsdSc and HsdSp synthetic target sites (Fig. 3). Compared to the plasmid lacking any HsdSc and HsdSp target sites (pPK15617), the conjugation efficiency of the plasmid containing one HsdSc target site (5′CAGN4CTG; pPK15621) was reduced 5-fold in Z. mobilis ZM4 (Fig. 3A). The plasmid containing one HsdSp target site (5′GAAGN7TCC; pPK15619) was more dramatically restricted, since the efficiency of conjugation into the parent strain was 4,900-fold less than that of the plasmid lacking any sites (pPK15617). Elimination of hsdSp restored the conjugation efficiency of pPK15619 to near that of the plasmid lacking any HsdSc and HsdSp target sites (pPK15617), indicating the specific role of the HsdRMSp system in restriction of the GAAGN7TCC sequence present on plasmid pPK15619. These results also suggest that the HsdRMSp restriction system is more active in Z. mobilis ZM4 than HsdMSc.
FIG 3.
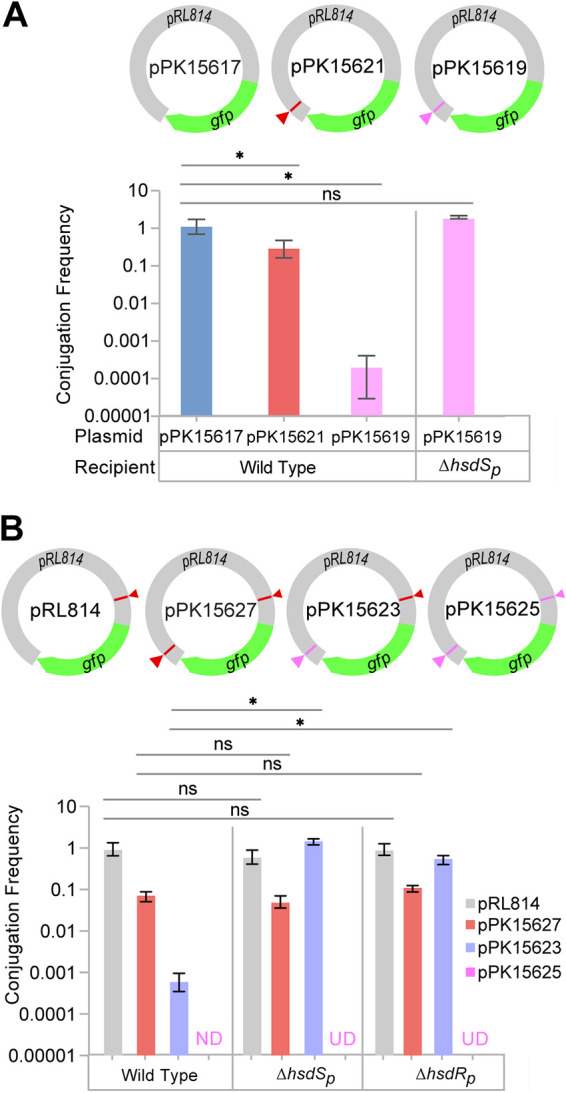
Functional relevance of the type I RM target sequences as measured by conjugation frequency. (A) The conjugation frequency of pPK15617 into Z. mobilis ZM4 (1.4 × 10−5 ± 0.5 × 10−5) served as a normalization factor for comparing the conjugation efficiency (y axis) of plasmids in wild-type and mutant ZM4 strains as indicated. (B) The conjugation frequency of pRL814 into Z. mobilis ZM4 (0.97 × 10−3 ± 0.5 × 10−3) served as a normalization factor for comparing the conjugation efficiency (y axis) of plasmids in wild-type and mutant ZM4 strains as indicated. When conjugation of a plasmid was below the limit of detection (0.00001), the sample is marked “ND” (not detected). When conjugation experiment of a plasmid was not done, it is marked “UD” (undetermined). Error bars represent the standard deviations of the conjugation frequency means obtained from three independent experiments. Statistical significance was determined using a paired Student's t test (*, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant). On the plasmids, red triangles represent restriction sites recognized by HsdSc and pink triangles represent restriction sites recognized by HsdSp.
We also measured the impact of additional synthetic HsdSc and HsdSp DNA sites, using the plasmid pRL814, which has one naturally occurring HsdSc target site in lacI (Fig. 3B). Adding a second HsdSc site to pRL814 (pPK15627) decreased the conjugation efficiency in the parent strain 12.6-fold, whereas adding a HsdSp site to pRL814 (pPK15623) decreased conjugation 1,950-fold. Further, the conjugation of a plasmid bearing two HsdSp target sites (pPK15625) could not be detected even at a frequency 10,000-fold below the conjugation frequency of the plasmid pRL814. Thus, each RM system is a barrier to efficient DNA uptake, and the newly discovered HsdRMSp seems to have the largest impact. Comparing the conjugation efficiency of pPK15627, which has two HsdSc target sites, to that of pPK15623, which has one HsdSc and one HsdSp target site, in strains lacking HsdSp (ΔhsdSp) or the plasmid-encoded restriction enzyme, HsdRp (ΔhsdRp), showed that the conjugation efficiency improved only for the plasmid with an HsdSp target site (Fig. 3B). These results indicate that the HsdRp endonuclease does not impact the HsdMSc system. Also, the lack of HsdRp imparts an equivalent mutant phenotype as HsdSp, a property expected for proteins that function as a complex.
Improvement of efficiency of conjugation of foreign genes into Z. mobilis ZM4 requires removal of hsdSc and hsdSp.
Our goal was to use the knowledge of Z. mobilis ZM4 RM systems to improve uptake of foreign DNA. Therefore, we tested the conjugation efficiency of plasmids containing heterologous DNA of interest into Z. mobilis ZM4 mutants lacking either or both type I RM systems. We utilized pRL814 as the parent plasmid, which has one HsdSc site. As a control for the relative size of the plasmid, we determined the frequency of conjugation into the parent Z. mobilis ZM4 of plasmid pPK15346, which contains the carotenoid-synthesizing genes crtI, crtB, and crtE from Rhodobacter sphaeroides 2.4.1 (35) and which lacks any known restriction sites in addition to the vector site (Fig. 4A). This plasmid mobilized as efficiently as pRL814 into Z. mobilis ZM4, showing that the size of the plasmid did not significantly impact the conjugation efficiency under these conditions.
FIG 4.
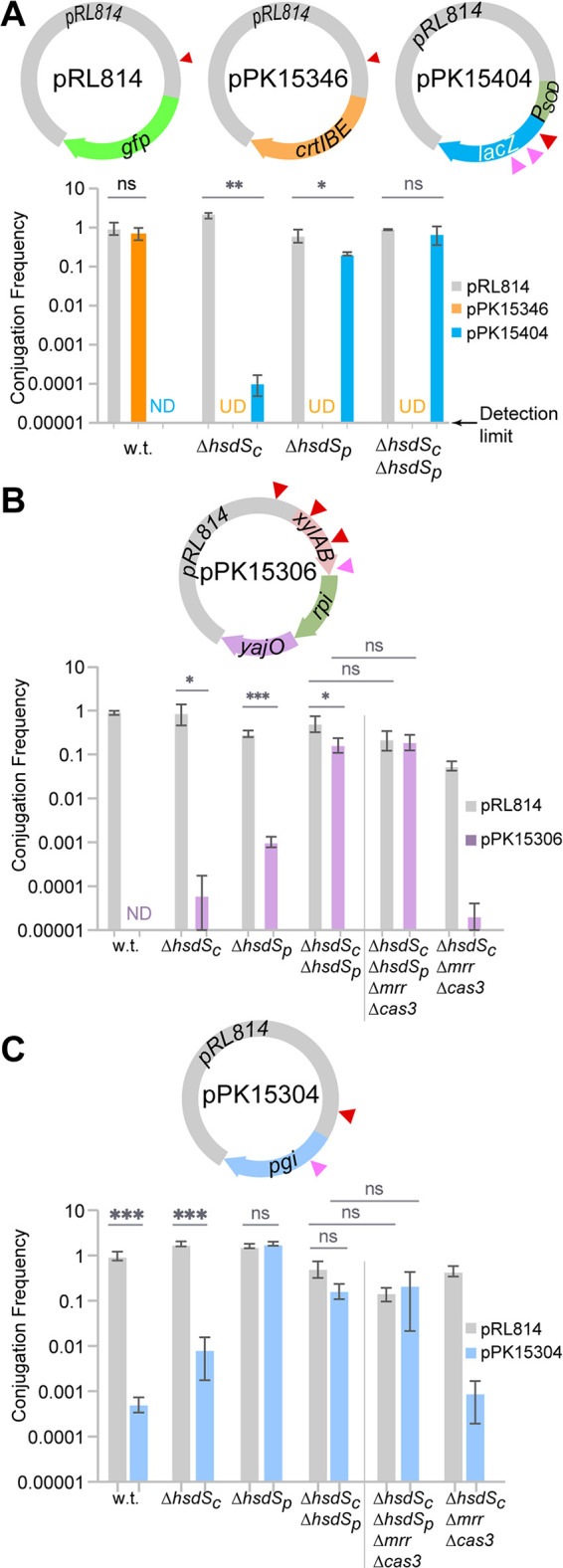
Role of HsdSc, HsdSp, Mrr, and Cas3 in restricting pPK15404, pPK15306, and pPK15304. (A) The frequency of conjugation of pRL814 into Z. mobilis ZM4 (0.78× 10−3 ± 0.09 × 10−3) served as a normalization factor for comparing the conjugation frequency (y axis) of the other plasmids in wild-type (w.t.) and mutant ZM4 strains. (B) The frequency of conjugation of pRL814 into Z. mobilis ZM4 (0.24 × 10−3 ± 0.08 × 10−3) served as a normalization factor for comparing the conjugation frequency (y axis) of plasmids in wild-type (w.t.) and mutant ZM4 strains as indicated. (C) The frequency of conjugation of pRL814 into Z. mobilis ZM4 (6 × 10−3 ± 0.6 × 10−3) served as a normalization factor for comparing the conjugation frequency (y axis) of plasmids in wild-type (w.t.) and mutant ZM4 strains. When conjugation of a plasmid was below the limit of detection (0.00001), the sample is marked “ND” (not detected). When conjugation experiment of a plasmid was not done, it is marked “UD” (undetermined). Error bars represent the standard deviations of the conjugation frequency means obtained from three independent experiments. Statistical significance was determined using a paired Student's t test (*, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant). On the plasmids, red triangles represent restriction sites recognized by HsdSc and pink triangles represent restriction sites recognized by HsdSp.
To evaluate restriction of other foreign genes in Z. mobilis ZM4, we measured the conjugation efficiency of pPK15404, pPK15306, and pPK15304 into Z. mobilis strains (Fig. 4A to C). Plasmid pPK15404 contains E. coli lacZ, which serves as a gene expression reporter in many bacteria and contains one HsdSc recognition site and two HsdSp sites. Plasmid pPK15306 contains a synthetic operon composed of E. coli xylA, xylB, rpi, and yajO, previously reported to direct xylose into a metabolic pathway (36–38), and contains three HsdSc sites and one HsdSp site. Plasmid pPK15304 contains E. coli pgi, involved in conversion of d-glucose 6-phosphate to d-fructose 6-phosphate in glycolysis and contains one recognition site for HsdSc and HsdSp. We found that these plasmids were severely restricted in Z. mobilis ZM4, since the conjugation frequency of pPK15404 and pPK15306 was below our detection limit (10−5) and the conjugation frequency of pPK15304 was 1,900 times lower than that of plasmid pRL814 (Fig. 4A to C). Deletion of hsdSc or hsdSp alone was not sufficient to overcome the restriction barrier for pPK15306, consistent with both types of target sites present on the plasmid. However, when both systems were deleted, the conjugation frequency was nearly identical to that of the vector control, indicating that the plasmids were no longer restricted. For plasmids pPK15304 and pPK15404, elimination of HsdSc had a small effect, whereas elimination of HsdSp had a much larger effect, since the conjugation frequency was similar to the vector control. These results show that elimination of the HsdMSc and HsdRMSp restriction systems in Z. mobilis ZM4 removes the restriction barrier for multiple sets of foreign genes and enables efficient uptake of heterologous DNA.
Mrr and Cas3 do not restrict pPK15306 and pPK15304.
Although we do not know the target sequence for Mrr and the target sequence for Cas3 is variably acquired as part of the defense mechanism (13, 14), we created mutants lacking one or both of these activities to determine any contribution to the restriction of foreign DNA by Z. mobilis ZM4. Deletion of either mrr or cas3 alone or in combination did not increase conjugation of pPK15306 or pPK15304 over the limit of detection for this assay (not shown). We also tested the effect of deleting mrr and cas3 in a ΔhsdSc strain, a background with increased conjugation efficiency, to rule out the possibility that small effects of mrr or cas3 could have been missed in our assay. However, we observed no improvement in conjugation with either plasmid in this strain (Fig. 4B and C). We also tested a strain lacking all four defense activities (ΔhsdSc ΔhsdSp Δmrr Δcas3 strain) as a recipient for conjugation experiments. This strain was as permissive in conjugating pPK15306 and pPK15304 as the ΔhsdSc ΔhsdSp strain (Fig. 4B and C), indicating there is no impairment in conjugation with this quadruple mutant. As a comparison, the doubling times of the wild-type, ΔhsdSc, ΔhsdSc ΔhsdSp, and ΔhsdSc ΔhsdSp Δmrr Δcas3 strains grown anaerobically in ZRMG medium (see Materials and Methods) at 30°C were 78.5 ± 0.7 min, 97.5 ± 3.5 min, 82.0 ± 2.8 min, and 95.5 ± 0.7 min, respectively. Although Mrr and Cas3 did not impact conjugation efficiency in our experiments, this quadruple mutant strain is available as a potential all-purpose recipient for engineering foreign DNA into Z. mobilis ZM4 in the future, because it would not require any prior knowledge of specific restriction sites in the foreign DNA.
DISCUSSION
RM systems provide a formidable barrier to entry of foreign DNA (8, 25, 39) and hinder genetic engineering. A thorough analysis of RM systems is a prerequisite to developing genetically tractable strains to promote DNA uptake by either conjugation or transformation. This study reports a comprehensive analysis of restriction systems of Z. mobilis ZM4 and the successful development of strains devoid of multiple restriction activities. A key advance was the discovery of a type I restriction system encoded in native plasmid pZM32 and its target site, which imparts a robust restriction barrier in Z. mobilis ZM4 but is not present in several other Z. mobilis strains.
A second type I RM system is encoded on a Z. mobilis ZM4 plasmid.
Our bioinformatic analysis confirmed the presence of previously known genes encoding subunits of chromosomally encoded HsdMSc system (21, 22, 25, 40), Mrr, a type IV restriction enzyme (21, 22, 25, 40), and the endonuclease, Cas3, of the type I F CRISPR-Cas system (7, 14). In addition, we also found previously unknown and unannotated genes for a complete type I RM system (hsdRMSp) on plasmid pZM32. The proteins of this type I RM system (HsdRMSp) are very distinct from the proteins of the previously annotated chromosomal type I RM system (HsdMSc); the HsdSc-HsdSp proteins and the HsdMc-HsdMp proteins share only 34% and 28% sequence similarity, respectively.
In this work, we showed that removal of both type I RM systems improved conjugation efficiency of heterologous genes in Z. mobilis ZM4. However, the most formidable barrier to improving conjugation efficiency was the newly identified plasmid encoded type I restriction system. As reported previously (22, 25) and confirmed here, elimination of the activity encoded by the HsdMSc system provided a small improvement in uptake of plasmids containing heterologous DNA. However, since most of the plasmids used in our studies had recognition sites for both the hsdMSc and the hsdRMSp systems, an optimal improvement in conjugation efficiency was observed when both activities were eliminated. Z. mobilis mutants lacking mrr have previously been shown to increase DNA uptake of the shuttle vector pBBR1MCS-3 (22), although we did not observe any increase with the plasmids used in our experiments. Since the recognition sequence for Mrr in Z. mobilis is unknown, it is possible that the plasmids used in this study lacked the sequence for Mrr restriction. Nevertheless, because future synthetic biology plasmid designs may have a target site for Mrr or the CRISPR-Cas3 system, we created a quadruple mutant strain (ΔhsdSc ΔhsdSp Δmrr Δcas3; PK15509) lacking all four defense systems of Z. mobilis as a suitable all-purpose platform strain for future genetic engineering of Z. mobilis ZM4.
An important advance was the finding that the restriction activity of the HsdMSc system in Z. mobilis ZM4 is less than that of the HsdRSMp system but that HsdMSc is still an active RM system that needs to be evaded for maximal DNA uptake. As reported previously (25) and confirmed here, no gene encoding a full-length HsdRc could be found in Z. mobilis ZM4. Since other bacteria are known to share a single HsdR with multiple HsdMS systems (6, 41, 42), we assessed if HsdRp is shared with HsdMSc system in Z. mobilis ZM4. Because elimination of HsdR did not affect the conjugation efficiency of a plasmid that is generally restricted by the HsdMSc system, this result suggests that HsdRp is not shared with the HsdMSc system in Z. mobilis ZM4. Rather, we found that HsdMc contains a conserved domain found in the N terminus of type I restriction enzyme R protein (HsdR_N). Recently, it has been proposed that such proteins evolved from a fusion of the N terminus of HsdR with HsdM of type I RM system (29). This class of fusion protein is proposed to retain the methylation activity but lack the motor and helicase domains responsible for reeling in DNA sequence that promote ATP-driven cleavage, suggesting that DNA cleavage occurs in closer proximity to the HsdS binding site than with a typical type I HsdR (29).
We have also demonstrated the usefulness of our previously published markerless genome modification method (5) to generate restriction deficient strains by sequentially deleting the four genes encoding the different defense systems from different regions of the genome without introduction of any permanent antibiotic resistance markers. Thus, the absence of any antibiotic resistance cassettes provides an excellent starting point for metabolic strain engineering. Further, the restriction systems can easily be reintroduced back into engineered strains if needed for industrial robustness. We also found that deletion of genes from one of the endogenous plasmids showed the same frequency as deletion of genes from chromosome. Thus, this method can be used for further modification of the endogenous plasmids.
Z. mobilis ZM4 ZMO1005 possibly encodes a CcrM-like methyltransferase.
In addition to methylome sequencing analysis enabling identification of the target sequence for the plasmid-encoded type I RM system and confirming the sequence of the chromosomally encoded system (8, 43), we also found an additional methylated sequence indicative of the methyltransferase CcrM, which methylates the adenine of GANTC in alphaproteobacteria (44–46).
Bioinformatic analysis indicated that ZMO1005 encodes a protein with an N6-methyltransferase family domain and showed 60% similarity with CcrM, a class β-methyltransferase of Caulobacter sp. Additionally, ZMO1005 contains conserved motif IV (DLIFADPPYNLQLGG) and motif I (ILDPFFGVGTTGAAA) of class β-methyltransferases (33). ccrM is prevalent in alphaproteobacteria and plays a vital role in DNA replication, DNA repair, and gene regulation (32, 43, 47, 48). CcrM, DnaA, GcrA, CtrA, and SciP are five master regulators that together control cell cycle progression by modulating epigenetic changes (43, 46–48). BLASTP analysis of these proteins against Z. mobilis ZM4 genome indicates the presence of all except sciP in Z. mobilis ZM4. Overall, this analysis indicates that CcrM (ZMO1005) might play an important role in Z. mobilis ZM4, which needs to be explored.
Future prospect of genome engineering in Z. mobilis ZM4.
A primary focus of this work was to identify RM systems of Z. mobilis ZM4 and to create strains with improved ability to take up plasmids encoding heterologous metabolic functions. Indeed, we improved the conjugation efficiency of plasmids containing genes that encode β-galactosidase, a xylose utilization pathway (36–38), and phosphoglucoisomerase (49, 50). Given the utility of β-galactosidase as a widespread reporter in gene expression studies, our development of a strain lacking RM systems provides a new useful tool for analyzing promoter-specific gene expression in Z. mobilis ZM4. Similarly, the efficient introduction into Z. mobilis ZM4 of heterologous metabolic functions like xylose utilization (36) or phosphoglucoisomerase (49) provides a proof of concept for the usefulness of these strains for future metabolic engineering.
Conclusion.
This study illustrates the impact of Z. mobilis ZM4 RM systems on restricting foreign DNA. We conducted comprehensive bioinformatic, genetic, and high-throughput methylome sequence analyses to identify all RM systems of Z. mobilis ZM4. We created a strain lacking all restriction systems, which accepts all foreign genes tested so far. This work will therefore help accelerate genetic engineering of Z. mobilis ZM4 by eliminating restriction of heterologous DNA and improving DNA uptake efficiency.
MATERIALS AND METHODS
Materials.
All restriction endonucleases, Q5 polymerase, and Gibson assembly HiFi master mix were from New England Biolabs, Inc. GOTaq Flexi DNA polymerase was from Promega. Primers were obtained from Integrated DNA Technologies (IDT). A Sony MA900 fluorescence activated cell sorter (FACS) was used for sorting nonfluorescent cells from fluorescent cells, and an Azure C600 imager was used for screening of fluorescent colonies as previously described (5).
Strains, plasmids, and growth conditions.
Bacterial strains, primers, and plasmids used in this study are listed in Tables 2 to 4, respectively. Z. mobilis ZM4 and its derivatives were grown in ZRMG medium (1% yeast extract, 0.2% KH2PO4, 2% glucose) (5), and E. coli strains were grown in Luria-Bertani (LB) medium (51). Chloramphenicol was used at a final concentration of 120 μg/ml for Z. mobilis ZM4 and 20 μg/ml for E. coli strains. Spectinomycin was used at a final concentration of 120 μg/ml for Z. mobilis ZM4 and 50 μg/ml for E. coli strains. For growth of E. coli WM6026, 0.1 mM m-diaminopimelate (DAP) was added to liquid media and 0.15 mM DAP to solid media (5).
TABLE 2.
Strains
| Strain | Description | Source and/or reference |
|---|---|---|
| DH5α | E. coli F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 ϕ80dlacZΔM15 Δ (lacZYA-argF)U169 hsdR17(rK− mK+) λ− | Lab collection |
| WM6026 | E. colilacIqrrnB3 ΔlacZ4787 hsdR514 ΔaraBAD567 ΔrhaBAD568 rph-1 attλ::pAE12(ΔoriR6K-cat::Frt5) ΔendA::Frt uidA(ΔMluI)::pir attHK::pJK1006 (ΔoriR6K-cat::Frt5; trfA::Frt) ΔdapA::Frt | 67 |
| Z. mobilis ZM4 (PK15256) | Zymomonas mobilis ZM4 ATCC 31281 | GLBRC (31)a |
| PK15394 | Z. mobilis Δcas3 (zmo0681) | This work |
| PK15395 | Z. mobilis Δcas3 (zmo0681) Δmrr (zmo0028) | This work |
| PK15407 | Z. mobilis Δcas3 (zmo0681) Δmrr (zmo0028) ΔhsdSc (zmo1933) | This work |
| PK15410 | Z. mobilis ΔhsdSc (zmo1933) | This work |
| PK15509 | Z. mobilis Δcas3 (zmo0681) Δmrr (zmo0028) ΔhsdSc (zmo1933) ΔhsdSp (zmop32x028) | This work |
| PK15510 | Z. mobilis Δcas3 (zmo0681) Δmrr (zmo0028) ΔhsdRp (zmop32x025) | This work |
| PK15527 | Z. mobilis ΔhsdSc (zmo1933) ΔhsdSp (zmop32x028) | This work |
GLBRC, Great Lakes Bioenergy Research Center.
TABLE 3.
Primers
| Plasmid constructed | Plasmid/gene amplified | Primer | Sequence (5′–3′) |
|---|---|---|---|
| pPK15436 (pRL814-crtIEB) | pRL814 backbone | P1 | ATGTATATCTCCTTCTTAAAGTTAAAC |
| P2 | GCTTGATATCGAATTCCTG | ||
| crtBI of R. sphaeroides | P3 | TTTAAGAAGGAGATATACATATGCCCTCGATCTCGCCC | |
| P4 | TATTTATAATAGAAAGTAAAGACTAGATCGGGTTGGCCCG | ||
| crtE of R. sphaeroides | P5 | CCCGATCTAGTCTTTACTTTCTATTATAAATAAAGGAGACCTTTCATGAGGCACAAGATGGCGTTTGAACAGC | |
| P6 | GCAGGAATTCGATATCAAGCTCAGACGCGGGCCGCGAC | ||
| pPK15404 (pRL814-lacZ) | pRL814 backbone | P7 | CTTAAGGCCGGATCTTGCGGCCCC |
| P8 | TGGGATTACACATGGCATG | ||
| Promoter of sod of Z. mobilis | P9 | GAGGGGCCGCAAGATCCGGCCTTAAGGCTGGGAATAGCATTTCTC | |
| P10 | TCATGGTCATGATGTGATCTCCCAATTC | ||
| lacZ of E. coli MG1655 | P11 | AGATCACATCATGACCATGATTACGGATTCAC | |
| P12 | CCATGCCATGTGTAATCCCAGCGAAATACGGGCAGACATG | ||
| pPK15306 (pRL814-xylA, xylB, rpi and yajO) | pRL814 backbone | P13 | AATTTAGTATATCCTTTGTCGGGTAATTTTTTAATAATTGTTATCCGCTCACAATTG |
| P14 | TGGGATTACACATGGCATG | ||
| xylA of E. coli MG1655 | P15 | ATTCAATTGTGAGCGGATAACAATTATTAAAAAATTACCCGACAAAGGATATACTAAATTATGCAAGCCTATTTTGACC | |
| P16 | TTCTGTTAGTCTGCTTTGTTATTTGTCGAACAGATAATGG | ||
| P17 | ATTCAATTGTGAGCGGATAAC | ||
| xylB of E. coli MG1655 | P18 | GTTCGACAAATAACAAAGCAGACTAACAGAAGGAAGTAACACATGTATATCGGGATAGATCTTG | |
| P19 | TTTCCTTCGTTTGCCCTTTACGCCATTAATGGCAG | ||
| P20 | GTTCGACAAATAACAAAGCAGAC | ||
| rpi of E. coli MG1655 | P21 | ATTAATGGCGTAAAGGGCAAACGAAGGAAACGACAGGAAGTACATATATGACGCAGGATGAATTG | |
| P22 | GATTATAAAGTTAATTTGCGTCATTTCACAATGGTTTTGAC | ||
| P23 | ATTAATGGCGTAAAGGGC | ||
| yajO of E. coli MG1655 | P24 | CATTGTGAAATGACGCAAATTAACTTTATAATCAAGGAACAGAAACAGATGCAATACAACCCCTTAG | |
| P25 | CTCATCCATGCCATGTGTAATCCCATTATTTAAATCCTACGACAGGATG | ||
| P26 | CATTGTGAAATGACGCA | ||
| pPK15304 (pRL814-pgi) | pRL814 backbone | P27 | AATTGTTATCCGCTCACAATTG |
| P28 | TGGGATTACACATGGCATG | ||
| pgi of E. coli MG1655 | P29 | ATTGTGAGCGGATAACAATTAAAGTCACAATTCTCAAAATCAGAAG | |
| P30 | CCATGCCATGTGTAATCCCACGATGATTAACCGCGCCA | ||
| pPK15303-2 | pPK15303 backbone | P31 | CGGCCTGTTTAGCGCTCGGTCTTGCCTT |
| P32 | GGGCATAAACGTGCCGAGGATGACGATG | ||
| Upstream of mrr of Z. mobilis | P33 | TCCTCGGCACGTTTATGCCCGTGACAAC | |
| P34 | TTCAAAAAGCTATCGAGCCTTTTACTTAAAATAATC | ||
| Downstream of mrr of Z. mobilis | P35 | AGGCTCGATAGCTTTTTGAAAAAGCCGTTTC | |
| P36 | ACCGAGCGCTAAACAGGCCGCTAACCTAG | ||
| pPK15357 | pPK15303-2 with modified RBS of gfp | P37 | AAGATATTAGAAGGAGGTCAAAAATGAGCAAAGGAGAAGAACT |
| P38 | TTTTGACCTCCTTCTAATATCTTGTTAAACTAATTCTAGATGT | ||
| pPK15381 | pPK15357 backbone | P39 | GTGCCGAGGATGACGATG |
| P40 | AGCGCTCGGTCTTGCCTT | ||
| Upstream of hsdSc of Z. mobilis | P41 | CTCATCGTCATCCTCGGCACTTCATTGAGCGCAATCTG | |
| P42 | GGTCAAGGTACAGATCGCTTAGGCGATC | ||
| Downstream of hsdSc of Z. mobilis | P43 | AAGCGATCTGTACCTTGACCATGCAGCTG | |
| P44 | GCAAGGCAAGACCGAGCGCTTTGTGCCTGCACATGCTG | ||
| pPK15472 | Upstream of hsdSp of Z. mobilis | P45 | CTCATCGTCATCCTCGGCACGACGATGGAGCAAAGACAG |
| P46 | TAGTGCTCATTATTCCCTCAGTCCATTAGAC | ||
| Downstream of hsdSp of Z. mobilis | P47 | TGAGGGAATAATGAGCACTACTACCGATATCGTTG | |
| P48 | GCAAGGCAAGACCGAGCGCTATCACTTCCCCCGCCTGAG | ||
| pPK15474 | Upstream of hsdRp of Z. mobilis | P49 | CTCATCGTCATCCTCGGCACATGCGGGCGAACATGCCC |
| P50 | TGGCTATGGGATGCGTGCCAACGACCCC | ||
| Downstream of hsdRp of Z. mobilis | P51 | TGGCACGCATCCCATAGCCATTTCCGATTAC | |
| P52 | GCAAGGCAAGACCGAGCGCTACGATCAGTTGCTCGAAAAAG | ||
| pPK15380 | Upstream of cas3 of Z. mobilis | P53 | TGCATTATGATTTATTCAGAAATGATGGAAAATATTATGCATC |
| P54 | GATCTTCTCCCCGCAACCAATAA | ||
| Downstream of cas3 of Z. mobilis | P55 | TCTGAATAAATCATAATGCAGGCTCGGC | |
| P56 | CTCATCGTCATCCTCGGCACCGTTTATCGGCTTTGCTC | ||
| pPK15536 | pRL814 with HsdSp target sequence | P57 | GAGTTCCCCCGGGGGATCCCTAGTTCT |
| P58 | GCGCTTCCTGCAGGAATTCGATATCAAGCT |
TABLE 4.
Plasmids
| Plasmid | Descriptiona | Source and/or reference |
|---|---|---|
| pRL814 | Broad-host-range plasmid containing pBBR-1 origin of replication, lacIq gene, PT7A1-O34-gfp, mob, and spectinomycin resistance cassette | GLBRC (52)b |
| pPK15305 | pRL814 with pgi gene cassette replacing the gfp locus | This work |
| pPK15306 | pRL814 with xylAB, rpi, and yajO gene cassette inserted at the gfp locus | This work |
| pPK15346 | pRL814 with crtIEB cassette inserted at the gfp locus | This work |
| pPK15404 | pRL814 with lacZ cassette inserted at the gfp locus | This work |
| pPK15303 | Suicide plasmid; cat gfp p15A ori mob with 500-bp regions upstream and downstream of the ldh gene | 5 |
| pPK15303-2 | Suicide plasmid pPK15296 with 500-bp regions upstream and downstream of the mrr gene in place of ldh flanking regions | This work |
| pPK15343 | pPK15303-2 with optimized RBS for gfp | This work |
| pPK15357 | pPK15303-2 with optimized RBS for enhanced expression of gfp | This work |
| pPK15380 | Suicide plasmid pPK15357 with 500-bp regions upstream and downstream of the cas3 gene in place of mrr flanking regions | This work |
| pPK15381 | Suicide plasmid pPK15357 with 500-bp regions upstream and downstream of the hsdSc gene in place of mrr flanking regions | This work |
| pPK15472 | Suicide plasmid pPK15357 with 500-bp regions upstream and downstream of the hsdSp gene in place of mrr flanking regions | This work |
| pPK15474 | Suicide plasmid with 500-bp regions upstream and downstream of the hsdRp gene | This work |
| pPK15536 | pRL814 with target sequence of HsdSp (5′CGAAGCGCGAGTTCC3′) at the SmaI restriction site | This work |
| pPK15617 | pRL814 lacking HsdSc target sequence (5′GTCAGTGGGCTG3′; nt 364 to 375 from the translational start site of lacI) | This work |
| pPK15619 | pPK15617 with HsdSp target sequence (5′CGAAGCGCGAGTTCC3′) at the SmaI restriction site | This work |
| pPK15621 | pPK15617 with HsdSc target sequence (5′GTCAGTGGGCTG3′) at the SmaI restriction site | This work |
| pPK15623 | pRL814 with HsdSp target sequence (5′CGAAGCGCGAGTTCC3′) at the SmaI restriction site | This work |
| pPK15625 | pRL814 with two HsdSp target sequences, the first in lacI replacing HsdSc target sequence (5′CAGCCCACTGAC-3′) with the HsdSp target sequence (5′CGAAGCGCGAGTTCC3′) and the second at the SmaI restriction site (insertion of 5′CGAAGCGCGAGTTCC3′) | This work |
| pPK15627 | pRL814 with HsdSc target sequence (5′GTCAGTGGGCTG3′) at the SmaI restriction site | This work |
RBS, ribosome binding site.
GLBRC, Great Lakes Bioenergy Research Center.
Sample preparation for SMRT sequencing.
Cell lysis and genomic DNA isolation were done as described by using a MasterPure complete DNA and RNA extraction kit (Lucigen) and PacBio single-molecule real-time (SMRT) DNA sequencing. Strains were grown anaerobically in 50 ml ZRMG until late exponential phase and harvested by centrifugation at 6,000 × g at 4°C. For cell lysis, the cell pellet was resuspended in 10 ml TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8]), incubated with 5 μl lysozyme (30,000 U/μl; Lucigen) at 37°C and 30 min, followed by the addition of 100 μl tissue and cell lysis solution (Lucigen) premixed with 20 μl 10 mg/ml proteinase K and then incubated at 65°C for 30 min. High-molecular-weight genomic DNA was isolated from the lysed cell suspension by extracting (once or twice) with an equal volume of phenol-chloroform-isoamyl alcohol (24:24:1) and centrifuging at 13,000 × g at 4°C for 10 min to separate the phases. The aqueous phase was then extracted with an equal volume of chloroform-isoamyl alcohol (24:1) as described above and transferred to a tube containing ammonium acetate (to a final concentration of 0.75 M) and 2.5 volumes of absolute ethanol. Genomic DNA was spooled out and resuspended in 200 μl TE buffer. Thirty micrograms of DNA was used for SMRT DNA sequencing at the Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, MD.
Conjugation of plasmids into Z. mobilis ZM4.
A DAP auxotrophic donor, E. coli WM6026, was used to conjugate plasmids into Z. mobilis ZM4 or its mutant derivatives as described previously (5). A stable vector pRL814 or its derivatives, which contains a broad-host-range MobV type conjugation system and pBBR-1 origin of replication, were used for conjugation. CFU were determined after conjugation by plating 100 μl of appropriate 10-fold dilutions of conjugation mixtures onto ZRMG plates, with or without antibiotic. The conjugation frequency was determined by dividing the CFU measured from plates with antibiotic (total number of exconjugants) by the CFU from plates without antibiotic (total number of viable cells). The conjugation frequency from three independent experiments was used to determine the means and standard deviations. Conjugation frequency was determined by normalizing the frequency of one plasmid to that of a control plasmid. The limit of detection in this assay is a conjugation efficiency of 0.00001. GraphPad Prism was used to calculate means, standard deviations, and P values.
Construction of plasmids with the HsdMSc and HsdRMSp target sites.
pRL814-derived plasmids containing no, single, or multiple HsdMSc and HsdRMSp target sites were constructed at Genewiz, Inc., using site-directed mutagenesis as follows: (i) pPK15617 contains a deletion of the HsdSc recognition sequence (5′GTCAGTGGGCTG3′; nucleotides [nt] 364 to 375 from the translational start site of the lacI gene) from pRL814; (ii) pPK15621 contains an insertion of the HsdSc recognition sequence (5′GTCAGTGGGCTG3′) into pPK15617 at the SmaI restriction site; (iii) pPK15619 contains an insertion of the HsdSp recognition sequence (5′CGAAGCGCGAGTTCC3′) into pPK15617 at the SmaI restriction site; (iv) pPK15627 contains an insertion of a second HsdSc recognition sequence (5′GTCAGTGGGCTG3′) into pRL814 at the SmaI restriction site; (v) pPK15623 contains an insertion of the HsdSp recognition sequence (5′CGAAGCGCGAGTTCC3′) into pRL814 at the SmaI restriction site; and (vi) pPK15625 contains two HsdSp recognition sequences (5′CGAAGCGCGAGTTCC3′) in pRL814; one replaces the HsdSc recognition sequence (5′GTCAGTGGGCTG3′) in lacI, and the second is an insertion at the SmaI restriction site. The sequence of the conjugation mobilization element (mobA) and the regions of insertions or deletions of the HsdMSc and HsdRMSp target sites were confirmed by Sanger sequencing.
Cloning of heterologous genes into vector pRL814.
Cloning of heterologous genes into vector pRL814 (52) was achieved by PCR amplification of the vector and indicated genes followed by Gibson assembly using NEBuilder HiFi DNA assembly cloning kit. The Gibson assembly products were transformed into E. coli DH5α using a heat shock method (53). Plasmids were isolated using a plasmid extraction kit (Thermo Fisher Scientific) and confirmed by PCR and Sanger sequencing using primers specific to the DNA fragment junctions. DNA fragments were amplified using primers described in Table 3 for the plasmids listed in Table 4.
Deletion of genes.
Deletion of genes from the Z. mobilis ZM4 genome relied on a homologous recombination method (5) where in the first step, a suicide plasmid, pPK15534 (Table 4), containing 500 bp of DNA upstream and downstream of the target gene to be deleted was conjugated into the recipient Z. mobilis strain from the donor E. coli DAP auxotroph WM6026. Exconjugants that contained plasmid DNA recombined at either the upstream or the downstream position of the gene were selected on ZRMG solid medium containing 120 μg/ml chloramphenicol. The recombinant strains were grown without selection and sorted by FACS to enrich for nonfluorescent strains that had lost the plasmid. The desired deletion strain was identified by PCR assays with specific primers, and the deletion boundaries were confirmed by Sanger sequencing. pPK15357 was used to delete mrr, pPK15380 was used to delete cas3, pPK15381 was used to delete hsdSc, pPK15472 was used to delete hsdSp, and pPK15474 was used to delete hsdRp (Table 4).
Identifying conserved domains of restriction systems.
Z. mobilis ZM4 open reading frames (ORFs) (31) were searched for conserved domains using CDD-Search at NCBI (34, 54), which uses the CDD database or the SRA (Sequence Read Archive) database (55) to scan for a set of precalculated position-specific scoring matrices (PSSM; a unique identifier for domain) in a peptide. CDD-search uses RPS-blast, a variant of PSI-blast, to translate DNA sequence into peptide sequences for all six frames and performs a scan for a set of PSSM with each peptide sequence. These domains included the specificity-determining (HsdS) activity of the RM Type1_S-TRD-CR-like superfamily (30, 34); methyltransferase (HsdM) of the N6-methyltransferase superfamily (34, 56); the type I restriction endonuclease (HsdR) superfamily (34, 57); type IV restriction endonuclease activities of the Mrr type endonuclease superfamily and PD-(D/E)XK superfamily (58, 59); type II restriction endonuclease activities of the GIY-YIG superfamily (58, 60) and the HNH endonuclease superfamily (58, 61); and signature nucleases of CRISPR-Cas system-associated endonucleases, such as Cas3, Cas9, and Cas10 (13, 62). The complete genome sequence, GenBank entry GCA_003054575.1, was analyzed in segments of 8 to 10 kb as described for systematic screening for modification-dependent restriction endonucleases in E. coli (63). The CDD identifiers were then analyzed for domains using phmmer, InterPro, and UniProt databases (64).
Bioinformatic analysis of hsdMSc and hsdRMSp from different Z. mobilis strains.
To screen for hsdMSc and hsdRMSp in different Z. mobilis strains, a local tBLASTn search (65) function for hsdSc, hsdMc, hsdSp, hsdMp, and hsdRp of Z. mobilis strain ZM4 against 16 unique genome sequence assemblies obtained from the NCBI database was performed. Sequence comparison of hsdSc and hsdSp of Z. mobilis ZM4 was done using NCBI BLAST and Clustal Omega (66).
ACKNOWLEDGMENTS
We thank Isabel Askenasy, Erin Mettert, and Donna Bates for comments on the manuscript.
This work was funded in part by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC-07ER64494) and by the Center for Bioenergy Innovation, U.S. DOE Bioenergy Research Center, supported by the Office of Biological and Environmental Research in the DOE Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. DOE under contract DE-AC05-00OR22725.
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Contributor Information
Patricia J. Kiley, Email: pjkiley@wisc.edu.
Robert M. Kelly, North Carolina State University
REFERENCES
- 1.He MX, Wu B, Qin H, Ruan ZY, Tan FR, Wang JL, Shui ZX, Dai LC, Zhu QL, Pan K, Tang XY, Wang WG, Hu QC. 2014. Zymomonas mobilis: a novel platform for future biorefineries. Biotechnol Biofuels 7:101. 10.1186/1754-6834-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang SH, Fei Q, Zhang YP, Contreras LM, Utturkar SM, Brown SD, Himmel ME, Zhang M. 2016. Zymomonas mobilis as a model system for production of biofuels and biochemicals. Microb Biotechnol 9:699–717. 10.1111/1751-7915.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, He Q, Yang Y, Wang J, Haning K, Hu Y, Wu B, He M, Zhang Y, Bao J, Contreras LM, Yang S. 2018. Advances and prospects in metabolic engineering of Zymomonas mobilis. Metab Eng 50:57–73. 10.1016/j.ymben.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Kremer TA, LaSarre B, Posto AL, McKinlay JB. 2015. N2 gas is an effective fertilizer for bioethanol production by Zymomonas mobilis. Proc Natl Acad Sci U S A 112:2222–2226. 10.1073/pnas.1420663112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lal PB, Wells F, Y L, Ghosh IN, Landick R, Kiley PJ. 2019. A markerless method for genome engineering in Zymomonas mobilis. Front Microbiol 10:2216. 10.3389/fmicb.2019.02216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loenen WA, Dryden DT, Raleigh EA, Wilson GG. 2014. Type I restriction enzymes and their relatives. Nucleic Acids Res 42:20–44. 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong G, He M, Feng H. 2016. Functional characterization of CRISPR-Cas system in the ethanologenic bacterium Zymomonas mobilis ZM4. Adv Microbiol 6:178–189. 10.4236/aim.2016.63018. [DOI] [Google Scholar]
- 8.Lee JYH, Carter GP, Pidot SJ, Guerillot R, Seemann T, Goncalves da Silva A, Foster TJ, Howden BP, Stinear TP, Monk IR. 2019. Mining the methylome reveals extensive diversity in Staphylococcus epidermidis restriction modification. mBio 10:e02451-19. 10.1128/mBio.02451-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird A. 2020. The selfishness of law-abiding genes. Trends Genet 36:8–13. 10.1016/j.tig.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Koonin EV, Makarova KS, Wolf YI, Krupovic M. 2020. Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire. Nat Rev Genet 21:119–131. 10.1038/s41576-019-0172-9. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Buso L, Golparian D, Parkhill J, Unemo M, Harris SR. 2019. Genetic variation regulates the activation and specificity of restriction-modification systems in Neisseria gonorrhoeae. Sci Rep 9:14685. 10.1038/s41598-019-51102-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasu K, Nagaraja V. 2013. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77:53–72. 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarova KS, Wolf YI, Koonin EV. 2018. Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J 1:325–336. 10.1089/crispr.2018.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y, Han J, Wang B, Hu X, Li R, Shen W, Ma X, Ma L, Yi L, Yang S, Peng W. 2019. Characterization and repurposing of the endogenous type I-F CRISPR-Cas system of Zymomonas mobilis for genome engineering. Nucleic Acids Res 47:11461–11475. 10.1093/nar/gkz940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riley LA, Guss AM. 2021. Approaches to genetic tool development for rapid domestication of non-model microorganisms. Biotechnol Biofuels 14:30. 10.1186/s13068-020-01872-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dryden DTF, Roberts GA. 2021. DNA modification, restriction endonucleases: type I enzymes, p 72–76. In Jez J (ed), Encyclopedia of biochemistry, 3rd ed. Elsevier, Amsterdam, The Netherlands. 10.1016/B978-0-12-819460-7.00101-8. [DOI] [Google Scholar]
- 17.Rao DN, Dryden DT, Bheemanaik S. 2014. Type III restriction-modification enzymes: a historical perspective. Nucleic Acids Res 42:45–55. 10.1093/nar/gkt616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Aelst K, Toth J, Ramanathan SP, Schwarz FW, Seidel R, Szczelkun MD. 2010. Type III restriction enzymes cleave DNA by long-range interaction between sites in both head-to-head and tail-to-tail inverted repeat. Proc Natl Acad Sci U S A 107:9123–9128. 10.1073/pnas.1001637107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beaulaurier J, Schadt EE, Fang G. 2019. Deciphering bacterial epigenomes using modern sequencing technologies. Nat Rev Genet 20:157–172. 10.1038/s41576-018-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Y, Cohen-Karni D, Xu D, Chin HG, Wilson G, Pradhan S, Roberts RJ. 2010. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res 38:5527–5534. 10.1093/nar/gkq327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou S-l, Zhang K, You L, Zhao X-m, Jing X, Zhang M-h. 2012. Enhanced electrotransformation of the ethanologen Zymomonas mobilis ZM4 with plasmids. Eng Life Sci 12:152–161. 10.1002/elsc.201100106. [DOI] [Google Scholar]
- 22.Kerr AL, Jeon YJ, Svenson CJ, Rogers PL, Neilan BA. 2011. DNA restriction-modification systems in the ethanologen, Zymomonas mobilis ZM4. Appl Microbiol Biotechnol 89:761–769. 10.1007/s00253-010-2936-1. [DOI] [PubMed] [Google Scholar]
- 23.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DT. 2002. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol Cell 9:187–194. 10.1016/s1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 24.Johnston CD, Cotton SL, Rittling SR, Starr JR, Borisy GG, Dewhirst FE, Lemon KP. 2019. Systematic evasion of the restriction-modification barrier in bacteria. Proc Natl Acad Sci U S A 116:11454–11459. 10.1073/pnas.1820256116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu B, He M, Feng H, Zhang Y, Hu Q, Zhang Y. 2013. Construction and characterization of restriction-modification deficient mutants in Zymomonas mobilis ZM4. Chin J Appl Environ Biol 19:189–197. 10.3724/SP.J.1145.2013.00189. [DOI] [Google Scholar]
- 26.O'Connell Motherway M, O'Driscoll J, Fitzgerald GF, Van Sinderen D. 2009. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb Biotechnol 2:321–332. 10.1111/j.1751-7915.2008.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mermelstein LD, Papoutsakis ET. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage phi 3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 59:1077–1081. 10.1128/aem.59.4.1077-1081.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts RJ, Vincze T, Posfai J, Macelis D. 2015. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 43:D298–D299. 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bower EKM, Cooper LP, Roberts GA, White JH, Luyten Y, Morgan RD, Dryden DTF. 2018. A model for the evolution of prokaryotic DNA restriction-modification systems based upon the structural malleability of type I restriction-modification enzymes. Nucleic Acids Res 46:9067–9080. 10.1093/nar/gky760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao P, Tang Q, An X, Yan X, Liang D. 2011. Structure of HsdS subunit from Thermoanaerobacter tengcongensis sheds lights on mechanism of dynamic opening and closing of type I methyltransferase. PLoS One 6:e17346. 10.1371/journal.pone.0017346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S, Vera JM, Grass J, Savvakis G, Moskvin OV, Yang Y, McIlwain SJ, Lyu Y, Zinonos I, Hebert AS, Coon JJ, Bates DM, Sato TK, Brown SD, Himmel ME, Zhang M, Landick R, Pappas KM, Zhang Y. 2018. Complete genome sequence and the expression pattern of plasmids of the model ethanologen Zymomonas mobilis ZM4 and its xylose-utilizing derivatives 8b and 2032. Biotechnol Biofuels 11:125. 10.1186/s13068-018-1116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mouammine A, Collier J. 2018. The impact of DNA methylation in Alphaproteobacteria. Mol Microbiol 110:1–10. 10.1111/mmi.14079. [DOI] [PubMed] [Google Scholar]
- 33.Woodcock CB, Horton JR, Zhang X, Blumenthal RM, Cheng X. 2020. Beta class amino methyltransferases from bacteria to humans: evolution and structural consequences. Nucleic Acids Res 48:10034–10044. 10.1093/nar/gkaa446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A. 2020. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268. 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burger BT, Imam S, Scarborough MJ, Noguera DR, Donohue TJ. 2017. Combining genome-scale experimental and computational methods to identify essential genes in Rhodobacter sphaeroides. mSystems 2:e00015-17. 10.1128/mSystems.00015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M, Chou Y-C, Howe W, Eddy C, Evans K, Mohagheghi A. May2007. Zymomonas pentose-sugar fermenting strains and uses thereof. US patent 7223575B2.
- 37.Chou YC, Linger J, Yang S, Zhang M. 2015. Genetic engineering and improvement of a Zymomonas mobilis for arabinose utilization and its performance on pretreated corn stover hydrolyzate. J Biotechnol Biomater 5:179. 10.4172/2155-952X.1000179. [DOI] [Google Scholar]
- 38.Zhang Y, Vera JM, Xie D, Serate J, Pohlmann E, Russell JD, Hebert AS, Coon JJ, Sato TK, Landick R. 2019. Multiomic fermentation using chemically defined synthetic hydrolyzates revealed multiple effects of lignocellulose-derived inhibitors on cell physiology and xylose utilization in Zymomonas mobilis. Front Microbiol 10:2596. 10.3389/fmicb.2019.02596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costa SK, Donegan NP, Corvaglia AR, Francois P, Cheung AL. 2017. Bypassing the restriction system to improve transformation of Staphylococcus epidermidis. J Bacteriol 199:e00271-17. 10.1128/JB.00271-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou SL, Hong LF, Wang C, Jing X, Zhang MH. 2012. Construction of an unmarked Zymomonas mobilis mutant using a site-specific FLP recombinase. Food Technol Biotechnol 50:406–411. [Google Scholar]
- 41.Sitaraman R, Dybvig K. 1997. The hsd loci of Mycoplasma pulmonis: organization, rearrangements and expression of genes. Mol Microbiol 26:109–120. 10.1046/j.1365-2958.1997.5571938.x. [DOI] [PubMed] [Google Scholar]
- 42.Dybvig K, Sitaraman R, French CT. 1998. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc Natl Acad Sci U S A 95:13923–13928. 10.1073/pnas.95.23.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozdon JB, Melfi MD, Luong K, Clark TA, Boitano M, Wang S, Zhou B, Gonzalez D, Collier J, Turner SW, Korlach J, Shapiro L, McAdams HH. 2013. Global methylation state at base-pair resolution of the Caulobacter genome throughout the cell cycle. Proc Natl Acad Sci U S A 110:E4658–67. 10.1073/pnas.1319315110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malone T, Blumenthal RM, Cheng X. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol 253:618–632. 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 45.Fujimoto D, Srinivasan PR, Borek E. 1965. On the nature of the deoxyribonucleic acid methylases. Biological evidence for the multiple nature of the enzymes. Biochemistry 4:2849–2855. 10.1021/bi00888a041. [DOI] [PubMed] [Google Scholar]
- 46.Adhikari S, Curtis PD. 2016. DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol Rev 40:575–591. 10.1093/femsre/fuw023. [DOI] [PubMed] [Google Scholar]
- 47.Collier J, McAdams HH, Shapiro L. 2007. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci U S A 104:17111–17116. 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reisenauer A, Shapiro L. 2002. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J 21:4969–4977. 10.1093/emboj/cdf490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Felczak MM, Jacobson TB, Ong WK, Amador-Noguez D, TerAvest MA. 2019. Expression of phosphofructokinase is not sufficient to enable Embden-Meyerhof-Parnas glycolysis in Zymomonas mobilis ZM4. Front Microbiol 10:2270. 10.3389/fmicb.2019.02270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobson TB, Adamczyk PA, Stevenson DM, Regner M, Ralph J, Reed JL, Amador-Noguez D. 2019. H and (13)C metabolic flux analysis elucidates in vivo thermodynamics of the ED pathway in Zymomonas mobilis. Metab Eng 54:301–316. 10.1016/j.ymben.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 51.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 52.Ghosh IN, Martien J, Hebert AS, Zhang Y, Coon JJ, Amador-Noguez D, Landick R. 2019. OptSSeq explores enzyme expression and function landscapes to maximize isobutanol production rate. Metab Eng 52:324–340. 10.1016/j.ymben.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 54.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration. 2011. The sequence read archive. Nucleic Acids Res 39:D19–D21. 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chain PS, Comerci DJ, Tolmasky ME, Larimer FW, Malfatti SA, Vergez LM, Aguero F, Land ML, Ugalde RA, Garcia E. 2005. Whole-genome analyses of speciation events in pathogenic Brucellae. Infect Immun 73:8353–8361. 10.1128/IAI.73.12.8353-8361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bianco PR, Xu C, Chi M. 2009. Type I restriction endonucleases are true catalytic enzymes. Nucleic Acids Res 37:3377–3390. 10.1093/nar/gkp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pingoud A, Wilson GG, Wende W. 2014. Type II restriction endonucleases—a historical perspective and more. Nucleic Acids Res 42:7489–7527. 10.1093/nar/gku447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kosinski J, Feder M, Bujnicki JM. 2005. The PD-(D/E)XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics 6:172. 10.1186/1471-2105-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunin-Horkawicz S, Feder M, Bujnicki JM. 2006. Phylogenomic analysis of the GIY-YIG nuclease superfamily. BMC Genomics 7:98. 10.1186/1471-2164-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mehta P, Katta K, Krishnaswamy S. 2004. HNH family subclassification leads to identification of commonality in the His-Me endonuclease superfamily. Protein Sci 13:295–300. 10.1110/ps.03115604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1:e60. 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lutz T, Flodman K, Copelas A, Czapinska H, Mabuchi M, Fomenkov A, He X, Bochtler M, Xu SY. 2019. A protein architecture guided screen for modification dependent restriction endonucleases. Nucleic Acids Res 47:9761–9776. 10.1093/nar/gkz755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang H, Letunic I, Lopez R, Luciani A, Madeira F, Marchler-Bauer A, Mi H, Natale DA, Necci M, Nuka G, Orengo C, Pandurangan AP, Paysan-Lafosse T, Pesseat S, Potter SC, Qureshi MA, Rawlings ND, Redaschi N, Richardson LJ, Rivoire C, Salazar GA, Sangrador-Vegas A, Sigrist CJA, Sillitoe I, Sutton GG, Thanki N, Thomas PD, Tosatto SCE, Yong SY, Finn RD. 2019. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360. 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blodgett JA, Thomas PM, Li G, Velasquez JE, van der Donk WA, Kelleher NL, Metcalf WW. 2007. Unusual transformations in the biosynthesis of the antibiotic phosphinothricin tripeptide. Nat Chem Biol 3:480–485. 10.1038/nchembio.2007.9. [DOI] [PMC free article] [PubMed] [Google Scholar]