Significance
Over the last few years, there has been a great deal of scientific debate regarding whether the plague bacterium, Yersinia pestis, spread from a Western European reservoir during the second plague pandemic, or if it repeatedly came to Europe from Asia. Here, we make a synthesis of the available evidence, including genomes of ancient DNA and historical, archeological, and ecological information. We conclude that the bacterium most likely came to Europe from Asia several times during the second plague pandemic.
Keywords: Black Death, European plague, Yersinia pestis, molecular evolution, ecological epidemiology
Abstract
The second plague pandemic started in Europe with the Black Death in 1346 and lasted until the 19th century. Based on ancient DNA studies, there is a scientific disagreement over whether the bacterium, Yersinia pestis, came into Europe once (Hypothesis 1) or repeatedly over the following four centuries (Hypothesis 2). Here, we synthesize the most updated phylogeny together with historical, archeological, evolutionary, and ecological information. On the basis of this holistic view, we conclude that Hypothesis 2 is the most plausible. We also suggest that Y. pestis lineages might have developed attenuated virulence during transmission, which can explain the convergent evolutionary signals, including pla decay, that appeared at the end of the pandemics.
Researchers agree that the second plague pandemic was caused by Yersinia pestis (1–9), which arrived in Europe from Caffa transported by Genoese galleys on the Black Sea at the beginning of the Black Death (10). However, there is no consensus among researchers as to the origins of plague epidemics in Europe following the Black Death and ravaging Europe until the 19th century, as attested by historical documents (11).
The two main theories are that one or more plague reservoirs remained in Western Europe during the entire second plague pandemic (referred to in the following as Hypothesis 1) (3, 4, 8, 12) or the bacteria repeatedly invaded Europe from non–Western European reservoir(s) during the same period (referred to in the following as Hypothesis 2) (6, 7, 9, 11, 13). Here, we assess these two hypotheses using a broad spectrum of evidence, including historical and archeological, genetic, evolutionary, and ecological information.
Results and Discussion
Assessment of the Two Hypotheses.
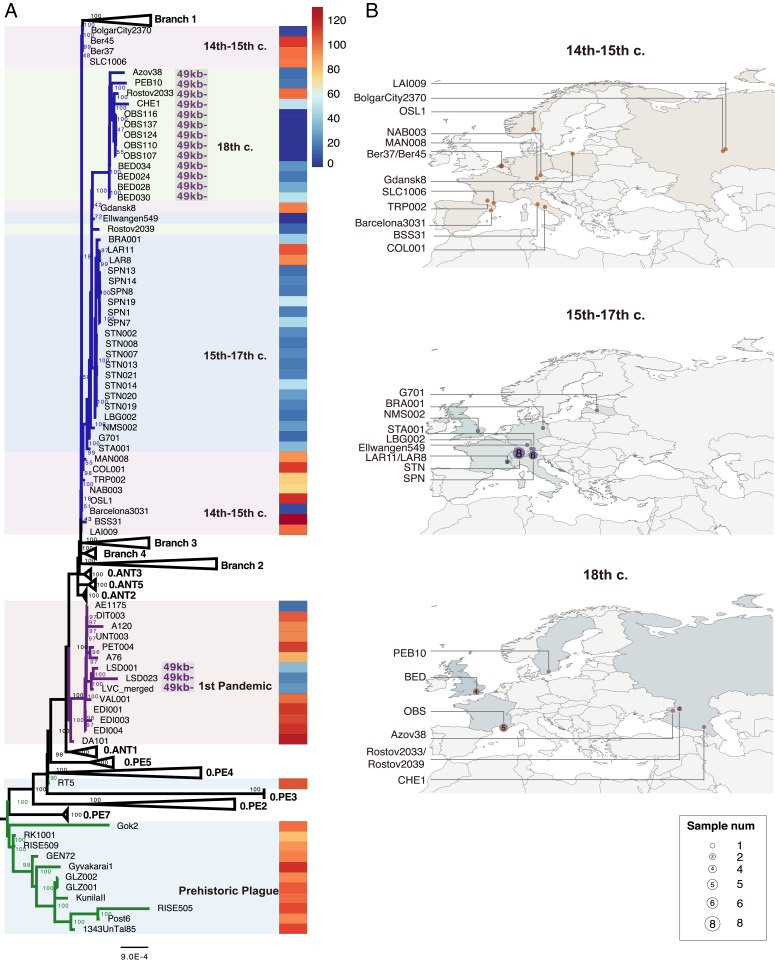
For the purpose of understanding the evolution of the plague bacteria, more than 100 ancient Y. pestis genomes have been published to date. The last 17 were recently reported during a short period by four distinct research groups (7–9, 12). Using most of the ancient genomes (criteria for exclusion are described in Methods), along with 499 modern ones, we present here the most updated phylogeny (Fig. 1).
Fig. 1.
Phylogeny and archaeological site locations of ancient genomes. (A) A maximum likelihood phylogeny was obtained with 574 genomes of Y. pestis (including 75 ancient genomes) involved based on 12,608 SNPs. The numbers at each node indicate the bootstrap values of 1,000 replicates. Branches highlighted in blue correspond to the second pandemic, which is subdivided in three groups: the 14th to 15th century group, which also includes the Black Death and the Pestis secunda (1,357 to 1,366) strains; the 15th to 17th century group; and the 18th century group (which also includes the BED genomes for homogeneity). Branches in purple correspond to the first Pandemic, and branches in green correspond to the prehistoric plague. The ratio between the depth of pla and that of the entire pPCP1 plasmid for all ancient genomes is shown in the rightmost heatmap, with a color scale ranging from 0 (dark blue) to 130+ (dark red). (B) Geographic distribution of the three waves during the second pandemic.
The updated phylogeny confirms the almost clonal nature of the Black Death strains in comparison to all other lineages of the second plague pandemic, including the strains from the Pestis secunda [Ber37 and Ber 45, The Netherlands (6), and BolgarCity2370, Russia (3)], which are placed on Branch 1 [see also London-Ind6330, United Kingdom (3)], as well as to all other strains, which are placed on the post–Black Death branch. There is general agreement that the post–Black Death branch was hosted in a novel wild rodent reservoir—either in Europe or outside Europe (3–8, 12, 14). The original hypothesis (Hypothesis 1) claims that such a plague reservoir existed in Western Europe (15), perhaps in the Alps (16). However, a newer hypothesis (Hypothesis 2) claims that the plague reservoir was in Asia, possibly close to Eastern Europe (6, 7, 9, 11, 13).
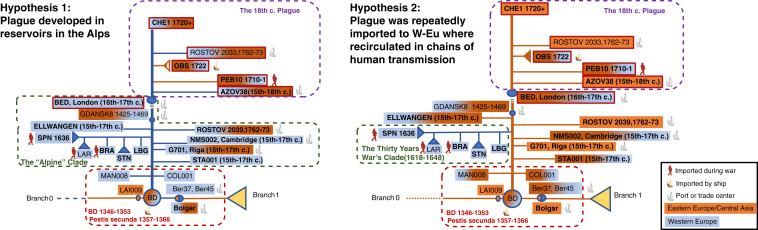
In order to more easily view the phylogeny from the second plague pandemic and to better contrast the evidence for the two hypotheses, we generated two schematic figures (Fig. 2) and a table (Table 1).
Fig. 2.
Schematic comparison between the two main hypotheses for the interpretation of the Y. pestis phylogeny of the second plague pandemic. Historic and evolutionary information is included in the schematic figures. In addition to the symbols explained in the figure, we outlined in red the strains showing the 49-kb deletion. Pla decay (meaning both, full, or partial absence of the pla gene) is indicated by the names in bold.
Table 1.
Main differences between the two competing hypotheses proposed to explain the phylogeny of Y. pestis of the second plague pandemic; genomic and evolutionary, historical and archaeological, and ecological arguments are considered
| Hypothesis 1 | Hypothesis 2 | |
| Main differences | ||
| Origin of the outbreaks | Plague established in Western European reservoirs (for example, marmots in the Alps) (3, 4, 8) | Plague was repeatedly imported from Eastern European or Asian reservoirs (6, 7, 9, 11, 13) |
| Transmission | Mediated by rats infected by wild rodents, as in China during the Third Pandemic (3) | Imported by rats, humans, and goods and subsequently spread by chains of human transmission, as in Europe during the Third Pandemic (17, 22) |
| Vector | Xenopsylla cheopis and other rodent fleas | Any ectoparasite, including Pulex irritans, and body lice (14, 30) |
| Supporting information | ||
| Population genomics | Western European strains (the Alpine clade) are basal in the subbranch (although of different eras). A model proposes Western European strains as ancestral sources for the transmissions (SI Appendix, Fig. S1) | The oldest (LAI009) and the most recent strains, in addition to Bolgar, are all from Eastern Europe (Russia) as well as all the strains of the 18th century; the majority of the Western European strains in the phylogeny come from ports |
| History and archaeology | A hypothesis (16) suggests that the plagues from the 16th century in the Alps were not imported by major trade centers | Multiple historical records assert that plague was imported for outbreaks associated with the Black Death strains (partially reviewed in refs. 6, 7, 11, 13) for BRA (4), SPN (in the Alps), PEB10 (7), and OBS (5). Multiple records of importation are historically attested (18), particularly in harbors |
| Climate | No climatic signal in support, although four datasets of climatic proxies were from the Alps (13) | Strong signals of climate-driven introductions of plague from Asia (13) |
| Evolution | Y. pestis developed in Western European reservoir strains pla+/pla− and Delta49kb, possibly as a form of adaptation to the local host (rodents) | Y. pestis developed pla+/pla− and Delta49kb strains possibly as a form of adaptation to the new host (humans) and/or new vectors (fleas or body lice) |
Hypothesis 1 is supported by a phylogenetic analysis based on the currently available ancient genomes, which infers high posterior probability for a Western European source of the transmissions on the post–Black Death branch (SI Appendix, Fig. S1). However, as the dataset includes 41 ancient genomes from Western Europe against only 8 strains from Eastern Europe (including Gdansk and Riga), the proposed origins from Western Europe are likely to be biased toward a European reservoir due to a size-effect bias. Notably, the most basal genome LAI009 (4) (the Black Death’s lineage), Bolgar (at the root of Branch 1), and the most recent genome [CHE1 (7)] all originated from Western Russia, implying that they might have been closer to a putative Asian or Eastern European reservoir. This continuity does represent strong evidence in support of Hypothesis 2.
Using only genomic data, Hypothesis 1 might be seen as the most parsimonious hypothesis since it proposes an internal source for all western Eurasian outbreaks. However, for two locations (Pestbacken, Sweden 1710 [PEB10] and Marseille, France 1722 [OBS]), an origin from the Ottoman Empire is historically and archaeologically well supported (7). Thus, Hypothesis 1 needs to account for a back and forth spread, which reintroduced plague on two occasions to the Ottoman Empire and back again to Western Europe. Notably, none of the strains from the 18th century appear to have originated in Western Europe according to historical sources (7, 9).
Hypothesis 1 assumes the existence of a wild rodent plague reservoir in the Alps, which is not supported by ecological evidence (13). Instead, a study of more than 7,000 historical plague outbreaks and 15 tree-ring datasets (four of which from the Alps) found climatic signals in support of frequent reimportations of plague from Asia into Eastern and Western European harbors (13).
Intriguingly, only a few genotyped strains are nodes on the backbone of the post–Black Death branch: the strains of the Black Death itself, the strain from Gdansk 1425 to 1469, and the strains from London (BED, 16th to 17th century). While the strains of the Black Death were notoriously imported into Western Europe from the Mongol Empire via Caffa in Crimea (10), both Gdansk and London were very active harbors also in historical times and were very often hit by plague. Interestingly, Y. pestis was also recovered from a rat found in Gdansk. Although the genome is partial due to the different single-nucleotide polymorphism (SNP) profile, it is clear that the strain from the rat could not have infected the victim (Gdansk8) (9). Being a port, Gdansk may indeed have hosted diverse importations of infected rats in the period from 1425 to 1469, as it happened in European harbors during the third pandemic (17).
Hypothesis 2 is consistent with the ecological as well as with the historical evidence (Fig. 2 and Table 1). The only Western European subcluster, the “Alpine cluster” formed by LBG (Landsberg, Germany), STN (Stans, Switzerland), BRA (Brandenburg, Germany), LAR (Lariey, French Alps), and SPN (San Procolo a Naturno, Italian Alps), may naturally be explained by the circulation of soldiers and troops in Europe during the Thirty Years War (1618 to 1648), which made up human chains of transmission with historically documented epidemic events (12, 18, 19). For three strains (SPN from the Italian Alps, LAR from the French Alps, and BRA from Northern Germany), the relationship with the time of the Thirty Years War is historically and archaeologically documented (4, 7, 12). Human chains of transmission, which do not require the presence of rats to start and sustain an epidemic, might explain the circulation of the plague within Europe over long periods of time. They might be due to interpersonal contacts, crowding, infected parasites in clothes or goods (14, 20, 21), or contact with infected pets or fur (6). Several chains of human transmission within Europe could be reconstructed for cases of the last century (17, 22) as well as for the second pandemic (23, 24).
Two Convergent Mutations.
To better understand the evolution of Y. pestis, we examined two more mutations, which were recently discovered in ancient strains. In the most recent subclade of the second pandemic, starting with BED, there is a 49-kb deletion with unknown function. This deletion was also present in the last lineage of the first pandemic and, in both cases, might have accounted for the decline of the pandemic (4, 7, 25). We found the same mutation in the Rostov 2033 strain in the 18th century clade (Figs. 1 and 2). By contrast, a second strain found in the same cemetery in Rostov (Rostov 2039) has a different SNP pattern and lacks the chromosomal deletion.
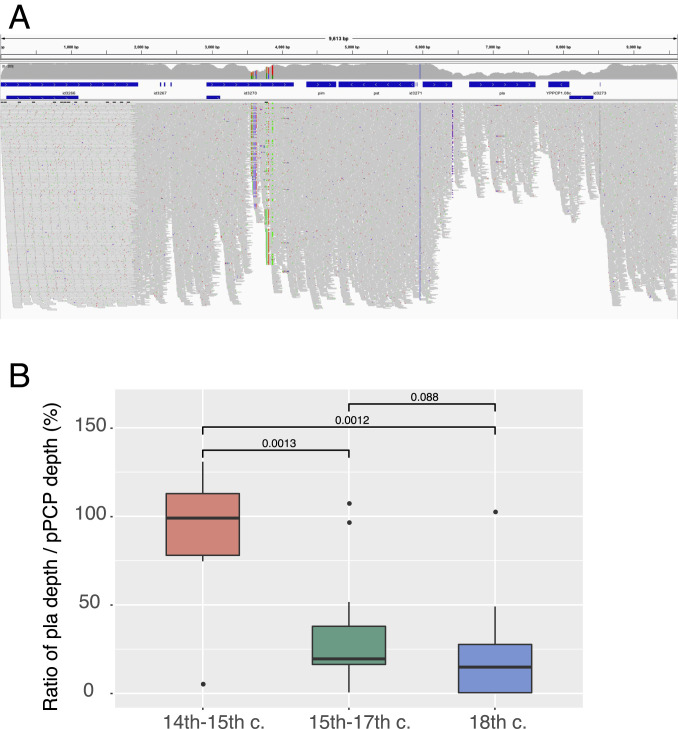
Another mutation, the depletion of the pla gene on the plasmid pPCP1, has recently been proposed as the cause of the disappearance of the second plague pandemic in the 18th century (8) given that the pla gene is an important virulence factor of Y. pestis. We checked for the presence of the pla+/pla− plasmids in all published ancient strains. The ratio in coverage depth between pla and the whole pPCP1 plasmid indicates the status of pla loss in an organism (Fig. 3). If the depth of pla is significantly lower than that of pPCP1, it might properly be concluded that the pla gene was lost in some pPCP1 plasmids. Our analyses show that the ratio of pla in the Black Death and post–Black Death genomes appears to be different when compared with the prehistoric and the first pandemic lineages (Fig. 1). We have also checked randomly selected modern Y. pestis genomes in different lineages, and their depth of pla and pPCP1 are quite consistent, indicating no other pla loss in modern plagues. By contrast, the generalized depletion of pla extensively observed during the post–Black Death era and at the end of the first pandemic (Fig. 1) seems to be consistent. Given that the sequencing data were generated by several different research groups, a systemic error during sequencing is unlikely.
Fig. 3.
The decay of the pla gene. (A) Depth plot of the pPCP1 plasmid in strain CHE1 using Integrative Genomics Viewer. The annotated genes of the pPCP1 plasmid are marked with blue bars. The average sequencing depth of whole pPCP1 plasmid is 195.65x, while the average sequencing depth of the pla region is 96.04x. (B) Group-wise comparison of the ratio between the depth of pla and that of whole pPCP1 plasmid among three waves of the second pandemic. Boxplots depict the upper, median, and lower quartiles of the ratios; individual dots indicate outliers that lie outside of 1.5 times the interquartile range; and vertical lines indicate the range of all ratios except for outliers. The P values of group-wise comparison using the Wilcoxon test are labeled on the top, two of which are statistically significant (P < 0.05). Data of B are provided in Dataset S4.
It seems that full pla strains were slightly depleted at the end of the second pandemic (8), with the same phenomenon at the end of the first pandemic. Notably, however, Rostov2033, one of the most recent genomes of the second pandemic, shows full read pPCP1 plasmids, whereas CHE, the most recent historical strain, shows very slight pla decay (Fig. 3). This observation is not fully in agreement with the proposed hypothesis that pla depletion contributed to the end of the pandemic. An alternative explanation for this phenomenon (8) is that the differences observed in the full pla plasmids might be due to different forms of plague. In particular, bubonic plague and pneumonic plague need the pla gene to develop, whereas primary septicemic plague does not (8). It seems that plague existed in all three forms, at least from the time of the first pandemic; however, this does not add any specific evolutionary information to the observed variability.
The Evolution of the pla Gene.
We propose an evolutionary hypothesis for the presence of lineages with pla decay. One of the optimized survival strategies for an emerging pathogen is to balance its virulence to the main host with its transmission strategy. This trade-off hypothesis was previously demonstrated for Y. pestis (26, 27). This mechanism would allow the bacterium to reduce virulence and enhance the time of survival of the host and, consequently, of the pathogen (28). After experiencing the Black Death and successive waves, the pla decay strains might have attempted to acquire a fitness advantage, reducing their virulence by increasing the time to death. Indeed, we observe among the victims only pla+/pla− mixed strains, whereas pla− lineages might have survived longer in the host population, providing a milder form of the illness. The Eastern European/Asia clade of the 18th century (including CHE1) further lost the 49-kb region, which can be the result of an extension of a virulence attenuated pattern. Such events of attenuated virulence might have occurred multiple times in the Y. pestis evolutionary history and left out host-adapted lineages, such as for 0.PE2 and 0.PE4 (29). Therefore, the possible virulence reduction caused by pla decay and loss of the 49-kb region is not necessarily the reason for the extinction of plague at the end of the first and second pandemics but might be the result of a form of adaptation to a new host, which may be the wild rodent in the putative Western European reservoir (Hypothesis 1), a new host in the Asian reservoir, or the human host (Hypothesis 2) as well as their vectors. We observed that the newly published strains from Lariey [French Alps (12)] do not show pla decay in contrast to other Alpine lineages (SPN). This evidence might exclude the hypothesis of an adaptation to a host in a Western European reservoir. Thus, we tentatively propose that this mechanism of pla decay would support the presence of human-to-human transmission chains mediated by human ectoparasites (fleas and body lice) during plague pandemics in Europe, the plausibility of which has previously been demonstrated (17, 22, 30), while the vector competence was supposed to be low (31).
Conclusion
Altogether, the most consistent interpretation of the current information is in support of Hypothesis 2. This implies that there must have existed a reservoir outside of Western Europe [with the Ottoman Empire, Persia, and Central Asia as possible candidates (7, 9)]. Such a reservoir could then feed, with multiple introductions, the second plague pandemic outbreaks in Western Europe along Northern and Southern trade routes (6, 32). In addition, there might have been continuous recirculation of plague in (Western and Eastern) Europe with the movement of goods, travelers, and troops. The recirculation of plague within Western Europe, mediated by trade and travel, might mimic the presence of a local reservoir [humans cannot be regarded as reservoir hosts, as plague significantly increases mortality beyond natural values (33–35)] and assure the long-lasting re-emergence of epidemics on the continent. Such recirculation of plague could explain the collateral branches on the phylogenetic tree—an example of which may be the Thirty Years War Clade (1618 to 1648) evidenced in Fig. 2. Additional ancient strains from Asia and Eastern Europe and more accurate dating and historical contextualization will provide further evidence to clarify the phylogeny and evolution of the Y. pestis pathogen.
Methods
SNP Calling and Evaluation.
All raw reads of 111 published ancient genomes were downloaded from NCBI SRA and EBI ENA databases. For the clones from each sampling site, only one genome with the highest quality (sequencing depth) as reported in corresponding publications was chosen, and genomes covering less than 20% of the chromosome length of the CO92 assembly (GCF_000009065.1) were excluded from further analysis, which resulted in a dataset with a total of 75 ancient genomes (Dataset S1). We trimmed and quality filtered raw reads using Trimmomatic v0.38 (36), and reads shorter than 30 bp and below a quality score of 20 were discarded. Subsequently, the filtered reads were mapped against the CO92 assembly with BWA mem model (v0.7.17) (37), and the aligned reads were extracted from bam files using SAMtools (v1.9) (38) view command (-bF 4); then, different runs of the same sample were merged using SAMtools merge command. Sequences with more than 10 soft and hard clipped alignments were filtered out by samclip, and duplicates were removed using Picard’s MarkDuplicates module. SNP calling was performed using the UnifiedGenotyper of the Genome Analysis Toolkit (GATK v3.8) (39) under the “EMIT_ALL_CONFIDENT_SITES” option with a minimum confidence threshold 10; a vcf file for every ancient genome was produced, and SNPs that were close to each other by less than 20 bp were excluded.
A total of 499 modern genomic assemblies of Y. pestis available in NCBI GenBank database on October 19, 2020, were downloaded (Dataset S2) and then aligned to the CO92 assembly using NASP’s convert_external_genome command (40), which was based on MUMmer’s nucmer and delta-filter modules (v3.23) (41). A FASTA file one-to-one position aligned with a reference FASTA for every modern genome was created.
All vcf files and aligned FASTA files were used to aggregate sample calls into matrices with NASP’s matrix module. For ancient genomes, an SNP would be called when supported by ≥3 reads and ≥90% allele frequency. After manually validating the calls for ancient genomes with notably longer branch lengths (SPN strains, BSS31, and SLC1006) according to their published SNP lists, we got a final dataset of 12,608 polymorphic loci (Dataset S3).
Phylogenetic Analyses and Geographic Extent of Sampled Sites.
A FASTA file, concatenated of all SNP sites, was used to generate a maximum likelihood tree with 1,000 fast bootstrap replicates using IQ-TREE (v1.6.5) (42) with the option -m MFP+ASC to infer the best substitution model and account for ascertainment bias correction. Then, FigTree (v1.4.3) was used to visualize the generated tree. The packages of ggplot2, maptoos, and maps in R (v3.6.1) were used to mark the archaeological site locations of samples from the second plague pandemic. The longitude and latitude for each site was taken from the website mapcoordinates (https://www.mapcoordinates.net/en).
Phylogenetic Analyses with Calculation of MCMC Posterior Probability.
A maximum likelihood tree for 47 ancient genomes during the second pandemic was rebuilt using RAxML (v8.2.11) (43) with 100 replicates and GTRGAMMA model. It was rerooted to strain LAI009 and transformed into newic format using FigTree. We used the ReorderData function in evobiR (v1.1, R package) to match the source records (country names) to the order of tips on the phylogenetic tree. Then, the make.simmap function in phytools (v0.7–70, R package) (44) was used to perform stochastic source mapping based on ARD model and the tip states on the tree, with 10,000 generations of MCMC sampling every 100 generations.
pPCP1 and pla Analysis
Samtools depth command was used to count the depth of whole pPCP1 plasmid and pla gene for each sample from bam files. The packages of ggplot2 and ggsignif were used to obtain the boxplots and group-wise comparisons (Wilcoxon test) of the ratio between the depth of pla and that of whole pPCP1 plasmid among three subclades of the second pandemic. The coverage plots of pPCP1 in CHE1 (Fig. 3) were visualized using Integrative Genomics Viewer (v2.8.11) (45).
Supplementary Material
Acknowledgments
This work was supported by the National Key Program for Infectious Diseases of China (Grant numbers 2018ZX10101003 and 2018ZX10714-002), Key Scientific and Technology Project of Inner Mongolia Autonomous Region (Grant number 2021ZD0006), and the University of Ferrara under the Bando per il finanziamento della ricerca scientifica “Fondo per l’Incentivazione alla Ricerca” 2020. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Katharine Rose Dean is thanked for constructive comments on the manuscript, including improving the language. Two anonymous reviewers on an earlier version of this paper are greatly thanked for comments and suggestions that helped us to improve the paper.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101940118/-/DCSupplemental.
Data Availability
Publicly available genomes are listed in Dataset S1 and Dataset S2. All other study data are included in the article and/or supporting information.
References
- 1.Haensch S., et al., Distinct clones of Yersinia pestis caused the black death. PLoS Pathog. 6, e1001134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos K. I., et al., A draft genome of Yersinia pestis from victims of the Black Death. Nature 478, 506–510 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spyrou M. A., et al., Historical Y. pestis genomes reveal the European Black Death as the source of ancient and modern plague pandemics. Cell Host Microbe 19, 874–881 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Spyrou M. A., et al., Phylogeography of the second plague pandemic revealed through analysis of historical Yersinia pestis genomes. Nat. Commun. 10, 4470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bos K. I., et al., Eighteenth century Yersinia pestis genomes reveal the long-term persistence of an historical plague focus. eLife 5, e12994 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Namouchi A., et al., Integrative approach using Yersinia pestis genomes to revisit the historical landscape of plague during the Medieval Period. Proc. Natl. Acad. Sci. U.S.A. 115, E11790–E11797 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guellil M., et al., A genomic and historical synthesis of plague in 18th century Eurasia. Proc. Natl. Acad. Sci. U.S.A. 117, 28328–28335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Susat J., et al., Yersinia pestis strains from Latvia show depletion of the pla virulence gene at the end of the second plague pandemic. Sci. Rep. 10, 14628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morozova I., et al., New ancient Eastern European Yersinia pestis genomes illuminate the dispersal of plague in Europe. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piazza M. D., Cronaca, Giuffrida A., Ed. (Renzo Mazzone, Palermo, 1980). [Google Scholar]
- 11.Bramanti B., Stenseth N. C., Walløe L., Lei X., Plague: A disease which changed the path of human civilization. Adv. Exp. Med. Biol. 918, 1–26 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Seguin-Orlando A., et al., No particular genomic features underpin the dramatic economic consequences of 17th century plague epidemics in Italy. iScience 24, 102383. 10.1016/j.isci.2021.102383. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid B. V., et al., Climate-driven introduction of the Black Death and successive plague reintroductions into Europe. Proc. Natl. Acad. Sci. U.S.A. 112, 3020–3025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbieri R., Drancourt M., Raoult D., The role of louse-transmitted diseases in historical plague pandemics. Lancet Infect. Dis. 21, e17–e25 10.1016/S1473-3099(20)30487-4. (2021). [DOI] [PubMed] [Google Scholar]
- 15.Ell S. R., Immunity as a factor in the epidemiology of medieval plague. Rev. Infect. Dis. 6, 866–879 (1984). [DOI] [PubMed] [Google Scholar]
- 16.Carmichael A. G., Plague persistence in Western Europe: A hypothesis. Medieval Globe 1, 8 (2014). [Google Scholar]
- 17.Bramanti B., Dean K. R., Walløe L., Chr Stenseth N., The third plague pandemic in Europe. Proc. Biol. Sci. 286, 20182429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alfani G., Plague in seventeenth-century Europe and the decline of Italy: Aan epidemiological hypothesis. Eur. Rev. Econ. Hist. 17, 408–430 (2013). [Google Scholar]
- 19.Lammert G., Geschichte der Seuchen, Hungers-und Kriegsnoth zur Zeit des dreissigjährigen Krieges (J. F. Bergmann, 1890). [Google Scholar]
- 20.Krauer F., Schmid B. V., Mapping the plague through natural language processing. medRxiv [Preprint] (2021) https://www.medrxiv.org/content/10.1101/2021.04.27.21256212v1. Accessed 30 April 2021. [DOI] [PubMed]
- 21.Barbieri R., et al., Unbiased lexicometry analyses illuminate plague dynamics during the second pandemic. bioRxiv [Preprint] (2021) https://www.biorxiv.org/content/10.1101/2021.05.06.442897v1. Accessed 6 May 2021.
- 22.Dean K. R., Krauer F., Schmid B. V., Epidemiology of a bubonic plague outbreak in Glasgow, Scotland in 1900. R. Soc. Open Sci. 6, 181695 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whittles L. K., Didelot X., Epidemiological analysis of the Eyam plague outbreak of 1665–1666. Proc. Biol. Sci. 283, 20160618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alfani G., Bonetti M., A survival analysis of the last great European plagues: The case of Nonantola (Northern Italy) in 1630. Popul. Stud. (Camb.) 73, 101–118 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Keller M., et al., Ancient Yersinia pestis genomes from across Western Europe reveal early diversification during the First Pandemic (541–750). Proc. Natl. Acad. Sci. U.S.A. 116, 12363–12372 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinnebusch B. J., Jarrett C. O., Bland D. M., “Fleaing” the plague: Adaptations of Yersinia pestis to its insect vector that lead to transmission. Annu. Rev. Microbiol. 71, 215–232 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Cui Y., et al., Evolutionary selection of biofilm-mediated extended phenotypes in Yersinia pestis in response to a fluctuating environment. Nat. Commun. 11, 281 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lathem W. W., Price P. A., Miller V. L., Goldman W. E., A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science 315, 509–513 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Sebbane F., Uversky V. N., Anisimov A. P., Yersinia pestis plasminogen activator. Biomolecules 10, 1554 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dean K. R., et al., Human ectoparasites and the spread of plague in Europe during the Second Pandemic. Proc. Natl. Acad. Sci. U.S.A. 115, 1304–1309 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miarinjara A., Bland D. M., Belthoff J. R., Hinnebusch B. J., Poor vector competence of the human flea, Pulex irritans, to transmit Yersinia pestis. Parasit. Vectors 14, 317 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bramanti B., Namouchi A., Schmid B. V., Dean K. R., Stenseth N. C., Reply to Barbieri et al.: Out of the Land of Darkness: Plague on the fur trade routes. Proc. Natl. Acad. Sci. U.S.A. 116, 7622–7623 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samia N. I., et al., Dynamics of the plague-wildlife-human system in Central Asia are controlled by two epidemiological thresholds. Proc. Natl. Acad. Sci. U.S.A. 108, 14527–14532 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Begon M., Epizootiologic parameters for plague in Kazakhstan. Emerg. Infect. Dis. 12, 268–273 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nilsson P., et al., The genome of the great gerbil reveals species-specific duplication of an MHCII Gene. Genome Biol. Evol. 12, 3832–3849 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv (2013) https://arxiv.org/abs/1303.3997. Accessed 26 May 2013.
- 38.Li H., et al., The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van der Auwera G. A., et al., From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinf. 43, 111011–111033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahl J. W., et al., NASP: An accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb. Genom. 2, e000074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurtz S., et al., Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 45.Robinson J. T., et al., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available genomes are listed in Dataset S1 and Dataset S2. All other study data are included in the article and/or supporting information.