Abstract
Size‐exclusion chromatography (SEC) is a widely adopted method for the isolation of extracellular vesicles (EVs) from complex samples. SEC can efficiently remove high‐abundant proteins, while often requires multiple fractionation operation using diversified column settings. In this study, we aim to establish a simplified SEC method to acquire high quality EVs. In comparison of all three cross‐linked Sepharose resins with the sample types of FBS and human serum (HS), CL‐6B and CL‐4B showed superior performance in regular SEC to CL‐2B in terms of significantly narrower EV and protein peaks, higher resolutions and EV purity. By increasing their bed volumes to 20 ml, the resolutions of CL‐6B and CL‐4B columns could be significantly improved, while the CL‐6B column had the best performance with higher particle yields and tighter EV peaks. With the CL‐6B 20 ml column, we further established a simplified dichotomic SEC method that only requires two bulk elutions to acquire EVs in the Eluate 1 and proteins in the Eluate 2. We further justified that such CL‐6B columns were reusable for at least 10 consecutive times, and the dichotomic SEC was applicable to EV isolations from HS and FBS‐free supernatants of fluorescently labelled and unlabelled SW620 cells. The proteomics analysis implicated that although the two methods had dissimilar abilities in removing different co‐isolating contaminant proteins from EVs, the dichotomic SEC and ultracentrifugation could isolate EVs from human plasma with comparable purity. This dichotomic SEC has its intriguing potential to be used for EV preparation toward clinical testing and/or basic research.
Keywords: clinical application, dichotomic size‐exclusion chromatography, extracellular vesicles, human plasma/serum, simplified
1. INTRODUCTION
Extracellular vesicles (EVs) are cell‐derived compartments or particles that have diversified biological or pathological functions. EVs could be roughly classified as exosomes and microvesicles (MVs) according to their biogenesis and characteristics (Crescitelli et al., 2013; Raposo & Stoorvogel, 2013). In general, exosomes are EVs with a diameter size of 30–200 nm, which are formed intracellularly along with the maturation of multivesicular endosomes (MVE) (Ciferri et al., 2021; Doyle & Wang, 2019; Thery et al., 2018). If not considering the intracellular origin, EVs with the similar size to exosomes can be collectively called small EVs (sEVs) (Kowal et al., 2016; Thery et al., 2018). After the membrane of MVE fuses with the plasma membrane of a cell, exosomes can be released (Thery et al., 2018). Different from exosomes, MVs are bigger EVs with typical sizes ranged from 500 to 1000 nm in diameter. MVs are largely formed through direct budding of the plasma membrane, followed by the extracellular release. However, EVs tend to have far more complex subpopulations. For example, Kowal et al have found through proteomics analyses that different EV subtypes isolated from human dendritic cells share common biomarkers such as flotillin and heat‐shock 70‐kDa protein (HSP70), while there is also a set of five proteins that have differently relative abundances in distinct populations (Kowal et al., 2016). Differential functions and characteristics among all EV subpopulations are still unclear. But, it is widely accepted that EVs are of promising values in the diagnosis, prognosis and treatment of various human diseases, as they carry tissue and/or organ derived biomolecules, such as proteins, RNAs, DNAs and lipids (Ayers et al., 2019; Boelens et al., 2014; Guo et al., 2016; Li et al., 2013; Xu et al., 2016).
EV biomarker researches are facing a number of challenges especially in standardizing the isolation and characterization methods toward clinical applications (Soekmadji et al., 2020). In addition, sample collection, storage, analytical methods and sample size are of significance in the validation stages of any EV biomarkers (Clayton et al., 2019; Soekmadji et al., 2020). As emphasized by the minimal information for studies of EVs 2018 (MISEV2018) (Thery et al., 2018) and other position papers from International Society for EVs (ISEV) (Lener et al., 2015; Mateescu et al., 2017; Russell et al., 2019), isolation of purified EVs is one of the most important and difficult prerequisites prior to any subsequent analyses. Especially, isolation of EVs from complex body fluids is often in a dilemma between purity and ease of operation that are both critical for the real‐world clinical applications (Soekmadji et al., 2020). Among over 30 types of biofluids, plasma/serum are one of the most widely used sample types for biomarker discovery and disease diagnosis (Witwer et al., 2013). However, there are a variety of biomolecules or biological nanoparticles in plasma/serum that have similar size and/or density to EVs, primarily including lipoproteins, subcellular organelles, cellular debris and viral particles (Mathieu et al., 2019). Among them, low/very low/high density lipoproteins (LDL, VLDL, HDL) represent ones of the most common contaminants in EV isolation as there are ∼1016 lipoproteins/ml in plasma (Simonsen, 2017). MISEV2018 recommended that several categories of protein markers could be analysed and reported to justify EV purity, including those showing the presence of EVs (Categories 1 and 2), and the depletion of commonly co‐isolating contaminants (Category 3) (Thery et al., 2018). In addition, the particle per protein unit ratio (particle/protein ratio) is also an important parameter to estimate EV purity (Dong et al., 2020; Thery et al., 2018). Furthermore, Tian et al. reported another indirect way to estimate EV purity via the ratio of Triton X‐100 disruptable particles to total particles using nano‐flow cytometry (nFCM) (Tian et al., 2020).
In current opinions, ultracentrifugation (UC), especially density‐gradient UC is one of the most accepted methods for EV isolation with high purity. But UC cannot be easily used in laboratory medicine as it is often time‐consuming, with low particle recovery rate and requires major instruments (Tian et al., 2020). Here, the particle recovery rate has been defined as the ratio of the recovered particles to the input particles (Tian et al., 2020). The filtration‐ or precipitation‐ based methods suffer from either severe protein contamination or introduction of exogenous chemical materials. In contrast, size‐exclusion chromatography (SEC) offers an important addictive‐free choice to bypass the need of major instruments while isolating EVs with the purity comparable to UC (Thery et al., 2018; Xu et al., 2016).
SEC uses polymer to form porous stationary phase in a column that allows particles of different sizes to be differentially eluted by gravity‐flow (Boing et al., 2014) or using liquid chromatography systems (Baranyai et al., 2015). Although it is known that chylomicron, LDL and VLDL cannot be completely removed, SEC can efficiently separate EVs from most of high‐abundant protein contaminations in body fluid such as albumin (Monguio‐Tortajada et al., 2019).
Most studies with the SEC isolation of EVs require the collection of approximately 26 eluent fractions, and some of the fractions can be pooled for EV analyses (Benedikter et al., 2017; Ludwig et al., 2019; Stranska et al., 2018; Takov et al., 2019). Indeed, for clinical applications, we believe that a simplified isolation protocol using gravity‐flow SEC for EV isolation is of phenomenal demands. To address this question, three aspects of SEC need to be evaluated. First, the selection of resins needs to be sufficiently addressed. Cross‐linked (CL) Sepharose based materials are among the mostly used SEC resins in EV isolation (Monguio‐Tortajada et al., 2019); however, comparison of these materials has not been fully addressed. Second, optimization of the bed volume on EV isolation should be performed. The bed volume of 10 ml is the most popular setting in SEC; while regarding columns with the same internal diameter, it has been shown that increased bed volume may lead to better separation performance (Lane et al., 2019). But, such a hypothesis has not been tested in gravity‐flow columns with bed volume greater than 10 ml so far per our knowledge at least according to peer‐reviewed publications. Third, with the above‐mentioned resin and bed volume combinations, the isolation protocol should be as easy as possible that can be operated by even under‐trained personnel in laboratory medicine.
In this study, we stepwise addressed the above‐mentioned three questions as summarized in Figure 1. Specifically, we compared the performance of all three Sepharose CL resins, namely CL‐2B, CL‐4B, and CL‐6B, followed by the bed volume optimization. With these optimized conditions, we demonstrated that the dichotomic SEC separation with 2 bulk elution steps was sufficient for the isolation of EVs from foetal bovine serum (FBS), human serum (HS) and FBS‐free cell culture supernatants with high purity and particle recovery rate. Such a simplified EV isolation method has advantages to be potentially used in clinical settings.
FIGURE 1.
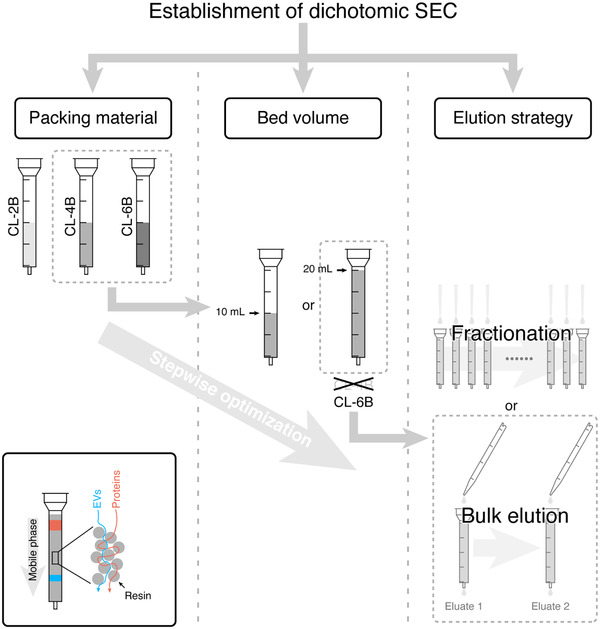
Workflow of developing a simplified SEC method for EV isolation. The principle of the SEC in EV isolation is summarized in the bottom‐left panel
2. MATERIAL AND METHODS
2.1. FBS
FBS (Lot#: 2014014), purchased from Biological Industries, Israel, were centrifuged sequentially at 300 × g for 10 min at room temperature (RT), and 17000 × g for 20 min at 4°C to remove debris. Supernatants were then filtered by 0.22 μm Minisart High Flow PES Syringe Filters (Sartorius, Guangzhou, China) and aliquoted for storage at ‐80°C.
2.2. Human serum (HS)
Collection of human blood samples for this study was approved by the Ethics Committee of the First Affiliated Hospital of Jinan University. Four healthy donors (3 females and 1 male, aged 24–32, normal diet) were included. Samples from Donor 1 was prepared and aliquoted for the evaluation of the performance of different resins and bed volumes, as well as the proteomic comparison of dichotomic SEC and UC. Serum from Donor 2–4 were employed for the validation of the dichotomic SEC method. For every donor, peripheral blood was drawn from the median cubital veins or basilic veins with a 22‐gauge needle (Improve Medical, Guangzhou, China) and Improvacuter vacutainers with no additive (Improve Medical). Blood samples were kept upright at RT for 1‐1.5 h to allow blood coagulation. Coagulated human blood samples were then processed following the identical protocol for aliquoting FBS as described above.
2.3. SEC column packing and equilibration
SEC columns were freshly packed the day before use. Sepharose CL‐2B, CL‐4B or CL‐6B resins (all from GE Healthcare, Shanghai, China) were thoroughly resuspended by gently inverting for over 2 min. To prepare SEC columns with the bed volumes of 10 ml and 20 ml, 13.5 ml and 27 ml resins were loaded into the Econo‐Pac chromatography columns (Bio‐Rad Laboratories, Inc., Guangzhou, China), respectively. Columns were prepared and kept standing still for overnight at 4°C, allowing resins to settle and reach the specified bed volume per gravity. Prior to EV isolation, the upper bed support provided along with the Econo‐Pac chromatography column was added on the top of the bed to prevent it from being disturbed or running dry. Columns were then equilibrated by 2 times of bed volume of PBS (0.22 μm filtered, similar hereinafter) before sample loading. The last 1 ml of PBS dripped out from SEC columns was collected for the detection of background particles and proteins.
2.4. EV isolation with regular SEC
HS and FBS samples were thawed at RT, followed by the centrifugation at 17000 × g, 10 min at 4°C to remove aggregates generated during freeze‐thaw. Supernatant was then loaded into SEC columns and fractionated according to Boing et al with minor modifications (Boing et al., 2014). Specifically, 1 ml of sample was loaded on the top of the equilibrated SEC column to allow its dripping out fully by gravity flow. For fractionation, 500 μl PBS was added for each fraction and the eluate was collected. The eluate of the first 500 μl PBS elution was counted as Fraction 1. A total number of 26 and 52 fractions were collected for 10 ml and 20 ml columns, respectively. All fractions were stored at ‐80°C for downstream analyses.
2.5. Dichotomic SEC for EV isolation
For dichotomic SEC, 2 bulk elutions with PBS at different combinations (7.5 ml and 18.5 ml, 8 ml and 18 ml, 8.5 ml and 17.5 ml, 9 ml and 17 ml, or 9.5 ml and 16.5 ml) were performed immediately after the sample (1 ml) being completely dripped out of the column (Figure 4(a)). Sample flow‐through and the eluate of the first bulk elution were collected as the Eluate 1. The second bulk elution was then collected as the Eluate 2. The Eluate 1 and 2 were respectively concentrated by using an Amicon Ultra‐4 filters with 30 kDa molecular weight cut off (MWCO) membranes (Merck Millipore Ltd., Guangzhou, China) via centrifuging at 5000 × g with a 35° fixed‐angle rotor.
FIGURE 4.
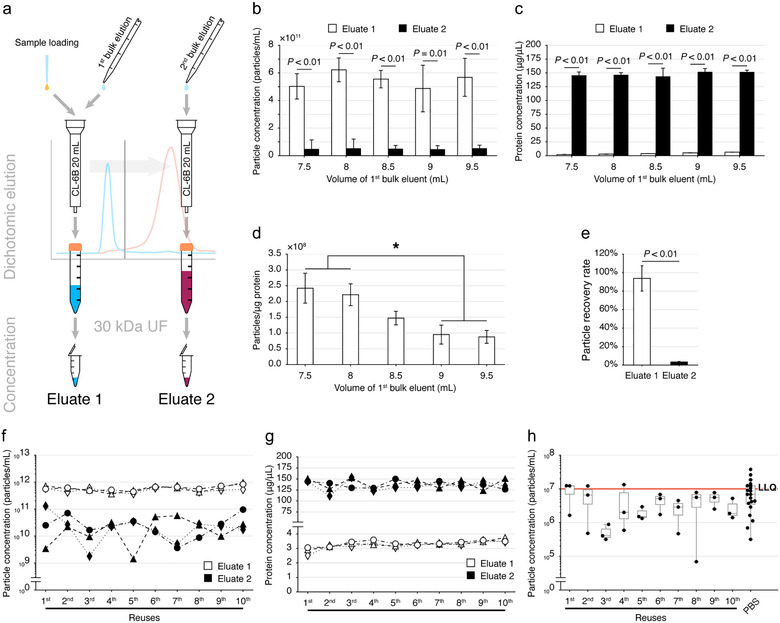
Feasibility of dichotomic SEC for EV isolation from FBS. (a) Schematic diagram of the dichotomic SEC strategy for EV isolation. (b,c) Comparison of the particle (b) and protein (c) concentration recovered in the Eluate 1 and 2 with different bulk eluent volumes. Data are shown as mean±SD, n = 3, Student's t‐test. (d) Particle to protein ratio of Eluate 1. Data are shown as mean±SD, n = 3. *Significant difference (P < 0.05), n = 3, Tukey's multiple comparisons. (e) The particle recovery rate. SEC‐derived particles were counted by NTA and loaded into the SEC column, the particles in the Eluate 1 and 2 were quantified with NTA to calculate the particle recovery rate. Data are shown as mean±SD, n = 3. (f,g) Particle yield (f) and protein concentration (g) comparisons in SEC columns reused for 10 consecutive times, column n = 3. (h) Particle leftover determination. The last 1 ml of equilibrating PBS dripped out from SEC columns was collected for the detection of background particles with NTA. LLQ: lower limit of quantification of NTA. One‐way ANOVA followed by Dunnett's multiple comparison against the PBS only group was performed, column n = 3
2.6. SEC column cleaning and regeneration
For experiments evaluating the performance of reused columns, we performed column cleaning and regeneration following the manufacturer's instruction of the Sepharose resins provided by GE Healthcare (Instruction 71‐7098‐00 AD). Specifically, used SEC columns were cleaned with 2 × bed volume of the cleaning solution (0.1% Triton X‐100 in distilled water, 0.22 μm filtered), followed by a wash with 2 × bed volume of PBS for regeneration. Columns should be re‐equilibrated with 1 × bed volume of PBS upon next use.
2.7. Nanoparticle tracking analysis (NTA)
Particle size and concentration were determined by the NanoSight NS300 analyzer (Malvern, Shanghai, China) equipped with a 488 nm blue laser and sCMOS camera as we previously described with minor modifications (Guo et al., 2016). Briefly, samples were diluted for ≥20 times with PBS and loaded into the instrument. For each sample, videos of 3 random views were captured with the instrumental parameters set as follows: temperature controller, on; temperature, 25°C; camera level, 14; capture duration, 60 s. Focusing was adjusted daily using an abundant sample. Videos were analysed with the following parameters: viscosity, water (0.89 cP); detection threshold, 3. The particle size and mean concentration computed from the 3 videos was reported, using the NanoSight NTA 3.4 software (Malvern). Particles larger than 60 nm in diameter were analysed based on the particle size sensitivity analyses reported by Bachurski et al. and others (Bachurski et al., 2019; Holcar et al., 2020).
2.8. Transmission electron microscopy (TEM)
TEM was performed as Thery et al described with minor modifications (Thery et al., 2006). In brief, 20 μl of the Eluate 1 isolated from HS of Donor 3 was fixed by adding equal volume of 4% (w/v) paraformaldehyde. 10 μl of the fixed sample was absorbed by a Formvar‐carbon coated TEM grid, followed by another fixation with 1% (w/v) glutaraldehyde. Grids were stained with uranyl‐oxalate solution (4% uranyl acetate, 0.0075 M oxalic acid, pH 7) and dried in methyl cellulose‐UA (0.4% uranyl acetate, 1.8% methyl cellulose). Finally, grids were observed with a Tecnai G2 transmission electron microscope (ThermoFisher Scientific, Shanghai, China) operated at 100 kV.
2.9. Immunoblotting
Protein concentrations were determined by the BCA protein assay (Thermo), and immunoblotting (IB) analyses were performed as we previously described (Zhang et al., 2017).
For IB on the multiple fractions acquired from regular SEC samples, two sample loading strategies were employed. For fractions with sufficient proteins, 20 μg protein was loaded. For fractions with less than 20 μg protein per 32 μl, 32 μl of samples was used. All samples were respectively adjusted to the volume of 32 μl with PBS, followed by the addition of 8 μl of the 5 × sample loading buffer (Fude Biological, Hangzhou, China). After being boiled for 10 min, samples were loaded into the NuPAGE 10% Bis‐Tris Gel (1.5 mm × 15 well, Thermo). Electrophoresis was performed at 200 V for 35 min in 1 × NuPAGE MOPS SDS Running Buffer (Thermo), followed by the PVDF membrane (Thermo) transfer at 20 V constant for 1 h in 1 × NuPAGE Transfer Buffer (Thermo) with 10% methanol. Membranes were blocked with 5% milk in Tris buffered saline with 0.2% Tween (TBST) and incubated with primary antibodies at 4°C overnight. Primary antibodies included rabbit anti‐apolipoprotein A‐I/ApoA1 pAb (Cat: 10686‐T52, SinoBiological, Beijing, China; 1:10000), rabbit anti‐integrin β3 mAb (Cat: 13166S, Cell Signaling Technology, Shanghai, China; 1:500), rabbit anti‐syntenin mAb (Cat: ab133267, Abcam, Shanghai, China; 1:1000), rabbit anti‐TSG101 mAb (Cat: MABC649, Merck; 1:2000), rabbit anti‐CD9 mAb (Cat: ab92726, Abcam; 1:1000), rabbit anti‐CD63 mAb (Cat: ab134045, Abcam; 1:2000), rabbit anti‐albumin mAb (Cat: ab207327, Abcam; 1:2000), and mouse anti‐ApoB mAb (Cat: SC‐393636, Santa Cruz Biotechnology, Shanghai, China; 1:5000). For secondary antibody staining, membranes were washed for 3 times with TBST (5 min each time), and incubated with the secondary antibody for 1 h at RT. HRP‐conjugated secondary antibodies included anti‐rabbit IgG and anti‐mouse IgG (both from CST; 1:2000). Finally, membranes were washed 3 times with TBST (5 min each time) prior to the incubation with Clarity Western ECL Substrate (Bio‐Rad Laboratories, Inc.). Images were taken by using a 5200 Multi imager with 590 nm filter (Tanon Science & Technology co., Shanghai, China).
For IB on the Eluate 1 and the Eluate 2 from the dichotomic SEC, considering the abundance of target protein, 5, 10, 20 or 30 μg protein/lane was loaded, respectively. Coomassie blue R‐250 staining analysis was used to verify the loading of the same amount of protein calculated based on the BCA results.
All IB raw images with short and long exposures, along with the corresponding bright field were summarized in Supplementary Figure S1.
2.10. Dichotomic SEC experiments on SW620 cell culture supernatants
SW620 cells were maintained at 37°C in complete Dulbecco's modified Eagle's medium (cDMEM; Thermo) containing 10% FBS, 1 mM sodium pyruvate (Thermo), 1% penicillin/streptomycin (Thermo), and 10 μg/ml ciprofloxacin as we previously described (Guo et al., 2016). To prepare supernatants for EV analyses, SW620 cells were seeded in 75 cm2 flasks at 1.5 × 107 cells/flask, and cultured for 48 h. Cells were washed in‐flask twice with PBS and rinsed with DMEM, followed by culturing in 10 ml DMEM without FBS for 24 h to allow EV secretion. The supernatant was then collected and sequentially centrifuged using a GL‐21 M centrifuge (Cence, Changsha, China) at 220 × g for 10 min and 15000 × g for 30 min at RT to remove cells and debris, respectively. After being filtered by a 0.22 μm filter, the supernatant was concentrated to ∼1 ml using a 30 kDa MWCO ultrafiltration (UF) device (Merck). The retentate was ready for SEC separation. Twenty‐one flasks with ∼210 ml supernatant were prepared for the EV experiments, including assays on particle and protein concentration, IB and EV purity.
2.11. Fluorescent labelling of SW620 cells
For fluorescent labelling experiments, eight 75 cm2 flasks of adherent cells were washed in‐flask twice with PBS. A final concentration of 5 μM 5(6)‐Carboxyfluorescein diacetate N‐succinimidyl ester (CFDA‐SE; Sigma‐Aldrich, Shanghai, China) in PBS was then added in‐flask at 1 ml per 1 × 107 cells. For the unlabelled group (8 flasks), PBS was used to mimic the CFDA‐SE labelling. Flasks were then incubated for 10 min at 37°C. Staining was then terminated by adding 5 × volume of cDMEM. Cells were washed in‐flask twice with PBS and rinsed with DMEM, followed by culturing in 10 ml DMEM without FBS for 24 h to allow EV secretion. Supernatants for EV analyses were then prepared with the same procedure as described above.
2.12. The nFCM analysis
The nFCM (NanoFCM Inc., Xiamen, China) analysis was performed to evaluate the measurement of particle size, fluorescence intensity, and EV purity. As EVs will be lysed by Triton X‐100, the EV purity can be indirectly reflected by the fraction of Triton X‐100 disrupted particles to the total particles (Tian et al., 2020). A silica nanosphere cocktail (Cat. S16M‐Exo, NanoFCM Inc.) that contained a mixture of 68 nm, 91 nm, 113 nm and 155 nm beads was employed as the particle size standards. SW620 EVs were mixed with Triton X‐100 to reach a final concentration of 1% (v/v), and placed on ice for 1 h. Diluted samples were then mixed with equal amount of the 1% (v/v) FITC‐labelled 250 nm silica nanosphere (Cat. S08211, NanoFCM Inc.), which served as the counting standard for flow rate calibration (Mao et al., 2020). The instrumental parameters of the nFCM analysis were set as follows: Laser, 10 mW, 488 nm; SS Decay, 10%; Sampling pressure, 0.8 kPa; Samp Period, 100 μs; Time to record, 1 min.
2.13. Fluorometry
Culture supernatants from CFDA‐SE labelled and unlabelled SW620 cell were subjected to the dichotomic SEC. The Eluate 1 and the Eluate 2 were respectively concentrated to 100 μl via UF. Next, 70 μl of the concentrated Eluate 1 or Eluate 2 were loaded into a 96 well‐plate. The fluorescence intensity (FI) was measured using a Victor X5 multilabel plate reader (PerkinElmer, Turku, Finland). Samples were excited with 488 nm laser, while the emitted light was recorded through a 535/25 nm bandpass filter.
2.14. Proteomics analysis on EVs
2.14.1. Platelet‐free plasma
A number of EV proteomics studies have been published including Karimi et al (Karimi et al., 2018) and Lane et al (Lane et al., 2019), etc., and most of these studies used the sample type of platelet‐free plasma (PFP). To make better inter‐study comparison, we here used the same sample type of PFP for the proteome comparison. To prepare PFP, peripheral blood from Donor 1 was drawn from the median cubital veins or basilic veins with a 22‐gauge needle and BD vacutainers with K2EDTA (BD). The first 3 ml of blood was discarded. Immediately after collection, tubes were softly inverted for 8–10 times. Blood samples were centrifuged twice at 2500 × g, 15 min at RT to collect PFP in supernatants. PFP was centrifuged at 15000 × g, 30 min at RT and filtered by 0.22 μm Millex®‐ GP Millipore Express PES Membrane Filter Units (Merck) and stored at ‐80°C in aliquots for further use. Prior to EV isolation with dichotomic SEC or UC, PFP were thawed at RT and centrifuged at 17000 × g, 10 min at 4°C to remove aggregates generated during freeze‐thaw.
2.14.2. Ultracentrifugation
To compare with the dichotomic SEC, EVs isolated through UC was prepared as we described with minor modifications (Guo et al., 2016; Zhang et al., 2017). In brief, thawed PFP (2 ml) was diluted with PBS to 28 ml, followed by centrifugation at 100000 × g, 2 h at 4°C using the Optima L‐100 XP Ultracentrifuge with an SW 32Ti rotor (Beckman Coulter, Inc., CA, USA). The pellet was resuspended and washed with 26 ml PBS and centrifuged again at 100000 × g, 2 h at 4°C. The supernatant was discarded, and the pellet (EVs) was resuspended with 50 μl of PBS.
2.14.3. Protein extraction
EVs isolated by using dichotomic SEC and UC were respectively mixed with 4% SDS lysis buffer contained 4 mM PMSF and 4 × protease inhibitor cocktail (Roche, Shanghai, China) to reach a final concentration of 1% SDS. After being incubated on ice for 30 min, lysates were centrifuged at 17000 × g for 30 min at 4°C. Supernatants were collected, and protein concentrations were measured by BCA kits.
2.14.4. Protein digestion
In‐solution protein digestion was performed as we described previously (Guo et al., 2016; Lu et al., 2019; Tang et al., 2018; Zhang et al., 2017). Briefly, 200–250 μg of EV lysates were reduced in 8 M urea containing 50 mM dithiothreitol (DTT) at 37°C for 1 h, and alkylated in 150 mM iodoacetamide (IAA) at RT for 30 min. Proteins were then transferred into a 30 kDa ultracentrifugal filter (Millipore) and centrifugally (12000 × g) washed sequentially with 8 M urea twice, and 50 mM triethylammonium bicarbonate (TEAB) for five times. Trypsin was then added at a mass ratio of 1:30, and proteins were digested for 12 h at 37°C. Peptides were collected, and desalted by using the MonoTip C18 Pipette Tips (GL Sciences, Tokyo, Japan). Peptides were then dried with a cold‐trap speed vacuum. Prior to the mass spectrometry (MS) analysis, peptides were reconstituted by deionized water containing 0.1% (v/v) formic acid.
2.14.5. Data‐independent acquisition mass spectrometry
Peptides were analysed in a data‐independent acquisition (DIA) mode by the liquid chromatography (LC) MS, equipped with an EASY‐nanoLC 1200 HPLC system and Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo) as we previously described with minor modifications (Lu et al., 2019; Tang et al., 2018; Zhang et al., 2017). For LC separation, 1 μg of peptide was loaded into an analytical column (Omics High Resolution, 0.1 mm × 50 cm) and separated with a 130 min gradient and online DIA MS analysis. The DIA MS parameters were set as follows. (1) MS parameters: detector type, orbitrap; orbitrap resolution, 120000; scan range, 350–1200 m/z; RF lens, 30%; AGC target, 1000000; maximum injection time, 100 ms. (2) MS/MS parameters: isolation mode, quadrupole; activation type, HCD; collision energy, 33%; detector type, orbitrap; orbitrap resolution, 50000; mass range, normal; scan range: 200–2000 m/z; AGC target, 100000; maximum injection time, 86 ms. All MS raw data are available in iProX (Ma et al., 2019) (accession number: PXD026034).
2.14.6. Database searches and protein quantification
Spectronaut Pulsar version 14.10 (Biognosys, Switzerland) was employed for database searches against the Swiss‐Prot Human fasta database (downloaded on Feb 24, 2021, 20396 entries). The DIA raw files of EVs isolated by using UC and the dichotomic SEC were processed with the Spectronaut directDIA experimental analysis workflow. Search parameters included: enzyme, trypsin; max peptide length, 52; min peptide length, 9; missed cleavages, 2; fixed modifications, carbamidomethyl (C); variable modifications, acetyl (N‐term), deamidation (NQ), oxidation (M); data filtering, Qvalue; precursor Qvalue cutoff, 0.01; protein Qvalue cutoff, 0.01; quantity MS‐level, MS2; cross run normalization, true; normalization strategy, global normalization.
2.15. Gene ontology analysis
The DAVID tool (Database for Annotation, Visualization and Integrated Discovery, DAVID; http://david.abcc.ncifcrf.gov/) was used for the gene ontology (GO) cellular components analysis on the EV proteomes (Huang da et al., 2009).
2.16. Data analyses and statistics
Data trimming, distribution and cumulative curves fitting, peak detection and full width at half maximum (FWHM) calculation were performed using in‐house scripts written in Python version 3.7.4.
The resolution (Rs) of a column was calculated by Equation 1:
| (1) |
where Vproteins and VEVs represent the retention volume of the protein and EV peaks, respectively. The Rs was calculated according to the fitted distribution curves.
The particle recovery rate was defined as R = Ni/N, where N denotes the particle number used as the source material and Ni denotes the particle number recovered by each isolation method (Tian et al., 2020).
The Shapiro‐Wilk test was employed for the normality test. The Student's t‐test, and One‐way ANOVA tests were performed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA). Significant difference was accepted when P < 0.05.
3. RESULTS
3.1. Comparison of Sepharose CL beads for EV isolation with SEC
While packing SEC columns with Sepharose CL beads (CL‐2B, CL‐4B and CL‐6B) at the bed volume of 10 ml, 26 fractions were collected in each analysis. We first used FBS to optimize the resin selection. In general, we found that all resins had the same feature to elute FBS particles largely in the early fractions (Figure 2(a‐c)). With NTA analysis, the size distribution of particles in all fractions were primarily within the diameter range of 60–200 nm (Supplementary Figure S2). Meanwhile, most proteins were eluted in the late fractions (Figure 2(a‐c) and Supplementary Figure S3(a‐c)). As compared with the CL‐4B or CL‐6B columns, the CL‐2B column showed a longer particle peak tail and a broader protein peak in the fitted distribution curve. With HS from Donor 1, we reproduced similar distribution pattern observed in the FBS experiments (Figure 2(a‐c)). In both FBS and HS experiments, the particle peaks of CL‐4B and CL‐6B columns showed normal‐like distributions, while the CL‐2B column exhibited non‐normal distribution. Particularly, in HS experiments, the data point of cumulative 50% of particles appeared in Fraction 7 with the CL‐6B column, which was close to the centre of the particle peak (Figure 2(c)). In case of using the CL‐4B column, such a data point located at Fraction 8, deviated from the centre of particle peak of Fraction 6 (Figure 2(b)).
FIGURE 2.
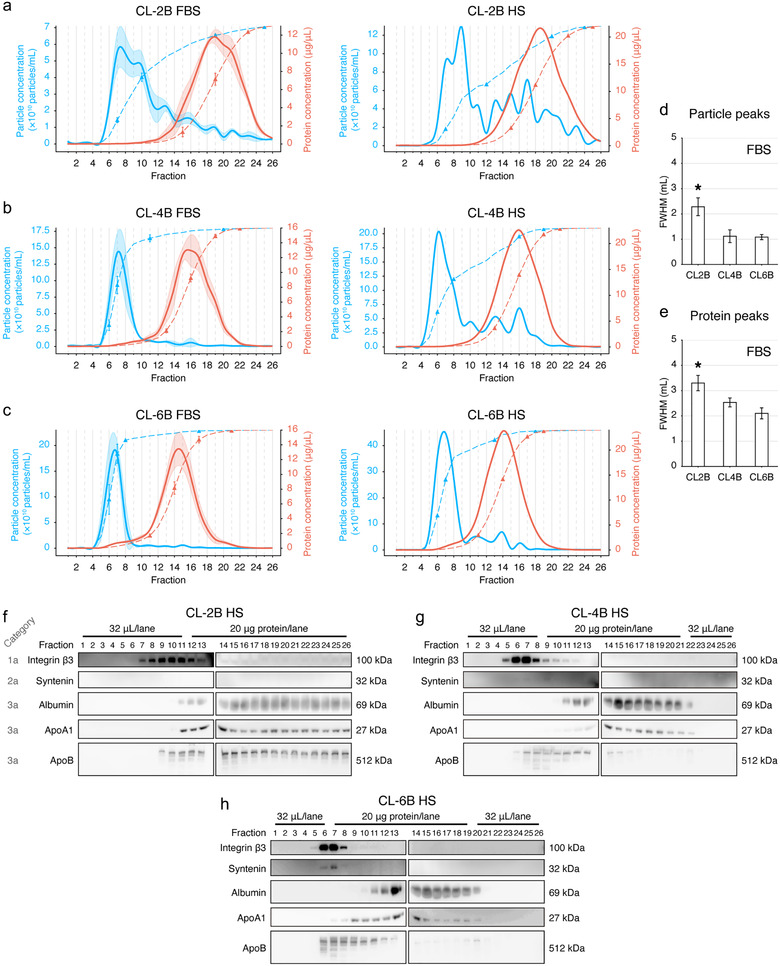
Resin optimization for SEC columns. (a,b,c) Fraction distribution of particle (blue) and protein (red) concentrations. Optimization using FBS (n = 3) and verification with human serum (HS) from Donor 1 are shown. FBS or HS samples were separated using the SEC columns with the bed volume of 10 ml that were packed with the resins of CL‐2B (a), CL‐4B (b) and CL‐6B (c), respectively. A total of 26 fractions were collected in each experiment, 500 μl of PBS was used for eluting each fraction. The dashed lines indicate the cumulative percentage. The 10%, 50%, 90% and 99% data points were labelled with triangles on each cumulative curve. The transparent shades illustrate the SD of each fraction. (d,e) Statistical comparison of the full width at half maximum (FWHM) of the particle (d) and protein (e) peaks in the FBS experiment. Data are shown as the mean±SD. *Significant difference, P < 0.05, Tukey's multiple comparisons test. (f,g,h) Immunoblotting of protein markers of EVs and contaminants for all SEC fractions using CL‐2B (f), CL‐4B (g) and CL‐6B (h) columns in the HS experiment. The sample loading volumes or sample loading protein amounts are labelled
When performing EV isolation from FBS, the CL‐2B columns had the FWHMs of 2.3±0.4 ml for particle peaks (Figure 2(d)) and 3.3±0.3 ml for protein peaks (Figure 2(e)), respectively. The FWHMs of CL‐2B columns were significantly greater than those of the CL‐4B and CL‐6B columns (Figure 2(d&e)), while CL‐4B and CL‐6B columns had no significant different FWHMs in terms of the particle (P = 0.99) (Figure 2(d)) and the protein (P = 0.15) peaks (Figure 2(e)), respectively. However, no significant difference was observed among the Rs of the CL‐2B (1.1 ± 0.2), the CL‐4B (1.4 ± 0.2) and the CL‐6B (1.5 ± 0.1) columns (P = 0.08).
We employed the protein markers suggested by MISEV2018 to estimate the purity of EVs isolated from HS. For the CL‐2B 10 ml column, the EV Category 1a marker integrin β3 was enriched in Fractions 7–13, which was overlapped with the contaminants of albumin (Category 3a, Fractions 11–26), ApoA1 (Category 3a, Fractions 11–26) and ApoB (Category 3a, Fractions 9–26) (Figure 2(f)).
For the CL‐4B 10 ml columns, integrin β3‐rich fractions (Fractions 5–10) were largely separated from albumin‐rich fractions (Fractions 11–22) and ApoA1‐rich fractions (Fractions 11–22) (Figure 2(g)). However, the VLDL marker of ApoB was still co‐enriched with the EV‐rich fractions at Fractions 6–10 (Figure 2(g)).
For the CL‐6B 10 ml columns, integrin β3‐rich fractions (Fractions 5–8) also tended to deviate from the albumin‐rich fractions (Fractions 10–20) and ApoA1‐rich fractions (Fractions 8–20) (Figure 2(h)). ApoB still appeared as a major contaminant of EVs as it was enriched at Fractions 6–13 (Figure 2(h)). Although the Category 1b marker of syntenin was undetectable or ambiguous in the CL‐2B (Figure 2(f)) or CL‐4B (Figure 2(g)) experiments, it was observed to be enriched in the Fractions 6 and 7 in the CL‐6B experiment (Figure 2(h)).
To be noted, the peak fractions of integrin β3 in the IB experiments (Figure 2(f‐g)) were all corresponding to the NTA particle peaks shown in Figure 2(a‐c), suggesting that particle concentrations estimated by NTA tended to be correlated to EV concentrations.
Taking these results together, Sepharose CL‐4B and CL‐6B were superior to CL‐2B in EV isolation with SEC; thus CL‐2B was not considered in the subsequent experiments.
3.2. Optimization of the CL‐4B and CL‐6B columns with increased bed volumes
We next proportionally increased the bed volume of SEC columns to 20 ml with total fraction number of 52 (500 μl per fraction).
Here, we still started from the optimization procedure using FBS. Through quantification, the FBS‐derived EV peaks were detected at the elution volumes of 6.9 ± 0.2 ml (Figure 3(a)) and 6.8 ± 0.1 ml (Figure 3(b)) using the CL‐4B and CL‐6B columns, respectively. In addition, their protein peaks were respectively located at the elution volumes of 16.0 ± 0.2 ml (Figure 3(a), Supplementary Figure S3(d)) and 14.8 ± 0.1 ml (Figure 3(b), Supplementary Figure S3(e)). Such a distribution pattern could be reproduced with HS from Donor 1. We observed that the data point of cumulative 50% of particles was located at the centre of CL‐6B particle peak (Fraction 13) (Figure 3(b)), while this data point was at Fraction 17 (∼4 fractions away from the centre) in the CL‐4B experiment (Figure 3(a)).
FIGURE 3.
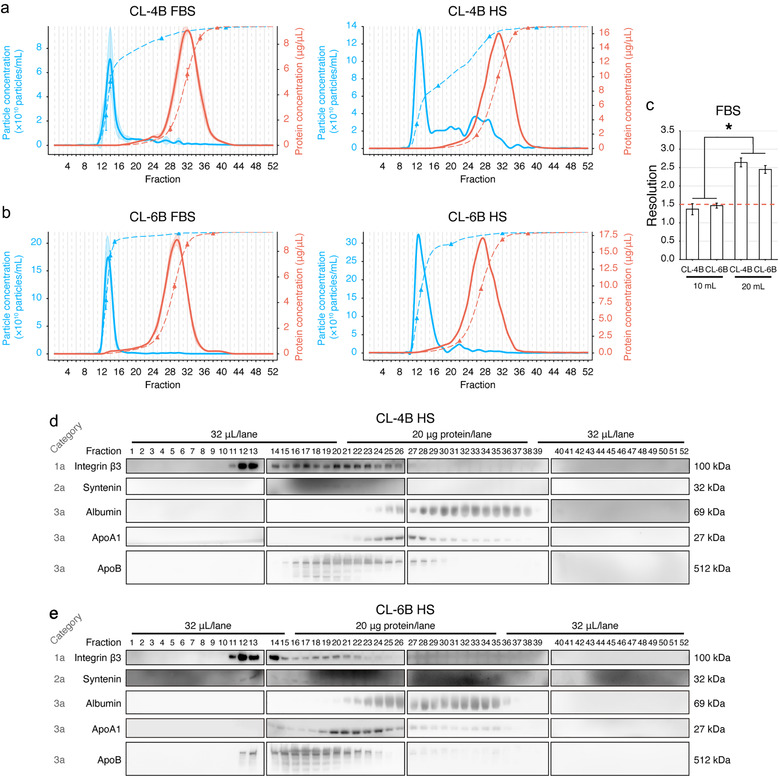
Optimization on the resin bed volume of SEC columns. (a,b) Fraction distribution of particle (blue) and protein (red) concentrations of CL‐4B (a) and CL‐6B (b) columns with the bed volume of 20 ml. Optimization using FBS (n = 3) and verification with human serum (HS) from Donor 1 are shown. A total of 52 fractions were collected in each experiment, 500 μl of PBS was used for eluting each fraction. The dashed lines indicate the cumulative percentage. The 10%, 50%, 90% and 99% data points were labelled with triangles on each cumulative curve. The transparent shades illustrate the SD of each fraction. (c) Statistical comparison of the resolution (Rs) in EV isolated from FBS. The dashed red line indicates the threshold of baseline separation (Rs = 1.5). *Significant difference (P < 0.05), n = 3, Tukey's multiple comparisons. (d,e) Immunoblotting of protein markers of EVs and contaminants for all SEC fractions using CL‐4B (d) and CL‐6B (e) columns in the HS experiment. The sample loading volumes or sample loading protein amounts are labelled
For both resins, increasing the bed volume from 10 ml to 20 ml significantly improved the Rs (Figure 3(c)). Especially, we found that the Rs of columns with the bed volume of 10 ml did not reach the baseline separation threshold of Rs = 1.5 (Barth, 2019). In contrast, the Rs values of CL‐4B and CL‐6B columns with the bed volume of 20 ml were 2.6 ± 0.1 and 2.5 ± 0.1, respectively, although they had no significantly difference (P = 0.25) (Figure 3(c)).
Per IB analysis, when using the CL‐4B 20 ml column to separate HS, integrin β3 was peaked in the Fraction 12 and 13, while abundant integrin β3 could still be observed in Fractions 14–26 (Figure 3(d)), indicating that EVs were eluted continuously and abundantly even after Fraction 16 (elution volume of 8 ml) until Fraction 26 (elution volume of 13). In the CL‐6B 20 ml experiment, integrin β3 was primarily enriched in the Fractions 11–14, although faint bands of integrin β3 could still be observed in Fractions 15–25 (Figure 3(e)). Albumin started to be eluted from Fraction 24 and Fraction 21 in the CL‐4B (Figure 3(d)) and CL‐6B (Figure 3(e)) experiments, respectively. ApoA1 started to be eluted from Fraction 22 and Fraction 20 in the CL‐4B (Figure 3(d)) and CL‐6B (Figure 3(e)) experiments, respectively. These fractions were distant from the particle peaks in either CL‐4B or CL‐6B experiments. However, there were still abundant ApoB co‐enriched in integrin β3‐rich fractions in both CL‐4B and CL‐6B experiments.
In addition, when summing up the recovered particles in all fractions from the above conditions, CL‐4B 20 ml column yielded significantly less particles as compared with other columns (Supplementary Figure S4).
With the above results, the CL‐6B column with bed volume of 20 ml was adopted for the subsequent analyses considering its relatively more concentrated particle peaks, low level of contaminants, and high level of EV yield.
3.3. Evaluation of the dichotomic SEC for EV isolation with FBS
Using the CL‐6B column with bed volume of 20 ml, we next simplified the isolation workflow with FBS into a dichotomic manner, i.e. separating EVs and proteins into two eluates (Eluate 1 and Eluate 2) with two consecutive bulk elutions (Figure 4(a)). The 30 kDa MWCO UF devices were used to concentrate the eluates to 200 μl.
We compared the performance of 7.5, 8, 8.5, 9 and 9.5 ml of PBS as the first bulk elution for EV acquisition. The eluent volumes of the second bulk elution were correspondingly set as 18.5, 18, 17.5, 17 and 16.5 ml. As such, the total eluent volume was the same of 26 ml for each SEC separation. In all bulk elution combinations, the Eluate 1s showed a significantly higher level of particle concentration (Figure 4(b)) and a significantly lower level of protein concentration (Figure 4(c)) compared to that in Eluate 2s. No significant difference was observed among the particles recovered in Eluate 1s (P = 0.64) or Eluate 2s (P = 0.9997) with different bulk elution combinations (Figure 4(b)). Similarly, no significant difference was observed among the proteins recovered in different Eluate 2s (P = 0.63) (Figure 4(c)).
Parallel to the significantly augmented protein concentration in Eluate 1s (Figure 4(c)), the particle to protein ratios decreased along with the increased bulk elution volumes (Figure 4(d)). The particle to protein ratios of Eluate 1s in the 7.5 ml and the 8 ml groups were significantly higher than either the 9 ml or the 9.5 ml groups (Figure 4(d)). Thus, Eluate 1s with 7.5 ml and 8 ml bulk elutions had sufficient ability to separate particles from proteins in FBS. For better particle yields, the Eluate 1 with 8 ml of bulk elution was used for subsequent analyses. With such a condition, we further validated that particles could be well recovered (93.8±13.8)% in the Eluate 1, while (3.3 ± 0.6)% particles could be observed in the Eluate 2 (Figure 4(e)).
We next tested whether the reused column would affect the particle isolation performance. Three SEC columns were employed to isolate particles from the same volume (1 ml) of FBS for 10 consecutive times, followed by cleaning and regeneration between each adjacent reuses. An eluate with PBS was used for determining the leftover particles in each reused column. In every column, particles (Figure 4(f)) and proteins (Figure 4(g)) were predominantly separated into the Eluate 1 and the Eluate 2, respectively. Comparing columns reused for different times, no significantly different particle or protein concentrations were observed among either Eluate 1s or Eluate 2s (all P > 0.05 by Dunnett's multiple comparisons test), respectively. No leftover particles were observed between reuses as all PBS eluates from regenerated columns had no significant difference from the PBS background (P = 0.61) (Figure 4(h)). Protein concentrations were all below the limit of detection of BCA for all PBS background obtained from regenerated columns, showing that no protein leftover in reused columns.
At this point, an SEC‐based dichotomic method for EV isolation was established by combining the CL‐6B column with the bed volume of 20 ml and 30 kDa UF for concentrating the eluates.
3.4. Application of the dichotomic SEC method to HS
We next tested the feasibility of isolating EVs from HS using this dichotomic SEC. We provided a video record to demonstrate the procedure and operation of this method, showing that it took ∼22 min starting from sample (1 ml) loading to obtain the Eluate 1 (Supplementary Video 1). Per NTA, much higher particle concentration (approximately 1 order of magnitude higher) could be acquired from HS (Figure 5(a)) than FBS (Figure 4(b)) with the same 1 ml sample inputs. The particle concentration in Eluate 1s ((3.4 ± 1.8) × 1012 particles/ml) was significantly higher than Eluate 2s ((2.1 ± 0.3) × 1011 particles/ml) (Figure 5(a)). The protein concentrations in the Eluate 2 were at ∼240 μg/μl level, which were significantly higher than that in the Eluate 1 (∼3.3 μg/μl level; Figure 5(b)). In Eluate 1s, the dichotomic SEC yielded a particle/protein ratio of (1.2 ± 0.9) × 109 particles/μg protein.
FIGURE 5.
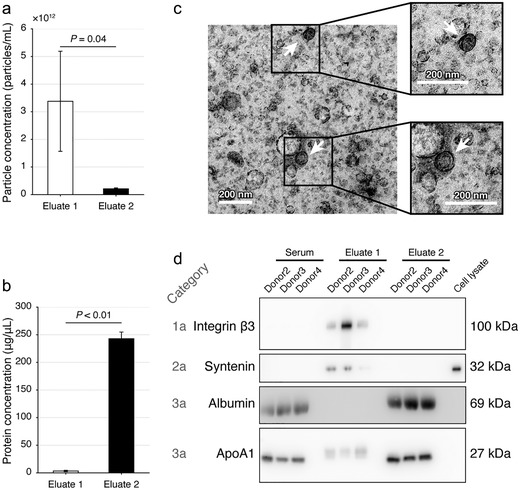
Evaluation of the dichotomic SEC method using human serum. (a, b) The particle (a) and protein (b) concentrations of the Eluate 1 and 2 with the dichotomic SEC method. Data are shown with mean±SD, donor n = 3, Student's t‐test. (c) Representative transmission electronic microscopy images of the Eluate 1. Scale bar = 200 nm. (d) Immunoblotting verification of the protein markers. The cell lysate was acquired from SW620 cells
With TEM, we observed that a variety of small cup‐shaped particles with ∼200 nm in diameter could be visually confirmed in Eluate 1 (Figure 5(c)). With IB, remarkable enrichment of integrin β3 and syntenin was observed in the Eluate 1 as compared with HS or Eluate 2 (Figure 5(d)). Albumin and ApoA1 were notably depleted from Eluate 1s as compared with HS or Eluate 2s (Figure 5(d)).
These NTA, TEM, particle/protein ratio and protein marker results justified that enriched EVs were obtained in the Eluate 1 from HS.
3.5. Evaluation of dichotomic SEC with SW620 cell culture supernatants
We next applied this dichotomic SEC method to FBS‐free cell culture supernatants, a less complex but equally important biofluid in both clinical and basic researches. As compared with the Eluate 2, the Eluate 1 had significantly higher level of particles (P < 0.01) (Figure 6(a)) and significantly lower level of proteins (P < 0.01) (Figure 6(b)).
FIGURE 6.
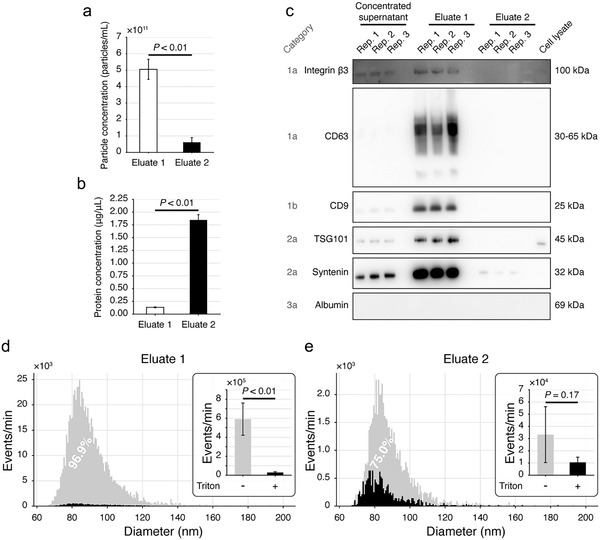
Evaluation of the dichotomic SEC method with SW620 cell culture supernatants. (a,b) The particle (a) and protein (b) concentrations of the two eluates. (c) Immunoblotting verification of the protein markers. (d,e) Size distribution and purity of the particles in Eluate 1 (d) and Eluate 2 (e) determined by nFCM. Triton X‐100 treatment was used to estimate the purity of EVs. All data were acquired from 3 biological replicates. Data are shown as mean±SD, Student's t‐test
IB results showed that the Eluate 1 enriched remarkably higher level of EV markers including integrin β3, CD63, TSG101, syntenin and CD9 compared to the Eluate 2 (Figure 6(c)). No albumin was observed in the Eluate 1, the Eluate 2 or SW620 cell lysate (Figure 6(c) and Supplementary Figure S1).
For estimating EV purities, we prepared SW620 supernatants, followed by the determination of Triton X‐100 disruptable particles in the Eluate 1 and the Eluate 2 using nFCM. The particle concentrations measured by NTA were (5.0 ± 0.8) × 1011 particles/ml and (6.6 ± 2.7) × 1010 particles/ml (n = 3) in Eluate 1s and Eluate 2s, respectively. With nFCM, a SiNP cocktail containing 68, 91, 113 and 155 nm beads was employed to fit a standard curve for particle size determination (Supplementary Figure S5). Most particles in the Eluate 1 (Figure 6(d)) and the Eluate 2 (Figure 6(e)) were distributed between sizes of 65 to 120 nm. After being treated with 1% Triton X‐100, > 95% particles in the Eluate 1 were disrupted and the number of events detected by the nFCM decreased significantly (P < 0.01) (Figure 6(d)). Meanwhile, ∼75% of the particles in Eluate 2 could also be lysed by the Triton X‐100 (Figure 6(e)). Such a significant event loss through Triton X‐100 treatment in the Eluate 1 indicated its high purity of EVs.
3.6. Application of the dichotomic SEC to isolate fluorescent EVs
CFDA‐SE labelled SW620 cells were used for releasing fluorescent EVs, followed by the EV isolation with the dichotomic SEC. Over 96% of cells could be labelled with 5 μM of CFDA‐SE (Supplementary Figure S6(a)), while we observed no significant cell viability changes comparing labelled and unlabelled cells (Supplementary Figure S6(b)).
Through nFCM analysis, fluorescent EVs mostly appeared as peaks with lower than the green fluorescence intensity (FI) of 250 and above the fluorescence baseline (Figure 7(a)). Interestingly, we observed that the fluorescence baseline of the Eluate 2 was much higher than the Eluate 1. CFDA‐SE was a known protein labelling dye, such baseline elevation in the Eluate 2 echoed that proteins were mostly enriched in this eluate (Figure 7(a)).
FIGURE 7.
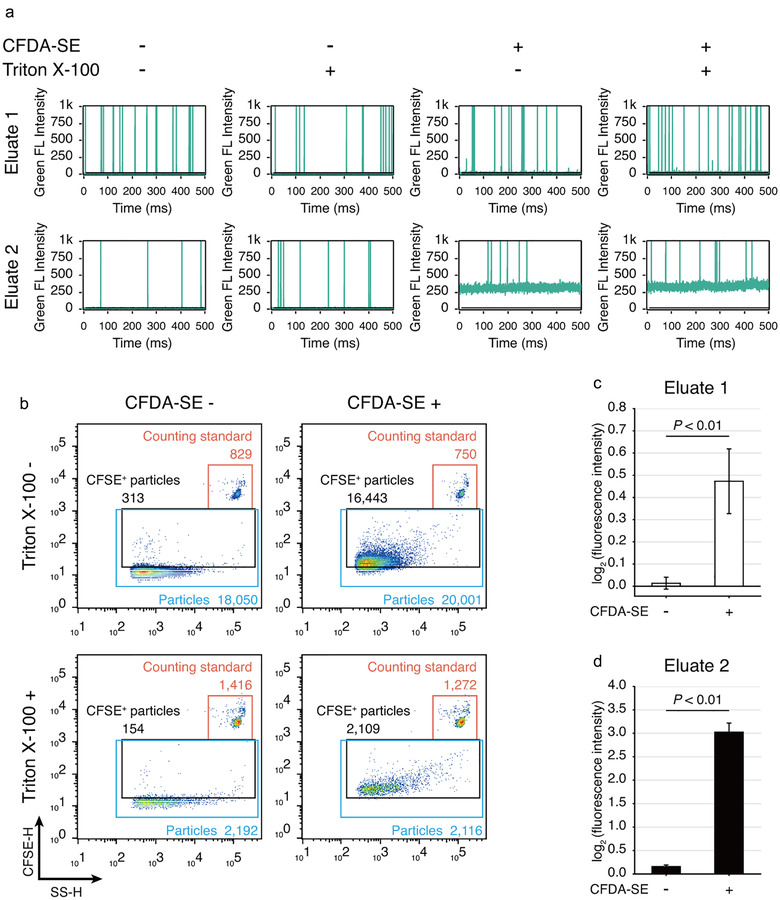
Application of the dichotomic SEC method to isolate fluorescent EVs released from CFDA‐SE labelled SW620 cells. (a) Representative green fluorescence burst traces obtained from nFCM. Culture supernatant was harvested from CFDA‐SE labelled SW620 cells and subjected to EV isolation. Each sample was treated in parallel with or without Triton X‐100. (b) Representative scatter plots of nFCM analysis on the Eluate 1. (c,d) Statistical analysis of the fluorescence intensity of Eluate 1 (c) and Eluate 2 (d) measured by a fluorometer. Data are shown as mean±SD, n = 3, Student's t‐test
As compared with the CFDA‐SE‐unlabelled group, 95.5% particles in the Eluate 1 isolated from supernatants of CFDA‐SE‐labelled cells showed positive fluorescent signal (Figure 7(b)). In addition, 92.4% of such fluorescent particles could be disrupted by Triton X‐100 (Figure 7(b)). With fluorometry, we confirmed that CFDA‐SE significantly increased the FI of the Eluate 1 (Figure 7(c)) and the Eluate 2 (Figure 7(d)).
3.7. Proteome comparison of EVs isolated by dichotomic SEC and UC
As no method can isolate completely “pure” EVs, proteomic analysis may help to holistically illustrate the quality of EV isolation. Here, EVs isolated from PFP by the dichotomic SEC (SEC‐EVs) and UC (UC‐EVs) were subjected to the proteomics profiling and quantification via DIA‐MS analyses (Figure 8).
FIGURE 8.
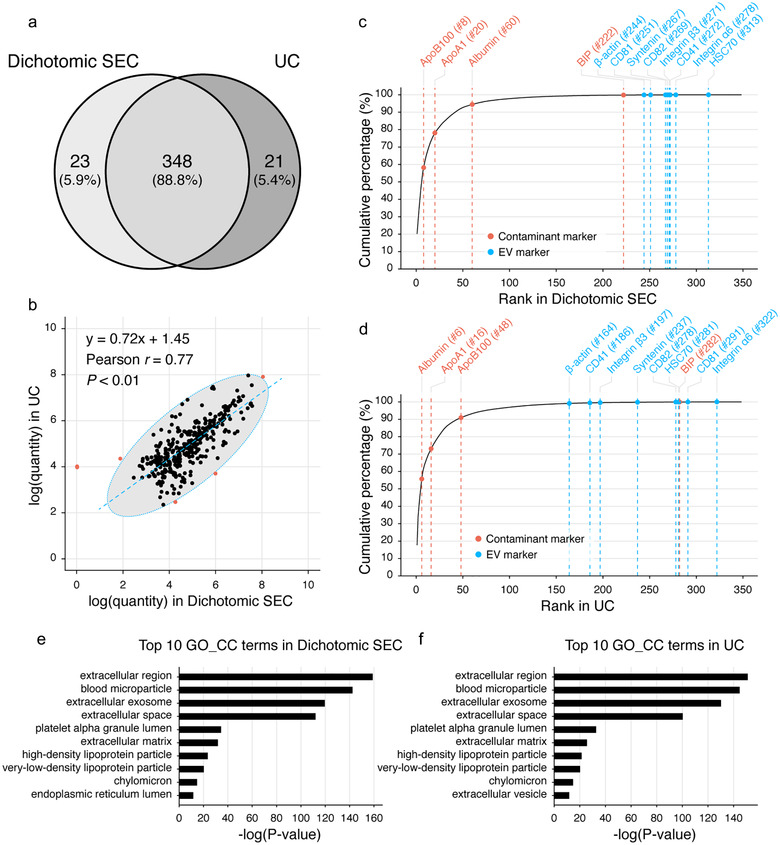
Proteomics comparison of plasma EVs isolated by the dichotomic SEC and UC. (a) Venn diagram comparison of the EV proteomes. (b) Correlation of protein quantities. Outliers (red dots) are determined and excluded through the confidence ellipse which contains 99.4% (three times of standard deviations, 3σ) of all data points. (c,d) Cumulative curves of the protein quantities of EVs isolated with the dichotomic SEC (c) and UC (d). Proteins are ranked and aligned based on their quantity values from high to low. Protein names of representative protein markers of EVs (blue) and common contaminants (red), together with their respective ranks are labelled above each curve. (e,f) Top 10 Gene ontology cellular component (GO_CC) terms enriched by the DAVID tool
We identified and quantified a total of 392 protein groups in either SEC‐EVs or UC‐EVs (Figure 8(a), Supplementary Table S1). Among them, 88.8% (348/392) protein groups were detected in both EVs, while 5.9% (23/392) and 5.4% (21/392) protein groups could only be found in SEC‐EVs or UC‐EVs, respectively. When analysing the 348 overlapped proteins, SEC‐EV and UC‐EV protein quantities were significantly correlated with a Pearson r of 0.77 (P < 0.01) (Figure 8(b)).
We then plotted cumulative curves of protein quantities for each EV proteome, and the proteins were ranked from high to low per quantities (Figure 8(c) and (d)). A total of 12 protein markers of EVs or contaminants listed in MISEV2018 could be quantified in both proteomes (Figure 8(c) and (d)). Among them, EV markers including CD81, CD82, integrin α6, integrin β3, CD41, syntenin, heat shock protein HSC70 and β‐actin shared similar ranking between the SEC‐EVs and UC‐EVs (Figure 8(c) and (d)). Regarding the contaminant markers, we observed that ApoB100 was one of the major co‐isolated contaminants in SEC‐EVs (rank #8, taking over 2.9% of the total protein quantities; Figure 8(c)), while UC‐EVs had less ApoB100 contamination (rank #48, taking over 0.24% of total protein quantities; Figure 8(d)). However, albumin (rank #6, taking over 4.7% of all protein quantities; Figure 8(d)) appeared to be a major contaminant in UC‐EVs, while SEC‐EVs had much less albumin contamination (rank #60, taking over 0.16% of total protein quantities; Figure 8(c)). As to other co‐isolating contaminants, similar amounts of ApoA1 (rank #20 in SEC‐EVs and #16 in UC‐EVs) and BIP (rank #222 in SEC‐EVs and #282 in UC‐EVs) were observed.
Per GO_CC enrichment, the identified proteins in SEC‐EVs and UC‐EVs were enriched in highly similar terms (Figure 8(e) and (f)). The top 10 GO_CC terms of proteins identified from SEC‐EVs (Figure 8(e)) and UC‐EVs (Figure 8(f)) were almost identical and targeting EV properties. These proteomics results implicated that although with difference, the dichotomic SEC and UC could isolate EVs from human plasma with comparable purity.
4. DISCUSSION
In this study, as discussed below, we addressed several key questions leading to the establishment of a dichotomic SEC method for EV separation toward clinical applications.
We focused on serum samples in the optimization stage as they represented as one of the most complex sample types that were used in laboratory medicine. Indeed, the resin evaluation has been widely emphasized by numerous groups for EV isolations from plasma/serum. For example, Boing et al have found that in platelet‐free human plasma, Sepharose CL‐2B based SEC can remove most albumin from EVs, although the isolation efficiency is inadequate (Boing et al., 2014). Benedikter et al have found that in cell culture supernatants with EV‐depleted FBS, EVs are enriched in Fractions 5–10/11, and proteins are eluted in Fractions 13–20 with CL‐4B columns (Benedikter et al., 2017). Baranyai et al have shown that albumin in plasma can be efficiently removed by using CL‐4B or Sephacryl S‐400 columns (Baranyai et al., 2015). Until very recently, Ter‐Ovanesyan et al proposed that CL‐6B could also be used for the SEC EV isolations from human plasma (Preprint available from: https://doi.org/10.1101/2020.10.13.337881). In this study, we found that both CL‐4B and CL‐6B were superior to CL‐2B in separation of EVs from proteins in both FBS and HS.
It is known that the pore size and bead diameter of resins have great influences on the fractionation performance of SEC columns (Monguio‐Tortajada et al., 2019). Here, the pore size of CL‐2B is 75 nm, larger than that of CL‐4B (42 nm) or CL‐6B (24 nm), while these resins have similar bead diameters (CL‐6B: 40–165 μm; CL‐4B: 45–165 μm; CL‐2B: 60–200 μm). Per SEC theory, EVs under the size of 75 nm in diameter will be retained by the CL‐2B resins, which may result in the tailing of the EV peaks acquired from such a column. Hence, the smaller pore sizes may serve as a possible factor contributing to the better performance of CL‐4B and CL‐6B. Such a hypothesis is partially favoured by Lane et al, implicating that the CL‐4B column has tighter elution profile and increased particle yield than the CL‐2B column in SEC on cell culture supernatants, while similar particle/protein ratios are observed in the EV fractions of the two types of columns (Lane et al., 2019). They also proposed that such difference could be driven by the smaller pore size of CL‐4B (Lane et al., 2019).
We next justified that whether the increased bed volume could further improve the performance of CL‐4B and CL‐6B columns. According to the rate theory of chromatography (McNaught & Wilkinson, 1997), an increase in the number of theoretical plates will lead to an increase in retention time and broadening the bands simultaneously. Using columns with the same inner radius, the bed volume increase is equivalent to the augmented number of theoretical plates. In this study, we showed that by increasing the bed volume from 10 ml to 20 ml, the Rs values of both CL‐4B and CL‐6B columns in the FBS experiments could be significantly improved to > 1.5. Together with purity estimate through IB analysis on HS EVs, we confirmed that CL‐6B columns had better performance than CL‐4B in separating EVs and proteins. In accordance, Ter‐Ovanesyan et al reported that for CL‐2B, CL‐4B or CL‐6B columns, increasing the bed volumes from 10 ml to 20 ml could remove more albumin (Preprint available from: https://doi.org/10.1101/2020.10.13.337881). In this optimization stage, our data highlighted that CL‐6B 20 ml columns performed well with the complex biofluids such as serum. But, for other sample types with different complexities or specific downstream applications, goal‐oriented adjustments may still be needed.
We further proved that the dichotomic SEC method with the resin of 20 ml CL‐6B and bulk elution could acquire EVs from FBS or HS. As proposed by numerous reports, the early fractions of SEC are often not collected as the EV peaks mainly appeared in the midway fractions. However, even with the same column setting, there are multiple collection and/or pooling choices for EV isolation. For example, with CL‐4B columns, #7‐9 (Lane et al., 2019) and #5‐10/11 (Benedikter et al., 2017) have been used for EV isolation. While with CL‐2B columns, #6‐8 (Lane et al., 2019) or only the #4 (Ludwig et al., 2019) fraction have been collected for EVs. With the commercialized qEV column, the #7‐9 (Tian et al., 2020), #7‐10 (Stranska et al., 2018) or #7‐12 (Takov et al., 2019) fractions have been pooled. The dichotomic SEC method proposed in this study could simplify the SEC procedure by avoiding multiple fractionation and pooling operations. Such an ease‐to‐use feature allows increased reproducibility in clinical settings, while acquiring high quality EVs with high particle recovery rate. Comparably, Tian et al have shown that qEV can achieve the particle recovery rate of 64.7±13.1%, whereas UC and UF can yield the particle recovery rates of 39.6±4.6% and 36.9±2.4%, respectively (Tian et al., 2020). In addition, we showed that most particles isolated by the dichotomic SEC had the sizes ranged 60–200 nm in diameter as determined by NTA, which were in the size range of sEVs.
Equally important is that the robustness of this dichotomic method is notable. First, we showed that the performance of acquiring EVs were not significantly affected upon bulk elution with 7.5‐9.5 ml of PBS. Second, we showed that the CL‐6B column could be reused for at least 10 times with no observable particle and protein leftover or declined performance. Indeed, to our knowledge, there is no report specifically addressing the recyclability of SEC columns for EV isolation. In addition, this feature is useful for the EV preparation, for example in the field of cell‐free therapy (Lai et al., 2011).
Furthermore, we demonstrated that the dichotomic SEC was applicable to the EV isolation from cell culture supernatants. We showed that both fluorescent EVs and protein particles could be distinguished by nFCM. It is known that CFDA‐SE will be cleaved to generate carboxyfluorescein succinimidyl ester (CFSE) post entering cells. CFSE binds to protein amine groups via its succinimidyl ester group, and becomes stably fluorescent. CFSE‐labelled cells have been widely used for the preparation of fluorescent EVs (Tian et al., 2013). To be noted, we found with the nFCM analysis that the Eluate 2 became highly fluorescent, indicating that CFSE‐labelled protein particles were enriched in Eluate 2. This further justified the successful separation of fluorescent EVs from proteins with this dichotomic SEC method.
It should be emphasized that the dichotomic SEC method cannot avoid known co‐isolating protein contaminants in regular SEC for EV isolation. For example, Karimi et al used a 10 ml CL‐2B column to separate plasma EVs, in which severe lipoprotein contaminations were detected (Karimi et al., 2018). Such an observation was comparable to our results showing that CL‐2B 10 ml columns had remarkably poorer performance than CL‐4B and CL‐6B columns. The IB results shown in this study also implicated that lipoproteins could not be completely removed from the particle fractions with any SEC columns tested. Interestingly, Karimi et al further performed proteomics analysis on purified plasma EVs with combination of UC, density cushion and SEC (Karimi et al., 2018). Comparing proteomics results from Karimi et al (Karimi et al., 2018) and us, the EV‐relevant GO_CC terms were overlapped in general, while the contamination terms of VLDL, HDL and chylomicron were found in our data, but avoided by Karimi et al, suggesting their better EV purity. In this study, we also provided a proteomics comparison of the dichotomic SEC‐EVs and UC‐EVs. The top 10 GO_CC terms of the two proteomic datasets were almost identical, with only 1 term difference. We could also show that approximately 2.9% SEC‐EV proteins and only 0.24% of UC‐EV proteins were ApoB100. However, UC‐EV proteins had 4.9% albumin contamination, while it was only 0.16% for SEC‐EV proteins. These results suggest that the dichotomic SEC and UC have dissimilar abilities in removing different co‐isolating contaminant proteins in EV isolations. Using the purity estimation method reported by Tian et al (Tian et al., 2020), we could show that a 96.9% purity could be achieved when using dichotomic SEC to isolate EVs from cell culture supernatants. However, we could not do such a purity estimation on serum samples, as lipoproteins could be disrupted by Triton X‐100 into small particles as pointed out by Tian et al (Tian et al., 2020).
With the dichotomic SEC to isolate EVs from HS, we showed that the particle/protein ratio was (1.2 ± 0.9) × 109 particles/μg protein. There are comparable results reported by other groups. Brennan et al showed that the qEV for HS EV isolation could obtain the particle/protein ratios ranged 108∼109 particles/μg protein (Brennan et al., 2020). Yang et al performed regular SEC on HS, and they observed the particle/protein ratios to be ranged from 7 × 108∼1 × 109 particles/μg protein (Yang et al., 2021). Takov et al found that the particle/protein ratio of ∼1.9 × 108 particles/μg protein could be achieved from the EV isolation from rat plasma using the qEVorigenal SEC columns (Takov et al., 2019).
In conclusion, we successfully established a simplified and robust dichotomic SEC method to separate EVs and proteins from FBS, human serum and FBS‐free cell culture supernatants. This dichotomic SEC method, characterized according to MISEV2018 recommendations, only requires two bulk elutions of loaded samples to acquire EVs in the Eluate 1 and proteins in the Eluate 2, which does not need multiple fractionation and pooling operations. The keys to this method are the selection of CL‐6B as the column resin, the increase of bed volume to 20 ml, and the bulk elution volume of 8 ml. Such a method has its intriguing potential to be used for the EV preparation toward clinical testing and/or basic researches.
CONFLICT OF INTEREST
No potential competing interest was reported by the authors.
AUTHOR CONTRIBUTIONS
Yizhi Cui and Tong Wang conceived the idea and directed the entire study. Yizhi Cui and Jiahui Guo designed and performed most of the experiments. Caihong Wu contributed to the experiments for resin selection, and nFCM analysis. Xinyi Lin performed proteomics and contributed to immunoblotting; Jian Zhou and Jiayi Zhang contributed to purity evaluations; Wenting Zheng performed the validation experiments with HS samples. Yizhi Cui and Jiahui Guo analysed the data. Yizhi Cui, Tong Wang and Jiahui Guo wrote the paper. All authors reviewed and approved the manuscript.
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGEMENTS
This work was supported by the National Key R&D Program of China to T.W. (2020YFE0202200); National Science and Technology Major Project of China to T.W. (2018ZX10732‐101‐002); the National Natural Science Foundation of China (31800692 to Y.C., 81973353 to T.W.); the Fund of Innovation and Entrepreneurship Leading Team Project of Guangzhou to T.W. (201809010009); the Guangdong Key Project for Research and Development to T.W. (2016B020238002); the Special Fund of Foshan Summit Plan to Y.C. (2019C020).
Guo, J., Wu, C., Lin, X., Zhou, J., Zhang, J., Zheng, W., Wang, T., & Cui, Y. (2021). Establishment of a simplified dichotomic size‐exclusion chromatography for isolating extracellular vesicles toward clinical applications. Journal of Extracellular Vesicles, 10, e12145. 10.1002/jev2.12145
Contributor Information
Tong Wang, tongwang@email.jnu.edu.cn.
Yizhi Cui, cuiyizhi01@email.jnu.edu.cn.
REFERENCES
- Ayers, L., Pink, R., Carter, D. R. F., & Nieuwland, R. (2019). Clinical requirements for extracellular vesicle assays. Journal of Extracellular Vesicles, 8(1), 1593755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachurski, D., Schuldner, M., Nguyen, P. H., Malz, A., Reiners, K. S., Grenzi, P. C., Babatz, F., Schauss, A. C., Hansen, H. P., Hallek, M., & von Strandmann, E. P. (2019). Extracellular vesicle measurements with nanoparticle tracking analysis ‐ An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. Journal of Extracellular Vesicles, 8(1), 1596016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranyai T., Herczeg K., Onódi Z., Voszka I., Módos K., Marton N., Nagy G., Mäger I., Wood M. J., El Andaloussi S., Pálinkás Z., Kumar V., Nagy P., Kittel Á., Buzás E. I., Ferdinandy P., Giricz Z. (2015). Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLOS ONE, 10(12), e0145686. 10.1371/journal.pone.0145686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, H. G. (2019). Chromatography Fundamentals, Part VIII: The Meaning and Significance of Chromatographic Resolution.
- Benedikter, B. J., Bouwman, F. G., Vajen, T., Heinzmann, A. C. A., Grauls, G., Mariman, E. C., Wouters, E. F. M., Savelkoul, P. H., Lopez‐Iglesias, C., Koenen, R. R., Rohde, G. G. U., & Stassen, F. R. M. (2017). Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies. Scientific Reports, 7(1), 15297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens, M. C., Wu, T. J., Nabet, B. Y., Xu, B., Qiu, Y., Yoon, T., Azzam, D. J., Twyman‐Saint Victor, C., Wiemann, B. Z., Ishwaran, H., Brugge, P. J. T., Jonkers, J., Slingerland, J., & Minn, A. J. (2014). Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell, 159(3), 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boing, A. N., van der Pol, E., Grootemaat, A. E., Coumans, F. A., Sturk, A., Nieuwland, R. (2014). Single‐step isolation of extracellular vesicles by size‐exclusion chromatography. Journal of Extracellular Vesicles, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, K., Martin, K., FitzGerald, S. P., O'Sullivan, J., Wu, Y., Blanco, A., Richardson, C., & Gee, M. M. Mc (2020). A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Scientific Reports, 10(1), 1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri, M. C., Quarto, R., & Tasso, R. (2021). Extracellular vesicles as biomarkers and therapeutic tools: From pre‐clinical to clinical applications. Biology (Basel), 10(5), 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, A., Boilard, E., Buzas, E. I., Cheng, L., Falcon‐Perez, J. M., Gardiner, C., Gustafson, D., Gualerzi, A., Hendrix, A., Hoffman, A., Jones, J., Lässer, C., Lawson, C., Lenassi, M., Nazarenko, I., O'Driscoll, L., Pink, R., Siljander, P. R. ‐ M., Soekmadji, C., … Nieuwland, R. (2019). Considerations towards a roadmap for collection, handling and storage of blood extracellular vesicles. Journal of Extracellular Vesicles, 8(1), 1647027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescitelli R., Lässer C., Szabó T. G., Kittel A., Eldh M., Dianzani I., Buzás E. I., Lötvall J. (2013). Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. Journal of Extracellular Vesicles, 2(1), 20677. 10.3402/jev.v2i0.20677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, L., Zieren, R. C., Horie, K., Kim, C. J., Mallick, E., Jing, Y., Feng, M., Kuczler, M. D., Green, J., Amend, S. R., Witwer, K. W., de Reijke, T. M., Cho, Y ‐ K., Pienta, K. J., & Xue, W. (2020). Comprehensive evaluation of methods for small extracellular vesicles separation from human plasma, urine and cell culture medium. Journal of Extracellular Vesicles, 10(2), e12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, L. M., & Wang, M. Z. (2019). Overview of Extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells, 727, 8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J., Cui, Y., Yan, Z., Luo, Y., Zhang, W., Deng, S., Tang, S., Zhang, G., He, Q. Y., & Wang, T. (2016). Phosphoproteome characterization of human colorectal cancer SW620 cell‐derived exosomes and new phosphosite discovery for C‐HPP. Journal of Proteome Research, 15(11), 4060–4072. [DOI] [PubMed] [Google Scholar]
- Holcar M., Ferdin J., Sitar S., Tušek‐Žnidarič M., Dolžan V., Plemenitaš A., Žagar E., Lenassi M. (2020). Enrichment of plasma extracellular vesicles for reliable quantification of their size and concentration for biomarker discovery. Scientific Reports, 10(1), 21346. 10.1038/s41598-020-78422-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da, W., Sherman, B. T., & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4(1), 44–57. [DOI] [PubMed] [Google Scholar]
- Karimi N., Cvjetkovic A., Jang S. C., Crescitelli R., Hosseinpour Feizi M. A., Nieuwland R., Lötvall J., Lässer C. (2018). Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cellular and Molecular Life Sciences, 75(15), 2873–2886. 10.1007/s00018-018-2773-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal, J., Arras, G., Colombo, M., Jouve, M., Morath, J. P., Primdal‐Bengtson, B., Dingli, F., Loew, D., Tkach, M., & Thery, C. (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proceedings of the National Academy of Sciences of the United States of America, 113(8), E968‐E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, R. C., Chen, T. S., & Lim, S. K. (2011). Mesenchymal stem cell exosome: A novel stem cell‐based therapy for cardiovascular disease. Regen Med, 6(4), 481–492. [DOI] [PubMed] [Google Scholar]
- Lane, R. E., Korbie, D., Trau, M., & Hill, M. M. (2019). Optimizing size exclusion chromatography for extracellular vesicle enrichment and proteomic analysis from clinically relevant samples. Proteomics, 19(8), 1800156. [DOI] [PubMed] [Google Scholar]
- Lener T., Gimona M., Aigner L., Börger V., Buzas E., Camussi G., Chaput N., Chatterjee D., Court F. A., Portillo H. A. d., O'Driscoll L., Fais S., Falcon‐Perez J. M., Felderhoff‐Mueser U., Fraile L., Gho Y. S., Görgens A., Gupta R. C., Hendrix A., Hermann D. M., Hill A. F., Hochberg F., Horn P. A., Kleijn D. d., Kordelas L., Kramer B. W., Krämer‐Albers E., Laner‐Plamberger S., Laitinen S., Leonardi T., Lorenowicz M. J., Lim S. K., Lötvall J., Maguire C. A., Marcilla A., Nazarenko I., Ochiya T., Patel T., Pedersen S., Pocsfalvi G., Pluchino S., Quesenberry P., Reischl I. G., Rivera F. J., Sanzenbacher R., Schallmoser K., Slaper‐Cortenbach I., Strunk D., Tonn T., Vader P., Balkom B. W. M. v., Wauben M., Andaloussi S. E., Théry C., Rohde E., Giebel B. (2015). Applying extracellular vesicles based therapeutics in clinical trials – an ISEV position paper. Journal of Extracellular Vesicles, 4(1), 30087. 10.3402/jev.v4.30087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T, Yan, Y, Wang, B, Qian, H, Zhang, X, Shen, L, Wang, M, Zhou, Y, Zhu, W, Li, W, Wenrong, Xu (2013). Exosomes derived from human umbilical cord mesenchymal stem cells alleviate liver fibrosis. Stem Cells and Development, 22(6), 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Zhang J., Lian X., Sun L., Meng K., Chen Y., Sun Z., Yin X., Li Y., Zhao J., Wang T., Zhang G., He Q. (2019). A hidden human proteome encoded by ‘non‐coding’ genes. Nucleic Acids Research, 47(15), 8111–8125. 10.1093/nar/gkz646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, N., Razzo, B. M., Yerneni, S. S., & Whiteside, T. L. (2019). Optimization of cell culture conditions for exosome isolation using mini‐size exclusion chromatography (mini‐SEC). Experimental Cell Research, 378(2), 149–157. [DOI] [PubMed] [Google Scholar]
- Ma, J., Chen, T., Wu, S., Yang, C., Bai, M., Shu, K., Li, K., Zhang, G., Jin, Z., He, F., Hermjakob, H., & Zhu, Y. (2019). iProX: An integrated proteome resource. Nucleic Acids Resource, 47(D1), D1211–D1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, C., Xue, C., Wang, X., He, S., Wu, L., & Yan, X. (2020). Rapid quantification of pathogenic Salmonella Typhimurium and total bacteria in eggs by nano‐flow cytometry. Talanta, 217, 121020. [DOI] [PubMed] [Google Scholar]
- Mateescu B., Kowal E. J. K., van Balkom B. W. M., Bartel S., Bhattacharyya S. N., Buzás E. I., Buck A. H., de Candia P., Chow F. W. N., Das S., Driedonks T. A. P., Fernández‐Messina L., Haderk F., Hill A. F., Jones J. C., Van Keuren‐Jensen K. R., Lai C. P., Lässer C., di Liegro I., Lunavat T. R., Lorenowicz M. J., Maas S. L. N., Mäger I., Mittelbrunn M., Momma S., Mukherjee K., Nawaz M., Pegtel D. M., Pfaffl M. W., Schiffelers R. M., Tahara H., Théry C., Tosar J. P., Wauben M. H. M., Witwer K. W., Nolte‐‘t Hoen E. N. M. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA – an ISEV position paper. Journal of Extracellular Vesicles, 6(1), 1286095. 10.1080/20013078.2017.1286095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, M., Martin‐Jaular, L., Lavieu, G., Thery, C.Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell‐to‐cell communication. Nature Cell Biology 2019, 21(1), 9–17. [DOI] [PubMed] [Google Scholar]
- McNaught, A. D., & Wilkinson, A. (1997) Compendium of chemical terminology, vol. 1669 (1259). Blackwell Science Oxford;. [Google Scholar]
- Monguio‐Tortajada, M., Galvez‐Monton, C., Bayes‐Genis, A., Roura, S., & Borras, F. E. (2019). Extracellular vesicle isolation methods: Rising impact of size‐exclusion chromatography. Cellular and Molecular Life Sciences, 76(12), 2369–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo, G., & Stoorvogel, W. (2013). Extracellular vesicles: Exosomes, microvesicles, and friends. Journal of Cell Biology, 200(4), 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A. E., Sneider, A., Witwer, K. W., Bergese, P., Bhattacharyya, S. N., Cocks, A., Cocucci, E., Erdbrugger, U., Falcon‐Perez, J. M., Freeman, D. W., Gallagher, T. M., Hu, S., Huang, Y., Jay, S. M., Kano, S ‐ I., Lavieu, G., Leszczynska, A., Llorente, A. M., Lu, Q., … Vader, P. (2019).Biological membranes in EV biogenesis, stability, uptake, and cargo transfer: An ISEV position paper arising from the ISEV membranes and EVs workshop. Journal of Extracellular Vesicles, 8(1), 1684862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen, J. B. (2017). What Are We Looking At? Extracellular Vesicles, Lipoproteins, or Both? Circulation Research, 121(8), 920–922. [DOI] [PubMed] [Google Scholar]
- Soekmadji, C., Li, B., Huang, Y., Wang, H., An, T., Liu, C., Pan, W., Chen, J., Cheung, L., Manuel Falcon‐Perez, J., Gho, Y. S., Holthofer, H. B., Le, M. T. N., Marcilla, A., O'Driscoll, L., Shekari, F., Shen, T. L., Torrecilhas, A. C., Yan, X., … Zheng, L. (2020). The future of extracellular vesicles as theranostics ‐ an ISEV meeting report. Journal of Extracellular Vesicles, 9(1), 1809766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranska, R., Gysbrechts, L., Wouters, J., Vermeersch, P., Bloch, K., Dierickx, D., Andrei, G., Snoeck, R. (2018). Comparison of membrane affinity‐based method with size‐exclusion chromatography for isolation of exosome‐like vesicles from human plasma. Journal of translational medicine, 16(1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takov, K., Yellon, D. M., & Davidson, S. M. (2019). Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size‐exclusion chromatography: Yield, purity and functional potential. Journal of Extracellular Vesicles, 8(1), 1560809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, S., Deng, S., Guo, J., Chen, X., Zhang, W., Cui, Y., Luo, Y., Yan, Z., He, Q. Y., Shen, S., & Wang, T. (2018). Deep coverage tissue and cellular proteomics revealed IL‐1beta can independently induce the secretion of TNF‐associated proteins from human synoviocytes. Journal of Immunology, 200(2), 821–833. [DOI] [PubMed] [Google Scholar]
- Thery, C., Amigorena, S., Raposo, G., & Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current Protocols in Cell Biology, Chapter 30(1), 3–22. [DOI] [PubMed] [Google Scholar]
- Thery, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., Antoniou, A., Arab, T., Archer, F., GK, A ‐ S., Ayre, D. C., Bach, J ‐ M., Bachurski, D., Baharvand, H., Balaj, L., Baldacchino, S., Bauer, N. N., Baxter, A. A., Bebawy, M., … Zuba‐Surma, E. K. (2018).Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, T., Zhu, Y. L., Hu, F. H., Wang, Y. Y., Huang, N. P., Xiao, Z. D. (2013). Dynamics of exosome internalization and trafficking. Journal of Cellular Physiology, 228(7), 1487–1495. [DOI] [PubMed] [Google Scholar]
- Tian, Y., Gong, M., Hu, Y., Liu, H., Zhang, W., Zhang, M., Hu, X., Aubert, D., Zhu, S., Wu, L., & Yan, X. (2020).Quality and efficiency assessment of six extracellular vesicle isolation methods by nano‐flow cytometry. Journal of Extracellular Vesicles, 9(1), 1697028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer K. W., Buzás E. I., Bemis L. T., Bora A., Lässer C., Lötvall J., Nolte‐‘t Hoen E. N., Piper M. G., Sivaraman S., Skog J., Théry C., Wauben M. H., Hochberg F. (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of Extracellular Vesicles, 2(1), 20360. 10.3402/jev.v2i0.20360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R., Greening, D. W., Zhu, H. J., Takahashi, N., Simpson, R. J. (2016). Extracellular vesicle isolation and characterization: Toward clinical application. Journal of Clinical Investigation, 126(4), 1152–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Wang, Y., Wei, S., Zhou, C., Yu, J., Wang, G., Wang, W., & Zhao, L. (2021). Extracellular vesicles isolated by size‐exclusion chromatography present suitability for RNomics analysis in plasma. Journal of translational medicine, 19(1), 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Lu, S., Zhou, Y., Meng, K., Chen, Z., Cui, Y., Shi, Y., Wang, T., & He, Q. Y. (2017). Motile hepatocellular carcinoma cells preferentially secret sugar metabolism regulatory proteins via exosomes. Proteomics, 17(13‐14). [DOI] [PubMed] [Google Scholar]
- Zhang, W., Chen, X., Yan, Z., Chen, Y., Cui, Y., Chen, B., Huang, C., Zhang, W., Yin, X., He, Q. Y., He, F., & Wang, T. (2017). Detergent‐insoluble proteome analysis revealed aberrantly aggregated proteins in human preeclampsia placentas. Journal of Proteome Research, 16(12), 4468–4480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information


