Summary
The metabolic inflammation (meta-inflammation) of obesity is characterized proinflammatory macrophage infiltration into adipose tissue. Catalysis by deoxyhypusine synthase (DHPS) modifies the translation factor eIF5A to generate a hypusine (Hyp) residue. Hypusinated eIF5A (eIF5AHyp) controls the translation of mRNAs involved in inflammation, but its role in meta-inflammation has not been elucidated. Levels of eIF5AHyp were found to be increased in adipose tissue macrophages from obese mice and in murine macrophages activated to a proinflammatory M1-like state. Global proteomics and transcriptomics revealed that DHPS deficiency in macrophages altered the abundance of proteins involved in NF-κB signaling, likely through translational control of their respective mRNAs. DHPS deficiency in myeloid cells of obese mice suppressed M1 macrophage accumulation in adipose tissue and improved glucose tolerance. These findings indicate that DHPS promotes the post-transcriptional regulation of a subset of mRNAs governing inflammation and chemotaxis in macrophages and contributes to a proinflammatory M1-like phenotype.
Graphical Abstract
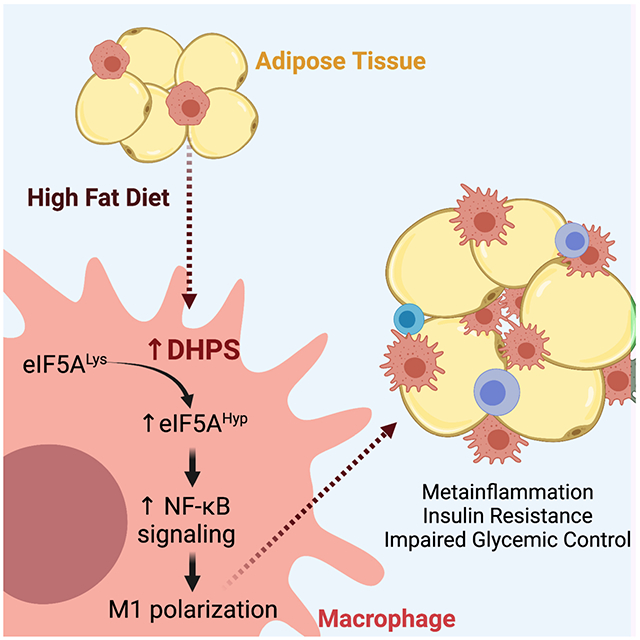
eTOC Blurb (In Brief)
Anderson-Baucum et al. show that eIF5A hypusination, catalyzed by deoxyhypusine synthase (DHPS), is a feature of macrophages that reside in adipose tissue of obese mice. The authors show that deletion of the Dhps gene in macrophages blunts their proinflammatory characteristics, resulting in improved insulin sensitivity and glucose homeostasis.
Introduction
Macrophages are involved in a diversity of functions ranging from tissue remodeling and organ patterning to removal of dead cells and repair of injured tissue (McNelis and Olefsky, 2014; Murray, 2017, 2017; Murray et al., 2014). Systemic signals and locally secreted stimuli can activate macrophages into specialized phenotypes (polarization). Traditional categorization of macrophage polarization describes two distinct phenotypes: M1 macrophages, which exhibit a proinflammatory phenotype and M2 macrophages, which are associated with tissue repair and anti-inflammatory responses (Murray, 2017; Peterson et al., 2018). In the setting of obesity, an increased presence of M1 macrophages has been identified in a number of metabolically-active tissues including the liver, pancreatic islet, muscle, and adipose tissue (Amano et al., 2014; Lumeng et al., 2007; Peterson et al., 2018; Ying et al., 2019; Zheng et al., 2016). Of particular importance are adipose tissue macrophages, which may comprise up to 50% of total adipose cells in obesity and drive insulin resistance, metabolic dysfunction, as well as a state of systemic inflammation (meta-inflammation) (Amano et al., 2014; Lumeng et al., 2007; Peterson et al., 2018; Zheng et al., 2016).
In this study, we explored the significance and mechanism whereby eukaryotic translation initiation factor 5A (eIF5A) governs meta-inflammation. EIF5A is a highly conserved protein among eukaryotes and is the only protein known to be post-translationally modified at a conserved lysine residue to generate the amino acid hypusine (Hyp) (Caraglia et al., 2013; Henderson and Hershey, 2011; Wolff et al., 2007). The hypusine modification is critical for eIF5A function and is catalyzed by the sequential action of two enzymes: deoxyhypusine synthase (DHPS) for transfer of a 4-aminobutyl moiety of the polyamine spermidine to Lys50 residue of eIF5A and deoxyhypusine hydroxylase (DOHH) for hydroxylation of the deoxyhypusine residue (Park et al., 2010). EIF5AHyp functions in mRNA translation during cellular processes that include proliferation, apoptosis, and cell differentiation (Gregio et al., 2009; Levasseur et al., 2019; Padgett et al., 2021; Robbins et al., 2010; Saini et al., 2009; Schuller et al., 2017). A role for eIF5AHyp has also been implicated in the translation of mRNAs involved in inflammation (de Almeida et al., 2014; Gobert et al., 2020; Maier et al., 2010). Although global Dhps gene disruption generates an embryonic-lethal phenotype in mice (Nishimura et al., 2012), the physiological roles of eIF5AHyp have been investigated by utilizing pharmacologic inhibitors of DHPS in mice and cell lines and conditional knockout mice (Colvin et al., 2013; Gobert et al., 2020; Levasseur et al., 2019; Maier et al., 2010; Padgett et al., 2021; Puleston et al., 2019). However, to date, assessment of the role of DHPS in macrophages in vivo in the setting of meta-inflammation has not been undertaken. In this study, we generated myeloid-specific Dhps knockout mice using the Lyz2-Cre driver strain of mice to investigate the role of DHPS in the context of meta-inflammation and macrophage migration as seen in obesity.
Results
DHPS and eIF5AHyp levels are increased in adipose tissue macrophages of obese mice
To assess if changes in DHPS and eIF5AHyp levels are associated with obesity and adipose tissue inflammation, we performed multi-color flow cytometry and immunoblot analyses of immune cells isolated from the stromal vascular fraction (SVF) of epididymal adipose tissue from C57BL/6J male mice fed a low-fat diet (LFD, 10% calories from fat) or a high-fat diet (HFD, 60% calories from fat) for 16 weeks. Flow cytometry analysis of SVF revealed that 16 weeks of HFD feeding increased numbers of total immune cells (CD45+) and the fraction of total immune cells that are positive for eIF5AHyp (Fig. 1A and B), compared to LFD controls. This increase in eIF5AHyp was confirmed by immunoblot analysis (Fig. 1C) and was found to be consistent with an increase in DHPS (Fig. 1C), the enzyme responsible for the rate-limiting step in eIF5A hypusination. We next performed flow cytometry to assess the levels of eIF5AHyp specifically in adipose tissue macrophages, the cells known to exacerbate adipose inflammation. Sixteen weeks of HFD feeding increased the percentage of proinflammatory (M1-like) macrophages (CD45+CD11b+F4/80+iNOS+) in the SVF (Fig. 1D), and increased the percentage of these macrophages producing eIF5AHyp (Fig. 1E). These increases in cell populations were specific to the SVF, since we did not observe similar increases in the spleen (Fig. 1D and E). Because eIF5AHyp is a factor that affects the rate of mRNA translation, we performed polyribosome profiling of SVF cells to assess translational alterations associated with HFD feeding. RNAs associated with polyribosomes are typically actively translated, whereas those associated with monoribosomes are translationally reduced (Evans-Molina et al., 2013). As shown in Fig. 1F, an increase of mRNAs associated with polyribosomes and an elevated polyribosome-to-monoribosome (P/M) ratio was detected in the HFD versus the LFD group, consistent with an increase of mRNA translation.
Figure 1. DHPS is required for the proinflammatory M1-like phenotype.
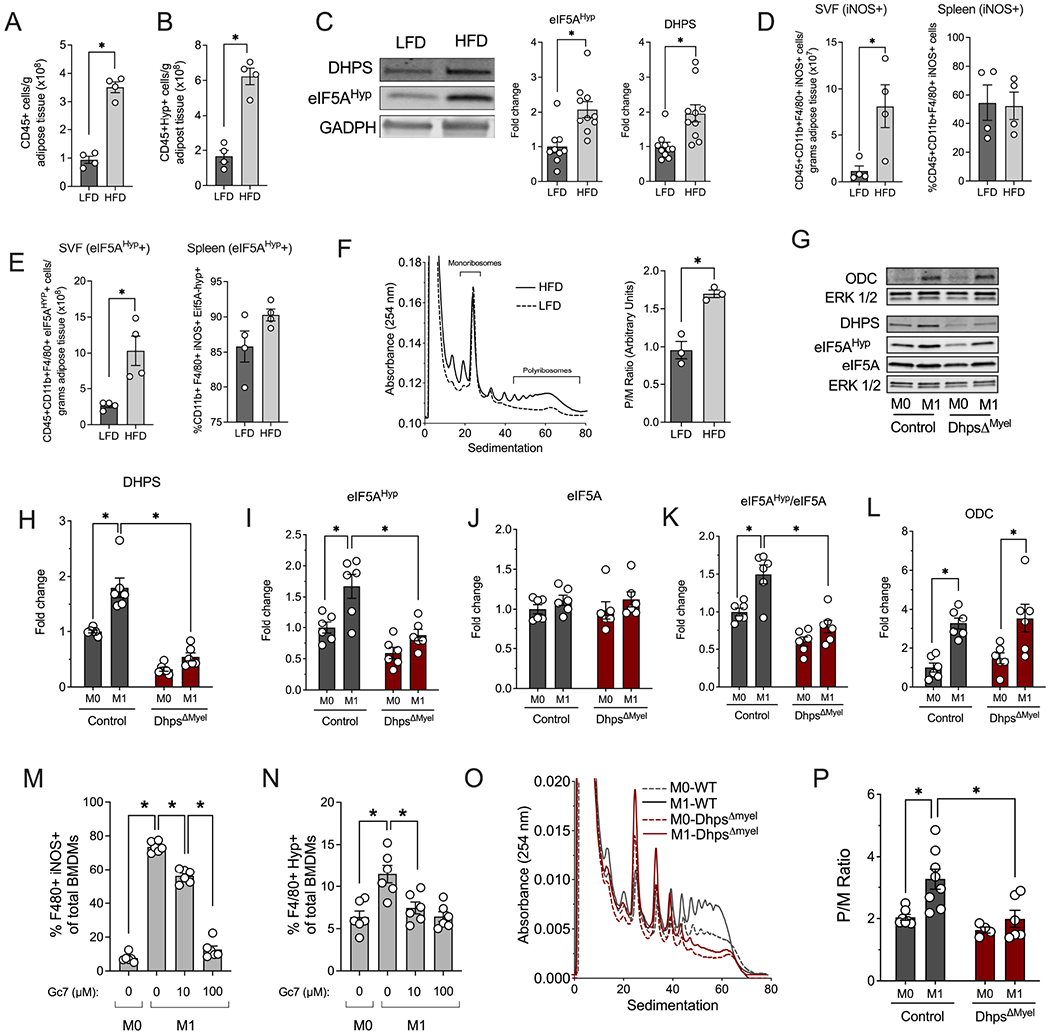
Immune cells from the adipose stromal vascular fraction (SVF) were isolated from male C57BL6/J mice (n=10 per group) fed a low fat diet (LFD, 10% kcal from fat) or a high fat diet (HFD, 60% kcal from fat) for 16 wks and samples were analyzed by flow cytometry using specific antibodies, immunoblotting or polyribosome profiling. (A) Quantification of CD45+ cells by flow cytometry and (B) Quantification of CD45+ eIF5AHyp+ cells by flow cytometry; (C) Representative immunoblot and quantification of DHPS and eIF5AHyp levels relative to GAPDH control; (D) Quantification of CD45+ CD11b+ F4/80+I NOS+ cells in the SVF or spleen by flow cytometry; (E) Quantification of CD45+ CD11b+ F4/80+ eIF5AHyp+ cells in the SVF or spleen by flow cytometry; (F) Representative polyribosome profile and quantification of the polyribosome/monoribosome (P/M) ratio; (G-R) Unpolarized (M0) or M1-polarized macrophages from control (combination of DhpsLoxp/Loxp and DhpsLoxp/+) and DhpsΔMyel mice (n=6-8 per group) were subjected to immunoblotting, flow cytometry and polyribosome profiling analysis. (G) Representatives immunoblots; Quantification of (H) DHPS, (I) eIF5AHyp, and (J) Total eIF5A relative to GAPDH; (K) eIF5AHyp levels relative to total eIF5A levels; (L) ODC levels relative to GAPDH; (M) Quantification of F4/80+ iNOS+ cells; (N) Quantification of F4/80+ eIF5AHyp+ cells. (O) Representative polyribosome profile and (P) quantification of P/M ratio. *p<0.05 by t-test or ANOVA. Data are presented as mean ± SEM.
DHPS activity in macrophages is increased during M1 polarization
To clarify the role of DHPS in macrophage phenotypes, we next performed studies in vitro using bone marrow-derived macrophages (BMDMs) from C57BL/6J mice. Similar to the observations from SVF cells of HFD-fed mice, DHPS and eIF5AHyp levels were increased upon M1 macrophage polarization (LPS+IFN-γ) in BMDMs from C57BL/6J mice (Fig. 1G–K). Additionally, ornithine decarboxylase (ODC), a rate-limiting enzyme in polyamine biosynthesis was also increased under these conditions (Fig. 1G and L). To correlate if M1 formation was dependent upon DHPS activity, we next utilized a small molecule inhibitor of DHPS, N1-guanyl-1,7-diaminoheptane (Gc7) (Lee et al., 1995). For these studies, we performed multi-color flow cytometry to assess eIF5AHyp levels and macrophage phenotypes. M1 polarization (LPS, IFN-γ) of BMDMs led to increased pro-inflammatory macrophage formation (F4/80+iNOS+) (Fig. 1M) with an attendant increase in eIF5AHyp levels (Fig. 1N). Treatment with increasing concentrations of Gc7 (10, 100 μM) led to a dose-dependent reduction in F4/80+iNOS+ cells with an expected decrease in eIF5AHyp levels (Fig. 1M–N).
Prior studies showed that inhibition of DHPS with the small molecule Gc7 also appeared to block the formation of M2-like macrophages (Puleston et al., 2019). We therefore performed flow cytometry and immunoblot analyses to assess eIF5AHyp levels following M2 polarization (using IL4 incubation). When BMDMs are alternatively polarized to the M2 state, there is the expected increase in M2 macrophages (F4/80+CD206+) (Suppl. Fig. S1A) but without any change in eIF5AHyp levels (Suppl. Fig. S1B). These findings were confirmed by immunoblot analyses (Suppl. Fig. S1C), wherein no changes in either DHPS or eIF5AHyp levels were observed. Accordingly, upon M2 polarization, treatment with increasing concentrations of Gc7 (10, 100 μM) had no effect on eIF5AHyp levels (Suppl. Fig. S1B). These findings notwithstanding, increasing concentrations of Gc7 (10, 100 μM) still reduced the formation of F4/80+CD206+ M2 macrophages (Suppl. Fig. S1A) in agreement with prior studies (Puleston et al., 2019). These findings suggest an effect of Gc7 on M2 polarization that may be independent of DHPS inhibition. Collectively, the data in Suppl. Fig S1A–C suggest that treatment with Gc7 may only reflect its effect on DHPS under specific conditions (M1 polarization), and that genetic deletion approaches are needed to clarify the role of DHPS, especially in macrophage phenotypes and meta-inflammation.
DHPS activity governs mRNA translation in macrophages upon M1polarization
To clarify the role of DHPS in M1 macrophage function and polarization, we generated myeloid lineage-specific Dhps knockout mice (DhpΔMyel). DhpsLoxp/Loxp mice (Levasseur et al., 2019) were crossed with mice expressing Cre recombinase under control of the endogenous Lyz2 promoter. The Cre transgene was carried as heterozygous in these studies to ensure that phenotypes were not related to deletion of Lyz2. DhpsΔMyel mice were born at the expected Mendelian ratios. BMDMs from DhpsΔMyel and their littermate controls (a combination of DhpsLoxp/+ and DhpsLoxp/Loxp) were isolated and polarized to the M1 phenotype. As shown in Figure 1G–L, DhpsΔMyel BMDMs showed the expected reduction in DHPS and eIF5AHyp levels, while levels of ODC were unchanged compared with controls. By polyribosome profiling, we observed that Dhps deficiency resulted in a relative decrease in the total RNA associated with polyribosomes in M1-polarized BMDMs (Fig. 1O), which was reflected in the reduced P/M ratio (Fig. 1P). This finding is suggestive of a reduction in mRNA translation initiation. Conversely, Dhps deficiency did not significantly affect the polyribosome profile or the P/M ratio in unpolarized (M0) BMDMs (Fig. 1O and P).
To determine if DHPS affects the translation of genes encoding proinflammatory cytokines and chemokines during M1 polarization, we evaluated the production of a set of cytokines and chemokines in DhpsΔMyel versus control BMDMs. Although the secretion of TNF-α, IL-6, IFN-γ-induced protein 10 (IP-10), IL-1α, monocyte chemoattractant protein 1 (MCP-1) and macrophage inflammatory protein-2 (MIP-2) were not altered by Dhps deletion (Suppl. Fig. S1D) during M1 polarization, secreted MIP-1α (p<0.05) and IL-1β (p=0.09) were decreased or trended to decrease in DhpsΔMyel BMDMs compared to control BMDMs (Fig. 2A and B). Because eIF5AHyp plays a critical role in mRNA translation, we next tested if the translation of mRNAs encoding MIP-1α (Ccl3) and IL-1β (Il1b) were reduced. RNA was isolated from polyribosome profiling sedimentation fractions and reversed-transcribed for real-time PCR analysis. Deletion of Dhps did not affect total levels of Ccl3 and Il1b transcript levels. However, there was a leftward shift in these transcripts in their polyribosome profiles compared to control BMDMs (Fig. 2C and D), indicating a translation initiation block in DhpsΔMyel BMDMs.
Figure 2. eIF5AHyp regulates mRNA translation initiation.
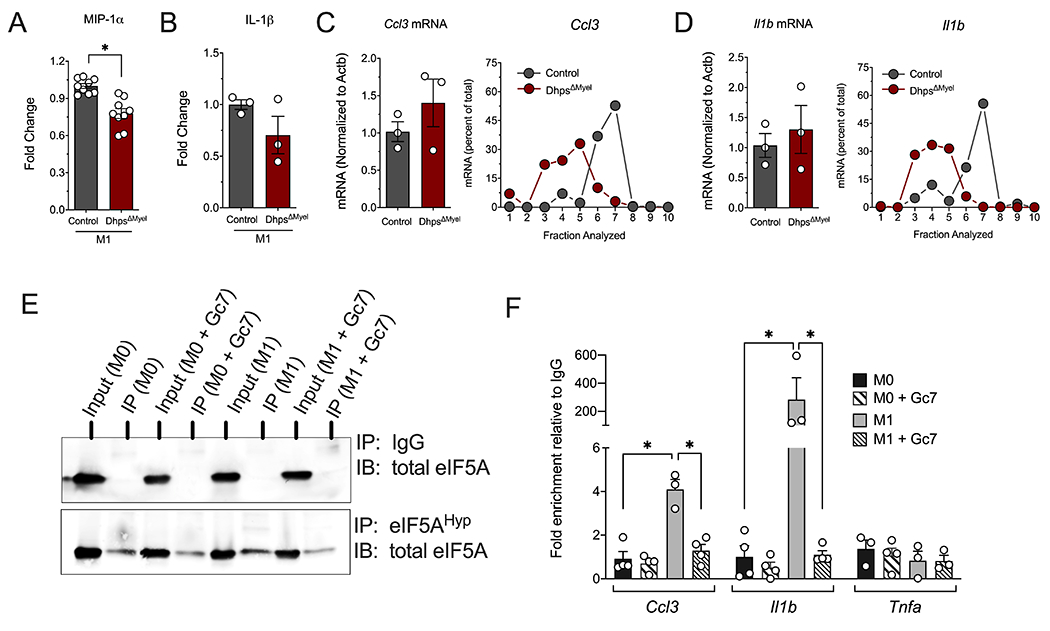
Bone marrow-derived macrophages (BMDMs) from control (combination of DhpsLoxp/Loxp and DhpsLoxp/+) and DhpsΔMyel mice (n=3-10 mice per group) were M1-polarized with LPS + IFN-γ and subjected to flow cytometry and polyribosome profiling. (A) Levels of MIP-1α and (B) IL-1β in media from M1-polarized BMDMs. The mRNA from fractions taken during polyribosome profiling were isolated and quantified for (C) Ccl3 and (D) Il1b; (E) Representative immunoblot of total eIF5A after immunoprecipitation with anti-eIF5AHyp antibody or control IgG in RAW 264.7 cell lysate; (F) Quantification of RNA for Ccl3, Il1b, and Tnfa after RNA immunoprecipitation with anti-eIF5AHyp antibody in RAW 264.7 cell lysate (n=4 independent experiments). *p<0.05 by ANOVA. Data are presented as mean ± SEM.
eIF5AHyp interacts with target mRNAs during translation
If eIF5AHyp directly controls the translation of Ccl3 and Il1b, we surmised that these mRNAs should be bound directly by eIF5AHyp, but not by unhypusinated eIF5A. To test this possibility, we performed RNA immunoprecipitation (RIP) assays using anti-eIF5AHyp antibody or control IgG from the RAW 264.7 mouse macrophage cell line, then performed quantitative RT-PCR on the recovered RNA. As shown in the immunoblots in Fig. 2E, immunoprecipitation with anti-eIF5AHyp antibody resulted in recovery of eIF5AHyp from RAW 264.7 cell lysate, whereas control IgG showed no recovery. We utilized the anti-eIF5A antibody in similar immunoprecipitations from RAW 264.7 cells under M0 and M1 polarization conditions and with and without Gc7 to inhibit hypusination. Fig. 2F shows that under M0 conditions, with or without Gc7, no recovery of Ccl3 or Il1b was observed over IgG control. Under M1 conditions, however, significant recovery of both Ccl3 and Il1b was observed, and this recovery was completely abrogated upon co-incubation with Gc7. By contrast, under no condition was recovery of Tnfa, the mRNA encoding TNF-α, observed. These data support the hypothesis that eIF5AHyp is directly and specifically bound to Ccl3 and Il1b mRNAs.
DHPS governs macrophage migration in a zebrafish model
MIP-1α (encoded by Ccl3) is a chemokine that is important in immune cell migration to sites of injury (Charmoy et al., 2010). We therefore asked if macrophage migration is impaired in the absence of DHPS function. We next performed a tailfin injury assay in zebrafish (Suppl. Fig. S2A), a model organism that allows for rapid testing of biologic processes in vivo. We employed the transgenic zebrafish line Tg(mpeg:EGFP)gl22 (Ellett et al., 2011) in which macrophages are labeled with green fluorescent protein. Tail fin injury results in increased macrophage migration to the site of injury within 6 hours, as observed by GFP+ cells migrating to the site of injury (Suppl. Fig. S2B and C). Treatment of zebrafish larvae with either the DHPS inhibitor Gc7 or a morpholino directed against dhps (to reduce DHPS levels) resulted in a significant decrease in the number of GFP+ macrophages migrating to the injury site as compared with control larvae (Suppl. Fig. S2B and C). These results suggest that inhibition or reduction of DHPS reduces macrophage migration.
DHPS promotes post-transcriptional regulation of inflammatory signaling in macrophages
To define the identity and functional properties of the DhpsΔMyel macrophages under polarized conditions, we differentiated BMDMs and subjected them to M1 polarization (LPS+IFN-γ) or M2 polarization (IL4) and performed multiplex tandem mass tag (TMT) LC-MS/MS-based proteomics analysis (Fig. 3A). A total of 9652 proteins were identified in our analysis. Notably, we identified DHPS as a protein significantly downregulated in DhpsΔMyel macrophages (Fig. 3B). We next identified proteins that were altered in control macrophages as a result of M1 polarization, then among those we identified proteins that were significantly altered in DhpsΔMyel macrophages (p<0.05); this strategy allowed us to focus analysis on those proteins that were acutely differentially regulated by DHPS under conditions of M1 polarization. Fig. 3C shows the heat map of 111 proteins altered in control macrophages by M1 polarization that are differentially regulated in DhpsΔMyel macrophages. Application of KEGG Pathway Analysis showed that these proteins clustered in pathways affecting broadly (a) nitric oxide signaling (cGMP-PKG) (Francis et al., 2010), (b) infection/inflammation (influenza A, legionellosis), and (c) meta-inflammation (adipocytokine signaling) (Fig. 3D)—all pathways consistent with a role for DHPS in promoting M1 macrophage polarization. Notably, there were several proteins whose levels changed in DhpsΔMyel macrophages such that their aggregate effect would be to reduce NF-κB nuclear translocation and pro-inflammatory signaling; these include interleukin 17 receptor A (IL-17RA) (Feng et al., 2019), serine/threonine kinase 11 (STK11/LKB1) (Liu et al., 2015), tripartite motif containing 13 (TRIM13) (Huang and Baek, 2017), PARP1, and NF-κB inhibitor α (IκBα) among others (Fig. 3C, red outline).
Figure 3. Dhps deletion diminishes adoption of an M1-like proteomic signature.
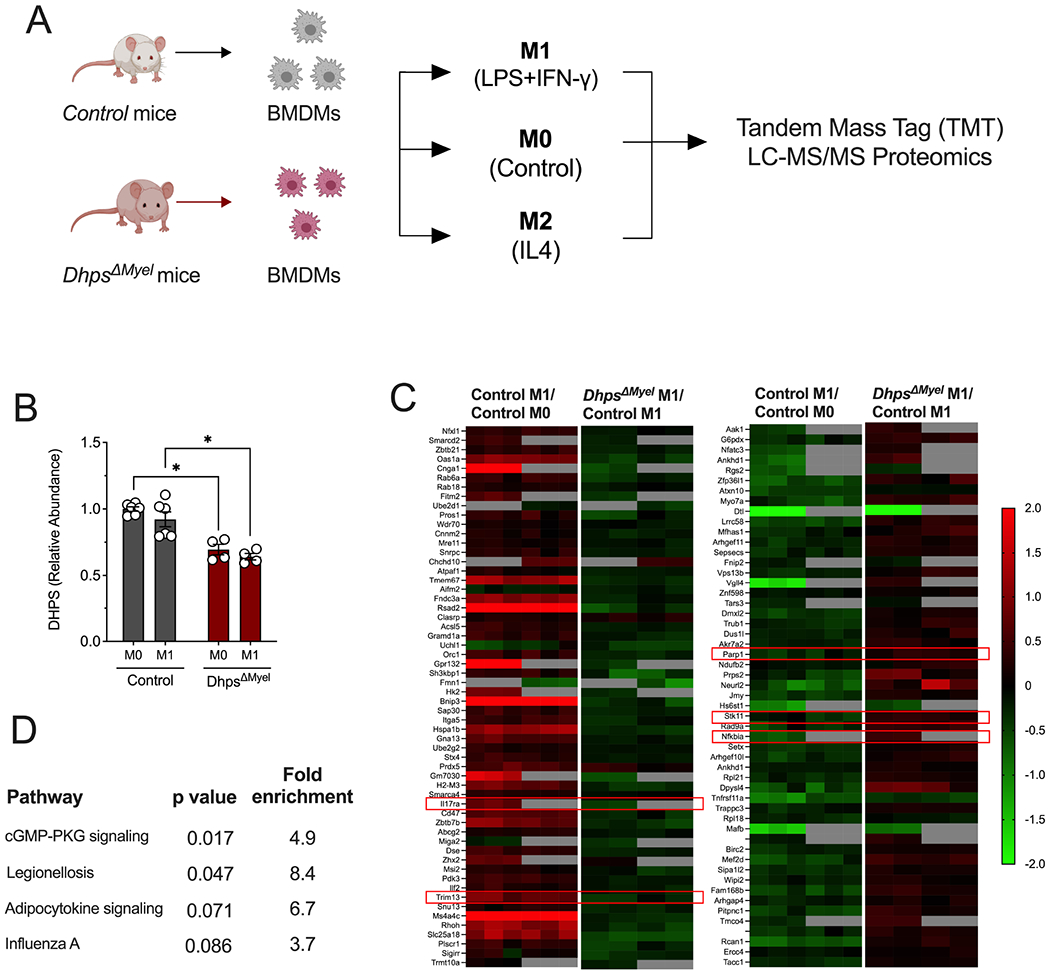
Bone marrow-derived macrophages (BMDMs) from control (combination of DhpsLoxp/Loxp and DhpsLoxp/+) mice (n=6) and DhpsΔMyel mice (n=4) were unpolarized (M0) or M1-polarized with LPS + IFN-γ, and multiplex tandem mass tag (TMT) LC-MS/MS-based proteomics analysis was performed. (A) Schematic of experimental design; (B) Quantification of DHPS levels from proteomics; (C) Heatmaps of significantly differentially regulated proteins (identified by their coding genes) in control cells upon M1 polarization, and those that are differentially regulated in DhpsΔMyel macrophages upon M1 polarization. Red boxes highlight key inflammatory molecules discussed in the text; (D) KEGG Pathway Analysis of proteins differentially regulated in DhpsΔMyel macrophages upon M1 polarization. *p<0.05 by ANOVA. Data are presented as mean ± SEM.
In the setting of M2 polarization (IL4), where we observed no significant changes in eIF5AHyp levels (Suppl. Fig S1), proteomics analysis revealed 53 proteins regulated in DhpsΔMyel macrophages that were altered in response to IL4 incubation (Suppl. Fig. S3); however, KEGG pathway analysis showed no clustering of these proteins, and no clear pathways emerged that would suggest consistent alterations in macrophage signaling or function. These findings emphasize that the role of DHPS in macrophage polarization likely pertains primarily to the M1 state.
DHPS promotes an M1-like transcriptomic signature
The link to NF-κB next led us to interrogate the transcriptional consequences of Dhps depletion in macrophages. Control and DhpsΔMyel BMDMs polarized under M1 conditions as above were harvested and subjected to RNA sequencing. Hierarchical clustering analysis of the transcriptomes indicated that the replicates of each group clustered together and there was no overlap between groups (Suppl. Fig. S4A). When using a fold-change (FC) >2 and a false discovery rate (FDR) <0.05, only 20 genes reached differential expression threshold (volcano plot, Suppl. Fig. S4B). On the one hand, this result is consistent with the role of DHPS in regulating the activity of a translation factor, where changes in gene expression would not be expected in the immediate-early period following cell polarization; on the other, the stringency of these cut-off parameters might belie early changes in gene expression that might have occurred subsequent to inhibition of NF-κB signaling. Using less stringent FC >2 and p<0.05, we identified a total of 70 genes that were differentially expressed (volcano plot, Suppl. Fig. S4C). Genes representing the signatures of M1 and M2 BMDM phenotypes were compiled from published resources (Gensel et al., 2017; Murray, 2017; Murray et al., 2014), evaluated in the transcriptomes, and represented by heat maps (Suppl. Fig. S4D). Dhps deficiency induced a higher level of expression of several components of an M2 macrophage gene signature (CD163, Retnla, Trem26, Itgb3, Tgfb1 and Itgax) and decreased the expression of several components of an M1 macrophage gene signature, which are known NF-κB targets, including Tlr2, Fcgr3, Il-27, Ccl2, Fcgr1, Cxcl1 and Ptges (Suppl. Fig. S4D). In parallel, we determined that Dhps deficiency induced a differential expression of genes associated with anti-inflammatory and tissue repair responses, including Col1a2, Timp3 and Fstl1 (Suppl. Fig. S4D). Finally, we compared all 111 significantly altered proteins in response to LPS+IFN-γ to genes that were significantly altered, and identified only 7 genes that showed changes consistent with the alterations in proteins (Dhps, Nipsnap1, Lrrc45, Hmgxb4, Pes1, Fcgr3, and Zmiz2); these data emphasize that protein alterations in DhpsΔMyel macrophages likely occurs at the post-transcriptional level. Overall, our transcriptomics data show that DhpsΔMyel BMDMs respond to M1-polarizing conditions with an altered gene expression profile that was biased toward downregulation of NF-κB targets and upregulation of anti-inflammatory signatures.
Dhps governs M1 macrophage infiltration into adipose tissue during meta-inflammation
To test a role for DHPS in obesity-induced meta-inflammation and glucose intolerance, DhpsΔMyel mice and littermate controls were fed either a LFD or a HFD for 5 weeks. At the end of the feeding period, both HFD-fed control and DhpsΔMyel mice gained body weight attributable to increases in fat mass compared to LFD mice (Fig. 4A and B). HFD-fed DhpsΔMyel mice exhibited improved glucose tolerance after an intraperitoneal glucose bolus compared to HFD-fed control mice (Fig. 4C and D). This improvement in glucose tolerance in HFD-fed DhpsΔMyel mice was coincident with an improvement in insulin sensitivity based on an insulin tolerance test (Fig. 4E and F). Next, we examined proinflammatory macrophage infiltration into the adipose tissue of HFD mice. There were no differences in the total number of macrophages in the adipose tissue in HFD-fed DhpsΔMyel mice compared to HFD-fed control mice (Fig. 4G and H). However, the adipose tissue of HFD-fed DhpsΔMyel mice had significantly fewer M1-like macrophages (iNOS+F4/80+ cells) compared to the tissue of HFD-fed control mice (Fig. 4G and I).
Figure 4. Dhps deletion reduces proinflammatory macrophage accumulation in adipose tissue and improves glucose tolerance during meta-inflammation.
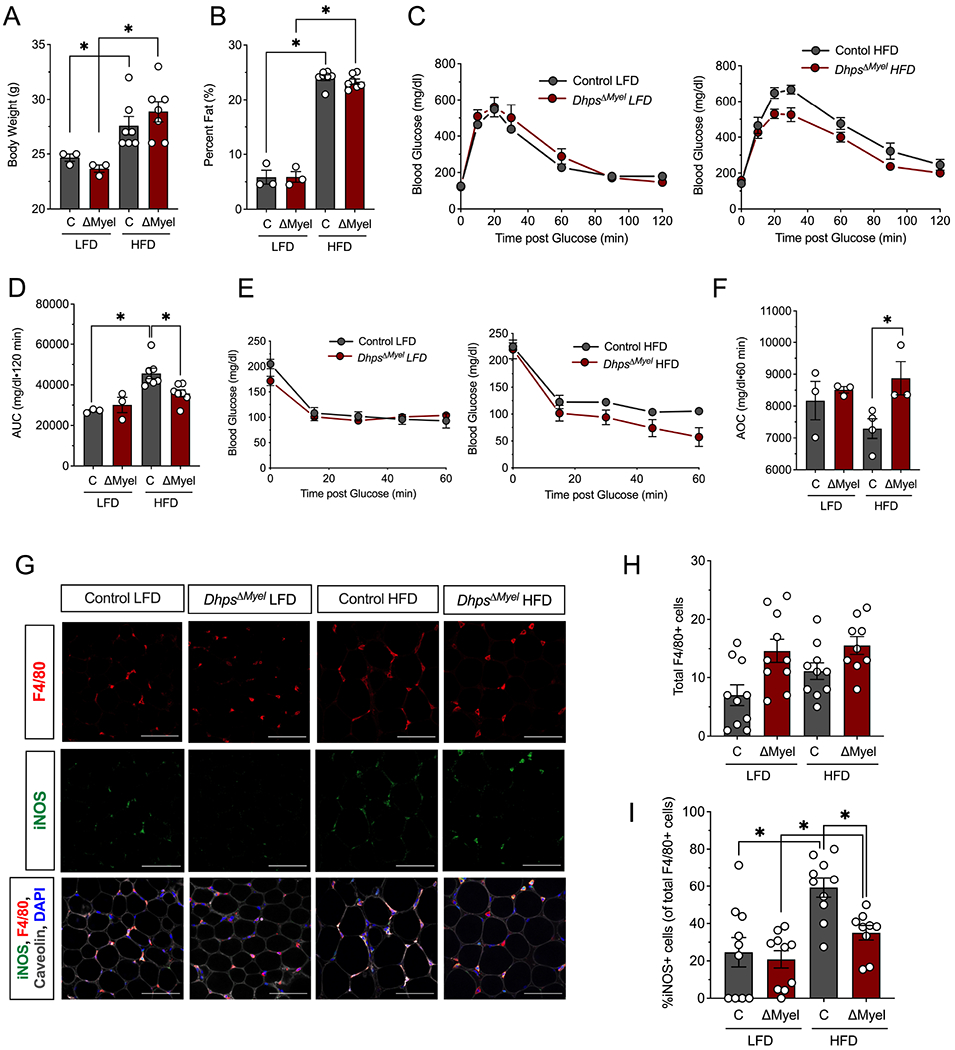
Control (combination of DhpsLoxp/Loxp and DhpsLoxp/+) and DhpsΔMyel male mice were fed a low fat diet (LFD, 10% kcal from fat) or a high fat diet (HFD-60% kcal from fat) (n=10 per group) for 5 weeks. (A) Body weight at end of study. (B) Percentage body fat at end of study as determined by EchoMRI. (C) Glucose tolerance tests (GTTs) at end of study (n=7 mice per group); (D) AUC analysis of GTTs. (E) Insulin tolerance tests (ITTs) at end of study; (F) AOC analysis of ITTs. (G) Representative immunofluorescence micrographs of epididymal adipose tissue stained for the proteins indicated (scale bar = 100 μm); (H) Quantification of the average number of F4/80+ cells per field of view (4 fields of view from each of 9-10 animals); (I) Quantification of the percent of adipose tissue resident M1 macrophages (iNOS+/F4/80+ cells) to total macrophages (F4/80+ cells) (n=3-7) *p<0.05 by ANOVA. Data are presented as mean ± SEM.
Discussion
The known functions of eIF5A depend on the hypusine modification at the conserved Lys50 residue. Hypusine is formed by the consecutive actions of the enzymes DHPS and DOHH, utilizing the polyamine spermidine as the amine donor (Park and Wolff, 2018). In the context of mammalian disease pathogenesis, our prior studies suggest a role for these proteins in the translation of mRNAs that govern cellular proliferation in diabetes and autoimmunity (Colvin et al., 2013; Levasseur et al., 2019). Notably, most of the prior published studies of DHPS/eIF5AHyp in mammalian systems have utilized either silencing of these proteins in cell lines/yeast or use of inhibitors of DHPS, the latter of which are known to cause effects unrelated to hypusine formation (Landau et al., 2010; Oliverio et al., 2014). Hence, our studies reported here represent a fundamental understanding of these proteins in mammals by way of targeted genetics.
Macrophages can be broadly classified as M1 (or “classically activated”) and M2 (or “alternatively activated”), although this somewhat simplistic classification scheme belies a more complicated gradient that exists between these phenotypes. M1 macrophages, driven by canonical TLR4/LPS signaling, are defined as proinflammatory and produce proinflammatory cytokines (e.g. IL-6, TNF-α, IL-1β, IL-12p40) and chemokines (e.g. CCL3, CCL4, CCL5) (Chawla et al., 2011; McLaughlin et al., 2017). A major transcription factor driving M1 polarization is NF-κB, whose activation and nuclear translocation drives the transcription of genes encoding many of these cytokines and chemokines (Wang et al., 2014). Our proteomics data are consistent with a post-transcriptional role for DHPS in the alteration of NF-κB regulators (IL17RA, STK11/LKB1, TRIM13, PARP1, and IκBα), which then drive the gene expression responses that we observed in our transcriptomics data. We believe these post-transcriptional changes are likely controlled at the level of mRNA translation, and the polyribosome profiling data with Ccl3 and Il1b (both NF-κB targets) are consistent with translational regulation.
Adipose tissue macrophages from lean mice express mostly genes associated with an M2 phenotype, including Ym1, Arginase 1 and I1-10 (Lumeng et al., 2007; Orr et al., 2012); however, these genes are downregulated with an attendant increase in Tnfa and Nos2 in adipose tissue macrophages (M1) from obese mice (Lumeng et al., 2007, 2008). A potential role for eIF5AHyp in proinflammatory M1 macrophages was originally suggested in studies utilizing the DHPS inhibitor Gc7 in macrophage-derived cell lines (de Almeida et al., 2014), which demonstrated reduced TNF-α production in response to Gc7 incubation. Consistent with that study, our studies showed that deletion of Dhps enhanced expression of genes associated with an M2 phenotype, including Cd163, Itgax, Itgb3 and Retnla, as well as tissue repair, Col1a2 and Timp3. Our studies suggest that under inflammatory conditions, Dhps deletion shifts macrophages away from an M1-like phenotype, which appears to be consistent with an effect of NFκ B suppression.
One key finding in our study was that Dhps deletion in macrophages led to a defect in the translation initiation of the mRNAs encoding IL-1β (Il1b) and MIP-1α (Ccl3) and to a loss of their binding to eIF5AHyp. IL-1β and MIP-1α are proinflammatory cytokines produced by adipocytes, adipose tissue macrophages, and other immune cells (DiPietro et al., 1998; Maurer and von Stebut, 2004; Stienstra et al., 2010; Wen et al., 2011). Polyproline-containing motifs have been suggested to serve as targets for facilitation of translation by eIF5AHyp in yeast (Gutierrez et al., 2013). Whereas IL-1β has consecutive proline residues, this is not the case with MIP-1α. Therefore, polyproline structural features may not be predictive of targets for eIF5AHyp in mammals. The significant decrease of MIP-1α production by Dhps deletion in M1 macrophages suggests that this chemokine may play a key role in accumulation of macrophages in adipose tissue of obese mice. We demonstrate that DHPS is necessary for the migration of macrophages to the tail injury site in zebrafish. In this regard, during obesity, accumulation of macrophages in the adipose tissue is mostly mediated by recruitment. However proliferation of resident macrophages also occurs (Amano et al., 2014; Zheng et al., 2016). Further studies will be necessary to elucidate if DHPS/eIF5AHyp induces infiltration or proliferation of macrophages within adipose tissue of obese mice.
Two recent studies (Nakamura et al., 2021; Puleston et al., 2019) suggest that inhibition of DHPS with Gc7 reduces the alternative (M2-like) activation of macrophages through changes in the levels of mitochondrial OXPHOS proteins, a result seemingly at odds with the findings in our study. In our proteomics, we found no clear evidence of alterations in mitochondrial OXPHOS proteins. Our findings are consistent with those Wilson and colleagues (Gobert et al., 2020), which interrogated DhpsΔMyel macrophages in the context of microbial infection. However, in line with Puleston et al., we show here that Gc7 does decrease M2 macrophage formation, but it does so without any change in eIF5AHyp levels. A key difference between these studies and ours is our use of genetic Dhps deletion rather than Gc7. First, as acknowledged in the report by Puleston et al., Gc7 may have effects independent of its intended target (Landau et al., 2010; Oliverio et al., 2014). Gc7 resembles natural polyamines and therefore may inhibit or augment other biologic processes that polyamines engage (independently of hypusine formation), such as stabilization of nucleic acid structure, protection from oxidative damage and nucleic acid depurination, and regulation of ion channels (Casero et al., 2018). For example, polyamines, particularly putrescine, tempers M1 macrophage polarization via changes in chromatin structure; as such, reduction in polyamines (by disruption of the gene encoding ODC) aggravates M1 macrophage polarization and inflammation in a mucosal inflammation model (Hardbower et al., 2017). Second, it is possible that the outcome of enzyme inhibition may differ from genetic deletion of the enzyme, since the former may not allow time to establish cellular compensation for other vital biologic processes of an enzyme. It is noteworthy that myeloid-specific deletion of Dhps in our studies did not result in the absence of such myeloid cells nor in the alteration of total numbers of such cells in adipose tissue.
Limitations of study
Some limitations of our study should be acknowledged. First, we deleted Dhps from the inception of Lyz2 activation during development; as noted above, the chronic nature of this deletion may have provided sufficient time for compensatory mechanisms to occur, such that other acute roles of DHPS/eIF5AHyp are not evident in our studies. Second, it remains to be determined if the phenotype observed here can be unequivocally linked to the action/inaction of eIF5AHyp, although the direct interaction of eIF5Hyp with Ccl3 and Il1b mRNAs is supportive of this notion. It is possible that DHPS might have other functions apart from its catalytic activity, and these functions would have been removed from our genetic deletion studies. Definitive resolution of this issue, however, must await studies of eIF5A deletion and/or mutation of the Lys50 residue, the target of hypusination. Despite these limitations, our data suggest that DHPS/eIF5AHyp, which lie downstream of the polyamines, may function discretely from the polyamines and could serve as targets to independently modulate macrophage activation.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Raghavendra G. Mirmira (mirmira@uchicago.edu).
Materials Availability
There are restrictions to the availability of anti-hypusine antibody for flow cytometry use due to the requirement for a materials transfer agreement. More information and requests for resources and reagents should be directed to Dr. Raghavendra Mirmira (mirmira@uchicago.edu).
Data and Code Availability
RNA sequencing data have been deposited in GEO and all proteomics data have been deposited in Proteome Exchange and all are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All primary data presented in this paper have been deposited as a unified Prism 9.1.1 file at Mendeley and are publicly available as of the date of publication, and all original immunoblot images are provided in a single PowerPoint file at Mendeley and are also available as of the date of publication. The DOI is listed in the key resources table. Original microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-ODC | Lisa Shantz, Pennsylvania State University College of Medicine | N/A |
| mouse anti-DHPS | Santa Cruz | Cat# sc-365077, RRID:AB_10846806 |
| rabbit anti-eIF5AHyp | Mirmira laboratory | N/A |
| mouse anti-eIF5A | BD Pharmigen | Cat# 611977, RRID:AB_399398 |
| rabbit anti-BiP | Cell Signaling | Cat# 3183, RRID: AB_10695864 |
| Mouse anti-Hsp90 | Enzo | Cat# SPA-830J, RRID: AB_1505637 |
| rabbit anti-cleaved caspase-3 | Cell Signaling | Cat# 9664, RRID: AB_2070042 |
| Mouse anti-phosho-histone-H3 | Millipore | Cat# 05-598, RRID: AB_309832 |
| rabbit anti-PCNA | Santa Cruz | Cat# sc-7907, RRID: AB_2160375 |
| rabbit anti-ERK 1/2 | Santa Cruz | Cat# sc-94, RRID: AB_2140110 |
| mouse anti-caveolin | Novus | Cat# NB100-615, RRID:AB_10003431 |
| rabbit anti-iNOS | Novus | Cat# NBP1-33780, RRID: AB_10004114 |
| mouse anti-GADPH | Novus | Cat# NB600-502, RRID: AB_10077682 |
| rat anti-F4/80 | Abcam | Cat# Ab6640, RRID: AB_1140040 |
| PE anti-mouse CD206 | Biolegend | Cat# 141706, RRID: AB_10895754 |
| FITC anti-mouse F4/80 | Biolegend | Cat# 123108, RRID: AB_893502 |
| PECy7 anti-mouse CD45 | Biolegend | Cat# 109829 |
| APC Cy7 anti-mouse CD11b | Biolegend | Cat# 101225, RRID: AB_830641 |
| APC anti-mouse iNOS | Invitrogen | Cat# 175920-82, RRID: AB_2573244 |
| chicken anti-GFP | Aves labs | Cat# GFP-1020, RRID: AB_10000240 |
| anti-chicken secondary antibody | Molecular probes | Cat# A-11040, RRID: AB_2534097 |
| Anti-rabbit PE Dazzle | Invitrogen | Cat# A11036 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RPMI Medium 1640 (1X) | Gibco | 11875-093 |
| PBS | Gibco | 14190-144 |
| HEPES Buffer | Corning | 25-060-CI |
| Collagenase | Sigma | C2-22 |
| Collagenase | Sigma | C7657 |
| Penicilin Streptomycin | Thermo Fisher Scientific | 15140-122 |
| Heat inactivated FBS | Gibco | 10082-147 |
| EDTA | Fisher Scientific | BP120-500 |
| Ammonium chloride | Fisher Scientific | A661-500 |
| Potassium bicarbonate | Fisher Scientific | P184-500 |
| rmM-CSF recombinant mouse | R&D Systems | 416-ML |
| rmIFN-γ recombinant mouse | Prospec | Cyt-358-b |
| rmIL-4 recombinant mouse | R&D Systems | 404-ML |
| Lipopolysaccharides from Escherichia coli 0111:B4 | Sigma Aldrich | L2630 |
| rhIFN-γ recombinant human | Prospec | CYT-206b |
| DAPI | Thermo Fisher Scientific | D1306 |
| Sucrose | Fisher Scientific | BP220-212 |
| Gc7 | Biosearch Technologies | G1000 |
| Acid acetic | Sigma Aldrich | A6283 |
| 1-Phenyl-2-thiourea | Acros organics | 207250250 |
| Formaldehyde | Fisher Scientific | 04042-500 |
| PIPES | Sigma Aldrich | P6757-500 |
| MgSO4 | Fisher Scientific | BP213-1 |
| EGTA | Fisher Scientific | 02783-100 |
| TOPRO-3 | Thermo Fisher Scientific | T3605 |
| Urea | Sigma | U0631 |
| 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) | Sigma | H3375 |
| Acetonitrile, HPLC grade | J.T. Baker | 9829-03 |
| Acetonitrile anhydrous | Sigma | 271004 |
| Ammonium hydroxide solution | Sigma | 338818 |
| BCA Protein Assay Kit | Thermo Scientific Pierce | A53225 |
| DTT | Thermo Fisher Scientific | 20291 |
| Iodoacetamide | Thermo Fisher Scientific | 90034 |
| EDTA disodium salt solution 0.5M | Sigma | E7889 |
| Formic acid | Sigma | 33015 |
| HPLC Grade Water | J.T. Baker | 4218-03 |
| Hydroxylamine Solution 50% | Sigma | 467804 |
| Lysyl Endopeptidase | Wako Chemicals | 129-02541 |
| PMSF (Phenylmethylsulfonyl fluoride) | Roche | 10837091001 |
| Reversed phase tC18 SepPak | Waters | 186002319 |
| Sequencing grade modified trypsin | Promega | V5117 |
| Sodium chloride solution; 5M | Sigma | S5150 |
| TMTpro 16plex reagent kit | Thermo Fisher Scientific | A44520 |
| Tris (hydroxymethyl)aminomethane hydrochloride pH 8.0; 1M | Sigma | T2694 |
| Trifluoroacetic acid | Sigma | 91707 |
| Commercial Resources | ||
| Normal Chow diet | Harlan Labs | 2018S |
| Low fat diet | Research Diets | D12450B |
| High fat diet (60%) | Research Diets | D12492 |
| Mouse cytokine/chemokine magnetic bead Milliplex Map Kit | Millipore | MCYTOMAG-70K-PMX |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | 4368814 |
| Experimental Models: Organisms and strains | ||
| C57BL/6 mice | Jackson Laboratories | 000664 |
| Lyz2 promoter driven-Cre recombinase mouse | Jackson Laboratories | 004781 |
| DhpsLoxp/Loxp mice | Raghavendra G Mirmira, deposited into Jackson Laboratories | 034895 |
| DhpsΔMyel mice | Raghavendra G. Mirmira | N/A |
| Wildtype (AB) Zebrafish | Zebrafish International Resource Center | ZL1 |
| Tg(mpeg1:GFP) Zebrafish | Zebrafish International Resource Center | ZL9940 |
| Experimental Models: Cell lines and Tissue | ||
| Mouse: RAW264.7 | ATCC | TIB-71 |
| Oligonucleotides | ||
| Ccl3 (mouse) | Applied Biosystems | Taqman-Assay ID Mm00441259_g1, Cat #4453320 |
| Il1b (mouse) | Applied Biosystems | Taqman-Assay ID Mm00434228_m1, Cat 4453320 |
| Morpholino | ||
| Dhps MO GGTTATGGATGTAAATCCGGCTTTT | Gene Tools, LLC | N/A |
| Deposited data | ||
| RNA Sequencing data | GEO | Accession: GSE144614 |
| Proteomics data | Proteome Exchange | PXD026266 |
| All other data in this manuscript | Mendeley Data | Mirmira, Raghavendra (2021), “Deoxyhypusine Synthase promotes a pro inflammatory macrophage phenotype”, Mendeley Data, V1, doi: 10.17632/kz5rwgpgwc.1 (http://dx.doi.org/10.17632/kz5rwgpgwc.1) |
| Software and Algorithms | ||
| Image Studio Software | LI-COR | Version 3.1.4 |
| Prism Software | GraphPad, Inc. | Version 9.1 |
| STAR 2.3.0 | Alexander Dobin | https://github.com/alexdobin/STAR |
| ngsutils | Marcus Breese and Yunlong Liu | https://github.com/ngsutils/ngsutils |
| EdgeR package | Bioconductor | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| MS-GF+ | Omics Group, Pacific Northwest National Lab | https://omics.pnl.gov/software/ms-gf |
| DAVID | Laboratory of Human Retrovirology and Immunoinformatics | https://david.ncifcrf.gov/tools.jsp |
| mzRefinery | Omics Group, Pacific Northwest National Lab | https://omics.pnl.gov/software/mzrefinery |
| MASIC | Omics Group, Pacific Northwest National Lab | https://omics.pnl.gov/software/masic |
| Decon2LS_V2 | Omics Group, Pacific Northwest National Lab | https://omics.pnl.gov/software/decontools-decon2ls |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mice used in these studies were male and were maintained under protocols approved by the University of Chicago and Indiana University institutional animal care and use committees. Mice were housed in a standard 12 h light:dark cycle with a temperature range between 18-22 °C. Mice were fed a normal chow diet (16% kcal from fat; Harlan Labs; 2018S), low fat diet (10% kcal from fat; Research Diets; D12450B), or high fat diet (60% kcal from fat D12492) as indicated. Weight and percentage of fat were determined using EchoMRI™ (EchoMRI LLC). Health status was checked daily by veterinary technicians. C57BL6/J mice were purchased from Jackson Laboratories. Mice with Loxp sites flanking exons 2 to 7 of the Dhps gene (DhpsLoxp/+ mice) on the C57BL6/J background were generated previously by our group (Levasseur et al., 2019) and are available from Jackson Laboratories. Mice harboring the Lyz2 promoter driven-Cre recombinase were purchased from Jackson Laboratories.
Zebrafish
Zebrafish (D. rerio) were maintained under protocols approved by the University of Chicago and Indiana University or institutional animal care and use committees. Zebrafish were maintained at 28.5°C in a recirculating aquaculture system enclosed in a cabinet and subjected to a 14 h light/10 h dark cycle. Tg(mpeg1:EGFP) zebrafish (Ellett et al., 2011) were obtained from the Zebrafish International Resource Center (ZIRC). Embryos bearing the Tg(mpeg1:GFP) transgene were collected at spawning, then maintained at 28.5 °C in petri dishes filled with egg water supplemented with 1-Phenyl-2-thiourea (Acros) to 0.003% to prevent pigmentation. At 3 days post fertilization, transgenic zebrafish were genotyped by epifluorescence using a Leica M205FA dissecting microscope.
RAW264.7 Murine Leukemia Macrophage Cell Line
RAW264.7 is a male murine leukemia macrophage-derived cell line that was established from a tumor induced by a murine leukemia virus (Ralph and Nakoinz, 1977). RAW264.7 cells were maintained at 37 °C and 5% CO2 atmosphere, and in RPMI 1640 medium supplemented with 2 mM L-glutamine and 10% fetal bovine serum, 100 units/mL penicillin, and 100 mg/ mL streptomycin.
Bone Marrow-Derived Murine Macrophages
Bone marrow-derived macrophages (BMDMs) were isolated and polarized as described previously (Ying et al., 2013). Briefly, femurs were isolated from mice and placed in complete medium (RPMI with 10% heat inactivated FBS, 10 mM HEPES, 100 units/ml penicillin, and 100 mg/ml streptomycin). The femurs were cleaned by removing muscle tissue and rinsed in complete medium. The ends of the femur were cut and the bone marrow was flushed with 10 mL complete medium using a 23g needle. After centrifugation (at 450 xg, 5 minutes, 4 °C), the cells were resuspended in complete medium supplemented with 10 ng/mL M-CSF (R&D biotech). At day 6-7, BMDMs from male control and DhpsΔMyel mice were cultured in RPMI media containing 10% fetal bovine serum and 10 ng/ml M-CSF (R&D Systems). On day 7, the BMDMs were polarized to the M1 state with media containing 10 ng/ml LPS (Sigma Aldrich) and 25 ng/ml IFN-γ (Prospec), or to the M2 state with media containing 10 ng/ml IL-4 (R&D Systems). For immunoblot, cytokines measurement, flow cytometry analyses, transcriptomics, and proteomics, cells were polarized for 16 h. For polyribosome profiling, the macrophages were polarized for 4-6 h.
Murine Stromal Vascular Cells
Upon euthanasia of male mice, epididymal adipose tissue was dissected and minced in 3 ml of isolation buffer (1:1 PBS:HBSS, 1% FBS, pen/strep, and 10 mM HEPES). Tissue was then digested with collagenase (1 mg/ml final concentration) for 40 min while shaking at 37 °C. For the last 10 minutes of digestion, EDTA was added at a final concentration of 2 mM. Cells were centrifuged at 500 xg for 10 min at 4 °C to separate stromal vascular cells (pellet) from adipocytes (supernatant). The stromal vascular fraction cell pellet was incubated with red blood cell lysis buffer (155 mM ammonium chloride, 10 mM potassium bicarbonate, 0.1 mM EDTA) for 5 min at room temperature. Stromal vascular fraction cells were washed twice in PBS prior to lysis for immunoblot analysis or polyribosome profiling.
METHOD DETAILS
Glucose Tolerance Tests and Insulin Tolerance Tests
Intraperitoneal glucose tolerance tests (GTTs) were performed as previously described (Evans-Molina et al., 2009). Briefly, after an overnight fast mice were injected intraperitoneally with glucose at 2 g/kg. Blood was sampled from the tail vein at 0, 10, 20, 30, 60, 90, and 120 min, and glucose was measured using an AlphaTRAK 2 glucometer (Zoetis). Insulin tolerance testing was performed on random fed mice following an intra- peritoneal injection of insulin at 0.75 U/kg. Blood was sampled from the tail vein at 0, 15, 30, 45, and 60 min, and glucose was measured in the manner described above.
Flow Cytometry
Cells were incubated with CD16/CD32 Fc block prior to staining. To stain for surface antigens, cells were incubated with antibodies against F4/80 (BM-8, Biolegend), CD45 (Biolegend) or CD11b (Biolegend) for 30 min. For intracellular staining the cells were permeabilized using BD Cytofix/Cytoperm (BD Pharmigen) and incubated with antibodies for CD206 (Biolegend) and/or iNOS (Invitrogen). The anti-eIF5AHyp antibody was developed in-house and used at a 1:75 dilution. All other antibodies were used at a 1:100 dilution. Cells were analyzed on the LSR Fortessa cytometer (BD). Data were analyzed using FlowJo software (Tree Star).
Immunoblotting
Western blotting was performed as previously described (Maier et al., 2010). Briefly, whole cell extracts of cells were prepared in a buffer containing SDS, and 5 μg extract was resolved by electrophoresis on a 4%–20% SDS-polyacrylamide gel, and and blotted onto a PVDF membrane. The blots were blocked and probed with the following primary antibodies with overnight incubation at 4 °C (1:1000 – 1:5000 dilution): rabbit anti-ODC (from Lisa Shantz, Pennsylvania State University College of Medicine), mouse anti-DHPS (Santa Cruz), rabbit anti-eIF5AHyp (MilliporeSigma), mouse total anti-eIF5A (BD Biosciences), and rabbit anti-ERK 1/2 (Santa Cruz). LiCor anti-rabbit or anti-mouse (1:10000) secondary antibodies were used for visualization and quantification. Immunoblots were visualized using the LiCor Odyssey system (LiCor Biosciences) and quantitated using Odyssey Imaging software (LiCor Biosciences).
Immunofluorescence
Epididymal adipose tissue from mice fed a NCD or HFD were fixed in 4% paraformaldehyde, paraffin embedded, and sectioned onto glass slides. The sectioned adipose tissue was stained using the following antibodies: iNOS (1:200, Novus), Caveolin (1:200, Novus), F4/80 (1:200, Abcam) and DAPI (Thermo Fisher), and counter-stained with following secondary antibodies: goat anti-guinea pig (Alexa 488; 1:400), donkey anti-rabbit (Alexa-568; 1:250) and donkey anti-mouse (Alexa-647; 1:300). All images were acquired using a Zeiss LSM800 confocal microscope (Carl Zeiss).
Polyribosome profiling and RT-PCR
The polyribosome profiling was performed in BMDM as previously described (Templin et al., 2014; Tersey et al., 2012). Briefly, 6h after polarization the cells were lysed and centrifuged through a 10-50% sucrose gradient and fractionated using a piston gradient fractionator (Bio-Comp Instruments). The RNA absorbance at 254 nm was continuously measured using an in-line UV monitor and the fractions were collected. Total RNA from the fractions was reverse transcribed and subjected to qPCR. Polyribosome-to-monoribosome (P/M) ratios were determining by calculating the area under the curve corresponding to the polyribosome peaks (more than two ribosomes) and dividing it by the area under the curve for the monoribosome (80S) peak. Primers for Ccl3 and Il1b were used for quantitative RT-PCR. Taqman primers for Ccl3 and Il1b were obtained from Applied Biosystems (Foster City, CA).
Cytokines quantification
BMDMs were M1 polarized for 16h, as described above. Conditioned media was collected and spun for 10min at 12,000rpm to remove cell debris. Levels of secreted TNF-α, IL-6, IP-10, IL-1α, IL-1β, MCP-1, MIP-1α, and MIP-2 for the DhpsΔMyel macrophages were quantified using a mouse cytokine/chemokine magnetic bead Milliplex Map Kit and a Luminex 200 instrument, per manufacturers’ instructions. Values were normalized to the average value (in pg/ml) of control cells treated on the same day and presented as fold increase.
RNA immunoprecipitation (RIP) assay
RIP assay was performed according to the manufacturer’s protocol of the Magna RIP Kit (EMD Millipore). Briefly, RAW264.7 cells were treated with 100 μM Gc7, 10 ng/ml LPS + 25 ng/ml IFN-γ, or 100 μM GC7 +10 ng/ml LPS + 25 ng/ml IFN-γ overnight. After treatments, cells were harvested, lysed in 100 μl RIP lysis buffer, then incubated with magnetic protein A/G beads conjugated to the antibodies (rabbit anti-eIF5AHyp, rabbit IgG) at 4 °C overnight. After six washes, the immunoprecipitate was treated with proteinase K solution and RNA was extracted using phenol/chloroform/isoamyl alcohol, and levels of Ccl3, Il1b, and Tnfa mRNAs were assayed by SYBR Green I-based quantitative RT-PCR. Primers (forward and reverse) were: Ccl3: 5’-AAGGTCTCCACCACTGCCCTTG and 5’-CTCAGGCATTCAGTTCCAGGTC; Il1b: 5’-AACCTGCTGGTGTGTGACGTTC and 5’-CAGCACGAGGCTTTTTTGTTGT; Tnfa: 5’-AGCACAGAAAGCATGATCCGCG and 5’-GACTTTCTCCTGGTATGAGATAGC.
Tandem mass tag (TMT) LC-MS/MS proteomics and data analysis
Control, M1 and M2 BMDMs from WT control (n=6) and DhpsΔMyel (n=4) mice were obtained as described above and cell pellets were dissolved in 8 M urea in 50 mM TRIS-HCI, pH 8.0. Samples were incubated for 15 min at 6°C in the Thermomixer R (Eppendorf) and protein concentration was measured by BCA Protein Assay. Before next 1 hour incubation at 37 °C, 250 mM dithiothreitol solution was added to a final concentration of 5 mM to reduce disulfide bonds. Free thiol groups were alkylated with 10 mM iodoacetamide for 45 min at 25°C in the dark. Samples were diluted 4-fold with 50 mM Tris-HCI, pH 8.0 to lower urea concentration to 2 M. Proteins were digested with Lys-C (Wako) at 1:50 enzyme:substrate ratio at 25 °C for 2 h. After the Lys-C digestion step, proteins were digested with 1:10 trypsin:protein ratio for 14 h at 25 °C. The digestion reaction was stopped by acidifying the samples with 100% formic acid to a final concentration of 1% formic acid. Peptides were desalted by solid phase extraction using Sep-Pak tC18 96-well Plate (25 mg Sorbent per Well; Waters) and dried in a SpeedVac vacuum concentrator.
Desalted peptides were dissolved in 100 mM HEPES, pH 8.5, quantified by BCA assay (Thermo Fisher Scientific) and equal amounts (50 μg) of peptides were labeled with 16-plex TMT reagent (Thermo Fisher Scientific) as previously described (Mertins et al., 2018). Samples (total = 30) were randomized in two sets of 16-plex reagents. For normalization between the two sets, one channel of each set was occupied with a reference sample comprised of a pooled aliquot of individual samples. Peptides were fractionated into 96 fractions with high pH reverse phase chromatography and further concatenated into 24 fractions (Mertins et al., 2018). Each fraction was dried in a vacuum centrifuge and dissolved in 3% acetonitrile, 0.1 % formic acid before being loaded into trapping column (4 cm x 150 μm i.d. packed in-lab with 5 μm Jupiter C18 particle, Phenomenex) connected to Waters NanoAcquity liquid chromatography system. Separation was performed on a 70 cm reversed-phase column (75 μm i.d. 3 μm packed in-lab Jupiter C18 particle, Phenomenex). Peptides were eluted at a flow rate 300 nL/min using a gradient of acetonitrile (solvent B) in water (solvent A) both containing 0.1% formic acid, which ramped to 8%-12%-30%-45%-95% B at 2-20-75-97-100 min, respectively. Eluting peptides were directly analyzed online on an orbitrap mass spectrometer (Q Exactive HF-X, Thermo Fisher Scientific). MS spectra were collected from 300 to 1800 m/z with 60,000 mass resolution. The top 16 most intense parent ions with 2 to 6 charges were submitted to high-energy collision dissociation with normalized collision energy of 30. Tandem mass spectra were collected at 45,000 mass resolution for each parent ion, which were then dynamically excluded for 45 seconds.
RNA-sequencing and analysis
RNA sequencing and analysis was performed as previously described (Levasseur et al., 2019). Purified total RNA was first evaluated for its quantity and quality using a Bioanalyzer 2100 (Agilent). All RNA samples had an RNA Integrity Number (RIN) value of eight or higher. cDNA was synthesized with poly-dT priming using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara Clontech Laboratories Inc.), followed by cDNA shearing with an AFA sonicator (Covaris) and size selection with AMPure beads (Beckman Coulter). Barcoded cDNA library was then prepared using the Ion Plus Fragment Library Kit (Thermo Fisher Scientific). Average size of library insert was approximately 110 bp. Pooled libraries were applied to ion sphere particles (ISP) with template preparation and amplification using Ion OneTouch 2, followed by ISP loading onto a PI chip and sequencing on Ion Proton semiconductor (Life Technologies).
Zebrafish treatments and tail fin injury
Morpholino (MO) was purchased from Gene Tools, LLC. For knockdown of dhps, 4 ng of dhps MO (targeting the splice site at the junction of exon 2 and exon 3) was injected in one-cell stage embryos (Mastracci et al., 2015). After sorting, embryos were immobilized and anesthetized with tricaine, and then subjected to tail-fin injury, where the distal end of the tailfin was excised using a sharp scalpel. For Gc7 incubation experiments, the embryos were treated for 6h with 20 mM Gc7 or vehicle (0.5 mM acetic acid) and then fixed with 3% formaldehyde in a PEM buffer (0.1 M PIPES, 1 mM MgSO4, 2 mM EGTA, pH 7) at 4°C overnight. For whole mount immunofluorescence staining, the following dilutions of antibodies were used: 1:200 chicken anti-GFP (Aves Labs) primary antibody, 1:500 anti-chicken secondary antibody (Molecular Probes) and 1:500 TOPRO-3 (Thermo Fisher) for DNA staining. Images were collected with a LSM700 confocal microscope (Zeiss) or an A1 confocal microscope (Nikon).
QUANTIFICATION AND STATISTICAL ANALYSIS
Proteomics analysis and quantification
Collected spectra were converted into peak lists using Decon2LS_V2 (Jaitly et al., 2009) combined with mzRefinery (Gibbons et al., 2015), both using default parameters. Peptides were then identified with MS-GF+ (Kim and Pevzner, 2014) by searching spectra against mouse reference proteome database from Uniprot Knowledgebase, downloaded on January 29, 2021. The searching parameters included: (1) parent ion mass tolerance ≤ 20 ppm, (2) tryptic digestion in at least one of the termini, (3) maximum of 2 trypsin missed cleavage site, (4) cysteine carbamidomethylation and N-terminal/lysine TMT addition as static modifications, (5) methionine oxidation as variable modification. Identifications were filtered MSGF probability ≤ 1.0E −9 and 1.0E −10 at peptide-spectrum match and protein level, respectively, to keep the false-discovery rate <1%. The quantitative information of the identified peptides was obtained by extracting the TMT reporter ion intensities using MASIC (Monroe et al., 2008). Proteomics data were processed in MatLab R2019 and all samples met quality control analysis based on evaluation of Pearson correlation between samples in the same group, percent of missing peptide abundance measures (of 161,221 peptides), and Median Absolute Deviation relative peptide abundance (Matzke et al., 2011).
RNA sequencing
A Phred quality score (Q score) was used to measure the quality of sequencing reads. More than 90% of the sequencing reads reached Q30 (99.9% base call accuracy). The sequencing data were first assessed using FastQC for quality control (Babraham Bioinformatics). All sequenced libraries were mapped to the mouse genome (UCSC mm10) using STAR RNA-seq aligner (Dobin et al., 2013). The reads distribution across the genome was assessed using bamutils (from ngsutils) (Breese and Liu, 2013). Uniquely mapped sequencing reads were assigned to mm10 refGene genes using featureCounts (Liao et al., 2014). Differential expression (DE) analyses were performed using edgeR v3.22.3 implemented in the Bioconductor package (McCarthy et al., 2012) to identify differentially expressed mRNAs between DhpsΔMyel and control mice. Biological coefficients of variation between the samples were estimated using an empirical Bayes approach under the assumption that the data follows a negative binomial distribution. A total of 10,552 mRNAs remained after filtering low expression transcripts and used in the differential expression analysis by edgeR. Statistical significance was defined as P value ≤ 0.05 and a two-fold change (FC) of expression level between comparison of DhpsΔMyel and control mice. The heat map and locus-by-locus volcano plot were performed using R package.
Image Processing and Quantification
Immunofluorescence in adipose tissue sections was analyzed by acquiring Z stacks images using an LSM700 confocal microscope (Zeiss). From each tissue section, 5-6 fields of view were randomly selected, and the number of iNOS+ and F4/80+ cells were counted in each field of view. All image analyses were performed in a blinded fashion and independently by two individuals.
Statistical analysis:
For proteomics data, statistical comparisons were made with a standard paired t-test using a Bonferroni correction for multiple test correction to compare M0, M1, and M2 within Dhps knockout and control mice. A standard two-tailed t-test was used to compare between the Dhps knockout and Control mice (Bramer et al., 2019). Differentially abundant proteins were submitted to functional enrichment analysis using DAVID (Huang et al., 2009). Statistical analysis for other data was performed using GraphPad Prism 9.1 software. To compare data from two independent groups/conditions, a standard paired or unpaired two-tailed student’s t-test was performed. For experiments involving 3 or more independent groups/conditions, data were analyzed by one-way ANOVA followed by a Bonferroni multiple test correction. Results were considered significant when p<0.05. The number of replicates (including number of animals used in each experiment) are indicated in the figures and/or figure legends. All data are expressed as mean ± SEM.
Supplementary Material
Highlights.
eIF5AHyp is enriched in adipose tissue macrophages in obesity
Proinflammatory macrophages augment DHPS and eIF5AHyp production
DHPS supports the production of proteins that promote NFκB signaling in macrophages
Dhps deletion in macrophages of obese mice improves insulin sensitivity and glycemia
Acknowledgements
The authors thank K. Orr, K. Randhave, J. Nelson, and M. Gritsenko for technical assistance, and Dr. D. Leclerc for helpful discussions. This work was supported by NIH grants R01 DK060581 and R01 DK124906 (to RGM), U01 127786 (to RGM, TOM, BJWB, and CEM), R01 DK093954 and R01 DK127308 (to CEM), F32 DK104501 and T32 DK064466 (to EAB), R01 DK121987-01A1 and JDRF Career Development Award 5-CDA-2016-194-A-N (to TLM). This work utilized core services supported by NIH grants P30 DK097512 (to Indiana University) and P30 DK020595 (to the University of Chicago). Proteomics analyses were performed in the Environmental Molecular Sciences Laboratory located at Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the DOE under Contract DE-AC05-76RLO 1830. The content is solely the responsibility of the authors and does not represent the official views of the funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- de Almeida OP, Toledo TR, Rossi D, Rossetto Dde B, Watanabe TF, Galvao FC, Medeiros AI, Zanelli CF, and Valentini SR, (2014). Hypusine modification of the ribosome-binding protein eIF5A, a target for new anti-inflammatory drugs: understanding the action of the inhibitor GC7 on a murine macrophage cell line. Curr Pharm Des 20, 284–292. [DOI] [PubMed] [Google Scholar]
- Amano SU, Cohen JL, Vangala P, Tencerova M, Nicoloro SM, Yawe JC, Shen Y, Czech MP, and Aouadi M, (2014). Local Proliferation of Macrophages Contributes to Obesity-Associated Adipose Tissue Inflammation. Cell Metab. 19, 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramer LM, Stratton KG, White AM, Bleeker AH, Kobold MA, Waters KM, Metz TO, Rodland KD, and Webb-Robertson B-JM, (2019). P-Mart: Interactive Analysis of Ion Abundance Global Proteomics Data. J. Proteome Res 18, 1426–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breese MR, and Liu Y, (2013). NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics 29, 494–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraglia M, Park MH, Wolff EC, Marra M, and Abbruzzese A, (2013). eIF5A isoforms and cancer: two brothers for two functions? Amino Acids 44, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero RA, Murray Stewart T, and Pegg AE, (2018). Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat. Rev. Cancer 18, 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmoy M, Brunner-Agten S, Aebischer D, Auderset F, Launois P, Milon G, Proudfoot AEI, and Tacchini-Cottier F, (2010). Neutrophil-Derived CCL3 Is Essential for the Rapid Recruitment of Dendritic Cells to the Site of Leishmania major Inoculation in Resistant Mice. PLoS Pathog. 6, e1000755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Nguyen KD, and Goh YPS, (2011). Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol 11, 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin SC, Maier B, Morris DL, Tersey SA, and Mirmira RG, (2013). Deoxyhypusine synthase promotes differentiation and proliferation of T helper type 1 (Th1) cells in autoimmune diabetes. J. Biol. Chem 288, 36226–36235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPietro LA, Burdick M, Low QE, Kunkel SL, and Strieter RM, (1998). MIP-1alpha as a critical macrophage chemoattractant in murine wound repair. J. Clin. Invest 101, 1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR, (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F, Pase L, Hayman JW, Andrianopoulos A, and Lieschke GJ, (2011). mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans-Molina C, Robbins RD, Kono T, Tersey SA, Vestermark GL, Nunemaker CS, Garmey JC, Deering TG, Keller SR, Maier B, et al. (2009). Peroxisome Proliferator-Activated Receptor γ Activation Restores Islet Function in Diabetic Mice through Reduction of Endoplasmic Reticulum Stress and Maintenance of Euchromatin Structure. Mol. Cell. Biol 29, 2053–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans-Molina C, Hatanaka M, and Mirmira RG, (2013). Lost in translation: endoplasmic reticulum stress and the decline of β-cell health in diabetes mellitus. Diabetes Obes Metab 15Suppl 3, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Zheng C, Zhou Z, Xiong H, Feng F, Xie F, and Wu Z-D, (2019). IL-17A neutralizing antibody attenuates eosinophilic meningitis caused by Angiostrongylus cantonensis by involving IL-17RA/Traf6/NF-μB signaling. Exp. Cell Res 384, 111554. [DOI] [PubMed] [Google Scholar]
- Francis SH, Busch JL, Corbin JD, and Sibley D, (2010). cGMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol. Rev 62, 525–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensel JC, Kopper TJ, Zhang B, Orr MB, and Bailey WM, (2017). Predictive screening of M1 and M2 macrophages reveals the immunomodulatory effectiveness of post spinal cord injury azithromycin treatment. Sci. Rep 7, 40144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons BC, Chambers MC, Monroe ME, Tabb DL, and Payne SH, (2015). Correcting systematic bias and instrument measurement drift with mzRefinery. Bioinformatics 31, 3838–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert AP, Finley JL, Latour YL, Asim M, Smith TM, Verriere TG, Barry DP, Allaman MM, Delagado AG, Rose KL, et al. (2020). Hypusination Orchestrates the Antimicrobial Response of Macrophages. Cell Rep. 33, 108510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregio APB, Cano VPS, Avaca JS, Valentini SR, and Zanelli CF, (2009). eIF5A has a function in the elongation step of translation in yeast. Biochem Biophys Res Commun 380, 785–790. [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Shin B-S, Woolstenhulme CJ, Kim J-R, Saini P, Buskirk AR, and Dever TE, (2013). eIF5A Promotes Translation of Polyproline Motifs. Mol. Cell 51, 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardbower DM, Asim M, Luis PB, Singh K, Barry DP, Yang C, Steeves MA, Cleveland JL, Schneider C, Piazuelo MB, et al. (2017). Ornithine decarboxylase regulates M1 macrophage activation and mucosal inflammation via histone modifications. Proc. Natl. Acad. Sci. U. S. A 114, E751–E760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A, and Hershey JW, (2011). Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci 108, 6415–6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, and Baek S-H, (2017). Trim13 Potentiates Toll-Like Receptor 2-Mediated Nuclear Factor kB Activation via K29-Linked Polyubiquitination of Tumor Necrosis Factor Receptor-Associated Factor 6. Mol. Pharmacol 91, 307–316. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA, (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, and Smith RD, (2009). Decon2LS: An open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics 10, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, and Pevzner PA, (2014). MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun 5, 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau G, Bercovich Z, Park MH, and Kahana C, (2010). The role of polyamines in supporting growth of mammalian cells is mediated through their requirement for translation initiation and elongation. J Biol Chem 285, 12474–12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Park MH, and Folk JE, (1995). Diamine and triamine analogs and derivatives as inhibitors of deoxyhypusine synthase: synthesis and biological activity. J Med Chem 38, 3053–3061. [DOI] [PubMed] [Google Scholar]
- Levasseur EM, Yamada K, Piñeros AR, Wu W, Syed F, Orr KS, Anderson-Baucum E, Mastracci TL, Maier B, Mosley AL, et al. (2019). Hypusine biosynthesis in β cells links polyamine metabolism to facultative cellular proliferation to maintain glucose homeostasis. Sci. Signal 12, eaax0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W, (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhang W, Zhang M, Zhu H, Moriasi C, and Zou M-H, (2015). Liver kinase B1 suppresses lipopolysaccharide-induced nuclear factor κB (NF-κB) activation in macrophages. J. Biol. Chem 290, 2312–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, Bodzin JL, and Saltiel AR, (2007). Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest 117, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, DelProposto JB, Westcott DJ, and Saltiel AR, (2008). Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes 57, 3239–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Ogihara T, Trace AP, Tersey SA, Robbins RD, Chakrabarti SK, Nunemaker CS, Stull ND, Taylor CA, Thompson JE, et al. (2010). The unique hypusine modification of eIF5A promotes islet beta cell inflammation and dysfunction in mice. J Clin Invest 120, 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastracci TL, Robertson MA, Mirmira RG, and Anderson RM, (2015). Polyamine biosynthesis is critical for growth and differentiation of the pancreas. Sci. Rep 5, 13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MM, Waters KM, Metz TO, Jacobs JM, Sims AC, Baric RS, Pounds JG, and Webb-Robertson B-JM, (2011). Improved quality control processing of peptide-centric LC-MS proteomics data. Bioinformatics 27, 2866–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer M, and von Stebut E, (2004). Macrophage inflammatory protein-1. Int. J. Biochem. Cell Biol 36, 1882–1886. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, and Smyth GK, (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin T, Ackerman SE, Shen L, and Engleman E, (2017). Role of innate and adaptive immunity in obesity-associated metabolic disease. J. Clin. Invest 127, 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNelis JC, and Olefsky JM, (2014). Macrophages, Immunity, and Metabolic Disease. Immunity 41, 36–48. [DOI] [PubMed] [Google Scholar]
- Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, Clauser KR, Clauss TR, Shah P, Gillette MA, et al. (2018). Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc 13, 1632–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe ME, Shaw JL, Daly DS, Adkins JN, and Smith RD, (2008). MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput. Biol. Chem 32, 215–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, (2017). Macrophage Polarization. Annu. Rev. Physiol 79, 541–566. [DOI] [PubMed] [Google Scholar]
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. (2014). Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity 41, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Kurihara S, Takahashi D, Ohashi W, Nakamura Y, Kimura S, Onuki M, Kume A, Sasazawa Y, Furusawa Y, et al. (2021). Symbiotic polyamine metabolism regulates epithelial proliferation and macrophage differentiation in the colon. Nat. Commun 12, 2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Lee SB, Park JH, and Park MH, (2012). Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development. Amino Acids 42, 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliverio S, Corazzari M, Sestito C, Piredda L, Ippolito G, and Piacentini M, (2014). The spermidine analogue GC7 (N1-guanyl-1,7-diamineoheptane) induces autophagy through a mechanism not involving the hypusination of eIF5A. Amino Acids 46, 2767–2776. [DOI] [PubMed] [Google Scholar]
- Orr JS, Puglisi MJ, Ellacott KL, Lumeng CN, Wasserman DH, and Hasty AH, (2012). Toll-like receptor 4 deficiency promotes the alternative activation of adipose tissue macrophages. Diabetes 61, 2718–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett LR, Robertson MA, Anderson-Baucum EK, Connors CT, Wu W, Mirmira RG, and Mastracci TL, (2021). Deoxyhypusine synthase, an essential enzyme for hypusine biosynthesis, is required for proper exocrine pancreas development. FASEB J. 35, e21473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, and Wolff EC, (2018). Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J. Biol. Chem 293, 18710–18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Nishimura K, Zanelli CF, and Valentini SR, (2010). Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids 38, 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KR, Cottam MA, Kennedy AJ, and Hasty AH, (2018). Macrophage-Targeted Therapeutics for Metabolic Disease. Trends Pharmacol. Sci 39, 536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston DJ, Buck MD, Klein Geltink RI, Kyle RL, Caputa G, O’Sullivan D, Cameron AM, Castoldi A, Musa Y, Kabat AM, et al. (2019). Polyamines and eIF5A Hypusination Modulate Mitochondrial Respiration and Macrophage Activation. Cell Metab. 30, 352–363.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph P, and Nakoinz I, (1977). Antibody-dependent killing of erythrocyte and tumor targets by macrophage-related cell lines: enhancement by PPD and LPS. J. Immunol 1950119, 950–954. [PubMed] [Google Scholar]
- Robbins RD, Tersey SA, Ogihara T, Gupta D, Farb TB, Ficorilli J, Bokvist K, Maier B, and Mirmira RG, (2010). Inhibition of deoxyhypusine synthase enhances islet beta cell function and survival in the setting of endoplasmic reticulum stress and type 2 diabetes. J Biol Chem 285, 39943–39952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, and Dever TE, (2009). Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Wu CC-C, Dever TE, Buskirk AR, and Green R, (2017). eIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 66, 194–205.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stienstra R, Joosten LAB, Koenen T, van Tits B, van Diepen JA, van den Berg SAA, Rensen PCN, Voshol PJ, Fantuzzi G, Hijmans A, et al. (2010). The Inflammasome-Mediated Caspase-1 Activation Controls Adipocyte Differentiation and Insulin Sensitivity. Cell Metab. 12, 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templin AT, Maier B, Tersey SA, Hatanaka M, and Mirmira RG, (2014). Maintenance of Pdx1 mRNA Translation in Islet β-Cells During the Unfolded Protein Response. Mol. Endocrinol 28, 1820–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tersey SA, Nishiki Y, Templin AT, Cabrera SM, Stull ND, Colvin SC, Evans-Molina C, Rickus JL, Maier B, and Mirmira RG, (2012). Islet β-Cell Endoplasmic Reticulum Stress Precedes the Onset of Type 1 Diabetes in the Nonobese Diabetic Mouse Model. Diabetes 61, 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Liang H, and Zen K, (2014). Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol 5, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Gris D, Lei Y, Jha S, Zhang L, Huang MT-H, Brickey WJ, and Ting JP-Y, (2011). Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol 12, 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff EC, Kang KR, Kim YS, and Park MH, (2007). Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids 33, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W, Cheruku PS, Bazer FW, Safe SH, and Zhou B, (2013). Investigation of Macrophage Polarization Using Bone Marrow Derived Macrophages. J. Vis. Exp 50323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W, Lee YS, Dong Y, Seidman JS, Yang M, Isaac R, Seo JB, Yang B-H, Wollam J, Riopel M, et al. (2019). Expansion of Islet-Resident Macrophages Leads to Inflammation Affecting β Cell Proliferation and Function in Obesity. Cell Metab. 29, 457–474.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Yang Q, Cao J, Xie N, Liu K, Shou P, Qian F, Wang Y, and Shi Y, (2016). Local proliferation initiates macrophage accumulation in adipose tissue during obesity. Cell Death Dis. 7, e2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited in GEO and all proteomics data have been deposited in Proteome Exchange and all are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All primary data presented in this paper have been deposited as a unified Prism 9.1.1 file at Mendeley and are publicly available as of the date of publication, and all original immunoblot images are provided in a single PowerPoint file at Mendeley and are also available as of the date of publication. The DOI is listed in the key resources table. Original microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-ODC | Lisa Shantz, Pennsylvania State University College of Medicine | N/A |
| mouse anti-DHPS | Santa Cruz | Cat# sc-365077, RRID:AB_10846806 |
| rabbit anti-eIF5AHyp | Mirmira laboratory | N/A |
| mouse anti-eIF5A | BD Pharmigen | Cat# 611977, RRID:AB_399398 |
| rabbit anti-BiP | Cell Signaling | Cat# 3183, RRID: AB_10695864 |
| Mouse anti-Hsp90 | Enzo | Cat# SPA-830J, RRID: AB_1505637 |
| rabbit anti-cleaved caspase-3 | Cell Signaling | Cat# 9664, RRID: AB_2070042 |
| Mouse anti-phosho-histone-H3 | Millipore | Cat# 05-598, RRID: AB_309832 |
| rabbit anti-PCNA | Santa Cruz | Cat# sc-7907, RRID: AB_2160375 |
| rabbit anti-ERK 1/2 | Santa Cruz | Cat# sc-94, RRID: AB_2140110 |
| mouse anti-caveolin | Novus | Cat# NB100-615, RRID:AB_10003431 |
| rabbit anti-iNOS | Novus | Cat# NBP1-33780, RRID: AB_10004114 |
| mouse anti-GADPH | Novus | Cat# NB600-502, RRID: AB_10077682 |
| rat anti-F4/80 | Abcam | Cat# Ab6640, RRID: AB_1140040 |
| PE anti-mouse CD206 | Biolegend | Cat# 141706, RRID: AB_10895754 |
| FITC anti-mouse F4/80 | Biolegend | Cat# 123108, RRID: AB_893502 |
| PECy7 anti-mouse CD45 | Biolegend | Cat# 109829 |
| APC Cy7 anti-mouse CD11b | Biolegend | Cat# 101225, RRID: AB_830641 |
| APC anti-mouse iNOS | Invitrogen | Cat# 175920-82, RRID: AB_2573244 |
| chicken anti-GFP | Aves labs | Cat# GFP-1020, RRID: AB_10000240 |
| anti-chicken secondary antibody | Molecular probes | Cat# A-11040, RRID: AB_2534097 |
| Anti-rabbit PE Dazzle | Invitrogen | Cat# A11036 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RPMI Medium 1640 (1X) | Gibco | 11875-093 |
| PBS | Gibco | 14190-144 |
| HEPES Buffer | Corning | 25-060-CI |
| Collagenase | Sigma | C2-22 |
| Collagenase | Sigma | C7657 |
| Penicilin Streptomycin | Thermo Fisher Scientific | 15140-122 |
| Heat inactivated FBS | Gibco | 10082-147 |
| EDTA | Fisher Scientific | BP120-500 |
| Ammonium chloride | Fisher Scientific | A661-500 |
| Potassium bicarbonate | Fisher Scientific | P184-500 |
| rmM-CSF recombinant mouse | R&D Systems | 416-ML |
| rmIFN-γ recombinant mouse | Prospec | Cyt-358-b |
| rmIL-4 recombinant mouse | R&D Systems | 404-ML |
| Lipopolysaccharides from Escherichia coli 0111:B4 | Sigma Aldrich | L2630 |
| rhIFN-γ recombinant human | Prospec | CYT-206b |
| DAPI | Thermo Fisher Scientific | D1306 |
| Sucrose | Fisher Scientific | BP220-212 |
| Gc7 | Biosearch Technologies | G1000 |
| Acid acetic | Sigma Aldrich | A6283 |
| 1-Phenyl-2-thiourea | Acros organics | 207250250 |
| Formaldehyde | Fisher Scientific | 04042-500 |
| PIPES | Sigma Aldrich | P6757-500 |
| MgSO4 | Fisher Scientific | BP213-1 |
| EGTA | Fisher Scientific | 02783-100 |
| TOPRO-3 | Thermo Fisher Scientific | T3605 |
| Urea | Sigma | U0631 |
| 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) | Sigma | H3375 |
| Acetonitrile, HPLC grade | J.T. Baker | 9829-03 |
| Acetonitrile anhydrous | Sigma | 271004 |
| Ammonium hydroxide solution | Sigma | 338818 |
| BCA Protein Assay Kit | Thermo Scientific Pierce | A53225 |
| DTT | Thermo Fisher Scientific | 20291 |
| Iodoacetamide | Thermo Fisher Scientific | 90034 |
| EDTA disodium salt solution 0.5M | Sigma | E7889 |
| Formic acid | Sigma | 33015 |
| HPLC Grade Water | J.T. Baker | 4218-03 |
| Hydroxylamine Solution 50% | Sigma | 467804 |
| Lysyl Endopeptidase | Wako Chemicals | 129-02541 |
| PMSF (Phenylmethylsulfonyl fluoride) | Roche | 10837091001 |
| Reversed phase tC18 SepPak | Waters | 186002319 |
| Sequencing grade modified trypsin | Promega | V5117 |
| Sodium chloride solution; 5M | Sigma | S5150 |
| TMTpro 16plex reagent kit | Thermo Fisher Scientific | A44520 |
| Tris (hydroxymethyl)aminomethane hydrochloride pH 8.0; 1M | Sigma | T2694 |
| Trifluoroacetic acid | Sigma | 91707 |
| Commercial Resources | ||
| Normal Chow diet | Harlan Labs | 2018S |
| Low fat diet | Research Diets | D12450B |
| High fat diet (60%) | Research Diets | D12492 |
| Mouse cytokine/chemokine magnetic bead Milliplex Map Kit | Millipore | MCYTOMAG-70K-PMX |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | 4368814 |
| Experimental Models: Organisms and strains | ||
| C57BL/6 mice | Jackson Laboratories | 000664 |
| Lyz2 promoter driven-Cre recombinase mouse | Jackson Laboratories | 004781 |
| DhpsLoxp/Loxp mice | Raghavendra G Mirmira, deposited into Jackson Laboratories | 034895 |
| DhpsΔMyel mice | Raghavendra G. Mirmira | N/A |
| Wildtype (AB) Zebrafish | Zebrafish International Resource Center | ZL1 |
| Tg(mpeg1:GFP) Zebrafish | Zebrafish International Resource Center | ZL9940 |
| Experimental Models: Cell lines and Tissue | ||
| Mouse: RAW264.7 | ATCC | TIB-71 |
| Oligonucleotides | ||
| Ccl3 (mouse) | Applied Biosystems | Taqman-Assay ID Mm00441259_g1, Cat #4453320 |
| Il1b (mouse) | Applied Biosystems | Taqman-Assay ID Mm00434228_m1, Cat 4453320 |
| Morpholino | ||
| Dhps MO GGTTATGGATGTAAATCCGGCTTTT | Gene Tools, LLC | N/A |
| Deposited data | ||
| RNA Sequencing data | GEO | Accession: GSE144614 |
| Proteomics data | Proteome Exchange | PXD026266 |
| All other data in this manuscript | Mendeley Data | Mirmira, Raghavendra (2021), “Deoxyhypusine Synthase promotes a pro inflammatory macrophage phenotype”, Mendeley Data, V1, doi: 10.17632/kz5rwgpgwc.1 (http://dx.doi.org/10.17632/kz5rwgpgwc.1) |
| Software and Algorithms | ||
| Image Studio Software | LI-COR | Version 3.1.4 |
| Prism Software | GraphPad, Inc. | Version 9.1 |
| STAR 2.3.0 | Alexander Dobin | https://github.com/alexdobin/STAR |
| ngsutils | Marcus Breese and Yunlong Liu | https://github.com/ngsutils/ngsutils |
| EdgeR package | Bioconductor | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| MS-GF+ | Omics Group, Pacific Northwest National Lab | https://omics.pnl.gov/software/ms-gf |
| DAVID | Laboratory of Human Retrovirology and Immunoinformatics | https://david.ncifcrf.gov/tools.jsp |
| mzRefinery | Omics Group, Pacific Northwest National Lab | https://omics.pnl.gov/software/mzrefinery |
| MASIC | Omics Group, Pacific Northwest National Lab | https://omics.pnl.gov/software/masic |
| Decon2LS_V2 | Omics Group, Pacific Northwest National Lab | https://omics.pnl.gov/software/decontools-decon2ls |


