Significance
The function of tissues is controlled by complex biological processes, in which transcriptional mechanisms play a fundamental role. The transcriptional coactivator PGC-1α is a prototypical regulator of adaptive responses evoked by environmental and internal cues (e.g. exercise-adaptation in skeletal muscle). Here, we found that PGC-1α-RNA–binding is central to assemble multiprotein complexes controlling gene transcription. Interestingly, PGC-1α-RNA–binding also regulates chromatin recruitment in transcriptionally active liquid-like nuclear condensates. Collectively, our findings unravel mechanistic insights into the regulation of complex transcriptional networks by PGC-1α, which we demonstrate to be essential to enhance the expression of key genes controlling skeletal muscle oxidative metabolism.
Keywords: gene transcription, transcriptional coactivator, chromatin, liquid–liquid phase separation, RNA-binding protein
Abstract
Plasticity of cells, tissues, and organs is controlled by the coordinated transcription of biological programs. However, the mechanisms orchestrating such context-specific transcriptional networks mediated by the dynamic interplay of transcription factors and coregulators are poorly understood. The peroxisome proliferator–activated receptor γ coactivator 1α (PGC-1α) is a prototypical master regulator of adaptive transcription in various cell types. We now uncovered a central function of the C-terminal domain of PGC-1α to bind RNAs and assemble multiprotein complexes including proteins that control gene transcription and RNA processing. These interactions are important for PGC-1α recruitment to chromatin in transcriptionally active liquid-like nuclear condensates. Notably, such a compartmentalization of active transcription mediated by liquid–liquid phase separation was observed in mouse and human skeletal muscle, revealing a mechanism by which PGC-1α regulates complex transcriptional networks. These findings provide a broad conceptual framework for context-dependent transcriptional control of phenotypic adaptations in metabolically active tissues.
Complex biological programs are highly regulated at the transcriptional level in the nucleus, involving the dynamic assembly of large multiprotein complexes at enhancers and gene promoters in a context-dependent manner (1, 2). While great strides have been made to understand the molecular underpinnings of the general transcriptional machinery, the mechanisms integrating environmental stimuli with specific transcriptional programs remain poorly understood. The peroxisome proliferator–activated receptor-γ coactivator-1α (PGC-1α) is a prototypical transcriptional coactivator controlling context-specific transcriptional networks (3). The specificity achieved by PGC-1α requires its selective recruitment to regulatory elements on the genome (4), in which it forms a multiprotein complex containing different transcription factors (TFs) and additional transcriptional regulators with enzymatic activity (5, 6). Accordingly, PGC-1α can control highly specific transcriptional programs coordinating distinct processes such as the fasting response in the liver, nonshivering thermogenesis in brown adipose tissue, and the adaptive response to endurance exercise in skeletal muscle (7–9). However, the molecular mechanisms by which PGC-1α integrates multiple signaling pathways into a spatiotemporal transcriptional output are enigmatic.
PGC-1α is highly conserved between mouse and human with 94% identity. This protein is intrinsically disordered for the large majority of its amino acids and contains only a single folded domain, an RNA recognition motif (RRM). Three sequence features are present in the intrinsically disordered part, an LXXLL motif, typically involved in nuclear receptor binding (10), and two arginine–serine-rich (RS) regions. The segment of residues 564 through 797 of the mouse protein, comprising the two RS and the RRM, is generally termed the C-terminal domain (CTD) (11). Deletion of the CTD of PGC-1α blunts its positive effects on gene expression, which was originally attributed to impaired RNA processing (11). A direct coupling of transcription to RNA processing was supported by PGC-1α interactions with splicing factors and components of the RNA polymerase 2 complex via the CTD and a minigene splicing assay (11). However, the impact of PGC-1α on RNA binding, processing, and, ultimately, gene transcription remains unclear. Therefore, the main aim of this study was to elucidate the mechanistic underpinning that defines the dynamic transcriptional activity of PGC-1α. We therefore performed a comprehensive interrogation of the PGC-1α-containing transcriptional complex and defined the impact of these interactions with RNAs and other proteins on subnuclear sequestration and ultimately transcriptional regulation.
Results
PGC-1α Function Is Impaired by Deletion of the CTD.
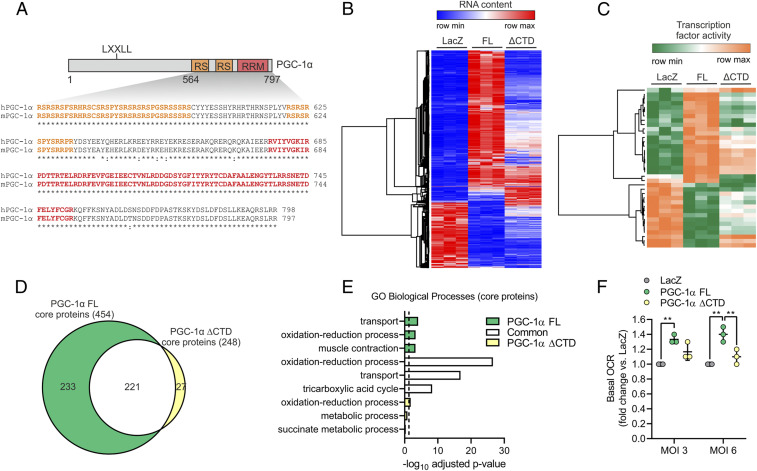
To investigate the function of PGC-1α CTD containing the well-conserved RS regions and RRM (Fig. 1A), we overexpressed either full-length (FL) PGC-1α or a truncated protein lacking the CTD (ΔCTD) in C2C12 myotubes. Both PGC-1α FL and ΔCTD exhibited comparable levels of overexpression at the RNA and protein level in a context of very low expression of endogenous PGC-1α (SI Appendix, Fig. S1 A and B). Moreover, deletion of the CTD of PGC-1α did not affect protein half-life compared to FL protein (SI Appendix, Fig. S1C). Transcriptome analysis revealed a large number of differentially expressed genes (DEG) upon overexpression of PGC-1α FL, whereas overexpression of ΔCTD induced a drastically different set of DEG (Fig. 1B and SI Appendix, Fig. S2A, and Dataset S1). Based on Gene Ontology (GO) analysis, the vast majority of DEG regulated by both PGC-1α FL and ΔCTD were related to metabolic pathways, while the function of the ΔCTD-dependent DEG and those that are exclusively controlled by the CTD is less clearly defined (SI Appendix, Fig. S2B). We next leveraged our transcriptomic data to infer TFs regulated in a PGC-1α CTD-dependent manner by using Integrated System for Motif Activity Response Analysis (ISMARA) (12). This analysis strongly suggests that the CTD of PGC-1α plays a central role in modulating the activity of a wide range of TFs such as ATF3, CEBPB, STAT2, and NFAT5 among others (Fig. 1C and Dataset S2). Interestingly, PGC-1α–induced activation of the nuclear receptor ERRα, known to bind at the N-terminal domain, was also predicted to be blunted in PGC-1α ΔCTD by ISMARA analysis, suggesting that CTD-specific features and mechanisms play a critical regulatory function.
Fig. 1.
PGC-1α function is impaired by deletion of the CTD. (A) Illustration of mouse PGC-1α (UniProt ID: O70343) with the CTD aligned against the human protein (UniProt ID: Q9UBK2). (B and C) Heat maps with DEG (B) and predicted changes in TF activity (C) induced by overexpression of LacZ (control), PGC-1α FL, or ΔCTD in C2C12 myotubes. (D and E) Overlap (D) and GO analysis (E) of core proteins regulated by PGC-1α FL or ΔCTD in C2C12 myotubes. The dashed line represents GO statistical cutoff (adjusted P value < 0.05). (F) Basal oxygen consumption rate (OCR) of C2C12 myotubes transduced with PGC-1α FL or ΔCTD at two different multiplicity of infections (MOIs). Values are mean ± SD; **P < 0.01.
The biological impact of the deletion of PGC-1α CTD was further investigated via whole-proteome analysis of skeletal muscle cells. Proteome remodelling mediated by PGC-1α FL was reduced in PGC-1α ΔCTD, although the effect was milder compared to that observed at the transcriptome level (SI Appendix, Fig. S2 C and D and Dataset S3). Next, we integrated the transcriptomic and proteomic datasets to identify a set of “core proteins,” (e.g., exhibiting consistent PGC-1α–dependent up- or down-regulation of transcripts and proteins). We found that deletion of the CTD of PGC-1α drastically impaired the expression of such core proteins, which are primarily involved in transport processes (e.g., TIMM17A, ABCD1, SLC25A20, and MCUR1) and oxidative metabolism (e.g., LDHB, ACADM, MDH2, and OGDH) (Fig. 1 D and E). Importantly, even though many core proteins in the overlap between PGC-1α FL and ΔCTD are also involved in oxidative phosphorylation and tricarboxylic acid cycle (Fig. 1E and SI Appendix, Fig. S2 E and F), only overexpression of PGC-1α FL significantly increased basal oxygen consumption of skeletal muscle cells (Fig. 1F). Altogether, these data demonstrate that both the transcriptional and functional output of PGC-1α are highly dependent on the CTD.
The PGC-1α CTD Mediates RNA-Dependent Protein–Protein Interactions.
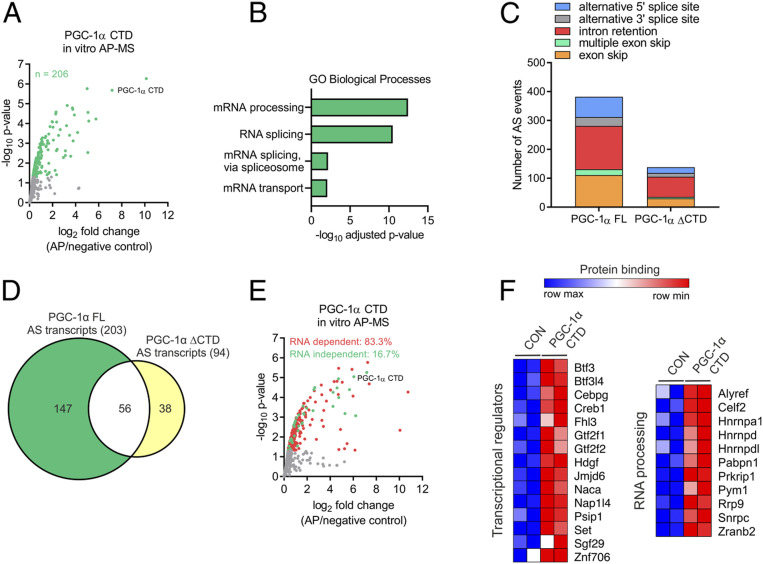
Mechanistically, the CTD of PGC-1α represents a potential platform for protein–protein and protein–RNA interactions. By implementing an in vitro affinity purification followed by mass spectrometry (in vitro AP-MS) assay, we identified over 200 nuclear proteins interacting directly or indirectly with the CTD of PGC-1α (Fig. 2A and Dataset S4). Most of PGC-1α CTD-interacting proteins were associated with the regulation of RNA processing and gene transcription (Fig. 2B and Dataset S4). Consistently, analysis of our transcriptomic data demonstrated that PGC-1α FL regulates RNA processing events (Fig. 2C and Dataset S5), corresponding to the alternative splicing of 203 transcripts (Fig. 2D). In contrast, over 60% of the alternative splicing capacity was lost when the CTD of PGC-1α was deleted (Fig. 2 C and D). These data support and further expand previous findings proposing a role of PGC-1α in RNA processing via CTD-mediated protein–protein interactions (11, 13). It is, however, unknown to what extent direct RNA binding to the CTD of PGC-1α is involved in this process. We therefore carried out in vitro AP-MS in the absence and presence of RNase A, which revealed that 83% of PGC-1α CTD protein–protein interactions rely on RNA (Fig. 2E and SI Appendix, Fig. S3 A and B and Dataset S4). Many of the RNA-dependent interacting proteins are involved in skeletal muscle function, but a large number is involved in RNA transcription and processing, implying a direct link between the binding of RNAs and splicing factors to the CTD of PGC-1α (Fig. 2F and SI Appendix, Fig. S3C). These findings indicate that the CTD of PGC-1α regulates gene transcription and RNA processing in an RNA binding–dependent manner.
Fig. 2.
Most PGC-1α CTD protein interactions depend on RNA binding. (A) Volcano plot with green dots representing proteins interacting with the CTD of PGC-1α. (B) GO analysis of PGC-1α CTD interactome. (C and D) Alternative splicing (AS) analysis of transcriptomic data showing the number of AS events (C) and transcripts (D) regulated by PGC-1α FL or ΔCTD in C2C12 myotubes. (E) Volcano plot with red and green dots representing PGC-1α CTD RNA-dependent and -independent protein interactions, respectively. (F) Heat maps showing PGC-1α CTD RNA-dependent protein interactions with proteins regulating gene transcription and RNA processing.
PGC-1α Chromatin Recruitment Is Regulated by RNA Binding at the CTD.
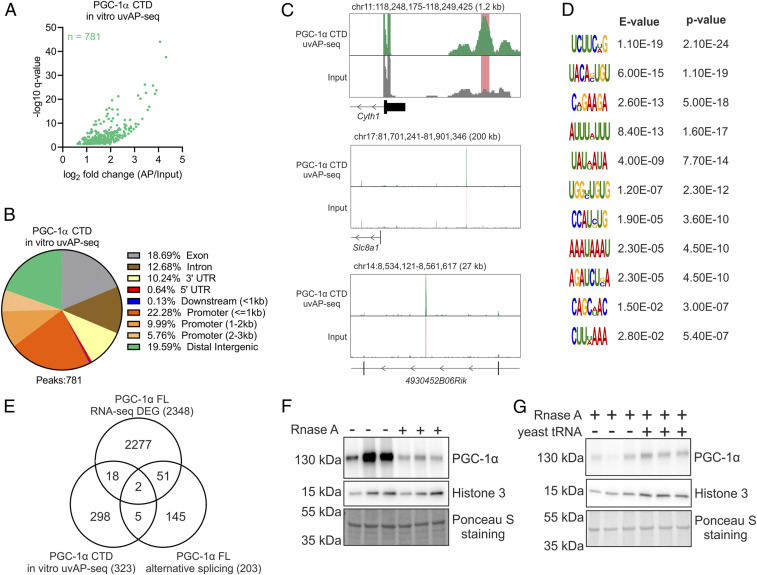
We next implemented in vitro cross-linking affinity purification followed by sequencing (in vitro uvAP-seq) to investigate the RNA-binding capacity of the PGC-1α CTD, which revealed direct interactions with a large number of RNAs (Fig. 3A and Dataset S6). Interestingly, although PGC-1α can interact with protein-coding RNAs (e.g., Ptges, Cntnap5b, Mm2pr, and Slco5a1), the vast majority of bound RNAs were transcribed at regulatory elements such as promoters and distal intergenic regions (Fig. 3 B and C), supporting and expanding previous reports of nonpolyadenylated RNAs binding to PGC-1α (14). Motif discovery analysis uncovered 11 motifs enriched among RNAs bound by the CTD (Fig. 3D). Such a diversity of RNA motifs has recently been found to be a common characteristic of a large number of RNA-binding proteins (RBPs) (15), including PGC-1α FL in primary hepatocytes (16). Overall, our results demonstrate that PGC-1α is a bona fide RBP, with a large promiscuity for binding several types of RNAs harboring different recognition motifs.
Fig. 3.
PGC-1α chromatin recruitment is regulated by RNA binding at the CTD. (A) Volcano plot with green dots representing RNAs interacting with the CTD of PGC-1α. (B) Annotation of in vitro AP-seq peaks representing PGC-1α CTD bound RNAs. (C) Genome browser track view of representative peaks located at promoter (Top), distal intergenic (Middle), and intronic regions (Bottom). (D) Motif discovery analysis of PGC-1α CTD bound RNAs. (E) Overlap of transcripts that are differentially expressed (DEG), alternative-spliced, and bound by PGC-1α. (F and G) Protein content of PGC-1α FL bound to chromatin fractions in the absence and presence of 1 mg/mL of RNase A (F) or following the addition of 5 µg/µL of yeast tRNA (G). Immunoblots are representative of three independent experiments, each performed in triplicate.
Previous studies have linked PGC-1α CTD function to RNA processing (11) (e.g., for the stabilization of hepatic metabolic transcripts) (16). Integration of our RNA sequencing (RNA-seq) and in vitro uvAP-seq datasets, however, revealed that over 90% of protein-coding RNAs bound by PGC-1α were neither DEG nor alternatively spliced, while 97% of DEG were neither alternatively spliced nor bound by PGC-1α (Fig. 3E). A total of 18 genes were both differentially expressed and bound by the CTD of PGC-1α (e.g., Supv3l1, Osbpl2, Pdss1, and Clybl), while only Ndufaf5 and Cluh were additionally alternatively spliced. While the small overlap between RNA binding and differential gene expression was also observed in the liver (16), our results indicate that RNA processing is not the main function of RNA binding to the CTD. PGC-1α function has been proposed to be modulated by binding of noncoding RNAs at DNA regulatory elements (14, 17), although comparison of our RNA binding and PGC-1α FL chromatin immunoprecipitation sequencing (ChIP-seq) data (4) (Dataset S7) demonstrates a low correlation between RNA- and chromatin-binding profiles (SI Appendix, Fig. S4 A and B). Notably, similar binding profiles have been recently reported for many other RBPs (15, 18). Therefore, RNA interactions with the CTD of PGC-1α at the site of genomic recruitment (e.g., the reported interaction with enhancer RNAs) (14) seems to be a less-frequent event. To define how RNA binding modulates PGC-1α function, we measured its recruitment to chromatin in the absence and presence of RNase A via subcellular fractionation. This experiment revealed that the absence of nuclear RNAs strongly decreases the amount of chromatin-bound PGC-1α FL (Fig. 3F). Importantly, subsequent reloading of nuclei with yeast transfer RNA (tRNA) was sufficient to restore chromatin-bound PGC-1α FL levels (Fig. 3G). We furthermore found that deletion of the RRM of PGC-1α significantly impaired the activation both PPARβ/δ and ERRα in human embryonic kidney 293 (HEK293) cells, while deletion of the entire CTD exacerbated this effect (SI Appendix, Fig. S4 C and D). This suggests that CTD-specific mechanisms controlling transcriptional activation by PGC-1α might be conserved between different cell types, though future studies will be required to dissect potential cell type–specific mechanisms. Our data therefore demonstrate that beside regulating protein–protein interactions, RNA binding at the CTD of PGC-1α is a crucial mechanism controlling its dynamic recruitment to chromatin and transcriptional activation.
The CTD of PGC-1α Regulates Its Localization in Nuclear Condensates.
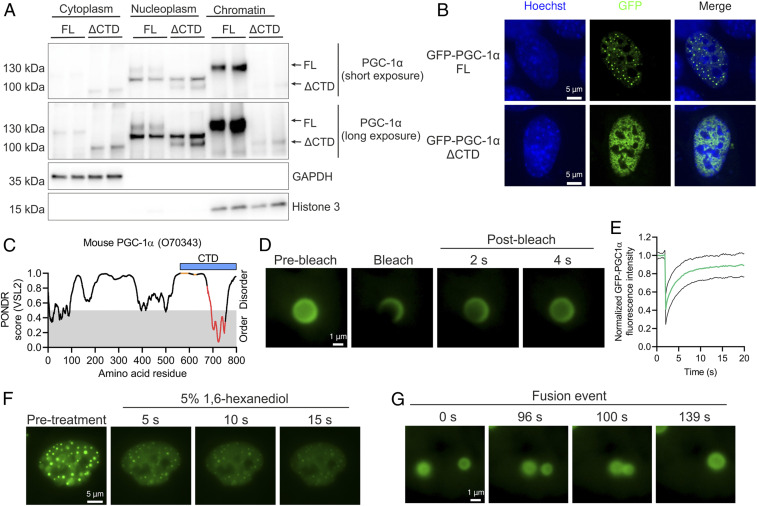
While residual levels of PGC-1α FL in cytoplasm and nucleoplasm of C2C12 myotubes were observed, we found that most of the protein was strongly bound to chromatin (Fig. 4A). In stark contrast, chromatin binding of PGC-1α ΔCTD was markedly reduced, which was associated with a shift toward higher relative levels in the nucleoplasm and cytoplasm compared to the FL protein (Fig. 4A). Intriguingly, both endogenous and GFP-fused PGC-1α FL proteins form characteristic nuclear foci, originally defined as nuclear speckles (11, 17). We now observed that deletion of the CTD of GFP-fused PGC-1α completely abolished its localization in nuclear foci in C2C12 myoblasts (Fig. 4B). Together, our data indicate that a large fraction of PGC-1α foci represent a chromatin-rich subcompartment such as chromatin condensates rather than nuclear speckles that are thought to reside in the nucleoplasm (19).
Fig. 4.
PGC-1α is localized within liquid-like nuclear condensates. (A) Subcellular fractionation of C2C12 myotubes expressing PGC-1α FL or ΔCTD. (B) Nuclear localization of GFP–PGC-1α FL or ΔCTD fusion proteins transfected in C2C12 myoblasts. (C) Predictor of Natural Disordered Regions (PONDR) analysis of mouse PGC-1α protein, with orange dots, red dots, and blue bar representing RS domains, RRM, and the CTD region under investigation, respectively. (D and E) Live-cell imaging (D) and quantification (E) of GFP–PGC-1α FL FRAP experiments in C2C12 myoblasts. Green and black lines denote mean and SD of FRAP quantification, respectively. (F) Live-cell imaging of 5% 1,6-hexanediol treatment of C2C12 myoblasts transfected with GFP–PGC-1α FL. (G) Live-cell imaging of a GFP–PGC-1α FL droplet fusion event in C2C12 myoblasts. Immunoblots and microscopy images are representative of at least three independent experiments each in triplicate.
Chromatin condensates are membrane-less organelles formed via liquid–liquid phase separation that play a critical regulatory role in gene expression (20, 21). Interestingly, the nuclear foci containing PGC-1α resemble nuclear condensates formed by TFs and transcriptional coregulators (22, 23). The formation of liquid-like droplets is often driven by the presence of intrinsically disordered regions (IDRs) on proteins and by multivalent protein and RNA interactions of RBPs (24). PGC-1α is largely disordered (Fig. 4C and SI Appendix, Fig. S5A), yet our findings demonstrate that its N-terminal IDRs alone are insufficient to mediate the formation of nuclear foci (Fig. 4B). Conversely, in presence of the RRM, PGC-1α is found in foci (Fig. 4B), implying that RNA binding plays an important role in localizing PGC-1α within nuclear foci. To assess potential liquid-like properties of PGC-1α foci, we carried out fluorescence recovery after photobleaching (FRAP) analysis that revealed a rapid fluorescence recovery characteristic of liquid-like condensates (Fig. 4 D and E). We additionally studied the physical properties of PGC-1α foci by using the aliphatic alcohol 1,6-hexanediol, which resulted in a dissolving of foci within seconds (Fig. 4F and SI Appendix, Fig. S5B). Finally, live-cell imaging–based detection of fusion events between different foci (Fig. 4G and SI Appendix, Fig. S5C and Movie S1) further evidence the liquid-like properties of PGC-1α–containing condensates.
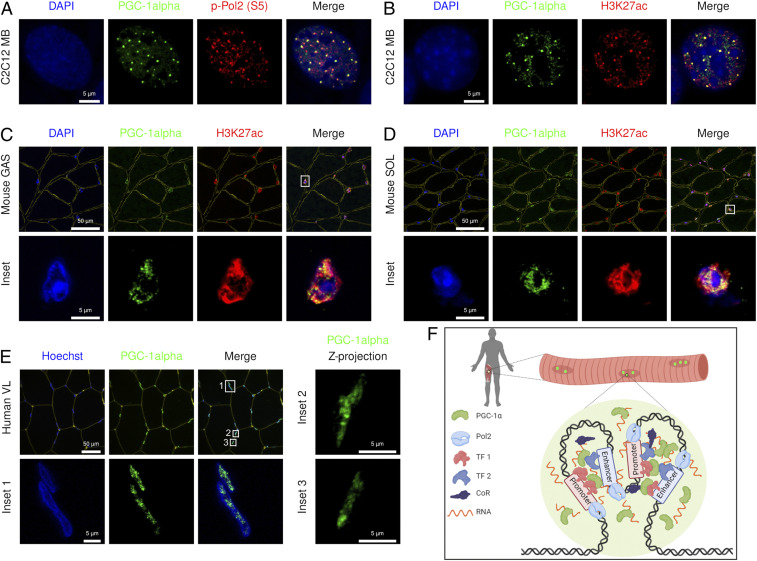
PGC-1α Nuclear Condensates Are Sites of Active Transcription in Mouse and Human Skeletal Muscle.
It is tempting to speculate that PGC-1α, as a strong transcriptional coactivator, might be partitioned in subcompartments favoring active gene expression. Consistent with this hypothesis, we found that PGC-1α condensates exhibit an enrichment of RNA polymerase 2 phosphorylated at S5 of its CTD repeat (p-Pol2 [S5]) and histone 3 acetylated at K27 (H3K27ac) in skeletal muscle myoblasts, thus reflecting active transcription at enhancers and promoters (Fig. 5 A and B). Deletion of the RRM of PGC-1α resulted in a mixed population of cells comprising foci-negative and -positive nuclei, with little colocalization with H3K27ac, as observed in PGC-1α ∆CTD-expressing cells (SI Appendix, Fig. S6A). Using immunofluorescence staining of PGC-1α (SI Appendix, Fig. S6B), we also found transcriptionally active, PGC-1α–containing condensates in nuclei from mouse skeletal muscle, both in the glycolytic and oxidative muscles gastrocnemius and soleus, respectively (Fig. 5 C and D). Importantly, further supporting our cell and mouse data, we found that PGC-1α was also present in distinct nuclear condensates in human skeletal muscle (Fig. 5E). Collectively, these findings suggest an important function of nuclear condensates to sequester transcriptionally active multiprotein complexes that contain chromatin-recruited PGC-1α at enhancers and promoters, which might be important for the highly orchestrated control of transcriptional networks encoding biological program of metabolic tissue plasticity (e.g., those observed in skeletal muscle).
Fig. 5.
PGC-1α condensates compartmentalize active transcription. (A and B) Images of transfected GFP-PGC-1α FL costained with DAPI, p-Pol2 (S5), and H3K27ac in C2C12 myoblasts (MB). (C and D) Images of endogenous PGC-1α costained with laminin (yellow), DAPI, and H3K27ac in mouse skeletal muscle (GAS: gastrocnemius, SOL: soleus). (E) Images of endogenous PGC-1α costained with wheat germ agglutinin (yellow) and Hoechst in human skeletal muscle (VL: vastus lateralis). (F) Model in which PGC-1α is compartmentalized within active transcriptional condensates via multivalent protein and RNA interactions at the CTD in a tissue- and context-specific manner (prepared with Biorender). Microscopy images are representative of at least three independent samples or experiments each in triplicate.
Discussion
Physical inactivity, which impairs skeletal muscle metabolism and function, can accelerate the development of a wide spectrum of diseases, including type 2 diabetes, cancer, and nonalcoholic fatty liver disease (25). Transcriptional coregulators have been heralded as master controllers of complex transcriptional networks modulating energy metabolism (26, 27), but the underlying molecular mechanisms remain elusive. Notably, this has limited the development of therapeutic strategies to modulate transcriptional coregulators in the context of metabolic diseases, in which PGC-1α is a highly attractive target. Our results reveal key mechanistic insights into the transcriptional regulation of energy metabolism by PGC-1α, a prototypical member of the coactivator protein superfamily. We now demonstrate that the highly conserved CTD of PGC-1α fulfils a crucial role in controlling gene expression by compartmentalizing multiprotein complexes of transcriptional regulators within liquid-like nuclear condensates.
The N-terminal domain of PGC-1α was originally discovered to be essential for transcriptional activation in the GAL4-UAS reporter system (5, 28). Here, we demonstrate that the CTD of PGC-1α regulates its transcriptional output via both RNA binding–dependent and –independent mechanisms, thus controlling downstream effects on energy metabolism. RNA binding regulates this process by mediating key protein–protein interactions with transcriptional regulators, including several TFs (e.g., BTF3 and CEBPG) and components of transcriptional coregulator complexes (e.g., SGF29 and JMJD6). Remarkably, the assembly of such multiprotein complexes on the chromatin is dynamically regulated by PGC-1α–RNA binding, which might facilitate the spatiotemporal adaptive response to stimuli such as exercise and diet (3). Accordingly, chromatin-bound RBPs have recently emerged as key regulators of gene transcription at active promoters and enhancers (15, 18). It should be noted that PGC-1α ∆RRM retains some transcriptional activity, which suggests RNA-independent mechanisms by which the CTD of PGC-1α regulates gene transcription. Indeed, although only very few of such factors have been found, RNA-independent PGC-1α CTD protein interactors include transcriptional regulators such as MEF2D and BRD3. In addition, other CTD domains of PGC-1α (e.g., SR domains) contribute to RNA binding (13, 17), suggesting the presence of multiple nonexclusive RNA-dependent and -independent mechanisms. Overall, our data demonstrate that the capacity of the CTD of PGC-1α to form specific multiprotein complexes and to strengthen chromatin binding is a key mechanism to fully activate TFs, including N-terminal interacting proteins such as ERRα and PPARβ/δ. It appears that such properties might allow the CTD of PGC-1α to regulate distinct transcriptional networks. In line with this idea, it has been reported that PGC-1α isoforms lacking the CTD, including the RS regions and RRM motif, regulate different transcriptional programs and biological processes in skeletal muscle cells compared to the FL protein (29, 30).
We have also uncovered an additional layer of complexity by which the CTD of PGC-1α modulates gene transcription that implicates its localization into specific liquid-like nuclear condensates. Both RNAs and RBPs play a key role in the formation of biomolecular condensates and the modulation of their biophysical properties (24, 31). Although the localization of PGC-1α in nuclear condensates appears to be partially dependent on its RRM (e.g., RNA-independent mechanisms), our data suggest that RNA binding is critical for the proper compartmentalization of active transcription. Importantly, the presence of nuclear condensates containing PGC-1α in mouse and human myonuclei in situ strongly implies an important physiological role in the regulation of skeletal muscle function. Therefore, our findings suggest that PGC-1α might represent an integral component of such membrane-less organelles able to control context- and/or tissue-specific transcriptional networks (Fig. 5F). Since nuclear condensate formation is already observed in the sedentary muscle, future studies will aim at interrogating how the payload of these condensates, including proteins of the transcription machinery, but also DNA regulatory elements, is affected by different states (e.g., exercise training, aging, and obesity) and their associated signaling pathways mediating specific posttranslational modification on the CTD of PGC-1α (e.g., phosphorylation, acetylation, methylation, or ubiquitination, all of which have been found on the CTD) (3).
Collectively, our findings imply that modulation of PGC-1α nuclear condensate formation represents an attractive strategy to regulate the function of this transcriptional coactivator in exercise or disease contexts. Notably, the establishment of abnormal liquid-like condensates has recently been found to be associated with the development of disorders such as cancer and neurodegenerative diseases (32). Hence, the mechanism controlling PGC-1α function discovered in this work might have important implications to the development of strategies to improve energy metabolism under pathological conditions. Indeed, targeting RBPs (33) and transcriptional condensates (34) with pharmacological compounds is emerging as a promising therapeutic approach, thus opening avenues in the treatment of metabolic diseases. Ultimately, the mechanistic insights discovered in this study could represent a cornerstone for a conceptual framework that describes how transcriptional coregulators modulate complex transcriptional programs defining the phenotype of metabolically active tissues. Future studies will reveal how this targeted partitioning intersects with similar processes that have been described for general and basal transcriptional regulators. Thereby, a more global understanding of dynamic transcription and a link to extra- and intracellular cues and signaling pathways will be achieved.
Materials and Methods
Humans.
Human skeletal muscle biopsies from young healthy men (n = 3) were obtained in a previously published study (35). The current study thus relies on the appropriate ethical approval (Regional Ethics Committee for Copenhagen: H-16040740) and complied with the ethical guidelines of the Declaration of Helsinki II as reported in the previous study. Written informed consent was obtained from all subjects prior to enrollment in the study.
Animals.
Male C57BL/6JRj mice were housed in a conventional facility with a 12-h night/day cycle at 23 °C, with free access to food and water. All experiments were approved by the veterinary office of the canton Basel-Stadt, Switzerland.
Cell Culture.
C2C12 myoblasts were grown in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (growth medium). To induce differentiation, growth medium of about 90% confluent myoblasts was changed to DMEM supplemented with 2% horse serum (differentiation medium). Experiments using C2C12 myotubes were performed after 4 d of differentiation. Cells were maintained at 37 °C, 95% O2, and 5% CO2.
Generation of DNA Plasmids and Transfections.
Mouse GFP-PGC-1α FL plasmid (Addgene, #4) was used as template for PCR amplification of a ΔCTD fragment containing amino acids 1 through 564 of PGC-1α. SalI and BamHI restriction sites were added to the 5′ and 3′ end of the insert, respectively. The destination plasmid GFP–PGC-1α FL was digested with SalI and BamHI, following which the PGC-1α ΔCTD insert was ligated. Deletion ΔRRM of mouse PGC-1α was performed with the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, # E0554S), using the plasmid cloning DNA (pcDNA)-f:PGC1 plasmid (Addgene, # 1026) as template. Primers to generate the PGC-1α ΔRRM construct were designed with NEBaseChanger (http://nebasechanger.neb.com). Plasmid sequences were corroborated via Sanger sequencing.
Transfection of C2C12 myoblasts was performed using Optimal-Minimal Essential Medium (Opti-MEM) (Thermo Fisher Scientific, #31985070) and Lipofectamine 2000 (Thermo Fisher Scientific, #11668019) following manufacturer’s instructions. A total 24 h after seeding, cells were transfected with 0.5 μg of EGFP–PGC-1α FL (Addgene, #4) or ΔCTD for 24 h.
Reporter Gene Assay.
Reporter gene assays were performed in 96-well plates using 2 × 104 HEK293 cells per well grown in growth medium without antibiotics. Cells were transfected using Opti-MEM (Thermo Fisher Scientific, #31985070) and polyethylenimine (Polysciences, # 23966). Plasmids and polyethylenimine were diluted in Opti-MEM, following which they were mixed in a 1:3 ratio of ug DNA:ug polyethylenimine and incubated for 20 min at room temperature before adding to the cells. Cells were transfected 24 h after seeding with 5 ng pRL-SV40 (Promega, #E2231), 25 ng pPPRE ×3-TK-luc (Addgene, #1015), 25 ng pERRE-luc (gift of Dr. Junichi Sadoshima), 10 ng pBABE puro PPARδ (Addgene, #8891), 10 ng pERRα (gift of Dr. Vincent Giguère), 60 ng PGC-1α FL (Addgene, #1026), and 60 ng PGC-1α ΔRRM or 60 ng PGC-1α ΔCTD (Addgene, #1030). The total amount of plasmid DNA was kept constant at 100 ng per well by using the control plasmid pAdenoX-LacZ. A total 48 h after transfection, firefly and Renilla luciferase activities were measured with Dual-Glo Luciferase Assay System (Promega, # E2920) following manufacturer’s instructions. Renilla luciferase activity was used for normalization.
Generation of Adenoviral Vectors.
Adenovirus vectors were generated with the Adeno-X Adenoviral System 3 following manufacturer’s instructions (Takara, #632267). Briefly, mouse PGC-1α FL and ΔCTD were PCR-amplified from the pcDNA-f:PGC1 (Addgene, # 1026) and pcDNA-f:PGC1(delta CTD) (Addgene, #1030) plasmid vectors, respectively. N-terminal HA and FLAG tags were introduced during PCR amplification, with the amplicon subcloned into the pAdenoX-ZsGreen1 vector to generate HA-Flag–PGC-1α FL and ΔCTD adenoviruses. The LacZ gene was also subcloned into the pAdenoX-ZsGreen1 vector, which was used to generate a LacZ control adenovirus. All plasmids were corroborated via Sanger sequencing. Adenoviruses were produced and amplified in Adeno-X 293 cells (Takara, # 632271), while titter was determined by fluorescence-activated cell sorting.
Adenovirus Transduction.
Cells were transduced with HA-Flag–PGC-1α FL, ΔCTD, or LacZ adenovirus at multiplicity of infection (MOI) 2 to 6. Adenoviruses were prepared in the corresponding medium, and cells were transduced for 4 h. Next, cells were washed once with phosphate-buffered saline (PBS) and then incubated in adenovirus-free medium for a total of 48 h.
RNA Purification and QPCR.
Cells were collected in TRI Reagent (Sigma #T9424), following which RNA was purified and reverse-transcribed using Direct-zol RNA MiniPrep (Zymo Research, #R2050) and iScript complementary DNA (cDNA) Synthesis Kit (Bio-Rad, #1708891), respectively. Relative changes in messenger RNA (mRNA) content were quantified by qPCR on a StepOnePlus system (Applied Biosystems) using Fast SYBR Green Master Mix (Thermo Fisher Scientific, #4385612). The ΔΔCT method was used for analysis, with TATA-binding protein (Tbp) as endogenous control.
RNA-seq and Alternative Splicing Analysis.
Libraries were prepared with TruSeq Stranded mRNA Library Kit (Illumina, #20020595), pair-end sequencing was performed using the HiSEq. 2500 (Illumina), and data were analyzed on the Galaxy platform (https://usegalaxy.eu/). Reads were trimmed with Trim Galore! (Galaxy version 0.4.3.1), and quality was assessed using FastQC (Galaxy version 0.72+galaxy1). Reads were aligned to the mm10 version of the mouse genome using STAR (Galaxy version 2.7.2b), while strand specificity and read counting were performed with Infer Experiment (Galaxy version 2.6.4.1) and featureCounts (Galaxy version 1.6.4+galaxy1), respectively. Next, we used DESeq2 (Galaxy version 2.11.40.6) for differential expression analysis (q value < 0.01 and fold change ≥ 2), and the resulting data were annotated with Annotate DESeq2/DEXSeq output tables (Galaxy version 1.1.0). Overlap between different datasets was determined with Venny (version 2.1, https://bioinfogp.cnb.csic.es/tools/venny/). GO analysis was performed with Database for Annotation, Visualization and Integrated Discovery (DAVID) 6.8 (https://david.ncifcrf.gov/), with significance defined as adjusted (Benjamini) P value < 0.05. TF activity analysis was achieved with ISMARA (https://ismara.unibas.ch/mara/), in which z value > 2 was considered significant. We used DESeq2 normalized counts to generate heat maps and for hierarchical clustering using Morpheus (https://clue.io/morpheus). Finally, in order to identify alternative splicing events, the SplAdder workflow was employed (36). We produced splicing graphs for all samples belonging to a comparison we were interested in. Afterward we called splicing events (exon_skip, intron_retention, alt_3prime, alt_5prime, and mult_exon_skip) with the SplAdder test procedure using default parameters.
Protein Extraction and Immunoblotting.
Protein was extracted with protein lysis buffer (50 mM Tris⋅HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA (pH 8.0), 5% Glycerol, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate (SDS), 1 mM nicotinamide, and 1X Halt Protease Inhibitor Mixture [Thermo Fisher Scientific, #87786]), as previously described (37).
Immunoblotting was performed with precast (Bio-Rad, #4561096) or home-made SDS-polyacrylamide gels, as previously described (37), and proteins were detected with a primary antibody to PGC-1α (Santa Cruz Biotechnology, #sc-518025), α-tubulin (Cell Signaling Technology, #2144S), histone 3 (Abcam, #ab1791), GAPDH (Cell Signaling Technology, #2118S), and HA (Sigma, #11867423001). If required, Ponceau S (Sigma, #P7170-1L) staining was used as loading control. Secondary antibodies for mouse (Agilent, #P0260), rabbit (Agilent, #P0399), and rat (Jackson ImmunoResearch, #112-035-003) were used. Antibody binding was detected using enhanced chemiluminescence horseradish peroxidase (HRP) substrate detection kit for standard (Thermo Fisher Scientific, #32106), medium (Thermo Fisher Scientific, #34076), or high (Thermo Fisher Scientific, #34095) sensitivity.
Protein Half-Life.
Protein half-life was measured by treating cells with DMSO as control or 100 μg/mL of cycloheximide (Sigma, #C4859) for 30 or 60 min. Next, cells were collected for protein extraction and immunoblotting.
MS Analysis of Whole-Cell Proteome.
Cells were lysed in 80 μL of lysis buffer (1% sodium deoxycholate, 0.1 M Tris, 10 mM TCEP, and pH = 8.5) using 10 cycles of sonication (Bioruptor, Diagnode). Samples were reduced for 10 min at 95 °C and alkylated at 15 mM chloroacetamide for 30 min at 37 °C. Proteins were digested by incubation with sequencing-grade modified trypsin (1/50 weight/weight; Promega, V5113) for 12 h at 37 °C. Tryptic digests were acidified (pH < 3) using TFA and cleaned up using iST cartridges (PreOmics, P.O.00027) according to the manufacturer’s instructions. Samples were dried under vacuum and stored at −20 °C.
Sample aliquots comprising 25 μg of peptides were labeled with isobaric tandem mass tags (TMT 10-plex, Thermo Fisher Scientific, 90110), as described previously (38). Shortly, peptides were resuspended in 20 μL labeling buffer (2 M urea, 0.2 M Hepes, and pH 8.3), and 5 μL of each TMT reagent were added to the individual peptide samples followed by a 1 h incubation at 25 °C, shaking at 500 rpm. To quench the labeling reaction, 1.5 μL aqueous 1.5-M hydroxylamine solution was added, and samples were incubated for another 10 min at 25 °C shaking at 500 rpm followed by pooling of all samples. The pH of the sample pool was increased to 11.9 by adding 1 M phosphate buffer (pH 12) and incubated for 20 min at 25 °C shaking at 500 rpm to remove TMT labels linked to peptide hydroxyl groups. Subsequently, the reaction was stopped by adding 2 M hydrochloric acid until a pH < 2 was reached. Finally, peptide samples were further acidified using 5% TFA, desalted using Sep-Pak Vac 1cc (50 mg) C18 cartridges (Waters, WAT054960) according to the manufacturer’s instructions, and dried under vacuum.
TMT-labeled peptides were fractionated by high-pH reversed-phase separation using a XBridge Peptide BEH C18 column (3.5 µm, 130 Å, and 1 mm × 150 mm; Waters, 186003562) on an Agilent 1260 Infinity high-performance liquid chromatography (HPLC) system. Peptides were loaded on column in buffer A (20 mM ammonium formate in water, pH 10) and eluted using a two-step linear gradient from 2 to 10% in 5 min and then to 50% buffer B (20 mM ammonium formate in 90% acetonitrile, pH 10) over 55 min at a flow rate of 42 µl/min. Elution of peptides was monitored with a ultraviolet (UV) detector (215 and 254 nm), and a total of 36 fractions were collected, pooled into 12 fractions using a postconcatenation strategy as previously described (39), and dried under vacuum.
Dried peptides were resuspended in 0.1% aqueous formic acid and subjected to liquid chromatography (LC)-MS/MS analysis using a Q Exactive HF Mass Spectrometer fitted with an EASY-nLC 1000 (Thermo Fisher Scientific) and a custom-made column heater set to 60 °C. Peptides were resolved using a reversed-phase (RP)-HPLC column (75 μm × 30 cm) packed in-house with C18 resin (ReproSil-Pur C18–AQ, 1.9 μm resin; Dr. Maisch, r119.aq.) at a flow rate of 0.2 μL/min−1. The following gradient was used for peptide separation: from 5 to 15% B over 10 min to 30% B over 60 min to 45% B over 20 min to 95% B over 2 min followed by 18 min at 95% B. Buffer A was 0.1% formic acid in water and buffer B was 80% acetonitrile and 0.1% formic acid in water.
The mass spectrometer was operated in data-dependent acquisition mode with a total cycle time of ∼1 s. Each MS1 scan was followed by high-collision dissociation (HCD) of the 10 most abundant precursor ions with dynamic exclusion set to 30 s. For MS1, 3e6 ions were accumulated in the Orbitrap over a maximum time of 100 ms and scanned at a resolution of 120,000 full width at half maximum (FWHM) (at 200 m/z). MS2 scans were acquired at a target setting of 1e5 ions, maximum accumulation time of 100 ms, and a resolution of 30,000 FWHM (at 200 m/z). Singly charged ions and ions with an unassigned charge state were excluded from triggering MS2 events. The normalized collision energy was set to 35%, the mass isolation window was set to 1.1 m/z, and one microscan was acquired for each spectrum.
The acquired raw-files were converted to the mascot generic file (mgf) format using the msconvert tool (part of ProteoWizard, version 3.0.4624 [2013-6-3]) and searched using MASCOT against a murine database (consisting of 49,434 forward- and reverse-protein sequences downloaded from Uniprot on 11/24/2014) and 390 commonly observed contaminants. The precursor ion tolerance was set to 10 ppm, and fragment ion tolerance was set to 0.02 Da. The search criteria were set as follows: full tryptic specificity was required (cleavage after lysine or arginine residues unless followed by proline), three missed cleavages were allowed, and carbamidomethylation (C) and TMT6plex (K and peptide N terminus) were set as fixed modification and oxidation (M) as a variable modification. Next, the database search results were imported into the Scaffold Q+ software (version 4.3.2, Proteome Software Inc.), and the protein false discovery rate was set to 1% based on the number of decoy hits. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. Acquired reporter ion intensities in the experiments were employed for automated quantification and statistical analysis using a modified version of our in-house developed SafeQuant R script version 2.3 (38). This analysis included adjustment of reporter ion intensities, global data normalization by equalizing the total reporter ion intensity across all channels, summation of reporter ion intensities per protein and channel, calculation of protein abundance ratios, and testing for differential abundance using empirical Bayes-moderated t-statistics. The calculated P values were corrected for multiple testing using the Benjamini−Hochberg method, with significance defined as q value < 0.05 and fold change ≥ 1.2. Overlap between different datasets was determined with Venny (version 2.1, https://bioinfogp.cnb.csic.es/tools/venny/). GO analysis was performed with DAVID 6.8 (https://david.ncifcrf.gov/), with significance defined as adjusted (Benjamini) P value < 0.05.
Assessment of Oxygen Consumption.
Basal oxygen consumption was measured using the Seahorse XF Cell Mito Stress Test Kit (Agilent, #103015-100) on a Seahorse XF96 Analyzer (Agilent) according to the manufacturer’s instructions.
Subcellular Fractionation.
Subcellular fractionation was performed as previously described (40, 41), with the following modifications. Cells, on 10-cm plates, were washed twice with ice-cold PBS and collected into 5 mL ice-cold PBS. Samples were centrifuged for 2 min at 500 g at 4 °C, and the cell pellet was resuspended in 500 μL of ice-cold cytoplasmic lysis buffer (0.15% Nonidet P-40, 10 mM Tris⋅HCl [pH 7.0], 150 mM NaCl, and 1X Halt Protease Inhibitor Mixture [Thermo Fisher Scientific, #87786]). Following 5-min incubation on ice, samples were homogenized by using a glass Donce homogenizer with 20 strokes with a tight pestle on ice. The resulting cell lysate was layered onto 1,250 μL of ice-cold sucrose buffer (10 mM Tris⋅HCl [pH 7.0], 150 mM NaCl, 25% sucrose, and 1X Halt Protease Inhibitor Mixture) and centrifuged for 10 min at 16,000 g at 4 °C. The supernatant containing the cytoplasmic fraction was transferred to a prechilled tube and snap-frozen in liquid nitrogen. Nuclei pellet was washed once with 1 mL of nuclei wash buffer (0.1% Triton X-100, 1 mM EDTA, and 1X Halt Protease Inhibitor Mixture in PBS) and centrifuged for 1 min at 1,150 g at 4 °C. The nuclei pellet was resuspended in 200 μL of glycerol buffer (20 mM Tris⋅HCl [pH 8.0], 75 mM NaCl, 0.5 mM EDTA, 50% glycerol, 0.85 mM DTT, and 1X Halt Protease Inhibitor Mixture), after which 200 μL of nuclei lysis buffer (1% Nonidet P-40, 20 mM Hepes [pH 7.5], 300 mM NaCl, 1 M urea, 0.2 mM EDTA, 1 mM DTT, and 1X Halt Protease Inhibitor Mixture) was added. Samples were mixed by pulsed vortex and incubated on ice for 2 min. Following centrifugation for 2 min at 18,500 g at 4 °C, the supernatant containing the nucleoplasm fraction was transferred to a prechilled tube and snap-frozen in liquid nitrogen. The chromatin pellet was incubated with 50 μL of 1X RQ1 DNase Reaction Buffer with 30U of RQ1 RNase-Free DNase (Promega, # M6101) for 10 min at 37 °C. Next, samples were placed on ice, and 50 μL of storage buffer (10 mM Tris [pH 7.4], 1 mM EDTA, 25 mM NaCl, 10% glycerol, and 1X Halt Protease Inhibitor Mixture) were added, following which sonication was performed using Bioruptor Plus (Diagenode) at 4 °C for 15 min with 30 s on and 30 s off. Finally, samples were centrifuged at 16,100 g for 10 min at 4 °C, and the supernatant containing the chromatin was transferred to a prechilled tube and snap frozen in liquid nitrogen. Samples were prepared for immunoblotting as described above.
RNase A and Yeast tRNA Treatment of Nuclei.
Nuclei were isolated as described above, washed once with 1 mL of ice-cold PBS, and centrifuged for 1 min at 1,150 g at 4 °C. The pellet was resuspended in 500 μL of 0.05% Tween-20 in PBS and incubated on ice for 10 min to permeabilize the nuclei. Next, samples were centrifuged for 1 min at 1,150 g at 4 °C and washed once with 1 mL of ice-cold PBS. Samples were resuspended in either 100 μL of PBS with or without 1 mg/mL of RNase A (Sigma, #R4642) and were incubated for 15 min at 37 °C. Next, samples were centrifuged at 2,300 g for 10 min at 4 °C, and chromatin fraction was extracted as described above.
For tRNA treatments, all samples were digested with RNase A, following which samples were washed twice with 1 mL of ice-cold PBS. Nuclei pellets were resuspended in 100 µl of PBS containing 1 U/μL RNasin Ribonuclease Inhibitors (Promega, # N2615) with or without 5 µg/µl of yeast tRNA (Thermo, #AM7119) and were then incubated for 15 min at 37 °C. Subsequently, 1 mL of nuclei wash buffer (0.1% Triton X-100, 1mM EDTA, and 1X Halt Protease Inhibitor Mixture in PBS) was added, and samples were centrifuged for 1 min at 1,150 g at 4 °C. Chromatin fraction was extracted as described above in the Subcellular Fractionation section.
In Vitro AP-MS.
C2C12 myotubes were transduced with PGC-1α FL at MOI 3 and nuclear extract was prepared with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, #78835). Pull-down and negative control comprised 350 μg of protein from nuclear extract, while 12 μg of recombinant N-terminal His-Tag CTD of mouse PGC-1α (US Biological Life Sciences, #156296) was added only to pull-down samples. Samples were incubated overnight at 4 °C with rotation, following which protein–protein complexes were purified with Pierce His Protein Interaction Pull-Down Kit (Thermo Fisher Scientific, #21277) following manufacturer’s instructions. RNA-dependent interactions were assessed by treating nuclear extracts with 1 U/μL RNasin Ribonuclease Inhibitors (Promega, # N2615) as control or 1 mg/mL of RNase A (Sigma, #R4642) for 15 min at 37 °C before adding the recombinant protein for overnight incubation.
Eluted proteins were incubated with four volumes of 100% trichloroacetic acid on ice for 10 min. Samples were then centrifuged at 18,500 g for 5 min, and the protein pellet was washed twice with 200 μL cold acetone. The final protein pellet was resuspended in 40 μL Gua buffer (2 M Guanidinium-HCl, 0.1 M Ammonium bicarbonate, and 5 mM TCEP), sonicated 10 times with Vial Tweeter ultrasonicator (Hielscher), and incubated for 10 min at 95 °C, followed by alkylation of proteins with 15 mM chloroacetamide for 30 min at 37 °C. Next, guanidium-HCl was diluted below 0.4 M with 0.1 M ammonium bicarbonate prior to adding 0.5 μg trypsin and incubated for 12 h at 37 °C shaking at 300 rpm. Tryptic digest was acidified (pH < 3) using TFA and desalted using C18 reverse phase spin columns (Microspin, The Nest Group, Inc., #SEM SS18V) according to the manufacturer’s instructions. Peptides were dried under vacuum and stored at −20 °C.
Dried peptides were resuspended in 0.1% aqueous formic acid and subjected to LC-MS/MS analysis using a Orbitrap Fusion Lumos Mass Spectrometer fitted with an EASY-nLC 1200 (Thermo Fisher Scientific) and a custom-made column heater set to 60 °C. Peptides were resolved using a RP-HPLC column (75 μm × 36 cm) packed in-house with C18 resin (ReproSil-Pur C18–AQ, 1.9 μm resin; Dr. Maisch, r119.aq.) at a flow rate of 0.2 μL/min−1. The following gradient was used for peptide separation: from 5% B to 12% B over 5 min to 35% B over 40 min to 50% B over 15 min to 95% B over 2 min followed by 18 min at 95% B. Buffer A was 0.1% formic acid in water, and buffer B was 80% acetonitrile and 0.1% formic acid in water.
The mass spectrometer was operated in DDA mode with a cycle time of 3 s between master scans. Each master scan was acquired in the Orbitrap at a resolution of 120,000 FWHM (at 200 m/z), and a scan range from 375 to 1,500 m/z followed by MS2 scans of the most intense precursors in the linear ion trap at “Rapid” scan rate with isolation width of the quadrupole set to 1.4 m/z. Maximum ion injection time was set to 50 ms (MS1) and 35 ms (M S2) with an automatic gain control target set to 1e6 and 1e4, respectively. Only peptides with charge state 2 to 5 were included in the analysis. Monoisotopic precursor selection was set to Peptide, and the Intensity Threshold was set to 5e3. Peptides were fragmented by HCD with collision energy set to 35%, and one microscan was acquired for each spectrum. The dynamic exclusion duration was set to 30 s.
The acquired raw-files were imported into the Progenesis QI software (version 2.0, Nonlinear Dynamics Limited), which was used to extract peptide precursor ion intensities across all samples applying the default parameters. The generated mgf-file was searched using MASCOT against a murine database (consisting of 33,968 forward- and reverse-protein sequences downloaded from Uniprot on 20180710) and spiked with the sequence of his-tagged Ppargc1a and 392 commonly observed contaminants using the following search criteria: full tryptic specificity was required (cleavage after lysine or arginine residues, unless followed by proline); three missed cleavages were allowed; carbamidomethylation (C) was set as fixed modification; oxidation (M) and acetyl (Protein N-term) were applied as variable modifications; and mass tolerance of 10 ppm (precursor) and 0.6 Da (fragments) were applied. The database search results were filtered using the ion score to set the false discovery rate to 1% on the peptide and protein level, respectively, based on the number of reverse-protein sequence hits in the dataset. Quantitative analysis results from label-free quantification were processed using the SafeQuant R package version 2.3.2 (38) (https://github.com/eahrne/SafeQuant/) to obtain peptide relative abundances. This analysis included global data normalization by equalizing the total peak/reporter areas across all LC-MS runs, data imputation using the knn algorithm, and summation of peak areas per protein and LC-MS/MS run, followed by calculation of peptide abundance ratios. Only isoform-specific peptide ion signals were considered for quantification. To meet additional assumptions (normality and homoscedasticity) underlying the use of linear regression models and t tests, MS intensity signals were transformed from the linear to the log-scale. The summarized peptide expression values were used for statistical testing of between-condition, differentially abundant peptides. Here, empirical Bayes-moderated t tests were applied, as implemented in the R/Bioconductor limma package (http://bioconductor.org/packages/release/bioc/html/limma.html), with significance defined as P value < 0.05 and fold change ≥ 1.2. Overlap between different datasets was determined with Venny (version 2.1, https://bioinfogp.cnb.csic.es/tools/venny/). GO analysis was performed with DAVID 6.8 (https://david.ncifcrf.gov/), with significance defined as adjusted (Benjamini) P value < 0.05. Heat maps were generated with Morpheus (https://clue.io/morpheus).
In Vitro uvAP-Seq.
Cells were transduced as for in vitro AP-MS. Total RNA was then extracted with Direct-zol RNA MiniPrep kit (Zymo Research, #R2050), and 5 μg of RNA were used for input, pull-down, or negative control samples. Pull-down samples were prepared by incubating 5 μg of RNA with 12 μg of recombinant N-terminal His-Tag CTD of mouse PGC-1α (US Biological Life Sciences, #156296) for 30 min at 37 °C with shaking at 500 rpm in 500 μL of PBS with RNasin Ribonuclease Inhibitors (Promega, #N261B). Negative control samples comprised 5 μg of RNA combined with HisPur Cobalt Resin (see details below), which were incubated as described for pull-down samples. RNA–protein interactions were crosslinked by UV irradiating (5 mJ/cm2 at 250 nm UV wavelength) opened tubes with samples on ice, following which PBS with RNasin Ribonuclease Inhibitors was added to 1 mL final volume. RNA–protein complexes were purified with Pierce His Protein Interaction Pull-Down Kit (Thermo Fisher Scientific, #21277) with the following modifications. HisPur Cobalt Resin was washed five times with wash solution and once with PBS in a 1.5-mL tube. Cross-linked samples were combined with HisPur Cobalt Resin and were incubated for 1 h at 4 °C with rotation in 1.5-mL tubes. Following incubation, HisPur Cobalt Resin containing the immobilized bait protein–RNA complexes were washed five times with wash buffer by centrifugation at 1,250 × g for 1 min. RNA was then partially digested by resuspending washed HisPur Cobalt Resin in 1 mL of PBS containing 10 μL 1:1,500 diluted Ambion RNase I (Thermo Fisher Scientific, #AM2294), following incubation at 37 °C for 3 min with shaking at 700 rpm and then washed five times as described above. Subsequently, DNA was digested by incubating pull-down, negative control, and input samples for 30 min at 37 °C with shaking at 800 rpm with 0.05 U/µL RQ1 RNase-Free DNase in 1X RQ1 RNase-Free DNase Reaction Buffer (Promega, # M6101). RNA was eluted from HisPur Cobalt Resin by incubating samples in a final volume of 100 µL PBS with 0.4 mg/mL of Proteinase K (Macherey-Nagel, #740506) and incubated for 2 h at 65 °C with shaking at 800 rpm. Finally, RNA was purified with Direct-zol RNA MiniPrep kit and stored at −80 °C.
Purified RNA from input and pull-down samples was used to prepare libraries with Stranded Total RNA Prep with Ribo-Zero Plus kit (Illumina, # 20040525). Of note, the amount of RNA recovered from negative control samples was not sufficient for library preparation. Single-end sequencing was performed using the NextSEq. 500 (Illumina), and data were analyzed on the Galaxy platform (https://usegalaxy.eu/). Reads were trimmed with Trim Galore! (Galaxy version 0.4.3.1), and quality was assessed using FastQC (Galaxy version 0.72+galaxy1). Reads were aligned to the mm10 version of the mouse genome using STAR (Galaxy version 2.7.2b), and duplicated reads were removed with MarkDuplicates (Galaxy version 2.18.2.2). Next, we used MACS2 callpeak (Galaxy version, #2.1.1.20160309.6) for peak calling (q value < 0.05 and fold change ≥ 1.5), while ChIPseeker (Galaxy version, #1.18.0+galaxy1) was used to annotate peaks. FASTA files from significant peaks were generated with Extract Genomic DNA (Galaxy version, #3.0.3), and DNA was converted to RNA with RNA/DNA (Galaxy version, # 1.0.2). FASTA files were used for motif discovery with Discriminative Regular Expression Motif Elicitation (DREME) (Version 5.1.1, https://meme-suite.org/), with E-value threshold < 0.05. Normalized BAM files were generated with bamCoverage (Galaxy version 3.0.2.0), and data were visualized on the Integrated Genome Browser-9.1.4 (42) to generate representative genome browser figures. The overlap between in vitro uvAP-seq and PGC-1α ChIP-seq data (Gene Expression Omnibus [GEO] accession: GSE51178) was performed with bedtools Intersect intervals (Galaxy version, #2.29.0), while overlap with RNA-seq and alternative splicing data were defined with Venny (version 2.1, https://bioinfogp.cnb.csic.es/tools/venny/).
Confocal Microscopy.
Cells were seeded at 2 × 104 cells per well on glass coverslips in 24-well plates and transfected as described in the Generation of DNA Plasmids and Transfections section. Cells were fixed with 4% formaldehyde in PBS for 15 min, washed three times with PBS, and incubated for 15 min with 1 µg/mL Hoechst 33342 (Thermo Fisher Scientific, #H3570) in PBS. Next, cells were washed three times with PBS and mounted in 5 µl of ProLong Gold Antifade Mountant (Thermo Fisher Scientific, #P36930).
For immunofluorescence staining of cells, samples were fixed as described in the paragraph above and permeabilized with 0.1% Triton X-100 in PBS for 5 min, washed three times with PBS, and blocked with 10% goat serum in PBS for 30 min. Next, cells were incubated for 1 h with primary antibodies (dilution in 10% goat serum in PBS) to p-Pol2 CTD repeat (S5) (1:1,000 dilution; Abcam, #ab5408), H3K27ac (1:2,000; Abcam, #ab4729), and PGC-1α (1:1,000 dilution; Merck, #ST1202). Following three washes with PBS, cells were incubated for 1 h with secondary antibodies (1:1,000 dilution in 10% goat serum in PBS) conjugated to Alexa Fluor 568 (Thermo Fisher Scientific, #A-21124, #A-11011 or #A-11004). Cells were then washed three times with PBS and mounted in 5 µl of ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, # P36931). All immunofluorescence staining steps were performed at room temperature.
Mouse skeletal muscle samples (gastrocnemius and soleus) were mounted in optimal cutting temperature medium (Tissue-Tek O.C.T) at resting length, snap frozen in liquid nitrogen-cooled isopentane, and stored at −80 °C. Muscle sections (10 μm) were cut at −22 °C on a cryostat (Leica, CM1950), collected on microscope slides, and stored at −80 °C. Muscle sections were fixed in ice-cold 4% formaldehyde in PBS for 10 min, washed with PBS, blocked, and permeabilized in PBS containing 10% goat serum and 0.4% triton X-100 for 1 h. Sections were then incubated overnight at 4 °C with primary antibody (dilution in 10% goat serum in PBS) to PGC-1α (1:100 dilution; Merck, #ST1202), H3K27ac (1:400 dilution; Abcam, #ab4729), and laminin 2α (1:200, Abcam, #ab11576). Next, sections were washed four times for 10 min in PBS and incubated for 1 h at room temperature with secondary antibodies (dilution in 10% goat serum in PBS) conjugated to Alexa Fluor 488 (1:200 dilution, Thermo Fisher Scientific, #A21131), Alexa Fluor 647 (1:200 dilution, Jackson, #711-605-152), and Cy3 (1:400 dilution; Jackson, #112-165-143). Sections were then washed four times for 10 min in PBS and mounted with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, # P36931) and coverslips (VWR, #631-0147).
Human skeletal muscle biopsies from vastus lateralis were taken at rest, cut (8-µm thickness), and mounted on positively charged glass slides. Cryosections were fixed in ice-cooled 4% paraformaldehyde diluted in PBS for 10 min and washed three times for 5 min with an immune-buffer solution (IB = 0.25% bovine serum albumin (BSA), 50 mM glycine, 0.033% saponin, and 0.05% sodium azide diluted in PBS). Sections were permeabilized with 0.1% Triton-X100 for 30 min followed by three washes for 5 min with IB. The sections were then blocked for 1 h in a blocking solution containing 2% BSA and 10% goat serum (Gibco, #16210-064) followed by overnight incubation at 4 °C with primary antibody to PGC-1α (1:300 dilution; Merck, #ST1202) diluted in IB. The next day, sections were washed three times for 10 min in IB and incubated in blocking buffer with Alexa Fluor 568 anti-mouse (1:200 dilution) for 2 h. To stain the glycoproteins of the sarcolemma, sections were incubated with Alexa Fluor 488 conjugated with wheat germ agglutinin diluted in PBS (1:200) during 10 min at 37 °C (Invitrogen, #W11261). Sections were then incubated with Hoechst 33342 (Invitrogen, #H3570) followed by three washes for 10 min in IB and mounting in Vectashield antifade mounting medium (Vector Laboratories, #H-1000).
C2C12 cells and mouse skeletal muscle samples were imaged with an Olympus SpinSR CSU-W1 confocal microscope equipped with UPLAPO 100XOHR objective lens and Hamamatsu Flash4 version 3 scientific complementary metal–oxide–semiconductor (sCMOS) camera. Z-stack images at 100× magnification were taken using Olympus cellSens Dimension software. Human skeletal muscle samples were images with a Zeiss 780 system with a Zeiss Plan-Apochromat 63× 1.4 NA objective. The system was driven by Zeiss Zen Black 2012.
Live-Cell Imaging.
Cells were seeded at 2 × 104 cells per chamber on an 8-well chamber slide (ibidi, #80826) and transfected as described in the Generation of DNA Plasmids and Transfections section. Live-cells imaging was performed using FEI MORE microscope, with cells maintained at 37 °C and 5% CO2. As the light source for GFP imaging, a spectraX light-emitting diode with 470/24-nm excitation and 517/20-nm emission filter was used. Images were recorded with Hamamatsu OCRA flash 4.0 cooled sCMOS at 100× (numerical aperture 1.4, oil) magnification with Live Acquisition 2.5 software, while postprocessing and deconvolution were carried out with Fiji 1.52p and Huygens Professional 19.10 software, respectively.
Cells were imaged before and after treatment with 5% 1,6-Hexanediol (Sigma, #240117) and diluted in growth medium for up to 60 s.
FRAP was performed with a 488-nm laser and 470/24-nm filter with laser power set to 100% (dwell time: 0.962 ms/µm2, line overlapping 75%, region of interest (ROI) loop count: 10, exposure time: 20 ms, and cycle time: 185 ms). A total 10 images were obtained prebleaching, while 399 images were obtained every 185 ms postbleaching. Intensities of FRAP regions were extracted with Fiji 1.52p software tool. FRAP Data were background corrected and analyzed with the online tool EasyFRAP-web (https://easyfrap.vmnet.upatras.gr/).
ChIP-Seq Analysis.
Previously published PGC-1α ChIP-seq data (GEO accession: GSE51178) (4) was analyzed on the Galaxy platform (https://usegalaxy.eu/) for comparison with the datasets generated in this study. Reads were trimmed with Trim Galore! (Galaxy version 0.4.3.1), and quality was assessed using FastQC (Galaxy version 0.72+galaxy1). Reads were aligned to the mm10 version of the mouse genome using Bowtie2 (Galaxy Version 2.3.4.3+galaxy0), and low-quality reads (phred < 20) were filtered out with Filter (Galaxy Version 2.4.1). Next, we used MACS2 callpeak (Galaxy version, #2.1.1.20160309.6) for peak calling (q value < 0.05 and fold change ≥ 2), while ChIPseeker (Galaxy version, #1.18.0+galaxy1) was used to annotate peaks. Normalized BAM files were generated with bamCoverage (Galaxy version 3.0.2.0).
Statistics.
All qPCR, immunoblotting, reporter gene assays, confocal microscopy, and live-cell imaging experiments were performed at least three independent times with similar results. FRAP quantification was performed with a total of 50 foci from independent experiments. Number of replicates per experiment is indicated in the figure legend when appropriate.
For qPCR, oxygen consumption, and reporter gene assays data analysis, statistical significance was determined with one-way ANOVA with Tukey’s post hoc test using GraphPad Prism (version 8.0). Significance was considered with a P value < 0.05. Values are expressed as mean ± SD.
RNA-seq, ChIP-seq, mass spectrometry, in vitro AP-MS, and in vitro AP-seq experiments were performed once with two or three biological replicates as indicated in figure legends. Statistical analysis of these experiments is described above in their corresponding sections.
Supplementary Material
Acknowledgments
We thank C. Beisel (Genomics Facility Basel, ETH Zürich), P. Demougin (Life Sciences Training Facility, Biozentrum), and K. Schleicher (Imaging Core Facility, Biozentrum) for technical help. We thank Prof. Erik A. Richter from the Molecular Physiology Section at University of Copenhagen for his contribution on the human experiments. We thank the Core Facility for Integrated Microscopy at University of Copenhagen for assistance with imaging of human skeletal muscle samples. This work was supported by the Novartis Foundation for Medical-Biological Research (J.P.-S.), Research Fund of the University of Basel (J.P.-S.), the Swiss National Science Foundation, the European Research Council Consolidator Grant 616830-MUSCLE_NET, Swiss Cancer Research Grant KFS-3733-08-2015, the Swiss Society for Research on Muscle Diseases, SystemsX.ch, the Novartis Stiftung für Medizinisch-Biologische Forschung, and the University of Basel (C.H.). C.H.-O. is supported by a research grant from the Danish Diabetes Academy, which is funded by the Novo Nordisk Foundation (Grant NNF17SA0031406).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105951118/-/DCSupplemental.
Data Availability
RNA-seq and in vitro AP-seq data are available at the Gene Expression Omnibus under accession number GSE156594. Whole-proteome analysis and in vitro AP-MS data are available at the Proteomics Identification Database (PRIDE) under accession number PXD026725. All additional data are included in the article and/or supporting information, while unique materials generated in this study are available from the corresponding authors on request.
References
- 1.Haberle V., Stark A., Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 19, 621–637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mouchiroud L., Eichner L. J., Shaw R. J., Auwerx J., Transcriptional coregulators: Fine-tuning metabolism. Cell Metab. 20, 26–40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kupr B., Handschin C., Complex coordination of cell plasticity by a PGC-1α-controlled transcriptional network in skeletal muscle. Front. Physiol. 6, 325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baresic M., Salatino S., Kupr B., van Nimwegen E., Handschin C., Transcriptional network analysis in muscle reveals AP-1 as a partner of PGC-1α in the regulation of the hypoxic gene program. Mol. Cell. Biol. 34, 2996–3012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puigserver P., et al., Activation of PPARgamma coactivator-1 through transcription factor docking. Science 286, 1368–1371 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Wallberg A. E., Yamamura S., Malik S., Spiegelman B. M., Roeder R. G., Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell 12, 1137–1149 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Lin J., et al., Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418, 797–801 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Puigserver P., et al., A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92, 829–839 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Yoon J. C., et al., Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413, 131–138 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Savkur R. S., Burris T. P., The coactivator LXXLL nuclear receptor recognition motif. J. Pept. Res. 63, 207–212 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Monsalve M., et al., Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6, 307–316 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Balwierz P. J., et al., ISMARA: Automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 24, 869–884 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho H., et al., Transcriptional coactivator PGC-1α contains a novel CBP80-binding motif that orchestrates efficient target gene expression. Genes Dev. 32, 555–567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguilo F., et al., Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1α. Cell Rep. 14, 479–492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Nostrand E. L., et al., A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavares C. D. J., et al., Transcriptome-wide analysis of PGC-1α-binding RNAs identifies genes linked to glucagon metabolic action. Proc. Natl. Acad. Sci. U.S.A. 117, 22204–22213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long J., et al., Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Invest. 126, 4205–4218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao R., et al., Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178, 107–121.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spector D. L., Lamond A. I., Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3, a000646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., Sharp P. A., A phase separation model for transcriptional control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson B. A., et al., Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boija A, et al., Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855 e1816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabari B. R., et al., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y., Protter D. S., Rosen M. K., Parker R., Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Booth F. W., Roberts C. K., Thyfault J. P., Ruegsegger G. N., Toedebusch R. G., Role of inactivity in chronic diseases: Evolutionary insight and pathophysiological mechanisms. Physiol. Rev. 97, 1351–1402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Malley B. W., Qin J., Lanz R. B., Cracking the coregulator codes. Curr. Opin. Cell Biol. 20, 310–315 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiegelman B. M., Heinrich R., Biological control through regulated transcriptional coactivators. Cell 119, 157–167 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Knutti D., Kaul A., Kralli A., A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol. Cell. Biol. 20, 2411–2422 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez-Redondo V., et al., Peroxisome proliferator-activated receptor γ coactivator-1 α isoforms selectively regulate multiple splicing events on target genes. J. Biol. Chem. 291, 15169–15184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruas J. L., et al., A PGC-1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 151, 1319–1331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roden C., Gladfelter A. S., RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 22, 183–195 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alberti S., Dormann D., Liquid-liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Wu P., Inhibition of RNA-binding proteins with small molecules. Nat. Rev. Chem. 4, 441–458 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Klein I. A., et al., Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henríquez-Olguin C., et al., Cytosolic ROS production by NADPH oxidase 2 regulates muscle glucose uptake during exercise. Nat. Commun. 10, 4623 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kahles A., Ong C. S., Zhong Y., Rätsch G., SplAdder: Identification, quantification and testing of alternative splicing events from RNA-Seq data. Bioinformatics 32, 1840–1847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pérez-Schindler J., Summermatter S., Santos G., Zorzato F., Handschin C., The transcriptional coactivator PGC-1α is dispensable for chronic overload-induced skeletal muscle hypertrophy and metabolic remodeling. Proc. Natl. Acad. Sci. U.S.A. 110, 20314–20319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahrné E., et al., Evaluation and improvement of quantification accuracy in isobaric mass tag-based protein quantification experiments. J. Proteome Res. 15, 2537–2547 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., et al., Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11, 2019–2026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer A., Churchman L. S., A detailed protocol for subcellular RNA sequencing (subRNA-seq). Curr. Protoc. Mol. Biol. 120, 4.29.1–4.29.18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kustatscher G., Wills K. L., Furlan C., Rappsilber J., Chromatin enrichment for proteomics. Nat. Protoc. 9, 2090–2099 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freese N. H., Norris D. C., Loraine A. E., Integrated genome browser: Visual analytics platform for genomics. Bioinformatics 32, 2089–2095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and in vitro AP-seq data are available at the Gene Expression Omnibus under accession number GSE156594. Whole-proteome analysis and in vitro AP-MS data are available at the Proteomics Identification Database (PRIDE) under accession number PXD026725. All additional data are included in the article and/or supporting information, while unique materials generated in this study are available from the corresponding authors on request.